Abstract
Acquired drug resistance is a major factor limiting the effectiveness of targeted cancer therapies. Targeting tumors with kinase inhibitors induces complex adaptive programs that promote the persistence of a fraction of the original cell population, facilitating the eventual outgrowth of inhibitor-resistant tumor clones. We show that the addition of a newly identified CDK7/12 inhibitor, THZ1, to targeted therapy enhances cell killing and impedes the emergence of drug-resistant cell populations in diverse cellular and in vivo cancer models. We propose that targeted therapy induces a state of transcriptional dependency in a subpopulation of cells poised to become drug tolerant, which THZ1 can exploit by blocking dynamic transcriptional responses, remodeling of enhancers and key signalling outputs required for tumor cell survival in the setting of targeted therapy. These findings suggest that the addition of THZ1 to targeted therapies is a promising broad-based strategy to hinder the emergence of drug-resistant cancer cell populations.
Keywords: Targeted therapy, resistance, CDK7, THZ1, cancer
Introduction
Large-scale genomic profiling efforts have facilitated the characterization of molecular alterations in cancers and aided the development of targeted kinase inhibitors for a wide array of cancer types. However, resistance to these targeted therapies invariably develops and limits their clinical efficacy(1–3). Resistance often emerges following an initial period of drug responsiveness via clonal evolution of the cancer cell population. This may entail acquisition of treatment-refractory mutations in the original target(4, 5), reactivation of key downstream effectors of the targeted pathway(6, 7), activation of alternative signalling pathways(8, 9), or cell state changes(10) which render the cell population indifferent to the original therapy. The emergence of acquired resistance is facilitated by the rapid induction of a complex network of pro-survival and pro-proliferative pathways upon exposure to targeted therapy(11–13), collectively promoting the persistence of a fraction of the original population in a drug-tolerant state(14) and permitting the eventual outgrowth of resistant clones.
We hypothesized that repression of the transcriptional changes that underlie the adaptive pro-survival and pro-proliferative responses induced by targeted therapy would interfere with the establishment of the drug tolerant state, resulting in improved therapeutic efficacy. This would be advantageous clinically as it would circumvent having to anticipate, elucidate, and target the myriad of potential drug resistance mechanisms that might arise in a particular patient. To test our hypothesis, we employed a novel transcriptional repressor, THZ1(15). THZ1 is a covalent CDK7 inhibitor, which additionally targets CDK12 at higher doses(15). CDK7 is a key regulator of the cell cycle(16–18), and together with CDK12, regulates RNA polymerase II (RNAPII)-mediated transcription(19–23). Prior studies have identified subsets of cancers with marked sensitivity to THZ1 monotherapy (T-cell acute lymphoblastic leukemia(15), MYCN-dependent neuroblastoma(24), small cell lung cancer(25), and triple negative breast cancer(26)). Vulnerability to THZ1 has been shown to be mediated by a strong dependency on specific oncogenes regulated by super-enhancer elements, and their corresponding transcriptional circuits (i.e., RUNX in acute lymphoblastic leukemia, MYCN in neuroblastoma, and in the case of triple negative breast cancer a core set of genes termed the “Achilles cluster” (15, 24, 26)). Importantly, THZ1 has been shown to be typically well-tolerated in mouse models, suggesting a therapeutic index for this agent in combination therapy approaches(15, 24, 25),(26). Here, we sought to test whether there may be a basis for therapeutic synergy among THZ1 and targeted cancer therapies in a variety of oncogene-driven cancer models, and specifically, whether transcriptional CDK inhibition may be a novel therapeutic approach to impede the ability of cancer cells to persist in the setting of targeted cancer therapy. For ease of reading, we use the term targeted therapy here to refer to small molecule inhibitors of receptor tyrosine kinases and additional signal transduction kinases (e.g. MEK1/2, BRAF). THZ1 as an inhibitor of CDK7 could also be classified as targeted therapy, although we do not refer to it as such herein.
Results
THZ1 suppreses the emergence of resistant cell populations in vitro
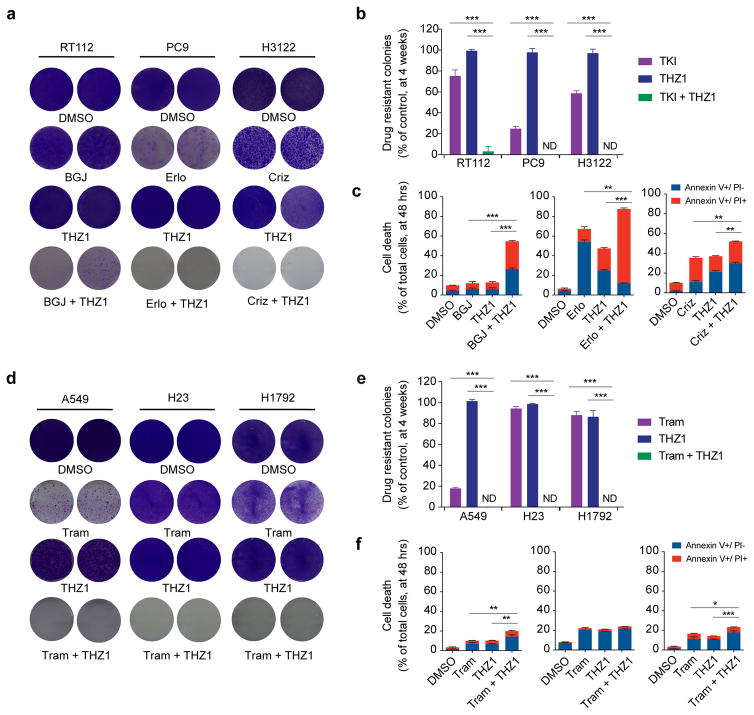
To determine whether THZ1 can suppress the emergence of resistant cancer cell populations, we first performed colony formation assays in vitro in a panel of well-established oncogene-dependency models with diverse receptor-tyrosine kinase dependencies and lineages (Fig. 1a,b, and Supplementary Fig. 1a, Supplementary Fig. 2a). Of note, despite low nanomolar potency of targeted agents in the models employed (Supplementary Fig. 1a), resistant cell populations were readily detected by four weeks of kinase inhibitor treatment, underscoring the need for strategies that further enhance the efficacy of targeted therapies (Fig. 1a,b and Supplementary Fig. 2a). The models employed exhibited variable sensitivity to THZ1 monotherapy; however, in the majority of models high-dose THZ1 (e.g. 1 μM or higher) eliminated all or nearly all cells at 96 hours (Supplementary Fig. 1b) as THZ1 acts as a general suppressor of transcription at high doses akin to actinomycin(15). To avoid general toxicity associated with high-dose THZ1 and to assess for therapeutic synergy with targeted kinase inhibitors we employed sub-lethal low nanomolar doses, corresponding approximately to the IC50 of THZ1 (Supplementary Fig. 1a). This dose of THZ1 approximated the minimal dose which produced a robust response in combination with targeted kinase inhibitors. At these sub-lethal doses colony formation at four weeks with THZ1 treatment alone was comparable to control. Colony formation with targeted tyrosine kinase inhibition alone had variable effect with up to 90% reduction in colony formation compared to control depending on the cellular model, however yielded resistant colonies in all models (Fig. 1a,b and Supplementary Fig. 2a). In contrast, combination treatment with THZ1 and targeted kinase inhibition, yielded few, or no detectable colonies. Combination treatment also significantly enhanced cell death at an early time point (48 hours) compared to either single agent alone (Fig. 1c, and Supplementary Fig. 2b). We observed an equally striking effect when treating with THZ1 in combination with a MEK-inhibitor (trametinib) in KRAS-mutant NSCLC or gastric cancer cellular models, and with THZ1 in combination with a BRAF-inhibitor (vemurafenib) in a BRAF-mutant melanoma model (Fig. 1d–f, Supplementary Fig. 1a,b and Supplementary Fig. 2a). Additionally, colony formation results at four weeks were maintained in long-term assays with no detectable drug-resistant colonies noted in combination-treated wells at three months (Supplementary Fig. 2c). We considered the specificity of the observed synergy by performing colony formation assays with THZ1 in combination with kinase inhibitors not targeting the dependency of the cellular model tested (e.g., A549, a KRAS-dependent model, was treated with crizotinib (a MET/ALK inhibitor) instead of trametinib), and we found that combination treatment with a mismatched kinase inhibitor plus THZ1 had no effect on colony outgrowth (Supplementary Fig. 2d). Taken together, these data suggest that THZ1 broadly has the ability to prevent resistance emergence to targeted kinase inhibition in diverse genetic contexts and lineages.
