Abstract
The MADS-box transcription factors play essential roles in many physiological and biochemical processes of plants, especially in fruit ripening. Here, a tomato MADS-box gene, SlCMB1, was isolated. SlCMB1 expression declined with the fruit ripening from immature green to B + 7 (7 days after Breaker) fruits in the wild type (WT) and was lower in Nr and rin mutants fruits. Tomato plants with reduced SlCMB1 mRNA displayed delayed fruit ripening, reduced ethylene production and carotenoid accumulation. The ethylene production in SlCMB1-RNAi fruits decreased by approximately 50% as compared to WT. The transcripts of ethylene biosynthesis genes (ACS2, ACS4, ACO1 and ACO3), ethylene-responsive genes (E4, E8 and ERF1) and fruit ripening-related genes (RIN, TAGL1, FUL1, FUL2, LoxC and PE) were inhibited in SlCMB1-RNAi fruits. The carotenoid accumulation was decreased and two carotenoid synthesis-related genes (PSY1 and PDS) were down-regulated while three lycopene cyclase genes (CYCB, LCYB and LCYE) were up-regulated in transgenic fruits. Furthermore, yeast two-hybrid assay showed that SlCMB1 could interact with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1, respectively. Collectively, these results indicate that SlCMB1 is a new component to the current model of regulatory network that regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening.
Introduction
Fruit ripening has always been the focus of scientific research, which is mainly due to not only the uniqueness of this biological process but also the important role that fruits provide nutrition for animal and human1,2. The biological study of fruit ripening have resulted in significant gains in knowledge over recent years. Fruit ripening is a complex physiological and biochemical process with many metabolic changes (e.g. color, flavor, texture, aroma and nutrition) which are regulated by genetic regulators, external signals and endogenous hormones3,4. Typically, two classes of fruits have been identified as the non-climacteric fruits (e.g. pepper and strawberry) and the climacteric fruits (eg. pear and tomato). In climacteric fruits, ethylene biosynthesis and respiration increased remarkably at the beginning of fruit ripening and the burst in ethylene production is necessary for normal ripening of fruits, while in non-climacteric fruits, these changes are not observed3.
Ethylene is an important plant endogenous hormone which plays significant regulatory roles in the growth and development of plants, such as flavor generation, fruit ripening, leaf senescence, and other programmed signals of senescence and defense5–9. It is well known that ethylene is a key regulatory factor at the beginning of ripening and is necessary for the process of fruit ripening10,11. To date, ACS (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE) and ACO (1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE) are identified as two kinds of key enzymes in ethylene biosynthesis12–14. Alexander et al. (2002) have reported that fruit ripening and ethylene biosynthesis are markedly repressed when the expression of SlACS2 was suppressed significantly in SlACS2-RNAi lines. Moreover, some studies have shown that exogenous ethylene can evidently induce the accumulation of SlACS2 transcripts15–17. It has been revealed that the transcripts of SlACO1 and SlACO3 are markedly accumulated when tomato fruit ripening is triggered15,18. In addition, previous studies manifest that SlACO1 positively regulates tomato fruit ripening2,19, when the expression of SlACO1 was suppressed in transgenic tomato fruits, the biosynthesis of endogenous ethylene was decreased and the storage ability of tomato fruits increased20. In addition to ethylene synthesis, ethylene response and perception are also essential to the ripening of fruits. E4 and E8, which are induced by ethylene, are generally considered as two classical genes of ethylene perception and response21. Many studies have shown that the accumulation of E4 transcripts is significantly induced by exogenous ethylene22,23. Also the expression of E4 is inhibited when the biosynthesis of ethylene is suppressed24. E8, which takes part in the feedback regulation of ethylene biosynthesis, is a fruit ripening-specific expression gene and is activated when fruit ripening is triggered25. After being characterized, the E8 promoter is widely used as a fruit-specific promoter to drive the transcripts of exogenous genes to study their function in transgenic tomato26,27.
Tomato is generally used as an excellent model plant for fruit ripening study, not only because of several desirable attributes, such as short life cycle, small genome size, efficient stable transformation, high-density genetic maps and the completion of tomato genome sequence28–30, but also the existence of lots of well-characterized ripening tomato mutants, such as Green ripe (Gr), never ripe (Nr), color nonripening (cnr) and ripening inhibitor (rin), have been found and studied31–35. These superiority and mutants of tomato help us to reveal the mechanism of fruit ripening36. The rin mutant tomato plants exhibit enlarged sepals, an altered inflorescence architecture and inhibited fruit ripening due to the absence of two functional MADS-box genes, SlMADS-MC and SlMADS-RIN. Loss one of these MADS-box gene, the SlMADS-RIN, results in failure of fruit ripening, whereas loss of the other, SlMADS-MC, the phenotypes of enlarged sepals and altered inflorescence architecture are observed32.
The MADS-box genes, encoding DNA-binding transcription factors, which have been characterized from the genome of plants, animals and fungi, play significant roles in numerous biological processes37. In tomato, at least 36 functional MADS-box transcriptional factors have been characterized and analyzed38. Among tomato MADS-box genes, many of these, such as TAGL1 (TOMATO AGAMOUS LIKE1), FUL1(FRUITFULL1), FUL2, SlFYFL and SlMADS1, have been studied and identified to be involved in fruits development or/and ripening. Overexpression of the TAGL1 gene leads to enhanced lycopene in tomato fruits and the transition of sepals into succulent fruit organs, but when the transcripts of TAGL1 are suppressed using RNAi approach, the transgenic tomato fruits display the phenotype of reduced pericarp thickness, altered starch accumulation and ripening inhibition39–41. Recently, a new study reported that TAGL1 inhibits cuticle development and lignin biosynthesis42. Tomato FUL1 and FUL2 genes are the homolog of the Arabidopsis FRUITFULL (FUL) gene, Shima et al. have reported that there is functional redundancy between FUL1 and FUL2 and these two proteins could form heterodimers with RIN to regulate fruit ripening through regulating the expression of ripening-related genes43. Moreover, tomato FRUITFULL (FUL1 and FUL2) homologs and TAGL1 also are able to form high order complexes with RIN to regulate fruit ripening44. SlFYFL and SlMADS1 are two MADS-box genes that participate in fruit ripening, overexpression of SlFYFL leads to the phenotypes of inhibited fruit ripening45 and suppression of SlMADS1 results in ripening inhibition46.
The fruit ripening regulatory network requires a number of regulators that regulate each other or/and the expression of other ripening-related genes to successfully complete the fruit ripening process. Although many regulators of MADS-box family have been reported to participate in tomato fruit ripening, there are still so many ripening-related MADS-box genes need to be investigated. This will contribute to enriching the fruit ripening regulatory network and to further revealing the mechanism of fruit ripening regulation. Recently, a new study by Soyk, S. et al. showed that mutation of the SEP MADS-box gene, J2 (accession no. Solyc04g005320) results in longer inflorescence in tomato (S. pimpinellifolium)47,48. The result of sequence alignment showed this J2 gene which has been named as SlCMB1 (accession no. XM_004237013) by the NCBI database is the investigated gene in our study. Interestingly, this gene was also found to be involved in tomato fruit ripening in addition to its roles in the architecture development in our study. To comprehensively investigate the diversified functions of MADS-box genes, SlCMB1 was isolated from the wild type tomato (Solanum lycopersicon Mill. cv. Ailsa Craig). RNAi repression of SlCMB1 was carried out to study the exact role of SlCMB1 in fruit ripening of tomato, and our results showed that suppression of SlCMB1 results in inhibited ethylene biosynthesis and reduced carotenoid accumulation during tomato fruit ripening. In addition, we analyzed the SlCMB1-RNAi lines at molecular and physiology levels. This paper enhanced our insights into the roles of SlCMB1 playing in multiple biological processes.
Results
Molecular characterization of SlCMB1
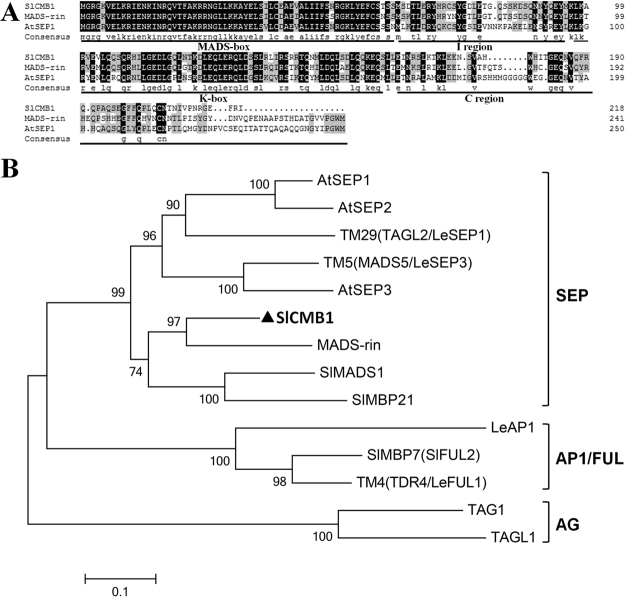
Based on the bioinformatics analysis of tomato MADS-box transcription factor family in our laboratory and the sequence on the NCBI (National Center for Biotechnology Information) web site, a tomato MADS-box gene was cloned from the wild type tomato (Solanum lycopersicon Mill. cv. Ailsa Craig). We named this gene as SlCMB1 (accession number: XM_004237013) following the existing name at the NCBI database. The nucleotide sequence analyses indicated that SlCMB1 contained a 717-bp ORF encoding a protein with 238 amino acids and this protein had an estimated molecular mass of 27.5 kDa (pI 8.62). Multiple alignment result showed that SlCMB1 had the typical MADS-box domains (i.e. the MADS domain, the K domain and the I domain) and the C-terminal region of SlCMB1 had significant difference to other known MADS-box proteins (Fig. 1A)38,49. In addition, phylogenetic analysis displayed that SlCMB1 belongs to the SEPALLATA (SEP) clade and showed higher similarity with SlMADS-RIN compared with other functional MADS-box proteins (Fig. 1B). Moreover, the result of promoter analysis showed that an 8 bp cis-regulatory element, ERE motif (ATTTCAAA) (Supplementary Fig. S1), which is an ethylene-responsive element was found at position −1844 in the promoter region of SlCMB1 gene, indicating that SlCMB1 gene might play an essential regulatory role in the process of fruit ripening.
