Significance
Natural killer T (NKT) cells are selected by CD1d molecules in the thymus. Two homologous genes in the mouse genome encode for CD1d proteins. The two CD1d isoforms are not equivalent in their lipid antigen presentation capabilities, affecting the development and the T cell antigen receptor repertoire of NKT cells.
Keywords: CD1, MHC, NKT
Abstract
MHC class I-like CD1 molecules have evolved to present lipid-based antigens to T cells. Differences in the antigen-binding clefts of the CD1 family members determine the conformation and size of the lipids that are presented, although the factors that shape CD1 diversity remain unclear. In mice, two homologous genes, CD1D1 and CD1D2, encode the CD1d protein, which is essential to the development and function of natural killer T (NKT) cells. However, it remains unclear whether both CD1d isoforms are equivalent in their antigen presentation capacity and functions. Here, we report that CD1d2 molecules are expressed in the thymus of some mouse strains, where they select functional type I NKT cells. Intriguingly, the T cell antigen receptor repertoire and phenotype of CD1d2-selected type I NKT cells in CD1D1−/− mice differed from CD1d1-selected type I NKT cells. The structures of CD1d2 in complex with endogenous lipids and a truncated acyl-chain analog of α-galactosylceramide revealed that its A′-pocket was restricted in size compared with CD1d1. Accordingly, CD1d2 molecules could not present glycolipid antigens with long acyl chains efficiently, favoring the presentation of short acyl chain antigens. These results indicate that the two CD1d molecules present different sets of self-antigen(s) in the mouse thymus, thereby impacting the development of invariant NKT cells.
CD1 molecules are glycoproteins that adopt an MHC class I-like structure (1). In contrast to the polymorphic MHC molecules that present peptides, CD1 molecules exhibit limited polymorphism and are ideally suited to present lipid-based antigens (2). Based on sequence homology, function, and tissue distribution, the CD1 family has been classified into three distinct groups: group I, includes CD1a, -b, and -c; group II consists of CD1d, and group III is composed of CD1e (3). The number of CD1 isoforms expressed in any given mammal varies considerably, however. For example, humans express all five CD1 isoforms, rabbits lack CD1c, and mice express only the CD1d protein. However, the basis for such varied usage of genes within the CD1 locus across mammals is largely unknown, but presumably reflects important functional differences related to pathogen-driven evolutionary pressures.
Lipid antigen-loaded CD1d molecules are recognized by a population of innate-like T lymphocytes, the natural killer T (NKT) cells. Two broad classes of NKT cells, termed type I and type II, have been defined on the basis of T cell antigen receptor (TCR) expression and antigen reactivity (4). Type I NKT cells or invariant NKT cells (iNKT) typically express a TCR that is the product of a canonical rearrangement between the Vα14 gene segment (Vα24 in human) and frequently the Jα18 gene segment, with a complementarity determining region (CDR)-3α invariant at the amino acid level (5, 6). In mice, the Vα14 invariant chain is coexpressed with a limited set of Vβ chains, predominantly Vβ8.2, Vβ7, and Vβ2, with variable CDR3β loop sequences (7). Essentially all of these cells are responsive to the prototypical antigen α-galactosylceramide (αGC) when presented by CD1d molecules, irrespective of their TCRβ repertoire diversity (8–10). How variation in CD1d itself impacts on type I NKT cell repertoire and function remains unclear, however.
Upon antigenic stimulation, type I NKT cells respond rapidly by producing Th1, Th2, and Th17 cytokines, and can also be cytotoxic (11, 12). Type I NKT cells can be activated by bacterial ligands, and sense changes in host lipid metabolism through recognition of transiently expressed endogenous lipids (13–15). Chronic stimulation by these “autoantigens” leads some type I NKT cells to produce cytokines at steady state, which, in turn, influences the development and activation of surrounding cells (16, 17). Thus, their unique properties establish type I NKT cells as important regulators of immune responses and tissue homeostasis (18).
In mice, two genes encoding the CD1d isoform have been identified, CD1D1 and CD1D2. These two genes are located on chromosome 3 and are approximately 9 kb apart, arranged in opposite transcriptional orientation (19). The two genes share 95% sequence homology with each other, and are likely the product of gene duplication (2, 20). Mouse CD1d molecules are expressed primarily by cells of the hematopoietic lineage, including B and T cells, macrophages, and dendritic cells (21, 22). At the transcriptional level, CD1D1 is expressed at higher levels than CD1D2 in all tissues tested except the thymus, where the mRNA levels appear to be equal for both genes (19). Of the 17 amino acid differences between CD1d1 and CD1d2 proteins, 9 occur within the antigen-binding cleft, which includes a Cys168→Trp168 mutation. The cysteine 168 residue participates in an intradomain disulfide bond that is highly conserved in classical and nonclassical MHC I molecules (1, 19, 23), and is thought to be critical for the folding of MHC-I (24, 25). The impact of this mutation on CD1d2 structure and function is unknown. Interestingly, the bovine CD1d gene (26) and the primordial CD1-encoding gene found in chickens (27) also contain an amino acid change at position 168. This change was previously shown to affect the nature of the antigens presented by these molecules (2). Intriguingly, rats possess only one CD1D gene, which, based on sequence homology, more resemble mouse CD1D1 than CD1D2, with a cysteine at position 168 (28). It remains unclear what advantage, if any, the gene duplication event that led to the existence of CD1D2 in the mouse might be.
The development of type I NKT cells requires expression of CD1d by cortical thymocytes (29) and is impaired in mice lacking CD1d expression. Interestingly, even though some targeting strategies led to the disruption of both CD1D genes (30, 31), one particular strain of CD1D-deficient mice, which was generated by using embryonic stem cells of 129/Sv origin and has been largely distributed in the scientific community and used in multiple studies, disrupted only the CD1D1 gene (32). Furthermore, sequencing of the CD1D2 gene from C57BL/6 mice revealed a frame-shift mutation at the beginning of the fourth exon encoding the α3 domain, thereby abolishing CD1d2 protein expression in this strain (22). Altogether, these results contributed to the assumption that CD1d2 molecules do not play any role in the development of type I NKT cells. Earlier studies that identified type I NKT cells by using costaining for TCRβ chains and the NK1.1 marker concurred with this notion (33). However, it remains unknown whether the CD1D2 gene is mutated in other strains of mice and whether its gene product might play a role in the selection and function of iNKT cells.
Here we report that CD1d2 molecules are expressed in the thymus of CD1D1−/−-deficient mice (32), where they select functional type I NKT cells. High-throughput sequencing of CD1D gene transcripts from the thymus of BALB/c mice showed a transcript ratio of CD1D1:CD1D2 close to 1:1, supporting the possibility for a role of CD1d2 in shaping the development of iNKT cells. The CD1d2 crystal structures revealed that CD1d2 adopts an overall architecture similar to CD1d1 and that the Cys168Trp mutation does not collapse the antigen-binding groove. However, the CD1d2 A′-pocket was markedly restricted in size, thereby favoring the loading of lipid antigens with shorter acyl-chain length, as shown by the structure of CD1d2 in complex with a truncated acyl chain (C10) analog of α-GC. Collectively, our results demonstrate that CD1d2 is expressed in the thymus of some mouse strains, where it likely presents a different repertoire of self-antigens than CD1d1 and can thereby impact the selection of iNKT cells and their function.
Results
Vα14-Jα18 TCR Usage in Conventional CD4+ and CD8+ T Cells from CD1D1−/− Mice.
