Significance
Analysis of structural and functional properties of cytosolic malate dehydrogenase (cMDH) orthologs of five genera of marine molluscs adapted to an ∼60 °C range of temperatures reveals the role of protein flexibility in adaptation to temperature. Molecular dynamics simulation (MDS) analysis reveals protein-wide as well as local adaptation in flexibility. Sequence regions involved in binding and catalysis show significant interspecific, temperature-related differences in flexibility. MDS analysis is shown to provide a powerful means of examining adaptive change in protein evolution at different temperatures.
Keywords: adaptation, amino acid sequence, evolution, malate dehydrogenase, molecular dynamics simulations
Abstract
Orthologous proteins of species adapted to different temperatures exhibit differences in stability and function that are interpreted to reflect adaptive variation in structural “flexibility.” However, quantifying flexibility and comparing flexibility across proteins has remained a challenge. To address this issue, we examined temperature effects on cytosolic malate dehydrogenase (cMDH) orthologs from differently thermally adapted congeners of five genera of marine molluscs whose field body temperatures span a range of ∼60 °C. We describe consistent patterns of convergent evolution in adaptation of function [temperature effects on KM of cofactor (NADH)] and structural stability (rate of heat denaturation of activity). To determine how these differences depend on flexibilities of overall structure and of regions known to be important in binding and catalysis, we performed molecular dynamics simulation (MDS) analyses. MDS analyses revealed a significant negative correlation between adaptation temperature and heat-induced increase of backbone atom movements [root mean square deviation (rmsd) of main-chain atoms]. Root mean square fluctuations (RMSFs) of movement by individual amino acid residues varied across the sequence in a qualitatively similar pattern among orthologs. Regions of sequence involved in ligand binding and catalysis—termed mobile regions 1 and 2 (MR1 and MR2), respectively—showed the largest values for RMSF. Heat-induced changes in RMSF values across the sequence and, importantly, in MR1 and MR2 were greatest in cold-adapted species. MDS methods are shown to provide powerful tools for examining adaptation of enzymes by providing a quantitative index of protein flexibility and identifying sequence regions where adaptive change in flexibility occurs.
Temperature has a strong imprint on protein evolution: Orthologous proteins of species evolved at different temperatures commonly exhibit temperature-adaptive differences in function and structural stability (1–6). One of the central concepts used to link structure and function in the context of adaptation to temperature has been protein “flexibility,” qualitatively defined as the ease with which a protein can alter its conformation during function or in response to a change in temperature (3, 6–9). In vivo studies of intact cells using neutron scattering methods (10) and in vitro and in silico analyses of individual proteins using a variety of physical and biochemical techniques have provided evidence for higher flexibility in cold-adapted proteins (1–9). These intrinsic differences in flexibility are hypothesized to lead to similar flexibilities at normal cell temperatures—i.e., to the conservation of “corresponding states” of structure that represent an optimal balance between stability and flexibility (1, 3). This hypothesis is closely tied to the concept that proteins exist in an ensemble of conformational microstates within which the distribution of functional and nonfunctional (or less functional) conformations varies with temperature (1, 11, 12).
Measuring rates of heat denaturation by orthologous proteins is the most common experimental approach that has been used for comparing structural stabilities. Although the differences in denaturation rate observed among orthologs are consistent with adaptation, these differences are at best only an indirect measure of flexibility. Furthermore, heat denaturation data cannot elucidate how flexibility varies among different regions of the protein, especially regions involved in binding and catalysis, where reversible changes in protein conformation may be especially critical. Thermal distortion of these regions could have negative effects on function; thus, adaptive variation in flexibility in these regions might be of particular selective importance in adaptation to temperature. This prediction is based in part on the fact that binding and catalysis commonly involve large changes in enzyme conformation (e.g., ref. 13); thus, thermal perturbation of these conformational changes could be a critical selective factor in adaptation to temperature (14). A related aspect of protein flexibility pertains to the catalytic rate constant (kcat). For certain enzymes—e.g., members of the 2-hydroxy-acid dehydrogenase family to which malate dehydrogenase (MDH) and lactate dehydrogenase (LDH) belong—the active site-specific rate of the reaction—the kcat value—depends on the rate of conformational changes during catalysis (9, 13). The higher kcat values of orthologs of cold-adapted species relative to those of warm-adapted species have been conjectured to reflect a higher intrinsic level of enzyme flexibility (7, 9).
The goals of the present study were to provide clearer and more quantitative linkages between temperature-adaptive changes in kinetic properties, thermal stability, and structural flexibility. For this analysis, we examined orthologs of a single, well-studied protein, the cytosolic paralog of MDH (cMDH) (EC 1.1.1.37), from five genera of marine molluscs, within each of which congeners adapted to different temperatures occur. The species selected for this study have widely different body temperatures, from lows <0 °C for north temperate species to highs near ∼55 °C for tropical species from rocky intertidal habitats (Fig. S1). This group of species seemed likely to provide a good study system for elucidating adaptive changes and for revealing possible patterns of convergent evolution among the species of the different genera.
To go beyond the analytical approaches used in most previous studies of temperature-driven protein evolution, we used molecular dynamics simulation (MDS) methods to characterize global protein flexibility, as measured by the root mean square deviation (rmsd) of the movement of backbone atoms, and residue-specific flexibility, as measured by the root mean square fluctuation (RMSF) of each residue in these ∼330-residue enzymes (15–19). Our criterion for quantifying flexibility was thus the amount of change (measured in angstroms) in movement throughout the entire protein (rmsd) and by individual amino acid residue (RMSF) atoms that occurs when temperature is increased. In our RMSF analysis, we were particularly interested in comparing interspecific differences in the regions of the enzyme that undergo large changes in conformation during function and serve key roles in ligand binding and catalysis (20). We conjectured that the thermal sensitivities of the flexibility of these regions would provide insights into interortholog differences in temperature effects on function—notably, binding of cofactor (NADH), as indexed by the Michaelis–Menten constant (KMNADH). Ligand binding events are known to be critical in adaptation of enzymes to temperature (1, 2); thus, thermal distortion of ligand binding sites or perturbation of the conformational changes that accompany ligand binding could be reflected in the RMSF characteristics of the regions of the enzyme that are critical for catalytic activity. Our study exploits MDS approaches in comparisons of orthologs of differently adapted congeners from several genera, and it illustrates the utility of these in silico approaches in the study of protein evolution.
