Significance
Before the emergence of oxygenic photosynthesis and the accumulation of oxygen on Earth, life was essentially composed of anaerobic microorganisms. However, very little is known about which metabolisms were present at the time. Anaerobic carbon fixation through the Wood–Ljungdahl pathway is believed to be among the most ancient, and still plays a pivotal role in modern ecosystems. However, its origin and evolutionary history has been disputed. We analysed the distribution and phylogeny of carbon monoxide dehydrogenase/acetyl-CoA synthase, the main enzymatic complex of the pathway in thousands of bacterial and archaeal genomes. We show that this complex was already at work in the last universal common ancestor and has been remarkably conserved in microorganisms over more than 3.5 billion years.
Keywords: Wood–Ljungdahl pathway, evolution, methanogens, acetogens, LUCA
Abstract
Carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) is a five-subunit enzyme complex responsible for the carbonyl branch of the Wood–Ljungdahl (WL) pathway, considered one of the most ancient metabolisms for anaerobic carbon fixation, but its origin and evolutionary history have been unclear. While traditionally associated with methanogens and acetogens, the presence of CODH/ACS homologs has been reported in a large number of uncultured anaerobic lineages. Here, we have carried out an exhaustive phylogenomic study of CODH/ACS in over 6,400 archaeal and bacterial genomes. The identification of complete and likely functional CODH/ACS complexes in these genomes significantly expands its distribution in microbial lineages. The CODH/ACS complex displays astounding conservation and vertical inheritance over geological times. Rare intradomain and interdomain transfer events might tie into important functional transitions, including the acquisition of CODH/ACS in some archaeal methanogens not known to fix carbon, the tinkering of the complex in a clade of model bacterial acetogens, or emergence of archaeal–bacterial hybrid complexes. Once these transfers were clearly identified, our results allowed us to infer the presence of a CODH/ACS complex with at least four subunits in the last universal common ancestor (LUCA). Different scenarios on the possible role of ancestral CODH/ACS are discussed. Despite common assumptions, all are equally compatible with an autotrophic, mixotrophic, or heterotrophic LUCA. Functional characterization of CODH/ACS from a larger spectrum of bacterial and archaeal lineages and detailed evolutionary analysis of the WL methyl branch will help resolve this issue.
Among the six carbon fixation pathways encountered in prokaryotes (1), the Wood–Ljungdahl (WL) (or reductive acetyl-CoA) pathway has been proposed to be one of the oldest (2). It consists of two branches, the methyl (Eastern) and the carbonyl (Western) (3, 4) (Fig. 1A). In the methyl branch, CO2 is progressively reduced to methyl (-CH3), whereas in the carbonyl branch, a CO2 molecule is reduced to CO (carbonyl moiety), which is combined with the methyl coming from the methyl branch and CoA to form acetyl-CoA (Fig. 1A). This complex reaction is carried out by a single multimeric, bifunctional, and oxygen-sensitive enzyme complex called carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS). In addition, CODH/ACS can also function in reverse in some bacteria and archaea to carry out acetyl-CoA degradation to CO2 and tetrahydromethanopterin/tetrahydrofolate (H4MPT/THF)-bound methyl, e.g., from lactate in Archaeoglobus fulgidus (5).
Fig. 1.
(A) The reactions of the WL pathway. In the methyl branch, CO2 is progressively reduced to formyl/formate (-CHO, HCOOH), methenyl (-CH), methylene (-CH2), and eventually methyl (-CH3). There are two nonhomologous versions of the methyl branch, each using a different cofactor to which the reduced carbon compounds are bound, tetrahydromethanopterin (H4MPT) or tetrahydrofolate (THF), which are commonly associated with methanogenic Archaea and acetogenic Bacteria, respectively. The carbonyl branch reduces a CO2 molecule to CO (carbonyl moiety), which is combined with the methyl coming from the methyl branch and CoA to form acetyl-CoA. This reaction is achieved through the CODH/ACS enzymatic complex. [H] denotes one reducing equivalent (=1e−+1H+). (B) Nomenclature and function of the CODH/ACS subunits. (C) Organization of the CODH/ACS cluster in Bacteria and Archaea with indication of homologous subunits. The genes in yellow correspond to the homologous accessory proteins CooC and AcsF, which are responsible for nickel insertion.
Five different subunits constitute the CODH/ACS complex, and their coding genes are often found in a cluster (Fig. 1 B and C). Four of them are homologous between Bacteria and Archaea, but follow a different and somewhat confusing terminology. In Archaea, they are called CdhA (α-subunit), CdhC (β-subunit), CdhD (δ subunit), and CdhE (γ subunit), while in Bacteria, their respective homologs are called AcsA (β), AcsB (α), AcsD (δ), and AcsC (γ). In addition, there exists a subunit exclusive to Archaea called CdhB (ε-subunit), and one exclusive to Bacteria (AcsE). For the remainder of this work we shall refer to components by the names of their archaeal homologs.
Functional studies have dissected the role of each subunit in the overall reaction in Bacteria and Archaea. CdhA catalyzes the first step: the reduction of CO2 to CO or the reverse reaction, CO oxidation into CO2. CdhA has a homolog called CooS that performs the same reaction, but is not part of the CODH/ACS cluster (6). In Methanosarcina, the archaeal-specific subunit CdhB has been shown to bind to CdhA and suggested to provide the cofactor-binding sites for FAD during FAD-dependent CO oxidation, or FAD/FADH2-mediated activity regulation during autotrophic or acetotrophic growth (7). CdhC catalyzes the second step: condensation of CO (carbonyl moiety) with a methyl moiety and CoA to form acetyl-CoA. CdhD–CdhE form a corrinoid iron–sulfur complex that provides the methyl for this second step of the reaction. In Archaea, this complex alone suffices to transfer the methyl moiety bound to H4MPT from the methyl branch to a corrinoid cofactor, which is used by CdhC (8). Conversely, in Bacteria the CdhD–CdhE subcomplex contains an extra subunit, AcsE, which catalyzes the transfer of the methyl moiety bound to tetrahydrofolate (THF) (from the methyl branch) to the corrinoid (9). In this work, we will refer to CdhABC as the oxidoreductase module and CdhDE/AcsE as the methyltransferase module (Fig. 1B).
