Abstract
tRNA maturation and quality control are crucial for proper functioning of these transcripts in translation. In several organisms, defective tRNAs were shown to be tagged by poly(A) or CCACCA tails and subsequently degraded by 3′-exonucleases. In a deep-sequencing analysis of tRNA 3′-ends, we detected the CCACCA tag also in Escherichia coli. However, this tag closely resembles several 3′-trailers of tRNA precursors targeted for maturation and not for degradation. Here, we investigate the ability of two important exonucleases, RNase R and RNase T, to distinguish tRNA precursors with a native 3′-trailer from tRNAs with a CCACCA tag. Our results show that the degrading enzyme RNase R breaks down both tRNAs primed for degradation as well as precursor transcripts, indicating that it is a rather nonspecific RNase. RNase T, a main processing exonuclease involved in trimming of 3′-trailers, is very inefficient in converting the CCACCA-tagged tRNA into a mature transcript. Hence, while both RNases compete for trailer-containing tRNA precursors, the inability of RNase T to process CCACCA tails ensures that defective tRNAs cannot reenter the functional tRNA pool, representing a safeguard to avoid detrimental effects of tRNAs with erroneous integrity on protein synthesis. Furthermore, these data indicate that the RNase T-mediated end turnover of the CCA sequence represents a means to deliver a tRNA to a repeated quality control performed by the CCA-adding enzyme. Hence, originally described as a futile side reaction, the tRNA end turnover seems to fulfill an important function in the maintenance of the tRNA pool in the cell.
Keywords: tRNA decay, tRNA maturation, tRNA quality control, RNase T, RNase R
INTRODUCTION
Transfer RNAs (tRNAs) play a crucial role as adapter molecules that translate the genetic information of mRNA into protein sequence. These essential transcripts are synthesized as precursor molecules that are subjected to a series of processing events, ranging from removal of 5′-leader and 3′-trailer regions, addition of the 3′-terminal CCA end, excision of introns (if present) to modification of certain nucleosides and (in eukaryotes) export of the mature tRNA into the cytoplasm (Betat et al. 2010; Hopper 2013).
In E. coli, many of the tRNA maturation steps are well-characterized. The 5′-leader cleavage of the precursor tRNA is achieved by RNase P, and multiple endo- and exoribonucleases are involved in the removal of the 3′-trailer. The CCA triplet is already encoded in all tRNA genes. Hence, the CCA-adding enzyme is not required for maturation, but contributes to efficient cell growth by restoring damaged CCA ends (Deutscher et al. 1977; Zhu and Deutscher 1987; Lizano et al. 2008). In the 3′-trailer processing, the initial endonucleolytic cleavage is performed by RNase E (Li and Deutscher 2002), followed by the subsequent action of exonucleolytic RNases T, PH, D, II, and BN to trim the tRNA 3′-trailer to its mature terminus (Reuven and Deutscher 1993). At present, the detailed mechanism regarding the concerted action of the participating RNases is not fully understood. Evidently, most of these RNases seem to have redundant activities, as an E. coli strain lacking up to four of these five RNases is still viable (Padmanabha and Deutscher 1991; Kelly and Deutscher 1992; Reuven and Deutscher 1993). However, RNase T and RNase PH are described as the primary enzymes responsible for short 3′-trailer removal (Li and Deutscher 1996).
RNase T is a member of the DEDD 3′–5′ exonuclease superfamily and was initially discovered in the context of tRNA CCA end turnover in E. coli (Deutscher et al. 1984; Deutscher and Marlor 1985). In contrast to RNase PH and other exonucleases involved in tRNA 3′-end processing, RNase T is the only enzyme that efficiently (and repeatedly) removes the terminal A residue of the CCA terminus, leaving a tRNA with an incomplete and nonfunctional 3′-CC end which is subsequently repaired by the CCA-adding enzyme (Deutscher et al. 1977, 1984). However, a functional relevance of this futile cycle has not been described so far. RNase T is single-strand specific and strongly inhibited by 3′-terminal C residues in the trailer sequence (Zuo and Deutscher 2002). The unusual substrate specificity has been elucidated mechanistically, and a series of crystal structures unraveled that a substrate RNA containing a 3′-terminal CMP induces a conformational change within the binding pocket that moves the scissile phosphate away from the active site (Hsiao et al. 2011, 2012).
Another E. coli exonuclease acting on tRNA 3′-ends is RNase R. However, this RNase is not involved in tRNA processing, but degradation. So far, this enzyme is the only known exoribonuclease able to digest complex secondary structures in vitro if a 3′-overhang of at least 7 nucleotides (nt) is present (Vincent and Deutscher 2006, 2009a,b; Matos et al. 2009). It has been suggested that RNase R is involved in tRNA quality control and decay (Li et al. 2002; Wilusz et al. 2011).
In general, several control mechanisms monitoring functional and structural integrity of tRNAs have been identified. Unstable tRNA precursors are prone to short poly(A) tailing by poly(A) polymerase, rendering these transcripts susceptible for degradation (Li et al. 2002; Mohanty et al. 2012; Mohanty and Kushner 2013). tRNAs lacking intact sugar–phosphate backbone have been shown to be deprived of efficient CCA addition, suggesting a role of the CCA-adding enzyme in kinetically controlled proofreading (Dupasquier et al. 2008). Moreover, the participitation of eukaryotic, bacterial, and archaeal CCA-adding enzymes in tRNA quality control has been extended toward an unprecedented proofreading mechanism for tRNAs with unstable or damaged acceptor stems (Wilusz et al. 2011).
In a crystallographic analysis, an archaeal CCA-adding enzyme was shown to initiate an on-enzyme refolding of tRNA substrates with structurally unstable acceptor stems. A torque-like motion against the 3′-end induces an isomerization step within the acceptor stem, presenting the terminal A residue as an alternative discriminator position, which in turn enables a second round of CCA incorporation (Wilusz et al. 2011; Betat and Mörl 2015; Kuhn et al. 2015). Such a 3′-CCACCA tag facilitates subsequent tRNA degradation by exonucleolytic activities (Wilusz et al. 2011). In E. coli, the aforementioned RNase R has been suggested as the responsible nuclease for the CCACCA-facilitated tRNA decay (Matos et al. 2009; Vincent and Deutscher 2006, 2009b; Wilusz et al. 2011).
As tRNA maturation and decay are opposing events, they have to be discriminated carefully in order to maintain cell homeostasis intact. However, when all of these tRNA pathways are active simultaneously within the cell, the question arises as to how E. coli exoribonucleases discriminate between a precursor tRNA (that carries the trailer sequence downstream from the encoded CCA end) and an unstable tRNA tagged for degradation by the CCA-adding enzyme (tRNA–CCACCA).
Using deep sequencing tailored to specifically resolve the 3′ tRNA end, we identified a small but significant portion of CCACCA-tagged tRNAs in E. coli. Hence, we examined the in vitro activities of RNase T and RNase R, two major contributors to tRNA maturation and tRNA degradation. Our data suggest that the discrimination between the CCACCA degradation tag and a highly similar trailer sequence is guided by the substrate specificity of RNase T, allocating this RNase to an indirect contributor to quality control. With its unique activity in the repeated turnover of the terminal A residue of regular CCA ends, RNase T induces a permanent monitoring of correct tRNA structures.
