Abstract
Type 1 diabetes (T1D) is a chronic disease characterized by an autoimmune-mediated destruction of insulin-producing pancreatic β cells. Environmental factors such as viruses play an important role in the onset of T1D and interact with predisposing genes. Recent data suggest that viral infection of human islets leads to a decrease in insulin production rather than β cell death, suggesting loss of β cell identity. We undertook this study to examine whether viral infection could induce human β cell dedifferentiation. Using the functional human β cell line EndoC-βH1, we demonstrate that polyinosinic-polycytidylic acid (PolyI:C), a synthetic double-stranded RNA that mimics a byproduct of viral replication, induces a decrease in β cell–specific gene expression. In parallel with this loss, the expression of progenitor-like genes such as SOX9 was activated following PolyI:C treatment or enteroviral infection. SOX9 was induced by the NF-κB pathway and also in a paracrine non–cell-autonomous fashion through the secretion of IFN-α. Lastly, we identified SOX9 targets in human β cells as potentially new markers of dedifferentiation in T1D. These findings reveal that inflammatory signaling has clear implications in human β cell dedifferentiation.
Keywords: Inflammation, Virology
Keywords: Beta cells, Diabetes, NF-kappaB
EndoC-βH1 cells as a model of human β cell dedifferentiation in type 1 diabetes.
Introduction
Type 1 diabetes (T1D) is an autoimmune disease whose etiology is not fully understood. It is a chronic disease in which insulin-producing pancreatic β cells undergo autoimmune attack. The increased incidence of T1D indicates that environmental factors are important mediators in the pathogenesis. Among these exogenous factors, a number of epidemiological and experimental results suggest that viral infection by enteroviruses such as coxsackieviruses or echoviruses may contribute to the development of T1D (1–4). Indeed, enteroviral proteins have been observed in pancreatic islets from T1D patients (5–7). Moreover, experimental models showed that infection with a coxsackievirus (CVB4E2) isolated from a patient with diabetic ketoacidosis (8) induces diabetes in mice (9). Additionally, coxsackievirus infection gives rise to diabetes in hyperglycemic mice engrafted with human islets (10). Lastly, genetic and experimental studies showed that rare variants of genes involved in recognition of dsRNA, a byproduct of replication of most viruses, protect against T1D (1, 4). However, a final link between viral infection and the onset of T1D is still lacking. Furthermore, mechanisms involved in the alteration of human β cell function following viral infection are unknown. For example, recent data indicate that hyperglycemic mice rescued by engrafted human islets became diabetic upon infection with coxsackieviruses. Importantly, diabetes was not due to human β cell death, but instead, to decreased insulin production, suggesting β cell loss of identity (10).
Rodent models have been useful to increase our understanding of how viral infections could contribute to the development of T1D in humans (1–4). For instance, CVB5 Faulkner and MCA strains, which had been adapted to enter rodent insulinoma cells such as MIN6 cells (11), were shown to disrupt insulin granule stores in this cell line (12). However, several differences exist between humans and rodents with respect to enteroviral infection. For example, coxsackievirus and adenovirus receptor (CAR) is expressed in human β cells, while it is not or barely detectable in murine β cells (10, 13). Moreover, the β cell’s sensitivity to different coxsackievirus strains varies between species (14). Thus, studying how enteroviral infection could lead to T1D in humans is of major importance. Therefore, the use of the functional human β cell line EndoC-βH1 (15) represents a suitable tool to better understand how viral infection could contribute to the development of T1D in humans.
Enteroviral, and indeed most viral, replication results in the formation of dsRNAs (16). In the present study, we treated EndoC-βH1 cells with polyinosinic-polycytidylic acid (PolyI:C). PolyI:C is a synthetic dsRNA recognized in the cytoplasm by dsRNA pattern recognition receptors (PRRs) that mimics, at least in part, the effects of viral infection on β cells (16, 17). We demonstrated that human β cells dedifferentiate following PolyI:C treatment and enteroviral infection. The expression of β cell–specific genes decreased, while progenitor-like genes such as SOX9 were activated. SOX9 was induced in an NF-κB–dependent manner, at least in part in a paracrine non–cell-autonomous fashion through the secretion of cytokines such as IFN-α. Finally, we discovered SOX9 targets in human β cells that represent potentially new marks of human β cell dedifferentiation in T1D.
Results
Human β cells dedifferentiate upon PolyI:C treatment.
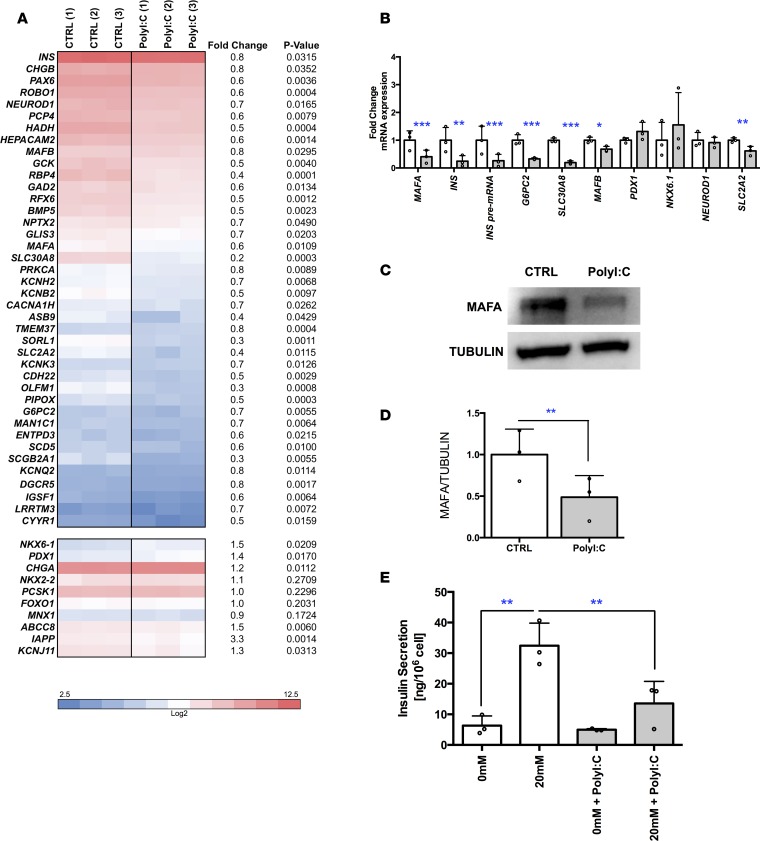
We first asked whether PolyI:C could disturb β cell identity. To this end, we transfected EndoC-βH1 cells with PolyI:C and performed global transcriptomic analyses 24 hours later. Mock-transfected cells (incubated with Lipofectamine alone) were used as control. As expected (18), PolyI:C treatment induced the expression of PRRs such as TLR3, IFIH1, and DDX58 that recognize dsRNA, demonstrating the sensitivity of EndoC-βH1 cells to PolyI:C (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97732DS1). PolyI:C treatment also induces the expression of B2M (β-2 microglobulin) (Supplemental Figure 1C). Interestingly, PolyI:C treatment decreased the expression of many genes related to β cell identity or function (Figure 1A). For example, the expression of the transcription factor MAFA decreased following PolyI:C treatment (Figure 1B), which was further confirmed at the protein level (Figure 1, C and D for quantification). INS mRNA levels also decreased, as was the case for its pre-mRNA, suggesting a decrease in INS transcription by PolyI:C treatment (Figure 1B). We also observed a decrease in the expression of genes linked to β cell function, such as G6PC2 and SLC30A8 (ZNT8) (Figure 1B). Finally, PolyI:C treatment decreased glucose-stimulated insulin secretion (Figure 1E).
Figure 1. Downregulation of human β cell identity in PolyI:C-transfected EndoC-βH1 cells.
EndoC-βH1 cells were either mock transfected (control) or transfected with PolyI:C and analyzed 24 hours later. (A and B) Heatmap from global transcriptomic analysis and RT-qPCR data represent downregulated β cell genes (n = 3). (C and D) Western blot analysis (representative of 3 independent experiments) and quantification of MAFA expression (n = 3). (E) EndoC-βH1 cells were either mock transfected (control, white) or transfected with PolyI:C (gray) and glucose-stimulated insulin secretion (GSIS) was performed 6 days later with 0 or 20 mM glucose. Insulin secreted in ng/106 cells is represented (n = 3). Data from RT-qPCR, Western blot, and GSIS analyses represent the mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 relative to BSA by 1-way ANOVA with Bonferroni’s correction.
