Abstract
Local protein synthesis in mature axons may play a role in synaptic plasticity, axonal arborization, or functional diversity of the circuit. To gain insight into this question, we investigated the axonal localization of translational regulators and associated mRNAs in five parallel olfactory circuits, four in the main olfactory bulb and one in the accessory olfactory bulb. Axons in all four main olfactory bulb circuits exhibited axonal localization of Fragile X granules (FXGs), structures that comprise ribosomes, mRNA, and RNA binding proteins including Fragile X mental retardation protein (FMRP) and the related protein FXR2P. In contrast, FXGs were not seen in axons innervating the accessory olfactory bulb. Similarly, axons innervating the main olfactory bulb, but not the accessory olfactory bulb, contained the FXG-associated mRNA Omp (olfactory marker protein). This differential localization was not explained by circuit-dependent differences in expression of FXG components or Omp, suggesting that other factors must regulate their axonal transport. The specificity of this transport was highlighted by the absence from olfactory axons of the calmodulin transcript Calm1, which is highly expressed in peripheral olfactory neurons at levels equivalent to Omp. Regulation of axonal translation by FMRP may shape the structure and function of the axonal arbor in mature sensory neurons in the main olfactory system but not in the accessory olfactory system.
Keywords: FXR2P, olfactory epithelium, olfactory marker protein, olfactory sensory neuron, RRID: AB_398856, RRID: AB_476964, RRID: AB_664696, RRID: AB_653928, RRID: AB_2149710, RRID: AB_306884, vomeronasal organ, vomeronasal sensory neuron
1 INTRODUCTION
The remarkable structural and functional diversity of neurons relies on precise cell type-dependent regulation of the proteome. One critical mechanism regulating neuronal protein expression is the transport of mRNA specifically into axons where this RNA is translated into proteins in response to appropriate stimuli. This local protein synthesis has been well-characterized in growing and regenerating axons (Korsak, Mitchell, Shepard, & Akins, 2016). More recent evidence has revealed that axonal translation also occurs in axons that are integrated into mature circuits, where local translation can support synaptic plasticity (Akins et al., 2017; Baleriola et al., 2014; Gumy et al., 2011; Shigeoka et al., 2016; Taylor et al., 2009; Younts et al., 2016). A key question is how differences in local axonal translation contribute to functional and structural diversity of neurons. Insight into this question can be gained by determining how similar classes of neurons differ in the axonal localization of identified mRNAs and the RNA binding proteins that associate with these mRNAs.
Fragile X granules (FXGs) are attractive candidates for supporting such diversity in the axonal arbor of mature neurons. These granules, which are only found in axons that have finished growing and have integrated into circuits, are composed of RNA binding proteins and associate with both ribosomes and mRNA (Akins, LeBlanc, Stackpole, Chyung, & Fallon, 2012; Akins et al., 2017; Christie, Akins, Schwob, & Fallon, 2009). FXG constituent proteins include Fragile X mental retardation protein (FMRP), a key regulator of activity-dependent protein synthesis in neurons. Dysregulation of this protein synthesis due to loss of FMRP leads to the autism-related disorder Fragile X syndrome (Bassell & Warren, 2008; Bhakar, Dölen, & Bear, 2012; Darnell & Klann, 2013). FXGs also contain the Fragile X related proteins FXR1P and FXR2P in a circuit-dependent manner (Christie et al., 2009). While all three Fragile X related proteins regulate protein synthesis in the somatodendritic domain of all neurons, these proteins only assemble into FXGs in axons of a stereotyped subset of neurons (Akins et al., 2012; Christie et al., 2009). FXGs thus reveal cell type-dependent mechanisms for regulating the axonal proteome.
A broad survey of FXG expression throughout the juvenile mouse brain suggested differential expression of FXGs between parallel peripheral olfactory circuits (Akins et al., 2012). FXGs were found in olfactory sensory neurons (OSNs) that express odorant receptors (ORs) and innervate the main olfactory bulb. In contrast, FXGs were not identified in the pheromone-detecting vomeronasal sensory neurons (VSNs) that innervate the accessory olfactory bulb (Akins et al., 2012). In addition to the two examined populations, there are also several additional classes of peripheral olfactory neurons that can be identified by their anatomical location in the nasal cavity as well as the signaling cascades that transduce chemosensory information (Figure 1). These include classes of OSNs that do not express ORs and that are thought to mediate stereotyped behaviors (Stowers & Logan, 2010). Whether FXGs are present in axons of these non-OR-expressing OSN subtypes, and hence may contribute to potential activity-dependent changes in axons of these cells, has not been determined. Investigating the differential utilization of axonal Fragile X proteins by parallel circuits promises to provide insight into the functional consequence of axonal translation.
FIGURE 1.
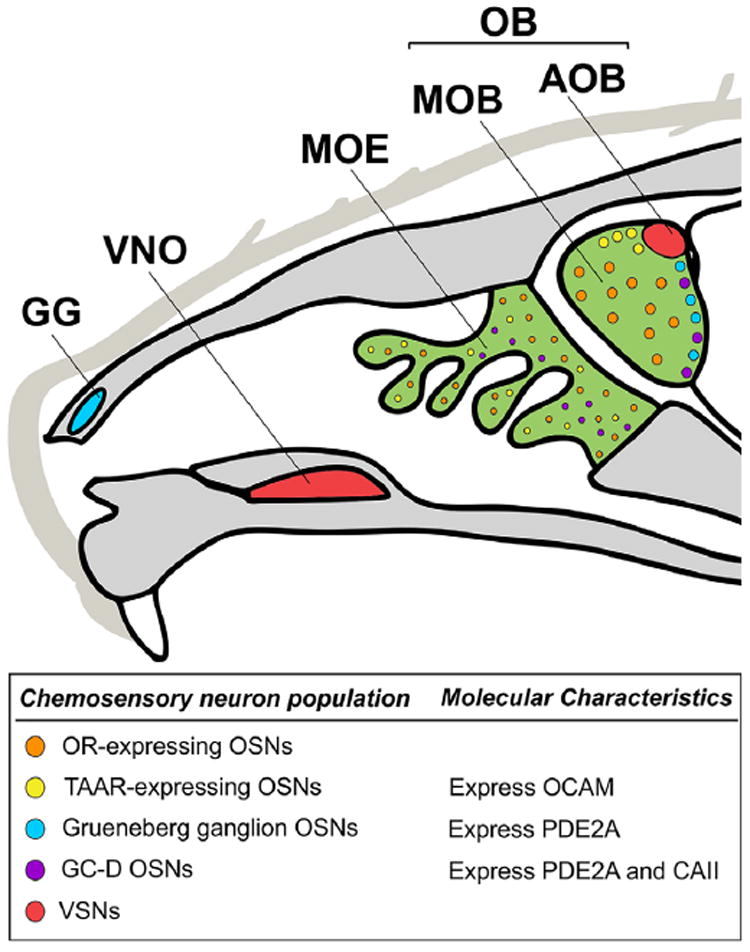
Overview of mouse olfactory anatomy. Neurons expressing chemosensory receptors are located in the main olfactory epithelium (MOE, green), Grueneberg ganglion (GG, blue), and vomeronasal organ (VNO, red). These neurons all project axons to the olfactory bulb (OB). Axons from the MOE and GG form glomeruli in the main olfactory bulb (MOB, green) whereas axons from the VNO form glomeruli in the accessory olfactory bulb (AOB, red). OSNs expressing odor receptors (ORs, orange) form glomeruli throughout the MOB. The subpopulation of OSNs in the MOE that express trace amine-associated receptors (TAARs, yellow) form glomeruli on the dorsal surface of the MOB. Both the OSNs from the MOE that express guanylyl cyclase D (GC-D, purple) and the OSNs from the GG form “necklace” glomeruli that encircle the posterior OB. In addition to their location within the MOB, the TAAR, GG, and GC-D OSNs can be identified through their unique molecular characteristics within the OSN population: TAAR OSNs express olfactory cell adhesion molecule (OCAM), GG and GC-D OSNs express phosphodiesterase 2A (PDE2A), and GCD OSNs express carbonic anhydrase II (CAII)
Within OSN axons, FXGs associate with the mRNA Omp, which encodes olfactory marker protein, a broadly conserved regulator of olfactory-induced intracellular signal transduction that is also implicated in olfactory plasticity (Buiakova et al., 1996; Kass, Moberly, Rosenthal, Guang, & McGann, 2013; Keller & Margolis, 1975; Lee, He, & Ma, 2011; Reisert, Yau, & Margolis, 2007). In these cells, FMRP regulates the axonal translation of OMP protein, but is not required for the axonal transport of the Omp mRNA (Akins et al., 2017). In addition to OSNs, OMP is also expressed by VSNs. However, whether VSN axons contain the Omp mRNA has not been explored to our knowledge. This question is of particular interest since axonal localization of Omp to VSN axons, which do not contain FXGs, would reveal cell type-dependent regulation of translation of the same mRNA. Conversely, if VSN axons do not contain Omp mRNA, this would suggest that these cell types differ in their requirement for axonally synthesized OMP protein.