RESULTS
CCACCA-extended tRNAs are detected in E. coli cells
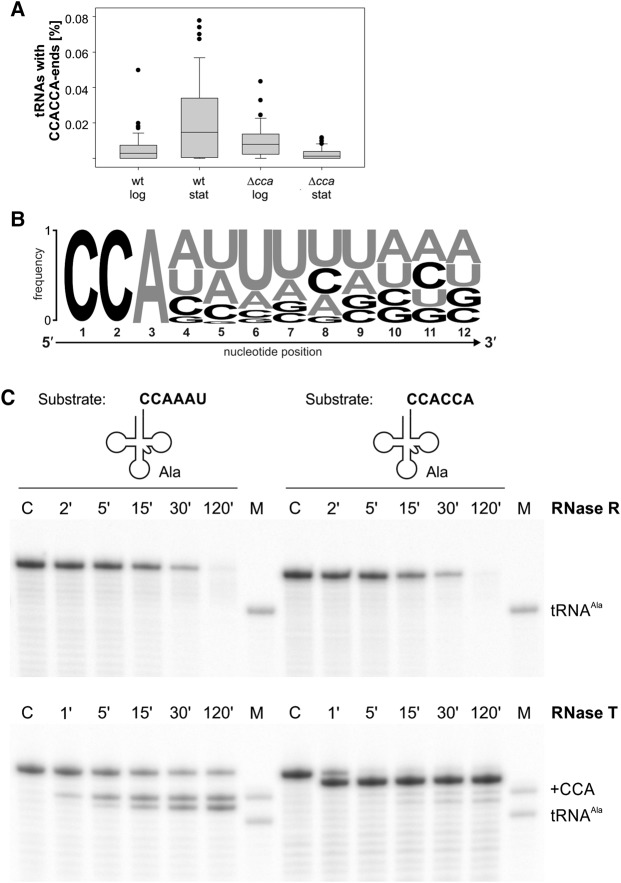
To address whether and at which conditions tRNA 3′-CCA ends are extended by an additional CCA triplet in E. coli in a manner similar to the observed 3′-terminal extensions in S. cerevisiae (Wilusz et al. 2011), we determined the 3′-termini of the tRNAs from E. coli CA244 cells at both exponential and stationary phase by means of deep sequencing. To elucidate the effect of the CCA-adding enzyme, as it is most likely extending the CCA termini, we also sequenced the tRNAome of the strain CA244 Δcca (lacking the CCA-adding enzyme).
The biological replicates showed high reproducibility (Supplemental Fig. S1A–D), allowing for combining the two sequencing data sets into a metaset. For each tRNA isoacceptor, the total mapped reads with CCACCA extensions are presented as a fraction of all reads of the corresponding tRNA isoacceptor. Under both growth conditions, we detected low but reproducible amounts of tRNAs with extended CCACCA ends. In the stationary phase, the amount of tRNA–CCACCA increased to 0.02% on average, with a maximum of 0.08% for some isoacceptors. These values are in the range previously observed in wildtype yeast (e.g., 0.01%–0.02% after 6 h of growth at 37°C) (Wilusz et al. 2011), while they were far less prevalent in the strain lacking the CCA-adding enzyme, supporting a role of this enzyme in generating the CCACCA extension (Fig. 1A). To rule out any methodological bias during library preparation, we permuted the second CCA motif and quantified the fractions carrying those extensions, i.e., CCACAC and CCAACC (Supplemental Fig. S1E, F). Thereby, we found only a marginal number of reads which scored one to three orders of magnitude lower than the tRNA–CCACCA reads. This supports the confidence in detecting the CCACCA extensions despite their low abundance.
FIGURE 1.
Abundance and recognition of tRNA CCACCA extensions as well as 3′-trailer sequences in E. coli. (A) Boxplots of the fraction of E. coli CA244 wt and Δcca tRNAs during exponential (log) and stationary (stat) growth carrying CCACCA extensions. Values are given as a fraction of respective tRNA subpopulations normalized to the total tRNA reads for each isoacceptor. (B) Nucleotide frequencies within the 3′-trailer region of the E. coli K12 tRNA genes starting from the CCA triplet (weblogo.berkeley.edu). Letter height correlates with abundance; A and U residues are highlighted in gray. (C) Radiolabeled E. coli tRNAAla with either its native 3′-trailer (CCAAAU, left) or the corresponding degradation tag (CCACCA, right) was incubated with either RNase R (upper panel, 20 ng µL−1) or RNase T (lower panel, 0.6 ng µL−1). Assays were carried out in a 40 µL total reaction volume with 5 µM tRNA substrate. C, control reactions without enzyme incubated for the maximal reaction time; M, size marker tRNAAla, 73 nt.
3′-Overhangs of tRNAs are differently recognized by E. coli RNases R and T
Correct 3′-processing is one of the essential steps to yield mature tRNAs. The processing machinery has to deal with an amount of different tRNA 3′-trailer sequences and distinguish them from degradation tags (Li and Deutscher 1996; Wilusz et al. 2011). To investigate the sequence distribution of the 3′-trailer region of all E. coli tRNA genes, we generated an RNA sequence logo that summarizes the overall position-specific abundance of individual nucleotides (Fig. 1B; Crooks et al. 2004). A and U residues occur with high frequency at the first positions downstream from the CCA triplet, whereas C and G residues are underrepresented.
To address how E. coli exoribonucleases discriminate between a tRNA precursor to be processed and an unstable tRNA that has been tagged for degradation, we investigated the substrate requirements of the two key RNases in tRNA degradation and maturation, focusing on the native trailer sequence, and compared it to the 3′-CCACCA degradation tag. To this end, we overexpressed and purified His-tagged versions of E. coli RNase T and RNase R using Ni2+ affinity chromatography followed by gel filtration (Supplemental Fig. S2). The activity of both enzymes was tested in a comparative in vitro analysis using radioactively labeled in vitro transcripts of either an E. coli precursor tRNAAla(GGC)–CCAAAU, carrying the native AU-rich trailer, or the corresponding tRNA with the CCACCA degradation tag. Whereas RNase R degrades both RNA sequences at equal efficiency, RNase T shows different and rather trailer-dependent activities (Fig. 1C). The latter removed the native trailer efficiently, and in about 50% of the transcripts, even the last A residue of the conserved CCA end was cut off—a frequently observed side reaction of this enzyme, described as tRNA 3′-end turnover (Deutscher et al. 1984, 1985; Zuo and Deutscher 2002; Hsiao et al. 2012). With tRNAAla(GGC)–CCACCA, however, RNase T only cuts off the terminal A residue of the CCACCA tag, which again corresponds to the side reaction on the standard CCA end (Fig. 1C). Hence, RNase T activity seems to depend much more on the composition of the 3′-trailer sequence than RNase R.