Figure 1.
Multiple sequence alignment and phylogenetic analysis of SlCMB1 with other MADS-box proteins. (A) Multiple sequence alignment of SlCMB1 with two other MADS-box proteins. The black part are the identical amino acids, and the gray are the similar amino acids. The MADS-box, K-box, I region, and C region are identified. (B) Phylogenetic analysis of the SlCMB1 with other known MADS-box proteins. SlCMB1 is in bold. Accession numbers of these proteins are listed as follows: AtSEP1 (AED92207.1), AtSEP2 (AEE73791.1), AtSEP3 (AEE30503.1), MADS-RIN (AF448522), SlMBP21 (NP_001275579), SlMADS1 (NP_001234380), LeAP1 (AAP83378.1/AY306154), TM5 (MADS5/LeSEP3) (NP_001234384/AY306153), TM29 (TAGL2/LeSEP1) (NP_001233911/AY306152), LeFUL1 (AY098732.1), LeFUL2 (NM_001307938.1), TAG1 (L26295.1), TAGL1 (AY098735.2).
Expression pattern analysis of SlCMB1
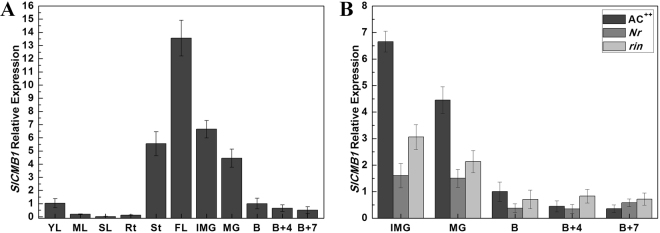
The tissue specificity of gene expression may be correlated with specific biological functions. Thus quantitative real-time PCR (qRT-PCR) was performed to analyze the transcripts of SlCMB1 in different tissues of the wild type tomato (Solanum lycopersicum (Mill. cv. Ailsa Craig AC++)) and ripening mutants (Nr and rin). Low expression levels of SlCMB1 were observed in roots, leaves and B (Breaker) to B + 7 (Breaker + 7d) fruits (Fig. 2A). Whereas, predominant expression was observed in flowers, stems, IMG and MG fruits (Fig. 2A,B), suggesting that SlCMB1 might play roles in the development of these tissues. Furthermore, the transcripts of SlCMB1 rapidly declined with fruit ripening in AC++ fruits, and a simlar expression pattern of SlCMB1 was observed in the mutant Nr and rin fruits (Fig. 2A,B). But the expression levels of SlCMB1 in Nr and rin fruits at IMG and MG stages were significantly less than in AC++ (Fig. 2B), suggesting that SlCMB1 expression may be impacted by SlMADS-RIN and/or ethylene(Fig. 2B).
Figure 2.
Expression patterns of SlCMB1 in different tissues of AC++ (Mill. cv. Ailsa Craig) and ripening-related mutant fruits. (A) The expression of SlCMB1 in YL, young leaves; ML, mature leaves; SL, senescent leaves; Rt, roots; St, stems; FL, flowers at anthesis; IMG, immature green fruits; MG, mature green fruits; B, breaker fruits; B + 4, fruits 4 days after breaker; B + 7, 7 days after breaker fruits in AC++. (B) Expression of SlCMB1 in different stages of AC++, Nr, and rin fruits. The SlCMB1 relative expression in B stage fruits of AC++ was set the control sample. Error bars indicate SE.
Generation of SlCMB1-RNAi Lines
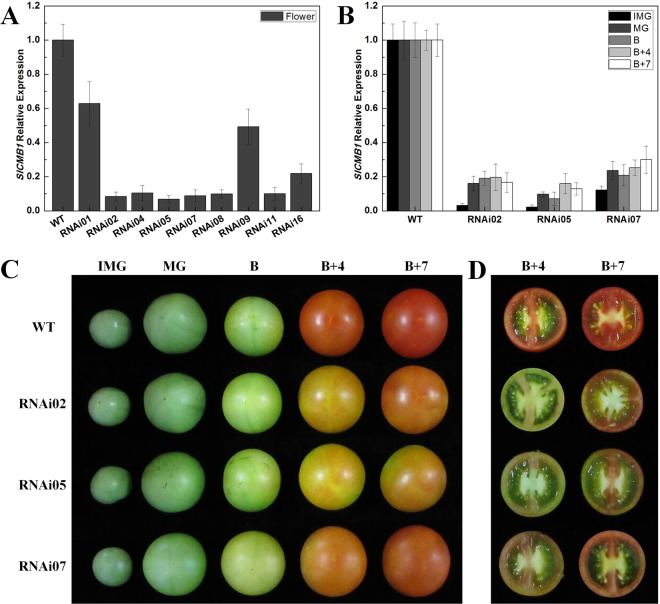
To further study the roles of SlCMB1 in tomato growth and development, an RNAi expression vector targeting the C-terminal specific fragment of SlCMB1 was generated (Supplementary Fig. S2) and transferred into the wild type tomato (AC++). Nine independent transgenic lines were confirmed by PCR with primers of NPT II (Supplementary Table S1), then total RNAs of these independent transgenic lines were extracted from flowers to investigate the relative expression of SlCMB1, respectively. The qRT-PCR results displayed that the transcript levels of SlCMB1 in six transgenic lines were significantly decreased by 90–94% compared with the wild-type (Fig. 3A). Later, three independent transgenic lines (RNAi02, RNAi05, RNAi07) which had the lowest accumulation of SlCMB1 transcripts were selected for further characterization.
Figure 3.
Phenotypes and expression analyses of SlCMB1 in RNAi lines. (A) The relative expression of SlCMB1 between the WT and all nine SlCMB1 RNAi lines. The tissue is flower at anthesis. (B) Relative expression of SlCMB1 in the WT and the three selected silencing lines from IMG to B + 7 fruits. The tissues are the fruits at different stages (C). Phenotypes of fruits at different stages from IMG to B + 7 stage in WT and RNAi lines. (D) Transverse sections of WT and SlCMB1 RNAi fruits at the B + 4 and B + 7 stage. WT, wild type. Data are the means ± SD of three independent biological replicates. The transcripts of SlCMB1 in the wild type are normalized to 1. Error bars indicate SE.
Expression profile analysis of ripening- and carotenoid-related genes in SlCMB1-RNAi fruits
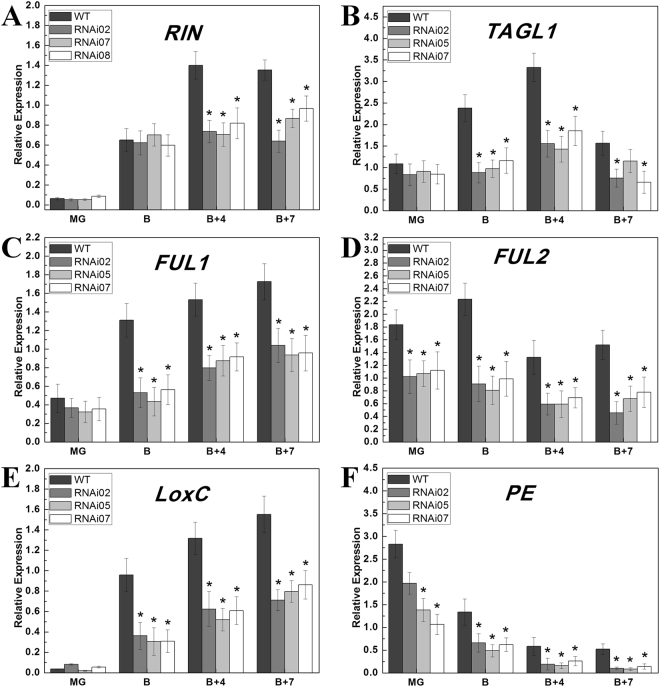
To verify the suppression of SlCMB1 in the selected transgenic lines, the total RNA was extracted from IMG, MG, B, B + 4 and B + 7 fruits of WT and the transgenic lines, respectively. The qRT-PCR result displayed that the transcripts of SlCMB1 in fruits at different stages of selected transgenic lines (RNAi02, RNAi05, RNAi07) were markedly reduced to roughly 5–30% of control levels (Fig. 3B). To confirm the specific suppression of SlCMB1, the expression of RIN was tested, because it is the most closely related (67.3% identity at nucleotide level) gene to SlCMB1 in tomato. Meanwhile, the result of multiple sequence alignment between RIN and SlCMB1 fragment showed that the 426 bp fragment of SlCMB1 using in our study was specific (Supplementary Fig. S3). Figure 4A showed that there was no significant difference of RIN expression in SlCMB1-RNAi fruits and the wild type at MG and B stage but its transcripts were reduced significantly at B + 4 and B + 7 stage in SlCMB1-RNAi fruits. These results indicated that the reduced expression of RIN at B + 4 and B + 7 stage of transgenic fruits was not caused by the construct of SlCMB1-RNAi vector but the suppression of SlCMB1 in tomato fruits.
