Abstract
Respiratory viral infections represent the most common cause of allergic asthma exacerbations. Amplification of type 2 immune response is strongly implicated in asthma exacerbation, but how virus infection boosts type 2 responses is poorly understood. We report a significant correlation between release of host double stranded DNA (dsDNA) following rhinovirus infection and exacerbation of type 2 allergic inflammation in humans. In a mouse model of allergic airway hypersensitivity, we show that rhinovirus infection triggers dsDNA release associated with neutrophil extracellular traps (NETs) formation (NETosis). We further demonstrate that inhibiting NETosis by blocking neutrophil elastase, or degrading NETs with DNase protects mice from type 2 immunopathology. Furthermore, injection of mouse genomic DNA alone is sufficient to recapitulate many features of rhinovirus-induced type 2 immune responses and asthma pathology. Thus, NETosis and its associated extracellular dsDNA contribute to the pathogenesis and may represent potential therapeutic targets of rhinovirus-induced asthma exacerbations.
Keywords: rhinovirus, asthma, exacerbation, NET, NETosis, neutrophil, neutrophil elastase, host DNA, dendritic cell
Introduction
Allergic asthma is a major and increasingly prevalent public health problem. Acute asthma exacerbations are a major unmet medical need. Respiratory virus infections (mostly rhinovirus [RV]) are strongly associated with asthma exacerbations1–3 and synergistic interactions between allergen exposure/sensitization and virus infections increase risk of exacerbation4,5, though mechanisms involved are poorly understood. Key aspects of RV-induced exacerbations are orchestrated by “type 2” immune cells, in particular adaptive type 2 helper T (TH2) cells3,6–9. Since viral infections induce immune responses with a predominant type 1 profile10,11, additional factors must participate in such type 2 immune-mediated exacerbation3.
Airway neutrophilia is increased during virus-induced exacerbations12–14 and neutrophils can undergo a process called NETosis that releases neutrophil extracellular traps (NETs) into the extracellular space. NETs contain double-stranded (ds)DNA, histone and non-histone proteins, including neutrophil elastase (NE) and myeloperoxidase (MPO), which regulate NET formation and are anti-microbial15,16. Some viruses can induce NETosis17 and host dsDNA, alone or in combination with antimicrobial components, can be sensed by the innate or adaptive immune system to boost type 2 immunity18–20. However, the potential role of NETs has never been investigated in virus-induced asthma exacerbations. We postulated that NETs might be released during RV-induced exacerbation to boost type 2 immune responses and drive exacerbation severity.
Here, we report that RV infection induces dsDNA release in humans, a phenomenon that is amplified in subjects with asthma and related to type 2 cytokine induction and exacerbation severity. In mice, RV infection is a potent inducer of NETs thereby promoting type 2 pathology. Thus, NETosis, by releasing dsDNA, plays a central functional role in exacerbation pathogenesis.
Results
RV-induced dsDNA is related to asthma exacerbation severity
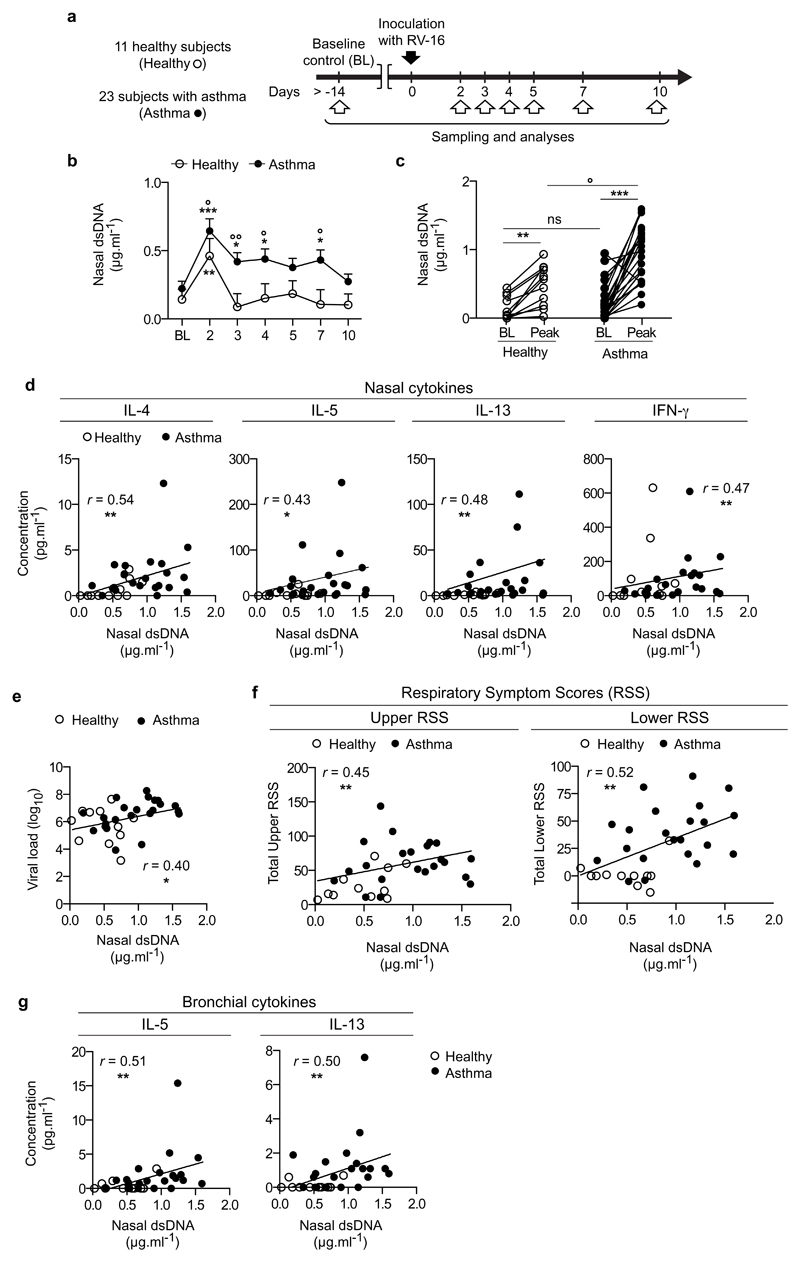
To determine whether host dsDNA is released during RV-induced asthma exacerbations and assess the relationship between RV infection, host dsDNA and type 2 immune responses, we analysed nasal lavage samples collected after RV-16 inoculation from 23 (who had lavage available) of 28 subjects with mild/moderate atopic asthma and 11 healthy control subjects who had previously undertaken a study investigating asthma exacerbation pathogenesis7,9 (Fig.1a and Supplementary Table 1). Since rhinoviruses are RNA viruses with no dsDNA intermediates, dsDNA detected in the airways after RV infection is most likely host-derived. Nasal dsDNA concentrations in the upper airways were increased on day 2 post RV inoculation (p.i.) compared to baseline both in healthy subjects and subjects with asthma, but to a higher extent in the latter group (Fig. 1b). On days 3, 4 and 7, dsDNA concentrations were induced compared to baseline in subjects with asthma but not healthy subjects, and were greater in subjects with asthma than in healthy subjects (Fig.1b). After infection, peak concentrations of dsDNA (i.e., the maximal concentrations of dsDNA detected during the infection for each subject) were increased in both groups of volunteers compared to baseline, but peak concentrations of dsDNA were higher in subjects with asthma compared to healthy subjects (Fig.1c). No differences were found between subjects with asthma treated with inhaled corticosteroid (ICS) or without ICS treatment (Supplementary Fig.1a-c).
Figure 1. Host dsDNA is released during human RV infection, is increased in RV-induced asthma exacerbation and correlates with type 2 cytokine production and exacerbation severity.
(a) Experimental outline. 11 healthy control and 23 subjects with asthma underwent sampling and analysis at baseline (BL) and 2, 3, 4, 5, 7 and 10 days after RV16 infection. (b) Concentrations of extracellular dsDNA in the acellular fraction of the nasal lavage fluid of subjects with asthma vs. healthy subjects at baseline and after RV16 infection, measured over time. (c) Comparison of baseline and peak (i.e., the maximal concentrations of dsDNA detected during the infection for each subject) levels of extracellular dsDNA, and between subjects with asthma and healthy subjects. (d) Correlation between peak levels of extracellular dsDNA and peak levels of the indicated nasal cytokines (i.e., the maximal levels of cytokines detected during the infection for each subject). (e) Correlation between peak concentrations of nasal dsDNA and peak viral load detected in the nose. (f) Correlation of peak levels of extracellular dsDNA with total upper and lower RSS. (g) Correlation of bronchial concentrations (on day 4 during infection) of IL-5 and IL-13 with peak concentrations of nasal dsDNA. (b-c) Asterisks (*) compare differences within the groups; circles (˚) compare differences between subjects with asthma and healthy subjects (b) at the indicated time points or (c) at the peak. The statistical significances of the differences between the baseline and the various times averages or the peak levels, and between the cohorts at the various time points were determined using contrasts between least square means estimated in the mixed model (described in the Online methods). (d-g) The correlation analysis used was nonparametric (Spearman’s correlation). Error bars indicate SEM. */˚, P<0.05; **/˚˚, P<0.01; ***/˚˚˚, P<0.001.
