Abstract
Revertant mosaicism, or “natural gene therapy”, is the phenomenon in which germline mutations are corrected by somatic events. In recent years, revertant mosaicism has been identified in all major types of epidermolysis bullosa, the group of heritable blistering disorders caused by mutations in the genes encoding epidermal adhesion proteins. Moreover, revertant mosaicism appears to be present in all patients with a specific subtype of recessive epidermolysis bullosa. We therefore hypothesized that revertant mosaicism should be expected at least in all patients with recessive forms of epidermolysis bullosa. Naturally corrected, patient-own cells are of extreme interest for their promising therapeutic potential, and their presence in all patients would open exciting, new treatment perspectives to those patients. To test our hypothesis, we determined the probability that single nucleotide reversions occur in patients’ skin using a mathematical developmental model. According to our model, reverse mutations are expected to occur frequently (estimated 216x) in each patient’s skin. Reverse mutations should, however, occur early in embryogenesis to be able to drive the emergence of recognizable revertant patches, which is expected to occur in only one per ~10,000 patients. This underestimate, compared to our clinical observations, can be explained by the “late-but-fitter revertant cell” hypothesis: reverse mutations arise at later stages of development, but provide revertant cells with a selective growth advantage in vivo that drives the development of recognizable healthy skin patches. Our results can be extrapolated to any other organ with stem cell division numbers comparable to skin, which may offer novel future therapeutic options for other genetic conditions if these revertant cells can be identified and isolated.
Introduction
Revertant mosaicism (RM), or “natural gene therapy”, is the phenomenon in which the effect of germline mutations is corrected by somatic mutational events, and hence constitutes a modifier of disease. RM was first reported in Lesch-Nyhan syndrome in 1988 [1], and subsequently in several other genetic syndromes [2,3]. In 1997, RM was first reported in a genetic skin condition, epidermolysis bullosa (EB) [4], the group of heritable blistering disorders caused by mutations in the genes encoding the components of the epidermal-dermal adhesion complex [5]. While long considered an extraordinary phenomenon, RM has been identified in all major types of EB in recent years (Table 1) [4–19]. Moreover, in a Dutch study RM appeared to be present in all patients with the generalized intermediate subtype of junctional EB (formerly: non-Herlitz junctional EB) on clinical examination, and could be proven at the DNA level in 60% of patients with this EB type [11]. RM has also been shown to be an important disease-modifier in ichthyosis with confetti, another genetic skin disorder, caused by mutations in the KRT1 or KRT10 genes [20,21]. These findings have led to the conclusion that, instead of being extraordinary, RM seems to be rather common in EB and led us to hypothesize that RM is present in all patients with EB. In the light of the exciting progress made on revertant cell therapy in recent years [22], boosted especially by the combination with the induced pluripotent stem cell approach [23], as well as the recent successful regeneration of an entire human epidermis from exogenously corrected epidermal stem cells [24], the presence of RM in all EB patients would have important implications for future revertant cell therapy development. In this study, we therefore sought to obtain proof for our hypothesis by employing a mathematical developmental model of the skin. Our results indicate that revertant cells should be present in the skin of all EB patients, but they need a significant selective growth advantage to be able to grow out to clinically recognizable healthy skin patches.
Table 1. Revertant mosaicism in epidermolysis bullosa.
| Major EB type | EB subtype | OMIM | Corrected gene | OMIM | References |
|---|---|---|---|---|---|
| EB simplex | EBS, generalized severe | 131760 | KRT14 | 148066 | [6] |
| EBS, autosomal recessive K14 | 601001 | KRT14 | 148066 | [7] | |
| Junctional EB | JEB, generalized intermediate (AR) | 226650 | COL17A1 | 113811 | [4,8–12] |
| LAMB3 | 150310 | [13] | |||
| Dystrophic EB | Recessive DEB, generalized severe | 226600 | COL7A1 | 120120 | [14–16] |
| Recessive DEB, generalized intermediate | 226600 | COL7A1 | 120120 | [16,17]] | |
| Dominant DEB | 131750 | COL7A1 | 120120 | [16] | |
| Kindler syndrome | - | 173650 | FERMT1 | 607900 | [18,19] |
EB, epidermolysis bullosa; EBS, epidermolysis bullosa simplex; JEB, junctional epidermolysis bullosa; DEB, dystrophic epidermolysis bullosa; AR, autosomal recessive.
Results and discussion
We used a simplified mathematical developmental model, in which we focused on single nucleotide corrections of nonsense mutations in recessive types of EB (REB), the dominant type of reversion mutations observed in EB [25]. We also focused on the probability that reverse mutations occur until adulthood is reached, not the probability that reverse mutations occur during adult life, because, as discussed later in more detail, the revertant patches that we observe in EB patients develop before adulthood. In this model, the number of basal keratinocytes (BKs), n, starting with a single embryonic ectodermal progenitor cell, increases exponentially in the epidermis, n = 2y, with each generation, y. The probability that reverse mutations occur until adulthood then depends on four factors: (1) the total number of BKs in an adult human body (estimated to be 36×109, ~235), (2) the number of mitoses required to obtain this number (36×109–1), (3) the probability of a nucleotide alteration per nucleotide per mitosis (~1×10−9) [26–28], and (4) the number of target nucleotides that are able to correct the germline mutation when altered (assumed 6 in a recessive model, ignoring the possibility of a nonsense codon being changed into one of the other two nonsense codons). See Table 2 and Methods section at the end of this manuscript for details on the mathematical calculations and quantitative estimates used in this study.[29–35] Our model shows that, during the total number of mitoses, the probability P that at least one reverse mutation occurs approaches 1 and reverse mutations are expected to occur 216 times in an average adult human body. This indicates that, indeed, the occurrence of reverse mutations should not be considered extraordinary, but rather an event that can be expected with mathematical certainty in REB patients’ skin carrying mutations that are correctable by single nucleotide mutations. This finding corresponds well to the results of a recent study that demonstrated a strong correlation between the high population incidence of basal cell carcinomas and the high number of basal stem cell mitoses in the skin, which was attributed to stochastic events of numerous randomly occurring somatic mutations [36].
