Significance
Known regulators of Wallerian degeneration appear to function locally in distal axons after axotomy to drive axon destruction. How competence to undergo axon death is established, and whether different subsets of neurons use alternate genetic programs to execute axon death, remains unclear. In this study, we show the transcription factor Pebbled (Peb) is required primarily in glutamatergic, but not cholinergic neurons, for appropriate axon degeneration after axotomy. Analysis of Peb mutant phenotypes also revealed an axon death phenotype where axon shafts initially broke into large fragments, but then failed to disintegrate. Peb is unique as a transcription factor shown to regulate Wallerian degeneration, and our analysis identifies a genetically accessible step in axon death signaling in vivo.
Keywords: Wallerian degeneration, axons, axon death
Abstract
Genetic studies of Wallerian degeneration have led to the identification of signaling molecules (e.g., dSarm/Sarm1, Axundead, and Highwire) that function locally in axons to drive degeneration. Here we identify a role for the Drosophila C2H2 zinc finger transcription factor Pebbled [Peb, Ras-responsive element binding protein 1 (RREB1) in mammals] in axon death. Loss of Peb in Drosophila glutamatergic sensory neurons results in either complete preservation of severed axons, or an axon death phenotype where axons fragment into large, continuous segments, rather than completely disintegrate. Peb is expressed in developing and mature sensory neurons, suggesting it is required to establish or maintain their competence to undergo axon death. peb mutant phenotypes can be rescued by human RREB1, and they exhibit dominant genetic interactions with dsarm mutants, linking peb/RREB1 to the axon death signaling cascade. Surprisingly, Peb is only able to fully block axon death signaling in glutamatergic, but not cholinergic sensory neurons, arguing for genetic diversity in axon death signaling programs in different neuronal subtypes. Our findings identify a transcription factor that regulates axon death signaling, and peb mutant phenotypes of partial fragmentation reveal a genetically accessible step in axon death signaling.
Neurons are connected over long distances by their axons, which can extend over more than a meter in humans. Maintenance of axon integrity is essential for sustained neural circuit function because axon breakage can block nervous system signal propagation. Axon loss is a hallmark of nervous system injuries, such as traumatic brain injury and spinal cord injury (1–3), is a unifying feature of neurodegenerative diseases (4), and is strongly correlated with functional loss in patients (5).
Wallerian degeneration (WD; axotomy) serves as a useful model to study basic aspects of axon biology, and to identify axon death signaling molecules. Severed axons, after a defined latent phase, undergo explosive fragmentation and are ultimately cleared by surrounding phagocytes (6–8). The discovery of the slow WD (WldS) mutant mouse, where severed distal axons survived for weeks after axotomy, radically changed our view of axonal biology (9). The observed long-term survival of distal severed axon fibers in WldS animals demonstrated that axon degeneration is a controlled process, and that under some conditions, axons could survive for weeks without a cell body (10–13). A growing number of studies across several species support the notion that the competence to undergo degeneration is likely a genetically programmed event: in the rock lobster, distal severed axons have been found to survive for a year after transection in vivo, and remain capable of evoked release at neuromuscular junctions (14); fragments of Aplysia axons can survive in vitro for extended periods of time without degeneration (15); and in Caenorhabditis elegans, most distal severed axons never degenerate (16). Despite these surprising observations, to our knowledge, nothing is known about transcriptional mechanisms that regulate the competence of axons to degenerate.
In axons that do undergo WD, the execution of degeneration is driven by axon death signaling molecules. Drosophila dSarm (sterile α, ARM, and TIR domain protein) was the first endogenous molecule shown to actively promote axon death (17). Sarm1 functions in a conserved role in mammals (17, 18), where it has been proposed to act as an NAD+ hydrolase that drives axonal degeneration through promoting metabolic catastrophe (19, 20). Drosophila dSarm is similarly capable of NAD+ hydrolysis (20), but requires signaling downstream through the BTB/BACK domain molecule Axundead to execute axon death in vivo (21). The E3 ubiquitin ligase Highwire/Phr1 also modulates axon death signaling (22, 23) through a mechanism that appears to involve regulating levels of the NAD+ biosynthetic molecule dNmnat/Nmnat2 (22, 24). In both Drosophila and mammals, all neurons tested thus far have been strongly protected by loss-of-function mutations in dSarm/Sarm1 (17, 18, 25), which suggests that axon death signaling molecules are engaged to drive destruction in a wide array of, or perhaps all, neuronal subtypes.
In this study, we present the identification and characterization of a role for Pebbled (Peb), a transcription factor, in axon degeneration. peb mutants show two predominant axon death-defective phenotypes: (i) severed distal axons are fully preserved morphologically, or (ii) the axon shaft breaks into large fragments (partially fragmented axons, PFAs) that fail to disintegrate further, lingering in the nervous system for weeks. The PFA phenotype in peb mutants is not observed in control or axon death mutants, and therefore defines a genetically accessible step in axon death signaling. Surprisingly, while PFAs form in all neurons, the ability of peb mutations to completely suppress axon degeneration was only observed in glutamatergic neurons, and not in cholinergic neurons, arguing that Peb functions to differentially modulate competence to undergo axon degeneration in distinct subsets of neurons.
Results
Isolation of a Mutant That Suppresses WD.
To identify novel endogenous regulators of WD, we performed an X chromosome-based F2 forward genetic screen in glutamatergic sensory neurons in the adult Drosophila wing using mosaic analysis using a repressible cell marker (MARCM) (26). Flies were bred to generate neuronal MARCM clones using the OK371-GAL4 driver, and axons were severed by surgical removal of the distal half of the wing. Animals were aged for 7 d postaxotomy (dpa), after which wings were dissected from the animal and axon death was quantified and imaged in the proximal wing vein. Neurons with cell bodies that were proximal to the injury site remained healthy and uninjured, and served as internal controls, represented by “cb” (cell body) in the upper right corner of each figure panel.
