Abstract
Background and Purpose
Acid‐sensing ion channels (ASICs) play an important role in synaptic plasticity and learning, as well as in nociception and mechanosensation. ASICs are involved in pain and in neurological and psychiatric diseases, but their therapeutic potential is limited by the lack of ligands activating them at physiological pH.
Experimental Approach
We extracted, purified and determined the structure of a bisbenzylisoquinoline alkaloid, lindoldhamine, (LIN) from laurel leaves. Its effect on ASIC3 channels were characterized, using two‐electrode voltage‐clamp electrophysiological recordings from Xenopus laevis oocytes.
Key Results
At pH 7.4 or higher, LIN activated a sustained, proton‐independent, current through rat and human ASIC3 channels, but not rat ASIC1a or ASIC2a channels. LIN also potentiated proton‐induced transient currents and promoted recovery from desensitization in human, but not rat, ASIC3 channels.
Conclusions and Implications
We describe a novel ASIC subtype‐specific agonist LIN, which induced proton‐independent activation of human and rat ASIC3 channels at physiological pH. LIN also acts as a positive allosteric modulator of human, but not rat, ASIC3 channels. This unique, species‐selective, ligand of ASIC3, opens new avenues in studies of ASIC structure and function, as well as providing new approaches to drug design.
Abbreviations
- ASIC
acid‐sensing ion channel
- GMQ
2‐guanidine‐4‐methylquinazoline
Introduction
Acid‐sensing ion channels (ASICs) belong to the degenerin/epithelial Na+ channel superfamily (Kellenberger and Schild, 2002). The six identified ASIC subunits (ASIC1a, ASIC1b, ASIC2a, ASIC2b, ASIC3 and ASIC4) encoded by four genes (ACCN1–ACCN4) (Wemmie et al., 2006) can form homotrimeric or heterotrimeric cation‐selective ion channels (Benson et al., 2002; Askwith et al., 2004; Sherwood et al., 2011). ASIC1a, ASIC2a, ASIC2b and ASIC4 isoforms are found in the CNS (Akopian et al., 2000; Alvarez de la Rosa et al., 2003; Wemmie et al., 2003), while ASIC3 and ASIC1b isoforms are predominantly expressed in the peripheral nervous system (Chen et al., 1998; Babinski et al., 1999). ASICs are implicated in neurotransmission, synaptic plasticity, learning, ischaemia and neuronal cell death and contribute to perception of acid‐mediated inflammatory or post‐operative pain (Wemmie et al., 2002, 2003; Xiong et al., 2004; Gao et al., 2005; Yagi et al., 2006; Friese et al., 2007; Deval et al., 2008, 2011; Yen et al., 2009).
ASIC subunits demonstrate high‐sequence conservation between different mammalian species (rat, mouse and human) and isoforms (rat isoforms show 45–80% sequence similarity). The crystal structure of chicken ASIC1 at 1.9 Å resolution showed that the three‐dimensional architecture of ASIC subunits form trimeric functional complexes. Each ASIC subunit consists of relatively short intracellular N‐terminus and C‐terminus and two transmembrane domains (TM1 and TM2) connected by a large cysteine‐rich extracellular domain (Jasti et al., 2007; Gonzales et al., 2009).
Homomeric channels composed of ASIC1a or ASIC3 subunits are activated by protons at pH just below 7.0, while those composed of ASIC1b and especially ASIC2a require much lower pH for activation (Lingueglia et al., 1997; Waldmann et al., 1997a; Hesselager et al., 2004; Lin et al., 2008). In response to fast decrease in extracellular pH, all homomeric ASICs generate a transient, quickly inactivating current. In addition, channels composed of ASIC3 subunits respond to pH changes from 7.4 to 6.5 or lower by slowly developing a sustained current, which lasts for the duration of the acidic stimulation (Osmakov et al., 2014). The nature of this sustained component is not well understood.
