Abstract
Significance: Developing evidence in the literature suggests that sirtuin 5 (SIRT5) may be involved in metabolic reprogramming, an emerging hallmark of cancer by which neoplastic cells reconfigure their metabolism to support the anabolic demands of rapid cell division. SIRT5 is one of the seven members of the nicotinamide adenine dinucleotide-dependent sirtuin family of lysine deacetylases. It removes succinyl, malonyl, and glutaryl groups from protein targets within the mitochondrial matrix and other subcellular compartments. SIRT5 substrates include a number of proteins integral to metabolism.
Recent Advances: New work has begun to elucidate the roles of SIRT5 in glycolysis, tricarboxylic acid cycle, fatty acid oxidation, nitrogen metabolism, pentose phosphate pathway, antioxidant defense, and apoptosis.
Critical Issues: In this study, we summarize biological functions of SIRT5 reported in normal tissues and in cancer and discuss potential mechanisms whereby SIRT5 may impact tumorigenesis, particularly focusing on its reported roles in metabolic reprogramming. Finally, we review current efforts to target SIRT5 pharmacologically.
Future Directions: The biological significance of SIRT5 has been elucidated in the context of only an extremely small fraction of its targets and interactors. There is no doubt that further studies in this area will provide a wealth of insights into functions of SIRT5 and its targets in normal and neoplastic cells. Antioxid. Redox Signal. 28, 677–690.
Keywords: : SIRT5, sirtuins, desuccinylase, demalonylase, deglutarylase, metabolic reprogramming
Introduction
Sirtuins modulate cellular metabolism and other processes in response to fluctuations in cellular energy demands. Originally identified in yeast, the silent information regulator 2 genes (sirtuins) are highly conserved proteins present in species ranging from bacteria to humans. The seven mammalian sirtuin proteins (SIRT1–7) are nicotinamide adenine dinucleotide (NAD+)-dependent lysine deacetylases implicated in regulating diverse aspects of cell biology, including metabolism, genome stability, and gene expression, among others (11). Sirtuin activity is regulated, in part, by alterations in cellular NAD+ levels; NAD+ levels undergo modest fluctuations under normal conditions and are reported to change dramatically under conditions of nutrient deprivation, obesity, and aging (4).
Despite their conserved catalytic domains, sirtuins are generally nonredundant in their functions, in part, due to differences in their subcellular localizations, biochemical targets, and enzymatic activities. Although sirtuins were originally characterized as deacetylases, a substantial body of literature has demonstrated that all sirtuins are capable of removing alternative lysine modifications, a topic discussed in depth subsequently. SIRT1, 6, and 7 are predominantly nuclear under most conditions, and SIRT2 generally resides in the cytosol, but undergoes nuclear translocation during mitosis (11, 99). These nonmitochondrial sirtuins participate in diverse cellular processes and have been extensively discussed in other recent reviews (12, 24, 35, 37, 40, 42, 46, 54).
SIRT3, SIRT4, and SIRT5 are primarily present in the mitochondrial matrix and possess distinct biochemical activities and substrate specificities (42). SIRT3 represents the major deacetylase activity in mitochondria, playing prominent roles in promoting fatty acid oxidation (FAO), tricarboxylic acid (TCA) cycle activity, and the mitochondrial unfolded protein response, among other processes, while suppressing levels of reactive oxygen species (ROS) (33, 42, 53). Among its multiple roles in ROS management, SIRT3 deacetylates and activates superoxide dismutase (SOD)2 and isocitrate dehydrogenase (IDH)2, an enzyme involved in regenerating the antioxidant glutathione (GSH). SIRT3 is implicated in tumor suppression, and elevated SIRT3 activity partially explains ROS suppression associated with the longevity-promoting intervention calorie restriction (31, 78, 89).
SIRT4 is a very weak deacetylase that inhibits the pyruvate dehydrogenase complex (PDC) by removing lipoamide modifications from the PDC E2 subunit (60). SIRT4 also deacetylates and inhibits malonyl-coenzyme A (CoA) decarboxylase, thereby reducing FAO (44). SIRT4 was reported to adenosine diphosphate (ADP)-ribosylate glutamate dehydrogenase (GDH) to block glutaminolysis, a role thought to be important for SIRT4 function in suppressing tumorigenesis (28, 39, 66). Very recently, SIRT4 was shown to remove multiple derivatives of methylglutaryl (methylglutaryl, hydroxymethylglutaryl, and 3-methylglutaryl groups) from lysine residues to regulate leucine metabolism, thereby inhibiting insulin secretion (1).
Like SIRT4, SIRT5 also possesses very weak deacetylase activity. Instead, SIRT5 targets lysine succinylation, malonylation, and glutarylation (Ksucc, Kmal, and Kglu) groups on its target proteins (16, 42, 70, 73, 74, 80, 94). The published roles of SIRT5 implicate this enzyme in the regulation of mitochondrial metabolism and other cellular processes, some of them extramitochondrial. Emerging literature highlights an involvement of SIRT5 in oncogenesis, the major focus of our discussion.
General phenotypes of malignancy have been conceptualized in the form of established hallmarks, such as uncontrolled cell cycle progression, genome instability, and metastatic potential, and the more recently described emerging hallmarks, including altered metabolism. Sirtuins act as major regulators of many of these hallmarks (11, 29). Roles for SIRT1, SIRT3, and SIRT6 in malignancy have been extensively characterized, and an emerging literature has begun to elucidate roles for SIRT4 and SIRT7 in cancer. Functions of SIRT5 in cancer have been less well defined (11, 42). In this study, we review the enzymatic activities and functions of SIRT5 in normal cell metabolism and in cancer metabolic reprogramming.
A Brief Primer on Sirtuin Biochemistry
Sirtuin family members share a core catalytic domain, but vary in their flanking N- and C-terminal sequences (11). Sirtuin core domains differ at specific amino acids—differences that help confer distinct enzymatic activities. Structural analysis reveals that the sirtuin catalytic domain comprises two subdomains, a highly conserved Rossmann fold typically found in NAD+-consuming enzymes and a more variable zinc-binding finger (20, 65). These two subdomains are connected by several homologous, but structurally variant, loops that collectively form a binding pocket for an acylated lysine and NAD+ (117). This binding pocket aligns the acyl-lysine and the ribose moieties of NAD+ with key amino acids responsible for catalyzing the sirtuin reaction (18). Although not directly involved in catalysis, the zinc finger is integral for maintaining proper enzyme structure and function (10).
Sirtuin-mediated deacetylation is initiated by the cleavage and release of nicotinamide (NAM) derived from NAD+ and the formation of an intermediate between ADP-ribose and the acyl moiety. Upon resolution of this intermediate, the newly deacetylated lysine and 2′-O-acyl-ADP-ribose are released (117). A product of the sirtuin reaction, NAM, inhibits sirtuin catalytic activity in a noncompetitive manner (5). NAM is recycled back to NAD+ via a salvage pathway, the major source of NAD+ in mammalian cells (81).
SIRT5 Possesses Unique Biochemical Activities
SIRT5 remains a somewhat mysterious protein, in part, because SIRT5 enzymatic activity is unique compared with that of other sirtuin proteins. SIRT5 was initially characterized as a lysine deacetylase, although it possesses very weak deacetylase activity (71). In 2011, the crystal structure of SIRT5 bound to a substrate peptide was solved, revealing that SIRT5 possesses a larger binding cavity compared with other sirtuins, with three unique active site amino acid residues, Ala86, Tyr102, and Arg105 (16, 74). It was established that SIRT5 efficiently removes acidic acyl groups—succinyl, malonyl, and glutaryl moieties—from lysine residues (16, 42, 70, 73, 74, 80, 94). Similar to phosphorylation, succinylation, malonylation, or glutarylation of a lysine residue (Ksucc, Kmal, and Kglu) renders it negatively charged at physiologic pH (74, 94).
Recently, it was reported that SIRT7, like SIRT5, is capable of desuccinylating histones (16, 47, 73, 74). However, since SIRT7 is not thought to localize to the mitochondria and since cellular Ksucc, Kmal, and Kglu levels globally increase in response to loss of SIRT5, current evidence supports the view that SIRT5 function is nonredundant with other sirtuins in the mitochondria and likely elsewhere in the cell as well (16, 47, 70, 73, 84, 94, 112).
Ksucc, Kmal, and Kglu are structurally quite similar (74). Succinyl-, malonyl-, and glutaryl-CoA, the precursors to Ksucc, Kmal, and Kglu, respectively, are derived from metabolites succinate, malonate, and glutarate (34). Succinyl-CoA is a 4-carbon mitochondrial TCA cycle intermediate (11a). Malonyl-CoA is a three-carbon product of acetyl-CoA carboxylation in the cytosol, involved in fatty acid synthesis (96). Mitochondrial malonyl-CoA is generated by ACSF3 and the major source of mitochondrial protein lysine malonylation (6). Glutaryl-CoA, a product of tryptophan and lysine catabolism, comprises four carbons and is predominantly mitochondrial (79). How these moieties become conjugated to lysines remains incompletely defined and a subject of active investigation.
Acetylation of nuclear and cytosolic lysines is catalyzed by lysine acetyltransferase (KAT) enzymes (ketoacyl coenzyme A thiolase) that employ acetyl-CoA as the source of acetyl groups. In addition to acetyl-CoA, the human KAT p300 has been found to accommodate propionyl-CoA as well as larger moieties such as crotonyl-, butyryl-, hydroxyisobutyryl-, succinyl-, and glutaryl-CoA (83, 84, 94). It is highly likely that acylation of many SIRT5 substrates occurs in a nonenzymatic manner. Acyl-CoAs possess a highly reactive thioester bond and function as acyl carrier species. Under chemical conditions in the mitochondrial matrix, the acyl groups can be conjugated to lysine side chains in an enzyme-independent mechanism, as in the case of mitochondrial lysine succinylation (38). Importantly, succinyl-CoA was recently found to exhibit much greater lysine acylation propensity than acetyl-CoA (101).
