Abstract
Using multiple drugs to kill cancer cells can decrease drug resistance development. However, this approach is frequently limited by the bioavailability and toxicity of the combined agents and delivery at ratios to specific locations that synergistically kill the cancer cells. Loading the individual agents into a nanoparticle that releases the drugs at synergizing ratios at a single location is one approach to resolve this concern. Celecoxib and Plumbagin are two drugs that were identified from a screen to synergistically kill melanoma cells compared to normal cells. Combined use of these agents by traditional approaches was not possible due to poor bioavailability and toxicological concerns. This study details the development of a nanoliposomal-based agent containing Celecoxib and Plumbagin, called CelePlum-777, which is stable and releases these drugs at an optimal ratio for maximal synergistic killing efficacy. CelePlum-777 was more effective at killing melanoma than normal cells and inhibited xenograft melanoma tumor growth by up to 72% without apparent toxicity. Mechanistically, the drug combination in CelePlum-777 led to enhanced inhibition of melanoma cell proliferation mediated by decreasing levels of key cyclin important for cancer cell proliferation and survival, which was not observed with the individual agents. Thus, a novel nanoparticle based drug has been developed containing Celecoxib and Plumbagin that lacks toxicity and deliverers the agents at a synergistically killing drug ratio to kill cancer cells.
Keywords: Plumbagin, Celecoxib, Nanoliposomes, COX-2, STAT3, Cyclins, Tumor development
INTRODUCTION
Despite the development of targeted pathway inhibitors and immune system modulators in melanoma, recurrent resistant disease development remains a problem (1). One emerging research area to address this issue has been to screen for pharmacological agents that synergize with one another to decrease the occurrence of resistant disease (2). However, this approach is frequently hindered by the individual drugs having unique solubility, bioavailability, toxicity profiles and limiting drug compatibility (3, 4). Furthermore, the mechanism of action of the drug combination causing enhanced efficacy has generally been difficult to ascertain (5). Nanotechnology is one approach being used to resolve these issues, which involves loading the drugs into a single nanoparticle at an optimized synergizing drug ratio (6). The nanoparticle simultaneously delivers the agents to tumor cells, releasing the drugs at the optimized synergizing ratio, and since there are two cooperating drugs, there can be decreased resistance development (6). A further advantage is that the FDA evaluates nanoparticles containing multiple active ingredients as a single drug product rather than two separate ones, which could significantly reduce costs and potentially move the agent more quickly to the clinic (7).
This study details the development of a nanoparticle containing Celecoxib and Plumbagin that were agents identified from a screen to synergistically kills melanoma cells. The non-steroidal anti-inflammatory drug Celecoxib inhibits cyclooxygenase-2 (COX-2) activity (8). COX-2 is an inducible enzyme that plays an important role in the production of prostaglandin E2 (PGE2) (8). COX-2 is overexpressed in carcinomas of the colon, breast, lung, prostate, cervix, stomach, and skin (9, 10). In melanoma cells, the key target modulated by Celecoxib is COX-2, which plays an important role in development of 50–70% of tumors (11). Celecoxib inhibits COX-2 activity thereby reducing the production of PGE2 (8), leading to a compensatory increase in COX-2 protein levels (12). PGE2 affects cellular proliferation, motility, invasiveness, angiogenesis, and promotes survival by inhibiting apoptosis (12). In addition, PGE2 is a tumor-inducing eicosanoid that promotes tumor growth and more invasive disease (13, 14). Celecoxib can have negative side effects depending on the dose and rate at which it is administered (15). Concentration of Celecoxib required to induce apoptosis of cultured cancer cells are high, ranging from 25 to 100 μmol/L, and use at these concentrations in animals, which would be equivalent to 200 mg per day, causes negative cardiovascular side effects (15, 16).
Plumbagin (5-hydroxy-2-methyl-1, 4-napthoquinone) is a quinoid isolated from the roots of the Plumbago zeylanica plant (17). Plumbagin retards the growth of various cancer types (18) and topical treatment inhibits ultraviolet radiation-induced squamous cell carcinomas in mice (19). Therapeutic use of Plumbagin has been limited due to toxicity, and poor aqueous solubility (19). Plumbagin has anti-bacterial, anti-fungal and anti-cancer properties by suppression of NF-κB, AKT/mTOR, STAT3, induction of ROS, p53, JNK and/or activation of the NRF2-ARE signaling pathway (20–23). Of these proteins, the key target modulated in cancers by Plumbagin is STAT3, which plays an important role in the development of 50–70% of many cancer types (20, 21, 23).
The COX-2 and STAT3 pathways inhibited by Celecoxib and Plumbagin respectively, are activated in 50 to 70% of melanoma patients, reducing cellular apoptosis, increasing proliferation and aiding an invasive phenotype (24, 25). An oral formulation of these agents is not feasible due to drug incompatibility involving solubility, bioavailability and toxicity (26–28). In this study, a single nanoparticle has been developed called CelePlum-777, containing Celecoxib and Plumbagin. Since the drugs are encapsulated in a lipid shell they are soluble when administered intravenously, the agents are released simultaneously from the nanoparticle at the synergizing ratio making them bioavailable at the optimized effective drug levels, and since the agents synergize, lower concentration of each would be required, thereby decreasing toxicity. CelePlum-777 inhibited xenograft melanoma tumor growth by up to 72% without apparent systemic toxicity. Mechanistically, the synergizing agents simultaneously inhibited COX-2 and STAT3 in melanomas, which enhanced the antiproliferative effect mediated by a more efficient reduction in key cyclins important in melanoma cell survival.
MATERIALS AND METHODS
Cell lines and culture conditions
Normal human primary melanocytes NHEM 558 was obtained Lonza in 2001, Walkersville, MD. Normal human fibroblast FF2441 and foreskin keratinocyte HFK was provided by Dr. Craig Myers between 2005–2006; Penn State College of Medicine, Hershey, PA. Mutant V600E-BRAF human melanoma cell line 1205 Lu was provided by Dr. Herlyn in 2003; Wistar Institute, Philadelphia, PA and UACC 903 was provided by Dr. Mark Nelson between 1995–1999; University of Arizona, Tucson, AZ. Wild type BRAF melanoma cell lines containing C8161.Cl9 was provided by Dr. Danny Welch in 2003; University of Kansas, Kansas City, KS) and MelJuSo was provided by Dr. Judith Johnson between 1995–1999; Institute for Immunology, Germany. Cell lines were maintained in a 37°C humidified 5% CO2 atmosphere incubator and periodically monitored for phenotypic and genotypic characteristics, and tumorigenic potential to validate and confirm cell line identity. Last assessment was within the last 6 months.
Screening to identify agents that synergize with Plumbagin
Plumbagin was screened together with in-house anticancer agents. For the initial screen, 5X103 UACC 903 melanoma cells were seeded into 96-well plates for 24 hours. Cells were then treated with 5 μmol/L of Plumbagin singly or in combination with the drug library at concentration ranging from 0.5 to 100 μmol/L for 72 hours. Cell viability was measured by MTS assay (Promega, Madison, WI) (29–31).
Synergy analysis when treating cultured cells with Plumbagin and Celecoxib dissolved in DMSO
UACC 903 cells were seeded into a 96-well plate at a density of 5X103 cells per well in 100 μL of media and grown for 48 hours. Cells were then treated with 30–50 μmol/L of Celecoxib (Sigma) or 5 μmol/L of Plumbagin (Sigma) singly or in combination for 72 hours. Cell viability was measured by MTS assay. Potential synergy between the drugs was assessed using the Chou-Talalay method to estimate the combination index (CI) using Calcusyn software (32, 33). The CI values of <0.9 were considered synergistic, >1.1 considered antagonistic, and values 0.9–1.1 considered as additive (32, 33).
Thermal stability of Plumbagin and Celecoxib
To determine whether these compounds would be stable during synthesis of nanoparticle, the thermal stability of Plumbagin and Celecoxib in DMSO solution was assessed at various time points at 60°C. 5X103 UACC 903 cells per well in 100 μL of media were seeded and grown in a 96-well plate for 48 hours and then treated with agents for 72 hours. Efficacy of the heat-treated compounds for killing UACC 903 melanoma cells was assessed by MTS assay.