Figure 4.
Relative expression of ripening-related genes in fruits of the wild type (WT) and SlCMB1-RNAi lines. The total RNA were extracted for the qRT-PCR assay from MG, B, B + 4 and B + 7 fruits of WT and RNAi lines. Three independent biological replications of each sample were used. (A) Expression of RIN in RNAi lines and the wild type. (B) Expression of TAGL1 in RNAi lines and the wild type. (C) Expression of FUL1 in RNAi lines and the wild type. (D) Expression of FUL2 in RNAi lines and the wild type. (E) Expression of LoxC in RNAi lines and the wild type. (F) Expression of PE in RNAi lines and the wild type. MG, mature green; B, breaker; B + 4, 4 days after B; and B + 7, 7 days after B. The significant differences were marked with the asterisks between the RNAi and WT fruits (P < 0.05). Error bars indicate SE.
Because the ripening time of SlCMB1-RNAi fruits was delayed 3 to 5 days (Table 1), so the expression of a number of known ripening-related genes were analyzed in SlCMB1-RNAi and the wild-type tomato fruits. The results showed that the transcripts of five ripening-related genes, RIN, TAGL1, FUL1, FUL2, LoxC (Lipoxygenase C) and PE (PECTINESTERASE) were significantly down-regulated in SlCMB1-RNAi fruits (Fig. 4A–F). These results suggested that SlCMB1 might regulate the fruit ripening of tomato through influencing the expression of ripening-related genes.
Table 1.
Days from anthesis to B stage for the wild type and SlCMB1-RNAi lines.
| Tomato Lines | Days |
|---|---|
| Wild type | 37.9 ± 0.66 |
| RNAi02 | 42.1 ± 0.68 |
| RNAi04 | 41.1 ± 0.82 |
| RNAi05 | 43.3 ± 0.51 |
| RNAi07 | 41.8 ± 0.73 |
| RNAi08 | 41.4 ± 0.68 |
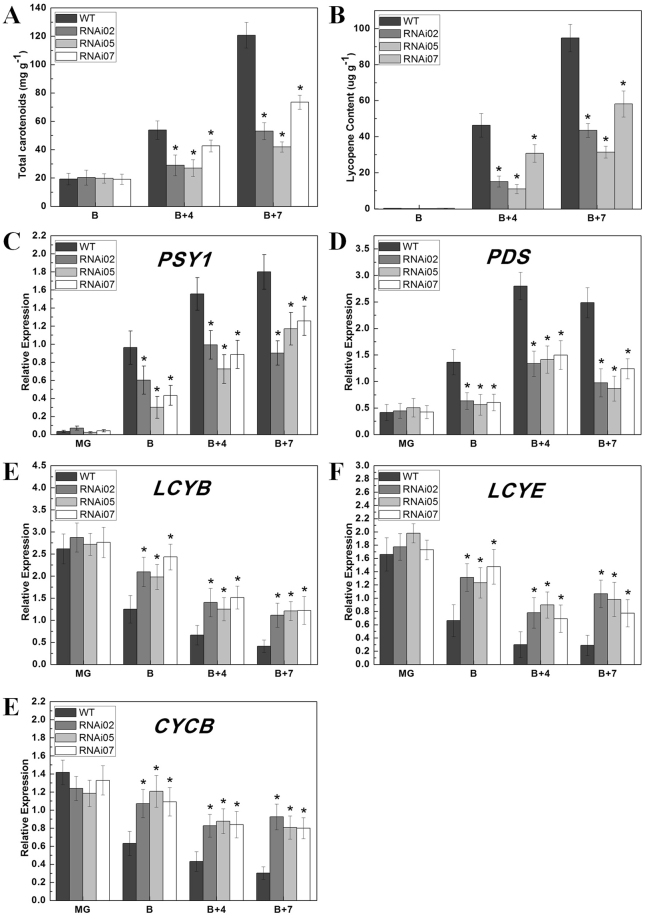
For the reduced carotenoid accumulation was observed in transgenic fruits, several known genes which have been reported to be involved in the carotenoid biosynthesis pathway were examined. It has been reported that PSY1 is the rate-limiting enzyme in the synthesis process of lycopene and is induced by ethylene50. In this study, the qRT-PCR results showed that its expression was dramatically down-regulated in B, B + 4 and B + 7 fruits of the SlCMB1-RNAi lines (Fig. 5B). Another enzyme, phytoene desaturase (PDS), which involved in the lycopene synthesis was also notablely down-regulated in SlCMB1-RNAi lines (Fig. 5C). Furthermore, other three lycopene cyclase genes, CYCB (chromoplast-specific lycopene cyclase), LCYB (lycopene β-cyclase) and LCYE (lycopene ε-cyclase), which participate in the cyclization of lycopene were significantly up-regulated in the SlCMB1-RNAi tomato fruits at different stages (Fig. 5D–F). These results indicate that suppression of SlCMB1 reduced the carotenoid accumulation and influenced the transcripts of related genes involving in the carotenoid biosynthesis pathway.
Figure 5.
Pigments accumulation and relative expression levels of the carotenoid synthesis related genes in the SlCMB1 RNAi and the wild-type (WT) fruits. (A) Analysis of carotenoid accumulation in B, B + 4 and B + 7 fruits of the SlCMB1-RNAi lines and the wild type. (B) Analysis of lycopene content in B, B + 4 and B + 7 fruits of the SlCMB1-RNAi lines and the wild type. (C) Expression of PSY1 in fruits of the wild type and transgenic lines. (D) Expression of PDS in fruits of the wild type and transgenic lines. (E) Expression of LCYB in fruits of the wild type and transgenic lines. (F) Expression of LCYE in fruits of the wild type and transgenic lines. (G) Expression of CYCB in fruits of the wild type and transgenic lines. MG, maturate green; B, breaker; B + 4, 4 days after B; B + 7, 7 days after B. Three independent biological repeats of each sample were used. Data are presented as means ± SD of at least three individual fruits. The significant differences were marked with the asterisks between the RNAi and WT fruits (P < 0.05). Error bars indicate SE.
Silencing of SlCMB1 inhibited fruit ripening and carotenoid accumulation
In the process of tomato fruit development and ripening, the time from pollination to fruit ripening was recorded. We found that the red color of wild type fruit was deeper than the SlCMB1-silenced fruit (Fig. 3C,D) and the ripening time of SlCMB1-silenced fruits was delayed 3 to 5 days as compared to the wild type (Table 1). Previous studies have reported that the accumulation of carotenoids is the main reason for the pigmentation change in tomato ripening fruits51. In our study, we extracted and examined the total carotenoids and lycopene in B, B + 4 and B + 7 fruits of the RNAi lines and the wild-type. As shown in Fig. 5A,B, the carotenoid and lycopene contents in B + 4 and B + 7 fruits of SlCMB1-RNAi lines were significantly lower than in the wild type tomato plants, indicatng that reduction of SlCMB1 transcripts inhibited fruit ripening and altered the carotenoid content of tomato fruits.
Repression of SlCMB1 reduced the production of ethylene and the transcripts of ethylene-related genes
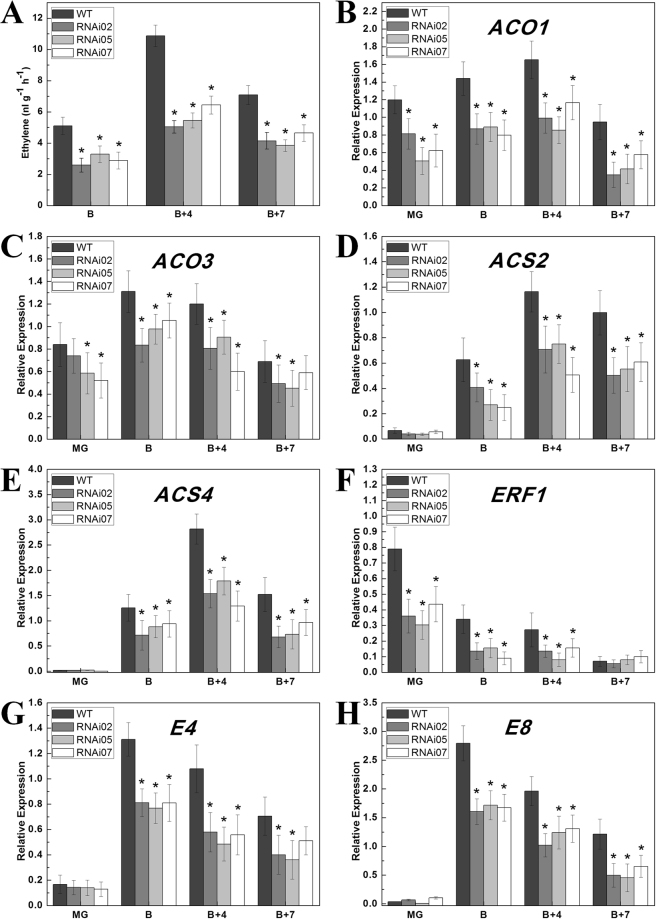
Fruit ripening and carotenoid accumulation of tomato were affected by the gas hormones ethylene52. To further explore the impacts of reduced SlCMB1 mRNA on ethylene biosynthesis, ethylene production was measured during the process of fruit ripening. The result showed that the ethylene production of wild-type fruits displayed a massive and rapid accumulation at B + 4 stage, and declined at B + 7 stage. SlCMB1-RNAi fruits exhibited a similar trend in production of ethylene similar to the wild type with the highest level of ethylene in the B + 4 stage. However, overall ethylene production in SlCMB1-RNAi fruits were approximately 50% lower when compared to the wild type in all stages (Fig. 6A).