To investigate these questions, we explored the extent to which axons of chemosensory neurons contain FXGs and the Omp mRNA. Consistent with past results, we found that OSNs contained FXGs in their axons. In contrast, FXGs were not found in VSN axons at any examined age. Expression in OSN axons was consistent across several subclasses of OSNs that mediate responses to distinct chemosensory cues, even those that mediate stereotyped behaviors. We also found the FXG-target Omp mRNA in OSN axons innervating the main olfactory bulb but not within VSN axons innervating the accessory olfactory bulb. Although Omp is a highly abundant mRNA, high mRNA levels are not sufficient for axonal transport in OSNs as we could not detect axonal localization in these cells of the calmodulin transcript Calm1, which is expressed in OSNs at levels similar to Omp and which localizes to axons of cortical neurons (Magklara et al., 2011; Taylor et al., 2009). Northern blot and PCR analyses indicated that the Omp transcript comprised the same sequence in OSN cell bodies, VSN cell bodies, and OSN axons, suggesting that differences in the Omp mRNA do not account for the cell type-specific axonal localization. These studies therefore reveal cell type-specific regulation of the axonal localization of both Fragile X proteins and mRNAs between parallel chemosensory circuits. These findings further suggest that local translation contributes to axonal plasticity in several OSN circuits that respond to diverse environmental cues and elicit distinct behavioral responses.
2 MATERIALS AND METHODS
2.1 Animals
All work with animals was performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Drexel University. C57BL/6 mice were obtained from Charles River and bred in-house. Male and female animals were used interchangeably since we observed no effect of sex on any measure. Mice were anesthetized by intraperitoneal injection with ketamine/xylazine/acepromazine. Following rapid decapitation, brains and nasal cavity were rapidly dissected and processed as detailed below.
2.2 Immunofluorescence
Brains and nasal cavities from 30 day old mice were embedded in OCT compound (Sakura Finetek), rapidly frozen, and stored at −80°C until cryosectioning. Slide-mounted coronal sections of OCT-embedded brains were prepared using a Leica 3050S cryostat at 20 μm and stained the same day. Tissue sections were fixed with room temperature PBS (0.1M phosphate, pH 7.4; 150 mM sodium chloride) containing 4% paraformaldehyde (PFA). Sections were then washed three times in PBS and incubated with blocking solution [PBST (10 mM phosphate buffer, pH 7.4, and 0.3% Triton X-100) and 1% blocking reagent (Roche Life Science)] for 30 min to occupy nonspecific binding sites. Sections were then treated with blocking solution containing primary antibodies (Table 1) overnight at room temperature. These sections were then washed for 5 min with PBST. Tissue was incubated with appropriate fluorescent secondary antibodies (Table 1) in blocking solution for 1 hr, washed for 5 min with PBST, slide mounted in NPG mounting medium (4% n-propylgallate, 80% glycerol, 5 mM phosphate pH 7.4), and coverslipped. Sections were imaged using a Leica SPE II confocal microscope.
TABLE 1.
Antibodies used in this study
| Primary antibody | Source | Target | RRID | Primary dilution | Secondary antibody |
|---|---|---|---|---|---|
| Mouse anti-FXR2P | Monoclonal 55 from BD Biosciences #611330 | Raised against human FXR2P amino acids 520–639; recognizes a single band of the appropriate size by Western in lysates from mouse brain; gives identical staining as seen with A42 | AB_398856 | 1:500 | Alexa-conjugated goat anti-mouse IgG2b (Life Technologies; 1:1,000) |
| Mouse anti-FXR2P | Monoclonal A42 from Sigma | Raised against human FXR2P amino acids 12–426; recognizes a single band of the appropriate size by Western and does not cross react with either FMRP or FXR1P (Zhang et al., 1995); recognizes a single band of the appropriate size by Western in lysates from mouse brain | AB_476964 | 1:500 | Alexa-conjugated goat anti-mouse IgG1 (Life Technologies; 1:1,000) |
| Goat anti-OMP | Polyclonal from Wako 544-10001 | Raised against purified natural Rat OMP; recognizes a single band at 19 kDa in mouse olfactory bulb extracts from wild type mice but not from OMP knockout mice (Buiakova et al., 1996) | AB_664696 | 1:1,000 | Alexa-conjugated donkey anti-goat (Life Technologies; 1:1,000) |
| Goat anti-PDE2A | Polyclonal from Santa Cruz Biotechnology #SC17227 | Raised against an N-terminal epitope of human PDE2A; Recognizes a single band of the appropriate size in Westerns of mouse olfactory epithelium; Stains the expected subset of OSNs in the olfactory epithelium (Hansen & Finger, 2008); Stains the expected subset of olfactory glomeruli (Figure 3) | AB_653928 | 1:1,000 | Alexa-conjugated donkey anti-goat (Life Technologies; 1:1,000) |
| Goat anti-OCAM | Polyclonal from R&D Systems #AF778 | Raised against recombinant mouse OCAM amino acids 20–700; Recognizes a single band of the appropriate length in mouse olfactory epithelial lysates; Recognizes the expected OSN populations in the olfactory epithelium (Murdoch & Roskams, 2008); Stained the expected subset of olfactory glomeruli (Zou et al., 2007) (Figure 3) | AB_2149710 | 1:100 | Alexa-conjugated donkey anti-goat (Life Technologies; 1:1,000) |
| Sheep anti-CAII | Polyclonal from Abcam #ab8953 | Raised against Human Carbonic Anhydrase II, purified from human erythrocytes. Recognizes a single band of the appropriate size in lysates from porcine retinal Müller glia cells (Merl, Ueffing, Hauck, & von Toerne, 2012); Stained the expected subset of olfactory glomeruli (Figure 3) | AB_306884 | 1:1,000 | Alexa-conjugated donkey anti-sheep (Life Technologies; 1:1,000) |
2.3 Quantification of FXG abundance
Confocal micrographs of FXR2P, CAII, OCAM, and PDE2A immunostaining collected without pixel saturation were processed using ImageJ. FXG abundance and glomerular area were quantified in glomeruli identified using a combination of their anatomical location and molecular phenotype as described in results. FXGs were identified based on their FXR2P signal. In brief, we used confocal images with a resolution of 1024 × 1024 pixels and 0.179 μm/pixel corresponding to an image of approximately 183 × 183 μm. These images were processed using the ImageJ rolling ball background subtraction command with a radius of 5 pixels, which subtracts from each pixel the mean intensity value of a circle of radius 5 centered on that pixel. We then identified structures that were more than 1.5 standard deviations brighter than the surrounding pixels to accentuate structures that were the approximate size of FXGs. Images were then autothresholded using the built-in Max Entropy setting. Macro-based FXG identification did not work for images with few to no FXGs. In these cases, granules were counted manually using previously described criteria (Akins et al., 2012; Christie et al., 2009). Graphs and statistical analyses were produced using Prism 6.0 (Graphpad). Quantifications are expressed as mean ± SEM.
2.4 In situ hybridization
Brains from 30-day-old mice were embedded in OCT compound (Sakura Finetek), rapidly frozen, and stored at −80°C until cryosectioning. Brains were sectioned at 20 μm, mounted on slides, and stored at −20°C until use. All subsequent steps were performed at room temperature unless otherwise noted. On the day of staining, slides were allowed to warm to room temperature, fixed in PBS containing 4% PFA for 10 min, and washed 3X in PBS. Sections were treated in 0.01M sodium citrate (pH 6) for 30 min at 75°C followed by incubation for 10 min in 0.2M HCl followed by 2 min in PBS containing 1% Triton X- 100. Sections were then rinsed twice for 1 min each in PBS before a 10-min equilibration in 2X SSC + 10% formamide. Sections were incubated overnight at 37°C in hybridization solution (10% dextran sulfate, 1 mg/ml E. coli tRNA, 2 mM Vanadyl Ribonucleoside, 200 μg/ml BSA, 2X SSC, 10% deionized formamide) containing 250 nM Stellaris fluorophore-labeled oligonucleotide probes (Biosearch Technologies; sequences in Supporting Information Table S1) along with the A42 anti-FXR2P antibody (Sigma; 1:500). Sections were then rinsed two times for 30 min each at 37°C in 2X SSC + 10% formamide followed by 2X SSC and then PBST. Sections were then incubated in blocking solution plus a secondary antibody for 1 hr to detect A42, washed three times in PBST, mounted in NPG, and imaged using a Leica SPE II confocal microscope.
