ABSTRACT
Helicobacter pylori infection and high dietary salt intake are risk factors for the development of gastric adenocarcinoma. One possible mechanism by which a high-salt diet could influence gastric cancer risk is by modulating H. pylori gene expression. In this study, we utilized transcriptome sequencing (RNA-seq) methodology to compare the transcriptional profiles of H. pylori grown in media containing different concentrations of sodium chloride. We identified 118 differentially expressed genes (65 upregulated and 53 downregulated in response to high-salt conditions), including multiple members of 14 operons. Twenty-nine of the differentially expressed genes encode proteins previously shown to undergo salt-responsive changes in abundance, based on proteomic analyses. Real-time reverse transcription (RT)-PCR analyses validated differential expression of multiple genes encoding outer membrane proteins, including adhesins (SabA and HopQ) and proteins involved in iron acquisition (FecA2 and FecA3). Transcript levels of sabA, hopA, and hopQ are increased under high-salt conditions, whereas transcript levels of fecA2 and fecA3 are decreased under high-salt conditions. Transcription of sabA, hopA, hopQ, and fecA3 is derepressed in an arsS mutant strain, but salt-responsive transcription of these genes is not mediated by the ArsRS two-component system, and the CrdRS and FlgRS two-component systems do not have any detectable effects on transcription of these genes. In summary, these data provide a comprehensive view of H. pylori transcriptional alterations that occur in response to high-salt environmental conditions.
KEYWORDS: outer membrane proteins, gene regulation, gastric cancer, diet, two-component signal transduction system, two-component regulatory systems
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that colonizes the stomach in about 50% of the world's population, resulting in a gastric inflammatory response. Most individuals colonized with H. pylori remain asymptomatic, but the presence of this organism increases the risk for peptic ulcer disease or gastric cancer (1, 2). In the absence of antibiotic therapy, H. pylori colonization can persist for life (3, 4). Several factors influence the clinical outcomes of H. pylori infection. These include variation among H. pylori strains in production of proteins that interact with host cells (5), genetic variation among human hosts (2, 6), and composition of the diet (7).
Human epidemiologic studies have demonstrated a link between high dietary salt intake and increased gastric cancer risk (7–10). In addition, experimental studies have demonstrated an increased risk for gastric cancer in animals fed a high-salt diet (11–14). The mechanisms by which a high-salt diet increases the risk of gastric cancer are unclear. One possibility is that a high-salt environment in the stomach modulates H. pylori gene expression (15–19). In previous proteomic studies, altered production of multiple H. pylori proteins has been observed in response to high-salt conditions (16, 19), but relatively little is known about the effects of high-salt conditions on H. pylori gene transcription.
In the current study, we analyzed the salt-responsive transcriptome of H. pylori using transcriptome sequencing (RNA-seq) methodology and detected salt-responsive regulation of 118 genes. We then focused further experiments on salt-responsive changes in the transcription of genes encoding outer membrane proteins (OMPs), which are potentially difficult to monitor using proteomic methods due to amino acid sequence relatedness among certain OMPs, as well as suboptimal detection of OMPs using methods such as two-dimensional difference in gel electrophoresis (2D-DIGE). We show that the transcription of sabA, hopA, and hopQ is upregulated in response to high-salt conditions, whereas transcription of fecA2 and fecA3 is downregulated in response to high-salt conditions. Two-component systems (TCSs), comprised of a sensor histidine kinase and a cognate response regulator, are commonly used by bacteria to detect and respond to environmental signals (reviewed in reference 20). Therefore, we analyzed the role of three TCSs in the observed transcriptional regulation: the ArsRS system (which is involved in acid-responsive gene transcription) (21–27), the FlgRS TCS (a pH-responsive TCS that primarily controls flagellar synthesis) (28, 29), and the CrdRS TCS (which responds to copper and nitrosative stress) (30, 31). We show that transcription of sabA, hopA, hopQ, and fecA3 is derepressed in an arsS mutant strain, but salt-responsive transcription of these genes is not mediated by the ArsRS TCS. These findings provide new insights into the effects of high-salt conditions on gene transcription in H. pylori.
RESULTS
Identification of differentially expressed genes.
To identify H. pylori genes differentially expressed in response to changes in environmental salt concentration, H. pylori was cultured for 6 h in brucella broth containing either 0.5% NaCl (BB-fetal bovine serum [FBS]-0.5%, routine conditions [see Materials and Methods for details]) or 1.25% NaCl (BB-FBS-1.25%, high-salt conditions). Quadruplicate analyses were performed for each condition. A previous study showed that growth of H. pylori in medium containing 1.25% NaCl was not substantially reduced compared to growth in medium containing 0.5% NaCl (15). Consistent with the previous results, the optical density at 600 nm (OD600) for the four cultures grown in BB-FBS-0.5% was 0.34 ± 0.04, and the OD600 for the four cultures grown in BB-FBS-1.25% was 0.29 ± 0.02. RNA was isolated and libraries were constructed and sequenced as described in Materials and Methods. For each gene, we calculated a transcript abundance ratio (i.e., transcript abundance under high-salt conditions divided by transcript abundance under routine conditions), based on the RNA-seq data. The mean ± standard deviation (SD) of all the calculated transcript abundance ratios (corresponding to 1,597 genes) was 1.02 ± 0.23. We designated differentially expressed genes as those with transcript abundance ratios that were >2 standard deviations above or below the mean (i.e., transcript abundance ratio of >1.46 or <0.69) and exhibiting false discovery rates (FDR) of <0.05. Based on these criteria, 65 genes were upregulated and 53 genes were downregulated in response to a high salt concentration (Table 1).
TABLE 1.
Differentially expressed genes identified by RNA-seq
| Gene expression | Gene no. |
Gene name or function | Ratiob | |
|---|---|---|---|---|
| B8 | 26695/J99a | |||
| Upregulated | HPB8_46 | HP1484 | Membrane protein | 1.53 |
| HPB8_73 | HP1459 | rluB, tRNA synthetase | 1.54 | |
| HPB8_104 | − | Predicted | 2.11 | |
| HPB8_111 | − | Predicted | 1.86 | |
| HPB8_188 | HP1426 | Predicted | 1.62 | |
| HPB8_193 | HP1288 | Predicted | 1.75 | |
| HPB8_243 | HP1240 | maf, septum formation | 1.46 | |
| HPB8_251 | − | Predicted | 2.22 | |
| HPB8_297 | HP1193 | tas1, aldoketoreductase | 1.49 | |
| HPB8_336 | HP1162 | Integral membrane protein | 1.50 | |
| HPB8_338 | HP1160 | Predicted | 1.85 | |
| HPB8_378 | HP1546 | Predicted | 1.73 | |
| HPB8_386 | HP1116 | Predicted | 1.48 | |
| HPB8_435 | HP0385 | Predicted | 1.53 | |
| HPB8_475 | HP1412 | Predicted | 1.47 | |
| HPB8_476 | jhp0955 | Predicted | 2.35 | |
| HPB8_477 | jhp0954 | Predicted | 1.76 | |
| HPB8_485 | HP0441 | virB4, ATPase | 1.55 | |
| HPB8_528 | jhp0936 | Predicted | 1.64 | |
| HPB8_529 | HP0996 | Predicted | 1.71 | |
| HPB8_530 | − | Predicted | 1.47 | |
| HPB8_555 | jhp1408 | Predicted | 1.62 | |
| HPB8_562 | − | Predicted | 1.75 | |
| HPB8_640 | HP0008 | Predicted | 1.60 | |
| HPB8_650 | HP0903 | ackA, acetate kinase | 1.63 | |
| HPB8_883 | HP0681 | Predicted | 1.49 | |
| HPB8_884 | HP0682 | Predicted | 1.61 | |
