Abstract
Objective
To decipher whether age-independent cardiovascular risk is associated with DNA methylation at CpG level and to determine whether these differential methylation signatures are associated with the incidence of cardiovascular events.
Approach and Results
We designed a two-stage, cross-sectional, epigenome-wide association study. Age-independent cardiovascular risk calculation was based on vascular age and on the residuals of the relationship between age and cardiovascular risk. Blood DNA methylomes from two independent populations were profiled using the Infinium HumanMethylation450 BeadChip. The discovery stage of these studies was performed in the REGICOR cohort (n=645). Next, we validated the initial findings in the Framingham Offspring Study (n=2,542). Eight CpGs located in four genes (AHRR, CPT1A, PPIF and SBNO2) and three intergenic regions showed differential methylation in association with age-independent cardiovascular risk (p-value≤1.17·10−7). These CpGs explained 12.01–15.16% of the variability of age-independent cardiovascular risk in REGICOR, and 7.51–8.53% in Framingham. Four of them were only related to smoking, three were related to smoking and BMI, and one to diabetes, triglycerides levels, and BMI (p-value≤7.81·10−4). Additionally, we developed methylation risk scores based on these CpGs and observed an association between these scores and cardiovascular disease incidence (HR=1.32; 95% CI: 1.16–1.51).
Conclusions
Age-independent cardiovascular risk was related to different DNA methylation profiles, with eight CpGs showing differential methylation patterns. Most of these CpGs were associated with smoking, and three of them were also related to BMI. Risk scores based on these differential methylation patterns were associated with cardiovascular events and could be useful predictive indices.
Keywords: epigenetics, epidemiology, cardiovascular disease risk factors, cardiovascular genomics, atherosclerosis
Subject codes: Cardiovascular disease, Epigenetics, Genetic, Association Studies, Epidemiology, Risk factors
INTRODUCTION
Coronary heart disease (CHD) is the leading cause of worldwide morbidity and mortality1,2 and atherosclerosis is its principal mechanism.3 Atherosclerosis development and progression is driven by age,4,5 and the presence of vascular risk factors (VRFs) such as smoking, diabetes, hypertension, and total, high-density lipoprotein (HDL) and low-density lipoprotein (LDL) cholesterol levels.6 The cumulative effect of aging and the presence of these VRFs is considered and summarized in the functions used to determine an individual’s cardiovascular risk (CVR).6
Atherosclerotic vascular burden is also influenced by heritable factors. Epigenetic signatures are heritable traits but they are also modifiable by age and environmental factors. The best-known epigenetic mechanism regulating gene expression, DNA methylation, is related to atherosclerotic traits7. Thus, it may be a mediator between aging, environmental factors, VRFs, and atherosclerosis. As both CVR and DNA methylation are highly determined by age, linking DNA methylation with age-independent CVR might contribute to understand the individual and cumulative effect of the confluence of VRFs on DNA methylation. Two approaches, the difference between vascular8,9 and chronological age (Δage) and the residuals of the correlation between age and CVR, have been used to calculate age-independent CVR.
We hypothesized that specific DNA methylation signatures are influenced by the confluence of VRFs and that, in turn, these signatures are associated with the progression of atherosclerosis and the incidence of cardiovascular events. Our aims were to identify CpGs showing differential methylation related to age-independent CVR and to determine the association of these differential methylation signatures with the presence of subclinical atherosclerosis and with the incidence of coronary or cardiovascular events.
MATERIALS AND METHODS
The online-only Data Supplement provides details on Materials and Methods. Very briefly, we performed an initial cross-sectional study through a two-stage epigenome-wide association study (EWAS) strategy, using two independent populations. Based on whole blood differential methylation profiles associated with age-independent CVR, we developed two methylation risk scores (MRSs) and assessed their association with subclinical atherosclerosis and with the incidence of clinical coronary and cardiovascular events.
RESULTS
The flowchart showing the study procedure is presented in Figure I in the online-only Data Supplement.
Estimate of the age-independent CVR in the study populations
We defined two approaches to determine age-independent CVR, the residuals and the Δage approach. Two of the 648 individuals with DNA methylation data in the REGICOR study were excluded after DNA methylation quality control analysis. We also excluded individuals younger and older than the suitable age range (35–79 years) to estimate CVR (n=67) and those individuals with missing values on the VRFs considered in the risk function (n=5). Finally, we estimated the CVR of 574 individuals (residuals approach) and the difference between vascular and chronological age of 465 individuals (Δage approach).
A descriptive analysis of the main sociodemographic and clinical characteristics of the individuals included in the discovery stage of the Δage approach is shown in Table 1; equivalent information for the individuals included in the residual approach is shown in Table I in the online-only Data Supplement.
Table 1. Descriptive characteristics of participants in the discovery stage (REGICOR cohort) in the whole population and across quartiles of the difference between vascular and chronological.
