Abstract
Escherichia albertii is an emerging member of the Enterobacteriaceae causing human and animal enteric infections. Antimicrobial resistance among enteropathogens has been reported to be increasing in the past years. The purpose of this study was to investigate antibiotic resistance and resistance genes in E. albertii isolated from Zigong city, Sichuan province, China. The susceptibility to 21 antimicrobial agents was determined by Kirby–Bauer disk diffusion method. The highest prevalence was tetracycline resistance with a rate of 62.7%, followed by resistance to nalidixic acid and streptomycin with a rate of 56.9 and 51.0%, respectively. All isolates were sensitive or intermediate susceptible to imipenem, meropenem, amoxicillin–clavulanic acid, and levofloxacin. Among 51 E. albertii isolates, 15 were extended-spectrum β-lactamase-producing as confirmed by the double disk test. The main β-lactamase gene groups, i.e., blaTEM, blaSHV, and blaCTX-M, were detected in17, 20, and 22 isolates, respectively. Furthermore, four colistin-resistant isolates with minimum inhibitory concentrations of 8 mg/L were identified. The colistin-resistant isolates all harbored mcr-1 and blaCTX-M-55. Genome sequencing showed that E. albertii strain SP140150 carried mcr-1 and blaCTX-M-55 in two different plasmids. This study provided significant information regarding antibiotic resistance profiles and identified the co-occurrence of β-lactamase and MCR-1 encoding genes in E. albertii isolates.
Keywords: Escherichia albertii, β-lactam, β-lactamases, ESBL, mcr-1
Introduction
Escherichia albertii is a gram-negative facultative rod bacterium belonging to a member of the Enterobacteriaceae. It was previously recovered from stool specimens of sick Bangladeshi children and was preliminarily identified as atypical eae-positive Hafnia alvei (Albert et al., 1991, 1992). In 2003, it was proposed as a new species, named E. albertii based on further genotypic and biochemical studies (Huys et al., 2003). E. albertii was reported to be the probable cause of death for redpoll finches (Carduelis flammea) in Alaska in 2004 (Oaks et al., 2010). In recent years, E. albertii was reported to be an emerging human enteropathogen associated with many sporadic infections and outbreaks in humans (Konno et al., 2012; Ooka et al., 2012, 2013; Asoshima et al., 2014; Murakami et al., 2014; Brandal et al., 2015; Inglis et al., 2015). Besides, it has been detected in water and raw meats of animal origin (Felfoldi et al., 2010; Maheux et al., 2014; Asoshima et al., 2015; Lindsey et al., 2015; Maeda et al., 2015; Wang et al., 2016), thus posing a high risk to public health.
Antimicrobials are one of the most successful forms of chemotherapy used in the treatment of infectious diseases by killing or inhabiting the growth of microorganisms. The most important and widely used antimicrobials are β-lactam drugs. In recent years, antimicrobial resistance among Gram-negative bacteria has been reported to be increasing (MacVane, 2017). In Enterobacteriaceae, resistance to the β-lactam is mediated by production of β-lactamase enzymes which inactivate the drugs by hydrolyzing the β-lactam ring (Bush and Bradford, 2016). Some clinically most important enzymes are as follows: (1) extended-spectrum β-lactamases (ESBLs), including SHV, TEM and CTX-M types; (2) carbapenemases, including class A (KPC types), class B metallo-β-lactamases (MBLs), and class D oxacillinases; and (3) the AmpC cephalosporinases (Bonomo, 2017).
Colistin is an antibiotic of last resort for the treatment of extensively drug-resistant Gram-negative bacteria (Kaye et al., 2016). In a recent study, the emergence of plasmid-mediated colistin resistance has been reported in E. coli, raising a great concern around the world (Liu et al., 2016). Thereafter, plasmid-mediated colistin resistance gene (mcr-1) has also been identified in other members of the Enterobacteriaceae from South America, Asia, Europe and Africa, suggesting that mcr-1 might be widespread (Olaitan et al., 2016; Al-Tawfiq et al., 2017).
To date, little is known about the antibiotic resistance of E. albertii. This study investigated the antibiotic resistance, identified the ESBL-producing and colistin-resistant isolates, and determined the distribution of β-lactamase genes and mcr-1 gene in E. albertii isolated from Zigong city, Sichuan province, China.
Materials and Methods
Bacterial Isolates
A total of 51 isolates of E. albertii were recovered from various samples collected in Zigong city, Sichuan province between 2013 and 2015, including diarrheal patient feces (3), healthy carrier feces (3), duck intestine (19), chicken intestine (18), chicken meat (3), duck meat (2), raw mutton (1), raw pork (1), and egret excrement (1). All isolates were confirmed to be E. albertii based on combination of diagnostic multiplex PCR, 16S rDNA sequencing, and multi-locus sequence typing (MLST) analysis as described previously (Wang et al., 2016) and stored at -80°C in Luria-Bertani (LB) medium (Oxoid, United Kingdom) with 30% (vol/vol) glycerol.