Results
Temperature Effects on KMNADH and Resistance to Heat Denaturation.
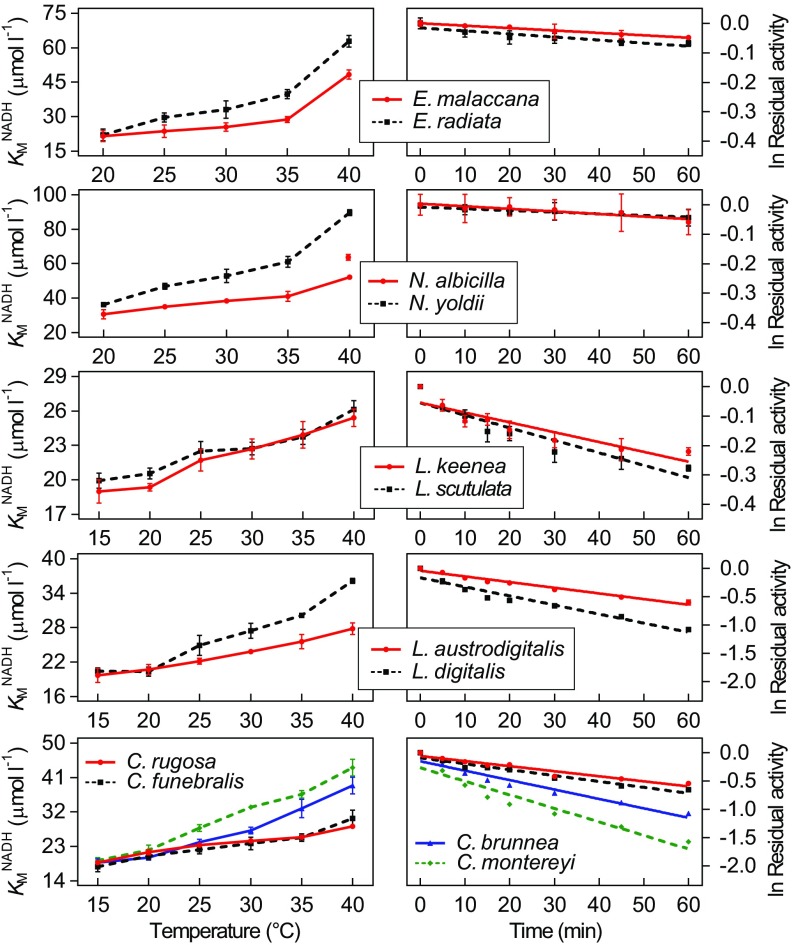
Fig. 1 illustrates effects of temperature on cofactor binding affinity (KMNADH) (Left) and heat-induced loss of activity (Right) for 12 cMDH orthologs of five sets of congeners. Field body temperatures of the 12 species range between ∼55 °C (Echinolittorina congeners) to slightly less than 0 °C (Chlorostoma congeners) (biogeographic distributions of the species are given in Fig. S1). Within each genus, orthologs of the more warm-adapted congener(s) exhibited lower values of KMNADH (an intrinsically higher cofactor binding affinity) at a common temperature of measurement, especially at higher temperatures, and the increase in KMNADH with increasing temperature of measurement was generally lower in warm-adapted orthologs as well. This pattern of variation in temperature/KM relationships is typical for many enzymes (1, 2).
Fig. 1.
Effects of temperature on cofactor binding affinity (KMNADH) (Left) and time-dependent loss of activity (Right) for 12 orthologs of cMDH from five genera of marine molluscs (mean ± SEM, n = 5, genus Echinolittorina; mean ± SEM, n = 3, genus Nerita; mean ± SEM, n = 3, genus Lottia; mean ± SEM, n = 3, genus Littorina; mean ± SEM, n = 3, genus Chlorostoma). Approximate upper lethal temperatures for the species are E. malaccana (55 °C), Echinolittorina radiata (52 °C), Nerita albicilla (50 °C), Nerita yoldii (50 °C), L. austrodigitalis (41 °C), L. digitalis (40 °C), Littorina keenae (48 °C), Littorina scutulata (45 °C), C. rugosa (∼45 °C), C. funebralis (42 °C), C. brunnea (36 °C), and C. montereyi (36 °C). (See Fig. S1 for biogeographic information and sources of thermal tolerance data. Slopes of rates of loss of activity are given in Table S1).
In parallel with these interspecific differences in thermal effects on function, the rates of heat denaturation were generally lowest in orthologs of the most warm-adapted species within a genus (Table S1 presents slopes of activity loss during heating). These intercongener differences are illustrated well by cMDHs of turban snails [genus Chlorostoma (formerly Tegula)] which occur at common sites, but at different heights along the subtidal to high intertidal transect. The subtidal species Chlorostoma montereyi is most heat-sensitive, followed by the low intertidal species Chlorostoma brunnea and the midzone intertidal species Chlorostoma funebralis and Chlorostoma rugosa. Orthologs of the tropical genera Echinolittorina and Nerita, which include species—e.g., Echinolittorina malaccana—whose body temperatures are among the highest recorded for eukaryotes (21, 22), have much higher thermal stabilities than orthologs of two temperate genera, Lottia and Chlorostoma, which generally occur either subtidally or in the mid- to low intertidal zone. Orthologs of the two temperate-zone Littorina congeners, which are found higher in the intertidal zone than congeners of Lottia and Chlorostoma, are of intermediate stability.