As reported in the original study9, the type 2 cytokines interleukin (IL)-5 and IL-13 were induced by RV infection in the nose of the 23 subjects with asthma included herein, and peak nasal concentrations were higher during infection in asthma than in healthy subjects (data not shown). One exception was IL-4 levels, which achieved statistical significance between baseline and the peak and between control and asthma groups in the original study9, but not in the current sub-groups (Supplementary Fig.1d). Nonetheless, peak concentrations of all three type 2 cytokines correlated with those of nasal dsDNA during infection (Fig.1d). Concentrations of the type 1 cytokine interferon (IFN)-γ, which were increased in both groups after RV infection but were not different between subjects with asthma and healthy subjects (data not shown), also correlated with dsDNA release (Fig.1d). As previously reported9, nasal viral loads on day 3 were higher in the subjects with asthma included herein, compared to healthy subjects (data not shown), and correlated with dsDNA concentrations (Fig.1e).
The 23 subjects with asthma included herein also displayed greater upper and lower respiratory symptom scores (RSS) compared to healthy subjects following inoculation with RV (ref. 9 and data not shown). Nasal concentrations of host dsDNA were positively correlated not only with cold symptom severity (total upper RSS), but, importantly, also with asthma exacerbation severity (total lower RSS) (Fig.1f) and the associated lower airway type 2 (bronchial mucosal lining fluid concentrations of IL-5 and IL-139) immune response (Fig.1g).
These results show that RV infection in subjects with asthma induces dsDNA release, which correlates with type 2 immune-mediated asthma exacerbation severity.
RV induces dsDNA in a mouse model of asthma exacerbation
To further investigate the relationships between RV-mediated release of host dsDNA and asthma exacerbation, we employed a clinically relevant mouse model (Fig.2a, Supplementary Fig.2a and Supplementary Fig.3a) able to recapitulate key aspects of RV-induced asthma exacerbation in humans9. House dust mite (HDM)-sensitized BALB/c mice that were challenged with HDM (called hereafter “allergic” mice) exhibited signs of allergic airway inflammation when compared to PBS-sensitized HDM-challenged controls, including higher numbers of total cells, eosinophils and lymphocytes in the bronchoalveolar lavage fluid (BALF), and higher serum concentrations of IgE and HDM-specific IgG2a and IgG1 (Fig.2b-c and Supplementary Fig.2b-d). Allergic mice inoculated with RV-1b displayed exacerbated features of asthma when compared to allergic mice inoculated with ultraviolet-inactivated RV-1b (UV) and RV–inoculated non-allergic mice, including increased numbers of total cells and numbers and percentages of eosinophils and lymphocytes in the BALF, higher serum concentrations of IgE, greater concentrations of BALF Muc5AC protein, enhanced airway inflammatory cell infiltration, mucus production and airway hyperreactivity (Fig.2b-g and Supplementary Fig.2b-g). Virus load was similar between RV-inoculated allergic and non-allergic animals (Supplementary Fig.2h).
Figure 2. RV-induced exacerbation of allergic airway inflammation and type 2 immune responses are associated with host dsDNA release in mice.
(a) Experimental outline. HDM-sensitized or PBS-injected mice were challenged intranasally (i.n.) with HDM prior to inoculation with RV1b or UV-inactivated RV1b (UV). (b) Total cell counts and differential immune cell counts and percentages in the bronchoalveolar lavage fluid (BALF). (c) Absolute quantification of serum levels of total IgE. (d) Relative quantification of Muc5AC protein in the BALF. (e) Inflammatory score estimated from hematoxylin and eosin staining of lung sections. (f) Percentage positivity of periodic acid Schiff (PAS)-stained goblet cells per total epithelial cells. (g) Measurement of dynamic airway resistance. (h-i) Gating strategy to analyse the recruitment of TH2 lymphocytes (SSClowCD3+CD4+ICOS+ST2+) to the lung of mice by flow cytometry. (j) Total numbers and percentage of TH2 lymphocytes among live cells in the lung. (k-l) Levels of TH2 cytokines in the supernatant of cells isolated from the lung (k) and the mediastinal lymph nodes (MLNs) (l) and restimulated ex vivo with HDM. (m) Time course analysis of extracellular dsDNA release in the BALF of HDM-naïve mice after RV1b inoculation. Asterisks (*) compare differences between the indicated time points vs. the baseline. (n) Concentration of extracellular dsDNA in the BALF of the indicated groups of mice. Data are (b-j;m-n) of 1 experiment representative of 3 independent experiments, each replicate containing 5 mice/group; (k-l) pooled from 3 independent experiments, each symbol representing the mean of 1 experiment in which LN cells from the 5 mice were pooled by group. Differences between multiple groups were estimated using a one-way ANOVA with Tukey’s post hoc test (b-d,g,j-n, data show mean + SD) or Kruskal-Wallis test (e-f, data show median + interquartile range). *, P<0.05; **, P<0.01; ***, P<0.001. AU, arbitrary unit.
Quantification of lung TH2 lymphocytes (defined here as singlet live, SSClowCD3+CD4+ICOS+ST2+ cells21, Fig.2h) revealed their higher numbers and percentages in the lungs of RV-infected allergic mice compared to uninfected allergic and infected non-allergic mice (Fig.2i-j and Supplementary Fig.3b). In addition, lung and mediastinal lymph node (MLN) cells isolated from RV-infected allergic mice produced more TH2 cytokines than cells isolated from uninfected allergic and infected non-allergic mice upon ex vivo HDM restimulation (Fig.2k-l and Supplementary Fig.3c-d). None of the TH2 cytokines were detectable in unstimulated cells (data not shown). These results confirm that RV-induced exacerbation of allergic airway inflammation is associated with a robust type 2 immune response in a clinically relevant context.
We next sought to investigate whether dsDNA was released following RV inoculation in our model. In naïve mice, RV induced an early burst of dsDNA release in the airways starting at 24 hours, peaking at day 2 and returning to baseline 4 days p.i. (Fig.2m). In the allergy setting, HDM challenge in combination with UV-treated virus inoculation induced low level of dsDNA. As in naïve mice, dsDNA was robustly increased in HDM-challenged RV-inoculated mice. Further induction was observed in RV HDM-sensitized, HDM-challenged (infected allergic) animals compared with both uninfected allergic and infected non-allergic mice (Fig.2n).
These results recapitulate our observations in humans (Fig.1b-c) that RV infection is able to trigger dsDNA release and that this is amplified in the airways of allergic subjects.
DNase protects against RV-induced exacerbation
To investigate whether dsDNA could drive type 2 immune-mediated asthma exacerbation, we treated mice with DNase (Fig.3a, Supplementary Fig.4a and Supplementary Fig.5a), abolishing dsDNA accumulation in the airways of mice inoculated with RV (Fig.3b and Supplementary Fig.4b).
Figure 3. DNase treatment prevents RV-induced type 2 mediated exacerbation of allergic airway inflammation.
(a) Experimental outline. Mice were treated with DNaseI by i.p. injection 4 hours before inoculation and 1 and 2 days p.i. and by the i.n. route 8 hours and 1 and 2 days p.i. (b) Levels of dsDNA in the BALF at day 1 p.i. (c) Total cell counts and differential immune cell counts and percentages in bronchoalveolar lavage fluid (BALF). (d) Absolute quantification of serum levels of total IgE. (e) Relative quantification of Muc5AC protein in the BALF. (f) Inflammatory score estimated from hematoxylin and eosin staining of lung sections. (g) Percentage positivity of periodic acid Schiff (PAS)-stained goblet cells per total epithelial cells. (h) Measurement of dynamic airway resistance. (i) Total number and percentage of TH2 lymphocytes (SSClowCD3+CD4+ICOS+ST2+) among live cells in the lungs of mice. (j,k) Levels of TH2 cytokines in the supernatant of cells isolated from the lung (j) and the mediastinal lymph nodes (MLNs) (k) and restimulated ex vivo with HDM. Data in (b-i) are from one experiment representative of 3 independent experiments with 6 (c-h) or 5 (b,i) mice/group. Data in (j-k) are pooled from 3 independent experiments, each symbol representing the mean of 1 experiment in which LN cells from the 6 mice were pooled by group. Differences between multiple groups were estimated using one-way ANOVA with Tukey’s post hoc test (b-e, h, i-k, data show mean + SD) or Kruskal-Wallis test (f-g, data show median + interquartile range). */˚, P<0.05. **/˚˚, P<0.01; ***/˚˚˚, P<0.001; circles (˚) compare DNase-treated vs. vehicle-injected counterparts. AU, arbitrary unit.