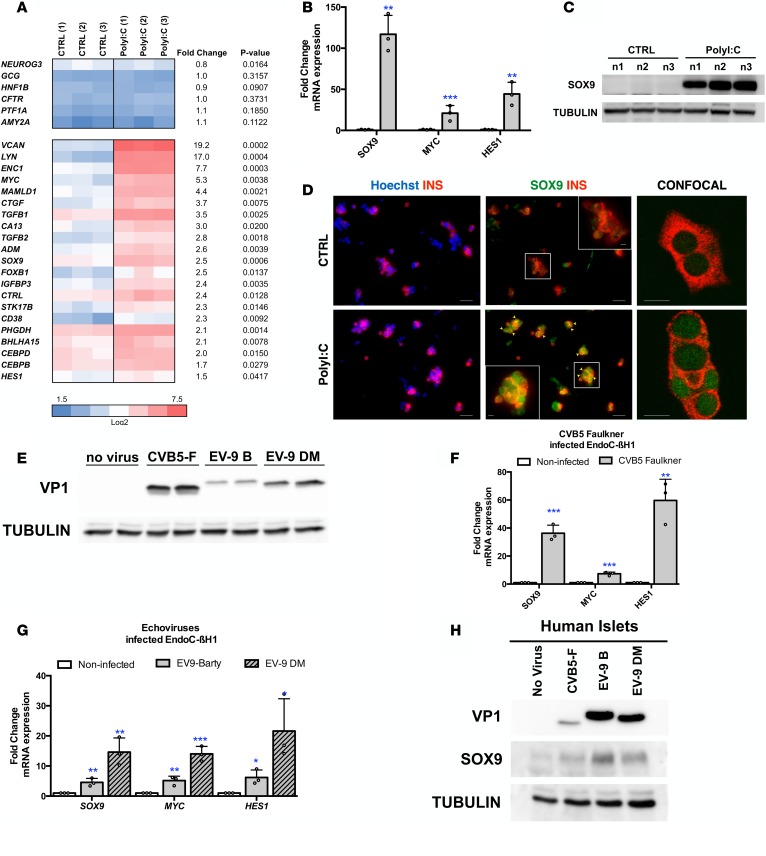
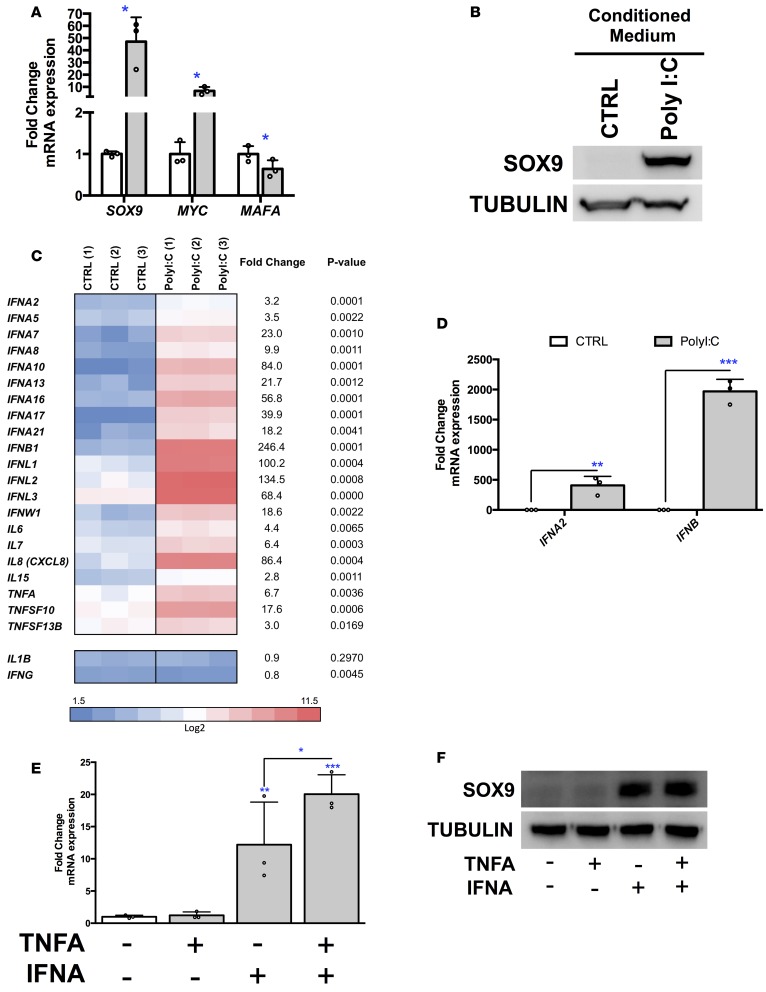
We next asked whether PolyI:C treatment activates the expression of genes normally not or barely expressed in pancreatic β cells. Global transcriptomic analysis indicated that expression of NEUROG3, an endocrine progenitor marker, GCG, an α cell marker, HNF1B and CFTR, 2 ductal markers, and PTF1A and AMY2A, 2 acinar markers, were not induced by PolyI:C (Figure 2A). VCAN (versican) that encodes a large extracellular matrix proteoglycan and the tyrosine-protein kinase LYN were highly induced (Figure 2A). It is also the case for the 2 β cell disallowed genes LRIG3 (leucine-rich repeats and immunoglobulin-like domains 3) and LMO4 (LIM domain only 4) (Supplemental Figure 2) (19, 20). Interestingly, the expression level of mRNA encoding the 3 transcription factors SOX9, HES1, and MYC, normally barely or not expressed in human β cells (primary and EndoC-βH1 cells), was increased upon treatment with PolyI:C (Figure 2A). Quantitative reverse transcription PCR (RT-qPCR) analyses further confirmed the induction of SOX9, HES1, and MYC mRNA upon PolyI:C treatment (Figure 2B). SOX9 induction was further validated at the protein level by Western blot (Figure 2C). Finally, transfection of PolyI:C into human islets induced SOX9 expression in insulin-positive β cells (Figure 2D).
Figure 2. PolyI:C induces the expression of SOX9, HES1, and MYC in human β cells.
(A–C) EndoC-βH1 cells were either mock transfected (CTRL) or transfected with PolyI:C and analyzed 24 hours later (n = 3). (A and B) Heatmap from global transcriptomic analysis and RT-qPCR data represent induced genes. (C) Western blot analysis of SOX9 expression (n = 3; n1, n2, and n3 correspond to 3 independent experiments). (D) Primary human islet cells were dissociated and either mock transfected (CTRL) or transfected with PolyI:C and analyzed 24 hours later by immunocytochemistry. Nuclei are stained with Hoechst 33342 stain (blue), insulin is in red, and SOX9 is in green. Arrowheads point to insulin+ (INS+) cells that stain positive for SOX9 following PolyI:C treatment. Scale bars: 25 μm. The inset shows a higher-magnification image (×2.1 magnification; scale bar: 5 μm). Representative images of 4 independent experiments. (E–G) EndoC-βH1 cells were infected with enteroviruses at 5 × 104 TCID50 and harvested 24 hours later. (E) Western blot analysis of VP1 expression (n = 3). (F and G) qPCR analyses of SOX9, MYC, and HES1 expression (n = 3). (H) Human islets were infected with enteroviruses at 5 × 107 TCID50 and harvested 24 hours later. Western blot analyses of VP1 and SOX9 expression are shown (representative images of 3 independent experiments). Data from RT-qPCR, Western blots, and immunofluorescence represent the mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 relative to control by Student’s t test. CVB5-F, coxsackievirus strain B5 Faulkner; EV-9 B, echovirus 9 Barty.
PolyI:C treatment induced a nearly 3-fold increase in the number of apoptotic cells (from 6.6% ± 2.8% to 15% ± 2%), indicating that PolyI:C treatment induces some β cell death in addition to dedifferentiation (Supplemental Figure 3, A and B). To exclude the possibility that dedifferentiation occurred in dying cells, we sorted annexin V/propidium iodide double-negative cells and performed RT-qPCR analyses. Remarkably, we observed similar genes down- and upregulated in live cells when compared with the bulk experiment (compare Supplemental Figure 3, C and D with Figures 1B and 2B).
By mining our transcriptomic data, we observed that PolyI:C treatment induced the expression of several genes linked to antiviral responses, coding for chemokines or for proteins related to immune response (Supplemental Figure 4) that were previously shown to be induced upon infection of human pancreatic islets by coxsackieviruses (10, 21). We thus tested whether infection with enteroviruses such as CVB5 and echovirus 9 (EV-9) upregulated the expression of pancreatic progenitor markers in EndoC-βH1 cells. CVB5 Faulkner, EV-9 Barty, and EV-9 DM strains efficiently infected EndoC-βH1 cells, as demonstrated by the detection of the major viral capsid protein VP1 (Figure 2E). Importantly, infection of EndoC-βH1 cells by all 3 virus strains induced SOX9, HES1, and MYC expression (Figure 2, F and G), as demonstrated for PolyI:C. Infection by all 3 virus strains also increased SOX9 levels in human pancreatic islets (Figure 2H).