Manufacture of CelePlum-777 containing Celecoxib and Plumbagin
Celecoxib and Plumbagin drugs alone or in combination at a 2.5:1, 5:1, 10:1 or 20:1 ratio were encapsulated into a nanoliposome called CelePlum-777 by combining L-α-Phosphatidylcholine (ePC) and 1,2-Dipalmitoyl-sn-Glycero-3-Phosphoethanolamine-N-[Methoxy(Polyethylene glycol)-2000] ammonium salt (DPPE-PEG-2000) in chloroform at 95:5 mol % for a final lipid concentration of 25 mg/mL (Avanti Polar Lipids). Solvent was removed and mixture dried under nitrogen gas followed by resuspension in sterile saline solution or water at 60°C with vortexing every 5 minutes over a 20-minute period followed by sonication and extrusion at 60°C through a 100-nm polycarbonate membrane using Avanti Mini Extruder (Avanti Polar Lipids). The particle size and zeta potential were measured using a Malvern Zetasizer (Malvern Instruments) (28).
Characterization of CelePlum-777
Drug encapsulation
Encapsulation of Celecoxib and Plumbagin singly or in combination at a ratio of 20:1 in the liposomal formulation was estimated by UV-visible spectrophotometry (SPECTRAmax M2 plate reader; Molecular devices) (34). Free drugs not incorporated into the nanoliposomes were separated using 10 kDa Centricon filter tube (Millipore). 1.0 mL of CelePlum-777 liposomal solution was taken in 10 kDa Centricon filter tube followed by centrifugation at 3,750 rpm for 30 minutes. Next, 0.5 mL of purified liposomal solution was combined with 0.5 mL of 1:1 ratio of chloroform and methanol to destroy the liposomal structure and release the drug into the solution. Following vortexing for 10 minutes, the precipitated lipids were separated following centrifugation at 10,000 rpm for 15 minutes. The supernatant was then used to measure the amount of each respective drug alone or in combination and concentrations were calculated from a standard curve of Plumbagin or Celecoxib ranging from 0.01 and 0.1 mg/mL. 1:1 ratio of chloroform and methanol was used as the reference blank. Percentage drug incorporated in the liposome was calculated as the free drug(s)/total drug(s)X100.
Stability
Stability of CelePlum-777 stored at 4°C was measured at various time intervals over 1 to 6 weeks. During this period, no aggregation or precipitation of the liposomes was observed. Assessing stability involved comparing size and zeta potential using the Malvern Zetasizer as well as assessing efficacy for killing UACC 903 melanoma cells using the MTS assay (28).
In vitro drug-release kinetics of CelePlum-777
The in vitro release kinetics of the compounds contained in CelePlum-777 was estimated at room temperature following dialysis through a molecular weight cut off 25 kDa membrane (Spectra Por, Los Angeles, CA) (35). 1.0 mL of purified CelePlum-777 solution in saline or water was placed into a dialysis membrane bag and suspended in 1 L of 10 mM reduced glutathione. 0.05 mL samples of the CelePlum-777 solution contained in the dialysis bag was removed at various time intervals (from 0.5 to 96 hours) and amount of Celecoxib and Plumbagin released at each time point estimated using UV-visible spectrophotometry as detailed in ref (34).
Assessment of cell viability, proliferation and apoptosis following treatment of CelePlum-777 in cultured melanoma cells
5X103 cells per well in 100 μL of media were seeded and grown in a 96-well plate for 48 or 72 hours respectively for those representing normal cells (NHEM 558-melanocytes, FF2441-fibroblast and HFK-keratinocytes) and melanoma cell lines (UACC 903, 1205 Lu, C8161.Cl9 and MelJuSo). Each cell line was treated with empty control liposome, Plumbagin liposome (5 μmol/L), Celecoxib liposome (100 μmol/L) or CelePlum-777 (containing 100 μmol/L Celecoxib + 5 μmol/L Plumbagin) for 24, 48, or 72 hours and viability measured by MTS assay (11, 36, 37). Rates of proliferation and apoptosis were measured by using a colorimetric cell proliferation ELISA BrdU kit (Roche diagnostics, Indianapolis, IN) or Apo-ONE Homogenous caspase-3/7 assay kit (Promega, Madison, WI), respectively. Data represent averages of at least 3 independent experiments; bars, S.E.M.
Western blot analysis
Cell lysates from UACC 903 or 1205 Lu melanoma cell lines treated with empty control liposome, 100 μmol/L Celecoxib liposome, 5 μmol/L Plumbagin liposome, or CelePlum-777 (containing 100 μmol/L Celecoxib + 5 μmol/L Plumbagin) for 6 or 24 hours were prepared in RIPA lysis buffer containing Halt Protease & Phosphatase Inhibitor Cocktail (Thermo Scientific) (29–31). Blots were probed with antibodies according to each supplier’s recommendations: antibodies to AKT (pan) (11E7), phospho AKT (S473), Cleaved PARP (Asp 214), phospho STAT3 (Y705), STAT3, cyclin H, cyclin B1, caspase 3, cyclin A2, cyclin E1, cyclin E2, cPLA2, and phospho cPLA2 (S505) from Cell Signaling Technology; cyclin D1, Erk2, p21, p27, COX-1, COX-2, alpha enolase and secondary antibodies conjugated with horseradish peroxidase from Santa Cruz Biotechnology. Immunoblots were developed using the enhanced chemiluminescence detection system or Super signal West Femto Chemiluminescent Substrate (Thermo Fisher Scientific).
Analysis of caspase-3/7 activity in total cell lysates
Caspase-3/7 activity in the cell lysates collected for Western blot analysis was used to determine the Apo-ONE homogenous caspase-3/7 activity assay kit (Promega Corporation) (36). In brief, 30–40 μg in 50 μL lysis buffer was incubated with caspase-3/7 substrate (R110-Z-DEVD dissolved in caspase-3/7 assay buffer) for 4 hours at 37°C with constant shaking in a light protected container. Amount of R110 released was determined using a SPECTRA max-M2 plate reader at a 485 nm excitation and 520 nm emission wavelengths. Average relative fluorescence units values from duplicate wells were plotted as a bar graph.
Tumorigenicity assessments
Animal experiments to assess the efficacy of drug treatment were performed according to protocols approved by the Institutional Animal Care and Use Committee at Penn State University. Tumor kinetics were measured by subcutaneous injection of 1X106 UACC 903 or 1205 Lu cells were injected above both left and right rib cages of 4–6 week-old female Athymic-Foxn1nu nude mice (Harlan Sprague Dawley) (28, 29). Six days later, when a fully vascularized tumor had formed, mice were randomly divided into 7 different groups and treated intravenously on alternate days for 3–4 weeks. Body weight in grams and dimensions of developing tumors in mm3 were measured on alternate days. Group 1 (Empty liposomes); Group 2 (Celecoxib liposome, 15 mg/Kg body weight); Group 3 (Plumbagin liposome, 0.75 mg/Kg body weight); Group 4 (Plumbagin liposome, 1.5 mg/Kg body weight); Group 5 (20:1 ratio of CelePlum-777; Celecoxib 15 mg/Kg + Plumbagin 0.75 mg/Kg body weight); Group 6 (10:1 ratio of CelePlum-777; Celecoxib 15 mg/Kg + Plumbagin 1.5 mg/Kg body weight) and Group 7 (10:1 ratio combined liposomes containing the individual agents of Celecoxib liposome; 15 mg/Kg + Plumbagin liposome; 1.5 mg/Kg body weight).
Size and time match tumors for analysis of biological processes regulating tumor development
2.5X106 of 1205 Lu cells were injected s.c. into nude mice, generating tumors of the same size developing at parallel time points. Six days later, mice were treated daily with liposomes containing Celecoxib at 15 mg/Kg body weight, Plumbagin at 1.5 mg/Kg body weight, or CelePlum-777 containing Celecoxib 15 mg/Kg + Plumbagin 1.5 mg/Kg body weight daily for up to 15 days. Empty liposome in saline was used as a vehicle control. Tumors were harvested at days 13 and 15 for comparison of rates of cellular proliferation, apoptosis, and vessel density by immunohistochemistry (38). Cell proliferation was measured using mouse anti-human Ki-67 antibody staining (Pharmigen). Apoptosis rates were measured using the terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) TMR Red Apoptosis kit (Roche). Vessel density indicative of angiogenesis was measured using a purified rat anti-mouse CD31 (PECAM-1) monoclonal antibody for immunostaining (Pharmingen). Numbers of Ki-67 or TUNEL stained cells were quantified as the percentage of total cells in tumors using the IP Lab imaging software program. Areas containing vessels were quantified and compared between tumor sections. For all tumor analyses, minimums of 4–6 different tumors with 4–6 fields per tumor section were analyzed.