RNase R has no pronounced specificity toward CCACCA
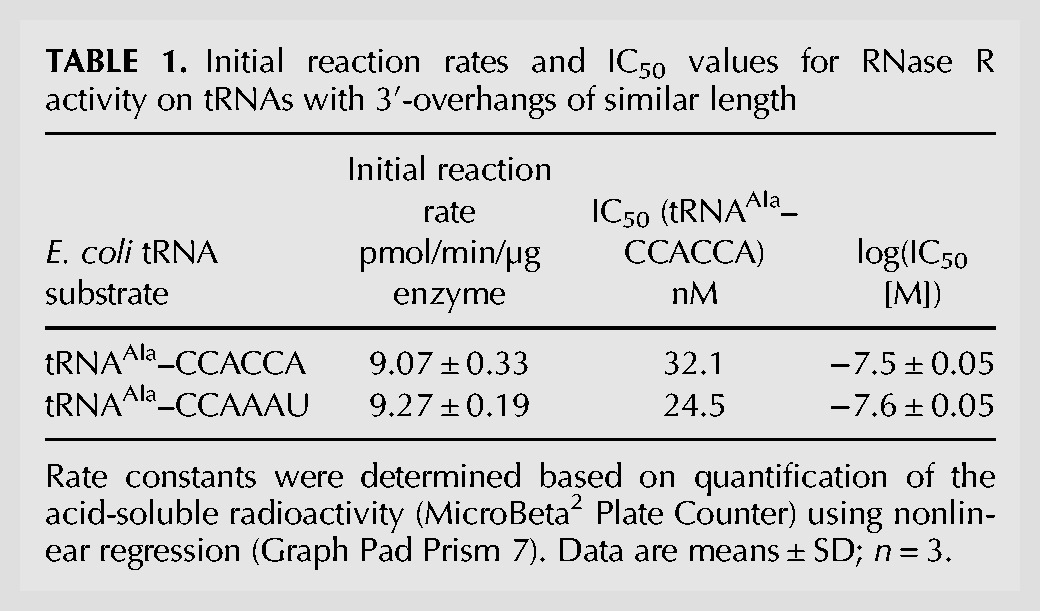
To further analyze RNase R-mediated degradation of tRNAs with native trailer or CCACCA tail, we determined the reaction rates by monitoring the release of radioactively labeled nucleotides from an internally 32P-labeled tRNA during degradation (Fig. 2A). It turned out that the initial reaction rates for the degradation of both tested tRNAs were identical within error margin (9.07 ± 0.33 pmol min−1 µg−1 for tRNAAla–CCACCA and 9.27 ± 0.19 pmol min−1 µg−1 for tRNAAla–CCAAAU, respectively; Table 1). A similar result was obtained in a competition analysis addressing the potential of tRNAAla–CCAAAU to interfere with tRNAAla–CCACCA degradation (Fig. 2B). Here, a constant amount of internally labeled tRNAAla–CCACCA was incubated with RNase R under nonsaturating conditions and increasing concentrations of nonlabeled tRNAAla–CCACCA or tRNAAla–CCAAAU, respectively. The IC50 values, representing the half maximal inhibitory concentration of the competing tRNA, are 24.5 nM for tRNAAla–CCAAAU compared to 32.1 nM for tRNAAla–CCACCA (Table 1). Because of these highly similar competitor effects, we reason that a tRNA with a CCACCA degradation tag will not be recognized preferentially by RNase R.
FIGURE 2.
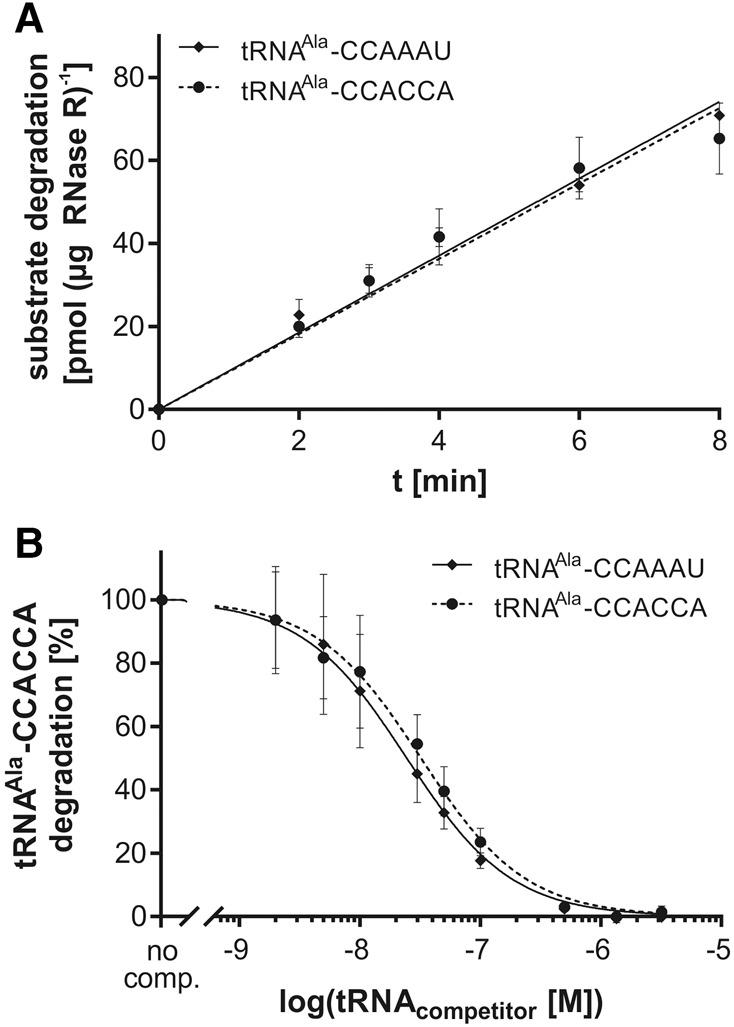
RNase R activity on tRNA substrates with 3′-overhangs of similar length. (A) Time-dependent degradation of internally labeled tRNAAla–CCACCA and tRNAAla–CCAAAU yield almost identical curves, implying that RNase R does not discriminate between the two 3′-overhangs; n = 3. (B) Competition studies with tRNAAla–CCACCA and tRNAAla–CCAAAU of the RNase R catalyzed reaction show identical inhibition curves, indicating that the enzyme accepts both substrate overhangs independent of their sequence composition. No comp., control reaction in the absence of competitor. Data are means ± SD; n = 3.
TABLE 1.
Initial reaction rates and IC50 values for RNase R activity on tRNAs with 3′-overhangs of similar length
RNase T discriminates between tRNA 3′-overhangs
The first analysis of the tRNA-maturating RNase T revealed that tRNAs with different 3′-trailer sequences were accepted as substrates to a different extent (Fig. 1C), and we detected a distinct product band pattern representing the processing activity of RNase T. Concerning the particular product formation, our data show that RNase T readily digested its natural precursor tRNA substrate, resulting in a mature tRNA–CCA (and the 3′-end turnover product tRNA–CC). In contrast, a tRNA–CCACCA was not recognized accordingly. However, the terminal AMP of this tag was instantly trimmed, while the following sequence CCACC was barely acted upon. This raises the question as to what extent RNase T is able to mature a tRNA that is actually primed for degradation. Therefore, we performed steady-state kinetics to evaluate the capability of RNase T to convert a tRNA–CCACCA substrate into a tRNA that is able to reenter the functional tRNA pool in the cell. Our initial tests showed that the sequence composition of the tRNA trailer has a considerable impact on the substrate preference (Fig. 3A) and, consequently, reaction velocity of RNase T. As a result, a tRNA substrate with a favored trailer sequence was rapidly degraded to the CCA end. However, on a transcript carrying a trailer sequence that is rather inefficiently removed (like the CCACCA tag), the degradation of the C residues represents the rate-limiting step, and the removal of the following A residue was considerably faster. As a consequence, after CC degradation, the enzyme efficiently removed also the terminal A residue of the CCA end. Hence, the trailer composition determines the nature of the main reaction products that are monitored in the kinetic analysis. For tRNA–CCAAAU (natural trailer), the main reaction product to be quantified was tRNA–CCA, and for tRNA–CCACCA (degradation tag), the main reaction product resulting from RNase T action was tRNA–CC (Fig. 3; Supplemental Fig. S3).