Figure 1. THZ1 in combination with targeted therapy enhances cell death and hinders the establishment of drug-resistant colonies in diverse oncogene-addicted cellular models.
a, Receptor tyrosine kinase-dependent cell lines, RT112 (FGFR), PC9 (EGFR) and H3122 (ALK) were treated with DMSO, the corresponding tyrosine kinase inhibitor (TKI: BGJ398 (BGJ), erlotinib (Erlo), or crizotinib (Criz)), THZ1, or THZ1 in combination with the corresponding TKI. Colony formation was assayed by crystal violet staining at 4 weeks. DMSO was stained by 1 week. Two representative wells from a minimum of three biological replicates are shown per condition. (RT112: BGJ398 1 μM, THZ1 100 nM, PC9: erlotinib 1 μM, THZ1 100 nM, H3122: crizotinib 250 nM, THZ1 50 nM.) Note: Colony formation for RT112 with THZ1 at 150 nM in combination with BGJ398 yielded no detectable colonies at 4 weeks (Supplementary Fig. 2f).
b, Quantification of colony formation in (a), shown as a percentage of the control. Mean (3 biological replicates) +/− standard deviation (SD) shown (* p-value < 0.05, ** < 0.005, *** < 0.0005, two-sided t-test). ND = not detectable.
c, Cell death analysis with cells treated as in (a) by flow cytometry with Annexin V/PI staining, following 48 hours of treatment. Mean (3 biological replicates) +/− SD shown (* p-value < 0.05, ** < 0.005, *** < 0.0005, two-sided t-test, comparing total cell death (Annexin V+/PI− or PI+)). Left panel: RT112, middle panel: PC9, right panel: H3122.
d, KRAS-mutant cell lines, A549, H23, H1792 were treated with DMSO, trametinib (Tram), THZ1, or a combination of THZ1 and trametinib. Colony formation was assayed by crystal violet staining at 4 weeks. DMSO was stained by 1 week. Two representative wells from a minimum of three independent biological replicates are shown per condition. (A549: trametinib 200 nM, THZ1 150 nM, H23: trametinib 500 nM, THZ1 100 nM, H1792: trametinib 500 nM, THZ1 500 nM).
e, Quantification of colony formation in (d), shown as a percentage of the control. Mean (3 biological replicates) +/− SD shown (* p-value < 0.05, ** < 0.005, *** < 0.0005, two-sided t-test). ND = not detectable.
f, Cell death analysis with cells treated as in (d) by flow cytometry with Annexin V/PI staining, following 48 hours of treatment. Mean (3 biological replicates) +/− SD shown (* p-value < 0.05, ** < 0.005, *** < 0.0005, two-sided t-test, comparing total cell death (Annexin V+/PI− or PI+)). Left panel: A549, middle panel: H23, right panel: H1792. Note: H23 had different drug response dynamics compared to the other cell lines tested, with the cell death in the combination-treated group ensuing close to the two-week time point.
THZ1 as an adjunct to targeted therapy may offer a more broadly applicable strategy to rational combination therapy approaches
We further compared our results with one of the prominent approaches for addressing resistance, namely rational combination therapy employing two or more kinase inhibitors to simultaneously target both the driver-oncogene and previously identified resistance mechanisms(3, 27–30). We tested rational combination therapies in RT112, PC9 and H3122 cells, using BGJ398, erlotinib, or crizotinib respectively, in combination with agents targeting known resistance mechanisms for these cell lines (8, 31–36). As expected, rational combination therapy decreased the proportion of cells surviving acute treatment at 96 hours (Supplementary Fig. 2e), and reduced the outgrowth of resistant clones with variable success at four weeks and three months (Supplementary Fig. 2f,g). Rational combination therapy with targeted kinase inhibition and MEK or PI3K inhibition conferred the greatest decrease in colony formation at three months across the three models. Approaches utilizing these rational combinations have however been challenging to translate clinically due to toxicity which has compromised the ability to assess their clinical efficay(37–40). In contrast to the rational combination therapy approaches we tested, THZ1 in combination with targeted therapy consistently yielded no detectable colonies across all tested backgrounds at time points up to three months (Supplementary Fig. 2f,g).
CDK7 or CDK12-deficient cells display enhanced sensitivity to targeted therapy
To confirm that the therapeutic effects noted with THZ1 are due to CDK7 and/or CDK12 inhibition, we tested whether genetic depletion of CDK7 or CDK12 mimics the effects of THZ1 treatment. We generated CDK7 or CDK12-deficient PC9 cells using CRISPR-Cas9 gene editing (Supplementary Fig. 3a,b). Both CDK7 and CDK12-deficient PC9 cells displayed enhanced sensitivity to erlotinib at 48 hours as compared to PC9 cells with a control RNA guide (CDK7_12_dummy) (Supplementary Fig. 3c). CDK12 depletion, however, had more modest effects. We furthermore performed colony formation assays with CDK7 or CDK12-deficient PC9 cells; however, the general cytotoxicity of CDK7 or CDK12 depletion precluded the performance of longer-term experiments. In an orthogonal approach we performed colony formation with THZ531, a recently described selective covalent CDK12 inhibitor(41). THZ531 demonstrated therapeutic synergy with BGJ398 in RT112, and erlotinib in PC9, corroborating the data from CDK12-deficient PC9 cells (Supplementary Fig. 3d,e). These findings suggest that the synergy noted with THZ1 in combination with targeted therapies is conferred through inhibition of CDK7 and CDK12.
THZ1 in combination with targeted therapy retards tumor growth and improves survival in vivo
To assess the efficacy and toxicity of targeted therapy in combination with THZ1 in vivo, we performed xenograft studies using cell-line models of FGFR-mutant bladder carcinoma (RT112) and EGFR-mutant NSCLC (PC9) (Fig. 2a, Supplementary Fig. 4a). Tumor-bearing mice were treated with a) vehicle, b) BGJ398 (RT112) or erlotinib (PC9), c) THZ1, or d) combination treatment with the appropriate targeted therapy and THZ1. THZ1 in combination with targeted therapy retarded tumor growth compared to THZ1 or targeted therapy alone (Supplementary Fig.4a), and significantly improved survival (Fig. 2a). Importantly, combination therapy was well tolerated, with no weight loss or behavioral changes observed (Supplementary Fig. 4d).
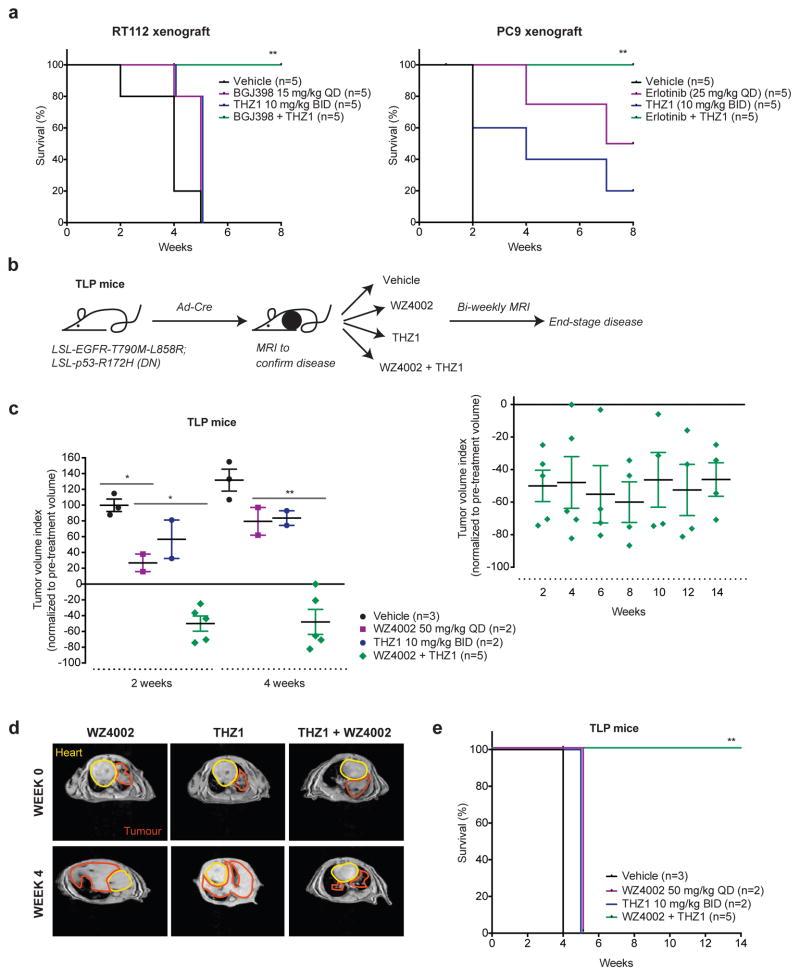
Figure 2. THZ1 in combination with targeted therapy increases survival in xenograft models and a novel EGFR-T790M-L858R LSL/−; p53-R172HLSL/− non-small cell lung cancer (NSCLC) genetically-engineered mouse model.
a, Xenografts of RT112 and PC9 tumors were treated with the indicated drugs for eight weeks (n=5 mice in each treatment group, equivalent to 10 tumors in each group). Survival over time is shown as a percentage for each treatment group. P-values are based on log-rank (Mantel-Cox) test analysis (* p-value < 0.05, ** < 0.005, *** < 0.0005).
b, Schematic of novel NSCLC genetically-engineered mouse model containing lox-stop-lox (LSL) EGFR-T790M-L858R and LSL p53-R172H dominant negative (DN) alleles (TLP mice). Mice were induced by intranasal administration at 6 weeks of age with Adenovirus-Cre recombinase. Upon determination of lung tumor growth by MRI, mice were randomized into treatment groups and imaged biweekly until end-stage disease to determine tumor response.
c, Tumor volume index, normalized to pre-treatment volume, for TLP mice treated with the indicated drugs at 2 and 4 weeks (left panel). Mean +/− standard error of the mean is shown (* p-value < 0.05, ** < 0.005, *** < 0.0005, two-sided t-test). Combination treated mice had long-term tumor-regression (right panel). Tumor volume index for combination-treated mice is shown up to 14 weeks.
d, Representative MRI images for mice treated with WZ4002, THZ1, or the combination of the two, pre-treatment and at week 4, showing significant tumor regression with combination treatment. Heart and tumor areas are drawn up and marked with yellow and red lines, respectively.
e, Survival curves for TLP mice treated with the indicated drugs. P-value determined by log-rank (Mantel-Cox) test analysis (* p-value < 0.05, ** < 0.005, *** < 0.0005).