The fact that four CODH/ACS subunits are homologous between Archaea and Bacteria would suggest that this enzyme was present before the divergence between these two domains of life, i.e., in the last universal common ancestor (LUCA). This has important implications in the debate about the physiology and ecology of the earliest microorganisms, in the most general sense formulated as an autotrophic versus heterotrophic LUCA (2, 10). However, previous analyses have reported conflicting results, by either indicating presence in the LUCA (10–12), or conversely concluding for no evidence to support that (13). Nonetheless, most of these studies focused on very few taxa (11–13), or studied only one or two subunits at a time (6, 13). Furthermore, the approach of analyzing bacterial and archaeal homologs together and not precisely identifying interdomain transfers might have complicated the interpretation of results, leading to conflicting conclusions regarding the presence or not of a CODH/ACS in the LUCA. Even the most recent and thorough analyses could only predict the methyltransferase module in LUCA (10), or CdhC in the ancestor of Archaea (14). These results might be inherent to the automated nature of these analyses, combined with a restricted taxonomic sampling or difficulties in correctly identifying orthologs from horizontal gene transfers. For instance, CdhA is often excluded from the set of genes inferred in the LUCA (10), because it is very difficult to distinguish it from CooS, which is subject to frequent interdomain transfers (6).
Surprisingly, while CODH/ACS has been generally thought to be a feature of most methanogenic archaea and some acetogenic, aceticlastic, carboxydotropic, sulfate-reducing, and anammox bacteria, the identification of CODH/ACS subunit homologs has led to predict the presence of the enzyme complex in a number of anaerobic uncultured lineages whose genomes were made recently available from both Archaea (15–21), and Bacteria (12, 22, 23). This suggests a larger distribution than previously known and provides a unique opportunity to understand the origin and evolutionary history of this ancient enzyme complex.
Here, we have carried out an exhaustive phylogenomic analysis of the whole CODH/ACS complex in the largest taxonomic representation to date. Our results allow us to dissect the paths that shaped the current distribution and diversity of CODH/ACS in both Bacteria and Archaea and to resolve the debate about its ancestry and its evolutionary implications. Our analysis also opens perspectives on some important functional and ecological aspects of CODH/ACS.
Results and Discussion
Remarkable Conservation and Expanded Distribution of the CODH/ACS Gene Cluster in both Bacteria and Archaea.
We carried out an exhaustive search for homologs of all six subunits (the four common ones plus the two domain-specific ones) in the genomes of 5,763 Bacteria and 639 Archaea, representative of the diversity of the two domains (Methods). In both Bacteria and Archaea, the five subunits of CODH/ACS usually formed a conserved gene cluster, indicating a potentially functional enzyme (Datasets S1 and S2). Clusters with fewer subunits were less common, and important examples are described further below.
The striking conservation of the CODH/ACS cluster allowed us to identify actual CdhA subunits from isolated CooS homologs present at different genomic locations, which are otherwise very difficult to separate based on sequence alone. Phylogenetic analysis of CdhA/CooS homologs (Fig. S1) indicates that CooS genes appeared early in bacterial evolution, possibly by duplication of CdhA in the Firmicutes, and have been subject to extensive intra and interdomain horizontal transfers and further duplications, consistent with previous analyses (6). The wide distribution of CooS highlights the widespread potential for anaerobic carboxydotrophy in both Bacteria and Archaea. Although it is unknown whether a CooS can substitute the function of CdhA, the presence of lone CdhA/CooS genes in a genome should not be treated as evidence for a functional WL pathway, or even a functional carbonyl branch (24).
Other than the presence of genes coding for the domain-specific subunits (AcsE for Bacteria and CdhB for Archaea), conserved sequence features also helped in the identification of archaeal and bacterial versions of the complex. For example, the bacterial-type CdhC contains a specific N-terminal domain that is not present in the archaeal counterpart (Fig. S2). Residues in this domain are known to form the hydrophobic channel connecting the active sites of CdhA and CdhC and also participate in the conformational change that allows the interaction of CdhC with the methyltransferase module (25). Conversely, the archaeal-type CdhD contains a specific N-terminal extension, which has no significant similarity to any sequence in current databases (Fig. S3).
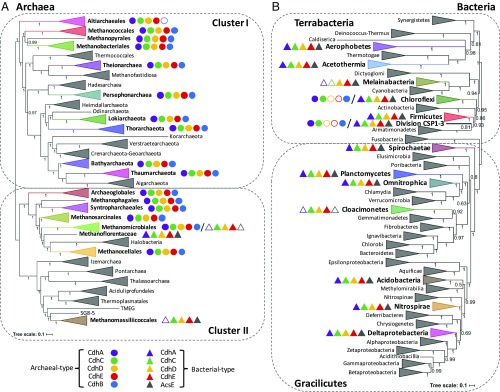
Mapping of CODH/ACS clusters on up-to-date reference phylogenies of the Archaea and Bacteria illustrates the wide distribution of this enzyme complex (Fig. 2 and Datasets S1 and S2). A complete and likely functional five-subunit enzyme complex appears to be a widespread characteristic of many novel archaeal lineages from various anaerobic ecosystems that have recently populated the archaeal tree, notably Cluster I Archaea (Fig. 2A) (26). Moreover, we report a complete CODH/ACS in the partial genome of an uncultured member of the Thaumarchaeota (RBG_16_49_8) (Fig. 2A and Dataset S1). This suggests a wider range of metabolic capabilities and role in biogeochemical cycles for this archaeal phylum, which is thought to fix carbon only through the 3-hydroxypropionate/4-hydroxybutyrate pathway (3, 27).
Fig. 2.
Distribution of complete or almost complete CODH/ACS complexes mapped on Bayesian reference phylogenies of Archaea (A) and Bacteria (B) based, respectively, on a concatenation of 41 markers (8,710-amino acid positions) (26), and from a concatenation of RNA polymerase subunits B, B′, and IF-2 (2,337-amino acid positions) (this work). Both trees were calculated in PhyloBayes, with the CAT+GTR+Γ4 model. Values at nodes represent Bayesian posterior probabilities. The scale bar represents the average number of substitutions per site. The archaeal and bacterial trees are rooted according to ref. 34, i.e., in Archaea between two large clades corresponding to cluster I and cluster II, and in Bacteria between two large clades roughly corresponding to Terrabacteria and Gracilicutes. Partial CODH/ACS clusters in one member of Hadesarchaea and one of Thaumarchaeota composed of only two of five subunits (CdhDE and CdhAB, respectively) have been omitted, but are included in Dataset S1. The clades corresponding to uncultured nanosized lineages in both Archaea [Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, Nanohaloarchaeota (DPANN)] and Bacteria [candidate phyla radiation (CPR)] were not included in these reference trees because they do not have CODH/ACS and their placement needs to be assessed with more in-depth analysis. For discussion and details on analyses, see main text and Methods.