Toxicity assessments
At the end of tumorigenicity assessment treatment, blood was collected from each euthanized animal in a serum separator tube with lithium heparin (BD Microtainer) following cardiac puncture and analyzed for levels of GLU (Glucose), BUN (Blood urea nitrogen), CREA (Creatinine), Phosphate, TP (Total Protein), CAL (Calcium), GLO (Globulin), ALT (Alanine aminotransferase), ALKP (Alkaline phosphatase), TBIL (Total bilirubin), CHOL (Total cholesterol), TRIG (Total triglyceride), AST (Aspartate aminotransferase) and AMY (Amylase) to demonstrate no vital organs toxicity. A portion of liver, heart, kidney, pancreas, spleen, intestine and stomach tissue from each animal was formalin-fixed and paraffin-embedded to examine changes in cell morphology and tissue organization following H&E staining (29–31).
Statistical Analysis
Statistical analysis was performed using Prism 4.0 GraphPad Software. One-way or Two-way Analysis of Variance (ANOVA) was used for group wise comparisons. For comparison between two groups, Student’s t test (2 tailed) was used. Results represent at least two to three independent experiments and are shown as averages ± S.E.M. Results with a P value less than 0.05 were considered significant. Sample sizes and number of times experiments were repeated are indicated in the figure legends. Number of asterisks in the figures indicates the level of statistical significance as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
RESULTS
Combining Celecoxib with Plumbagin led to synergistic killing of melanoma cells
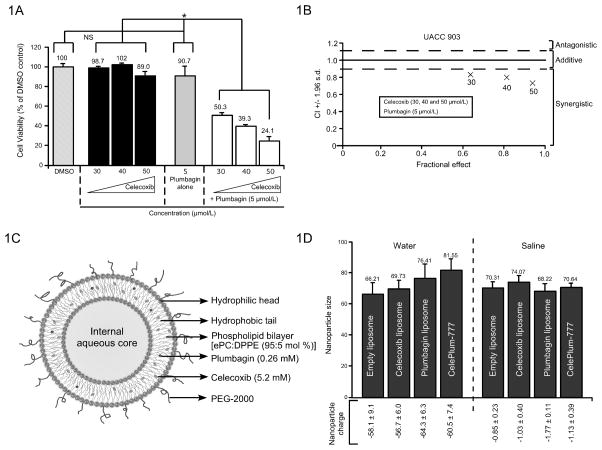
To identify an agent that synergizes with Plumbagin, a small library of compounds was screened to identify those that cooperatively to kill UACC 903 melanoma cells, which identified Celecoxib as a potential candidate (Data not shown). To further validate the initial screen, UACC 903 cells were treated with various concentration of Plumbagin (3–5 μmol/L) combined with 30, 40, 50 μmol/L Celecoxib for 72 hours. Simultaneous treatment with Plumbagin (3–5 μmol/L) or Celecoxib (30 to 50 μmol/L) killed UACC 903 cells while the single agents alone had a negligible effect (Fig. 1A & Supplemental Fig 1). To determine whether the killing effect was additive or synergistic, the Chou-Talalay method was used for determining the CI values using Calcusyn software (32). Using this approach, CI values <0.9 are considered synergistic, >1.1 are antagonistic, and values 0.9 to 1.1 are additive (32). CI values were 0.83, 0.80, and 0.74 for 30, 40, and 50 μmol/L of Celecoxib plus 5 μmol/L of Plumbagin, respectively, demonstrating a strong synergistic killing effect (Fig. 1B). Thus, treatment of cultured UACC 903 melanoma cells with Celecoxib and Plumbagin at ratios of 10:1 led to cooperatively synergistic inhibition. Based on this observation, ratios of 10:1 and 20:1 were selected for subsequent studies in cultured cells and in animals.
Figure 1. Development of synergistically acting CelePlum-777.
(1A). Celecoxib and Plumbagin inhibit the viability of UACC 903 cells in a cooperative manner as measured by MTS assay. Data represent averages of at least 3 independent experiments; bars, S.E.M. (1B). Calcusyn analysis suggest synergistic killing by Celecoxib and Plumbagin and CI values range from 0.74 to 0.83. (1C). Structure of CelePlum-777 showing predicted location of Celecoxib and Plumbagin within the liposome. (1D). Size and charge of CelePlum-777 dissolved in water or saline. Size and zeta potential of empty liposomes, Celecoxib or Plumbagin liposomes alone, or CelePlum-777. Data represent averages of at least 3 independent experiments; bars, S.E.M.
Development of a nanoliposome containing Celecoxib and Plumbagin at a drug ratio of 10:1 or 20:1 called CelePlum-777
Combining Celecoxib and Plumbagin for oral animal treatment is limited by solubility, bioavailability and toxicity (15, 39). Furthermore, solubilizing the agents in DMSO for intravenous administration would cause animal lethality and use for intraperitoneal or subcutaneous administration is not a viable option for human use. Therefore, encapsulation of both agents into a single nanoparticle was selected as an approach to overcome poor bioavailability, toxicity and solubility issues related to these agents (40). To create a viable nanoparticle containing both Celecoxib and Plumbagin certain feasibility assessment needed to be undertaken. These involved assessing compound thermal stability, identifying the optimal liposomal lipid composition and determining whether sufficient amount of each compound at the optimal synergizing ratio could be loaded into the lipid shell of the liposome. Agent thermostability was measured by heating Plumbagin or Celecoxib at 60°C overtime and then measuring efficacy for killing UACC 903 cells by MTS assay (Supplemental Fig. 2). Celecoxib could be heated for up to 30 minutes without decreasing efficacy (Supplemental Fig. S2A) and prolonged heating of Plumbagin did not affect its inhibitory activity (Supplemental Fig. S2B).
To identify the optimal lipid formulation to enable loading of sufficient quantities of Celecoxib or Plumbagin into liposomes, several formulations were evaluated and results assessed for size, zeta potential, membrane fluidity and surface hydration (28). A PEGylated liposomal system made of 95:5 mol % for ePC: DPPE PEG-2000 was selected to form a stable liposome ~70 nm in size. To demonstrate that sufficient quantities of Celecoxib and Plumbagin could be loaded into the lipid shell of the liposomes, individual Celecoxib liposomes were made containing 2, 3 and 4 mg of compound (Supplemental Table 1). Those made with 3 and 4 mg crashed, which set the maximal amount of Celecoxib that could be loaded. Liposomes containing 0.2, 0.4 and 0.8 mg of Plumbagin, were all viable (Supplemental Table 1). Based on these results, it was predicted that a liposomal formulation could be made containing 2 mg of Celecoxib and 0.1 mg of Plumbagin giving a ratio of 20:1 or 2 mg of Celecoxib and 0.2 mg of Plumbagin giving a ratio of 10:1 (Supplemental Table 1). The same lipid formulation was also found to be suitable for making a control empty liposome, one containing Celecoxib or Plumbagin alone and one containing both drugs at ratios of 10:1 or 20:1 was optimized to attain the best drug ratio for maximal potency and tested on the UACC 903 melanoma cell line (Supplemental Fig. 3). These liposomes were made and dissolved in saline or water and found to be viable, which is diagrammatically shown in Fig. 1C. Size of the liposomes in water ranged from 66 to 81 nm and zeta potential charge was −56 to −64 mV, while in saline the size ranged from 68 to 74 nm with a zeta potential charge of −0.9 to −1.8 mV (Fig. 1D).