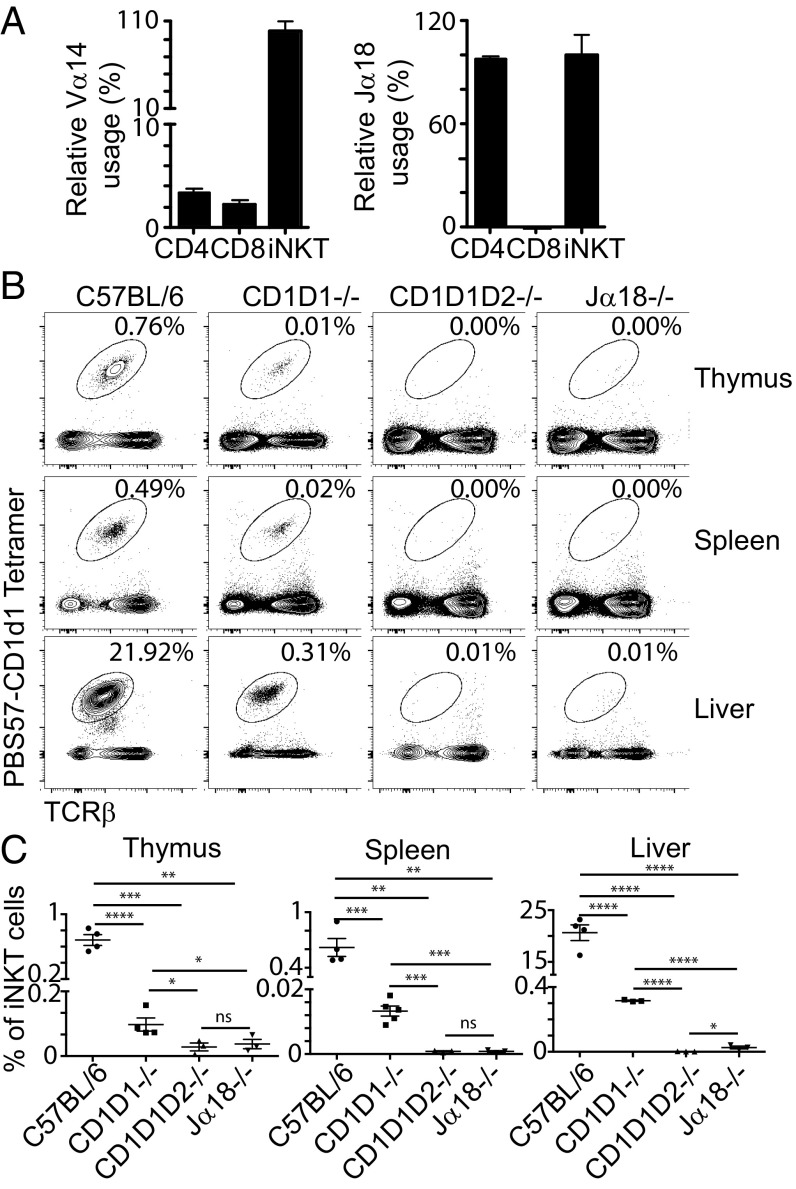
We wanted to determine the frequency of the Vα14–Jα18 rearrangement within conventional CD4+ and CD8+ TCRβ+ T cells. To ensure that CD4+ and CD8+ cell populations would be devoid of any potentially contaminating iNKT cells, we sorted cells from the spleens of C57BL/6 CD1D1−/− TCRCα+/− mice by flow cytometry. The frequency of gene usage was measured by quantitative PCR and normalized to the levels found in PBS57-CD1d tetramer+ TCRβ+ cells sorted from the spleen of CD1D1+/− TCRCα+/− mice. The strategy of amplification is depicted in SI Appendix, Fig. S1. As seen in Fig. 1A, 2–3% of conventional T cells use the Vα14 gene segment. Conventional splenic CD8+ T cells that use the Vα14 gene segment appear to only very rarely rearrange with the Jα18 gene segment, as revealed by the use of a Jα18-specific probe. Surprisingly, the situation appeared different for CD4+ T cells, in which the results suggested that all Vα14 segments were rearranged with Jα18, like in iNKT cells. Sequencing of the PCR product showed that it corresponded to the canonical Vα14–Jα18 iNKT rearrangement. These perplexing results suggested that, within the CD4+ T cells derived from CD1D1-deficient mice, Vα14 might have preferentially rearranged with Jα18, contradicting previous findings showing that Vα14 can rearrange with all Jα segments (34, 35). An alternative explanation could be provided if some residual iNKT cells were still present in the sample of the sorted CD4+ T cells from CD1D1−/− TCRCα+/− mice. Indeed, upon careful flow cytometry analysis involving the acquisition of several millions of events, we clearly detected a population of PBS57-CD1d1 tetramer+ cells in the thymus, spleen, and liver of CD1D1−/− mice (Fig. 1B). This population was absent from the organs of CD1D1/CD1D2−/− mice and greatly decreased in Jα18−/− mice, which lack the invariant Traj18 gene segment (36, 37). The few cells that were labeled in Jα18−/− mice likely correspond to the atypical iNKT cells previously identified in that strain (37, 38). These results contrast with previous experiments that examined the presence of NKT cells in CD1D1−/− mice by using TCRβ+ and NK1.1+ costaining and concluded that CD1d2 expression could not rescue NKT cell development (33). Instead, our results demonstrate that, even though the proportion of iNKT cells in CD1D1−/− mice is only 1–4% of the iNKT cells found in the organs of C57BL/6 (B6) mice (Fig. 1C), CD1D1−/− mice should not be considered as iNKT cell-deficient, and CD1d2 molecules can potentially select for iNKT cells.
Fig. 1.
Frequency of Vα14 usage and Vα14-Jα18 rearrangement usage among CD4+ and CD8+ T cells purified from the spleen of CD1D1−/− mice and frequency of iNKT cells in CD1D1−/− mice. (A) Relative frequency of Vα14 and Vα14-Jα18 usage in CD4+ and CD8+ T cells from CD1D1−/− mice. The frequency was normalized to the amount of the various PCR products found in iNKT cells sorted from the spleen of CD1D1+/− TCRα+/− mice, which was set at 100%. Results are shown as mean ± SD. (B) Cells from the thymus, spleen, and liver of 6–8-wk-old C57BL/6, CD1D1−/−, CD1D1D2−/−, and Jα18−/− mice were stained with CD1d1 tetramers loaded with PBS57 (PBS57-CD1d), and TCRβ mAbs and analyzed by flow cytometry (data representative of n ≥ 3 per group across two experiments). The percentage of iNKT cells in each sample is shown. (C) Summary of the data shown in B with percentage of iNKT cells in thymus, spleen, and liver of 6–8-wk-old C57BL/6, CD1D1−/−, CD1D1D2−/−, and Jα18−/− mice (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant).
The CD1D2 Sequence from CD1D1−/− Mice and the WT BALB/c Strain Is Functional.
B6 mice harbor a point mutation in exon 4 of CD1D2, which encodes the α3 domain (22). This frame-shift mutation introduces a stop codon, abolishing surface expression of the molecule. In the 129/sv strain, however, CD1D2 does not contain this mutation (22). To confirm that CD1D2 in the CD1D1−/− mice (32) is not the pseudogene found in the B6 mice, we designed primers that span part of exon 4 and the adjoining intronic area. Genomic DNA from CD1D1−/− mice, B6 mice, and another WT strain, BALB/c, was purified and used for amplification of the exon 4 fragment. Sequencing results confirmed that, whereas the B6-derived sequence contains a stop codon in exon 4, BALB/c and 129/Sv-derived CD1D1−/− mice had a functional CD1D2 sequence. These results demonstrate that no cross-over of the CD1 locus ever occurred during the backcross of the CD1D1−/− mice to B6 mice and that the CD1 locus is of 129/sv origin in these mice. Furthermore, these results suggest that CD1d2 molecules have the potential to be expressed in BALB/c mice.
CD1d2 Protein Is Expressed in the Thymus but Not the Periphery of CD1D1−/− Mice.
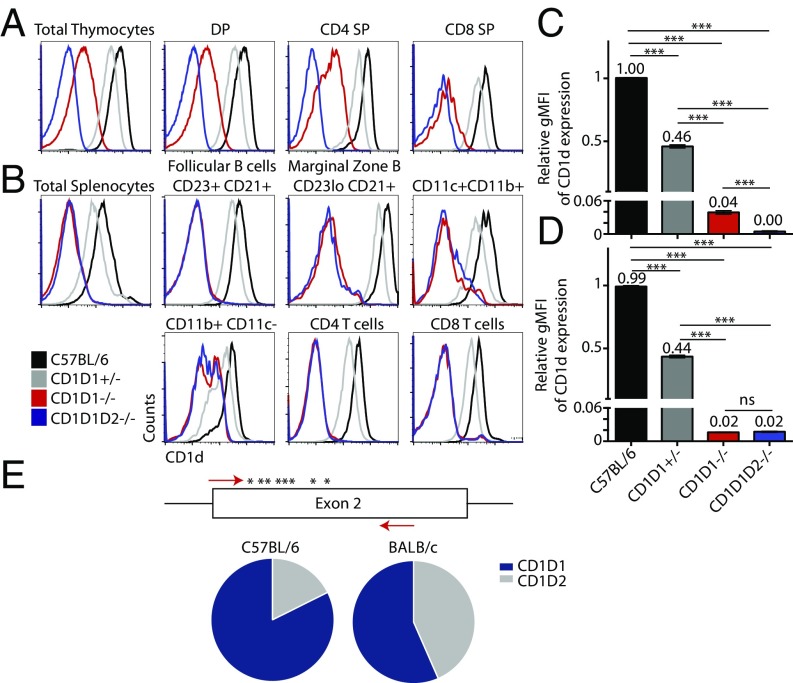
Because no serological reagent currently exists to discriminate CD1d1 from CD1d2 proteins, we first examined CD1d2 expression on cells derived from B6 CD1D1−/− mice. Controls were provided by staining cells from double-deficient CD1D1/CD1D2 mice (30, 33) and heterozygous CD1D1+/− mice generated by crossing CD1D1−/− mice with B6 mice. In this latter strain, the CD1d protein levels represent the combined gene activity of a single copy of CD1D1 B6 allele (because CD1D2 is mutated in B6) and a single copy of the functional CD1D2 allele from the KO mice (because CD1D1 has been knocked out in these mice and the CD1D2 allele is of 129/Sv origin). As seen in Fig. 2, we observed a low level of CD1d expression in the thymus of CD1D1−/− mice, attributable to CD1d2 expression (Fig. 2A). In particular, whereas CD1D1+/− thymocytes displayed haploinsufficient patterns, expressing half the amount of CD1d found on thymocytes from WT mice [as measured by geometric mean fluorescence intensity (gMFI)], the CD1D1−/− thymocytes expressed only 1/25 of the level expressed in WT mice (Fig. 2C). Interestingly, this expression was limited to the thymus, in agreement with previous results (33). In the periphery, we observed no protein expression of CD1d2 on whole splenocytes or individual splenic populations (Fig. 2 B and D).
Fig. 2.