Because the activation of the majority of ASICs requires a marked reduction in pH, protons are unlikely to play a significant role as ASIC activators under physiological or pathological conditions. It was therefore proposed that there are other ligands besides protons that can activate ASICs in vivo (Wemmie et al., 2013). Several ligands have been identified as potential activators of ASICs, including a synthetic compound 2‐guanidine‐4‐methylquinazoline (GMQ), which activates and potentiates ASIC3 at pH < 7.4 through an extracellular domain binding site distinct from the proton sensor (Yu et al., 2010). A classical ASIC inhibitor amiloride applied at high concentrations has been recently shown to activate a sustained current through ASIC3 channels under conditions of moderate acidification (Li et al., 2011). Low MW NMDA receptor channel blockers at submillimolar concentrations produced potentiation and/or inhibition of ASICs at pH of 6.5 and lower (Tikhonova et al., 2014). The spider toxin PcTx1 that binds to ASIC1a at pH 7.4 was proposed to increase affinity of the channel to protons and convert it to a desensitized state (Chen et al., 2005). Interestingly, PcTx1 acted as an agonist on chicken ASIC1a and as a positive allosteric modulator on rat ASIC1b (Chen et al., 2005, 2006; Baconguis and Gouaux, 2012). The coral snake toxin MitTx demonstrated isoform‐specific activation of ASICs, acting at nanomolar concentrations on ASIC1a and ASIC1b and at micromolar concentrations on ASIC3 (Bohlen et al., 2011). Among endogenous ligands, lysophosphatidylcholine (LPC) and arachidonic acid at micromolar concentrations and pH 7.4 were shown to evoke a constitutive depolarizing ASIC3‐mediated current and cause pain in rats (Marra et al., 2016). Collectively, despite availability of ligands that activate ASICs, none of them activate ASICs independently of protons (i.e. at pH > 7.4).
In this study, we identify the first low MW natural compound, lindoldhamine (LIN) (PubChem CID 10370752), which acts as an agonist and positive allosteric modulator of human and rat ASIC3 channels in a broad range of pH, including pH > 7.4, when proton activation is entirely absent. In contrast to typical low MW regulators of ASICs, which always include basic chemical groups (e.g. amidine or guanidinium) (Yu et al., 2010; Li et al., 2011), LIN is largely aromatic and carries no charge.
Methods
Isolation of the active component
LIN was purified from an acetic acid extract of Laurus nobilis leaves. The plant material was collected in the North Caucasus and stored at room temperature in dry conditions. Dried laurel leaves (2g) were crushed and extracted with 80 mL of 10% acetic acid containing 10 mM EDTA and 1 mM PMSF for protease inhibition. Extraction lasted for 18 h at 24°C with constant stirring and was followed by 20 min centrifugation at 10000× g. The supernatant was collected, lyophilized and stored at −20°C till the next stages of purification. LIN was purified by two consecutive runs of the supernatant through chromatography columns connected to HPLC. First, the supernatant components were separated on the reverse‐phase Luna C18 column (250 × 10 mm, 5 μm; Phenomenex, Torrance, USA) using a 60 min linear gradient of acetonitrile (0 to 60%) in 0.1% Trifluoroacetic acid (TFA) at a constant flow rate of 5 mL·min−1. In the second stage, the active component was isolated, using a Vydac C18 column (250 × 4.6 mm, 5 μm) with a 60 min 10 to 40% linear gradient of acetonitrile in 0.1% TFA at the constant flow rate of 1 mL·min−1. The eluted compounds were detected by absorbance at 210 and 280 nm.
Mass spectrometry
The purity of the chromatographic fractions and the determination of MW were performed using a Shimadzu LCMS‐2020 single quadrupole detector equipped with an electrospray ion source (ESI) coupled to a LC‐20A HPLC. Separation was achieved using Symmetry C18 column (Waters, Milford, USA, 75 × 4.6 mm, 3.5 μm) and 5 min 10 to 60% acetonitrile gradient. Spectra in the 300–2000 m/z range were collected in both positive and negative modes. Lab Solution 5.65 software was used for data acquisition and analysis.
NMR spectroscopy
NMR spectra were acquired in D2O, pH 3.0 at 14°C on the Bruker Avance III 600 MHz NMR spectrometer equipped with the 5 mm CPTXI cryoprobe (Bruker Corp., Billerica, MA, USA). The following spectra were used to elucidate the structure of lindoldhamine: 1H, 13C, two‐dimensional (2D) DQF‐COSY, ROESY (τ m = 75 and 200 m), 1H–13C HSQC, 1H–13C HMBC (J long = 4.5 and 7.0 Hz) and 1H–15N HMBC (J long = 7.0 Hz). Free induction decay (FID) resolution for indirect dimensions was at least 40 Hz for 13C and 30 Hz for 1H. Bruker TopSpin software was used for data acquisition and analysis. The raw NMR data are available for download from https://drive.google.com/open?id=0B‐FosxT8cujXQWhIUnh6dDNIY1E.