The distribution of CODH/ACS in members of the Asgard superphylum is interesting to note, as they have been proposed to be the archaeal clade from which eukaryotes emerged (28). In particular, the presence of CODH/ACS in a Lokiarchaeota genome was recently used to support a hydrogen-dependent and autotrophic ancestor of eukaryotes (18). Although we found CODH/ACS in additional Lokiarchaeota and Thorarchaeota genomes (Dataset S1), no homologs could be identified in any other Asgard genome, in particular the Heimdallarchaeota, currently indicated as the closest relatives of eukaryotes (28).
The Altiarchaeales are the only archaea which appear to consistently lack a CdhB homolog, in agreement with previous reports (15) (Fig. 2A and Dataset S1). As Altiarchaeales may form a deep branch in the archaeal phylogeny (Fig. 2A) (26), their reduced four-subunit cluster might represent an ancestral version of the archaeal enzyme, with CdhB appearing later. It was theorized that the role of the CdhB subunit is to bind FAD as an alternative electron donor or for regulation of the overall CODH reaction (7); thus its origin might be related to further fine tuning of the enzyme during archaeal diversification. Alternatively, the Altiarchaeales might have specifically lost or replaced their CdhB subunit. Experimental characterization of CODH/ACS in this archaeal lineage will help to elucidate these two alternative scenarios.
Finally, Methanomassiliicoccales and Methanoflorentaceae have a complete or almost complete bacterial-type CODH cluster and lack an archaeal-type CODH altogether (Fig. 2A and Dataset S1). However, only Methanoflorens stordalenmirensis has a genuine CdhA, while phylogenetic analysis suggests that all homologs in Methanomassiliicoccales are indeed CooS (Fig. S1). Some Methanomicrobiales also harbor a partial bacterial-type CdhCDE cluster (Fig. 2A and Dataset S1), which might be functional, fixing carbon from CO instead of CO2.
Concerning Bacteria, complete and functional CODH/ACS complexes appear to be present in a wide range of additional lineages of ecological significance (Fig. 2B and Dataset S2). Acetothermia are a dominant taxon in subsurface environments and were suggested as the earliest bacterial lineage to possess the WL pathway (12). Aerophobetes have been mainly found in brackish lakes and cold seep environments (22), where they have been hypothesized to function as carbon recyclers, either taking advantage of saline sediment organic matter or fixing carbon under nutrient-poor conditions (29). Division CSP1–3 has been indicated as an important member of the carbon fixing community in subsurface aquifer ecosystems (23). We found a divergent CODH/ACS complex in a member of Cloacimonetes, a lineage of uncultured bacteria initially identified from a wastewater treatment plant anaerobic digester (30), and which has since been found also in marine sediments (e.g., ref. 22). Interestingly, we report a partial and divergent CODH/ACS, including only the methyltransferase module (AcsE–CdhDE), possibly the remnant of a complete cluster, in Melainabacteria. As this phylum is a close relative of Cyanobacteria (31), this might provide information on the emergence of oxygenic photosynthesis and the Calvin cycle of carbon fixation. Finally, we confirmed the presence of an archaeal-type oxidoreductase module (CdhABC) in the Firmicute Desulforudis audaxviator (32), in addition to its original bacterial version. Such a cluster also exists as the only complex in two members of Dehalococcoidia (Chloroflexi), as previously noted (33) (Fig. 2B and Dataset S2).
The wide distribution of CODH/ACS clusters and their strong conservation begs the question of the evolutionary history of this complex in Archaea and Bacteria. To avoid artifacts that may be introduced by the large evolutionary distance between archaeal and bacterial homologs, as well as the presence of interdomain transfers that might bias phylogenetic analysis, we chose to analyze separately the bacterial and archaeal CODH/ACS clusters.
Evolution of CODH/ACS in Archaea.
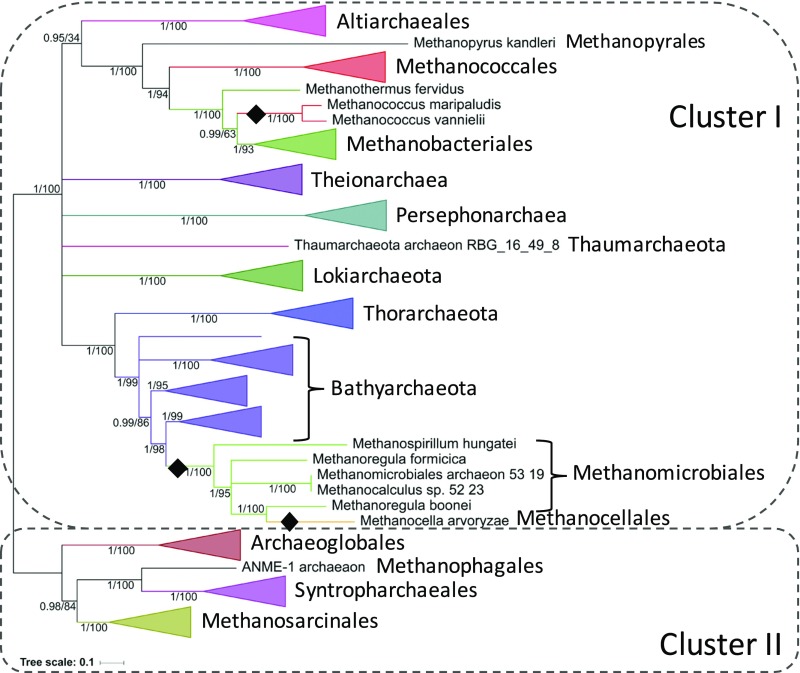
Single gene trees of the five individual subunits (CdhABCDE) did not show major incongruence and could be assembled into a concatenated dataset to increase the phylogenetic signal (Methods). Although not completely resolved, the resulting tree is largely consistent with the reference archaeal phylogeny, by recovering the monophyly of major orders and phyla and their relationships, as well as Cluster I and Cluster II Archaea (34) (Fig. 3). This suggests a mainly vertical inheritance of this enzyme, and by extension the likely presence of a five subunit CODH/ACS (CdhABCDE) in the ancestor of all present-day Archaea, significantly extending a recent analysis, which could only infer CdhC (14). Two likely intradomain transfer events can nevertheless be highlighted, which led to the replacement of the original CODH/ACS (indicated by black diamonds in Fig. 3). One of these likely occurred from Bathyarchaeota to some Methanomicrobiales and one Methanocellales and is interesting from an ecological point of view: at least Methanoregula boonei (35), Methanoregula formicica (36), and Methanocella arvoryzae (37) all require acetate as a carbon source for growth, and in principle do not utilize CODH/ACS to fix carbon. Their CODH/ACS could therefore be used in the catabolic direction or possibly to fix carbon under not yet explored conditions. On the other hand, one member of Methanomicrobiales (Methanospirillum hungatei) might be capable of carbon fixation, as it has been shown not to be dependent on acetate (38). Exploration of the function of these CODH/ACS complexes in both Methanomicrobiales and Bathyarchaeota will provide information on the direction of the reaction in these lineages.