FIGURE 3.
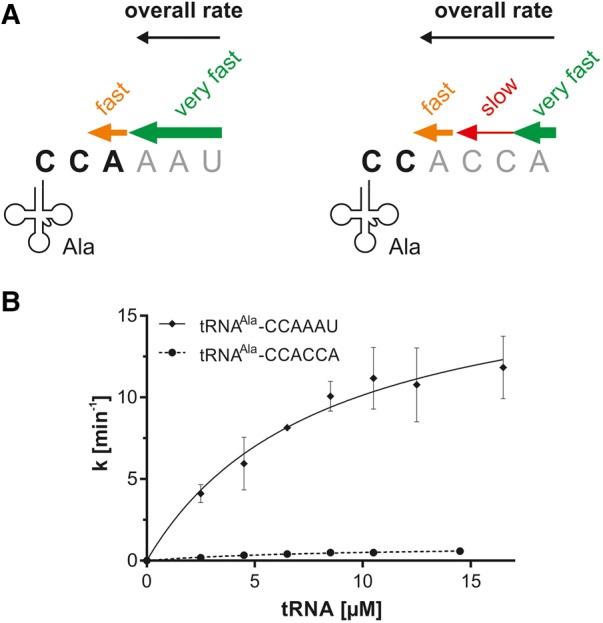
RNase T kinetics on tRNA substrates with 3′-overhangs of similar length. (A) Our initial experiments showed that tRNAs are degraded to different extents, depending on the sequence of the 3′-extension (reaction velocity is indicated by size and color of the individual arrows; experimental data are not shown). For the kinetic analysis, main reaction products able to reenter the functional tRNA pool were taken into account (3′-end in bold characters). tRNA–CCAAAU with the natural trailer (left) is rapidly degraded to the CCA end as the main reaction product (green arrow) that was analyzed in the kinetic analysis. The subsequent 3′-end turnover (removal of the terminal A residue, orange arrow) is slower and therefore was not considered. Individual bases of the degradation tag CCACCA are removed at different reaction velocities (right). Although the terminal A residue of the CCACCA sequence is also removed very fast (green arrow), degradation of the two following C residues is very slow (red arrow) and corresponds to the rate-limiting step of the reaction. The subsequent 3′-end turnover (removal of the terminal A residue of the resulting CCA end, orange arrow) is then removed very efficiently. Hence, for this tRNA, tRNA–CC represents the resulting main reaction product for the kinetic analysis. (B) Kinetic analysis of tRNAAla–CCAAAU trailer removal by RNase T. The CCACCA tag is nearly not converted. k is the reaction rate normalized to the total enzyme concentration. Data are means ± SD; n = 3–4. A different scale of the kinetics of the CCACCA removal is displayed in Supplemental Figure S2.
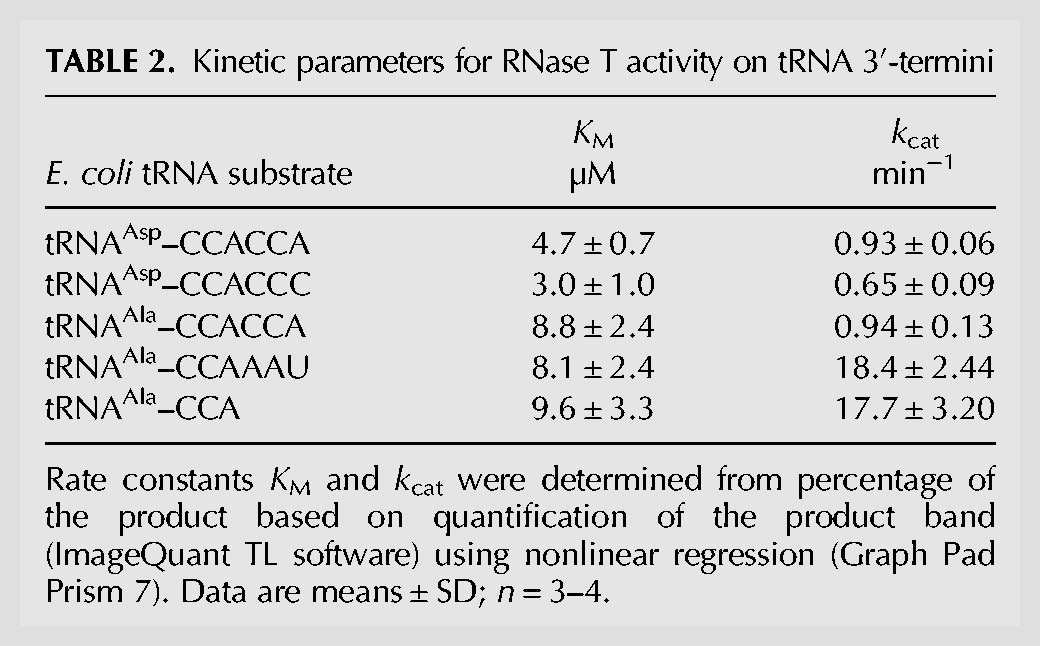
Increasing concentrations of tRNA substrates were incubated with RNase T and separated on high resolution denaturing polyacrylamide gels. Product formation was determined by densitometric quantification of the bands corresponding to the main reaction products tRNA–CCA and tRNA–CC, respectively. For comparison, E. coli precursor tRNAAsp(GUC)—one of three tRNAs with a native short trailer sequence devoid of A and U residues out of a total of 88 E. coli tRNAs—was included in the studies (tRNAAsp(GUC)–CCACCC or –CCACCA, respectively, Supplemental Fig. S3). For all tested tRNA substrates, we obtained apparent KM values in a comparable range from 3.0 to 8.8 µM (Table 2). In contrast, the sequence composition of the tRNA trailer had a considerable impact on the turnover number kcat (Table 2; Fig. 3). The conversion rate of the natural precursor tRNAAla–CCAAAU was 18.4 ± 2.44 min−1, a value that is almost 20-fold higher than the kcat measured for tRNAAla–CCACCA (0.94 ± 0.13 min−1). RNase T strongly discriminates between tRNA 3′-overhangs without C residues and the degradation tag CCACCA. As a result, RNase T-mediated conversion of a tRNA–CCACCA (labeled for degradation) into a mature tRNA is rather inefficient. Similarly, tRNAAsp–CCACCC, comprising a rare trailer sequence, is a poor substrate for RNase T (Table 2).