Electrophysiological recordings from Xenopus laevis oocytes
All animal care and experimental procedures were in agreement with the guidelines of ‘European convention for the protection of vertebrate animals used for experimental and other scientific purposes’ (Strasbourg, 18.III.1986). Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny et al., 2010; McGrath and Lilley, 2015). Oocytes were surgically removed from the ovarian tissue of X. laevis, under conditions of cold anaesthesia. The whole animal was cooled down to 2–4°C, and then a small part of the ovary (~200 oocytes) was removed through a small (less than 1 cm) incision in the abdominal wall, which then was sewn with a single stitch. For each animal, the interval between surgeries was at least 3 months. A total of 10 female X. laevis frogs were involved in this study, and they were reused for no more than five times each. Oocytes were defolliculated and injected with 2.5–10 ng of cRNA. cRNA transcripts were synthesized from the linearized cDNA templates using the HiScribe™ T7 High Yield RNA Synthesis Kit (NEB, Ipswich, USA) according to the protocol for capped transcripts supplied by the manufacturer. Human ASIC3 (hASIC3) cDNA (AF057711.1) was subcloned from EX‐Q0260‐B02 (GeneCopoeia, Inc., Rockville, USA) to pcDNA3.1 and linearized by NaeI (Promega, Madison, USA). Similarly, cRNAs were also synthesized from PCi plasmids containing rat ASIC3 (rASIC3), rat ASIC1a and ASIC2a isoforms. Integrity of ASIC genes was confirmed by DNA sequencing of the entire inserted fragments. After injection, the oocytes were kept for 2–3 days at 19°C and then up to 7 days at 15°C in ND‐96 medium supplemented with gentamycin (50 μg·mL−1) and containing (in mM): 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2 and 10 HEPES titrated to pH 7.4 with NaOH. Two‐electrode voltage‐clamp recordings were performed using the GeneClamp500 amplifier (Axon Instruments, Inverurie, UK) at the holding potential of −50 mV. The data were filtered at 20 Hz and digitized at 100 Hz using the L780 AD converter (L‐Card, Moscow, Russia). Microelectrodes were filled with 3 M KCl. The external solution was ND‐96 with pH adjusted to 8.5, 8.0, 7.8, 7.6, 7.4, 7.3 or 7.0. Proton‐activated currents through ASIC channels were elicited by application of ND‐96, in which 5 mM of HEPES was substituted with 10 mM acetic acid at pH 4.0 or 10 mM MES at pH < 7.0. Extracellular solutions were applied using a computer‐controlled valve system.
Data and statistical analysis
The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2015). In all experiments, the responses were recorded by a person not blinded to the treatment. Analysis of electrophysiological data was performed using OriginPro 8.6 software (OriginLab, Northampton, USA). Curves were fitted using the logistic equation: F(x) = A − A∕(1 + (x∕x0)n), where F(x) is the current amplitude at the ligand concentration x, A is the maximal current amplitude, x0 is the EC50 value and n is the Hill coefficient (slope factor). All data are presented as mean ± SEM. Significant differences between normalized data measurements were determined using the non‐parametric Kruskal–Wallis ANOVA test, with the significance level set at P < 0.05.
Materials
GMQ was obtained from Sigma‐Aldrich (Steinheim, Germany). Peptide APETx2 was also produced by the method of heterologous expression in Escherichia coli. Lyophilized LIN was dissolved in ND96 buffer corresponding to that used in the experiment. Fresh solutions of the compounds were made immediately before the experiments.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017).
Results
Isolation from L. nobilis
We tested a number of medicinal plant extracts and invertebrate venoms available in our laboratory and found that the acetic acid extract from laurel leaves produced strong activation of ASICs. The extract from L. nobilis leaves was separated by two‐stage reverse‐phase chromatography (see Methods), and its components were subsequently tested on human ASIC3 channels using electrophysiology. As a result, only one component of the extract (MW of 568.25 Da) was active (Supporting Information Figure S1). In L. nobilis leaves, this component is abundant and comprises ~0.03% of the weight of crude dried plants. Two‐electrode voltage‐clamp recordings from X. laevis oocytes expressing hASIC3 showed that at the holding membrane potential of −50 mV, this component induced a slowly increasing inward current at pH 7.8 or caused strong potentiation of ASIC‐mediated transient currents activated by protons (pH 5.5, Figure 1A). Solubility limit of LIN was 30 mM in H2O and 3 mM in the conditioning buffer ND96, and the latter was the maximum concentration we applied in electrophysiological experiments.
Figure 1.
