Abstract
We report our group’s performance for protein-protein complex structure prediction and scoring in Round 37 of the Critical Assessment of PRediction of Interactions (CAPRI), an objective assessment of protein-protein complex modeling. We demonstrated noticeable improvement in both prediction and scoring compared to previous rounds of CAPRI, with our human predictor group near the top of the rankings and our server scorer group at the top. This is the first time in CAPRI that a server has been the top scorer group. To predict protein-protein complex structures, we used both multi-chain template-based modeling (TBM) and our protein-protein docking program, LZ-erD. LZerD represents protein surfaces using 3D Zernike descriptors (3DZD), which are based on a mathematical series expansion of a 3D function. Because 3DZD are a soft representation of the protein surface, LZerD is tolerant to small conformational changes, making it well suited to docking unbound and TBM structures. The key to our improved performance in CAPRI Round 37 was to combine multi-chain TBM and docking. As opposed to our previous strategy of performing docking for all target complexes, we used TBM when multi-chain templates were available and docking otherwise. We also describe the combination of multiple scoring functions used by our server scorer group, which achieved the top rank for the scorer phase.
Keywords: CAPRI, protein docking prediction, protein-protein docking, protein structure prediction, computational methods, prediction accuracy, structure modeling, template-based modeling, protein-protein interaction, LZerD
Introduction
Detailed, atomic-level structures of protein-protein complexes are crucial for understanding the many biological processes mediated by protein-protein interactions. Despite the importance, the majority of protein-protein complex structures remain unsolved by experimental techniques such as X-ray crystallography and cryo-electron microscopy, in large part because such methods are difficult and resource-intensive. Computational protein complex modeling complements experimental techniques by producing structure models of protein-protein complexes. A substantial number of protein-protein complexes can be modeled using template-based modeling (TBM) or protein docking. Protein docking uses the tertiary structure of single proteins, which have much better structural coverage than protein complexes as well as higher availability of single-chain templates for TBM1–3. Many computational protein complex modeling methods have been developed, and an objective, community-wide evaluation, the Critical Assessment of Prediction of Interactions (CAPRI)4, was established in 20015. Since then, over 120 targets have been released for prediction. Our group has participated in CAPRI since 2009, participating in rounds 18–24 and 26–396. The core of our prediction pipeline is a protein-protein docking program named LZerD7–11 developed in our group. LZerD represents protein surfaces using 3D Zernike descriptors (3DZD)8;12;13, which are based on a mathematical series expansion of a 3D function. The 3DZD comprise a soft representation of the protein surface shape, making LZerD more tolerant to the conformational change that occurs on binding than similar shape-based docking methods.
In this work, we summarize our group’s performance in Round 37 of CAPRI. Our performance improved compared to previous rounds of CAPRI6 for both the predictor phase, in which we create models of a protein-protein complex, and the scorer phase, in which we re-rank the models of all predictors. Our human predictor group was among the best performing and our server scorer group reached the top rank, which is the first time a server group has been the top ranked scorer. We identified two major changes that led to our improved predictions. First and foremost, we found that we missed several easy cases in the previous CASP/CAPRI round, Round 306. Concretely, out of 25 targets assessed in Round 30, 7 targets we missed were classified as easy homodimers by the assessors4. Thus, for Round 37, we used multi-chain template-based modeling (TBM) for many targets, leading to several medium quality models and even one achieving high quality. In addition, we streamlined the docking and scoring procedure used when no multi-chain template was found. The use of both TBM and docking was central to our improved performance. For the scorer phase, our server group applied a straightforward combination of multiple scores to achieve the top rank.
Materials and Methods
Predictor procedure
The Kihara lab has two CAPRI groups, the Kihara human predictor group and the LZerD server predictor group. For Round 37 of CAPRI, the prediction procedure we used in CAPRI 306 was modified in several ways. Because we missed several easy homodimer cases in Round 30, we used template-based modeling (TBM) for complexes when applicable, rather than just for single chains. The overall procedure is to check for multi-chain templates and use multi-chain TBM if possible. Otherwise, single chains are modeled then complex models are created using the LZerD docking program and ranked using knowledge-based scoring functions. Prior to submission, for models from both TBM and docking, we rebuild the side-chains and relax the models using molecular dynamics to reduce atomic clashes. We will describe the shared steps of the pipeline, outlined in Figure 1, and then discuss the differences between the groups.
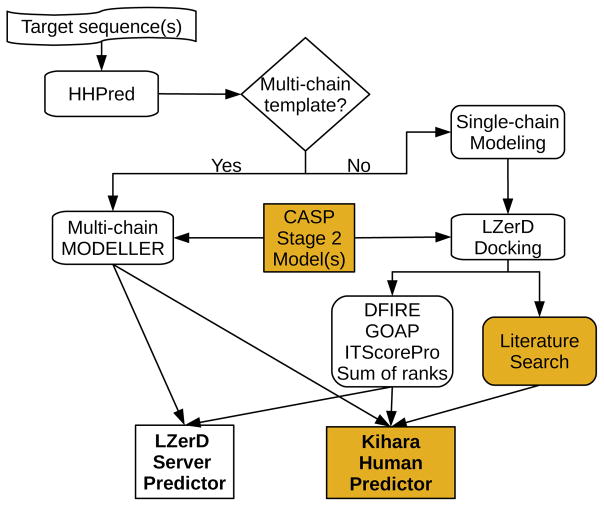
Figure 1.
Outline of predictor procedure. Filled boxes indicate procedures used only by the human group. First, the target sequence(s) are input into HHPred. If a multi-chain template is found, MODELLER is used to create complex models. Otherwise, single-chain models are created with MODELLER, which are used as input to LZerD docking. The docking poses produced by LZerD are chosen using the sum of ranks of three knowledge-based scoring functions: DFIRE, GOAP, and ITScorePro. The human predictor group has two differences: first, CASP Stage 2 server models are used as input to either MODELLER or LZerD; second, literature information is used to inform docking and/or model selection.