In control wings, injured axons typically undergo fragmentation by 12 h postaxotomy (hpa), degenerate fully by 24 hpa, and are cleared by surrounding glia after 5 dpa (26). We screened ∼2,000 independent X-chromosome mutant stocks, and identified one, 345x, that exhibited suppression of WD (Fig. 1A). While uninjured 345x mutant axons were similar in morphology to control, 60% of injured axons in 345x mutants remained partially or fully preserved at 7 dpa compared with 0% in wild-type control: 22% of severed 345x mutant axons remained morphologically intact, while 38% initiated fragmentation, but were maintained as PFAs (Fig. 1B). We defined PFAs as GFP+ axon fragments that spanned the imaging field-of-view, and were aligned linearly such that they appeared to represent the remnants of a previously intact axon. We rarely observed PFAs in control animals, and only early in the phase of axon degeneration (Fig. 1D), but never in dsarm, axundead, or highwire mutants (17, 21, 22). Therefore, 345x mutations result in an axon death phenotype, apparently specific to the execution phase of axonal destruction.
Fig. 1.
Mutation 345x causes defective WD in vivo. (A) Glutamatergic clones within the L1 anterior wing vein were labeled with mCD8::GFP. Uninjured and injured (7 dpa) are shown. (Scale bar, 10 μM.) Cell bodies (cb) of uninjured clones within each wing were counted and indicated in the upper right corner of each panel. Mutant 345x exhibited two axon death-defective phenotypes: either severed intact or PFAs. (B) Quantification of peb mutant phenotype. PFAs (gray) and severed intact axons (black). Daggers (†) and asterisks (*) represent PFA and severed intact significance, respectively (n > 30 wings). Two-way ANOVA, error = SEM (**,††P < 0.01; ***,†††P < 0.001; ****,††††P < 0.0001). (C) Quantification of total continuous GFP+ axon debris length, 7 dpa. Unpaired t test, error = SEM ****P < 0.0001. (D and E) Time-course data for both controls and 345x mutants over 14 dpa (n > 30 wings). (F) Schematic of the adult wing showing two imaging fields-of-view, proximal and distal to the injury site. (G and H) Severed intact axons were imaged and traced along the entirety of the wing (individual images, outlined in grey, stitched together, 63× magnification). Asterisks indicate cell bodies of uninjured clones (n = 14 wings, 27 severed intact axons). (I) Mitochondria (green) and glutamatergic neurons (red) of control and peb mutant clones imaged and indicated time points. (Scale bar, 10 μM; Insets, 1 μM.) (J) Quantification of mitochondria length at 8 hpa (control: n = 11 wings, 185 mitos. peb: n = 13 wings, 241 mitos. dsarm: n = 11 wings, 158 mitos.), 1 dpa (control: n = 10 wings, 211 mitos. peb: n = 16 wings 170 mitos. dsarm: n = 10 wings, 128 mitos), and 7 dpa (peb: n = 15 wings, 182 mitos. dsarm: n = 12 wings, 209 measurements). One-way ANOVA, Tukey’s multiple comparisons test, error = SEM, ****P < 0.0001. ctrl, control; n/a, not applicable; ns, not significant.
To distinguish PFAs from normal cellular debris produced by degenerating axons, we measured the length of all GFP+ axonal membrane debris from both control and 345x clones at 7 dpa. The sparse debris in controls averaged 0.9 μm in length, while 345x mutant axons fragments averaged 3.0 μm, ranging in size from 0.2 to 25.5 μm in length (Fig. 1C). Preserved intact axons and PFAs persisted in 345x mutants over time, as both phenotypes were observed out to 14 dpa (Fig. 1 D and E), although at reduced levels.
While one portion of a severed axon in 345x mutants was fully protected, it was plausible that PFAs might be generated at other positions along the axon. To explore this possibility, we scored axon morphological integrity with single-axon resolution along the entire length of individual severed intact axons in the wing vein of 345x mutants (Fig. 1 F and G). We found that when axons were preserved in the proximal wing, 100% of those axons remained morphologically intact along their entire length (Fig. 1H). We conclude that severed axons in 345x mutants fall into one of three phenotypic categories: (i) they are morphologically preserved along their entire length, (ii) they generate PFAs along their entire length, or (iii) they degenerate and are cleared normally.
Mitochondrial dysfunction is a hallmark in axon degeneration, and is observed after axotomy and in neurodegenerative diseases (27–29). In WldS-expressing axons, mitochondria persist after axotomy long after mitochondria in control animals have been destroyed (30). We therefore examined mitochondrial morphology before and after axotomy in control and 345x mutant clones. Using a GFP targeted to mitochondria, we found no significant difference in the average mitochondria length in uninjured neurons between control and 345x mutants (Fig. 1I). Just before the explosive fragmentation stage of the distal axon, at 8 h after injury, wild-type mitochondria began to decrease in size, with an average length of 0.36 μm. At 8 hpa, mitochondria of 345x mutants were significantly longer, with lengths averaging 0.9 μm (Fig. 1I). However, when we looked at injured axons 24 h after injury, mitochondria in debris of control axons and severed intact 345x mutant axons were indistinguishable (Fig. 1J). These data show that 345x mutant mitochondria exhibit a normal, albeit delayed, morphological response to axon injury within axon death-defective axons. This loss of mitochondria in 345x mutant axons that are morphologically intact is strikingly different from dsarm-null mutant (Fig. 1 I and J) or WldS-expressing severed axons (30). Surprisingly, 345x mutations are capable of preserving axon integrity despite significant depletion of mitochondria.