CD1D1−/− mice express low amounts of CD1d molecules on the surface of hematopoietic cells of the thymus, but not the periphery. (A) Flow-cytometric analysis of CD1d levels of thymic DP (CD4+CD8+), CD4, and CD8 subsets from C57BL/6 (black), CD1D1+/− (gray), CD1D1−/− (red), and CD1D1D2−/− (blue) mice. (B) Flow-cytometry analysis of CD1d levels of spleen T and B subsets derived from the aforementioned mouse strains. (C) Bars represent the relative gMFI of CD1d expression on total thymocytes, normalized to C57BL/6. Bars depict mean ± SEM of at least three mice per group across at least two experiments (***P < 0.001 by Student t test). (D) Bars represent the relative gMFI of CD1d expression on total splenocytes, normalized to C57BL/6. Bars depict mean ± SEM of at least three mice per strain across at least two experiments. (E) Ratio of CD1D1/CD1D2-encoding transcripts in the thymus of B6 and BALB/c mice. Primers containing specific Ion Torrent tags were designed against segments identical in exon 2 of CD1D1 and CD1D2 genes. CD69−CD4+CD8+ preselection DP thymocytes were sorted from three independent B6 and three independent BALB/c mice, and mRNA was extracted and transformed into cDNA. CD1d exon sequence was amplified, and PCR products were sequenced by using high-throughput sequencing. Transcript identity was determined based on an 8-nt polymorphism (asterisk) that exists between the two genes in this exon. The ratio of CD1D1 and CD1D2 mRNA expression was significantly different between the two strains (P < 0.0001).
Early experiments with the use of RNase protection assays suggested that CD1D2-encoding transcripts were equally abundant compared with CD1D1 transcripts in BALB/c thymocytes (19). To verify these findings, we generated cDNA from B6 and BALB/c DP thymocytes and amplified a portion of exon 2 by using primers targeting identical sequences between CD1D1 and CD1D2 genes. This segment of DNA, however, contains eight nucleotide polymorphisms between the two genes. PCR products were sequenced by using high-throughput sequencing, and the relative expression of CD1D1 and CD1D2 transcripts was determined in each strain. The transcript ratio of CD1D1 to CD1D2 was overwhelmingly skewed toward CD1D1 in B6, with an expression ratio close to 5:1. This likely reflects the degradation of CD1D2-encoding mRNA as a result of nonsense-mediated mRNA decay (39). In BALB/c, however, the ratio of CD1D1:CD1D2 is close to 1:1 (Fig. 2E). Altogether, these results demonstrate that CD1d2 molecules can be expressed in the thymus of mice with a functional CD1D2 gene.
Development and Repertoire of CD1d2-Selected iNKT Cells.
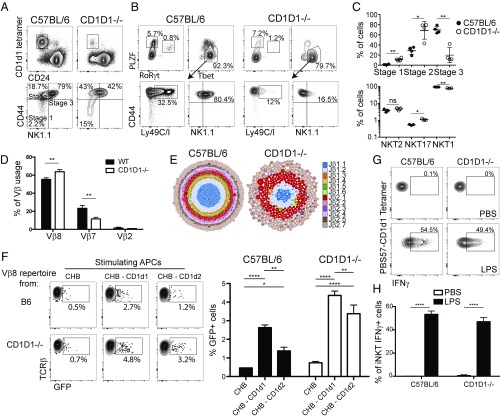
We enriched PBS57-CD1d1+ cells from the thymus of B6 and CD1D1−/− mice by using magnetic beads and analyzed the maturation of iNKT cells by using the cell-surface markers CD44 and NK1.1 to follow their maturation progress from stages 1 to 3 (40, 41). CD1d2-selected iNKT cells in CD1D1−/− mice appeared less mature compared with CD1d1-selected iNKT cells from B6 mice (Fig. 3A). Indeed, the majority of iNKT cells in the CD1D1−/− mice were of the more immature stage 1 (CD44−NK1.1−) or stage 2 (CD44+NK1.1−) phenotype compared with WT mice, in which approximately 80% of cells are the most mature stage 3 (CD44+NK1.1+) phenotype (Fig. 3C). However, the absolute number of iNKT stages detected in CD1D1−/− thymi was significantly lower than that observed in control thymi as a result of reduced total iNKT cell numbers in the CD1D1−/− mice. Because analysis of iNKT cells by staging is insufficient, as populations within each stage are heterogeneous (16, 42), we also characterized the CD1d2-selected iNKT cells by their expression of the transcription factors PLZF, RORγt, and Tbet to define terminally differentiated effector cells. Compared with the iNKT subset makeup found in the B6 thymus, iNKT cells in the thymus of CD1D1−/− mice showed a trending increase in the iNKT2 (PLZFhi, Tbetlo, RORγtlo) and iNKT17 (PLZFmid, Tbetlo, RORγthi) populations and a concomitant decrease in iNKT1 (PLZFlo, Tbethi, RORγtlo) cells (Fig. 3 B and C). Interestingly, even though the vast majority of the CD1d2-selected iNKT cells are of the iNKT1 phenotype (i.e., PLZFlo Tbethi), these cells are mostly NK1.1− and lack expression of the NK receptors Ly49C/I, in striking contrast with the same population of iNKT1 cells in B6 mice (Fig. 3B). This might have contributed to their lack of detection in previous studies (33).
Fig. 3.
Development and repertoire of CD1d2-selected iNKT cells. PBS57-CD1d1 tetramer-reactive thymocytes were enriched from C57BL/6 and pooled CD1D1−/− thymi (≥5 mice per sample) by using MACS beads. Enriched cells were stained with indicated markers and assessed by flow cytometry to characterize specific stages of iNKT-cell development (A) (data representative of n ≥ 3 per group) or functional subsets of iNKT cells as defined by the expression of the transcription factors PLZF, RoRγt, and Tbet (B). (C) Graphs depict mean ± SEM of A and B (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant). (D) PBS57-CD1d1 tetramer-reactive thymocytes were enriched by MACS beads from individual B6 or pooled CD1D1−/− thymi (≥5 mice per sample). Enriched cells were stained with PBS57-CD1d1 tetramers, TCRβ, Vβ8, Vβ7, and Vβ2 mAbs and assessed by flow cytometry to characterize Vβ usage (data representative of n ≥ 3 samples per group). Bars depict mean ± SEM of percentage of iNKT cells expressing each Vβ (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant). (E) Visual representation of Vβ8 CDR3 sequences from sorted iNKT cells of B6 (Left) and CD1D1−/− (Right) mice. Each CDR3 sequence is represented by a dot, with the size of the dot proportional to the number of times this CDR3 sequence was found with the sample, and each Jβ is represented by a color. Vβ8 rearrangements were amplified by PCR with a V-specific primer and a C-specific reverse primer followed by high-throughput sequencing using the Ion Torrent platform. Sequence analysis was performed with in-house software, and gene identity was assigned on the basis of sequence alignment with published sequences (International ImMunoGeneTics Information System). (F) Vβ8 rearrangements from B6 and CD1D1−/− iNKT cells were amplified and cloned into retroviral plasmids. Retroviruses were produced and used to transduce Vα14-expressing hybridoma. Hybridoma were sorted for similar TCR expression and transduced with NFAT-GFP reporting construct. Antigen presentation assay using the B cell lymphoma CHB cells transfected or not with CD1D1- or CD1D2-expressing constructs was performed. The percentage of GFP+ hybridoma cells from triplicate cultures was recorded by using flow cytometry after 18 h of cultures and quantified (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by two-way ANOVA). Results are representative of three independent experiments. (G) Intracellular IFN-γ expression of liver iNKT cells 7 h after LPS injection. iNKT cells were identified by using PBS57-CD1d1 tetramer and TCRβ mAbs and stained intracellularly for IFN-γ. Plots are representative of at least three mice per group. (H) Bars depict mean ± SEM of percentage of responding iNKT cells within each strain or condition (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant).
Next, we examined the Vβ repertoire expressed by iNKT cells selected on CD1d1 in B6 mice or on CD1d2 in CD1D1−/− mice by flow cytometry. Similar to what has been reported previously for the CD1d1-selected iNKT repertoire (43, 44), CD1d2-selected iNKT cells were enriched for Vβ8+ and Vβ7+ cells compared with conventional T cells. However, a small but consistent increase in the proportion of cells using the Vβ8 gene segment, concomitant to a reduction in Vβ7 gene usage, was observed for CD1d2-selected iNKT cells compared with CD1d1-selected iNKT cells (Fig. 3D). To further compare the TCR repertoires of CD1d1-selected and CD1d2-selected iNKT cells, we sorted PBS57-CD1d1 tetramer+ cells from the thymi of two C57BL/6 mice and more than 30 pooled CD1D1−/− thymi. Total RNA was extracted, and cDNA was synthesized. We amplified Vβ8.1/8.2 rearrangements by using specific Vβ8.1/2 and Cβ primers, and the PCR fragments were sequenced by using next-generation sequencing. Fig. 3E presents a visual depiction of all sequences found in the B6 and CD1D1−/− Vβ8+ iNKT repertoires, where every dot represents a unique CDR3β sequence, the size of the dot represents how many times this sequence was found, and the color represents which Jβ was used. As expected, CD1d1-selected iNKT cells expressed a polyclonal repertoire with no overrepresentation of any particular sequence (45). Although the CD1d2-selected iNKT cell repertoire was also polyclonal, it appeared to be much more limited, with fewer clones and fewer Jβs represented. In fact, when comparing the overlap between the two repertoires, we observed that only 16% of the sequences from CD1D1−/− iNKT cells were contained within the B6 repertoire. Thus, the majority of Vβ8.1/8.2 TCR sequences found in the iNKT cells from CD1D1−/− mice appear unique to these iNKT cells. To further explore the antigen reactivity of CD1d1 vs. CD1d2-selected iNKT TCR repertoires, we cloned libraries of Vβ8.1/8.2 TCR sequences into retroviral plasmids. We then transduced a TCR-negative T hybridoma with the Vα14-Jα18 TCRα chain and the two different Vβ8 libraries derived from B6 or CD1D1−/− iNKT cells. The hybridoma was further transduced with a NFAT-GFP construct, allowing for measurement of TCR stimulation on a per-cell basis. Hybridomas expressing Vβ8 TCR repertoires derived from B6 or CD1D1−/− mice were cultivated overnight in the presence of the CHB lymphoma that had been previously transfected (or not) with CD1d1- or CD1d2-expressing constructs. As seen in Fig. 3F, approximately 2% of the hybridoma expressing the B6 iNKT cell repertoire expressed GFP upon stimulation by CD1d-expressing APCs. The proportion of autoreactive cells was increased to 4–5% of the cells when the repertoire derived from CD1D1−/− iNKT cells was interrogated in a similar fashion (Fig. 3F). Cell sorting of these GFP+ cells and subsequent restimulation with CD1d-expressing APCs demonstrated the autoreactivity of these cells toward CD1d-expressing APCs. Altogether, these data suggest that the TCR repertoire of iNKT cells found in CD1D1−/− mice is more autoreactive than the TCR repertoire expressed by B6-derived iNKT cells. This might be a result of the nature of the CD1d2 molecules themselves, alterations in the self-lipid repertoire, and/or their low levels of expression in the thymus of CD1D1−/− mice (Fig. 2).