Figure 6.
Determination of ethylene production and the expression of ethylene-related genes in the wild type (WT) and the SlCMB1-RNAi fruits. (A) Production of ethylene in WT and SlCMB1-RNAi fruits. B, B + 4 and B + 7 fresh fruits were sealed in air-tight jars and then the 1 mL headspace gas was sampled 24 h later. The date represent means from at least three biological repeats. (B–E) Expression of four ethylene biosynthesis-related genes (ACO1, ACO3, ACS2 and ACS4) in fruits of SlCMB1-RNAi and the wild type. (F–H) Expression of three ethylene responsive genes (ERF1, E4 and E8) in WT and SlCMB1 RNAi fruits. The total RNA were extracted for qPCR assay from MG (mature green), B (breaker), B + 4 (4 days after B) and B + 7 (7 days after B) fruits of WT and RNAi lines. Three replications for each sample were used. The significant differences were marked with the asterisks between the RNAi and WT fruits (P < 0.05). Date are the means ± SD of three independent biological replicates. Error bars indicate SE.
The ethylene production was affected by the transcripts of the ethylene synthesis and response genes. Given that the reduction of ethylene production in the SlCMB1-RNAi lines, multiple crucial genes involved in the ethylene biosynthesis and response were detected in fruits of the wild-type and RNAi lines at different stages. The results showed that four crucial ethylene biosynthesis genes (ACO1, ACO3, ACS2 and ACS4) and three ethylene response genes (E4, E8 and ERF1) were notably down-regulated in SlCMB1-RNAi fruits, especially in B and B + 4 fruits (Fig. 6B–H). These results suggested that down-regulation of SlCMB1 reduced the ethylene production and the transcripts of ethylene biosynthesis and response genes.
SlCMB1 could interact with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1
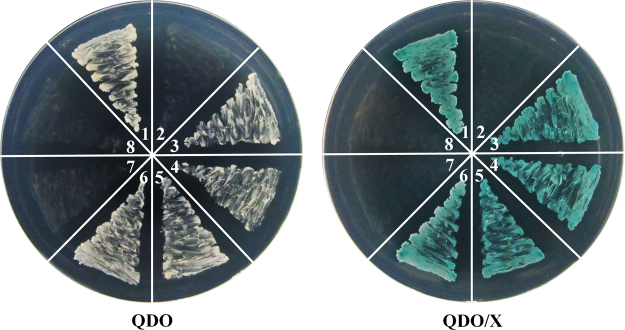
To confirm the existence of interaction between SlCMB1 and other ripening-related proteins, SlMADS-RIN, SlMADS1, SlAP2a and TAGL1 were preferentially selected for the yeast two hybrid assays. The ORF (open reading frames) of these four genes were amplified, respectively. The ORF of SlCMB1 was cloned into the pGADT7 vector to be as the prey and the ORF of SlMADS-RIN, SlMADS1, SlAP2a and TAGL1 was cloned into the pGBKT7 vector to be as the bait, respectively. Self activation of pGBKT7-RIN, pGBKT7-SlMADS1, pGBKT7-SlAP2a and pGBKT7-TAGL1 were tested and the results were negative (Supplementary Fig. S5). Furthermore, the empty prey and bait vector containing the construct of each prey and bait were used as the negative controls, respectively. Figure 7 displayed that the yeast could grow on the selective medium (QDO) and turn blue on the plate containing the X-α-gal indicator (QDO/X), indicating that SlCMB1 can interact with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1, respectively.
Figure 7.
Yeast two-hybrid assay of SlCMB1 with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1. QDO, SD medium lacking Trp, Leu, His, and adenine. QDO/X, SD medium lacking Trp, Leu, His, and adenine with X-a-Gal. 1. pGBKT7-53 & pGADT7-T (positive control); 2. pGBKT7-Lam & pGADT7-T (negative control); 3. pGADT7-SlCMB1 & pGBKT7-RIN; 4. pGADT7-SlCMB1 & pGBKT7-SlMADS1; 5. pGADT7-SlCMB1 & pGBKT7-SlAP2a; 6. pGADT7-SlCMB1 & pGBKT7-TAGL1; 7. pGADT7 (empty bait vector control); 8. pGBKT7 (empty prey vector control).
Discussion
In this study, we characterized a SEP MADS-box gene, SlCMB1, which is recently reported to be involved in the regulation of tomato (S. pimpinellifolium) inflorescence architecture48 and named it following the existing name on the NCBI web site. It is known that MADS-box genes always play diverse roles in different developmental processes, such as MC (MACROCALYX), J (JOINTLESS) and SlMBP21 and so on48,53–57. In order to further investigate other potential roles of SlCMB1 in the development of tomato, we isolated this gene from Solanum lycopersicon (Mill. cv. Ailsa Craig) and investigated this gene. SlCMB1 had a higher homology with RIN, the key regulator of fruit ripening, at the amino acid level. The promoter analysis result displayed that the ethylene-responsive element, ERE-motif (ATTTCAAA) (Supplementary Fig. S1)58,59, was found in the promoter region of SlCMB1 gene. Expression profile of SlCMB1 showed that its transcripts rapidly declined with fruit ripening in the wild type fruits, and its expression pattern in Nr and rin mutant fruits was similar with that in AC++ fruits (Fig. 2A,B). These results indicated that SlCMB1 may involve in fruit ripening and/or ethylene biosynthesis.
In plants, ethylene plays a very important role in fruit ripening, and the biosynthesis pathway of ethylene has been well studied60–62. Two patterns of ethylene production systems, the autoinhibitory (system 1) and the autocatalytic (system 2), have been identified. In the system 1, basal ethylene is produced in the immature fruits and vegetative organs, while in the system 2 ethylene production is greatly increased at the beginning of flower senescence and climacteric fruit ripening17,63. ACS (ACC synthase) and ACO (ACC oxidase) are the critical rate-limiting enzymes in the process of ethylene biosynthesis. In tomato, co-suppression of SlACS2 and SlACS4 using antisense approach could reduce the ethylene biosynthesis of system 2 and the fruit ripening is inhibited in the transgenic lines64. In addition, SlACS2 plays a significant role in the transition of ethylene biosynthesis system (from system 1 to system 2)17. It has been reported that SlACO1 and SlACO3 express at the beginning of fruit ripening, the transcripts peak of SlACO1 at B + 3 stage and then falls back to B stage levels, whereas SlACO3 expresses transiently during ripening, the synthesis of ACO1 may be the first step of ethylene synthesis, after that the transcripts of ACS genes are induced by produced ethylene and then more ACC are produced18. In our study, the transcripts of SlACS2, SlACS4, SlACO1 and SlACO3 were significantly decreased in SlCMB1-silenced fruits (Fig. 6B–E). Moreover, the production of ethylene in SlCMB1-silenced fruits is significantly lower than the wild type (Fig. 6A). These results suggested that SlCMB1 may contribute to ethylene biosynthesis by promoting the expression of ethylene synthesis genes during tomato fruit ripening.
It has been known that E4, E8 and ERF1 are important ethylene-responsive genes in the process of fruit ripening23,25. In the mutations which block fruit ripening, the transcript of E4 is suppressed with the inhibited high-level ethylene biosynthesis23. E8 is reported to involve in ethylene biosynthesis in the process of fruit ripening25. The ethylene-responsive gene ERF1 which is initiated by ethylene is reported as an immediate target for EIN365. In this study, the transcripts of these three genes (E4, E8 and ERF1) were dramatically reduced in the SlCMB1-RNAi fruits compared with the wild type (Fig. 6F–H). Down-regulation of these ethylene response genes indicated that suppression of SlCMB1 impacts ethylene synthesis and fruit ripening.