| HPB8_897 | HP0696 | acxB | 1.65 | |
| HPB8_898 | HP0697 | acxC | 1.60 | |
| HPB8_929 | HP0724 | dcuA | 1.86 | |
| HPB8_930 | HP0725 | sabA | 1.62 | |
| HPB8_976 | HP0767 | Predicted | 1.58 | |
| HPB8_1017 | HP0809 | fliL | 1.49 | |
| HPB8_1088 | HP0876 | frpB1 | 1.62 | |
| HPB8_1089 | HP0877 | ruvC | 1.52 | |
| HPB8_1092 | HP0882 | Predicted | 1.53 | |
| HPB8_1124 | HP0461 | Predicted | 1.81 | |
| HPB8_1139 | HP1022 | Predicted | 1.75 | |
| HPB8_1140 | HP1022 | Predicted | 1.47 | |
| HPB8_1154 | HP1036 | folK | 1.60 | |
| HPB8_1176 | HP1057 | Predicted | 1.46 | |
| HPB8_1213 | HP1093 | Predicted | 1.54 | |
| HPB8_1293 | HP0269 | miaB, tRNA methylation | 1.46 | |
| HPB8_1336 | HP0229 | hopA | 1.73 | |
| HPB8_1347 | HP0219 | Predicted | 1.51 | |
| HPB8_1352 | HP0213 | gidA, division protein | 1.49 | |
| HPB8_1408 | HP0157 | arok | 1.51 | |
| HPB8_1430 | HP0135 | Predicted | 1.74 | |
| HPB8_1452 | HP0113 | Predicted | 1.55 | |
| HPB8_1453 | HP0112 | fucA, fucose metabolism | 1.63 | |
| HPB8_1491 | − | Predicted | 1.62 | |
| HPB8_1514 | HP0054 | hypAVM, DNA methylase | 1.77 | |
| HPB8_1561 | HP0427 | Predicted | 1.85 | |
| HPB8_1562 | HP0426 | Predicted | 1.48 | |
| HPB8_1563 | HP0426 | Predicted | 1.95 | |
| HPB8_1588 | HP0036 | Predicted | 1.55 | |
| HPB8_1600 | HP0025 | hopD | 1.48 | |
| HPB8_1609 | HP0016 | Predicted | 1.55 | |
| HPB8_1610 | HP0015 | Predicted | 1.55 | |
| HPB8_1653 | − | Predicted | 2.29 | |
| HPB8_1655 | HP1564 | metQ | 1.57 | |
| HPB8_1669 | HP1541 | mfd, DNA transcription | 1.58 | |
| HPB8_1701 | HP1399 | rocF | 1.66 | |
| HPB8_p0001 | − | Predicted | 1.81 | |
| HPB8_p0002 | jhp0828 | Predicted | 1.74 | |
| HPB8_p0003 | − | Predicted | 1.47 | |
| Downregulated | HPB8_106 | HP1435 | pspA, signal peptide protease | 0.59 |
| HPB8_159 | HP1320 | rpsJ | 0.60 | |
| HPB8_160 | HP1319 | rplC | 0.56 | |
| HPB8_162 | HP1317 | rplW | 0.67 | |
| HPB8_170 | HP1309 | rplN | 0.68 | |
| HPB8_195 | HP1286 | Secreted protein | 0.53 | |
| HPB8_226 | HP1254 | bioC, biotin synthesis | 0.63 | |
| HPB8_234 | HP1246 | rpsF | 0.68 | |
| HPB8_235 | HP1245 | ssb, DNA binding protein | 0.67 | |
| HPB8_236 | HP1244 | rpsR | 0.65 | |
| HPB8_299 | HP1192 | Secreted motility protein | 0.54 | |
| HPB8_334 | HP1163 | fixS | 0.66 | |
| HPB8_394 | HP1100 | porA | 0.67 | |
| HPB8_439 | HP0389 | sodB | 0.67 | |
| HPB8_743 | HP0549 | murI, glutamate racemase | 0.57 | |
| HPB8_761 | HP0565 | Predicted | 0.46 | |
| HPB8_786 | HP0588 | oorD, oxoglutarate ferrodoxin oxidoreductase | 0.66 | |
| HPB8_800 | HP0601 | flaA | 0.52 | |
| HPB8_829 | HP0630 | mda66, drug activity modulator | 0.37 | |
| HPB8_842 | HP0641 | Predicted | 0.39 | |
| HPB8_843 | HP0641 | 3-Hydroxyacid dehydrogenase | 0.39 | |
| HPB8_844 | HP0642 | frxA, NAD(P)H-flavin oxidoreductase | 0.38 | |
| HPB8_921 | HP0715 | Predicted ABC transporter | 0.65 | |
| HPB8_925 | HP0719/720 | Predicted | 0.62 | |
| HPB8_938 | HP0731 | Predicted | 0.67 | |
| HPB8_939 | HP0733 | Predicted | 0.60 | |
| HPB8_958 | HP0751 | flaG | 0.67 | |
| HPB8_1010 | HP0802 | ribA, riboflavin biosynthesis | 0.67 | |
| HPB8_1015 | HP0807 | fecA2 | 0.54 | |
| HPB8_1016 | HP0808 | acpS, ACP synthase | 0.67 | |
| HPB8_1020 | HP0812 | Predicted | 0.51 | |
| HPB8_1023 | HP0815 | motA, flagellar motor | 0.66 | |
| HPB8_1061 | HP0851 | Predicted integral membrane protein | 0.59 | |
| HPB8_1108 | HP0474 | modB, molybdenum transporter | 0.65 | |
| HPB8_1111 | HP0472 | horE | 0.56 | |
| HPB8_1122 | HP0463 | hsdM, restriction enzyme | 0.63 | |
| HPB8_1146 | HP1028 | Predicted | 0.64 | |
| HPB8_1238 | HP0326 | neuA3, CMP-N-acetylneuraminic acid synthetase | 0.65 | |
| HPB8_1239 | HP0325 | flgH, flagellar basal ring | 0.64 | |
| HPB8_1247 | HP0318 | Predicted | 0.50 | |
| HPB8_1266 | HP0296 | rplU | 0.68 | |
| HPB8_1271 | HP0291 | pheA, predicted chorismate mutase | 0.65 | |
| HPB8_1307 | HP0256 | Predicted | 0.64 | |
| HPB8_1338 | HP0227 | hopM/omp5 | 0.56 | |
| HPB8_1346 | HP0220 | nifS | 0.63 | |
| HPB8_1378 | HP0189 | Predicted | 0.67 | |
| HPB8_1450 | HP0115 | flaB | 0.56 | |
| HPB8_1492 | HP0073 | ureA | 0.67 | |
| HPB8_1493 | HP0072 | ureB | 0.68 | |
| HPB8_1511 | HP0057 | Predicted | 0.41 | |
| HPB8_1525 | HP1354 | DNA methyltransferase | 0.67 | |
| HPB8_1526 | HP1355 | nadC, NAD synthesis | 0.64 | |
| HPB8_1551 | HP1382 | nucG endonuclease | 0.64 | |
| HPB8_1702 | HP1400 | fecA3 | 0.39 | |
Corresponding genes in the H. pylori 26695 and J99 genomes are shown (63, 64). The gene name for H. pylori J99 is listed if the gene is absent in H. pylori 26695. A minus indicates the absence of the corresponding gene in H. pylori 26695 and J99.
Ratios of transcript abundance (high-salt versus routine conditions) were calculated as described in Materials and Methods.
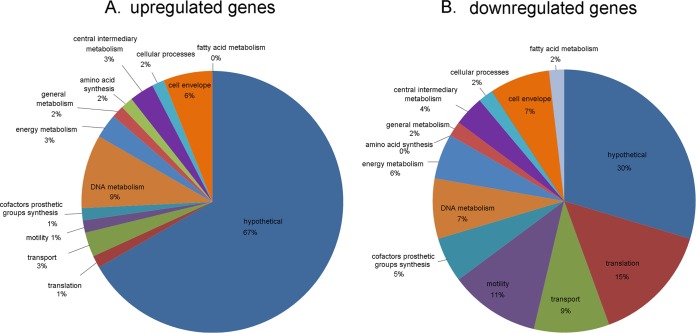
Figure 1 provides a graphical summary of the assigned functions of genes upregulated (Fig. 1A) or downregulated (Fig. 1B) in response to high-salt conditions. The majority of salt-responsive genes have no known functions (hypothetical proteins). The functions of the remaining genes are classified into the categories of protein translation, transport, motility, cofactor synthesis, and OMPs. Specifically, the list (Table 1) includes genes encoding OMPs (sabA, hopA, hopD, horE, hopM) and specialized OMPs involved in iron transport (fecA3, fecA2, frpB1), as well as genes encoding proteins involved in acetone metabolism (acxB, acxC), motility (flaA, flaB, flgH, motA, flaG), and acid resistance (e.g., ureA, ureB, rocF). Genes involved in motility (11%), translation (15%), and transport (9%) comprise a third of the genes for which transcript levels are decreased in bacteria grown under high-salt conditions (Fig. 1B). In comparison, genes involved in motility (1%), translation (1%), and transport (3%) comprise only 5% of the genes for which transcription is upregulated in bacteria grown under high-salt conditions (Fig. 1A).
FIG 1.
Functional classifications of differentially expressed genes. The pie charts show the proportions of differentially expressed genes identified in the RNA-seq analysis that were upregulated (n = 65) (A) or downregulated (n = 53) (B) in response to high-salt conditions.
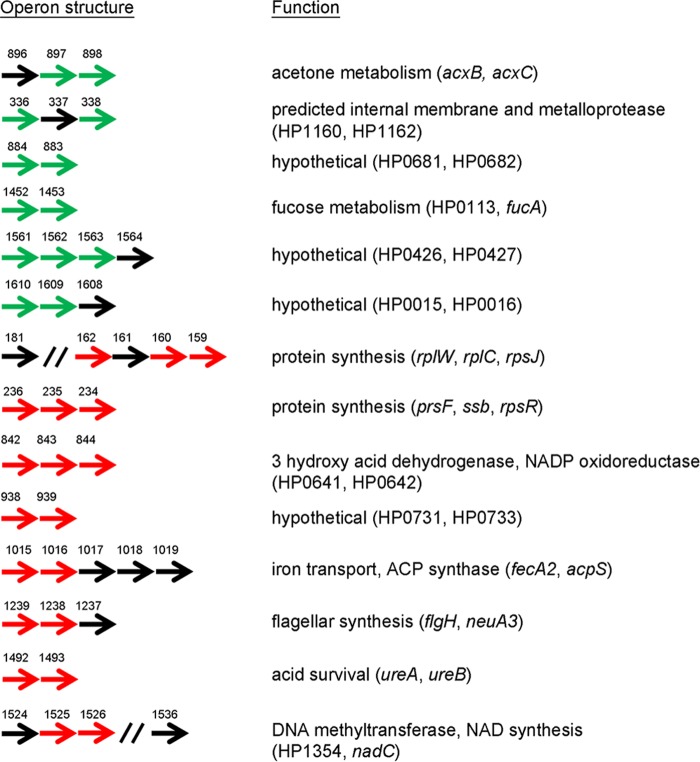
Many of the salt-responsive genes listed in Table 1 are located next to each other in the genome. Based on the operon map published for H. pylori strain 26695 (32), it is predicted that 13 of the 65 identified upregulated genes are transcribed in six different operons, and 20 of the 53 downregulated genes are transcribed in eight different operons (Fig. 2). Two operons (HPB8_159 to HPB8_181, and HPB8_234 to HPB8_236) encode proteins involved in protein synthesis. Other operons are involved in acetone metabolism (HPB8_896 to HPB8_898), fucose metabolism (HPB8_1452 to HPB8_1453), acid survival (HPB8_1492 to HPB8_1493), flagellar synthesis (HPB8_1237 to HPB8_1239), and iron transport and acyl carrier protein metabolism (HPB8_1015 to HPB8_1016). The functions of genes in four of the operons are not known.
FIG 2.
Mapping of salt-responsive genes to operons. Among the differentially expressed genes listed in Table 1, multiple genes were mapped to the same operons. Thirteen of the genes upregulated in response to high-salt conditions were mapped to six operons, and 20 of the downregulated genes were mapped to eight operons (32). Upregulated genes are shown in green, and downregulated genes are shown in red. The repressed gene HPB8_170 is not shown for the HPB8_181 to HPB8_162 operon. The numbers above the genes are the gene numbers in the H. pylori B8 genome. Corresponding gene numbers in H. pylori reference strain 26695 (HP numbers) are listed in the right column when appropriate. Functions associated with each operon are also shown on the right.