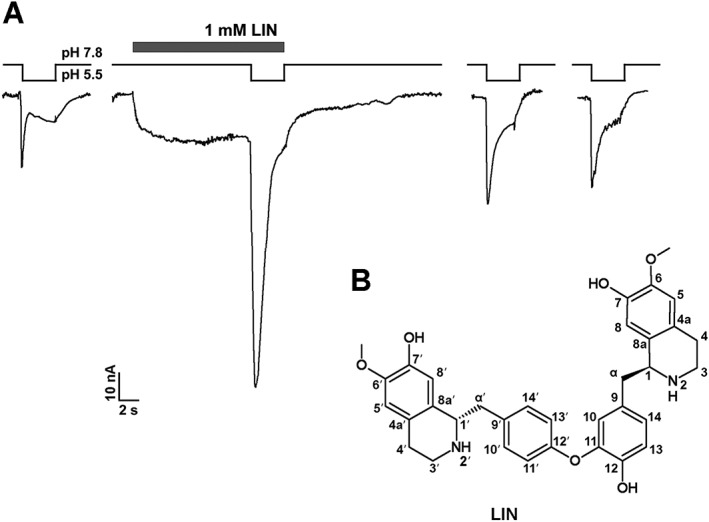
Effects of LIN on hASIC3 and its structure. (A) Activating and potentiating effects of LIN, extracted from L. nobilis leaves, on human ASIC3 channels. LIN (1mM) was first applied at pH 7.8, and then pH was changed to 5.5 (mean for LIN‐induced steady current was 46.5 ± 7.6%, and mean for LIN‐potentiation of transient current was 253 ± 31% of pH 5.5 induced current). The whole‐cell currents shown in the Figure were recorded from the same cell. (B) Chemical formula of LIN. Atoms are numbered in agreement with earlier work (Guha et al., 1979).
Structure of the active component
1H NMR spectrum of the compound showed 11 protons in the aromatic region (7.1–6.2 ppm, Supporting Information Table S1 and Figure S2), in which two signals had an observable splitting constant (7.133 and 6.841 ppm, 2H each, J = 8.3 Hz) and seven singlets of 1H each. In the aliphatic region, there were two methoxy singlets (3.657 and 3.867 ppm, 3H and 3H), two overlapped protons near 4.6 ppm and 12 overlapped protons of high multiplicity in the range of 3.4–2.0 ppm, making the total of 31 protons in the NMR spectrum (Supporting Information Figure S2). 13С NMR, together with 2D 13C HSQC/HMBC spectra, showed 10 carbon resonances in the aliphatic region (two signals near ~24 ppm, four signals near ~38 and four signals at ~55 ppm) and 22 carbon signals in aromatic carbons (111–163 ppm), making the total of 32 carbon resonances. Two of the carbon resonances (130.78 and 117.49) were connected in HSQC with two aromatic doublets of 2H each (7.133 and 6.841); consequently, these resonances were part of the symmetric phenyl ring, making the total number of 34 carbon atoms per molecule. At least one nitrogen was observed in the 15N HMBC spectrum, and at least two oxygens were detected in the methoxy groups. All of the acquired 1H/13C NMR spectra were pH‐independent in the range of pH 2.0–6.8 (Supporting Information Figure S2), suggesting that ionogenic groups are not present in the molecule.
Further analysis of 1H/13C chemical shifts, 13C HMBC, COSY and ROESY connectivities (Supporting Information Table S1) suggested that the molecule consists of two similar fragments with only one ROESY NMR contact between them (10–11′13′ contact in Supporting Information Table S1). The structures of both fragments were established as coclaurines (CID 160487) connected via the ether bond. Instead of the expected doublets, the 13H and 14H protons had a singlet, likely due to redundant chemical shifts of these atoms. The ether bond 11C–O–12′C was supported by the ROESY contact and characteristic 13C chemical shifts of carbons 11C and 13C (143.20 and 146.09, Supporting Information Table S1). The resulting formula for this compound is C34H36N2O6 (calculated mass 569.2646, five protons are invisible in D2O), in good agreement with the MW of 569.25 predicted by ESI+ MS. As a result, our NMR experiments determined the chemical structure of the active component as a bisbenzylisoquinoline alkaloid, LIN ((1R)‐1‐[[4‐[2‐hydroxy‐5‐[[(1R)‐7‐hydroxy‐6‐methoxy‐1,2,3,4‐tetrahydroisoquinolin‐1‐yl]methyl]phenoxy] phenyl]methyl]‐6‐methoxy‐1,2,3,4‐tetrahydroisoquinolin‐7‐ol) (PubChem CID 10370752) (Figure 1B). Although this bisbenzylisoquinoline alkaloid was first described in 1976 (Lu and Chen, 1976), the corresponding NMR data have not been available. Thus, our Supporting Information Table S1 represents the first complete NMR signal LIN assignment by means of modern high‐resolution 2D NMR. By measuring [α]D with a PerkinElmer Model 241 polarimeter, we determined LIN stereochemistry that turns out to be the same as proposed previously (Lu and Chen, 1976).