The 345x Mutation Maps to the C2H2 Zinc Finger Transcription Factor Pebbled/RREB1.
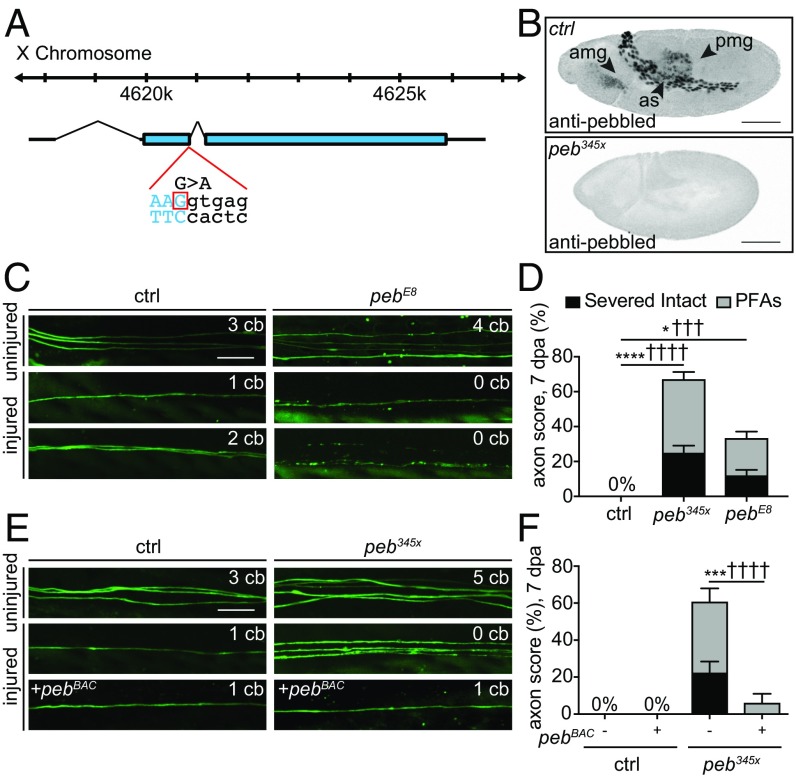
345x mutants were homozygous-lethal, suggesting the mutation affected an essential gene. This observation was supported by the fact that lethality associated with 345x cosegregated with the axon death phenotype during backcrossing over five generations. To identify the gene affected by the 345x mutation, we used genome-wide sequencing to identify mutations in the backcrossed 345x mutant stock, and found only one mutation that remained in 345x compared with the original isolate, which was a mutation in the pebbled (peb) gene that resulted in the loss of a splice donor site in exon 2 and generated a premature stop codon (Fig. 2A). In a parallel set of experiments, we screened a collection of X-chromosome deficiency lines that had been recombined onto an FRT chromosome with the MARCM approach. In total, these deficiencies removed 38.7% of X chromosome genes, and two nonoverlapping deficiency lines led to a suppression of axon death (Fig. S1). The first deficiency uncovered the E3 ubiquitin ligase Highwire, a known axon death signaling molecule (22, 23). The second overlapped with the genetic region to which we mapped 345x, and included the peb gene.
Fig. 2.
345x encodes the transcription factor pebbled. (A) The 345x mutation in exon 2 of the pebbled gene. (B) Pebbled in control embryos localized to the AS and midgut (anterior AM, posterior PM). peb345x mutant embryos lacked Peb protein. (Scale bars, 100 μm.) (C and D) Representative images and quantifications showing pebE8 mutant allele phenocopied peb345x mutants (n > 30 wings). (E and F) Representative images and quantifications showing genomic pebBAC rescued defective axon death back to control levels (n > 30 wings). (Scale bar, 10 μm.) Two-way ANOVA, error = SEM (*,†P < 0.05; ***,†††P < 0.001, ****,††††P < 0.0001). amg, anterior midgut; as, amnioserosa; ctrl, control; pmg, posterior midgut.
Peb [Ras-responsive element binding protein 1 (RREB1) in mammals] is a conserved transcription factor that contains 14 C2H2 zinc finger domains (31). In Drosophila embryos, Peb is expressed in amnioserosa (AS), anterior and posterior midgut (AM and PM, respectively), trachea, and in the peripheral nervous system and imaginal discs during later stages (32–34). Loss of Peb function leads to defects in embryonic germband retraction and dorsal closure of the embryonic epidermis, resulting in lethality. Peb also plays important roles in axon guidance in photoreceptor cells in the developing Drosophila visual system (35, 36), but roles for Peb/RREB1 in axon death have not been previously reported. We performed immunohistochemistry (IHC) of early-stage embryos with α-Peb antibodies and confirmed that Peb was localized to the AS, and both the AM and PM (Fig. 2B) (32). Consistent with our prediction that the 345x mutations would lead to a loss of Peb protein product, we found that embryos homozygous for 345x lacked α-Peb immunoreactivity (Fig. 2B). IHC analyses in wing tissues during later stages of development show Peb is detectable only within the nuclei of developing and mature neurons and not present in the cytoplasm of the cell body and axon (Fig. S3).