Twenty-nine of the 118 salt-responsive genes identified in this study encode proteins that were differentially abundant in previous proteomic analyses of H. pylori grown under high- or low-salt conditions (Table 2). Four of the differentially expressed genes correspond to salt-responsive proteins previously identified by 2D-DIGE analysis (16) of bacterial lysates from either H. pylori strain 26695 or strain 7.13. Twenty-six correspond to salt-responsive proteins in H. pylori strain 7.13 that were identified using iTRAQ, MudPIT, or surface biotinylation techniques (19). The effect of high-salt conditions on protein levels (increased or decreased) matched the direction of change observed in RNA-seq experiments for 27 of the 29 proteins (Table 2).
TABLE 2.
Differentially expressed genes corresponding to differentially abundant proteins detected by proteomic methods
| Gene no. in B8 genome | Gene no. in 26695/J99 genome | Gene name or function | Differential expression detected by: |
|||||
|---|---|---|---|---|---|---|---|---|
| 2D-DIGEa,c | MudPIT 26695b,c | MudPIT 7.13b,c | iTRAQ 26695b,c | iTRAQ 7.13b,c | Biotinylation 26695b,c | |||
| HPB8_106 | HP1435 | pspA | Yes | |||||
| HPB8_162d | HP1317 | rplW | Yes | |||||
| HPB8_188d | HP1426 | Predicted | Yes | |||||
| HPB8_195 | HP1286 | Secreted protein | Yes | |||||
| HPB8_234 | HP1246 | rpsF | Yes | |||||
| HPB8_235 | HP1245 | ssb | Yes | |||||
| HPB8_394 | HP1100 | porA | Yes | |||||
| HPB8_439 | HP0389 | sodB | Yes | |||||
| HPB8_475 | HP1412 | Predicted | Yes | Yes | ||||
| HPB8_555 | jhp1408 | Predicted | Yes | |||||
| HPB8_761 | HP0565 | Predicted | Yes | Yes | ||||
| HPB8_800 | HP0601 | flaA | Yes | Yes | ||||
| HPB8_829 | HP0630 | mda66 | Yes | Yes | Yes | |||
| HPB8_844 | HP0642 | frxA | Yes | Yes | Yes | |||
| HPB8_897 | HP0696 | acxB | Yes | |||||
| HPB8_938 | HP0731 | Predicted | Yes | |||||
| HPB8_958 | HP0751 | flaG | Yes | Yes | ||||
| HPB8_1010 | HP0802 | ribA | Yes | |||||
| HPB8_1017 | HP0809 | fliL | Yes | |||||
| HPB8_1247 | HP0318 | Predicted | Yes | |||||
| HPB8_1271 | HP0291 | pheA | Yes | |||||
| HPB8_1336 | HP0229 | hopA | Yes | Yes | ||||
| HPB8_1430 | HP0135 | Predicted | Yes | |||||
| HPB8_1450 | HP0115 | flaB | Yes | |||||
| HPB8_1493 | HP0072 | ureB | Yes | |||||
| HPB8_1600 | HP0025 | hopD | Yes | Yes | ||||
| HPB8_1701 | HP1399 | rocF | Yes | |||||
| HPB8_1702 | HP1400 | fecA3 | Yes | Yes | Yes | Yes | Yes | |
| HPB8_p0002 | jhp0828 | Predicted | Yes | |||||
Loh et al., 2012 (16).
Voss et al., 2015 (19).
“Yes” indicates that differential expression was detected at the protein level in the indicated study.
In two cases, the direction of change in response to high-salt conditions (i.e., upregulation or downregulation) was nonconcordant when RNA-seq results were compared with proteomic results.
Validation of RNA-seq data.
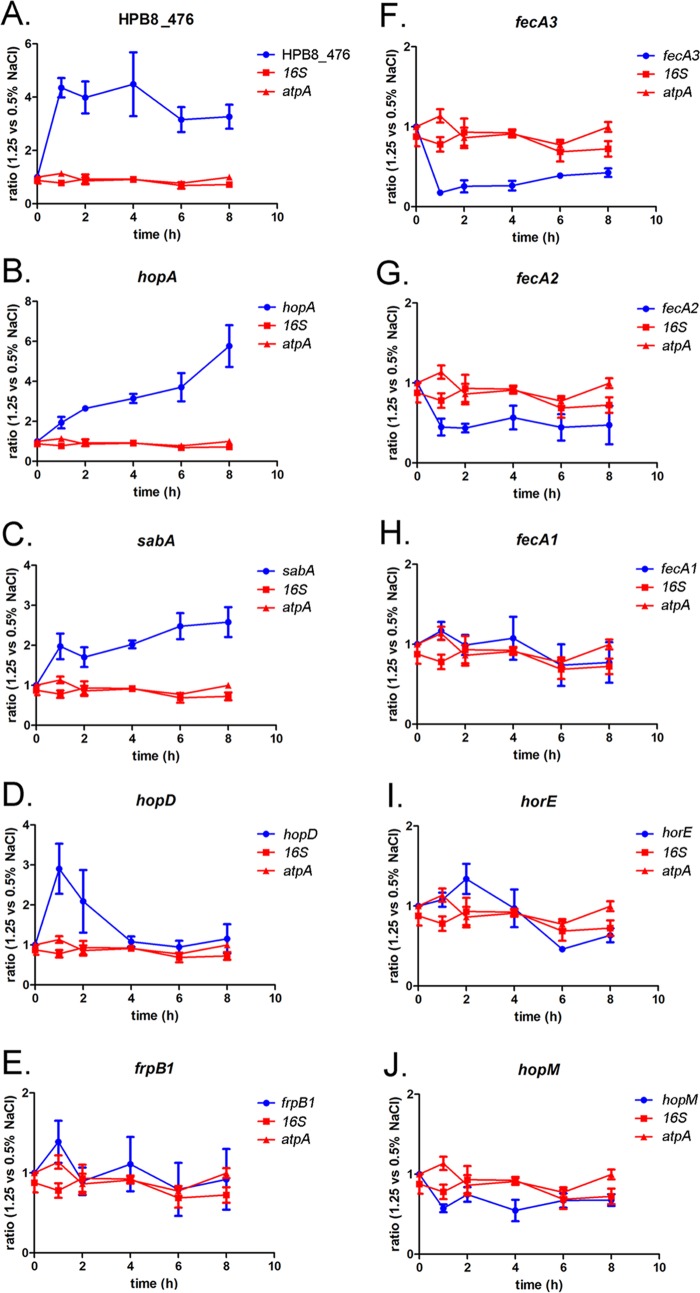
We next undertook studies to validate the RNA-seq results for several of the differentially expressed genes, using real-time reverse transcription (RT)-PCR. We analyzed RNA from broth cultures of H. pylori 7.13 grown for multiple time periods (0, 1, 2, 4, 6, and 8 h) in BB-FBS-0.5% or BB-FBS-1.25% medium. RT-PCR analysis confirmed that the transcription of HPB8_476 (which exhibited the highest level of change in the RNA-seq experiments) was upregulated in the presence of elevated levels of NaCl. In contrast, there were no significant differences in atpA or 16S rRNA transcript levels.
We next focused on eight genes encoding OMPs (hopA, sabA, hopD, frpB1, fecA3, fecA2, horE, and hopM) that were differentially expressed in the RNA-seq experiments. Three of these (fecA3, hopA, and hopD) encode proteins that were previously reported to be altered at the proteomic level in response to alterations in salt concentration (upregulated levels of HopA and HopD and downregulated levels of FecA3 under high-salt conditions) (19). As shown in Fig. 3, hopA (panel B) and sabA (panel C) transcript levels were increased at multiple time points after exposure of the bacteria to high-salt conditions. Transcript levels of hopD increased at early time points (1 h and 2 h) after bacterial exposure to high-salt conditions, followed by a return to basal levels (Fig. 3D). We confirmed the downregulated expression of fecA2 and fecA3 in response to growth under high-salt conditions (Fig. 3F and G). Consistent with the RNA-seq data, we did not detect any salt-responsive changes in a third fecA gene (fecA1) in response to high-salt conditions (Fig. 3H). Minimal differences in frpB1, HPB8_1111 (horE), or HPB8_1338 (hopM) transcript levels were detected (Fig. 3E, I, and J) when bacteria grown under the two conditions were compared. Thus, the RT-PCR experiments confirmed differential expression of multiple OMP-encoding genes that were identified by RNA-seq, and the results were also consistent with previous proteomic results. Possible reasons for discordance between some of the RNA-seq data and RT-PCR results are considered in Discussion.
FIG 3.
Real-time RT-PCR analysis of differentially expressed genes encoding OMPs. Real-time RT-PCR experiments were performed to validate the differential expression of salt-responsive genes encoding OMPs. H. pylori strains were cultured for 1, 2, 4, 6, and 8 h in medium containing 0.5% NaCl or 1.25% NaCl (BB-FBS-0.5% or BB-FBS-1.25%). RNA isolated from these cultures was processed as described in Materials and Methods and used to generate cDNA, which was used in the real-time RT-PCR analyses. Transcript abundance was calculated using the ΔΔCT method, with each transcript signal normalized to the abundance of the gyrB internal control. At each of the indicated time points, the normalized transcript signals obtained for H. pylori grown in BB-FBS-1.25% were compared to the normalized transcript signals obtained for H. pylori grown in BB-FBS-0.5% to calculate a ratio of transcript abundance. (A to J) Effect of high-salt conditions on the expression of the indicated genes. Transcript levels of 16S rRNA and atpA were monitored as controls. (A) HPB8_476 is a control gene that exhibited the greatest magnitude of change in the RNA-seq experiments in response to high-salt conditions. (H) fecA1 was not differentially expressed in RNA-seq experiments. Each panel depicts results obtained with RNA derived from at least four independent sets of biological samples (i.e., four bacterial cultures grown under high-salt conditions and four cultures grown under routine conditions). The mean and standard error of the mean are reported.