Physiochemical and stability characterization of CelePlum-777
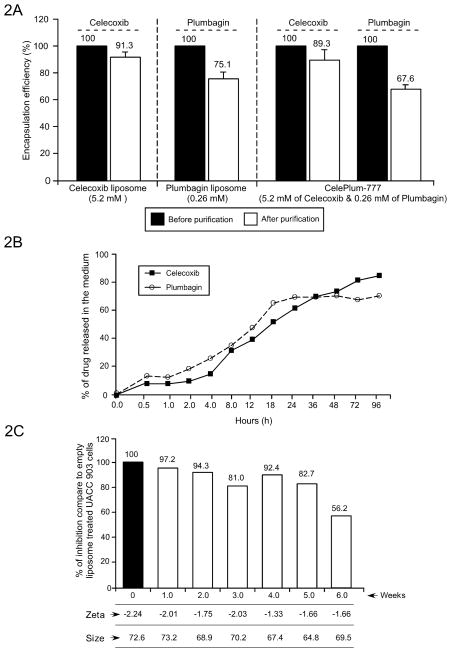
The entire drug was solubilized during liposome manufacture but a certain amount was loosely bound to the exterior lipids of the nanoparticle. To measure the proportion encapsulated in the lipid shell, free loosely bound or free drug was removed by Centricon Ultra centrifugation followed by lysis of the liposomes. The amount of each drug in the lipid shell was then estimated by determining the quantity present at the absorption maxima of the compound extrapolated from a standard curve of Celecoxib or Plumbagin ranging from 0.01 and 0.1 mg/mL. Encapsulation for Celecoxib or Plumbagin in liposomes containing each agent alone was 91 and 75%, respectively (Fig. 2A). Encapsulation of Celecoxib and Plumbagin in CelePlum-777 liposomes was 89.3 % and 67.6%, respectively (Fig. 2A). To demonstrate that the nanoparticles releases the drugs at similar ratios to maintain the 20:1 or 10:1 synergistic killing ratio, drug release kinetics was examined by dialyzing CelePlum-777 in 10 mM glutathione (GSH) over 96 hours, the liposome lysed and drugs remaining estimated from a standard curve (Fig. 2B). Celecoxib and Plumbagin release occurred at a steady rate over 96 hours (Fig. 2B) and since the drugs were loaded at 10:1 and 20:1 ratios, the similar release kinetics suggested that dugs would be freed from the nanoparticle at a rate that would maintain the synergistic effects of the agents. Long-term storage of CelePlum-777 was measured by storage in sterile saline at 4°C and sampled over 6 weeks to assess size, zeta potential, and efficacy for killing UACC 903 melanoma cells. Size and charge remained consistent over 6-weeks. For up to 5 weeks CelePlum-777 retained killing efficacy above 80%, but at week 5, killing efficacy decreased to 56% (Fig. 2C). Thus, CelePlum-777 can be stored at 4°C for up to 5 weeks without changes in the physiochemical or killing properties.
Figure 2. Characterization of drug encapsulation, stability, and release kinetics of CelePlum-777.
(2A). Drug encapsulation efficiency. Nanoliposomes containing Celecoxib alone (5.2 mM), Plumbagin alone (0.26 mM), or the combination (5.2 mM Celecoxib and 0.26 mM Plumbagin), Plumbagin encapsulation alone was 75.1%, Celecoxib encapsulation alone was 91.3% and the encapsulation of both agents was 67.6% and 89.3%, respectively. Data represent averages of at least 3 independent experiments; bars, S.E.M. (2B). Drug release kinetics of CelePlum-777. 71% of Celecoxib and 69% of Plumbagin was released from CelePlum-777 over 96 hours. (2C). Stability of CelePlum-777. CelePlum-777 was stored at 4°C and stability measured over weekly assessing size, charge, and cancer cell killing efficacy indicating stability for up to 5-weeks. Data represent averages of at least 2 independent experiments.
Comparing killing efficacy of CelePlum-777 to nanoliposomes containing individual drugs
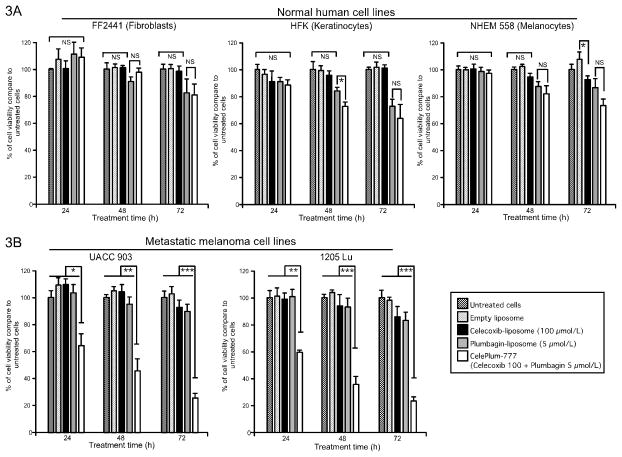
Efficacy of 20:1 ratio of CelePlum-777 for killing normal cells (NHEM 558-melanocytes, FF2441-fibroblast and HFK-keratinocytes) (Fig. 3A) and melanoma cells UACC 903 or 1205 Lu (Fig. 3B) were compared to empty liposomes or those containing only Celecoxib or Plumbagin for 24, 48, and 72 hours by MTS assay. For normal cell lines (the fibroblasts, keratinocytes or melanocytes), a general observation was that at least one of the single agents led to a decrease in cell viability that was equivalent to the killing observed with the drug combination (Fig. 3A). Only for keratinocytes at the 48-hour time point was there a slight difference in cell killing for the drug combination that was statistically significant from the Plumbagin treatment on its own. Since this only occurred at the 48-hour time point and not at the other two times, this was attributed to a slight variance in experimental outcome. Therefore, one of the two drugs deceased normal cell proliferation alone or in combination with the other agent and the killing was not synergistic. In the case of the melanoma cell lines, the combined agents were in every case significantly more effective in killing the melanoma cells than both of the agents on their own, suggesting co-operation in a synergistic fashion. Thus, there was no synergistic killing of the normal cell but it did occur for the melanoma cell lines where the killing for the combined drugs was clearly synergistic (Fig. 3A). In contrast, melanoma cells treated with CelePlum-777 were more effectively killed than those treated with empty liposomes or those containing Celecoxib or Plumbagin alone (Fig. 3B). Comparable results were also obtained with C8161.Cl9 (wild type BRAF) and MelJuSo (wild type BRAF and NRAS mutation) melanoma cell lines suggesting that synergistic killing occurred independently of the BRAF mutation status (Supplemental Fig. 4).
Figure 3. Efficacy of CelePlum-777 compared to nanoliposomes containing individual drugs.
Normal (3A) or melanoma cell lines (3B) were treated with empty liposome, Celecoxib liposome (100 μmol/L), Plumbagin liposome (5 μmol/L), or CelePlum-777 (100 μmol/L Celecoxib + 5 μmol/L Plumbagin) for 24, 48, and 72 hours and cell survival assessed by MTS assay. Data represent averages of at least 3 independent experiments; bars, S.E.M.
CelePlum-777 inhibited melanoma tumor growth with negligible toxicity
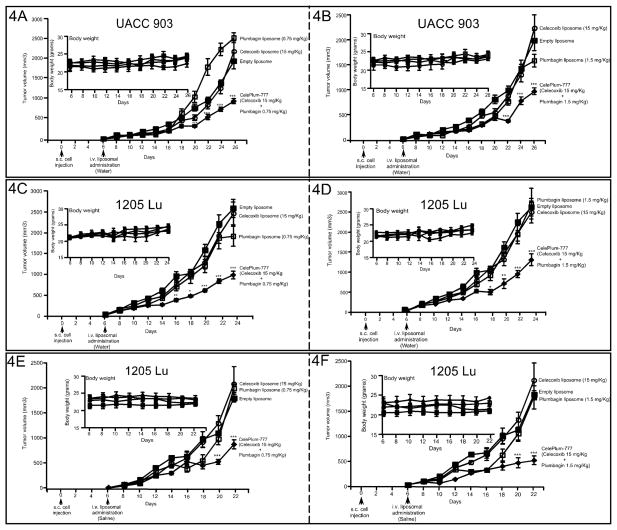
To determine whether CelePlum-777 synergistically inhibited melanoma tumor development, nude mice containing vascularized tumors were treated intravenously with 20:1 ratio of CelePlum-777 (15 mg/Kg Celecoxib + 0.75 mg/Kg Plumbagin) or 10:1 CelePlum-777 (15 mg/Kg Celecoxib + 1.5 mg/Kg Plumbagin) based on body weight, on alternate days. CelePlum-777 administered in water decreased tumor growth by up to 62% in both UACC 903 (Figs. 4A & 4B) and 1205 Lu (Figs. 4C & 4D) xenografts (P < 0.001, two-way analysis of variance). Resuspending CelePlum-777 in saline (Figs. 4E & 4F) did not alter the tumor inhibitory efficacy leading to tumor inhibition by up to 72%. CelePlum-777 treatment at either the 10:1 or 20:1 drug ratio did not alter animal body weight, suggesting negligible toxicity (Figs. 4A to 4F; insets). No significant changes in serum parameters indicative of major organ toxicity were observed (Supplemental Table 2). Analysis of H & E stained tissue sections comparing control or CelePlum-777 treated mice showed no changes in cellular morphology or architecture of liver, heart, kidney, spleen, and intestine (Supplemental Fig. 5). Collectively, these results suggested that CelePlum-777 inhibits xenografted melanoma tumor growth without significant organ related toxicity.