DNase-treated RV-infected allergic mice had reduced features of allergic asthma exacerbation compared to vehicle-treated counterparts, with lower numbers of total cells, numbers and percentages of eosinophils and lymphocytes in the BALF, concentrations of serum IgE, concentrations of BALF Muc5AC protein, airway inflammatory cell infiltration, mucus production and airway hyperreactivity (Fig.3c-h and Supplementary Fig.4c-f). DNase treatment did not affect BALF neutrophil numbers and viral replication (Supplementary Fig.4d,g), suggesting that reduction of exacerbated disease was not due to a difference in viral loads. Serum concentrations of HDM-specific IgG2a and IgG1 remained unchanged upon DNase treatment (Supplementary Fig.4h).
Assessing specifically type 2 immune responses, DNase treatment also reduced numbers and percentages of lung TH2 lymphocytes (Fig.3i and Supplementary Fig.5a-b), as well as TH2 cytokine production following ex vivo HDM restimulation of lung and MLN cells (Fig.3j-k and Supplementary Fig.5c-d).
Thus, DNase treatment substantially reduced type 2 immune responses and RV-induced exacerbation of allergic airway inflammation, consistent with the idea that extracellular DNA contributes to boosting of type 2 immune responses and exacerbation of asthma by RV.
dsDNA is sufficient to boost type 2 immune responses
Next, we investigated whether exogenous dsDNA, when injected alone, could recapitulate features of virus-driven exacerbation of type 2 immune responses and asthma pathology (Fig.4a, Supplementary Fig.6a and Supplementary Fig.7a). Administration of dsDNA from mouse splenocytes recapitulated many, but not all, of the immune and pathological features seen after RV infection. When compared to vehicle-treated allergic mice, dsDNA-treated allergic mice exhibited greater numbers of total cells, numbers and percentages of eosinophils and lymphocytes in the BALF, concentrations of serum IgE, airway inflammatory cell infiltration and lung and MLN-associated type 2 immune responses (Fig.4b-c,e,h-j, Supplementary Fig.6b-d and Supplementary Fig.7b-d). However, dsDNA-treated allergic mice did not exhibit greater mucus production or enhanced airway hyperactivity compared to vehicle-treated allergic mice (Fig.4d,f-g and Supplementary Fig.6e). Similar to RV infection, dsDNA also did not increase serum concentrations of HDM-specific IgG2a and IgG1 above concentrations seen in allergic mice (Supplementary Fig.6f). In non-allergic mice, dsDNA treatment did not interfere with any aspect of lung inflammation assessed in this model (Supplementary Fig.6 and Supplementary Fig.7).
Figure 4. Host dsDNA is sufficient to exacerbate type 2 immune responses in allergic mice.
(a) Experimental outline. Mice were injected i.t. with 10μg of endogenous dsDNA 24h, 36h, 48h and 72h after the last HDM challenge. (b) Total cell counts and differential immune cell counts and percentages in bronchoalveolar lavage fluid (BALF). (c) Absolute quantification of serum levels of total IgE. (d) Relative quantification of Muc5AC protein in the BALF. (e) Inflammatory score estimated from hematoxylin and eosin staining of lung sections. (f) Percentage positivity of periodic acid Schiff (PAS)-stained goblet cells per total epithelial cells. (g) Measurement of dynamic airway resistance. (h) Total number and percentage of TH2 lymphocytes (SSClowCD3+CD4+ICOS+ST2+) among live cells in the lung. (i,j) Levels of TH2 cytokines in the supernatant of cells isolated from the lung (i) and the mediastinal lymph nodes (MLNs) (j) and restimulated ex vivo with HDM. Data are (b-h) of 1 experiment representative of 3 independent experiments with 5 mice/group; (i-j) pooled from 3 independent experiments, each symbol representing the mean of 1 experiment in which LN cells from the 6 mice were pooled by group. Differences between multiple groups were estimated using one-way ANOVA with Tukey’s post hoc test (b-d, g, h-j, data shown mean + SD) or Kruskal-Wallis test (e-f, data show median + interquartile range). */˚, P<0.05. **/˚˚, P<0.01; ***/˚˚˚, P<0.001; circles (˚) compare dsDNA-injected vs. vehicle-injected counterparts. ns, not significant; AU, arbitrary unit.
Thus host dsDNA release, as observed after RV infection, is sufficient to exacerbate many features of type 2-mediated allergic inflammation.
DNase inhibits monocyte-derived dendritic cell recruitment
During TH2 sensitization, host dsDNA acts preferentially on dendritic cells (DCs) to boost type 2 immune responses19,20, and monocyte-derived DCs (mo-DCs) are recruited to the lung and subsequently to the MLN upon allergen challenge to maintain TH2 cell-mediated immunity to HDM22. We thus reasoned that DNase may suppress type 2 immune-mediated exacerbations by inhibiting mo-DC recruitment in our model.
Numbers of mo-DCs (defined here as singlet live, CD11c+MHC-IIhighCD103-CD11b+CD64+ cells22) and also CD11b+ conventional DCs (cDC2, defined here as singlet live, CD11c+MHC-IIhighCD103-CD11b+CD64-MAR-1- cells22), the DC subset responsible for priming TH2 responses to HDM23,24, were evaluated on day 3 p.i. (Fig.5a). As expected in allergic mice22, numbers of mo-DCs recruited to the lung were higher compared to PBS-sensitized controls (Fig.5b-c). However, there was no recruitment of mo-DCs to the MLNs (Fig.5e-f). Following RV infection alone, numbers of mo-DCs were higher in lungs and MLNs compared to UV-inoculated controls, and numbers were further increased in infected allergic mice (Fig.5c,f). Numbers of cDC2 were also higher in the lungs of allergic mice and in RV infected mice compared to non-allergic/infected control mice, but, unlike mo-DCs, cDC2 numbers were not higher in RV-infected allergic mice compared to UV-inoculated allergic controls (Fig.5d). In the MLNs numbers of cDC2 were also increased in RV-inoculated as compared to UV-inoculated allergic mice (Fig.5g).
Figure 5. DNase treatment inhibits monocyte-derived dendritic cell recruitment during RV-induced exacerbation of allergic airway inflammation.
(a) Experimental outline. (b-d) Quantification of lung pro-TH2 dendritic cell (DC) populations. (b) Gating strategy to identify DC subsets present in the lung. We defined the monocyte-derived DC subset (mo-DC) as singlet living CD11c+MHCIIhighCD103-CD11b+CD64+ cells and the conventional CD11b+ DC subset (CD11b+cDC2) as CD11c+MHCIIhighCD103-CD11b+CD64-MAR- cells. (c,d) Total numbers of moDCs (c) and CD11b+cDC2 (d) among live cells in the lung are shown. (e-g) Quantification of MLN pro-TH2 DC populations. (e) Gating strategy to identify DC subsets present in the MLN. (f,g) Total numbers of moDCs (f) and CD11b+cDC2 (g) among live cells in the MLN are shown. (h-j) Levels of CCL2 (h), CCL7 (i) and CCL12 (j) in the acellular fraction of BALF. (b-j) Data are of 1 experiment representative of 2 independent experiments with 5 mice/group. Differences between multiple groups were estimated using one-way ANOVA with Tukey’s post hoc test. Error bars indicate SD. */˚, P<0.05. **/˚˚, P<0.01; ***/˚˚˚, P<0.001; circles (˚) compare DNase-treated vs. vehicle-injected counterparts. ns, not significant.
We then investigated whether mo-DC recruitment to the lung and MLNs was affected by DNase treatment and found that DNase-treated RV-infected allergic mice had lower numbers of mo-DCs in the lung and MLNs compared to vehicle-treated counterparts (Fig.5c,f). Conversely, DNase treatment of RV-infected allergic mice did not reduce numbers of cDC2 in the lung or MLNs compared to vehicle-treated counterparts (Fig.5d,g).
During respiratory syncytial virus (RSV) infection, mo-DC recruitment is driven by the chemokines CCL2, CCL7 and CCL1225. One day p.i., BALF concentrations of CCL2, CCL7 and CCL12 were increased in RV-infected allergic mice compared to UV-inoculated controls (Fig.5h-j). Moreover, concentrations of CCL7 and CCL12 were higher in RV-inoculated allergic mice than in RV-inoculated non-allergic mice (Fig.5i-j). DNase treatment substantially decreased concentrations of CCL2 and CCL7, but not CCL12, in DNase-treated RV-infected allergic and non-allergic mice as compared to vehicle-treated counterparts.
Taken together, our results are concordant with the hypothesis that RV-induced extracellular dsDNA, by promoting CCL2 and CCL7 secretion and boosting mo-DC recruitment, contributes to RV-induced exacerbation of allergic airway inflammation.
RV-induced NETs mediate asthma exacerbation pathogenesis
We next investigated mechanisms underlying dsDNA release during RV-induced asthma exacerbations. Virus-induced asthma exacerbations are associated with neutrophil recruitment to the airways12,13 and viruses are able to prime neutrophils to extrude NETs26, mainly composed of dsDNA. We therefore postulated that RV infection could induce NETs thus providing a source of dsDNA to contribute to exacerbation of allergic asthma.