2.5 Northern blotting
For Northern blotting, the vomeronasal epithelium (VNO), olfactory epithelium (OE), main olfactory bulbs (MOBs), prefrontal cortex (Ctx), and cerebellum (Cbm) were rapidly dissected from 2 to 3-month-old mice and flash frozen in liquid nitrogen and stored until the RNA was extracted with TRIzol reagent. Nine animals were pooled for the VNO sample, three for the OE and OB, and two for the Ctx and Cbm. 20 μg of RNA was loaded per lane. Since the OE expresses Omp at high levels, 2 μg of OE RNA was combined with 18 μg of Ctx RNA (pilot studies indicated no detectable Omp in the cortex sample). 20 μg of sample RNA or 9 μg of Millenium RNA ladder (Ambion AM7151) were each mixed with 1X MOPS buffer [20 mM 3-morpholinopropane-1-sulfonic acid (MOPS), 5 mM sodium acetate, and 1 mM ethylene-diamine-tetraacetic acid (EDTA)], 4% PFA, 5 M formamide, and RNA loading dye (800 mM urea, 2 mM Tris-HCl, 100 μM EDTA, 0.005% xylene cyanol, and 0.005% bromophenol blue) denatured at 65°C for 5 min and immediately removed to ice. RNA was run on a 1.5% agarose gel (containing 1X MOPS and 3% PFA) in MOPS buffer at 40 volts for 6 hr, rinsed twice in 20X SSC (3 M sodium chloride, 0.3 M sodium citrate) for 15 min each, then transferred to nylon membrane overnight via capillary action using 20X SSC.
2.6 Membrane-based detection of total RNA
The following incubations were performed at room temperature with gentle rocking. The membrane was rinsed in 6X SSC for 5 min, dried on blotting paper for 5 min, then crosslinked with UV light (wavelength 254 nm, 1.5 J over 1 min). To identify total RNA, the membrane was stained for 5 min with 0.2% methylene blue (Sigma M9140-25G) in 300 mM sodium acetate. This signal was then imaged with a Gel Doc XR+ system (Bio-Rad) using standard bright field settings. The membrane was then destained in 0.2X SSC 1% sodium dodecyl sulfate (SDS) for 15 min and washed twice in deionized water for 5 min.
2.7 Preparation of Omp Northern probe
The full-length mouse Omp sequence comprising 5′ UTR, coding sequence, and 3′ UTR was cloned from pCMV-Sport6.1-OMP (Open Biosystems) into pCRBlunt II-TOPO (Invitrogen 450245). Antisense RNA probe was prepared using biotin-labeled uridine with the Thermo Fisher MEGAscript T7 Transcription Kit AM1334 as per manufacturer’s instructions. Immediately before use, Omp probe was denatured at 70°C for 5 min and transferred to ice.
2.8 Membrane-based detection of Omp mRNA
The following incubations were performed with gentle rocking at room temperature unless otherwise stated. Following total RNA detection, membranes were incubated for 1 hr in preheated DIG EasyHyb (Roche Life Science 11603558001) at 68°C. The membrane was then incubated overnight at 68°C in EasyHyb solution containing 100 ng/ml of biotinylated Omp probe. The membrane was washed twice for 5 min each in 2X SSC containing 0.1% SDS followed by two 15 min washes in preheated 0.1X SSC 0.1% SDS at 68°C. The membrane was then stained with horseradish peroxidase (HRP)-conjugated streptavidin (Thermo Fisher N100, diluted in EasyHyb, final concentration 250 ng/ml) for 1 hr. The membrane was washed twice for 5 min in 2X SSC containing 0.1% SDS and twice for 5 min in 0.1X SSC 0.1% SDS. The membrane was then incubated for 5 min without rocking in SuperSignal West Femto Maximum Sensitivity Substrate (Pierce 34095, prepared per manufacturer’s instructions). The membrane was then imaged on a Bio-Rad Gel Doc XR+ using the standard chemiluminescent protocol.
2.9 PCR
Tissue was dissected from 75-day-old mice, and RNA was prepared using Trizol (Thermo Fisher). 1 μg of RNA from each tissue was converted to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). PCR from each cDNA sample was performed as indicated in Results using OneTaq (New England Biolab) for 30 cycles as per the manufacturer directions. Primers, designed to specifically target the Omp sequence using NCBI Primer-BLAST, were as follows: OMP 6 forward, ATTCCCTGACGCTGGTGGTAG; OMP 608 reverse, AAGGAGATCCAGGCAAGGGA; OMP 506 forward, TGGAGCCTGCCAACCTAAAG; OMP 2119 reverse, AGGGCACACAGTCTTTATTGTGA. PCR products were size fractionated using agarose gel electrophoresis. The resulting gels were then imaged using a Bio-Rad Gel Doc XR+ using the standard ethidium bromide protocol.
2.10 Figure preparation
Images used in figures were adjusted for brightness, contrast, and gamma using Photoshop CS6 13.0.6 (Adobe). Figures were prepared using Corel Draw X7 (Corel).
3 RESULTS
3.1 FXGs are found in OSN axons but not VSN axons
As a first step toward understanding potential roles for axonal translation in peripheral olfactory neurons, we explored the potential differential expression of FXGs between axons in the main and accessory olfactory systems. To identify FXGs, we immunostained for FXR2P followed by standardized image processing and analysis (Akins et al., 2012, 2017; Christie et al., 2009). We have previously shown that FXGs are found throughout life in OSN axons innervating the main olfactory bulb, both in the olfactory nerve layer as well as in their glomerular targets (Akins et al., 2012, 2017; Christie et al., 2009). Consistent with these past studies, we found that FXGs were present in OSN axons innervating glomeruli in the main olfactory bulb (Figure 2a).
FIGURE 2.
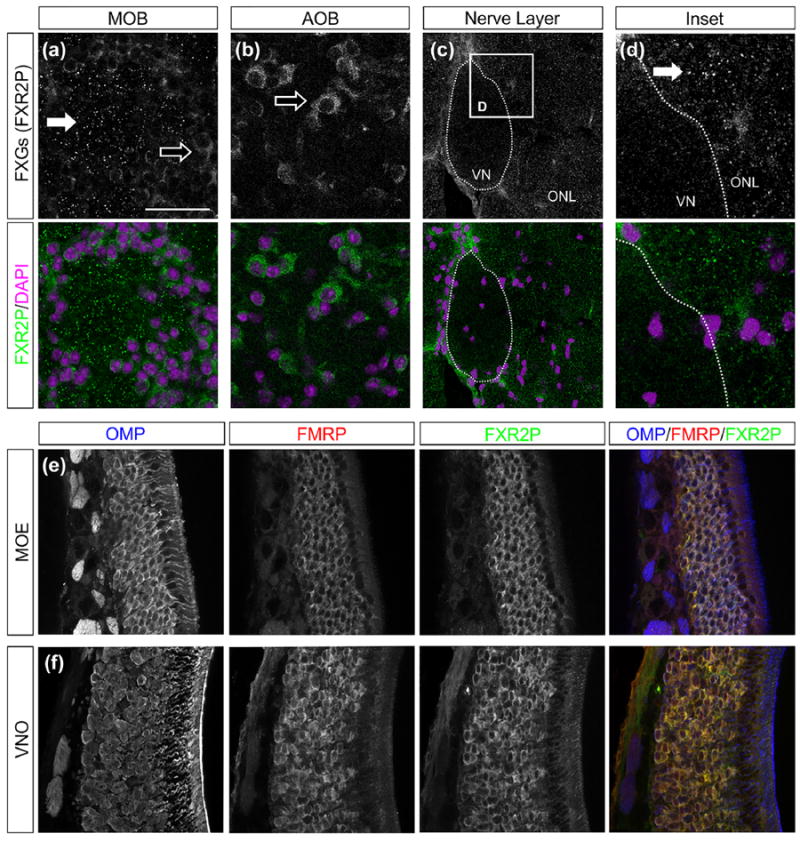
FXGs localize to OSN axons but not VSN axons. (a–d) FXGs were identified by immunostaining for FXR2P (green) and location in the olfactory bulb was determined by identifying cells using somatic FXR2P signal as well as DAPI (magenta). (a) In glomeruli of the main olfactory bulb (MOB), FXR2P is expressed in the cell bodies of periglomerular cells (open arrow) and in FXGs of OSN axons (solid arrow). (b) In glomeruli of the accessory olfactory bulb (AOB), FXR2P is only expressed in periglomerular cells (open arrow). (c, d) FXGs are expressed in OSN axons coursing through the olfactory nerve layer (ONL) of the MOB, but not VSN axons coursing through the vomeronasal nerve (VN). (e, f) OSNs in the main olfactory epithelium (MOE) as well as VSNs in the vomeronasal organ (VNO) express OMP protein (blue), FMRP (red), and FXR2P (green). Scale bar: 40 μm in a, b; 60 μm in c; 15 μm in d; 55 μm in e, f
FXGs were not previously identified in axon terminals of VSNs in a broad survey of FXG expression throughout the brain of 15-day-old mice, the age of peak FXG expression in most circuits in the mouse brain (Akins et al., 2012). However, in contrast to axons in the rest of the brain, mouse OSNs exhibit peak FXG expression in 30-day-old mice that continues throughout life (Akins et al., 2017; Christie et al., 2009). We therefore asked whether VSN axons contain FXGs and similarly exhibit peak expression at ages later than 15 days old. However, we did not detect FXGs in VSN axon terminals in the accessory olfactory bulb in either 30 day old mice (Figure 2b) or in adult mice (not shown). This lack of FXG localization in terminals could potentially reflect inefficient axonal transport of these complexes in VSNs. To investigate this possibility, we asked whether FXGs are found in more proximal portions of VSN axons by examining the vomeronasal nerve as it courses along the medial aspect of the main olfactory bulb. We were unable to detect FXGs in the vomeronasal nerve at any position (Figure 2c,d). Importantly, in all sections in which we did not detect FXGs in VSN axons, we could readily detect FXGs in nearby OSN axons in both the olfactory nerve layer and glomeruli within the main olfactory bulb. The inability to detect FXGs in VSN axons therefore did not reflect technical limitations. Instead, our findings indicate that FXGs are specifically localized to axons of OSNs but not VSNs.