Table 2. Quantitative estimates and calculated values in our developmental model of the skin.
| Quantitative values | Explanation | References | |
|---|---|---|---|
| 6 complexes | Average number of rete ridges-dermal papillae complexes per mm skin length | [29] | |
| 0.070 mm | Average height of rete ridges-dermal papillae complexes | [30] | |
| 1.348 mm | DEJ length corresponding to 1 mm of skin length | Calculated in this study | |
| 1.817 mm2 | DEJ surface corresponding to 1 mm2 of skin surface | Calculated in this study | |
| 81.7% | Increase in dermo-epidermal contact due to rete ridges-dermal papillae complexes | Calculated in this study | |
| 10 μm | Diameter of circular base of basal keratinocytes | [31] | |
| 20,000 cells | Number of basal keratinocytes per mm2 skin | Used in this study | |
| 15,000 cells | Number of basal keratinocytes per mm2 skin | [32] | |
| 20,000–30,000 cells | Number of basal keratinocytes per mm2 skin | [33] | |
| 23,146 cells | Number of basal keratinocytes per mm2 skin | Calculated in this study | |
| 1.8 m2 | Average skin surface in adult human body | Used in this study | |
| 3.3x106 mm2 | Average total DEJ surface in adult human body | Calculated in this study | |
| 36x109 cells | Average total number of basal keratinocytes in adult human body | Calculated in this study | |
| (36x109–1) mitoses | Number of mitoses needed to obtain total number of basal keratinocytes | Calculated in this study | |
| 35.1 generations | Number of generations of cells needed to obtain total number of basal keratinocytes | Calculated in this study | |
| 1x10-9 | Approximate per nucleotide point mutation rate per mitosis | [26–28] | |
| 6x109 nucleotides | Approximate number of nucleotides per genome | [34] | |
| 6 mutations | Expected number of novel point mutations per mitosis | Calculated in this study | |
| 216x109 mutations | Expected number of point mutations in basal keratinocytes of human body at adulthood | Calculated in this study | |
| 36x | Average point mutation frequency of each nucleotide in basal keratinocytes of human body at adulthood | Calculated in this study | |
| 105 | Total number of somatic mutations expected to have accumulated per BK at adulthood | Calculated in this study | |
| 6 nucleotides | Number of target nucleotides for a reverse mutation in REB patients with 2 nonsense mutations | Calculated in this study | |
| 6x10-9 | Probability of a reverse point mutation per mitosis | Calculated in this study | |
| 1.6x10-94 | Probability that no reverse mutation occurs during total (36x109–1) mitoses (Pnot) | Calculated in this study | |
| ~1 | Probability that at least one reverse mutation occurs during total number of mitoses (P = 1 –Pnot) | Calculated in this study | |
| 216 mutations | Expected number of reverse point mutations during (36x109–1) mitoses | Calculated in this study | |
| 100 mm2 | Minimal size of revertant patch to be clinically recognizable | [9], Fig 1 | |
| 2x106 cells | Number of basal keratinocytes required for a revertant patch | Calculated in this study | |
| 21 generations | Number of generations needed to obtain 2x106 basal keratinocytes | Calculated in this study | |
| 14th generation | Generation in which revertant cell should occur to obtain revertant patch of 100 mm2 in generation 35 | Calculated in this study | |
| 0.0001 | Probability P that at least one reverse mutation occurs in the first 14 generations | Calculated in this study | |
| 1/10,000 patients | Number of patients predicted to carry clinically recognizable revertant skin patch | Calculated in this study | |
| 1:1,000 | Long term proliferating epidermal stem cells:other basal keratinocytes ratio | [24,35] | |
| 156 | Expected number of reverse mutations in epidermal stem cells during adult life | Calculated in this study | |
DEJ, dermo-epidermal junction; REB, recessive epidermolysis bullosa
Knowing that revertant mutations should occur frequently in the skin of patients with REB, the next question is whether these revertant cells will be able to grow out to healthy, revertant skin patches that are clinically recognizable. To induce recognizable patches, i.e. patches covering at least 1 cm2 (corresponding to 2×106 revertant BKs) (Fig 1) [9], reverse mutations in our model should arise in the 14th cell generation the latest (214 = 16,384 BKs) (Methods section, Table 2). This number of cells is supposed to be reached before the 4th week of embryogenesis [37]. The probability P that at least one reverse mutation will have occurred in the first 14 generations is only ~0.0001. A clinically recognizable, healthy revertant skin patch should therefore be expected in only one per ~10,000 patients (~1/0.0001).
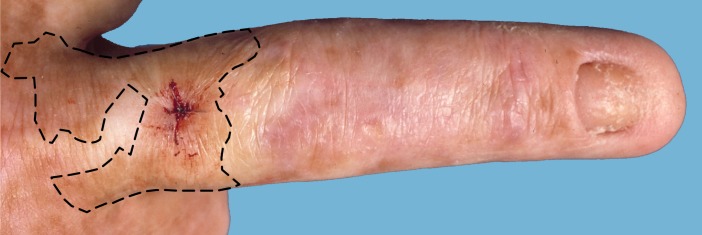
Fig 1. The smallest revertant patch observed in any of our patients.
Photograph of the revertant patch on the dorsal middle finger of patient EB093-01 (deceased) who had generalized intermediate junctional EB due to compound heterozygous COL17A1 mutations c.[3676C>T];[4319dup], p.[Arg1226*];[Gly1441Trpfs*14] [9]. Note stitched biopsy site for confirmation of type XVII collagen re-expression. The size of the patch was approximately 2 cm2 and it allowed him to wear a wedding ring. Based on this, we concluded revertant patches should be ~1 cm2 minimum in order to be clinically recognizable.
Evidently, this conclusion strongly contradicts our clinical observations that revertant patches are present in all Dutch patients with generalized intermediate junctional EB [11]. This underestimate can only be partly explained by the developmental model itself. First, the model ignores cell loss due to normal biologic processes, like apoptosis and differentiation. As the probability that reverse mutations occur depends on the number of mitoses needed to obtain final cell numbers, integrating cell loss will increase this probability by means of increasing the total number of mitoses. To get an impression of the effect of cell loss, we next used a conservative model in which we assumed that 90% of new cells in each generation would be lost, ny = 1.1y, as the exact cell loss rate is unknown. In this model 359×109 mitoses (255 generations) are required to obtain the 36×109 BKs of an adult human body; and 19.6×106 mitoses (152 generations) are required for the 2×106 cells of a 1 cm2 recognizable revertant patch, assuming identical growth and loss rates for mutant and revertant cells. The reverse mutation should then occur no later than in the 103rd generation, when the absolute minimal number of cells needed to obtain a revertant skin patch at adulthood is reached (18×103 BKs, 180×103 mitoses), which again is before the 4th week of embryonic development [37]. Loss of 90% of cells in each generation would thus increase the required number of mitoses ~10x compared to the initial model. The probability of recognizable patches (P = 0.0011) consequently also increases a factor 10. Hence, even with 90% cell loss in each generation, recognizable revertant patches could be expected in only one per ~909 patients (~1/0.0011) and cannot explain their much more frequent clinical appearance.
