Summary
Microbial or endogenous molecular patterns as well as pathogen functional features can activate innate immune systems. Whereas detection of infection by pattern recognition receptors has been investigated in details, sensing of virulence factors activities remains less characterized. In Drosophila, genetic evidences indicate that the serine protease Persephone belongs to a danger pathway activated by abnormal proteolytic activities to induce Toll signaling. However, neither the activation mechanism of this pathway nor its specificity has been determined. Here, we identify a unique region in the pro-domain of Persephone that functions as bait for exogenous proteases independently of their origin, type, or specificity. Cleavage in this bait region constitutes the first step of a sequential activation and licenses the subsequent maturation of Persephone to the endogenous cysteine cathepsin 26-29-p. Our results establish Persephone itself as an immune receptor able to sense a broad range of microbes through virulence factor activities rather than molecular patterns.
Keywords: innate immunity, danger signal, virulence factors, cysteine cathepsin, cathepsin 26-29-p, immune receptor, clip-serine protease, NF-κB pathway, bacterial proteases, fungal proteases
Graphical Abstract
Highlights
-
•
All pathogen-secreted proteases activate the danger-sensing arm of the Toll pathway
-
•
The protease Persephone is the immune sensor for microbial proteolytic activities
-
•
A sensitive region in Persephone zymogen functions as a bait for exogenous proteases
-
•
Bait-region hydrolysis primes maturation of Persephone by the host cathepsin 26-29-p
Innate immune systems are activated by microbial molecular patterns or pathogen functional features. Issa et al. show that the Drosophila Toll pathway senses pathogen proteases through a hydrolysis-sensitive region localized in the Persephone pro-domain. Cleavage of this bait region primes maturation of Persephone and activation of the pathway by the host cathepsin 26-29-p.
Introduction
The metazoan innate immune system has developed numerous strategies to control fungal and bacterial infections, ranging from physical barriers to a sophisticated array of molecules and cells that function to suppress or prevent microbial invasion. Initial recognition of microbial invaders is mediated by a set of germline-encoded pattern recognition receptors (PRRs) that sense highly conserved pathogen-associated molecular patterns (PAMPs) not present in the host (Medzhitov and Janeway, 2002). Engagement of PRRs activates intracellular signaling pathways leading to the expression of pro-inflammatory cytokines, chemokines, and soluble antibacterial effector molecules. PRRs also recognize endogenous molecules or damage-associated molecular patterns (DAMPs) released during tissue or cellular damage resulting from infection or tissue injury (Bianchi, 2007). Innate immune system can also be activated by pathogen functional features acting as danger signals such as toxins or enzymatic activity of virulence factors. Whereas the activation of PRRs by microbial or endogenous molecular patterns has been characterized in structural detail, the sensing of danger signals remains less well understood (Yin et al., 2015).
In Drosophila melanogaster, two evolutionarily conserved signaling pathways, Toll and immune deficiency (IMD), control the expression of anti-microbial peptides (AMPs) following immune challenge (Hoffmann, 2003). The IMD pathway, activated by diaminopimelic acid-containing peptidoglycan (DAP-type PGN) common to most Gram-negative bacteria, regulates expression of a set of AMPs, among which is Diptericin, via the NF-κB transcription factor Relish (Kleino and Silverman, 2014). On the other hand, the Toll pathway is activated by lysine-containing peptidoglycan (Lys-type PGN) found in Gram-positive bacteria and by β-glucans characteristic of fungal cell walls and activates a different set of AMPs, including the antifungal peptide Drosomycin (Drs), through dorsal-related immune factor (DIF), another NF-κB-like transcription factor (Valanne et al., 2011). Of note, sensing of these PAMPs occurs upstream of the receptor Toll, which functions as a receptor for the cytokine Spaetzle (Spz) in Drosophila (Levashina et al., 1999, Weber et al., 2003). Circulating PRRs, e.g., peptidoglycan recognition protein (PGRP)-SA for Lys-PGN or glucan-binding protein (GNBP) 3 for β-1-3 glucans, sense infection in the hemolymph and activate a serine protease referred to as modular serine protease (ModSP). Activation of ModSP triggers the sequential activation of the Clip-serine proteases (Clip-SPs) Grass and Spaetzle-processing enzyme (SPE) (Buchon et al., 2009, El Chamy et al., 2008, Gobert et al., 2003, Gottar et al., 2006). The latter processes the Spz precursor to generate an active Toll ligand. Of note, a second cascade, the so-called danger signal cascade, can independently activate SPE and the Toll receptor. Rather than PRRs, it is dependent on the Clip-SP Persephone (Psh) and has been shown to be activated by some bacterial or fungal proteases (El Chamy et al., 2008, Gottar et al., 2006, Ligoxygakis et al., 2002). However, neither the activation mechanism of this arm of the Toll pathway nor its specificity has been determined. Microbial proteases play a central role in the host colonization and in the control of the immune system. Various examples have emerged across species showing that during close host-pathogen co-evolution, immune systems developed the mean to sense this danger (Chavarría-Smith et al., 2016, Cheng et al., 2015, de Zoete et al., 2011, LaRock et al., 2016, Turk, 2007). However, due to the high variety of protease enzymatic specificities, such systems are able to detect only a limited number of proteases.
Clip-SPs such as Psh belong to the chymotrypsin family and are expressed and secreted as inactive zymogens with a regulatory N-terminal pro-domain or “Clip domain” connected to the catalytic domain by a 23–92 amino acid linker (Veillard et al., 2016). Their activation relies on a proteolytic cleavage immediately upstream of the catalytic domain. The new N terminus, which contains the consensus sequence I/V-V-G-G-, folds into the enzymatic active site and triggers the catalytic activity (Hedstrom, 2002, Veillard et al., 2016). Because Clip-SPs are organized in cascade, their activation site matches the specificity of the upstream serine protease. Thus, a majority of Clip-SPs present an arginyl or lysyl residue in P1 position upstream of the activation site (Schechter and Berger nomenclature) to be processed by proteases with trypsin-like specificity (Ross et al., 2003, Veillard et al., 2016). Strikingly, Psh differs from most proteases and Clip-SPs in that it contains an unusual histidine in P1 position of the activation site, a residue that can be accepted by the substrate-binding sites of very few proteases. Because the zymogen of Psh (pro-Psh) is sensitive to hydrolysis by the Beauveria bassiana protease Pr1A, it has been suggested that pro-Psh could directly be activated by microbial proteases (Gottar et al., 2006). However, the restricted specificity needed to activate Psh is hard to reconcile with the structural and enzymatic diversities of secreted microbial proteases thought to be detected by the danger arm of the Toll pathway. This apparent contradiction prompted us first to determine the nature of the proteases sensed by this arm of the Toll pathway and then to investigate in more detail the molecular mechanism of its activation.
Results
Psh-Dependent Toll Pathway Activation Correlates with the Presence of Microbial Extracellular Proteases
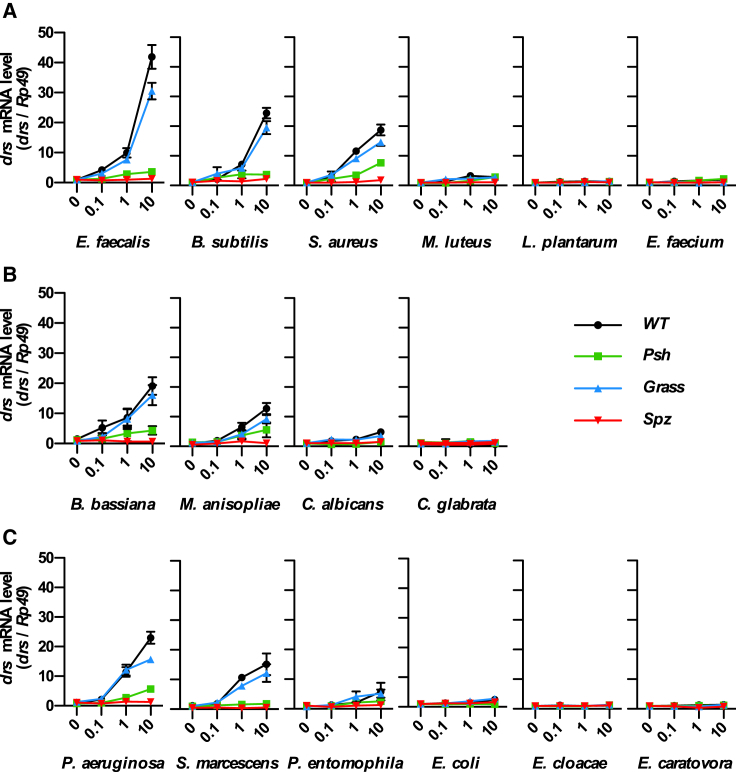
Previous studies have used an array of challenges to monitor the Psh-dependent activation of Toll, ranging from septic injuries with bacteria to natural infections with enthomopathogenic fungi, injection of purified proteases, or ectopic expression of exogenous proteases (El Chamy et al., 2008, Gottar et al., 2006). As a result of this heterogeneity, we do not have a clear picture of the pathogens that activate the Psh pathway. Thus, we used a standardized screen with a unique mode of infection (septic injury by pricking) and a defined microorganism load (OD600 from 0.1 to 10) to monitor the activation of either the PRR or the danger arm of the Toll pathway by an array of pathogenic microorganisms. Flies, either wild-type or mutants for Spz (required for both arms), Grass (required for PRR arm only), or Psh (required for danger arm only), were challenged with a panel of infectious microorganisms, and expression of the AMP Drs, a marker of Toll activation, was measured 16 hr post-infection. For the Gram-positive bacteria Enterococcus faecalis, Staphylococcus aureus, and Bacillus subtilis and the fungi B. bassiana and Metarhizium anisopliae, but also the Gram-negative bacteria Pseudomonas aeruginosa and Serratia marcescens, we observed a dose-dependent activation of the Toll pathway, which was abolished in spzrm7 and psh1 null mutants but only weakly affected in Grasshrd null mutant flies (Figure 1). By contrast, the Gram-positive bacteria Micrococcus luteus, Lactobacillus plantarum, and Enterococcus faecium and the fungi Candida albicans and Candida glabrata did not induce significant drs expression under these conditions, although they have been shown to activate Toll through the PAMP pathway when inoculated at high concentrations (Gobert et al., 2003, Gottar et al., 2006, Lemaitre et al., 1997). These results indicate that the Psh pathway can be activated by a range of different microorganisms, including Gram-negative bacteria.