Taken together, these data indicate that dedifferentiation occurs in dsRNA-treated human β cells, as is the case following infection with enteroviruses.
The NF-κB subunit RELA mediates the induction of SOX9 by PolyI:C in EndoC-βH1 cells.
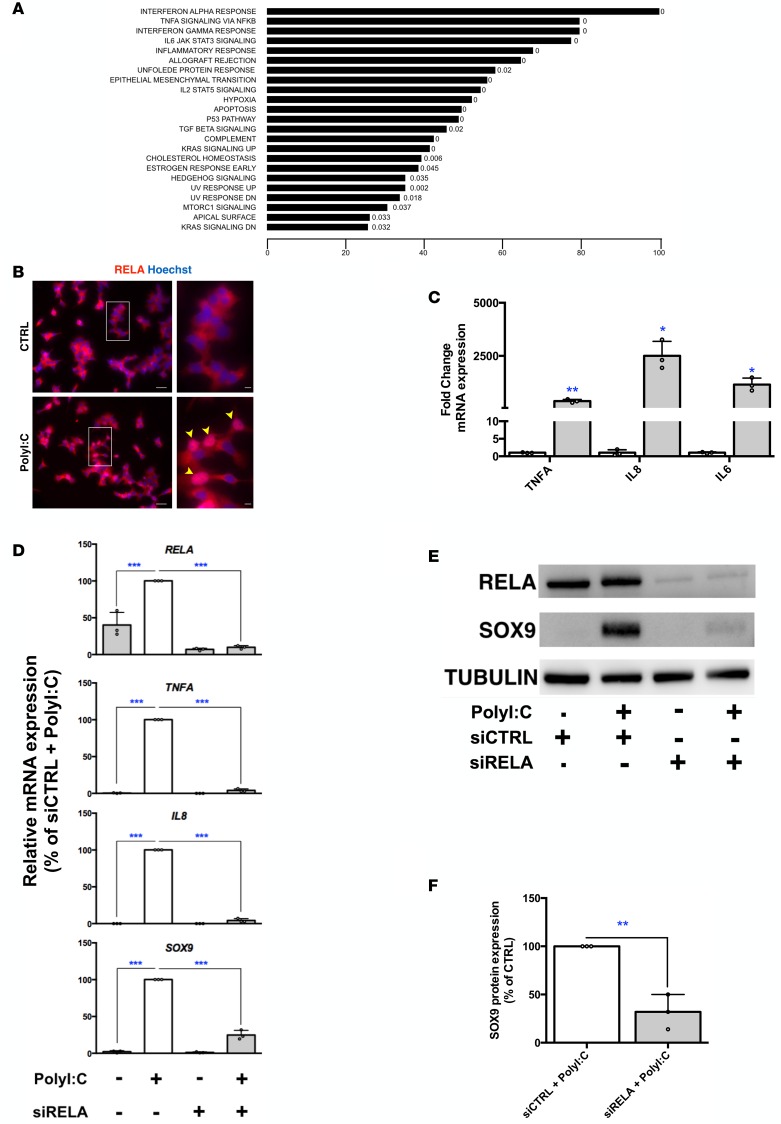
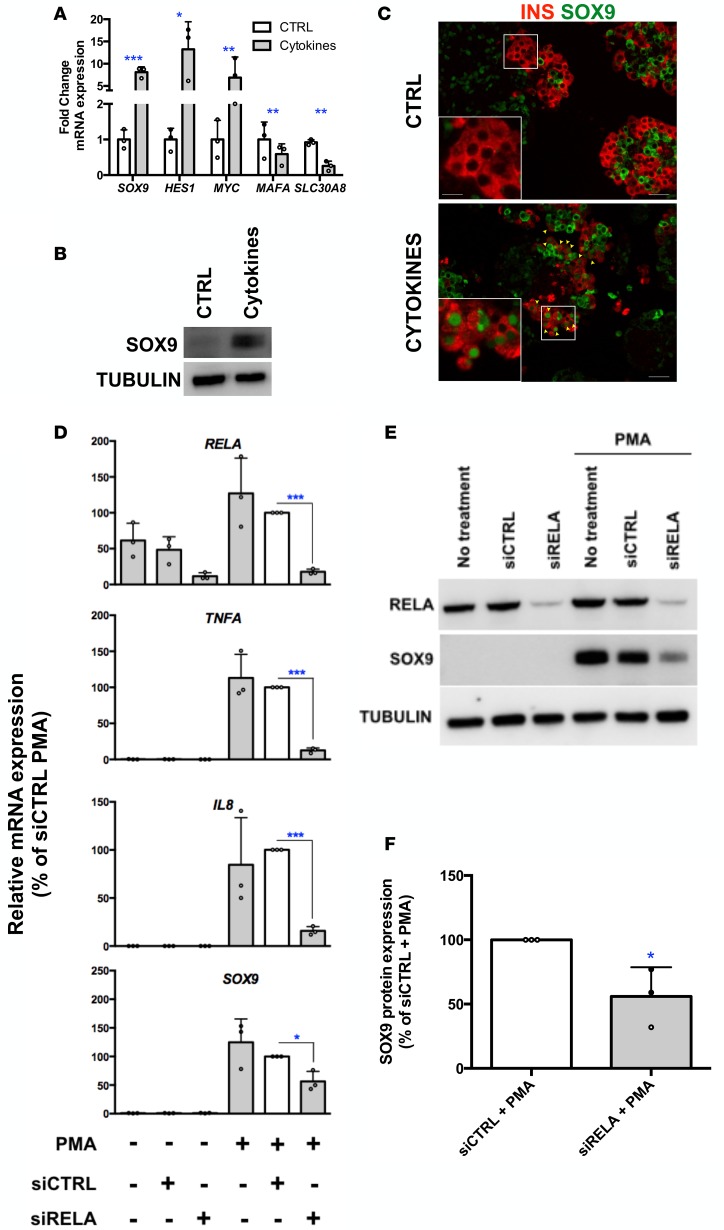
To determine the mechanisms involved in β cell dedifferentiation induced by PolyI:C, we searched for pathways regulated by PolyI:C using gene set enrichment analysis (GSEA). The NF-κB pathway was listed among the top regulated pathways (Figure 3A). PolyI:C treatment induced the nuclear translocation of RELA, a major subunit of NF-κB (Figure 3B) and upregulated the expression of NF-κB downstream targets such as TNFA, IL8, and IL6 (Figure 3C). Moreover, efficient siRNA-mediated downregulation of RELA (Figure 3, D and E) reduced TNFA, IL8 (Figure 3D), and IL6 (data not shown) induction by PolyI:C. Remarkably, RELA knockdown weakened the induction of SOX9 by PolyI:C, as measured at the mRNA (Figure 3D) and protein levels (Figure 3, E and F). To strengthen our hypothesis that RELA is largely responsible for the induction of SOX9 upon PolyI:C treatment, we tested the effect of additional NF-κB activators. Incubation of EndoC-βH1 cells with a cocktail of proinflammatory cytokines (TNF-α + IL-1β + IFN-γ), which are involved in β cell dysfunction and death through NF-κB in T1D-related models (22), induced SOX9 expression both at the mRNA and protein levels (Figure 4, A and B). Treatment of human islets with cytokines also induced SOX9 expression in β cells (Figure 4C). PMA, another potent NF-κB activator that induces TNFA and IL8 expression in EndoC-βH1 cells (Figure 4D), increased SOX9, HES1, and MYC expression while decreasing MAFA and SLC30A8 levels (Figure 4, D and E and Supplemental Figure 5A). SOX9 induction by PMA treatment was prevented when the NF-κB subunit RELA was downregulated with siRNA (Figure 4, D–F) or inhibited using ammonium pyrrolidinedithiocarbamate (PDTC), a potent NF-κB inhibitor (Supplemental Figure 5, A and B).
Figure 3. The NF-κB pathway is involved in SOX9 induction by PolyI:C.
(A) Gene set enrichment analysis of global transcriptomic data representing pathways regulated following PolyI:C treatment with the corresponding FDR q values. The x axis represents the percentage of regulated genes in the corresponding pathway. (B) Immunostaining for RELA indicates that PolyI:C treatment (24 hours) induces the nuclear translocation of RELA. Scale bars: 25 μm. Arrowheads point to nuclear RELA localization. The insets show higher-magnification images (×3 magnification; scale bars: 5 μm). n = 3, representative images of 3 independents experiments. (C) EndoC-βH1 cells were either mock transfected (CTRL) or transfected with PolyI:C and the NF-κB targets TNFA, IL8, and IL6 were analyzed 24 hours later by qPCR (n = 3). (D–F) EndoC-βH1 cells were transfected with control nontarget siRNA (siCTRL) or siRNA targeting RELA (siRELA). They were next either mock transfected or transfected with PolyI:C 24 hours later, RNA was prepared for RT-qPCR (n = 3) (D), and proteins for Western blot analyses (n = 3, representative blot of 3 independent experiments) (E and F). Data from immunofluorescence, RT-qPCR, and Western blots represent the mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01; and ***P < 0.001 relative to control by Student’s t test or ANOVA with Bonferroni’s correction for multiple comparisons.