Antibiotic Susceptibility Testing
Susceptibility to antimicrobials was determined by Kirby–Bauer disk diffusion method on Mueller Hinton agar (MHA). The antibiotics used in the study included imipenem (IMP), meropenem (MEM), piperacillin (PRL), ampicillin/sulbactam (SAM), amoxicillin/clavulanic acid (AMC), cefepime (FEP), cefuroxime (CXM), cephalothin (KF), ceftriaxone (CRO), aztreonam (ATM), kanamycin (K), streptomycin (S), gentamicin (CN), nalidixic acid (NA), levofloxacin (LEV), norfloxacin (NOR), ciprofloxacin (CIP), trimethoprim/sulfamethoxazole (SXT), tetracycline (TE), furadantin (F), and chloramphenicol (C) (Oxoid, United Kingdom). The inoculated plates were incubated for 24 h aerobically at 37°C. The diameters of the zones of inhibition was interpreted according to the Clinical Laboratory Standards Institute (CLSI) guidelines (CLSI, 2016).
The minimum inhibitory concentration (MIC) of colistin was determined by broth microdilution method recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group1.
Identification of the ESBL-Producing Isolates
The double disk test was performed to confirm the ESBL phenotype. It was carried out on MHA with two pairs of disks (ceftazidime + ceftazidime/clavulanic acid and cefotaxime + cefotaxime/clavulanic acid) (BD Diagnostics, United States). The results were interpreted as recommended by the CLSI (CLSI, 2016). E. coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as the quality control strains.
Detection of the β-Lactamase Gene Groups
Escherichia albertii isolates were inoculated on LB agar and incubated overnight at 37°C. A colony was suspended in 50 μl of sterilized distilled water and boiled for 10 min. The cell suspension was centrifuged at 10,000 × g for 5 min, and the supernatant was used as template DNA. PCR was performed to screen the main β-lactamase gene groups, i.e., blaTEM, blaSHV, blaCTX-M, blaKPC, and blaNDM genes. All primers and PCR conditions used in this study were presented in Supplementary Table S1. Each reaction tube contained 10 μl of master Mix (Qiagen, Germany), 0.5 μM of forward and reverse primers, and 1 μl of template DNA, and was made up to a total volume of 20 μl with sterile distilled water.
Detection and Sequencing of the mcr-1 Gene
All 51 isolates were subjected to PCR for the presence of mcr-1 gene using primers described previously (Supplementary Table S1). The expected PCR products were sequenced using the ABI 3730 Automated DNA Analyzer (Applied Biosystems, United States).
The mcr-1 sequences obtained in this study have been deposited in GenBank under accession numbers: KX765477–KX765480.
Plasmid Profiling
Plasmid DNA profiles of all 51 E. albertii isolates were analyzed using the S1-nuclease pulsed-field gel electrophoresis (PFGE) method. Briefly, the bacterial cells embedded in agarose were lysed by SDS/proteinase K, and then were digested with 8 U S1-nuclease at 37°C for 30 min. Finally, each sample was resolved by PFGE in a Chef-Mapper (Bio-Rad, United States) at 14°C, with a switch time 2.16 to 54.17 s at 6 V/cm for 18 h. Each DNA band visualized was considered as a unit length of linear plasmid. The approximate size of each plasmid was determined by comparing profiles with XbaI-digested DNA from Salmonella serotype Braenderup strain H9812 (Bai et al., 2017).
Mating Experiments
Four of the mcr-1-positive isolates were selected for conjugation experiments. Filter conjugation was carried out using E. coli J53 (sodium azide-resistant) as the recipient. The donor and recipient were grown on LB medium to an optical density at 600 nm of 0.5, mixed equally, and then incubated on sterilized filter paper for 4 h. The filter was then resuspended in LB medium, and dilutions were plated on M-H agar containing sodium azide (150 μg/mL and colistin (4 μg/mL) to select for transconjugants. Mobilization efficiency was calculated as the number of transconjugant colonies divided by the number of donor colonies (Wang et al., 2011).
Whole-Genome Sequencing
Genomic DNA was isolated from an overnight culture using the Wizard Genomic DNA purification kit (Promega, United States) according to the manufacturer’s instructions. Total DNA obtained was subjected to quality control by agarose gel electrophoresis and quantified by Qubit (Life Technologies, United States). The complete genome was sequenced by single molecule real-time (SMRT) technology using the Pacific Biosciences (PacBio) sequencing platform performed at the Beijing Novogene Bioinformatics Technology, Co., Ltd. (McCarthy, 2010). The filtered reads were assembled to generate one contig without gaps using SMRT Analysis 2.3.0 (Berlin et al., 2015). The protein-coding sequences (CDSs) were predicted using GeneMarkS (Besemer et al., 2001). ARDB (Antibiotic Resistance Genes Database)2 was used to search for antimicrobial resistance genes (Liu and Pop, 2009).