Figure 1.
Psh-Dependent Activation of the Toll Pathway
Wild-type w1118 flies and Grasshrd (Grass), Spaetzlerm7 (Spz), and psh1 (Psh) mutant flies were immune challenged by septic injury with three different inocula diluted in PBS (OD600 = 0.1, 1, and 10) of Gram-positive bacteria (A), fungi (B), or Gram-negative bacteria (C). PBS (OD600 = 0) was used as control. Flies were collected 16 hr after challenge and drs gene expression was monitored by qRT-PCR in total RNA extracts. Ribosomal protein 49 (Rp49) mRNA was used as reference gene. Data represent means ± SEs of three independent experiments, each containing three groups of ten flies (five males and five females). Results were normalized to the value obtained for w1118 control flies.
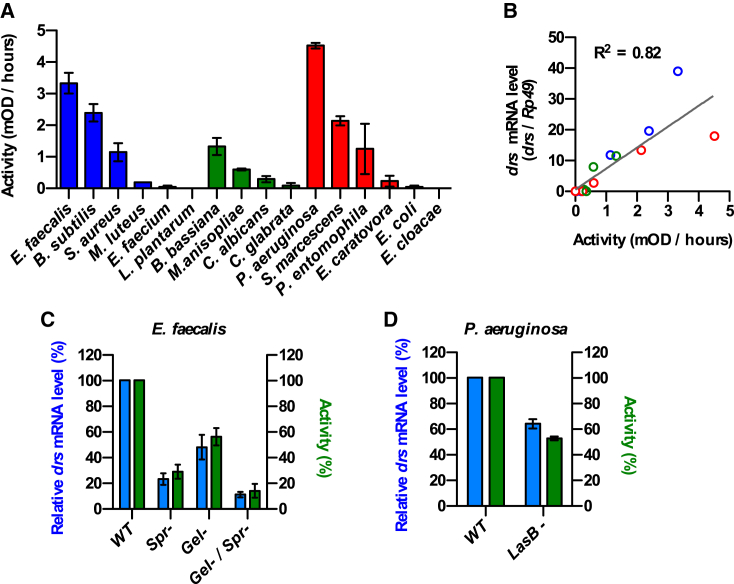
We next used AzoDye collagen, a non-specific protease substrate, to determine the level of extracellular proteolytic activity released by each microorganism in the culture medium. The highest activities were measured in the case of E. faecalis and P. aeruginosa cultures, while at the opposite no activity could be detected for Enterobacter cloacae or L. plantarum (Figure 2A). Remarkably, we observed a strict correlation between the levels of secreted proteolytic activity and drs expression in vivo (Figure 2B). E. faecalis strains mutant for the metalloproteinase gelatinase GelE and the serine protease SprE secreted significantly less proteolytic activity, and this correlated with reduced expression of drs in infected flies (Figure 2C). Similar results were obtained with a P. aeruginosa strain mutant for the elastase LasB (Figure 2D). Altogether, these results reveal that extracellular proteolytic activity is associated with activation of the Toll pathway, independently of the type, structure, specificity, or origin of the secreted proteases.
Figure 2.
Psh-Dependent Toll Pathway Activation versus Microbial Proteolytic Activity
(A) Extracellular proteolytic activity of Gram-positive bacteria (blue), fungi (green), and Gram-negative bacteria (red) grown to early stationary phase was determined on the non-specific substrate AzoDye collagen (15 mg/mL) in 0.1 M TrisHCl buffer (pH 8). Data represent means ± SEs of three independent experiments.
(B) Psh-dependent induction of drs expression upon immune challenge (OD600 = 10) from Figure 1 was correlated to the respective proteolytic activity determined on AzoDye collagen.
(C and D) Extracellular proteolytic activity and Psh-dependent induction of drs expression were determined under the same conditions as above for E. faecalis serine protease E (SprE) and gelatinase (GelE) null mutant strains (C) or for the P. aeruginosa elastase (LasB) null mutant (D) and expressed relative to the values obtained from the respective parental strains.
Data represent means ± SEs of three independent experiments.
Processing of pro-Psh by Secreted Microbial Proteases
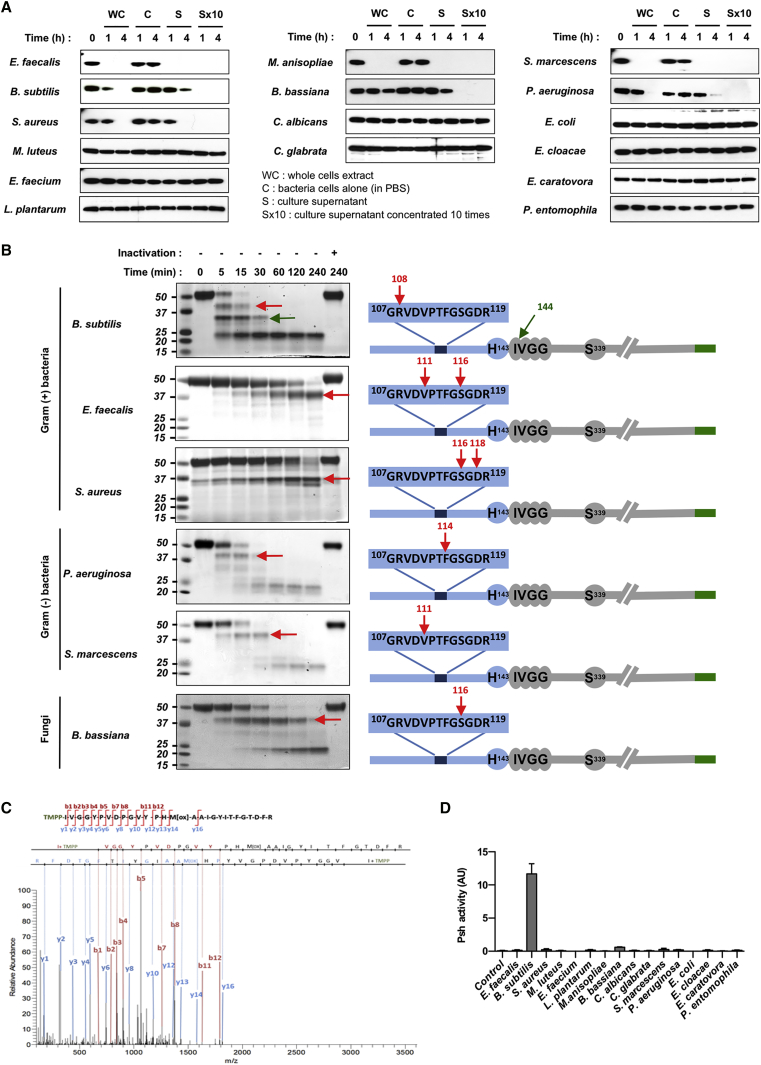
Because pro-Psh has been shown to be sensitive to hydrolysis by the B. bassiana protease Pr1A, we expressed a recombinant form of pro-Psh (rpro-Psh) carrying a C-terminal hexa-histidine Tag in S2 cells. Rpro-Psh was purified from the cell culture supernatant by affinity chromatography, incubated with bacterial and fungal preparations, and analyzed by western blot using an anti-HisTag antibody. As expected, no hydrolysis of rpro-Psh was observed after incubation with microorganisms devoid of extracellular proteolytic activity (Figure 3A). In contrast, incubation with cell-free culture media of pathogens secreting proteases led to rpro-Psh hydrolysis. However, no hydrolysis product could be observed, probably because of the instability of the tag or of the activity of co-secreted carboxy-peptidase(s).
Figure 3.
Hydrolysis of rpro-Psh by Microbial Extracellular Proteases
(A) rpro-Psh labeled with a C-terminal HisTag (0.2 μg/μL) was incubated at 29°C with whole-cell cultures of microorganisms grown to early stationary phase and microorganism cells suspended only in PBS, cell-free culture medium (supernatant), or the same medium (supernatant) concentrated ten times. After 1 or 4 hr, 1 μg aliquots were removed and the reaction was stopped at 100°C for 5 min. Residual rpro-Psh was visualized by western blot with an anti-6HisTag antibody. Representative results of at least two independent experiments.
(B) rpro-Psh was incubated under the same conditions with cell-free media (supernatant). At various time points, 5 μg of proteins were removed and the reaction was stopped at 100°C for 5 min. Controls were performed after pre-inactivation of the media for 5 min at 100°C. Hydrolysis products were visualized by Coomassie blue staining after SDS-PAGE electrophoresis. Identifications of neo-N-terminal peptides were determined by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) analysis after in-gel protein N-terminal chemical derivatization method using TMPP reagent. Arrows indicate identified N-terminal extremities.
(C) Characterization of TMPP-derivatized peptide of the expected N-terminal extremity of the catalytic domain by MS/MS fragmentation spectrum. The corresponding MS/MS fragmentation table is presented as Table S4.
(D) Cell-free supernatant of S2 cells expressing rpro-Psh (200 μL) was incubated in 0.1 M TrisHCl buffer (pH 8) with 200 μL of cell-free medium from microorganism cultures (final volume, 600 μL). After 1 hr, proteolytic activity of the generated rpro-Psh hydrolysis products was determined on the fluorogenic substrate Z-Arg-AMC for 30 min at 29°C in 0.1 M TrisHCl buffer (pH 8) supplemented with 5 mM CaCl2.
A time course analysis on SDS-PAGE gel using Coomassie blue staining revealed a high variability in the early hydrolysis profiles of rpro-Psh incubated in media conditioned by different pathogens. While E. faecalis, S. aureus, or B. bassiana generated stable products, other microorganisms, such as P. aeruginosa or S. marcescens, seemed to sequentially degrade rpro-Psh (Figure 3B). Identification of the N-terminal extremities of the hydrolysis products by mass spectrometry after in-gel labeling with TMPP (Ayoub et al., 2015) revealed that only B. subtilis was able to release the expected N-terminal extremity of the activated form of the Psh catalytic domain, with a cleavage after the His143 (Figures 3B, 3C, and S1). Furthermore, the Psh catalytic domain released by B. subtilis was active when incubated with Z-Arg-AMC, a fluorogenic substrate (Figure 3D). Incubation of rpro-Psh with the B. subtilis purified protease subtilisin resulted in cleavage of rpro-Psh at the extremity of the clip domain, generating a proteolytic active form of Psh. Identical results were obtained when the B. subtilis subtilisin was incubated with a catalytically inactive mutant version of pro-Psh containing a substitution of Ser338 in the protease active site (Figure S2). This confirms that release of the Psh catalytic domain by subtilisin is direct and does not implicate an auto-processing event.
