Abstract
Neurodevelopmental disorders (NDDs) represent a diverse group of syndromes characterized by abnormal development of the central nervous system and whose symptomatology includes cognitive, emotional, sensory, and motor impairments. The identification of causative genetic defects has allowed for creation of transgenic NDD mouse models that have revealed pathophysiological mechanisms of disease phenotypes in a neural circuit- and cell type-specific manner. Mouse models of several syndromes, including Rett syndrome, Fragile X syndrome, Angelman syndrome, Neurofibromatosis type 1, etc., exhibit abnormalities in the structure and function of dopaminergic circuitry, which regulates motivation, motor behavior, sociability, attention, and executive function. Recent advances in technologies for functional circuit mapping, including tissue clearing, viral vector-based tracing methods, and optical readouts of neural activity, have refined our knowledge of dopaminergic circuits in unperturbed states, yet these tools have not been widely applied to NDD research. Here, we will review recent findings exploring dopaminergic function in NDD models and discuss the promise of new tools to probe NDD pathophysiology in these circuits.
Graphical abstract
Introduction
In the last decade, the widespread adoption of technologies for functional circuit mapping in animal models has greatly enhanced our ability to understand the input-output relationships between populations of neurons and determine their function in vivo. These include techniques for the visualization, reconstruction, and analysis of intact circuits across micro- and macroscales. Examples include serial section electron microscopy [1,2], the Brainbow toolkit [3,4] and intersectional labelling strategies [5,6], improved neuroinformatic tools for neurite tracing [7], tissue clearing [8,9], light sheet microscopy [10,11], and serial two-photon tomography [12,13]. Additionally, optogenetic [14] and chemogenetic [15] actuators, genetically encoded indicators of neuronal activity [16,17], and advanced in vivo imaging modalities [18–23] have allowed for the functional deconstruction of genetically defined circuits in order to probe their contributions to complex behaviors. The development of viral vectors that can deliver transgenes in a pathway- and cell type-specific manner [24–28] or broadly transduce neurons across the CNS [29] have greatly facilitated efforts to anatomically and functionally characterize complex neurobiological systems in both basal and disease states.
New tools for ‘connectomic’ or circuit-centered research that can survey large scale functional connectivity patterns are particularly well suited to the study of neurodevelopmental disorders (NDDs), such as autism spectrum disorder (ASD), where diverse genetic and environmental insults during neurodevelopment can vastly perturb circuit architecture and physiology across brain areas [30,31]. While the neural substrates of ASD symptomatology are multifaceted, mesencephalic dopamine systems, consisting of A8 retrorubral, A9 nigrostriatal, and A10 mesocorticolimbic projections [32], represent circuits of interest given their potential contribution to several common ASD symptoms, including perseverant interests, stereotyped movements, impaired attention and executive function, and difficulty with social interactions [33]. Several recent studies implicate these circuits in behavioral phenotypes observed in rodent NDD disease models, including Angelman syndrome (AS), Rett syndrome (RS), fragile X syndrome (FXS), neurofibromatosis type 1 (NF1), etc. (Table 1), yet widespread adoption of new tools for functional circuit mapping has yet to occur in these models. In this review, we will highlight common patterns of cellular and circuit level phenotypic variation across NDD mouse models and discuss the promise of recent neurotechnological advances such as whole brain tissue clearing and gene delivery by systemic viral vectors to further elucidate NDD pathophysiology in dopaminergic circuits.
Table 1.
Diverse behavioral, synaptic, and cellular phenotypes are observed in nigrostriatal and mesocorticolimbic pathways in mouse models of neurodevelopmental syndromes and ASD candidate genes.
| Neurodevelopmental Syndromes | |||
|---|---|---|---|
| Syndrome | Mouse Model | Major Findings | Citation |
| 15q11–13 Duplication Syndrome | Ube3a-2× | Triplication of Ube3a synergizes with seizures to reduce expression of the glutamatergic synapse organizer Cbln1, impairs glutamatergic transmission in VTA neurons, and leads to loss of sociability. | [99] |
| 16p11.2 Deletion Syndrome | 16p11.2+/− | Mice carrying a homologous chromosomal deletion to 16p11.2 (7F3) exhibit abnormal synaptic signaling and increased numbers of dopamine D2 receptor (D2R)-expressing medium spiny neurons (MSNs) in the striatum, fewer D1 receptor (D1R)-expressing neurons in the cortex, locomotor hyperactivity, and deficits in motor control. | [157] |
| Angelman Syndrome | Ube3am−/p+ | Ube3am−/p+ mice display enhanced electrically evoked dopamine release in the NAc and reward seeking but decreased sensitivity to drugs that increase dopamine overflow. | [100] |
| TH-Cre∷Ube3am−/p+, TH-Cre∷Ube3aFLOX/p+ | Loss of maternal Ube3a in tyrosine hydroxylase (TH)-expressing neurons enhanced optical self-stimulation via increased GABA release from dopaminergic terminals in the NAc. | [101] | |
| Fragile X Syndrome | Fmr1−/y | Fmr1−/y mice are more sensitive to the rewarding effects but less sensitive to the motor effects of cocaine compared to wildtype; the number of TH-expressing neurons in reduced in the SNc but not VTA of these mice. | [158] |
| Fmr1−/y | Locomotor sensitization, conditioned place preference, and synaptic changes in the NAc following repeated cocaine is reduced in Fmr1−/y mice. | [159] | |
| Neurofibromatosis Type 1 | Nf1+/−:Nf1FLOX∷GFAP-Cre:Nf1FLOX/FLOX | Mice with one non-functional Nf1 allele in all somatic cells and complete Nf1 knockout in glial fibrillary acid protein (GFAP)-expressing cells display reduced striatal dopamine and TH expression in vivo and reduced dopaminergic neurite outgrowth in vitro. | [47] |
| Nf1+/−, TH-Cre∷Nf1FLOX/FLOX, GFAP-Cre∷GFAPFLOX/FLOX | Knockout of Nf1 in TH or GFAP-expressing cells is associated with reduced dopamine content in the hippocampus and deficits in spatial working memory. | [46] | |
| Rett Syndrome | Mecp2+/−, Mecp2−/y | SNc neurons exhibit decreased somal size, dendrite count, and striatal dopamine release in Mecp2+/− mice and symptomatic Mecp2−/y males. | [52] |
| Mecp2+/− | Mecp2+/− display aberrant motor coordination and motor skill learning secondary to reduced striatal dopamine content, down-regulation of tyrosine hydroxylase expression, and dopamine D2 receptor (D2R) up-regulation. | [102] | |
| Mecp2S421A | Loss of MeCP2 phosphorylation at position 421 results in accelerated amphetamine sensitization and changes in MSN excitability in the NAc. | [160] | |
| Dlx5/6-Cre∷DMecp2FLOX/y | Conditional knockout of Mecp2 in the striatum phenocopies Mecp2+/− mice in dopamine deregulation and motor dysfunction. | [161] | |
| Non-Syndromic ASD Genes | |||
| Gene (Product) | Mouse Model | Major Findings | Citation |
| Cntnap4 (CNTNAP4) | Cntnap4−/− | Loss of Cntnap2 results in enhanced dopamine release in the NAc and dorsal striatum through a presynaptic mechanism and results in excessive grooming. | [53] |
| Nlgn1 (Neuroligin-1) | Nlgn1−/− | Nlgn1−/− display reduced GluN2A-containing NMDA receptor currents and glutmatergic inputs in dopamine D1 receptor (D1R) and D2R-expressing striatal medium spiny neurons (MSNs), respectively. | [162] |
| Nlgn2 (Neuroligin-2) | Nlgn2 miR knockdown | Striatal knockdown of Nlgn2 results in downregulation of dopaminergic synapses and upregulation of GABAergic synapses. | [163] |
| Nlgn3 (Neuroligin-3) | Nlgn3−/−, Nlgn3R451C | Both Nlgn3 knockout mice and mice modeling the R451C polymorphism demonstrate enhanced repetitive motor routines by impairing inhibitory transmission onto D1R-expressing MSNs in the NAc. | [164] |
| Shank3 (SHANK3) (Note: loss of SHANK3 is also seen in Phelan McDermid Syndrome) | Shank3 shRNA knockdown | Shank3 knockdown via short hairpin RNA (shRNA) in the VTA impairs excitatory synapse maturation, reduces dopaminergic neuron excitability via increased inhibitory tone, reduces social preference, and can be rescued an mGluR1 agonist or optogenetic stimulation of dopaminergic neurons | [98] |
| Shank3b−/− | Loss of Shank3b alters the development of excitatory inputs to medium spiny neurons of the dorsomedial striatum, which can be rescued by chemogenetic inhibition of corticostriatal inputs. | [165] | |
| Shank3bfx/fx | Loss of Shank3b in a conditional knock-in model results in abnormal motor, social and exploratory behaviors; repetitive grooming; and synaptic changes in the striatum. These deficits are rescued with germline re-activation of Shank3 expression. | [166] | |
Elucidating abnormal patterns of dopaminergic connectivity in NDD models
Dopaminergic projection neurons are a heterogenous population whose function, activity, neurotransmitter content, and pattern of connectivity varies with brain region and connection target [34–36]. For example, efferents arising from the midbrain ventral tegmental area (VTA) project throughout the extended amygdala [including the nucleus accumbens (NAc)], hippocampus, and prefrontal cortex (the mesocorticolimbic pathway) and have been widely studied for their role in cognition, reinforcement, and motivation [37,38], while dopaminergic populations in the substantia nigra pars compacta (SNc) project primarily to the dorsal striatum (nigrostriatal pathway) and are critical for the selection and execution of motor programs and habitual behavior [39,40]. Other populations outside the mesencephalon include those in the dorsal raphe nucleus (DRN)/ventral periaqueductal gray area (vPAG) that affect social behavior, nociception, and arousal [41–43] and tuberoinfundibular projections from the hypothalamic arcuate nucleus to the median eminence that regulate prolactin release [44].