Fig. 3.
Bayesian phylogeny based on a concatenation of the five subunits of the archaeal-type CODH/ACS (CdhABCDE, 2,063-aa positions), calculated by PhyloBayes, with the CAT+GTR+Γ4 model. Black diamonds indicate instances of intradomain transfers (see main text for discussion). Cluster I and cluster II refer to the clades identified in the reference archaeal phylogeny in Fig. 2A. Values at nodes are posterior probabilities and bootstrap supports calculated by maximum likelihood in IQTree under the TEST option (100 replicates). Nodes below 0.95 posterior probability support were collapsed. Scale bar represents the average number of substitutions per site. For details on analyses, see Methods.
The rare instances of bacterial-type CODH/ACS in a few archaea belonging to Methanomicrobiales, Methanoflorentaceae, and Methanomassiliicoccales (Fig. 2A and Dataset S1) are likely due to one or two ancient interdomain transfer events (Fig. S4). Methanomicrobiales are CO2 or formate-dependent hydrogenotrophic methanogens; Methanoflorentaceae are hydrogenotrophic methanogens but their carbon source is unknown (probably CO2); and Methanomassiliicoccales are methyl-dependent hydrogenotrophic methanogens. Contrary to M. stordalenmirensis, which acquired a complete bacterial type and is expected to perform the reaction normally, the inability of these Methanomicrobiales and Methanomassiliicoccales to fix carbon can be explained by the fact that they never acquired CdhA during these transfers. Instead, they only possess an independently acquired CooS, which might only be able to function in the oxidative direction (Fig. S1). However, this raises the question of what the function of the bacterial type CODH/ACS complex would be in these lineages. Funneling of acetyl-CoA into acetoclastic methanogenesis, as is the case of some Methanosarcinales, seems unlikely as none of those organisms is known to utilize acetate in that manner. Nonetheless, the consistent lack of CdhA opens another possibility, namely, carbon fixation by direct integration of CO through CdhC.
Modularity and Tinkering of CODH/ACS in Bacteria.
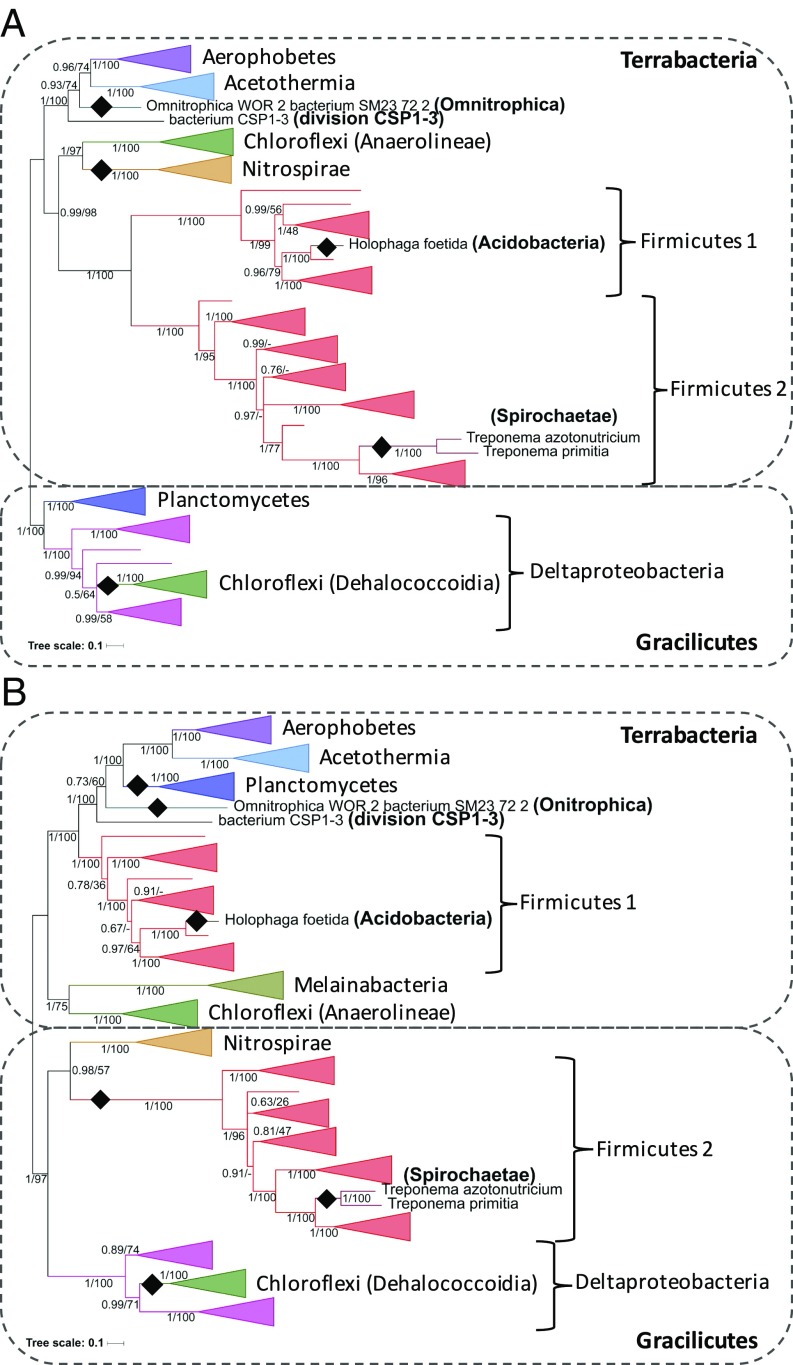
Compared to Archaea, the evolution of CODH/ACS follows a more complex path in Bacteria. The oxidoreductase module (CdhAC), and the methyltransferase module (AcsE–CdhDE) showed, in fact, partly different evolutionary histories and therefore their phylogenies had to be analyzed separately (Methods) (Fig. 4, respectively). Both are overall congruent with the reference phylogeny, notably by forming two clades roughly corresponding to the Terrabacteria and Gracilicutes groups (Fig. 4). This suggests that a five subunit CODH/ACS (AcsE–CdhACDE) would have been present in the ancestor of all present-day Bacteria. However, a few discrepancies are nevertheless visible in the evolutionary history of the two modules, due to both recent and more ancient transfers (indicated by black rectangles in Fig. 4).