By contrast, no proteolytic activity could be detected for rpro-Psh incubated with the media conditioned by the other pathogens, confirming the unique capacity of the B. subtilis to effectively and directly activate Psh (Figure 3D). Strikingly, however, the first hydrolysis products generated during incubation with all other microbial conditioned media always corresponded to cleavage(s) in the same region (G107-G118) of the Clip domain (Figure 3B). This indicates that this short sequence is highly susceptible to hydrolysis, independently of the specificity of the attacking protease. This region may thus act as bait for microbial proteases.
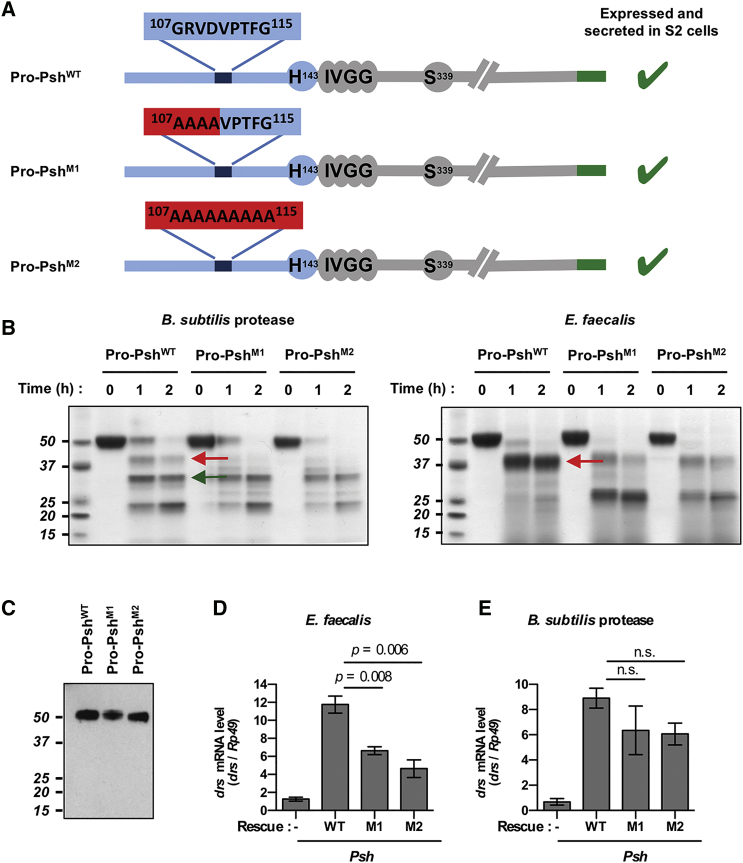
Cleavage in a Bait Region of the Clip Domain Is Essential for the Activation of Psh by Microbial Proteases
We hypothesized that cleavage in the “bait region” of the Clip domain may constitute the first step of a Psh sequential activation. To test this hypothesis, we constructed pro-Psh mutants in which this hypothetic bait region was substituted partially (rpro-PshM1) or totally (rpro-PshM2) by alanyl residues (Figure 4A). In vitro, these mutations significantly reduced the cleavages in the bait region by both E. faecalis conditioned medium and B. subtilis subtilisin. However, they did not affect the release of the specific active Psh catalytic domain upon incubation with B. subtilis subtilisin (Figure 4B). Interestingly, E. faecalis proteases reported their activity within the catalytic domain, with a cleavage after the Gly202. In parallel, wild-type rpro-Psh, rpro-PshM1, and rpro-PshM2 were expressed in psh1 null mutant flies under the control of a fat body-specific promoter (Figure 4C). Expression of wild-type rpro-Psh restored Toll pathway activation upon B. subtilis subtilisin injection or E. faecalis infection (Figures 4D and 4E). Expression of rpro-PshM1 and rpro-PshM2 also allowed almost full restoration of the Toll pathway inducibility to the B. subtilis subtilisin in the psh1 mutant background. However, neither of the mutant forms rescued drs expression after E. faecalis infection (Figure 4D). These results demonstrate the essential function of the bait region for Psh activation by extracellular microbial proteases and confirm the unique mode of action of B. subtilis subtilisin.
Figure 4.
Psh Inactivation by Mutations in the Bait Region
(A) Structure of pro-Psh mutants with partial (pro-PshM1) or total substitution (pro-PshM2) of the bait region by alanyl residues.
(B) Purified mutant or wild-type rpro-Psh (0.2 μg/μL) was incubated for 1 or 2 hr at 29°C with cell-free E. faecalis culture medium supernatant or with purified B. subtilis protease (1 nM). Following electrophoresis, hydrolysis products were visualized by Coomassie blue staining. Red arrows indicate fragments resulting from hydrolysis in the bait region and the green arrow indicates the active catalytic domain.
(C–D) Wild-type pro-Psh, pro-PshM1, and pro-PshM2 were expressed under the control of the fat body Yolk driver in psh1 mutant flies.
(C) Secretion of wild-type and mutant pro-Psh in the blood was determined by western blot using an anti-6HisTag antibody.
(D and E) drs mRNA levels were monitored by qPCR in psh1 mutant flies (psh1; yolk-GAL4/+) expressing the wild-type rpo-Psh (WT; psh1; yolk-GAL4/UAS-pro-Psh), the M1 mutant (M1; psh1; yolk-GAL4/UAS-pro-PshM1), and the M2 mutant (M2; psh1; yolk-GAL4/UAS-proPshM2) for 24 hr at 25°C following E. faecalis challenge (OD600 = 1) (D) or 16 hr at 25°C following B. subtilis protease injection (E).
Data represent means ± SEs of three independent experiments, each containing three groups of ten flies (five males and five females). p values obtained from Student’s t test are indicated on the graphs.
A Cysteine Protease Inhibitor Blocks Psh-Dependent, but Not Grass-Dependent, Toll Pathway Activation
The results above suggest a sequential activation of pro-Psh involving an endogenous protease that would cleave the remaining amino acids of the Psh pro-domain at His143. Among the proteases secreted into the Drosophila hemolymph, serine proteases do not possess the specificity needed to accept a histidine in P1 position, and various approaches performed in the laboratory did not reveal immune function for the two matrix-metalloproteases in Toll pathway activation. Extracellular cysteine proteases, particularly cathepsins, represent an interesting alternative and have been suggested in the past to participate in insect immunity (Fujimoto et al., 1999, Serbielle et al., 2009).
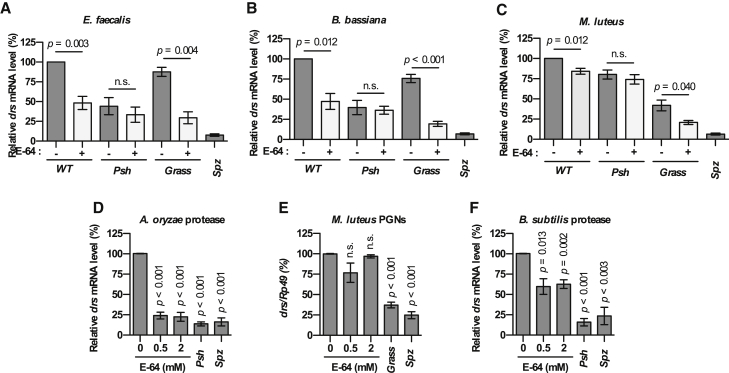
In order to evaluate their implication in Toll pathway activation, we injected the cysteine protease inhibitor E-64 into the body cavity of wild-type flies 2 hr before immune challenge and monitored drs expression (Figure 5). E-64 blocked Toll pathway activation in response to E. faecalis or B. bassiana as efficiently as Psh inactivation (Figures 5A and 5B). However, it only had a limited impact following M. luteus infection (Figure 5C). E-64 totally suppressed the Psh-dependent Toll pathway activation following microbial challenge when injected in Grasshrd mutant flies, but had no additional impact when injected into Psh null mutant flies. Moreover, E-64 totally blocked activation of the Toll pathway in response to purified A. oryzae protease in wild-type flies, but did not affect the response to injection of purified M. luteus PGNs (Figures 5D and 5E). Importantly, E-64 injection had only a marginal impact on the activation of Toll pathway upon injection of B. subtilis subtilisin (Figure 5F). Altogether, these data reveal that a cysteine protease inhibited by E-64 participates in the activation of pro-Psh by microbial proteases unable to directly cleave at His143 as B. subtilis subtilisin does.
Figure 5.
The Cysteine Protease Inhibitor E-64 Blocks Psh-Dependent Induction of drs Expression upon Immune Challenge
(A–C) A total of 18.4 nL of PBS alone or containing 2 mM of the cysteine protease inhibitor E-64 were injected into w1118 (WT), psh1 (Psh), and Grasshrd flies (Grass). spzrm7 mutant flies (Spz) were used as control. After 2 hr, flies were immune challenged for 24 hr by septic injury with E. faecalis (OD600 = 1) (A), by natural infection with B. bassiana (B), or by septic injury with M. luteus (OD600 > 200) (C).
(D–F) The E-64 inhibitor was injected at 0.5 or 2 mM into w1118 flies and Grasshrd, psh1, and spzrm7 mutant flies were used as control. After 2 hr, flies were challenged for 16 hr with A. oryzae protease (D), for 24 hr with M. luteus peptidoglycans (E), and for 16 hr with B. subtilis protease (F). Flies were collected and drs gene expression was monitored by qRT-PCR in total RNA extracts. Ribosomal protein 49 (Rp49) mRNA was used as reference gene. Results were normalized to the value obtained with the control conditions.
Data represent means ± SEs of three independent experiments, each containing three groups of ten flies (five males and five females). p values obtained from Student’s t test are indicated on the graphs.