Figure 4. CelePlum-777 treatment synergistically inhibited melanoma tumor growth.
Vascularized xenografts of UACC 903 (A and B) or 1205 Lu (C, D, E, and F) melanoma cells were treated intravenously on alternate days with liposomes containing single agents (Celecoxib 15 mg/Kg or Plumbagin 0.75 or 1.5 mg/Kg body weight), or CelePlum-777 (Celecoxib 15 mg/Kg + Plumbagin 0.75 or 1.5 mg/Kg body weight) for 3 to 4 weeks. The line graph indicates tumor volume (mm3) and inset body weight. Data represent experiments of 3 mice per group, containing two tumors per mouse; bars, S.E.M.
Tumor inhibitory efficacy of CelePlum-777 was next compared to nanoliposomes containing only Celecoxib or Plumbagin that were then combined at the synergizing 10:1 ratio. The purpose was to demonstrate that CelePlum-777 was as effective as the combined liposomes containing the individual agents. CelePlum-777 or liposomes containing Celecoxib and Plumbagin at the 10:1 ratio led to similar levels of tumor inhibition (Supplemental Fig. 6).
CelePlum-777 led to enhanced inhibition of melanoma cell proliferation
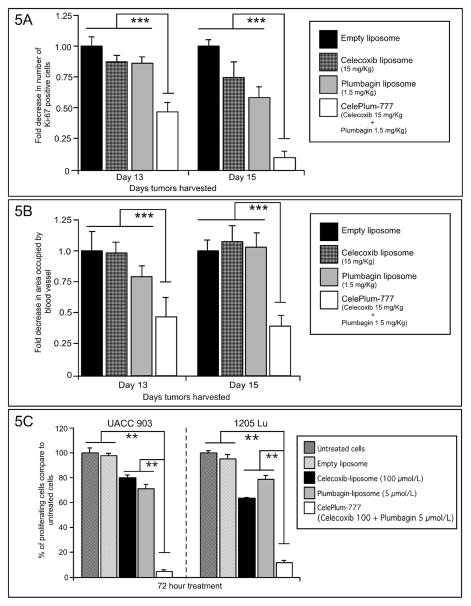
To identify the underlying mechanism by which CelePlum-777 synergistically inhibited melanoma tumor growth, an established published approach was used (38, 41). It involved quantifying the rates of cell proliferation (using Ki-67 staining), apoptosis (using TUNEL staining), and tumor angiogenesis (using CD31 staining) occurring in time and size matched tumors treated with 10:1 ratio of CelePlum-777 compared with control-exposed animals. Size and time matched tumors at days 13 and 15 were compared to identify statistically quantifiable differences in cell proliferation, apoptosis or vascular development between CelePlum-777 treatments compared to liposomes containing the individual drugs. At day 13, a statistically significant reduction of approximately 50% in proliferating cells (Fig. 5A) and vascular development (Fig. 5B) was observed (P < 0.001, two-way analysis of variance). In contrast, there was no significant change in TUNEL positive cells in CelePlum-777 treated tumors compared with individual controls (Supplemental Fig. 7). To further confirm the growth inhibition of cultured cells after treatment with CelePlum-777 compared to controls, the rates of cell proliferation was measured after 72 hours of treatment. Treatment with liposomes containing Celecoxib or Plumbagin reduced melanoma cell proliferation by 20 to 35%, while nanoparticles containing both agents decreased cell survival by 90–95% (Fig. 5C). Thus, CelePlum-777 cooperatively decreased the proliferative potential of melanoma cells compared to the individual agents in both tumors and cultured cells, suggesting this process led to the synergistic effects on tumor development. Apoptosis was only evident in cultured cells and not tumors, suggesting it was not key in the tumor inhibitory process.
Figure 5. CelePlum-777 treatment consistently led to enhanced inhibition of tumor and cultured cell proliferation.
Size and time matched xenografted tumors were removed from mice on days 13 and 15, following treatment from day 6 with liposomes containing Celecoxib at 15 mg/Kg body weight, Plumbagin at 1.5 mg/Kg body weight, or CelePlum-777 containing Celecoxib 15 mg/Kg + Plumbagin 1.5 mg/Kg body weight. Tumor sections were immunostained for Ki-67 (5A) or CD31 (5B) to assess proliferation and vascular development, respectively. Images were quantified and plotted as fold difference in cells expressing Ki-67 or area occupied by blood vessel compared with controls. Data was obtained from three to four tumors, with four to five fields averaged per tumor. Data represent averages of at least 3 independent experiments; bars, S.E.M. Rates of cell proliferation in cultured cells (5C) following CelePlum-777 treatment for 72 hours reduced melanoma cell proliferation compared to liposomes containing Celecoxib or Plumbagin alone.
CelePlum-777 inhibited the activity of STAT3 and COX-2 to co-operatively decrease key cyclins levels to retard melanoma cell proliferation
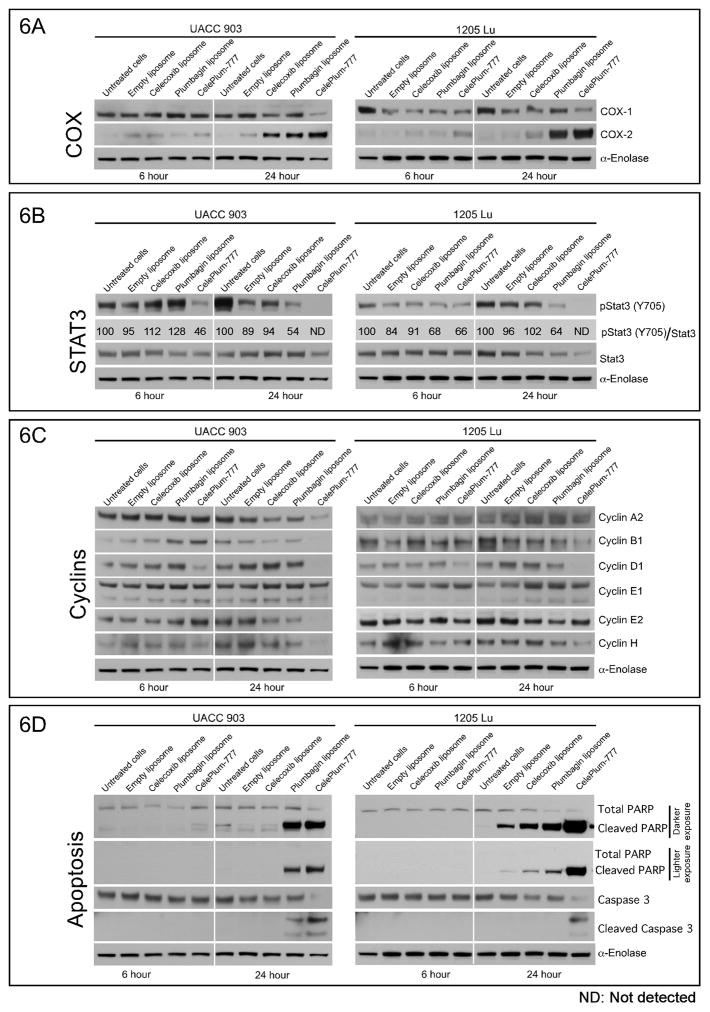
To identify the synergistic effects mediated by CelePlum-777 on the signaling pathways compared to Celecoxib or Plumbagin alone, cells were treated with liposomes containing each agent alone or CelePlum-777. CelePlum-777 had negligible effects on the signaling of other pathways including those of the AKT and cPLA2 signaling pathways (Supplemental Table 3). The drug combination consistently led to increased COX-2 levels. For UACC 903 cells at the 24-hour time point, Celecoxib and Plumbagin increased COX-2 levels and the effect for the combination treatment appeared to be additive. It is unclear why Plumbagin increased COX-2 levels but this also occurred for the 1205 Lu cell line at 24-hour. Celecoxib did increase COX-2 levels in 1205 Lu cells but to a significantly lesser extent. Thus, these data suggest that Plumbagin might also be inhibiting COX-2 signaling, which in turn increases COX-2 levels but this is purely speculative. However, it is accepted that when Celecoxib binds to and inhibits the activity of COX-2 there is a compensatory increase in protein expression to alleviate the inhibition(11, 42–45). COX-2 assays following treatment with Celecoxib have shown reduced activity when the protein is inhibited with Celecoxib and there is this compensatory increase in COX-2 protein levels (11). Therefore, the increase in protein expression has been used to indicate effective inhibition of COX-2 activity and is a compensatory response to inhibition (11, 42–45) (Fig. 6A and Supplemental Table 3).