ASIC activation by LIN
The ability of LIN to activate ASICs was studied using two‐electrode voltage clamp of X. laevis oocytes expressing different ASIC subunits. No LIN‐activated currents were recorded from uninjected oocytes (Supporting Information Figure S3). At the extracellular pH ≥ 7.4, LIN caused sustained inward currents through human and rat homomeric ASIC3 channels (Figure 2). The slow inward current activation kinetics in human ASIC3 (hASIC3) channels (τ on = 848 ± 454 ms, n = 6, at 1 mM of LIN, Figure 2B) were similar to the kinetics of the sustained current activated by protons (τ on = 869 ± 348 ms, n = 5, pH 6.0). At pH 7.8, the amplitude of the LIN‐activated currents through hASIC3 channels increased with LIN concentration but did not reach its maximal value at the highest applied LIN concentration of 3 mM (Figure 2E). Fitting with the logistic equation approximated the maximal amplitude of the LIN‐activated current (I max) as 122.6 ± 22.4% of the control current in response to the drop of pH to 5.5, the apparent half‐maximal concentration (EC50) of 1.53 ± 0.83 mM and the Hill coefficient (n H) of 0.93 ± 0.16 (n = 6). Increasing the extracellular pH to 8.5 resulted in significant enhancement of LIN‐induced activation (Figure 2F), indicating that it did not require protons. In the presence of 1 μM of the APETx2 toxin, a well‐known ASIC3‐selective inhibitor, the amplitude of the LIN‐induced currents was markedly reduced (Figure 3A, B). This inhibition produced by the recombinant APETx2 was slightly weaker but comparable with the previously reported inhibition of proton‐induced currents by APETx2 obtained from a natural source (~80%, Diochot et al., 2004). Therefore, the inhibition by APETx‐2 of LIN‐activated currents, illustrated in Figure 3, strongly supports their ASIC3‐specific origin.
Figure 2.
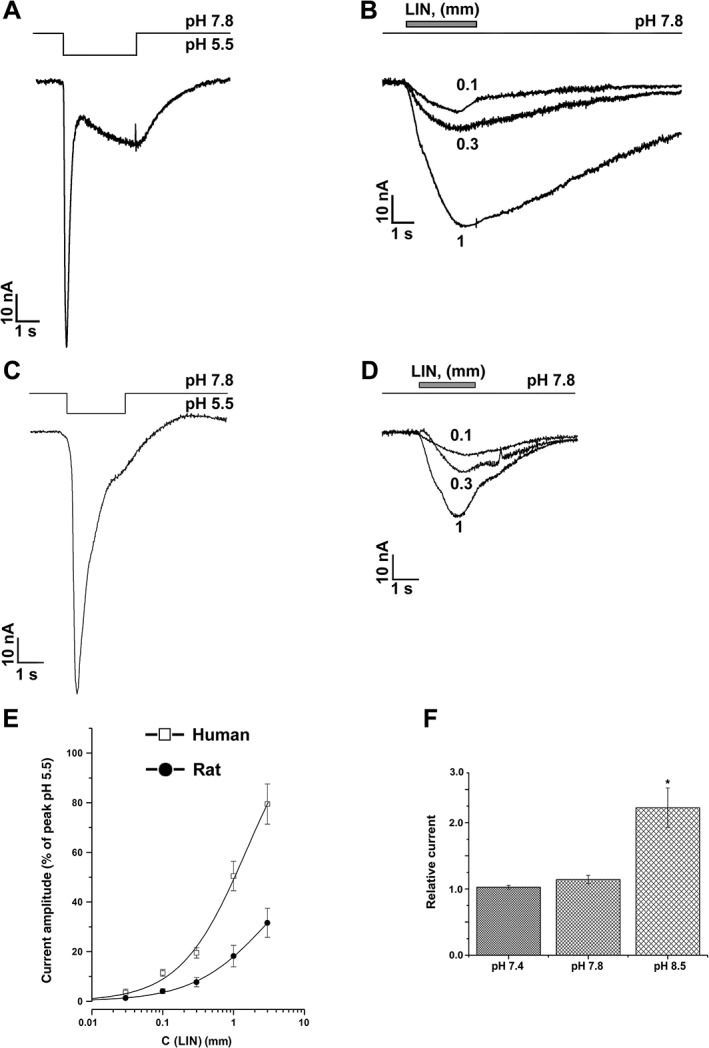
Activation of human and rat ASIC3 channels by LIN. Whole‐cell currents recorded from (A, B) human or (C, D) rat ASIC3 channels in response to the (A, C) pH 5.5 stimulus or to (B, D) 1 mM LIN application at the constant pH 7.8. Single‐exponential fitting of current recovery after termination of LIN application gave the τ off time constants of 4.42 ± 0.02 s (0.1 mM LIN), 6.61 ± 0.04 s (0.3 mM LIN) and 11.13 ± 0.07 s (1 mM LIN) for hASIC3 and 2.7 ± 0.2 s (0.1 mM LIN), 2.48 ± 0.09 s (0.3 mM LIN) and 2.02 ± 0.02 s (1 mM LIN) for rASIC3. (E) Dose–response curves for hASIC3 and rASIC3 activation by LIN. Data presented are mean ± SEM (n = 5). Solid lines through the points fit with the logistic equation. (F) Amplitude of 1 mM LIN‐induced hASIC3 current measured at pH 7.4, 7.8 and 8.5. Data shown are mean ± SEM; n = 5. *P < 0.05, significantly different from current amplitudes at pH 7.8; Kruskal–Wallis ANOVA.