Mesencephalic dopaminergic neurons in mice arise from a pool of progenitors in the midbrain floor plate under the control of numerous signaling molecules, including sonic hedgehog, WNT1, engrailed 1 and 2, fibroblast growth factor-8, etc., undergo radial migration to their final positions in either the VTA or SNc by embryonic day 13.5, and exhibit extensive axonal outgrowth along the anteroposterior and dorsoventral axes with synaptogenesis in downstream targets continuing into postnatal development [45]. Consequently loss of NDD-associated genes, such as EN2, MECP2, CNTNAP2, and NF1, produce hypo- or hyperdopaminergic behavioral phenotypes, such as abnormal motor, cognitive, or social behavior, in mouse models by perturbing neuronal maturation, migration, or neurite outgrowth [46–53]. Efforts to understand these phenotypes would benefit from a comprehensive ultrastructural understanding of how specific NDD-associated genetic changes alter dopaminergic circuit architecture and function and inform new therapeutic strategies, such as whole brain gene therapy or genome editing, to help ameliorate NDD symptomatology.
Several recent viral vector-based labeling methods are likely to greatly enhance our understanding of the input-output relationship between dopaminergic efferent and afferent connections in NDD models (Table 2A). This toolkit includes a new adenoassociated viral (AAV) vector for retrograde labeling (AAV2-retro) [26] (in addition to existing retrograde labeling vectors [24,25,27]), intersectional strategies to target individual neuronal projections and their inputs (INTERSECT [5], TRIO [6]), mGRASP for fluorescent labeling of connections between synaptic partners [54,55], and a single cell projection mapping via RNA barcoding (MAP-seq [56]). The recently developed brain-penetrant AAV PHP.eB can efficiently deliver viral transgenes to the CNS after peripheral administration (Figure 1A–B) [29,57], including Brainbow reagents [58] for multicolor labeling via stochastic expression of fluorescent proteins (XFPs) [57] and genetically encoded calcium indicators (GECIs; [59,60]). This tool should also prove useful for non-invasive delivery of optogenetic [14] or chemogenetic [15] tools, AAV-optimized CRISPR-Cas9 effectors for genome editing (e.g. [61–63]), and therapeutic transgenes across large brain volumes. Additionally, PHP.eB can deliver the cargo of interest co-administered with a titratable inducer vector for controlled sparseness while maintaining high viral transgene copy number [57], which is beneficial for effective neurite tracing with methods such as mGRASP [55] or Brainbow [58] (Figure 1C–D). This method is also likely to benefit sensors that need sparse expression to reduce background fluorescence [64–66].
Table 2.
Selected recent advances in neurotechnologies for structural analysis of circuit architecture in rodent models.
| A. Viral Vectors and Vector-Based Labeling Methods | ||
|---|---|---|
| Tool | Summary | Citation |
| rAAV2-retro | A recombinant AAV2 variant for retrograde targeting of projection neurons that can be used for both functional and tracing studies. Efficient retrograde transduction was observed in many cortical and subcortical regions. | [26] |
| PHP.eB and PHP.S | AAV9-based vectors for efficient CNS (PHP.eB) or PNS (PHP.S) transduction after peripheral (intravenous or retro-orbital) virus administration. Can be used with a titratable inducer vector for controlled sparseness of multicolor labels that preserves color diversity; inducers involve use of tetracycline-controlled transactivator (tTA) or Cre-dependent Flp-based constructs. | [29,57] |
| TRIO | Tracing the Relationship Between Input and Output; A combinatorial two-vector system that maps the input-output relationship of a population of neurons. In this method, canine adenovirus-2 (CAV-2) [25] is used to deliver a Flp recombinase transgene to axons in a specific projection terminal field for retrograde transduction of the cell bodies; Flp-dependent RVdG [24] component vectors are later delivered to the cell bodies for monosynaptic retrograde tracing of inputs. Cre-dependent Flp can be used for cell type-specific targeting using a Cre driver line (cTRIO). Cannot be used for functional studies due to lethality of RVdG. | [6] |
| INTERSECT | INTronic Recombinase Sites Enabling Combinatory Targeting; A two-component system that allows for functional projection targeting using Flp- and Cre-dependent viral vectors via axonal targeting in a downstream region using replication incompetent herpes simplex virus (HSV) [27] carrying a Cre-dependent Flp recombinase transgene. Either Cre- and Flp-ON and -OFF strategies can be used. | [5] |
| MAP-Seq | Multiplexed Analysis of Projections by Sequencing; allows for parallel mapping of single neuron axonal arbors via recovery of RNA barcodes in from terminal fields after delivery of AAV viral barcode libraries to the cell body. Does not distinguish fibers of passage, so downstream regions must be chosen carefully for RNA recovery and sequencing. | [56] |
| mGRASP | Mammalian GFP Reconstitution Across Synaptic Partners; A method for fluorescently labeling synaptic connections that employs AAV-mediated delivery of synapse-targeted split GFP fragments in genetically defined pre- and post-synaptic neuronal partners. Cre-ON and Cre-OFF strategies can be used for studying microcircuits. | [54,55] |
| B. Large Volume Imaging Modalities | ||
| Light Sheet Microscopy (LSM) | Originally developed over 100 years ago, LSM illuminates the sample with a thin sheet of light and detects the emitted fluorescent signal with an orthogonally arranged detection objective. Variants include CLARITY optimized LSM (COLM) for use in cleared tissue [76], SPED (Spherical Aberration-assisted Extended Depth of Field) LSM that improves scan speed via extended depth of field [68], and an adaptive LSM that integrates multiple fields of view with 10 degrees of freedom that are autonomously adjusted in real time for improved spatial resolution and image quality [11]. | [10,11,68] |
| High-Speed Volumetric STP Tomography | High-speed Volumetric Serial Two-Photon Tomography; A high speed imaging platform based on Serial Two-Photon Tomography (STP) [13] that creates 3D reconstructions of neuronal axonal arbors via the integration of fast volumetric 2-photon microscopy and a vibrating microtome to image bright, sparsely labelled neurons in cleared samples embedded in gelatin. Includes computational tools for the registration and visualization of large (up to 100 TB) data sets, although labeling must be sufficiently sparse to prevent neurite reconstruction errors when axons from different neurons are closely positioned. | [12] |
| C. Tissue Clearing Methods | ||
| CLARITY | Hydrogel-based clearing method that utilizes 4% SDS for lipid removal after sample has been embedded in an acrylamide-bisacrylamide gel and cross-linked with formaldehyde. Clearing can be accelerated with electrophoresis at the expense of tissue integrity. Compatible with immunolabeling and endogenous fluorescence. The EDC-CLARITY variant is compatible with HCR (hybridization chain reaction) probes for bulk RNA labeling. | [75,76,86] |
| PACT PACT-deCAL | Passive CLARITY Technique; A passive CLARITY-based clearing method for rapid clearing of thick sections that employs 8% SDS as the detergent. Compatible with immunolabeling, endogenous fluorescence, smFISH (single molecule), and smHCR probes for single and bulk RNA labeling. Produces reversible expansion of tissue and can be used with RIMS (Reflective Index Matching Solution), a non-viscous mounting medium that decreases the refractive index of the sample for better optical access. PACT-deCAL uses EDTA/EGTA to decalcify samples for bone clearing. | [71,78,85,167] |
| PARS | Perfusion Assisted Agent Release In Situ; An active CLARITY-based clearing method that involves intracranial and/or transcardial perfusion of reagents for whole body clearing. | [71,78] |
| SWITCH | System-Wide Control of Interaction Time and Kinetics of Chemicals; A fixation and clearing method that exploits the pH dependence of glutaraldehyde-tissue gel formation for uniform fixation prior to delipidation with SDS. This method provides added tissue integrity for multiplexed immunolabeling. Not compatible with smFISH or smHCR probes. | [84] |
| uDISCO | A whole-body clearing method based on 3DISCO; it utilizes dehydration with tert-butanol followed by delipidation with diphenyl ether for fast sample clearing. Maintenance of endogenous fluorescence is improved relative to 3DISCO and other solvent based methods, whereby fluorescence deteriorates within several days after clearing. Shrinks tissues by approximately 40% for faster LSM imaging. | [79] |
| Sca/eS | An improved version of Sca/eA2 [168] that achieves tissue transparency via partial delipidation and hyperhydration via urea, sorbitol, glycerol, and Triton X-100. Preserves endogenous fluorescence and limits expansion better than most other methods, although large0 sample clearing can take several weeks. A simplified protocol Sca/eSQ can be used in thick (<500 micron) sections. | [72] |
| CUBIC | Clear, Unobstructed, Brain/Body Imaging Cocktails and Computational Analysis; A clearing method based on Sca/eA2 that uses urea, aminoalcohols, TRITON X-100, and high sucrose concentrations. Maintains endogenous fluorescence, can be perfused for whole body clearing, produces reversible tissue expansion, and exhibits superior decolorization (i.e. loss of the heme chromophore) relative to other techniques. | [73,74] |
| Tissue Expansion Methods for High Resolution Microscopy | ||
| ExM | Expansion Microscopy; A tissue expansion technology whereby the fixed and permeabilized sample is embedded in a superabsorbent hydrogel containing sodium acrylate and acrylamide, cross-linked with N-N′-methylenebisacrylamide, and digested with a protease to produce a 4.5-fold sample expansion. Newer variants display improved protein retention (proExM) and are compatible with immunolabeling, smFISH, and smHCR (ExFISH). | [80–82] |
| ePACT | Expansion PACT; Variant of the PACT tissue clearing method that utilizes a superabsorbent hydrogel and enzymatic digestion to increase sample size up to 5-fold for high resolution imaging with preserved endogenous fluorescence. | [78] |
| MAP | Magnified Analysis of Proteome; a hydrogel-based clearing method that expands tissue without the use of enzymatic digestion via treatment with high acrylamide concentrations (up to 20%) prior to SDS treatment. Compatible with immunolabeling but not RNA detection. | [83] |
Figure 1. Broadly transducing AAVs permit brainwide transgene expression and facilitate neurite tracing.