Figure 4. Potent NF-κB activators induce SOX9 expression in human β cells.
(A and B) EndoC-βH1 cells were treated with proinflammatory cytokines (TNF-α, IL-1β, and IFN-γ). (A) SOX9, HES1, MYC, MAFA, and SLC30A8 mRNAs were analyzed 72 hours later (n = 3). (B) SOX9 protein was analyzed 72 hours later (n = 3, representative Western blot of 3 independent experiments). (C) Human islets were exposed to IL-1β, TNF-α, and IFN-γ for 72 hours. Islets were harvested prior to immunofluorescence staining. Insulin is in red and SOX9 is in green. Scale bars: 25 μm. Arrowheads point to insulin+ (INS+) cells that stain positive for SOX9 following cytokine treatment. The insets show higher-magnification images (×2.4 magnification; scale bar: 5 μm). n = 2, representative images of 2 independent experiments. (D–F) EndoC-βH1 cells were transfected with control nontarget siRNA (siCTRL) or siRNA targeting RELA (siRELA). They were next treated with PMA for 8 hours. RNA was prepared for RT-qPCR (n = 3) (D) and proteins for Western blot analyses (E and F) (n = 3). Data from RT-qPCR and Western blot represented as the mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 relative to control by Student’s t test.
These data indicate that PolyI:C activates the NF-κB pathway through RELA, which in turn is necessary for SOX9 induction in human β cells.
PolyI:C-treated EndoC-βH1 cells secrete factors that disrupt β cell identity of neighboring cells.
In models of T1D, CD8+ T cells, endothelial cells, and macrophages secrete proinflammatory cytokines that presumably induce β cell dysfunction and death (22). β Cells themselves also secrete proinflammatory cytokines following viral infection or cytokine exposure (refs. 21, 22 and Supplemental Figure 4). We wondered whether factors secreted by β cells following PolyI:C transfection could disrupt neighboring β cell identity in a paracrine non–cell-autonomous fashion. To test this hypothesis, we incubated fresh (non–PolyI:C-transfected) EndoC-βH1 cells with the conditioned medium of PolyI:C-treated EndoC-βH1 cells. Interestingly, SOX9 (both mRNA and protein levels) increased following incubation with conditioned medium from PolyI:C-treated cells (Figure 5, A and B). This induction was not due to PolyI:C that remained present in the conditioned medium, as (a) addition of PolyI:C to the culture medium (without transfection) did not modulate gene expression in naive EndoC-βH1 cells even at high concentration (Supplemental Figure 6A), and (b) treatment of conditioned medium from PolyI:C-transfected cells with RNAse-A did not modify its ability to induce SOX9 expression in naive cells (Supplemental Figure 6B). Taken together, these observations indicate that SOX9 induction by conditioned medium was exclusively due to secreted factors.
Figure 5. SOX9 is also induced in a paracrine non–cell-autonomous fashion.
(A and B) EndoC-βH1 cells were incubated with conditioned medium from mock- (CTRL) or PolyI:C-transfected cells. SOX9, MYC, and MAFA mRNA (n = 3) (A) and SOX9 protein (n = 3; representative Western blot of 3 independent experiments) (B) were analyzed 72 hours later. (C and D) EndoC-βH1 cells were either mock transfected (CTRL) or transfected with PolyI:C and analyzed 24 hours later. (C) Heatmap from global transcriptomic analysis represents upregulated IFN, IL, and TNF family members (n = 3). (D) RT-qPCR analysis of IFNA2 and IFNB1 expression (n = 3). (E and F) EndoC-βH1 cells were treated with TNF-α and IFN-α. SOX9 mRNA (n = 3) (E) and protein (n = 3; representative Western blot of 3 independent experiments) (F) were analyzed 72 hours later. Data from RT-qPCR and Western blots represent the mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01; and ***P < 0.001 relative to control by Student’s t test or ANOVA with Bonferroni’s correction for multiple comparisons.
We next searched for the secreted factor(s) that induce SOX9 expression in a paracrine non–cell-autonomous fashion. Further mining our microarray data indicated that PolyI:C treatment upregulated the expression of many secreted cytokines. This is, for example, the case for IFN, IL, and TNF family members (Figure 3C and Figure 5, C and D). On the other hand, the expression of IL1B and IFNG was not induced by PolyI:C (Figure 5C). We thus tested whether selected cytokines induced by PolyI:C activate SOX9 expression. It was not the case for TNF-α (Figure 5, E and F). In contrast, IFN-α induced SOX9 expression both at the mRNA and protein levels. Moreover, coincubation of IFN-α with TNF-α further increased SOX9 expression (Figure 5E). Note that conditioned medium from PolyI:C-treated cells induced a 46.3 ± 18-fold increase in SOX9 mRNA, while IFN-α plus TNF-α increased SOX9 mRNA by only 20.3 ± 2.3-fold. Thus, there must be additional factors that contribute to the change in SOX9 mRNA.
These data indicate that PolyI:C treatment induces the production of secreted factors such as IFN-α and TNF-α that could induce SOX9 expression in a non–cell-autonomous fashion.
SOX9 targets in dedifferentiated human β cells.
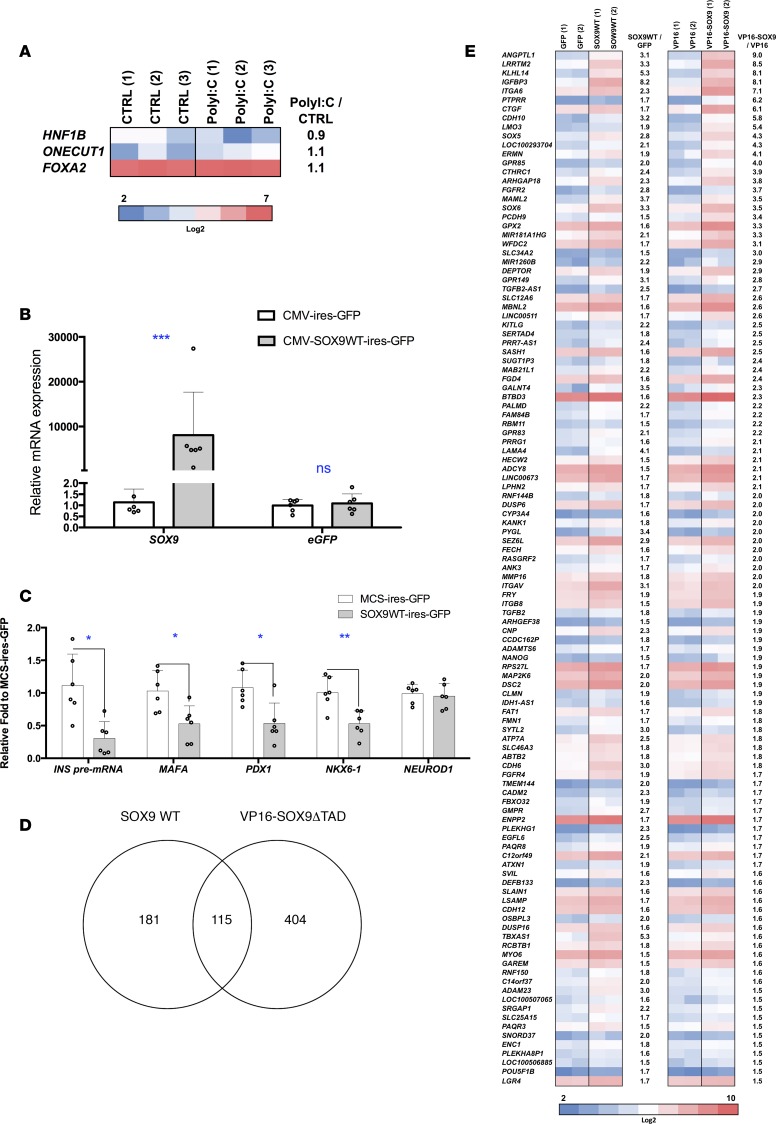
At this point, our data indicated that inflammatory signals induced human β cell dedifferentiation and SOX9 could represent an important transcription factor in this process. To further define the role of SOX9 in β cells, we searched for its target genes. Previous data described HNF1B, ONECUT1, and FOXA2 as SOX9 targets in mouse pancreatic duct cells (23). None of them was induced by PolyI:C in EndoC-βH1 cells (Figure 6A). To define novel SOX9 targets, we overexpressed SOX9 in EndoC-βH1 cells. We transfected EndoC-βH1 cells with either an MCS-ires-GFP (control) or a SOX9-ires-GFP plasmid and sorted GFP+ cells by FACS (Figure 6B). SOX9 overexpression induced a downregulation of INS presplicing mRNA and several β cell–related transcription factors such as MAFA, PDX1, and NKX6-1 (Figure 6C). This expression pattern resembles the one observed in transgenic mice with ectopic expression of SOX9 in β cells (24). We also overexpressed in EndoC-βH1 cells a constitutively trans-activating SOX9 mutant (VP16-SOX9ΔTAD) in which the SOX9 transactivation domain (TAD) was replaced by VP16 that increases SOX9 transcriptional activity (Supplemental Figure 7) (25). We performed global transcriptomic analyses and detected 115 genes upregulated by both wild-type SOX9 (SOX9WT) and VP16-SOX9ΔTAD constructs (Figure 6, D and E). We next crossed the list of genes induced by WT and mutant super transactivator form SOX9 with one of the genes induced following PolyI:C treatment. We obtained a list of 25 genes induced by both SOX9 and PolyI:C (Figure 7A) and selected 4 of them (CTGF, LRRTM2, DEPTOR, and MAML2). We confirmed by RT-qPCR their upregulation following SOX9WT and VP16-SOX9ΔTAD transfection (Figure 7B) and following PolyI:C treatment (see below). Moreover, when SOX9 expression was blunted using siRNA (Figure 7, C and D), CTGF, LRRTM2, DEPTOR, and MAML2 induction by PolyI:C was decreased (Figure 7E). Sox9 induction has recently been observed in dedifferentiated β cells from nonobese diabetic (NOD) mice (26). Interestingly, we found that 10 out of 25 genes induced by SOX9 overexpression and PolyI:C treatment have been described as induced in dedifferentiated β cells from NOD mice and Ctgf, Lrrtm2, Deptor, and Maml2 belonged to this list (Supplemental Figure 8).