| N=465 | Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | P-trend | |
|---|---|---|---|---|---|---|
| N=116 | N=116 | N=116 | N=116 | |||
| Age* | 58.5(9.32) | 59.3(10.8) | 60.6(9.20) | 60.0(8.29) | 54.2(7.29) | <0.001 |
| Sex, female, n (%) | 241(51.8) | 61(52.6) | 50(43.1) | 56(48.3) | 56(48.3) | 0.709 |
| FRESCO CVR (%)†‡ | 1.85 [0.87;3.21] | 0.76 [0.43;1.97] | 1.96 [0.88;3.11] | 1.97 [1.27;3.79] | 2.31 [1.58;3.54] | <0.001 |
| Vascular age* | 63.2(10.3) | 55.6(11.6) | 62.7(9.13) | 66.4(8.33) | 68.3(6.68) | <0.001 |
| Δage*‡ | 4.70(7.06) | −3.68(2.69) | 2.06(1.18) | 6.41(1.28) | 14.1(4.25) | <0.001 |
| Residuals from age/CVR (%)† | −0.56 [−1.78;0.46] | −1.24 [−2.38;−0.61] | −0.62 [−1.85;−0.21] | −0.03 [−2.11;0.60] | 0.83 [−0.59;1.98] | <0.001 |
| Total cholesterol, mg/dl* | 209(35.4) | 194(31.3) | 208(29.3) | 211(33.8) | 222(40.6) | <0.001 |
| LDL cholesterol, mg/dl* | 136(31.2) | 116(25.0) | 137(24.1) | 141(28.5) | 150(35.6) | <0.001 |
| HDL cholesterol, mg/dl* | 54.2(12.7) | 64.0(13.9) | 54.4(10.5) | 49.7(8.93) | 48.4(10.4) | <0.001 |
| Triglycerides, mg/dl† | 83.0 [63.0;115] | 66.0 [53.8;80.5] | 79.5 [59.8;104] | 90.5 [70.8;124] | 108 [82.8;146] | <0.001 |
| Cholesterol treatment, n (%) | 89(19.1) | 22(19.0) | 23(19.8) | 24(20.7) | 20(17.2) | 0.792 |
| SBP, mmHg* | 127(17.8) | 123(17.3) | 125(15.5) | 129(16.6) | 132(20.4) | <0.001 |
| DBP, mmHg* | 76.8(9.89) | 74.3(9.19) | 75.1(9.24) | 77.5(9.38) | 80.5(10.5) | <0.001 |
| HTN, n (%)‡ | 165(35.5) | 29(25.0) | 29(25.0) | 46(39.7) | 61(52.6) | <0.001 |
| HTN treatment, n (%) | 90(19.4) | 14(12.1) | 18(15.5) | 24(20.7) | 34(29.3) | 0.001 |
| Glucose, mg/dl* | 94.0(13.7) | 91.1(9.44) | 92.7(9.53) | 93.9(11.4) | 98.4(20.5) | <0.001 |
| Diabetes, n (%)‡ | 12(2.58) | 0(0.00) | 0(0.00) | 2(1.72) | 10(8.62) | <0.001 |
| Diabetes treatment, n (%) | 8(1.72) | 0(0.00) | 0(0.00) | 2(1.72) | 6(5.17) | 0.001 |
| Smoking status, n (%) | <0.001 | |||||
| Current smokers | 83(17.8) | 5(4.31) | 4(3.45) | 17(14.7) | 57(49.1) | - |
| Former 1–5 years | 23(4.95) | 3(2.59) | 9(7.76) | 6(5.17) | 5(4.31) | - |
| Former >5 years | 122(26.2) | 41(35.3) | 32(27.6) | 32(27.6) | 16(13.8) | - |
| Never smokers | 237(51.0) | 67(57.8) | 71(61.2) | 61(52.6) | 38(32.8) | - |
| BMI, kg/m2‡ | 26.5(3.94) | 25.1(3.64) | 26.4(3.13) | 27.2(4.34) | 27.4(4.18) | <0.001 |
| Obesity, n (%)‡ | 83(17.9) | 9(7.83) | 19(16.4) | 27(23.5) | 28(24.1) | <0.001 |
Mean (Standard deviation).
Median (Interquartile range).
FRESCO CVR, FRESCO cardiovascular risk; Δage, difference between vascular and chronological age; LDL, Low-Density Lipoprotein; HDL, High-Density Lipoprotein; SBP, Systolic Blood Pressure; DBP, Diastolic Blood Pressure; HTN, Hypertension, defined as previous treatment or SBP≥140mmHg or DBP≥90mmHg; Diabetes, defined as previous treatment or glycemia≥126 mg/dl; BMI, Body Mass Index; Obesity, defined as BMI≥30kg/m2.
In the Framingham Offspring Cohort, after the DNA methylation quality control and exclusion of duplicates, we excluded individuals not within the valid age range (n=209) and those with missing values for any VRF used in the risk function (n=161). Therefore, from 2,542 individuals remaining after the quality control, we calculated the CVR of 2,172 participants and the difference between vascular and chronological age of 1,823 participants. The main sociodemographic and clinical characteristics of these individuals are shown in Tables II and III in the online-only Data Supplement.