To confirm that the mutation of peb was responsible for the axon death phenotype, we assayed axon death in adult wing glutamatergic neurons with a second peb allele, pebE8 (37). We found that pebE8 mutant clones phenocopied the 345x mutant phenotype, although the phenotype was slightly weaker: in pebE8 mutant MARCM clones 12% of severed axons remained morphologically intact, and 21% formed PFAs (Fig. 2 C and D). We next crossed a bacterial artificial chromosome (BAC) containing the genomic copy of peb into 345x mutants, and found that reintroduction of a wild-type copy of pebBAC into 345x mutants completely rescued the axon death phenotypes (i.e., both axon degeneration, the production of PFAs, and clearance of axonal debris) to control levels (Fig. 2 E and F). Addition of pebBAC had no effect on uninjured axons (Fig. S2). Furthermore, we found that the pebBAC rescued the lethality of the 345x mutation. We conclude that 345x is a mutation in the peb gene, and henceforth refer to it as peb345x.
Human RREB1 Can Functionally Substitute for Pebbled.
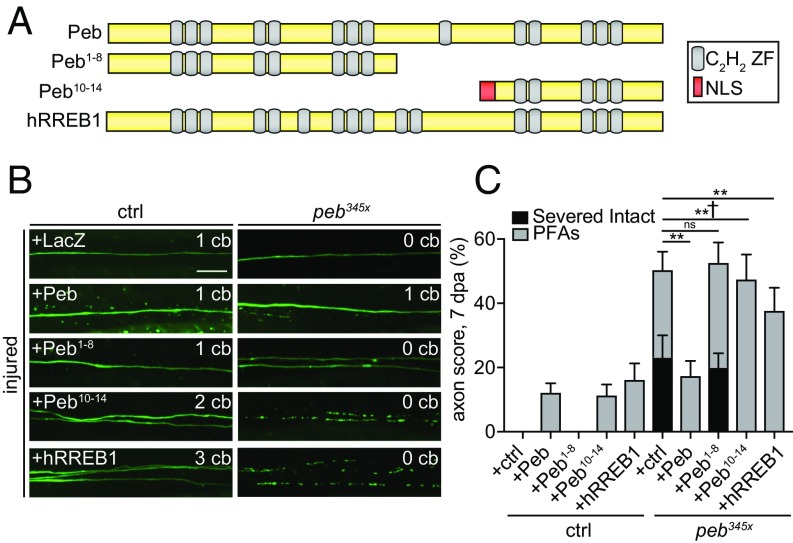
To explore how Peb regulates axon death, we drove expression of cDNA constructs encoding full-length Peb, truncated Peb containing zinc fingers 1–8 or 10–14, and the human homolog, Ras-responsive element binding protein 1, (Peb, Peb1−8, Peb10−14, and hRREB1, respectively) in control and peb345x clones, and quantified axon degeneration 7 dpa (Fig. 3A). Overexpression of full-length Peb using the GAL4/UAS system (and driving expression with the OK371-GAL4 driver) was sufficient to completely rescue the blockade of axon degeneration observed in peb345x clones: 100% of mutant axons initiated axon fragmentation (Fig. 3 B and C). Expression of the C-terminal zinc finger domains 10–14 (in Peb10−14) was sufficient to rescue the axon death defect in peb mutants. In contrast, expression of N-terminal Peb1−8 failed to rescue the axon protective phenotype. This defines the key domains essential for Peb function in prodegenerative signaling, and provides additional support for a transcriptional role for Peb, consistent with previous work demonstrating that C-terminal domains are required for Peb to bind DNA in vitro (31). Expression of hRREB1 was also sufficient to fully rescue the blockade of axon degeneration (Fig. 3 B and C and Fig. S2), arguing that Peb and hRREB1 exhibit similar properties with respect to target gene regulation. However, while the initiation of axon fragmentation was strongly rescued, 17% of the GFP+ injured axons persisted as PFAs in peb345x background rescued with Peb. This approximates the percentage of axons that form PFAs in peb345x mutant clones expressing a UAS-LacZ control rescue construct, and we observed PFAs in peb345x mutants rescued with hRREB1 expression. Additionally, expression of either Peb or hRREB1 in control animals led to the production of PFAs, which are only ever rarely observed in the earliest phase of normal axon degeneration (Fig. 3 B and C). These data suggest Peb expression levels may need to be fine-tuned for proper rescue of execution of axon death and destruction of PFAs, with too much or too little Peb resulting in PFA formation.
Fig. 3.
The Peb C-terminal domain or hRREB1 rescue axon death defects in peb mutants. (A) Schematic of Pebbled protein. Pebbled contains 14 C2H2 zinc finger domains (gray ovals). N-terminal Peb1−8 and C-terminal Peb10−14 (with NLS added) truncated proteins and hRREB1 are shown. (B and C) Representative images and quantification of Peb and hRREB1 rescue of peb clones, 7 dpa (Scale bar, 10 μm.) (n > 30 wings). Two-way ANOVA, Tukey’s multiple comparisons test, error = SEM (†P < 0.05; **P < 0.01; ctrl, control; ns, not significant).
Peb Loss Can Fully Preserve Severed Axons in Glutamatergic, but Not Cholinergic Neurons.