Multi-chain template-based modeling
The sequences for all query proteins were input into HHPred14 to search for templates. In the case of homo-oligomers, a candidate template was used if the number of chains in its biological unit matched the number of chains provided by the organizers. In the case of hetero-oligomers, a candidate template was used if it was a hit for all sequences and had the correct number of chains. A hit was defined as a template that was found within top 10 ranks in a HHPred search and has at least a sequence identity of 10%. Most of the templates found had over 30% sequence identity to the query. If an appropriate multi-chain template was found, it was used to create a model of the complex with MODELLER15. Table 1 lists whether each target was modeled using TBM or docking. If no multi-chain template could be found, single-chain models were created and docking was performed (described in more detail below). Table S1 lists the multi-chain templates and single-chain templates used for each target.
Table 1.
Per-target predictor performance summary for CAPRI Round 37.
| Target | Oligo | Technique | Int. | LZerD hits | Kihara hits | Groups with hits |
|---|---|---|---|---|---|---|
|
|
|
|
|
|
||
| T110 | 3 | 3zpe | 1 | 10/5** | 10/7** | 22/18** |
| T111 | 3 | 4d0u | 1 | 10*** | 10*** | 27/23***/4** |
| T112 | 3 | 2vtw, 2ium | 1 | 0 | 0 | 19 |
| T113 | 1+1 | Docking | 1 | 0 | 1 | 6 |
| T114 | 2 | 4q95 | 1 | 0 | 0 | 0 |
| T115 | 1 | Docking | - | Canceled | Canceled | Canceled |
| T116 | 2 | 3d36, 4q20, 4u7o, 4gcz, 4bix | 1 | 0 | 0 | 0 |
| T117 | 2x(1+1) | Docking | 1 | 0 | 5 | 1 |
| 2 | 0 | 0 | 0 | |||
| 3 | 0 | 0 | 0 | |||
| 4 | 0 | 0 | 0 | |||
| T118 | 8 | 3r1m | 1 | 1** | 1** | 24/22** |
| 2 | 1** | 1** | 24/13***/11** | |||
| 3 | 1*** | 1*** | 24/18***/4** | |||
| T119 | 2 | 3ox4, 5br4 | 1 | 5 | 5 | 22/5** |
| T120 | 1+1 | 4dh2 | 1 | 10 | 10/1** | 20/7** |
“Oligo”: oligomeric state. A single number indicates a homomeric complex and a plus sign (+) indicates a heteromeric complex, e.g. 1+1 is a heterodimer and 2x(1+1) is a heterotetramer. T115 was canceled because it was found to be a monomer. “Technique”: a PDB code is the template used to model the complex structure with template-based modeling (TBM), “Docking” means single chains were modeled using TBM and the complex was modeled using docking. “Int.”: unique interface in a multimeric complex.
indicates high quality models and
indicates medium quality models; e.g., 6/1***/4** means six acceptable or better models, of which one was high and four were medium quality.
CAPRI model qualities defined previously30. Models with clashes or low sequence identity are excluded. “LZerD hits” and “Kihara hits”: number of models of each quality in the 10 submitted models. For T118, we only submitted 1 model instead of 10. “Groups with hits”: number of groups with at least one model of each quality.
Protein-protein docking with LZerD
To dock protein complexes, we use LZerD, a pairwise protein-protein docking program developed in our lab7–9;11. LZerD represents the surface shape of a protein using 3D Zernike descriptors (3DZD)8;12;13, based on a mathematical series expansion of a 3D function (in this case, the protein surface). The 3DZD are a soft representation of the surface shape, which confers tolerance to the conformational changes associated with binding. In our earlier study, LZerD was shown to have superior performance to other shape-based docking methods on unbound docking7. Protein-protein docking with LZerD involves creating patches on the surface of each input protein and then matching the patches between two proteins. To create patches, points are evenly spread on the surface of the protein and 3DZD are computed for the sphere centered at each point. In addition, the surface normal is computed for each point. Matching patches between the receptor and ligand proteins are found using geometric hashing. Each pair of matching patches is evaluated using a shape score, which consists of four terms: the correlation of the 3DZD vectors, the angle between the surface normals, the buried surface area, and the excluded volume (representing atomic clash). Patch pairs with a good shape score are used to create tens to hundreds of thousands of candidate docking poses, which can be further evaluated using other scoring functions.
The docking poses produced by LZerD were ranked using the LZerD shape score, then the top 20,000 were clustered with an RMSD cutoff of 4.0 Å. Next, the number of clusters was further reduced to 9999 using the shape score. Rather than the pre-filtering process used in CAPRI Round 306, all cluster centers were scored using DFIRE16, GOAP17, and ITScorePro18. Then, the rank by each scoring function was computed and the ranks of each decoy were summed. For example, a decoy that is rank 1 by all three scoring functions would have a rank sum of 3. The ten decoys with the lowest rank sum were chosen. This procedure was used to rank LZerD decoys in the prediction phase.
Model refinement
After the decoys to be submitted are selected, the structures were relaxed using molecular dynamics (MD) simulation to reduce the number of atom clashes. CHARMM19 was used for MD simulation with the CHARMM22 force field and an implicit solvent model, FACTS20. For the entire simulation, all Cα atoms were restrained with a harmonic constraint of 100 kcal/mol/Å2. The complex was minimized with 500 steps of the steepest descent algorithm followed by 1000 steps of the adopted basis Newton-Raphson algorithm. Finally, the structure was equilibrated for 20 ps using a temperature of 100 K, 2 fs timestep, and fixed covalent hydrogen bond lengths.