Identification and Characterization of 26-29-p, a Cathepsin Involved in Psh-Dependent Toll Pathway Activation
We next monitored activation of the Toll pathway in fly lines in which the seven Drosophila cathepsins or the bleomycin hydrolase, all known to be sensitive to E-64, were inactivated by transposon insertion or RNA interference. The flies harboring a P-element inserted in the gene encoding the 26-29-protease (26-29-p: CG8947) showed reduced drs expression following infection with E. faecalis, M. luteus, or B. bassiana (Figures S3A–S3C). This reduction was comparable to that observed in psh1 mutant flies. CG8947 mutant flies also displayed an increased susceptibility to infections by E. faecalis and B. bassiana (Figures S3D and S3E). Remarkably, while most cysteine cathepsins are intracellular and present a short and unstructured inhibitory pro-fragment, cathepsin 26-29-p contains both a signal peptide and a 26 kDa N-terminal pro-domain of unknown function and has been found in the circulating system of various insects (Fujimoto et al., 1999, Serbielle et al., 2009).
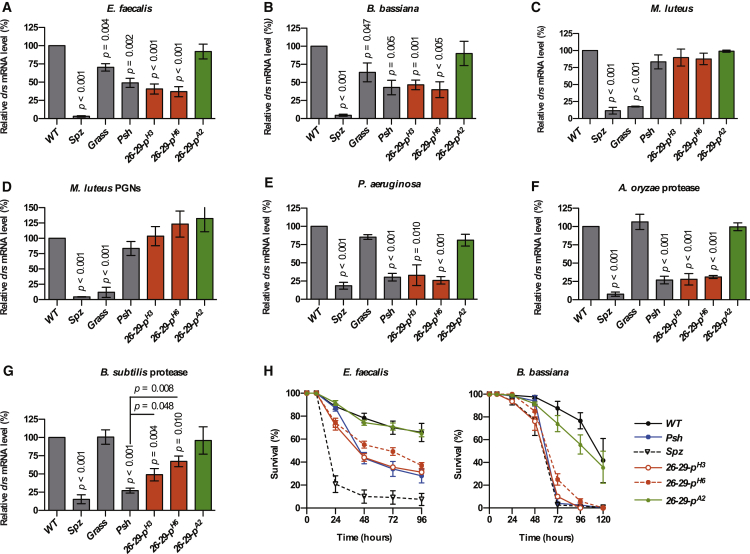
To characterize the function of the cathepsin 26-29-p, we generated additional mutant alleles by imprecise excision of the P-element inserted in the first exon of the 26-29-p gene. We obtained two new alleles, 26-29-pH3 and 26-29-pH6, carrying deletions of the 26-29-p coding region as well as a revertant 26-29-pA2 resulting from a clean excision of the P-element (Figure S4). Toll pathway activation was fully restored in the 26-29-pA2 flies, confirming that the phenotype we observed was caused by the insertion in the 26-29-p gene. 26-29-pH3 and 26-29-pH6 null mutant flies showed reduced activation of the Toll pathway 24 hr after immune challenge with E. faecalis or B. bassiana, similar to psh1 mutants (Figures 6A and 6B). This reduction was not observed upon infection with M. luteus bacteria or following injection of purified M. luteus PGNs (Figures 6C and 6D). Moreover, activation of the Toll pathway was totally abolished in 26-29-pH3 and 26-29-pH6 flies in response to infection with Gram-negative P. aeruginosa bacteria or injection of purified protease from A. oryzae (Figures 6E and 6F). By contrast, deletions in the 26-29-p gene only partially reduced Toll pathway activation upon injection of the purified subtilisin from B. subtilis (Figure 6G). Finally, 26-29-pH3 and 26-29-pH6 mutants, but not 26-29-pA2 revertant flies, showed a similar increased susceptibility to infection by E. faecalis and B. bassiana as psh1 mutant flies (Figure 6H). We conclude that the 26-29-p cathepsin is required for activation of Psh by microbial proteases, with the exception of those that can directly activate it, such as B. subtilis subtilisin.
Figure 6.
Cathepsin 26-29-p Is Required for Psh Sequential Activation
Two null mutant alleles of the 26-29-p gene (26-29-pH3 and 26-29-pH6) were generated by imprecise P-element excision resulting in deletions spanning transcriptional start site and the first two exons of the gene. A clean excision restoring expression of wild-type 26-29-p was used as control (26-29-pA2) (Figure S4).
(A–G) Flies of the indicated genotype were challenged by septic injury with E. faecalis (OD600 = 1) (A), M. luteus (OD600 > 200) (C), or P. aeruginosa (OD600 = 1) (E); by natural infection with B. bassiana (B); or by injection of M. luteus peptidoglycans (D), A. oryzae protease (F), or B. subtilis protease (G). After 24 hr at 29°C (16 hr for the purified proteases), flies were collected and drs gene expression was monitored by qRT-PCR in total RNA extracts. Ribosomal protein 49 (Rp49) mRNA was used as reference gene. Results were normalized to the value obtained with the wild-type flies. Data represent means ± SEs of three independent experiments, each containing three groups of ten flies (five males and five females). p values obtained from one-way ANOVA test are indicated on the graphs.
(H) Survival of adult flies challenged with E. faecalis by septic injury (OD600 = 1) or with B. bassiana by natural infection. Data represent means ± SEs of three independent experiments, each containing three groups of 20 flies (10 males and 10 females). Log-rank statistical analyses are presented as Table S5.
Sequential Activation of Psh by Microbial Proteases and Cathepsin 26-29-p
To confirm these genetic results, we expressed a recombinant form of the pro-cathepsin 26-29-p carrying a C-terminal hexa-histidine Tag in Drosophila S2 cells. In agreement with its proposed extracellular function, rpro-cathepsin 26-29-p was not detected in the cell extract but was exclusively found in the culture media (Figure S5A). To activate rpro-cathepsin 26-29-p, we incubated the concentrated supernatant with pepsine at an acidic pH following the common procedure for recombinant pro-cathepsin activation (Brömme et al., 2004). Under these conditions, rpro-cathepsin 26-29-p was processed and the proteolytic activity of its generated active form could be followed on the fluorogenic substrate Z-FR-AMC (Figures S5B and S5C).
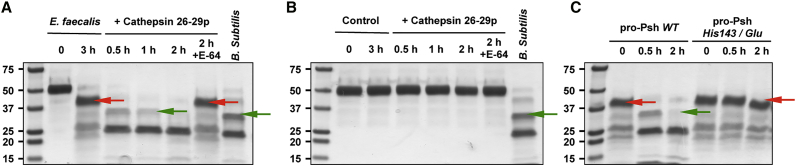
We then incubated the activated cathepsin 26-29-p extract with the full-length rpro-Psh or after its pre-processing into the bait region by E. faecalis culture media (Figures 7A and 7B). Remarkably, under our conditions, cathepsin 26-29-p had no effects on the full-length pro-Psh but cleaved the pre-processed form following a hydrolysis pattern similar to the one already observed after incubation with B. subtilis. Indeed, mass spectrometry analysis confirmed that cathepsin 26-29-p released the active form of Psh with a first cleavage at the extremity of the Clip domain after the His143. In agreement with these results, cathepsin 26-29-p was not able to cleave the partially processed rpro-Psh mutant containing a substitution of His143 in the same conditions (Figure 7C).
Figure 7.
Sequential Activation of rpro-Psh by Bacterial Proteases and Cathepsin 26-29-p
(A and B) Purified rpro-Psh was incubated in 0.1 M Tris buffer (pH 8) with (A) or without (B) E. faecalis culture supernatant at 29°C. After 3 hr, the partially processed rpro-Psh was incubated with the pre-activated cathepsin 26-29-p for 0.5–2 hr at 29°C in 0.2 M sodium acetate buffer (pH 5.5). The generated Psh hydrolysis products were then visualized by Coomassie blue staining after SDS-PAGE electrophoresis. Incubation under the same conditions with cathepsin 26-29-p pre-inactivated with E-64 was used as control. For comparison, hydrolysis products generated by B. subtilis subtilisine generated as previously described are presented. The N-terminal extremities of the hydrolysis products of interest were determined by N-terminal labeling and mass spectrometry. Red arrows indicate N-terminal extremities localized in the bait region, and the green arrows show the expected N-terminal extremity of the active form of Psh.
(C) Alternatively, the experiment was repeated with rpro-Psh His143/Glu.
Altogether with the genetic data, these results confirmed the function of cathepsin 26-29-p in the sequential activation of Psh during infections.
Discussion
A Pathway for Sensing an Array of Microbial Proteolytic Activities
Two theories were proposed in the late 1980s and early 1990s to account for activation of innate immunity by a limited number of receptors. Heralded by C. Janeway and P. Matzinger, these theories postulated that innate immunity was activated by non-self and danger signals, respectively (Janeway, 1989, Matzinger, 1994). During infections, damage to host tissues and cells leads to the release of intracellular molecules that can activate innate immunity upon binding PRRs (e.g., activation of TLR3, 7, 8, and 9 by self DNA or RNA or of the C-type lectin receptor DNGR1 by F-actin) (Ahrens et al., 2012, Zelenay and Reis e Sousa, 2013). Conserved from flies to mammals, damage-induced responses are important for triggering tissue repair, but they are delayed and occur when pathogen progression already impacts the host integrity (Allen and Wynn, 2011, Iwasaki and Medzhitov, 2015, Srinivasan et al., 2016). Of note, danger can be sensed before damage occurs, through monitoring the activity of microbial molecules associated with pathogenesis. Sometimes referred to as effector-triggered immunity, this strategy of surveillance focuses on toxins or virulence factors. For example, pore-forming toxins activate NLRP3 and the inflammasome in mammals, while the cytotoxic necrotizing factor-1 from E. coli modifies the enzyme Rac2, triggering its interaction with IMD and induction of AMPs (Boyer et al., 2011, Martinon et al., 2009). Defense reactions can also be activated in numerous hosts upon sensing microbial proteases (e.g., Chavarría-Smith et al., 2016, Cheng et al., 2015, de Zoete et al., 2011, Turk, 2007). Notably, in human type 2 immune response is activated by allergens such as the dust mite cysteine cathepsin or by excreted proteases from multicellular parasites by a yet unknown mechanism (Medzhitov and Janeway, 2002).