Discovery stage of the EWAS
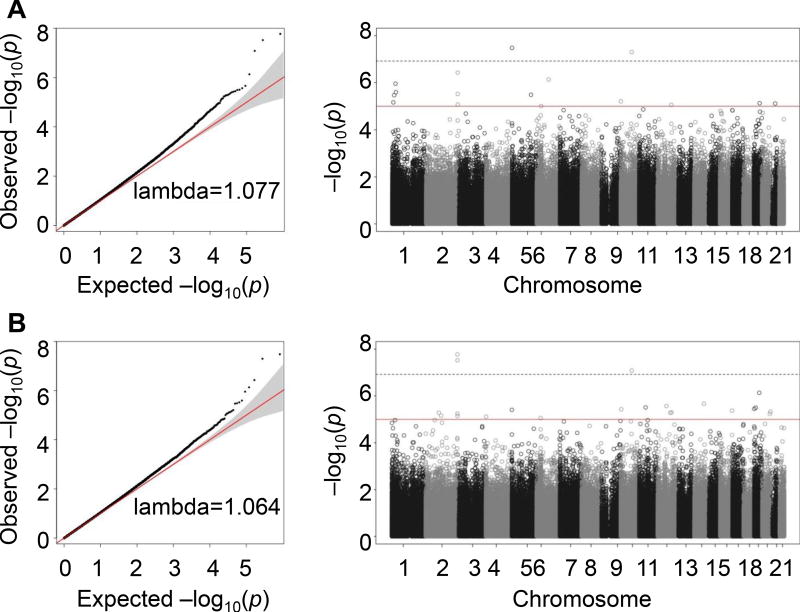
As we hypothesized that methylation is influenced by the convergence of VRFs other than age, we studied their association through an epigenome-wide strategy. We identified 29 differentially methylated CpGs in association with age-independent CVR using the residuals approach and 24 using the Δage approach, defining an arbitrary p-value threshold <1·10−5. Of these, 48 were unique differentially methylated CpGs in association with age-independent CVR (Tables IVA and IVA in the online-only Data Supplement). These CpGs were located in 32 genes and 16 intergenic regions. The Manhattan and QQ plots are shown in Figure 1.
Figure 1.
QQ plots and Manhattan plots of the association between methylation and age-independent cardiovascular risk in the discovery stage of the model adjusted for sex, age, 395 cell type, and surrogate variables. Plots are classified according to the approach used to describe age-independent CVR (residuals from age/CVR association, A; or difference between vascular and chronological age, B).
Validation stage and metaanalysis
We assessed the association between the age-independent CVR and the initially identified 48 CpGs in the Framingham population. Seven CpGs (cg00574958, cg05575921, cg05951221, cg12057156, cg12547807, cg18608055, cg27537125) were significant (p-value <1.04·10−3; 0.05/48) in both approaches, and one additional (cg19939077) was significant only in the residuals approach (Tables VA and VB in the online-only Data Supplement).
By metaanalyzing the results observed in the REGICOR and the Framingham studies, we validated eight of the 48 initially discovered CpGs in seven differential loci (Table 2), using a p-value threshold <1.17·10−7 that was defined by the Bonferroni correction for multiple comparisons (0.05/427,948). Two of these CpGs were located in the same locus (cg05951221 and cg21566642), therefore seven independent loci were identified. The model adjusted for age, sex, cell type, and surrogate variables was considered as the main analysis.
Table 2. Differentially methylated CpGs associated with age-independent CVR in the discovery, validation and metaanalysis stage using (A) the Δage approach and (B) the residuals approach, and the model adjusted for age, sex, cell type and surrogate variables.
For those CpGs located in intergenic regions, the closest genes are given.
| A. Δage approach | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Features | Discovery stage (REGICOR) |
Validation stage (Framingham) |
Metaanalysis (Discovery + Validation) |
|||||||||
|
| ||||||||||||
| CpG | Chr* | Position (bp) |
Gene | Coeff* | SE* | P-value | Coeff* | SE* | P-value | Coeff* | SE* | P-value |
| cg12547807 | 1 | 9473751 | NA; (LOC100506022) | −2.04·10−2 | 4.53·10−3 | 6.83·10−6 | −1.60·10−2 | 2.23·10−3 | 6.91·10−13 | −1.69·10−2 | 2.00·10−3 | <2·10−16 |
| cg27537125 | 1 | 25349681 | NA; (MIR4425) | −2.13·10−2 | 4.38·10−3 | 1.10·10−6 | −8.46·10−3 | 2.35·10−3 | 3.09·10−4 | −1.13·10−2 | 2.07·10−3 | 4.20·10−8 |
| cg05951221 | 2 | 233284402 | NA; (AC068134.6; ALPPL2) | −2.74·10−2 | 6.14·10−3 | 8.40·10−6 | −1.07·10−2 | 3.48·10−3 | 2.02·10−3 | −1.48·10−2 | 3.03·10−3 | 1.05·10−6 |
| cg21566642 | 2 | 233284661 | NA; (AC068134.6; ALPPL2) | −3.45·10−2 | 6.79·10−3 | 3.71·10−7 | −9.73·10−3 | 3.57·10−3 | 6.46·10−3 | −1.51·10−2 | 3.16·10−3 | 1.78·10−6 |
| cg05575921 | 5 | 373378 | AHRR | −3.98·10−2 | 7.19·10−3 | 3.30·10−8 | −1.66·10−2 | 3.01·10−3 | 3.15·10−8 | −2.01·10−2 | 2.78·10−3 | 4.64·10−13 |
| cg19939077 | 10 | 81108060 | PPIF | −1.40·10−2 | 5.59·10−3 | 1.24·10−2 | −7.12·10−3 | 2.75·10−3 | 9.71·10−3 | −8.46·10−3 | 2.47·10−3 | 6.14·10−4 |
| cg00574958 | 11 | 68607622 | CPT1A | −2.01·10−2 | 5.02·10−3 | 6.21·10−5 | −1.76·10−2 | 2.88·10−3 | 9.27·10−10 | −1.82·10−2 | 2.50·10−3 | 2.82·10−13 |
| cg18608055 | 19 | 1130866 | SBNO2 | −1.12·10−2 | 4.10·10−3 | 6.23·10−3 | −9.40·10−3 | 1.73·10−3 | 5.18·10−8 | −9.67·10−3 | 1.59·10−3 | 1.20·10−9 |
| B. Residuals approach | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Features | Discovery stage (REGICOR) |