Enzyme-independent acylation was recently shown to occur within the cytosol as well (41, 102); acylation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and pyruvate kinase muscle isozyme M2 (PKM2) by malonyl-CoA was found to be catalytically independent (41). Consistently, SIRT5 was shown to target GAPDH and PKM2 (59, 70, 104, 106). Thus, many lysine modifications targeted by SIRT5 may simply occur as a passive by-product of cellular metabolism, rather than as an actively enzyme-catalyzed event.
SIRT5 possesses a well-defined mitochondrial localization sequence (MLS) and the majority of SIRT5 proteins localize to the mitochondrial matrix (61, 64). However, a significant fraction of SIRT5 is cytosolic and a modest portion is even nuclear (64, 73). Mechanisms of SIRT5 localization to nonmitochondrial compartments have not been clearly elucidated. In humans, alternative splicing of the Sirt5 mRNA yields two dominant isoforms, SIRT5iso1 and SIRT5iso2, both of which contain an N-terminal MLS, but differ at their C-termini (61). Several other SIRT5 isoforms are reported in the NCBI database (14, 22). Based on studies using ectopically expressed green fluorescent protein (GFP)-tagged SIRT5, one report found that while both SIRT5 isoforms localized to mitochondria, only SIRT5iso1 was present in the cytoplasm. Most studies of SIRT5 have focused on elucidating functions of SIRT5iso1, the only reported form of SIRT5 present in mice (14, 61, 73).
Unbiased mass spectrometry-based proteomic surveys have proven extremely valuable in defining cellular substrates of, and potential functions for, SIRT5. This work has been greatly facilitated by the availability of a viable SIRT5-deficient mouse model (53, 73, 113). These mice showed greatly increased tissue Ksucc, Kmal, and Kglu levels, but little change in Kac, confirming the in vitro characterization of SIRT5 activity (73, 74, 94, 113). Collectively, these studies revealed that SIRT5 targets hundreds of protein substrates, typically on multiple lysines per substrate protein. A large majority of these modified proteins localize to the mitochondrial matrix, with many involved in cellular metabolism, particularly in the TCA cycle, urea cycle, and FAO (70, 73, 74, 80, 94). However, in the cytosol, SIRT5 was shown to act on multiple glycolytic enzymes as well as proteins associated with the 40S and 60S ribosomal subunits, to name a few (61, 73). Ksucc, Kmal, and Kglu sites were also identified on histones, suggesting a potential role for SIRT5 in transcriptional regulation (73, 74, 94, 107).
Ksucc and Kglu levels are highest in mitochondria, where succinyl-CoA, as a TCA cycle intermediate, and glutaryl-CoA are particularly abundant. By contrast, Kmal is especially prominent in the cytosol (94, 107). Pathway analysis of Ksucc has identified potential roles for SIRT5 in regulating glycolysis, keto acid breakdown, redox homeostasis, cardioblast proliferation, acid thiol ligation, and protein translation (16, 73, 80, 94). Similarly, examining Kmal enrichment revealed proteins that modulate glycolysis and gluconeogenesis, retinoid X receptor inhibition, and estrogen biosynthesis (16, 70), and Kglu sites predominantly occurred on proteins involved in aerobic respiration, redox homeostasis, and the TCA cycle (16, 94). All three marks are enriched on proteins involved in FAO (1, 16, 70, 73, 74, 94). The diversity of SIRT5 targets and interacting partners indicates that SIRT5 is functionally important in multiple subcellular compartments, and not only in the mitochondrial matrix.
SIRT5 Function in Organismal Homeostasis
SIRT5 is expressed in most tissues, with the highest SIRT5 levels in the heart and brain (61, 68). Despite the widespread expression of SIRT5 and unique enzymatic activities, germline SIRT5 deficiency is well tolerated in mice under basal conditions (53, 85, 113). One study showed that SIRT5-deficient animals on the C57BL/6 background were born at a sub-Mendelian ratio (113), although this effect had not been observed in previous work using 129 background mice (53). Sirt5 knockouts (KOs) show perturbed FAO and mildly depressed ketogenesis (80) and elevated blood ammonia levels following a prolonged fast (113). Sirt5 KOs showed marked cardiac protein hypersuccinylation and develop mild cardiac dysfunction and hypertrophy with age under basal conditions (85). SIRT5 protects from myocardial and cerebral injury in response to ischemia (7, 67). In humans, the presence of a single-nucleotide polymorphism in the SIRT5 promoter correlates with reduced Sirt5 mRNA expression and gene expression changes, suggestive of aging in different brain regions (25).
SIRT5 Expression in Cancer
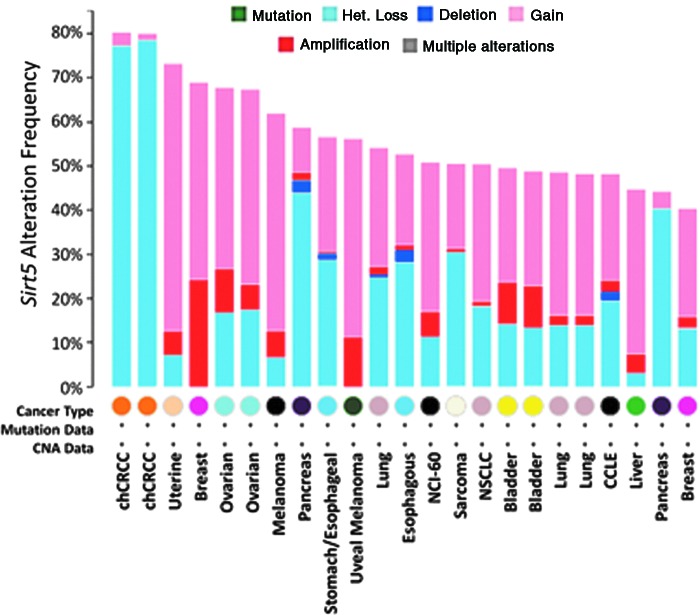
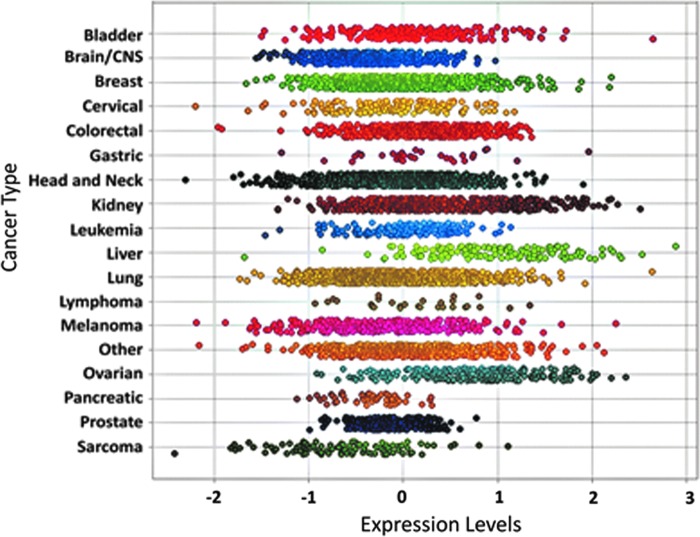
In humans, the Sirt5 gene is found on the highly unstable cytogenetic band on chromosome 6p23 (58, 93). Likewise, gain or loss of the Sirt5 gene locus occurs frequently across a range of cancers, although often in the context of nonfocal genomic events resulting in alterations in many flanking genes in addition to SIRT5 (Fig. 1) (23, 58, 69, 93). Sirt5 mRNA expression is detectable across a wide variety of cancers (Fig. 2).
FIG. 1.
SIRT5 gene alteration across multiple cancer types. The frequency of SIRT5 gene alteration was measured across cancer types in The Cancer Genome Atlas (cBioportal) on April 15, 2017. The following genetic alterations were queried: copy number gain (low-level gene gain), amplification (high-level gene amplification), het loss (heterozygous deletion), and deletion (homozygous deletion). Cancer types with alteration frequency greater than 40% are shown (69).
FIG. 2.
SIRT5 mRNA expression across multiple cancer types. SIRT5 expression across >30,000 cancer samples using the Oncomine powertools Oncomine Gene Expression Browser (82). Normalized linear expression plotted for all cancer samples profiled by Affymetrix U133A, U133A 2.0, and U133 Plus 2.0 arrays in the Oncomine database. Samples stratified by cancer type as indicated. Courtesy of Dr. Scott Tomlins, Department of Pathology at the University of Michigan, Ann Arbor, MI.
A number of studies have shown that in specific tissues, Sirt5 mRNA expression tends to be elevated in tumors relative to the corresponding normal tissue type, an issue discussed in more depth later in the review, suggesting a potential tumor-promoting role for SIRT5 in certain contexts (36, 50, 56, 91). For example, Sirt5 mRNA and protein expression levels are frequently elevated in human lung cancers and serve as a predictor of recurrence, rendering SIRT5 a candidate biomarker for poor survival in non small cell lung cancer (NSCLC) (50, 56). Additionally, tumor tissues from patients suffering from Waldenstrom's macroglobulinemia, a B cell malignancy, show elevated Sirt5 mRNA levels compared with B-lymphocytes from healthy donors (91). Likewise, Sirt5 mRNA expression was markedly increased in grade III estrogen receptor-negative/progesterone receptor-positive invasive breast tumors (36).
In contrast, Sirt5 mRNA expression was significantly reduced in endometrial carcinoma, another hormone-responsive cancer, compared with benign endometria. Sirt5 mRNA levels were also lower in head and neck squamous cell carcinoma compared with corresponding noncancerous tissue (3, 43). For the remainder of this review, we discuss known functions of SIRT5 in normal and malignant tissues, focusing on SIRT5 interactors and targets that may be most relevant in cancer.
SIRT5 in Normal and Cancer Metabolism
Cancer cells reprogram their metabolism to support the anabolic demands of cell growth (29). Malignant cells require amplified production of nucleosides, amino acids, and lipids for synthesis of RNA, DNA, and membranes. Additionally, these cells require mechanisms for efficient ROS elimination (11). A particularly notable aspect of metabolic reprogramming involves rewiring of glucose and glutamine metabolism. For example, glycolysis becomes a major source of ATP synthesis in many cancer cells and provides precursors for numerous anabolic processes and for antioxidant defense (57).