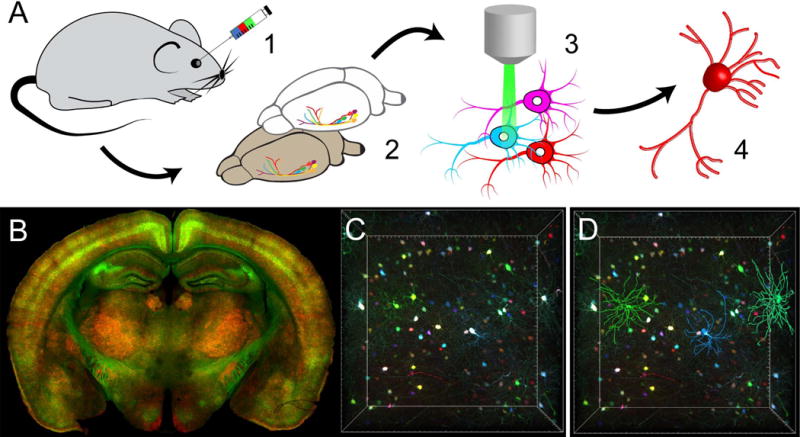
A. Workflow for multicolor labelling with PHP.eB and neurite tracing. Viral particles carrying red, green, or blue XFP transgenes with or without a titratable inducer are systemically introduced via retro-orbital injection (1). Following transduction (4–8 weeks), brains are fixed and cleared (2). Tissue samples can then be imaged with light sheet or confocal microscopy (3) prior to neurite tracing (4). B. Brainwide transduction of neurons (green) or astrocytes (red) using cell type-specific promoters (hSyn1 and GFAP, respectively) and gene regulatory elements following retro-orbital injection of PHP.eB (1×1012 viral genomes/mouse). C–D. Sparsely labelled striatal neurons were successfully traced after transduction by PHP.eB multicolor XFPs.
The utility of viral vector-based mapping tools has been improved by microscopic techniques for rapid imaging of large samples, such as light sheet microscopy [11,67,68] or high-speed volumetric serial two-photon (STP) tomography [12] (Table 2B), and tissue clearing protocols that render biological samples optically transparent for analysis of intact circuits in whole brains or thick slices (Figure 2) [8,9]. Several tissue clearing strategies have been recently described or refined (Table 2C); these include immersion clearing with high refractive index (RI) solutions (SeeDB [69], FRUIT [70], RIMS [71]), clearing via hyperhydration (Sca/eS [72], CUBIC [73,74]), hydrogel embedding followed by detergent delipidation (CLARITY [75,76], PARS [77,78], PACT [77,78]), and solvent-based clearing methods (uDISCO [79]). Clearing methods that build upon water-absorbent CLARITY hydrogels to create hyperabsorbant hydrogels have also been implemented to facilitate high resolution imaging of small structures, such as individual dendrites or neurites (ExM [80–82], ePACT [78], and MAP [83]). Hydrogel-based methods preserve endogenous fluorescence while maintaining compatibility with tools for proteomic analysis [76,78,82–84], RNA profiling (smFISH or smHCR probes [71,81,85,86]), and time-stamped fluorescent readouts of neuronal activity (e.g. ArcTRAP [87,88]).
Figure 2. Visualization of intact circuits using hydrogel-based clearing methods.
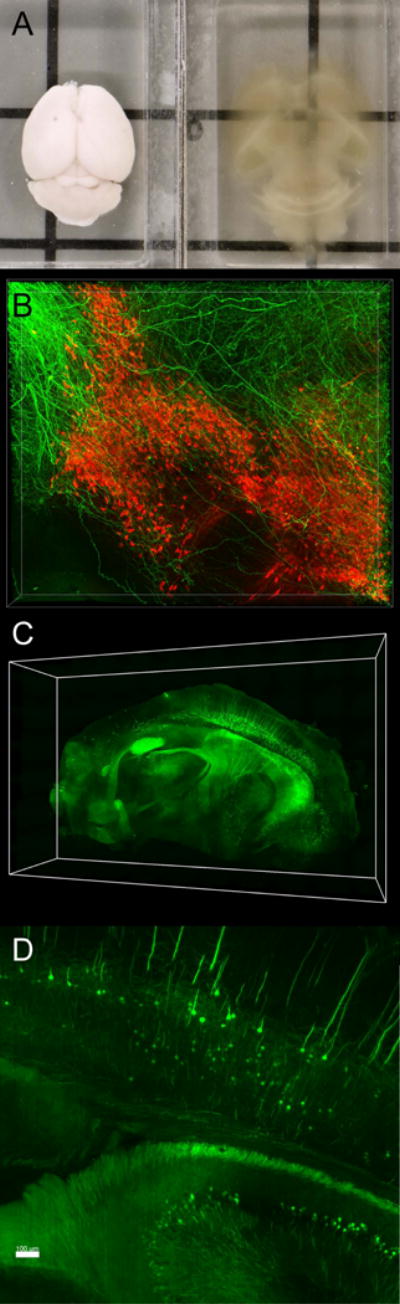
A. A mouse brain before and after clearing with PARS and long-term storage in RIMS (adapted from [71]). The sample demonstrates moderate tissue expansion due to acrylamide embedding. B. Confocal image of dopaminergic neurons in the SNc (red) and cholinergic afferents from the pedunculopontine tegmental nucleus (green) visualized in a 1mm-thick PACT-cleared section. C–D. Whole brain imaging of fluorescently-labelled cells in Thy1-eYFP mice using light sheet microscopy.
Several recent studies have successfully integrated these technologies to probe the structure of dopaminergic and related circuits in healthy mice. For example, retrograde labeling, tissue clearing, and LSM have been used to parse SNc subcircuit connectivity and function [89], identify an anatomically distinct projection to the posterior striatum [90] that preferentially encodes novel cue information rather than reward prediction errors [91], and refine our knowledge of cholinergic inputs to the SNc and VTA [92]. An input-output analysis of VTA connections using TRIO uncovered a novel projection from the anterior cingulate cortex to the lateral NAc that produces behavioral reinforcement using an optogenetic intracranial self-stimulation paradigm [93].
Bridging the gap between synaptic function and neural circuit dynamics in NDD models
One common feature amongst NDDs is that the causative genes (e.g. FMR1 in FXS, UBE3A in AS, MECP2 in RS, NF1 in NF1, EN1 and EN2, SHANK genes, etc.) affect synapse formation, maintenance, and plasticity in rodent models [94,95]. As such, there have been considerable efforts to characterize synapse function in dopaminergic circuits in these mice. For example, reduction in SHANK-3, an excitatory synapse scaffolding protein whose loss of function is associated with Phelan McDermid Syndrome (also called 22q13 deletion syndrome; see [96] for a review) and some non-syndromic ASD cases [97], via delivery of a short hairpin RNA (shRNA) into the VTA impairs maturation of excitatory synapses and reduces dopaminergic neuron excitability and social preference via increased inhibitory tone [98]. Mice modeling 15q11–13 Duplication Syndrome (where Ube3A protein levels are increased three-fold) exhibit a loss of sociability due to downregulation of the glutamatergic synapse organizer CBLN1 in the VTA [99]. Altered neurotransmitter content or release from VTA or SNc neurons in downstream targets has been reported in mouse models of AS [100,101], NF1 [46,47], and RS [52,102] and in Cntnap2 knockout mice [53]. While there is a large body of research delineating the role of synaptic or microcircuit deficits in NDD models [103–106], less is known about how those changes alter population dynamics or neuron ensemble activity to produce behavioral phenotypes; improvements in optical tools to monitor neural activity across multiple spatial scales [59,107–109] should help bridge this divide.
Understanding how networks of interconnected neurons encode and translate relevant environmental stimuli into a motivated behavior requires a high throughput readout of neuron firing with single-cell resolution. Metal electrodes or electrode arrays are a robust tool to measure spiking with high temporal precision and can be coupled with optogenetic tools to manipulate activity or infer cell identity (i.e. opto-tagging, [110]). Opto-tagging has been used to monitor diverse populations across the CNS (e.g. cortical interneurons [111,112], AgRP neurons in the arcuate nucleus [113], dopaminergic neurons in the VTA [114], etc.), yet this technique is limited in the number of neurons it can sample and may not be suitable for all populations due to the challenges in efficiently and accurately opto-tagging (e.g. cortical pyramidal neurons), as well as genetically similar populations that are too sparse or dense to be reliably identified. In contrast, optogenetic stimulation of dopaminergic circuitry during blood oxygen level dependent contrast (BOLD) fMRI imaging can approximate mesocorticolimbic or nigrostriatal network activity in rodents [115–117], yet this technique lacks both cellular resolution and temporally precise neurophysiological readouts.