Figure 6. Genes induced in EndoC-βH1 cells by SOX9.
(A) EndoC-βH1 cells were either mock transfected (CTRL) or transfected with PolyI:C and analyzed 24 hours later. Heatmap from global transcriptomic analysis represents previously described SOX9 target genes (n = 3). (B–E) EndoC-βH1 cells were transfected with MCS-ires-GFP, SOX9WT-ires-GFP, VP16-ires-GFP, or VP16-SOX9ΔTAD-ires-GFP plasmid. GFP+ cells were sorted by FACS 48 hours later and RNAs were prepared (n = 2–6). (B and C) RT-qPCR analyses (n = 6). (D and E) Global transcriptomic analyses with Venn diagram and heatmap that represent genes upregulated (>1.5-fold) by SOX9WT-ires-GFP and VP16-SOX9ΔTAD-ires-GFP (n = 2). Data from RT-qPCR represent the mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001 relative to control by Student’s t test. ns, not significant.
Figure 7. Potentially new SOX9 targets in dedifferentiated human β cells.
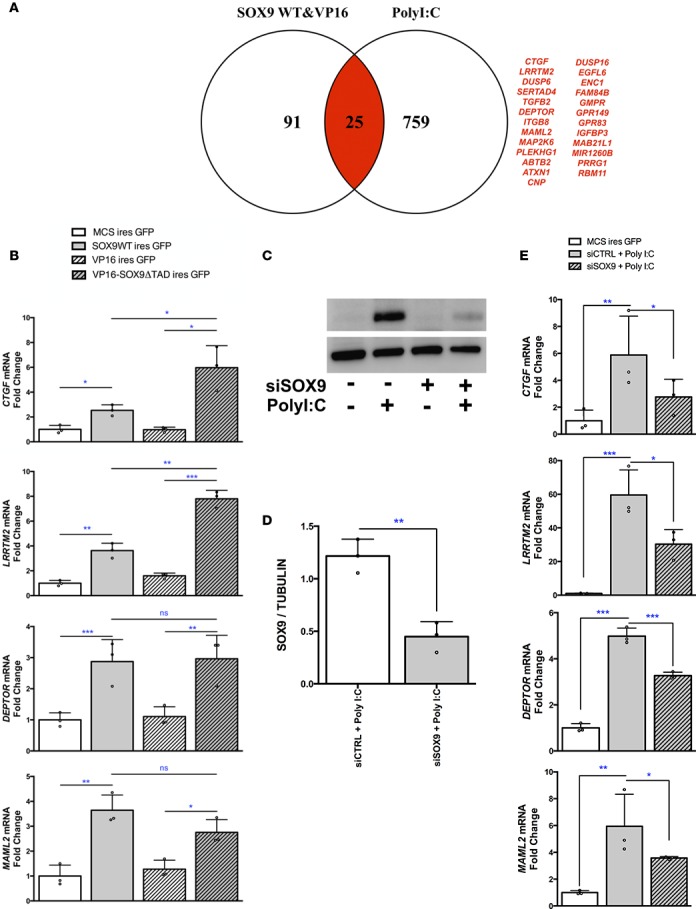
(A) Venn diagram with genes upregulated (>2-fold) in EndoC-βH1 cells treated with PolyI:C and following ectopic expression of SOX9/VP16-SOX9ΔTAD. Genes upregulated by both PolyI:C and SOX9/VP16-SOX9ΔTAD are shown in red. (B) EndoC-βH1 cells were transfected with MCS-ires-GFP, SOX9WT-ires-GFP, VP16-ires-GFP, or VP16-SOX9ΔTAD-ires-GFP plasmid. GFP+ cells were sorted by FACS 48 hours later and RNA was prepared for RT-qPCR analyses (n = 3). (C–E) EndoC-βH1 cells were transfected with control nontarget siRNA (siCTRL) or siRNA targeting SOX9 (siSOX9). They were next either mock transfected or transfected with PolyI:C. Twenty-four hours later, proteins were prepared for Western blot analyses (n = 3) (C and D) and RNA for RT-qPCR (n = 3) (E). Data from RT-qPCR and Western blot represent the mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01; and ***P < 0.001 relative to control by Student’s t test or ANOVA with Bonferroni’s correction for multiple comparisons. ns, not significant.
Taken together, our data show that SOX9 and its targets represent potentially new markers of human β cell dedifferentiation.
Discussion
The concept of loss of β cell identity or dedifferentiation as a cause of diabetes mellitus is gaining prominence. Dedifferentiation has been observed in several models where β cells were challenged with environmental cues such as glucotoxicity, pregnancy, and hypoxia (24, 27, 28). Recent data also suggest that β cell dedifferentiation occurs in T1D. For example, in humans, the frequency of degranulated chromogranin A+/hormone– cells increases in pancreata from T1D patients (29). Moreover, in NOD mice, a subpopulation of degranulated β cells resists immunological attacks while decreasing β cell markers and gaining pancreatic progenitor traits (26). We undertook the present study to elucidate the mechanisms involved in human β cell dedifferentiation in T1D.
Enteroviral infections have been associated with the development of T1D, but our knowledge of the way enteroviruses act on human β cells and induce diabetes is limited. A number of rodent models have been useful to better understand mechanisms of enterovirus-induced β cell dysfunction. However, several differences exist between human and rodent β cells in response to viral assault. As access to primary human islet preparations is limited, we recently developed a functional human β cell line by lentiviral transduction of 2 RIP-regulated oncogenes (SV40T and hTERT) (15), and validated in the present study this line to model β cell infection by viruses. We demonstrated that infection of EndoC-βH1 cells with enteroviruses induced the expression of the major viral capsid protein VP1. Moreover, treatment of EndoC-βH1 cells with PolyI:C induced a gene expression profile that resembles the one previously observed following infection of human pancreatic islets by coxsackieviruses (10, 21).
We observed marks of dedifferentiation following PolyI:C treatment characterized by a decreased expression of β cell–specific genes such as INS, MAFA, and SLC30A8. Such decrease in gene expression was independent of β cell death, which corroborates recent data that indicate that enteroviral infection of human islets results in a loss of insulin production (10). Interestingly, we observed that glucose-stimulated insulin secretion was diminished following PolyI:C treatment. In parallel with the decrease in β cell–specific genes and function, we observed a marked increase of progenitor markers such as SOX9, HES1, and MYC as additional marks of dedifferentiation. It is noteworthy that the expression of the pancreatic endocrine progenitor marker NEUROG3 as well as ALDH1A3 that are both increased in β cell dedifferentiation in type 2 diabetes (T2D) (28, 30, 31) were not induced in our model, suggesting alternative mechanisms of β cell dedifferentiation following viral infection.