Fig. 4.
Bayesian phylogeny based on a concatenation of (A) the two oxidoreductase module subunits (CdhAC, 1,248-aa positions) and (B) the three methyltransferase module subunits (AcsE-CdhDE, 965-aa positions) calculated in PhyloBayes, with the CAT+GTR+Γ4 model. Terrabacteria and Gracilicutes refer to the clades identified in the reference bacterial phylogeny in Fig. 2B. Black diamonds indicate instances of intradomain transfers: recent ones from Firmicutes to Acidobacteria, Spirochaetae, and from Deltaproteobacteria to Dehalococcoidia; and three more ancient ones from Chloroflexi (Anaerolineae) to Nitrospirae, and from Aerophobetes/Acetothermia to Plactomycetes/Omnitrophica. The direction of these transfers was deduced by comparing the reference and gene trees to identify misplaced clades (recipients). Values at nodes are posterior probabilities and bootstrap supports calculated by maximum likelihood. Scale bars represent the average number of substitutions per site. For discussion and details on analyses, see main text and Methods.
An interesting discrepancy concerns the Firmicutes: while they are monophyletic in the oxidoreductase module CdhAC tree (Fig. 4A), they are not in the tree based on the methyltransferase module AcsE–CdhDE (Fig. 4B), where a clade of Firmicutes (Firmicutes 2) is misplaced within the Gracilicutes clade and might have replaced their original copy with one obtained from the Nitrospirae. These different evolutionary histories might be linked to differences in physiology, energy conservation, and pathways related to the WL carbonyl branch, as observed in the model acetogens Moorella thermoacetica (Firmicutes 1), and Acetobacterium woodii (Firmicutes 2). In fact, M. thermoacetica (Firmicutes 1) and A. woodii (Firmicutes 2) belong to the groups of Ech and Rnf acetogens, respectively, according to their nonhomologous primary energy conservation complexes (39). The Proteobacterial homolog of Rnf, called Rsx, forms part of a system that senses oxidative stress (40, 41). Moreover, in a phylogeny of the Firmicutes with molecular dating, the divergence of Firmicutes 1 and Firmicutes 2 clades was shown to occur close to that of Bacilli, the main aerobic lineage of the Firmicutes, at 2,400 Ma, around the time of the Great Oxygenation Event (GOE) (42). Because Deltaproteobacteria and Nitrospirae might have also emerged after the GOE (43, 44), it could be speculated that Firmicutes 2 replaced their original methyltransferase module with one acquired from these lineages producing a CODH/ACS complex with higher tolerance to oxygen. Experimental work is required to explore this hypothesis.
Concerning the presence of archaeal-type CdhABC clusters in Dehalococcoidia (Chloroflexi) and D. audaxviator (Firmicutes) (Fig. 2B and Dataset S2), phylogenetic analysis suggests that these arose from a single horizontal transfer of the entire complex from an undetermined member of Methanogens class I to Bacteria and then subsequent transfers among bacterial lineages (Fig. S5A). Interestingly, a second acquisition of a CdhDE/AcsE module from Deltaproteobacteria led to the emergence of unique cases of hybrid archaeal/bacterial CODH/ACS enzyme complexes, including one with six subunits in D. audaxviator (Fig. S5 B and C). This shows that tinkering of a hybrid bacterial/archaeal CODH/ACS is a rare but possible event and may have been driven for stoichiometric and reaction optimization reasons, as D. audaxviator is extremely limited in its carbon sources (32). It will be extremely interesting to experimentally characterize these atypical CODH/ACS enzyme complexes, in particular the relative role of the two redundant AcsE (bacterial) and CdhB (archaeal) subunits.
A CODH/ACS in LUCA?
Current genomic data indicate that functional CODH/ACS complexes are much more widely distributed in Bacteria and Archaea than previously known. Such an extended sampling allowed us to clarify the evolutionary history of the CODH/ACS complex, which had been left unclear or partially resolved by previous analyses.
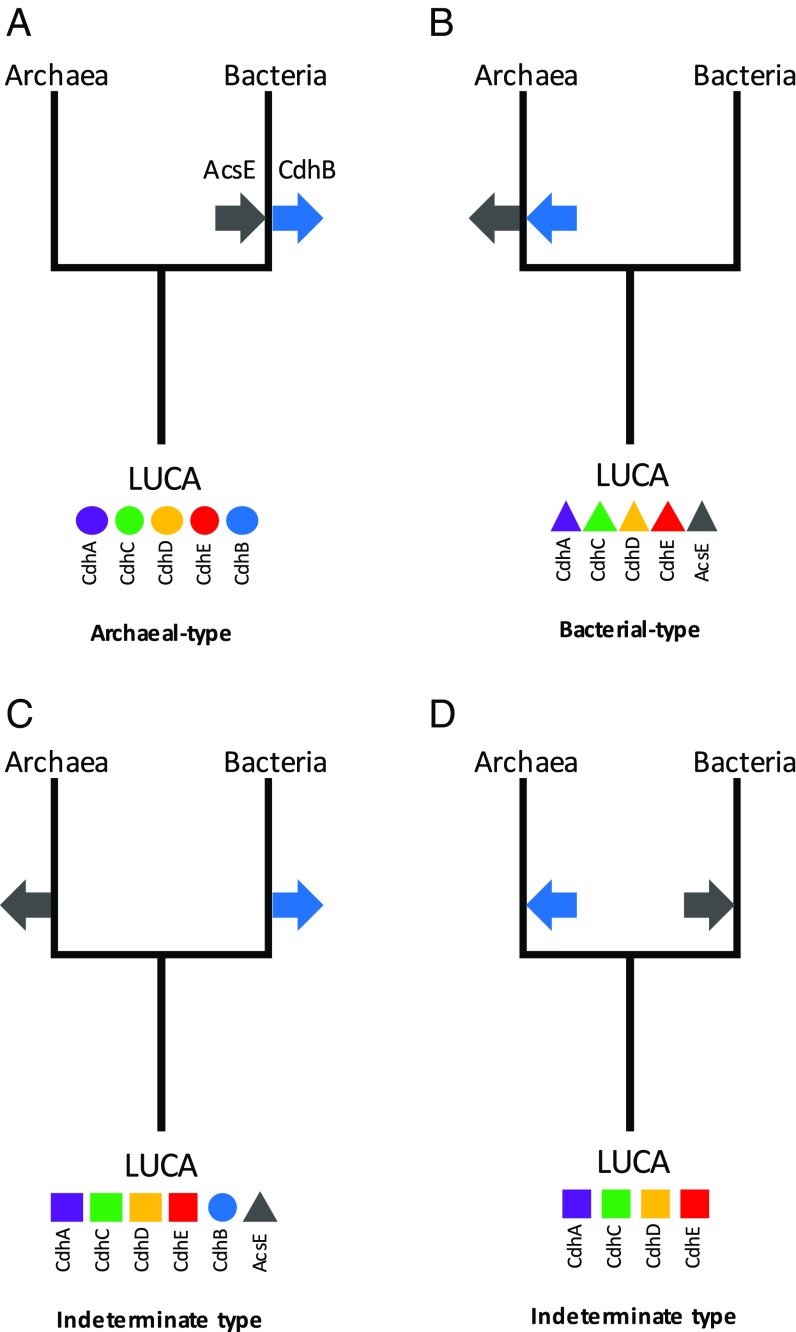
The striking conservation and largely vertical inheritance of CODH/ACS subunits strongly argues for the presence of a five-subunit complex in the ancestor of each domain (CdhABCDE in Archaea and AcsE–CdhACDE in Bacteria). By extension, this allows us to confidently infer the presence of a functional enzyme complex in the LUCA, consisting of at least the four common subunits CdhACDE. Nonetheless, the precise nature of the enzyme complex in the LUCA remains open to speculation, with four possible scenarios (Fig. 5). In the first scenario, LUCA had a modern archaeal-type five-subunit CODH/ACS, while Bacteria would have acquired AcsE and lost CdhB (Fig. 5A). In the second scenario, LUCA had a five-subunit bacterial version, and Archaea would have acquired CdhB and lost AcsE (Fig. 5B). In the third scenario, LUCA had a six-subunit CODH/ACS, similar to the present-day situation observed in Desulforudis, while AcsE would have been lost in Archaea and CdhB in Bacteria (Fig. 5C). In the fourth scenario, LUCA would have possessed a version of the complex with only the four common subunits (CdhACDE), while the specific archaeal (CdhB) and bacterial (AcsE) subunits were subsequent inventions (Fig. 5D).