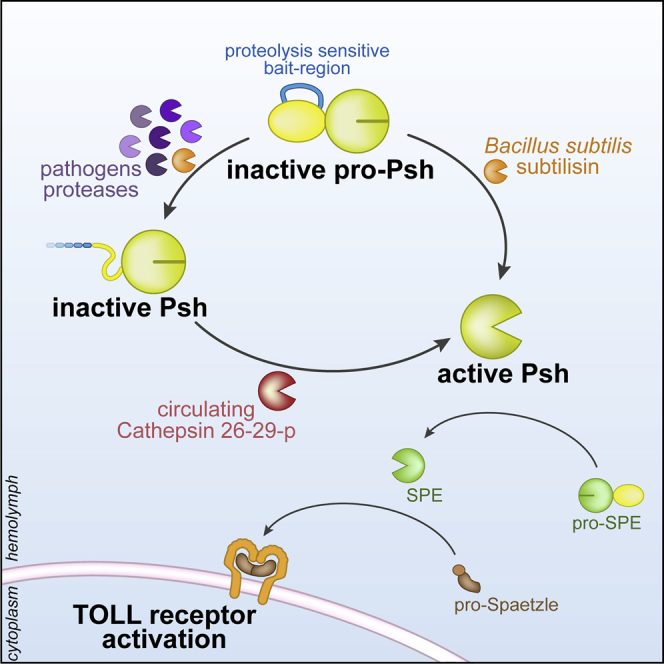
One critical aspect of innate immunity is to reconcile sensing of an immense range of potential microbial inducers with a restricted number of receptors (Janeway, 1989). We now have a reasonable understanding of the way this is achieved for PRRs. However, up to now it has remained unclear how this occurs in the context of virulence factors, which often target specific host molecules. This is particularly true for pathogen proteases due to their high enzymatic specificities. The identification of a critical region in the pro-domain of Psh, functioning as bait for microbial proteases independently of their origin, type, or specificity, provides for the first time an example of an innate immunity receptor able to sense a broad range of microbes through virulence factors, rather than molecular patterns.
Under physiological conditions, our previous studies have shown that the serine protease inhibitors of the serpin family such as Necrotic (Nec) control the residual activation of the danger arm by endogenous proteases since Nec null mutant flies display a constitutive and Psh-dependent activation of the Toll pathway (Levashina et al., 1999, Ligoxygakis et al., 2002). However, while we focused our study on exogenous proteases, we can hypothesize that the lack of specificity of the Psh activation mechanism allows the sensing of abnormal concentration of endogenous proteases in the hemolymph. Indeed, a Psh-dependent activation of the Toll pathway was observed after overexpression of an active form of the Grass serine protease in the hemolymph as well as after necrosis triggering (El Chamy et al., 2008, Ming et al., 2014).
A Unique Mode of Activation among Serine Proteases
The mechanism solved here involves sequential activation of Psh, with an initial cleavage in the bait region by microbial proteases licensing the subsequent maturation of Psh to the endogenous cathepsin 26-29-p. Since peptide bond hydrolysis is irreversible, proteolytic enzymes are tightly regulated at the transcriptional and post-translational levels (Khan and James, 1998). Two-step processes are frequently observed for activation of zymogens belonging to the chymotrypsin family as they allow strict spatial (i.e., activation of neutrophil and mast cell serine proteases in the azurophilic granules) or temporal regulation (i.e., activation of plasmin and thrombin during fibrinolysis and coagulation, respectively) (Caughey, 2016, Collen, 1999, Korkmaz et al., 2008, Wood et al., 2011). However, in most cases, sequential activation involves two highly specific cleavage sites not compatible with the sensing of a broad range of proteases. Hence, the long bait region highly sensitive to proteolysis described here constitutes an original strategy to detect exogenous proteases independent of their specificities.
Interestingly, the mode of activation of Psh is reminiscent of the mechanism of inhibition by α2-macroglobulin (α2-M), a non-specific inhibitor targeting both self- and non-self-proteases and clearing them from the tissue fluids (Garcia-Ferrer et al., 2017, Goulas et al., 2017). Indeed, α2-M contains a 25 amino acid-long bait region, which is also sensitive to all classes of proteases. Upon cleavage of this bait region, α2-M undergoes a structural rearrangement, thus trapping the target protease(s) (Marrero et al., 2012).
Overall, the proposed model is evocative of the guard system in plants, where structural modification of host proteins by pathogen effectors (here, Psh) is sensed by guard receptors (here, the cathepsin 26-29-p) to trigger an appropriate immune response (Jones and Dangl, 2006).
An Essential Immune Function for a Circulating Cathepsin
Cysteine cathepsins have long been known to participate in intracellular protein turnover inside the endosome/lysosome compartments. However, it has now been shown that in specific physiological or pathological conditions they can be addressed to alternative intracellular localizations or even extracellular space (Brix et al., 2008). Remarkably, we visualized the recombinant cathepsin 26-29-p only in the cell supernatant, but not in the cellular lysate. This observation corroborates previous studies that described the presence of cysteine 26-29-p homologs in the hemolymph of the Lepidopteran Manduca sexta and the flesh flies Sarcophaga peregrina (Fujimoto et al., 1999, Serbielle et al., 2009). This specific extracellular localization could be due to the 26 kDa N-terminal pro-domain of unknown function that is unique among cysteine cathepsins.
In mammals, cysteine cathepsins are involved in both adaptive and innate immunity. In addition to their main role in the degradation of phagocytized microbes in the phago-lysosomes, cathepsins are also essential for the regulation of MHC class II-dependent antigen presentation (Sadegh-Nasseri and Kim, 2015). In the innate immune system, cathepsins are mandatory for addressing TLR7 and TLR9 to the reticulum and for the post-translational processing of several cytokines (e.g., IL-8 or TNF-alpha) (Ewald et al., 2008, Ha et al., 2008, Ohashi et al., 2003, Park et al., 2008). Of note, cathepsin C and cathepsin L participate in the activation of serine proteases in the context of immune responses (i.e., neutrophil serine proteases and granzymes for cathepsin C and complement protease C3 for cathepsine L) (Hamon et al., 2016, Liszewski et al., 2013).
In addition, cysteine cathepsins are found extracellularly in inflammatory conditions, although the biological significance of this observation is still unclear. Associated with deleterious effects, such as degradation of the extracellular matrix or basal membrane components leading to the loss of tissue integrity, extracellular cathepsins may represent a bystander event rather than a specific response to infection and have even been considered as potential therapeutic targets in chronic inflammatory diseases (Vasiljeva et al., 2007). Our results described for the first time the implication of a circulating cysteine cathepsin in the model organism Drosophila melanogaster innate immune system. They suggest that the immune function of extracellular cysteine cathepsins in mammals is a topic that deserves further attention.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-His (C-term) Antibody | Invitrogen | #P/N 46-0693 |
| Bacterial and Virus Strains | ||
| Escherichia coli ATCC23724 | ATCC | ATCC23724 |
| Enterobacter cloacae | H. Monteil lab (University Louis Pasteur, Strasbourg) | N/A |
| Staphylococcus aureus RN6390 | H. Monteil lab (University Louis Pasteur, Strasbourg) | N/A |
| Serratia marcescens Db11 20C2 | Nehme et al., 2007 | 20C2 |
| Bacillus subtilis | J. Millet lab (Pasteur institute of Paris) | N/A |
| Erwinia carotovora Ecc15 | Basset et al., 2000 | N/A |
| Enterococcus faecalis OG1RF | E. Murray lab (University of Texas) Kawalec et al., 2005 | TX4002 |
| E. faecalis gelE- | E. Murray lab (University of Texas) Kawalec et al., 2005 | TX5264 |
| E. faecalis sprE- | E. Murray lab (University of Texas) Kawalec et al., 2005 | TX5243 |
| E. faecalis gelE- and sprE- | E. Murray lab (University of Texas) Kawalec et al., 2005 | TX5128 |
| Enterococcus faecium | E. Murray lab (University of Texas) Kawalec et al., 2005 | N/A |
| Pseudomonas aeruginosa PA14 | F.M. Ausubel lab; Liberati et al., 2006 | N/A |
| Pseudomonas aeruginosa; Elastase null mutant | F.M. Ausubel lab; Liberati et al., 2006 | ID31939 |
| Pseudomonas aeruginosa; Protease IV null mutant | F.M. Ausubel lab; Liberati et al., 2006 | ID37740 |
| Pseudomonas entomophila | B. Lemaitre lab (University of Lausanne) | N/A |
| Micrococcus luteus ATCC4698 | ATCC | ATCC4698 |
| Lactobacillus plantarum | W.J. Lee lab (Seoul National University) | N/A |
| Beauveria Bassiana 80.2 | Gottar et al., 2006 | N/A |
| Metarhizium anisopliae V275 | Gottar et al., 2006 | N/A |
| Candida glabrata | Gottar et al., 2006 | N/A |
| Candida albicans | Isolated by M. Koenig (CHU Strasbourg-Hautepierre) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| rpro-PshWT | This paper | N/A |
| rpro-PshM1 | This paper | N/A |
| rpro-PshM2 | This paper | N/A |
| rpro-Psh Ser339/Ala | This paper | N/A |
| rpro-Psh His144/Glu | This paper | N/A |
| rpro-Psh His144/Glu: Ser339/Ala | This paper | N/A |
| Protease, from Bacillus Sp. | Sigma | #P5985 |
| Protease, from Aspergillus oryzae | Sigma | #P6110 |
| Peptidoglycan from Micrococcus luteus | Sigma | #53243 |
| Pepsine from porcine gastric mucosa | Sigma | #P6887 |
| Azo dye-impregnated collagen | Sigma | #A4341 |
| Z-Arg-MCA | Sigma-aldrich | #C8022 |
| Z-Phe-Arg-AMC | Bachem | l-1160 |
| (N-succinimidyloxycarbonyl-methyl)tris-(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP) | Ayoub et al., 2015 | N/A |
| Porcine trypsin | Promega | #V5111 |
| Deposited Data | ||
| Images of gels and blots | Mendeley data | https://doi.org/10.17632/mzgnrcftzv.1 |
| Experimental Models: Cell Lines | ||
| Drosophila S2 cells | Invitrogen | #R69007 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster w1118 | El Chamy et al., 2008 | N/A |
| D. melanogaster psh1 | El Chamy et al., 2008 | N/A |
| D. melanogaster grasshrd | El Chamy et al., 2008 | N/A |
| D. melanogaster spzrm7 | El Chamy et al., 2008 | N/A |
| D. melanogaster C564-gal4 driver | Bloomington stock center | #6982 |
| D. melanogaster 26-29-pKG00154 | Bloomington stock center | #13051 |
| D. melanogaster 26-29-pH3 | This paper | N/A |
| D. melanogaster 26-29-pH6 | This paper | N/A |
| D. melanogaster 26-29-pA2 | This paper | N/A |
| D. melanogaster UAS-pro-PshWT | This paper | N/A |
| D. melanogaster UAS-pro-PshM1 | This paper | N/A |
| D. melanogaster UAS-pro-PshM2 | This paper | N/A |
| D. melanogaster ATT-3B VK00033 | Bloomington stock center | #24871 |
| P(SUPor-P)26-29-pKG00154 | Bloomington stock center | #13051 |
| P(TRIP.HMS00725)attP2 | Bloomington stock center | #32932 |
| P(TRIP.HMS02491)attP2 | Bloomington stock center | #42655 |
| P(TRIP.HMS00910)attP2 | Bloomington stock center | #33955 |
| Mi(MIC)CG11459MI08810 | Bloomington stock center | #50488 |
| P(EPgy2)CtsB1EY03339 | Bloomington stock center | #15434 |
| P(TRIP.GL00551)attP2 | Bloomington stock center | #36591 |
| P(SUPor-P)CG1440KG04580 | Bloomington stock center | #13977 |
| Recombinant DNA | ||
| pMT/V5-His vector | Invitrogen | #V412020 |
| pJM1345: rpro-PshWT in pMT/V5-His | this paper | N/A |
| pJM1681: rpro-PshM1 in pMT/V5-His | this paper | N/A |
| pJM1682: rpro-PshM2 in pMT/V5-His | this paper | N/A |
| pUAST-attB vector | Bischof et al., 2007 | N/A |
| pJM1692: pro-PshWT in pUAST-attB | this paper | N/A |
| pJM1693: pro-PshM1 in pUAST-attB | this paper | N/A |
| pJM1694: pro-PshM2 in pUAST-attB | this paper | N/A |
| pJM1674: rpro-Psh Ser339/Ala in pMT/V5-His | this paper | N/A |
| pJM1675: rpro-Psh His144/Glu in pMT/V5-His | this paper | N/A |
| pJM1676: rpro-Psh His144/Glu: Ser339/Ala in pMT/V5-His | this paper | N/A |
| pJM1689: rpro-cathepsin 26-29-p in pMT/V5-His | this paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 05 | GraphPad Software | N/A |
| Mascot algorithm v2.5.1 | Matrix Science | N/A |
| Scaffold software | Proteome Software | N/A |
| OASIS online application | Yang et al., 2011 | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Florian Veillard (f.veillard@ibmc-cnrs.unistra.fr).