In addition, we tested THZ1 in combination with the covalent T790M-mutant-EGFR selective inhibitor, WZ4002(42), in a novel EGFR-T790M-L858R LSL/−; p53-R172HLSL/− (TLP) genetically-engineered mouse model (GEMM) of NSCLC (Fig. 2b). L858R is an activating mutation of EGFR, T790M is a gatekeeper mutation which confers decreased sensitivity to first and second generation EGFR inhibitors, and p53-R172H is a dominant negative p53 mutation that is found in 38% of EGFR-mutant NSCLC and is associated with more advanced, aggressive disease(43). Upon detectable tumor-burden by Magnetic Resonance Imaging (MRI) mice were randomized into treatment groups (Fig. 2b). Thereafter tumor growth was evaluated by biweekly MRI. Treatment with WZ4002 resulted in initial response at two weeks (p=0.0117, two-tailed t-test); however, tumors rapidly developed resistance and rebounded by four weeks, reaching end-stage disease by five weeks of treatment, emphasizing the aggressive nature of this EGFR-mutant, p53-mutant GEMM. In stark contrast, combined THZ1-WZ4002 treatment resulted in a dramatic response with extensive long-term tumor regression (Fig. 2c, d). Mice in the combination arms continued to have significant tumor regression at 14 weeks of treatment (Fig. 2c, right panel). Furthermore, combination-treated mice had 100% survival vs. 0% survival for single-agent treated mice at 14 weeks (p=0.0019, log-rank test) (Fig. 2e), and no overt toxicity was evident in the combination-treated animals despite long-term treatment (Supplementary Fig. 4d).
We further tested THZ1 in combination with crizotinib in a previously published GEMM of EML4-ALK NSCLC(44) (Supplementary Fig. 4b,c). Mice in the combination treatment arm had significant tumor regression compared to crizotinib-treated mice (p=0.013, two-tailed t-test). In this model however, mice treated with THZ1 (or combination treatment) developed complications secondary to twice daily intraperitoneal injections (i.e., scarring, ascites, peritonitis) with longer-term dosing. The increased complications may be related to differences in the background, as C57BL/6 (TLP model) and Nu/Nu mice (xenograft studies) tolerated THZ1 administration well compared to mice on a mixed genetic background (EML4-ALK model).
Consistent with prior in vivo studies (15, 24, 25),(26) we show that intraperitoneal administration of THZ1 10 mg/kg BID is sufficient to saturate CDK7 binding in TLP GEMM tumor nodules and liver (Supplementary Fig. 4e).
THZ1 impedes the engagement of targeted therapy-induced transcriptional programs that promote tumor cell survival
To investigate the mechanisms by which THZ1 may enhance cell death and suppress resistance emergence in combination with targeted therapy we examined gene expression by RNA-seq in six independent cellular models representing diverse oncogenic dependencies and lineages (RT112, PC9, H3122, A375, N87, A549). Cells were treated for 48 hours with vehicle, THZ1, the corresponding targeted therapy, or combined THZ1 and targeted therapy treatment. Treatment with targeted kinase inhibitors induced extensive, but variable degrees of change in gene expression across the cellular models studied (Fig. 3a and Supplementary Fig. 5a). Consistent with prior work targeted therapy up-regulated the expression of genes involved in pro-survival programs, including NF-κB-, STAT- and TGF-β-driven transcription programs(12, 13, 45, 46) (Supplementary Fig. 5b). Cell lines had gene expression changes suggesting MAPK reactivation, including downregulation of negative regulators of the MAPK pathway (i.e., DUSP, SPRED and SPRY family members) (Supplementary Fig. 5b). We also noted FRA1 (FOSL1) downregulation consistent with activation of the previously described tumor secretome(11), and upregulation of stemness factors, such as Wnt/Hedgehog and SOX family members. Downregulation of cell cycle genes and upregulation of cell senescence programs further suggested transition to a quiescent cell state (Supplementary Fig. 5b). Importantly, the specific genes altered were generally distinct between the cell lines but highlighted programs serving similar functions, many of which have previously been implicated in drug-resistance(11–13, 31, 47–49). The original characterization of drug tolerant persisters(14) was performed at later time points, thus we confirmed that gene expression changes induced at 48 hours were stable by comparing with gene expression following seven days of treatment with targeted therapy. Indeed we found that the transcriptional programs induced and repressed at 48 hours were largely maintained at seven days, suggesting that this earlier time point is reflective of the drug tolerant state (Supplementary Fig. 5c).
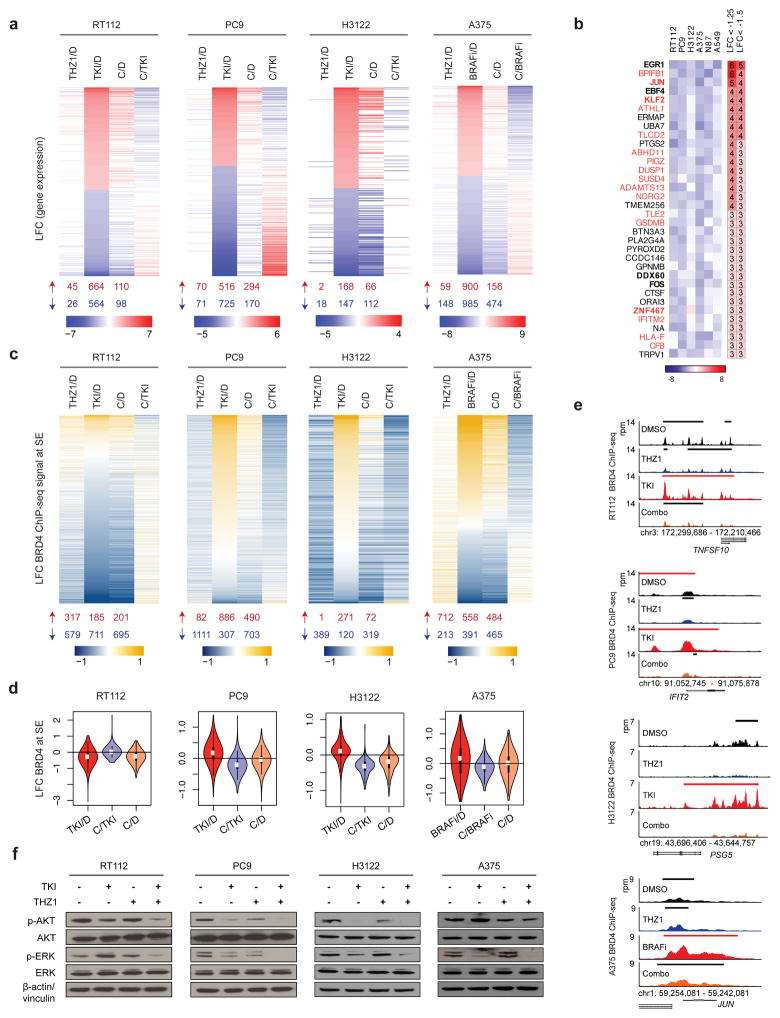
Figure 3. THZ1 attenuates targeted therapy-induced transcriptional and enhancer remodeling.
a, Differentially expressed genes at 48 hours following treatment with THZ1, TKI or BRAFi, or THZ1 in combination with TKI or BRAFi (Combination, C) compared to DMSO control (D), filtered by genes that were up- or downregulated greater than or equal to 1.5 log2-fold change (LFC) of read counts per million with targeted kinase inhibition, or less than or equal to −1.5 LFC, respectively. Column four shows LFC of combination-treated versus targeted therapy-treated cells (C/TKI, C/BRAFi). Heat maps show averaged values for 3 biological replicates per condition. The number of upregulated and downregulated genes (LFC ≥1.5, and ≤ −1.5, respectively) for each condition is summarized below each column, in red and blue respectively. LFC values between −1.5 and 1.5 are shown in white.
b, Heat map showing most downregulated genes in combination-treated cells versus targeted therapy-treated cells across all six cell lines. Only genes whose expression was downregulated less than −1.5 LFC were considered, and only genes affected in a minimum of three cell lines were included. Genes in bold are transcription factors. Genes in orange are EGR1 targets. Columns on the right indicate the number of cell lines with LFC<−1.25, and LFC<−1.5 for each respective gene.
c, LFC of BRD4 ChIP-seq signal at super-enhancer regions (SE) following 48-hour treatment with THZ1, targeted-therapy (TKI, BRAFi) or THZ1 in combination with targeted therapy (C) compared to DMSO control (D). The fourth column shows combination treatment compared to targeted therapy treatment (C/TKI). The heat map plots the union of SE regions identified in DMSO-treated cells and targeted therapy-treated cells. The number of upregulated and downregulated regions is summarized below each column, in red and blue respectively.
d, Violin plots showing the distribution of LFC of BRD4 ChIP-seq density at super-enhancer regions plotted in (c), for TKI compared to DMSO control (D), combination treatment compared to targeted therapy treatment (C/TKI), and combination compared to DMSO control (C/D).
e, BRD4 gene tracks for control-, THZ1-, targeted therapy (TKI)-, and combination (Combo)-treated cells for TNFSF10 (RT112), IFIT2 (PC9), PSG5 (H3122), and JUN (A375). Signal of ChIP-seq occupancy is in reads per million (rpm). Black bars indicate typical enhancers and red bars super-enhancers.
f, Immunoblot analysis for AKT and ERK activity for cells treated for 24 hours with control, the corresponding kinase inhibitor (BRAFi for A375), THZ1, or THZ1 in combination with the kinase inhibitor, at the doses used in colony formation assays.