Second, we assumed that all BKs are equal and need to be revertant. In reality, a hierarchy among BKs is recognized: long-term proliferating epidermal stem cells produce clonogenic transient amplifying cells that provide the other (transient amplifying) BKs to the epidermal proliferating units, which collectively form skin patches [35,38]. Only a limited number of epidermal stem cells thus need to be revertant in order to acquire recognizable revertant patches. This critical number of revertant stem cells per mm2 needed for recognizable patches is unknown, but it is undoubtedly smaller than the number used before. To estimate the possible effect of BK hierarchy, we assumed an epidermal stem cell:non-stem cell BK ratio of 1:1,000, based on the review by Strachan and Ghadially [35] and the recent experimental study by Hirsch et al. [24], who showed that the number of stem cells in their skin transplants was approximately 1.8×103 per cm2, i.e. approximately 1:1,000 to the number of 2×106 BKs that we calculated to be present per cm2 skin. This reduces the number of mitoses in which reverse mutations can occur 1,000-fold. Consequently, the probability P that at least one reverse mutation occurs decreases substantially to 0.19 (as compared to ~1 in the initial model). As the ratio of cells in the entire body skin vs. a minimal recognizable patch will be left unchanged—both are reduced 1,000-fold by considering epidermal stem cells—the probability of a recognizable patch will also be left unchanged, i.e. one per ~10,000 patients. In other studies, the stem cell:non-stem cell BKs ratio has been estimated to be up to 1:10 [36,39], but even if stem cell numbers in the skin are this high, the probability of the occurrence of recognizable revertant skin patches would not the altered. Hence, integrating stem cells in the model cannot explain the discrepancy between our clinical observations and the predictions following from the model.
Third, we used the average human somatic cell mutation rate (1×10−9/nt/mitosis) [27], as data on the actual mutation rate for BKs are not available, which could be substantially higher. However, only a ~100,000-fold increase in mutation rate to at least 1×10−4 per nucleotide per mitosis could explain the occurrence of reverse mutations in the 14th cell generation in every patient. A recent study performing ultra-deep sequencing of 74 cancer genes in 234 normal sun-exposed skin biopsies from human eyelids identified 2–6 somatic mutation per megabase per cell (12,000–36,000 mutations per diploid genome) [40]. We calculated the average number of somatic mutations to be approximately 105 per BK at adulthood (see Methods section). Assuming these BKs self-renew monthly to maintain homeostasis for 60 years (720 mitoses), this would accumulate to approximately 4,425 somatic mutations in BKs in aged skin. Although the data by Martincorena et al. might thus suggest a higher mutation rate in human skin than the average human mutation rate, most mutations in their study had a UV-signature, indicating that such a higher mutation rate is likely attributable to environmental factors rather than to an intrinsically higher mutation rate. In addition, their results were obtained on entire epidermis, making it hard to compare to our calculations for BKs only. One could obviously argue that environmental factors play a major role in the occurrence of reverse mutations in human skin and indeed UV-exposure might induce reverse mutations during life. Interestingly though, many reverse mutations do not have a UV-signature [11], reducing the probability that they occur due to environmental factors and supporting our hypothesis that they should arise early during embryonic development.
The three points mentioned above may explain a small portion of the discrepancy between our clinical observations and the predictions from the model. However, another, more likely explanation why revertant cells grow out to recognizable patches in more-than-expected patient numbers is that revertant cells possess a strong selective growth advantage over their non-revertant neighbors, equivalent to what is seen for tumor cells carrying driver mutations in certain cancer-related genes [40]. If revertant cells are able to survive longer or to go through more cycles of cell divisions, reverse mutations would be allowed to occur at later stages of development, when their occurrence is more likely (Fig 2). In hematologic diseases involving hematopoietic bone marrow stem cells, such a growth advantage is believed to explain the ability to detect reverse mutations and clinical improvement in the first place [41–46]. As skin is also a continuously self-renewing organ, similar mechanisms may be active in skin as in the bone marrow. Additionally, growth potential differences between wild-type and mutant cell lineages have been shown for other diseases like Wiskott-Aldrich syndrome and neurofibromatosis type I [47,48].
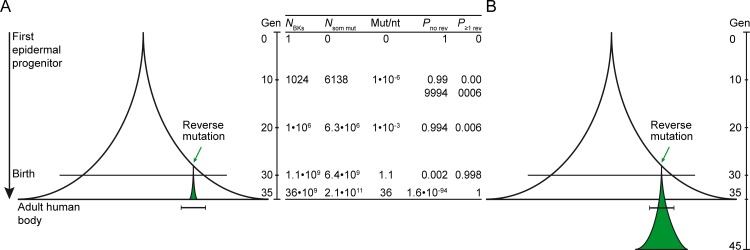
Fig 2. Selective growth advantage may explain the occurrence of clinically recognizable revertant patches.
(A) Schematic representation of the “bell-shaped” increase in cell number according to the n = 2y model starting with a single progenitor. In the later generations, the probability of a reverse mutation approaches 1 asymptotically. However, a reverse mutation that occurs in later generations cannot result in a clinically recognizable patch since the revertant cell cannot go through the required number of mitoses. (B) We therefore propose that revertant cells have a selective growth advantage, e.g. they possess the ability to go through more generations than their mutant neighbors. This would allow reverse mutations to occur at later stages and still result in visible patches. Green area: revertant area. Horizontal bar: size of minimal clinically recognizable patch (≥ 1 cm2). Gen, generation of cells. NBKs, number of basal keratinocytes. Nsom mut, expected total number of new somatic mutations. Mut/Nt, average number of mutations per nucleotide. Pno rev, probability of no reverse mutation. P≥1 rev, probability of at least one reverse mutation.
Several arguments support a selective growth advantage of revertant cells in REB skin. First, revertant patches are not confined to the lines of epidermal development, the lines of Blaschko [49,50]. Rather, they appear to develop in one Blaschko-line segment and then expand centrifugally into adjacent segments (Fig 3). This observation indicates that reverse mutations occur when epidermal formation from the Blaschko-lines is completed, i.e. after the end of the 4th week [37]. This is in line with our conclusion that reverse mutations likely occur in later stages when the number of epidermal cells has reached the critical amount to allow their occurrence with a higher probability.
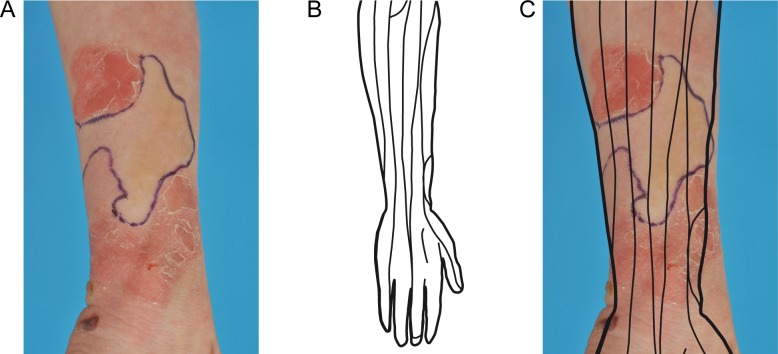
Fig 3. A selective growth advantage of revertant cells is supported by the patterns of the revertant skin patches.