The complete genome sequences of SP140150 isolate are available at GenBank under the accession numbers: CP025676–CP025679.
Ethics Statement
Samples were collected and detected as part of the infectious disease surveillance program led by National Institute for Communicable Disease Control and Prevention, China CDC and implemented by Zigong Center for Disease Control and Prevention. The study was approved by the ethics committee of National Institute for Communicable Disease Control and Prevention, China CDC, according to the medical research regulations of National Health and Family Planning Commission of the People’s Republic of China.
Results
Antibiotic Resistance of E. albertii Isolates
Antimicrobial resistance in E. albertii isolates was determined against 21 antibiotics. The highest prevalence was tetracycline resistance with a rate of 62.7% (32/51), followed by resistance to nalidixic acid and streptomycin with a rate of 56.9% (29/51) and 51.0% (26/51), respectively. Resistance rate to cefuroxime, piperacillin, and chloramphenicol was 45.1, 43.1, and 41.2%, respectively. Lower resistance was observed for ampicillin/sulbactam, cefepime, cephalothin, ceftriaxone, aztreonam, kanamycin, gentamicin, norfloxacin, ciprofloxacin, trimethoprim/sulfamethoxazole, and furadantin with a rate ranging from 17.6 to 39.2%. All isolates were sensitive or intermediate susceptible to imipenem, meropenem, amoxicillin–clavulanic acid, and levofloxacin (Figure 1).
FIGURE 1.
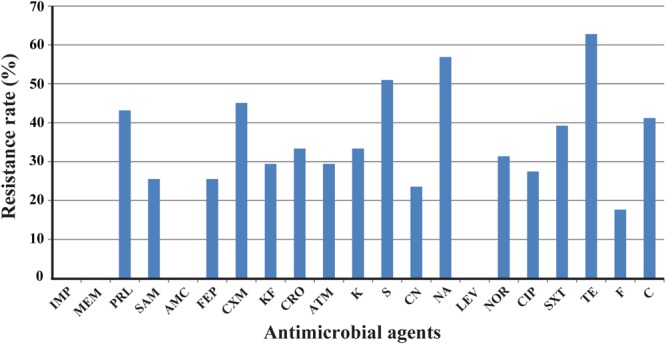
Frequency of antimicrobial resistance of 51 Escherichia albertii isolates. IMP, imipenem; MEM, meropenem; PRL, piperacillin; SAM, ampicillin/sulbactam; AMC, amoxicillin/clavulanic acid; FEP, cefepime; CXM, cefuroxime; KF, cephalothin; CRO, ceftriaxone; ATM, aztreonam; K, kanamycin; S, streptomycin; CN, gentamicin; NA, nalidixic acid; LEV, levofloxacin; NOR, norfloxacin; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; TE, tetracycline; F, furadantin; C, chloramphenicol.
Ten isolates (19.6%) were resistant to one antimicrobial agent, while the majority exhibited resistance to two or more antimicrobials tested. All isolates from diarrheal patients and healthy carriers were susceptible to imipenem, meropenem, piperacillin, cefepime, aztreonam, and levofloxacin (Supplementary Table S2).
ESBL-Producing E. albertii Isolates
Among 51 E. albertii isolates, 15 isolates from four sources, i.e., six isolates from duck intestine, six from chicken intestine, two from chicken meat and one from raw mutton samples, were ESBL-producing as confirmed by the double disk test. All isolates recovered from diarrheal patients and healthy carriers were non-ESBL-producing (Supplementary Table S2).
All ESBL-producing E. albertii isolates were resistant to piperacillin, cefuroxime, cephalothin, ceftriaxone, aztreonam, tetracycline and chloramphenicol, whereas resistance to streptomycin and nalidixic acid were observed in 14 of the 15 ESBL-producing isolates. ESBL-producing E. albertii isolates have higher resistance rate than non-ESBL-producing isolates, as expected (Table 1).
Table 1.
Comparison of the resistance rates of ESBL-producing and of non-ESBL-producing E. albertii isolates.