Analysis of transcriptional profiles of hopQ, vacA-like genes, and cagA.
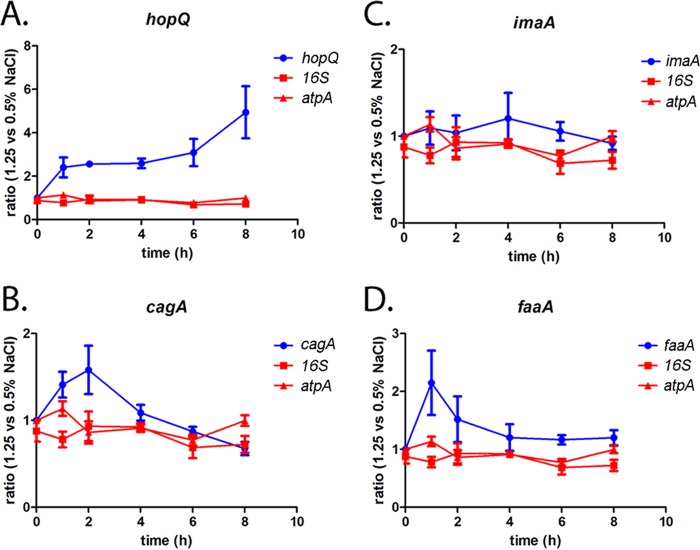
Several OMPs (HopQ, ImaA, and FaaA) and CagA were previously shown to undergo changes in abundance in response to H. pylori growth under high-salt conditions, based on proteomic analysis (16, 19), but the corresponding genes did not meet the criteria for being differentially expressed in the current RNA-seq analysis. To resolve this apparent discrepancy, we analyzed salt-responsive transcription of these genes by real-time PCR over an 8-h time course. As shown in Fig. 4A, growth of H. pylori in BB-FBS-1.25% resulted in increased hopQ transcript levels compared to growth in BB-FBS-0.5%, and this difference was detected at multiple time points. In the case of cagA (Fig. 4B), an initial increase in transcript levels was detected after bacterial exposure to high-salt conditions (1 h and 2 h), but the transcript levels subsequently returned to baseline levels (Fig. 4B). No significant changes in imaA or faaA transcription were detected in response to alterations in salt concentration (Fig. 4C and D).
FIG 4.
hopQ transcript levels are increased in response to high salt concentrations. Previous proteomic experiments (16, 19) detected differential abundance of CagA, HopQ, ImaA, and FaaA when bacteria grown under low-salt conditions or routine conditions were compared with bacteria grown in medium containing higher salt concentrations. H. pylori was cultured for the indicated times in BB-FBS-0.5% or BB-FBS-1.25% medium. Salt-dependent changes in the transcript levels of these genes were monitored using real-time RT-PCR. Transcript abundance was calculated using the ΔΔCT method, with each transcript level normalized to the abundance of the gyrB internal control. At each of the indicated time points, the normalized transcript levels for H. pylori grown in BB-FBS-1.25% were compared to the normalized transcript levels for H. pylori grown in BB-FBS-0.5% to calculate a ratio of transcript abundance. (A to D) Effect of increased salt concentrations on transcription of the indicated genes (hopQ, cagA, imaA, and faaA). The effects of high-salt conditions on expression of atpA and 16S rRNA were monitored as controls. Each panel depicts results obtained with RNA derived from four independent experiments, using H. pylori grown in either BB-FBS-0.5% or BB-FBS-1.25% medium. The mean and standard error of the mean are reported.
Salt-responsive changes in hopQ and sabA detected using promoter-reporter fusions.
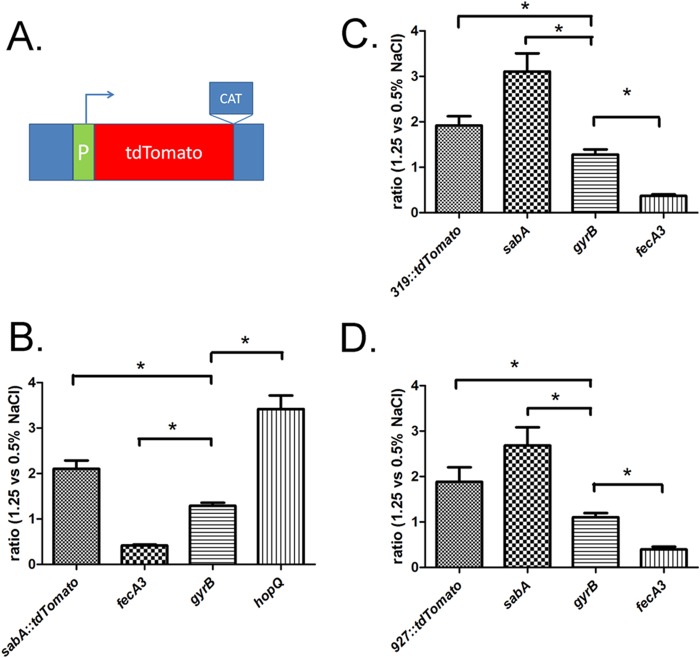
H. pylori 7.13 contains two identical copies of hopQ (designated HPB8_319 and HPB8_927) (33). To analyze the effects of environmental salt concentration on transcription of the individual hopQ alleles, we generated strains in which either the HPB8_319 or HPB8_927 open reading frame (ORF) was deleted and replaced with an ORF encoding the fluorescent protein tdTomato. In the resulting strains, transcriptional expression of tdTomato is driven by the promoters of HPB8_319 or HPB8_927 (Fig. 5A). As a positive control, we used the same approach to generate a transcriptional fusion in which tdTomato transcription is driven by the sabA promoter. The original goal of these experiments was to detect the tdTomato protein fluorometrically, but the intensity of the fluorescent signals was insufficiently strong for reliable quantification using a fluorescent plate reader. Therefore, we investigated salt-responsive expression of the transcriptional fusions at the transcriptional level, using primers specific for tdTomato. As shown in Fig. 5, an H. pylori strain containing the sabA promoter-tdTomato fusion (Fig. 5B) grown in BB-FBS-1.25% medium showed increased levels of tdTomato transcript compared to the same strain grown in BB-FBS-0.5% medium. Increased levels of tdTomato transcript were also detected in strains harboring the HPB8_319 (panel C) or HPB8_927 (panel D) promoter-tdTomato fusions, indicating that both 7.13 hopQ alleles are responsive to increased salt conditions. As internal controls, we monitored the expression of sabA, hopQ, fecA3, and gyrB for each strain, where appropriate, using primers specific for these genes. The expression of these control genes was consistent with the salt-responsive transcriptional trends previously detected in the wild-type 7.13 strain (Fig. 3). These results provide additional evidence indicating that hopQ and sabA are salt-responsive genes and indicate that the salt-responsive expression of these genes is mediated by DNA sequences upstream of the ATG initiation sites.
FIG 5.
Experiments with transcriptional reporters show that transcription of sabA and hopQ is upregulated in response to high-salt conditions. (A) Schematic representation of the key components of the transcriptional reporters. Each of the transcriptional fusion constructs contains a DNA sequence (including the promoter [P] and 5′-untranslated region [UTR]) upstream of the translational start site of the gene of interest fused to the tdTomato ORF. Immediately downstream of the ORF is an antibiotic selection marker (chloramphenicol acetyltransferase [CAT]) that confers resistance to chloramphenicol. The sabA reporter is designated sabA::tdTomato, and the hopQ reporters (for HPB8_319 and HPB8_927) are designated 319::tdTomato and 927::tdTomato, respectively. These transcriptional fusion constructs were introduced into the H. pylori chromosome as described in Materials and Methods. The introduction of each specific gene::tdTomato reporter resulted in deletion of the corresponding ORF (sabA or one of the copies of hopQ). Each H. pylori reporter strain was cultured for 6 h in BB-FBS-0.5% or BB-FBS-1.25% medium. RNA was isolated and transcript levels were quantified as described in Materials and Methods. All transcript data were normalized to 16S rRNA, and the normalized data then were analyzed to compare gene expression in H. pylori cultures grown in BB-FBS-0.5% or BB-FBS-1.25% medium. (B to D) Salt-responsive changes in strains harboring the sabA::tdTomato (B), 319::tdTomato (C), or 927::tdTomato (D) transcriptional reporters are shown. In experiments with the sabA::tdTomato reporter strain, transcript levels of fecA3, gyrB, and hopQ were monitored in parallel. In experiments with the hopQ 319::tdTomato and 927::tdTomato reporter strains, the expression of sabA, gyrB, and fecA3 was similarly monitored. Each panel depicts results from five independent experiments using RNA derived from H. pylori cultures grown in BB-FBS-0.5% (n = 5) or BB-FBS-1.25% (n = 5) medium. The mean and standard error of the mean are reported. Significant differences (P < 0.05, Student's t test) in expression between the indicated samples are indicated by an asterisk.
Effect of ArsS, CrdS, and FlgS mutation on sabA, hopQ, and hopA expression.