Changes in gene expression induced by targeted therapy were significantly attenuated in the combination-treated arm; a finding that was most prominent in RT112, PC9, and A375, however consistent across all six models (Fig. 3a, and Supplementary Fig. 5a,b,d). Interestingly, combination treatment attenuated the expression of genes upregulated by targeted therapy, as well as of those downregulated by targeted therapy (i.e., these genes were less repressed in the combination treated arm). Supervised analyses further corroborated that THZ1 is interfering with the establishment of the adaptive responses to targeted therapy (Supplementary Fig. 5b). We noted, for instance, a diminished upregulation of NF-κB/Interferon pathway members, such as TNFSF10, MX1 and MX2 in RT112 combination-treated cells compared to BGJ398-treated cells. Lastly, in contrast to the gene expression changes noted with targeted therapy, treatment with low-dose THZ1 alone did not alter gene expression extensively (Fig. 3a, and Supplementary Fig. 5a, b, e).
We next considered which genes were most perturbed across all six models by combination treatment compared to targeted therapy alone, and found that only 34 genes were downregulated across multiple cell lines (three or more lines with ≤ − 1.5 log2-fold change). This small subset of most perturbed genes was enriched for transcription factors (e.g. EGR1, JUN, KLF2, FOS and their target genes) (Fig. 3b). These findings suggest that targeted therapy may induce a state that is dependent on the activity of these transcription factors, and is consistent with the finding that THZ1-sensitive cell lines have overexpression of oncogenic transcription factors and factors involved in the regulation of RNAPII-driven transcription(15). To further investigate the significance of EGR1 downregulation in the adaptive response to targeted therapy, we generated EGR1-deficient PC9 cells and performed colony formation assays with vehicle, erlotinib, THZ1 or combination therapy. Knockdown of EGR1 resulted in significantly diminished colony formation following erlotinib treatment compared to sg dummy PC9 cells, thus partly phenocopying the effect of THZ1 in combination with erlotinib (Supplementary Fig. 6a,b). EGR1 knockdown however had no effect on colony formation with THZ1 treatment alone or combination treatment. These findings further corroborate the importance of EGR1, and EGR1-dependent programs, for the adaptive rewiring to targeted therapies.
Considering shared upregulated genes across the six models with combination treatment versus targeted therapy alone we noted an enrichment for histone proteins (Supplementary Fig. 7a). CDK7 inhibition has been shown to impair the 3′-end processing of histone mRNAs, leading to aberrant polyadenylation(22), which may result in an artifactual gain of histone mRNA signal in the RNAseq analysis.
THZ1 prevents the rapid and dynamic remodeling of the enhancer landscape elicited by targeted therapies
Given that tumor cells acquire enhancers and super-enhancers at genes that control tumor cell identity46–49, and that THZ1 has been shown to disproportionally perturb super-enhancer-driven transcription8–10, we asked whether targeted therapy induces changes in the enhancer landscape in our models and whether THZ1 has an impact on enhancer remodeling. To address this we performed ChIP-seq for BRD4, a member of the bromodomain and extraterminal domain family of transcriptional coactivators and elongation factors(50). BRD4 localizes at enhancers and promoters and is known to be rapidly and dynamically regulated in response to such factors as NF-κB pathway activation(51), and most recently in response to inhibition by MEK1/2 in triple-negative breast cancer(52).
We found that targeted therapy led to a redistribution of BRD4 occupancy, and the addition of THZ1 attenuated the gain and loss of BRD4 signal elicited by targeted therapy at super-enhancers, as well as typical enhancers (Fig. 3c,d,e and Supplementary Fig. 8a,b,c). In line with our transcriptome findings, changes in BRD4 signal density induced by targeted therapy varied greatly across models (Supplementary Fig. 8c), and these changes were largely maintained at seven days (Supplementary Fig. 8d). Furthermore, super-enhancer associated genes had higher expression than typical enhancers, and changes in the enhancer landscape paralleled those in gene expression (Supplementary Fig. 9a,b). Interestingly, THZ1 monotherapy also induced changes in BRD4 signal density that varied considerably across models (Fig. 3c and Supplementary Fig. 8a,c), however these changes did not correspond to significant effects on transcription or cell viability. Taken together these findings suggest that THZ1 impinges on the ability of tumor cells to efficiently redistribute transcription factors and remodel enhancers that allow them to escape targeted kinase inhibition.
Repression of the adaptive responses to targeted therapy by THZ1 results in more complete ERK suppression
The adaptive responses to targeted therapy have previously been shown to re-activate key survival pathways, such as AKT/PI3K and ERK/MAPK(31, 32, 34, 53, 54), therefore we performed a targeted immunoblotting analysis in a subset of cellular models to consider activation of these key survival pathways. We found that combination treatment with THZ1 and targeted therapy resulted in enhanced AKT pathway suppression in receptor-tyrosine kinase dependent models, and more complete ERK suppression in all cell lines tested (Fig. 3f, Supplementary Fig. 10a). These data, along with the finding that MAPK pathway repressors, such as DUSPs, are downregulated with targeted therapy (Supplementary Fig. 5b), suggests that the transcriptional reprogramming engaged by targeted therapy converges in part on MAPK reactivation. Downregulation or deletion of MAPK negative regulators leading to MAPK reactivation has previously been shown to confer resistance to FGFR and ALK inhibition in lung cancer (32, 53), BRAF inhibition in melanoma(55), MET inhibition in gastric cancer(56), and to MEK inhibition more broadly(57). To further consider the role of MAPK in our models we knocked down two well-described negative regulators of MAPK, NF1 and SPRED2, in PC9 cells. NF1 or SPRED2 knockdown resulted in an observable rescue from erlotinib and to a lesser degree erlotinib plus THZ1 treatment (Supplementary Fig. 10b–e). We further examined ERK activity in NF1 or SPRED2-deficient PC9 cells following treatment with vehicle, erlotinib, THZ1 or erlotinib plus THZ1 (Supplementary Fig. 10f). NF1 and SPRED2-deficient cells displayed greater ERK activity following erlotinib or combination treatment, compared to sg dummy PC9 cells. Combination treatment nevertheless resulted in comparatively enhanced ERK suppression compared to erlotinib treatment alone. Cumulatively these findings suggest that ERK activity is a key component of the cellular survival program in the context of targeted therapy, and that repression of the adaptive responses to targeted therapy, by THZ1, more fully suppresses this component.
Discussion
A growing body of evidence suggests that tumors are able to evade targeted cancer therapies by an extensive repertoire of resistance mechanisms(7, 11, 34). This poses a significant therapeutic challenge, as it may not be feasible to target the multitude of potentially relevant escape pathways in each individual patient. Recent studies indicate that targeted therapies acutely elicit pro-survival and pro-proliferative responses, which promote the persistence of a drug-tolerant population and facilitate resistance emergence (11–14). We hypothesized that we may be able to interfere with the adaptive reprogramming response and enhance the efficacy of targeted cancer therapy by employing a novel transcriptional repressor, THZ1.
THZ1 is a covalent CDK7/12 inhibitor with reported single agent activity in T-cell acute lymphoblastic leukemia(15), MYCN-dependent neuroblastoma(24), small cell lung cancer(25), and triple negative breast cancer(26), and recently in combination with BH3 mimetic drugs in peripheral T-cell lymphomas(58). Here we show that THZ1’s mode of action may also be leveraged to prevent resistance emergence to targeted therapies. THZ1 in combination with targeted therapies resulted in significantly enhanced cell death and decreased resistance emergence in cellular cancer models and led to significant tumor regression and increased survival in vivo in xenograft models and immunocompetent genetically-engineered mouse models. Notably, these findings were broadly applicable across diverse oncogenic dependencies and lineages, suggesting that CDK7/12 inhibition may be a promising broad-based strategy for enhancing the effectiveness of targeted therapies.
As noted, THZ1 is a covalent inhibitor of both CDK7 and CDK12, and we cannot from our study conclude the extent to which the inhibition of CDK7 or CDK12 contributes to the noted therapeutic effect. CDK7 and CDK12-specific compounds are in development and will aid in determining the contribution of each to the noted synergy. Here we show that genetic depletion of CDK7 or CDK12 resulted in enhanced sensitivity to erlotinib in PC9 cells, and additionally that a novel covalent CDK12-specific compound, THZ531(41), as an adjunct to targeted therapy, results in comparable synergy to THZ1 in colony formation studies. These findings suggest roles for both CDK7 and CDK12 in the noted therapeutic synergy. CDK7-specific compounds were not tested as none are currently published.