Figure 3.

LIN‐induced current inhibition by ASIC3‐specific inhibitor APETx2. (A) Whole‐cell currents recorded from human ASIC3 channels in response to 1 mM LIN application alone (black trace) or at presence of 1 μM APETx2 at the constant pH 7.8. (B) Inhibition of 1 mM LIN‐induced hASIC3 current measured at pH 7.8 by 1 μM APETx2. Data shown are mean ± SEM; n = 5. *P < 0.05, significantly different from current amplitudes at pH 7.8; Kruskal–Wallis ANOVA.
LIN activation of rat ASIC3 (rASIC3) channels was weaker than hASIC3 channels but was similarly far from saturation at 3 mM LIN (Figure 2C, D). Fitting the rASIC3 activation curve with the logistic equation yielded the I max and EC50 values of 65.3 ± 13.3% and 3.2 ± 1.5 mM, n H of 0.82 ± 0.07 (n = 5) respectively (Figure 2E). The average current recovery time constant after termination of LIN application (τ off) for rASIC3 (2.3 ± 0.4 s, n = 7) was three times smaller than that for hASIC3 (6.1 ± 2.9 s, n = 5), suggesting higher LIN affinity to hASIC3. No significant activation of rASIC1a or rASIC2a by LIN was detected at up to 1 mM LIN concentrations (Supporting Information Figure S4).
Comparison of LIN with another ASIC3 activator, GMQ
To compare the potency of LIN with that of the known ASIC3 activator GMQ, we measured their activity in the same experiment (Figure 4). When applied simultaneously with the pH step from 7.8 to 7.0, 1 mM GMQ activated inward currents through human and rat ASIC3 channels with the 133 ± 14% (n = 5) and 106 ± 5% (n = 5) amplitudes of their control values in the absence of GMQ respectively. Strikingly, at the same 1 mM concentration, LIN activated human and rat ASIC3‐mediated currents with the amplitudes of 285 ± 15% (n = 5) and 139 ± 8% (n = 5) of the control respectively. Therefore, LIN‐induced activation of ASIC3 channels occurred not only without assistance of protons, it was also much more potent than activation by the well‐known ligand GMQ.
Figure 4.