| Antibiotic | Non-ESBL-producing (N = 36) Resistant [n (%)] | ESBL-producing (N = 15) Resistant [n (%)] |
|---|---|---|
| Imipenem | 0 (0) | 0 (0) |
| Meropenem | 0 (0) | 0 (0) |
| Piperacillin | 7 (19.4) | 15 (100) |
| Ampicillin/sulbactam | 6 (16.7) | 7 (46.7) |
| Amoxycillin/clavulanic acid | 0 (0) | 0 (0) |
| Cefepime | 0 (0) | 13 (86.7) |
| Cefuroxime | 8 (22.3) | 15 (100) |
| Cephalothin | 0 (0) | 15 (100) |
| Ceftriaxone | 2 (5.6) | 15 (100) |
| Aztreonam | 0 (0) | 15 (100) |
| Kanamycin | 6 (16.7) | 11 (73.3) |
| Streptomycin | 12 (33.3) | 14 (93.3) |
| Gentamicin | 7 (19.4) | 5 (33.3) |
| Nalidixic acid | 15 (41.7) | 14 (93.3) |
| Levofloxacin | 0 (0) | 0 (0) |
| Norfloxacin | 6 (16.7) | 10 (66.7) |
| Ciprofloxacin | 4 (11.1) | 11 (73.3) |
| Sulfamethoxazole | 12 (33.3) | 8 (53.3) |
| Tetracycline | 17 (47.2) | 15 (100) |
| Furadantin | 0 (0) | 9 (60.0) |
| Chloramphenicol | 6 (16.7) | 15 (100) |
Distribution of β-Lactamase Genes
The main β-lactamase gene groups (blaTEM, blaSHV, blaCTX-M, blaKPC, and blaNDM) were screened by PCR. Eight E. albertii isolates did not contained any β-lactamase from gene groups tested and none was positive for blaKPC and blaNDM. The blaTEM, blaSHV, and blaCTX-M were detected in 17 (33.3%), 20 (39.2%), and 22 (43.1%) isolates, respectively. All 15 ESBL-producing isolates contained 1–3 β-lactamase genes (Supplementary Table S2).
mcr-1-Positive and Colistin-Resistant E. albertii Isolates
Four out of 51 isolates (one from raw mutton, one from raw chicken meat, and two from chicken intestine) were positive for MCR-1 encoding gene by PCR. Sequencing analysis showed that the four nucleotide sequences are identical to the first reported mcr-1 sequence in plasmid pHNSHP45 (GenBank accession number KP347127).
The MICs of colistin of all 51 E. albertii isolates were determined by broth microdilution method. The four mcr-1-positive isolates were colistin-resistant with MIC of 8 mg/L. The MICs of colistin of all mcr-1-negative isolates were less than 4 mg/L.
The colistin resistance genes of all four mcr-1-positive isolates were successfully transferred by conjugation into sodium azide resistant E. coli J53. The transfer frequencies of isolates SP140128, SP140089, SP140149, and SP140150 were similar (4.2 × 10-3, 8.8 × 10-4, 3.8 × 10-4, and 1.3 × 10-3, respectively).
Co-occurrence of blaCTX-M and mcr-1 Genes in E. albertii Isolates
Except one isolate T150248 from healthy carrier, plasmids ranging in size from 36 to 283 kbp were identified by S1-nuclease-based PFGE in 50 E. albertii isolates. Among which, 23 isolates harbored one plasmid; 13 harbored two plasmids; 10 harbored three plasmids; three harbored four plasmids; and one isolate harbored five plasmids (Supplementary Table S2). Two different size plasmids (56 and 113 kbp) were identified in all four colistin-resistant isolates, and an additional 45 kbp plasmid was present in three out of four colistin-resistant isolates (Table 2).
Table 2.
Antimicrobial resistance profiles, β-lactamase genes, and plasmid content of colistin-resistant E. albertii isolates.
| Isolate | Origin | Antibiotic resistance profile | β-Lactamase gene∗∗ | Plasmids (kb)∗∗∗ |
|---|---|---|---|---|
| SP140128 | Mutton | PRL, SAM∗, FEP, CXM, KF, CRO, ATM, K, S, NA, NOR, CIP, SXT∗, TE, F∗, C | blaCTX-M-55 | 113, 56 |
| SP140089 | Chicken meat | PRL, SAM∗, FEP, CXM, KF, CRO, ATM, K, S, NA, NOR, CIP∗, SXT∗, TE, F, C | blaCTX-M-55 | 113, 56, 45 |
| SP140149 | Chicken intestine | PRL, SAM∗, FEP, CXM, KF, CRO, ATM, K, S, NA, LEV∗, NOR, CIP, SXT∗, TE, F∗, C | blaCTX-M-55 | 113, 56, 45 |
| SP140150 | Chicken intestine | PRL, SAM∗, FEP, CXM, KF, CRO, ATM, K, S, NA, NOR, CIP, SXT∗, TE, F, C | blaCTX-M-55 | 113, 56, 45 |
PRL, piperacillin; SAM, ampicillin/sulbactam; FEP, cefepime; CXM, cefuroxime; KF, cephalothin; CRO, ceftriaxone; ATM, aztreonam; K, kanamycin; S, streptomycin; NA, nalidixic acid; LEV, levofloxacin; NOR, norfloxacin; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; TE, tetracycline; F, furadantin; C, chloramphenicol. ∗Intermediate sensitivity to antibiotics. ∗∗blaCTX-M-55 type was determined by sequencing. ∗∗∗Plasmid sizes were estimated by S1-nuclease PFGE method.