Validation stage (Framingham) |
Metaanalysis (Discovery+ Validation) |
|||||||||
|
| ||||||||||||
| CpG | Chr* | Position (bp) |
Gene | Coeff* | SE* | P-value | Coeff* | SE* | P-value | Coeff* | SE* | P-value |
| cg12547807 | 1 | 9473751 | NA; (LOC100506022) | −4.16 | 9.84·10−1 | 2.39·10−5 | −6.77 | 1.27 | 9.80·10−8 | −5.14 | 7.78·10−1 | 4.01·10−11 |
| cg27537125 | 1 | 25349681 | NA; (MIR4425) | −5.30 | 1.20 | 1.07·10−5 | −4.74 | 1.23 | 1.15·10−4 | −5.03 | 8.60·10−1 | 5.14·10−9 |
| cg05951221 | 2 | 233284402 | NA; (AC068134.6; ALPPL2) | −8.90 | 1.58 | 1.68·10−8 | −6.62 | 1.90 | 4.98·10−4 | −7.97 | 1.21 | 5.20·10−11 |
| cg21566642 | 2 | 233284661 | NA; (AC068134.6; ALPPL2) | −8.81 | 1.59 | 3.01·10−8 | −5.90 | 1.98 | 2.85·10−3 | −7.67 | 1.24 | 6.08·10−10 |
| cg05575921 | 5 | 373378 | AHRR | −8.51 | 1.84 | 3.92·10−6 | −7.08 | 1.74 | 4.60·10−5 | −7.75 | 1.26 | 8.73·10−10 |
| cg19939077 | 10 | 81108060 | PPIF | −7.43 | 1.39 | 8.21·10−8 | −5.80 | 1.62 | 3.35·10−4 | −6.74 | 1.05 | 1.50·10−10 |
| cg00574958 | 11 | 68607622 | CPT1A | −3.27 | 1.43 | 2.28·10−2 | −7.01 | 1.55 | 5.74·10−6 | −5.00 | 1.05 | 2.00·10−6 |
| cg18608055 | 19 | 1130866 | SBNO2 | −4.83 | 1.04 | 3.55·10−6 | −4.62 | 1.03 | 7.19·10−6 | −4.72 | 7.32·10−1 | 1.11·10−10 |
CpG, CpG ID; Chr, chromosome location; Position (bp), basepair genomic position; NA, non-annotated gene; Coeff, regression coefficient; SE, standard error of the regression.
Four of those CpGs were validated in both approaches used to estimate the age-independent CVR (cg05575921, located within AHRR; cg18608055, within SBNO2; cg27537125, in an intergenic region close to a non-coding RNA [MIR4425]; and cg12547807, in another intergenic region). Apart from this CpG site, the residuals approach provided three CpGs (cg05951221 and cg21566642, located in the same intergenic region close to the gene ALPP2 and the long non-coding RNA ACO68134.6, and cg19939077, within PPIF). The Δage approach contributed one additional CpGs (cg00574958, located within CPT1A).
Seven of the eight identified CpGs were found to be hypomethylated in association with smoking status (cg05575921, cg05951221, cg12547807, cg18608055, cg19939077, cg21566642, and cg27537125) (Table 3 and Table VI in the online-only Data Supplement). Three of them (cg12547807, cg18608055, cg19939077) were also hypomethylated in association with BMI. The eighth CpG (cg00574958) was hypomethylated in association with diabetes, BMI, and triglycerides levels.
Table 3. Metaanalysis of the association between the methylation level in the identified CpGs and vascular risk factors in the REGICOR and the Framingham studyies.
The association between differential methylation sites and BMI was evaluated in the total population; associations with SBP and DBP and with lipid levels were assessed in non-treated individuals for hypertension and for hypercholesterolemia, respectively.
| CpG | Total population | Individuals non-treated for hypertension |
Individuals non-treated for hypercholesterolemia |
Statistics | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Smoking | Diabetes | BMI† | SBP† | DBP† | Total cholesterol |
HDL cholesterol† |
TG† | ||
| cg00574958 | 1.35·10−3 | −3.22·10−3* | −4.97·10−4* | −2.33·10−5 | 6.76·10−6 | −2.30·10−5 | −5.56·10-5 | −5.52·10−5* | Coefficient |
| 8.35·10−4 | 8.36·10−4* | 5.57·10−5* | 3.01·10−5 | 5.04·10−5 | 1.17·10−5 | 2.62·10−5 | 6.91·10−6* | SE† | |
| 1.07·10−1 | 1.15·10−4* | <2·10−16* | 4.38·10−1 | 8.93·10−1 | 4.95·10−2 | 3.36·10−2 | 1.33·10−15* | P-value | |
| cg05575921 | −1.60·10−1* | −6.53·10−3 | 5.68·10−4 | 6.56·10−5 | 9.27·10−5 | 1.18·10−5 | −1.20·10−4 | −3.58·10−5 | Coefficient |
| 3.89·10−3* | 3.86·10−3 | 2.57·10−4 | 1.28·10−4 | 2.14·10−4 | 5.12·10−5 | 1.13·10−4 | 2.98·10−5 | SE† | |
| <2·10−16* | 9.03·10−2 | 2.68·10−2 | 6.08·10−1 | 6.66·10−1 | 8.18·10−1 | 2.89·10−1 | 2.31·10−1 | P-value | |
| cg05951221 | −8.46·10−2* | −6.29·10−3 | −1.31·10−4 | 3.08·10−4 | −3.84·10−4 | 1.03·10−4 | −1.95·10−4 | −3.60·10−5 | Coefficient |