(A) Dorsal forearm of an 8-year old boy (EB134-01) with generalized intermediate junctional EB due to the compound heterozygous COL17A1 mutations c.[1260del];[3496_3497del], p.[Thr421Leufs*72];[Ser1166Leufs*6] showing a revertant patch (indicated by blue line). (B) The lines of Blaschko on the dorsal forearm as deduced by Blaschko and Happle [49,50]. (C) The lines of Blaschko projected on the revertant patch on the dorsal forearm of patient EB134-01. The revertant patch has clearly exceeded the boundaries of the Blaschko lines and has grown into adjacent Blaschko line segments. The parents of patient EB134-01 have given written informed consent (as outlined in PLOS consent form) to publish this photograph.
Second, a segmental phenotype due to somatic second-hit mutations has not been reported in carriers of REB mutations. As there are many more target nucleotides for gene silencing than for mutation correction, the probability that the wild-type allele is silenced in carriers far exceeds the probability of reverse mutations in REB patients’ skin [27]. Although carriers may not all have been meticulously evaluated for small areas of increased skin blistering propensity, the lack of reports on a segmental phenotype in carriers suggests a significant growth disadvantage of cells harboring second-hit mutations.
Third, the expanding revertant patches seen in ichthyosis with confetti due to multiple correction events in either the KRT1 or KRT10 genes seem to support this hypothesis of a growth advantage or greater fitness of corrected skin cells over their mutant neighbors [20,21,51,52]. While normal epidermal stem cell units are considered to colonize up to 3 mm in diameter [35], patches in ichthyosis with confetti due to KRT10 mutations, which first appear around the age of 6 years, are reported to grow up to 4 cm in diameter [20].
In our model, the 29th cell generation is the first where the number of mitoses is so high that more than one reverse mutation is expected to occur. Revertant cells need another 21 generations to acquire the 2×106 cells of a minimal recognizable patch. They would thus need to go through at least 15 (21 vs. 6) more generations than their mutant neighbors. Future studies, perhaps using in vivo imaging techniques in new luminescent and fluorescent genetic animal models [53], need to clarify whether the revertant growth advantage is indeed of this order of magnitude. Two different mechanisms may act synergistically in providing the enormous revertant growth advantage: first, revertant cells may survive longer and, second, may possess the ability to go through more cell divisions.
Despite their likely growth advantage, the majority of patients do not report expansion of revertant patches in adulthood, and existing revertant patches do not expand into blistered areas at their borders. This suggests a limited time frame where the growth advantage can be exerted, with an end point before adulthood. In a child with Kindler syndrome the revertant patch expanded relatively to body size until the age of 8 years [18]. This indicates that revertant patches may have the ability to expand at least until this age. Similar observations have been made in ichthyosis with confetti (IWC type I) due to KRT10 frameshift mutations, in which multiple revertant patches arise during childhood and stop expanding before adulthood [20]. In IWC type II caused by KRT1 frameshift mutations, on the other hand, patches usually arise at a later age [21], indicating that the time window of revertant growth advantage is limited, but likely varies per gene or even mutation.
Why revertant patches lose their ability to expand further after a certain time point and are not able to cover the entire body with new, healthy skin and thus cure the disease, is unknown. In the study by Hirsch et al., exogenously corrected epidermal stem cells were cultured to produce 0.85 m2 of new epidermis to treat a child with junctional EB due to a homozygous LAMB3 mutation [24]. From this study, it would be expected that corrected stem cells would have the ability to expand in vivo as well, but this is not what is seen in adult EB patients. In their review, Colom and Jones state that cells with carcinogenic driver mutations likely have a short-term advantage over their neighbors, after which their growth is constrained and they revert to homeostatic behavior. If this is true, a similar mechanism could be active in revertant cells [54]. Also, age-related changes in the balance between proliferation and differentiation gene expression programs, for instance due to epigenetic changes at the epidermal differentiation complex locus (1q21) or of proliferation and differentiation gene networks [55–58], could play a role. Further studies comparing the genomic and epigenomic constitution of revertant and non-revertant cell lineages could help solve this issue.
In addition to existing patches not expanding further after a certain age, we have neither witnessed the occurrence of new revertant patches in adult EB patients nor has this been reported in the literature. Of note, one patient claimed that his revertant patch had occurred “during life”, but long-term photographic could only confirm the presence of the same patch throughout follow-up, leaving the question unanswered whether this is a true example of a new revertant patch occurring later in life [13]. This is particularly remarkable when the occurrence of new mutations during adulthood is considered (see Methods section). Analogous to the calculations used to obtain the frequency of reverse mutations during embryonic development until adulthood, it can be estimated that at least 156 new reverse mutations will occur in epidermal stem cells alone during adult life (Table 2). The lack of new revertant patches arising during adulthood thus supports the hypothesis that the growth advantage of revertant cells decreases during life and has a limited time window.
Altogether, our developmental model predicts that REB patients with mutations that are correctable by single point reversions carry numerous reverse mutations in their skin. Our observation of revertant patches in at least all the patients with one junctional REB subtype seems to form the clinical proof for this prediction. The question remains as to whether it is also true in other REB subtypes, but this prediction should encourage clinicians to scrutinize every EB patient for revertant patches (i.e. by asking the patients about the presence of skin areas that never blister, clinical examination and a ballpoint rub test on suspected patches [16]), as their presence offers exciting future therapeutic prospects [22,23]. To produce clinically recognizable revertant patches, reverse mutations should, however, arise so early during embryogenesis that that is expected to occur in only one per ~10,000 patients. We therefore postulate the “late-but-fitter revertant cell” hypothesis: reverse mutations arise at later stages of development, but confer a strong selective growth advantage to the revertant cells that drives their development into revertant patches. Our results indicate that revertant cells should also be present in the affected tissues in other genetic diseases, provided their stem cell division numbers are comparable to skin (and bone marrow), like the colorectal tissues [36]. Table 3 shows all genetic conditions in which RM has been identified to date [1,20,21,41,46,59–75]. Next generation sequencing techniques have opened unprecedented opportunities for the detection of low-grade somatic mosaicism and are perfectly suited to prove whether this hypothesis is indeed true [76]. If ways can be found to subsequently identify and isolate these cells, this could offer novel opportunities for revertant cell therapy in other genetic diseases too.