Consistent with prior studies we noted that high-dose THZ1 or genetic depletion of CDK7 or 12 is cytotoxic. Kwiatkowski et al have previously shown that high-dose THZ1 results in complete CDK7 inhibition, leading to global downregulation of steady-state mRNA levels by 12 hours with a concomitant dramatic loss of cell viability, while low-dose THZ1 only downregulates a subset of transcripts(15). This subset of preferentially downregulated transcripts largely consists of oncogenes whose high-level expression is driven by super-enhancers, as well as additional genes that form the core regulatory circuitry of these oncogenes. In the current study, we employed lower doses of THZ1 (~IC50), at which transcription was modestly affected by monotherapy (Supplementary Fig. 5e) and cell viability effects were minimal, suggesting incomplete target inhibition. Our data suggest that this partial CDK7/12 inhibition is sufficient to hinder enhancer remodeling and the establishment of novel transcriptional circuits elicited by targeted therapy. Thus, akin to models intrinsically sensitive to THZ1, where low-dose THZ1 disrupts the transcription of key transcription factors thus interfering with core pre-existing oncogenic circuits, we believe that THZ1 in the setting of targeted therapy disrupts the establishment of novel pro-survival circuits, possibly by lowering levels of transcription factors key to the reprogramming process.
Consistent with prior studies we found that targeted therapies acutely induce extensive transcriptional changes that support drug-resistance emergence(11–13, 31, 47, 48). Our data suggest that although similar adaptive responses are engaged across diverse models (e.g., NF-κB/STAT pathway activation), the specific molecules involved in the adaptive reprogramming response vary based on the specific cellular context (e.g., IGFBP5 was upregulated by BGJ398 in RT112, while IGFBP2 was upregulated by trametinib in A549). These results are consistent with the extensive heterogeneity noted in the adaptive kinome response to lapatinib in ERBB2-dependent breast cancer cell lines(59), and further highlight the advantages in preventing the adaptive reprogramming response as a whole, rather than targeting individual tumor type-specific components.
We additionally found that targeted cancer therapies elicit a rapid and dynamic remodeling of the enhancer landscape across a spectrum of solid cancer models, supporting transcriptional programs that facilitate resistance emergence. Acute remodeling of enhancers, has been shown to occur in response to pro-inflammatory stimuli, leading to rapid inflammatory gene activation(51, 60), and recently in response to MEK1/2 inhibition in triple-negative breast cancer models(52). Our mechanistic studies across multiple cellular cancer models suggest that CDK7/12 inhibition, by THZ1, prevents active enhancer formation at genes promoting resistance emergence in response to targeted therapy and impedes the engagement of the transcriptional programs and signaling outputs that characterize the drug tolerant state. Curiously, we found that THZ1 monotherapy, at the low doses employed in the study, had widely varying effects on BRD4 distribution at enhancers across cellular models, yet did not lead to significantly altered transcriptional outputs or decreased cellular viability.
Our work builds conceptually on prior studies that have suggested that blocking the adaptive reprogramming elicited by targeted therapy may have a therapeutic impact in cancer models. Several studies have shown that HDAC inhibitors can suppress, in vitro, the emergence of drug resistance(7, 14). Furthermore, HDAC inhibitors in combination with targeted therapies, have been tested in several early phase clinical trials in solid cancers (61–67). However, the majority have unfortunately shown minimal, or no, response, emphasizing the need for additional strategies to counter resistance. In addition, Stuhlmiller et al.(59) have shown that BET bromodomain inhibition can suppress the transcription of lapatinib-induced kinases in ERBB2-dependent breast cancer cell lines, preventing downstream SRC/FAK signalling and AKT reactivation, and arresting growth in vitro. These findings were recently extended to triple-negative breast cancer models, showing that pharmacological inhibition of BRD4 can prevent resistance emergence to MEK inhibition(52). Further in vitro and in vivo studies are necessary to delineate differences in the mechanism of action of BET inhibition, compared to CDK7/12 inhibition, in the context of therapeutic synergy with targeted therapies in diverse clinical settings.
Cancer cells that are highly dependent on transcription for maintenance of their oncogenic state, so-called transcriptionally addicted cells, have previously been shown to be highly susceptible to THZ1 monotherapy(15, 24, 25). Here, we propose a model whereby targeted therapy induces a state of acquired transcriptional addiction, in a subpopulation of cells poised to become drug tolerant, that is thereby highly vulnerable to THZ1 treatment. This provides a potentially powerful therapeutic intervention that obviates the need to anticipate or elucidate the myriad of drug resistance mechanisms that might arise in a particular patient.
Methods
Cell lines
RT112, PC9, NCI-H3122, N87, OE19, NCI-H2077, NCI-H1975, HCC827, EBC-1, NCI-H1703, A549, NCI-H23, NCI-H1792, and GSU were cultured in RPMI media, and A375 and NCI-H2009 were cultured in DMEM. Both types of media were supplemented with 10% FBS, and penicillin/streptomycin/L-glutamine. All cell lines were cultured at 37 °C in a humidified chamber in the presence of 5% CO2. Cell lines were obtained from ATCC, Sigma-Aldrich, or collaborating labs primarily in 2014. A375, N87, EBC-1, GSU, and NCI-H2009 were obtained in 2009. Cell lines were not authenticated. Cells were not passaged for more than 6 months. Cell lines used for RNAseq/ChIP-seq studies and for animal studies were mycoplasma tested and negative (MycoAlert PLUS, Lonza).
Cell viability assays
1500 cells were seeded in 96-well plates, allowed to adhere overnight, and then incubated with media containing vehicle or drug as indicated for 96 h. Following 96 h, cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability assay (Promega). Plates were read on a Tecan Infinite M200 Pro plate reader. All conditions were tested in triplicate, unless otherwise noted. Drug curves and IC50 values were generated using GraphPad Prism 6 (GraphPad Software).
Colony formation assays
100 000 cells were seeded in 6-well plates, allowed to adhere overnight, and then incubated with media containing vehicle or drug as indicated for four weeks, unless otherwise noted. Media (and drug) were replaced weekly. At 4 weeks plates were fixed with 1% paraformaldehyde and then stained with 0.1% crystal violet as previously described (http://medicine.yale.edu/lab/kim/resources/protocols/cell/crystal_violet_stain.aspx) to assess colony formation. Control wells were stained by one week. Results were quantified using an ImageJ Colony Area PlugIn(68). Values of less than 1% on the quantification were considered as not detectable (ND). Drug doses were as follows: RT112: BGJ398 1 μM, THZ1 100 nM (Figure 1) and 150 nM (Supplementary Figure 2f,g), PC9: erlotinib 1 μM, THZ1 100 nM, NCI-H3122: crizotinib 250 nM, THZ1 50 nM, A549: trametinib 200 nM, THZ1 150 nM, NCI-H23: trametinib 500 nM, THZ1 100 nM, NCI-H1792: trametinib 500 nM, THZ1 500 nM, NCI-H2077: BGJ398 1 μM, THZ1 10 nM, NCI-H1975: WZ4002 1 μM, THZ1 500 nM, HCC827: erlotinib 50 nm, THZ1 75 nM, N87: lapatinib 100 nM, THZ1 25 nM, OE19: lapatinib 150 nM, THZ1 125 nM, EBC-1: crizotinib 10 nM, THZ1 50 nM, NCI-H1703: imatinib 1 μM, THZ1 30 nM, GSU: trametinib 10 nM, THZ1 200 nM, NCI-H2009: trametinib 50 nM, THZ1 25 nM, A375: vemurafenib 1 μM, THZ1 50 nM.
Apoptosis/cell death analysis
100 000 cells were seeded in 6-well plates, allowed to adhere overnight, and then incubated with media containing vehicle or drug as indicated for 24 h or 48 h. Cell death was quantified using the Alexa Fluor 488 Annexin V/Dead Cell Apoptosis kit for flow cytometry (Invitrogen), according to the manufacturer’s protocol. All conditions were assayed in triplicate. Data were acquired using a BD LSRFortessa X-20 (BD Biosciences), and analyzed in FlowJo.
Xenograft tumor studies
Xenograft studies were approved by the Dana Farber Cancer Institute Animal Care and Use Committee. RT112, PC9, and A549 xenograft models were established by subcutanoues (s.c.) injection of 2 × 106 cells in Matrigel (Corning) into both flanks of nude mice (NU/NU, #088 Charles River) when animals were 8–10 weeks of age. The xenograft studies were powered to include 5 mice (10 tumors per treatment group) providing 82% power to detect an underlying difference in survival between 70% and 10% at eight weeks in Fisher’s exact test at a one-sided 0.05 level. When tumors reached between 100–200 mm3, as measured by caliper, mice were randomized to four groups of five female mice each, for each cell line: 1) vehicle, 2) BGJ398 (RT112), erlotinib (PC9) or trametinib (A549), 3) THZ1, or 4) combination treatment with THZ1 plus BGJ398 (RT112), erlotinib (PC9) or trametinib (A549). The animals were randomized to treatment using simple randomization by cage. Investigators were not blinded to group allocation. The following dosing regimens were employed: BGJ398 15 mg/kg once daily (QD) by oral gavage, erlotinib 25 mg/kg QD by oral gavage, trametinib 2.5 mg/kg QD by oral gavage, and THZ1 10 mg/kg twice daily (BID) by intraperitoneal (i.p.) injection. BGJ398 was dissolved in PEG300, erlotinib was dissolved in 0.5% methylcellulose and 0.4% Tween 80, and THZ1 was dissolved in 10% DMSO in D5W. Caliper measurements were then performed weekly and continued for eight weeks. A549 xenografts had severe ulcerations therefore were excluded from the study. RT112 xenografts had one mouse in the combination-treated group that was censored at week 5 (found dead, cause not known, tumor size small). PC9 xenografts had one mouse in the erlotinib-treated arm censored at week 1 due to ulceration.