Alternatively, genetically encoded calcium indicators (GECIs; e.g. the GCaMP6 family of proteins [16]) provide cell type-specific fluorescent readouts of neuron activity during behavior that is stable over months of testing and is scalable [21]. Using two-photon mesoscopes with wide field of view objectives [108] or random access scanning strategies [107] to image through large cranial windows in head-fixed mice, researchers can record the calcium dynamics of hundreds to thousands of neurons at once. Bulk calcium signals can also be measured across superficial cortical areas using a wide field fluorescence macroscope featuring a 12 mm field of view, which has been used to assess global representations of motivated behavior in multiple cell types [59]. While these tools have been optimized for relatively superficial (< 1mm deep) structures, several tools should help extend the depth of non-invasive optical access, such as three-photon microscopy [118], the implementation of axially elongated Bessel foci [119], photoacoustic tomography [120], and guidestar-assisted wavefront engineering techniques to limit optical scattering [121], such as time reversal of ultrasound encoded light (TRUE) [122,123].
Several recent technologies have provided optical access to deep brain areas in behaving mice for activity measurements in bulk or with single-cell resolution. For bulk measurements, fiber photometry [124] and TEMPO [125] allow for quantification of calcium or voltage sensor dynamics, respectively, using implanted optical fibers in order to correlate activity of genetically defined populations with behavioral events. Calcium imaging via implanted gradient index microendoscopes (GRIN lenses) provides single cell resolution at depths >4mm below the skull surface [126]. While two-photon GRIN lens imaging is most commonly performed in head-fixed mice [127], several strategies such as 2-photon fiberscopes [128,129] and miniaturized head-mounted 2-photon microscopes [130,131] have been developed for freely moving behavior. Head-mounted miniaturized epifluorescence microscopes [132] are also available and have been more widely adopted for use in behaving animals. Single cell calcium dynamics have been imaged via GRIN lens in the VTA [133], SNc [134], and interconnected regions, including the dorsal striatum [134], lateral hypothalamus [134,135], medial preoptic area [136], medial prefrontal cortex [137,138], bed nucleus of the stria terminalis [133], hippocampus [139,140], etc. Additionally, chronic imaging windows have permitted monitoring of sparsely labelled SNc axons in the dorsal striatum, which revealed distinct temporal and spatial encoding of reward and motor signals [39]. While several groups employ cortical two-photon calcium imaging in NDD models, including RS [141] and FXS [142] mice, analysis of deeper structures has not been reported to date.
Considerations and future outlook
The identification of causative genetic defects in neurodevelopmental syndromes and subsequent creation of transgenic mouse models has greatly enhanced our understanding of the developmental perturbations that produce synaptic, cellular, and behavioral phenotypes in these mice. While several recent studies examining dopaminergic circuitry have uncovered pathophysiological mechanisms underlying aberrant social interactions, positive reinforcement, stereotyped behavior, etc., few studies have employed new technologies for functional circuit mapping in NDD models. This may be due to several factors; first, given that phenotype expression is dependent on genetic background in many mouse models, such as NF1 [143], it will be important to continue identifying and developing minimal gene regulatory elements (promoters, enhancers, miRNA binding sites) that can be accommodated within well-tolerated viral capsids for cell type-specific targeting without the need to cross mice to Cre or Flp driver lines. Several cell type-specific promoters have been developed to target different cell populations in the CNS, including catecholaminergic (tyrosine hydroxylase promoter), serotonergic (FEV), Purkinje (PCP2) [144], and forebrain GABAergic (mDlx5/6) neurons [145], although they vary in leakiness and promoter size, which can limit packageable transgene size due to AAV carrying capacity of 4.7 kb [146]. Second, many of these techniques require specialized equipment, reagents, or expertise that makes implementation challenging. Several helpful imaging, tissue clearing, and data analysis protocols have recently been published [54,76,78,133,147–149] that can help guide potential users.
To effectively integrate measures of neural activity with comprehensive dopaminergic connectomes in mouse models obtained with such tools for precise structural and functional analysis of intact circuits, several advances will be required. First, we will need better computational methods for automated detection, segmentation, and tracing of individual fluorescently labelled neurons in whole cleared brains. This task is currently labor intensive and works poorly for neurons with complex morphology, such as catecholaminergic neurons with large axonal arbors that traverse several mm of brain tissue. Recent successes in overcoming these challenges include the reconstruction of single projection neurons in the claustrum, which branch extensively throughout the entire cerebrum [150]. Second, we will need improved tools for converting neural activity states into fluorescent labels that can be superimposed upon neuronal reconstructions. Several technologies show promise, such as CaMPARI [151] and iTANGO [152], which provide light timestamped indicators of intracellular calcium or dopaminergic neurotransmission, respectively. Hybridization chain reaction (HCR) probes for single-cell, multiplexed RNA detection have been validated for hydrogel-based clearing methods [86,153] and could allow for medium throughput identification of projection- or activity-dependent changes in gene activity in mutant and wildtype mice. At this time, only PACT/PARS [71], EDC-CLARITY [86], and Ex-FISH [81] have been demonstrated to be compatible with RNA profiling, yet clearing methods are advancing rapidly and will likely be useful for a broader range of applications in the future.
When examining the role of functional circuit mapping technologies in elucidating dopaminergic connectivity in NDD models, one cannot neglect the ontogeny of these circuits. Several methods have been used to clear mouse embryos at various stages of development (reviewed by [154]), yet it is difficult to employ viral vector-based tracing and labeling techniques in the developing mouse. It is thus of great interest to identify AAVs that cross the blood-placenta barrier and selectively target the developing embryonic nervous system. AAV selection platforms, such as CREATE (Cre recombination–based AAV targeted evolution), which has been used to develop vectors that efficiently target the central (PHP.B/PHP.eB) or peripheral (PHP.S) nervous systems [29,57] when given systemically, could yield new vectors for in utero transgene delivery. Additionally, tools for large volume functional imaging of developing organisms, such as a two-beam light sheet microscope with adaptive optics and automated cell tracking [11], have been applied to early embryonic mice [155,156], yet new methods to maintain optical access within the amnion will be necessary to image and track post-implantation fetal cells.
Going forward, we anticipate that continued technological advances can yield progressively more precise and comprehensive functional and connectomic maps of dopaminergic circuitry across development. As these tools become more widely adopted by NDD researchers, we will likely gain newfound understanding of how functional and structural abnormalities synergize to produce behavioral and cognitive phenotypes in mouse models and reveal putative mechanisms of disease symptomatology in human populations. Ultimately these discoveries can inform the creation of behavioral and pharmacological therapies that target circuit- or cell-type specific mechanisms of disease in order to benefit the health of affected children and adults.
Highlights.
Dopaminergic dysfunction is common in many neurodevelopmental disorders.
New circuit mapping tools have refined our knowledge of these circuits at baseline.
When used in animal models, these tools will likely elucidate NDD pathophysiology.
Acknowledgments
We would like to acknowledge support from the Children’s Tumor Foundation (Young Investigator Award 2016-01-006 to JER), as well as the Heritage Medical Research Institute (VG) and the Tianqiao and Chrissy Chen Institute for Neuroscience at Caltech. We would like to thank Jennifer Treweek, Benjamin Deverman, Ken Chan, Min Jang, Alon Greenbaum, and Ryan Cho for histological images used in the manuscript figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: None
References
- 1.Joesch M, Mankus D, Yamagata M, Shahbazi A, Schalek R, Suissa-Peleg A, Meister M, Lichtman JW, Scheirer WJ, Sanes JR. Reconstruction of genetically identified neurons imaged by serial-section electron microscopy. Elife. 2016:5. doi: 10.7554/eLife.15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vazquez-Reina A, Kaynig V, Jones TR, Roberts M, et al. Saturated reconstruction of a volume of neocortex. Cell. 2015;162(3):648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Weissman TA, Pan YA. Brainbow: New resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics. 2015;199(2):293–306. doi: 10.1534/genetics.114.172510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuriel S, Gudes S, Draft RW, Binshtok AM, Lichtman JW. Multispectral labeling technique to map many neighboring axonal projections in the same tissue. Nat Methods. 2015;12(6):547–552. doi: 10.1038/nmeth.3367. [DOI] [PubMed] [Google Scholar]
- 5⋆.Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, Zalocusky KA, et al. Targeting cells with single vectors using multiple-feature boolean logic. Nat Methods. 2014;11(7):763–772. doi: 10.1038/nmeth.2996. This study introduced Flp- and Dre-dependent DIO constructs to enable intersectional viral targeting strategies. The resulting technology, INTERSECT, allows for pathway specific targeting via Cre-dependent retrograde delivery (with HSV vectors) of Flp-dependent constructs. Exclusion targeting is also possible depending on the starting orientation of the exons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6⋆⋆.Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, Luo L. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature. 2015;524(7563):88–92. doi: 10.1038/nature14600. Schwarz and colleagues introduced TRIO (‘tracing the relationship between input and output’) and cTRIO (‘cell-type-specific TRIO’) methods for input-output mapping of inputs to a population of neurons that project to a starter area. Using this method, starter cells are selected based on a known projection target (in this case, the olfactory bulb, auditory cortex, hippocampus, cerebellum, and medulla) of a region of interest (the locus coeruleus, LC). A Cre-dependent retrograde canine adenovirus vector expressing Cre-dependent Flp recombinase was injected into each starter location in DBH-Cre mice, which express Cre recombinase in noradrenergic neurons Flp-dependent RVDg vectors were then used to map inputs to the LC based on starter location. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acciai L, Soda P, Iannello G. Automated neuron tracing methods: An updated account. Neuroinformatics. 2016;14(4):353–367. doi: 10.1007/s12021-016-9310-0. [DOI] [PubMed] [Google Scholar]