MDS: Temperature-Correlated Differences in Global and Localized Flexibility.
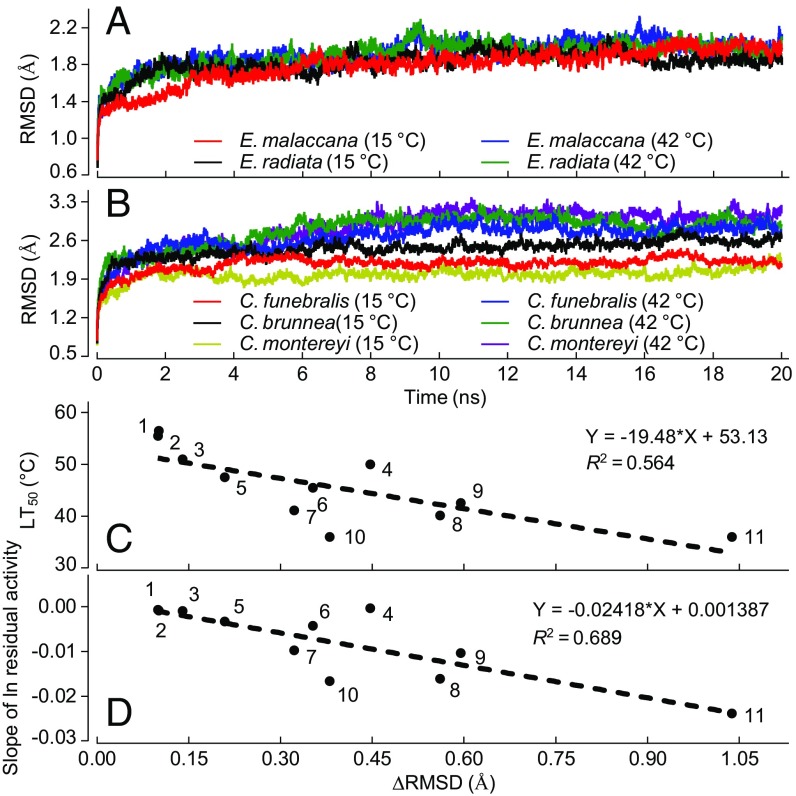
MDS procedures were used to evaluate global flexibility of structure and localized flexibility of individual amino acid residues throughout the ∼333-residue sequence. rmsd analysis reflected the average amount of movement of backbone atoms throughout the entire protein structure. As shown in Fig. 2A and Tables S2 and S3, at a low simulation temperature of 15 °C, there was relatively little interspecific variation in rmsd. For all sets of congeners, rmsd values ranged between 2.549 Å (C. brunnea) and 1.885 Å (E. malaccana) at 15 °C. The only significant difference within a genus was found between C. brunnea and C. montereyi (P = 0.0119). However, when the simulation temperature was increased to 42 °C—a temperature commonly experienced by congeners of Echinolittorina and Nerita, but several degrees greater than the thermal ranges of congeners of Littorina, Lottia, and Chlorostoma—significant differences (P < 0.05) in rmsd were found among genera and congeners. Thus, the increase in rmsd, ΔRMSD, when simulation temperature was increased from 15 to 42 °C was inversely related to the thermal tolerance limits (LT50) of the species (Fig. 2B) and, as expected from the thermal denaturation relationships shown in Fig. 1, positively correlated with the rate of denaturation of the cMDH orthologs (Fig. 2C). In summary, it was the change in rmsd with rising temperature, not the value of rmsd per se at a relatively low temperature (15 °C), that provided an index of differences in global protein stability among orthologs.
Fig. 2.
MDS analysis of backbone atom movements in cMDHs. (A and B) The rmsd of backbone atom positions for cMDHs from Echinolittorina malaccana, E. radiata, C. funebralis, C. brunnea, and C. montereyi at simulation temperatures of 15 and 42 °C (n = 3). (C) The ΔRMSD (the difference of rmsd value between 42 and 15 °C) over the equilibration state (10–20 ns) vs. lethal temperature (LT50) for cMDH orthologs of 11 species of molluscs. (D) The ΔRMSD vs. the rate of thermal denaturation for 11 cMDH orthologs. Species are numbered as follows: 1, E. malaccana; 2, E. radiata; 3, N. albicilla; 4, N. yoldii; 5, L. keenae; 6, L. scutulata; 7, L. austrodigitalis; 8, L. digitalis; 9, C. funebralis; 10, C. brunnea; and 11, C. montereyi. The relationship between ΔRMSD and LT50 or rate of thermal denaturation was analyzed by a least-squares linear regression analysis model.