The absence of FXGs in VSN axons could reflect lack of expression of FMRP and FXR2P by the VSNs. To test this possibility, we investigated the expression of these proteins in both OSN cell bodies in the main olfactory epithelium as well as in VSN cell bodies in the vomeronasal organ. As expected, we detected both FMRP and FXR2P in OSN cell bodies (Figure 2e). We also detected expression of both FMRP and FXR2P in VSN cell bodies (Figure 2f). The absence of FXGs in vomeronasal axons therefore does not reflect lack of expression by VSNs of the FXG constituent proteins FMRP or FXR2P.
3.2 FXGs localize to axons of multiple classes of OSNs mediating distinct chemosensory modalities
OSNs are subdivided into several subpopulations that detect different classes of chemical cues. These subpopulations can be identified based both on their molecular phenotype as well as the anatomical location of their glomerular targets within the olfactory bulb (Figure 1). The largest subpopulation comprises those that detect volatile odorant molecules using ORs. These OSNs innervate glomeruli throughout the olfactory bulb and, based on this anatomical location, are the cells whose axons were analyzed above (Figure 2). Whether FXGs are also found in other subpopulations of OSNs has not been investigated.
We first asked whether FXGs are found in axons of OSNs that detect biogenic amines using trace amine-associated receptors (TAARs) rather than ORs. Axons of TAAR-expressing OSNs contain OCAM (olfactory cell adhesion molecule) and innervate the dorsal olfactory bulb (Johnson et al., 2012). In contrast, other classes of OR-expressing OSNs that express axonal OCAM innervate the ventral olfactory bulb (Treloar, Gabeau, Yoshihara, Mori, & Greer, 2003). We therefore asked whether dorsal glomeruli innervated by OCAM-positive axons contain FXGs. We could readily detect FXGs in these glomeruli that are innervated by TAAR-expressing OSNs (Figure 3a). Further, FXG abundance in this population of OSNs was indistinguishable from that seen in the population of OR-expressing OSNs (Figure 3d). TAAR-expressing OSNs thus contain FXGs in their axons.
FIGURE 3.
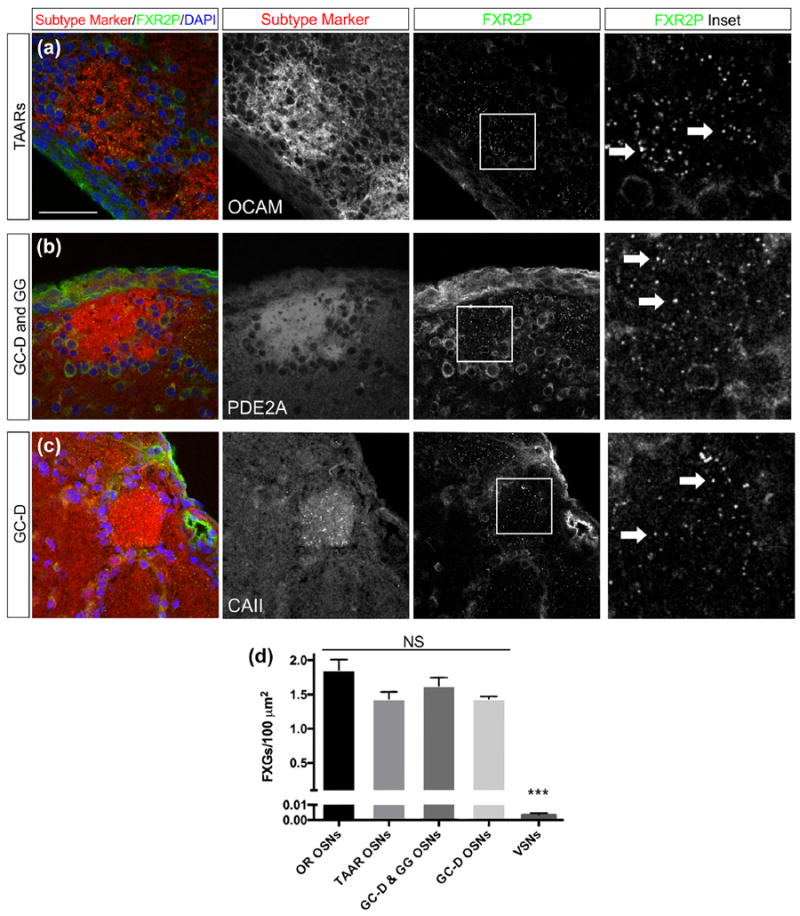
FXGs localize to axons of multiple OSN subtypes. OSN subtypes were identified in the main olfactory bulb by immunostaining for molecular markers (red): OCAMfor TAAR-expressing OSNs (a), PDE2A for the mixed population of GC-D-expressing OSNs and OSNs from the Grueneberg ganglion (GG) (b), and CAII for GC-D-expressing OSNs (c). Axons of all OSN subtypes examined expressed FXGs, indicated by FXR2P (green, arrows) (a–c). (d) FXG abundance differed among the five olfactory neuron subtypes (ANOVA, p < .0001). The OSN subtypes did not differ from each other (pairwise Tukey’s post hoc comparisons, p > .05). All OSN subtypes differed from VSNs (pairwise Tukey’s, p < .0001). FXG densities as granules per 100 μm2: OR OSNs: 1.9 ± 0.06 (n = 750 images, five mice); TAARs: 1.5 ± 0.05 (n = 331 images, five mice); GC-D: 1.4 ± 0.07 (n = 264 images, five mice); GG & GC-D: 1.6 ± 0.07 (n = 313 images, five mice); VSNs: 0.0036 ± 0.0014 (n = 45 images, four mice). Scale bar: 50 μm; 15 μm in inset images
We next asked whether FXGs are found in axons of OSNs that innervate “necklace” glomeruli in the caudal olfactory bulb. These OSNs include two subclasses, both of which express the cyclic nucleotide metabolism protein PDE2A (phosphodiesterase 2A) (Juilfs et al., 1997; Liu, Fraser, & Koos, 2009). One of these subclasses also expresses guanylyl cyclase-D (GC-D) and mediates detection of learned social cues such as foods eaten by other members of the same species (Munger et al., 2010). The second subclass of PDE2A-expressing OSNs are located in the rostral nasal cavity in the Grueneberg ganglion, rather than in the main olfactory epithelium. Grueneberg ganglion OSNs detect several stimuli including cues from stressed members of the same species as well as temperature (Brechbühl, Klaey, & Broillet, 2008; Schmid, Pyrski, Biel, Leinders-Zufall, & Zufall, 2010). FXGs were readily detected in PDE2A-containing necklace glomeruli (Figure 3b). Further, FXGs were as abundant in these OSNs as in the OR- and TAAR-expressing OSNs (Figure 3d).
To determine if FXGs are found at equivalent levels in both populations of necklace glomeruli, we assessed FXG abundance in glomeruli receiving input from GC-D OSNs. The axons of these OSNs are unique in the OB as the only ones that contain carbonic anhydrase II (CAII) (Hu et al., 2007). We thus performed double immunofluorescence for CAII and FXR2P and assessed FXG abundance in the GC-D glomeruli. We found that FXGs are as abundant in GC-D glomeruli as they are in the total population of necklace glomeruli as well as in glomeruli innervated by OR- and TAAR-expressing OSNs (Figure 3d). Unfortunately, we were not able to identify immunostaining conditions that allowed us to selectively label axons of Grueneberg ganglion OSNs. However, the observation that all necklace glomeruli, including those specifically identified as GC-D glomeruli, contain FXGs at equivalent levels suggests that axons of Grueneberg ganglion neurons also contain a similar abundance of FXGs. Together with the data presented above, our findings indicate that FXGs are found in axons of multiple classes of OSNs that mediate detection of a wide variety of chemosensory cues.