- 8.Treweek JB, Gradinaru V. Extracting structural and functional features of widely distributed biological circuits with single cell resolution via tissue clearing and delivery vectors. Curr Opin Biotechnol. 2016;40:193–207. doi: 10.1016/j.copbio.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162(2):246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chhetri RK, Amat F, Wan Y, Hockendorf B, Lemon WC, Keller PJ. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Methods. 2015;12(12):1171–1178. doi: 10.1038/nmeth.3632. [DOI] [PubMed] [Google Scholar]
- 11⋆.Royer LA, Lemon WC, Chhetri RK, Wan Y, Coleman M, Myers EW, Keller PJ. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat Biotechnol. 2016;34(12):1267–1278. doi: 10.1038/nbt.3708. The authors developed an adaptive light sheet microscope to address challenges caused by the spatial and temporal heterogeneity of live specimens. Improvements include the development of a multi-view imaging method that employs four orthogonally placed optical arms (two for light sheet illumination and two for detection) with 10 digitally adjustable degrees of freedom and new software (AutoPilot) for real-time image quality monitoring and autonomous microscope adjustment to improve spatial resolution and image quality. [DOI] [PubMed] [Google Scholar]
- 12.Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, Chandrashekar J. A platform for brain-wide imaging and reconstruction of individual neurons. Elife. 2016;5:e10566. doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9(3):255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18(9):1213–1225. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth BL. Dreadds for neuroscientists. Neuron. 2016;89(4):683–694. doi: 10.1016/j.neuron.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin MZ, Schnitzer MJ. Genetically encoded indicators of neuronal activity. Nat Neurosci. 2016;19(9):1142–1153. doi: 10.1038/nn.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girven KS, Sparta DR. Probing deep brain circuitry: New advances in in vivo calcium measurement strategies. ACS Chem Neurosci. 2017;8(2):243–251. doi: 10.1021/acschemneuro.6b00307. [DOI] [PubMed] [Google Scholar]
- 19.Peron S, Chen TW, Svoboda K. Comprehensive imaging of cortical networks. Curr Opin Neurobiol. 2015;32:115–123. doi: 10.1016/j.conb.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Yang W, Yuste R. In vivo imaging of neural activity. Nat Methods. 2017;14(4):349–359. doi: 10.1038/nmeth.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji N, Freeman J, Smith SL. Technologies for imaging neural activity in large volumes. Nat Neurosci. 2016;19(9):1154–1164. doi: 10.1038/nn.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu D, Xu J, McMahon MT, van Zijl PC, Mori S, Northington FJ, Zhang J. In vivo high-resolution diffusion tensor imaging of the mouse brain. Neuroimage. 2013;83:18–26. doi: 10.1016/j.neuroimage.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gozzi A, Schwarz AJ. Large-scale functional connectivity networks in the rodent brain. Neuroimage. 2016;127:496–509. doi: 10.1016/j.neuroimage.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Callaway EM, Luo L. Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J Neurosci. 2015;35(24):8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junyent F, Kremer EJ. Cav-2–why a canine virus is a neurobiologist’s best friend. Curr Opin Pharmacol. 2015;24:86–93. doi: 10.1016/j.coph.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 26⋆.Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, Huang CC, et al. A designer aav variant permits efficient retrograde access to projection neurons. Neuron. 2016;92(2):372–382. doi: 10.1016/j.neuron.2016.09.021. The authors performed several rounds of selection to identify AAV-2 capsids that provided retrograde transduction in projections from the striatum to the substantia nigra pars reticulata and projections from deep cerebellar nuclei to the cerebellar cortex. The resulting AAV2-retro virus was shown to provide efficient, Cre-dependent retrograde transduction several loci, including cortical and subcortical structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraefel C, Marconi P, Epstein AL. Herpes simplex virus type 1 (hsv-1)-derived amplicon vectors for gene transfer and gene therapy. Methods Mol Biol. 2015;1254:295–316. doi: 10.1007/978-1-4939-2152-2_21. [DOI] [PubMed] [Google Scholar]
- 28.Salganik M, Hirsch ML, Samulski RJ. Adeno-associated virus as a mammalian DNA vector. Microbiol Spectr. 2015;3(4) doi: 10.1128/microbiolspec.MDNA3-0052-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29⋆⋆.Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu WL, Yang B, Huber N, Pasca SP, Gradinaru V. Cre-dependent selection yields aav variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34(2):204–209. doi: 10.1038/nbt.3440. Deverman and colleagues developed a platform for AAV capsid selection (Cre recombination-based AAV targeted evolution or CREATE) that they used to discover AAV9 variants that broadly transduced the CNS after peripheral (intravenous or retro-orbital) injection. The resulting PHP.B vector transduced cells in multiple brain regions, including the motor and visual cortices, striatum, thalamus, hippocampus, and cerebellum, better than AAV9 with reduced gene transfer to the liver. Additionally, PHP.B was also able to transduce human differentiated cortical neurons and astrocytes derived from induced pluripotent stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YC, Frei JA, Kilander MB, Shen W, Blatt GJ. A subset of autism-associated genes regulate the structural stability of neurons. Front Cell Neurosci. 2016;10:263. doi: 10.3389/fncel.2016.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nomi JS, Uddin LQ. Developmental changes in large-scale network connectivity in autism. Neuroimage Clin. 2015;7:732–741. doi: 10.1016/j.nicl.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kizer JS, Palkovits M, Brownstein MJ. The projections of the a8, a9 and a10 dopaminergic cell bodies: Evidence for a nigral-hypothalamic-median eminence dopaminergic pathway. Brain Res. 1976;108(2):363–370. doi: 10.1016/0006-8993(76)90192-x. [DOI] [PubMed] [Google Scholar]
- 33.Fuccillo MV. Striatal circuits as a common node for autism pathophysiology. Front Neurosci. 2016;10:27. doi: 10.3389/fnins.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardozo Pinto DF, Lammel S. Viral vector strategies for investigating midbrain dopamine circuits underlying motivated behaviors. Pharmacol Biochem Behav. 2017 doi: 10.1016/j.pbb.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulin JF, Zou J, Drouin-Ouellet J, Kim KY, Cicchetti F, Awatramani RB. Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep. 2014;9(3):930–943. doi: 10.1016/j.celrep.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogt Weisenhorn DM, Giesert F, Wurst W. Diversity matters - heterogeneity of dopaminergic neurons in the ventral mesencephalon and its relation to parkinson’s disease. J Neurochem. 2016;139(Suppl 1):8–26. doi: 10.1111/jnc.13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasser HM, Calu DJ, Schoenbaum G, Sharpe MJ. The dopamine prediction error: Contributions to associative models of reward learning. Front Psychol. 2017;8:244. doi: 10.3389/fpsyg.2017.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wise RA. Forebrain substrates of reward and motivation. J Comp Neurol. 2005;493(1):115–121. doi: 10.1002/cne.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howe MW, Dombeck DA. Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature. 2016;535(7613):505–510. doi: 10.1038/nature18942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gremel CM, Lovinger DM. Associative and sensorimotor corticobasal ganglia circuit roles in effects of abused drugs. Genes Brain Behav. 2017;16(1):71–85. doi: 10.1111/gbb.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthews GA, Nieh EH, Vander Weele CM, Halbert SA, Pradhan RV, Yosafat AS, Glober GF, Izadmehr EM, Thomas RE, Lacy GD, Wildes CP, et al. Dorsal raphe dopamine neurons represent the experience of social isolation. Cell. 2016;164(4):617–631. doi: 10.1016/j.cell.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Sugam JA, Lowery-Gionta EG, McElligott ZA, McCall NM, Lopez AJ, McKlveen JM, Pleil KE, Kash TL. Mu opioid receptor modulation of dopamine neurons in the periaqueductal gray/dorsal raphe: A role in regulation of pain. Neuropsychopharmacology. 2016;41(8):2122–2132. doi: 10.1038/npp.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho JR, Treweek JB, Robinson JE, Xiao C, Bremner LR, Greenbaum A, Gradinaru V. Dorsal raphe dopamine neurons modulate arousal and promote wakefulness by salient stimuli. Neuron. 2017 doi: 10.1016/j.neuron.2017.05.020. in press. [DOI] [PubMed] [Google Scholar]