Table 3. Revertant mosaicism in genetic diseases other than epidermolysis bullosa.
| Disease | OMIM | Corrected gene | OMIM | References |
|---|---|---|---|---|
| Lesch-Nyhan syndrome | 300322 | HPRT | 308000 | [1] |
| Ichthyosis with confetti, KRT10 | 609165 | KRT10 | 148080 | [20] |
| Ichthyosis with confetti, KRT1 | 609165 | KRT1 | 139350 | [21] |
| Fanconi anemia, complementation group A | 227650 | FANCA | 607139 | [41] |
| Dyskeratosis congenita type 1 | 127550 | TERC | 602322 | [46] |
| Duchenne muscular dystrophy | 310200 | DMD | 300377 | [59] |
| Myotonic dystrophy | 160900 | DMPK | 605377 | [60] |
| Tyrosinemia type I | 276700 | FAH | 613871 | [61] |
| Bloom syndrome | 210900 | RECQL3 | 604610 | [62] |
| Adenosine deaminase deficiency | 102700 | ADA | 608958 | [63] |
| Hereditary motor and sensory neuropathy type 1A | 118220 | PMP22 duplication | 601097 | [64] |
| X-linked severe combined immunodeficiency | 300400 | IL2RG | 308380 | [65] |
| Fanconi anemia, complementation group C | 227645 | FANCC | 613899 | [66] |
| Wiskott-Aldrich syndrome | 301000 | WAS | 300392 | [67] |
| X-linked hypohidrotic ectodermal dysplasia with immunodeficiency | 300291 | NEMO | 300248 | [68] |
| Omenn syndrome | 603554 | RAG1 | 179615 | [69] |
| T-cell immunodeficiency | 610163 | CD3-zeta (CD247) | 186780 | [70] |
| Fanconi anemia, complementation group N | 610832 | PALB2 | 610355 | [71] |
| Fanconi anemia, complementation group I | 609053 | FANCI | 611360 | [72] |
| Leukocyte adhesion deficiency type 1 | 116920 | ITGB2 | 600065 | [73] |
| Autosomal recessive severe combined immunodeficiency | 600802 | JAK3 | 600173 | [74] |
| Autosomal recessive severe combined immunodeficiency | 608971 | IL7R | 146661 | [75] |
Methods
Determining the occurrence of reverse mutations in basal keratinocytes
We have used an exponential model in which the total number of basal keratinocytes (BKs), n, starting with a single embryonic ectodermal progenitor cell, doubles with every generation of cells, y, according to n = 2y. The total number of BKs, ntotal, in an average adult human body is a function of the total body dermo-epidermal junction (DEJ) surface SDEJ and the number of BKs per unit DEJ. The SDEJ is not identical to the human skin surface Sskin, as it is greatly increased by the presence of rete ridges and dermal papillae. Assuming an average of 6 rete ridges-dermal papillae complexes per mm skin length (Lskin) with an average height of 0.070 mm [28,29], the DEJ can be described as a sinus function with amplitude 0.035 mm and period 12π (Table 2):
The exact length of the line described by this sinus, i.e. the actual DEJ length LDEJ, is calculated by solving the integral of the function (Mathematica, Wolfram Research Inc., Champaign, IL)
Using a = 0 and b = 1 mm Lskin reveals that 1 mm of Lskin corresponds to 1.348 mm LDEJ. Consequently, since the “finger like appearance” of dermal papillae resembles two interfering sinus waves [77], 1 mm2 Sskin corresponds to (1.348)2 ≈ 1.817 mm2 SDEJ. Rete ridges thus theoretically expand the dermal-epidermal surface contact by 81.7%.
The number of BKs was measured to be between 15,000 and 20,000 to 30,000 per mm2 skin [33,33]. Assuming that BKs have a circular base of diameter ~10 μm [30] and surface ~78.5 μm2, approximately 12,739 BKs are expected per mm2 SDEJ (1 mm2 SDEJ / 78.5×10−6 mm2 per BK) and 23,146 BKs per mm2 Sskin. Taken together, we decided to use 20,000 BKs per mm Sskin in further calculations.
Assuming a total human adult body surface of 1.8 m2 (corresponding to 3.3×106 mm2 SDEJ) the total number of BKs in an adult human body ntotal = 1.8×106 mm2 × 20,000 BKs/mm2 = 36×109. This number is reached in the 35th generation, after a total of 36×109–1 mitoses. Given a per nucleotide point mutation rate of approximately 1×10−9 per mitosis [26–28] and 6×109 nucleotides per diploid genome [34], ~6 novel point mutations may be expected in the daughter cell’s genome in each cell division. In the 35th generation of cells, the total expected number of new point mutations that have occurred in the body’s BKs is approximately 6 × total number of mitoses = 216×109, indicating that each single nucleotide is expected to be mutated on average 36 times in the human basal skin layer at adulthood. In total, 3.6×1012 somatic mutations are expected to have accumulated in the body’s BKs at adulthood, which equates to an average of 105 somatic mutations per BK.
Alteration of one of the nucleotides of a nonsense codon will usually change the nonsense into a sense codon. If nonsense mutations are present on both alleles, there are therefore 6 target nucleotides for a reversion, thereby ignoring the small probability that the nonsense codon will be changed into one of the other two possible nonsense codons. For frame-shift and splice-site mutations, the number of potential target nucleotides for reversion events are usually unknown but theoretically numerous. For missense mutations, this number is probably less than 6. We therefore decided to use the number of 6 target nucleotides of nonsense codons in our calculations. A larger or smaller number of target nucleotides would not affect the order of magnitude of the probability of the occurrence of reverse mutations. The probability of a reverse point mutation per mitosis in case of 6 target nucleotides is thus prev = 6 × 1×10−9 = 6×10−9, The probability that no reverse mutation will occur during the total number of mitoses until the 35th generation (Pnot) is
Therefore, the probability P that at least one reverse mutation occurs during these mitoses is
The expected number of reverse mutations in 36×109–1 mitoses is (36×109–1) × 6×10−9 = 216.
Determining the occurrence of clinically recognizable revertant skin patches
One cm2 of revertant Sskin corresponds to 100 mm2 × 20,000 BKs/mm2 = 2×106 revertant BKs. According to the 2y model, it takes 21 generations to obtain this number from one single revertant ancestor. This implies that a reverse mutation should have arisen at the latest in the y = 35–21 = 14th generation, corresponding to a stage of 214 = 16,384 BKs. Considering that in early embryonic stages rete ridges and dermal papillae are not yet developed [37], the reverse mutation should have arisen at the embryonic stage where the embryo’s body surface was less than 2 mm2. For comparison, a 4-week embryo measures approximately 5 mm in length and, if compared to a cylinder with height = 5 mm and radius = 0.5 mm, its body surface is in the order of 17 mm2, indicating that a reverse mutation should arise before the 4th week of embryonic development in order to give rise to a clinically recognizable revertant skin patch. The probability Pnot that a reverse mutation does not arise in the first 14 generations is
and the probability P that at least one reverse mutation arises in the first 14 generations is
Therefore, a clinically recognizable revertant skin patch is expected once in only ~1/0.0001 = 10,000 patients.
Determining the occurrence of reverse mutations in epidermal stem cells during adult life
The number of long-term proliferating epidermal stem cells (ESCs) in an adult human body was estimated earlier to be approximately 36×106 (total number of BKs / 1,000). Assuming every ESC divides asymmetrically every month to maintain homeostasis, analogous to the self-renewal time of the entire epidermis, every ESC would go through 12 × 60 = 720 mitoses until the age of 78 years, totaling to 720 mitoses × 36×106 ESCs = 26×109 mitoses in the entire human body. Assuming the daughter cells would remain as ESCs in the basal layer, 26×109 mitoses × 6 mutations/mitosis = 156×109 new somatic mutations are expected to accumulate, equivalent to 26 mutations per nucleotide (156×10 9 mutations / 6×109 nucleotides per diploid genome). As there are 6 candidate nucleotides for reverse mutations, 6 × 26 = 156 reverse mutations are expected in ESCs during adult life.