SIRT5 Promotes Glycolysis and Regulates Carbon Flux into Mitochondria
Glucose is transported into the cell by glucose transport proteins, particularly GLUT1 (57, 88). SIRT5 depletion in H1299 NSCLC cells led to a decrease in GLUT1 mRNA and protein levels (57). Glycolysis ultimately results in the conversion of glucose to either pyruvate (via pyruvate kinase, including the PKM2 isoform) or lactate (via lactate dehydrogenase, LDHA, or LDHB) (11a). SIRT5 was shown to demalonylate GAPDH and other enzymes in the glycolytic cascade in mouse liver, resulting in increased GAPDH activity and elevated glycolytic flux (70). An independent proteomic survey in 293T cells confirmed that SIRT5 physically interacts with GAPDH (59).
In the final step of glycolysis, PKM2 catalyzes conversion of phosphoenolpyruvate to pyruvate, and PKM2 suppression impairs conversion of glucose into pyruvate (13). One report found that SIRT5 desuccinylates PKM2 at a single lysine residue in A549 NSCLC and 293T cells, suppressing PKM2 activity. SIRT5-dependent inhibition of PKM2 in these cells led to reduced glycolytic flux, reduced mitochondrial respiration, and enhancement of cell proliferation and protection against oxidative insult. Additionally, the interaction between PKM2 and SIRT5 was enhanced by increased cellular ROS levels. This suggests a potential feedback mechanism, whereby ROS induces SIRT5 to restrict carbon entry into the TCA cycle, thereby minimizing further ROS production (106, 118). Yet another report found that desuccinylation of PKM2 by SIRT5 maintains its cytosolic localization in a proinflammatory environment (104), further demonstrating the context-dependent impact of SIRT5 activity.
Elevated lactic acid levels foster a microenvironment that suppresses antitumoral immune response (8, 63). SIRT5 depletion in H1299 NSCLC cells correlates with reduced LDHA mRNA and protein levels through mechanisms that remain to be elucidated (57). LDHA consumes pyruvate to produce lactate, suppressing conversion of pyruvate to acetyl-CoA by PDC, which would otherwise fuel the TCA cycle. SIRT5 desuccinylates the PDC E1α subunit at multiple sites, suppressing overall PDC enzymatic activity in 293T cells. SIRT5-deficient liver mitochondria show increased pyruvate-dependent respiration (73). However, conflicting results from HepG2 cells and mouse cardiac tissue suggest that the impact of SIRT5 on PDC and mitochondrial respiration may be context dependent (9, 85).
Although it is a very weak deacetylase, cytosolic SIRT5 has been reported to inhibit PDC through deacetylation and deactivation of signal transduction and activator of transcription 3 (STAT3) in A549 lung cancer cells (71, 108). Upon phosphorylation by interleukin-6-stimulated Janus kinases, cytosolic STAT3 undergoes rapid nuclear translocation to bind and activate the promoters of hundreds of progrowth and prosurvival genes. In contrast, acetylation promotes mitochondrial translocation of STAT3, which has been shown to drive RAS-dependent tumorigenesis (111). Mitochondrial STAT3 binds and activates PDC, correlating with elevated mitochondrial membrane potential (MMP) (86, 115) and ATP synthesis in PC3 prostate cancer cells (100, 108). In PC3 cells, deacetylation of cytosolic STAT3 by SIRT5 inhibited its mitochondrial translocation and interaction with PDC, thereby reducing pyruvate conversion into acetyl-CoA, lowering the MMP, and inhibiting ATP production. Two of the three Kac sites of STAT3, K685 and K707, are also conserved in STAT1 and STAT5b, indicating that SIRT5 may regulate the mitochondrial activity of multiple STAT family members (108).
In summary, SIRT5 regulates several aspects of glucose metabolism across multiple subcellular compartments. In the cytosol, it promotes glycolysis via effects on GAPDH and likely other glycolytic enzymes and it suppresses PKM2 activity in noninflammatory conditions (70, 106). Similarly, SIRT5 inhibits pyruvate entry into the TCA cycle by deacetylating cytosolic STAT3 and inhibiting its mitochondrial translocation, thereby indirectly reducing PDC function (45). Mitochondrial SIRT5 also directly desuccinylates and inhibits PDC (73). Overall, these roles for SIRT5 in glucose metabolism, on balance, favor metabolic reprogramming and a tumor-promoting cellular metabolism (Fig. 3).
FIG. 3.
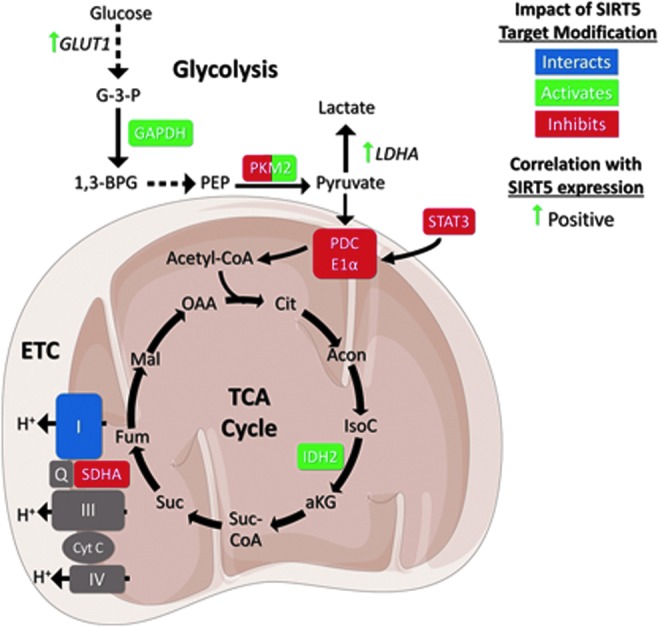
SIRT5 regulates glycolysis, the TCA cycle, and the ETC. SIRT5 impacts activities and expression levels of enzymes involved in glycolysis, the TCA cycle, and the ETC. See text for details. Figures were produced using material from Servier Medical Art (www.servier.com). αKG, alpha ketoglutarate; Acetyl-CoA, acetyl-coenzyme A; Acon, aconitate; Cit, citrate; Cyt C, cytochrome C; ETC, electron transport chain; Fum, fumarate; G-3-P, glucose 3 phosphate; GAPDH, glyceraldehyde phosphate dehydrogenase; GLUT1, glucose transporter 1; IDH2, isocitrate dehydrogenase 2; I–IV, complex I–IV; IMM, inner mitochondrial membrane; IsoC, isocitrate; LDHA, lactate dehydrogenase A; Mal, malate; MM, mitochondrial matrix; OAA, oxaloacetate; PDCE1α, pyruvate dehydrogenase complex E1α subunit; PEP, phosphoenolpyruvate; PKM2, pyruvate kinase muscle isozyme 2; Q, Quinolone; SDH/complex II, succinate dehydrogenase; STAT3, signal transducer and activator of transcription 3; Suc, succinate; Suc-CoA, succinyl-coenzyme A; TCA cycle, tricarboxylic acid cycle.
SIRT5 Regulation of the TCA Cycle and Electron Transport Chain
SIRT5 also regulates multiple TCA cycle enzymes. SIRT5 inhibits the conversion of succinate to fumarate by desuccinylating succinate dehydrogenase subunit A (SDHA) in 293T cells (73). SDHA, also known as Complex II of the electron transport chain, catalyzes conversion of succinate to fumarate, coupled to reduction of NAD+ to NADH and the generation of ubiquinol from ubiquinone (11a). SIRT5-depleted 293T cells, or SIRT5-deficient liver mitochondria, show increased succinate-dependent respiration (73). Contradictory results have been reported using distinct approaches for measuring Complex II activity and mitochondrial respiration (115). SIRT5 also desuccinylates IDH2, increasing enzyme activity and upregulating conversion of isocitrate to alpha-ketoglutarate (α-KG) in 293T cells. SIRT5 depletion induces increased IDH2 succinylation and impaired IDH2 activity (118). SIRT5 was also found to interact with the complex I subunit NDUFA4 (Fig. 3) (59), although the functional significance of this interaction has not been elucidated. Cytochrome c (Cyt C) was found to be deacetylated by SIRT5 in vitro, a topic we discuss subsequently (86).
In recent years, mutations in IDH1 and IDH2 have been identified as driver events in a number of cancer types, particularly including gliomas and acute myeloid leukemia (AML) (15). These mutations cause IDH enzymes to produce a novel oncometabolite, (R)-2-hydroxyglutarate (2-HG). A large literature documents the ability of 2-HG to inhibit α-KG-dependent dioxygenases, particularly those involved in DNA and histone demethylation, in turn leading to widespread epigenetic dysregulation and promoting tumorigenesis.
Li et al. identified a novel role for 2-HG in inhibiting the TCA cycle enzymes, SDH and fumarate hydratase, resulting in increased cellular succinate levels and widespread increased protein succinylation, predominantly not only on mitochondrial proteins but also in the cytosol and the nucleus. Functionally, this hypersuccinylation was associated with mitochondrial dysfunction and accumulation of the antiapoptotic protein B cell lymphoma-2 (BCL-2). In cells expressing mutant IDH1, enforced SIRT5 expression relieved 2-HG-induced protein hypersuccinylation, reversing mitochondrial dysfunction and apoptotic resistance. Xenograft tumor formation by cells expressing mutant IDH was impaired by SIRT5 overexpression (45). These findings imply a tumor suppressive role for SIRT5 specifically in the context of mutant IDH1.
Taking these findings altogether, SIRT5 controls the production of several intermediates in the TCA cycle and electron transport. SIRT5 desuccinylates and inhibits SHDA, restricting TCA cycling by decreased production of fumarate as well as electron transport (73). By contrast, SIRT5 promotes the production of TCA cycle intermediate α-KG by activating IDH2; however, in glioma cells bearing the IDH1 R123H mutant, SIRT5 relieves the oncogenic effects of 2-HG accumulation (45, 118).