| 3.60·10−3* | 3.57·10−3 | 2.38·10−4 | 1.23·10−4 | 2.06·10−4 | 5.00·10−5 | 1.10·10−4 | 2.92·10−5 | SE† | |
| <2·10−16* | 7.85·10−2 | 5.81·10−1 | 1.23·10−2 | 6.18·10−2 | 3.92·10−2 | 7.71·10−2 | 2.17·10−1 | P-value | |
| cg12547807 | −1.75·10−2* | −1.28·10−4 | −3.65·10−4* | 6.64·10−5 | −4.03·10−5 | 1.81·10−5 | 4.47·10−5 | −1.71·10−5 | Coefficient |
| 1.52·10−3* | 1.51·10−3 | 1.01·10−4* | 5.12·10−5 | 8.58·10−5 | 2.09·10−5 | 4.63·10−5 | 1.22·10−5 | SE† | |
| <2·10−16* | 9.32·10−1 | 2.95·10−4* | 1.94·10−1 | 6.38·10−1 | 3.87·10−1 | 3.34·10−1 | 1.61·10−1 | P-value | |
| cg18608055 | −1.31·10−2* | 7.88·10−4 | −5.45·10−4* | 4.47·10−5 | −2.70·10−5 | 5.67·10−5 | 8.90·10−5 | −1.95·10−5 | Coefficient |
| 1.83·10−3* | 1.83·10−3 | 1.22·10−4* | 6.13·10−5 | 1.03·10−4 | 2.55·10−5 | 5.67·10−5 | 1.50·10−5 | SE† | |
| 8.37·10−13* | 6.66·10−1 | 7.99·10−6* | 4.66·10−1 | 7.93·10−1 | 2.60·10−2 | 1.17·10−1 | 1.93·10−1 | P-value | |
| cg19939077 | −1.22·10−2* | −1.54·10−3 | −4.05·10−4* | −5.68·10−7 | 1.14·10−4 | 2.32·10−5 | −1.10·10−4 | −2.33·10−5 | Coefficient |
| 1.59·10−3* | 1.59·10−3 | 1.06·10−4* | 5.38·10−5 | 9.01·10−5 | 2.20·10−5 | 4.93·10−5 | 1.30·10−5 | SE† | |
| 1.8·10−14* | 3.34·10−1 | 1.42·10−4* | 9.92·10−1 | 2.04·10−1 | 2.92·10−1 | 2.58·10−2 | 7.32·10−2 | P-value | |
| cg27537125 | −1.09·10−1* | −5.76·10−3 | −2.09·10−4 | 3.16·10−4 | −3.19·10−4 | 1.03·10−4 | −1.84·10−4 | −2.08·10−5 | Coefficient |
| 3.77·10−3* | 3.79·10−3 | 2.52·10−4 | 1.31·10−4 | 2.20·10−4 | 5.18·10−5 | 1.16·10−4 | 3.06·10−5 | SE† | |
| <2·10−16* | 1.28·10−1 | 4.07·10−1 | 1.60·10−2 | 1.47·10−1 | 4.64·10−2 | 1.12·10−1 | 4.96·10−1 | P-value | |
| cg21566642 | −1.84·10−2* | −3.52·10−5 | −4.38·10−5 | 5.92·10−5 | −5.52·10−5 | 1.34·10−5 | −1.51·10−5 | 6.12·10−7 | Coefficient |
| 1.17·10−3* | 1.17·10−3 | 7.79·10−5 | 4.10·10−5 | 6.87·10−5 | 1.64·10−5 | 3.66·10−5 | 9.63·10−6 | SE† | |
| <2·10−16* | 9.76·10−1 | 5.74·10−1 | 1.48·10−1 | 4.22·10−1 | 4.12·10−1 | 6.81·10−1 | 9.49·10−1 | P-value | |
Significant associations (p-value<7.81·10−4 = 0.05/(8 CpGs*8 VRFs))
BMI, Body Mass Index; SBP, Systolic Blood Pressure; DBP, Diastolic Blood Pressure; HDL, High-Density Lipoprotein; TG, Triglycerides; SE, Standard Error.
The identified CpGs explained 15.16% of the variability of the age-independent CVR estimated through the residuals approach and 12.01% of the variability of the CVR assessed through the Δage approach in REGICOR (excluding cg21566642 to avoid redundant information already provided by cg00574958). In the Framingham population, these eight CpGs explained 7.51% and 8.53% of the variability of the CVR from the residuals and the Δage approach, respectively.
We analyzed whether genetic variants located close to the validated CpGs were associated with CHD. Data on CHD have been contributed by CARDIoGRAMplusC4D investigators and were downloaded from www.cardiogramplusc4d.org. We did not find any CVD-associated SNP located 10,000 bp downstream or upstream of the identified CpGs.
Development of methylation risk scores and evaluation of their association with subclinical atherosclerosis and clinical events
We developed methylation risk scores (MRSs) based on the identified CpGs, one for each population used in the residuals and Δage studies (REGICOR, n=574 and n=465, and Framingham, n=2,172 and n=1,823, respectively). We used seven CpGs (excluding cg21566642 to avoid redundant information).
1. Association of methylation risk scores with subclinical atherosclerosis
The association between the MRSs and arterial stiffness measurements in the REGICOR population is shown in Table 4A. Measurements related to arterial stiffness were available for 534 individuals included in the EWASs. Both MRSs were directly associated with arterial distensibility coefficient and inversely with pulse wave velocity. In model 2, the association with those parameters was only found in the left common carotid artery.
Table 4. Association of the MRSs with (A) subclinical atherosclerosis in the REGICOR cohort (N=534) and (B) CVD or CHD incidence in the Framingham Offspring Cohort.