Genetically-engineered EGFR-p53-mutant NSCLC mouse model
The study was approved by the Dana Farber Cancer Institute Animal Care and Use Committee. Mice (both male and female) bred to contain conditional EGFR-T790M-L858R lox-stop-lox (LSL) allele and the p53-dominant negative R172H LSL allele to a final genotype of EGFR- T790M-L858RLSL/−; p53-R172HLSL/− maintained on a mixed background, were induced at 6 weeks of age with Adenovirus-Cre recombinase by intranasal administration (69) to allow for Cre-mediated recombination of lox-stop-lox modified mutant-EGFR and p53 alleles. Upon clinical signs of disease magnetic resonance imaging (MRI) was performed to establish pre-treatment tumor burden in the lungs (generally 16–20 weeks of age). Mice were imaged using a 7 Tesla BioSpec (Bruker Biospin) optimized for image requisition of pulmonary parenchyma and vessels in mice. Animals were anesthetized with 2% isoflurane IsoFlo; Abbott) in 100% oxygen via a nose cone. Respiratory and cardiac gating was applied to minimize motion artifacts during imaging. 24 slices (1 mm) were collected. Tumor volume per animal was quantified manually, based on a minimum of eight consecutive axial image sequences, using the 3D Slicer. Upon determination of the pre-treatment volume, mice were randomized (by simple randomization) into treatment groups as follows: 1) vehicle, 2) WZ4002 (covalent T790M-mutant-EGFR selective inhibitor, 50 mg/kg QD by oral gavage) 3) THZ1 (10 mg/kg, BID, i.p.) or 4) THZ1+WZ4002. WZ4002 was dissolved in 5% N-methylpyrrolidone, and THZ1 in 10% DMSO in D5W. Pharmacokinetics properties of THZ1 are provided in Kwiatkowski et al.(15), and Wang et al.(26) Investigators were not blinded to group allocation. Mice were imaged biweekly by MRI until end-stage disease to determine tumor volume. Mice weights and signs of toxicity were monitored daily during the course of treatment. End-stage disease was reached when animals acquired clinical symptoms secondary to their lung tumors, in accordance with Dana Farber Cancer Institute Animal Care and Use Committee regulations.
Genetically-engineered EML4-ALK NSCLC mouse model
This study was approved by the Dana Farber Cancer Institute Animal Care and Use Committee. A previously described GEMM of NSCLC with doxycycline-inducible EML4-ALK was employed(70). Upon determination of the pre-treatment volume by MRI (as described above for the TLP GEMM), mice were randomized into treatment groups as follows: 1) vehicle, 2) crizotinib (50 mg/kg QD by oral gavage) 3) THZ1 (10 mg/kg, BID, i.p.) or 4) THZ1+crizotinib. Investigators were not blinded to group allocation. Crizotinib was dissolved in 5% hydroxypropyl methylcellulose, and THZ1 in 10% DMSO in D5W. Mice were imaged at two weeks and at four weeks by MRI to determine tumor volume. Mice weights and signs of toxicity were monitored daily during the course of treatment. End-stage disease was reached when animals acquired clinical symptoms secondary to their lung tumors, in accordance with Dana Farber Cancer Institute Animal Care and Use Committee regulations.
RNA-seq analysis
RNA was isolated from RT112, PC9, H3122, A375, N87, and A549 following treatment with DMSO, the appropriate targeted therapy, THZ1, or targeted therapy in combination with THZ1 at the doses used in colony formation assays (see above relevant section for dosing). Cells were harvested following 48 hours of treatment. For the targeted therapy arm cells were also harvested following 7 days of treatment. Cell number was determined and total RNA was isolated using the RNeasy micro kit (Qiagen). Ambion® ERCC RNA Spike-In Mix (Life technology) was added to total RNA. cDNA libraries were prepared using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer’s instructions. Library integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent). Sequencing was performed on the HiSeq 2000 platform (Illumina) to a minimum depth of 30 million reads per sample.
QC-passed reads were aligned to the human reference genome (hg19) using bowtie2(71) and quantified as gene-level read counts using RSEM(72). Genes assigned less than five reads in all samples were removed. For each cell line analyzed at given time point and conditions (untreated, THZ1-treated, targeted therapy-treated, and combination-treated), read count data was post-processed by removing lowly-expressed genes, applying data normalization, and calculating differential expression. Only genes expressing over ten reads in at least three samples were retained. Read counts were normalized to log-counts-per-million values with the voom transformation(73). Expression changes for each gene in treated cells compared to untreated controls was determined using the limma package(74) as log2-transformed fold change and a multiple-testing adjusted p-value. Heatmap visualization was performed using R. Log2-transformed fold changes were not scaled and were colored on a blue-red scale.
EGR1 target genes were defined based on JASPAR Predicted Transcription Factor Targets(75) (http://amp.pharm.mssm.edu/Harmonizome/dataset/JASPAR+Predicted+Transcription+Factor+Targets).
Chromatin Immunoprecipitation
RT112, PC9, H3122, A375, N87, and A549 cells were treated for 48 hours with vehicle, targeted therapy, THZ1, and targeted therapy in combination with THZ1 at the doses employed in colony formation assays (see above relevant section for dosing). Cells were crosslinked for 10 min at room temperature by the addition of one-tenth of the volume of 11% formaldehyde solution (11% formaldehyde, 50mM HEPES pH 7.5, 100mM NaCl, 1mM EDTA pH 8.0, 0.5mM EGTA pH 8.0) to the growth media followed by 5 min quenching with 2.5 M glycine. Cells were washed twice with PBS, the supernatant was aspirated and cells collected, and the cell pellet was flash frozen in liquid nitrogen. Frozen crosslinked cells were stored at −80°C. 50 μL of Dynal magnetic beads (Sigma) were blocked with 0.5% BSA (w/v) in PBS. Magnetic beads were bound with 10 μg of the indicated antibody. For BRD4 occupied regions, we performed ChIP-seq experiments using a Bethyl antibody (cat# A301-985A100). For H3K27Ac occupied regions, we performed ChIP-seq experiments using an Abcam antibody (cat# AB4729, lot#GR183922-1). Crosslinked cells were lysed with lysis buffer 1 (50mM HEPES pH 7.5, 140mM NaCl, 1mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100), pelleted and resuspended in lysis buffer 2 (10 mM TrisHCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). The subsequent pellet was resuspended in and sonicated in sonication buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 1 mM EGTA, 0.1% Na-deoxycholate, 0.1% SDS, and 1% Triton X-100). Lysates were sonicated for 4 minutes (1-second ‘ON’ and 4-seconds ‘OFF’) at 40% amplitude on a QSonica Sonicator on ice. Sonicated lysates were cleared and incubated overnight at 4°C with magnetic beads bound with antibody to enrich for DNA fragments bound by the indicated factor. Beads were washed two times with sonication buffer, one time with sonication buffer with 500 mM NaCl, one time with LiCl wash buffer (10 mM TrisHCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate) and one time with TE buffer. DNA was eluted in elution buffer (50 mM TrisHCl pH 8.0, 10 mM EDTA, 1% SDS). Cross-links were reversed overnight. RNA and protein were digested using RNase A and Proteinase K, respectively, and DNA was purified with phenol chloroform extraction and ethanol precipitation.
ChIP-Seq Analysis
Illumina sequencing libraries were generated and data was processed according to(76). In brief, libraries were generated following the Illumina TruSeqTM DNA Sample Preparation v2 kit protocol with minor changes. All ChIP-Seq data sets were aligned using bowtie to build NCBI36/hg19 of the human genome with -p 4 --best -k 2 -m 2 --sam -l 40. Wiggle files for gene tracks were created using MACS(77) 1.4.2 with options –w –S –space=50 to count reads in 50bp bins. These were divided by the number of treatment reads to normalize to mapped-reads-per-million, and were displayed in the UCSC genome browser.
Super-enhancers were identified using BRD4 ChIP-Seq and the ROSE algorithm (https://bitbucket.org/young_computation/rose/). Enhancer constituents were identified using MACS with input control and with two parameter sets: --keep-dup=1 –p 1e-9 and –keep-dup=all –p 1e-9. The collapsed union of these regions was used as input for ROSE with parameters –s 12500 –t 1000 and input control. Super-enhancers identified in the DMSO and targeted therapy conditions were collapsed to capture both baseline and acquired super-enhancers upon targeted therapy treatment. The list of combined DMSO and targeted therapy super-enhancers were associated with expressed genes by finding the single expressed Ensembl transcript whose transcription start site was nearest the center of the super-enhancer.
Densities of H3K27Ac or BRD4 ChIP-Seq reads (Figure 3c,d and Supplementary Figure 7 and 8) were calculated using bamToGFF (https://github.com/BradnerLab/pipeline). Super-enhancers identified in DMSO or targeted therapy conditions described above were treated as one bin (-m 1), reads were extended to be 200bp (default) and the reads-per-million (-r) normalized density (-d) of reads was calculated therein. These RPM-normalized density values were log2-normalized after addition of one pseudocount, and log2 values were used for fold-change calculations.