SIRT5 Promotes FAO
FAO is initiated when fatty acids of varying lengths are converted into fatty acyl-CoA intermediates, which are then transported into mitochondria by way of the carnitine shuttle. For very long fatty acyl-CoA (VLCFA) substrates, very long-chain acyl-CoA dehydrogenase (VLCAD) converts these intermediates to enoyl-CoA. Enoyl-CoA hydratase (ECHA) is a subunit of the mitochondrial trifunctional (MTF) complex that catalyzes conversion of enoyl-CoA to ketoacyl- and acetyl-CoA to break down fatty acids for energy (11a). SIRT5, together with SIRT3, promotes FAO by deacetylating VLCAD in mice, increasing its activity and enhancing conversion of acyl-CoA to enoyl-CoA (Fig. 4).
FIG. 4.
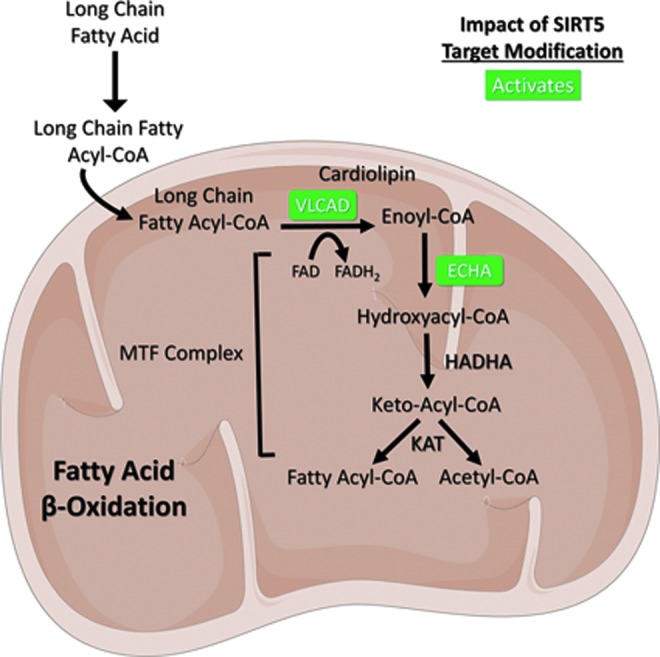
SIRT5 promotes fatty acid β-oxidation. SIRT5 desuccinylates VLCAD and ECHA to activate their enzymatic activities. See text for details. Figures were produced using material from Servier Medical Art (www.servier.com). ECHA, enoyl-coenzyme A hydratase; HADH, hydroxyacyl-coenzyme A dehydrogenase; KAT, lysine acetyltransferase; MTF, mitochondrial trifunctional; VLCAD, very long chain acyl-coenzyme A dehydrogenase.
SIRT5-mediated desuccinylation works alongside SIRT3-mediated deacetylation to upregulate VLCAD function by stabilizing its association with flavin adenine dinucleotide, a redox cofactor required for VLCAD activity. Notably, SIRT3 and SIRT5 target overlapping, highly evolutionarily conserved lysine residues, K298/K299 and K507. Indeed, K298/K299 lie within the active site of VLCAD, and K507 occurs within the inner mitochondrial transmembrane domain, potentially affecting VLCAD localization to this organelle. Cooperative deacetylation of VLCAD by SIRT3 and SIRT5 is required for VLCAD-cardiolipin substrate binding, increasing FAO flux through the regulation of mitochondrial membrane remodeling (95, 115).
SIRT5 also desuccinylates and activates ECHA, thereby promoting the final step in lipid metabolism. Consistently, SIRT5-deficient myocardium shows impaired FAO (7). Overall, SIRT5 promotes robust ATP generation through enhancing the breakdown of very long-chain fatty acid molecules, an event that can yield upward of 70 ATP molecules per VLCFA to support tissue energy demands (11a).
SIRT5 Regulates Nitrogen Metabolism
Glutaminolysis involves the conversion of glutamine to glutamate and ammonia by glutaminase enzymes (GLS1 and GLS2) and subsequently of glutamate to α-KG by GDH. A by-product of this pathway is ammonia, which is highly neurotoxic if it accumulates to an appreciable degree (11a). SIRT5 desuccinylates and is proposed to inhibit GLS2, thereby reducing production of glutamate and ammonia from glutamine (Fig. 5). In MDA-MB-231 breast cancer cells, SIRT5 was shown to interact with GLS2. Indeed, inhibition of SIRT5 by either genetic KD or a novel SIRT5 inhibitor, MC3482, correlated with increased GLS2 succinylation and elevated cellular glutamate and ammonia levels in MDA-MB-231 cells and mouse myoblast C2C12 cell lines. Overexpression of SIRT5 reversed these phenotypes. Ectopic expression of SIRT5 reduced expression of autophagy and mitophagy proteins; consistently, ammonia is known to promote expression of factors involved in cellular mitophagy and autophagy (55, 76). Importantly, however, the impact of SIRT5 on GLS2 biochemical activity has not been directly measured (76).
FIG. 5.
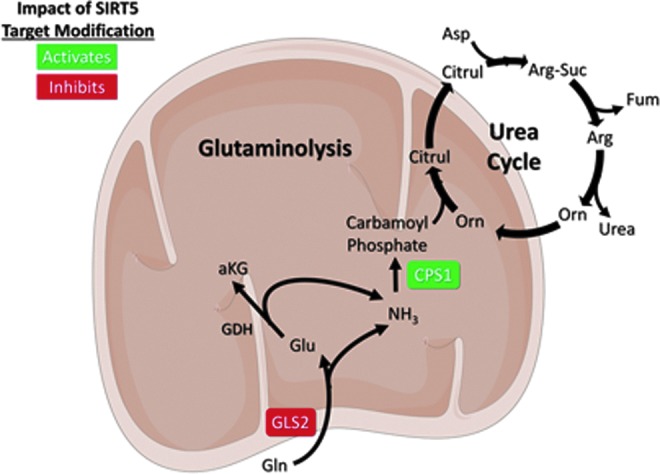
SIRT5 downregulates glutaminolysis and promotes ammonia detoxification. SIRT5 modifies CPS1 to increase its activity. SIRT5 suppresses ammonia production via proposed regulation of GLS2. See text for details. Figures were produced using material from Servier Medical Art (www.servier.com). αKG, alpha ketoglutarate; Arg, arginine; Arg-Suc, arginosuccinate; Asp, aspartate; Citrul, citrulline; CPS1, carbamoyl phosphate synthetase 1; Fum, fumarate; GDH, glutamate dehydrogenase; Gln, glutamine; GLS2, glutaminase 2; Glu, glutamate; NH3, ammonia; Orn, ornithine.
Ammonia is neutralized via conversion to urea in the urea cycle, a process that occurs primarily in the liver and kidney. Carbamoyl phosphate synthetase 1 (CPS1) converts ammonia to carbamoyl phosphate to initiate formation of urea for excretion and represents the rate-limiting step in urea biosynthesis (11a). SIRT5 was originally reported to activate urea cycle function via deacetylation of CPS1. SIRT5 associates with CPS1 in mitochondria, and CPS1 activity is decreased in Sirt5 KO liver tissues (68). Multiple studies have since determined that SIRT5 also desuccinylates and deglutarylates CPS1 to activate it (94, 113). These findings demonstrate that SIRT5 regulates cellular ammonia levels, both at the source of ammonia production, by potentially inhibiting GLS2, and by promoting the detoxifying conversion of ammonia to urea (Fig. 5).
SIRT5 Activates the Pentose Phosphate Pathway and Antioxidant Defense
Hyperactive glutaminolysis drives rapid production of glutamate and aspartate, which, in turn, promotes amino acid and nucleic acid anabolism (11a, 68). The sugar backbone of DNA, ribose-5-phosphate, is generated by the pentose phosphate pathway (PPP). The PPP is initiated through conversion of glucose-6-phosphate (G6P) from glycolysis to 6-phosphogluconolactone by G6P dehydrogenase, an enzyme activated by SIRT5-mediated deglutarylation (Fig. 6) (118). The activity of the enzymes in the PPP is coupled to reduction of two molecules of NADP+ into NADPH, a major reducing equivalent within the cell (11a).
FIG. 6.
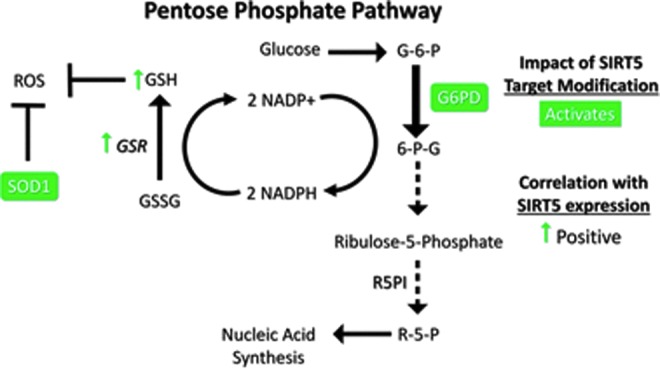
SIRT5 promotes ROS detoxification via SOD1 and the pentose phosphate pathway. SIRT5 desuccinylates SOD1 and deglutarylates G6PD, activating both enzymes. Figures were produced using material from Servier Medical Art (www.servier.com). 6-G-P, 6-phosphogluconate; G-6-P, glyceraldehyde-6-phosphate; G6PD, glucose-6-phosphate dehydrogenase; GSH, glutathione; GSR, glutathione reductase; GSSG, oxidized glutathione; NADP/NADPH, nicotinamide adenine dinucleotide phosphate; R-5-P, ribose 5 phosphate; R5PI, ribulose 5 phosphate isomerase; ROS, reactive oxygen species; SOD1, superoxide dismutase 1.
NADPH production by the PPP plays a major role in suppressing accumulation of ROS, toxic by-products of oxygen metabolism that can react with and damage cellular macromolecules (11a). NADPH is required for regeneration of the reduced form of GSH, a potent cellular ROS scavenger, from its oxidized form by glutathione reductase (GSR) (2). SIRT5 plays a role in ROS elimination by stimulating NADPH production via the PPP to regenerate GSH (118). Likewise, manipulation of SIRT5 expression revealed a positive correlation between SIRT5 and GSR mRNA levels in NSCLC cells (56). Furthermore, Sirt5 KO mouse embryonic fibroblasts (MEFs) exhibited reduced GSH levels and NADPH production and increased sensitivity to oxidative stress (Fig. 6) (118). Elevated GSH is also implicated in conferring a multidrug resistance (MDR) phenotype in cancers (see Regulation of Apoptotic Resistance by SIRT5) (2, 40a).