In human nasal lavages, the NET-associated protein NE16,27 was induced by RV infection only in subjects with asthma, and there was a non-significant trend towards increased NE during infection in subjects with asthma compared to healthy subjects (Fig.6a). In addition, nasal concentrations of NE were positively correlated with those of dsDNA at day 4 p.i. (Fig.6b). Thus, NETs induced by RV infection are a likely source of dsDNA in RV-infected human airways. In our mouse model, NE concentrations were also increased in the BALF of RV-inoculated mice at day 1 p.i., were greater in RV-infected allergic mice than in both uninfected allergic and infected non-allergic mice (Fig.6c) and correlated with dsDNA concentrations (Fig.6d).
Figure 6. NETs released during RV infection promote type 2 immune-mediated exacerbation of allergic airway inflammation.
(a) Nasal baseline (BL) and day 4 p.i. levels of neutrophil elastase (NE) in subjects with asthma vs. healthy subjects. The statistical significances of the differences between the baseline and the peak, and between the groups at the various time points were determined using contrasts between least square means estimated in the mixed model (described in the Online methods). (b) Correlation between nasal levels of NE and dsDNA levels at day 4 p.i. in human samples. The correlation analysis used was nonparametric Spearman’s correlation. (c) Levels of NE in the bronchoalveolar lavage fluid (BALF) of the indicated groups of mice. Differences between groups were estimated using a Welch's ANOVA (compensating for the heterogeneity in the variances between the groups) with Tukey’s post hoc test. (d) Correlation between BALF NE levels and levels of dsDNA in mice. The correlation analysis used was parametric Pearson’s correlation. (e) Experimental outline for experiments shown in f-i. Mice were treated with neutrophil-depleting anti-Ly6G antibodies (α-Ly6G) 24 h prior to RV1b inoculation. (f,g) Total number of neutrophils (f) and levels of extracellular dsDNA (g) in the BALF of indicated groups of mice. (h,i) Quantification of NETs by high resolution confocal scanning microscopy (citrullinated histone H3 [Cit-H3], red, MPO [green] and DNA [DAPI, blue]) (h) and levels of Cit-H3 protein by Western Blot (i) in the lungs of the indicated groups of mice. Insets in h magnified on the right side. GAPDH was used as a loading control in i. (j) Experimental outline for experiments shown in k-o. Mice were treated by i.p. injection of the NE inhibitor (NEi; GW311616A) 12 hours before RV1b inoculation and every 12 hours after. (k) RV1b viral RNA in the lung tissue of mice. (l) Total and differential immune cell counts in BALF. (m) Measurement of dynamic airway resistance. (n) Total number and percentage of TH2 lymphocytes (SSClow, CD4+, CD3+, ICOS+, ST2+) among living cells in the lung. (o) Levels of TH2 cytokines in the supernatant of cells isolated from the mediastinal lymph node (MLN) and stimulated ex vivo with HDM. (c-o) Differences between groups were estimated using a Welch's ANOVA (compensating for the heterogeneity in the variances between the groups) with Tukey’s post hoc test. Data are (c-n) 1 experiment representative of 2 independent experiments with 5 mice/group; (o) pooled from 2 independent experiments, each symbol representing the mean of 1 experiment in which LN cells from the 5 mice were pooled by group. Blot images in i have been cropped and full-length blots can be found in Supplementary Fig.9e,f. Data show mean + SD. */˚, P<0.05; **/˚˚, P<0.01; ***/˚˚˚, P<0.001; circles (˚) compare α-Ly-6G- or NEi-treated vs. vehicle-injected counterparts. ns, not significant. Scale bar in h =100μm, except in magnified areas=10µm.
To assess the dependency of dsDNA release on neutrophil recruitment to the airways of RV-infected allergic mice, neutrophils were depleted by treating mice intraperitoneally (i.p.) with anti-Ly6G neutralizing antibodies (α-Ly6G) prior to RV inoculation (Fig.6e and Supplementary Fig.9a). α-Ly6G treatment blocked RV-induced neutrophil recruitment to the airways (Fig.6f), and decreased BALF dsDNA to concentrations observed in UV-inoculated allergic mice (Fig.6g).
To unambiguously test whether RV can induce NETs in the lungs of mice, identification and relative quantification of NET formation were performed in our model of RV-induced asthma exacerbation. NETs were identified by immunofluorescence as areas staining triple positive for DNA, citrullinated histone H3 (Cit-H3) and MPO (a NET-associated protein16) and were semi-quantified by Western blots of Cit-H3 in lungs of mice28. NETs were abundantly present in lungs of RV-infected mice at day 2 p.i., regardless of their allergic status (Fig.6h,i, Supplementary Fig.8 and Supplementary Fig.9a-f). NETs and Cit-H3 were virtually absent in lungs of UV-inoculated allergic and non-allergic mice. Blocking neutrophil recruitment with α-Ly6G abolished NET and Cit-H3 protein detection in the lungs without affecting eosinophil numbers (Fig.6h,i and Supplementary Fig.9), demonstrating that neutrophils, but not eosinophils, represented the source of extracellular traps and dsDNA. To corroborate this further, we assessed neutrophil recruitment and dsDNA concentrations in eosinophil-deficient ΔdblGATA BALB/c mice29 (Supplementary Fig.10a). The number of neutrophils and dsDNA were not affected by eosinophil deficiency (Supplementary Fig.10b,c). These data confirmed that dsDNA release in RV-infected allergic mice was dependent on neutrophil-, but not eosinophil-derived extracellular traps.
Finally, to formally assess the contribution of NETs to RV-induced asthma exacerbation and avoid non-specific effects of DNase, we used a specific inhibitor of NETosis (GW311616A; a neutrophil elastase inhibitor, NEi16,27) in our mouse model (Fig.6j and Supplementary Fig.11a). We confirmed that NEi reduced RV-induced NETs in the airways of infected mice (Supplementary Fig.11b-d) without affecting BALF neutrophil numbers and viral load (Fig.6k,l). NEi-treated RV-infected allergic mice displayed a reduction in several features of type 2-mediated exacerbation of allergic airway inflammation when compared to vehicle-treated counterparts, including less BALF total cells, eosinophils and lymphocytes (Fig.6l), airway hyperreactivity (Fig.6m), numbers and percentages of lung TH2 cells and release of TH2 cytokines from HDM re-stimulated MLN cells (Fig.6n,o). We conclude that NETs are induced by RV in vivo, and that targeting NETosis recapitulates the effects of DNase treatment (Fig.3) and allows protection of mice from RV-induced exacerbation of allergic airway inflammation.
Discussion
The nature of the signals that amplify TH2 responses during virus-induced asthma exacerbation remains a critical question in asthma research. Building on previous studies reporting that dsDNA released from dying host cells is responsible for developing type 2 immune responses following alum-mediated sensitization19,20, we show here that host-derived dsDNA released during RV infection is amplified in subjects with asthma undergoing RV-induced asthma exacerbations, and that dsDNA levels are strongly related to both exacerbation symptom severity and RV-induced type 2 cytokine release. Similar increases in airway levels of dsDNA are also found in RV-infected mice.
Here we developed a clinically relevant mouse model of RV-induced exacerbation of allergic airway inflammation that recapitulates many features of RV-induced asthma exacerbation in humans. Indeed, even though RV1b does not naturally infect mice and its capability to replicate is limited to 1 to 2 days6, features of allergic asthma exacerbations in the mouse model are of substantial duration and are completely dependent on virus replication, like in humans. Moreover, the timing of dsDNA and exacerbation responses is similar in mice and humans, with a peak of dsDNA release on day 2, and asthma exacerbation detectable at days 3-4 (as quantified by the respiratory scores in humans9 and airway hyperreactivity and allergic airway inflammation in mice).
…Exogenous DNase administration abolished all relevant exacerbation outcomes in this model, revealing a crucial functional role played by dsDNA or dsDNA-containing structures in RV-induced exacerbation of type 2-mediated allergic airway inflammation.
…Two major sources of extracellular host DNA in tissues are NETs and necrotic cells. When injected in the absence of virus infection, dsDNA from splenocytes recapitulated many but not all of the exacerbating effects of RV on asthma-related outcomes in mice, suggesting that more complex structures containing dsDNA, rather than dsDNA alone, were responsible for the full aggravating effects. This assumption was concordant with the fact that the commercially available DNase that we used degrades not only dsDNA but also DNA-associated proteins19,30–33. We therefore postulated that NETs, which contain both dsDNA and proteins/peptides, were the main source of airway dsDNA elicited by RV infection. This hypothesis is supported by several observations. First, RV1b robustly induced NETs in RV-infected animals. Second, NET inhibition in RV-infected mice had the same beneficial effects as DNase on the characteristic features of asthma. Third, Ly6G blocking antibody almost completely abolished neutrophil recruitment and NET formation and decreased levels of dsDNA to levels observed in non-infected allergic mice. Fourth, the use of eosinophil-deficient ΔdblGATA mice rules out a potential contribution of eosinophils as a source of extracellular traps and dsDNA. Finally, experimental RV16 infection in humans induces NE release correlated with the nasal levels of dsDNA.
…However, there is significant dsDNA release in non-infected HDM-exposed mice in the absence of NETs. We speculate that in this case HDM induces dsDNA release by other mechanisms, likely related to HDM-induced tissue damage34, and this dsDNA is derived from necrotic airway resident cells rather than NETs.