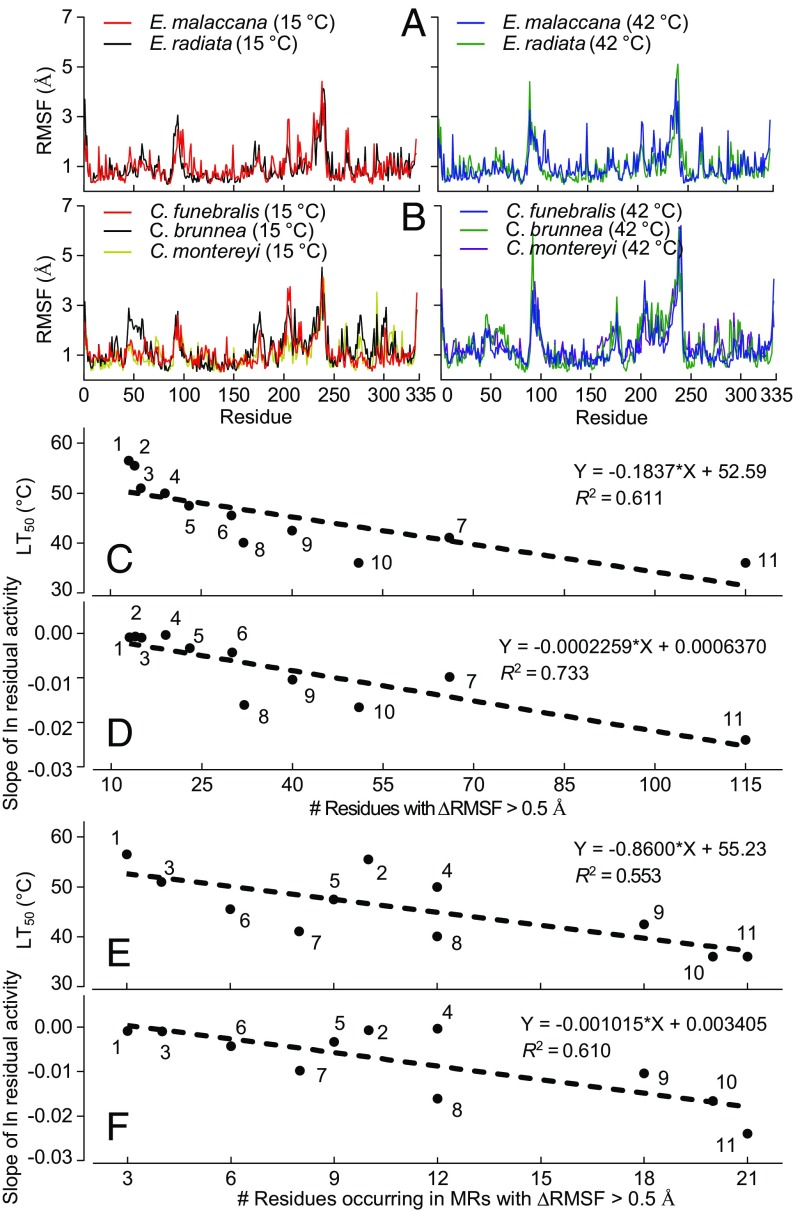
Tables S2 and S3 summarize the averaged-across-sequence RMSF values obtained at 15 and 42 °C for 11 of the 12 species for which data are given in Fig. 1. With the exception of the cMDHs of the congeners of Chlorostoma, there were few significant interspecific differences (P < 0.05) in temperature effects on average RMSF. However, there are two further ways of analyzing the RMSF data that lead to clearer insights into temperature effects on enzyme flexibility and adaptive differences in stability among orthologs. First, a residue-by-residue analysis showed that there was a large amount of residue-specific variation in individual residue movement across the sequence (Fig. 3). Data at simulation temperatures of 15 and 42 °C are presented for the sequences of the species occurring at highest (Echinolittorina congeners) and lowest (Chlorostoma congeners) field temperatures. In these four orthologs, as in the orthologs of all 11 species so examined, the highest RMSF values were found in two regions of the enzyme known to play important roles in function: residues 91–101, which comprise the catalytic loop region that folds onto the catalytic site during ligand binding, and residues 230–245, which are involved in catalysis (19, 22). In the closely related 2-hydroxy-acid dehydrogenase LDH, the homologous regions are described as “mobile regions” (MRs) by Coquelle et al. (23) and “major movers” by Gerstein and Chothia (13). We use the abbreviation MR in the discussion to follow and include residues 90–105 in MR1 and residues 230–245 in MR2.
Fig. 3.
MDS analysis of side chain movements (RMSF) in molluscan cMDHs. RMSF spectra for congeners of Echinolittorina (A) and Chlorostoma (B) at simulation temperatures of 15 and 42 °C (n = 3). Values for average RMSF across the full sequence are given in Table S1. (C and D) Number of residues showing a change in RMSF (ΔRMSF) > 0.5 Å vs. the species’ lethal temperature (LT50) (C) and the rate of thermal denaturation (D) for the cMDH orthologs. (E and F) Number of residues occurring in MR regions (residues 90–105 and 230–245) showing ΔRMSF > 0.5 Å vs. LT50 (E) and the rate of thermal denaturation (F) for the cMDH orthologs. The numbers assigned to each species are given in the Fig. 2 legend. The relationships between the number of residues showing significant change and the LT50 or rate of thermal denaturation were analyzed by least-squares linear regression.
A second metric for comparing the RMSF responses of orthologs is shown in Table 1 and Fig. 3 C and D. A value of ΔRMSF >0.5 Å was used as the threshold value of ΔRMSF that indicates a significant change in structural movements (24). The specific residues exhibiting this degree of change in RMSF are listed in Table 1. The number of residues showing a ΔRMSF > 0.5 Å correlated strongly with enzyme thermal stability. These interortholog differences indicated that increases in temperature not only cause larger average ΔRMSD in cold-adapted orthologs (Fig. 2), but also lead to larger increases in the number of residues that fluctuate in position by > 0.5 Å in the cold-adapted proteins. Residues located in MR1 and MR2 also show significant interspecific differences (Fig. 3 E and F and Table 1). In cold-adapted orthologs, there were generally a larger number of residues in these functionally important regions exhibiting a ΔRMSF > 0.5 Å. Interortholog variation was also noted in the relative effects of increasing temperature on MR1 vs. MR2; for some orthologs, MR1 was more sensitive than MR2, whereas for other orthologs, the opposite pattern was observed (Table 1).
Table 1.