Experimental Model and Subject Details
Fly strains
All Drosophila melanogaster were reared at 25°C on standard cornmeal-agar medium in a 12-hour light-dark cycle. w1118 flies were used as wild-type control throughout the experiments. psh1, Grasshrd and spzrm7 mutant flies have been described previously (El Chamy et al., 2008). The C564-gal4 driver (#6982) is from Bloomington stock center. Cathepsin mutant flies are described in Table S3. The 26-29-pH3, 26-29-pH6 and the 26-29-pA2 mutants were created by excision of the P-element 26-29-pKG00154 from Bloomington stock center #13051 (Figure S4). For psh rescue experiments, wild-type pro-psh, pro-pshM1 and pro-pshM2 were cloned in pUAST-ATTB vector and injected in ATT-3B VK00033 fly lines from Bloomington stock center #24871.
Bacterial stocks
Escherichia coli (ATCC23724), Enterobacter cloacae (kind gift of H. Monteil), Serratia marcescens (20C2; described in Nehme et al., 2007), Bacillus subtilis (kind gift from J. Millet, Pasteur Institute of Paris) and Erwinia carotovora (Ecc15) were grown in Luria-Bertani broth (LB). Staphylococcus aureus (RN6390), Enterococcus faecalis (OG1RF), Pseudomonas aeruginosa (PA14) and Pseudomonas entomophila (kind gift of B. Lemaitre) were grown in Bushnell Haas Broth (BHB). Micrococcus luteus (4698) was grown in Tryptic Soy Broth (TSB). Lactobacillus plantarum (kind gift of WJ. Lee) was grown in deMedan, Rogosa and Sharpe broth (MRS). Fungi Beauveria Bassiana (80.2) and Metarhizium anisopliae (V275) were grown in modified TKI broth (described in Lohse et al., 2014) and Candida glabrata and Candida albicans (isolated by Pr M. Koenig, CHU Strasbourg-Hautepierre) in Sabouraud Broth. Enterococcus faecium and E. faecalis gelE- and sprE- null mutants are a kind gift from E. Murray (University of Texas) and have been described in Kawalec et al. (2005). P. aeruginosa Elastase (ID31939) and Protease IV (ID37740) null mutants are a kind gifts from F.M. Ausubel and are described in Liberati et al. (2006).
Method Details
Expression of wild-type rpro-Psh, pro-Psh mutants and cathepsin 26-29-p
The coding sequence of pro-Psh was amplified by PCR using the cDNA clone GH12385 (DGRC) as template. KpnI and XhoI sites were introduced on 5′ and 3′ respectively of the psh cDNA using the following primers: 5′-GGGGGGTACCAAGATGCCATTGAAGTGGTCCCTGC-3′ and 5′-GGGGCTCGAGCACCCGATTGTCCGGCCAGA-3′ with Phusion High-Fidelity (New England Biolabs) and sub-cloned into KpnI-XhoI sites (New England Biolabs) of the pMT-V5-HisA vector (Invitrogen). The ligated product was transformed into chemically competent E. coli DH5α cells (Invitrogen). Appropriate insertion of the psh gene into the pMT-V5-HisA vector was verified by DNA sequencing (GATC Biotech sequencing center). Plasmid DNA named pJM1345 was extracted from these transformed E. coli DH5α cells using a Plasmid purification kit (QIAGEN). Psh mutants were obtained by PCR-directed mutagenesis using pJM1345 as template and are described in Tables S1 and S2. The Psh constructs were also digested by KpnI-PmeI restriction enzymes and sub-cloned into the pUAST-attB vector (described in Bischof et al., 2007). Similarly, the coding sequence of cathepsin 26-29-p was amplified by PCR using the cDNA clone pRE18380 (DGRC) and primers T7/IMU1347 (Table S2) and sub-cloned into EcoRI-ApaI of the pMT-V5-His expression vector.
Immune challenge
For infection by septic injury, flies were injured with a thin tungsten needle previously dipped in a microorganism suspension diluted in PBS at the indicated concentrations. Flies were challenged by natural infection with B. bassiana. 18.4 nL of a solution of B. subtilis (P5985; Sigma-Aldrich) or A. oryzae (P6110; Sigma-Aldrich) proteases, diluted at (1:2000) in PBS or 9.2 nL of a sonicated suspension of M. luteus peptidoglycans (5mg/ml; Sigma-Aldrich) were injected into the fly body cavity (Nanoject II apparatus; Drummond Scientific). When needed, 18.4 nL of E-64 (Sigma) diluted in PBS at 0.5 or 2 mM were injected 2 hours before immune challenge.
Quantitative RT-PCR
RNA was isolated with the NucleoSpin 96 RNA Kit (Macherey-Nagel) according to the manufacturer’s instructions. RNA was reversed transcribed by using the iScriptcDNA Synthesis Kit (Bio-Rad). Analysis of RNA expression was performed by real-time quantitative RT-PCR by using the iTaq SYBR Green Kit (Bio-Rad). Ribosomal protein 49 (Rp49) mRNA was used for normalization. The primers were as follows: for drs forward, 5′-CGTGAGAACCTTTTCCAATATGATG-3′ and reverse, 5′-TCCCAGGACCACCAGCAT-3′; Rp49 forward, 5′-GACGCTTCAAGGGACAGTATCTG-3′ and reverse, 5′-AAACGCGGTTCTGCATGAG-3′; 26-29-p forward, 5′-CGCAGGCTTGGCTTTCTCAG-3′ and reverse, 5′- GGCGTACGGAATGTACAGGG-3′.
Azo-collagen Assay
Bacterial and fungal cultures were centrifuged for 15 min at 5,000 g. 200 μL of cell free culture supernatant were then incubated for 8 hours at 29°C under constant shacking in 0.1 M TrisHCl buffer pH 8 (final volume: 600 μl) with 0.15 mg/ml of AzoDye-collagen (Sigma). The reaction was stopped by adding 200 μL of 3 M glycine, pH 3 and the AzoDye-collagen fibers were harvested by centrifugation (10 min at 15,000 g). The absorbance at 520 nm of the clear supernatant (200 μl) was determined in a 96-well plate using a spectrophotometer LB940 (BERTHOLD Technologies). Controls were done in the same conditions with pre-inactivated supernatant (10 min at 100°C).
Expression and purification of wild-type and mutated forms of rpro-Psh
Drosophila S2 cells were maintained at 25°C in Schneider’s medium (Biowest) supplemented with 10% FCS (Thermo Scientific). A total of 2 × 106 S2 cells were co-transfected with each plasmid of interest (1 μg) and with puromycin-selection plasmid (0.1 μg) by calcium phosphate precipitation and selected in the presence of puromycin (0.1 μg/ml). S2 cells stably transfected were then grown in Insect-Xpress medium (Biowhittaker) supplemented with 1% GlutaMAX-1 (gibco) and 1% Pen Strep (gibco). Wild-type or mutant rpro-Psh expression was then induced with Cu2SO4 at 0.5 M for 3 days at 25°C. Cultures were harvested by centrifugation at 1,500 g for 5 min and the cell-free supernatant dialyzed 2 times against 4 L of Ni-Sepharose binding buffer (20 mM sodium phosphate buffer, 500 mM NaCl, 20 mM imidazole, pH 7.4), applied to a 10 mL pre-equilibrated Ni2+-Sepharose 6 Fast Flow matrix (GE Healthcare, Pittsburgh, PA) and bound proteins were eluted with binding buffer supplemented with 500 mM imidazole. Eluted fractions containing the protein of interest were pooled and dialyzed against 0.1 M Tris-buffer pH 8. Protein concentrations of the final samples were determined by BCA Assay (Sigma).