Several publications have suggested that host dsDNA exerts its effects mainly on innate immune cells through a STING-TBK1-IRF3-dependent signalling pathway19,20,35–38. In this study, in agreement with previous ones19,20, we propose that NET-derived dsDNA acts as an adjuvant to promote recruitment of pro-TH2 DCs to the lung and to the MLNs. More specifically, our data support that RV infection, by inducing NETs, is able to boost mo-DC recruitment during the allergen challenge, which has been shown to maintain specifically TH2 cell-mediated immunity against HDM22. Another report suggested that mo-DC recruitment was driven after RSV infection to preferentially induce TH1 immune responses25, suggesting that the functional specialization of mo-DCs during RV infection, most probably originates from the TH2 microenvironment. However, in non-sensitized mice, the release of dsDNA during RV infection increases mo-DC recruitment without promoting type 2 immune responses, suggesting the need to provide two separate signals, one provided by the TH2 microenvironment and the other provided by the virus, to allow mo-DCs to drive the exacerbation of TH2-mediated adaptive immune responses.
In addition to acting on mo-DCs, there are other ways by which dsDNA could contribute to exacerbation of allergic airway inflammation. An earlier study has demonstrated that host dsDNA released from dying cells and complexed with antimicrobial peptide is able to directly stimulate CD4+ T cells to induce TH2 cell differentiation and activation18. Several others reported that NETs and antimicrobial peptides are both released during respiratory viral infection39,40, suggesting that in addition to the role played by mo-DCs, a complex of dsDNA and antimicrobial peptides released during the NETosis process could interact with CD4+ T cells and exacerbate type 2 immune responses.
Given that dsDNA alone only partially recapitulates the exacerbating effects of RV infection, it is reasonable to postulate that other NET-associated molecules may contribute to virus-induced exacerbation of allergic airway inflammation. Among these molecules, the antimicrobial peptide LL37 has been reported to facilitate the transport of self-dsDNA into antigen-presenting cells35,41 making it a possible partner for helping dsDNA to boost type 2 responses. In addition, other studies have shown that NE can contribute to airway hyperresponsiveness and airway inflammation in rodent models42,43 and inhibition of NE can attenuate RV-induced airway hyperreactivity in our model. These data, together with the fact that dsDNA alone, as opposed to RV1b inoculation, did not increase airway hyperreactivity, support the hypothesis that NET-derived NE may contribute to AHR during exacerbation. The other NET-associated proteins rather seem to have inhibitory functions on type 2 immunity. Indeed, Cathepsin G stimulated type I IFN production by plasmacytoid DCs, which stimulated antiviral responses and autoimmunity but dampened type 2 responses44,45. Moreover, NE has also been reported to attenuate DC maturation and function, resulting in reduced allostimulatory ability or inactivation of regulatory T cells46–48. Further experiments will be required to determine the respective roles of these NET-associated-proteins/peptides.
NETs, by releasing dsDNA and antimicrobial peptides, are beneficial defence structures that can help the host to fight against infections26,49,50. However, several studies have revealed that NETs could exert adverse effects characterized by massive influx of neutrophils into the lung when their regulation is altered40,49,51. In line with these studies, our data confirm that massive neutrophil recruitment and excessive NET deposits associated with substantial dsDNA release in the airway of allergic mice after RV infection promotes exacerbation of type 2 immune responses. As other major causes of asthma exacerbation (e.g., bacterial infection and cigarette smoke40,52,53) are related with neutrophil recruitment and NETosis, it would be interesting to investigate the impact of NETs and associated dsDNA on the exacerbation of type 2 immune response in these conditions as well.
A remaining question is how RV infection drives NETosis. Since neutrophils are not a target for RV replication54, it is plausible that the virus itself is not able to promote NETosis and that secondary signals are needed during RV infection to drive this process in vivo. Investigating the signal(s) induced by the virus to promote NETosis is a very interesting perspective for future research. As dsDNA starts to be released 24 hours after RV inoculation in mice and NETs are clearly present at 48 hours, it is plausible that the secondary signals activating NETosis are released from RV-infected respiratory epithelial cells, the primary site of RV replication54.
In conclusion, our findings identify a novel important role for NETs and NET-associated dsDNA in virus-induced asthma exacerbation. Our data also suggest controlling the release of NETs (for example with inhibitors of NETosis) or speeding up their clearance (for example with DNase) as novel potential therapeutic options for treatment of virus-induced exacerbation of type 2 immune responses.
Online Methods
In vivo human model of RV-induced allergic asthma exacerbation
28 mild to moderate atopic subjects with asthma and 11 non-atopic, non-asthmatic healthy volunteers without a recent viral illness or serum neutralizing antibodies to RV-16 (RV16) at screening were recruited to a previously reported study investigating the pathogenesis of RV induced asthma exacerbations9. Subjects with asthma were excluded if they had severe disease, a recent asthma exacerbation or current symptoms of allergic rhinitis. Clinical characteristics of healthy volunteers recruited by strict inclusion/exclusion criteria have been previously reported9, but are reproduced for clarity in Table 1. Clinical characteristics for the 28 subjects diagnosed with asthma were also previously published9, but because nasal lavage samples for measuring dsDNA for the present studies were only remaining for 23 out of the 28 previously reported subjects with asthma, the clinical characteristics of these 23 are resumed in Table 1.
All volunteers were successfully infected with RV16 (100 TCID50) diluted in 250μl of 0.9% saline, via atomizer into both nostrils and respiratory symptoms scores were recorded and nasal lavages were performed at baseline and on days 2, 3, 4, 5, 7 and 10 post RV16 inoculation as reported9.
Nasal (IL-4, IL-5, IL-13 and IFN-γ) and bronchial (IL-5 and IL-13) soluble mediators were measured using Meso Scale Discovery array platform (Rockville, MD) in the fluid lining the nasal or bronchial mucosa sampled two weeks prior to infection (baseline) and on days 2, 3, 4, 5, 7 and 10 (nasal) and day 4 (bronchial) during infection, as reported9. Virus loads were measured by qPCR in the nasal lavage at baseline and on days 2, 3, 4, 5, 7 and 10 during infection as reported9. Neutrophil elastase (NE) was assessed by ELISA (Abcam, Cambridge, UK) in the nasal lavage at baseline and on day 4 during infection. All subjects gave written informed consent and the St. Mary’s Hospital Ethic Committee approved the study.
Measurement of double-stranded DNA
Double-stranded DNA (dsDNA) was measured in the acellular fraction of nasal lavages (human studies) and BALF (mouse studies) using Quant-iT PicoGreen dsDNA reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Mice and viruses
WT BALB/c mice were purchased from Harlan Laboratories and maintained in specific pathogen-free conditions at Imperial College London (UK). Mice aged 6-8 weeks were used for each experiment. All animal experiments were reviewed and approved by the Animal Welfare and Ethical Review Board (AWERB) within Imperial College London and approved by the UK Home Office in accordance with the Animals Act 1986 and the ARRIVE guidelines. Minor-group RV serotype-1b (RV1b) were grown in Ohio HeLa cells (European Collection of Cell Cultures). Viruses were obtained from the American Type Culture Collection and were passaged five times in HeLa cells prior purification6. Viruses were titrated on HeLa cells by standard methods and were inactivated by UV-light exposure at 1.200mJ/cm2 for 30 minutes. WT BALB/c and eosinophil-deficient ΔdblGATA (C.Cg-Gata1tm6Sho/J) BALB/c mice were purchased from The Jackson Laboratory, and mice were housed and bred in specific pathogen-free facilities at the University of Liège (Belgium). Mice aged 6-8 weeks were used and experiments were approved by the IACUC of the University of Liège (approval number 1593).
Mouse model of RV-induced exacerbation of allergic airway inflammation
The experimental protocol is shown in Fig. 2a. Lightly isoflurane-anesthetized WT BALB/c mice were sensitized by intra-tracheal (i.t) exposure to crude House Dust Mite extracts (HDM; Dermatophagoides farinae; lot numbers: 248039 and 189262; Lenoir, NC) (100μg in 50μl phosphate-buffered saline [PBS]) at day -1. Ten days after sensitization, anesthetized mice were dosed i.n. with 10μg of HDM in 50μl PBS once daily on two consecutive days (-2 and -1) to trigger allergic airway inflammation. Control non-allergic mice received 50 µl PBS i.n. during sensitization phase and were challenged with HDM as for sensitized mice. One day after the last challenge (day 0), allergic and non-allergic mice were inoculated i.n. with 50 μl (2.5 x 106 TCID50/mL) RV serotype 1b (RV1b). Infection control groups were dosed with 50μl UV-inactivated RV1b (UV). Mice were sacrificed at different time points (at day 1, 2, 4 and 9) post inoculation (p.i.) for end-point analyses. Parameters of airway allergy, including cytology of the bronchoalveolar lavage fluid (BALF), lung histology, and total IgE serum levels were assessed as described previously55. The extent of peribronchial inflammation was estimated by a score calculated by quantification of peribronchial inflammatory cell layers surrounding the diameter-matched bronchi in lung sections stained with hematoxylin and eosin, as described previously56. Mucus production was quantified as percentage of periodic acid-Schiff-stained goblet cells per total epithelial cells in randomly selected bronchi, as described56. Four randomly selected sections were analyzed per mouse lung, and the average value was used as individual score for each mouse. Airway hyper-responsiveness was evaluated at day 3 post RV1b inoculation as enhanced pause (PenH) in response to nebulized challenge with increasing doses of methacholine (Sigma-Aldrich, St Louis, MO) using an unrestrained whole-body plethysmography system (Electromedsystems, Bordon, UK) as previously described57. PenH is displayed as average values for a 5 min log period post-methacholine challenge.