In addition to its function in the PPP, SIRT5 possesses other roles in ROS management, via desuccinylation and activation of SOD1, a key antioxidant enzyme in ROS detoxification. NSCLC cells expressing a succinylation mimetic mutant form of SOD1 exhibited reduced SOD1 activity and cell viability. Conversely, cells co-overexpressing WT SOD1 and SIRT5 exhibited augmented SOD1 function and decreased levels of cellular ROS (Fig. 6), with enhanced xenograft formation ability (50). Consistently, an independent study in Regulation of Apoptotic Resistance by SIRT5 human neuroblastoma cells showed that SIRT5 overexpression was protective against hydrogen peroxide-induced damage (48). SIRT5 protein expression declined upon induction of oxidative stress in H92c rat cardiomyocytes, and SIRT5 depletion sensitized cells to oxidative stress-induced death (51). Mechanisms of SIRT5 downregulation in this context have not been elucidated. Although SIRT3 had previously been considered to play a dominant role in cellular antioxidant defense among the sirtuins, these recent studies identify SIRT5 as a major factor in this process as well.
Regulation of Apoptotic Resistance by SIRT5
Several studies have demonstrated that SIRT5 promotes apoptotic resistance in normal and cancer cells. Like several other sirtuins (92, 98, 103), SIRT5 is reported to deacetylate forkhead box O3A (FOXO3A) (105) despite the weak deacetylase activity of SIRT5 (71). FOXO3A is a transcription factor that promotes expression of genes involved in antioxidant defense and translocates to the nucleus upon deacetylation by SIRT5. Notably, this effect relieves elevated ROS levels and proapoptotic signaling induced upon treatment of NSCLC cells with cigarette smoke extract (105).
The antiapoptotic role of SIRT5 in rat cardiomyocytes in response to oxidative stress mentioned previously likely relates to the observation that SIRT5 directly interacts with the B cell lymphoma-XL (BCL-XL) antiapoptotic protein. While the specific impact of SIRT5 on BCL-XL remains unclear, this interaction may help explain the prosurvival functions of SIRT5 described in many contexts (Fig. 7) (51).
FIG. 7.
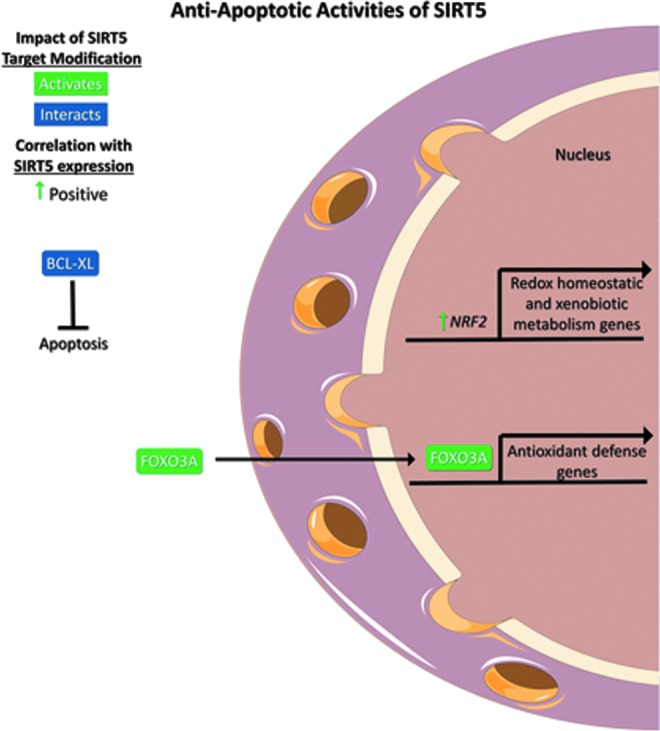
SIRT5 promotes apoptotic resistance. SIRT5 expression positively correlates with the expression of NRF2, a regulator of redox homeostatic and xenobiotic metabolism genes. SIRT5 deacetylates and promotes the nuclear translocation of FOXO3A, a transcriptional regulator of antioxidant defense genes. SIRT5 also interacts with BCL-XL, which inhibits apoptosis. Figures were produced using material from Servier Medical Art (www.servier.com). BCL-XL, B cell lymphoma-XL; FOXO3A, forkhead box O3A; NRF2, nuclear factor erythroid-2-related factor 2.
Survival of photoreceptor cells in mice was also found to be SIRT5 and NAD+ dependent (49). Similarly, SIRT5 was also shown to protect MEFs from mitophagic mitochondrial fragmentation by way of regulating the localization, expression, and dynamics of the profragmentation dynamin-related protein 1. Mitochondrial fragmentation initiates the mitophagic degradation of the organelle and can initiate the apoptotic cascade (26). This link between SIRT5 and mitophagy is consistent with the finding that experimental manipulation of SIRT5 levels in cultured breast cancer cells or mouse myoblasts affects expression of mitophagy-related genes (76). Although the precise mechanism of SIRT5 activity remains to be elucidated in these settings, collectively, these data reveal a role for SIRT5 as an antimitophagy factor.
Primary or secondary resistance to genotoxic chemotherapy is a major impediment to cancer treatment and cure. In vitro, SIRT5 depletion reduces cell viability and sensitizes multiple NSCLC cell lines to genotoxic chemotherapeutics, including cisplatin (CDDP), fluorouracil, and bleomycin. In cultured NSCLC cells, experimental manipulation of SIRT5 levels led to corresponding changes in nuclear factor erythroid-2-related factor 2 (NRF2) expression through mechanisms yet to be defined. NRF2 regulates expression of genes involved in cellular redox homeostasis and xenobiotic metabolism, including chemotherapy drugs. A significant positive correlation was identified between SIRT5 and NRF2 (NRE2L2) mRNA levels in NSCLC patients (Fig. 7). In these clinical samples, SIRT5 levels also correlated with expression of a variety of ATP-dependent drug efflux pump transcripts, which promote an MDR phenotype, conferred by increased drug expulsion (56).
SIRT5 is also a negative regulator of the tumor suppressor SUN domain-containing protein 2 (SUN2), a protein that enhances CDDP sensitivity of lung cancer cells. Most NSCLC cells express low levels of SUN2 in comparison with normal lung tissue, and elevated levels of SUN2 protein within the inner nuclear membrane of lung cancer are associated with improved patient survival. SIRT5 suppresses proapoptotic activity of SUN2 and desensitizes NSCLC cells to chemotherapeutics, although the mechanism of SIRT5 activity against SUN2 has not yet been identified (50). Overall, SIRT5 plays roles in resistance to multiple chemotherapeutics, especially CDDP.
Although many reports have identified prosurvival functions of SIRT5, in other contexts, SIRT5 may facilitate cell death. One report found that the impact of SIRT5 on apoptosis was localization dependent in both normal human neurons and neuroblastoma cells. Nuclear and cytosolic SIRT5 supported cell survival, whereas mitochondrial SIRT5 enhanced cell death. Importantly, the fraction of SIRT5 localized to mitochondria was greater in neuroblastoma cells than in neurons (75).
As mentioned previously, SIRT5 overexpression in IDH mutant cells relieved accumulation of the antiapoptotic protein BCL-2 that normally accompanies excess 2-HG production, thereby resensitizing IDH mutant cells to cell death signaling (45). Cyt C, the major initiator of the intrinsic apoptotic cascade, was shown to be deacetylated and stimulated by SIRT5 in vitro, although a role for SIRT5 in deacetylating Cyt C in vivo could not be demonstrated (86). SIRT5 was also found to promote necroptosis, a cell death pathway characterized by the formation of the RIPK3 complex. SIRT5 depletion protected L929 cells from tumor necrosis factor-induced necroptosis (77). Collectively, these findings support a proapoptotic role for SIRT5 in both normal and cancer cells.
Pharmacologic Targeting of SIRT5
SIRT5 is largely dispensable in nonmalignant tissues and cells (53, 85, 113), yet it plays a progrowth and prosurvival role in several cancer types (48, 51, 56, 57, 105), rendering it a potentially attractive drug target. Although many SIRT5 inhibitors have been described, these agents have not been evaluated for their clinical anticancer activity in cells or in whole animals. The majority of these inhibitors lack selectivity for SIRT5; the SIRT5 inhibitors, suramin, thiobarbiturates, NAM, cambinol, GW5074, and Nɛ-carboxyethyl-thiourea-lysine, have all demonstrated in vitro inhibition of other sirtuins at comparable potencies (21, 30, 32, 62, 87, 90, 97, 109, 114). By contrast, a SIRT5 target covalent inhibitor linked to a cyclic pentapeptide inhibited SIRT5 in vitro deacetylase activity at a 50% inhibitory concentration (IC50) of 7.5 μM and SIRT1/2/3/6 at IC50 values between 200 and 1000 μM (52). Moreover, MC3482 demonstrated ∼40% inhibition of SIRT5 desuccinylase activity in MDA-MB-231 cells at a concentration of 50 μM, but was found to have no significant impact on SIRT1 or SIRT3 activities. The effects of MC3482 treatment on the remaining sirtuins have not been reported, and the mechanism for SIRT5 selectivity of MC3482 has yet to be defined (76).
Collectively, these studies have provided insights to support the development of new, more selective, and potent SIRT5 inhibitors. Current efforts to achieve SIRT5-selective molecules aim at targeting the substrate-binding pocket, which contains the SIRT5-specific residues Tyr102, Arg105, and Ala86. Large-scale screening methods are underway to identify highly selective compounds (110). Recently, a microdroplet-based approach has been applied to the identification of SIRT5 inhibitors; this technique allows the use of a longer, near-native peptide substrate derived from the SDHA protein sequence, enhancing throughput and potentially reducing the false discovery rate (27). SIRT5 inhibitors may eventually prove to be effective anticancer therapeutics, either as single agents or to sensitize cancer cells to established genotoxic therapies, although potential cardiac toxicity represents a significant concern given data from the Sirt5 KO mouse model (85, 113).