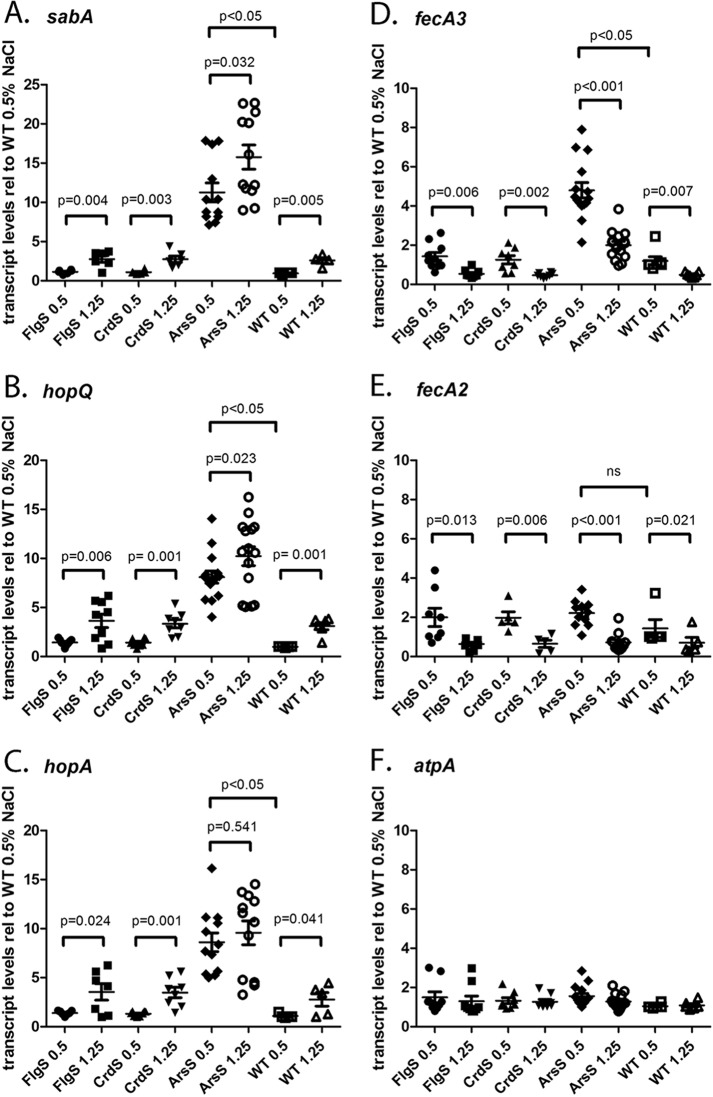
We next investigated a possible role for the ArsRS, CrdRS, or FlgRS TCSs in the regulation of salt-responsive OMPs. To do this, we analyzed transcript levels of salt-responsive genes encoding OMPs in strains harboring mutations in genes for ArsS, CrdS, or FlgS sensor kinases. We first analyzed the effects of the sensor kinase mutations in H. pylori strains cultured in medium containing 0.5% NaCl. Consistent with the known involvement of the ArsRS TCS in the repression of sabA (22, 34), we detected an increase in sabA transcript levels (about 12-fold, P < 0.05) in the ArsS mutant compared to the wild-type strain (Fig. 6A). Similar derepression of hopQ (9-fold, P < 0.05) (Fig. 6B), hopA (9-fold, P < 0.05) (Fig. 6C), and fecA3 (6-fold, P < 0.05) (Fig. 6D) was observed in the ArsS mutant compared to the wild-type strain, suggesting that the ArsRS TCS represses the expression of these genes. There was a smaller, nonsignificant difference in transcript levels for fecA2 (1.5-fold) (Fig. 6E) in the ArsS mutant compared to the wild-type strain. Mutations to either the CrdS or FlgS sensor kinases did not alter the basal level of hopQ, hopA, sabA, fecA3, or fecA2.
FIG 6.
ArsS regulates sabA, hopQ, hopA, and fecA3 expression. Strains containing insertional mutations in genes encoding ArsS, CrdS, or FlgS sensor kinases were cultured in BB-FBS-0.5% or BB-FBS-1.25% medium for 6 h. Transcript abundance was calculated using the ΔΔCT method, with each transcript signal normalized to the abundance of the gyrB internal control. The normalized transcript signals obtained for each sensor kinase mutant grown in BB-FBS-0.5% and BB-FBS-1.25% media were then compared to the normalized transcript signals obtained for wild-type H. pylori 7.13 grown in BB-FBS-0.5% medium (relative levels reported on the y axis). Each panel depicts the transcript levels for each strain and growth condition relative to that of the wild-type strain grown in BB-FBS-0.5% medium. The expression levels of the indicated genes (sabA, hopQ, hopA, fecA3, fecA2, and atpA) are shown. Each panel depicts results from at least seven independent biological sets of H. pylori strains grown in BB-FBS-0.5% and BB-FBS-1.25% media. Symbols indicate relative transcript abundance in the indicated H. pylori strains, grown under high-salt or routine conditions. The mean and standard error of the mean are reported. Statistical differences and corresponding P values are shown for comparisons of each strain grown under high-salt conditions and routine conditions (Student's t test). In addition, analyses of variance (ANOVA), followed by Dunn's multiple-comparison test, were conducted to examine statistical differences in the transcript levels expressed by mutant strains grown in BB-FBS-0.5% medium and the levels expressed by the WT strain grown in BB-FBS-0.5% medium.
We then examined the effects of the ArsS, CrdS, and FlgS kinase mutations on the ability of H. pylori strains to regulate sabA, hopQ, hopA, fecA3, and fecA2 transcript levels in response to salt. As shown in Fig. 6A to C, growth of wild-type H. pylori strain 7.13 under high-salt conditions resulted in ∼3-fold induction of sabA, hopQ, and hopA transcripts compared to that of wild-type strains grown at 0.5% NaCl. A similar effect of salt on expression of sabA, hopQ, and hopA was detected in H. pylori strains containing either the FlgS or CrdS mutations. Growth of the ArsS mutant strain under high-salt conditions resulted in detectable increases in transcription of sabA, hopQ, and hopA, but the magnitude of the increase was lower than that which was observed in the wild-type strain. Compared to the ArsS mutant strain grown in BB-FBS-0.5%, transcript levels in the ArsS mutant strain grown in BB-FBS-1.25% increased by 1.6-fold for sabA (P = 0.032), 1.5-fold for hopQ (P = 0.023), and 1.35-fold for hopA (P = 0.541). Since sabA, hopQ, and hopA are highly expressed in the arsS mutant strain, the magnitude of salt induction for these transcripts is likely to be less than what is observed for the wild-type strain and FlgS or CrdS mutants. Repression of both fecA3 (Fig. 6D) and fecA2 (Fig. 6E) transcripts in the FlgS, CrdS, and ArsS sensor kinases was observed when the strains were grown under high-salt conditions, compared to when the strains were grown under routine conditions. Taken together, these results indicate that the ArsRS TCS regulates basal transcription but does not affect salt-regulated transcription of the genes encoding the OMPs SabA, HopQ, HopA, FecA3, and FecA2.
DISCUSSION
In this study, we used RNA-seq technology to define the salt-responsive transcriptome of H. pylori. We identified 65 H. pylori genes that were upregulated and 53 genes that were downregulated in response to high-salt conditions. Among the differentially expressed genes, multiple genes were mapped to 14 operons (based on comparison to the operon map of H. pylori strain 26695) (32), and the remaining 85 genes are predicted to be monocistronically transcribed. Within each of these 14 operons, the differentially expressed genes identified by RNA-seq were all upregulated or downregulated in the same manner. Functions associated with the identified operons include acetone metabolism (35, 36), acid survival (37, 38), flagellar synthesis (39, 40), and iron transport (41). Genes associated with these functions have been documented to affect H. pylori colonization in animal models (35, 40, 42, 43). The effect of salt on these operons may therefore serve to promote H. pylori colonization under elevated salt conditions.
Previous proteomic studies (16, 19) detected differential abundance of 29 of the corresponding encoded proteins when H. pylori strains were cultured under high-salt and normal-salt conditions. High-salt conditions had a similar effect on gene transcription and protein levels (i.e., upregulated or downregulated) for 27 of the 29 proteins. The higher number of differentially expressed genes detected by RNA-seq in comparison to the number of differentially abundant proteins detected in proteomic experiments presumably reflects limited sensitivity of the proteomic methods. Posttranscriptional changes potentially account for instances where differential abundance of proteins was detected in proteomic experiments and corresponding differences were not detected in RNA-seq experiments.
Comparative analysis of transcription in H. pylori cultures grown for 6 h under either high-salt conditions or routine conditions allowed us to detect differences in steady-state levels of transcripts, which are relevant to the previously published proteomic data (19). Exposure of H. pylori to high-salt conditions probably results in additional transcriptional changes that are transient and would not have been detected by RNA-seq at the 6-h time point. Consistent with expectations, RT-PCR experiments revealed transcriptional changes in multiple genes that were detectable within 1 h after exposure of the bacteria to high-salt conditions and persisted over a longer time course (up to 8 h after exposure to high-salt conditions). The magnitude of the differences in transcript levels detected by RNA-seq was relatively low (<2-fold) for most genes. Similarly, high-salt conditions caused changes that were relatively small in magnitude in RT-PCR experiments at multiple time points in the current study, as well as in previous proteomic experiments (16, 19).
An examination of the functional groupings associated with up- or downregulated genes suggests a coordinated transcriptional response when the bacteria are subjected to elevated salt concentrations. For example, motility genes accounted for a larger percentage (11%) of downregulated genes than upregulated genes (1%). The downregulated genes include genes encoding the flagellin subunits (FlaA, FlaB) (44, 45) and genes required for flagellar rotation (motA) (46). Consistent with this, H. pylori motility decreases when the bacteria are grown under high-salt conditions, compared to routine conditions (19). The decrease in H. pylori motility may represent an adaptation that favors H. pylori attachment to host cells under conditions of elevated salt concentrations. In addition to alterations in genes associated with motility, changes in transcription of genes associated with protein translation and nutrient transport were detected. Genes involved in translation and transport account for a larger proportion (15% [translation] and 9% [transport]) of downregulated genes than the corresponding percentages (1% and 3%) for each category among the upregulated genes. After 6 h, the density of cultures grown under high-salt conditions (OD600 = 0.29 ± 0.02) was slightly lower than that of cultures grown under routine conditions (OD600 = 0.34 ± 0.04). Thus, the decrease in translation may reflect an adjustment of H. pylori growth in response to stress conditions. Three of the differentially expressed genes encode proteins involved in iron uptake. Curiously, two of these iron transporters (FecA2 and FecA3) (41), which in Escherichia coli are involved in ferric citrate transport (47), are downregulated in response to high-salt conditions, whereas RNA-seq studies suggested that FrpB1, an iron binding protein involved in iron acquisition when hemoglobin is an iron source (48), is upregulated in response to high-salt conditions. The different effects of high-salt conditions on the expression of these iron transporters (upregulated expression of frpB1 and downregulated expression of fecA2 and fecA3 under high-salt conditions) may reflect a strategy in which mechanisms for iron acquisition are altered under conditions of salt stress.