Previous studies showed that four MADS-box proteins, RIN, FUL1, FUL2, and TAGL1, are essential to fruit ripening40,43,66. Recently, the molecular and biochemical studies indicate that RIN involves in ethylene biosynthesis by increasing the transcripts of ethylene biosynthesis genes and ethylene signaling genes such as ACS2 and ACS436,67,68. Further more, RIN could directly regulate the expression of FUL1 by binding to its promoter69 and another MADS-box gene, TAGL1, which activity in ripening is executed through direct activation of ACS2 that is reported to be a target of MADS-RIN36,70. In this study, the expression levels of these four MADS-box genes, RIN, TAGL1, FUL1 and FUL2, were down-regulated in the SlCMB1-RNAi fruits (Fig. 4A–D). RIN is the key regulator in tomato fruit ripening and acts upstream of ethylene biosynthesis, when it is repressed, fruit ripening and ethylene biosynthesis will be delayed32. In this study, although there was no significant difference of RIN expression in SlCMB1-RNAi fruits at MG and B stage, the remarkable reduced RIN mRNA in tomato plants were observed in transgenic fruits at B + 4 and B + 7 stage (Fig. 4A) suggesting that there might be direct or indirect regulatory relationship between SlCMB1 and RIN. In addition to these genes described above, the transcripts of other one ripening-related gene, LoxC, and a cell wall metabolism gene, PE, were also inhibited in SlCMB1-RNAi fruits (Fig. 4E,F). Moreover, phenotype analysis showed that fruit ripening in SlCMB1-silenced lines was delayed (Fig. 3C,D; Table 1). These results indicated that repression of SlCMB1 impacts the transcripts of ripening-related genes and delays the fruit ripening.
In this study, suppression of SlCMB1 resulted in delayed fruit ripening (Fig. 3C,D, Table 1) and reduced ethylene production (Fig. 6A). This is an interesting and new finding about this gene after it is reported to be involved in the regulation of inflorescence development48. This new finding will be contribute to helping us to further study the roles of SlCMB1 in the process of tomato growth and development. Up to now, a number of MADS-box genes have been reported to play multiple roles in the growth and development of plants, such as SlMADS1, SlMBP21 and TAGL141,48,54,55,70–74. For example, SlMADS1 (Solyc03g114840) was recently reported to play essential roles in the inflorescence development of tomato after it was found to be as a negative regulatory factor in tomato fruit ripening46,48. Further more, co-repression of the tomato FRUITFULL homologues (FUL1 and FUL2) results in the delayed fruit ripening and the decreased transcripts of ripening- and ethylene-related genes43. Similar alteration of fruit ripening is also observed in rin, Nr, nor, and Cnr mutant: delayed fruit ripening and reduced ethylene production29,32,33,35. According to our results and previous studies, we can speculate that SlCMB1 may be involved in the regulation of ethylene synthesis and fruit ripening, possibly through affecting the expression of ethylene synthesis, ethylene response and ripening-related genes.
The red pigmentation in tomato ripening fruits is mainly made up of β-carotene (5–40%) and lycopene (70–90%) which represent most of total carotenoids conferring the orange color and the red color, respectively75,76. Up to now, it is well known that the defects of carotenoid biosynthesis results in the reduction of carotenoid accumulation in the ripening-deficient mutant fruits77. The decreased expression of SlCMB1 leads to dramatically reduced carotenoid and lycopene content (Fig. 5A,B), explaining partly why SlCMB1-silenced fruits do not completely turn red at the same stage compared with the wild type (Fig. 3C,D). Moreover, the displayed orange-yellow or orange SlCMB1-silenced fruits (B + 4 and B + 7) and decreased carotenoid and lycopene content also implies increased β-carotene accumulation.
It is reported that PSY1, PDS, LCYE, LCYB and CYCB are the major enzymes and PSY1 is the rate-limiting enzyme in the carotenoid biosynthesis pathway. PSY1 catalyzes the conversion of geranylgeranyl diphosphate (GGPP) to phytoenethe, and PDS catalyzes the conversion of phytoene to ζ-carotene. In the carotenoid biosynthetic pathway, cyclization of lycopene forms two branches: one branch results in β-carotene and xanthophylls which catalyzed by two chloroplast and chromoplast lycopene β-cyclases, LCYB and CYCB, and the other results in α-carotene and xanthophyll catalyzed by LCYE and LCYB78. Furthermore, CYCB, a major enzyme in the cyclization of lycopene, is reported to be responsible for the transition from lycopene to β-carotene79. It is reported that the relative content of β-carotene and lycopene in tomato fruits during normal ripening is mediated by increased PSY1 transcripts and reduced expression of CYCB, in which these two effects are regulated partly by ethylene due to the induction of ethylene to PSY151,79,80. In this study, the expression of PSY1, which is induced by ethylene and is a crucial regulator of carotenoids biosynthesis during fruit ripening, was significantly down-regulated in response to reduced SlCMB1 (Fig. 5B). PDS, another regulator of carotenoids biosynthesis, was also notably inhibited in transgenic fruits (Fig. 5C), whereas the transcripts of CYCB, LCYB and LYCE were significantly increased in SlCMB1-RNAi fruits compared to the wild type (Fig. 5D–F). The change in expression of these genes is consistent with the decreased carotenoid and lycopene content. Previous studies have shown that RIN could interact with the major limiting enzyme PSY1 which is a direct target of RIN to control the pigment accumulation in carotenoid pathways during fruit ripening36,69. In this study, the expression of RIN was suppressed in the fruits of SlCMB1-RNAi lines at B + 4 and B + 7 stage (Fig. 4A) and the yeast two-hybrid assay showed that SlCMB1 can interact with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1, respectively (Figs 7; S5). These results can explain, on molecular and protein level, why the SlCMB1-silenced fruits exhibited a kind of light orange or orange phenotype at B + 4 and B + 7 stage (Fig. 3C,D). Analogously, previous studies showed that repression of the MADS-box gene TAGL1 leads to reduced ethylene production and increased β-carotene accumulation during fruit ripening40. Moreover, suppression of a tomato AP2/ERF gene, SlAP2a, results in reduced carotenoid accumulation through altering carotenoid pathway flux81,82. Based on previous investigations, we could speculate that SlCMB1 may play a significant role in regulation of carotenoid synthesis away from the lycopene and flux toward the β-carotene in SlCMB1-RNAi fruits, possibly through impacting ethylene biosynthesis or signal transduction or through regulating the expression of PSY1 by interacting with SlMADS-RIN.
Recent years, a growing number of transcription factor family, especially the MADS-box transcription factors, have been characterized and identified to play important regulatory roles in fruit ripening. It has been reported that MADS-box proteins can form homodimers, heterodimers, or higher-order protein complexes with other proteins to regulate plant growth and development83–85. Among the MADS-box proteins, SlMADS-RIN, a classical regulatory factor of fruit ripening, involves in the ethylene synthesis, ethylene response and ethylene perception in tomato66. Previous reports have shown that SlMADS-RIN can bind to SlACS2 and SlACS4 and associates with their promoters36,67,69. What is more, SlMADS-RIN also indirectly influences SlACO1 expression through binding to the promoter of a homeobox gene, HB-1, which generates an interaction with the promoter of SlACO136,86. Recent studies have shown that the ethylene-responsive genes E8 which could be induced by ethylene in fruit ripening is reported to be the direct target of SlMADS-RIN36,87. In addition, SlMADS1, SlAP2a and TAGL1 are the significant regulators in the process of fruit ripening. The MADS-box protein, SlMADS1, is reported to inhibit ethylene biosynthesis and influences fruit ripening as a negative regulator by interacting with SlMADS-RIN46. When SlAP2a, a member of the AP2/ERF superfamily, was repressed by RNAi approach in tomato, the transgenic lines displayed shorter ripening time and altered carotenoid accumulation81,82. TAGL1, another MADS-box gene, is reported to be as a positive regulator to involve in the regulation of fruit ripening and fleshy fruit expansion and suppression of TAGL1 in tomato results in yellow-orange fruits with thiner pericarps, decreased carotenoids and delayed ripening40,70. In our study, the yeast two-hybrid assays showed that SlCMB1 could interact with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1, respectively (Figs 7; S5). Previous studies have reported that the tomato FRUITFULL homologues (FUL1 and FUL2) act in fruit ripening via forming heterodimers with MADS-RIN43. The yeast three-hybrid assays have displayed that FUL1, TAGL1 and RIN could form higher order complexs88. Wang, S. et al. thought that there might be higher order complexes between FUL1, FUL2, MADS-RIN and TAGL1 in tomato fruit ripening89. Similarly, higer order complexs may also exists among SlCMB1, SlMADS-RIN, SlMADS1, SlAP2a and TAGL1 in the process of tomato fruit ripening. So we can speculate that SlCMB1 may increase the activity of SlMADS-RIN and TAGL1 and/or reduce the activity of SlMADS1 and SlAP2a through forming dimers or higher-order protein complexes to directly or indirectly regulate the expression of related genes such as ACO1, ACS2, ACS4 and E8, and finally the ethylene biosynthesis is increased and the fruit ripening is promoted.
In summary, the MADS-box transcription factor SlCMB1 plays an essential role in the process of fruit ripening acting as a positive regulator by modulating the ethylene biosynthesis and response and carotenoid accumulation through interacting with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1. SlCMB1 is a new member of the regulatory network of fruit ripening. Additionally, our results manifest that the higher levels of the SlCMB1 regulatory cascades in tomato fruit ripening await being discovered, such as identification of upstream regulatory factors, direct or indirect downstream targets and the interaction between these new regulatory components. We can believe that these follow-up works will contribute to adding more new components to enrich the ripening regulatory network and will bring a deeper understanding to the fruit ripening regulatory mechanism.