Residues with a significantly different value of the change in ΔRMSF between 42 and 15 °C in cMDHs from E. malaccana, E. radiata, N. albicilla, N. yoldii, L. keenae, L. scutulata, L. austrodigitalis, L. digitalis, C. funebralis, C. brunnea, and C. montereyi
| Species | Residue |
| E. malaccana | 91 92 106 107 109 149 170 222 234 282 290 325 33 |
| E. radiata | 32 91 92 97 98 99 100 188 189 237 239 240 244 300 |
| N. albicilla | 92 212 214 215 216 217 220 222 229 230 231 236 248 264 302 |
| N. yoldii | 22 26 44 62 90 91 92 197 205 232 233 234 235 236 237 238 239 240 255 |
| L. keenae | 1 14 15 16 42 43 44 45 46 47 48 89 91 92 93 94 95 96 97 188 235 242 262 |
| L. scutulata | 11 42 43 44 45 61 62 97 98 99 100 106 200 201 204 212 214 215 216 217 218 219 220 221 222 225 229 230 231 297 |
| L. austrodigitalis | 1 3 15 25 26 32 47 93 106 109 110 121 125 129 130 131 141 142 166 168 169 174 176 177 179 187 188 189 190 202 203 204 205 206 207 208 209 211 212 221 225 229 233 234 236 237 238 240 241 261 262 264 265 276 292 293 300 301 302 303 306 307 310 318 331 332 |
| L. digitalis | 22,170 180 188 197 205 216 219 220 221 222 224 230 231 232 233 234 235 236 237 238 239 240 241 259 264 292 293 294 329 330 331 |
| C. funebralis | 1 2 31 32 33 42 44 45 48 49 75 76 89 91 92 93 96 97 98 99 100 101 107 171 172 173 176 203 212 228 232 233 234 236 237 238 239 240 241 333 |
| C. brunnea | 62 63 64 65 89 90 91 92 93 95 96 97 98 99 100 176 197 199 200 205 206 208 209 210 214 215 216 217 218 219 220 221 222 224 225 226 227 228 230 231 232 234 235 236 237 238 239 240 289 297 299 |
| C. montereyi | 1 13 14 16 17 18 19 26 28 34 45 48 49 50 51 52 53 54 55 56 57 59 60 61 65 66 67 69 89 91 92 93 94 95 96 97 98 134 157 158 159 161 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 197 201 202 203 204 209 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 231 232 233 234 235 236 237 238 239 240 241 244 245 246 247 248 249 251 252 256 272 277 278 281 282 284 298 299 301 327 328 329 330 331 |
A ΔRMSF >0.5 Å is used as the threshold for defining a significant difference. The residues occurring in MR1 (residues 90–105) are shown in bold, and those in MR2 (residues 230–245) are underlined.
Discussion
cMDH undergoes large changes in conformation and structural compactness during ligand binding. Birktoft et al. (25) reported a change in solvent-accessible surface area of ∼4% during coenzyme binding, and Chapman et al. (20) reported a further 9.7% increase in protein compactness when substrate binding occurs. Displacements of structural elements central to binding and catalysis may involve movements >15 Å (26). We hypothesized that the functional properties associated with these large changes in protein conformation during formation of the ternary (enzyme–cofactor–substrate) complex would be inherently sensitive to temperature and subject to natural selection for adaptive change. As shown by data in Fig. 1, cofactor binding affinity, as indexed by KMNADH, is perturbed by changes in experimental temperature for all orthologs, but distinct interspecific patterns are shown. The orthologs of warm-adapted congeners generally show smaller increases in KMNADH with rising measurement temperature than more cold-adapted orthologs, especially at the upper end of the range of measurement temperatures. In parallel with the results of the kinetic studies, resistance to heat denaturation is almost always higher in orthologs of warm-adapted congeners. The similar patterns of adaptive variation found among congeners provide an illustration of convergent evolution at the protein level (2).
To develop a mechanistic explanation for the interspecific differences in effects of temperature on KMNADH, we employed MDS approaches to investigate whether the movement of the protein backbone throughout the enzyme was differentially sensitive to a rise in temperature from 15 to 42 °C between orthologs (ΔRMSD analysis) and if specific regions of the enzyme known to be critical for binding and catalysis showed differential responses as well (ΔRMSF analysis). The rmsd analyses shown in Fig. 2 and summarized in Tables S2 and S3 show that overall stability of cMDH structure is reflected in the extent to which the increase in simulation temperature altered average amounts of protein backbone movements throughout the subunit structure. At 15 °C, the orthologs exhibited relatively similar values for rmsd and no consistent trends related to adaptation temperature were evident. However, despite similar rmsd values at 15 °C for the molluscan cMDHs, when the simulation temperature was increased to 42 °C, the amount of change in rmsd (ΔRMSD) was inversely related to adaptation temperature, as indexed by the thermal tolerances of the species (Fig. 2C) and resistance of the orthologs to denaturation (Fig. 2D). In summary, in silico ΔRMSD analyses allow a precise measurement of protein thermal stability and may provide a more accurate measure of evolutionary differences in stability than in vitro heat denaturation experiments, which can be influenced by factors like protein aggregation in addition to intrinsic stability of the protein per se (9).
A more detailed analysis of structural stability relationships was gleaned from RMSF analyses, which allowed a residue-by-residue measurement of temperature effects on structural movements. Mobility differences noted through RMSF analysis mirrored those obtained through measurement of temperature factors (B factors) in crystallographic studies of protein structure (20). RMSF spectra (Fig. 3) showed similar patterns among orthologs. In agreement with other studies of MDH and LDH, the largest amounts of structural fluctuation occurred in regions known to be involved in ligand binding and catalysis—notably, the catalytic loop region (residues 91–101 in the numbering system that includes the N-terminal methionine residue) and the catalytic region (residues 233–245). Substrate binding occurred from residues 93–99 within the catalytic loop. In both of these functionally important regions, there is a high degree of conservation of sequence among orthologs in both LDHs and MDHs (2, 27, 28). Based on the high degree of sequence identity in these two key regions, we conjecture that the adaptive differences in thermal stability of NADH binding are likely to be due to sequence differences outside of MR1 and MR2. Thus, we propose that the sites of this adaptive variation in flexibility lie principally in regions of the sequence that influence the conformational stabilities of the MRs themselves and/or the energy changes that occur during formation of the ternary complex—notably, the collapse of the catalytic loop during cofactor binding.