We focused on the induction of SOX9, a transcription factor expressed in multipotent pancreatic progenitors and duct cells that contribute to the development of endocrine and exocrine cells of the adult pancreas (32). Healthy β cells do not express SOX9 (32), while aberrant SOX9 expression has been observed in islets of db/db mice, a model of T2D (33) and in mouse β cells following activation of the Hedgehog signaling pathway (34) or under hypoxic conditions (24). Remarkably, ectopic expression of SOX9 itself led to β cell dedifferentiation as revealed by a decrease in β cell–specific genes both in mice (24) and in humans (the present study).
There are around 106 islets dispersed in the human pancreas. The way by which viruses could induce an efficient decrease in functional β cell mass and β cell dedifferentiation in this islet setting remains obscure. One hypothesis is that following infection, viruses would replicate in a recurrent fashion in the human islets, and over weeks or months the viruses would infect β cells. This hypothesis is supported by the fact that signs of viral infection such as expression of the viral capsid protein VP1 are detected in the islets of the majority of brain-dead organ donors with T1D, i.e., potentially years after infection (35). Our data support an additional hypothesis, in which viral infection would activate the NF-κB and interferon regulatory factor (IRF) pathways in a limited number of β cells within an islet that would induce the production and secretion of NF-κB and IRF downstream cytokines. Such proinflammatory cytokines would next act in a paracrine fashion on neighboring β cells to induce their dedifferentiation, creating a vicious cycle (36, 37). Such a hypothesis is supported by our data that demonstrate that (a) PolyI:C treatment induces the production of cytokines such as IFN-α and TNF-α by EndoC-βH1 cells; and (b) the conditioned medium of PolyI:C-treated EndoC-βH1 cells induces SOX9 expression when added to naive β cells, as is the case when the cells are treated with IFN-α and TNF-α. We thus propose that within islets, viral infection could induce dedifferentiation in a paracrine non–cell-autonomous fashion, which would represent a mechanism to increase the process of human β cell dedifferentiation following viral infection.
Our data indicate that PolyI:C alters β cell identity and gives rise to a partial progenitor-like gene expression profile. In particular, PolyI:C induces the expression of the progenitor marker SOX9 in an NF-κB–dependent manner. We searched for SOX9 targets in human β cells. It was previously demonstrated that HNF1B, ONECUT1, and FOXA2 were SOX9 targets in a mouse pancreatic duct cell line (23). None of these genes were induced (neither by PolyI:C treatment nor SOX9 overexpression), which may be due to species (mouse vs. human) or cell type (duct vs. β cells) differences. By mining transcriptomic data obtained from SOX9 overexpression or PolyI:C treatment in EndoC-βH1 cells, we discovered potentially new SOX9 targets such as CTGF, DEPTOR, MAML2, and LRRTM2. Interestingly, by analyzing transcriptomic data from NOD mouse β cells that survive immunological attack (26), we discovered that the expression of Sox9 and its 4 above-described targets was induced, further supporting SOX9 and its targets as markers of β cell dedifferentiation. In chondrocytes, CTGF was recently demonstrated as a direct target of SOX9, with a role in differentiation (38). In the pancreas, CTGF is involved in β cell proliferation and regeneration (39–41). Following immunological attack, as observed during T1D progression, dedifferentiated cells could represent a pool of progenitor-like cells that can eventually proliferate and regenerate through SOX9 and CTGF. These dedifferentiation/redifferentiation cycles may lead to continuous turnover of β cells that are again targeted by the immune system or they could lead to a residual subpopulation of β cells, as observed in T1D patients with a disease duration of 50 years or longer (termed “medalists”) (42). Understanding whether dedifferentiated cells can redifferentiate into β cells and developing strategies to protect them from immune attacks may identify new therapeutic targets to replenish a sufficient amount of insulin-positive cells in vivo.
Methods
Culture of EndoC-βH1 cells and treatment.
The human pancreatic β cell line EndoC-βH1 was cultured in 5.6 mM glucose Dulbecco’s Modified Eagle’s Medium (DMEM, Thermo Fisher Scientific) supplemented with 2% BSA fraction V (Roche), 50 μM β-mercaptoethanol (Sigma-Aldrich), 5.5 μg/ml transferrin (Sigma-Aldrich), 6.7 ng/ml Na2SeO3, 10 mM nicotinamide (Calbiochem), and 100 μg/ml streptomycin and 100 U/ml penicillin (Thermo Fisher Scientific). Cells were seeded at 9 × 104 cells/cm2 on plates coated with 1.2% Matrigel containing 3 μg/ml fibronectin (both Sigma-Aldrich) and cultured at 37°C and 5% CO2 as previously described (15).
EndoC-βH1 cells were transfected with 1 μg/ml of PolyI:C (InvivoGen) using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) as previously described (43). The following compounds were used for treatment: 100 ng/ml PMA (Santa Cruz Biotechnology); 2,000 U/ml IFN-γ (R&D Systems); 1,100 U/ml TNF-α (R&D Systems); 1,000 U/ml IL-1β (R&D Systems); 2,000 U/ml IFN-α (Roche); and 10 μM PDTC (Santa Cruz Biotechnology).
Glucose-stimulated insulin secretion.
Five days after PolyI:C transfection, EndoC- βH1 cells were starved in DMEM containing 0.5 mM glucose. After a 24-hour starvation, cells were washed twice and then preincubated in Krebs-Ringer bicarbonate HEPES buffer (KRBH) containing 0.2% fatty acid–free BSA and 0 mM glucose for 1 hour. Insulin secretion was measured following a 40-minute incubation with KRBH containing 0.2% fatty acid–free BSA and 0 mM or 20 mM glucose. Insulin secretion and intracellular insulin were measured by ELISA as previously described (15).
Human islets and treatment.
Human islets were obtained from 7 donors. On the day of receiving the samples, approximately 100 human islets were handpicked for each condition, washed in PBS, and treated with 1 ml Accutase (PAA Laboratories) for 5 minutes at 37°C. Partial dissociation of the human islets was achieved with slow pipetting and Accutase was removed after a centrifugation step. The cells were seeded on Matrigel/fibronectin–coated coverslips and cultured for 24 hours in the same medium as used for EndoC-βH1 cells and transfected with PolyI:C as described above.
Assessment of cell death.
Apoptotic and necrotic cell death was determined following annexin-V/propidium iodide staining (Biolegend). AnnexinV and propidium iodide double-negative cells were considered alive and sorted using a FACSAria III cell sorter (BD Bioscience).
RNA isolation, reverse transcription, and qPCR.
An RNeasy Micro kit (Qiagen) was used to extract total RNA from EndoC-βH1 cells. A First-Strand cDNA kit (Thermo Fisher Scientific) was used to synthesize cDNA. RT-qPCR was performed using Power SYBR Green mix (Applied Biosystems) with a QuantStudio 3 analyzer. Custom primers were designed with IDT Primer-Quest online software and the efficiency was determined for each with a serial dilution of cDNA samples from EndoC-βH1 cells. PPIA or ACTB transcript levels were used for normalization of each sample. Probe sequences are listed in Supplemental Table 1.
Gene expression analysis.
EndoC-βH1 cells were isolated, RNA extracted, and gene expression assessed using the Affymetrix 2.0ST gene chip. Microarray data and all experimental details are available in the NCBI’s Gene Expression Omnibus (GEO) database (accession GSE104196). GSEA software and the Biological Process database of the Gene Ontology Consortium were used for gene expression comparisons (44).
siRNA transfection.
EndoC-βH1 cells were passaged and transfected using Lipofectamine RNAiMAX (Life Technologies) 24 hours later. SMARTpool siRNA for human RELA or SOX9, or ON-TARGETplus nontargeting control pool (siCTRL) were used (Dharmacon, GE healthcare Life Sciences) at a final concentration of 80 nM. Briefly, siRNA and Lipofectamine RNAiMAX were combined in OptiMEM and applied to the cells. Medium was replaced 2.5 hours later with fresh EndoC-βH1 culture medium.
Enterovirus infection.
For infection, 7.5 × 105 EndoC-βH1 cells were incubated with 5 × 104 of 50% tissue culture infectious dose (TCID50) CVB5 Faulkner, EV-9 Barty, or DM for 2 hours in infection medium (1× Hanks’ medium without FBS, 0.44% NaHCO3) as described previously (12). Twenty-four hours after infection, cells were harvested for RNA preparation or protein analysis. Five hundred human islets were infected with 5 × 107 TCID50/ml for 2 hours in islet culture medium (RPMI plus 5.5 mM glucose and 10% FBS). Twenty-four hours after infection, islets were harvested for protein analysis.
Preparation of conditioned medium.
EndoC-βH1 cells were transfected with or without PolyI:C and media were changed 2.5 hours later. Conditioned media were recovered after 24 hours, filtered, incubated with or without RNAse-A (1 mg/ml, Qiagen) for 30 minutes at 37°C, and added to untreated (naive) EndoC-βH1 cells.
Immunostaining and immunoblotting.