- 44.Grattan DR. 60 years of neuroendocrinology: The hypothalamo-prolactin axis. J Endocrinol. 2015;226(2):T101–122. doi: 10.1530/JOE-15-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bodea GO, Blaess S. Establishing diversity in the dopaminergic system. FEBS Lett. 2015;589(24 Pt A):3773–3785. doi: 10.1016/j.febslet.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Anastasaki C, Woo AS, Messiaen LM, Gutmann DH. Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum Mol Genet. 2015;24(12):3518–3528. doi: 10.1093/hmg/ddv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown JA, Emnett RJ, White CR, Yuede CM, Conyers SB, O’Malley KL, Wozniak DF, Gutmann DH. Reduced striatal dopamine underlies the attention system dysfunction in neurofibromatosis-1 mutant mice. Hum Mol Genet. 2010;19(22):4515–4528. doi: 10.1093/hmg/ddq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genestine M, Lin L, Durens M, Yan Y, Jiang Y, Prem S, Bailoor K, Kelly B, Sonsalla PK, Matteson PG, Silverman J, et al. Engrailed-2 (en2) deletion produces multiple neurodevelopmental defects in monoamine systems, forebrain structures and neurogenesis and behavior. Hum Mol Genet. 2015;24(20):5805–5827. doi: 10.1093/hmg/ddv301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brielmaier J, Matteson PG, Silverman JL, Senerth JM, Kelly S, Genestine M, Millonig JH, DiCicco-Bloom E, Crawley JN. Autism-relevant social abnormalities and cognitive deficits in engrailed-2 knockout mice. PLoS One. 2012;7(7):e40914. doi: 10.1371/journal.pone.0040914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kouwenhoven WM, Veenvliet JV, van Hooft JA, van der Heide LP, Smidt MP. Engrailed 1 shapes the dopaminergic and serotonergic landscape through proper isthmic organizer maintenance and function. Biol Open. 2016;5(3):279–288. doi: 10.1242/bio.015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown JA, Diggs-Andrews KA, Gianino SM, Gutmann DH. Neurofibromatosis-1 heterozygosity impairs cns neuronal morphology in a camp/pka/rock-dependent manner. Mol Cell Neurosci. 2012;49(1):13–22. doi: 10.1016/j.mcn.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gantz SC, Ford CP, Neve KA, Williams JT. Loss of mecp2 in substantia nigra dopamine neurons compromises the nigrostriatal pathway. J Neurosci. 2011;31(35):12629–12637. doi: 10.1523/JNEUROSCI.0684-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karayannis T, Au E, Patel JC, Kruglikov I, Markx S, Delorme R, Heron D, Salomon D, Glessner J, Restituito S, Gordon A, et al. Cntnap4 differentially contributes to gabaergic and dopaminergic synaptic transmission. Nature. 2014;511(7508):236–240. doi: 10.1038/nature13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng L, Kwon O, Lee B, Oh WC, Kim J. Using mammalian gfp reconstitution across synaptic partners (mgrasp) to map synaptic connectivity in the mouse brain. Nat Protoc. 2014;9(10):2425–2437. doi: 10.1038/nprot.2014.166. [DOI] [PubMed] [Google Scholar]
- 55.Kim J, Zhao T, Petralia RS, Yu Y, Peng H, Myers E, Magee JC. Mgrasp enables mapping mammalian synaptic connectivity with light microscopy. Nat Methods. 2011;9(1):96–102. doi: 10.1038/nmeth.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, Zador AM. High-throughput mapping of single-neuron projections by sequencing of barcoded rna. Neuron. 2016;91(5):975–987. doi: 10.1016/j.neuron.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu W, Sánchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, Gradinaru V. Engineered aavs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 2017 doi: 10.1038/nn.4593. in press). ⋆⋆ The authors used the CREATE platform to further select two new PHP.B variants for gene delivery after systemic exposure: PHP.eB, an enhanced capsid with 1.8–2.5 fold improved CNS targeting when compared to PHP.B, and PHP.S, which has high peripheral nervous system tropism without crossing the blood brain barrier. They also created a two-component tTA-TRE system that allows for dose-dependent reduction in transduced cell density when a co-administered tTA vector is titrated downward. This approach does not diminish copy number, allowing for high transgene expression in a sparse population of cells. Existing and new cell type-specific promoters for transgene targeting were also tested with PHP.eB, allowing for effective transduction of diverse cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the brainbow toolbox. Nat Methods. 2013;10(6):540–547. [PubMed] [Google Scholar]
- 59⋆.Allen WE, Kauvar IV, Chen MZ, Richman EB, Yang SJ, Chan K, Gradinaru V, Deverman BE, Luo L, Deisseroth K. Global representations of goal-directed behavior in distinct cell types of mouse neocortex. Neuron. 2017;94(4):891–907. e896. doi: 10.1016/j.neuron.2017.04.017. The authors developed a wide field fluorescence microscope (field of view: 12mm) for large scale cortical imaging of different cell types during motivated behavior. This is the first published study in which a broadly transducing, brain penetrant AAV vector (PHP.eB) was used to express GCaMP across a large cortical volume after peripheral administration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hillier D, Fiscella M, Drinnenberg A, Trenholm S, Rompani SB, Raics Z, Katona G, Juettner J, Hierlemann A, Rozsa B, Roska B. Causal evidence for retina-dependent and -independent visual motion computations in mouse cortex. Nat Neurosci. 2017 doi: 10.1038/nn.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. A multifunctional aav-crispr-cas9 and its host response. Nat Methods. 2016;13(10):868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using crispr-cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fine EJ, Appleton CM, White DE, Brown MT, Deshmukh H, Kemp ML, Bao G. Trans-spliced cas9 allows cleavage of hbb and ccr5 genes in human cells using compact expression cassettes. Sci Rep. 2015;5:10777. doi: 10.1038/srep10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, Saulnier JL, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11(8):825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, Schnitzer MJ. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350(6266):1361–1366. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flytzanis NC, Bedbrook CN, Chiu H, Engqvist MK, Xiao C, Chan KY, Sternberg PW, Arnold FH, Gradinaru V. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and caenorhabditis elegans neurons. Nat Commun. 2014;5:4894. doi: 10.1038/ncomms5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Osten P, Margrie TW. Mapping brain circuitry with a light microscope. Nat Methods. 2013;10(6):515–523. doi: 10.1038/nmeth.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomer R, Lovett-Barron M, Kauvar I, Andalman A, Burns VM, Sankaran S, Grosenick L, Broxton M, Yang S, Deisseroth K. Sped light sheet microscopy: Fast mapping of biological system structure and function. Cell. 2015;163(7):1796–1806. doi: 10.1016/j.cell.2015.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ke MT, Fujimoto S, Imai T. Seedb: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16(8):1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 70.Hou B, Zhang D, Zhao S, Wei M, Yang Z, Wang S, Wang J, Zhang X, Liu B, Fan L, Li Y, et al. Scalable and dii-compatible optical clearance of the mammalian brain. Front Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158(4):945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, Miyawaki A. Scales: An optical clearing palette for biological imaging. Nat Neurosci. 2015;18(10):1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 73.Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, Ukai H, Ueda HR. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159(4):911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 74.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157(3):726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 75.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497(7449):332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced clarity for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9(7):1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang HH, St-Pierre F, Sun X, Ding X, Lin MZ, Clandinin TR. Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell. 2016;166(1):245–257. doi: 10.1016/j.cell.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, Bjorkman PJ, et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc. 2015;10(11):1860–1896. doi: 10.1038/nprot.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan C, Cai R, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P, Plesnila N, Dichgans M, Hellal F, Erturk A. Shrinkage-mediated imaging of entire organs and organisms using udisco. Nat Methods. 2016;13(10):859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- 80.Chen F, Tillberg PW, Boyden ES. Optical imaging. Expansion microscopy. Science. 2015;347(6221):543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang JB, Marblestone A, Church GM, Raj A, et al. Nanoscale imaging of rna with expansion microscopy. Nat Methods. 2016;13(8):679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, DeGennaro EM, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34(9):987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ku T, Swaney J, Park JY, Albanese A, Murray E, Cho JH, Park YG, Mangena V, Chen J, Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016;34(9):973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, McCue M, et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163(6):1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shah S, Lubeck E, Schwarzkopf M, He TF, Greenbaum A, Sohn CH, Lignell A, Choi HM, Gradinaru V, Pierce NA, Cai L. Single-molecule rna detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development. 2016;143(15):2862–2867. doi: 10.1242/dev.138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell. 2016;164(4):792–804. doi: 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87⋆.Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang AC, Jennings JH, Adhikari A, Halpern CH, Witten IB, et al. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell. 2016;165(7):1776–1788. doi: 10.1016/j.cell.2016.05.010. These studies integrated ArcTRAP and CLARITY technologies, which allowed for identification of behavioral ensembles associated with rewarding (cocaine) or aversive (footshock) stimuli using a fast-acting formulation of tamoxifen (4-hydroxytamoxifen) to induce labeling of recently activated neurons. The paper also introduced a data processing workflow of cleared samples for image registration to an anatomical atlas, cell identification, annotation, and quantification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via trap: Targeted recombination in active populations. Neuron. 2013;78(5):773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89⋆.Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, Deisseroth K. Intact-brain analyses reveal distinct information carried by snc dopamine subcircuits. Cell. 2015;162(3):635–647. doi: 10.1016/j.cell.2015.07.014. Lerner and colleagues used TRIO in combination with CLARITY and CLARITY-optimized light sheet microscopy (COLM) to characterize inputs to dorsomedial and dorsolateral striatum-projecting dopaminergic neurons in the SNc and found evidence for parallel nigrostriatal circuits along the mediolateral axis. These findings were supported by qualitative differences in electrophysiological properties of each projection type and bulk calcium responses via fiber photometry during appetitive and aversive stimuli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90⋆.Menegas W, Bergan JF, Ogawa SK, Isogai Y, Umadevi Venkataraju K, Osten P, Uchida N, Watabe-Uchida M. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. Elife. 