TABLE 2.
Kinetic parameters for RNase T activity on tRNA 3′-termini
RNase T catalysis includes the tRNA 3′-end turnover
Our assays with RNase T revealed an efficient formation of the product tRNA–CC, containing an incomplete CCA terminus (as shown exemplarily in Fig. 4A), which represents a typical and unique feature of RNase T that is described as tRNA end turnover (Deutscher et al. 1984, 1985). Since kinetic parameters allow estimating how RNase T acts on functional and mature tRNAs ending with the CCA triplet compared to their precursor counterparts, we included this activity in our kinetic analysis. The turnover of the model substrate tRNAAla(GGC)–CCA was investigated under steady-state conditions as described above (Fig. 4B). Notably, the apparent kinetic parameters demonstrate that the turnover of the terminal AMP was in a similar range as that of its corresponding tRNA with the common trailer sequence (tRNA–CCAAAU), regarding both KM (9.6 ± 3.2 µM) and kcat (17.7 ± 3.20 min−1, Table 2). Hence, these data corroborate that both RNase T-catalyzed reactions, i.e., the tRNA–CCA end turnover and the trimming of AU-rich 3′-trailer sequences, are highly efficient and proceed at similar rates in E. coli.
FIGURE 4.
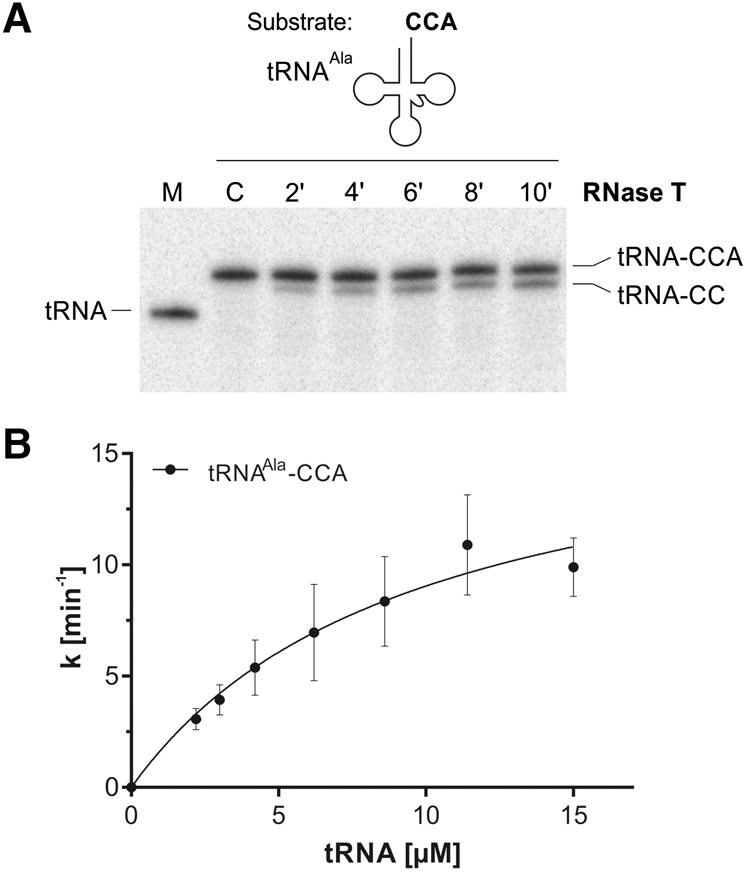
RNase T-catalyzed tRNA end turnover. (A) Time-dependent 3′-end turnover of a 5′-end labeled tRNAAla transcript ending with the CCA triplet. RNase T specifically catalyzes the removal of the terminal A residues and leaves the CC sequence intact. C, control reactions incubated without enzyme for the maximum reaction time; M, size marker tRNAAla, 73 nt. (B) Apparent kinetics of tRNA 3′-end turnover. Increasing amounts of tRNAAla–CCA were incubated with RNase T under steady-state conditions and product formation was determined by quantifying the intensity of the tRNAAla–CC band. Michaelis–Menten parameters were determined using GraphPadPrism software (Table 2); k is the reaction rate normalized to the total enzyme concentration. Data are means ± SD; n = 3.
DISCUSSION
As adapters connecting mRNA sequence with amino acid sequence in translation, tRNAs fulfill an essential role in the cell. Accordingly, these transcripts are subjected to a rigorous quality control and nonfunctional tRNAs are subsequently removed from the cellular tRNA pool. In E. coli, it was demonstrated that poly(A) polymerase is involved in this process by adding short poly(A) tails to defective tRNAs (Li et al. 2002; Mohanty et al. 2012). In 2011, a new tag sequence, CCACCA, for the specific decay of tRNAs was described (Wilusz et al. 2011). CCA-adding enzymes from all kingdoms of life are able to add a second CCA triplet to the 3′-end of a tRNA and triage it for degradation (Wilusz et al. 2011; Betat and Mörl 2015). While RNase R of E. coli is able to degrade such transcripts (Vincent and Deutscher 2006), it was unclear how the cell discriminates between a tRNA tagged for decay and a tRNA precursor that carries a 3′-trailer of similar length and composition that is subjected to final maturation. Furthermore, and similar to S. cerevisiae (Wilusz et al. 2011), our deep sequencing-based analysis proves a small but persistent occurrence of CCACCA-tagged tRNAs, suggesting that the mechanism is common also in E. coli (Fig. 1A). To shed light on this conundrum, we investigated the substrate specificity of the two main exonucleases acting on tRNA 3′-ends in tRNA decay and maturation, RNase R and RNase T.
The degrading exoribonuclease RNase R does not discriminate between a tRNA tagged for degradation and a precursor tRNA
RNase R is a nonspecific, 3′–5′-processive exonuclease that is able to degrade extensively structured RNAs (Cheng and Deutscher 2002). The major substrate requirement for RNase R activity is a single-stranded 3′-overhang of at least 7 nt that serves as a binding site for the enzyme to initiate degradation (Vincent and Deutscher 2006, 2009b). In a tRNA carrying a CCACCA tail as a tag for degradation, this sequence including the unpaired discriminator position fulfills this prerequisite. Hence, this tag or trailer sequences of similar length in a tRNA precursor should both represent proper substrates for RNase R-mediated degradation. The corresponding time series with a tRNA tagged with CCACCA and a tRNA precursor with a 6-nt long 3′-trailer (Figs. 1C and 2A) indicate no specificity of this enzyme toward the CCACCA-tagged tRNA. This is further corroborated by the competition experiment with tRNA–CCACCA (tag) and tRNA–CCAAAU (trailer) transcripts (Fig. 2B). As reported earlier, however, when tested on an RNA duplex with a homopolymeric 3′-overhang of 10 nt, RNase R exhibits a certain preference for A residues (Vincent and Deutscher 2006). Yet, this might be the case for longer 3′-overhangs, but we found that changes of 2–3 nt within the 7 nt overhang do not have an impact on RNase R activity.
Taken together, these observations suggest that RNase R, which exhibits almost no sequence specificity, recognizes both trailer and degradation tags of tRNAs and does not discriminate against transcripts to be processed into functional tRNAs and those marked for degradation. Hence, RNase R may have a considerable impact in the interplay between tRNA maturation and degradation.