Human prediction and server prediction
Here we describe the differences in the prediction procedures used by the LZerD server predictor group and the Kihara human predictor group. The overall procedure is outlined in Figure 1. The procedures unique to the human group are shown in filled boxes. In both groups, the first step is to search for multi-chain templates. For the server group, a multi-chain template is used as input to MODELLER. For the human group, CASP Stage 2 server models were available due to the concurrent CAPRI and CASP rounds. In cases with a multi-chain template, the human predictor group used the CASP server models in two ways. First, the CASP server model(s) and the multi-chain template were both used as as input to MODELLER. Second, the CASP server model(s) were superimposed onto the template and the superimposed CASP server models were saved into a new file for submission. Table S1 lists the CASP server models used for each target.
For cases where a multi-chain template was not found (T113, T115, and T117), both groups used single-chain modeling followed by docking. For T113B (CASP ID: T0885), a clear single-chain template was not detected using HHPred, so the server group used a model created using I-TASSER21. In docking cases, the server group selected docking decoys using the sum of ranks described above. The human group also used the sum of ranks but considered other information as well. For T113 and T115, the rank by PRESCO22;23 was also considered for human predictions. In addition, the human group also considered literature information where available to guide docking and model selection.
Scorer procedure
The scorer procedure used in CAPRI 37 is outlined in Figure 2. For each target, the 600–1300 candidate docking poses submitted by the predictor groups were scored using DFIRE16, GOAP17, and ITScorePro18. The rank by each scoring function was computed and the ranks for all three scores were combined into a rank sum (if a docking pose is rank 1 by all three scoring functions, its rank sum is 3). All scorer docking poses were also clustered with a 4 Å RMSD cutoff. The cluster member with the lowest rank sum was chosen to be the cluster representative. For the server group, the ten cluster representatives with the lowest rank sum were chosen; this is effectively equivalent to selecting the ten models with the lowest rank sum while skipping models that are very similar (e.g. in the same cluster). For the human group, the cluster representatives from the ten largest clusters were chosen. Prior to submission, we rebuilt the side-chains for the selected models and performed the MD relaxation protocol described above.
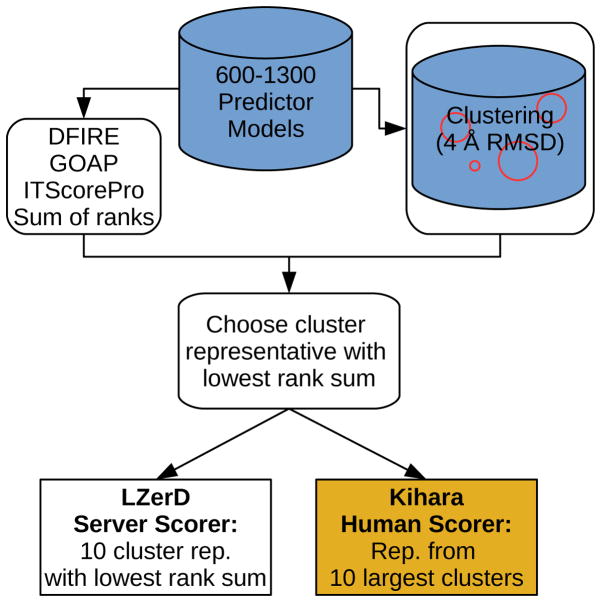
Figure 2.
Outline of scorer procedure. The 600–1300 models submitted by the predictors for the scorer round are scored using three knowledge-based scoring functions: DFIRE, GOAP, and ITScorePro. The rank sum (sum of the rank by each scoring function) is computed for each model. The models are also clustered with a 4 Å RMSD cutoff. Cluster representatives are chosen using the lowest rank sum. The LZerD server scorer group submits the 10 cluster representatives with the lowest rank sum. The Kihara human scorer group submits the cluster representatives from the 10 largest clusters.
Results and Discussion
Predictor results
Table 1 shows a summary of the results from the predictor phase from Round 37. CAPRI Round 37 contained targets T110-T120 which were selected from CASP12 (CASP IDs are listed in Table S1). Because target T115 was canceled, a total of ten targets were evaluated. This table includes two targets, T114 and T116, where no groups produced acceptable models. T114 did not have an appropriate multi-chain template and it was difficult to compute the alignment with the single-chain template. T116 had various multi-chain templates but there were large conformational changes. In addition, Round 37 included targets with more than two chains (T110, T111, T112, T117, and T118) which may have more than one interface. T118 is a homo-octamer with D4 symmetry; thus, there are 3 unique interfaces (one from the C4 portion and two between the C4 halves). In contrast, T112 is a homo-trimer with C3 symmetry and has only 1 unique interface. The CAPRI evaluation classifies target quality as the best quality achieved by all interfaces; thus, medium quality for interfaces 1 and 2 and high quality for interface 3 counts as medium quality for the target.
According to the CAPRI assessors’ slides presented at the CASP12 evaluation meeting held at Gaeta, Italy, on December 12, 2017 (http://predictioncenter.org/casp12/docs/presentations/CASP12_CAPRI_Lensink.pdf), our human group had hits for six targets out of ten, including one target with high quality models and three targets with medium quality models. The best group also had six targets with hits and medium quality for three targets but they achieved high quality for two targets. The LZerD server predictions were nearly as good, with five total hits, of which one was high and two were medium quality. In the assessors’ slides (page 42), compared to the other participants, our human predictor group (Kihara) tied for fourth out of 48 predictor groups and our server predictor group (LZerD) tied for tenth. Compared to previous rounds, the performance of our groups was noticeably better.