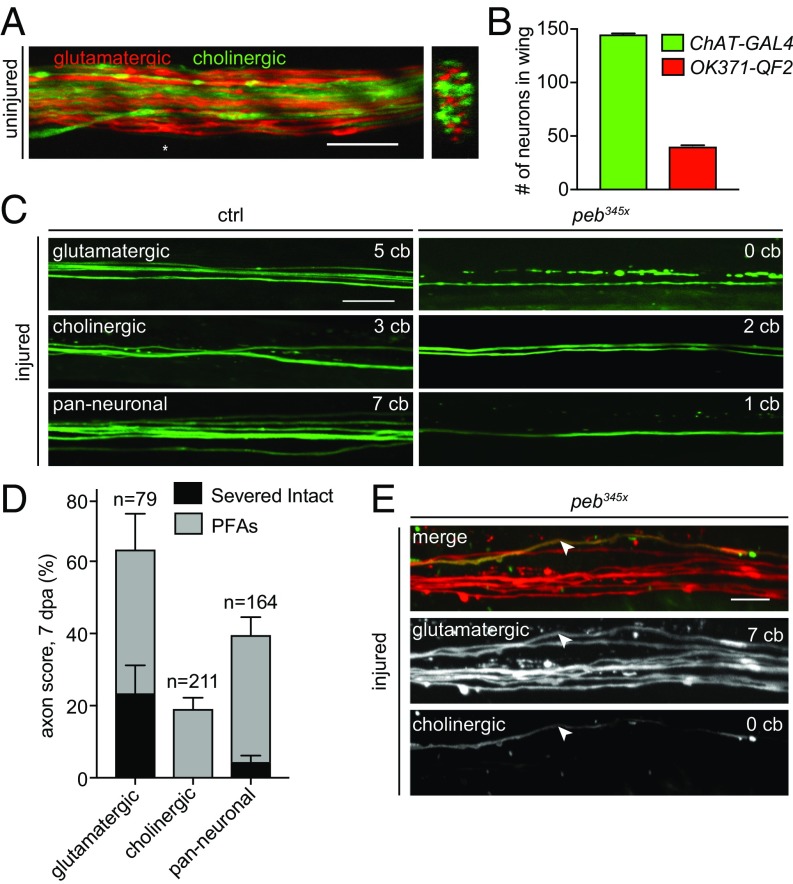
There are ∼250 sensory neurons in the Drosophila wing that send axonal projections to the thoracic ganglion in the CNS (38). Most cells fall into two subsets based on neurotransmitter profiles: cholinergic (∼145 cells labeled by ChAT-GAL4) and glutamatergic neurons (∼40 cell labeled by vGlut-QF2 or OK371-GAL4) (Fig. 4 A and B) (26). Because the peb345x mutation was identified and characterized in glutamatergic neurons, we wished to determine whether it also regulated axon death signaling in cholinergic neurons. Approximately 23% of severed peb345x glutamatergic clones failed to undergo axon degeneration at 7 dpa, and ∼40% of severed axons formed PFAs (Fig. 4 C and D). When we assayed cholinergic peb345x clones, we found a complete lack of protection from axon degeneration: 100% of all neurons began fragmenting (Fig. 4 C and D). Of severed axons, 19% form PFAs, indicating that peb also regulates PFA formation at some level in cholinergic neurons compared with control clones that underwent normal axon death (Fig. S4). In axotomy assays with the pan-neuronal driver nSyb-GAL4, we observed only 4% of severed mutant clones remaining intact, and there was no change in PFAs compared with peb345x glutamatergic clones. Control pan-neuronal clones had a normal degenerative response to injury. Given that glutamatergic neurons represent ∼20% of the nSyb-GAL4+ cells, we suspected the only cells protected in this background by the peb345x mutation were the glutamatergic neurons. Indeed, after screening many hundred clones of injured axons where we simultaneously labeled cholinergic neurons (with ChAT-GAL4) and glutamatergic neurons (with vGlut-QF2), we only found a single severed, intact axon that expressed ChAT-GAL4, and it was also positive for vGlut-QF2 and therefore also glutamatergic (Fig. 4E). The observation that only glutamatergic neurons can be fully protected by peb345x mutations, while PFAs appear with equal frequency in both cholinergic and glutamatergic neurons, further supports the notion that the initiation (i.e., fragmentation) and execution (i.e., full degradation) of axons are genetically separable and differentially regulated by Peb levels. Moreover, ability of peb mutants to completely block axon death is limited in the wing to glutamatergic sensory neurons.
Fig. 4.
Pebbled axon death defect observed only in glutamatergic neurons. (A) Cholinergic neurons (green) and glutamatergic neurons (red) in the adult wing vein. An asterisk (*) marks position of orthogonal view. (Scale bar, 10 μm.) (B) Number of cholinergic and glutamatergic neurons in the wing vein using ChAT-GAL4 and vGlut-QF2. (C) Axon death in glutamatergic, cholinergic, or all neurons. (D) Axon protection at 7 dpa (n > 30 wings per genotype. Glut = 79 injured neurons, chol = 211 injured neurons, all = 164 injured neurons). (E) Severed intact peb clone double labeled with OK371-QF2 (red) and ChAT-GAL4 (green), indicated by arrowhead. (Scale bar, 10 μm.)
peb Mutations Do Not Block dSarm-Induced Axon Death, Although dsarm Mutations Can Enhance peb Mutant Phenotypes in Axon Death.