3.3 Selective localization of Omp mRNA to OSN axons
Because OSNs and VSNs differ in their axonal localization of FXGs, which contain RNA binding proteins and associate with both ribosomes and mRNA (Akins et al., 2017), we asked whether they also differ in their axonal mRNA content. As a first step toward addressing this question, we investigated the axonal localization of the mRNA encoding OMP, a broadly conserved protein that is expressed in all of the chemosensory subsystems investigated here. Omp mRNA localizes to OSN axons, where it associates with FXGs (Akins et al., 2017; Ressler, Sullivan, & Buck, 1994; Vassar et al., 1994; Wensley et al., 1995). We therefore asked whether we could detect Omp mRNA in VSN axons. We performed in situ hybridization for the Omp mRNA combined with immunofluorescence for FXR2P and OMP. Consistent with past studies, we readily detected Omp mRNA in OSN axons in the main olfactory bulb (Figure 4a; 0.6 ± 0.14 puncta/100 μm2 in the MOB, n = 4 animals). As expected from these past studies, Omp mRNA in these glomeruli was always found in axons containing OMP protein and frequently colocalized with FXGs. In contrast, we could not detect Omp mRNA granules in VSN axons in the accessory olfactory bulb (Figure 4b; 0 puncta observed in four animals). The in situ hybridization approach that we used is highly sensitive (Batish, van den Bogaard, Kramer, & Tyagi, 2012), suggesting that our failure to detect Omp in VSN axons reflects an absence of this message from these axons. Further, we could readily detect Omp mRNA in OSN glomeruli adjacent to the accessory olfactory bulb in the same sections. Together, these data indicate that Omp mRNA is selectively localized to axons in OSNs but not VSNs.
FIGURE 4.
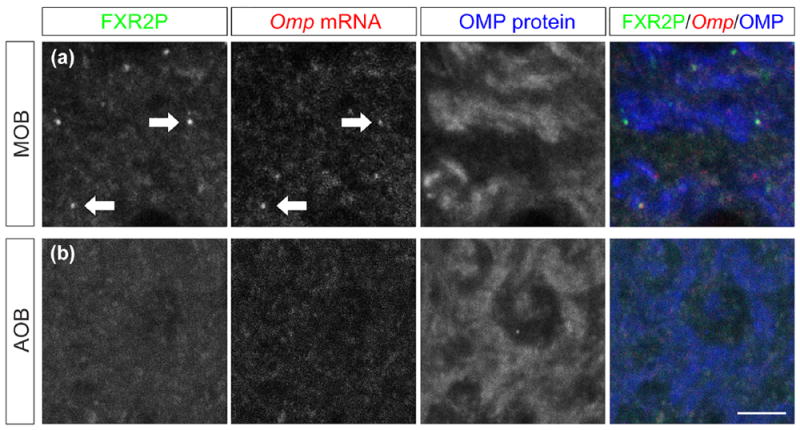
Omp mRNA localizes to axons of OSNs but not VSNs. (a) OSN axons in the main olfactory bulb (MOB) contain FXGs (green, indicated by FXR2P immunostaining, arrows), Omp mRNA (red, indicated by Omp in situ hybridization, arrows), and OMP protein (blue, indicated by OMP immunostaining). (b) VSN axons in the accessory olfactory bulb (AOB) do not contain FXGs or Omp mRNA, but do contain OMP protein. Scale bar: 5 μm
3.4 Differential axonal transport of Omp mRNA does not reflect cell type-specific transcript variants
To begin to address the molecular basis for the cell type-dependent axonal transport, we asked whether the axonal transport of Omp mRNA in OSNs but not VSNs reflected differences in the transcript between these two cell types. Omp mRNA has been shown to be expressed from a single exon with no splicing events (Danciger, Mettling, Vidal, Morris, & Margolis, 1989). However, we tested whether the axonal mRNA may reflect rare alternative transcript forms that were not detected in these past studies. Such forms may arise from previously unknown splicing events or usage of alternative polyadenylation sites and would most likely be different sizes. Therefore, we first asked whether there was a difference in Omp transcript length between the two cell types by Northern blot. Consistent with previous findings (Berghard, Buck, & Liman, 1996), we could detect no difference in Omp transcript length between RNA preparations from olfactory epithelium and vomeronasal organ (Figure 5a).
FIGURE 5.
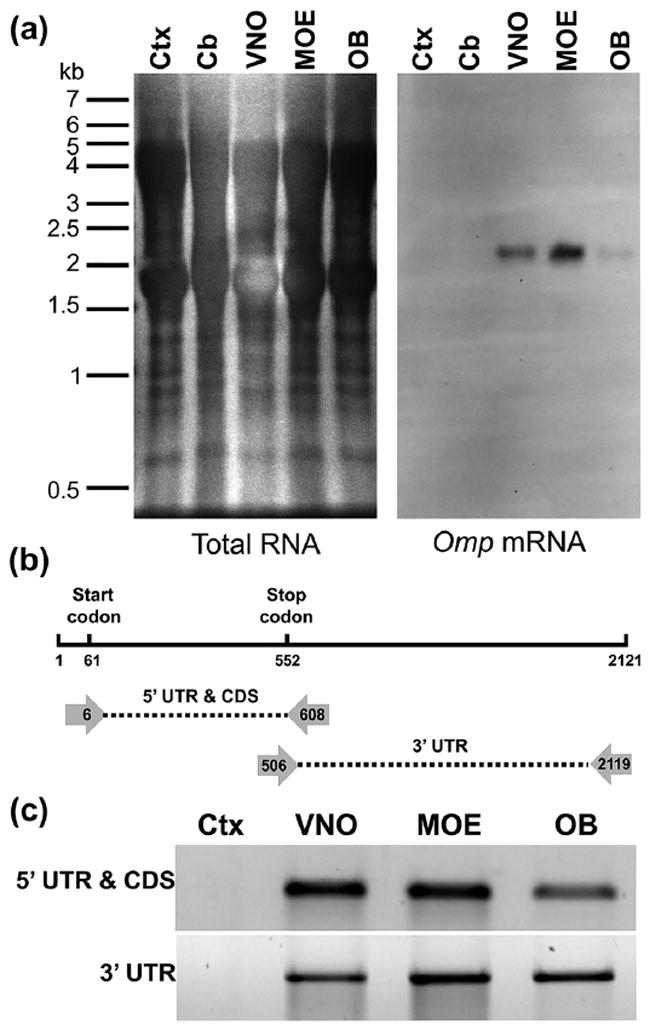
Omp transcripts are equivalent in VSN cell bodies, OSN cell bodies, and OSN axons. (a) Northern blot of RNA from cortex (Ctx), cerebellum (Cb), vomeronasal organ (VNO), main olfactory epithelium (MOE), and olfactory bulb (OB). Omp mRNA is a single size in the VNO (containing VSN cell bodies), MOE (containing OSN cell bodies), and OB (containing axons of both VSNs and OSNs). (b) Schematic of the Omp mRNA and primer locations for detecting possible alternative splicing at a higher resolution. UTR, untranslated region; CDS, coding sequence. (c) PCR using the primer pairs indicated in (b) produce products of equivalent size using cDNA prepared from MOE, VNO, and OB
We next asked whether the two tissues express Omp mRNA that differs in sequence, despite having the same length. We first performed PCR using primers that uniquely amplify the Omp 5′ UTR and coding sequence from the mouse genome (Figure 5b). As expected from the observation that the OMP protein is the same length in olfactory epithelium and vomeronasal organ, we could detect no differences in this portion of the transcript using cDNA prepared from these two tissues (Figure 5c). We next asked whether there were differences in the length of the 30 UTR between the two tissues using primers that uniquely amplify this portion of the transcript (Figure 5b). As with the 5′ UTR and coding sequence, we could detect no differences in the size of the 3′ UTR in cDNA prepared from either tissue. Since the primers used for these studies recognize unique sequences in the mouse genome, these findings indicate that the OSN and VSN Omp transcripts have the same sequences. The differences in axonal transport of the Omp mRNA between OSNs and VSNs therefore do not reflect the expression of tissue-specific forms of this transcript.
Our results do not rule out the possibility that axonal Omp mRNA represents a specific form of this transcript that is expressed at low levels in OSNs and not at all in VSNs. To test this possibility, we asked whether the axonal Omp mRNA differed from the somatic form. The Northern blot revealed that the Omp mRNA found in OSN axons in the olfactory bulb was an equivalent size to the Omp mRNA found in both OSN and VSN cell bodies (Figure 5a). Further, the PCR-based approach revealed that both the axonal and somatic mRNAs comprised the same sequences (Figure 5c). Taken together, these results indicate that the Omp mRNA is the same in OSN cell bodies, VSN cell bodies, and OSN axons.
3.5 The highly-expressed Calm1 mRNA does not localize to OSN axons
The localization of Omp mRNA to axons could merely be a byproduct of its high expression level in OSNs, although these axons do not contain the relatively abundant OSN mRNA encoding the G protein Gαolf (Wensley et al., 1995). To further test whether high abundance of an mRNA is sufficient for axonal localization of that mRNA, we asked whether the Calm1 mRNA encoding calmodulin localizes to OSN axons. Calm1 was chosen as a candidate as OSNs express Calm1 mRNA at levels comparable to those seen for Omp mRNA and nearly twice that seen for the Gαolf mRNA (Magklara et al., 2011). Further, the Calm1 transcript localizes to axons of cultured cortical neurons (Taylor et al., 2009) and is an FMRP target (Darnell et al., 2011). Since the in situ hybridization approach we used here has the potential for single molecule RNA sensitivity (Batish et al., 2012), we reasoned that we should be able to detect axonal Calm1 mRNA if OSNs transport this mRNA into axons. As expected, we could readily detect Calm1 mRNA in OSN cell bodies in the olfactory epithelium (Figure 6c–f). In the olfactory bulb, Calm1 mRNA was detected in mitral and tufted cells but not periglomerular or granule cells (Figure 6g–j), consistent with prior reports (Biffo, Goren, Khew-Goodall, Miara, & Margolis, 1991). However, we were not able to detect Calm1 mRNA in either olfactory nerve layer or glomeruli (Figure 6g,h) in sections from four different mice. In contrast, Omp mRNA is evident in OSN axons in olfactory nerve layer and glomeruli (Figure 6k) but not in mitral or tufted cells (Figure 6l,m). Therefore, the Calm1 transcript does not localize to OSN axons. Taken together with the results discussed above, these findings demonstrate that high transcript abundance is not sufficient for axonal localization of mRNAs and therefore reveal exquisite selectivity of axonal mRNA transport in olfactory axons.