Acknowledgments
We thank Lude Franke and Ruud Scheek for their constructive mathematical help and advice. We also thank the patients/parents/families for allowing us to use the published photographs to underpin our results. Many thanks go out to Jackie Senior, scientific editor at the Genetics Department of the University Medical Center Groningen, for carefully editing this manuscript.
Data Availability
All relevant data are contained within the paper.
Funding Statement
This study was supported in part by the Netherlands Organization for Health Research and Development (ZonMw) grant 92003541 to PCvdA, the Priority Medicines Rare Diseases (E-RARE) grant 113301091 from ZonMw to AMGP, and the Vlinderkind (Dutch Butterfly Child) Foundation to MFJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yang TP, Stout JT, Konecki DS, Patel PI, Alford RL, Caskey CT. Spontaneous reversion of novel Lesch-Nyhan mutation by HPRT gene rearrangement. Somat Cell Mol Genet. 1988;14: 293–303. [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003;40: 721–728. doi: 10.1136/jmg.40.10.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson RP. Somatic gene mutation and human disease other than cancer: an update. Mutat Res. 2010;705: 96–106. doi: 10.1016/j.mrrev.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 4.Jonkman MF, Scheffer H, Stulp R, Pas HH, Nijenhuis M, Heeres K, et al. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell. 1997;88: 543–551. [DOI] [PubMed] [Google Scholar]
- 5.Fine JD, Bruckner-Tuderman L, Eady RA, Bauer EA, Bauer JW, Has C, et al. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J Am Acad Dermatol. 2014;70: 1103–1126. doi: 10.1016/j.jaad.2014.01.903 [DOI] [PubMed] [Google Scholar]
- 6.Smith FJ, Morley SM, McLean WH. Novel mechanism of revertant mosaicism in Dowling-Meara epidermolysis bullosa simplex. J Invest Dermatol. 2004;122: 73–77. doi: 10.1046/j.0022-202X.2003.22129.x [DOI] [PubMed] [Google Scholar]
- 7.Schuilenga-Hut PH, Scheffer H, Pas HH, Nijenhuis M, Buys CH, Jonkman MF. Partial revertant mosaicism of keratin 14 in a patient with recessive epidermolysis bullosa simplex. J Invest Dermatol. 2002;118: 626–630. doi: 10.1046/j.1523-1747.2002.01715.x [DOI] [PubMed] [Google Scholar]
- 8.Darling TN, Yee C, Bauer JW, Hintner H, Yancey KB. Revertant mosaicism: partial correction of a germ-line mutation in COL17A1 by a frame-restoring mutation. J Clin Invest. 1999;103: 1371–1377. doi: 10.1172/JCI4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasmooij AM, Pas HH, Deviaene FC, Nijenhuis M, Jonkman MF. Multiple correcting COL17A1 mutations in patients with revertant mosaicism of epidermolysis bullosa. Am J Hum Genet. 2005;77: 727–740. doi: 10.1086/497344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonkman MF, Pasmooij AM. Revertant mosaicism—patchwork in the skin. N Engl J Med. 2009;360: 1680–1682. [DOI] [PubMed] [Google Scholar]
- 11.Pasmooij AM, Nijenhuis M, Brander R, Jonkman MF. Natural gene therapy may occur in all patients with generalized non-Herlitz junctional epidermolysis bullosa with COL17A1 mutations. J Invest Dermatol. 2012;132: 1374–1383. doi: 10.1038/jid.2011.477 [DOI] [PubMed] [Google Scholar]
- 12.Kowalewski C, Bremer J, Gostynski A, Wertheim-Tysarowska K, Wozniak K, Bal J, et al. Amelioration of junctional epidermolysis bullosa due to exon skipping. Br J Dermatol. 2016;174: 1375–1379. doi: 10.1111/bjd.14374 [DOI] [PubMed] [Google Scholar]
- 13.Pasmooij AM, Pas HH, Bolling MC, Jonkman MF. Revertant mosaicism in junctional epidermolysis bullosa due to multiple correcting second-site mutations in LAMB3. J Clin Invest. 2007;117: 1240–1248. doi: 10.1172/JCI30465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almaani N, Nagy N, Liu L, Dopping-Hepenstal PJ, Lai-Cheong JE, Clements SE, et al. Revertant mosaicism in recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2010;130: 1937–1940. doi: 10.1038/jid.2010.64 [DOI] [PubMed] [Google Scholar]
- 15.Pasmooij AM, Garcia M, Escamez MJ, Nijenhuis AM, Azon A, Cuadrado-Corrales N, et al. Revertant mosaicism due to a second-site mutation in COL7A1 in a patient with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2010;130: 2407–2411. doi: 10.1038/jid.2010.163 [DOI] [PubMed] [Google Scholar]
- 16.Kiritsi D, Garcia M, Brander R, Has C, Meijer R, Jose Escámez M, et al. Mechanisms of natural gene therapy in dystrophic epidermolysis bullosa. J Invest Dermatol. 2014;134: 2097–2104. doi: 10.1038/jid.2014.118 [DOI] [PubMed] [Google Scholar]
- 17.Van den Akker PC, Nijenhuis M, Meijer G, Hofstra RM, Jonkman MF, Pasmooij AM. Natural gene therapy in dystrophic epidermolysis bullosa. Arch Dermatol. 2012;148: 213–216. doi: 10.1001/archdermatol.2011.298 [DOI] [PubMed] [Google Scholar]
- 18.Lai-Cheong JE, Moss C, Parsons M, Almaani N, McGrath JA. Revertant mosaicism in kindler syndrome. J Invest Dermatol. 2012;132: 730–732. doi: 10.1038/jid.2011.352 [DOI] [PubMed] [Google Scholar]
- 19.Kiritsi D, He Y, Pasmooij AM, Onder M, Happle R, Jonkman MF, et al. Revertant mosaicism in a human skin fragility disorder results from slipped mispairing and mitotic recombination. J Clin Invest. 2012122; 1742–1746. doi: 10.1172/JCI61976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choate KA, Lu Y, Zhou J, Choi M, Elias PM, Farhi A, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330: 94–97. doi: 10.1126/science.1192280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choate KA, Lu Y, Zhou J, Elias PM, Zaidi S, Paller AS, et al. Frequent somatic reversion of KRT1 mutations in ichthyosis with confetti. J Clin Invest. 2015;125: 1703–1707. doi: 10.1172/JCI64415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gostyński A, Pasmooij AM, Jonkman MF. Successful therapeutic transplantation of revertant skin in epidermolysis bullosa. J Am Acad Dermatol. 2014;70: 98–101. doi: 10.1016/j.jaad.2013.08.052 [DOI] [PubMed] [Google Scholar]