Hydrolysis assay of rpro-Psh
Bacteria and fungi were grown to early stationary phase and centrifuged (5,000 g, 15 min). The cellular fraction was diluted in an equal volume of PBS and the cell-free medium was concentrated 10 times (CorningR Spin-X UF concentrators, Sigma). 20 μL of purified rpro-Psh (0.4 μg/μl in 0.1 M Tris buffer, pH 8) was incubated with 20 μL of the various microbial preparations. After 1 or 4 hours at 29°C, 1 μg of rpro-Psh was taken and the reaction was stopped at 100°C for 5 min in the presence of NuPAGE LDS Sample buffer (Invitrogen). Samples were electrophoresed for 2 hours at 100 V on a NuPAGE 4%–12% Bis-Tris Gel (Invitrogen) and resolved proteins electro-blotted onto nitrocellulose membrane (2 hours at 30 V). Non-specific binding sites were blocked with a 5% skim milk solution and membranes were then incubated with the monoclonal Anti-6His C-term antibody (Invitrogen) followed by the anti-mouse IgG-peroxidase conjugate (Sigma). Proteins of interest were visualized with the Chimioluminescent Reagent substrate Covalight (Covalab).
Alternatively, rpro-Psh was incubated under the same conditions with microbial cell-free medium. At various time points, 5 μg of proteins were removed and the reaction was stopped at 100°C for 5 min in the presence of NuPAGE LDS Sample buffer and subjected to SDS-PAGE electrophoresis as above. Proteins were then stained with SimplyBlue SafeStain (Invitrogen) and N-terminal extremities of the hydrolysis products of interest were determined by N-terminal labeling and mass spectrometry (see below).
Activity assay of rpro-Psh
Cell free supernatant of S2 cells expressing rPro-Psh (200 μl) was incubated in 0.1 M Tris buffer, pH 8 with B. subtilis protease (1 nM), with A. oryzae protease (100 nM) or with 200 μL of cell free medium of microbial culture (final volume: 600 μl). After 1 hour, proteolytic activity of the generated rpro-Psh hydrolysis products was determined on the fluorogenic substrate Z-Arg-AMC (Sigma Aldrich) for 30 min at 29°C in 0.1 M Tris buffer pH 8 supplemented with 5 mM CaCl2 (λex = 350 nm; λem = 460 nm).
Expression and activation of rpro-cathepsin 26-29-p
S2 cells were stably transfected with the expression plasmid of rpro-cathepsin 26-29-p as described previously. Expression of rpro-cathepsin 26-29-p was assessed in S2 cells lysate and cells culture supernatant by western blot using the monoclonal Anti-6His C-term antibody. To activate rpro-cathepsin 26-29-p, cells culture media was concentrated 20 times on Amicon centrifugal filter (Millipore) and then incubated with pepsine (0.002 mg/ml) in 0.1 M glycine buffer, pH 3. After incubation at 37°C, rpro-cathepsin 26-29-p processing was followed by western blot with the monoclonal Anti-6His C-term antibody. Activity of the generated hydrolysis products was assessed at 37°C in 0.1 M sodium acetate buffer, ph 5.5 on the fluorogenic substrate Z-Phe-Arg-AMC (Bachem) (λex = 350 nm; λem = 460 nm).
In vitro sequential activation of rpro-Psh
Purified rpro-Psh was incubated as previously described in 0.1 M Tris buffer, pH 8 with or without E. faecalis culture supernatant at 29°C. After 3 hours, the partially processed rpro-Psh was incubated with the pre-activated cathepsin 26-29-p for 30 min to 2 hours at 29°C in 0.2 M sodium acetate buffer, pH 5.5. The generated hydrolysis products were then visualized after SDS-PAGE electrophoresis using the SimplyBlue SafeStain (Invitrogen) and N-terminal extremities of the hydrolysis products of interest were determined by N-terminal labeling and mass spectrometry (see below). Controls were performed in presence of E-64 (0.2 mM). To confirm the capacity of cathepsin 26-29-p to release the N-terminal extremity of the active form of Psh, the experiment was repeated in the same conditions with rpro-Psh His143/Glu.
In-Gel N-Terminal Protein Derivatization strategy
Unless otherwise specified, all chemicals were obtained from Sigma Aldrich (St. Louis, MO). Using an automated robot platform (Massprep station, Waters), the gel slices containing protein samples were washed twice in 25 mM NH4HCO3 and CH3CN. The cysteine residues where subsequently reduced in 10 mM (tris(2-carboxyethyl)phosphine) at room temperature and then alkylated with 30 mM iodoacetamide. After dehydration with CH3CN, (N-succinimidyloxycarbonyl-methyl)tris-(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP) was added at a molar ratio of 200:1 (quantities of protein were evaluated based on 1D gel intensity band). Then 50 μL of the reaction buffer (100 mM Tris-HCl, pH 8.2) was added in each well (1H). Selective N-terminal TMPP derivatization is achieved by a careful control of reaction pH at 8.2. Residual derivatizing reagent was quenched by adding a solution of 0.1 M hydroxylamine at room temperature for 1 hour. The gel slices were then washed three times in 25 mM NH4HCO3 and CH3CN before dehydration with CH3CN. Enzymatic digestion was performed in-gel overnight at 37°C using porcine trypsin (Promega, Madison, WI, USA). Peptides were suspended in 10 μL of 1% CH3CN, 0.1% HCO2H in H2O.
Peptides were analyzed on a nanoUPLC-system (nanoAcquity, Waters) coupled to a quadrupole-Orbitrap hybrid mass spectrometer (Q-Exactive Plus, Thermo Scientific, San Jose, CA). Sample was concentrated/desalted on a Symmetry C18 precolumn (0.18 × 20 mm, 5 μm particle size; Waters) using a mobile phase composed of 99% solvent A (0.1% HCO2H in H2O) and 1% solvent B (0.1% HCO2H in CH3CN) at a flow rate of 5 μl/min for 3 minutes. Afterward, peptides were eluted at a flow rate of 450 nL/min using a UPLC separation column (BEH130 C18, 200 mm x 75 μm, 1.7 μm particle size; Waters) maintained at 60°C with the following gradient: from 1% to 50% B in 50 minutes.
The Q-Exactive Plus was operated in positive ion mode with source temperature set to 250°C and spray voltage to 2.0 kV. Spectra were acquired through automatic switching between full MS and MS/MS scans. Full scan MS spectra (300-1800 m/z) were acquired at a resolution of 70,000 at m/z 200 with an automatic gain control (AGC) fixed at 3 × 106 ions and a maximum injection time set to 50 ms, the lock-mass option being enabled (polysiloxane, 445.12002 m/z). Up to 10 most intense precursors (with a minimum of 2 charges) per full MS scan were isolated using a 2 m/z window and MS/MS spectra were acquired at a resolution of 17,500 at m/z 200 with an AGC fixed at 1 × 105 and a maximum injection time set to 100 ms. Peptide fragmentation was performed using higher energy collisional dissociation (HCD), with normalized collision energy being set to 27 and dynamic exclusion of already fragmented precursors being set to 10 s. The peptide match selection option was turned on. The system was fully controlled by XCalibur software (v3.0.63; Thermo Fisher Scientific).
Peak lists in MGF format were generated using the MSConvert algorithm of ProteoWizard software (v3.0.6090; http://proteowizard.sourceforge.net/). Searched against a SwissProt protein database combining Drosophila melanogaster (TaxID 7227) using Mascot algorithm v2.5.1 (Matrix science, London, UK). Mass tolerance was set to 5 ppm in MS mode and 0.07 Da in MS/MS mode, a maximum of one trypsin-missed cleavage was tolerated. Oxidation of methionine residues and carbamidomethylation of cysteine residues and TMPP of N-terminal peptide were considered as variable modifications. To gather and validate the identifications obtained, Scaffold software was used and spectra of labeled peptides were then carried out for manual validation (parameters Ion Score ≥ 0 and Ion -Identity Score = −40).
Quantification and Statistical Analysis
GraphPad Prism 05 (GraphPad Software) was used for mean and standard error calculations and Student’s t test or One-way ANOVA test. Log-rank analyses of survival assay were performed with the OASIS online application (Yang et al., 2011).
Data and Software Availability
Original images of gels and blots have been deposited to Mendeley Data and are available at https://doi.org/10.17632/mzgnrcftzv.1.
Acknowledgments
We thank Professors J.A. Hoffmann and J.L. Imler for critical reading of the manuscript. This work was supported by Centre National de la Recherche Scientifique, the Labex NetRNA (ANR-10-LABEX-0036_NETRNA), and a European Research Council Advanced Grant (AdG_20090506 “Immudroso,” to J.-M.R.) and benefits from funding from the state managed by the French National Research Agency as part of the Investments for the Future program. N.I. was supported by a fellowship from the region Alsace and from the Labex NetRNA. This work was also supported financially by the French Proteomic Infrastructure ProFI (ANR-10-INBS-08-03).
Author Contributions
F.V., J.-M.R., and N.M. designed the study, interpreted the results, and wrote the paper. N.I., E.L., and F.V. performed most of the experiments. N.G., C.S.-R., and A.V.D. designed, performed, and interpreted the mass spectrometry analysis.
Declaration of Interests
The authors declare no competing interests.
Published: February 15, 2018
Footnotes
Supplemental Information includes five figures and five tables and can be found with this article online at https://doi.org/10.1016/j.molcel.2018.01.029.