The data in Table 1 provide a basis for interpreting the sensitivities of NADH binding to temperature. MR1 and MR2 both show a general increase in flexibility, as indexed by the number of residues showing ΔRMSF values >0.5 Å, as adaptation temperature decreases. Thus, for the most warm-adapted species, E. malaccana, only three residues in the two MRs showed significant change. In contrast, for the most cold-adapted species, C. montereyi, 21 residues in MR1 and MR2 showed significant change. These differences reflect the trends for total number of residues shown in Fig. 3. We interpret the effects of rising temperature on KMNADH to reflect increases in the fraction of binding-incompetent conformations of the enzyme as temperature rises, with cold-adapted orthologs showing the greatest increase. This interpretation is consistent with the concept of conformational microstates (11) and their role in governing protein activity. Only a fraction of the possible suite of microstates will have the geometry required for ligand (here, NADH) recognition, and, as temperature rises, the fraction of binding-competent microstates decreases. The increase in binding-incompetent microstates with rising temperature is conjectured to be indexed by the number of residues in MR1 and MR2 that show ΔRMSF values > 0.5 Å. Moreover, as shown in Table 1, residues lying adjacent to one or both of the MRs also showed significant changes in ΔRMSF with rising temperature in cold-adapted species. This suggests that larger fractions of sequence in catalytically important regions of the enzyme become more mobile in these species as temperature rises. The MR2 of the ortholog of C. montereyi, a cold-adapted species, provided an illustration of this expanded region of high mobility; several sites in the sequence bordering each side of MR2 showed enhanced flexibility with rising temperature.
Differences in flexibility of the MRs may also underlie temperature-adaptive interortholog differences in kcat values. Whereas we did not measure the kcat values of the cMDH orthologs in this study, previous work on cMDH and LDH has shown that kcat varies inversely with evolutionary adaptation temperature (2, 9). Lower energy costs for the rate-determining conformational changes that occur during catalysis have been conjectured as responsible for this trend. The observed interspecific differences in flexibility of the catalytically important MRs of the enzyme are consistent with this hypothetical mechanism.
Identifying the specific amino acid substitutions that are responsible for the interortholog differences in ΔRMSF in MR1 and MR2 can be done with varying degrees of confidence. The clearest cause–effect linkage is for the two congeners of Lottia, whose cMDHs differ by only a single substitution: At position 291, the serine in the ortholog of the more heat-tolerant species, Lottia austrodigitalis, is replaced by a glycine in the ortholog of Lottia digitalis (27). Even though this site in the sequence falls well outside of MR1 and MR2, it appears to affect the temperature sensitivity (ΔRMSF) of both MRs. The differences in ΔRMSF of the MRs, which may be due to the occurrence of additional hydrogen bonds in the cMDH of L. austrodigitalis, appear to provide a good illustration of the effects of transmitted conformational effects through large regions of the protein, in keeping with the analysis of Gerstein and Chothia (13) and others (28). In congeners of Echinolittorina, only two differences in sequence were found (positions 48 and 114) (21). Thus, as in the case of the congeners of Lottia, differences in flexibility of MRs may be influenced by amino acid substitutions that fall outside of MR1 and MR2.
In summary, our combined use of in vitro methods (kinetic experiments and thermal stability analyses) and in silico MDS analysis has provided further insights into the mechanisms by which orthologous proteins become adapted for function in widely different thermal conditions. Most prior studies that used MDS methods to examine temperature effects on proteins compared distantly related species (7, 29–32); our work, with 12 differently adapted congeners belonging to five molluscan genera, allowed a multilineage analysis of convergent adaptation in flexibility. We show that protein flexibility as indexed by ΔRMSD and ΔRMSF is a common outcome of adaptive changes in amino acid sequence and that regions involved in large conformational changes during binding and catalysis are especially important features of the adaptive response. Our data support the microstate ensemble model of protein structure (11), which postulates rapid and temperature-sensitive interconversion of conformational microstates and a shift toward an ensemble with fewer ligand-binding-competent microstates with high or low extremes of temperature (1). It has been conjectured that the intensity of natural selection on proteins evolving at different temperatures may be governed by the types and magnitudes of conformational changes that are necessary for function (14). If this is the case, then the types of flexibility-modulating adaptations described for cMDHs may be common among many, but possibly not all, classes of enzymes. Investigations of other sets of orthologous proteins from species adapted to widely different ranges of temperature seem warranted to test the generality of the observations made in this and earlier studies (2). In vitro and in silico studies of these “naturally occurring mutants” could complement studies of laboratory-created mutants (29) and yield deeper insights into the processes of protein evolution across the proteome and provide a better understanding of how changes in sequence translate into adaptive modifications of protein function and structure.
Materials and Methods
Determination of KMNADH and Rates of Heat Denaturation.
Enzyme preparation involved ammonium sulfate precipitation of heat-treated homogenates (heating removed the thermally labile mitochondrial paralog, mMDH) to isolate a cMDH-rich protein fraction (27). Dialyzed aliquots of the ammonium sulfate-precipitated proteins were used for assays. Binding of NADH to cMDH was indexed by the apparent Michaelis–Menten constant, KMNADH (27). KMNADH values were calculated from the initial velocities of the reaction at each NADH concentration by using Prism software (Version 5.0; GraphPad Software). Thermal stabilities were measured as described by Fields et al. (33). Dialyzed enzyme preparations were heated at 42.5 °C for different periods of time. Aliquots were removed and assayed for residual cMDH activity. See SI Materials and Methods for more details.