- 23.Umegaki-Arao N, Pasmooij AM, Itoh M, Cerise JE, Guo Z, Levy B, et al. Induced pluripotent stem cells from human revertant keratinocytes for the treatment of epidermolysis bullosa. Sci Transl Med. 2014;26: 264ra164. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch T, Rothoeft T, Teig N, Bauer JW, Pellegrini G, De Rosa L, et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551: 327–332. doi: 10.1038/nature24487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasmooij AM, Jonkman MF, Uitto J. Revertant mosaicism in heritable skin diseases: mechanisms of natural gene therapy. Discov Med. 2012;14: 167–179. [PubMed] [Google Scholar]
- 26.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci USA. 2010;107: 961–968. doi: 10.1073/pnas.0912629107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328: 636–639. doi: 10.1126/science.1186802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reish RG, Zuhaili B, Bergmann J, Aflaki P, Koyama T, Hackl F, et al. Modulation of scarring in a liquid environment in the Yorkshire pig. Wound Repair Regen. 2009;17: 806–816. doi: 10.1111/j.1524-475X.2009.00546.x [DOI] [PubMed] [Google Scholar]
- 30.Giangreco A, Goldie SJ, Failla V, Saintigny G, Watt FM. Human skin aging is associated with reduced expression of the stem cell markers beta1 integrin and MCSP. J Invest Dermatol. 2010;130: 604–608. doi: 10.1038/jid.2009.297 [DOI] [PubMed] [Google Scholar]
- 31.Koehler MJ, Zimmermann S, Springer S, Elsner P, König K, Kaatz M. Keratinocyte morphology of human skin evaluated by in vivo multiphoton laser tomography. Skin Res Technol. 2011;17: 479–486. doi: 10.1111/j.1600-0846.2011.00522.x [DOI] [PubMed] [Google Scholar]
- 32.Bergstresser PR, Pariser RJ, Taylor JR. Counting and sizing of epidermal cells in normal human skin. J Invest Dermatol. 1978;70: 280–284. [DOI] [PubMed] [Google Scholar]
- 33.Potten CS. Biological dosimetry of local radiation accidents of skin: possible cytological and biochemical methods. Br J Radiol Suppl. 1986;19: 82–85. [PubMed] [Google Scholar]
- 34.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431: 931–945. doi: 10.1038/nature03001 [DOI] [PubMed] [Google Scholar]
- 35.Strachan LR, Ghadially R. Tiers of clonal organization in the epidermis: the epidermal proliferation unit revisited. Stem Cell Rev. 2008;4: 149–157. doi: 10.1007/s12015-008-9020-6 [DOI] [PubMed] [Google Scholar]
- 36.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347: 78–81. doi: 10.1126/science.1260825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loomis CA, Koss T, Chu D. Embryology In: Bolognia JL, Jorizzo JL, Rapini RP, editors. Dermatology 2nd edition Vol. 1. London: Elsevier Health Sciences; 2008. pp. 37–47. [Google Scholar]
- 38.Ghadially R. 25 Years of Epidermal Stem Cell Research. J Invest Dermatol. 2012;132: 797–810. doi: 10.1038/jid.2011.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen UB, Lowell S, Watt FM. The spatial relationship between stem cells and their progeny in the basal layer of human epidermis: a new view based on whole-mount labelling and lineage analysis. Development. 1999;126: 2409–2418. [DOI] [PubMed] [Google Scholar]
- 40.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348: 880–6. doi: 10.1126/science.aaa6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waisfisz Q, Morgan NV, Savino M, de Winter JP, van Berkel CG, Hoatlin ME, et al. Spontaneous functional correction of homozygous fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat Genet. 1999;22: 379–383. doi: 10.1038/11956 [DOI] [PubMed] [Google Scholar]
- 42.Wada T, Schurman SH, Otsu M, Garabedian EK, Ochs HD, Nelson DL, et al. Somatic mosaicism in Wiskott—Aldrich syndrome suggests in vivo reversion by a DNA slippage mechanism. Proc Natl Acad Sci USA. 2001;98: 8697–8702. doi: 10.1073/pnas.151260498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gross M, Hanenberg H, Lobitz S, Friedl R, Herterich S, Dietrich R, et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98: 126–135. doi: 10.1159/000069805 [DOI] [PubMed] [Google Scholar]
- 44.Soulier J, Leblanc T, Larghero J, Dastot H, Shimamura A, Guardiola P, et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood. 2005;105: 1329–1336. doi: 10.1182/blood-2004-05-1852 [DOI] [PubMed] [Google Scholar]
- 45.Hamanoue S, Yagasaki H, Tsuruta T, Oda T, Yabe H, Yabe M, et al. Myeloid lineage-selective growth of revertant cells in Fanconi anaemia. Br J Haematol. 2006;132: 630–635. doi: 10.1111/j.1365-2141.2005.05916.x [DOI] [PubMed] [Google Scholar]
- 46.Jongmans MC, Verwiel ET, Heijdra Y, Vulliamy T, Kamping EJ, Hehir-Kwa JY, et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am J Hum Genet. 2012;90: 426–433. doi: 10.1016/j.ajhg.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konno A, Wada T, Schurman SH, Garabedian EK, Kirby M, Anderson SM, et al. Differential contribution of Wiskott-Aldrich syndrome protein to selective advantage in T- and B-cell lineages. Blood. 2004;103: 676–678. doi: 10.1182/blood-2003-05-1739 [DOI] [PubMed] [Google Scholar]
- 48.Roehl AC, Mussotter T, Cooper DN, Kluwe L, Wimmer K, Högel J, et al. Tissue-specific differences in the proportion of mosaic large NF1 deletions are suggestive of a selective growth advantage of hematopoietic del(+/-) stem cells. Hum Mutat. 2012;33: 541–550. doi: 10.1002/humu.22013 [DOI] [PubMed] [Google Scholar]
- 49.Blaschko A. Die Nervenverteilung in der Haut in ihrer Beziehung zu den Erkrankungen der Haut. Wien & Leipzig: Braumüller; 1901. [Google Scholar]
- 50.Happle R, Fuhrmann-Rieger A, Fuhrmann W. How are the Blaschko lines arranged on the scalp? Hautarzt. 1984;35: 366–369. [PubMed] [Google Scholar]
- 51.Burger B, Spoerri I, Schubert M, Has C, Itin PH. Description of the natural course and clinical manifestations of ichthyosis with confetti caused by a novel KRT10 mutation. Br J Dermatol. 2012;166: 434–439. doi: 10.1111/j.1365-2133.2011.10639.x [DOI] [PubMed] [Google Scholar]
- 52.Hotz A, Oji V, Bourrat E, Jonca N, Mazereeuw-Hautier J, Betz RC, et al. Expanding the Clinical and Genetic Spectrum of KRT1, KRT2 and KRT10 Mutations in Keratinopathic Ichthyosis. Acta Derm Venereol. 2016;96: 473–478 doi: 10.2340/00015555-2299 [DOI] [PubMed] [Google Scholar]