Fig. 5.
Alternative scenarios for the nature of the CODH/ACS enzyme complex in LUCA and the transition to bacterial and archaeal types in the two domains (see main text for discussion). Inward facing arrows indicate emergence of a new subunit, and outward facing arrows indicate subunit loss.
This last scenario raises the important issue of whether such an ancestral complex possessed the capability to transfer methyl moieties into the WL carbonyl branch, particularly from the cofactors utilized in one of the two different methyl branch versions of the WL pathway (H4MPT and THF). As mentioned in the Introduction, while the archaeal CdhDE subcomplex contains a methyltransferase activity, in Bacteria the methyltransferase activity is carried out by the specific subunit AcsE. Therefore, if the four-subunit CODH/ACS in LUCA behaved like the archaeal type, the methyltransferase activity would have been embedded in the CdhDE subcomplex. Conversely, if CODH/ACS in LUCA behaved like the bacterial type, it can be asked whether AcsE could have been absent without compromising the activity of the enzyme. It has been recently shown in the Chloroflexi member Dehalococcoides mccartyi that the THF-corrinoid methyltransferase activity is present, despite the absence of an AcsE homolog. It was proposed that the activity could either be embedded in the rest of the CODH/ACS complex or performed by a different enzyme altogether (45). Furthermore, a CdhCDE (without AcsE) subcomplex of bacterial origin has been retained in Methanomicrobiales (see prior discussion) and is probably active. Therefore, it could be speculated that AcsE arose in Bacteria to more efficiently link the THF methyl branch with the carbonyl branch of the WL pathway. In any case, regardless of whether it resembled the modern bacterial or archaeal type, a four-subunit CdhACDE complex would have been sufficient for a functional WL carbonyl branch in the LUCA.
Another open question is the source of the methyl moiety, and by inference the lifestyle of the LUCA. In fact, a methyl moiety transferred from the H4MPT or THF version of the WL methyl branch, implying a fully carbon fixing/autotrophic lifestyle, is just one possibility. Alternatively, the ancestral CODH/ACS complex might have been independent of either methyl branch version, and AcsE or CdhDE might have been able to transfer methyl moieties from other sources e.g., methylamines, dimethylsulfide, methanol, methyl-CoM, chloromethane, etc., pointing to a mixotrophic LUCA. In fact, AcsE and CdhDE belong in the family of corrinoid methyltransferases, which includes methionine synthase (MetH), the H4MPT:CoM methyltransferase complex (MtrA-H) of methanogens, aromatic ether O-demethylase (MtvABC), the methylamine:CoM methyltransferases (Mtt, Mtb), methanol methyltransferase (Mta), and chloromethane:THF methyltransferase (CmuAB) (46, 47). All these enzymes are catalytically analogous and structurally very similar, composed of a TIM barrel and conserved active site residues. Some of them are also known to be promiscuous in their methyl source (48, 49). CODH/ACS in LUCA might have likewise been promiscuous in its use of methyl sources, as ancestral enzymes often have been found to possess a broader range of specificities (50, 51).
Finally, we would like to underline the fact that an ancestral presence of CODH/ACS does not by itself resolve the lifestyle of the LUCA. The ancient CODH/ACS might also have originally been unable to fix carbon and operated only catabolically (either alone or with a reverse methyl branch like for example in Archaeoglobales), consistently with a heterotrophic LUCA. Alternatively, it could have also switched back and forth from anabolic to catabolic function, depending upon the H2 levels, similarly to some present-day acetogens (39).
Characterization of CODH/ACS complexes from a larger spectrum of Bacteria and Archaea might eventually resolve this issue. At the same time, detailed evolutionary study of the two WL methyl branch versions will elucidate their antiquity and link with CODH/ACS.
Methods
Homology Searches.