The most significant difference compared to Round 30 was the use of template-based modeling (TBM) for complexes, which we used for eight targets. Five of the six targets that we successfully modeled used multi-chain TBM. One TBM case, T112, was successfully modeled by other groups but not by ours. Our submissions had fnat of 0.5, meeting the threshold for high quality, but L-RMSD 11 Å and I-RMSD 4.8 Å, exceeding the cutoffs of 10 Å and 4 Å, respectively, for acceptable. Furthermore, we had complete or partial success for both targets where we used docking (T113 and T117). For T113, the human group had one acceptable hit and was one of only six groups to get hits (Table 1). For T117, we had acceptable hits for one of the four interfaces (Figure 4C); however, the other interfaces were incorrect. Most notably, our human group was the only group in the experiment to have any hits for T117.
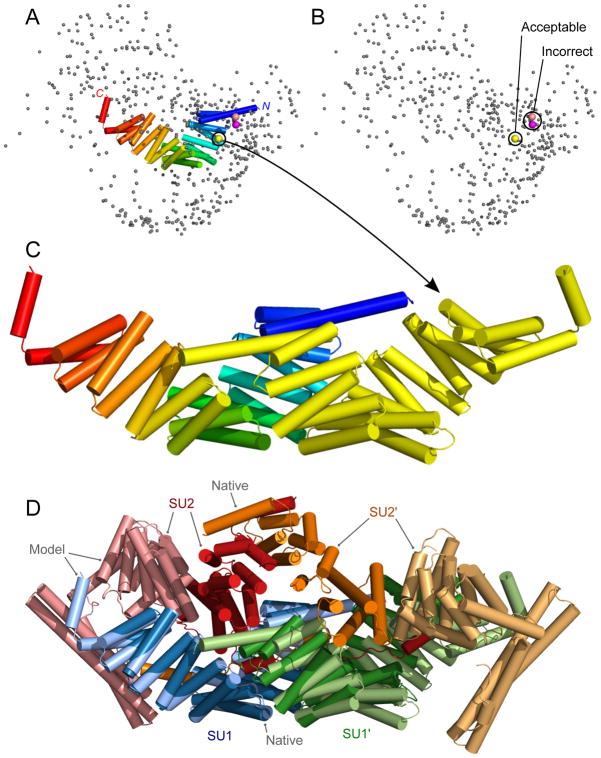
Figure 4.
Selection of dimer models for T117A/T0903 (SU1). A: Each sphere shows the center of mass for the N-terminus (residues 15-111) of the ligand (the docked chain). The yellow, salmon, and pink spheres were selected due to proximity to the concave face of the N-terminus (blue) of the receptor structure (shown as cartoon). B: As in panel A with the receptor structure removed for clarity. The yellow sphere represents an acceptable model while the salmon and pink spheres were incorrect. C: Cartoon representation of the model represented by the yellow sphere (acceptable: fnat 0.3, I-RMSD 2.2 Å, L-RMSD 11.1 Å). The receptor structure is colored from blue to red for N- to C-terminus while the ligand is shown in yellow. D: The tetramer model overlaid on the native structure (PDB ID: 5a7d). Dark colors: native; light colors: model. Blue and green: SU1; red and orange: SU2. The SU1 dimer model is the same as panel C. SU2 (light red and light orange) was incorrectly docked to the sides of the SU1 dimer.
Modeling of T117
T117 is a heterotetramer of inscuteable and partner of inscuteable (Pins) from D. melanogaster, which are involved in asymmetric cell division24. Pins is a component of a force generating complex, which connects to microtubules and generates pulling forces, and inscuteable is an adaptor protein that recruits the force generating complex containing Pins to sites of polarization24. The target consists of two copies of the tetratricopeptide-repeat (TPR) domain of Pins (referred to as SU1; 382 residues) and two copies of the asymmetric domain of inscuteable (referred to as SU2; 341 residues). In the crystal structure (PDB ID: 5a7d), SU1 forms a homodimer, and SU2 binds to SU1 at the outside of the complex (Figure 3). Predicting the structure of this complex was difficult because the two domains are large and they have an intertwined assembly: the SU1 homodimer forms a tunnel structure and the N-terminus of SU2 starts at one end of the tunnel, passes through the tunnel in an extended conformation (Figure 3B), and finally forms a folded domain on the outside of the other end of the tunnel. Moreover, there was no appropriate template that covered the whole complex. For the human prediction, we have successfully modeled a subcomplex using extensive biological knowledge.
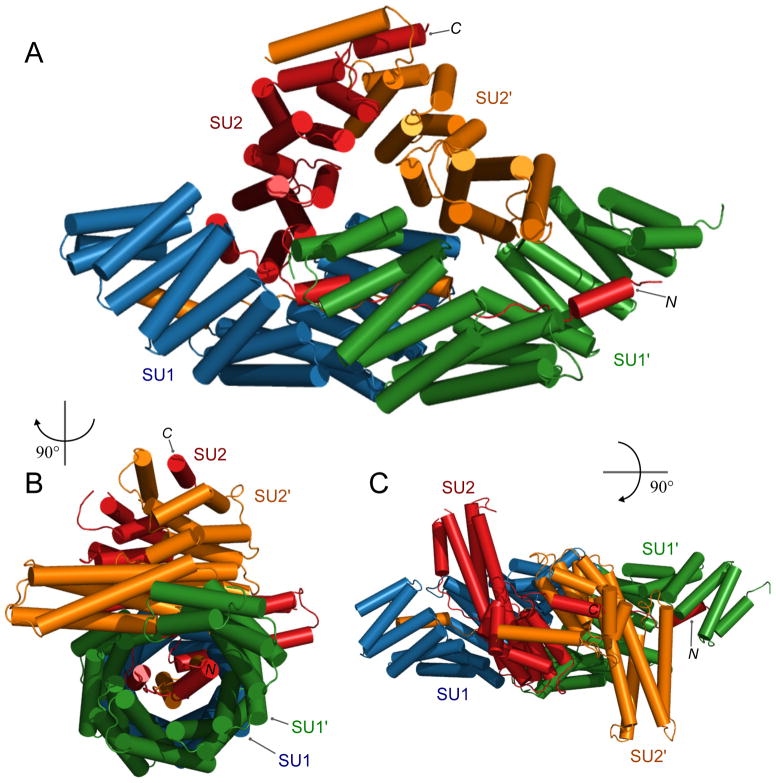
Figure 3.