CD1d2-Selected iNKT Cells Are Functional.
To test whether CD1d2-selected iNKT cells were functional in vivo, we injected 2 μg of the prototypic antigen αGC, which has a 26-carbon–long fatty acid chain, i.v. to B6 and CD1D1−/− mice and harvested livers from the animals 90 min later to interrogate IL-4 and IFN-γ production within iNKT cells (44). Unlike B6 controls, in which 90% of iNKT cells were activated as detected by IFN-γ intracellular cytokine staining, no CD1D1−/−-derived iNKT cells were responsive to injected antigen, resembling PBS solution-injected controls. These results probably reflect the absence of the CD1d molecule expression in the peripheral tissues required to present the antigen. However, iNKT cells can also be activated in a TCR-independent manner in response to inflammatory cytokines secreted by activated antigen-presenting cells (46, 47). Therefore, we injected 40 μg of LPS or PBS solution control to B6 or CD1D1−/− mice and harvested their livers 7 h after injection. Cells were stained intracellularly for IFN-γ directly ex vivo without further stimulation and/or addition of Brefeldin A. Although the activation of iNKT cells through LPS is less robust than through TCR engagement by the αGC/CD1d complex, the results clearly demonstrated that CD1d1-selected as well as CD1d2-selected iNKT cells could produce IFN-γ under these conditions (Fig. 3 G and H). These results demonstrate that CD1d2-selected iNKT cells are functional and capable of secreting cytokines upon stimulation. Furthermore, these results support the idea that the maintenance of iNKT cell effector function is TCR independent and does not require tonic signaling mediated by CD1d molecules.
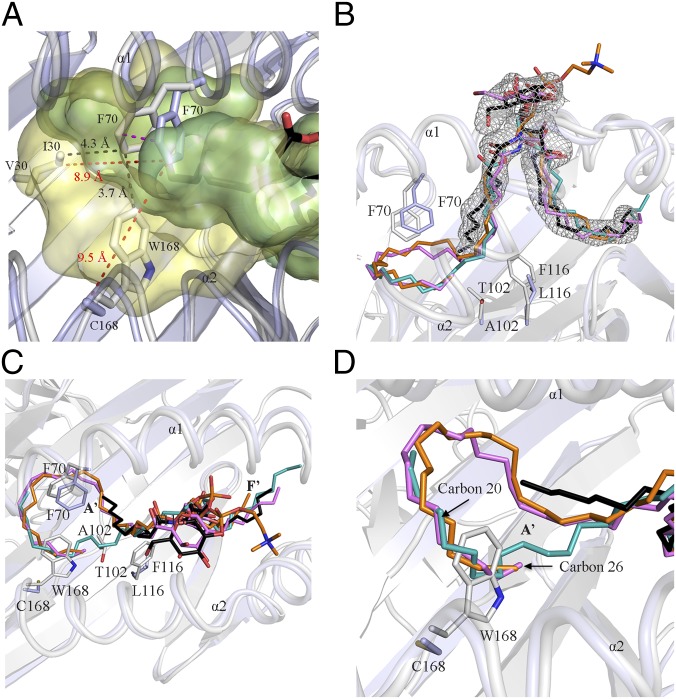
Overall Architecture of CD1d2.
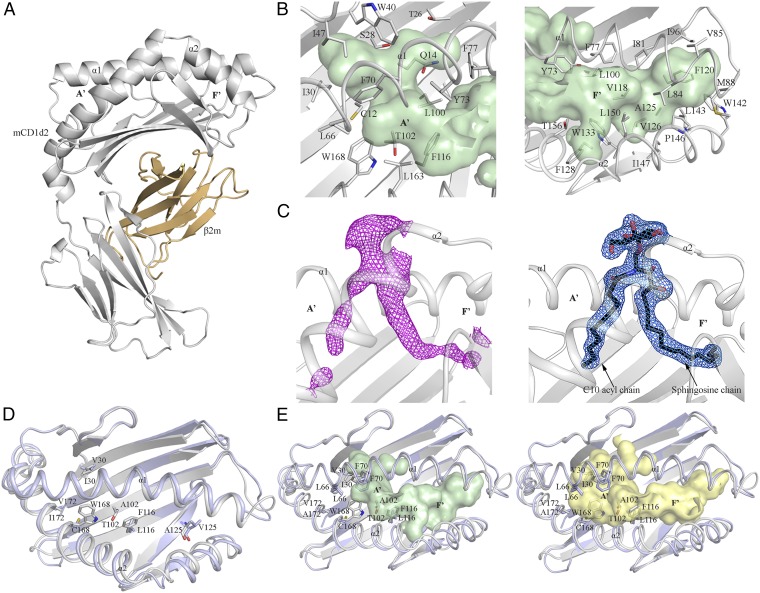
To gain insight into the detailed architecture of the CD1d2 molecule, we expressed and purified CD1d2 from the supernatant of insect cells and subsequently determined its structure. CD1d2 adopted a standard MHC-I fold, like CD1d1 (Fig. 4 and SI Appendix, Table S1). Similar to the overall architecture of other CD1 molecules (48–51), the α1- and α2-helices are positioned atop of a six antiparallel β-strands platform to form a deep hydrophobic antigen-binding groove (∼1,300 Å3) that accommodates lipid-based antigens (Fig. 4A). Following the nomenclature adopted for mouse CD1d1 (3), the antigen-binding cleft is composed of two distinct A′- and F′-pockets (Fig. 4 A and B) that differ in size and architecture. The contour of the A′-portal (∼550 Å3) is delineated by a number of bulky hydrophobic residues (W40, F70, Y73, F77, F116, and W168) with residues Y73 and F77 separating the A′-pocket from the F′-pocket (Fig. 4B). The F′-pocket (750 Å3) is shaped by an extensive network of hydrophobic residues. Here, residues L100, V118, F120, A125, F128, and L143 form the floor of the pocket and a number of key hydrophobic residues from the α1- and α2-helices (Y73, F77, I81, L84, V85, M88, W142, L143, P146, I147, L150, and T156) define the overall shape of the F′-pocket (Fig. 4B, Right).
Fig. 4.
CD1d2 crystal structure. (A) Cartoon representation of the CD1d2 crystal structure. CD1d2 and β2m are colored in gray and light orange, respectively. (B) Close-up views of the A′- (Left) and F′-pockets (Right) of CD1d2. Both pockets are colored in pale green and shown as surface representations. (C, Left) Unbiased Fo-Fc electron density map (magenta) contoured at 2.0σ level of the αGC (C10) lipid Ag. (C, Right) 2Fo-Fc electron density map (marine) contoured at 0.8σ level of the αGC (C10) lipid Ag. The lipid Ag is shown as black sticks. For clarity, only the α1- and α2-helices of CD1d2 are colored in light gray and shown as cartoon representations. (D) Superimposition of the α1- and α2-domains of the crystal structures of CD1d2 (gray) and CD1d1 (PDB ID code 1CD1; light blue) (48). The polymorphic residues between CD1d2 and CD1d1 are shown as sticks. For clarity, only the residues located inside the binding groove are shown. (E) Surface representation of the binding pocket of CD1d2 (light green) and CD1d1 (pale yellow). A superimposition of the α1- and α2-domains of CD1d2 (gray) and CD1d1 (light blue) is also shown.
The CD1d2 crystal structure also revealed clear separate unaccounted and continuous electron density within the A′- and F′-portals to protrude out of the binding cleft (SI Appendix, Fig. S2). This density suggested the presence of an endogenous bound lipid containing a head group that was acquired from the Hi5 insect cells during the recombinant expression of CD1d2. Although the precise identity of the endogenous ligand is undetermined, we modeled in the unbiased electron density the common self-membrane phospholipid phosphatidic acid (PA). This enabled us to evaluate the approximate length of the acyl chains that the endogenous lipid may contain. PA consisting of two acyl chains composed of 10 and 18 carbons could accommodate the continuous electron density that was visible in the A′- and F′-pockets, respectively (SI Appendix, Fig. S2).