2015;4:e10032. doi: 10.7554/eLife.10032. The authors used RVDg tracing, clearing with CLARITY, and whole brain light-sheet microscopy to map inputs to VTA and SNc neurons projecting to different projection sites. Projection-dependent rabies tracing was achieved via retrograde expression of the rabies cognate receptor (TVA) through the retrograde activity of AAV5. The authors found that most starter sites produced similar patterns of inputs from the ventral striatum, except for the posterior striatum; this region produced relatively more inputs from the globus pallidus, subthalamic nucleus, and zona incerta. Additionally, the authors created an automated workflow for image acquisition, atlas registration, neuron segmentation, and quantification of labelled cells in cleared brains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91⋆.Menegas W, Babayan BM, Uchida N, Watabe-Uchida M. Opposite initialization to novel cues in dopamine signaling in ventral and posterior striatum in mice. Elife. 2017:6. doi: 10.7554/eLife.21886. Menengas et al. compared bulk calcium activity across striatal subclasses using fiber photometry during a classical conditioning task. While ventral striatal neurons showed activation only in response to rewards and reward-predictive cues, the posterior tail of the striatum, a region their group had previously demonstrated to be an anatomically distinct subclass via rabies mapping in CLARITY-cleared brains, showed strong activation to novel cues regardless or reward state, as well as responses to aversive and neutral stimuli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao C, Cho JR, Zhou C, Treweek JB, Chan K, McKinney SL, Yang B, Gradinaru V. Cholinergic mesopontine signals govern locomotion and reward through dissociable midbrain pathways. Neuron. 2016;90(2):333–347. doi: 10.1016/j.neuron.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93⋆⋆.Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, Luo L. Circuit architecture of vta dopamine neurons revealed by systematic input-output mapping. Cell. 2015;162(3):622–634. doi: 10.1016/j.cell.2015.07.015. This study features cTRIO mapping of inputs to VTA dopaminergic (with DAT-Cre mice) and GABAergic (with GAD2-Cre mice). While inputs to the medial NAc-projecting dopaminergic neurons were biased towards the medial NAc shell, lateral NAc-projecting neurons received more inputs from the dorsal striatum, NAc core, and anterior cingulate cortex. The authors also performed cTRIO using membrane tethered GFP in the VTA and noticed similar medial-to-lateral differences in dopaminergic neuron axonal arborization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kwan V, Unda BK, Singh KK. Wnt signaling networks in autism spectrum disorder and intellectual disability. J Neurodev Disord. 2016;8:45. doi: 10.1186/s11689-016-9176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4:3. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harony-Nicolas H, De Rubeis S, Kolevzon A, Buxbaum JD. Phelan mcdermid syndrome: From genetic discoveries to animal models and treatment. J Child Neurol. 2015;30(14):1861–1870. doi: 10.1177/0883073815600872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, Orteschi D, Collins JS, Zollino M, Visconti P, Dupont B, et al. Prevalence of shank3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet. 2013;21(3):310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bariselli S, Tzanoulinou S, Glangetas C, Prevost-Solie C, Pucci L, Viguie J, Bezzi P, O’Connor EC, Georges F, Luscher C, Bellone C. Shank3 controls maturation of social reward circuits in the vta. Nat Neurosci. 2016;19(7):926–934. doi: 10.1038/nn.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krishnan V, Stoppel DC, Nong Y, Johnson MA, Nadler MJ, Ozkaynak E, Teng BL, Nagakura I, Mohammad F, Silva MA, Peterson S, et al. Autism gene ube3a and seizures impair sociability by repressing vta cbln1. Nature. 2017 doi: 10.1038/nature21678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Riday TT, Dankoski EC, Krouse MC, Fish EW, Walsh PL, Han JE, Hodge CW, Wightman RM, Philpot BD, Malanga CJ. Pathway-specific dopaminergic deficits in a mouse model of angelman syndrome. J Clin Invest. 2012;122(12):4544–4554. doi: 10.1172/JCI61888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berrios J, Stamatakis AM, Kantak PA, McElligott ZA, Judson MC, Aita M, Rougie M, Stuber GD, Philpot BD. Loss of ube3a from th-expressing neurons suppresses gaba co-release and enhances vta-nac optical self-stimulation. Nat Commun. 2016;7:10702. doi: 10.1038/ncomms10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kao FC, Su SH, Carlson GC, Liao W. Mecp2-mediated alterations of striatal features accompany psychomotor deficits in a mouse model of rett syndrome. Brain Struct Funct. 2015;220(1):419–434. doi: 10.1007/s00429-013-0664-x. [DOI] [PubMed] [Google Scholar]
- 103.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Monteiro P, Feng G. Shank proteins: Roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18(3):147–157. doi: 10.1038/nrn.2016.183. [DOI] [PubMed] [Google Scholar]
- 105.Piochon C, Kano M, Hansel C. Ltd-like molecular pathways in developmental synaptic pruning. Nat Neurosci. 2016;19(10):1299–1310. doi: 10.1038/nn.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mullins C, Fishell G, Tsien RW. Unifying views of autism spectrum disorders: A consideration of autoregulatory feedback loops. Neuron. 2016;89(6):1131–1156. doi: 10.1016/j.neuron.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sofroniew NJ, Flickinger D, King J, Svoboda K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. Elife. 2016:5. doi: 10.7554/eLife.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stirman JN, Smith IT, Kudenov MW, Smith SL. Wide field-of-view, multi-region, two-photon imaging of neuronal activity in the mammalian brain. Nat Biotechnol. 2016;34(8):857–862. doi: 10.1038/nbt.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen JL, Voigt FF, Javadzadeh M, Krueppel R, Helmchen F. Long-range population dynamics of anatomically defined neocortical networks. Elife. 2016:5. doi: 10.7554/eLife.14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anikeeva P, Andalman AS, Witten I, Warden M, Goshen I, Grosenick L, Gunaydin LA, Frank LM, Deisseroth K. Optetrode: A multichannel readout for optogenetic control in freely moving mice. Nat Neurosci. 2011;15(1):163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim H, Ahrlund-Richter S, Wang X, Deisseroth K, Carlen M. Prefrontal parvalbumin neurons in control of attention. Cell. 2016;164(1–2):208–218. doi: 10.1016/j.cell.2015.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kvitsiani D, Ranade S, Hangya B, Taniguchi H, Huang JZ, Kepecs A. Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature. 2013;498(7454):363–366. doi: 10.1038/nature12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mandelblat-Cerf Y, Ramesh RN, Burgess CR, Patella P, Yang Z, Lowell BB, Andermann ML. Arcuate hypothalamic agrp and putative pomc neurons show opposite changes in spiking across multiple timescales. Elife. 2015:4. doi: 10.7554/eLife.07122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Adamantidis AR, Tsai HC, Boutrel B, Zhang F, Stuber GD, Budygin EA, Tourino C, Bonci A, Deisseroth K, de Lecea L. Optogenetic interrogation of dopaminergic modulation of the multiple phases of reward-seeking behavior. J Neurosci. 2011;31(30):10829–10835. doi: 10.1523/JNEUROSCI.2246-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van Den Berge N, Albaugh DL, Salzwedel A, Vanhove C, Van Holen R, Gao W, Stuber GD, Ian Shih YY. Functional circuit mapping of striatal output nuclei using simultaneous deep brain stimulation and fmri. Neuroimage. 2017;146:1050–1061. doi: 10.1016/j.neuroimage.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Decot HK, Namboodiri VM, Gao W, McHenry JA, Jennings JH, Lee SH, Kantak PA, Jill Kao YC, Das M, Witten IB, Deisseroth K, et al. Coordination of brain-wide activity dynamics by dopaminergic neurons. Neuropsychopharmacology. 2017;42(3):615–627. doi: 10.1038/npp.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferenczi EA, Zalocusky KA, Liston C, Grosenick L, Warden MR, Amatya D, Katovich K, Mehta H, Patenaude B, Ramakrishnan C, Kalanithi P, et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351(6268):aac9698. doi: 10.1126/science.aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ouzounov DG, Wang T, Wang M, Feng DD, Horton NG, Cruz-Hernandez JC, Cheng YT, Reimer J, Tolias AS, Nishimura N, Xu C. In vivo three-photon imaging of activity of gcamp6-labeled neurons deep in intact mouse brain. Nat Methods. 2017;14(4):388–390. doi: 10.1038/nmeth.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lu R, Sun W, Liang Y, Kerlin A, Bierfeld J, Seelig JD, Wilson DE, Scholl B, Mohar B, Tanimoto M, Koyama M, et al. Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat Neurosci. 2017;20(4):620–628. doi: 10.1038/nn.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang LV, Yao J. A practical guide to photoacoustic tomography in the life sciences. Nat Methods. 2016;13(8):627–638. doi: 10.1038/nmeth.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Horstmeyer R, Ruan H, Yang C. Guidestar-assisted wavefront-shaping methods for focusing light into biological tissue. Nat Photonics. 2015;9:563–571. doi: 10.1038/nphoton.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ruan H, Jang M, Yang C. Optical focusing inside scattering media with time-reversed ultrasound microbubble encoded light. Nat Commun. 2015;6:8968. doi: 10.1038/ncomms9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Y, Lai P, Ma C, Xu X, Grabar AA, Wang LV. Optical focusing deep inside dynamic scattering media with near-infrared time-reversed ultrasonically encoded (true) light. Nat Commun. 2015;6:5904. doi: 10.1038/ncomms6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, Tye KM, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157(7):1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marshall JD, Li JZ, Zhang Y, Gong Y, St-Pierre F, Lin MZ, Schnitzer MJ. Cell-type-specific optical recording of membrane voltage dynamics in freely moving mice. Cell. 2016;167(6):1650–1662 e1615. doi: 10.1016/j.cell.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barretto RP, Schnitzer MJ. In vivo optical microendoscopy for imaging cells lying deep within live tissue. Cold Spring Harb Protoc. 2012;2012(10):1029–1034. doi: 10.1101/pdb.top071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ. In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J Neurophysiol. 2004;92(5):3121–3133. doi: 10.1152/jn.00234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Myaing MT, MacDonald DJ, Li X. Fiber-optic scanning two-photon fluorescence endoscope. Opt Lett. 2006;31(8):1076–1078. doi: 10.1364/ol.31.001076. [DOI] [PubMed] [Google Scholar]