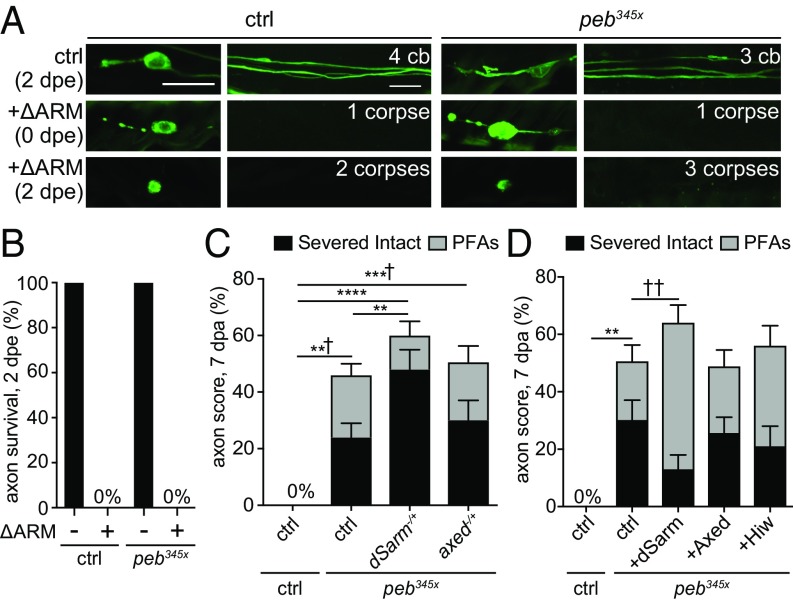
Activation of Sarm1 signaling potently drives axon death in mammalian and Drosophila neurons (19). To determine whether Peb acts downstream of dSarm, we crossed peb345x mutants into a background expressing a gain-of-function version of dSarm (dSarmΔARM) (21) and assayed for suppression of axon death. Loss of Peb function was not sufficient to block dSarmΔARM-induced axon death (Fig. 5 A and B), arguing that Peb does not act genetically downstream of dSarm, and demonstrating that constitutive activation of dSarm is sufficient to eliminate the appearance of PFAs. We next sought to determine whether peb mutants exhibited any dominant genetic interactions with components of the axon death signaling cascade, including dsarm (17), axundead (21), or highwire (22). We crossed loss-of-function mutations of dsarm or axed into the peb345x background, and found that loss of a single copy of dsarm was sufficient to double the number of intact axons in peb345x animals, while loss of a single copy of axed had no effect (Fig. 5C). Removal of one copy of dsarm or axed in control clones had no effect on axon degeneration (Fig. S4). In addition to the increase in fully protected axons, loss of one copy of dsarm also reduced the number of PFAs in peb345x mutants (Fig. 5C). Reciprocally, we found that overexpression of dSarm in a peb345x background led to a partial suppression of the ability of peb345x to fully protect axons, and an increase in the number of PFAs. Manipulation of Axed or Hiw had no effect (Fig. 5D). Overexpression of dSarm, Axed, and Hiw in control clones had no effect on axon degeneration (Fig. S4). These data indicate the neuroprotective effects of peb345x mutants are sensitive to dSarm levels.
Fig. 5.
The neuroprotective effects of peb345x mutants are sensitive to dSarm levels. (A and B) dSarmΔARM (ΔARM) induced axon death in uninjured control and peb345x clones at 2 d posteclosion (dpe) (n > 15 wings). (Scale bar, 10 μm.) (C) Quantifications of axon death 7 dpa in control and peb345x clones when one copy of dsarm or axed was removed (n > 30 wings). (D) Quantifications of axon death at 7 dpa in control and peb345x clones expressing dSarm, Axed, or Hiw (n > 30 wings). Two-way ANOVA, error = SEM (*,†P < 0.05; **,††P < 0.01; ***,†††P < 0.001; ****,††††P < 0.0001). ctrl, control.
Discussion
Distal axons separated from their cell bodies can survive in a functionally competent state for days to weeks after axotomy in multiple species (14, 21, 39). It therefore seems plausible that some axons are programmed to degenerate while others are not. In this study, we identify the transcription factor Pebbled (Peb)/RREB1 as an essential modulator of axon death in Drosophila. Through epistatic analysis, we can place peb upstream of dSarm or in a separate, parallel pathway, which ultimately converges on axon death. The simplest interpretation of our data is that Peb regulates axon death signaling in glutamatergic axons at the transcriptional level. Human RREB1 can rescue axon death phenotypes associated with loss of peb, arguing for strong conservation of the binding properties of Peb and hRREB1, and implying that RREB1 may play similar roles in axon biology in mammals.
Pebbled appears to identify a step in the axon death signaling cascade. Loss of Pebbled function resulted in the appearance of three axon phenotypes after injury: (i) full morphological preservation with a slightly delayed loss of mitochondria; (ii) the generation of PFAs that linger in the nervous system for weeks, but which also lose mitochondria after a short delay; or (iii) apparently normal axon death signaling and clearance. The cell biology of axon preservation in peb mutants is unique, and implies that peb mutants identify a genetically accessible step in axon death signaling. PFAs have not been observed in other axon death mutants (i.e., dsarm, hiw, or axed), as these mutants all fully block axon degeneration after axotomy (17, 21, 23). Understanding the nature of PFA production compared with normal axon degeneration is an important goal for future study. In the case of dsarm mutants, in addition to the axon shaft maintaining complete integrity, mitochondria also appeared well-preserved. That was not the case in peb mutants, where mitochondria degenerated after only a short delay, and preserved axons were severely depleted of mitochondria for the duration of their extended survival. Interestingly, the phenotype of individual peb mutant axons is consistent along the entirety of the axon shaft: we never observed an axon that generated PFAs in one portion, but was fully protected elsewhere. Unraveling the molecular basis of this all-or-none type of phenotypic expression is of great interest for the future. Finally, while some PFAs are observed in control animals immediately after the initiation of axon fragmentation, they quickly undergo explosive degeneration and are cleared. From these data, we conclude that Peb functions both at the initial phase of axon breakage into smaller fragments, and subsequently during the phase of explosive degeneration.