FIGURE 6.
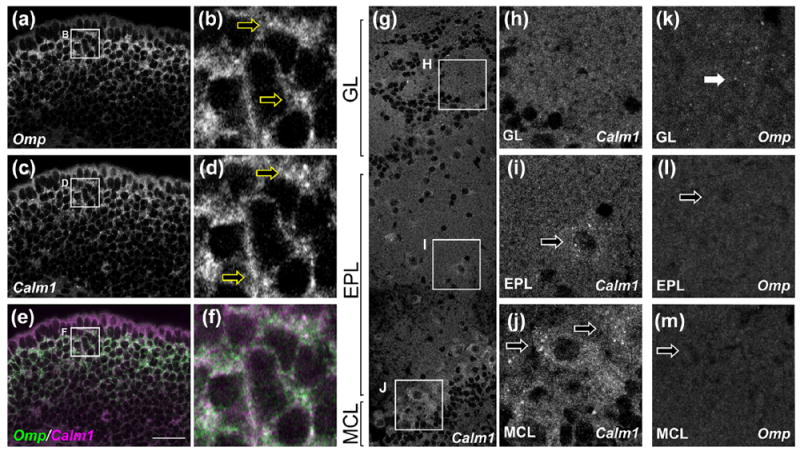
Calm1 mRNA does not localize to OSN axons. (a–f) In the main olfactory epithelium, OSN cell bodies contain both Omp mRNA (a, b, green in e, f, arrows in b) and Calm1 mRNA (c, d, red in e, f, arrows in d). (g–j) In the olfactory bulb, Calm1 mRNA was not found in OSN axons (g, h), but was seen in cell bodies of tufted cells (g, i) and mitral cells (g, j). (k–m) In contrast, Omp mRNA was found in OSN axons (k) but was not seen in resident olfactory bulb cells including tufted (l) and mitral (m) cells. Olfactory bulb layers and cell bodies were determined based on DAPI signal (not shown). GL, glomerular layer; EPL, external plexiform layer; MCL, mitral cell layer. Scale bar: 25 μm (a, c, e); 5 μm (b, d, f); 40 μm (g); 15 μm (h–m)
4 DISCUSSION
In this study, we demonstrate selective axonal localization in olfactory neurons of the Omp mRNA as well as translational regulator proteins in FXGs. These molecules are constituents of OSN axons but are not found in VSN axons either in the vomeronasal nerve or in the accessory olfactory bulb. We cannot rule out the possibility that the Omp mRNA is present in a supramolecular complex in VSN axons that prevents detection of this RNA by in situ hybridization. However, such a complex would differ from those localized to OSN axons, which do not completely mask the Omp mRNA from detection. Therefore, whether or not Omp mRNA exists in such VSN complexes, our inability to detect either Omp mRNA or FXGs in VSN axons, reveals fundamental differences in the control of the axonal proteome between OSNs and VSNs. In contrast to the cell type-dependent localization in axons, the Fragile X related proteins are found in the cell bodies of both OSNs and VSNs in non-FXG complexes. These proteins are therefore likely to regulate somatic translation in both OSNs and VSNs, while regulating axonal translation selectively in the OSNs. Our findings suggest that FMRP-dependent axonal translation is a feature of all OSNs, since FXGs are present in all four OSN subclasses we examined here. Here, we discuss the potential regulation of axonal RNA transport and its functional implications in the olfactory system.
4.1 Selectivity of axonal transport
The coordinate cell type-dependent localization of Omp mRNA and FXGs suggests that similar mechanisms regulate the axonal transport of mRNA and FXGs. This axonal localization to OSN axons could be the result of active transport or of passive processes such as diffusion. For example, Omp mRNA and FXGs may be absent from VSN axon terminals simply because VSN axons are much longer than OSN axons. However, we did not observe FXGs in VSN axons at any point in the vomeronasal nerve. Further, FXGs are found in terminals of Grueneberg ganglion OSN axons, which are substantially longer than VSN axons. This concept also does not easily fit with observations that FXGs in hippocampus are expressed in commissural axons but not the comparatively shorter Schaffer collaterals (Akins et al., 2012). Moreover, the lack of OSN axonal localization of the Calm1 mRNA, which is expressed at equivalent levels to the Omp mRNA, indicates that axonal localization does not depend solely on transcript abundance, which would be the case if passive mechanisms predominated. Localization of translational machinery to OSN axons must therefore reflect active transport.
Active RNA localization relies on specific recognition of sequence motifs by RNA binding proteins associated with motor proteins. One such RNA motif is the zip code that together with the RNA binding protein Imp1/Zbp1 mediates axonal transport of the mRNA encoding β-actin but not the one encoding γ-actin (Bassell et al., 1998; Kislauskis, Zhu, & Singer, 1994; Ross, Oleynikov, Kislauskis, Taneja, & Singer, 1997). A similar system is likely operating in OSNs since the Omp mRNA but not Calm1 mRNA is transported into axons. However, the Omp transcript does not contain a sequence that is obviously similar to the β-actin zip code. These findings thus suggest that OSNs regulate axonal RNA transport using a parallel system.
4.2 Differential axonal transport between OSNs and VSNs
The molecular basis for the differential axonal mRNA transport between OSNs and VSNs could reflect differences in either the mRNAs or in the axonal transport machinery. Our findings rule out that cell type-dependent differences in the Omp sequence underlie differential axonal transport of this transcript between OSNs and VSNs. We cannot, however, eliminate the possibility that the differential transport reflects differences in the Omp transcript abundance between the two cell types, since Omp is expressed at higher levels in the olfactory epithelium than it is in the vomeronasal organ (Figure 5). Indeed, axonal mRNA transport is influenced by the abundance of particular transcripts. For example, the β-actin and GAP-43 mRNAs compete for binding to Zbp1 and axonal transport in dorsal root ganglion neurons (Donnelly et al., 2013). A similar competition for binding to axonal transport machinery may result in other transcripts being axonally localized in VSN axons. However, VSNs also do not transport FXGs into axons, suggesting that RNA granules may be selectively excluded from VSN axons. Taken together, these findings suggest a fundamental difference in axonal transport of RNA and RNA binding proteins between OSNs and VSNs.
Determining how VSNs and OSNs differ in the mechanisms that regulate axonal mRNA transport would be facilitated by the identification of the factors that guide this transport in OSNs. Several lines of evidence suggest that FXGs are not the complexes that govern axonal RNA transport in OSNs. Omp mRNA localization to OSN axons is independent of FXGs and their constituent proteins FMRP and FXR2P, since axonal Omp transcript levels are unchanged in either Fmr1 or Fxr2 knockout animals (Akins et al., 2017). Further, both FMRP and FXR2P are ubiquitously expressed in neurons, including in VSNs (Figure 2f), which do not express FXGs (Akins et al., 2012). FXR2P may instead regulate precise localization within the axonal arbor, since subaxonal distribution of FXR2P is affected by myristoylation (Stackpole, Akins, & Fallon, 2014). These findings suggest that axonal RNA transport in OSNs is regulated by RNA binding proteins other than FMRP and FXR2P.
Alternatively, the cell type-dependent axonal mRNA localization in OSNs but not VSNs might reflect differences in the microtubule cytoskeleton along which mRNA-containing granules are transported. For example, VSN axons contain the microtubule-bundling protein Tau while OSN axons do not contain Tau (Viereck, Tucker, & Matus, 1989). Changes in cytoskeletal organization arising from absence of Tau have been proposed to underlie axonal RNA localization in OSNs (Wensley et al., 1995). However, this seems unlikely as Tau is expressed by other populations of FXG-containing neurons, including hippocampal dentate granule cells (Busciglio, Ferreira, Steward, & Cáceres, 1987). An additional possibility is that there are cell type-specific patterns of posttranslational microtubule modifications, which can affect the affinity of motor proteins for the microtubule cytoskeleton (Verhey & Gaertig, 2007). Future studies will address whether differential axonal transport between OSNs and VSNs reflects differences in the composition of transport granules or in the cytoskeletal pathways along which these granules travel.