A local database of genome sequences from 639 Archaea and 5,763 Bacteria was assembled, representing all genomes (one per species) present at the National Center for Biotechnology Information (NCBI) as of June 2016, and included some retroactively added archaea recently shown to possess a CODH/ACS, some genomes manually downloaded and curated from the Joint Genome Institute/Integrated Microbial Genomes (JGI/IMG (52) and the ggkbase (ggkbase.berkeley.edu/). Homology searches for the six subunits of CODH/ACS (CdhABCDE and AcsE) were performed as follows: from the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology page of each enzyme, five sequences from both Bacteria and Archaea, preferably from Swissprot and well annotated, were used to create a preliminary hidden Markov model (HMM) profile. This was used to perform hidden Markov model-based searches with HMMer (53) against the local genome database of Bacteria and Archaea separately. The first 20–30 hits in descending e-value order from each domain were used to create new domain-specific HMM profiles for each subunit (12 in total). In the new search, all hits with e-values lower than e−4 were kept and subjected to preliminary phylogenetic analysis to remove distant homologs, nearly identical copies arising from recent duplications and/or composite metagenomes. Synteny was manually checked, and only complete or almost-complete syntenic clusters (three to five subunits) were retained for further evolutionary analysis. Additional complete clusters present in some lineages resulting from recent duplications were not considered. For the analysis of CdhA/CooS, the same procedure was used, with the difference that homology searches against the local genome databases of Bacteria and Archaea were performed with both HMM profiles created for bacterial and archaeal CdhA, to find distant homologs.
Phylogenomic Analysis.
Single subunit datasets were aligned with MUSCLE (54) with default parameters and trimmed with BMGE 1.1 (55) using the BLOSUM30 substitution matrix to select unambiguously aligned positions. To check for phylogenetic congruence, single gene trees were calculated by IQTree under the TEST option with 100 bootstrap replicates (56) and collapsed at nodes showing <80% bootstrap support. These trees were tested for congruence against a strictly bifurcating tree of a blind concatenation, by performing an Internode Certainty (IC) test (57) in RaxML (58). Sequences responsible for incongruences at order level or above (in Archaea) or phylum level (in Bacteria) were removed and the procedure was repeated until no further incongruence was found. Incongruences were either due to horizontal gene transfer or to artifacts due to fast evolving or partial sequences. This led to three concatenated datasets with no more than 25% missing taxa per gene: CdhABCDE for Archaea (2,065-aa positions) and AcsE-CdhDE and CdhAC for Bacteria (962-aa positions and 1,250-aa positions, respectively). An additional four datasets were created to investigate interdomain and intradomain transfers that gave rise to hybrid clusters: AcsE–CdhDE and CdhC for Bacteria and interdomain transfers to Archaea (949- and 645-aa positions, respectively), CdhABC for Archaea and interdomain transfers to Bacteria (1,282-aa positions), and AcsE–CdhDE for Bacteria and intradomain hybrid transfers (955-aa positions). Finally, a dataset for CdhA/CooS from both Bacteria and Archaea was created (420-aa positions). Phylogenies from all concatenated datasets were inferred in IQTree under the TEST option with 100 bootstrap replicates and in PhyloBayes under the CAT+GTR+Γ4 model (with the exception of the Bacteria+interdomain transfers single-gene CdhC dataset where LG+Γ4 was used). For Bayesian analyses, four independent Markov chain Monte Carlo chains were run until convergence and checked by sampling every two cycles with a 25% burn-in. Support at nodes was evaluated by posterior probability values.
For the reference phylogeny of Archaea, we used a supermatrix of 41 genes (8,710-aa positions) from ref. 26. For the reference phylogeny of Bacteria, we used a concatenation of RNA polymerase subunits B, B′, and IF-2 (2,337-aa positions). Bayesian phylogenies were constructed in PhyloBayes (59) under the CAT+GTR+Γ4 model. Trees were rooted according to ref. 34.
Supplementary Material
Acknowledgments
We thank Alexander Probst for feedback on an earlier version of the paper, and two anonymous reviewers whose comments helped improve clarity and correctness of the manuscript. P.S.A. is supported by a PhD fellowship from Paris Diderot University and by funds from the PhD Programme “Frontières du Vivant (FdV)–Programme Bettencourt.” G.B. is a recipient of a Roux-Cantarini fellowship from the Institut Pasteur. S.G. acknowledges funding from the French National Agency for Research Grant ArchEvol (ANR-16-CE02-0005-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716667115/-/DCSupplemental.
References
- 1.Fuchs G. Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 2.Peretó J. Out of fuzzy chemistry: From prebiotic chemistry to metabolic networks. Chem Soc Rev. 2012;41:5394–5403. doi: 10.1039/c2cs35054h. [DOI] [PubMed] [Google Scholar]
- 3.Berg IA, et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 4.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Möller-Zinkhan D, Thauer RK. Anaerobic lactate oxidation to 3 CO2 by Archaeoglobus fulgidus via the carbon monoxide dehydrogenase pathway: Demonstration of the acetyl-CoA carbon-carbon cleavage reaction in cell extracts. Arch Microbiol. 1990;153:215–218. [Google Scholar]
- 6.Techtmann SM, et al. Evidence for horizontal gene transfer of anaerobic carbon monoxide dehydrogenases. Front Microbiol. 2012;3:132. doi: 10.3389/fmicb.2012.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong W, et al. Structure of the α2ε2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarbonylase/synthase complex. Proc Natl Acad Sci USA. 2008;105:9558–9563. doi: 10.1073/pnas.0800415105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grahame DA, DeMoll E. Partial reactions catalyzed by protein components of the acetyl-CoA decarbonylase synthase enzyme complex from Methanosarcina barkeri. J Biol Chem. 1996;271:8352–8358. doi: 10.1074/jbc.271.14.8352. [DOI] [PubMed] [Google Scholar]
- 9.Kung Y, et al. Visualizing molecular juggling within a B12-dependent methyltransferase complex. Nature. 2012;484:265–269. doi: 10.1038/nature10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss MC, et al. The physiology and habitat of the last universal common ancestor. Nat Microbiol. 2016;1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 11.Lindahl PA, Chang B. The evolution of acetyl-CoA synthase. Orig Life Evol Biosph. 2001;31:403–434. doi: 10.1023/a:1011809430237. [DOI] [PubMed] [Google Scholar]
- 12.Takami H, et al. A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS One. 2012;7:e30559. doi: 10.1371/journal.pone.0030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becerra A, Rivas M, García-Ferris C, Lazcano A, Peretó J. A phylogenetic approach to the early evolution of autotrophy: The case of the reverse TCA and the reductive acetyl-CoA pathways. Int Microbiol. 2014;17:91–97. doi: 10.2436/20.1501.01.211. [DOI] [PubMed] [Google Scholar]