To formally demonstrate that CD1d2 presents lipid antigens of shorter acyl-chain length, we conducted lipid loading experiments with a truncated analog of αGC in which the acyl chain was 10 carbon atoms in length [αGC (C10)]. Although we could not load recombinant CD1d2 with αGC, the αGC (C10) form readily complexed with CD1d2. We then subsequently determined the structure of CD1d2-αGC (C10; SI Appendix, Table S1), in which the C10 acyl chain was clearly observed within the A′-pocket of CD1d2. The structure of CD1d2-αGC (C10) was very similar to that of CD1d2 bound to endogenous lipids, and the mode of αGC (C10) presentation by CD1d2 was very similar to that of CD1d1-αGC. Accordingly, CD1d2 can present αGC (C10) in a manner analogous to CD1d1 presentation of α-GC.
Comparison of the CD1d2 and CD1d1 Crystal Structures.
CD1d2 and CD1d1 share high amino acid sequence identity (93%; SI Appendix, Fig. S3) that is reflected by a very similar α1- and α2-domain architecture with an rmsd of 0.9 Å over 178 Cα atoms (Fig. 4D). Thus, the presence of the Cys168Trp polymorphism did not lead to gross conformational changes in the CD1d2 architecture. The majority of the polymorphisms between CD1d1 and CD1d2 are evenly distributed over the α2-helix and the six β-sheet platform, whereas the α1-helix sequence is conserved between CD1d1 and CD1d2 (Fig. 4D and SI Appendix, Fig. S3). Ten of these polymorphic residues are located on the molecular surface of CD1d2 (SI Appendix, Fig. S4A). The remaining six residues (I30, T102, F116, W168, and A172 in CD1d2) are predominantly buried within the A′-pocket of CD1d2 (Fig. 4D and SI Appendix, Fig. S4B). Interestingly, residues I30, T102, F116, and W168 are directly implicated in remolding the overall architecture of the A′-portal relative to CD1d1 (Figs. 4E and 5A and SI Appendix, Fig. S4B). The substitution of V30, L116, and C168 in CD1d1 by the bulkier hydrophobic I30, F116, and W168 residues in CD1d2, respectively, contributes to a reduction of the A′-pocket volume of CD1d2 by ∼400 Å3 compared with CD1d1, and also serves to prevent the collapse of the A′-pocket as a result of the lack of the intermolecular disulfide bond. Here, to accommodate the endogenous lipid and αGC (C10), the side chain of F70 was repositioned in close vicinity of I30 and W168 (∼4 Å distance), and consequently this triad of residues contributed to restrict the size of the A′-portal (Figs. 4E and 5A). Accordingly, the structural rearrangement incurred by these polymorphic residues may impact the overall repertoire of lipids that can be presented by CD1d2 compared with CD1d1. Modeling of longer acyl chains in the A′-pocket was sterically disfavored by the architecture of the CD1d2 groove (Fig. 5 C and D). Indeed, the A′-pocket of CD1d2 favors the loading of shorter sized lipids compared with that of CD1d1, as evidenced by the CD1d2-αGC(C10) structure. In the CD1d2 A′-pocket, residues T102 and F116 also impacted the overall position of the acyl chain of the endogenous lipid (Fig. 5B). By contrast, the F′-pockets of CD1d2 and CD1d1 are highly conserved (Fig. 4D) whereby only A125 (CD1d2) was replaced by V125 (CD1d1; Fig. 4 D and E). Thus, molecular insights into the antigen-binding cleft of CD1d2 show that lipid antigens with long acyl chains are not as efficiently presented compared with CD1d1 molecules.
Fig. 5.
Structural comparison between CD1d1 and CD1d2. (A) Close-up view of a superimposition of the A′-pockets of CD1d1 and CD1d2 shown in yellow and light green, respectively. (B and C) Superimposition of CD1d-phosphatidylcholine (orange; PDB ID code 1ZHN) (71), CD1d-dioleoyl-PA (teal; PDB ID code 4MX7), CD1d-αGC (C25; violet; PDB ID code 3HE6) (9), and CD1d2-αGC (C10; black). (D) Close-up view of A. The α1- and α2-domains of the CD1d2 (gray) and CD1d1 (light blue; PDB ID code 1ZHN) are shown in each image.
Differential Antigen Presentation by CD1d1 and CD1d2 Molecules.
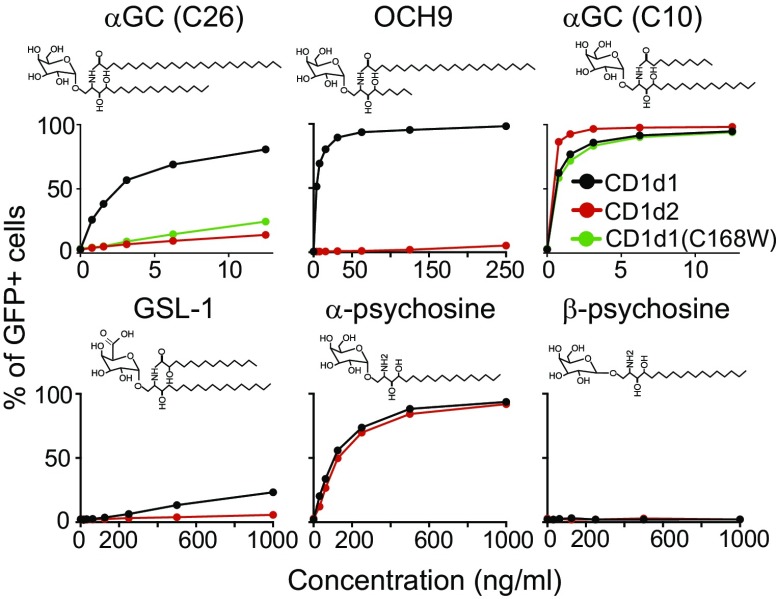
To test the antigen-presentation ability of CD1d1 and CD1d2 molecules, we cultivated hybridoma cells expressing a library of iNKT TCRs (expressing different TCRβ chains) derived from B6 thymic iNKT cells with B cell lymphoma CHB cells expressing similar levels of CD1d1 or CD1d2 molecules. Increasing concentrations of various antigens were added to the cultures, and the percentage of GFP+ hybridoma cells was assessed after 18 h. As seen in Fig. 6, CD1d2-expressing CHB cells poorly stimulated the hybridomas compared with CD1d1-expressing cells when cultivated in the presence of glycolipid antigens with long acyl chains, including the prototypical iNKT antigen αGC (C26), the αGC variant with a shorter sphingosine chain OCH9 (C24), and the Sphingomonas-derived antigen GSL-1 (C14). In striking contrast, a version of αGC with a 10-carbon acyl chain, or the α-psychosine antigen, which lack an acyl chain (52, 53), equivalently stimulated the hybridoma when cultivated with CD1d1- or CD1d2-expressing cells. The β-anomeric form of psychosine did not stimulate the hybridoma when cultivated with either APCs. CHB cells expressing CD1d1 molecules with the Cys168→Trp168 mutation were also poor presenters of αGC (C26) but could present αGC (C10) as efficiently as WT CD1d1 molecules (Fig. 6). Altogether, these results suggest that CD1d2 molecules are poor presenters of long acyl chain glycolipid antigens but can present certain antigens with short acyl chains as efficiently as CD1d1 molecules. These results demonstrate that both CD1d molecules in mice are not equivalent and can selectively present different lipid antigens to iNKT cells.
Fig. 6.
Differential antigen presentation by CD1d1 and CD1d2 molecules. Hybridoma-expressing Vβ8+ iNKT TCRs derived from C57BL/6 iNKT cells were cultivated for 18 h with CHB cells stably transfected with CD1D1 (black), CD1D2 (red), or the CD1D1 mutant in which the cysteine 168 was replaced by a tryptophan (green) and the indicated glycolipid antigens. Graphs depict the percentage of GFP+ hybridoma as a function of antigen concentration. Data represent the mean of duplicates for each condition and are representative of three independent experiments.
CD1d2-αGC-C10 Tetramer+ Cells.