Figure 6. CelePlum-777 decreased STAT3 and COX-2 activity leading to an enhanced decrease in key cyclin levels.
A, B, and C. UACC 903 or 1205 Lu melanoma cells were treated with empty liposome, Celecoxib liposome (100 μmol/L), Plumbagin liposome (5 μmol/L), or CelePlum-777 (100 μmol/L Celecoxib + 5 μmol/L Plumbagin) for 6 and 24 hours. Western blotting measure changes in protein expression of (6A) COX-2, (6B) STAT3 (6C) Cyclins and (6D) Apoptosis signaling pathways. Quantified pSTAT3 levels were normalized over STAT3 levels using ImageJ software.
The drug combination significantly decreased the protein levels of pSTAT3 (Y705) at the 24-hour time points (Fig. 6B), and that of certain cyclins (Fig. 6C & Supplemental Table 3) in both UACC 903 and 1205 Lu cells. Results of the scanned and quantified pSTAT3 levels were normalized over STAT3 levels show that for UACC 903 cells there is a pSTAT3 range from 95 to 128 in phosphorylation levels at 6 hours, which is likely just normal variation with this type of analysis and suggests it is probably not changing. At 24 hours, STAT3 phosphorylation levels are not detectable for the drug combination, while the range is 89 to 54 for the control and single agent treatments. Clearly this change is significant. Furthermore, the results for the 1205 Lu cell line likewise show a similar trend at 6 hours and also show no detectable STAT3 phosphorylation at 24 hours for the drug combination compared to the individual drug controls. In both melanoma cell lines, cyclins B1, D1 and H were cooperatively decreased after 24 hours of exposure to the drug combination. In addition to reductions of these cyclins in UACC 903 cells, cyclin A2, E1, and E2 were also decreased in one but not both cell lines. Since cyclins are key to the functioning of the cyclin-cyclin dependent kinase complex in melanoma cells, reductions in the levels of these proteins, which were more prominent following treatment with CelePlum-777 than with Celecoxib or Plumbagin alone, could account for the reduction of cell proliferation seen in cultured cells and in animal tumors. Increase in cleaved caspase-3 and PARP protein levels in cultured cells at late time points suggest this phenomenon might be restricted to cells growing in these conditions and not to tumors in the in vivo environment (Fig. 6D). In addition, CelePlum-777 caused significant cultured cell death, which consequently induced caspase-3/7 activity. Similar results were observed for protein lysates derived from cultured UACC 903 and 1205 Lu cells treated with agent alone or CelePlum-777 and analyzed for Apo-ONE homogenous caspase-3/7 activity (Supplemental Fig. 8). This was not observed in tumors (Figs. 5 &6), suggesting it did not occur in vivo but was a cell culture only phenomenon.
DISCUSSION
Malignant melanoma is an aggressive form of skin cancer due to its high metastatic nature and drug resistance (46). Despite the approval of V600E-BRAF inhibitors, Zelboraf (Vemurafenib), Tafinlar (Dabrafenib); and a Mek inhibitor, Mekinist (Trametinib) by the FDA for treating patients with MAPK signaling cascade activation (47). These targeted therapy approaches are hindered by drug resistance eventually leading to more aggressive disease development (48). Studies have shown that resistance develops by compensatory reactivation of MAPK signaling (49). Therefore, the development of novel agents are needed to decrease the possibility of resistance development and one approach to accomplish this objective is to develop single nanoparticle based agents containing multiple synergistically acting drugs (6, 50, 51).
Nanotechnology has the potential to delay drug resistance by delivering multiple synergizing drugs to a tumor to kill the cells more effectively than single agents (6, 7). Use of a single nanoparticle containing two synergizing drugs is an attractive therapeutic strategy because it has numerous merits over conventional therapy, including being considered as a single drug product by the FDA (6, 7, 50, 52). Combination pharmacological agent therapies can be limited because drugs can have different solubility, bioavailability and toxicity profiles, which creates complications in dosing and scheduling optimization (53). This study details the development of a nanoliposomal-based agent containing Celecoxib and Plumbagin, called CelePlum-777, which is stable and releases these drugs at an optimal ratio for maximal synergistic killing efficacy. Combining Celecoxib with Plumbagin in animals or humans is not possible because of poor bioavailability and intravenous administration in DMSO is lethal necessitating the development of a clinically viable approach (54). Encapsulating both agents at a synergistically killing drug ratio in a nanoparticle resolved these issues, since both were solubilized in a single vehicle that could be administered intravenously (40). The nanoparticle loaded and delivered the two drugs at synergizing ratios to inhibit tumor development at a significantly lower concentrations than required when using the single agents (52, 55). Furthermore, the probability of developing recurrent resistant disease would be less than if a single agent was used (52, 56). A final added advantage is that a nanoparticle delivery system has the added potential or extending the drug circulation half-life and promoting accumulation in the tumor due to the Enhanced Permeability and Retention (EPR) effect (57).
Very little is known regarding the development and efficacy of nanoparticles containing multiple drugs targeting key pathways important in melanoma development (52, 55). Typically, lower concentrations of synergizing drugs are also needed for efficacy, which occurred with CelePlum-777. Celecoxib induces cultured cancer cells apoptosis at 25 to 100 μmol/L, which is equivalent to 200 mg per day in animals but this concentration cause negative cardiovascular side effects (15, 16). Plumbagin has toxic effects making the use of low concentration important (19). CelePlum-777 enabled use of lower concentrations of each agent, but retained efficacy due to the synergism observed with the drug combination. CelePlum-777 had a negligible killing effect on normal cells and did not cause apparent systemic toxicity in mice.
CelePlum-777 synergistically inhibited xenografted melanoma tumor development by targeting COX-2 and STAT3, which are important pathways in melanoma development (8, 11, 24, 25, 58). These signaling pathways are constitutively activated in the majority of melanomas, functioning to reduce cellular apoptosis, increase proliferation and aid the invasive processes to promote melanoma metastasis development (8, 11, 24, 25, 58). It is important to note that CelePlum-777 or the individual drugs had negligible effects on proteins and pathways reported to be regulated by these agents in other cancer types, including AKT, NFkB, cPLA2, and COX-1 (8, 11, 24, 25, 58). These pathways were not inhibited or involved in the synergistic inhibition mediated by CelePlum-777. In contrast, modulating COX-2 and STAT3 led to consistent decrease in cyclins that are key to the functioning of the cyclin-cyclin dependent kinase complex in melanoma cells (59). Cyclin D1, a regulatory protein transcriptionally activated by the STAT3 signaling pathway, can promote cellular proliferation and metastasis when overexpressed (60). Cyclin A2, cyclin B1 and cyclin H are all factors that can drive cells through G2/M transition (61–64) and decreased levels seem to contribute to the arrest of cells in the G2/M phase of the cell cycle. Targeting multiple pathways in this manner might more effectively treat cancer without the development of recurrent resistant disease, making this a significant development (24, 65–68).
In summary, CelePlum-777 is a nanoparticle containing Celecoxib and Plumbagin that functions synergistically to reduce melanoma tumor growth with negligible systemic toxicity. It has a unique mechanism of action by simultaneously targeting COX-2 and STAT3 pathways, leading to decreased levels of several key cyclins important in melanoma development, which might more effectively treat cancer without development of recurrent resistant disease (69).
Supplementary Material
Acknowledgments
Grant support: NIH grants R01 CA-136667-02, RO1 CA-1138634-02, RO1 CA-127892-01A and The Foreman Foundation for Melanoma (Gavin Robertson), The Geltrude Foundation (Gavin Robertson), The Penn State Melanoma and Skin Cancer Center (Raghavendra Gowda), J. Roland, Mary R Elizabeth A Gilbert Memorial Trust (Raghavendra Gowda), The James Paul Sutton Medical Research Fund (Raghavendra Gowda), The Penn State Chocolate Tour Fund (Raghavendra Gowda & Gavin Robertson).