Data deposition
RNA-Seq and ChIP-Seq data have been submitted to the NCBI Gene Expression Omnibus under accession number GSE89129.
Immunoblotting
Cells were lysed in RIPA buffer (Roche) containing protease inhibitors (Roche) and Phosphatase Inhibitor Cocktails I and II (CalBioChem). Protein concentrations were determined using a Bradford assay (Bio-Rad). Proteins were separated by SDS gel electrophoresis using NuPAGE 4–12% Bis-Tris gels (Life Technologies) in MOPS buffer. Resolved protein was transferred to nitrocellulose membranes, blocked in 10% milk and probed with primary antibodies recognizing AKT (9272S), p-AKT (4060P), ERK (4695S), p-ERK (4370S), CDK12 (11973), tubulin (3873S), EGR1 (4153S) (all from Cell Signalling Technology), CDK7 (sc-723, Santa Cruz), SPRED2 (ab153700, Abcam), NF1 (sc-67, Santa Cruz), actin (A5441, Sigma-Aldrich) and vinculin (V9131, Sigma-Aldrich) in 5% milk or 3% bovine serum albumin as recommended by the manufacturer. After incubation with the appropriate secondary antibody (Pierce anti-mouse IgG/IgM (31444, Thermo Scientific) and anti-rabbit IgG (31460, Thermo Scientific)), blots were imaged on film.
Pull down/Immunoprecipitation (IP) experiments
TLP mice were treated for 72 hours with vehicle or THZ1 (10 mg/kg BID, i.p.), and livers and tumors were harvested 6 hours following the last treatment. Tumor and liver tissues were homogenized and lysed as previously described(25) and incubated with bio-THZ1 for pulldown as previously described(15). CDK7 target engagement was analyzed by immunoblotting.
CRISPR-CAS
CDK7/CDK12 experiments
Target sequences for CRISPR interference were designed using the sgRNA designer (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design) and CRISPR Design tool (http://crispr.mit.edu), provided by the Broad Institute, MIT and Feng Zhang lab, MIT, respectively. Off-target effects were considered using http://www.genome-engineering.org. A non-targeting sgRNA from the Gecko library v2 was used as a dummy sgRNA for control (78).
Sequences were as follows:
dummy guide 5′ATCGTTTCGCTTAACGGCG3′;
CDK7 sgRNA#1 5′TGTGATGCAAAGGTATTCCA3′;
CDK7 sgRNA#2 5′ATACACATCAGGTTGTAACC3′;
CDK7 sgRNA#3 5′TGAGAAGCTGGACTTCCTTG3′;
CDK12 sgRNA#1 5′GCTTGTGCTTCGATACCAAG3′;
CDK12 sgRNA #2 5′GCTCCCAGACTGGAATTAAG3′;
CDK12 sgRNA #3 5′GTAGGAGTCATAATTGCTCG3′;
Lenti CRISPRv2 vectors were cloned as previously described(78, 79). Briefly, HEK-293T cells were transduced with lentiCRISPRv2 using X-treme Gene 9 (Roche) according to the manufacturer’s instructions. On day 2, PC9 cells were seeded, and allowed to adhere overnight. On day 3 the supernatant of transduced HEK293T cells was collected and added to the PC9 cells through a 0.45 μm filter. Supernatant from transduced HEK293T cells was again collected and added to PC9 cells on day 4. On day 5, puromycin (1 mg/ml) was added to select infected cells (for four days).
NF1/SPRED2/EGR1 experiments
Oligonucleotides coding for guide RNAs that target the NF1 and SPRED2 genes were chosen from the Avana library(80) and cloned into lentiGuide-Puro two vector system using established methods(78). The sequence for the oligonucleotides are as follows (dummy guide was as above):
NF1_sg1 5′ GATATATCCAAAGACG 3′;
NF1_sg2 5′GGTGGAATGGGTCCAGGCCG3′;
NF1_sg3 5′ TCTTTAGTCGCATTTCTACC3′;
SPRED2_sg1 5′ACCAGAGATGACTCCAGCGG3′;
SPRED2_sg2 5′ AGGTTGCTCTCTCTTCTGAG3′;
SPRED2_sg3 5′CAAAGGCTCGGGCATCAGCA3′;
EGR1_sg1 5′CGGCCAGTATAGGTGATGGG3′
EGR2_sg2 5′AAGGCCTTAATAGTAGACAG3′;
EGR3_sg3 5′GAGTGAGGAAAGGATCCGAA3′;
RT-PCR
Total cellular RNA was isolated from cells using an RNeasy Mini Kit (Qiagen) and 1.0 μg was then reverse transcribed to cDNA using the High Capacity RNA to c-DNA kit (Life Technologies). Quantitative PCR reactions were performed on an ABI Prism 7300 platform (Life Technologies). CDK7 expression was checked using the following forward primer: 5′-GGGACAGTTTGCCACCGTTT-3′ and reverse primer: 5′-ATGTCCAAAAGCATCAAGGAGAC-3′. CDK12 expression was checked using the following forward primer: 5′-GAGGAGGCAGCAGAGAAGAG-3′ and reverse primer: 5′-TAAAAGTTGCAGCAAGGCGG-3′. CDK7 and CDK12 primers were designed using Primer 3 software. Relative gene expression was normalized to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the following forward primer: 5′-TTAGGAAAGCCTGCCGGTGACTAA-3′ and reverse primer: 5′-AAAGCATCACCC GGAGGAGAAATC-3′(81).
Statistical analysis
Data are expressed as mean +/− standard deviation. Statistical significance was determined using Student’s t-test, Mann-Whitney U test, or one-way ANOVA. For survival analyses log-rank test (Mantel-Cox) was used. Statistical analyses were performed in Prism 6 (GraphPad Software). Significance was set at P = 0.05.
Supplementary Material
Significance.
CDK7/12 inhibition prevents active enhancer formation at genes promoting resistance emergence in response to targeted therapy and impedes the engagement of transcriptional programs required for tumor cell survival. CDK7/12 inhibition in combination with targeted cancer therapies may serve as a novel therapeutic paradigm for enhancing the effectiveness of targeted therapies.
Footnotes
Conflict of interest disclosure statement: N.S.G, and N.K. are inventors on a patent application covering THZ1, licensed to Syros Pharmaceuticals, which was co-founded by N.S.G and R.A.Y. M.M. is a consultant/SAB chair for Origimed and holds equity in the company. M.M. gets research support from Bayer and patent royalties from LabCorp.
References
- 1.Ramos P, Bentires-Alj M. Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene. 2014 doi: 10.1038/onc.2014.314. [DOI] [PubMed] [Google Scholar]
- 2.Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. Drug resistance in cancer: an overview. Cancers. 2014;6:1769–92. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nature reviews Cancer. 2013;13:714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 4.Chell V, Balmanno K, Little AS, Wilson M, Andrews S, Blockley L, et al. Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene. 2013;32:3059–70. doi: 10.1038/onc.2012.319. [DOI] [PubMed] [Google Scholar]
- 5.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–42. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Mikse O, Liao RG, Li Y, Tan L, Janne PA, et al. Ligand-associated ERBB2/3 activation confers acquired resistance to FGFR inhibition in FGFR3-dependent cancer cells. Oncogene. 2014 doi: 10.1038/onc.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettazzoni P, Viale A, Shah P, Carugo A, Ying H, Wang H, et al. Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer research. 2015;75:1091–101. doi: 10.1158/0008-5472.CAN-14-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–72. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer cell. 2014;26:207–21. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 13.Blakely CM, Pazarentzos E, Olivas V, Asthana S, Yan JJ, Tan I, et al. NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell reports. 2015;11:98–110. doi: 10.1016/j.celrep.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–20. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher RP, Morgan DO. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–24. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 17.Larochelle S, Merrick KA, Terret ME, Wohlbold L, Barboza NM, Zhang C, et al. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Molecular cell. 2007;25:839–50. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schachter MM, Merrick KA, Larochelle S, Hirschi A, Zhang C, Shokat KM, et al. A Cdk7-Cdk4 T-loop phosphorylation cascade promotes G1 progression. Molecular cell. 2013;50:250–60. doi: 10.1016/j.molcel.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Molecular cell. 2009;34:387–93. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drapkin R, Le Roy G, Cho H, Akoulitchev S, Reinberg D. Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6488–93. doi: 10.1073/pnas.93.13.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Molecular and cellular biology. 2009;29:5455–64. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nature structural & molecular biology. 2012;19:1108–15. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelso TW, Baumgart K, Eickhoff J, Albert T, Antrecht C, Lemcke S, et al. Cyclin-dependent kinase 7 controls mRNA synthesis by affecting stability of preinitiation complexes, leading to altered gene expression, cell cycle progression, and survival of tumor cells. Molecular and cellular biology. 2014;34:3675–88. doi: 10.1128/MCB.00595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–39. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, et al. Targeting Transcriptional Addictions in Small Cell Lung Cancer with a Covalent CDK7 Inhibitor. Cancer cell. 2014;26:909–22. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, et al. CDK7-Dependent Transcriptional Addiction in Triple-Negative Breast Cancer. Cell. 2015;163:174–86. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong LN, Davies MA. Targeted therapy for melanoma: rational combinatorial approaches. Oncogene. 2014;33:1–9. doi: 10.1038/onc.2013.34. [DOI] [PubMed] [Google Scholar]