Materials and Methods
Plant materials and growth conditions
In this study, the near-isogenic tomato line, Solanum lycopersicum (Mill. cv. Ailsa Craig AC++), was used as the wild type. The wide type and transgenic tomato plants were planted in the greenhouse under the standard conditions as follows: 25 °C for 16 h (day) and 18 °C for 8 h (night). The tomato flowers were tagged at anthesis. The days and fruit color post-anthesis (DPA) was used to differentiate the ripening days of tomato fruits. In the wild type, we defined 20 DPA as the immature green, 35 DPA as the mature green that the fruit is green and shiny and no obvious color change is observed. The 38 DPA tomato fruits which color of fruits change from green to yellow was characterized as breaker (B) fruits. Besides, the material of B + 4 (4 days after breaker) fruits and B + 7 (7 days after breaker) fruits were also used in our study. All the needed samples were collected and immediately frozen in liquid nitrogen and then stored at −80 °C until being used.
Total RNA extraction, isolation and sequence analysis of SlCMB1
The total RNA from all WT and transgenic tomato plants tissues was extracted using the RNAiso Plus reagent (Takara, China) following the instructions of manufacturer. In order to synthesize the first strand cDNA, 1 μg total RNA samples which were digested with the Dnase I (Promega, USA) was used to perform the reverse transcription using the M-MLV reverse transcriptase (Promega) with the oligo(dT)20 primer according to the manufacturer’s protocol.
The full length of SlCMB1 gene was cloned using 1–2 μL cDNA with primers SlCMB1-Full-F and SlCMB1-Full-R (Supplementary Table S1). The DNA A-Tailing kit (Takara) was used to tail the amplified products. After that, the tailed products were cloned into the pMD18-T vector (Takara). The Escherichia coli JM109 transformation was performed to pick out the positive clones and confirmed by sequencing. Multiple sequence alignment was performed for comparison with other MADS-box proteins by DNAMAN (Version 6.0). The phylogenetic tree was constructed by MEGA(Version 5.2) according to the neighbor-joining bootstrap method as follows: bootstrap analysis of 1,000 replicates, pairwise deletion and poisson model. Moreover, 3500 bp nucleotide sequence upstream of the initiation codon ATG of predicted ORF of SlCMB1 gene was used to perform the promoter analysis on the Plant CARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
RNAi vector construction of SlCMB1 and plant transformation
In order to study the function of SlCMB1 in the process of tomato fruits development, a RNAi vector of SlCMB1 was constructed to down-regulate its expression. A 426-bp 3′ special fragment of SlCMB1 was amplified using primers SlCMB1-i-F and SlCMB1-i-R (Supplementary Table S1) which was tailed with the KpnI/HindIII and the XhoI/XbaI restriction enzyme sites at the 5′ end of primers, respectively. The enzyme digestion was performed using the HindIII/XbaI and KpnI/XhoI enzyme to digest the amplified SlCMB1 fragments, respectively. Then the digested products were ligated into the sense orientation at the HindIII/XbaI restriction enzyme site and into the antisense orientation at the KpnI/XhoI restriction enzyme site of pHANNIBAL plasmid, respectively. After being digested with SpeI/SacI, the expression unit what we needed, including the 35 S promoter of the cauliflower mosaic virus, the specific fragment of SlCMB1 in the antisense orientation, the PDK intron, specific fragment of SlCMB1 in the sense orientation, and the OCS terminator, was subcloned into the pBIN19 vector at the SacI and XbaI restriction site to obtain the RNAi vector for SlCMB1 gene silencing (Supplementary Fig. S2).
The generated binary plasmids which were verified by restriction digest analysis and by sequencing were transferred into the Agrobacterium tumefaciens LBA4404 strain, and Agrobacterium tumefaciens-mediated transformation was carried out according to the approach described by Chen et al.90. The primers NPTII-F and NPTII-R (Supplementary Table S1) were used to detect the transgenic plants. The selected positive transgenic lines were used for the subsequent investigation.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA extraction and reverse transcription were performed as described above. Then 5 times RNase/DNase-free water was used to dilute the synthesized cDNAs for the qRT-PCR analysis. The qRT-PCR was carried out on the CFX96TM Real-Time System (C1000TM Thermal Cycler, Bio-Rad) according to the manufacturer instructions. All the qRT-PCR reactions were performed in the 10 μL total sample volume including 5 μL SYBR Premix Go Taq (Promega, China), 0.25 μL each primer (10 mM), 3 μL nuclease-free water and 1.5 μL diluted cDNA. The reaction conditions were carried out as follows: 95 °C for 5 min, followed by 41 cycles of 95 °C for 15 s and 60 °C for 35 s. After the qRT-PCR cycles, Melt curve analysis of each qRT-PCR sample was performed to confirm each primer specificity: 95 °C for 1 min followed by a constant increase (0.25 °C per 1 s) of the temperature between 65 °C and 95 °C. The results of melt curve analysiss showed that only one product of each gene existed. The NTC (no template control) and NRT (no reverse transcription control) experiments were also carried out for each gene analysis. Tomato SlCAC91 was used as the internal standards. The 2−∆∆CT method92 was used to analyze the relative quantification of specific mRNA levels. The expression levels of ethylene synthesis and response genes, ripening-related genes and carotenoid synthesis genes were detected in fruits. All the primers of related genes used for qRT-PCR analysis are listed in Supplementary Table S2.
Pigments content determination
For the total carotenoid extraction, the sample (1 g) of each line was cut from the same area of pericarp around the equator of fruits at B, B + 4 and B + 7 stage. After being triturated in the liquid nitrogen, 10 mL hexane:acetone (60:40, v/v) was used and then the total carotenoids of each sample was extracted. Then the extract was centrifuged for 5 min at 4000 g, the absorbance of the samples was immediately examined at 450 nm. The carotenoid content of each samples was calculated as follows: total carotenoid (mg/mL) = 4 × (absorbancy at 450 nm) × 10 mL/1 g93,94.
The lycopene extraction was performed according to the method described by Fish, W. W. et al.95. 0.4 to 0.6 g sample of each line was cut from the same area of pericarp around the equator of fruits at B, B + 4 and B + 7 stage. The 50 mL centrifuge tubes were used in this experiment. Three independent experiments of each sample were carried out. After being triturated in the liquid nitrogen, 20 mL 0.05% (w/v) BHT in acetone: 95% ethanol: hexane (1:1:2, v/v) was used. Then the 50 mL centrifuge tubes were laid on a container that contained ice and shaked in a orbital shaker to mix for 15 min at 180 rpm. After shaking was finished, the ice deionized water (3 mL) were added into each tube, and then the samples were shaken to mix for another 5 min at 180 rpm. After 5 min of shaking, the tubes were left for 5 min at room temperature to allow for phase separation. The supernatant (hexane layer) was used to measure the absorbance at 503 nm. The quartz cuvette was 1 cm path length and the hexane solvent was used as the blank control. The carotenoid content of each samples was calculated as follows: Lycopene (mg/kg) = (A503 × 31.2)/g tissue95.
Ethylene measurements
The B, B + 4, B + 7 fruits of transgenic lines and the wild type tomato were used and put in a open jars (100 mL) for 3 h to minimize the impact of ethylene that induced by the wound of fruits picking. After being sealed, the jars were stored at the room temperate for 24 h. Then 1 mL of the headspace gas of each sample was injected into the gas chromatograph (Hewlett-Packard 5890 series) using the flame ionization detector. Each sample was normalized for fruit weight compared to the ethylene standards which concentration had been known81. Three independent experiments were performed for each sample.
Yeast two-hybrid assay
The MATCHMAKER GAL4 Two-Hybrid System III was used to perform the yeast two-hybrid assay following the method described by the manufacturer (Clontech). The PCR experiments were performed to amplify the open reading frame (ORF) of SlCMB1 using the primers SlCMB1(Y2H)F/R (Supplementary Table S1). The products were cloned into the EcoR I/BamH I site of the pGADT7 vector to generate the pGADT7-SlCMB1 vector (Supplementary Fig. S4B). Meanwhile, the ORFs of SlMADS-RIN, SlMADS1, SlAP2a and TAGL1 were also amplified using the primers SlMADS-RIN(Y2H)-F/R, SlMADS1(Y2H)-F/R, SlAP2a(Y2H)-F/R and TAGL1(Y2H)-F/R (Supplementary Table S1), respectively. After being digested by BamHI and EcoRI, the products were linked into the EcoRI/BamHI site of the vector pGBKT7 to generate the pGBKT7-RIN, pGBKT7-SlMADS1, pGBKT7-SlAP2a and pGBKT7-TAGL1 (Supplementary Fig. S4A), respectively. The pGADT7-SlCMB1 vector was transferred into Y187 and the pGBKT7-RIN, pGBKT7-SlMADS1, pGBKT7-SlAP2a and pGBKT7-TAGL1 were also transferred into Y2Hgold, respectively. The Y187 with prey (pGADT7-SlCMB1) was plated on SD (synthetic dropout) medium without Leu. The Y2HGold with the pGBKT7-RIN, pGBKT7-SlMADS1, pGBKT7-SlAP2a and pGBKT7-TAGL1 bait was plated on the SD medium without Trp (SDO), respectively. In parallel, the self-activation experiments of pGBKT7-RIN, pGBKT7-SlMADS1, pGBKT7-SlAP2a and pGBKT7-TAGL1 were performed on the SD medium with no Trp, His and adenine (TDO) (Supplementary Fig. S5), respectively. Then, the Y2HGold with bait (pGBKT7-RIN, pGBKT7-SlMADS1, pGBKT7-SlAP2a and pGBKT7-TAGL1) and the Y187 with prey (pGADT7-SlCMB1) were cultured together in the 2 × YPDA medium at 200 rpm for 24 h, respectively. The cultures were plated on the SD medium with no Leu and Trp (DDO) to select the diploids containing the prey and bait vectors, simultaneously. 3 to 5 days later, the fresh diploid cells were cultured on the SD medium which lacked adenine, His, Leu and Trp, with the indicator X-a-Gal (QDO/X) to confirm whether SlCMB1 could interact with SlMADS-RIN, SlMADS1, SlAP2a and TAGL1 or not, respectively. All plates were incubated at 30 °C for 3–5 d. The empty bait and prey vector containing the construct of each bait and prey were used as negative controls, respectively. In parallel, positive controls were also cultured. Three independent repetition of these experiments were performed with fresh transformants.