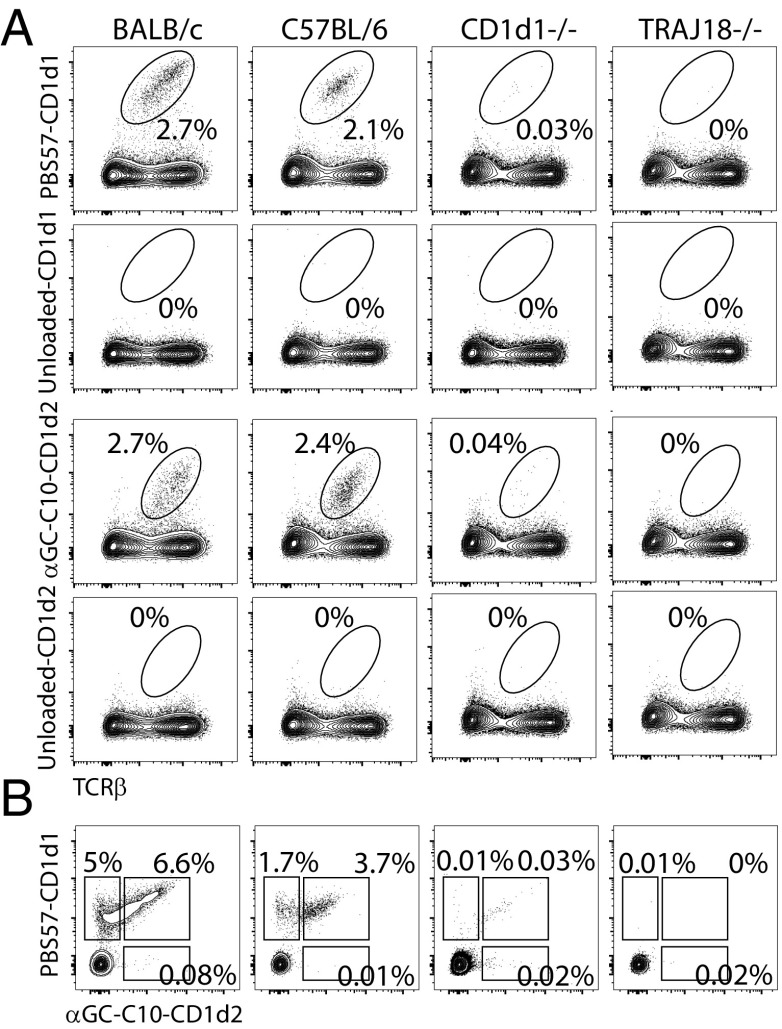
Having established that CD1d2 molecules preferentially present glycolipid antigens with short acyl chains, we loaded soluble CD1d2 monomers with αGC(C10) and generated CD1d2 tetramers. Thymocytes from BALB/c, B6, CD1D1−/−, and Jα18−/− mice were independently stained by using the PBS57-CD1d1 or the αGC(C10)-CD1d2 tetramers (Fig. 7A). The percentage of cells stained in each thymus was essentially identical between tetramers, even though αGC(C10)-CD1d2 tetramers stained with lower intensity compared with PBS57-CD1d1 tetramers. Costaining with saturating concentrations of CD1d1 and CD1d2 tetramers loaded with PBS57 or αGC(C10), respectively, were initially unsuccessful because no CD1d2 signal was detected in these instances (data not shown). However, sequential staining with the two tetramers could be achieved if CD1d2 tetramers were used first. With the use of this strategy, we did not detect a significant population of αGC(C10)-CD1d2-specific cells (Fig. 7B). Instead, a fraction of iNKT cells was stained with both tetramers, whereas some PBS57-CD1d1 tetramer+ cells were not labeled with the αGC(C10)-CD1d2 tetramer. Interestingly, cells labeled with the αGC(C10)-CD1d2 tetramer expressed the highest levels of TCR on their surface (Fig. 7B), suggesting that the αGC(C10)-CD1d2 binds only to the highest avidity iNKT TCRs.
Fig. 7.
αGC(C10)-CD1d2 tetramer staining. (A) Thymocytes from BALB/c, C57BL/6, CD1D1−/− and Jα18−/− mice were stained with CD1d1 tetramers loaded (or not) with PBS57 or CD1d2 tetramers loaded (or not) with αGC (C10) and TCRβ mAbs. The percentage of tetramer+ is shown. Data are representative of two independent experiments. (B) Thymocytes from the aforementioned mouse strains were first stained with αGC (C10)-CD1d2 tetramers, washed, and subsequently stained with PBS57-CD1d1 tetramers. Data are representative of three independent experiments. Simultaneous staining with both tetramers did not allow for the detection of the fluorophore associated with the CD1d2 tetramer.
Discussion
Peptide-presenting major histocompatibility molecules are encoded by the most polymorphic genes in the genome. Most of this polymorphism involves residues that are lining the antigen-binding groove of MHC molecules (54). At the population level, these polymorphisms provide various possible binding solutions to the myriad of antigenic peptides that need to be presented for immunosurveillance by T cells. By contrast, lipid-binding CD1 molecules are essentially monomorphic, with the number and complexity of CD1-encoding genes varying greatly between species. The functional consequence of expressing multiple CD1 proteins presumably reflects pathogen-driven evolutionary pressure. The mouse genome contains two genes, CD1D1 and CD1D2, which encode for the CD1d molecules. A collection of observations had contributed to the misleading idea that CD1D2 might not play any relevant role in the biology of lipid-reactive T cells. By contrast, we now show that, in the 129/Sv and BALB/c mouse strains, the CD1D2 gene is functional, and, in BALB/c mice, is expressed equivalently to the CD1D1 gene at the mRNA level in the thymus. The reason why CD1d2 protein expression was detected in the thymus of CD1D1−/− mice (albeit at low levels) but not in splenocytes is uncertain. Early experiments examining the pattern of CD1d expression in whole organs or in cell lines suggested that it was controlled, at least in part, at the transcriptional level (19, 55). Our current understanding of CD1D transcriptional regulation remains limited. The minimal promoter region within 200 bp from the transcriptional start site of CD1D1 has been identified and was shown to be regulated by the Ets family of transcription factors in B and myeloid cells (55). However, examination of the upstream DNA sequence of CD1D2 revealed that it is highly conserved with CD1D1, including the previously identified Ets binding site. It is possible that thymocytes express unique combinations of Ets transcription factors that allow for the transcription of CD1d2. Lineage-specific regulatory elements could also differentially control the expression of CD1D2 and CD1D1 in different cell types. What advantage, if any, differential CD1d2 expression between the thymus and other tissues might provide remains to be determined. Irrespective of its low level of expression in the thymus of CD1D1−/− mice, CD1d2 molecules are clearly capable of selecting some iNKT cells in these mice. Consequently, mice lacking only the CD1D1 gene (32) should not be considered as NKT cell-deficient.
Interestingly, the phenotype and repertoire of the CD1d2-selected iNKT cells in CD1D1−/− mice was not equivalent to the CD1d1-selected iNKT cells in B6 mice. The iNKT cells in the thymus of CD1D1−/− mice were somewhat enriched for the iNKT2 and iNKT17 subsets compared with the subset composition encountered in age-matched B6 mice. Furthermore, the thymic iNKT1 subset in CD1D1−/− mice expressed little to no NK1.1 molecules, which usually characterize this subset in B6 mice. Similarly, the Vβ repertoire of CD1d2-selected iNKT cells was enriched for usage of the Vβ8 chain with a concomitant decrease in Vβ7 usage compared with the CD1d1-selected iNKT cells. Previous reports showed that changes in Vβ selection occurred in mice expressing lower or higher levels of CD1d, and reflected changes in positive and negative selection according to Vβ affinities for self-ligand presented by CD1d molecules (56, 57). The TCR Vβ domain influences the thymic selection of iNKT cells by contributing to the avidity of iNKT TCR binding to CD1d complexed with endogenous ligand(s) (8). In heterozygote CD1D+/− mice that express half the levels of CD1d compared with WT mice, Vβ7+ iNKT cells were overrepresented with a concomitant decrease in Vβ8 usage, suggesting that Vβ7-containing iNKT TCRs have higher affinity for the self-ligand(s)–CD1d complexes that are responsible for the positive selection of these cells. By contrast, in CD1D1−/− mice, the expression level of CD1d2 was only 4% of the level of CD1d observed in WT mice, and yet we observed a preferential enrichment of Vβ8+ iNKT cells. Sequencing of the CDR3 region of these Vβ8+ iNKT cells demonstrated poor overlap with the repertoire of B6 Vβ8+ iNKT cells. Furthermore, the CD1d2-selected Vβ8 iNKT repertoire tended to be more autoreactive when reexpressed in an in vitro system. The reason(s) for these differences remains unclear. It is possible that the low level of CD1d2 expression contributed to the selection of iNKT cells expressing TCR of the highest avidity, which would otherwise be negatively selected in WT animals. Alternatively, the presentation of a different set of self-antigen(s) responsible for the positive selection of iNKT cells between CD1d1 and CD1d2 could contribute to these differences in repertoire and phenotype. These two possibilities are not exclusive. Future experiments aimed at manipulating the expression level of CD1d2 to match the levels observed for CD1d1 should be informative in that matter.
One aspect that determines the function of a CD1 protein is the size and shape of the antigen-binding groove. For example, human CD1b has a large antigen-binding groove that can accommodate ligands of as much as 80 carbon atoms in length (49). In contrast, the human CD1a isoform has a unique architecture that appears to favor “headless” antigens, with a restricted A′-pocket that prefers lipids of shorter acyl-chain length (C18–C20) (50, 58). Our work resonates with the CD1a molecule in that CD1d2 favors the presentation of shorter length lipid ligands in comparison with CD1d1. Despite high homology between the two CD1d molecules, changes in amino acids at position I30, T102, F116, and especially W168 restrict the size of the A′-pocket of CD1d2. The Trp-168 blocks part of the A′-pocket so that long fatty acid chain glycolipids will not fit. In agreement with the structural data, APCs engineered to express the CD1d2 molecules or the C168W CD1d1 mutant were poor presenters of common glycosphingolipids, including the prototypical antigen αGC, which consist of fatty acid with a length of 24–26 carbons. Interestingly, comparable observations had been previously reported whereby the equivalent cysteine residue in the bovine CD1d and the ancestral chicken CD1-2 are replaced by a tryptophan and phenylalanine, respectively (26, 27). These restricted the size of the A′- pocket of the two isoforms and reduced their capacity to bind long-chain lipid-based antigens (26, 27). Similarly, CD1d2 molecules efficiently presented only glycolipid antigens with short or no acyl chains. Of interest, α-psychosine, which contains a single lipid chain and has been proposed to possibly play a role in the selection of iNKT cells (52), was presented well by both CD1d molecules. Although the lipid portions of antigens are buried in the CD1d groove, the composition of the fatty acids, including the length, the degree of unsaturation, and the presence of other modifications, strongly influences antigenic potency (59–61). As such, it is reasonable to hypothesize that the balance between the various antigen species that might be presented by CD1d1 and/or CD1d2 in the thymus possibly affects the tuning of the iNKT TCR repertoire to different ligands. CD1d2 might also be important in shaping the repertoire of other CD1d-restricted T cells.
Methods
Mice.