Tetrameric structure of inscuteable and partner of inscuteable (Pins). A: SU1 (blue) and SU1′ (green) are Pins; SU2 (red) and SU2′ (orange) are inscuteable. The N- and C-termini of SU2 are labeled. B: as in panel A; rotated 90° left about the y-axis. C: as in panel A; rotated 90° forward about the x-axis. PDB ID: 5a7d.
For the server prediction, no heterotetrameric template was detected using HHPred; thus, a default topology of AABB (dimer of homodimers) was used. AABB was used as a default topology because the CASP instructions were to submit two copies of SU1 in one file and two copies of SU2 in another file such that the concatenation would form a tetramer. For SU1, the top two templates detected by HHPred were 4wndA and 4a1sA. 4wndA is the N-terminal tetratricopeptide-repeat (TPR) domain of G-protein-signaling modulator 2 from H. sapiens (67% identical). 4a1sA is the TPR domain of Pins from D. melanogaster (99% identical). Under 4wndA, the HHPred results also listed 3sf4, which contains three copies of the TPR domain of G-protein-signaling modulator 2 that form two distinct interfaces. Therefore, two different SU1 homodimers were modeled by superimposing two copies of 4a1sA onto each of the two interfaces in 3sf4 (first: chains A and B; second: chains A and C). While CASP server models were not generally available in time for the CAPRI server prediction deadline, they were available for this complex. For SU2, we chose the CASP server model Baker-RosettaServer-TS5 because it had rank 1 by GOAP and rank 4 by ITScorePro. Next, we docked SU2 homodimers using M-ZDOCK25 with C2 symmetry. The top SU2 homodimer from M-ZDOCK (ranked by ITScorePro18) was docked using LZerD against the two SU1 homodimers. For each SU1 homodimer, the top five heterotetramers by ITScorePro were selected. None of the models submitted by the server group achieved acceptable quality.
For the human prediction, we used the information that SU1 is a tetratricopeptide-repeat (TPR) protein24. A literature search for TPR proteins revealed a TPR dimer structure with an interlocking N-terminus (PDB ID: 2xpi26). Although this TPR dimer structure was not similar enough to use as a multi-chain template, it provided two critical structural insights: that the topology would be BAAB (symmetrical dimer of heterodimers) rather than AABB (dimer of homodimers) and that the TPR protein (SU1) would dimerize using its N-terminus. M-ZDOCK25 was used for symmetrical docking of SU1. Rather than selecting decoys using a scoring function, homodimers were manually selected using visual inspection. Three homodimer models were chosen that were consistent with an N-terminal interaction (residues 15-111; blue region of the cartoon) on the concave face (Figure 4A–B). Separately, SU2 was docked to SU1 using LZerD and ranked with ITScorePro. The top 10 SU1/SU2 heterodimers were superimposed onto the three selected SU1 homodimers to create 30 candidate tetramers. Finally, these 30 tetramers were re-ranked with ITScorePro and the top 10 were chosen for submission. One of the manually selected SU1 dimers achieved acceptable quality (Figure 4C), but the location of SU2 was not correct (Figure 4D).
Comparison of human and server predictor performance
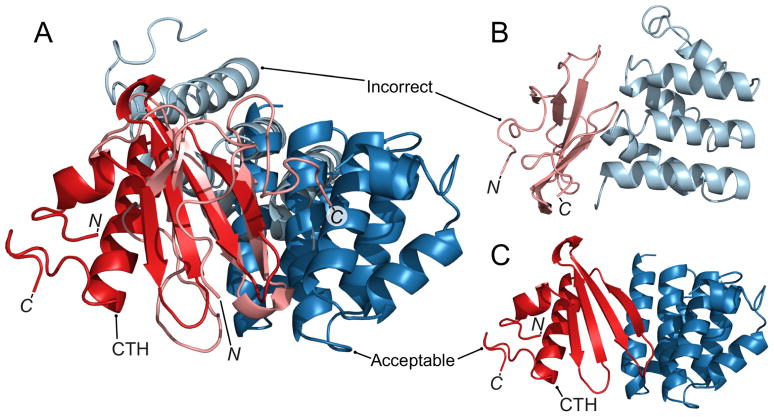
The difference in ranking between the LZerD server predictor group and the Kihara human predictor group is due to two targets (T113 and T120); specifically, improvement from no hits to 1 acceptable hit for T113 and from 10 acceptable hits to 1 medium hit for T120. For T113, the single-chain model of chain A (CASP ID T0884) used for the server submission has a relatively high proportion of coil (Figure 5B, light red) and RMSD of 11.4 Å to the native structure, likely due to a low (11%) sequence identity template. The CASP server model of chain A used for the human submission has a higher proportion of secondary structure (Figure 5C, dark red) and has a better RMSD of 7.9 Å to the native structure. Both models have a similar global fold, sharing a 4-strand antiparallel β sheet, but the sheet is less well-defined in the server model (Figure 5B, light red). In addition, the C-terminus of the server model appears to partially obstruct the binding site (Figure 5A). Thus, errors in the single-chain model of T113A used for the server submission prevented successful docking.
Figure 5.