4.3 Potential implications for OSN plasticity
The functional consequences of axonal translation in OSNs are unclear (Dubacq, Fouquet, & Trembleau, 2014). One possibility is that FXGs are involved in axon outgrowth as the peripheral olfactory system incorporates new OSNs throughout life (Schwob et al., 2017). However, OSNs do not express FXGs until after outgrowth when they have integrated into olfactory bulb circuits (Akins et al., 2017). Throughout the brain, FXG expression correlates with periods of robust plasticity and, in the adult hippocampus, is not dependent on ongoing neurogenesis (Akins et al., 2017; Christie et al., 2009). These findings suggest that FXG-dependent local translation in OSN axons supports the plasticity or maintenance, rather than formation, of olfactory circuits. Local dendritic translation underlies long term modification of synaptic strength at many synapses (Bramham & Wells, 2007), and presynaptic translation is required for long term depression at GABAergic synapses in the hippocampus (Younts et al., 2016). Local translation in either OSN axons or mitral cell dendrites may modulate synaptic signaling between these neurons, since synapses between OSNs and mitral cells exhibit plasticity that comprises both presynaptic and postsynaptic components (Ennis, Linster, Aroniadou-Anderjaska, Ciombor, & Shipley, 1998; Mutoh, Yuan, & Knöpfel, 2005; Tyler, Petzold, Pal, & Murthy, 2007). Of particular interest, the OSN to mitral cell synapse exhibits presynaptic metabotropic glutamate receptor-mediated long term depression (Mutoh et al., 2005). FMRP is a critical regulator of the postsynaptic manifestation of this plasticity at other synapses in the brain (Bassell & Warren, 2008; Bear, Huber, & Warren, 2004; Huber, Gallagher, Warren, & Bear, 2002). Further, in addition to synaptic plasticity, OSNs exhibit remarkable structural plasticity, with a high rate of activity-dependent synapse formation and elimination (Cheetham, Park, & Belluscio, 2016). To our knowledge, none of these forms of synaptic or axonal plasticity have been investigated at terminals of non-OR-expressing OSNs in the main olfactory bulb or at VSN terminals in the accessory olfactory bulb. While the non-OR OSN subsystems are generally thought to mediate innate responses, the behavioral output that arises from activation of these systems can be affected by context and past experience (Munger et al., 2010; Saraiva et al., 2016). Whether these behavioral differences reflect modifications to the connections between OSNs and their targets in the olfactory bulb is not known. Our findings suggest that FMRP-dependent regulation of axonal translation by odor exposure may support rapid experiencedependent modification of axonal structure and synaptic release properties in all classes of OSNs.
OMP modulates odor-induced cAMP signaling in OSNs (Reisert et al., 2007). While this function of OMP is best understood in OSN sensory cilia within the olfactory epithelium, it seems likely that OMP plays a similar role in OSN axons as well. OSN axons contain signal transduction machinery components that mirror those localized to the sensory cilia (Barnea et al., 2004; Cherry & Davis, 1999; Menco, Tekula, Farbman, & Danho, 1994; Strotmann, Levai, Fleischer, Schwarzenbacher, & Breer, 2004; Treloar, Uboha, Jeromin, & Greer, 2005; Wekesa & Anholt, 1999; Zou et al., 2007). This cAMP signal transduction machinery is not equivalent in OSN and VSN axons as, for example, the phosphodiesterase PDE4A is much more highly expressed in OSN axons than in VSN axons (Cherry & Davis, 1999). These observations suggest cell type-dependent functions for axonal cAMP signaling in olfactory neurons. Consistent with such roles, the cAMP transduction machinery in OSN axons is functionally active, as application of odorant molecules directly to axons results in local cAMP synthesis (Maritan et al., 2009). While OSN axons are unlikely to encounter odorants in the olfactory bulb, these signal transduction cascades may be activated in response to other cues. Local synthesis of OMP would provide a way to modulate these axonal responses separately from modulation of the responses induced by odorants in OSN cilia.
4.4 Summary
In these studies, we have identified that even closely related neurons that process similar types of information, such as the OSNs and VSNs, differ in their use of axonal translation. These observations suggest that multiple classes of OSNs, even those that mediate stereotyped behaviors, exhibit activity-dependent regulation of the axonal proteome. The proteins that are synthesized within axons also vary within cell type across development and in response to both physiological and pathophysiological cues (Baleriola et al., 2014; Gumy et al., 2011; Kalinski et al., 2015; Shigeoka et al., 2016; Taylor et al., 2009; Willis et al., 2007). Axonal translation is thus regulated at several different levels in order to modulate the axonal proteome in a cell type-dependent manner. Differences among neuronal cell types in the transcripts that are translated in axons and the regulatory mechanisms that govern this translation likely contribute to the vast structural and functional diversity seen in the axonal compartment across neurons.
Supplementary Material
Additional Supporting Information may be found online in the supporting information tab for this article.
TABLE S1 In situ probe sequences for experiments depicted in the paper
Acknowledgments
We thank H. Berk-Rauch and G. Wenk for assistance with experiments, the members of the Akins lab for critical review of this work, and RY Korsak for help in preparing Figure 1. Pilot studies were carried out in the lab of JR Fallon.
Funding information
NIH/NIMH, Grant Number: R00MH90237; NIH/NICHD, Grant Number: R01HD052083 (JR Fallon).
Footnotes
AUTHOR CONTRIBUTION
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Acquisition of data, study concept and design, analysis and interpretation of data, drafting and critical revision of the manuscript for intellectual content: all authors. Obtained funding: MRA.
CONFLICT OF INTEREST
All authors declare that we have no conflicts of interest.
References
- Akins MR, Berk-Rauch HE, Kwan KY, Mitchell ME, Shepard KA, Korsak LIT, Fallon JR, et al. Axonal ribosomes and mRNAs associate with fragile X granules in adult rodent and human brains. Human Molecular Genetics. 2017 doi: 10.1093/hmg/ddw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins MR, LeBlanc HF, Stackpole EE, Chyung E, Fallon JR. Systematic mapping of fragile X granules in the mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. The Journal of Comparative Neurology. 2012;520(16):3687–3706. doi: 10.1002/cne.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell. 2014;158(5):1159–1172. doi: 10.1016/j.cell.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea G, O’Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science. 2004;304(5676):1468–1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Kosik KS, et al. Sorting of beta -actin mRNA and protein to neurites and growth cones in culture. The Journal of Neuroscience. 1998;18(1):251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(12):4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends in Neurosciences. 2004;27(7):370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Berghard A, Buck LB, Liman ER. Evidence for distinct signaling mechanisms in two mammalian olfactory sense organs. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(6):2365–2369. doi: 10.1073/pnas.93.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annual Review of Neuroscience. 2012;35(1):417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffo S, Goren T, Khew-Goodall YS, Miara J, Margolis FL. Expression of calmodulin mRNA in rat olfactory neuroepithelium. Molecular Brain Research. 1991;10(1):13–21. doi: 10.1016/0169-328X(91)90051-X. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: Transport, translation and function. Nature Reviews Neuroscience. 2007;8(10):776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Brechbühl J, Klaey M, Broillet M-C. Grueneberg ganglion cells mediate alarm pheromone detection in mice. Science. 2008;321(5892):1092–1095. doi: 10.1126/science.1160770. [DOI] [PubMed] [Google Scholar]
- Buiakova OI, Baker H, Scott JW, Farbman A, Kream R, Grillo M, Margolis FL, et al. Olfactory marker protein (OMP) gene deletion causes altered physiological activity of olfactory sensory neurons. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9858–9863. doi: 10.1073/pnas.93.18.9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busciglio J, Ferreira A, Steward O, Cáceres A. An immunocytochemical and biochemical study of the microtubule-associated protein Tau during post-lesion afferent reorganization in the hippocampus of adult rats. Brain Research. 1987;419(1–2):244–252. doi: 10.1016/0006-8993(87)90590-7. [DOI] [PubMed] [Google Scholar]
- Cheetham CEJ, Park U, Belluscio L. Rapid and continuous activity-dependent plasticity of olfactory sensory input. Nature Communications. 2016;7:10729. doi: 10.1038/ncomms10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JA, Davis RL. Cyclic AMP phosphodiesterases are localized in regions of the mouse brain associated with reinforcement, movement, and affect. The Journal of Comparative Neurology. 1999;407(2):287–301. doi: 10.1002/(SICI)1096-9861(19990503)407:2<287∷AIDCNE9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: A presynaptic fragile X granule expressed in a subset of developing brain circuits. The Journal of Neuroscience. 2009;29(5):1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danciger E, Mettling C, Vidal M, Morris R, Margolis F. Olfactory marker protein gene: Its structure and olfactory neuron-specific expression in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(21):8565–8569. doi: 10.1073/pnas.86.21.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nature Neuroscience. 2013;16(11):1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Darnell RB, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, Twiss JL, et al. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. The Journal of Neuroscience. 2013;33(8):3311–3322. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq C, Fouquet C, Trembleau A. Making scent of the presence and local translation of odorant receptor mRNAs in olfactory axons. Developmental Neurobiology. 2014;74(3):259–268. doi: 10.1002/dneu.22122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis M, Linster C, Aroniadou-Anderjaska V, Ciombor K, Shipley MT. Glutamate and synaptic plasticity at mammalian primary olfactory synapses. Annals of the New York Academy of Sciences. 1998;855(1):457–466. doi: 10.1111/j.1749-6632.1998.tb10606.x. [DOI] [PubMed] [Google Scholar]