Measurement of relative house dust mite-specific antibody titers
Plates were coated overnight with HDM (5µg.ml-1 in PBS) and were blocked with 1% BSA in PBS before the addition of serum samples that had been diluted 1:200 in blocking buffer. Plates were washed 6 times with PBS containing 0.005% Tween-20 before incubation with biotinylated anti-mouse IgG2a (RMG2a-62; Biolegend, London, UK) and IgG1 (RMG1-1; Biolegend) at a concentration of 1μg.ml-1 for 1h. Plates were washed an additional six times, septavidin-HRP (R&D Systems, Minneapolis, MN) was added for 40 min and the amount of bound HDM-specific IgG1 or IgG2a was determined using TMB substrate (Sigma-Aldrich). For relative quantification, we used as standard, serum (top standard diluted 1:100 in blocking buffer) pooled from mice sensitized i.t. with 100 μg HDM and challenged ten days later i.n. with 50μg HDM during three consecutive days. These mice were inoculated with RV1b one day after the last challenge and culled at day 4 p.i. in order to collect the serum.
Quantitative PCR analysis of viral RNA copies
Apical lobes from each mouse were excised and total RNA was extracted and purified from homogenized lung tissue by using an RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. 1μg of RNA was reverse-transcribed for cDNA synthesis using random hexamer as primers (Omniscript RT kit, Qiagen). RNA from BALF cells was also extracted and converted to cDNA by the same process. Quantification of viral RNA copies number was conducted using specific RV1b genomic primers and probe sequence as previously described6. Analysis was performed using the QuantiTect Probe PCR Master Mix (Qiagen) and the LightCycler® 480 Real-Time PCR system (Roche, Indianapolis, IN). For Absolute quantification, each gene was normalized to level of 18s and the exact copies of gene of interest were calculated using a standard curve generated by amplification of plasmid DNA. In the graphs, data were expressed as viral RNA copies number per μg of cDNA reaction.
Ex vivo restimulation of mediastinal lymph nodes cells and total lung cells with HDM
During endpoint analyses, mediastinal lymph node(MLN)s were homogenized and MLN cells were cultured in Click’s medium supplemented with 5% heat-inactivated fetal bovine serum (Sigma-Aldrich) at a density of 2.105 cells/well in a 96-well plate. Cells were cultivated and restimulated in vitro for 3 days with or without 30 μg.ml-1 HDM. For lung cells, after obtaining single-lung-cell suspensions (see below), total lung cells were cultured and restimulated in vitro with HDM as described above. TH2 cytokine production (IL-4, IL-5 and IL-13) was measured in the cell supernatants by ELISA (R&D System Duoset) according to manufacturers’ specifications.
BALF neutrophil elastase, chemokine and MUC5ac quantifications
For neutrophil elastase (NE) and chemokine quantification in the airways, BALF was obtained by lavaging the lung with 1.5 ml PBS supplemented with 5mM EDTA (ThermoFisher). The levels of CCL2, CCL12, CCL7 and NE was measured in the acellular fraction of BALF by ELISA (R&D System duoset and Abnova, Taiwan, China) according to manufacturers’ specifications.
Mucin MUC5ac was determined by semiquantitative Enzyme-Linked Immuno Sorbent Assay (ELISA). BALF was diluted in PBS and dried overnight in Maxisorp plates (Nunc). MUC5ac was detected using a biotinylated anti-MUC5ac antibody (Thermofisher, Waltham, MA) and detected with streptavidin-HRP (R&D system) and TMB substrate (Sigma-Aldricht).
Flow cytometry
To obtain single-lung-cell suspensions, lungs were perfused with PBS through the right ventricle, and the left lung lobe was crudely dissociated using the GentleMACS™ tissue dissociator (Miltenyi Biotech, Germany). After dissociation, lung cells were isolated by enzymatic digestion using liberase™ (20μg/mL; Roche) and DNase I (5mg 10-2 ml-1; Sigma-Aldrich), in a final volume of 3ml of RPMI per lung (left lobe) at 37°C for 40min with agitation.
Before surface staining, cells from the digested lung were incubated with anti-CD16/CD32 antibodies (2.4G2; BD Biosciences) to avoid non-specific binding. All staining reactions were performed at 4°C for 20 to 30 min. Flow cytometry analyses were performed on a BD LSRII digital flow cytometer (BD Biosciences, San Jose, CA). TH2 lymphocytes were isolated from lung cell suspension using a combination of the following fluorochrome-conjugated anti-mouse antibodies against CD4 (RM4-5, BD biosciences), CD3e (500A2, BD bioscience), ICOS (C398.4A; Biolegend) and T1/ST2 (DJ8; MDBiosciences, Zurich, Switzerland) in PBS containing 1% BSA and 5% NaN3. Different subsets of dendritic cells were detected by flow cytometry in cell suspensions of lung and in mediastinal lymph node using a combination of the following fluorochrome-conjugated anti-mouse antibodies against MHCII (M5/114; Biolegend), CD11c (N418; Biolegend), CD11b (M1/70; Biolegend), CD103 (2E7; Biolegend), CD64 (X54-5/7.1; Biolegend), FcεR1 (MAR-1, Biolegend), CCR7 (4B12; Biolegend). In all experiments, fixable Live/dead® marker Near-IR (ThermoFisher) was used to eliminate death cells from analysis. Optimal PMT voltages and antibodies titration were performed in order to properly separate negative and positive staining populations. Gating strategy was set up according to FMO control for all antibodies. Final analysis and graphical output were performed using FlowJo software (Tree Star, Ashland, OR).
In vivo DNase I treatment
The experimental protocol is shown in Fig. 3a. Mice were injected by i.p. with 1.000 IU of DNaseI from bovine pancreas (Sigma-Aldrich) in 200μl PBS 4 hours prior RV1b inoculation and at 1 and 2 days p.i., and treated i.n. with 500 IU DNase I 50 μl PBS 8 h, 1 and 2 days after RV1b inoculation. In this model, mice were sacrificed at day 1 and 4 p.i. for end-point analyses.
In vivo administrations of dsDNA
Endogenous dsDNA was extracted from mouse spleens, as previously described19. The experimental protocol is shown in Fig. 4a. Mice were administered with 10μg of endogenous dsDNA in 50μl PBS 24, 36, 48 and 72 h after the last HDM challenge. In this model, mice were sacrificed at day 1 and 4 p.i. for-end point analyses.
In vivo treatment with anti-Ly6G neutralizing antibodies
The experimental protocol is shown in Fig. 6e. Mice were injected i.p. with 500μg of neutrophil-depleting anti-Ly6G (α-Ly6G; 1A8; BioXCell, Lebanon, NH) or isotype control (2A3; BioXCell) antibodies in 200μl of PBS 24h before RV1b inoculation. Mice were euthanized 1 or 2 days after RV1b inoculation for BALF neutrophil and dsDNA analysis or NET detection, respectively (see below).
Western blotting
Lung tissues were homogenized in RIPA buffer supplemented with protease inhibitor cocktail (Complete, Roche). An equivalent amount of protein per sample was resolved on gradient gels (4–12% Bis-Tris Plus gels, Life Technologies) and electroblotted on Invitrolon PVDF membranes (Life technologies). The membranes were blocked for 1h at room temperature with 5% of dry milk in PBS and incubated overnight at 4°C with antibodies to histone H3 citrullinated (rabbit polyclonal anti-H3Cit; 1:1000, cat. No. ab5103, Abcam, Cambridge, UK) in Tris buffer saline-Tween 0,05% + BSA 5%. The membranes were then incubated during 2h at room temperature with appropriate HRP-conjugated secondary antibodies in Tris buffer saline-Tween 0,05% + BSA 5%. Equal loading was confirmed by probing for GAPDH (mouse monoclonal anti-GAPDH; 1:40,000, cat. No. AM4300, ThermoFisher).