To date all known axon death signaling molecules—dSarm, Hiw, and Axed—have been proposed to function locally in the axon to drive destruction, and their neuroprotective effects extend to both glutamatergic and cholinergic neurons (17, 21, 22). Based on its expression in the nuclei of wing sensory neuron precursors and mature neurons, and the fact it is a C2H2 zinc finger transcription factor, we propose that Peb functions at the transcriptional level to help establish and maintain competence to undergo axon degeneration. While Peb appears to be expressed broadly in wing sensory neurons, surprisingly, the ability of peb mutants to fully block axon fragmentation is restricted to glutamatergic neurons. How Peb selectively protects glutamatergic axons is unclear, but could modify axonal phenotypes through the JNK signaling cascade. During embryogenesis, in AS peb mutants show increased levels of AP-1 transcriptional activity downstream of the JNK signaling cascade (40), which in turn inhibits cytoskeletal rearrangements that allow for cell migration (32, 40). We have found a lack of evidence to support a role for JNK signaling in axon death (17, 21), but some data support a neuroprotective role for this pathway by controlling baseline levels of Nmnat (41). Peb has also been shown to negatively regulate nervy, the Drosophila homolog of mammalian MTG8 proto-oncogene (31); however, we observed no alterations in axon death when we overexpressed nervy in glutamatergic neurons.
The nuclear localization requirement of the carboxy terminal DNA binding zinc finger domains of Peb for rescue suggests that Peb is regulating injury-induced axon degeneration at the transcriptional level. We attempted to identify direct transcriptional targets of Peb by expressing a tagged version of Peb in the Drosophila embryonic cell line, GM2, and performing ChIP with antibodies specific to tagged Peb and subsequent deep sequencing (ChIP-seq). This approach successfully identified the one known target for Peb, the transcriptional regulator Nervy, whose overexpression did not block axon death, and several potential Peb targets (Table S1). We did not find evidence for direct binding of Peb to regions containing known axon death signaling genes (i.e., dsarm, axed, or hiw). This could indicate that Peb does not directly modulate axon death genes in vivo to exert its effects, although it remains unclear how similar Peb transcriptional activity in GM2 cells might be compared with neurons. Finally, it remains unclear why dsarm, but not axed mutations, modify peb mutant phenotypes, given that Axed signals downstream of dSarm in axon death. A deeper understanding of the molecular basis of axon degeneration, and the nature of PFAs, will likely be required to answer this question.
Our analysis of peb mutant phenotypes reveals features of the cell biology of axon death. Our discovery of PFAs in peb mutants implies that axon degeneration can be genetically dissected into distinct activation and execution phases, and we propose that PFAs represent activation, but failure to execute axon death. We found that either increased or decreased Peb levels could lead to the production of PFAs, arguing that fine-tuning of Peb levels is essential for appropriate execution of axon death. Furthermore, Peb modulation of PFA production is not limited to glutamatergic neurons, since we found PFAs in cholinergic neurons under both loss- and gain-of-function Peb conditions. Interestingly, Peb allows us to genetically separate mitochondrial loss from axon degeneration. We observed mitochondrial degeneration even in fully protected peb mutant axons, indicating that mitochondrial destruction must occur through a Peb-independent signaling pathway.
Methods
See SI Methods for more information and detailed descriptions of animal genotypes, EMS mutagenesis, transgenic constructs, the wing axotomy assay, subsequent wing microscopy, IHC procedures, antibodies used, cell culture techniques, ChIP methods, and deep sequencing analyses.
Supplementary Material
Acknowledgments
We thank all M.R.F. laboratory members and colleagues in the field for feedback, comments, and suggestions; Jaeda Coutinho-Budd for her technical assistance in confocal image acquisition well as her scientific input and skilled editorial assistance; Dr. Maninjay Atianand for assistance with ChIP; and Amy Sheehan for generating the 5XUAS::5XMyc vector. Deficiency lines screened in this study were generated through a collaboration with the laboratories of Dr. Chris Doe and Dr. Tory Herman from the University of Oregon. Sequencing was performed at the Bauer Core Facility at Harvard University. This work was supported by National Institute of Neurological Disorders and Stroke Grant R01 NS053538 (to M.R.F.). During the period of this study, M.R.F. was an Investigator with the Howard Hughes Medical Institute and J.E.L. was a collaborating PI.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1140.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715837115/-/DCSupplemental.
References
- 1.Kelley BJ, Farkas O, Lifshitz J, Povlishock JT. Traumatic axonal injury in the perisomatic domain triggers ultrarapid secondary axotomy and Wallerian degeneration. Exp Neurol. 2006;198:350–360. doi: 10.1016/j.expneurol.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Henninger N, et al. Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain. 2016;139:1094–1105. doi: 10.1093/brain/aww001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagg T, Oudega M. Degenerative and spontaneous regenerative processes after spinal cord injury. J Neurotrauma. 2006;23:264–280. doi: 10.1089/neu.2006.23.263. [DOI] [PubMed] [Google Scholar]
- 4.Adalbert R, Coleman MP. Review: Axon pathology in age-related neurodegenerative disorders. Neuropathol Appl Neurobiol. 2013;39:90–108. doi: 10.1111/j.1365-2990.2012.01308.x. [DOI] [PubMed] [Google Scholar]
- 5.Coleman MP, Perry VH. Axon pathology in neurological disease: A neglected therapeutic target. Trends Neurosci. 2002;25:532–537. doi: 10.1016/s0166-2236(02)02255-5. [DOI] [PubMed] [Google Scholar]