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Haarberg HE, Smalley KS. Resistance to Raf inhibition in cancer. Drug discovery today Technologies. 2014;11:27–32. doi: 10.1016/j.ddtec.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Held MA, Langdon CG, Platt JT, Graham-Steed T, Liu Z, Chakraborty A, et al. Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer discovery. 2013;3:52–67. doi: 10.1158/2159-8290.CD-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahan A, Miller JM. The solubility-permeability interplay and its implications in formulation design and development for poorly soluble drugs. The AAPS journal. 2012;14:244–51. doi: 10.1208/s12248-012-9337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley ST, Frank KJ, Fricker G, Brandl M. Biopharmaceutical classification of poorly soluble drugs with respect to “enabling formulations”. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2013;50:8–16. doi: 10.1016/j.ejps.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Jia J, Zhu F, Ma X, Cao Z, Li Y, Chen YZ. Mechanisms of drug combinations: interaction and network perspectives. Nature reviews Drug discovery. 2009;8:111–28. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 6.Brys AK, Gowda R, Loriaux DB, Robertson GP, Mosca PJ. Nanotechnology-based strategies for combating toxicity and resistance in melanoma therapy. Biotechnology advances. 2016 doi: 10.1016/j.biotechadv.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Gowda R, Jones NR, Banerjee S, Robertson GP. Use of Nanotechnology to Develop Multi-Drug Inhibitors For Cancer Therapy. Journal of nanomedicine & nanotechnology. 2013:4. doi: 10.4172/2157-7439.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawk ET, Viner JL, Dannenberg A, DuBois RN. COX-2 in cancer--a player that’s defining the rules. J Natl Cancer Inst. 2002;94:545–6. doi: 10.1093/jnci/94.8.545. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh N, Chaki R, Mandal V, Mandal SC. COX-2 as a target for cancer chemotherapy. Pharmacol Rep. 2010;62:233–44. doi: 10.1016/s1734-1140(10)70262-0. [DOI] [PubMed] [Google Scholar]
- 10.Higashi Y, Kanekura T, Kanzaki T. Enhanced expression of cyclooxygenase (COX)-2 in human skin epidermal cancer cells: evidence for growth suppression by inhibiting COX-2 expression. International journal of cancer Journal international du cancer. 2000;86:667–71. doi: 10.1002/(sici)1097-0215(20000601)86:5<667::aid-ijc10>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 11.Gowda R, Madhunapantula SV, Desai D, Amin S, Robertson GP. Simultaneous targeting of COX-2 and AKT using selenocoxib-1-GSH to inhibit melanoma. Mol Cancer Ther. 2013;12:3–15. doi: 10.1158/1535-7163.MCT-12-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–86. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 13.Greene ER, Huang S, Serhan CN, Panigrahy D. Regulation of inflammation in cancer by eicosanoids. Prostaglandins & other lipid mediators. 2011;96:27–36. doi: 10.1016/j.prostaglandins.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nachat-Kappes R, Pinel A, Combe K, Lamas B, Farges MC, Rossary A, et al. Effects of enriched environment on COX-2, leptin and eicosanoids in a mouse model of breast cancer. PloS one. 2012;7:e51525. doi: 10.1371/journal.pone.0051525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies NM, McLachlan AJ, Day RO, Williams KM. Clinical pharmacokinetics and pharmacodynamics of celecoxib: a selective cyclo-oxygenase-2 inhibitor. Clinical pharmacokinetics. 2000;38:225–42. doi: 10.2165/00003088-200038030-00003. [DOI] [PubMed] [Google Scholar]
- 16.Schonthal AH, Chen TC, Hofman FM, Louie SG, Petasis NA. Celecoxib analogs that lack COX-2 inhibitory function: preclinical development of novel anticancer drugs. Expert opinion on investigational drugs. 2008;17:197–208. doi: 10.1517/13543784.17.2.197. [DOI] [PubMed] [Google Scholar]
- 17.Hafeez BB, Zhong W, Fischer JW, Mustafa A, Shi X, Meske L, et al. Plumbagin, a medicinal plant (Plumbago zeylanica)-derived 1,4-naphthoquinone, inhibits growth and metastasis of human prostate cancer PC-3M-luciferase cells in an orthotopic xenograft mouse model. Mol Oncol. 2012 doi: 10.1016/j.molonc.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomathinayagam R, Sowmyalakshmi S, Mardhatillah F, Kumar R, Akbarsha MA, Damodaran C. Anticancer mechanism of plumbagin, a natural compound, on non-small cell lung cancer cells. Anticancer research. 2008;28:785–92. [PubMed] [Google Scholar]
- 19.Sand JM, Bin Hafeez B, Jamal MS, Witkowsky O, Siebers EM, Fischer J, et al. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone), isolated from Plumbago zeylanica, inhibits ultraviolet radiation-induced development of squamous cell carcinomas. Carcinogenesis. 2012;33:184–90. doi: 10.1093/carcin/bgr249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XQ, Yang CY, Rao XF, Xiong JP. Plumbagin shows anti-cancer activity in human breast cancer cells by the upregulation of p53 and p21 and suppression of G1 cell cycle regulators. European journal of gynaecological oncology. 2016;37:30–5. [PubMed] [Google Scholar]
- 21.Hafeez BB, Fischer JW, Singh A, Zhong W, Mustafa A, Meske L, et al. Plumbagin Inhibits Prostate Carcinogenesis in Intact and Castrated PTEN Knockout Mice via Targeting PKCepsilon, Stat3, and Epithelial-to-Mesenchymal Transition Markers. Cancer prevention research. 2015;8:375–86. doi: 10.1158/1940-6207.CAPR-14-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan W, Tu B, Liu YY, Wang TY, Qiao H, Zhai ZJ, et al. Suppressive Effects of Plumbagin on Invasion and Migration of Breast Cancer Cells via the Inhibition of STAT3 Signaling and Down-regulation of Inflammatory Cytokine Expressions. Bone research. 2013;1:362–70. doi: 10.4248/BR201304007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafeez BB, Jamal MS, Fischer JW, Mustafa A, Verma AK. Plumbagin, a plant derived natural agent inhibits the growth of pancreatic cancer cells in in vitro and in vivo via targeting EGFR, Stat3 and NF-kappaB signaling pathways. International journal of cancer Journal international du cancer. 2012;131:2175–86. doi: 10.1002/ijc.27478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005;24:315–27. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- 25.Becker MR, Siegelin MD, Rompel R, Enk AH, Gaiser T. COX-2 expression in malignant melanoma: a novel prognostic marker? Melanoma Res. 2009;19:8–16. doi: 10.1097/CMR.0b013e32831d7f52. [DOI] [PubMed] [Google Scholar]
- 26.Montaguti P, Melloni E, Cavalletti E. Acute intravenous toxicity of dimethyl sulfoxide, polyethylene glycol 400, dimethylformamide, absolute ethanol, and benzyl alcohol in inbred mouse strains. Arzneimittel-Forschung. 1994;44:566–70. [PubMed] [Google Scholar]
- 27.Sharma A, Mayhew E, Bolcsak L, Cavanaugh C, Harmon P, Janoff A, et al. Activity of paclitaxel liposome formulations against human ovarian tumor xenografts. International journal of cancer Journal international du cancer. 1997;71:103–7. doi: 10.1002/(sici)1097-0215(19970328)71:1<103::aid-ijc17>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 28.Gowda R, Madhunapantula SV, Sharma A, Kuzu OF, Robertson GP. Nanolipolee-007, a novel nanoparticle-based drug containing leelamine for the treatment of melanoma. Mol Cancer Ther. 2014;13:2328–40. doi: 10.1158/1535-7163.MCT-14-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuzu OF, Gowda R, Sharma A, Robertson GP. Leelamine mediates cancer cell death through inhibition of intracellular cholesterol transport. Mol Cancer Ther. 2014;13:1690–703. doi: 10.1158/1535-7163.MCT-13-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gowda R, Madhunapantula SV, Kuzu OF, Sharma A, Robertson GP. Targeting multiple key signaling pathways in melanoma using leelamine. Mol Cancer Ther. 2014;13:1679–89. doi: 10.1158/1535-7163.MCT-13-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gowda R, Madhunapantula SV, Desai D, Amin S, Robertson GP. Selenium-containing histone deacetylase inhibitors for melanoma management. Cancer biology & therapy. 2012;13:756–65. doi: 10.4161/cbt.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 33.Ashton JC. Drug combination studies and their synergy quantification using the Chou-Talalay method--letter. Cancer Res. 2015;75:2400. doi: 10.1158/0008-5472.CAN-14-3763. [DOI] [PubMed] [Google Scholar]