Among the 118 differentially expressed genes, 8 encode OMPs. We used real-time RT-PCR methods to validate the RNA-seq results for several of these OMP-encoding genes, and we also analyzed genes encoding salt-responsive OMPs identified in previous proteomic experiments (19). Real-time RT-PCR experiments confirmed that the expression of genes encoding the OMPs SabA, HopA, and HopQ is upregulated in response to high-salt conditions and that the expression of genes encoding the OMPs FecA2 and FecA3 is downregulated in response to high-salt conditions. Salt-induced increases in the abundance of HopA, HopD, and HopQ have previously been shown in proteomic studies (16, 19). Both SabA and HopQ are adhesins that mediate H. pylori interactions with gastric epithelial cells. SabA binds the sialyl-dimeric Lewis X glycosphingolipid (49). HopQ binds to human carcinoembryonic antigen-related cell adhesion molecules (CEACAM) and thereby promotes the translocation of the CagA effector protein into host cells (50, 51). Real-time RT-PCR experiments failed to validate the RNA-seq results for several OMP-encoding genes. We speculate that false-positive results for several genes could have arisen in the RNA-seq analysis due to nucleotide sequence relatedness among genes encoding H. pylori OMPs.
Since TCSs often mediate responses to environmental stimuli, we examined a possible role of three TCSs in modulating salt-responsive gene expression in H. pylori. Previous work (22, 34) identified the acid-responsive ArsRS TCS as a regulator of sabA expression and showed that the ArsR response regulator binds a DNA fragment encompassing nucleotides −20 bp upstream and +38 bp downstream of the sabA transcriptional start site. The ArsRS TCS is activated when the sensor kinase ArsS phosphorylates the response regulator ArsR in response to low pH signals (21, 23, 25, 27, 52, 53). In the case of sabA, the activated ArsR binds to a region of DNA surrounding the transcriptional start site of sabA and blocks transcription (34). In addition, a mutation to ArsS results in the derepression of sabA expression (22). Examination of the DNA sequences upstream of the translational start sites of sabA, hopQ (HPB8_319, HPB8_927), hopA, and to a lesser extent fecA3 revealed considerable sequence relatedness, both upstream and downstream of the transcriptional start sites of the respective genes (see Fig. S1 in the supplemental material). The relatedness of regions upstream of sabA and fecA3 was mainly limited to sequences upstream of the fecA3 transcriptional start site. Given the degree of similarity between sequences upstream of sabA and regions upstream of several other salt-responsive genes encoding OMPs, including the region upstream of sabA that is bound by ArsR, we postulated that the ArsRS system would regulate hopQ, hopA, and fecA3 expression. Consistent with this, the ArsS mutant strain expressed higher levels of hopA, hopQ, and fecA3 than the wild-type strain. Recently, inactivation of ArsS has also been demonstrated to result in the derepression of genes encoding the OMPs SabB, LabA (HopD), and HopZ, with ArsR binding to the promoter regions of these genes (54). Our current finding that ArsRS regulates hopA, hopQ, and fecA3 suggests a key role of the ArsRS TCS in the regulation of OMP expression in H. pylori. Besides the ArsRS TCS, H. pylori utilizes two additional TCSs to sense environmental changes: the CrdRS TCS responds to copper and nitrosative stress (30, 31), and FlgRS is a pH-responsive TCS that primarily controls flagellar synthesis (28, 29, 55). Mutations to either the CrdRS or FlgRS TCS had no effect on sabA, hopQ, hopA, fecA2, or fecA3 expression. Similarly, high-salt conditions stimulated increased sabA, hopQ, and hopA transcription and decreased fecA2 and fecA3 transcription in the FlgS, CrdS, and ArsS mutants. We noted a decrease in the magnitude of salt induction of sabA, hopQ, and hopA transcripts in the ArsS mutant compared to that of the wild-type strain, which can be attributed to the fact that all three transcripts are expressed at close to maximal levels in the ArsS mutant. Thus, although an intact ArsRS TCS system represses expression of sabA, hopQ, hopA, and fecA3, the salt-responsive expression of these genes is not dependent on the ArsRS TCS. Further studies will be required to define the mechanisms by which these genes are regulated in response to changes in environmental salt concentrations.
Within the human stomach, H. pylori is likely to encounter large fluctuations in salt concentration due to variations in dietary salt intake. The transcriptional changes described in this study presumably enhance the ability of the bacteria to tolerate these changes. In addition to short-term transcriptional alterations in response to changes in salt concentration, a recent study revealed proteomic and genomic changes in H. pylori strains isolated from Mongolian gerbils fed a high-salt diet for 4 months (56). Thus, high-salt stress can lead to the adaptation and selection of H. pylori strains with enhanced fitness for growth in a high-salt environment. Collectively, the short-term and long-term effects of a high-salt environment on H. pylori probably result in an increased ability of the bacteria to tolerate a high-salt environment and may also alter the interactions of the bacteria with host cells (e.g., by increased expression of adhesins and effector proteins), which is potentially relevant to the increased risk of gastric cancer observed in persons who consume a high-salt diet.
MATERIALS AND METHODS
Bacterial culture methods.
H. pylori strain 7.13 was grown at 37°C in ambient air supplemented with 5% CO2 on Trypticase soy agar plates containing 5% sheep blood or in modified brucella broth containing 5% fetal bovine serum (FBS) (i.e., BB-FBS). When required, streptomycin (25 μg/ml) or chloramphenicol (5 μg/ml) was added to the culture medium. Escherichia coli was grown on Luria-Bertani medium in the presence of ampicillin (50 μg/ml), chloramphenicol (25 μg/ml), or streptomycin (25 μg/ml).
RNA isolation.
To prepare H. pylori RNA samples for RNA-seq analysis, H. pylori was grown to an OD600 of ∼0.5 in BB-FBS (25 ml) containing 0.5% NaCl, and the bacteria were pelleted by centrifugation. The bacteria were then resuspended and inoculated into brucella broth containing either 0.5% NaCl (i.e., BB-FBS-0.5%, routine conditions) or 1.25% NaCl (i.e., BB-FBS-1.25%, high-salt conditions) at an initial OD600 of 0.15. The cultures were grown for 6 h, and then the bacteria were pelleted and resuspended in RNAlater (Ambion) for 40 min. The cell suspensions were centrifuged at 3,500 × g, supernatants were decanted, and the pellets were stored at −80°C. Total RNA was prepared as previously described (56) using TRIzol reagent. Contaminating DNA was removed by digesting the RNA with RQ1 RNase-free DNase (Promega), and the samples were subjected to a cleanup step using RNeasy columns (Qiagen). Each RNA sample was eluted in 100 μl of water. H. pylori cultures for real-time RT-PCR analysis were grown using the same methods as described above, except that the cultures were grown for 1, 2, 4, 6, and 8 h before being harvested for RNA isolation.
Preparation of RNA-seq library and sequence analysis.
The total RNA quality was assessed using a 2100 Bioanalyzer (Agilent). At least 200 ng of DNase-treated total RNA (RNA integrity number greater than 6) was used to generate rRNA-depleted/mRNA-enriched libraries using TruSeq Ribo-Zero bacterial RNA kits (Illumina). Library quality was assessed using the 2100 Bioanalyzer (Agilent), and libraries were quantitated using KAPA library quantification Kits (KAPA Biosystems). Pooled libraries were subjected to 75-bp paired-end sequencing according to the manufacturer's protocol (Illumina HiSeq 3000). Bcl2fastq2 conversion software (Illumina) was used to generate demultiplexed Fastq files. A total of eight independent RNA-seq libraries from four sets of H. pylori cells grown under routine or high-salt conditions were sequenced. The number of sequence reads for each sample ranged from 24 to 33 million.
RNA-seq data were trimmed to remove all bases below Q3, and adapter sequences were removed using FastQ quality control software (FaQCs) (57). Trimmed sequences were aligned to the H. pylori B8 reference genome (33) (GenBank accession number GCA_000196755.1), which is nearly identical to the strain 7.13 genome, using Burrows-Wheeler alignment (BWA) (58) and default alignment options. Read counts were generated using HT-Seq v.0.6.1p1 (59) and default options. Transcripts associated with a total of 1,597 H. pylori genes were identified by RNA-seq. The EdgeR (60) package for R was used to analyze count files. Data from individual samples were normalized within EdgeR and analyzed using the generalized linear model (GLM).
To ensure that the RNA-seq results represented sequences corresponding to mRNA instead of contaminating DNA, we analyzed RNA-seq data for 14 putative nontranscribed intergenic regions in comparison to protein-encoding genes. The coverage for individual genes or regions was calculated and tabulated for all samples, using samtools mpileup. The average coverage for the entire genome was about 1,000× (maximum count, 598,758; median, 1,607; mean, 5,761, based on reads aligning to individual genes). The average coverage for the intergenic regions was 3.8 (ranging from 1 to 11), whereas the coverage for a panel of selected regulatory genes (predicted to be transcribed at low levels) ranged from 217 to 2,140 (see Table S1 in the supplemental material). The low coverage of putative noncoding regions provided evidence that the preparations used for RNA-seq analysis contained very low levels of contaminating DNA.
To identify differentially expressed genes, ratios of transcript abundance (number of sequence reads from bacteria grown under high-salt conditions divided by the number of sequence reads from bacteria grown under routine conditions) were calculated for each gene. The mean ± SD of all the calculated transcript abundance ratios (corresponding to 1,597 genes) was 1.02 ± 0.23. We designated differentially expressed genes as those exhibiting transcript abundance ratios that were >2 SDs above or below the mean (i.e., transcript abundance ratio of >1.46 or <0.69) and exhibiting false discovery rates (FDR) of <0.05.
Real-time RT-PCR.