- 6.Waller A. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Philos Trans R Soc Lond. 1850;140:423–429. [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena S, Caroni P. Mechanisms of axon degeneration: From development to disease. Prog Neurobiol. 2007;83:174–191. doi: 10.1016/j.pneurobio.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Conforti L, Gilley J, Coleman MP. Wallerian degeneration: An emerging axon death pathway linking injury and disease. Nat Rev Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- 9.Lunn ER, Perry VH, Brown MC, Rosen H, Gordon S. Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve. Eur J Neurosci. 1989;1:27–33. doi: 10.1111/j.1460-9568.1989.tb00771.x. [DOI] [PubMed] [Google Scholar]
- 10.Glass JD, Brushart TM, George EB, Griffin JW. Prolonged survival of transected nerve fibres in C57BL/Ola mice is an intrinsic characteristic of the axon. J Neurocytol. 1993;22:311–321. [Google Scholar]
- 11.Mack TG, et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald JM, et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 13.Martin SM, O’Brien GS, Portera-Cailliau C, Sagasti A. Wallerian degeneration of zebrafish trigeminal axons in the skin is required for regeneration and developmental pruning. Development. 2010;137:3985–3994. doi: 10.1242/dev.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parnas I, Dudel J, Atwood HL. Synaptic transmission in decentralized axons of rock lobster. J Neurosci. 1991;11:1309–1315. doi: 10.1523/JNEUROSCI.11-05-01309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benbassat D, Spira ME. The survival of transected axonal segments of cultured Aplysia neurons is prolonged by contact with intact nerve cells. Eur J Neurosci. 1994;6:1605–1614. doi: 10.1111/j.1460-9568.1994.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 16.Neumann B, Nguyen KCQ, Hall DH, Ben-Yakar A, Hilliard MA. Axonal regeneration proceeds through specific axonal fusion in transected C. elegans neurons. Dev Dyn. 2011;240:1365–1372. doi: 10.1002/dvdy.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osterloh JM, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J Neurosci. 2013;33:13569–13580. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science. 2015;348:453–457. doi: 10.1126/science.1258366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Essuman K, et al. The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron. 2017;93:1334–1343.e5. doi: 10.1016/j.neuron.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neukomm LJ, et al. Axon death pathways converge on axundead to promote functional and structural axon disassembly. Neuron. 2017;95:78–91.e5. doi: 10.1016/j.neuron.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 22.Xiong X, et al. The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 2012;10:e1001440. doi: 10.1371/journal.pbio.1001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babetto E, Beirowski B, Russler EV, Milbrandt J, DiAntonio A. The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 2013;3:1422–1429. doi: 10.1016/j.celrep.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilley J, Coleman MP. Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 2010;8:e1000300. doi: 10.1371/journal.pbio.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilley J, Orsomando G, Nascimento-Ferreira I, Coleman MP. Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 2015;10:1974–1981. doi: 10.1016/j.celrep.2015.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neukomm LJ, Burdett TC, Gonzalez MA, Züchner S, Freeman MR. Rapid in vivo forward genetic approach for identifying axon death genes in Drosophila. Proc Natl Acad Sci USA. 2014;111:9965–9970. doi: 10.1073/pnas.1406230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 29.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avery MA, et al. WldS prevents axon degeneration through increased mitochondrial flux and enhanced mitochondrial Ca2+ buffering. Curr Biol. 2012;22:596–600. doi: 10.1016/j.cub.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ming L, Wilk R, Reed BH, Lipshitz HD. Drosophila Hindsight and mammalian RREB-1 are evolutionarily conserved DNA-binding transcriptional attenuators. Differentiation. 2013;86:159–170. doi: 10.1016/j.diff.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Yip ML, Lamka ML, Lipshitz HD. Control of germ-band retraction in Drosophila by the zinc-finger protein HINDSIGHT. Development. 1997;124:2129–2141. doi: 10.1242/dev.124.11.2129. [DOI] [PubMed] [Google Scholar]
- 33.Wilk R, Reed BH, Tepass U, Lipshitz HD. The hindsight gene is required for epithelial maintenance and differentiation of the tracheal system in Drosophila. Dev Biol. 2000;219:183–196. doi: 10.1006/dbio.2000.9619. [DOI] [PubMed] [Google Scholar]
- 34.Pickup AT, Lamka ML, Sun Q, Yip MLR, Lipshitz HD. Control of photoreceptor cell morphology, planar polarity and epithelial integrity during Drosophila eye development. Development. 2002;129:2247–2258. doi: 10.1242/dev.129.9.2247. [DOI] [PubMed] [Google Scholar]
- 35.Oliva C, Sierralta J. Developmental biology. Dev Biol. 2010;344:911–921. doi: 10.1016/j.ydbio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Oliva C, et al. Hindsight regulates photoreceptor axon targeting through transcriptional control of jitterbug/Filamin and multiple genes involved in axon guidance in Drosophila. Dev Neurobiol. 2015;75:1018–1032. doi: 10.1002/dneu.22271. [DOI] [PubMed] [Google Scholar]
- 37.Strecker TR, Yip ML. Zygotic genes that mediate torso receptor tyrosine kinase functions in the Drosophila melanogaster embryo. Proc Natl Acad Sci USA. 1991;88:5824–5828. doi: 10.1073/pnas.88.13.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang Y, Bonini NM. Axon degeneration and regeneration: Insights from Drosophila models of nerve injury. Annu Rev Cell Dev Biol. 2012;28:575–597. doi: 10.1146/annurev-cellbio-101011-155836. [DOI] [PubMed] [Google Scholar]
- 39.Stengl M. Pigment-dispersing hormone-immunoreactive fibers persist in crickets which remain rhythmic after bilateral transection of the optic stalks. J Comp Physiol A Neuroethol. 1995;176:217–228. [Google Scholar]
- 40.Reed BH, Wilk R, Lipshitz HD. Downregulation of Jun kinase signaling in the amnioserosa is essential for dorsal closure of the Drosophila embryo. Curr Biol. 2001;11:1098–1108. doi: 10.1016/s0960-9822(01)00318-9. [DOI] [PubMed] [Google Scholar]
- 41.Walker LJ, et al. MAPK signaling promotes axonal degeneration by speeding the turnover of the axonal maintenance factor NMNAT2. eLife. 2017;6:e22540. doi: 10.7554/eLife.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.