- 34.Panwar P, Pandey B, Lakhera PC, Singh KP. Preparation, characterization, and in vitro release study of albendazole-encapsulated nanosize liposomes. International journal of nanomedicine. 2010;5:101–8. doi: 10.2147/ijn.s8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng S, Chang S, Lu J, Chen Z, Xie L, Nie Y, et al. Characterization of 9-nitrocamptothecin liposomes: anticancer properties and mechanisms on hepatocellular carcinoma in vitro and in vivo. PloS one. 2011;6:e21064. doi: 10.1371/journal.pone.0021064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madhunapantula SV, Hengst J, Gowda R, Fox TE, Yun JK, Robertson GP. Targeting sphingosine kinase-1 to inhibit melanoma. Pigment Cell Melanoma Res. 2012;25:259–74. doi: 10.1111/j.1755-148X.2012.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma A, Madhunapantula SV, Gowda R, Berg A, Neves RI, Robertson GP. Identification of Aurora Kinase B and WEE1 as Downstream Targets of (V600E)B-RAF in Melanoma. Am J Pathol. 2013 doi: 10.1016/j.ajpath.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahl JM, Cheung M, Sharma A, Trivedi NR, Shanmugam S, Robertson GP. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res. 2003;63:2881–90. [PubMed] [Google Scholar]
- 39.Rodrigues SV, Viana LM, Baumann W. UV/Vis spectra and solubility of some naphthoquinones, and the extraction behavior of plumbagin from Plumbago scandens roots in supercritical CO2. Analytical and bioanalytical chemistry. 2006;385:895–900. doi: 10.1007/s00216-006-0502-6. [DOI] [PubMed] [Google Scholar]
- 40.Alavijeh MS, Palmer AM. The pivotal role of drug metabolism and pharmacokinetics in the discovery and development of new medicines. IDrugs : the investigational drugs journal. 2004;7:755–63. [PubMed] [Google Scholar]
- 41.Sharma A, Sharma AK, Madhunapantula SV, Desai D, Huh SJ, Mosca P, et al. Targeting Akt3 signaling in malignant melanoma using isoselenocyanates. Clin Cancer Res. 2009;15:1674–85. doi: 10.1158/1078-0432.CCR-08-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munkarah AR, Genhai Z, Morris R, Baker VV, Deppe G, Diamond MP, et al. Inhibition of paclitaxel-induced apoptosis by the specific COX-2 inhibitor, NS398, in epithelial ovarian cancer cells. Gynecologic oncology. 2003;88:429–33. doi: 10.1016/s0090-8258(03)00084-2. [DOI] [PubMed] [Google Scholar]
- 43.Hinz B, Ramer R, Eichele K, Weinzierl U, Brune K. Up-regulation of cyclooxygenase-2 expression is involved in R(+)-methanandamide-induced apoptotic death of human neuroglioma cells. Molecular pharmacology. 2004;66:1643–51. doi: 10.1124/mol.104.002618. [DOI] [PubMed] [Google Scholar]
- 44.Na HK, Inoue H, Surh YJ. ET-18-O-CH3-induced apoptosis is causally linked to COX-2 upregulation in H-ras transformed human breast epithelial cells. FEBS letters. 2005;579:6279–87. doi: 10.1016/j.febslet.2005.09.094. [DOI] [PubMed] [Google Scholar]
- 45.Elrod HA, Yue P, Khuri FR, Sun SY. Celecoxib antagonizes perifosine’s anticancer activity involving a cyclooxygenase-2-dependent mechanism. Mol Cancer Ther. 2009;8:2575–85. doi: 10.1158/1535-7163.MCT-09-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12:349–61. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 47.Ugurel S, Rohmel J, Ascierto PA, Flaherty KT, Grob JJ, Hauschild A, et al. Survival of patients with advanced metastatic melanoma: The impact of novel therapies. European journal of cancer. 2016;53:125–34. doi: 10.1016/j.ejca.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 48.Solit D, Sawyers CL. Drug discovery: How melanomas bypass new therapy. Nature. 2010;468:902–3. doi: 10.1038/468902a. [DOI] [PubMed] [Google Scholar]
- 49.Shtivelman E, Davies MQ, Hwu P, Yang J, Lotem M, Oren M, et al. Pathways and therapeutic targets in melanoma. Oncotarget. 2014;5:1701–52. doi: 10.18632/oncotarget.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bregoli L, Movia D, Gavigan-Imedio JD, Lysaght J, Reynolds J, Prina-Mello A. Nanomedicine applied to translational oncology: A future perspective on cancer treatment. Nanomedicine : nanotechnology, biology, and medicine. 2016;12:81–103. doi: 10.1016/j.nano.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature nanotechnology. 2007;2:751–60. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 52.Parhi P, Mohanty C, Sahoo SK. Nanotechnology-based combinational drug delivery: an emerging approach for cancer therapy. Drug Discov Today. 2012;17:1044–52. doi: 10.1016/j.drudis.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 53.Hoelder S, Clarke PA, Workman P. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol Oncol. 2012;6:155–76. doi: 10.1016/j.molonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DiStefano V, Klahn JJ. Observations on the pharmacology and hemolytic activity of dimethyl sulfoxide. Toxicology and applied pharmacology. 1965;7:660–6. doi: 10.1016/0041-008x(65)90122-5. [DOI] [PubMed] [Google Scholar]
- 55.Hu CM, Aryal S, Zhang L. Nanoparticle-assisted combination therapies for effective cancer treatment. Therapeutic delivery. 2010;1:323–34. doi: 10.4155/tde.10.13. [DOI] [PubMed] [Google Scholar]
- 56.Liang XJ, Chen C, Zhao Y, Wang PC. Circumventing tumor resistance to chemotherapy by nanotechnology. Methods in molecular biology. 2010;596:467–88. doi: 10.1007/978-1-60761-416-6_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran MA, Gowda R, Sharma A, Park EJ, Adair J, Kester M, et al. Targeting V600EB-Raf and Akt3 using nanoliposomal-small interfering RNA inhibits cutaneous melanocytic lesion development. Cancer Res. 2008;68:7638–49. doi: 10.1158/0008-5472.CAN-07-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu F, Cao J, Wu J, Sullivan K, Shen J, Ryu B, et al. Stat3-Targeted Therapies Overcome the Acquired Resistance to Vemurafenib in Melanomas. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garrido C, Paco L, Romero I, Berruguilla E, Stefansky J, Collado A, et al. MHC class I molecules act as tumor suppressor genes regulating the cell cycle gene expression, invasion and intrinsic tumorigenicity of melanoma cells. Carcinogenesis. 2012;33:687–93. doi: 10.1093/carcin/bgr318. [DOI] [PubMed] [Google Scholar]
- 60.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–47. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 61.Yang CL, Liu YY, Ma YG, Xue YX, Liu DG, Ren Y, et al. Curcumin blocks small cell lung cancer cells migration, invasion, angiogenesis, cell cycle and neoplasia through Janus kinase-STAT3 signalling pathway. PloS one. 2012;7:e37960. doi: 10.1371/journal.pone.0037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lakshmanan I, Ponnusamy MP, Das S, Chakraborty S, Haridas D, Mukhopadhyay P, et al. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene. 2012;31:805–17. doi: 10.1038/onc.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai QS, Liu W, Wang XB, Lu N, Gong DD, Kong LY, et al. NCPMF-60 induces G2/M cell cycle arrest and apoptosis in human hepatocellular carcinoma HepG2 cells. Anti-cancer drugs. 2011;22:46–57. doi: 10.1097/CAD.0b013e3283405801. [DOI] [PubMed] [Google Scholar]
- 64.Horiuchi K, Umetani M, Minami T, Okayama H, Takada S, Yamamoto M, et al. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17278–83. doi: 10.1073/pnas.0608357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madhunapantula SV, Mosca PJ, Robertson GP. The Akt signaling pathway: An emerging therapeutic target in malignant melanoma. Cancer Biol Ther. :12. doi: 10.4161/cbt.12.12.18442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madhunapantula SV, Robertson GP. Is B-Raf a good therapeutic target for melanoma and other malignancies? Cancer Res. 2008;68:5–8. doi: 10.1158/0008-5472.CAN-07-2038. [DOI] [PubMed] [Google Scholar]
- 67.Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18:254–67. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie TX, Huang FJ, Aldape KD, Kang SH, Liu M, Gershenwald JE, et al. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66:3188–96. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- 69.Crawford S. Is it time for a new paradigm for systemic cancer treatment? Lessons from a century of cancer chemotherapy. Frontiers in pharmacology. 2013;4:68. doi: 10.3389/fphar.2013.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.