- 53.Ra H, González-González E, Uddin MJ, King BL, Lee A, Ali-Khan I, et al. Detection of non-melanoma skin cancer by in vivo fluorescence imaging with fluorocoxib A. Neoplasia. 2015;17: 201–207. doi: 10.1016/j.neo.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colom B, Jones PH. Clonal analysis of stem cells in differentiation and disease. Curr Opin Cell Biol. 2016;43: 14–21. doi: 10.1016/j.ceb.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 55.Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14: 753–63. doi: 10.1038/ncb2520 [DOI] [PubMed] [Google Scholar]
- 56.Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E. Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med. 2014;4: a015263 doi: 10.1101/cshperspect.a015263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abhishek S, Palamadai Krishnan S. Epidermal Differentiation Complex: a review on its epigenetic regulation and potential drug targets. Cell J. 2016;18: 1–6. doi: 10.22074/cellj.2016.3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Botchkarev VA. The molecular revolution in cutaneous biology: chromosomal territories, higher-order chromatin remodeling, and the çontrol of gene expression in keratinocytes. J Invest Dermatol. 2017;137: e93–e99. doi: 10.1016/j.jid.2016.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burrow KL, Coovert DD, Klein CJ, Bulman DE, Kissel JT, Rammohan KW, et al. Dystrophin expression and somatic reversion in prednisone-treated and untreated Duchenne dystrophy. CIDD Study Group. Neurology. 1991;41: 661–666. [DOI] [PubMed] [Google Scholar]
- 60.Brunner HG, Jansen G, Nillesen W, Nelen MR, de Die CE, Höweler CJ, et al. Brief report: reverse mutation in myotonic dystrophy. N Engl J Med. 1993;328: 476–480. doi: 10.1056/NEJM199302183280705 [DOI] [PubMed] [Google Scholar]
- 61.Kvittingen EA, Rootwelt H, Brandtzaeg P, Bergan A, Berger R. Hereditary tyrosinemia type I. Self-induced correction of the fumarylacetoacetase defect. J Clin Invest. 1993;91: 1816–1821. doi: 10.1172/JCI116393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellis NA, Lennon DJ, Proytcheva M, Alhadeff B, Henderson EE, German J. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am J Hum Genet. 1995;57: 1019–1027. [PMC free article] [PubMed] [Google Scholar]
- 63.Hirschhorn R, Yang DR, Puck JM, Huie ML, Jiang CK, Kurlandsky LE. Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat Genet. 1996;13: 290–295. doi: 10.1038/ng0796-290 [DOI] [PubMed] [Google Scholar]
- 64.Liehr T, Rautenstrauss B, Grehl H, Bathke KD, Ekici A, Rauch A, et al. Mosaicism for the Charcot-Marie-Tooth disease type 1A duplication suggests somatic reversion. Hum Genet. 1996;98: 22–28. [DOI] [PubMed] [Google Scholar]
- 65.Stephan V, Wahn V, Le Deist F, Dirksen U, Broker B, Müller-Fleckenstein I, et al. Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N Engl J Med. 1996;335: 1563–1567. doi: 10.1056/NEJM199611213352104 [DOI] [PubMed] [Google Scholar]
- 66.Lo Ten Foe JR, Kwee ML, Rooimans MA, Oostra AB, Veerman AJ, van Weel M, et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet. 1997;5: 137–148. [PubMed] [Google Scholar]
- 67.Ariga T, Yamada M, Sakiyama Y, Tatsuzawa O. A case of Wiskott-Aldrich syndrome with dual mutations in exon 10 of the WASP gene: an additional de novo one-base insertion, which restores frame shift due to an inherent one-base deletion, detected in the major population of the patient’s peripheral blood lymphocytes. Blood. 1998;92: 699–701. [PubMed] [Google Scholar]
- 68.Nishikomori R, Akutagawa H, Maruyama K, Nakata-Hizume M, Ohmori K, Mizuno K, et al. X-linked ectodermal dysplasia and immunodeficiency caused by reversion mosaicism of NEMO reveals a critical role for NEMO in human T-cell development and/or survival. Blood. 2004;103: 4565–4572. doi: 10.1182/blood-2003-10-3655 [DOI] [PubMed] [Google Scholar]
- 69.Wada T, Toma T, Okamoto H, Kasahara Y, Koizumi S, Agematsu K, et al. Oligoclonal expansion of T lymphocytes with multiple second-site mutations leads to Omenn syndrome in a patient with RAG1-deficient severe combined immunodeficiency. Blood. 2005;106: 2099–2101. doi: 10.1182/blood-2005-03-0936 [DOI] [PubMed] [Google Scholar]
- 70.Rieux-Laucat F, Hivroz C, Lim A, Mateo V, Pellier I, Selz F, et al. Inherited and somatic CD3zeta mutations in a patient with T-cell deficiency. N Engl J Med. 2006;354: 1913–1921. doi: 10.1056/NEJMoa053750 [DOI] [PubMed] [Google Scholar]
- 71.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39: 159–161. doi: 10.1038/ng1942 [DOI] [PubMed] [Google Scholar]
- 72.Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29: 211–218. doi: 10.1155/2007/151968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tone Y, Wada T, Shibata F, Toma T, Hashida Y, Kasahara Y, et al. Somatic revertant mosaicism in a patient with leukocyte adhesion deficiency type 1. Blood. 2007;109: 1182–1184. doi: 10.1182/blood-2007-08-039057 [DOI] [PubMed] [Google Scholar]
- 74.Ban SA, Salzer E, Eibl MM, Linder A, Geier CB, Santos-Valente E, et al. Combined immunodeficiency evolving into predominant CD4+ lymphopenia caused by somatic chimerism in JAK3. J Clin Immunol. 2014;34: 941–953. doi: 10.1007/s10875-014-0088-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bayer DK, Martinez CA, Sorte HS, Forbes LR, Demmler-Harrison GJ, Hanson IC, et al. Vaccine-associated varicella and rubella infections in severe combined immunodeficiency with isolated CD4 lymphocytopenia and mutations in IL7R detected by tandem whole exome sequencing and chromosomal microarray. Clin Exp Immunol. 2014;178: 459–469. doi: 10.1111/cei.12421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cohen AS, Wilson SL, Trinh J, Ye XC. Detecting somatic mosaicism: considerations and clinical implications. Clin Genet. 2015;87: 554–562. doi: 10.1111/cge.12502 [DOI] [PubMed] [Google Scholar]
- 77.Arao H, Obata M, Shimada T, Hagisawa S. Morphological characteristics of the dermal papillae in the development of pressure sores. J Tissue Viability. 1998;8: 17–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are contained within the paper.