One hundred nanograms of purified total RNA was reverse transcribed into cDNA using an iScript cDNA synthesis kit (Bio-Rad). Real-time RT-PCR was carried out on 1:20 dilutions of the cDNA preparations using an ABI real-time PCR machine, with SYBR green as the fluorochrome (iTaq universal SYBR mix; Bio-Rad). The abundance of each transcript was calculated using the ΔΔCT method. As controls, the expression of the housekeeping genes gyrB (DNA gyrase subunit B), atpA (encoding ATP synthase F1 α subunit), and 16S rRNA were monitored. The transcriptional profiles of the genes examined were similar if results were normalized to either 16S rRNA or gyrB (data not shown). The normalized transcript signals from high-salt conditions and routine conditions were then compared. The primer pairs used for real-time RT-PCR analysis are based on the published H. pylori B8 genome (33) and are listed in Table 3.
TABLE 3.
Primers used for real-time RT-PCR analyses
| Gene | Forward primer | Reverse primer |
|---|---|---|
| atpA | 5′-CTTCACGCAATTCGCTTCTG-3′ | 5′-AAGCCCTTAGCCCCAGCATA-3′ |
| fecA1 | 5′-ATGGTATGCGAACTACCGCC-3′ | 5′-TAGCGTTGCCCCACTTCAAT-3′ |
| fecA2 | 5′-TCTCGCACGGTGATTTCCAA-3′ | 5′-GCGCACCGAAATTTTAGGCA-3′ |
| fecA3 | 5′-ATGTGGGTATCCAAGCGCAA-3′ | 5′-TCTTGCTCGCTGAGTGATCC-3′ |
| frpB1 | 5′-TATTGCACCCCAAGCTTTTC-3′ | 5′-AAGGCTGTCTGTGGCTTCAT-3′ |
| gyrB | 5-CGTGGATAACGCTGTAGATGAGAGC-3′ | 5′-GGGATTTTTTCCGTGGGGTG-3′ |
| hopA | 5′-GATCCGATAAAAACCGCAAA-3′ | 5′-TAATTCAACGCCATGGTGAA-3′ |
| hopD | 5′-CAAGTGGGTGAATGGGACTT-3′ | 5′-GAATCGCGCTGATTTCATTT-3′ |
| hopM | 5′-CAATTATAGAACCACTGCAC-3′ | 5′-TATTCCAATTGTATCGTAGG-3′ |
| hopQ | 5′-AACTCTTGCGGCATCACTCTT-3′ | 5′-CCGATCTCAACGCTAAAAGC-3′ |
| horE | 5′-TTTTATGGGTGCGGGTTATC-3′ | 5′-GCCACCATAGGTGAGCAAAT-3′ |
| sabA | 5′-AAAGCATTCAAAACGCCAAC-3′ | 5′-CCCGCATAAAGACTCCAAAA-3′ |
| HPB8_476 | 5′-TAGCCTTGATCGGGTTTTTG-3′ | 5′-TAGCGGTTTGGAATTCCTTG-3′ |
| tdTomato | 5′-TCCCCGATTACAAGAAGCTG-3′ | 5′-CCCATGGTCTTCTTCTGCAT-3′ |
Generation of hopQ mutant strains.
H. pylori strain 7.13 contains two copies of the hopQ gene, designated HPB8_319 and HPB8_927 (33). To examine the expression of these two hopQ genes independently, we generated strains that contained a deletion of either HPB8_319 or HPB8_927. DNA constructs containing nucleotides 500 bp upstream of the translational start site of HPB8_319 or HPB8_927, followed by 500 bp downstream of the stop codons of these genes, were synthesized (Genscript) and cloned into pUC57, yielding plasmids pHPB8_319flank and pHPB8_927flank, respectively. The synthesized DNA in both pHPB8_319flank and pHPB8_927flank contained unique XbaI and SmaI restriction sites that separated the upstream and downstream regions of the targeted hopQ allele, and into this site, a selectable marker encoding kanamycin resistance was introduced, yielding plasmids pHPB8_319flank::kan and pHPB8_927flank::kan. The DNA sequences flanking HPB8_319 are different from those of HPB8_927; therefore, use of these plasmids allows targeted deletion of individual hopQ genes. The plasmids, which do not replicate in H. pylori, were used to transform H. pylori strain 7.13, and transformants were selected on brucella plates containing 10 μg/ml kanamycin. Strain HPB8_319-kan is a hopQ mutant strain that contains a kanamycin insertion in the hopQ HPB8_319 locus, while HPB8_927-kan contains a kanamycin insertion in the hopQ HPB8_927 locus.
Generation of sabA and hopQ promoter reporter strains.
To construct a sabA promoter reporter, a 1.03-kb fragment of DNA containing 517 bp upstream of the translational start and 509 bp downstream of the sabA translational stop codon was synthesized (Genscript). Restriction sites for XbaI and SmaI were introduced between the upstream and downstream regions. The synthesized DNA was introduced into pUC57, yielding plasmid pSabA-promXS. To allow introduction of a tdTomato cassette (Clontech) downstream of the sabA promoter, the tdTomato ORF was PCR amplified from plasmid ptdTomato (Clontech) with specific forward (5′-AGAATCTAGATGGTGAGCAAGGGCGAGGAGG-3′) and reverse (5′-ATTAGATCTCTACTTGTACAGCTCGTCCATG-3′) primers. These primers contain XbaI (forward primer) and BglII (reverse primer) restriction sites (underlined). The ∼1.5-kb PCR product was digested with XbaI/BglII and cloned into the XbaI/BamHI site of plasmid pAD-C (61), which contains a cat (chloramphenicol acetyltransferase) cassette, and the resultant plasmid was digested with XbaI-SmaI to release a 2.5-kb fragment containing the tdTomato ORF and cat cassette. The isolated tdTomato-cat DNA fragment was next ligated into XbaI/SmaI sites of pSabA-promXS, generating the sabA reporter pSabA-tdTomato. DNA sequencing was used to confirm the proper ligation of the sabA DNA with the tdTomato reporter. pSabA-tdTomato was introduced into H. pylori strain 7.13 by natural transformation, and transformants were selected on chloramphenicol plates.
A similar approach was used to generate hopQ-tdTomato reporter fusions. For this purpose, plasmids pHPB8_319flank and pHPB8_927flank used in the generation of the hopQ mutant (see above) were digested with XbaI and SmaI. The 2.5-kb XbaI-SmaI DNA fragment containing the tdTomato ORF and cat cassette was inserted into the digested pHPB8_319flank and pHPB8_927flank plasmids. The resultant plasmids (p319-tdTomato and p927-tdTomato) were sequenced to confirm the proper ligation of the tdTomato reporter. Plasmid p319-tdTomato was used to transform strain HPB8_927-kan, and plasmid p927-tdTomato was used to transform strain HPB8_319-kan, selecting for chloramphenicol resistance.
Generation of H. pylori sensor kinase mutants.
To generate mutants in ArsS, CrdS, and FlgS sensor kinases, H. pylori strain 7.13 was transformed with the plasmids p165Km1, p1364km1, and p244km36, respectively, as previously described (62). These plasmids contain cloned genes for arsS, crdS, and flgS from H. pylori strain J99 that were disrupted by insertion of a blunt-ended 1.2-kb kanamycin resistance cassette from pUC4K into unique BglII, Eco47III, and HindIII restriction sites in the arsS, crdS, and flgS DNA sequences, respectively (62). The resulting plasmids, which are unable to replicate in H. pylori, were then introduced into H. pylori strain 7.13 by natural transformation, and kanamycin-resistant transformants were selected. PCR analysis was performed in each case to confirm the appropriate insertion of the kanamycin resistance cassette into the desired site in the H. pylori chromosome.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants AI039657, AI118932, CA116087 from the NIH and by the Department of Veterans Affairs (merit review grant 2I01BX000627).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00626-17.