All four isolates harboring mcr-1 were positive for blaCTX-M group gene (blaCTX-M-55 type) and were ESBL-producing. The four colistin-resistant isolates showed indistinguishable PFGE patterns and they were all typed as sequence type (ST) 4479 (Supplementary Figure S1). They exhibited multi-drug resistance. All four isolates were resistant to 12 antibiotics tested in this study, i.e., piperacillin, cefepime, cefuroxime, cephalothin, ceftriaxone, aztreonam, kanamycin, streptomycin, nalidixic acid, norfloxacin, tetracycline, and chloramphenicol (Table 2).
Genome Features of E. albertii Strain SP140150 Harboring mcr-1
The completed genome sequence of SP140150 consists of a circular chromosome of 4,881,553 bp with an average GC content of 49.8% and three circular plasmids. Antimicrobial resistance genes were searched against these three plasmids. blaCTX-M-55 was identified in pEA-1, a plasmid of 129,356 bp in size with an average GC content of 51.9%. In addition, pEA-1 also carried chloramphenicol resistance gene cml, tetracycline resistance determinant tetA, streptomycin resistance genes aph6id and aph33ib, sulfonamide-resistant dihydropteroate synthase gene sul2, and multidrug efflux RND transporter OqxA and OqxB. None of any antimicrobial resistance genes was identified in pEA-2, the second plasmid of 57,110 bp in size with an average GC content of 46.1%. A 1626 bp CDS encoding MCR-1 was found located downstream of an insertion sequence ISApl1 in the third plasmid pEA-3. pEA-3 is 68,747 bp in size with an average GC content of 42.5%. pEA-3 possesses an IncFII-type backbone and contains 90 predicted CDSs encoding plasmid replication, maintenance and stability functions, and a type IV protein secretion system (Figure 2).
FIGURE 2.
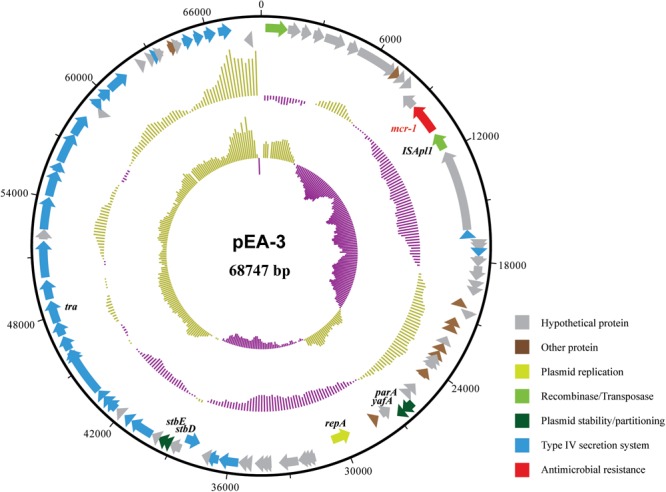
Structure of plasmid pEA-3 carrying mcr-1 from E. albertii strain SP140150. From outer circle to inner circle, each represents CDS, GC content and GC skew, respectively. The functions of corresponding CDSs are colored as indicated.
Discussion
Antimicrobial resistance is an increasingly encountered phenomenon among the Enterobacteriaceae and an alarming threat to global health (Laxminarayan et al., 2013). Resistance to β-lactams is primarily because of bacterially produced β-lactamases that are able to hydrolyze the β-lactam ring (Bush and Bradford, 2016). There are several reports of porin-mediated resistance in clinical isolates of enterobacteria, mainly affecting susceptibility to β-lactams (Martinez-Martinez et al., 1996; Poirel et al., 2004; Pavez et al., 2008; Perez et al., 2013).
Some E. albertii isolates in this study demonstrated resistance to four antimicrobials identified by the World Health Organization (WHO) as being of critical importance in the treatment of human infectious diseases, including piperacillin, ampicillin, cefotaxime, and cefepime (WHO, 2012). Furthermore, 29.4% of isolates demonstrated resistance to cephalothin that was identified by the WHO as highly important in human disease treatment (WHO, 2012).
Extended-spectrum β-lactamases are defined as enzymes produced by certain bacteria that are able to hydrolyze extended spectrum cephalosporin. TEM-group (exception of TEM-1 and TEM-2), SHV-group, and CTX-M-group β-lactamases are important types of ESBLs (Ghafourian et al., 2015). Among the three types, the prevalence of CTX-M is increasing in Enterobacteriaceae and predominates as a cause of extended spectrum cephalosporin resistance (Falagas and Karageorgopoulos, 2009). In the current study, the prevalence of blaCTX-M gene in E. albertii was in agreement with those reported in the other studies (Bora et al., 2014). Besides, blaCTX-M gene was mostly detected in isolates resistant to cefotaxime. Nineteen (37.3%) of E. albertii isolates carried two or three different β-lactamase gene groups, demonstrating the co-occurrence of these genes in various combinations (Shahid et al., 2011).