- 28.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nature biotechnology. 2012;30:679–92. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 29.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–6. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bivona TG, Doebele RC. A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nature medicine. 2016;22:472–8. doi: 10.1038/nm.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer discovery. 2012;2:934–47. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nature medicine. 2015;21:1038–47. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nature medicine. 2014;20:1027–34. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson FH, Johannessen CM, Piccioni F, Tamayo P, Kim JW, Van Allen EM, et al. A Functional Landscape of Resistance to ALK Inhibition in Lung Cancer. Cancer cell. 2015;27:397–408. doi: 10.1016/j.ccell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi N, Lucena-Araujo AR, Nakayama S, de Figueiredo-Pontes LL, Gonzalez DA, Yasuda H, et al. Dual ALK and EGFR inhibition targets a mechanism of acquired resistance to the tyrosine kinase inhibitor crizotinib in ALK rearranged lung cancer. Lung cancer. 2014;83:37–43. doi: 10.1016/j.lungcan.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer research. 2011;71:6051–60. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carter CA, Rajan A, Keen C, Szabo E, Khozin S, Thomas A, et al. Selumetinib with and without erlotinib in KRAS mutant and KRAS wild-type advanced nonsmall-cell lung cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2016;27:693–9. doi: 10.1093/annonc/mdw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soria JC, LoRusso P, Bahleda R, Lager J, Liu L, Jiang J, et al. Phase I dose-escalation study of pilaralisib (SAR245408, XL147), a pan-class I PI3K inhibitor, in combination with erlotinib in patients with solid tumors. Oncologist. 2015;20:245–6. doi: 10.1634/theoncologist.2014-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price KA, Azzoli CG, Krug LM, Pietanza MC, Rizvi NA, Pao W, et al. Phase II trial of gefitinib and everolimus in advanced non-small cell lung cancer. J Thorac Oncol. 2010;5:1623–9. doi: 10.1097/JTO.0b013e3181ec1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bedard PL, Tabernero J, Janku F, Wainberg ZA, Paz-Ares L, Vansteenkiste J, et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:730–8. doi: 10.1158/1078-0432.CCR-14-1814. [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, Kwiatkowski N, Olson CM, Dixon-Clarke SE, Abraham BJ, Greifenberg AK, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol. 2016;12:876–84. doi: 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi F, Kugawa S, Tateno H, Kokubu F, Fukuchi K. Analysis of EGFR, KRAS and P53 mutations in lung cancer using cells in the curette lavage fluid obtained by bronchoscopy. Lung cancer. 2012;78:201–6. doi: 10.1016/j.lungcan.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Akbay E, Mikse O, Tupper T, Cheng K, Wang Y, et al. Co-clinical trials demonstrate superiority of crizotinib to chemotherapy in ALK-rearranged non-small cell lung cancer and predict strategies to overcome resistance. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:1204–11. doi: 10.1158/1078-0432.CCR-13-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15535–40. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izumchenko E, Chang X, Michailidi C, Kagohara L, Ravi R, Paz K, et al. The TGFbeta-miR200-MIG6 pathway orchestrates the EMT-associated kinase switch that induces resistance to EGFR inhibitors. Cancer research. 2014;74:3995–4005. doi: 10.1158/0008-5472.CAN-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsdale R, Jorissen RN, Li FZ, Al-Obaidi S, Ward T, Sheppard KE, et al. The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Science signaling. 2015;8:ra82. doi: 10.1126/scisignal.aab1111. [DOI] [PubMed] [Google Scholar]
- 48.Della Corte CM, Bellevicine C, Vicidomini G, Vitagliano D, Malapelle U, Accardo M, et al. SMO gene amplification and activation of the hedgehog pathway as novel mechanisms of resistance to anti-epidermal growth factor receptor drugs in human lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:4686–97. doi: 10.1158/1078-0432.CCR-14-3319. [DOI] [PubMed] [Google Scholar]
- 49.Soucheray M, Capelletti M, Pulido I, Kuang Y, Paweletz CP, Becker JH, et al. Intratumoral Heterogeneity in EGFR-Mutant NSCLC Results in Divergent Resistance Mechanisms in Response to EGFR Tyrosine Kinase Inhibition. Cancer research. 2015;75:4372–83. doi: 10.1158/0008-5472.CAN-15-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. The Journal of biological chemistry. 2007;282:13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 51.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Molecular cell. 2014;56:219–31. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zawistowski JS, Bevill SM, Goulet DR, Stuhlmiller TJ, Beltran AS, Olivares-Quintero JF, et al. Enhancer Remodeling during Adaptive Bypass to MEK Inhibition Is Attenuated by Pharmacologic Targeting of the P-TEFb Complex. Cancer discovery. 2017;7:302–21. doi: 10.1158/2159-8290.CD-16-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malchers F, Ercanoglu MS, Schutte D, Castiglione R, Tischler V, Michels S, et al. Mechanisms of primary drug resistance in FGFR1 amplified lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017 doi: 10.1158/1078-0432.CCR-17-0478. [DOI] [PubMed] [Google Scholar]
- 54.Li F, Huynh H, Li X, Ruddy DA, Wang Y, Ong R, et al. FGFR-Mediated Reactivation of MAPK Signaling Attenuates Antitumor Effects of Imatinib in Gastrointestinal Stromal Tumors. Cancer discovery. 2015;5:438–51. doi: 10.1158/2159-8290.CD-14-0763. [DOI] [PubMed] [Google Scholar]
- 55.Shen CH, Kim SH, Trousil S, Frederick DT, Piris A, Yuan P, et al. Loss of cohesin complex components STAG2 or STAG3 confers resistance to BRAF inhibition in melanoma. Nature medicine. 2016;22:1056–61. doi: 10.1038/nm.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai AZ, Cory S, Zhao H, Gigoux M, Monast A, Guiot MC, et al. Dynamic reprogramming of signaling upon met inhibition reveals a mechanism of drug resistance in gastric cancer. Science signaling. 2014;7:ra38. doi: 10.1126/scisignal.2004839. [DOI] [PubMed] [Google Scholar]
- 57.Jing J, Greshock J, Holbrook JD, Gilmartin A, Zhang X, McNeil E, et al. Comprehensive predictive biomarker analysis for MEK inhibitor GSK1120212. Molecular cancer therapeutics. 2012;11:720–9. doi: 10.1158/1535-7163.MCT-11-0505. [DOI] [PubMed] [Google Scholar]
- 58.Cayrol F, Praditsuktavorn P, Fernando TM, Kwiatkowski N, Marullo R, Calvo-Vidal MN, et al. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat Commun. 2017;8:14290. doi: 10.1038/ncomms14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell reports. 2015;11:390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–71. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 61.Gray JE, Haura E, Chiappori A, Tanvetyanon T, Williams CC, Pinder-Schenck M, et al. A phase I, pharmacokinetic, and pharmacodynamic study of panobinostat, an HDAC inhibitor, combined with erlotinib in patients with advanced aerodigestive tract tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:1644–55. doi: 10.1158/1078-0432.CCR-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reguart N, Rosell R, Cardenal F, Cardona AF, Isla D, Palmero R, et al. Phase I/II trial of vorinostat (SAHA) and erlotinib for non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations after erlotinib progression. Lung cancer. 2014;84:161–7. doi: 10.1016/j.lungcan.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Dasari A, Gore L, Messersmith WA, Diab S, Jimeno A, Weekes CD, et al. A phase I study of sorafenib and vorinostat in patients with advanced solid tumors with expanded cohorts in renal cell carcinoma and non-small cell lung cancer. Invest New Drugs. 2013;31:115–25. doi: 10.1007/s10637-012-9812-z. [DOI] [PubMed] [Google Scholar]
- 64.Witta SE, Jotte RM, Konduri K, Neubauer MA, Spira AI, Ruxer RL, et al. Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:2248–55. doi: 10.1200/JCO.2011.38.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han JY, Lee SH, Lee GK, Yun T, Lee YJ, Hwang KH, et al. Phase I/II study of gefitinib (Iressa((R))) and vorinostat (IVORI) in previously treated patients with advanced non-small cell lung cancer. Cancer chemotherapy and pharmacology. 2015;75:475–83. doi: 10.1007/s00280-014-2664-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerber DE, Boothman DA, Fattah FJ, Dong Y, Zhu H, Skelton RA, et al. Phase 1 study of romidepsin plus erlotinib in advanced non-small cell lung cancer. Lung cancer. 2015;90:534–41. doi: 10.1016/j.lungcan.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bauman J, Verschraegen C, Belinsky S, Muller C, Rutledge T, Fekrazad M, et al. A phase I study of 5-azacytidine and erlotinib in advanced solid tumor malignancies. Cancer chemotherapy and pharmacology. 2012;69:547–54. doi: 10.1007/s00280-011-1729-2. [DOI] [PubMed] [Google Scholar]
- 68.Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PloS one. 2014;9:e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature protocols. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, Li D, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer research. 2010;70:9827–36. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016 doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology. 2016;34:184–91. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baranwal S, Wang Y, Rathinam R, Lee J, Jin L, McGoey R, et al. Molecular characterization of the tumor-suppressive function of nischarin in breast cancer. Journal of the National Cancer Institute. 2011;103:1513–28. doi: 10.1093/jnci/djr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.