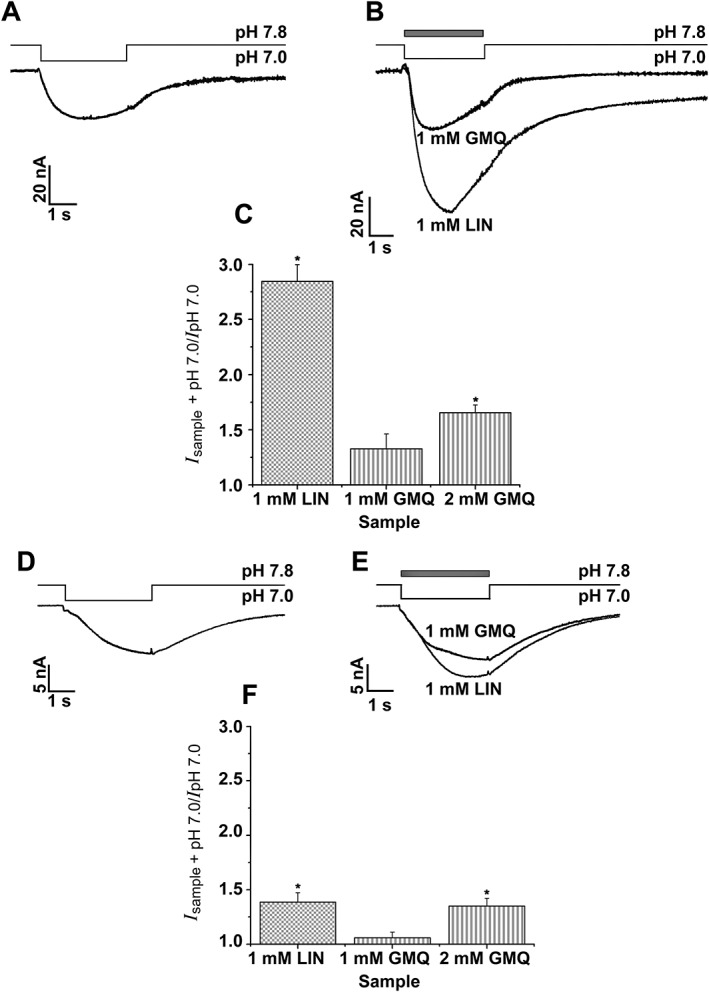
Comparison of LIN and GMQ effects on human and rat ASIC3 channels. Whole‐cell currents recorded from (A, B) human or (D, E) rat ASIC3 channels in response to 1 mM LIN and GMQ applications at the constant pH 7.0. The maximal current amplitudes for (C) hASIC3 and (F) rASIC3. Data shown are mean ± SEM; n = 5. *P < 0.05, significantly different from amplitudes of the control current at pH 7.0 and currents activated by LIN and GMQ; Kruskal–Wallis ANOVA.
The effect of LIN on proton‐induced currents
To study the effect of LIN on proton‐induced currents, we co‐applied a pH stimulus (Figure 5). LIN was applied at the concentration of 0.3 mM, which was sufficient to activate both hASIC3 and rASIC3 channels (Figure 2B–D). We measured the maximal current amplitude (Figure 5C) and the time, at which the current reaches its maximum (latency) (Figure 5D) and did not observe a proportional increase in values of these parameters with decreasing pH. For human and rat ASIC3, the most pronounced modulating effect was observed at pH 6.0–6.5, demonstrating that changes in both current amplitude and latency became smaller with increasing extracellular concentration of protons. At pH > 6.0, LIN induced a weak inhibition of proton‐induced rASIC3 currents and potentiation of hASIC3 currents. LIN therefore acted as a positive modulator only on hASIC3‐mediated proton‐induced currents.
Figure 5.
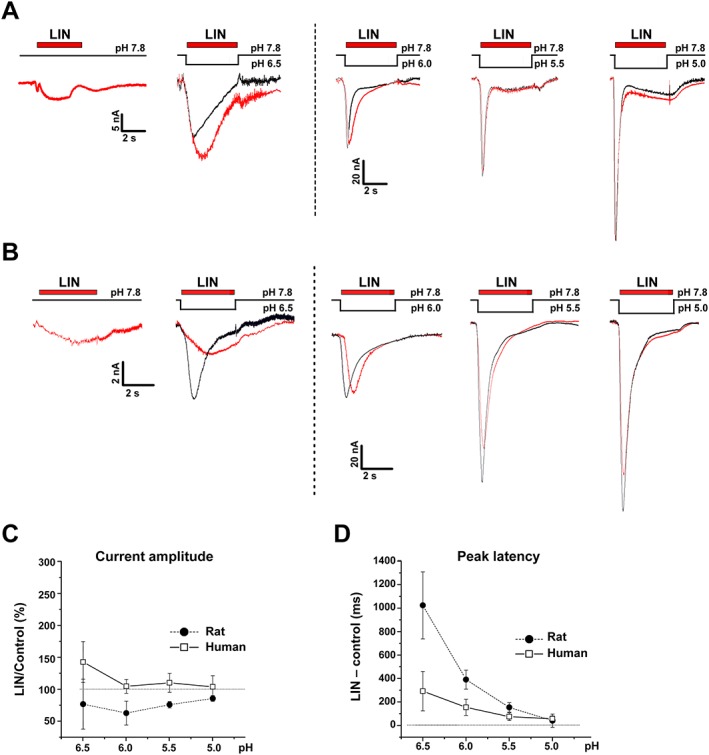
Effects of LIN co‐administered with acidic stimuli on ASIC3‐mediated currents. Whole‐cell currents recorded from (A) human or (B) rat ASIC3 channels held at pH 7.8 in response to different pH stimuli alone or co‐applied with 0.3 mM LIN. (C) Current amplitude and (D) latency time calculated for different pH stimuli. Data are mean ± SEM (n = 6).