The degradation tag CCACCA protects tRNAs from being converted into mature tRNAs by RNase T
In contrast to RNase R, the maturation enzyme RNase T shows a high sequence dependence in the removal of 3′-trailers. The time series experiment on a trailer-containing tRNA with CCAAAU overhang and a CCACCA-tagged tRNA show very different main reaction products (Fig. 1C). On the trailer tRNA, RNase T efficiently removes the AAU sequence as well as the terminal A residue of the CCA end and stops at the remaining CC residues. On the CCACCA-tagged tRNA, RNase T only removes the terminal A residue and stops at CCACC. These data together with our kinetic analysis are in agreement with the described C-effect of RNase T, where 3′-terminal C and CC positions dramatically block trailer removal, as they induce a disruptive conformational change within the binding pocket of the enzyme (Zuo and Deutscher 2002; Hsiao et al. 2011, 2012; Duh et al. 2015). Since RNase T action is nonprocessive and the enzyme frequently dissociates from its substrates (Zuo and Deutscher 2002), we presume that in vivo, the terminal AMP is readily reincorporated by CCA-adding enzyme, as previously observed with minihelix substrates containing 3′-CCACC (Kuhn et al. 2015).
The apparent KM values for RNase T catalysis are in a 3–9 µM range, indicating rather similar affinities to the different tRNA substrates (Table 2; Fig. 3). The kcat values, however, differ up to 28-fold, with the poorest turnover of tRNAAsp–CCACCC (trailer; kcat = 0.65 min−1), tRNAAsp–CCACCA (tag; kcat = 0.93 min−1), and tRNAAla–CCACCA (tag; kcat = 0.94 min−1). Hence, independent of the tRNA identity, the CCACCA tag is a very poor substrate for RNase T, and the only noticeable reaction is the removal of the terminal A residue of the CCACCA tag. Hence, this tag protects tRNAs selected for degradation from unwanted maturation and prevents such transcripts from reentering the pool of functional tRNAs in translation. In light of RNase T substrate specificity, it is quite surprising that the E. coli genome contains some tRNA genes with C-rich trailers at all, though at a rather low abundance (Fig. 1B). In the case of tRNAAsp, the other gene copies carry pyrimidine-rich trailers CCACTT and CCACTA, respectively, that might also interfere with RNase T-mediated maturation. However, as the sequences for the tRNA body are identical in these three genes, it is impossible to discriminate or quantify the processing efficiencies of the individual transcripts. Yet, as tRNA 3′-end processing is carried out by redundant activities in E. coli, it is likely that other RNases with different specificities are responsible for trimming of C-containing trailer sequences, such as RNase PH, RNase D, RNase BN, RNase II, or PNPase. As such tRNA precursors accumulate in RNase PH-deficient E. coli strains, it seems that RNase PH is the predominant activity responsible for an efficient removal of such trailers (Li and Deutscher 1996).
Our findings suggest that RNase T, as a major maturation enzyme in E. coli, is not involved in the recognition of tRNAs with a CCACCA degradation tag. In vivo, this discrimination against the C-rich tag is vitally important to prevent tRNAs tagged for degradation from transforming into mature transcripts that could reenter the translationally active tRNA pool of a cell. Accordingly, the CCACCA tag is not only a tag for degradation but also equally protects tRNAs from being trimmed by the processing enzyme RNase T.
tRNA quality control: an interplay between RNase R and RNase T
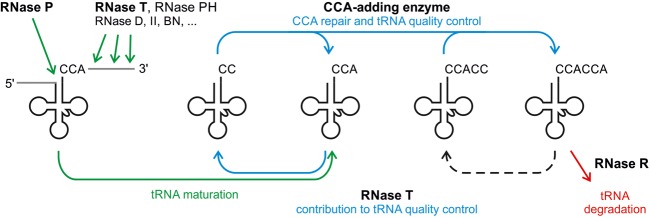
RNase T was originally described as a mere RNA processing enzyme (Reuven and Deutscher 1993; Li and Deutscher 1995; Li et al. 1998). However, our results, together with the reported scrutinizing function of the CCA-adding enzyme (Wilusz et al. 2011; Kuhn et al. 2015), indicate that RNase T is also contributing to the tRNA quality surveillance in E. coli in two ways (Fig. 5). First, it is the only RNase that does not stop at the CCA terminus, but readily removes the terminal A residue of this essential triplet. Such truncated CCA ends are no longer functional, but are restored by the CCA-adding enzyme, representing a process called tRNA 3′-end turnover (Fig. 4; Deutscher et al. 1984, 1985). The biological reason for this turnover remained rather unclear, but with the discovery of the quality control executed by the same enzyme (Wilusz et al. 2011), a new function can be assigned to it. As the CCA triplet is already encoded in all E. coli tRNA genes, the end turnover represents the sole opportunity to subject the tRNA pool to this type of quality control. In this scenario, RNase T activity guarantees that over time, a large part of the tRNA pool is repeatedly subjected to this quality control system.
FIGURE 5.
Interlocked quality control and processing of tRNAs in E. coli. After transcription, a set of endo- and exonucleases remove the 5′-leader and 3′-trailer sequences of the tRNA precursor (green). RNase T is a major processing enzyme in 3′-trailer removal. The resulting mature tRNA–CCA is repeatedly subjected to quality control (blue). RNase T removes the terminal A residue, resulting in tRNA–CC (end turnover). This truncated tRNA is then scrutinized by the CCA-adding enzyme. If the tRNA is intact, the enzyme completes the CCA end, releasing a functional tRNA. If the tRNA is damaged or unstable, the torque control mechanism of the CCA-adding enzyme leads to a refolding of the acceptor stem so that a second CCA triplet is added, and the tRNA is tagged by CCACCA for RNase R-mediated degradation (red). An occasional loss of the terminal A due to RNase T activity (dashed arrow) leads to a CCACC tag that is either reconverted into CCACCA (CCA-adding enzyme) or polyadenylated [poly(A) polymerase; not shown]. As a result, both products carry a 3′-tag long enough for being recognized by RNase R.
A second contribution of RNase T to tRNA quality control is based on its substrate specificity. While its preference for A-rich trailers might interfere with the poly(A) tail-mediated tRNA quality control described above, the inefficient removal of C residues ensures that CCACCA-tagged tRNAs are not further processed into mature transcripts that reenter the functional tRNA-pool. As RNase T efficiently removes 3′-terminal A residues, the CCACCA tail can be converted into a CCACC sequence (Fig. 1C). The discriminator base and this shortened tag represent only a 6-nt single strand and such a tRNA is a rather bad substrate for RNase R (Vincent and Deutscher 2006). Hence, this nonfunctional tRNA is not immediately degraded. As it is not recognized by the aminoacyl-tRNA synthetase, it remains uncharged and represents a rather inert tRNA transcript. Yet, its accumulation is not likely (and we did not observe it), as the second tRNA surveillance system, the poly(A) polymerase most likely sets in by adding a poly(A) tail (Li et al. 2002; Mohanty et al. 2012). As a consequence, the 3′-overhang is then long enough again for RNase R recognition and degradation. Hence, due to the individual substrate specificities of RNases T and R, such an elimination of CCACCA-tagged tRNAs represents a rather stochastic event. Polymerization (A-, CCA-, or CCACCA-addition) and degradation are constantly competing and running in parallel on the pool of tRNA substrates in the cell.