REFERENCES
- 1.Amieva MR, El-Omar EM. 2008. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Fox JG, Wang TC. 2007. Inflammation, atrophy, and gastric cancer. J Clin Invest 117:60–69. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atherton JC, Blaser MJ. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest 119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cover TL. 2016. Helicobacter pylori Diversity and gastric cancer risk. mBio 7:e01869-01815. doi: 10.1128/mBio.01869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLean MH, El-Omar EM. 2014. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 11:664–674. doi: 10.1038/nrgastro.2014.143. [DOI] [PubMed] [Google Scholar]
- 7.Cover TL, Peek RM Jr. 2013. Diet, microbial virulence, and Helicobacter pylori-induced gastric cancer. Gut Microbes 4:482–493. doi: 10.4161/gmic.26262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SA, Kang D, Shim KN, Choe JW, Hong WS, Choi H. 2003. Effect of diet and Helicobacter pylori infection to the risk of early gastric cancer. J Epidemiol 13:162–168. doi: 10.2188/jea.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsugane S. 2005. Salt, salted food intake, and risk of gastric cancer: epidemiologic evidence. Cancer Sci 96:1–6. doi: 10.1111/j.1349-7006.2005.00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsugane S, Sasazuki S. 2007. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 10:75–83. doi: 10.1007/s10120-007-0420-0. [DOI] [PubMed] [Google Scholar]
- 11.Bergin IL, Sheppard BJ, Fox JG. 2003. Helicobacter pylori infection and high dietary salt independently induce atrophic gastritis and intestinal metaplasia in commercially available outbred Mongolian gerbils. Dig Dis Sci 48:475–485. doi: 10.1023/A:1022524313355. [DOI] [PubMed] [Google Scholar]
- 12.Fox JG, Dangler CA, Taylor NS, King A, Koh TJ, Wang TC. 1999. High-salt diet induces gastric epithelial hyperplasia and parietal cell loss, and enhances Helicobacter pylori colonization in C57BL/6 mice. Cancer Res 59:4823–4828. [PubMed] [Google Scholar]
- 13.Gamboa-Dominguez A, Ubbelohde T, Saqui-Salces M, Romano-Mazzoti L, Cervantes M, Dominguez-Fonseca C, de la Luz Estreber M, Ruiz-Palacios GM. 2007. Salt and stress synergize H. pylori-induced gastric lesions, cell proliferation, and p21 expression in Mongolian gerbils. Dig Dis Sci 52:1517–1526. doi: 10.1007/s10620-006-9524-3. [DOI] [PubMed] [Google Scholar]
- 14.Gaddy JA, Radin JN, Loh JT, Zhang F, Washington MK, Peek RM Jr, Algood HM, Cover TL. 2013. High dietary salt intake exacerbates Helicobacter pylori-induced gastric carcinogenesis. Infect Immun 81:2258–2267. doi: 10.1128/IAI.01271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loh JT, Torres VJ, Cover TL. 2007. Regulation of Helicobacter pylori cagA expression in response to salt. Cancer Res 67:4709–4715. doi: 10.1158/0008-5472.CAN-06-4746. [DOI] [PubMed] [Google Scholar]
- 16.Loh JT, Friedman DB, Piazuelo MB, Bravo LE, Wilson KT, Peek RM Jr, Correa P, Cover TL. 2012. Analysis of Helicobacter pylori cagA promoter elements required for salt-induced upregulation of CagA expression. Infect Immun 80:3094–3106. doi: 10.1128/IAI.00232-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gancz H, Jones KR, Merrell DS. 2008. Sodium chloride affects Helicobacter pylori growth and gene expression. J Bacteriol 190:4100–4105. doi: 10.1128/JB.01728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gancz H, Merrell DS. 2011. The Helicobacter pylori ferric uptake regulator (Fur) is essential for growth under sodium chloride stress. J Microbiol 49:294–298. doi: 10.1007/s12275-011-0396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss BJ, Loh JT, Hill S, Rose KL, McDonald WH, Cover TL. 2015. Alteration of the Helicobacter pylori membrane proteome in response to changes in environmental salt concentration. Proteomics Clin Appl 9:1021–1034. doi: 10.1002/prca.201400176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pflock M, Kennard S, Delany I, Scarlato V, Beier D. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect Immun 73:6437–6445. doi: 10.1128/IAI.73.10.6437-6445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodwin AC, Weinberger DM, Ford CB, Nelson JC, Snider JD, Hall JD, Paules CI, Peek RM Jr, Forsyth MH. 2008. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology 154:2231–2240. doi: 10.1099/mic.0.2007/016055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh JT, Gupta SS, Friedman DB, Krezel AM, Cover TL. 2010. Analysis of protein expression regulated by the Helicobacter pylori ArsRS two-component signal transduction system. J Bacteriol 192:2034–2043. doi: 10.1128/JB.01703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcus EA, Sachs G, Wen Y, Scott DR. 2016. Phosphorylation-dependent and phosphorylation-independent regulation of Helicobacter pylori acid acclimation by the ArsRS two-component system. Helicobacter 21:69–81. doi: 10.1111/hel.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol 188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Servetas SL, Carpenter BM, Haley KP, Gilbreath JJ, Gaddy JA, Merrell DS. 2016. Characterization of key Helicobacter pylori regulators identifies a role for ArsRS in biofilm formation. J Bacteriol 198:2536–2548. doi: 10.1128/JB.00324-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta SS, Borin BN, Cover TL, Krezel AM. 2009. Structural analysis of the DNA-binding domain of the Helicobacter pylori response regulator ArsR. J Biol Chem 284:6536–6545. doi: 10.1074/jbc.M804592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spohn G, Scarlato V. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J Bacteriol 181:593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. 2009. The pH-responsive regulon of HP0244 (FlgS), the cytoplasmic histidine kinase of Helicobacter pylori. J Bacteriol 191:449–460. doi: 10.1128/JB.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung CL, Cheng HH, Hsieh WC, Tsai ZT, Tsai HK, Chu CH, Hsieh WP, Chen YF, Tsou Y, Lai CH, Wang WC. 2015. The CrdRS two-component system in Helicobacter pylori responds to nitrosative stress. Mol Microbiol 97:1128–1141. doi: 10.1111/mmi.13089. [DOI] [PubMed] [Google Scholar]
- 31.Waidner B, Melchers K, Stahler FN, Kist M, Bereswill S. 2005. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J Bacteriol 187:4683–4688. doi: 10.1128/JB.187.13.4683-4688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 33.Farnbacher M, Jahns T, Willrodt D, Daniel R, Haas R, Goesmann A, Kurtz S, Rieder G. 2010. Sequencing, annotation, and comparative genome analysis of the gerbil-adapted Helicobacter pylori strain B8. BMC Genomics 11:335. doi: 10.1186/1471-2164-11-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey VC, Acio CR, Bredehoft AK, Zhu L, Hallinger DR, Quinlivan-Repasi V, Harvey SE, Forsyth MH. 2014. Repetitive sequence variations in the promoter region of the adhesin-encoding gene sabA of Helicobacter pylori affect transcription. J Bacteriol 196:3421–3429. doi: 10.1128/JB.01956-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brahmachary P, Wang G, Benoit SL, Weinberg MV, Maier RJ, Hoover TR. 2008. The human gastric pathogen Helicobacter pylori has a potential acetone carboxylase that enhances its ability to colonize mice. BMC Microbiol 8:14. doi: 10.1186/1471-2180-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pflock M, Bathon M, Schar J, Muller S, Mollenkopf H, Meyer TF, Beier D. 2007. The orphan response regulator HP1021 of Helicobacter pylori regulates transcription of a gene cluster presumably involved in acetone metabolism. J Bacteriol 189:2339–2349. doi: 10.1128/JB.01827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott DR, Marcus EA, Weeks DL, Sachs G. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187–195. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 38.Sachs G, Weeks DL, Wen Y, Marcus EA, Scott DR, Melchers K. 2005. Acid acclimation by Helicobacter pylori. Physiology (Bethesda) 20:429–438. doi: 10.1152/physiol.00032.2005. [DOI] [PubMed] [Google Scholar]
- 39.Josenhans C, Vossebein L, Friedrich S, Suerbaum S. 2002. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol Lett 210:165–172. doi: 10.1111/j.1574-6968.2002.tb11176.x. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole PW, Lane MC, Porwollik S. 2000. Helicobacter pylori motility. Microbes Infect 2:1207–1214. doi: 10.1016/S1286-4579(00)01274-0. [DOI] [PubMed] [Google Scholar]
- 41.Danielli A, Romagnoli S, Roncarati D, Costantino L, Delany I, Scarlato V. 2009. Growth phase and metal-dependent transcriptional regulation of the fecA genes in Helicobacter pylori. J Bacteriol 191:3717–3725. doi: 10.1128/JB.01741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debowski AW, Walton SM, Chua EG, Tay AC, Liao T, Lamichhane B, Himbeck R, Stubbs KA, Marshall BJ, Fulurija A, Benghezal M. 2017. Helicobacter pylori gene silencing in vivo demonstrates urease is essential for chronic infection. PLoS Pathog 13:e1006464. doi: 10.1371/journal.ppat.1006464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eaton KA, Brooks CL, Morgan DR, Krakowka S. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun 59:2470–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leying H, Suerbaum S, Geis G, Haas R. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol Microbiol 6:2863–2874. doi: 10.1111/j.1365-2958.1992.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 45.Suerbaum S, Josenhans C, Labigne A. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol 175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dean GE, Macnab RM, Stader J, Matsumura P, Burks C. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol 159:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hussein S, Hantke K, Braun V. 1981. Citrate-dependent iron transport system in Escherichia coli K-12. Eur J Biochem 117:431–437. doi: 10.1111/j.1432-1033.1981.tb06357.x. [DOI] [PubMed] [Google Scholar]
- 48.Carrizo-Chavez MA, Cruz-Castaneda A, Olivares-Trejo Jde J. 2012. The frpB1 gene of Helicobacter pylori is regulated by iron and encodes a membrane protein capable of binding haem and haemoglobin. FEBS Lett 586:875–879. doi: 10.1016/j.febslet.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 49.Mahdavi J, Sonden B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, Altraja S, Wadstrom T, Kersulyte D, Berg DE, Dubois A, Petersson C, Magnusson KE, Norberg T, Lindh F, Lundskog BB, Arnqvist A, Hammarstrom L, Boren T. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Javaheri A, Kruse T, Moonens K, Mejias-Luque R, Debraekeleer A, Asche CI, Tegtmeyer N, Kalali B, Bach NC, Sieber SA, Hill DJ, Koniger V, Hauck CR, Moskalenko R, Haas R, Busch DH, Klaile E, Slevogt H, Schmidt A, Backert S, Remaut H, Singer BB, Gerhard M. 2016. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat Microbiol 2:16189. doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- 51.Koniger V, Holsten L, Harrison U, Busch B, Loell E, Zhao Q, Bonsor DA, Roth A, Kengmo-Tchoupa A, Smith SI, Mueller S, Sundberg EJ, Zimmermann W, Fischer W, Hauck CR, Haas R. 2016. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat Microbiol 2:16188. doi: 10.1038/nmicrobiol.2016.188. [DOI] [PubMed] [Google Scholar]
- 52.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. 2007. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J Bacteriol 189:2426–2434. doi: 10.1128/JB.01492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beier D, Frank R. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J Bacteriol 182:2068–2076. doi: 10.1128/JB.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acio-Pizzarello CR, Acio AA, Choi EJ, Bond K, Kim J, Kenan AC, Chen J, Forsyth MH. 2017. Determinants of the regulation of Helicobacter pylori adhesins include repeat sequences in both promoter and coding regions as well as the two-component system ArsRS. J Med Microbiol 66:798–807. doi: 10.1099/jmm.0.000491. [DOI] [PubMed] [Google Scholar]
- 55.Scott DR, Marcus EA, Wen Y, Singh S, Feng J, Sachs G. 2010. Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3, and NH4(+), is necessary for acid survival of Helicobacter pylori. J Bacteriol 192:94–103. doi: 10.1128/JB.00848-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loh JT, Gaddy JA, Algood HM, Gaudieri S, Mallal S, Cover TL. 2015. Helicobacter pylori adaptation in vivo in response to a high-salt diet. Infect Immun 83:4871–4883. doi: 10.1128/IAI.00918-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo CC, Chain PS. 2014. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinformatics 15:366. doi: 10.1186/s12859-014-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. 2014. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun 82:3457–3470. doi: 10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loh JT, Cover TL. 2006. Requirement of histidine kinases HP0165 and HP1364 for acid resistance in Helicobacter pylori. Infect Immun 74:3052–3059. doi: 10.1128/IAI.74.5.3052-3059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 64.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.