- Gumy LF, Yeo GSH, Tung Y-CL, Zivraj KH, Willis D, Coppola G, Fawcett JW, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Finger TE. Is TrpM5 a reliable marker for chemosensory cells? Multiple types of microvillous cells in the main olfactory epithelium of mice. BMC Neuroscience. 2008;9:115. doi: 10.1186/1471-2202-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhong C, Ding C, Chi Q, Walz A, Mombaerts P, Luo M, et al. Detection of Near-Atmospheric Concentrations of CO2 by an Olfactory Subsystem in the Mouse. Science. 2007;317(5840):953–957. doi: 10.1126/science.1144233. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(11):7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Tsai L, Roy DS, Valenzuela DH, Mosley C, Magklara A, Barnea G, et al. Neurons expressing trace amineassociated receptors project to discrete glomeruli and constitute an olfactory subsystem. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(33):13410–13415. doi: 10.1073/pnas.1206724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juilfs DM, Fülle H-J, Zhao AZ, Houslay MD, Garbers DL, Beavo JA. A subset of olfactory neurons that selectively express cGMP-stimulated phosphodiesterase (PDE2) and guanylyl cyclase-D define a unique olfactory signal transduction pathway. Proceedings of the National Academy of Sciences. 1997;94(7):3388–3395. doi: 10.1073/pnas.94.7.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski AL, Sachdeva R, Gomes C, Lee SJ, Shah Z, Houle JD, Twiss JL. mRNAs and protein synthetic machinery localize into regenerating spinal cord axons when they are provided a substrate that supports growth. The Journal of Neuroscience. 2015;35(28):10357–10370. doi: 10.1523/JNEUROSCI.1249-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass MD, Moberly AH, Rosenthal MC, Guang SA, McGann JP. Odor-specific, olfactory marker protein-mediated sparsening of primary olfactory input to the brain after odor exposure. The Journal of Neuroscience. 2013;33(15):6594–6602. doi: 10.1523/JNEUROSCI.1442-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Margolis FL. Immunological studies of the rat olfactory marker protein. Journal of Neurochemistry. 1975;24(6):1101–1106. doi: 10.1111/j.1471-4159.1975.tb03883.x. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. The Journal of Cell Biology. 1994;127(2):441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsak LIT, Mitchell ME, Shepard KA, Akins MR. Regulation of neuronal gene expression by local axonal translation. Current Genetic Medicine Reports. 2016;4(1):16–25. doi: 10.1007/s40142-016-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AC, He J, Ma M. Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. The Journal of Neuroscience. 2011;31(8):2974–2982. doi: 10.1523/JNEUROSCI.5067-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Fraser SE, Koos DS. Grueneberg ganglion olfactory subsystem employs a cGMP signaling pathway. The Journal of Comparative Neurology. 2009;516(1):36–48. doi: 10.1002/cne.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Lomvardas S, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145(4):555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritan M, Monaco G, Zamparo I, Zaccolo M, Pozzan T, Lodovichi C. Odorant receptors at the growth cone are coupled to localized cAMP and Ca2+ increases. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3537–3542. doi: 10.1073/pnas.0813224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menco BPM, Tekula FD, Farbman AI, Danho W. Developmental expression of G-proteins and adenylyl cyclase in peripheral olfactory systems. Light microscopic and freeze-substitution electron microscopic immunocytochemistry. Journal of Neurocytology. 1994;23(11):708–727. doi: 10.1007/BF01181645. [DOI] [PubMed] [Google Scholar]
- Merl J, Ueffing M, Hauck SM, von Toerne C. Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Müller cells. PROTEOMICS. 2012;12(12):1902–1911. doi: 10.1002/pmic.201100549. [DOI] [PubMed] [Google Scholar]
- Munger SD, Leinders-Zufall T, McDougall LM, Cockerham RE, Schmid A, Wandernoth P, Kelliher KR, et al. An olfactory subsystem that detects carbon disulfide and mediates food-related social learning. Current Biology. 2010;20(16):1438–1444. doi: 10.1016/j.cub.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, Roskams AJ. A Novel Embryonic Nestin- Expressing Radial Glia-Like Progenitor Gives Rise to Zonally Restricted Olfactory and Vomeronasal Neurons. Journal of Neuroscience. 2008;28(16):4271–4282. doi: 10.1523/JNEUROSCI.5566-07.2008. https://doi.org/10.1523/JNEUROSCI.5566-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H, Yuan Q, Knöpfel T. Long-term depression at olfactory nerve synapses. The Journal of Neuroscience. 2005;25(17):4252–4259. doi: 10.1523/JNEUROSCI.4721-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert J, Yau K-W, Margolis FL. Olfactory marker protein modulates the cAMP kinetics of the odour-induced response in cilia of mouse olfactory receptor neurons. The Journal of Physiology. 2007;585(3):731–740. doi: 10.1113/jphysiol.2007.142471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: Evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79(7):1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Molecular and Cellular Biology. 1997;17(4):2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva LR, Kondoh K, Ye X, Yoon K, Hernandez M, Buck LB. Combinatorial effects of odorants on mouse behavior. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(23):E3300–E3306. doi: 10.1073/pnas.1605973113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Pyrski M, Biel M, Leinders-Zufall T, Zufall F. Grueneberg ganglion neurons are finely tuned cold sensors. Journal of Neuroscience. 2010;30(22):7563–7568. doi: 10.1523/JNEUROSCI.0608-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, Hewitt Coleman J. Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. Journal of Comparative Neurology. 2017;525(4):1034–1054. doi: 10.1002/cne.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Holt CE, et al. Dynamic axonal translation in developing and mature visual circuits. Cell. 2016;166(1):181–192. doi: 10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackpole EE, Akins MR, Fallon JR. N-myristoylation regulates the axonal distribution of the Fragile X-related protein FXR2P. Molecular and Cellular Neuroscience. 2014;62:42–50. doi: 10.1016/j.mcn.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Logan DW. Olfactory mechanisms of stereotyped behavior: On the scent of specialized circuits. Current Opinion in Neurobiology. 2010;20(3):274–280. doi: 10.1016/j.conb.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann J, Levai O, Fleischer J, Schwarzenbacher K, Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. The Journal of Neuroscience. 2004;24(35):7754–7761. doi: 10.1523/JNEUROSCI.2588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. The Journal of Neuroscience. 2009;29(15):4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treloar HB, Gabeau D, Yoshihara Y, Mori K, Greer CA. Inverse expression of olfactory cell adhesion molecule in a subset of olfactory axons and a subset of mitral/tufted cells in the developing rat main olfactory bulb. The Journal of Comparative Neurology. 2003;458(4):389–403. doi: 10.1002/cne.10590. [DOI] [PubMed] [Google Scholar]
- Treloar HB, Uboha U, Jeromin A, Greer CA. Expression of the neuronal calcium sensor protein NCS-1 in the developing mouse olfactory pathway. The Journal of Comparative Neurology. 2005;482(2):201–216. doi: 10.1002/cne.20431. [DOI] [PubMed] [Google Scholar]
- Tyler WJ, Petzold GC, Pal SK, Murthy VN. Experience-dependent modification of primary sensory synapses in the mammalian olfactory bulb. The Journal of Neuroscience. 2007;27(35):9427–9438. doi: 10.1523/JNEUROSCI.0664-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nuñez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79(6):981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6(17):2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- Viereck C, Tucker RP, Matus A. The adult rat olfactory system expresses microtubule-associated proteins found in the developing brain. The Journal of Neuroscience. 1989;9(10):3547–3557. doi: 10.1523/JNEUROSCI.09-10-03547.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wekesa KS, Anholt RRH. Differential expression of G proteins in the mouse olfactory system. Brain Research. 1999;837(1–2):117–126. doi: 10.1016/S0006-8993(99)01630-3. [DOI] [PubMed] [Google Scholar]
- Wensley CH, Stone DM, Baker H, Kauer JS, Margolis FL, Chikaraishi DM. Olfactory marker protein mRNA is found in axons of olfactory receptor neurons. The Journal of Neuroscience. 1995;15(7):4827–4837. doi: 10.1523/JNEUROSCI.15-07-04827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Twiss JL, et al. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. Journal of Cell Biology. 2007;178(6):965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younts TJ, Monday HR, Dudok B, Klein ME, Jordan BA, Katona I, Castillo PE. Presynaptic protein synthesis is required for long-term plasticity of GABA release. Neuron. 2016;92(2):479–492. doi: 10.1016/j.neuron.2016.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, O’Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. The EMBO Journal. 1995;14(21):5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D-J, Chesler AT, Pichon CEL, Kuznetsov A, Pei X, Hwang EL, Firestein S. Absence of adenylyl cyclase 3 perturbs peripheral olfactory projections in mice. The Journal of Neuroscience. 2007;27(25):6675–6683. doi: 10.1523/JNEUROSCI.0699-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
TABLE S1 In situ probe sequences for experiments depicted in the paper