Fluorescence microscopy
To identify NETs from lung tissues, lungs were collected 2 days after RV infection without performing BALF and fixed 2 days with 4% paraformaldehyde in PBS. Lung tissues were paraffin-embedded and lung sections were cut (5 µm thick sections) for immunofluorescent staining. After deparaffinization and rehydration, tissue sections were boiled 20 minutes in 10mM Sodium Carbonate buffer for antigen retrieval. Lung section were permeabilized in PBS 0,5% triton X-100 and then, incubated with blocking buffer (2% of BSA and 2% of donkey serum in PBS) during 1 hour at room temperature (Sigma-Aldrich). For detection of NETs, samples were stained in blocking buffer with rabbit antibodies to histone H3 citrullinated (H3Cit; 1:100, cat. no. ab5103 abcam, Cambridge, UK) and with goat antibodies to MPO (1:40, cat .no. af3667, R&D system) during 2 hours at 37°C. After washing samples with PBS, secondary donkey anti-rabbit and anti-goat IgG antibodies conjugated with AlexaFluor 568 (1:200, polyclonal, ThermoFisher) and AlexaFluor 488 (1:200, polyclonal, ThermoFisher) were added in blocking buffer containing DAPI (1:1000; R&D system) and incubated 2h in the dark. Finally, samples were mounted with 10μl of ProLong Antifade reagent (ThermoFisher) on glass slides and stored at room temperature in the dark overnight. All samples were analyzed by fluorescent microscopy using standard filter sets. Controls were stained with secondary antibody only. Images were acquired on a LEICA TCS SP5 Confocal microscope.
In vivo treatment with neutrophil elastase inhibitor (NEi)
The experimental protocol is shown in Fig. 6j. Mice were injected i.p. with 50μg of neutrophil elastase inhibitor (GW311616A, AxonMedChem, Groningen, Netherlands) or vehicle control 12h before RV1b inoculation and every 12h until end point analyses. Mice were euthanized 2 or 4 days p.i. for NETs detection and assessment of allergic lung inflammation, respectively.
Statistical analyses
For the human studies, peak values were modeled using a linear mixed model after checking the normality of the residuals (Kolmogorov-Smirnov). Heterosedasticity between initial (s20) and peak (s2p) times on one hand, and correlation (r) between the successive measurements on the same experimental unit on the other hand, were modeled using these 3 variance components (i.e., s20, s2p and r) through an autoregressive model. Fixed factors were time (0 or peak), status (Asthma / ICS, Asthma / non-ICS) and healthy in some analyses or with a pooled asthma group versus the healthy one in other analyses. Post hoc comparisons were carried on using differences of least-square means. A similar model was used for the kinetics data, except that, due to the increased number of time points, more variance components needed to be estimated (variances at the various times and correlations between the various times). This was achieved using an unstructured variance-covariance matrix and estimating the corresponding 28 variance components. Differences were considered significant when the corresponding reported P values were <0,05. To study the correlation between the extracellular dsDNA level and nasal/bronchial cytokines or clinical symptom scores, standard least-square linear regressions were carried out. Coefficients of correlation (r) of Spearman (non-parametric analyses) were presented as measures of linear association for regression relationships. Analyses were performed using SAS 9.3. and Prism 6 (GraphPad Software).
For mouse experiments, respect of the assumptions of normal distribution of residuals and homoscedasticity was verified, and data were presented as mean + standard deviation (s.d.) for parametric analyses and as median + interquartile range for non-parametric analyses, as indicated in the figure legends. Coefficients of correlation (r) were presented as measures of linear association for regression relationships. Statistical analyses were performed using Prism 6 (GraphPad Software). One-way ANOVA with Tukey's post hoc tests for multiple comparisons, Kruskal-Wallis test or Welch's ANOVA were performed as mentioned in the respective figure legends. A P value less than 0.05 was considered significant.
In all figure panels depicting correlations, asterisks (*) indicate the level of significance of the coefficient of correlation. For human studies, in Figures 1b-c, and 6a, asterisks (*) compare differences within the groups (Healthy vs. Asthma), and circles (˚) compare differences between subjects with asthma and healthy subjects at the indicated time points (Figure 1b) or at the peak (Figure 1c, Figure 6a). For mouse studies, asterisks were used to indicate significant differences observed when comparing two groups of mice, except in Figure 3 and 5, where circles (˚) compare DNase-treated vs. vehicle-injected counterparts; in Figure 4, where circles (˚) compare dsDNA injected vs. vehicle-injected counterparts; in Figure 6, where circles (˚) compare α-Ly-6G- or NEi-treated vs. vehicle-injected counterparts. */˚P< 0.05, **/˚˚P< 0.01, ***/˚˚˚P< 0.001.
Supplementary Material
Acknowledgments
The authors thank all the CBS team from Imperial College London for animal management, M. Paulsen of the Flow Cytometry Core Facility from St Mary’s Campus (Imperial College London) for giving advice and C. Tytgat for secretarial assistance. The authors also thank S.Ormenese and JJ. Goval of the Cell Imaging Platform of the Groupe Interdisciplinaire de Génoprotéomique Appliquée (GIGA, Liège, Belgium) for help with confocal microscopy. M.T. is a postdoctoral fellow who has been supported by the Wallonie-Bruxelles International organization and by the European Academy of Allergy and Clinical Immunology. T.M. is a Research Associate of the F.R.S-FNRS and is supported by the Acteria Foundation. This work was supported by the European Research Council (ERC FP7 grant number 233015); a Chair from Asthma UK (number CH11SJ); the Medical Research Council Centre (grant number G1000758); National Institute of Health Research (NIHR) Biomedical Research Centre (grant number P26095); Predicta FP7 Collaborative Project (grant number 260895); and the NIHR Biomedical Research Centre at Imperial College London. SLJ is a NIHR Senior Investigator.
Abbreviations used
- BALF
bronchoalveolar lavage fluid
- cDC2
CD11b+ conventional DCs
- Cit-H3
citrullinated histone H3
- DC(s)
dendritic cell(s)
- dsDNA
double-stranded DNA
- HDM
house dust mite
- ICS
inhaled corticosteroids
- IFN
interferon
- IL
interleukin
- i.n.
intranasal(ly)
- i.p.
intraperitoneal(ly)
- MLN
mediastinal lymph node
- mo-DCs
monocyte-derived DCs
- MPO
myeloperoxidase
- NE
neutrophil elastase
- NEi
inhibitor of neutrophil elastase
- NET
neutrophil extracellular trap
- PBS
phosphate-buffered saline
- p.i.
post inoculation
- RSS
respiratory symptom score
- RSV
respiratory syncytial virus
- RV
rhinovirus
- TH2
adaptive type 2 helper T (cell)
- UV
ultraviolet-inactivated RV-1b
Footnotes
Author Contributions
M.T. made the initial observation by analysing human samples, performed most of the experiments and analyzed the data. M.T. designed the experiments with the help of M.R.E., R.S., T.M., F.B. and S.L.J. D.J.J. conducted the human experimental infection study, provided associated data and designed the supplementary Table 1. S.L.J. was the principle investigator for the human experimental infection study. M.T., D.S. and A.G performed the western blot experiments. D.S., Y.M.C, H.M. helped M.T. to process mouse samples in the laboratory. C.R. participated in confocal studies and experiments involving ΔdblGATA BALB/c mice. J.A. grew and purified virus. F.F. performed human and part of mouse statistical analyses. T-D.T. and V.P. contributed to experiments and discussion related to NET detection and inhibition. M.T. and T.M. designed the figures. All the authors provided feedback on the manuscript; M.T., T.M., F.B. and S.L.J. wrote the manuscript. T.M., F.B. and S.L.J. supervised the project; F.B. and S.L.J. secured funding.
Competing financial interests
DJJ has received support for travel expenses to attend Respiratory Conferences from AstraZeneca, Boehringer Ingelheim (UK), and GSK.
SLJ reports grants and/or personal fees from Centocor; Sanofi Pasteur; GSK; Chiesi; Boehringer Ingelheim; Novartis; grants, personal fees and shareholding from Synairgen; personal fees from Bioforce outside the submitted work; In addition, SLJ is involved in patents relating to use of interferon-β and interferon-λ for the treatment and prevention of virally-induced exacerbation in asthma and chronic pulmonary obstructive disease, and for induction of cross-reactive cellular responses against rhinovirus antigens.
The other authors declare they have no competing financial interests.