Carbapenem-resistant Enterobacteriaceae have been increasingly reported worldwide. The carbapenemases include NDMs, KPCs, OXA-48, and others (Tzouvelekis et al., 2012). KPCs are currently the most common cause of carbapenem resistance worldwide. The emergence of New Delhi metallo-β-lactamase (NDM-1) and its variants had raised a major public health concern. NDM-1 can hydrolyze a wide range of β-lactam antibiotics, including carbapenems (Khan et al., 2017). In China, Wang et al. (2015) has reported that Enterobacteriaceae remained susceptible to carbapenems. In the present study, none of the isolate was positive for blaKPC or blaNDM and all isolates demonstrated susceptibility to meropenem and imipenem.
Polymyxins are active against most members of the Enterobacteriaceae family, however, some are naturally resistant to polymyxins, like Proteus, Brucella, Legionella, Campylobacter, and Vibrio (Poirel et al., 2017). In additional to intrinsic resistance, mechanisms responsible for acquired resistance to polymyxins in Enterobacteriaceae have been identified, including genes encoding LPS-modifying enzymes; regulators of the PmrAB and PhoPQ two-component systems; the intrinsic regulator RamA (Poirel et al., 2017). Recently, Liu et al. (2016) had reported plasmid-mediated colistin resistance in E. coli. Thereafter, plasmid-mediated colistin resistance gene (mcr-1) has been proved to be widespread in other members of the Enterobacteriaceae (Olaitan et al., 2016). In this study, four genetically related E. albertii isolates were positive for mcr-1 and were colistin-resistant. When the mcr-1 harboring plasmid pEA-3 sequence was compared using BLASTn to the nucleotide database at NCBI (accessed 26.01.2018), several highly similar (99% identities with query coverage over 90%) mcr-1 harboring plasmids from E. coli, Cronobacter sakazakii or Salmonella enterica were identified, suggesting that mcr-1 has also spread to E. albertii.
Conclusion
The present study provides significant information regarding antibiotic resistance of E. albertii from human, animal, and raw retail meats for the first time. Co-occurrence of β-lactamase and MCR-1 encoding genes in E. albertii isolates were identified. Further epidemiological assessments on the drug resistance patterns of E. albertii and determination of the molecular resistance mechanisms are needed in the treatment and prevention of both human and animal infections.
Author Contributions
QL, HW, YM, JX, and YX designed the project; QL, HW, ZZ, XLiu, LZ, GY, XC, and JZ carried out the sampling work; YXu, XB, JW, XLi, NZ, SF, RF, and JL carried out the experiments and generated the data; XLi, JL, and YX analyzed the data and drafted the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by grants from the National Natural Science Foundation of China (81772152), the State Key Laboratory for Infectious Disease Prevention and Control (2015SKLID504), the National Basic Research Priorities Program of China (2015CB554201), and the Health and Family Planning Commission of Sichuan Province (150259).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00258/full#supplementary-material
Pulsed-field gel electrophoresis (PFGE) profiles of four mcr-1-positive E. albertii isolates. Genomes of E. albertii isolates were digested with Xba I and fragments separated on a 1% agarose gel using a CHEF-DR III PFGE apparatus, according to the protocol for E. coli O157:H7 from PulseNet, United States (http://www.cdc.gov/pulsenet/pathogens/index.html). Multi-locus sequence typing (MLST) was done according to the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
PCR primers used to screen the β-lactamase and MCR-1 encoding genes.
Antibiotic resistance patterns and prevalence of β-lactamase and MCR-1 encoding genes in E. albertii isolates.