EndoC-βH1 cells were cultured on Matrigel/fibronectin–coated coverslips and processed for RELA immunostaining as previously described (45). Human islets cultured on Matrigel/fibronectin–coated coverslips and treated with PolyI:C were processed for SOX9 (1/500; ab5535; Millipore) and insulin (1:1,000; Sigma-Aldrich). Handpicked human islets were treated with cytokines for 72 hours. Tissues were fixed in 10% formalin and processed for immunohistochemistry for SOX9 and insulin using a NxGen decloaking chamber (Biocare). The secondary antibodies were Alexa Fluor 488 goat anti-rabbit (1:400; Invitrogen) and Alexa Fluor 594 goat anti-mouse (1:400; Jackson ImmunoResearch).
For immunoblot assays, cells were lysed in RIPA buffer and sonicated. Equal amounts of protein (20 μg) were separated in a 4%–12% Bis-Tris gel and transferred onto a PVDF membrane using an iBLOT2 Dry Blotting System (Thermo Fisher Scientific). Membranes were immunoblotted with the following antibodies: MAFA (1/500; Gift from Alireza Rezania, Betalogics), SOX9 (1/500; ab5535; Millipore), VP1 (1/500; Gift from Merja Roivainen, Helsinki, Finland), RELA (1/250; sc8008; Santa Cruz Biotechnology), and tubulin (1/2,000; T9026; Sigma-Aldrich). Species-specific HRP-linked secondary antibodies (Cell Signaling Technology) were used for detection and visualization was performed on an ImageQuant LAS 4000 following ECL exposure (GE Healthcare).
Plasmid construction.
All retroviral vector plasmids used for transfection were derived from the pPRIG vector or its relatives (46, 47). For safety measures, retroviral psi (Ψ) packaging element was removed in each plasmid by an EheI digestion followed by religation (48). The pcDNA3-Flag-SOX9WT and the RCAS-Flag-SOX9ΔTAD plasmids were gifts from Philippe Jay and Pascal De Santa Barbara (both from INSERM-CNRS, Montpellier, France). The pPRIGΔΨ-FlagSOX9WT (referred to here as SOX9WT) was constructed following pcDNA3-FlagSOX9WT digestion with BamHI-EcoRI. The digested DNA was then subcloned into the pPRIGΔΨ bicistronic vector digested by the same enzymes. The pPRIGΔΨ-VP16-SOX9ΔTAD (referred to as VP16-SOX9ΔTAD) was constructed in 4 steps. First, the RCAS-Flag-SOX9ΔTAD was digested with ClaI and SalI and subcloned into the pPRIZΔΨ vector digested by the same enzymes. The pcDNA3-FlagSOX9WT plasmid was digested by NdeI-PasI and then subcloned into the pPRIZΔΨ-FlagSOX9ΔTAD vector digested by the same enzymes (referred as pPRIZΔψ-NWT-FlagSOX9ΔTAD). The multiple cloning site (MCS) of the pPRIGΔΨ-VP16-MCS plasmid was shortened by a AvaIII-SdaI digestion (as pPRIGΔψ-VP16-sMCS). Lastly, pPRIZΔψ-NWT-FlagSOX9ΔTAD was digested with SdaI–PstI and then subcloned into pPRIGΔψ-VP16-sMCS digested with PstI–MunI. Sequencing validated all developed plasmids. The plasmids used here are referred to as MCS-ires-GFP (CTRL), SOX9WT-ires GFP, VP16-ires-GFP, and VP16-SOX9ΔTAD-ires-GFP.
Plasmid transfection.
EndoC-βH1 cells were passaged and transfected using Lipofectamine 2000 (Life Technologies) 24 hours later. MCS-ires-GFP (CTRL), SOX9WT-ires GFP, VP16-ires-GFP, or VP16-SOX9ΔTAD-ires-GFP was used at a final of 1 μg/ml. Briefly, plasmid and Lipofectamine 2000 were combined in OptiMEM and applied to the cells. Three hours later, the medium was replaced with fresh EndoC-βH1 culture medium.
Luciferase assay.
SOX9 response element COL2A1 promoter and SOX9 binding motif were gifts from Laura Bridgewater (Brigham Young University, Provo, Utah, USA) and Marc van de Wetering (Hubrecht Institute, Utrecht, The Netherlands), respectively. Cells were cotransfected with a SOX9 response element–driven firefly luciferase reporter gene, with either MCS-ires-GFP (CTRL), SOX9WT-ires GFP, VP16-ires-GFP, or VP16-SOX9ΔTAD-ires-GFP and Renilla luciferase at a final concentration of 1 μg/ml. Firefly and Renilla luciferase activity was measured with a Dual Luciferase reporter assay system (Promega) following the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla luciferase activity to correct for variation in transfection efficiency.
Statistics.
Graphs were constructed by using Prism 6 software (GraphPad). Quantitative data are presented as the mean ± SD from 3 independent experiments. Statistical significance was estimated using a 1-tailed Student’s t test. A P value less than 0.05 was considered significant.
Study approval.
Human pancreata were obtained from brain-dead donors after informed consent was signed from next-of-kin, and processed for islet isolation according to the procedures approved by the ethics committee of the University of Pisa, Italy, the French Agency of Biomedicine, and King’s College Hospital NHS Foundation Trust (London, United Kingdom).
Author contributions
MO, KPK, MD, MS, OAC, and RS conceived and designed the experiments. MO, MD, AP, and KPK performed the experiments. P. Cattan, MB, PM, P. Choudhary, GCH, and SRB provided human islets. MO, KPK, MS, OAC, and RS wrote the manuscript
Supplementary Material
Acknowledgments
We would like to thank Delphine Bredel (INSERM U1016) for technical assistance and Anja Steffen and Undine Schubert for help on delivery of human islets. We also acknowledge the transcriptomic platform from the Cochin Institute for performing array hybridizations and Nicolas Glaser (INSERM U1016) for his help in further data analyses. This work was supported by the European Diabetes Research Programme in Cellular Plasticity Underlying the Pathophysiology of Type 2 Diabetes from EFSD-AstraZeneca (to R. Scharfmann), Fondation pour la Recherche Médicale (to M. Diedisheim and M. Oshima), and the Aide aux Jeunes Diabétiques (to M. Diedisheim). The Scharfmann laboratory is supported by The Foundation Bettencourt Schueller, belongs to the Laboratoire d’Excellence consortium Revive and to the Departement Hospitalo-Universitaire (DHU) Autoimmune and Hormonal disease. The M. Solimena laboratory is supported by the German Center for Diabetes Research (DZD e.V.), which is financed by the German Ministry for Education and Research. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (INNODIA, no. 115797). This Joint Undertaking receives support from the Union’s Horizon 2020 research and innovation program and the European Federation of Pharmaceutical Industries and Associations, JDRF, and The Leona M. and Harry B. Helmsley Charitable Trust (to R. Scharfmann, P. Marchetti, and M. Solimena). This work was also supported by the International Research Training Group “Immunological and Cellular Strategies in Metabolic Disease” (GRK 2251) of the German Research Foundation (DFG).
Version 1. 02/08/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(3):e97732. https://doi.org/10.1172/jci.insight.97732.
Contributor Information
Masaya Oshima, Email: oshima.masaya@gmail.com.
Klaus-Peter Knoch, Email: knoch-k@rcs.urz.tu-dresden.de.
Marc Diedisheim, Email: marc_diedisheim@yahoo.fr.
Antje Petzold, Email: antje.petzold@mailbox.tu-dresden.de.
Pierre Cattan, Email: pierre.cattan@sls.aphp.fr.
Marco Bugliani, Email: m.bugliani@ao-pisa.toscana.it.
Piero Marchetti, Email: piero.marchetti@med.unipi.it.
Pratik Choudhary, Email: pratik.choudhary@kcl.ac.uk.
Michele Solimena, Email: michele.solimena@tu-dresden.de.
Olivier Albagli-Curiel, Email: olivier.albagli-curiel@inserm.fr.