References
- 1.Johnston SL. The role of viral and atypical bacterial pathogens in asthma pathogenesis. Pediatr Pulmonol Suppl. 1999;18:141–143. [PubMed] [Google Scholar]
- 2.Johnston SL, et al. Community study of role of viral infections in exacerbations of asthma in 9-11 year old children. BMJ. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busse WW, Lemanske RF, Jr, Gern JE. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet. 2010;376:826–834. doi: 10.1016/S0140-6736(10)61380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green RM, et al. Synergism between allergens and viruses and risk of hospital admission with asthma: case-control study. Bmj. 2002;324:763. doi: 10.1136/bmj.324.7340.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray CS, et al. Study of modifiable risk factors for asthma exacerbations: virus infection and allergen exposure increase the risk of asthma hospital admissions in children. Thorax. 2006;61:376–382. doi: 10.1136/thx.2005.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett NW, et al. Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nat Med. 2008;14:199–204. doi: 10.1038/nm1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beale J, et al. Rhinovirus-induced IL-25 in asthma exacerbation drives type 2 immunity and allergic pulmonary inflammation. Sci Transl Med. 2014;6:256ra134. doi: 10.1126/scitranslmed.3009124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collison A, et al. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat Med. 2013;19:232–237. doi: 10.1038/nm.3049. [DOI] [PubMed] [Google Scholar]
- 9.Jackson DJ, et al. IL-33-dependent type 2 inflammation during rhinovirus-induced asthma exacerbations in vivo. Am J Respir Crit Care Med. 2014;190:1373–1382. doi: 10.1164/rccm.201406-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gern JE, Vrtis R, Grindle KA, Swenson C, Busse WW. Relationship of upper and lower airway cytokines to outcome of experimental rhinovirus infection. Am J Respir Crit Care Med. 2000;162:2226–2231. doi: 10.1164/ajrccm.162.6.2003019. [DOI] [PubMed] [Google Scholar]
- 11.Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol. 2012;12:295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Message SD, et al. Rhinovirus-induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL-10 production. Proc Natl Acad Sci U S A. 2008;105:13562–13567. doi: 10.1073/pnas.0804181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wark PA, et al. Neutrophil degranulation and cell lysis is associated with clinical severity in virus-induced asthma. Eur Respir J. 2002;19:68–75. doi: 10.1183/09031936.02.00226302. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, et al. Airway inflammation and illness severity in response to experimental rhinovirus infection in asthma. Chest. 2014;145:1219–1229. doi: 10.1378/chest.13-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 16.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schonrich G, Raftery MJ. Neutrophil Extracellular Traps Go Viral. Front Immunol. 2016;7:366. doi: 10.3389/fimmu.2016.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imanishi T, et al. Nucleic acid sensing by T cells initiates Th2 cell differentiation. Nat Commun. 2014;5:3566. doi: 10.1038/ncomms4566. [DOI] [PubMed] [Google Scholar]
- 19.Marichal T, et al. DNA released from dying host cells mediates aluminum adjuvant activity. Nat Med. 2011;17:996–1002. doi: 10.1038/nm.2403. [DOI] [PubMed] [Google Scholar]
- 20.McKee AS, et al. Host DNA released in response to aluminum adjuvant enhances MHC class II-mediated antigen presentation and prolongs CD4 T-cell interactions with dendritic cells. Proc Natl Acad Sci U S A. 2013;110:E1122–1131. doi: 10.1073/pnas.1300392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonhagen K, et al. ICOS+ Th cells produce distinct cytokines in different mucosal immune responses. Eur J Immunol. 2003;33:392–401. doi: 10.1002/immu.200310013. [DOI] [PubMed] [Google Scholar]
- 22.Plantinga M, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Janss T, et al. Interferon response factor-3 promotes the pro-Th2 activity of mouse lung CD11b+ conventional dendritic cells in response to house dust mite allergens. Eur J Immunol. 2016 doi: 10.1002/eji.201646513. [DOI] [PubMed] [Google Scholar]
- 24.Mesnil C, et al. Resident CD11b(+)Ly6C(-) lung dendritic cells are responsible for allergic airway sensitization to house dust mite in mice. PLoS One. 2012;7:e53242. doi: 10.1371/journal.pone.0053242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goritzka M, et al. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J Exp Med. 2015;212:699–714. doi: 10.1084/jem.20140825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenne CN, et al. Neutrophils recruited to sites of infection protect from virus challenge by releasing neutrophil extracellular traps. Cell Host Microbe. 2013;13:169–180. doi: 10.1016/j.chom.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Branzk N, et al. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol. 2014;15:1017–1025. doi: 10.1038/ni.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong SL, et al. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat Med. 2015;21:815–819. doi: 10.1038/nm.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu C, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caudrillier A, et al. Platelets induce neutrophil extracellular traps in transfusion-related acute lung injury. J Clin Invest. 2012;122:2661–2671. doi: 10.1172/JCI61303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs TA, et al. Extracellular DNA traps promote thrombosis. Proc Natl Acad Sci U S A. 2010;107:15880–15885. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakkim A, et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci U S A. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noges LE, White J, Cambier JC, Kappler JW, Marrack P. Contamination of DNase Preparations Confounds Analysis of the Role of DNA in Alum-Adjuvanted Vaccines. J Immunol. 2016;197:1221–1230. doi: 10.4049/jimmunol.1501565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan TK, et al. House dust mite-induced asthma causes oxidative damage and DNA double-strand breaks in the lungs. J Allergy Clin Immunol. 2016;138:84–96 e81. doi: 10.1016/j.jaci.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Chamilos G, et al. Cytosolic sensing of extracellular self-DNA transported into monocytes by the antimicrobial peptide LL37. Blood. 2012;120:3699–3707. doi: 10.1182/blood-2012-01-401364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lande R, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lande R, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 38.Ishii KJ, Akira S. Innate immune recognition of, and regulation by, DNA. Trends Immunol. 2006;27:525–532. doi: 10.1016/j.it.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Funchal GA, et al. Respiratory syncytial virus fusion protein promotes TLR-4-dependent neutrophil extracellular trap formation by human neutrophils. PLoS One. 2015;10:e0124082. doi: 10.1371/journal.pone.0124082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng OZ, Palaniyar N. NET balancing: a problem in inflammatory lung diseases. Front Immunol. 2013;4:1. doi: 10.3389/fimmu.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Romo GS, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koga H, et al. Inhibition of neutrophil elastase attenuates airway hyperresponsiveness and inflammation in a mouse model of secondary allergen challenge: neutrophil elastase inhibition attenuates allergic airway responses. Respir Res. 2013;14:8. doi: 10.1186/1465-9921-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki T, et al. Aerosolized human neutrophil elastase induces airway constriction and hyperresponsiveness with protection by intravenous pretreatment with half-length secretory leukoprotease inhibitor. Am J Respir Crit Care Med. 1996;153:1405–1411. doi: 10.1164/ajrccm.153.4.8616573. [DOI] [PubMed] [Google Scholar]
- 44.de Heer HJ, et al. Essential role of lung plasmacytoid dendritic cells in preventing asthmatic reactions to harmless inhaled antigen. J Exp Med. 2004;200:89–98. doi: 10.1084/jem.20040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skrzeczynska-Moncznik J, et al. DNA structures decorated with cathepsin G/secretory leukocyte proteinase inhibitor stimulate IFNI production by plasmacytoid dendritic cells. Am J Clin Exp Immunol. 2013;2:186–194. [PMC free article] [PubMed] [Google Scholar]
- 46.Maffia PC, et al. Neutrophil elastase converts human immature dendritic cells into transforming growth factor-beta1-secreting cells and reduces allostimulatory ability. Am J Pathol. 2007;171:928–937. doi: 10.2353/ajpath.2007.061043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roghanian A, Drost EM, MacNee W, Howie SE, Sallenave JM. Inflammatory lung secretions inhibit dendritic cell maturation and function via neutrophil elastase. Am J Respir Crit Care Med. 2006;174:1189–1198. doi: 10.1164/rccm.200605-632OC. [DOI] [PubMed] [Google Scholar]
- 48.Tateosian NL, et al. Neutrophil elastase treated dendritic cells promote the generation of CD4(+)FOXP3 (+) regulatory T cells in vitro. Cell Immunol. 2011;269:128–134. doi: 10.1016/j.cellimm.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Jenne CN, Kubes P. Virus-induced NETs--critical component of host defense or pathogenic mediator? PLoS Pathog. 2015;11:e1004546. doi: 10.1371/journal.ppat.1004546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yipp BG, Kubes P. NETosis: how vital is it? Blood. 2013;122:2784–2794. doi: 10.1182/blood-2013-04-457671. [DOI] [PubMed] [Google Scholar]
- 51.Hahn S, Giaglis S, Chowdhury CS, Hosli I, Hasler P. Modulation of neutrophil NETosis: interplay between infectious agents and underlying host physiology. Semin Immunopathol. 2013;35:439–453. doi: 10.1007/s00281-013-0380-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chrysanthopoulou A, et al. Neutrophil extracellular traps promote differentiation and function of fibroblasts. J Pathol. 2014;233:294–307. doi: 10.1002/path.4359. [DOI] [PubMed] [Google Scholar]
- 53.Grabcanovic-Musija F, et al. Neutrophil extracellular trap (NET) formation characterises stable and exacerbated COPD and correlates with airflow limitation. Respir Res. 2015;16:59. doi: 10.1186/s12931-015-0221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson DJ, Johnston SL. The role of viruses in acute exacerbations of asthma. J Allergy Clin Immunol. 2010;125:1178–1187. doi: 10.1016/j.jaci.2010.04.021. quiz 1188-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toussaint M, et al. Myeloid hypoxia-inducible factor 1alpha prevents airway allergy in mice through macrophage-mediated immunoregulation. Mucosal Immunol. 2013;6:485–497. doi: 10.1038/mi.2012.88. [DOI] [PubMed] [Google Scholar]
- 56.Marichal T, et al. Interferon response factor 3 is essential for house dust mite-induced airway allergy. J Allergy Clin Immunol. 2010;126:836–844 e813. doi: 10.1016/j.jaci.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Singanayagam A, et al. A short-term mouse model that reproduces the immunopathological features of rhinovirus-induced exacerbation of COPD. Clin Sci (Lond) 2015;129:245–258. doi: 10.1042/CS20140654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.