Sequencing of cMDH cDNA.
To obtain the data needed for the MDS analyses, we sequenced all of the cMDH orthologs that had not previously been sequenced. The protocols used for sequencing the cDNAs followed those of Fields et al. (33), Dong and Somero (27), and Liao et al. (21). The cDNA sequences were assembled by using DNAMAN software (Lynnon Biosoft), and the deduced amino acid sequences were aligned by using the ClustalX2 algorithm (34). See SI Materials and Methods for more details.
Molecular Modeling of cMDH cDNA.
By using the sequence data, 3D models were constructed by the I-TASSER server with a high C-score level (∼1.6). C-score is a confidence score for estimating the quality of predicted models by I-TASSER, which is typically in the range between −5 and 2, and a higher value signifies a model with a high confidence (35).
Molecular Dynamic Simulation of cMDH.
Computed 3D structures constructed as described above were used as the starting models of the simulations. Simulations were performed by using NAMD (Version 2.9) (36) with the CHARMM36 force field (37–39). Transferable intermolecular potential 3P water was used as the aqueous solution to create simulation conditions that more closely resembled the cellular environment (40). The solvated systems were subjected to energy minimization to remove energetically unfavorable contacts among water molecules and ions (steepest descent method, 5,000 steps). Each system was performed in the isobaric–isothermal (NPT) ensemble at 1 bar pressure and the set temperature by the Langevin Piston and Langevin Dynamic method, respectively. For all simulations, each system was assigned for 20 ns at 15 and 42 °C in triplicate. Trajectories of the structures were collected and stored every 0.002 ns during the simulation. See SI Materials and Methods for more details.
The VMD program was used to visualize and analyze the simulation trajectories (41). The rmsd of backbone atom positions and the RMSF for individual residues in all models were calculated and compared. The stabilized structure (10‒20 ns) was taken from the trajectory of the system to determine the movements of protein backbone and individual residue atoms. The averaged equilibrium rmsd of backbone atom positions and averaged equilibrium RMSF for individual residues were calculated and compared. The differences between the values obtained at 15 and 42 °C for rmsd (ΔRMSD) and RMSF (ΔRMSF) were calculated to provide an estimate of protein flexibility. Differences between species were analyzed by using one-way analysis of variance followed by the Tukey’s multiple comparisons test (P = 0.05) with GraphPad Prism software (Version 6.0).
Supplementary Material
Acknowledgments
We thank Drs. Brent Lockwood and Peter A. Fields for helpful suggestions on the manuscript. This work was supported by National Natural Science Foundation of China Grants 41776135 and 41476115; Nature Science Funds for Distinguished Young Scholars of Fujian Province, China Grant 2017J07003; and the State Key Laboratory of Marine Environmental Science.
Footnotes
The authors declare no conflict of interest.
Data deposition: The cMDH sequences of congeners of five genera of marine molluscs reported in this paper have been deposited in the GenBank database [accession nos. MF774406 (Echinolittorina malaccana), MF774407 (Echinolittorina radiata), MG256005 (Littorina keenae), MG256006 (Littorina scutulata), MF774409 (Nerita yoldii), MF774408 (Nerita albicilla), MG190821 (Chlorostoma montereyi), MG190820 (Chlorostoma brunnea), MG190818 (Chlorostoma funebralis), MG190819 (Chlorostoma rugosa), EU863452 (Lottia digitalis), and EU863454 (Lottia austrodigitalis)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718910115/-/DCSupplemental.
References
- 1.Somero GN, Lockwood BL, Tomanek L. Biochemical Adaptation: Response to Environmental Challenges from Life’s Origins to the Anthropocene. Sinauer Associates; Sunderland, MA: 2017. [Google Scholar]
- 2.Fields PA, Dong Y, Meng X, Somero GN. Adaptations of protein structure and function to temperature: There is more than one way to ‘skin a cat’. J Exp Biol. 2015;218:1801–1811. doi: 10.1242/jeb.114298. [DOI] [PubMed] [Google Scholar]
- 3.Feller G. Enzyme function at low temperatures in psychrophiles. In: Siddiqui KS, Thomas T, editors. Protein Adaptation in Extremophiles. Nova Science; Hauppauge, NY: 2008. pp. 35–69. [Google Scholar]
- 4.Feller G, Gerday C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat Rev Microbiol. 2003;1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 5.Dalhus B, et al. Structural basis for thermophilic protein stability: Structures of thermophilic and mesophilic malate dehydrogenases. J Mol Biol. 2002;318:707–721. doi: 10.1016/S0022-2836(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 6.Gerday C, et al. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 2000;18:103–107. doi: 10.1016/s0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
- 7.Olufsen M, Smalås AO, Moe E, Brandsdal BO. Increased flexibility as a strategy for cold adaptation: A comparative molecular dynamics study of cold- and warm-active uracil DNA glycosylase. J Biol Chem. 2005;280:18042–18048. doi: 10.1074/jbc.M500948200. [DOI] [PubMed] [Google Scholar]
- 8.Fields PA. Review: Protein function at thermal extremes: Balancing stability and flexibility. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:417–431. doi: 10.1016/s1095-6433(00)00359-7. [DOI] [PubMed] [Google Scholar]