MRSs were based on the residuals approach and the Δage approach in REGICOR and Framingham population, respectively. Model 1 was age- and sex-adjusted. Model 2 was adjusted for VRFs (age, sex, total and HDL cholesterol, diabetes, smoking status, SBP, and hypertensive treatment). (A) Subclinical atherosclerosis comprises arterial distensibility and pulse wave velocity for right and left carotid arteries. Both the coefficient and the p-value of each association are given. (B) Hazard ratios, confidence intervals (95%) and p-values of each association are given.
| A. Subclinical atherosclerosis | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Subclinical atherosclerosis |
MRS | Model 1 | Model 2 | |||
| Coeff | P-value | Coeff | P-value | |||
| Pulse wave velocity | Left | Residuals | −1.3·10−2 | 1.92·10−4 | −1.08·10−2 | 1.26·10−2 |
| Δage | −1.3·10−2 | 1.92·10−4 | −1.08·10−2 | 1.26·10−2 | ||
| Right | Residuals | −8.47·10−3 | 7.64·10−3 | −3.55·10−3 | 3.79·10−1 | |
| Δage | −8.47·10−3 | 7.63·10−3 | −3.55·10−3 | 3.79·10−1 | ||
| Distensibility coefficient | Left | Residuals | 2.59·10−2 | 1.59·10−4 | 2.36·10−2 | 5.75·10−3 |
| Δage | 2.59·10−2 | 1.59·10−4 | 2.36·10−2 | 5.74·10−3 | ||
| Right | Residuals | 1.83·10−2 | 3.7·10−3 | 9.42·10−3 | 2.4·10−1 | |
| Δage | 1.83·10−2 | 3.7·10−3 | 9.42·10−3 | 2.4·10−1 | ||
| B. CVD or CHD incidence | |||||
|---|---|---|---|---|---|
|
| |||||
| Incidence | MRS | Model 1 | Model 2 | ||
|
| |||||
| Hazard ratio and 95% CI |
P-value | Hazard ratio and 95% CI |
P-value | ||
| CVD (n=222) | Residuals | 1.32 (1.16;1.51) | 2.20·10−5 | 1.28 (1.09;1.51) | 3.09·10−3 |
| Δage | 1.33 (1.17;1.51) | 1.10·10−5 | 1.30 (1.10;1.53) | 1.74·10−3 | |
| CHD (n=94) | Residuals | 1.08 (0.88;1.32) | 4.84·10−1 | 0.97 (0.75;1.26) | 8.30·10−1 |
| Δage | 1.09 (0.89;1.34) | 4.17·10−1 | 0.99 (0.76;1.28) | 9.34·10−1 | |
Coeff, Linear Regression Coefficients; MRS, Methylation Risk Score; CVD, Cardiovascular Disease; CHD, Coronary-Heart Disease; CI, Confidence Intervals.
2. Association of methylation risk scores with CHD and CVD incidence
The associations between the MRSs and the incidence of clinical coronary (n=94) and cardiovascular (n=222) events in the Framingham population are shown in Table 4B. The median and interquartile range of the follow-up periods for CVD and CHD incidence were 7.66 (5.44; 9.89) and 7.87 (6.47; 9.26) years, respectively. Both MRSs were associated with higher CVR independently of the classical VRFs considered in the CVR function. The hazard ratios for CVD events per 1 standard deviation increase were similar in all the models of adjustment.
DISCUSSION
Here we identify seven loci (eight CpGs) showing differential DNA methylation associated with age-independent CVR, located in four genes (AHRR, CPT1A, PPIF and SBNO2) and three intergenic regions. All but one of these CpGs were associated with smoking, and three of them were also associated with BMI (two not previously reported). The eighth CpG was associated with BMI, diabetes and triglycerides. In addition, epigenetic risk scores based on the differentially methylated CpGs identified were associated with the incidence of CVD events.
A growing number of studies published in the last decade supports the role of DNA methylation in atherosclerosis and CVDs.7 The development of epigenome-wide techniques has allowed the identification of differentially methylated patterns across the genome that might not have been found through a candidate-gene methylation strategy. The published studies have analyzed the association between DNA methylation and atherosclerosis, mainly focused on either atherosclerotic and cardiovascular events or individual VRFs, but none of them have assessed the cumulative effect of several VRFs. To determine this cumulative effect, we estimated CVR using a validated risk function. Moreover, as age is a strong determinant of CVR,4,5 we estimated age-independent CVR by calculating the difference between vascular and chronological age and the residuals of the association between CVR and age.
Seven of the eight validated CpGs were associated with smoking, and three of them were additionally associated with BMI (two of them newly discovered: cg12547807 and cg19939077). Methylation at cg12547807, located in an intergenic region of chromosome 1, has been previously identified as related to smoking,10 as have both cg19939077 and cg18608055 (false discovery rate <0.05).10 cg19939077 was additionally described as differentially methylated in association with alcoholism11,12; it is located within PPIF, which encodes a protein of the peptidyl-prolyl cis-trans-isomerases, which have an important role in several CVDs.13
cg18608055 is located in SBNO2, which encodes a component of the pathways leading to the anti-inflammatory effect of IL-10.14 Previously identified as differentially methylated in association with BMI15,16 and serum C-reactive protein (CRP),17 it has also been reported to be associated with the cardiovascular biomarker GDF-15.18 The same study found it to be hypomethylated in individuals who had suffered a myocardial infarction (ncases=47, ncontrols=271), although this association was not statistically significant, probably due to the small population size. Similarly, our results suggest that hypomethylation of the cg18608055 is associated with an increased age-independent CVR.