References
- Albert M., Faruque S., Ansaruzzaman M., Islam M., Haider K., Alam K., et al. (1992). Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J. Med. Microbiol. 37 310–314. 10.1099/00222615-37-5-310 [DOI] [PubMed] [Google Scholar]
- Albert M. J., Alam K., Islam M., Montanaro J., Rahaman A., Haider K., et al. (1991). Hafnia alvei, a probable cause of diarrhea in humans. Infect. Immun. 59 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J. A., Laxminarayan R., Mendelson M. (2017). How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int. J. Infect. Dis. 54 77–84. 10.1016/j.ijid.2016.11.415 [DOI] [PubMed] [Google Scholar]
- Asoshima N., Matsuda M., Shigemura K., Honda M., Yoshida H., Hiwaki H., et al. (2014). Identification of Escherichia albertii as a causative agent of a food-borne outbreak occurred in 2003. Jpn. J. Infect. Dis. 67 139–140. 10.7883/yoken.67.139 [DOI] [PubMed] [Google Scholar]
- Asoshima N., Matsuda M., Shigemura K., Honda M., Yoshida H., Oda T., et al. (2015). Isolation of Escherichia albertii from raw chicken liver in Fukuoka city, Japan. Jpn. J. Infect. Dis. 68 248–250. 10.7883/yoken.JJID.2014.530 [DOI] [PubMed] [Google Scholar]
- Bai L., Wang L., Yang X., Wang J., Gan X., Wang W., et al. (2017). Prevalence and molecular characteristics of extended-spectrum beta-lactamase genes in Escherichia coli isolated from diarrheic patients in China. Front. Microbiol. 8:144. 10.3389/fmicb.2017.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C. S., Drake J. P., Landolin J. M., Phillippy A. M. (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33 623–630. 10.1038/nbt.3238 [DOI] [PubMed] [Google Scholar]
- Besemer J., Lomsadze A., Borodovsky M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29 2607–2618. 10.1093/nar/29.12.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo R. A. (2017). beta-lactamases: a focus on current challenges. Cold Spring Harb. Perspect. Med. 7:a025239. 10.1101/cshperspect.a025239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora A., Hazarika N. K., Shukla S. K., Prasad K. N., Sarma J. B., Ahmed G. (2014). Prevalence of blaTEM, blaSHV and blaCTX-M genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India. Indian J. Pathol. Microbiol. 57 249–254. 10.4103/0377-4929.134698 [DOI] [PubMed] [Google Scholar]
- Brandal L. T., Tunsjo H. S., Ranheim T. E., Lobersli I., Lange H., Wester A. L. (2015). Shiga toxin 2a in Escherichia albertii. J. Clin. Microbiol. 53 1454–1455. 10.1128/JCM.03378-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Bradford P. A. (2016). beta-lactams and beta-lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6:a025247. 10.1101/cshperspect.a025247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing, CLSI Supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Falagas M., Karageorgopoulos D. E. (2009). Extended-spectrum β-lactamase-producing organisms. J. Hosp. Infect. 73 345–354. 10.1016/j.jhin.2009.02.021 [DOI] [PubMed] [Google Scholar]
- Felfoldi T., Heeger Z., Vargha M., Marialigeti K. (2010). Detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary. Clin. Microbiol. Infect. 16 89–92. 10.1111/j.1469-0691.2009.02795.x [DOI] [PubMed] [Google Scholar]
- Ghafourian S., Sadeghifard N., Soheili S., Sekawi Z. (2015). Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr. Issues Mol. Biol. 17 11–21. 10.21775/cimb.017.011 [DOI] [PubMed] [Google Scholar]
- Huys G., Cnockaert M., Janda J. M., Swings J. (2003). Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 53 807–810. 10.1099/ijs.0.02475-0 [DOI] [PubMed] [Google Scholar]
- Inglis T. J., Merritt A. J., Bzdyl N., Lansley S., Urosevic M. N. (2015). First bacteraemic human infection with Escherichia albertii. New Microbes New Infect. 8 171–173. 10.1016/j.nmni.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye K. S., Pogue J. M., Tran T. B., Nation R. L., Li J. (2016). Agents of last resort: polymyxin resistance. Infect. Dis. Clin. North Am. 30 391–414. 10.1016/j.idc.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Khan A. U., Maryam L., Zarrilli R. (2017). Structure, genetics and worldwide spread of New Delhi metallo-beta-lactamase (NDM): a threat to public health. BMC Microbiol. 17:101. 10.1186/s12866-017-1012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T., Yatsuyanagi J., Takahashi S., Kumagai Y., Wada E., Chiba M., et al. (2012). Isolation and identification of Escherichia albertii from a patient in an outbreak of gastroenteritis. Jpn. J. Infect. Dis. 65 203–207. 10.7883/yoken.65.203 [DOI] [PubMed] [Google Scholar]
- Laxminarayan R., Duse A., Wattal C., Zaidi A. K., Wertheim H. F., Sumpradit N., et al. (2013). Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 13 1057–1098. 10.1016/S1473-3099(13)70318-9 [DOI] [PubMed] [Google Scholar]
- Lindsey R. L., Fedorka-Cray P. J., Abley M., Turpin J. B., Meinersmann R. J. (2015). Evaluating the occurrence of Escherichia albertii in chicken carcass rinses by PCR, Vitek analysis, and sequencing of the rpoB gene. Appl. Environ. Microbiol. 81 1727–1734. 10.1128/AEM.03681-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Pop M. (2009). ARDB–antibiotic resistance genes database. Nucleic Acids Res. 37 D443–D447. 10.1093/nar/gkn656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-Y., Wang Y., Walsh T. R., Yi L.-X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- MacVane S. H. (2017). Antimicrobial resistance in the intensive care unit: a focus on gram-negative bacterial infections. J. Intensive Care Med. 32 25–37. 10.1177/0885066615619895 [DOI] [PubMed] [Google Scholar]