- 9.Fields PA, Somero GN. Hot spots in cold adaptation: Localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc Natl Acad Sci USA. 1998;95:11476–11481. doi: 10.1073/pnas.95.19.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tehei M, et al. Adaptation to extreme environments: Macromolecular dynamics in bacteria compared in vivo by neutron scattering. EMBO Rep. 2004;5:66–70. doi: 10.1038/sj.embor.7400049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrabl JO, et al. The role of protein conformational fluctuations in allostery, function, and evolution. Biophys Chem. 2011;159:129–141. doi: 10.1016/j.bpc.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 13.Gerstein M, Chothia C. Analysis of protein loop closure. Two types of hinges produce one motion in lactate dehydrogenase. J Mol Biol. 1991;220:133–149. doi: 10.1016/0022-2836(91)90387-l. [DOI] [PubMed] [Google Scholar]
- 14.Lockwood BL, Somero GN. Functional determinants of temperature adaptation in enzymes of cold- versus warm-adapted mussels (genus Mytilus) Mol Biol Evol. 2012;29:3061–3070. doi: 10.1093/molbev/mss111. [DOI] [PubMed] [Google Scholar]
- 15.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 16.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 17.Smith LJ, Daura X, van Gunsteren WF. Assessing equilibration and convergence in biomolecular simulations. Proteins. 2002;48:487–496. doi: 10.1002/prot.10144. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi K, Salam MU. Comparing simulated and measured values using mean squared deviation and its components. Agron J. 2000;92:345–352. [Google Scholar]
- 19.Maiorov VN, Crippen GM. Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J Mol Biol. 1994;235:625–634. doi: 10.1006/jmbi.1994.1017. [DOI] [PubMed] [Google Scholar]
- 20.Chapman AD, Cortés A, Dafforn TR, Clarke AR, Brady RL. Structural basis of substrate specificity in malate dehydrogenases: Crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, α-ketomalonate and tetrahydoNAD. J Mol Biol. 1999;285:703–712. doi: 10.1006/jmbi.1998.2357. [DOI] [PubMed] [Google Scholar]
- 21.Liao ML, et al. Heat-resistant cytosolic malate dehydrogenases (cMDHs) of thermophilic intertidal snails (genus Echinolittorina): Protein underpinnings of tolerance to body temperatures reaching 55°C. J Exp Biol. 2017;220:2066–2075. doi: 10.1242/jeb.156935. [DOI] [PubMed] [Google Scholar]
- 22.Marshall DJ, Dong YW, McQuaid CD, Williams GA. Thermal adaptation in the intertidal snail Echinolittorina malaccana contradicts current theory by revealing the crucial roles of resting metabolism. J Exp Biol. 2011;214:3649–3657. doi: 10.1242/jeb.059899. [DOI] [PubMed] [Google Scholar]
- 23.Coquelle N, Fioravanti E, Weik M, Vellieux F, Madern D. Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environments. J Mol Biol. 2007;374:547–562. doi: 10.1016/j.jmb.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 24.Kovacic F, Mandrysch A, Poojari C, Strodel B, Jaeger KE. Structural features determining thermal adaptation of esterases. Protein Eng Des Sel. 2016;29:65–76. doi: 10.1093/protein/gzv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birktoft JJ, Rhodes G, Banaszak LJ. Refined crystal structure of cytoplasmic malate dehydrogenase at 2.5-A resolution. Biochemistry. 1989;28:6065–6081. doi: 10.1021/bi00440a051. [DOI] [PubMed] [Google Scholar]
- 26.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 27.Dong Y, Somero GN. Temperature adaptation of cytosolic malate dehydrogenases of limpets (genus Lottia): Differences in stability and function due to minor changes in sequence correlate with biogeographic and vertical distributions. J Exp Biol. 2009;212:169–177. doi: 10.1242/jeb.024505. [DOI] [PubMed] [Google Scholar]
- 28.Holland LZ, McFall-Ngai M, Somero GN. Evolution of lactate dehydrogenase-A homologs of barracuda fishes (genus Sphyraena) from different thermal environments: Differences in kinetic properties and thermal stability are due to amino acid substitutions outside the active site. Biochemistry. 1997;36:3207–3215. doi: 10.1021/bi962664k. [DOI] [PubMed] [Google Scholar]
- 29.Sharma R, Sastry GN. Deciphering the dynamics of non-covalent interactions affecting thermal stability of a protein: Molecular dynamics study on point mutant of Thermus thermophilus isopropylmalate dehydrogenase. PLoS One. 2015;10:e0144294. doi: 10.1371/journal.pone.0144294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papaleo E, et al. Protein flexibility in psychrophilic and mesophilic trypsins. Evidence of evolutionary conservation of protein dynamics in trypsin-like serine-proteases. FEBS Lett. 2008;582:1008–1018. doi: 10.1016/j.febslet.2008.02.048. [DOI] [PubMed] [Google Scholar]
- 31.Pasi M, Riccardi L, Fantucci P, De Gioia L, Papaleo E. Dynamic properties of a psychrophilic α-amylase in comparison with a mesophilic homologue. J Phys Chem B. 2009;113:13585–13595. doi: 10.1021/jp900790n. [DOI] [PubMed] [Google Scholar]
- 32.Du X, et al. Comparative thermal unfolding study of psychrophilic and mesophilic subtilisin-like serine proteases by molecular dynamics simulations. J Biomol Struct Dyn. 2017;35:1500–1517. doi: 10.1080/07391102.2016.1188155. [DOI] [PubMed] [Google Scholar]
- 33.Fields PA, Rudomin EL, Somero GN. Temperature sensitivities of cytosolic malate dehydrogenases from native and invasive species of marine mussels (genus Mytilus): Sequence-function linkages and correlations with biogeographic distribution. J Exp Biol. 2006;209:656–667. doi: 10.1242/jeb.02036. [DOI] [PubMed] [Google Scholar]
- 34.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacKerell AD, Jr, Feig M, Brooks CL., 3rd Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 39.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen WL, et al. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 41.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.