Taken together, this work suggests another layer of control to the tRNA surveillance system in E. coli. The tRNA 3′-end turnover catalyzed by RNase T and the CCA-adding enzyme is no longer regarded as a futile cycle, but represents an essential feature to scrutinize and monitor structural integrity of the tRNA pool in the cell. It is likely that a similar control mechanism exists in organisms of all three kingdoms of life. With two different degradation tags, poly(A) tail and CCACCA end, tRNAs seem to underlie a tight quality control. Yet, as the poly(A) tag can be efficiently removed by RNase T, it is very likely that such a tagged tRNA is not degraded, but can be processed into a mature transcript (Mohanty et al. 2012). In contrast, the CCACCA tag inhibits RNase T-mediated processing and keeps the tRNA on its path toward degradation. Thus, the combination of poly(A) polymerase, CCA-adding enzyme, and RNases with different substrate specificities represents an efficient interlocked control system that keeps the cell's tRNAs functional and intact.
MATERIALS AND METHODS
Growth conditions and RNA isolation
Escherichia coli CA244 wt and Δcca were grown at 37°C in LB medium (10 g L−1 NaCl, 10 g L−1 tryptone, and 5 g L−1 yeast extract). Cells were harvested in exponential (OD600 = 0.3) or stationary phase (OD600 = 3 for CA244 Δcca and OD600 = 5 for CA244 wt). Total RNA was isolated by hot-phenol extraction or TRIzol (Thermo Fisher Scientific). Full-length tRNA was purified by cutting corresponding bands from denaturing polyacrylamide gels and eluting overnight at 4°C with elution buffer (50 mM potassium acetate, 200 mM potassium chloride, and pH 7.0).
Deep sequencing of tRNAs
Total tRNA was 3′-dephosphorylated with T4 PNK (Thermo Fisher Scientific) for 45 min at 37°C and purified with the Clean and Concentrator Kit (Zymo Research). A sequencing adapter was ligated to the 3′-end of tRNAs using T4 RNA ligase 2 (NEB) for 2.5 h at 22°C. For preparation of the sequencing library, which contains 3′-terminal tRNA parts, the sample was subjected to random alkaline fragmentation for 20 min at 95°C in 100 mM NaHCO3 containing 2 mM EDTA pH 9.2. Subsequently, RNA fragments were 5′-phosphorylated with T4 PNK (NEB) and a second adapter was ligated to the 5′-termini with T4 RNA ligase 1 (NEB) at 22°C overnight. Using a primer complementary to the 3′-adapter, the RNAs were reverse transcribed into cDNA with Revert Aid H Minus RT (Thermo Fisher Scientific) for 1 h at 44°C. The RNA was degraded and the cDNA containing both adapters was amplified by PCR with Pfu DNA polymerase (Thermo Fisher Scientific). Amplicons in the range of 130–140 bp (i.e., 10–20 nt of the 3′-tRNA ends plus 120 nt adapters) were gel-purified and analyzed on 2100 Bioanalyzer (Agilent) and Qubit 3.0 (Thermo Fisher Scientific). cDNA libraries were sequenced on a HiSeq2000 (Illumina) machine. Sequenced reads were trimmed with fastx-toolkit (0.0.13.2; quality threshold: 20) and adapters were cleaved using cutadapt (1.2.1; minimal overlap: 1 nt). tRNA sequences were downloaded from the genomic tRNA database (Escherichia coli K12) and 10 nt upstream of the CCA tails were used for mapping of the sequencing reads. tRNA read counts were normalized by the depth of each sequencing library as read per million of mapped reads (rpM) (Mortazavi et al. 2008).
Cloning, overexpression, and purification of recombinant RNases
Coding regions of RNase T and RNase R were amplified by PCR from the genomic DNA of E. coli BL21 and inserted into the XhoI and NdeI sites of pET28a(+). Proteins were overexpressed with an N-terminal His-tag in E. coli BL21(DE3) pLys. Cells were grown at 37°C in 1.0–1.2 L LB supplemented with 34 μg mL−1 chloramphenicol and 30 μg mL−1 kanamycin to an OD600 of 0.8–1.0. Expression was induced by adding IPTG to a final concentration of 1 mM for 3 h at 37°C for RNase R or overnight at 18°C for RNase T expression. Cells were harvested and the pellet was stored at −80°C until further use.
For purification of the recombinant enzymes, all steps were performed at 4°C. The frozen pellet was thawed on ice and resuspended in binding buffer 25 mM Tris–HCl (pH 7.8), 300 mM KCl for RNase T, or 10 mM Tris–HCl (pH 7.6), 500 mM KCl, 10% glycerol for RNase R. The buffers for RNase T and RNase R were supplemented with 1 mg mL−1 lysozyme, 1 mM DTT and 0.1 mM PMSF or mini complete protease inhibitor cocktail from Roche, respectively. Cells were disrupted by sonication; the lysate was centrifuged at 16,000g for 1 h at 4°C. The resulting supernatant was applied to a HisTrapFF column using ÄKTA pure system (Amersham Biosciences) in the presence of 10 mM imidazole in the buffer, washed with 50 mM and eluted at 500 mM imidazole. The protein containing fractions were pooled and applied to a HiLoad 16/60 Superdex 200 (RNase R) or Superdex 75 column (RNase T). Fractions containing the target protein were pooled, separated into aliquots and stored at −80°C (RNase R) or at −20°C in the presence of 40% glycerol (RNase T). The purity of the protein preparations was assessed on 12.5% SDS–polyacrylamide gels and stained with Coomassie Brilliant Blue R-250 (Bio-Rad). Protein concentration was determined according to Bradford (Bradford 1976).
Preparation of tRNA substrates for RNases
tRNA substrates were in vitro transcribed as described previously (Schürer et al. 2002; Mörl et al. 2005). The 3′-dephosphorylated RNA was 5′-monophosphorylated in the presence of 1 mM ATP using T4 polynucleotide kinase to resemble the in vivo constellation. Free nucleotides were removed using either G25 columns (GE Healthcare) or preparative 10% PAGE. Radioactive labeling was carried out either internally by adding equimolar [α-32P] NTPs during transcription or at the 5′-end with [γ-32P] ATP (Hartmann Analytic) during 5′-monophosphorylation. The E. coli tRNA substrates tested were tRNAAla(GGC)–CCA, 5′-GGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCA-3′, tRNAAsp(GUC)–CCA, 5′-GGAGCGGUAGUUCAGUCGGUUAGAAUACCUGCCUGUCACGCAGGGGGUCGCGGGUUCGAGUCCCGUCCGUUCCGCCA-3′, with their respective 3′-trailer composition as designated in the main text.
RNase activity assays
All assays were performed at 37°C in three to four independent replicates. Prior to the reaction, RNA substrates were heated at 65°C for 5 min by shaking at 600 rpm and allowed to slowly cool down to room temperature.