Methylation status at cg05575921 has been associated with smoking,10,19 obesity,20 and triglycerides,21 but we found no relationship to BMI or lipid levels. This CpG is located within AHRR, the smoking-associated gene that is most epigenetically regulated.22 Its transcription factor is involved in the metabolism of toxins from cigarette smoke.23 cg27537125, cg05951221, and cg21566642 are also identified in association with smoking.10,19 The last two are located within an intergenic CGI of chromosome 2 (chr2:233,283,397–233,285,959). This CGI should be further analyzed in future studies assessing the association between age-independent CVR and epigenomic regions instead of sites.
Differential methylation at cg00574958 has been associated with metabolic syndrome,24 diabetes,25,26 BMI,16,27 and lipid levels19,21, in agreement with our findings. It is located within intron 1 of CPT1A, which encodes a key enzyme in the regulation of mitochondrial fatty acid oxidation.28
We hypothesized that VRFs may lead to vascular dysfunction, and thus epigenetic risk scores will be inversely associated with arterial distensibility and positively associated with pulse wave velocity. Remarkably, the associations between arterial wall properties and MRSs were contrary to that hypothesis. Engelen and coworkers previously reported a positive association between carotid distensibility and smoking in a healthy population (n=3,601), including data from 24 centers worldwide.29 In our study, smoking was related to six of the seven CpGs used in the development of the MRSs, which could explain the unexpected result.
We did observe an association between the age-independent CVR epigenetic risk scores and the incidence of clinical CVD, supporting the consistency of our approach and our results. These associations remained significant when adjusting for classical VRFs considered in the FRESCO risk function (i.e. age, sex, total and HDL cholesterol, diabetes, smoking status, SBP and hypertensive treatment). They could be the consequence of a synergistic effect of classical VRFs on DNA methylation. Also, as six of the CpGs used to build the MRS were related to smoking, it could be that the observed associations reflect the smoking status more precisely than the applied questionnaires, which consider it as a dichotomous instead of a continuous variable.
Finally, our rationale was that differential DNA methylation was the result of exposure to VRFs accumulated as age-independent CVR. It could act as a mediator, but not a causal determinant, of the association between CVR and CVD. Although we cannot discard the causal association between DNA methylation and CVD, we report that genetic variability in the identified loci was not associated with CHD and calls into question any causal role. The main strength of this study is the dual-approach design to identify the age-independent CVR. Both methods presented consistent and similar results in all the associations. Regarding the EWASs, we used powerful statistics by fitting models based on robust linear regression, adjusted for confounding variables and further non-measured confounding (surrogate variables). Not only did we use methylation data processed according to standardized protocols, but we validated our first findings in a large independent population. Lastly, we showed the potential clinical importance of our results by applying them to create age-independent CVR epigenetic risk scores, then evaluated them as predictors of clinical cardiovascular events.
Some limitations of our study should be noted. First, our results are based on European origin populations and cannot be extrapolated to other ethnic groups. Additionally, we could not adjust our analysis for genetic variability between the two populations of the study because we do not have genome-wide genetic data in the REGICOR population. Second, due to the cross-sectional study design and the potential bi-directional relationship between CVR and DNA methylation, we cannot infer causality of the association between these two variables. Finally, EWASs have several limitations, such as the type of sample used (peripheral blood circulating cells), the population size, or the data analysis and interpretation.
In summary, we identified seven loci showing differential methylation in relation to age-independent CVR. Six of these loci were associated with smoking. Moreover, three of them were associated with BMI, two of them for the first time (to our knowledge). We also showed that age-independent CVR epigenetic risk scores based on differential methylation patterns are related to the incidence of clinical cardiovascular events. They may also have potential value as research tools to unravel atherosclerosis mechanisms.
Supplementary Material
HIGHLIGHTS.
We identified eight differentially methylated CpGs in association with age-independent cardiovascular risk.
Seven of those CpGs were related to smoking three of them also with BMI (two of them newly associated with BMI). The eighth CpG was also associated with BMI as well as diabetes and triglycerides.
Risk scores based on these differential methylation patterns were related to clinical cardiovascular events. This association was independent of age, sex and classical vascular risk factors.
Acknowledgments
To Elaine M. Lilly, PhD, for her critical reading and revision of the English text.
Sources of funding: This project was funded by the Carlos III Health Institute–European Regional Development Fund (ERDF) [FIS PI15/00051, CIBERCV, CIBERESP]; and the Government of Catalonia through the Agency for Management of University and Research Grants [2014SGR240]. S.S-B. was funded by the Instituto de Salud Carlos III-Fondos FEDER (IFI14/00007). A.F-S. was funded by the Spanish Ministry of Economy and Competitivity (BES-2014-069718). The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195 and HHSN268201500001I). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Additional funding for SABRe was provided by Division of Intramural Research, NHLBI, and Center for Population Studies, NHLBI.
Abbreviations
- CGI
CpG island
- CHD
coronary heart disease
- CpG
5'-cytosine-phosphate-guanine-3'
- CVD
cardiovascular disease
- CVR
cardiovascular risk
- EWAS
epigenome-wide association study
- MRS
methylation risk score
- REGICOR
REgistre GIroní del COR
- VRF
vascular risk factor
Footnotes
Disclosures: The authors declare they do not have any disclosures.