- Maeda E., Murakami K., Sera N., Ito K., Fujimoto S. (2015). Detection of Escherichia albertii from chicken meat and giblets. J. Vet. Med. Sci. 77 871–873. 10.1292/jvms.14-0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheux A. F., Boudreau D. K., Bergeron M. G., Rodriguez M. J. (2014). Characterization of Escherichia fergusonii and Escherichia albertii isolated from water. J. Appl. Microbiol. 117 597–609. 10.1111/jam.12551 [DOI] [PubMed] [Google Scholar]
- Martinez-Martinez L., Hernández-Allés S., Albertí S., Tomás J. M., Benedi V. J., Jacoby G. A. (1996). In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum-cephalosporins. Antimicrob. Agents Chemother. 40 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. (2010). Third generation DNA sequencing: pacific biosciences’ single molecule real time technology. Chem. Biol. 17 675–676. 10.1016/j.chembiol.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Murakami K., Etoh Y., Tanaka E., Ichihara S., Horikawa K., Kawano K., et al. (2014). Shiga toxin 2f-producing Escherichia albertii from a symptomatic human. Jpn. J. Infect. Dis. 67 204–208. 10.7883/yoken.67.204 [DOI] [PubMed] [Google Scholar]
- Oaks J., Besser T., Walk S., Gordon D., Beckmen K., Burek K., et al. (2010). Escherichia albertii in wild and domestic birds. Emerg. Infect. Dis. 16 638–646. 10.3201/eid1604.090695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan A. O., Chabou S., Okdah L., Morand S., Rolain J. M. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16 289–290. 10.1016/S1473-3099(15)00540-X [DOI] [PubMed] [Google Scholar]
- Ooka T., Seto K., Kawano K., Kobayashi H., Etoh Y., Ichihara S., et al. (2012). Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 18 488–492. 10.3201/eid1803.111401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka T., Tokuoka E., Furukawa M., Nagamura T., Ogura Y., Arisawa K., et al. (2013). Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerg. Infect. Dis. 19 144–146. 10.3201/eid1901.120646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavez M., Neves P., Dropa M., Matté M. H., Grinbaum R. S., De Araújo M. R. E., et al. (2008). Emergence of carbapenem-resistant Escherichia coli producing CMY-2-type AmpC β-lactamase in Brazil. J. Med. Microbiol. 57 1590–1592. 10.1099/jmm.0.2008/002774-0 [DOI] [PubMed] [Google Scholar]
- Perez K. L., Alam M. J., Castillo A., Taylor T. M. (2013). Antibiotic resistance and growth of the emergent pathogen Escherichia albertii on raw ground beef stored under refrigeration, abuse, and physiological temperature. J. Food Prot. 76 124–128. 10.4315/0362-028X.JFP-12-277 [DOI] [PubMed] [Google Scholar]
- Poirel L., Héritier C., Spicq C., Nordmann P. (2004). In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J. Clin. Microbiol. 42 3831–3833. 10.1128/JCM.42.8.3831-3833.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Jayol A., Nordmann P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30 557–596. 10.1128/CMR.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Singh A., Sobia F., Rashid M., Malik A., Shukla I., et al. (2011). blaCTX-M, blaTEM, and blaSHV in Enterobacteriaceae from North-Indian tertiary hospital: high occurrence of combination genes. Asian Pac. J. Trop. Med. 4 101–105. 10.1016/S1995-7645(11)60046-1 [DOI] [PubMed] [Google Scholar]
- Tzouvelekis L., Markogiannakis A., Psichogiou M., Tassios P., Daikos G. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25 682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li Q., Bai X., Xu Y., Zhao A., Sun H., et al. (2016). Prevalence of eae-positive, lactose non-fermenting Escherichia albertii from retail raw meat in China. Epidemiol. Infect. 144 45–52. 10.1017/S0950268815001120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Xiong Y., Lan R., Ye C., Wang H., Ren J., et al. (2011). pO157_Sal, a novel conjugative plasmid detected in outbreak isolates of Escherichia coli O157:H7. J. Clin. Microbiol. 49 1594–1597. 10.1128/JCM.02530-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wang H., Yu Y., Xu X., Sun Z., Lu J., et al. (2015). Antimicrobial resistance monitoring of gram-negative bacilli isolated from 15 teaching hospitals in 2014 in China. Zhonghua Nei Ke Za Zhi 54 837–845. 10.3760/cma.j.issn.0578-1426.2015.10.005 [DOI] [PubMed] [Google Scholar]
- WHO (2012). Critically Important Antimicrobials for Human Medicine. Geneva: World Health Organization. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pulsed-field gel electrophoresis (PFGE) profiles of four mcr-1-positive E. albertii isolates. Genomes of E. albertii isolates were digested with Xba I and fragments separated on a 1% agarose gel using a CHEF-DR III PFGE apparatus, according to the protocol for E. coli O157:H7 from PulseNet, United States (http://www.cdc.gov/pulsenet/pathogens/index.html). Multi-locus sequence typing (MLST) was done according to the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
PCR primers used to screen the β-lactamase and MCR-1 encoding genes.
Antibiotic resistance patterns and prevalence of β-lactamase and MCR-1 encoding genes in E. albertii isolates.