Discussion
Although substantial literature now links many sirtuins to cancer, roles for SIRT5 in neoplasia have only begun to be addressed. There are several challenges to the study of SIRT5 in many contexts, including cancer. First, SIRT5 loss of function is associated with relatively mild phenotypes under basal conditions (53, 85, 113). Additionally, known functions of SIRT5 are pleiotropic and very context dependent (36, 45, 50, 56, 75, 77, 91, 104, 106). The fact that SIRT5 targets at least three distinct lysine modifications implies that this sirtuin likely exerts differing biological effects in different cell types, or even in the same cell type in differing physiological states, depending on the presence of these modifications. Finally, the effects of SIRT5 and its target modifications on hundreds of reported targets of SIRT5 remain to be defined (16, 42, 70, 73, 74, 80, 94). Still, a growing body of work has begun to elucidate the prosurvival and protumorigenic roles for SIRT5 in cancer cells, including promotion of metabolic reprogramming, antioxidant defense, and resistance to chemotherapy-induced cell death.
In the context of metabolic reprogramming, SIRT5 stimulates glycolysis and restricts TCA cycling and electron transport (57, 70, 73, 106, 108, 118). SIRT5 reduces cellular ROS by promoting NADPH production to support GSH regeneration, and by stimulating SOD1 antioxidant activity, thereby promoting cell survival (50, 106, 118). Conversely, specifically in the setting of IDH mutant cancer cells, SIRT5 appears to play a tumor suppressive role by reversing BCL-2 upregulation and mitigating epigenetic dysregulation in response to elevated 2-HG levels (45). Thus, SIRT5 likely impacts cancer biology in a context-specific manner, in some situations, promoting cancer cell survival and proliferation (51, 56, 57) and, in others, functioning to restrict cancer growth (45, 77). Such Janus-faced activity is similar to roles described for other sirtuins in malignancy (24).
Clearly, much remains to be learned about the biology of SIRT5. In this regard, in addition to direct substrates of SIRT5, several key cellular regulators have shown to be impacted by SIRT5 via mechanisms that remain undefined. In particular, expression levels of Glut1, Ldha, Bcl-xl, Bcl-2, Sun2, Nrf2 (45, 50, 56, 106, 118), promitophagy genes (Bnip3, Pink1, and Park2), proautophagy genes (Map1lc3b, Gabarap, and Gabarapl2) (76), pro-MDR genes (Gclc, Gclm, Ho-1, Gsr, and Gpx2), and proxenobiotic genes (Nqo-1, Txn, Txnrd1, Mrp1/2, and Atp7a) (56) are all altered by manipulation of SIRT5 levels. Whether SIRT5 regulates expression of these genes via direct chromatin modification, or alternatively whether these effects represent indirect consequences of changes in mitochondrial and cellular physiology brought about by alterations in SIRT5 activity, represents an important open question in this area.
Hundreds of potential SIRT5 substrates have been identified, and likewise hundreds were suggested to associate with endogenous SIRT5 in immunoprecipitation/mass spectrometry studies (59). The biological significance of SIRT5 has been elucidated in the context of only an extremely small fraction of these targets and interactors. Among this handful of targets, it seems that no overall conclusions can be made at this point regarding the general activating or inhibitory effects of desuccinylation, demalonylation, or deglutarylation on SIRT5 targets. Similarly, given the current state of this field, it is challenging to correlate specific SIRT5 target modifications and the pro- or antitumorigenic functions of SIRT5 as a general matter. There is no doubt that further studies in this area will provide a wealth of insights into functions of SIRT5 and its targets in normal and neoplastic cells.
A key question revolves around potential cooperativity, redundancy, or antagonism of SIRT5 with other sirtuins, especially SIRT3 and SIRT4, and relationships between the Kac and target modifications of SIRT5, Ksucc, Kmal, and Kglu. In this regard, mitochondrial sirtuins share numerous substrates in common (60, 116). This is relevant since all three mitochondrial sirtuins depend on the presence of mitochondrial NAD+ for activity, thus raising the general question of how the activities of sirtuins may be differentially regulated. The observation that sirtuins show differing affinities for NAD+, and differing sensitivity to inhibition by NAM, may offer a partial answer to this question (19). Adding to the complexity of potential cross-talk among sirtuins, distinct lysine modifications may exert additive or even opposing effects on substrate function, especially since Kac is quite chemically distinct from major target modifications of SIRT5.
SIRT5 may represent an attractive pharmacological target for treatment of specific cancer types, with a potentially wide therapeutic window. In specific cancer types, including uterine, breast, cutaneous and uveal melanoma, lung, and lymphoma, the SIRT5 gene frequently shows increased copy number; likewise, SIRT5 mRNA and protein expression is elevated in multiple tumor types (Figs. 1 and 2). Development of SIRT5 inhibitors suitable for in vivo studies represents a significant area of current investigation in many laboratories.
Abbreviations Used
- 1,3-BPG
1,3-bisphosphoglycerate
- 293T
human epithelial kidney 293 SV40 large T antigen cell line
- 2-HG
(R)-2-hydroxyglutarate
- A549
human nonsmall cell lung adenocarcinoma cell line
- Acon
aconitate
- Ala86
alanine 86
- Arg
arginine
- Arg105
arginine 105
- Arg-Suc
arginosuccinate
- Asp
aspartate
- BCL-2
B cell lymphoma-2
- BCL-XL
B cell lymphoma-XL
- C2C12
mouse myoblast cell line
- CDDP
cisplatin
- Cit
citrate
- Citrul
citrulline
- CoA
coenzyme A
- CPS1
carbamoyl phosphate synthetase 1
- Cyt C
cytochrome C
- ECHA
enoyl-coenzyme A hydratase
- ETC
electron transport chain
- FAO
fatty acid oxidation
- FOXO3A
forkhead box O3A
- Fum
fumarate
- G-3-P
glucose-3-phosphate
- G6P
glucose-6-phosphate
- G6PD
glucose-6-phosphate dehydrogenase
- GAPDH
glyceraldehyde phosphate dehydrogenase
- GDH
glutamate dehydrogenase
- Gln
glutamine
- GLS
glutaminase
- Glu
glutamate
- GLUT1
glucose transporter 1
- GSH
glutathione
- GSR
glutathione reductase
- GSSG
oxidized glutathione
- H92c
rat embryonic cardiomyocyte cell line
- HADH
hydroxyacyl-coenzyme A dehydrogenase
- IC50
50% inhibitory concentration
- IDH
isocitrate dehydrogenase
- IMM
inner mitochondrial membrane
- IsoC
isocitrate
- Kac
acetyllysine
- KAT
lysine acetyltransferase
- Kglu
glutaryllysine
- Kmal
malonyllysine
- KO
knockout
- Ksucc
succinyllysine
- L929
mouse fibroblast cells
- LDHA/B
lactase dehydrogenase A/B
- Mal
malate
- MC3482
Sirt5 inhibitor
- MDA-MB
231 human metastatic breast cancer cell line
- MDR
multidrug resistance
- MEFs
mouse endothelial fibroblasts
- MLS
mitochondrial localization sequence
- MMP
mitochondrial membrane potential
- MTF
mitochondrial trifunctional
- NAD+
nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
- NADPH
reduced form of NADP+
- NAM
nicotinamide
- NDUFA4
complex I subunit
- NH3/NH4+
ammonia
- NRF2
nuclear factor erythroid-2-related factor 2
- NSCLC
nonsmall cell lung cancer
- OAA
oxaloacetate
- Orn
ornithine
- PC3
human prostate cancer cell line
- PDC
pyruvate dehydrogenase complex
- PDCE1α
pyruvate dehydrogenase complex E1α subunit
- PEP
phosphoenolpyruvate
- PKM2
pyruvate kinase muscle isozyme M2
- PPP
pentose phosphate pathway
- Q
quinolone
- R-5-P
ribose 5 phosphate
- R5PI
ribulose 5 phosphate isomerase
- ROS
reactive oxygen species
- SDHA
succinate dehydrogenase subunit A
- SIRT1–7
seven mammalian sirtuin proteins
- SIRT5
sirtuin 5
- SIRT5iso1/iso2
SIRT5 isoform 1/2
- Sirtuins
silent information regulator 2
- SOD
superoxide dismutase
- STAT3
signal transducer and activator of transcription
- Suc
succinate
- Suc-CoA
succinyl coenzyme A
- SUN2
SUN domain-containing protein 2
- TCA
tricarboxylic acid cycle
- Tyr102
tyrosine 102
- VLCAD
very long chain acyl-CoA dehydrogenase
- VLCFA
very long fatty acyl-CoA
- WT
wild-type
- α-KG
alpha ketoglutarate
Acknowledgments
The authors would like to thank Dr. Scott Tomlins for contributing Figure 2 and Drs. Hening Lin, Adam Stein, Surinder Kumar, and William Giblin for helpful discussions. L.R.B.-R. and A.H.G. are supported by NIH training grants 4T32HL007517 and T32GM113900, respectively. The Lombard laboratory is supported by awards from NIH (R01GM101171, 2R01HL114858, and R21AG053561), DoD (OC140123), the Glenn Foundation for Medical Research, the Melanoma Research Alliance, Love Your Melon/St. Baldrick's Foundation, the Harrington Discovery Institute, and the Allen H. Blondy Research Fellowship for Melanoma from the University of Michigan Cancer Research Committee.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, Wagner GR, Thompson JW, Madsen AS, Green MF, Sivley RM, Ilkayeva OR, Stevens RD, Backos DS, Capra JA, Olsen CA, Campbell JE, Muoio DM, Grimsrud PA, and Hirschey MD. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab 25: 838–855.e15, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balendiran GK, Dabur R, and Fraser D. The role of glutathione in cancer. Cell Biochem Funct 22: 343–352, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Bartosch C, Monteiro-Reis S, Almeida-Rios D, Vieira R, Castro A, Moutinho M, Rodrigues M, Graca I, Lopes JM, and Jeronimo C. Assessing sirtuin expression in endometrial carcinoma and non-neoplastic endometrium. Oncotarget 7: 1144–1154, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belenky P, Bogan KL, and Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci 32: 12–19, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, and Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem 277: 45099–45107, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Bowman CE, Rodriguez S, Selen Alpergin ES, Acoba MG, Zhao L, Hartung T, Claypool SM, Watkins PA, and Wolfgang MJ. The mammalian malonyl-CoA synthetase ACSF3 is required for mitochondrial protein malonylation and metabolic efficiency. Cell Chem Biol 24: 673–684.e4, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylston JA, Sun J, Chen Y, Gucek M, Sack MN, and Murphy E. Characterization of the cardiac succinylome and its role in ischemia-reperfusion injury. J Mol Cell Cardiol 88: 73–81, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, Kastenberger M, Bogdan C, Schleicher U, Mackensen A, Ullrich E, Fichtner-Feigl S, Kesselring R, Mack M, Ritter U, Schmid M, Blank C, Dettmer K, Oefner PJ, Hoffmann P, Walenta S, Geissler EK, Pouyssegur J, Villunger A, Steven A, Seliger B, Schreml S, Haferkamp S, Kohl E, Karrer S, Berneburg M, Herr W, Mueller-Klieser W, Renner K, and Kreutz M. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab 24: 657–671, 2016 [DOI] [PubMed] [Google Scholar]