References
- 1.Op de Beeck A, Eizirik DL. Viral infections in type 1 diabetes mellitus--why the β cells? Nat Rev Endocrinol. 2016;12(5):263–273. doi: 10.1038/nrendo.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hober D, Sauter P. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat Rev Endocrinol. 2010;6(5):279–289. doi: 10.1038/nrendo.2010.27. [DOI] [PubMed] [Google Scholar]
- 3.Hyöty H. Viruses in type 1 diabetes. Pediatr Diabetes. 2016;17 Suppl 22:56–64. doi: 10.1111/pedi.12370. [DOI] [PubMed] [Google Scholar]
- 4.Petzold A, Solimena M, Knoch KP. Mechanisms of beta cell dysfunction associated with viral infection. Curr Diab Rep. 2015;15(10):73. doi: 10.1007/s11892-015-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dotta F, et al. Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci USA. 2007;104(12):5115–5120. doi: 10.1073/pnas.0700442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krogvold L, et al. Detection of a low-grade enteroviral infection in the islets of langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes. 2015;64(5):1682–1687. doi: 10.2337/db14-1370. [DOI] [PubMed] [Google Scholar]
- 7.Richardson SJ, Leete P, Bone AJ, Foulis AK, Morgan NG. Expression of the enteroviral capsid protein VP1 in the islet cells of patients with type 1 diabetes is associated with induction of protein kinase R and downregulation of Mcl-1. Diabetologia. 2013;56(1):185–193. doi: 10.1007/s00125-012-2745-4. [DOI] [PubMed] [Google Scholar]
- 8.Yoon JW, Austin M, Onodera T, Notkins AL. Isolation of a virus from the pancreas of a child with diabetic ketoacidosis. N Engl J Med. 1979;300(21):1173–1179. doi: 10.1056/NEJM197905243002102. [DOI] [PubMed] [Google Scholar]
- 9.Gerling I, Nejman C, Chatterjee NK. Effect of coxsackievirus B4 infection in mice on expression of 64,000-Mr autoantigen and glucose sensitivity of islets before development of hyperglycemia. Diabetes. 1988;37(10):1419–1425. doi: 10.2337/diab.37.10.1419. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher GR, et al. Viral infection of engrafted human islets leads to diabetes. Diabetes. 2015;64(4):1358–1369. doi: 10.2337/db14-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Hello H, Ylipaasto P, Smura T, Rieder E, Hovi T, Roivainen M. Amino acids of Coxsackie B5 virus are critical for infection of the murine insulinoma cell line, MIN-6. J Med Virol. 2009;81(2):296–304. doi: 10.1002/jmv.21391. [DOI] [PubMed] [Google Scholar]
- 12.Knoch KP, et al. PTBP1 is required for glucose-stimulated cap-independent translation of insulin granule proteins and Coxsackieviruses in beta cells. Mol Metab. 2014;3(5):518–530. doi: 10.1016/j.molmet.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ylipaasto P, et al. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia. 2004;47(2):225–239. doi: 10.1007/s00125-003-1297-z. [DOI] [PubMed] [Google Scholar]
- 14.Roivainen M, et al. Mechanisms of coxsackievirus-induced damage to human pancreatic beta-cells. J Clin Endocrinol Metab. 2000;85(1):432–440. doi: 10.1210/jcem.85.1.6306. [DOI] [PubMed] [Google Scholar]
- 15.Ravassard P, et al. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80(10):5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyalwidhe JO, et al. Coxsackievirus-induced proteomic alterations in primary human islets provide insights for the etiology of diabetes. J Endocr Soc. 2017;1(10):1272–1286. doi: 10.1210/js.2017-00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8(12):911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pullen TJ, Rutter GA. When less is more: the forbidden fruits of gene repression in the adult β-cell. Diabetes Obes Metab. 2013;15(6):503–512. doi: 10.1111/dom.12029. [DOI] [PubMed] [Google Scholar]
- 20.Pullen TJ, Huising MO, Rutter GA. Analysis of purified pancreatic islet beta and alpha cell transcriptomes reveals 11β-hydroxysteroid dehydrogenase (Hsd11b1) as a novel disallowed gene. Front Genet. 2017;8:41. doi: 10.3389/fgene.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ylipaasto P, et al. Global profiling of coxsackievirus- and cytokine-induced gene expression in human pancreatic islets. Diabetologia. 2005;48(8):1510–1522. doi: 10.1007/s00125-005-1839-7. [DOI] [PubMed] [Google Scholar]
- 22.Eizirik DL, Mandrup-Poulsen T. A choice of death--the signal-transduction of immune-mediated beta-cell apoptosis. Diabetologia. 2001;44(12):2115–2133. doi: 10.1007/s001250100021. [DOI] [PubMed] [Google Scholar]
- 23.Lynn FC, Smith SB, Wilson ME, Yang KY, Nekrep N, German MS. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci USA. 2007;104(25):10500–10505. doi: 10.1073/pnas.0704054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puri S, Akiyama H, Hebrok M. VHL-mediated disruption of Sox9 activity compromises β-cell identity and results in diabetes mellitus. Genes Dev. 2013;27(23):2563–2575. doi: 10.1101/gad.227785.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamachi Y, Cheah KS, Kondoh H. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol Cell Biol. 1999;19(1):107–120. doi: 10.1128/MCB.19.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rui J, Deng S, Arazi A, Perdigoto AL, Liu Z, Herold KC. β Cells that resist immunological attack develop during progression of autoimmune diabetes in NOD mice. Cell Metab. 2017;25(3):727–738. doi: 10.1016/j.cmet.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonas JC, et al. Chronic hyperglycemia triggers loss of pancreatic beta cell differentiation in an animal model of diabetes. J Biol Chem. 1999;274(20):14112–14121. doi: 10.1074/jbc.274.20.14112. [DOI] [PubMed] [Google Scholar]
- 28.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Md Moin AS, Dhawan S, Shieh C, Butler PC, Cory M, Butler AE. Increased hormone-negative endocrine cells in the pancreas in type 1 diabetes. J Clin Endocrinol Metab. 2016;101(9):3487–3496. doi: 10.1210/jc.2016-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim-Muller JY, et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat Commun. 2016;7:12631. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cinti F, et al. Evidence of β-cell dedifferentiation in human type 2 diabetes. J Clin Endocrinol Metab. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seymour PA. Sox9: a master regulator of the pancreatic program. Rev Diabet Stud. 2014;11(1):51–83. doi: 10.1900/RDS.2014.11.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latreille M, et al. MicroRNA-7a regulates pancreatic β cell function. J Clin Invest. 2014;124(6):2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landsman L, Parent A, Hebrok M. Elevated Hedgehog/Gli signaling causes beta-cell dedifferentiation in mice. Proc Natl Acad Sci USA. 2011;108(41):17010–17015. doi: 10.1073/pnas.1105404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG. The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia. 2009;52(6):1143–1151. doi: 10.1007/s00125-009-1276-0. [DOI] [PubMed] [Google Scholar]
- 36.Cavallo MG, et al. Viral infection induces cytokine release by beta islet cells. Immunology. 1992;75(4):664–668. [PMC free article] [PubMed] [Google Scholar]
- 37.Kaminitz A, Stein J, Yaniv I, Askenasy N. The vicious cycle of apoptotic beta-cell death in type 1 diabetes. Immunol Cell Biol. 2007;85(8):582–589. doi: 10.1038/sj.icb.7100093. [DOI] [PubMed] [Google Scholar]
- 38.Oh CD, et al. SOX9 directly regulates CTGF/CCN2 transcription in growth plate chondrocytes and in nucleus pulposus cells of intervertebral disc. Sci Rep. 2016;6:29916. doi: 10.1038/srep29916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford LA, et al. Connective tissue growth factor (CTGF) inactivation leads to defects in islet cell lineage allocation and beta-cell proliferation during embryogenesis. Mol Endocrinol. 2009;23(3):324–336. doi: 10.1210/me.2008-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guney MA, et al. Connective tissue growth factor acts within both endothelial cells and beta cells to promote proliferation of developing beta cells. Proc Natl Acad Sci USA. 2011;108(37):15242–15247. doi: 10.1073/pnas.1100072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley KG, et al. Connective tissue growth factor modulates adult β-cell maturity and proliferation to promote β-cell regeneration in mice. Diabetes. 2015;64(4):1284–1298. doi: 10.2337/db14-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keenan HA, et al. Residual insulin production and pancreatic ß-cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes. 2010;59(11):2846–2853. doi: 10.2337/db10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marroqui L, et al. TYK2, a candidate gene for type 1 diabetes, modulates apoptosis and the innate immune response in human pancreatic β-cells. Diabetes. 2015;64(11):3808–3817. doi: 10.2337/db15-0362. [DOI] [PubMed] [Google Scholar]
- 44.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandra V, et al. Extracellular acidification stimulates GPR68 mediated IL-8 production in human pancreatic β cells. Sci Rep. 2016;6:25765. doi: 10.1038/srep25765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albagli-Curiel O, Lécluse Y, Pognonec P, Boulukos KE, Martin P. A new generation of pPRIG-based retroviral vectors. BMC Biotechnol. 2007;7:85. doi: 10.1186/1472-6750-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlier G, et al. Human fucci pancreatic Beta cell lines: new tools to study Beta cell cycle and terminal differentiation. PLoS One. 2014;9(9):e108202. doi: 10.1371/journal.pone.0108202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkegaard JS, et al. Xenotropic retrovirus Bxv1 in human pancreatic β cell lines. J Clin Invest. 2016;126(3):1109–1113. doi: 10.1172/JCI83573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.