Human and server models for T113. Light colors: Server submission model 1. Dark colors: Human submission model 3. A: Server submission model 1 and human submission model 3 aligned using the PyMOL command “super” on chain A (dark red and light red). For the server submission, chain A was created using the template 2b5uA (11% sequence identity, model has RMSD 11.4 Å to native). For the human submission, the CASP server model Zhang-Server TS1 was used (template information not provided, model has RMSD 7.9 Å to native). The chain A models share a similar 4-strand antiparallel β sheet but in the human model, the C-terminus forms a helix (labeled CTH) behind the β sheet while in the server model, the C-terminus is in front of the β sheet. B: The server submission model 1 (incorrect: fnat 0.02, I-RMSD 12 Å, L-RMSD 31 Å). C: The human submission model 3 (acceptable: fnat 0.1, I-RMSD 3.7 Å, L-RMSD 7.3 Å). CTH: the C-terminal helix of chain A.
For T120, our server submissions were just under the threshold for medium (fnat 0.3, I-RMSD 3.4 Å, and L-RMSD 5.9 Å); using CASP server models to create the human submissions yielded one model with L-RMSD of 4.9 Å, despite increasing the I-RMSD slightly. Therefore, the difference in ranking between the LZerD server group and the Kihara human group was primarily due to errors in single-chain models.
Scorer results
Table 2 shows a summary of the results from the scorer phase of Round 37. In terms of the scorer results, the performance of our groups was also better than in previous rounds. Our human scorer group had hits for three targets, including one high quality hit and one medium quality hit. The LZerD scorer group had hits for six targets, with one high quality and three medium quality. Remarkably, the selection performance of the LZerD scorer group was the best among all the participated groups. According to the assessor’s presentation at the CASP12 meeting, the LZerD scorer group was first while our human scorer group (Kihara) tied for tenth among participated scorer groups.
Table 2.
Per-target scorer performance summary for CAPRI Round 37
| Target | Oligo | Int. | S.LZerD hits | S.Kihara hits | S.Groups with hits |
|---|---|---|---|---|---|
|
|
|
|
|
||
| T110 | 3 | 1 | 10** | 10 | 16/14** |
| T111 | 3 | 1 | 10/7***/3** | 10/7***/3** | 16/15***/1** |
| T112 | 3 | 1 | 2 | 0 | 14 |
| T113 | 1+1 | 1 | 0 | 0 | 2 |
| T114 | 2 | 1 | 0 | 0 | 0 |
| T115 | 1 | - | Canceled | Canceled | Canceled |
| T116 | 2 | 1 | 0 | 0 | 0 |
| T117 | 2x(1+1) | 1 | 0 | 0 | 2 |
| 2 | 0 | 0 | 0 | ||
| 3 | 0 | 0 | 0 | ||
| 4 | 0 | 0 | 0 | ||
| T118 | 8 | 1 | 10** | 1** | 16/2***/14** |
| 2 | 10** | 6/5** | 16/11***/5** | ||
| 3 | 10/6***/4** | 2/1** | 16/13***/3** | ||
| T119 | 2 | 1 | 7 | 0 | 14 |
| T120 | 1+1 | 1 | 4/2** | 0 | 5/2** |
“Oligo”: oligomeric state. A single number indicates a homomeric complex and a plus sign (+) indicates a heteromeric complex, e.g. 1+1 is a heterodimer and 2x(1+1) is a heterotetramer. T115 was canceled because it was found to be a monomer. “Int.”: unique interface in a multimeric complex.
indicates high quality models and
indicates medium quality models; e.g., 6/1***/4** means six acceptable or better models, of which one was high and four were medium quality.
CAPRI model qualities defined previously30. Models with clashes or low sequence identity are excluded. “S.LZerD hits” and “S.Kihara hits”: number of models of each quality in the 10 submitted scorer models. “S.Groups with hits”: number of groups with at least one scorer model of each quality.
To understand the effectiveness of the rank sum of the three scoring functions used for the LZerD scorer group, in Table 3 we compared the performance of the rank sum and individual scores. The rank sum clearly performed better than using each score individually. Out of the six targets that had any hits and a native structure available at the time of writing (T110, T111, T112, T113, T119, and T120), the rank sum performed better than any individual score for all cases except T113, where GOAP found one acceptable model that was missed by the rank sum.
Table 3.
Performance of single and combined scoring functions
| Target | S.LZerD hits | DFIRE | GOAP | ITScorePro |
|---|---|---|---|---|
|
|
|
|
|
|
| T110 | 10** | 2/1** | 1/1** | 2/1** |
| T111 | 10/7***/3** | 1*** | 0 | 1*** |
| T112 | 2 | 0 | 0 | 0 |
| T113 | 0 | 0 | 1 | 0 |
| T114 | 0 | ? | ? | ? |
| T115 | Canceled | Canceled | Canceled | Canceled |
| T116 | 0 | 0 | 0 | 0 |
| T117 | 0 | 0 | 0 | 0 |
| T118 | 10** | ? | ? | ? |
| T119 | 7 | 4 | 2 | 1 |
| T120 | 4/2** | 2/1** | 4/1** | 0 |
S.LZerD hits were chosen using the sum of the ranks of DFIRE, GOAP, and ITScorePro. “?” indicates that the performance is unknown at the time of writing because the correct structure was not available. S.LZerD results for these targets were provided from the assessors.
Comparison of human and server scorer performance
The LZerD scorer group had better performance than the human scorer group. When reviewing the selections to determine the reason for the difference in performance, we discovered that for targets T112-T120, the human scorer selections were inadvertently selected from the smallest clusters rather than the largest clusters. Therefore, we looked at what the results would have been using the largest clusters and found that in some cases, we would have had better results. For T113, the largest cluster includes the acceptable model submitted by our human predictor group, but the fnat is very close to the cutoff of 0.1, so it is unclear whether it would have been assessed as acceptable. For T118, the largest clusters include 5 medium hits, somewhat improved over just 1 medium hit. For T112 and T119, the largest clusters for each target include 3 acceptable hits. For T120, none of the largest clusters contained hits. Overall, if we had applied the intended selection criteria of largest clusters for the human scorer group, we expect that we would have achieved five total hits with two medium-quality and one high-quality. Nevertheless, this is still less successful than the server scorer group (six total hits, four medium-quality, and one high-quality).