Statistical analysis
The mean values of data were measured from three replicates and ‘Standard Error’ of the means was calculated. Data were analyzed by Origin 8.0 software, and t test (SAS 9.2) was used for assessing significant differences among the means.
Electronic supplementary material
Acknowledgements
This work was supported by National Natural Science Foundation of China (no. 30600044, 31572129), and the Natural Science Foundation of Chongqing of China (cstc2015jcyjA80026).
Author Contributions
Guoping Chen and Zongli Hu designed and managed the research work and improved the manuscript. Jianling Zhang, Zongli Hu, Qiyuan Yao, Xuhu Guo, Vanluc Nguyen, Fenfen Li performed the experiments. Jianling Zhang wrote the manuscript and prepared all figures. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21672-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goff SA, Klee HJ. Plant volatile compounds: sensory cues for health and nutritional value? Science. 2006;311:815–819. doi: 10.1126/science.1112614. [DOI] [PubMed] [Google Scholar]
- 2.Giovannoni J. Molecular biology of fruit maturation and ripening. Annual review of plant biology. 2001;52:725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- 3.Giovannoni JJ. Genetic regulation of fruit development and ripening. The Plant Cell. 2004;16:S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa F, et al. Use of homologous and heterologous gene expression profiling tools to characterize transcription dynamics during apple fruit maturation and ripening. BMC plant biology. 2010;10:1. doi: 10.1186/1471-2229-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abeles, F., Morgan, P. & Saltveit, M. The role of ethylene in agriculture. (eds Frederick B. et al.) Ch. 9, 264–296. Ethylene in Plant Biology (Second Edition), (Academic press, 1992).
- 6.Nath, P., Trivedi, P. K., Sane, V. A. & Sane, A. P. Role of Ethylene in Fruit Ripening. (eds Nafees A. Khan) Ch. 8, 151-184. Ethylene action in plants, (Springer, 2006).
- 7.Dolan L. The role of ethylene in root hair growth in Arabidopsis. Journal of Plant Nutrition and Soil Science. 2001;164:141–145. doi: 10.1002/1522-2624(200104)164:2<141::AID-JPLN141>3.0.CO;2-Z. [DOI] [Google Scholar]
- 8.Flores F, et al. Role of ethylene in the biosynthetic pathway of aliphatic ester aroma volatiles in Charentais Cantaloupe melons. Journal of experimental botany. 2002;53:201–206. doi: 10.1093/jexbot/53.367.201. [DOI] [PubMed] [Google Scholar]
- 9.Zhou H-W, Dong L, Ben-Arie R, Lurie S. The role of ethylene in the prevention of chilling injury in nectarines. Journal of Plant Physiology. 2001;158:55–61. doi: 10.1078/0176-1617-00126. [DOI] [Google Scholar]
- 10.Abeles, F. B., Morgan, P. W. & Saltveit Jr, M. E. Roles and Physiological Effects of Ethylene in Plant Physiology: Dormancy, Growth, and Development. (eds Frederick B. et al.) Ch. 5, 120–181. Ethylene in Plant Biology (Second Edition), (Academic press, 1992).
- 11.Hiwasa K, et al. Ethylene is required for both the initiation and progression of softening in pear (Pyrus communis L.) fruit. Journal of experimental botany. 2003;54:771–779. doi: 10.1093/jxb/erg073. [DOI] [PubMed] [Google Scholar]
- 12.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annual review of plant physiology. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- 13.Zarembinski TI, Theologis A. Ethylene biosynthesis and action: a case of conservation. Plant Molecular Biology. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- 14.Oetiker JH, Olson DC, Shiu OY, Yang SF. Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum) Plant Molecular Biology. 1997;34:275–286. doi: 10.1023/A:1005800511372. [DOI] [PubMed] [Google Scholar]
- 15.Olson DC, White JA, Edelman L, Harkins RN, Kende H. Differential expression of two genes for 1-aminocyclopropane-1-carboxylate synthase in tomato fruits. Proceedings of the National Academy of Sciences. 1991;88:5340–5344. doi: 10.1073/pnas.88.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barry CS, et al. Differential expression of the 1‐aminocyclopropane‐1‐carboxylate oxidase gene family of tomato. The Plant Journal. 1996;9:525–535. doi: 10.1046/j.1365-313X.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- 17.Barry CS, Llop-Tous MI, Grierson D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiology. 2000;123:979–986. doi: 10.1104/pp.123.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. Journal of experimental botany. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- 19.Blume B, Grierson D. Expression of ACC oxidase promoter—GUS fusions in tomato and Nicotiana plumbaginifolia regulated by developmental and environmental stimuli. The Plant Journal. 1997;12:731–746. doi: 10.1046/j.1365-313X.1997.12040731.x. [DOI] [PubMed] [Google Scholar]
- 20.Hu Z-L, Chen X-Q, Chen G-P, Lü L-J, Donald G. The influence of co-suppressing tomato 1-aminocyclopropane-1-carboxylic acid oxidase I on the expression of fruit ripening-related and pathogenesis-related protein genes. Agricultural Sciences in China. 2007;6:406–413. doi: 10.1016/S1671-2927(07)60063-7. [DOI] [Google Scholar]
- 21.Lincoln JE, Cordes S, Read E, Fischer RL. Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proceedings of the National Academy of Sciences. 1987;84:2793–2797. doi: 10.1073/pnas.84.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lincoln JE, Fischer RL. Diverse mechanisms for the regulation of ethylene-inducible gene expression. Molecular and General Genetics MGG. 1988;212:71–75. doi: 10.1007/BF00322446. [DOI] [PubMed] [Google Scholar]
- 23.Xu R, Goldman S, Coupe S, Deikman J. Ethylene control of E4 transcription during tomato fruit ripening involves two cooperativecis elements. Plant Molecular Biology. 1996;31:1117–1127. doi: 10.1007/BF00040829. [DOI] [PubMed] [Google Scholar]
- 24.Lincoln JE, Fischer RL. Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiology. 1988;88:370–374. doi: 10.1104/pp.88.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kneissl ML, Deikman J. The tomato E8 gene influences ethylene biosynthesis in fruit but not in flowers. Plant Physiology. 1996;112:537–547. doi: 10.1104/pp.112.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesanakurti D, Kolattukudy PE, Kirti PB. Fruit-specific overexpression of wound-inducedtap1 under E8 promoter in tomato confers resistance to fungal pathogens at ripening stage. Physiologia plantarum. 2012;146:136–148. doi: 10.1111/j.1399-3054.2012.01626.x. [DOI] [PubMed] [Google Scholar]
- 27.Krasnyanski SF, Sandhu J, Domier LL, Buetow DE, Korban SS. Effect of an enhanced CaMV 35S promoter and a fruit-specific promoter on uida gene expression in transgenic tomato plants. In Vitro Cellular & Developmental Biology-Plant. 2001;37:427–433. doi: 10.1007/s11627-001-0075-1. [DOI] [Google Scholar]
- 28.Moore S, Vrebalov J, Payton P, Giovannoni J. Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. Journal of experimental botany. 2002;53:2023–2030. doi: 10.1093/jxb/erf057. [DOI] [PubMed] [Google Scholar]
- 29.Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Current opinion in plant biology. 2007;10:283–289. doi: 10.1016/j.pbi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Consortium TG. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barry CS, Giovannoni JJ. Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proceedings of the National Academy of Sciences. 2006;103:7923–7928. doi: 10.1073/pnas.0602319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrebalov J, et al. A MADS-Box Gene Necessary for Fruit Ripening at the Tomato Ripening-Inhibitor (Rin) Locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 34.Thompson AJ, et al. Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiology. 1999;120:383–390. doi: 10.1104/pp.120.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature genetics. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 36.Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. The Tomato MADS-box Transcription Factor RIPENING INHIBITOR Interacts with Promoters Involved in Numerous Ripening Processes in a COLORLESS NONRIPENING-Dependent Manner1[W][OA] Plant Physiology. 2011;157:1568–1579. doi: 10.1104/pp.111.181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messenguy F, Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21. doi: 10.1016/S0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 38.Hileman LC, et al. Molecular and Phylogenetic Analyses of the MADS-box Gene Family in Tomato. Molecular Biology and Evolution. 2006;23:2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- 39.Itkin M, et al. TOMATO AGAMOUS‐LIKE 1 is a component of the fruit ripening regulatory network. The Plant Journal. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- 40.Vrebalov J, et al. Fleshy Fruit Expansion and Ripening Are Regulated by the Tomato SHATTERPROOF Gene TAGL1. The Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giménez E, et al. Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS One. 2010;5:e14427. doi: 10.1371/journal.pone.0014427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gimenez, E. et al. TOMATO AGAMOUS1 and ARLEQUIN/TOMATO AGAMOUS-LIKE1 MADS-box genes have redundant and divergent functions required for tomato reproductive development. Plant molecular biology, 1–19 (2016). [DOI] [PubMed]
- 43.Shima Y, et al. Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Molecular Biology. 2013;82:427–438. doi: 10.1007/s11103-013-0071-y. [DOI] [PubMed] [Google Scholar]
- 44.Ito Y. Regulation of Tomato Fruit Ripening by MADS-Box Transcription Factors. Japan Agricultural Research Quarterly: JARQ. 2016;50:33–38. doi: 10.6090/jarq.50.33. [DOI] [Google Scholar]