RNase R assays were carried out using internally labeled [α-32P] tRNA in 90–100 µL reactions (unless stated otherwise) in 20 mM Tris–HCl buffer (pH 8.0) containing 100 mM KCl, 0.25 mM MgCl2 and 1 mM DTT (Vincent and Deutscher 2006). Concentrations of RNA substrate and enzyme were as indicated. The reaction was stopped by adding ice cold trichloroacetic acid to a final concentration of 6%–9% as well as glycogen (50–70 ng µL−1) as carrier. The activity was determined by quantifying the acid-soluble products (MicroBeta2, PerkinElmer). For qualitative analysis, the acid-precipitate was washed once in 70% ethanol, air-dried, resuspended in RNA loading dye (10 mM Tris–HCl [pH 7.6], 80% formamide, 0.25% bromophenolblue, 0.25% xylencyanol), resolved on a 10% PAGE and visualized on a PhosphorImager (GE Healthcare). In the RNase R competition study, a constant amount of internally labeled tRNAAla–CCACCA (50 nM) was incubated with 0.2 ng µL−1 RNase R and increasing amounts of nonlabeled tRNAAla–CCACCA or tRNAAla–CCAAAU. Reaction conditions were chosen so that substrate conversion was under nonsaturating conditions. Product formation was determined by quantifying the acid-soluble radioactivity.
RNase T assays were carried out using 5′-end labeled tRNA. The time course experiment was carried out in a total volume of 40 µL containing 20 mM Tris–HCl buffer (pH 8.0), containing 50 mM KCl, 10 mM MgCl2, and 1 mM DTT (Zuo and Deutscher 2002). Concentrations of RNA substrate and enzyme were as indicated. At various time intervals, aliquots of 4 µL were withdrawn and the reaction was terminated in 6%–9% ice cold trichloroacetic acid and 50–70 ng µL−1 glycogen. The acidic precipitate was washed once in 70% ethanol, air-dried, resuspended in RNA loading dye and resolved on a 10% PAGE. Michaelis–Menten kinetics were performed in a total reaction volume of 10 µL. Initial rates of RNase T activity were determined under steady-state conditions so that product formation was linear over time and did not exceed 20%. Quantification of the product bands was carried out using ImageQuant TL software (GE Healthcare), and kinetic parameters were determined by nonlinear regression using GraphPad Prism 7.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sonja Bonin, Tobias Friedrich, and Karoline Raulf for expert technical assistance, Murray P. Deutscher for the generous gift of E. coli KO strains, and Irina Chelysheva and Johannes Wagner for mapping the sequencing data. We thank Hans-Jörg Hofmann for valuable discussions. This work was supported by the Deutsche Forschungsgemeinschaft (MO 634/12-1 to M.M. and CZ 234/1-1 to A.C.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064436.117.
REFERENCES
- Betat H, Mörl M. 2015. The CCA-adding enzyme: a central scrutinizer in tRNA quality control. Bioessays 37: 975–982. [DOI] [PubMed] [Google Scholar]
- Betat H, Rammelt C, Mörl M. 2010. tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization. Cell Mol Life Sci 67: 1447–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem 277: 21624–21629. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Marlor CW. 1985. Purification and characterization of Escherichia coli RNase T. J Biol Chem 260: 7067–7071. [PubMed] [Google Scholar]
- Deutscher MP, Lin JJ, Evans JA. 1977. Transfer RNA metabolism in Escherichia coli cells deficient in tRNA nucleotidyltransferase. J Mol Biol 117: 1081–1094. [DOI] [PubMed] [Google Scholar]
- Deutscher MP, Marlor CW, Zaniewski R. 1984. Ribonuclease T: new exoribonuclease possibly involved in end-turnover of tRNA. Proc Natl Acad Sci 81: 4290–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Marlor CW, Zaniewski R. 1985. RNase T is responsible for the end-turnover of tRNA in Escherichia coli. Proc Natl Acad Sci 82: 6427–6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duh Y, Hsiao YY, Li CL, Huang JC, Yuan HS. 2015. Aromatic residues in RNase T stack with nucleobases to guide the sequence-specific recognition and cleavage of nucleic acids. Protein Sci 24: 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupasquier M, Kim S, Halkidis K, Gamper H, Hou YM. 2008. tRNA integrity is a prerequisite for rapid CCA addition: implication for quality control. J Mol Biol 379: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK. 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YY, Yang CC, Lin CL, Lin JL, Duh Y, Yuan HS. 2011. Structural basis for RNA trimming by RNase T in stable RNA 3′-end maturation. Nat Chem Biol 7: 236–243. [DOI] [PubMed] [Google Scholar]
- Hsiao YY, Duh Y, Chen YP, Wang YT, Yuan HS. 2012. How an exonuclease decides where to stop in trimming of nucleic acids: crystal structures of RNase T-product complexes. Nucleic Acids Res 40: 8144–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KO, Deutscher MP. 1992. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J Bacteriol 174: 6682–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn CD, Wilusz JE, Zheng Y, Beal PA, Joshua-Tor L. 2015. On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell 160: 644–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. 1995. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc Natl Acad Sci 92: 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. 1996. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 86: 503–512. [DOI] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. 2002. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA 8: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. 1998. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc Natl Acad Sci 95: 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Reimers S, Pandit S, Deutscher MP. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J 21: 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizano E, Scheibe M, Rammelt C, Betat H, Mörl M. 2008. A comparative analysis of CCA-adding enzymes from human and E. coli: differences in CCA addition and tRNA 3′-end repair. Biochimie 90: 762–772. [DOI] [PubMed] [Google Scholar]
- Matos RG, Barbas A, Arraiano CM. 2009. RNase R mutants elucidate the catalysis of structured RNA: RNA-binding domains select the RNAs targeted for degradation. Biochem J 423: 291–301. [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. 2013. Deregulation of poly(A) polymerase I in Escherichia coli inhibits protein synthesis and leads to cell death. Nucleic Acids Res 41: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR. 2012. Polyadenylation helps regulate functional tRNA levels in Escherichia coli. Nucleic Acids Res 40: 4589–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörl M, Lizano E, Willkomm DK, Hartmann RK. 2005. Production of RNAs with homogeneous 5′ and 3′ ends. In Handbook of RNA biochemistry (ed. Hartmann RK, et al. ), pp. 22–35. Wiley-VCH Verlag GmbH, Weinheim, Germany. [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Padmanabha KP, Deutscher MP. 1991. RNase T affects Escherichia coli growth and recovery from metabolic stress. J Bacteriol 173: 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven NB, Deutscher MP. 1993. Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. FASEB J 7: 143–148. [DOI] [PubMed] [Google Scholar]
- Schürer H, Lang K, Schuster J, Mörl M. 2002. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res 30: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. 2006. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem 281: 29769–29775. [DOI] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. 2009a. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J Mol Biol 387: 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. 2009b. The roles of individual domains of RNase R in substrate binding and exoribonuclease activity. The nuclease domain is sufficient for digestion of structured RNA. J Biol Chem 284: 486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. 2011. tRNAs marked with CCACCA are targeted for degradation. Science 334: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Deutscher MP. 1987. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J 6: 2473–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP. 2002. The physiological role of RNase T can be explained by its unusual substrate specificity. J Biol Chem 277: 29654–29661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.