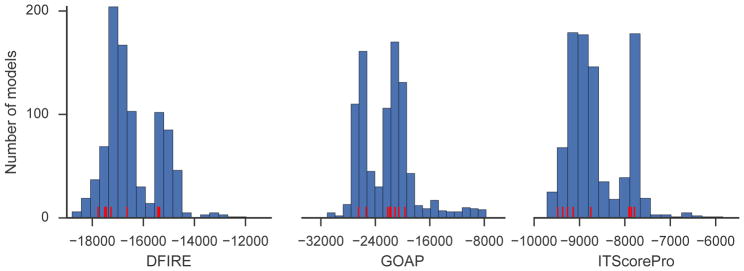
To further explore why the cluster size-based selection was less successful than using scoring functions, we examined the score distributions of the CAPRI scorer models and found that they are different from those for docking decoys generated by a single method such as LZerD. The score distributions for LZerD decoys have a single peak (i.e. are unimodal; see Figures 2 and 3 in our previous work6). In contrast, since the CAPRI scorer pools consist of models produced by different groups, the score distributions may have multiple peaks. For example, Figure 6 shows the distributions of three scores for the scorer decoys from target T120. The score distributions for T120 are bimodal (Figure 6) and several of the largest clusters fall in the peak with higher (i.e. worse) scores. This indicates that cluster size can be artificially inflated if there are many similar incorrect models; thus, the cluster size is not an ideal metric for the CAPRI scorer round.
Figure 6.
Distributions of scoring functions for the scorer models of T120 (contributed by multiple predictor groups). Red: the scores of the cluster centers of the 10 largest clusters.
Comparison of predictor and scorer performance
We also compared our performance for the prediction and the scoring categories. Most of the targets where our human predictor group was successful were also successfully selected by the LZ-erD scorer group. However, for T113, while the human predictor group submitted one acceptable model, the LZerD scorer group did not have any hits. Surprisingly, we found that the acceptable model from the human predictor group was in fact selected by the LZerD scorer group, but it did not meet the acceptable criteria due to having fnat 0.08, slightly lower than the cutoff for an acceptable model, 0.1. The differences between the models were introduced due to stochastic components of the side-chain rebuilding and relaxation procedure. Evidently, these differences led to a decrease in fnat for the model submitted by the LZerD scorer group.
Conclusion
For the recent Round 37 of CAPRI, we modified our prediction pipeline based on our analysis of Round 306. The most substantial change was the use of template-based complex modeling. However, for cases without a clear multi-chain template27, docking is still required. The improvement in our human predictor group rank to tied for 3rd in CAPRI Round 37 from 18th across CAPRI Rounds 28–35 was due to a combination of using template-based modeling when a multi-chain template was available and using docking when it was not. In future rounds, we may be able to further improve our performance by using additional template search methods, both for multi-chain and single-chain templates. Possible directions for improving template search include the use of an alternative sensitive search method28 or a structure-based search against local docking interface templates29, which were shown to perform well in this CAPRI/CASP round. Another lesson we learned was the inappropriateness of using clustering for scoring decoy sets, since decoys are collected from different research groups and thus the distribution is biased. To further improve decoy selection, we could develop a more sophisticated method that combines multiple features including statistical scoring functions as we used this time as well as other structure and energetic features of decoys.
Overall, computational protein complex modeling has made extraordinary progress across many rounds of CAPRI. Our lab’s protocol combines the strengths of TBM when complex template information is available and protein-protein docking when it is not. Clearly, the remaining challenges are complexes with neither multi-chain nor single-chain template structures and complexes with substantial conformational change upon binding. Complexes without clear single-chain templates may be modeled with continued improvements in template detection and ab initio folding while large conformational change can be addressed by using some combination of flexible docking and flexible refinement. We look forward to participating in future rounds of CAPRI to see how the community responds to these challenges.
Supplementary Material
Table S1: Multi-chain and single-chain templates used for CAPRI Round 37
Acknowledgments
We thank Charles Christoffer and Genki Terashi for helpful discussions. This work was supported by the National Institutes of Health (R01GM123055, R01GM097528) and the National Science Foundation (IIS1319551, DBI1262189, IOS1127027, DMS1614777).