References
- 1.GBD 2016 DALYs and HALE Collaborators SI. Abajobir AA, Abate KH, et al. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1260–1344. doi: 10.1016/S0140-6736(17)32130-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Global Status Report on Noncommunicable Diseases. 2014 http://www.who.int/nmh/publications/ncd-status-report-2014/en/
- 3.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 4.Barton M, Husmann M, Meyer MR. Accelerated Vascular Aging as a Paradigm for Hypertensive Vascular Disease: Prevention and Therapy. Can J Cardiol. 2016;32:680–686.e4. doi: 10.1016/j.cjca.2016.02.062. [DOI] [PubMed] [Google Scholar]
- 5.Dhingra R, Vasan RS. Age as a risk factor. Med Clin North Am. 2012;96:87–91. doi: 10.1016/j.mcna.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goff DC, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA Guideline on the Assessment of Cardiovascular Risk. Circulation. 2014;129:S49–S73. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Sanlés A, Sayols-Baixeras S, Subirana I, Degano IR, Elosua R. Association between DNA methylation and coronary heart disease or other atherosclerotic events: A systematic review. Atherosclerosis. 2017;263:325–333. doi: 10.1016/j.atherosclerosis.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 8.D’Agostino RB, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB. General Cardiovascular Risk Profile for Use in Primary Care. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 9.Groenewegen K, den Ruijter H, Pasterkamp G, Polak J, Bots M, Peters SA. Vascular age to determine cardiovascular disease risk: A systematic review of its concepts, definitions, and clinical applications. Eur J Prev Cardiol. 2016;23:264–274. doi: 10.1177/2047487314566999. [DOI] [PubMed] [Google Scholar]
- 10.Joehanes R, Just AC, Marioni RE, et al. Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Marioni RE, Hedman ÅK, et al. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philibert RA, Penaluna B, White T, Shires S, Gunter T, Liesveld J, Erwin C, Hollenbeck N, Osborn T. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics. 2014;9:1212–1219. doi: 10.4161/epi.32252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perrucci GL, Gowran A, Zanobini M, Capogrossi MC, Pompilio G, Nigro P. Peptidyl-prolyl isomerases: a full cast of critical actors in cardiovascular diseases. Cardiovasc Res. 2015;106:353–364. doi: 10.1093/cvr/cvv096. [DOI] [PubMed] [Google Scholar]
- 14.El Kasmi KC, Smith AM, Williams L, Neale G, Panopolous A, Watowich SS, Häcker H, Foxwell BMJ, Murray PJ. Cutting Edge: A Transcriptional Repressor and Corepressor Induced by the STAT3-Regulated Anti-Inflammatory Signaling Pathway. J Immunol. 2007;179:7215–7219. doi: 10.4049/jimmunol.179.11.7215. [DOI] [PubMed] [Google Scholar]
- 15.Ronn T, Volkov P, Gillberg L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;100:9440–9445. doi: 10.1093/hmg/ddv124. [DOI] [PubMed] [Google Scholar]
- 16.Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2016;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ligthart S, Marzi C, Aslibekyan S, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17:255. doi: 10.1186/s13059-016-1119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ek WE, Hedman AK, Enroth S, Morris AP, Lindgren CM, Mahajan A, Gustafsson S, Gyllensten U, Lind L, Johansson A. Genome-wide DNA methylation study identifies genes associated with the cardiovascular biomarker GDF-15. Hum Mol Genet. 2016;25:817–827. doi: 10.1093/hmg/ddv511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen A-K, Zeilinger S, Kastenmüller G, et al. Epigenetics meets metabolomics: an epigenome-wide association study with blood serum metabolic traits. Hum Mol Genet. 2014;23:534–545. doi: 10.1093/hmg/ddt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou Y-H, Hedman ÅK, Arnett DK, Fornage M, Pankow JS, Boerwinkle E. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24:4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekkers KF, van Iterson M, Slieker RC, et al. Blood lipids influence DNA methylation in circulating cells. Genome Biol. 2016;17:138. doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dertinger SD, Silverstone AE, Gasiewicz TA. Influence of aromatic hydrocarbon receptor-mediated events on the genotoxicity of cigarette smoke condensate. Carcinogenesis. 1998;19:2037–2042. doi: 10.1093/carcin/19.11.2037. [DOI] [PubMed] [Google Scholar]
- 24.Das M, Sha J, Hidalgo B, et al. Association of DNA Methylation at CPT1A Locus with Metabolic Syndrome in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study. In: Gaunt TR, editor. PLoS One. Vol. 11. 2016. p. e0145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kriebel J, Herder C, Rathmann W, et al. Association between DNA Methylation in Whole Blood and Measures of Glucose Metabolism: KORA F4 Study. PLoS One. 2016;11:e0152314. doi: 10.1371/journal.pone.0152314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW, Göring HHH, Cole SA, Comuzzie AG, Almasy L, Mahaney MC, Curran JE, Blangero J, Carless MA. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet. 2015;24:5330–5344. doi: 10.1093/hmg/ddv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aslibekyan S, Demerath EW, Mendelson M, et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity. 2015;23:1493–1501. doi: 10.1002/oby.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casals N, Zammit V, Herrero L, Fadó R, Rodríguez-Rodríguez R, Serra D. Carnitine palmitoyltransferase 1C: From cognition to cancer. Prog Lipid Res. 2016;61:134–148. doi: 10.1016/j.plipres.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Engelen L, Bossuyt J, Ferreira I, van Bortel LM, Reesink KD, Segers P, Stehouwer CD, Laurent S, Boutouyrie P. Reference Values for Arterial Measurements Collaboration. Reference values for local arterial stiffness. Part A: carotid artery. J Hypertens. 2015;33:1981–1996. doi: 10.1097/HJH.0000000000000654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.