The CD1D1−/− (32), CD1D1−/−CD1D2−/− (30), and Jα18(neo) (36) mice have been described previously and were backcrossed >10 times on the C57BL/6 background. C57BL/6 and BALB/c were purchased from Jackson Laboratories. All mice were used between 6 and 12 wk and were age-matched for each experiment. All mice were raised in a specific pathogen-free environment at the Biology Resource Center at National Jewish Health (NJH) and the Office of Laboratory Animal Research at the University of Colorado Denver (UCD). All animal procedures were approved by the NJH (protocol AS2780-10-16) and UCD [protocol B-64314(05)1E] Institutional Animal Care and Use Committees and were carried out in accordance with the approved guidelines.
Lymphocyte Isolation.
Single-cell suspensions were prepared from the thymus, spleen, and liver by manual disruption by using a syringe plunger. The liver was perfused with PBS solution and then cut into small pieces, disrupted with a syringe plunger, and liver lymphocytes were isolated by centrifugation by using a 33% (vol/vol) Percoll gradient (GE Healthcare).
High-Throughput Sequencing.
CD69−CD4+CD8+ preselection double-positive thymocytes were sorted by flow cytometry and washed twice in ice-cold PBS solution. Total RNA was extracted by using the RNeasy Mini Kit (Qiagen). cDNA was made by using SuperScript III Reverse Transcriptase (Invitrogen). The Vβ8-Cβ region was amplified by using specific Vβ8 (5′-ATTATTCATATGGTGCTGGC-3′) and Cβ (5′-CGATCTTGGGTGGAGTCACATTTCTC-3′) primers with required specific Ion Torrent tags. Purified PCR products were sent for high-throughput sequencing by using the Ion Torrent platform. Sequence analysis was done with in-house software, and gene identity was assigned on the basis of sequence alignment with published sequences (International ImMunoGeneTics Information System).
Alternatively, exon 2 of CD1d genes was amplified by PCR with Phusion high-fidelity polymerase (NEB) using specific primers (forward, 5′-TACACCTTCCGCTGCCT-3′ reverse, 5′-AGCTACTCGATAGACTTGAAAC-3′) with required specific Ion Torrent tags. Gel-purified PCR products were sequenced with the Ion Torrent platform. Sequence analysis was done with in-house software, and gene identity was assigned on the basis of sequence alignment with published sequences.
Quantitative PCR.
Total mRNA was extracted from sorted cells by using TRIzol solution (Invitrogen). RNA was treated with the DNA-free kit (Ambion) to remove contaminating DNA from RNA preparations. Reverse transcription was carried out by using the SuperScript III Kit (Invitrogen) following the manufacturer’s instructions. The amount of amplicon generated during the PCR was monitored with a DNA engine Opticon 2 apparatus (MJ Research) by using gene-specific primers and probes and Platinum Quantitative PCR SuperMix UDG (Invitrogen). The sequences of the primers and probes are as follows: Vα14 forward, 5-TACAGTGTGACCCCCGACAAC-3; Cα forward, 5-CCTCTGCCTGTTCACCGACTT-3; Cα reverse, 5-TGGCGTTGGTCTCTTTGAAG-3; Cα probe, 5 FAM-CTCCCAAATCAATGTGCCGAAAACCA-TAMRA-3; Vα14 probe, 5 FAM-CACCCTGCTGGATGACACTGCCAC-TAMRA-3; and Jα18 probe, 5 FAM-AGCTGGGACTCAGCTGATTGTCATACCTG-TAMRA-3.
Flow Cytometry.
PBS57-CD1d1 tetramer was obtained from the National Institutes of Health Tetramer Core Facility. The complete list of surface antibodies used is as follows. From BD Biosciences, anti-TCRβ (H57-597), anti-CD8α (53-6.7); from BioLegend, mCD1d (1B1), CD44 (IM7); from eBioscience, anti-B220 (RA3-6B2), anti-MHCII (I-A/I-E; M5/114.15.2), anti-CD4 (RM4-5), and anti-NK1.1 (PK1316). Surface antibody staining was done and then cells were fixed and permeabilized by using the FoxP3 buffer set (eBioscience). Fixed and permeabilized cells were incubated with intracellular antibodies including anti-PLZF (Mags.21F7; eBioscience), anti-Tbet (4B10; BioLegend), anti-RORγt (Q31-378; BD Biosciences), or anti–IFN-γ (XMG1.2, eBioscience). Cells were analyzed on a BD LSRFortessa device (BD Biosciences), and data were processed with FlowJo software (TreeStar).
Enrichment of CD1d Reactive Thymocytes.
Thymocytes were enriched for PBS57-CD1d reactive cells by incubating thymocyte cell suspensions with PE- or APC-conjugated PBS57-CD1d tetramers for 45 min at 4 °C, incubated with anti-PE or anti-APC magnetic microbeads (Miltenyi Biotec) for 15 min at 4 °C, followed by separation by using an autoMACS Pro Separator (Miltenyi Biotec) according to the manufacturer’s instructions. Cells were then later surface-stained and then intracellular-stained for flow cytometry.
In Vivo iNKT Cell Stimulation.
Mice were administered vehicle or 2 μg αGC (Funakoshi) by i.v. injection. Organs (spleen or liver) were harvested 90 min post injection. Cells were then later surface-stained and then intracellular-stained for flow cytometry without the use of Brefeldin A or further activation. Mice were administered PBS solution or 40 μg LPS (Sigma) by i.v. injection. Livers were harvested 7 h post injection. Cells were surface-stained and then intracellular-stained for flow cytometry.
TCRβ Library Preparation and T Cell Hybridoma.
The 58α−β− hybridoma cell line (62) was infected with hCD4-pA-GFP-NFAT-RV (63), and then cells positive for human CD4 were sorted by using an Aria cell sorter 2 d after transduction. Cells were next transduced with Vα14 iNKT TCRα chain-IRES-NGFR retrovirus, and cells positive for NGFR were sorted. Vβ8+ TCR rearrangements were amplified from total iNKT cells purified from the thymus of B6 or CD1D1−/− mice and cloned into retroviral plasmids as previously described (8). Cells were transduced with the retroviral TCRβ libraries, and TCRβ+ cells were sorted to obtain TCRαβ+ cells. TCR-reconstituted hybridomas (2.5 × 104 cells) were cultured for 18 h with antigens and CHB (5 × 105 cells) in 96-well round-bottomed plates. GFP expression on the hybridomas was analyzed by flow cytometry.
CHB Transfectants.
cDNA encoding CD1d1 and CD1d2 molecules were amplified from B6 or CD1D1−/− splenocytes by using the primers 5′-ATTTATCTCGAGCCACCATGCGGTACCTACC-3′ and 5′-ATTAAAGAATTCTCACCGGATGTCTTGATAAGCGC-3′ and cloned into retroviral plasmids. Plasmids were sequence-verified, and retroviruses were prepared as previously described (8) and used to transduce CD1d-negative CHB B cell lymphoma (64). Stably transduced cells were sorted for similar CD1d cell surface expression by flow cytometry by using an Aria cell sorter (BD Bioscience). The C168W CD1D1 mutant was generated by using QuikChange II Site-Directed Mutagenesis Kit following the manufacturer’s instructions. CHB cells expressing the C168W mutant were generated similarly.
Cloning, Expression, and Purification of CD1d2.
The mouse CD1d2/β2m gene (129/Sv sequence) harboring BirA and 6×-histidine tags was cloned into the baculovirus pFastBac Dual Expression vector as previously described (65). The CD1d2/β2m glycoprotein was expressed for 72 h at 23°C by using High Five expression insect cells. The baculovirus-expressed soluble CD1d2/β2m protein was then concentrated and buffer was exchanged against 20 mM Tris, pH 8.5, 500 mM NaCl, and purified by Ni-NTA. The protein was then dialyzed against 20 mM Tris, pH 8.5, 150 mM NaCl, and further purified by size-exclusion chromatography.
Crystallization and Structure Determination of CD1d2.
The purified CD1d2 protein and CD1d2-αGC (C10) complex (8–10 mg/mL) in 20 mM Tris, pH 8.5, 150 mM NaCl was crystallized in 26% PEG 3350, 0.1 M Bis-Tris, pH 6, 0.2 M CaCl2 at 20 °C by using the hanging-drop vapor-diffusion method. The crystals were flash-frozen before data collection in mother liquor containing 10–15% glycerol as a cryoprotectant. The crystals of CD1d2 were diffracted at the Australian Synchrotron facility (Melbourne, Australia). The dataset was processed and scaled by using the programs iMosflm and SCALA from the CCP4 suite, respectively (66–68). The crystals diffracted to 2.3-Å resolution and belonged to the space group of P21 with two molecules in asymmetric unit. The crystal structures of CD1d2 and CD1d2-αGC(C10) were determined by molecular replacement by using the Phaser-MR program (69) using the mouse CD1d1 as search model [Protein Data Bank (PDB) ID code 1CD1] (48). The model building and subsequent refinement cycles were performed with the program COOT (70) and the refinement program BUSTER 2.10 (https://www.globalphasing.com/buster/), respectively. The quality of the final structure was validated by using the PDB validation server from the PDB database (https://validate-rcsb-1.wwpdb.org/). The coordinates relating to the data reported in this study are deposited in the PDB database with ID codes 6BMH (CD1d2-αGalCer C10 structure) and 6BMK (CD1d2-PA C10 structure, endogenous).
Biotinylation of CD1d2.