- 14.Williams TA, et al. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc Natl Acad Sci USA. 2017;114:E4602–E4611. doi: 10.1073/pnas.1618463114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Probst AJ, et al. Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat Commun. 2014;5:5497. doi: 10.1038/ncomms6497. [DOI] [PubMed] [Google Scholar]
- 16.Lazar CS, Baker BJ, Seitz KW, Teske AP. Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches. ISME J. 2017;11:1118–1129. doi: 10.1038/ismej.2016.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwirichia R, et al. Metabolic traits of an uncultured archaeal lineage–MSBL1–From brine pools of the Red Sea. Sci Rep. 2016;6:19181. doi: 10.1038/srep19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF. Lokiarchaeon is hydrogen dependent. Nat Microbiol. 2016;1:16034. doi: 10.1038/nmicrobiol.2016.34. [DOI] [PubMed] [Google Scholar]
- 19.Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 2016;10:1696–1705. doi: 10.1038/ismej.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans PN, et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 21.Laso-Pérez R, et al. Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature. 2016;539:396–401. doi: 10.1038/nature20152. [DOI] [PubMed] [Google Scholar]
- 22.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 23.Hug LA, et al. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol. 2016;18:159–173. doi: 10.1111/1462-2920.12930. [DOI] [PubMed] [Google Scholar]
- 24.Giovannelli D, et al. Insight into the evolution of microbial metabolism from the deep-branching bacterium, Thermovibrio ammonificans. Elife. 2017;6:e18990. doi: 10.7554/eLife.18990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Can M, Armstrong FA, Ragsdale SW. Structure, function, and mechanism of the nickel metalloenzymes, CO dehydrogenase, and acetyl-CoA synthase. Chem Rev. 2014;114:4149–4174. doi: 10.1021/cr400461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adam PS, Borrel G, Brochier-Armanet C, Gribaldo S. The growing tree of archaea: New perspectives on their diversity, evolution and ecology. ISME J. 2017;11:2407–2425. doi: 10.1038/ismej.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pester M, Schleper C, Wagner M. The Thaumarchaeota: An emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol. 2011;14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaremba-Niedzwiedzka K, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Draft genome of an Aerophobetes bacterium reveals a facultative lifestyle in deep-sea anaerobic sediments. Sci Bull (Beijing) 2016;61:1176–1186. [Google Scholar]
- 30.Pelletier E, et al. “Candidatus Cloacamonas acidaminovorans”: Genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol. 2008;190:2572–2579. doi: 10.1128/JB.01248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Rienzi SC, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife. 2013;2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chivian D, et al. Environmental genomics reveals a single-species ecosystem deep within Earth. Science. 2008;322:275–278. doi: 10.1126/science.1155495. [DOI] [PubMed] [Google Scholar]
- 33.Wasmund K, et al. Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J. 2014;8:383–397. doi: 10.1038/ismej.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raymann K, Brochier-Armanet C, Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci USA. 2015;112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bräuer SL, Cadillo-Quiroz H, Ward RJ, Yavitt JB, Zinder SH. Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol. 2011;61:45–52. doi: 10.1099/ijs.0.021782-0. [DOI] [PubMed] [Google Scholar]
- 36.Yashiro Y, et al. Methanoregula formicica sp. nov., a methane-producing archaeon isolated from methanogenic sludge. Int J Syst Evol Microbiol. 2011;61:53–59. doi: 10.1099/ijs.0.014811-0. [DOI] [PubMed] [Google Scholar]
- 37.Sakai S, Conrad R, Liesack W, Imachi H. Methanocella arvoryzae sp. nov., a hydrogenotrophic methanogen isolated from rice field soil. Int J Syst Evol Microbiol. 2010;60:2918–2923. doi: 10.1099/ijs.0.020883-0. [DOI] [PubMed] [Google Scholar]
- 38.Ferry JG, Wolfe RS. Nutritional and biochemical characterization of Methanospirillum hungatii. Appl Environ Microbiol. 1977;34:371–376. doi: 10.1128/aem.34.4.371-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuchmann K, Müller V. Autotrophy at the thermodynamic limit of life: A model for energy conservation in acetogenic bacteria. Nat Rev Microbiol. 2014;12:809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- 40.Koo MS, et al. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biegel E, Schmidt S, González JM, Müller V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci. 2011;68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Letelier A, Olmedo-Alvarez G, Eguiarte LE, Souza V. Divergence and phylogeny of Firmicutes from the Cuatro Ciénegas basin, Mexico: A window to an ancient ocean. Astrobiology. 2012;12:674–684. doi: 10.1089/ast.2011.0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Battistuzzi FU, Hedges SB. A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol. 2009;26:335–343. doi: 10.1093/molbev/msn247. [DOI] [PubMed] [Google Scholar]
- 44.Thomas SH, et al. The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria. PLoS One. 2008;3:e2103. doi: 10.1371/journal.pone.0002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhuang W-Q, et al. Incomplete Wood-Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc Natl Acad Sci USA. 2014;111:6419–6424. doi: 10.1073/pnas.1321542111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vannelli T, Messmer M, Studer A, Vuilleumier S, Leisinger T. A corrinoid-dependent catabolic pathway for growth of a Methylobacterium strain with chloromethane. Proc Natl Acad Sci USA. 1999;96:4615–4620. doi: 10.1073/pnas.96.8.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee R, Ragsdale SW. The many faces of vitamin B12: Catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 48.Becher B, Müller V, Gottschalk G. The methyl-tetrahydromethanopterin: coenzyme M methyltransferase of Methanosarcina strain Gö1 is a primary sodium pump. FEMS Microbiol Lett. 1992;91:239–243. doi: 10.1128/jb.174.23.7656-7660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vepachedu VR, Ferry JG. Role of the fused corrinoid/methyl transfer protein CmtA during CO-dependent growth of Methanosarcina acetivorans. J Bacteriol. 2012;194:4161–4168. doi: 10.1128/JB.00593-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler LC, Lim SA, Marqusee S, Harms MJ. The thermostability and specificity of ancient proteins. Curr Opin Struct Biol. 2016;38:37–43. doi: 10.1016/j.sbi.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen IA, et al. IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Criscuolo A, Gribaldo S. BMGE (Block mapping and gathering with entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobert K, Salichos L, Rokas A, Stamatakis A. Computing the internode certainty and related measures from partial gene trees. Mol Biol Evol. 2016;33:1606–1617. doi: 10.1093/molbev/msw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 59.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.