- 45.Xie, Q. et al. Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Scientific reports4 (2014). [DOI] [PMC free article] [PubMed]
- 46.Dong T, et al. A Tomato MADS-box Transcription Factor, SlMADS1, Acts as a Negative Regulator of Fruit Ripening1[C][W] Plant Physiology. 2013;163:1026–1036. doi: 10.1104/pp.113.224436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SJ, Jiang K, Schatz MC, Lippman ZB. Rate of meristem maturation determines inflorescence architecture in tomato. Proceedings of the National Academy of Sciences. 2012;109:639–644. doi: 10.1073/pnas.1114963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soyk S, et al. Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell. 2017;169:1142–1155. doi: 10.1016/j.cell.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 49.Litt A, Irish VF. Duplication and Diversification in the APETALA1/FRUITFULL Floral Homeotic Gene Lineage: Implications for the Evolution of Floral Development. Genetics. 2003;165:821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartley GE, Viitanen P, Bacot K, Scolnik P. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. Journal of Biological Chemistry. 1992;267:5036–5039. [PubMed] [Google Scholar]
- 51.Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression) Plant Physiology. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng M, Yanofsky MF. Three ways to learn the ABCs. Current opinion in plant biology. 2000;3:47–52. doi: 10.1016/S1369-5266(99)00036-9. [DOI] [PubMed] [Google Scholar]
- 53.Nakano T, et al. MACROCALYX and JOINTLESS Interact in the Transcriptional Regulation of Tomato Fruit Abscission Zone Development. Plant Physiology. 2012;158:439–450. doi: 10.1104/pp.111.183731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu D, et al. The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. The Plant Journal. 2014;77:284–296. doi: 10.1111/tpj.12387. [DOI] [PubMed] [Google Scholar]
- 55.Li, N. et al. The MADS-box Gene SlMBP21 Regulates Sepal Size Mediated by Ethylene and Auxin in Tomato. Plant and Cell Physiology (2017). [DOI] [PubMed]
- 56.Szymkowiak EJ, Irish EE. Interactions between jointless and Wild-Type Tomato Tissues during Development of the Pedicel Abscission Zone and the Inflorescence Meristem. The Plant Cell. 1999;11:159–175. doi: 10.1105/tpc.11.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao L, et al. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature. 2000;406:910–913. doi: 10.1038/35022611. [DOI] [PubMed] [Google Scholar]
- 58.Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proceedings of the National Academy of Sciences. 1993;90:5939–5943. doi: 10.1073/pnas.90.13.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itzhaki H, Maxson JM, Woodson WR. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1)gene. Proceedings of the National Academy of Sciences. 1994;91:8925–8929. doi: 10.1073/pnas.91.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annual review of cell and developmental biology. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 61.Guo H, Ecker JR. The ethylene signaling pathway: new insights. Current opinion in plant biology. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Wang KL-C, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. The Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang S-H, Lu L-S, Wang NN, Charng Y-Y. Negative feedback regulation of system-1 ethylene production by the tomato 1-aminocyclopropane-1-carboxylate synthase 6 gene promoter. Plant Science. 2008;175:149–160. doi: 10.1016/j.plantsci.2007.11.004. [DOI] [Google Scholar]
- 64.Oeller PW, Min-Wong L. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254:437. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- 65.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Gene Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuste-Lisbona FJ, et al. Characterization of vegetative inflorescence (mc-vin) mutant provides new insight into the role of MACROCALYX in regulating inflorescence development of tomato. Scientific reports. 2016;6:18796. doi: 10.1038/srep18796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito Y, et al. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. The Plant Journal. 2008;55:212–223. doi: 10.1111/j.1365-313X.2008.03491.x. [DOI] [PubMed] [Google Scholar]
- 68.Fujisawa, M., Nakano, T. & Ito, Y. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC plant biology11 (2011). [DOI] [PMC free article] [PubMed]
- 69.Fujisawa M, et al. Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta. 2012;235:1107–1122. doi: 10.1007/s00425-011-1561-2. [DOI] [PubMed] [Google Scholar]
- 70.Itkin M, et al. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. The Plant Journal. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- 71.Roldan MVG, et al. Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Scientific reports. 2017;7:4402. doi: 10.1038/s41598-017-04556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giménez E, et al. Transcriptional Activity of the MADS Box ARLEQUIN/TOMATO AGAMOUS-LIKE1 Gene Is Required for Cuticle Development of Tomato Fruit. Plant Physiology. 2015;168:1036–1048. doi: 10.1104/pp.15.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gimenez E, et al. TOMATO AGAMOUS1 and ARLEQUIN/TOMATO AGAMOUS-LIKE1 MADS-box genes have redundant and divergent functions required for tomato reproductive development. Plant Molecular Biology. 2016;91:513–531. doi: 10.1007/s11103-016-0485-4. [DOI] [PubMed] [Google Scholar]
- 74.Garceau, D. C., Batson, M. K. & Pan, I. L. Variations on a theme in fruit development: the PLE lineage of MADS-box genes in tomato (TAGL1) and other species. Planta, 1–9 (2017). [DOI] [PubMed]
- 75.Burns J, Fraser PD, Bramley PM. Identification and quantification of carotenoids, tocopherols and chlorophylls in commonly consumed fruits and vegetables. Phytochemistry. 2003;62:939–947. doi: 10.1016/S0031-9422(02)00710-0. [DOI] [PubMed] [Google Scholar]
- 76.Alba R, et al. Transcriptome and Eelected Metabolite Analyses Reveal Multiple Points of Ethylene Control during Tomato Fruit Development. The Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fraser PD, Bramley PM. The biosynthesis and nutritional uses of carotenoids. Progress in lipid research. 2004;43:228–265. doi: 10.1016/j.plipres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 78.Hirschberg J. Carotenoid biosynthesis in flowering plants. Current opinion in plant biology. 2001;4:210–218. doi: 10.1016/S1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- 79.Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proceedings of the National Academy of Sciences. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alba R, et al. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. The Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung MY, et al. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. The Plant Journal. 2010;64:936–947. doi: 10.1111/j.1365-313X.2010.04384.x. [DOI] [PubMed] [Google Scholar]
- 82.Karlova R, et al. Transcriptome and Metabolite Profiling Show That APETALA2a Is a Major Regulator of Tomato Fruit Ripening. The Plant Cell Online. 2011;23:923–941. doi: 10.1105/tpc.110.081273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Favaro R, et al. Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Molecular Genetics and Genomics. 2002;268:152–159. doi: 10.1007/s00438-002-0746-6. [DOI] [PubMed] [Google Scholar]
- 84.Shchennikova AV, Shulga OA, Immink R, Skryabin KG, Angenent GC. Identification and Characterization of Four Chrysanthemum MADS-box Genes, Belonging to the APETALA1/FRUITFULL and SEPALLATA3 Subfamilies. Plant Physiology. 2004;134:1632–1641. doi: 10.1104/pp.103.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Folter S, et al. A Bsister MADS‐box gene involved in ovule and seed development in petunia and Arabidopsis. The Plant Journal. 2006;47:934–946. doi: 10.1111/j.1365-313X.2006.02846.x. [DOI] [PubMed] [Google Scholar]
- 86.Lin Z, et al. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. The Plant Journal. 2008;55:301–310. doi: 10.1111/j.1365-313X.2008.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin GZ, Wang YY, Cao BH, Wang WH, Tian SP. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. The Plant Journal. 2012;70:243–255. doi: 10.1111/j.1365-313X.2011.04861.x. [DOI] [PubMed] [Google Scholar]
- 88.Leseberg CH, et al. Interaction study of MADS-domain proteins in tomato. Journal of experimental botany. 2008;59:2253–2265. doi: 10.1093/jxb/ern094. [DOI] [PubMed] [Google Scholar]
- 89.Wang S, et al. Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. Journal of experimental botany. 2014;65:3005–3014. doi: 10.1093/jxb/eru137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen G, et al. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiology. 2004;136:2641–2651. doi: 10.1104/pp.104.041608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC plant biology. 2008;8:1. doi: 10.1186/1471-2229-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 93.Fray RG, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Molecular Biology. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- 94.Forth D, Pyke KA. The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. Journal of experimental botany. 2006;57:1971–1979. doi: 10.1093/jxb/erj144. [DOI] [PubMed] [Google Scholar]
- 95.Fish WW, Perkins-Veazie P, Collins JK. A Quantitative Assay for Lycopene That Utilizes Reduced Volumes of Organic Solvents. Journal of food composition and analysis. 2002;15:309–317. doi: 10.1006/jfca.2002.1069. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.