For the purpose of biotinylation, the purified CD1d2 was desalted and incubated eight parts of CD1d2 with one part of buffer A (100 mM Bicine, pH 8.3) and one part of buffer B (100 mM ATP, 100 mM MgOAc, 500 µM d-biotin) along with 2 μL of biotin protein ligase (1 mg/mL). The biotinylation reaction was carried out at room temperature overnight. The excess of free biotin was removed by size-exclusion chromatography by using a Superdex 200 10/30 gel filtration column.
Supplementary Material
Acknowledgments
We thank members of our laboratories for thoughtful discussions and critical comments on the manuscript; Chyung-Ru Wang for the CD1D1D2−/− mice used in this study; Paul B. Savage and Luc Teyton for the α- and β-psychosine antigens; the National Jewish Health flow cytometry facility, the Mucosal and Vaccine Research Colorado Flow Core, and the University of Colorado flow cytometry shared resource facility for assistance with cell sorting; the Center for Genes, Environment, and Health at National Jewish Health for sequencing; the National Institutes of Health (NIH) core facility for CD1d tetramers; the staff at the Australian Synchrotron for assistance with data collection; and the staff at the Monash Macromolecular crystallization facility. This work was supported by NIH Grants AI121761 and AI124076 (to L.G.); an American Association for Immunologists Careers in Immunology Fellowship (L.G.); Cancer Center Support Grant P30CA046934; the National Health and Medical Research Council; Worldwide Cancer Research Grant 16-1106 (to J.R.); and an Australian Research Council Future Fellowship FT160100074 (to J.L.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6BMH and 6BMK).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716669115/-/DCSupplemental.
References
- 1.Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: Structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol. 2013;31:529–561. doi: 10.1146/annurev-immunol-032712-095912. [DOI] [PubMed] [Google Scholar]
- 2.Zajonc DM. The CD1 family: Serving lipid antigens to T cells since the Mesozoic era. Immunogenetics. 2016;68:561–576. doi: 10.1007/s00251-016-0931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15:643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: What’s in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 5.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koseki H, et al. Homogenous junctional sequence of the V14+ T-cell antigen receptor alpha chain expanded in unprimed mice. Proc Natl Acad Sci USA. 1990;87:5248–5252. doi: 10.1073/pnas.87.14.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallevaey T, et al. T cell receptor CDR2 β and CDR3 β loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellicci DG, et al. Differential recognition of CD1d-α-galactosyl ceramide by the V β 8.2 and V β 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel O, et al. Vβ2 natural killer T cell antigen receptor-mediated recognition of CD1d-glycolipid antigen. Proc Natl Acad Sci USA. 2011;108:19007–19012. doi: 10.1073/pnas.1109066108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 13.Brigl M, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med. 2011;208:1163–1177. doi: 10.1084/jem.20102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gapin L. iNKT cell autoreactivity: What is ‘self’ and how is it recognized? Nat Rev Immunol. 2010;10:272–277. doi: 10.1038/nri2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gapin L, Godfrey DI, Rossjohn J. Natural killer T cell obsession with self-antigens. Curr Opin Immunol. 2013;25:168–173. doi: 10.1016/j.coi.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YJ, et al. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity. 2015;43:566–578. doi: 10.1016/j.immuni.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosby CM, Kronenberg M. Invariant natural killer T cells: Front line fighters in the war against pathogenic microbes. Immunogenetics. 2016;68:639–648. doi: 10.1007/s00251-016-0933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradbury A, Belt KT, Neri TM, Milstein C, Calabi F. Mouse CD1 is distinct from and co-exists with TL in the same thymus. EMBO J. 1988;7:3081–3086. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: Insights from comparative genomic analysis. Trends Immunol. 2003;24:412–418. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 21.Brossay L, et al. Mouse CD1 is mainly expressed on hemopoietic-derived cells. J Immunol. 1997;159:1216–1224. [PubMed] [Google Scholar]
- 22.Park SH, Roark JH, Bendelac A. Tissue-specific recognition of mouse CD1 molecules. J Immunol. 1998;160:3128–3134. [PubMed] [Google Scholar]
- 23.Porcelli SA. The CD1 family: A third lineage of antigen-presenting molecules. Adv Immunol. 1995;59:1–98. doi: 10.1016/s0065-2776(08)60629-x. [DOI] [PubMed] [Google Scholar]
- 24.Bjorkman PJ, et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 25.Warburton RJ, et al. Mutation of the alpha 2 domain disulfide bridge of the class I molecule HLA-A*0201. Effect on maturation and peptide presentation. Hum Immunol. 1994;39:261–271. doi: 10.1016/0198-8859(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, et al. Crystal structures of bovine CD1d reveal altered αGalCer presentation and a restricted A′ pocket unable to bind long-chain glycolipids. PLoS One. 2012;7:e47989. doi: 10.1371/journal.pone.0047989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zajonc DM, Striegl H, Dascher CC, Wilson IA. The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1. Proc Natl Acad Sci USA. 2008;105:17925–17930. doi: 10.1073/pnas.0809814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichimiya S, Kikuchi K, Matsuura A. Structural analysis of the rat homologue of CD1. Evidence for evolutionary conservation of the CD1D class and widespread transcription by rat cells. J Immunol. 1994;153:1112–1123. [PubMed] [Google Scholar]
- 29.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y-H, Chiu NM, Mandal M, Wang N, Wang C-R. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 31.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 32.Mendiratta SK, et al. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–477. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 33.Chen YH, et al. Expression of CD1d2 on thymocytes is not sufficient for the development of NK T cells in CD1d1-deficient mice. J Immunol. 1999;162:4560–4566. [PubMed] [Google Scholar]
- 34.Hager E, Hawwari A, Matsuda JL, Krangel MS, Gapin L. Multiple constraints at the level of TCRα rearrangement impact Vα14i NKT cell development. J Immunol. 2007;179:2228–2234. doi: 10.4049/jimmunol.179.4.2228. [DOI] [PubMed] [Google Scholar]
- 35.Bedel R, et al. Lower TCR repertoire diversity in Traj18-deficient mice. Nat Immunol. 2012;13:705–706. doi: 10.1038/ni.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawano T, et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Mutation of the Traj18 gene segment using TALENs to generate natural killer T cell deficient mice. Sci Rep. 2016;6:27375. doi: 10.1038/srep27375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uldrich AP, et al. A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat Immunol. 2011;12:616–623. doi: 10.1038/ni.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 40.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 41.Pellicci DG, et al. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195:835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gapin L. Development of invariant natural killer T cells. Curr Opin Immunol. 2016;39:68–74. doi: 10.1016/j.coi.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuda JL, et al. Natural killer T cells reactive to a single glycolipid exhibit a highly diverse T cell receptor β repertoire and small clone size. Proc Natl Acad Sci USA. 2001;98:12636–12641. doi: 10.1073/pnas.221445298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol. 2007;178:2706–2713. doi: 10.4049/jimmunol.178.5.2706. [DOI] [PubMed] [Google Scholar]
- 47.Vahl JC, et al. NKT cell-TCR expression activates conventional T cells in vivo, but is largely dispensable for mature NKT cell biology. PLoS Biol. 2013;11:e1001589. doi: 10.1371/journal.pbio.1001589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng Z, et al. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 49.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 50.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 51.Scharf L, et al. The 2.5 Å structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian α-linked glycosylceramides. Immunity. 2014;41:543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng S, et al. Psychosine variants as antigens for natural killer T cells. Chem Sci. 2017;8:2204–2208. doi: 10.1039/c6sc04218j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes AL, Hughes MK. Natural selection on the peptide-binding regions of major histocompatibility complex molecules. Immunogenetics. 1995;42:233–243. doi: 10.1007/BF00176440. [DOI] [PubMed] [Google Scholar]
- 55.Geng Y, Laslo P, Barton K, Wang CR. Transcriptional regulation of CD1D1 by Ets family transcription factors. J Immunol. 2005;175:1022–1029. doi: 10.4049/jimmunol.175.2.1022. [DOI] [PubMed] [Google Scholar]
- 56.Schümann J, Mycko MP, Dellabona P, Casorati G, MacDonald HR. Cutting edge: Influence of the TCR Vbeta domain on the selection of semi-invariant NKT cells by endogenous ligands. J Immunol. 2006;176:2064–2068. doi: 10.4049/jimmunol.176.4.2064. [DOI] [PubMed] [Google Scholar]
- 57.Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vβ bias of Vα14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med. 2006;203:1197–1207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birkinshaw RW, et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol. 2015;16:258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wun KS, et al. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 2011;34:327–339. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci USA. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossjohn J, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 62.Letourneur F, Malissen B. Derivation of a T cell hybridoma variant deprived of functional T cell receptor alpha and beta chain transcripts reveals a nonfunctional alpha-mRNA of BW5147 origin. Eur J Immunol. 1989;19:2269–2274. doi: 10.1002/eji.1830191214. [DOI] [PubMed] [Google Scholar]
- 63.Ise W, et al. CTLA-4 suppresses the pathogenicity of self antigen-specific T cells by cell-intrinsic and cell-extrinsic mechanisms. Nat Immunol. 2010;11:129–135. doi: 10.1038/ni.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haughton G, Arnold LW, Bishop GA, Mercolino TJ. The CH series of murine B cell lymphomas: Neoplastic analogues of Ly-1+ normal B cells. Immunol Rev. 1986;93:35–51. doi: 10.1111/j.1600-065x.1986.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 65.Patel O, et al. NKT TCR recognition of CD1d-α-C-galactosylceramide. J Immunol. 2011;187:4705–4713. doi: 10.4049/jimmunol.1100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 69.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giabbai B, et al. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: A molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.