References
- 1.Kihara D, Skolnick J. Microbial genomes have over 72% structure assignment by the threading algorithm PROSPECTOR Q. Proteins: Struct, Funct, Bioinf. 2004;55(2):464–473. doi: 10.1002/prot.20044. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Kihara D. Effect of using suboptimal alignments in template-based protein structure prediction. Proteins: Struct, Funct, Bioinf. 2011;79(1):315–34. doi: 10.1002/prot.22885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, Shen MY, Kelly L, Melo F, Sali A. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R, Grudinin S, Popov P, Neveu E, Lee H, Baek M, Park S, Heo L, Rie Lee G, Seok C, Qin S, Zhou HX, Ritchie DW, Maigret B, Devignes MD, Ghoorah A, Torchala M, Chaleil RA, Bates PA, Ben-Zeev E, Eisenstein M, Negi SS, Weng Z, Vreven T, Pierce BG, Borrman TM, Yu J, Ochsenbein F, Guerois R, Vangone A, Rodrigues JP, van Zundert G, Nellen M, Xue L, Karaca E, Melquiond AS, Visscher K, Kastritis PL, Bonvin AMJJ, Xu X, Qiu L, Yan C, Li J, Ma Z, Cheng J, Zou X, Shen Y, Peterson LX, Kim HR, Roy A, Han X, Esquivel-Rodríguez J, Kihara D, Yu X, Bruce NJ, Fuller JC, Wade RC, Anishchenko I, Kundrotas PJ, Vakser IA, Imai K, Yamada K, Oda T, Nakamura T, Tomii K, Pallara C, Romero-Durana M, Jiménez-García B, Moal IH, Férnandez-Recio J, Joung JY, Kim JY, Joo K, Lee J, Kozakov D, Vajda S, Mottarella S, Hall DR, Beglov D, Mamonov A, Xia B, Bohnuud T, Del Carpio CA, Ichiishi E, Marze N, Kuroda D, Roy Burman SS, Gray JJ, Chermak E, Cavallo L, Oliva R, Tovchi-grechko A, Wodak SJ. Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: a CASP-CAPRI experiment. Proteins: Struct, Funct, Bioinf. 2016;84(S1):323–348. doi: 10.1002/prot.25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janin J. Welcome to CAPRI: A Critical Assessment of PRedicted Interactions. Proteins Struct, Funct, Genet. 2002;47(3):257–257. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 6.Peterson LX, Kim H, Esquivel-Rodríguez J, Roy A, Han X, Shin WH, Zhang J, Terashi G, Lee M, Kihara D. Human and server docking prediction for CAPRI round 30–35 using LZerD with combined scoring functions. Proteins: Struct, Funct, Bioinf. 2017;85(3):513–527. doi: 10.1002/prot.25165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkatraman V, Yang YD, Sael L, Kihara D. Protein-protein docking using region-based 3D Zernike descriptors. BMC Bioinformatics. 2009;10:407. doi: 10.1186/1471-2105-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kihara D, Sael L, Chikhi R, Esquivel-Rodríguez J. Molecular surface representation using 3D Zernike descriptors for protein shape comparison and docking. Curr Protein Pept Sci. 2011;12(6):520–30. doi: 10.2174/138920311796957612. [DOI] [PubMed] [Google Scholar]
- 9.Li B, Kihara D. Protein docking prediction using predicted protein-protein interface. BMC Bioinformatics. 2012;13(1):7. doi: 10.1186/1471-2105-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquivel-Rodríguez J, Yang YD, Kihara D. Multi-LZerD: Multiple protein docking for asymmetric complexes. Proteins: Struct, Funct, Bioinf. 2012;80(7):1818–1833. doi: 10.1002/prot.24079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esquivel-Rodríguez J, Filos-Gonzalez V, Li B, Kihara D. Pairwise and Multimeric Protein-Protein Docking Using the LZerD Program Suite. In: Kihara D, editor. Protein Struct Predict, volume 1137 of Methods in Molecular Biology. Springer; New York, New York, NY: 2014. pp. 209–234. [DOI] [PubMed] [Google Scholar]
- 12.Canterakis N. 3D Zernike moments and Zernike affine invariants for 3D image analysis and recognition. In: Ersbøll BK, Johansen P, editors. Proc 11th Scand Conf Image Anal. 1999. pp. 85–93. Dansk Selskab for Automatisk Genkendelse af Mønstre. [Google Scholar]
- 13.Sael L, La D, Li B, Rustamov R, Kihara D. Rapid comparison of properties on protein surface. Proteins: Struct, Funct, Bioinf. 2008;73(1):1–10. doi: 10.1002/prot.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 16.Zhou HY, Zhou YQ. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11(11):2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Skolnick J. GOAP: A generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys J. 2011;101(8):2043–2052. doi: 10.1016/j.bpj.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang SY, Zou X. Statistical mechanics-based method to extract atomic distance-dependent potentials from protein structures. Proteins: Struct, Funct, Bioinf. 2011;79(9):2648–2661. doi: 10.1002/prot.23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 20.Haberthür U, Caflisch A. FACTS: Fast analytical continuum treatment of solvation. J Comput Chem. 2008;29(5):701–715. doi: 10.1002/jcc.20832. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, Kihara D. Detecting local residue environment similarity for recognizing near-native structure models. Proteins: Struct, Funct, Bioinf. 2014;82(12):3255–72. doi: 10.1002/prot.24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H, Kihara D. Protein structure prediction using residue- and fragment-environment potentials in CASP11. Proteins: Struct, Funct, Bioinf. 2016;84:105–117. doi: 10.1002/prot.24920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culurgioni S, Alfieri A, Pendolino V, Laddomada F, Mapelli M. Inscuteable and NuMA proteins bind competitively to Leu-Gly-Asn repeat-enriched protein (LGN) during asymmetric cell divisions. Proc Natl Acad Sci. 2011;108(52):20998–21003. doi: 10.1073/pnas.1113077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierce B, Tong W, Weng Z. M-ZDOCK: a grid-based approach for Cn symmetric multimer docking. Bioinformatics. 2005;21(8):1472. doi: 10.1093/bioinformatics/bti229. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010;29(21):3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aloy P, Ceulemans H, Stark A, Russell RB. The Relationship Between Sequence and Interaction Divergence in Proteins. J Mol Biol. 2003;332(5):989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Oda T, Lim K, Tomii K. Simple adjustment of the sequence weight algorithm remarkably enhances PSI-BLAST performance. BMC Bioinformatics. 2017;18(1):288. doi: 10.1186/s12859-017-1686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anishchenko I, Kundrotas PJ, Tuzikov AV, Vakser Structural templates for comparative protein docking. Proteins: Struct, Funct, Bioinf. 2015;83(9):1563–1570. doi: 10.1002/prot.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Méndez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: Current status of docking methods. Proteins: Struct, Funct, Bioinf. 2003;52(1):51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Multi-chain and single-chain templates used for CAPRI Round 37