Supplemental Information
References
- Ahrens S., Zelenay S., Sancho D., Hanč P., Kjær S., Feest C., Fletcher G., Durkin C., Postigo A., Skehel M. F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity. 2012;36:635–645. doi: 10.1016/j.immuni.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Allen J.E., Wynn T.A. Evolution of Th2 immunity: a rapid repair response to tissue destructive pathogens. PLoS Pathog. 2011;7:e1002003. doi: 10.1371/journal.ppat.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub D., Bertaccini D., Diemer H., Wagner-Rousset E., Colas O., Cianférani S., Van Dorsselaer A., Beck A., Schaeffer-Reiss C. Characterization of the N-terminal heterogeneities of monoclonal antibodies using in-gel charge derivatization of α-amines and LC-MS/MS. Anal. Chem. 2015;87:3784–3790. doi: 10.1021/ac504427k. [DOI] [PubMed] [Google Scholar]
- Basset A., Khush R.S., Braun A., Gardan L., Boccard F., Hoffmann J.A., Lemaitre B. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. USA. 2000;97:3376–3381. doi: 10.1073/pnas.070357597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M.E. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L., Magoc L., Dejardin S., Cappillino M., Paquette N., Hinault C., Charriere G.M., Ip W.K., Fracchia S., Hennessy E. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix K., Dunkhorst A., Mayer K., Jordans S. Cysteine cathepsins: cellular roadmap to different functions. Biochimie. 2008;90:194–207. doi: 10.1016/j.biochi.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Brömme D., Nallaseth F.S., Turk B. Production and activation of recombinant papain-like cysteine proteases. Methods. 2004;32:199–206. doi: 10.1016/s1046-2023(03)00212-3. [DOI] [PubMed] [Google Scholar]
- Buchon N., Poidevin M., Kwon H.M., Guillou A., Sottas V., Lee B.L., Lemaitre B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl. Acad. Sci. USA. 2009;106:12442–12447. doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey G.H. Mast cell proteases as pharmacological targets. Eur. J. Pharmacol. 2016;778:44–55. doi: 10.1016/j.ejphar.2015.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarría-Smith J., Mitchell P.S., Ho A.M., Daugherty M.D., Vance R.E. Functional and evolutionary analyses identify proteolysis as a general mechanism for NLRP1 inflammasome activation. PLoS Pathog. 2016;12:e1006052. doi: 10.1371/journal.ppat.1006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Li J.F., Niu Y., Zhang X.C., Woody O.Z., Xiong Y., Djonović S., Millet Y., Bush J., McConkey B.J. Pathogen-secreted proteases activate a novel plant immune pathway. Nature. 2015;521:213–216. doi: 10.1038/nature14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collen D. The plasminogen (fibrinolytic) system. Thromb. Haemost. 1999;82:259–270. [PubMed] [Google Scholar]
- de Zoete M.R., Bouwman L.I., Keestra A.M., van Putten J.P. Cleavage and activation of a Toll-like receptor by microbial proteases. Proc. Natl. Acad. Sci. USA. 2011;108:4968–4973. doi: 10.1073/pnas.1018135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Chamy L., Leclerc V., Caldelari I., Reichhart J.M. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat. Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald S.E., Lee B.L., Lau L., Wickliffe K.E., Shi G.P., Chapman H.A., Barton G.M. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y., Kobayashi A., Kurata S., Natori S. Two subunits of the insect 26/29-kDa proteinase are probably derived from a common precursor protein. J. Biochem. 1999;125:566–573. doi: 10.1093/oxfordjournals.jbchem.a022322. [DOI] [PubMed] [Google Scholar]
- Garcia-Ferrer I., Marrero A., Gomis-Rüth F.X., Goulas T. α2-macroglobulins: structure and function. Subcell. Biochem. 2017;83:149–183. doi: 10.1007/978-3-319-46503-6_6. [DOI] [PubMed] [Google Scholar]
- Gobert V., Gottar M., Matskevich A.A., Rutschmann S., Royet J., Belvin M., Hoffmann J.A., Ferrandon D. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- Gottar M., Gobert V., Matskevich A.A., Reichhart J.M., Wang C., Butt T.M., Belvin M., Hoffmann J.A., Ferrandon D. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas T., Garcia-Ferrer I., Marrero A., Marino-Puertas L., Duquerroy S., Gomis-Rüth F.X. Structural and functional insight into pan-endopeptidase inhibition by α2-macroglobulins. Biol. Chem. 2017;398:975–994. doi: 10.1515/hsz-2016-0329. [DOI] [PubMed] [Google Scholar]
- Ha S.D., Martins A., Khazaie K., Han J., Chan B.M., Kim S.O. Cathepsin B is involved in the trafficking of TNF-alpha-containing vesicles to the plasma membrane in macrophages. J. Immunol. 2008;181:690–697. doi: 10.4049/jimmunol.181.1.690. [DOI] [PubMed] [Google Scholar]
- Hamon Y., Legowska M., Hervé V., Dallet-Choisy S., Marchand-Adam S., Vanderlynden L., Demonte M., Williams R., Scott C.J., Si-Tahar M. Neutrophilic cathepsin C is maturated by a multistep proteolytic process and secreted by activated cells during inflammatory lung diseases. J. Biol. Chem. 2016;291:8486–8499. doi: 10.1074/jbc.M115.707109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom L. An overview of serine proteases. Curr. Protoc. Protein Sci. 2002;Chapter 21:10. doi: 10.1002/0471140864.ps2110s26. [DOI] [PubMed] [Google Scholar]
- Hoffmann J.A. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C.A., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kawalec M., Potempa J., Moon J.L., Travis J., Murray B.E. Molecular diversity of a putative virulence factor: purification and characterization of isoforms of an extracellular serine glutamyl endopeptidase of Enterococcus faecalis with different enzymatic activities. J. Bacteriol. 2005;187:266–275. doi: 10.1128/JB.187.1.266-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.R., James M.N. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A., Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2014;42:25–35. doi: 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz B., Moreau T., Gauthier F. Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie. 2008;90:227–242. doi: 10.1016/j.biochi.2007.10.009. [DOI] [PubMed] [Google Scholar]
- LaRock C.N., Todd J., LaRock D.L., Olson J., O’Donoghue A.J., Robertson A.A., Cooper M.A., Hoffman H.M., Nizet V. IL-1beta is an innate immune sensor of microbial proteolysis. Sci. Immunol. 2016;1:eaah3539. doi: 10.1126/sciimmunol.aah3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Reichhart J.M., Hoffmann J.A. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina E.A., Langley E., Green C., Gubb D., Ashburner M., Hoffmann J.A., Reichhart J.M. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- Liberati N.T., Urbach J.M., Miyata S., Lee D.G., Drenkard E., Wu G., Villanueva J., Wei T., Ausubel F.M. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P., Pelte N., Hoffmann J.A., Reichhart J.M. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297:114–116. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- Liszewski M.K., Kolev M., Le Friec G., Leung M., Bertram P.G., Fara A.F., Subias M., Pickering M.C., Drouet C., Meri S. Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity. 2013;39:1143–1157. doi: 10.1016/j.immuni.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse R., Jakobs-Schönwandt D., Patel A.V. Screening of liquid media and fermentation of an endophytic Beauveria bassiana strain in a bioreactor. AMB Express. 2014;4:47. doi: 10.1186/s13568-014-0047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero A., Duquerroy S., Trapani S., Goulas T., Guevara T., Andersen G.R., Navaza J., Sottrup-Jensen L., Gomis-Rüth F.X. The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew. Chem. Int. Ed. Engl. 2012;51:3340–3344. doi: 10.1002/anie.201108015. [DOI] [PubMed] [Google Scholar]
- Martinon F., Mayor A., Tschopp J. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.A., Jr. Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- Ming M., Obata F., Kuranaga E., Miura M. Persephone/Spätzle pathogen sensors mediate the activation of Toll receptor signaling in response to endogenous danger signals in apoptosis-deficient Drosophila. J. Biol. Chem. 2014;289:7558–7568. doi: 10.1074/jbc.M113.543884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme N.T., Liégeois S., Kele B., Giammarinaro P., Pradel E., Hoffmann J.A., Ewbank J.J., Ferrandon D. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 2007;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K., Naruto M., Nakaki T., Sano E. Identification of interleukin-8 converting enzyme as cathepsin L. Biochim. Biophys. Acta. 2003;1649:30–39. doi: 10.1016/s1570-9639(03)00152-3. [DOI] [PubMed] [Google Scholar]
- Park B., Brinkmann M.M., Spooner E., Lee C.C., Kim Y.M., Ploegh H.L. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Jiang H., Kanost M.R., Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Sadegh-Nasseri S., Kim A. Exogenous antigens bind MHC class II first, and are processed by cathepsins later. Mol. Immunol. 2015;68(2 Pt A):81–84. doi: 10.1016/j.molimm.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbielle C., Moreau S., Veillard F., Voldoire E., Bézier A., Mannucci M.A., Volkoff A.N., Drezen J.M., Lalmanach G., Huguet E. Identification of parasite-responsive cysteine proteases in Manduca sexta. Biol. Chem. 2009;390:493–502. doi: 10.1515/BC.2009.061. [DOI] [PubMed] [Google Scholar]
- Srinivasan N., Gordon O., Ahrens S., Franz A., Deddouche S., Chakravarty P., Phillips D., Yunus A.A., Rosen M.K., Valente R.S. Actin is an evolutionarily-conserved damage-associated molecular pattern that signals tissue injury in Drosophila melanogaster. eLife. 2016;5:e19662. doi: 10.7554/eLife.19662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B.E. Manipulation of host signalling pathways by anthrax toxins. Biochem. J. 2007;402:405–417. doi: 10.1042/BJ20061891. [DOI] [PubMed] [Google Scholar]
- Valanne S., Wang J.H., Rämet M. The Drosophila Toll signaling pathway. J. Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 2007;13:387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- Veillard F., Troxler L., Reichhart J.M. Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation. Biochimie. 2016;122:255–269. doi: 10.1016/j.biochi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Weber A.N., Tauszig-Delamasure S., Hoffmann J.A., Lelièvre E., Gascan H., Ray K.P., Morse M.A., Imler J.L., Gay N.J. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat. Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- Wood J.P., Silveira J.R., Maille N.M., Haynes L.M., Tracy P.B. Prothrombin activation on the activated platelet surface optimizes expression of procoagulant activity. Blood. 2011;117:1710–1718. doi: 10.1182/blood-2010-09-311035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.S., Nam H.J., Seo M., Han S.K., Choi Y., Nam H.G., Lee S.J., Kim S. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE. 2011;6:e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q., Fu T.M., Li J., Wu H. Structural biology of innate immunity. Annu. Rev. Immunol. 2015;33:393–416. doi: 10.1146/annurev-immunol-032414-112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S., Reis e Sousa C. Adaptive immunity after cell death. Trends Immunol. 2013;34:329–335. doi: 10.1016/j.it.2013.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.