- 129.Gobel W, Kerr JN, Nimmerjahn A, Helmchen F. Miniaturized two-photon microscope based on a flexible coherent fiber bundle and a gradient-index lens objective. Opt Lett. 2004;29(21):2521–2523. doi: 10.1364/ol.29.002521. [DOI] [PubMed] [Google Scholar]
- 130.Helmchen F, Fee MS, Tank DW, Denk W. A miniature head-mounted two-photon microscope. High-resolution brain imaging in freely moving animals. Neuron. 2001;31(6):903–912. doi: 10.1016/s0896-6273(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 131⋆⋆.Zong W, Wu R, Li M, Hu Y, Li Y, Li J, Rong H, Wu H, Xu Y, Lu Y, Jia H, et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat Methods. 2017 doi: 10.1038/nmeth.4305. Zong et al. report a new miniaturized head-mounted 2-photon microscope for use in behaving mice. The microscope employs a miniature collimator, scan lens, and custom MEMS (microelectromechanical system) scanning mirror within the head-mounted microscope body to achieve resonant scanning frequencies up to 40 Hz with a maximal field of view of 130 × 130 μm2. Fluorescent signals are relayed from the microscope to the photomultiplier tube detectors via a custom supple fiber bundle. The microscope was able to efficiently detect GCaMP signals in multiple cortical regions. [DOI] [PubMed] [Google Scholar]
- 132.Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nat Methods. 2011;8(10):871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Resendez SL, Jennings JH, Ung RL, Namboodiri VM, Zhou ZC, Otis JM, Nomura H, McHenry JA, Kosyk O, Stuber GD. Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat Protoc. 2016;11(3):566–597. doi: 10.1038/nprot.2016.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bocarsly ME, Jiang WC, Wang C, Dudman JT, Ji N, Aponte Y. Minimally invasive microendoscopy system for in vivo functional imaging of deep nuclei in the mouse brain. Biomed Opt Express. 2015;6(11):4546–4556. doi: 10.1364/BOE.6.004546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jennings JH, Ung RL, Resendez SL, Stamatakis AM, Taylor JG, Huang J, Veleta K, Kantak PA, Aita M, Shilling-Scrivo K, Ramakrishnan C, et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell. 2015;160(3):516–527. doi: 10.1016/j.cell.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McHenry JA, Otis JM, Rossi MA, Robinson JE, Kosyk O, Miller NW, McElligott ZA, Budygin EA, Rubinow DR, Stuber GD. Hormonal gain control of a medial preoptic area social reward circuit. Nat Neurosci. 2017;20(3):449–458. doi: 10.1038/nn.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Otis JM, Namboodiri VM, Matan AM, Voets ES, Mohorn EP, Kosyk O, McHenry JA, Robinson JE, Resendez SL, Rossi MA, Stuber GD. Prefrontal cortex output circuits guide reward seeking through divergent cue encoding. Nature. 2017;543(7643):103–107. doi: 10.1038/nature21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pinto L, Dan Y. Cell-type-specific activity in prefrontal cortex during goal-directed behavior. Neuron. 2015;87(2):437–450. doi: 10.1016/j.neuron.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of ca1 hippocampal place codes. Nat Neurosci. 2013;16(3):264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, Wei B, Veshkini M, La-Vu M, Lou J, Flores SE, et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature. 2016;534(7605):115–118. doi: 10.1038/nature17955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Banerjee A, Rikhye RV, Breton-Provencher V, Tang X, Li C, Li K, Runyan CA, Fu Z, Jaenisch R, Sur M. Jointly reduced inhibition and excitation underlies circuit-wide changes in cortical processing in rett syndrome. Proc Natl Acad Sci U S A. 2016;113(46):E7287–E7296. doi: 10.1073/pnas.1615330113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Goncalves JT, Anstey JE, Golshani P, Portera-Cailliau C. Circuit level defects in the developing neocortex of fragile x mice. Nat Neurosci. 2013;16(7):903–909. doi: 10.1038/nn.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Diggs-Andrews KA, Gutmann DH. Modeling cognitive dysfunction in neurofibromatosis-1. Trends Neurosci. 2013;36(4):237–247. doi: 10.1016/j.tins.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Leeuw CN, Korecki AJ, Berry GE, Hickmott JW, Lam SL, Lengyell TC, Bonaguro RJ, Borretta LJ, Chopra V, Chou AY, D’Souza CA, et al. Raav-compatible minipromoters for restricted expression in the brain and eye. Mol Brain. 2016;9(1):52. doi: 10.1186/s13041-016-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, Grimley JS, et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci. 2016;19(12):1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Grieger JC, Samulski RJ. Packaging capacity of adeno-associated virus serotypes: Impact of larger genomes on infectivity and postentry steps. J Virol. 2005;79(15):9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Amat F, Hockendorf B, Wan Y, Lemon WC, McDole K, Keller PJ. Efficient processing and analysis of large-scale light-sheet microscopy data. Nat Protoc. 2015;10(11):1679–1696. doi: 10.1038/nprot.2015.111. [DOI] [PubMed] [Google Scholar]
- 148.Pan C, Cai R, Quacquarelli FP, Gasemigharagoz A, Erturk A. Whole organ and organism tissue clearing by udisco. 2016 [Google Scholar]
- 149.Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, Ueda HR. Advanced cubic protocols for whole-brain and whole-body clearing and imaging. Nat Protocols. 2015;10(11):1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- 150.Reardon S. A giant neuron found wrapped around entire mouse brain. Nature. 2017;543(7643):14–15. doi: 10.1038/nature.2017.21539. [DOI] [PubMed] [Google Scholar]
- 151.Fosque BF, Sun Y, Dana H, Yang CT, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, Jayaraman V, et al. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347(6223):755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 152⋆⋆.Lee D, Creed M, Jung K, Stefanelli T, Wendler DJ, Oh WC, Mignocchi NL, Luscher C, Kwon HB. Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nat Methods. 2017 doi: 10.1038/nmeth.4234. Lee and colleagues engineered a light- and ligand-gated split tobacco etch virus (TEV) protease (iTANGO) that allows for fluorescent labelling of neurons endogenously exposed to dopamine in the presence of blue light. This elegant system combines the TANGO platform, which drives gene expression through the release of a cleaved transcription factor by TEV protease following G protein-coupled receptor activation; CRY2PHR, which interacts with its partner protein CIBN only in the presence of light; and AsLOV2, which undergoes a conformational change when exposed to the appropriate wavelength. The authors demonstrate that optogenetically evoked dopamine and motivated behavior can induce XFP expression during light exposure. [DOI] [PubMed] [Google Scholar]
- 153.Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016;92(2):342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kolesova H, Capek M, Radochova B, Janacek J, Sedmera D. Comparison of different tissue clearing methods and 3d imaging techniques for visualization of gfp-expressing mouse embryos and embryonic hearts. Histochem Cell Biol. 2016;146(2):141–152. doi: 10.1007/s00418-016-1441-8. [DOI] [PubMed] [Google Scholar]
- 155.Ichikawa T, Nakazato K, Keller PJ, Kajiura-Kobayashi H, Stelzer EH, Mochizuki A, Nonaka S. Live imaging and quantitative analysis of gastrulation in mouse embryos using light-sheet microscopy and 3d tracking tools. Nat Protoc. 2014;9(3):575–585. doi: 10.1038/nprot.2014.035. [DOI] [PubMed] [Google Scholar]
- 156.Stegmaier J, Amat F, Lemon WC, McDole K, Wan Y, Teodoro G, Mikut R, Keller PJ. Real-time three-dimensional cell segmentation in large-scale microscopy data of developing embryos. Dev Cell. 2016;36(2):225–240. doi: 10.1016/j.devcel.2015.12.028. [DOI] [PubMed] [Google Scholar]
- 157.Portmann T, Yang M, Mao R, Panagiotakos G, Ellegood J, Dolen G, Bader PL, Grueter BA, Goold C, Fisher E, Clifford K, et al. Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 d eletion syndrome. Cell Rep. 2014;7(4):1077–1092. doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fish EW, Krouse MC, Stringfield SJ, Diberto JF, Robinson JE, Malanga CJ. Changes in sensitivity of reward and motor behavior to dopaminergic, glutamatergic, and cholinergic drugs in a mouse model of fragile x syndrome. PLoS One. 2013;8(10):e77896. doi: 10.1371/journal.pone.0077896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Smith LN, Jedynak JP, Fontenot MR, Hale CF, Dietz KC, Taniguchi M, Thomas FS, Zirlin BC, Birnbaum SG, Huber KM, Thomas MJ, et al. Fragile x mental retardation protein regulates synaptic and behavioral plasticity to repeated cocaine administration. Neuron. 2014;82(3):645–658. doi: 10.1016/j.neuron.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Deng JV, Wan Y, Wang X, Cohen S, Wetsel WC, Greenberg ME, Kenny PJ, Calakos N, West AE. Mecp2 phosphorylation limits psychostimulant-induced behavioral and neuronal plasticity. J Neurosci. 2014;34(13):4519–4527. doi: 10.1523/JNEUROSCI.2821-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Su SH, Kao FC, Huang YB, Liao W. Mecp2 in the rostral striatum maintains local dopamine content critical for psychomotor control. J Neurosci. 2015;35(15):6209–6220. doi: 10.1523/JNEUROSCI.4624-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Espinosa F, Xuan Z, Liu S, Powell CM. Neuroligin 1 modulates striatal glutamatergic neurotransmission in a pathway and nmdar subunit-specific manner. Front Synaptic Neurosci. 2015;7:11. doi: 10.3389/fnsyn.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Uchigashima M, Ohtsuka T, Kobayashi K, Watanabe M. Dopamine synapse is a neuroligin-2-mediated contact between dopaminergic presynaptic and gabaergic postsynaptic structures. Proc Natl Acad Sci U S A. 2016;113(15):4206–4211. doi: 10.1073/pnas.1514074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, Sudhof TC. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158(1):198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Peixoto RT, Wang W, Croney DM, Kozorovitskiy Y, Sabatini BL. Early hyperactivity and precocious maturation of corticostriatal circuits in shank3b(−/−) mice. Nat Neurosci. 2016;19(5):716–724. doi: 10.1038/nn.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mei Y, Monteiro P, Zhou Y, Kim JA, Gao X, Fu Z, Feng G. Adult restoration of shank3 expression rescues selective autistic-like phenotypes. Nature. 2016;530(7591):481–484. doi: 10.1038/nature16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Greenbaum A, Chan KY, Dobreva T, Brown D, Balani DH, Boyce R, Kronenberg HM, McBride HJ, Gradinaru V. Bone clarity: Clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Sci Transl Med. 2017;9(387) doi: 10.1126/scitranslmed.aah6518. [DOI] [PubMed] [Google Scholar]
- 168.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14(11):1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]