- 9.Buler M, Aatsinki S-M, Izzi V, Uusimaa J, and Hakkola J. SIRT5 is under the control of PGC-1α and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB J 28: 3225–3237, 2014 [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarty SP. and Balaram H. Reversible binding of zinc in Plasmodium falciparum Sir2: structure and activity of the apoenzyme. Biochim Biophys Acta 1804: 1743–1750, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Chalkiadaki A. and Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer 15: 608–624, 2015 [DOI] [PubMed] [Google Scholar]
- 11a.Chandel NS. Navigating Metabolism. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2015. [Google Scholar]
- 12.Chang HC. and Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab 25: 138–145, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christofk HR, Vander Heiden MG, Wu N, Asara JM, and Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452: 181–186, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, and Sayers EW. GenBank. Nucleic Acids Res 44: D67–D72, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark O, Yen K, and Mellinghoff IK. Molecular pathways: isocitrate dehydrogenase mutations in cancer. Clin Cancer Res 22: 1837–1842, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, and Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.This reference has been deleted
- 18.Feldman JL, Dittenhafer-Reed KE, and Denu JM. Sirtuin catalysis and regulation. J Biol Chem 287: 42419–42427, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldman JL, Dittenhafer-Reed KE, Kudo N, Thelen JN, Ito A, Yoshida M, and Denu JM. Kinetic and structural basis for acyl-group selectivity and NAD(+) dependence in sirtuin-catalyzed deacylation. Biochemistry 54: 3037–3050, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finnin MS, Donigian JR, and Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol 8: 621–625, 2001 [DOI] [PubMed] [Google Scholar]
- 21.Fischer F, Gertz M, Suenkel B, Lakshminarasimhan M, Schutkowski M, and Steegborn C. Sirt5 deacylation activities show differential sensitivities to nicotinamide inhibition. PLoS One 7: e45098, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun 260: 273–279, 1999 [DOI] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, and Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giblin W, Skinner ME, and Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet 30: 271–286, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glorioso C, Oh S, Douillard GG, and Sibille E. Brain molecular aging, promotion of neurological disease and modulation by sirtuin 5 longevity gene polymorphism. Neurobiol Dis 41: 279–290, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guedouari H, Daigle T, Scorrano L, and Hebert-Chatelain E. Sirtuin 5 protects mitochondria from fragmentation and degradation during starvation. Biochim Biophys Acta 1864: 169–176, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Guetschow ED, Kumar S, Lombard DB, and Kennedy RT. Identification of sirtuin 5 inhibitors by ultrafast microchip electrophoresis using nanoliter volume samples. Anal Bioanal Chem 408: 721–731, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, and Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126: 941–954, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D. and Weinberg RA. Hallmarks of cancer: the next generation. Cell 144: 646–674, 2011 [DOI] [PubMed] [Google Scholar]
- 30.He B, Du J, and Lin H. Thiosuccinyl peptides as Sirt5-specific inhibitors. J Am Chem Soc 134: 1922–1925, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, and Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49: 186–199, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, and Bedalov A. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res 66: 4368–4377, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, and Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirschey MD. and Zhao Y. Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Mol Cell Proteomics 14: 2308–2315, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houtkooper RH, Pirinen E, and Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igci M, Kalender ME, Borazan E, Bozgeyik I, Bayraktar R, Bozgeyik E, Camci C, and Arslan A. High-throughput screening of Sirtuin family of genes in breast cancer. Gene 586: 123–128, 2016 [DOI] [PubMed] [Google Scholar]
- 37.Imai S. and Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol 24: 464–471, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.James AM, Hoogewijs K, Logan A, Hall AR, Ding S, Fearnley IM, and Murphy MP. Non-enzymatic N-acetylation of lysine residues by acetylCoA often occurs via a proximal s-acetylated thiol intermediate sensitive to glyoxalase II. Cell Rep 18: 2105–2112, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong SM, Lee A, Lee J, and Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem 289: 4135–4144, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing H. and Lin H. Sirtuins in epigenetic regulation. Chem Rev 115: 2350–2375, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Karwicka E. Role of glutathione in the multidrug resistance in cancer. Adv Cell Biol 2: 105–124, 2010 [Google Scholar]
- 41.Kulkarni RA, Worth AJ, Zengeya TT, Shrimp JH, Garlick JM, Roberts AM, Montgomery DC, Sourbier C, Gibbs BK, Mesaros C, Tsai YC, Das S, Chan KC, Zhou M, Andresson T, Weissman AM, Linehan WM, Blair IA, Snyder NW, and Meier JL. Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem Biol 24: 231–242, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S. and Lombard DB. Mitochondrial sirtuins and their relationships with metabolic disease and cancer. Antioxid Redox Signal 22: 1060–1077, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu ML, Hsu CM, and Yang MY. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol 34: 1847–1854, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, and Haigis MC. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell 50: 686–698, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, He X, Ye D, Lin Y, Yu H, Yao C, Huang L, Zhang J, Wang F, Xu S, Wu X, Liu L, Yang C, Shi J, He X, Liu J, Qu Y, Guo F, Zhao J, Xu W, and Zhao S. NADP(+)-IDH mutations promote hypersuccinylation that impairs mitochondria respiration and induces apoptosis resistance. Mol Cell 60: 661–675, 2015 [DOI] [PubMed] [Google Scholar]
- 46.Li L. and Bhatia R. The controversial role of Sirtuins in tumorigenesis—SIRT7 joins the debate. Cell Res 23: 10–12, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L, Liang J, Cheng Z, Shi L, Shang Y, and Yu W. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun 7: 12235, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang F, Wang X, Ow SH, Chen W, and Ong WC. Sirtuin 5 is anti-apoptotic and anti-oxidative in cultured SH-EP neuroblastoma cells. Neurotox Res 31: 63–76, 2017 [DOI] [PubMed] [Google Scholar]
- 49.Lin JB, Kubota S, Ban N, Yoshida M, Santeford A, Sene A, Nakamura R, Zapata N, Kubota M, Tsubota K, Yoshino J, Imai S, and Apte RS. NAMPT-mediated NAD(+) biosynthesis is essential for vision in mice. Cell Rep 17: 69–85, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, Jin W, Huang HH, and Chen X. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun 441: 191–195, 2013 [DOI] [PubMed] [Google Scholar]
- 51.Liu B, Che W, Zheng C, Liu W, Wen J, Fu H, Tang K, Zhang J, and Xu Y. SIRT5: a safeguard against oxidative stress-induced apoptosis in cardiomyocytes. Cell Physiol Biochem 32: 1050–1059, 2013 [DOI] [PubMed] [Google Scholar]
- 52.Liu J, Huang Y, and Zheng W. A selective cyclic peptidic human SIRT5 inhibitor. Molecules 21: 1217, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, and Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lombard DB. and Zwaans BM. SIRT3: as simple as it seems? Gerontology 60: 56–64, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lozy F. and Karantza V. Autophagy and cancer cell metabolism. Semin Cell Dev Biol 23: 395–401, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu W, Zuo Y, Feng Y, and Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol 35: 10699–10705, 2014 [DOI] [PubMed] [Google Scholar]
- 57.Lv XB, Liu L, Cheng C, Yu B, Xiong L, Hu K, Tang J, Zeng L, and Sang Y. SUN2 exerts tumor suppressor functions by suppressing the Warburg effect in lung cancer. Sci Rep 5: 17940, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahlknecht U, Ho AD, Letzel S, and Voelter-Mahlknecht S. Assignment of the NAD-dependent deacetylase sirtuin 5 gene (SIRT5) to human chromosome band 6p23 by in situ hybridization. Cytogenet Genome Res 112: 208–212, 2006 [DOI] [PubMed] [Google Scholar]
- 59.Marcon E, Jain H, Bhattacharya A, Guo H, Phanse S, Pu S, Byram G, Collins BC, Dowdell E, Fenner M, Guo X, Hutchinson A, Kennedy JJ, Krastins B, Larsen B, Lin ZY, Lopez MF, Loppnau P, Miersch S, Nguyen T, Olsen JB, Paduch M, Ravichandran M, Seitova A, Vadali G, Vogelsang MS, Whiteaker JR, Zhong G, Zhong N, Zhao L, Aebersold R, Arrowsmith CH, Emili A, Frappier L, Gingras AC, Gstaiger M, Paulovich AG, Koide S, Kossiakoff AA, Sidhu SS, Wodak SJ, Graslund S, Greenblatt JF, and Edwards AM. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat Methods 12: 725–731, 2015 [DOI] [PubMed] [Google Scholar]
- 60.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, and Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159: 1615–1625, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsushita N, Yonashiro R, Ogata Y, Sugiura A, Nagashima S, Fukuda T, Inatome R, and Yanagi S. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells 16: 190–202, 2011 [DOI] [PubMed] [Google Scholar]
- 62.Maurer B, Rumpf T, Scharfe M, Stolfa DA, Schmitt ML, He W, Verdin E, Sippl W, and Jung M. Inhibitors of the NAD(+)-dependent protein desuccinylase and demalonylase Sirt5. ACS Med Chem Lett 3: 1050–1053, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miao P, Sheng S, Sun X, Liu J, and Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life 65: 904–910, 2013 [DOI] [PubMed] [Google Scholar]
- 64.Michishita E, Park JY, Burneskis JM, Barrett JC, and Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16: 4623–4635, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Min J, Landry J, Sternglanz R, and Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell 105: 269–279, 2001 [DOI] [PubMed] [Google Scholar]
- 66.Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, Nishimura J, Hata T, Takemasa I, Mizushima T, Doki Y, Mori M, and Ishii H. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer 113: 492–499, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morris-Blanco KC, Dave KR, Saul I, Koronowski KB, Stradecki HM, and Perez-Pinzon MA. Protein kinase C epsilon promotes cerebral ischemic tolerance via modulation of mitochondrial Sirt5. Sci Rep 6: 29790, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakagawa T, Lomb DJ, Haigis MC, and Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137: 560–570, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Network TCGAR. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, and Verdin E. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell 59: 321–332, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.North BJ, Schwer B, Ahuja N, Marshall B, and Verdin E. Preparation of enzymatically active recombinant class III protein deacetylases. Methods 36: 338–345, 2005 [DOI] [PubMed] [Google Scholar]
- 72.This reference has been deleted
- 73.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, and Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 50: 919–930, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, and Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics 10: M111.012658, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pfister JA, Ma C, Morrison BE, and D'Mello SR. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS One 3: e4090, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T, Schutkowski M, Pellegrini L, Sansone L, Villanova L, Runci A, Pucci B, Morgante E, Fini M, Mai A, Russo MA, and Tafani M. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy 11: 253–270, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Preyat N, Rossi M, Kers J, Chen L, Bertin J, Gough PJ, Le Moine A, Rongvaux A, Van Gool F, and Leo O. Intracellular nicotinamide adenine dinucleotide promotes TNF-induced necroptosis in a sirtuin-dependent manner. Cell Death Differ 23: 29–40, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiu X, Brown K, Hirschey MD, Verdin E, and Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12: 662–667, 2010 [DOI] [PubMed] [Google Scholar]
- 79.Rao KS, Albro M, Dwyer TM, and Frerman FE. Kinetic mechanism of glutaryl-CoA dehydrogenase. Biochemistry 45: 15853–15861, 2006 [DOI] [PubMed] [Google Scholar]
- 80.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, and Verdin E. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab 18: 920–933, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Revollo JR, Grimm AA, and Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem 279: 50754–50763, 2004 [DOI] [PubMed] [Google Scholar]
- 82.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, and Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9: 166–180, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, Roeder RG, and Allis CD. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell 58: 203–215, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sabari BR, Zhang D, Allis CD, and Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol 18: 90–101, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sadhukhan S, Liu X, Ryu D, Nelson OD, Stupinski JA, Li Z, Chen W, Zhang S, Weiss RS, Locasale JW, Auwerx J, and Lin H. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc Natl Acad Sci U S A 113: 4320–4325, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, and Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 382: 790–801, 2008 [DOI] [PubMed] [Google Scholar]
- 87.Schuetz A, Min J, Antoshenko T, Wang CL, Allali-Hassani A, Dong A, Loppnau P, Vedadi M, Bochkarev A, Sternglanz R, and Plotnikov AN. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure 15: 377–389, 2007 [DOI] [PubMed] [Google Scholar]
- 88.Schwartzenberg-Bar-Yoseph F, Armoni M, and Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 64: 2627–2633, 2004 [DOI] [PubMed] [Google Scholar]
- 89.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, and Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143: 802–812, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suenkel B, Fischer F, and Steegborn C. Inhibition of the human deacylase Sirtuin 5 by the indole GW5074. Bioorg Med Chem Lett 23: 143–146, 2013 [DOI] [PubMed] [Google Scholar]
- 91.Sun JY, Xu L, Tseng H, Ciccarelli B, Fulciniti M, Hunter ZR, Maghsoudi K, Hatjiharissi E, Zhou Y, Yang G, Zhu B, Liu X, Gong P, Ioakimidis L, Sheehy P, Patterson CJ, Munshi NC, O'Connor OA, and Treon SP. Histone deacetylase inhibitors demonstrate significant preclinical activity as single agents, and in combination with bortezomib in Waldenstrom's macroglobulinemia. Clin Lymphoma Myeloma Leuk 11: 152–156, 2011 [DOI] [PubMed] [Google Scholar]
- 92.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, and Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest 119: 2758–2771, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takeshita A, Naito K, Shinjo K, Sahara N, Matsui H, Ohnishi K, Beppu H, Ohtsubo K, Horii T, Maekawa M, Inaba T, and Ohno R. Deletion 6p23 and add(11)(p15) leading to NUP98 translocation in a case of therapy-related atypical chronic myelocytic leukemia transforming to acute myelocytic leukemia. Cancer Genet Cytogenet 152: 56–60, 2004 [DOI] [PubMed] [Google Scholar]
- 94.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, and Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab 19: 605–617, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taylor WA, Mejia EM, Mitchell RW, Choy PC, Sparagna GC, and Hatch GM. Human trifunctional protein alpha links cardiolipin remodeling to beta-oxidation. PLoS One 7: e48628, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci 62: 1784–1803, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trapp J, Meier R, Hongwiset D, Kassack MU, Sippl W, and Jung M. Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins). ChemMedChem 2: 1419–1431, 2007 [DOI] [PubMed] [Google Scholar]
- 98.Vahtola E, Louhelainen M, Merasto S, Martonen E, Penttinen S, Aahos I, Kyto V, Virtanen I, and Mervaala E. Forkhead class O transcription factor 3a activation and Sirtuin1 overexpression in the hypertrophied myocardium of the diabetic Goto-Kakizaki rat. J Hypertens 26: 334–344, 2008 [DOI] [PubMed] [Google Scholar]
- 99.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, and Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 20: 1256–1261, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Visavadiya NP, Keasey MP, Razskazovskiy V, Banerjee K, Jia C, Lovins C, Wright GL, and Hagg T. Integrin-FAK signaling rapidly and potently promotes mitochondrial function through STAT3. Cell Commun Signal 14: 32, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wagner GR, Bhatt DP, O'Connell TM, Thompson JW, Dubois LG, Backos DS, Yang H, Mitchell GA, Ilkayeva OR, Stevens RD, Grimsrud PA, and Hirschey MD. A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab 25: 823–837.e8, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wagner GR. and Hirschey MD. A prob(e)able route to lysine acylation. Cell Chem Biol 24: 126–128, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang F, Nguyen M, Qin FX, and Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 6: 505–514, 2007 [DOI] [PubMed] [Google Scholar]
- 104.Wang F, Wang K, Xu W, Zhao S, Ye D, Wang Y, Xu Y, Zhou L, Chu Y, Zhang C, Qin X, Yang P, and Yu H. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1beta production and to prevent DSS-induced colitis in mice. Cell Rep 19: 2331–2344, 2017 [DOI] [PubMed] [Google Scholar]
- 105.Wang Y, Zhu Y, Xing S, Ma P, and Lin D. SIRT5 prevents cigarette smoke extract-induced apoptosis in lung epithelial cells via deacetylation of FOXO3. Cell Stress Chaperones 20: 805–810, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xiangyun Y, Xiaomin N, Linping G, Yunhua X, Ziming L, Yongfeng Y, Zhiwei C, and Shun L. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget 8: 6984–6993, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, and Zhao Y. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics 11: 100–107, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu YS, Liang JJ, Wang Y, Zhao XJ, Xu L, Xu YY, Zou QC, Zhang JM, Tu CE, Cui YG, Sun WH, Huang C, Yang JH, and Chin YE. STAT3 undergoes acetylation-dependent mitochondrial translocation to regulate pyruvate metabolism. Sci Rep 6: 39517, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang L-L, He Y-Y, Chen Q-L, Qian S, Wang Z-Y. Design and synthesis of new 9-substituted norharmane derivatives as potential Sirt5 inhibitors. J Heterocycl Chem 54: 1457–1466, 2016 [Google Scholar]
- 110.Yang L, Ma X, He Y, Yuan C, Chen Q, Li G, and Chen X. Sirtuin 5: a review of structure, known inhibitors and clues for developing new inhibitors. Sci China Life Sci 60: 249–256, 2017 [DOI] [PubMed] [Google Scholar]
- 111.Yu H, Pardoll D, and Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9: 798–809, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu J, Haldar M, Mallik S, and Srivastava DK. Role of the substrate specificity-defining residues of human SIRT5 in modulating the structural stability and inhibitory features of the enzyme. PLoS One 11: e0152467, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, Lin H, Schoonjans K, and Auwerx J. Metabolic characterization of a Sirt5 deficient mouse model. Sci Rep 3: 2806, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zang W, Hao Y, Wang Z, and Zheng W. Novel thiourea-based sirtuin inhibitory warheads. Bioorg Med Chem Lett 25: 3319–3324, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y, Bharathi SS, Rardin MJ, Lu J, Maringer KV, Sims-Lucas S, Prochownik EV, Gibson BW, and Goetzman ES. Lysine desuccinylase SIRT5 binds to cardiolipin and regulates the electron transport chain. J Biol Chem 292: 10239–10249, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Y, Bharathi SS, Rardin MJ, Uppala R, Verdin E, Gibson BW, and Goetzman ES. SIRT3 and SIRT5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-CoA dehydrogenase. PLoS One 10: e0122297, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao K, Harshaw R, Chai X, and Marmorstein R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc Natl Acad Sci U S A 101: 8563–8568, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou L, Wang F, Sun R, Chen X, Zhang M, Xu Q, Wang Y, Wang S, Xiong Y, Guan KL, Yang P, Yu H, and Ye D. SIRT5 promotes IDH2 desuccinylation and G6PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep 17: 811–822, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]