LIN potentiation of the transient hASIC3 currents
To further study the effect of LIN on the proton‐activated ASIC currents, we pre‐incubated cells with LIN for 15 s before applying the pH stimulus (pH drop from 7.8 to 5.5). LIN had a strong potentiating effect on hASIC3 currents resulted peak current amplitudes more than double that of the control (Figure 6A). The maximum current potentiation was observed at 100 μM LIN, while further increase in LIN concentration led to a reduction of potentiation and a delay in the peak response latency (Figure 6A, B). The reduced potentiation of the peak current at LIN concentrations higher than 100 μM was accompanied by an increase in the baseline current amplitude (Figure 6A, traces to the right), likely to be due to an enhanced contribution of LIN‐induced proton‐independent activation (Figure 2), suggesting that the two actions of LIN are mutually exclusive but additive in reaching the maximum net hASIC3 current. Fitting of the current amplitude with the logistic equation in the range of 0.1–300 μM LIN concentrations (EC50 = 3.77 ± 0.48 μM, n H = 1.12 ± 0.13, n = 7) (Figure 6B) demonstrated that LIN is nearly 1000 times more effective as a potentiator of the hASIC3 peak current in response to acidification than the proton‐independent activator at high pH. Notably, the lack of leftward or rightward shifts in the pH activation curve (Figure 6C) indicated that LIN did not alter sensitivity of hASIC3 to protons: pH50 = 6.04 ± 0.05 for control and pH50 = 6.03 ± 0.04 in presence of 30 μM LIN (n = 5). Under the same conditions, instead of potentiation observed for hASIC3‐mediated currents, LIN induced a weak inhibition of rASIC3‐mediated currents (Figure 6B, D).
Figure 6.
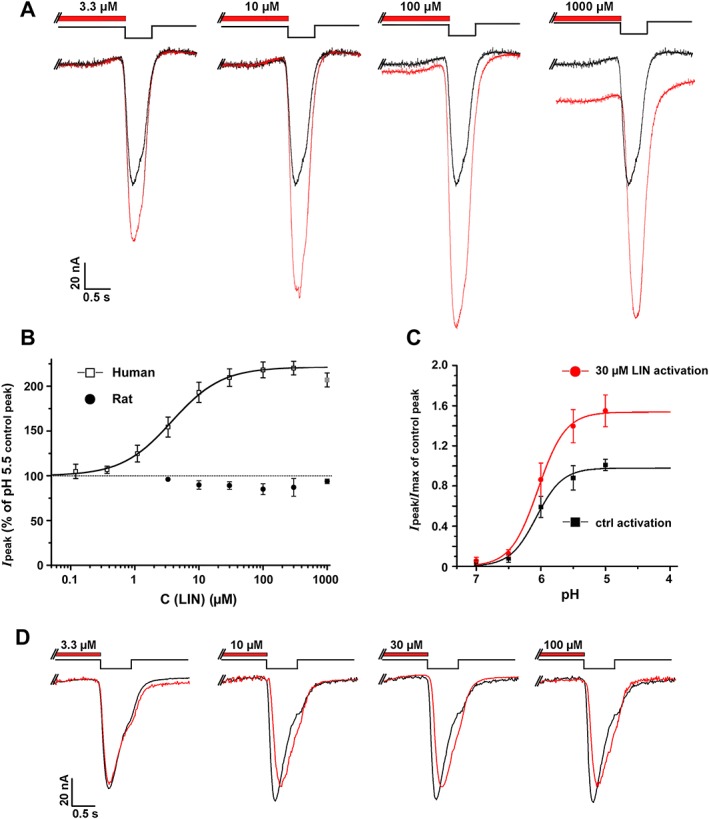
Effects on ASIC3‐mediated currents of LIN pre‐incubation. (A) Whole‐cell currents recorded from human ASIC3 channels held at pH 7.8 in response to different pH 5.5 stimulus, with (red) or without (black) 15 s pre‐incubation with LIN at different concentrations. (B) Peak current amplitude for hASIC and rASIC at different concentrations of pre‐incubated LIN. The dose–response curve for hASIC3 is fitted with the logistic equation. Data are mean ± SEM (n = 7). (C) pH dependence of hASIC3 activation in the absence and presence of 30 μM LIN. hASIC3 held at pH of 7.8 was activated by various acidic pH stimuli. The peak current amplitudes are plotted as a function of pH and fitted with the logistic equation (pH50 = 6.04 ± 0.05 and n H = 2.15 ± 0.46 for control vs. pH50 = 6.03 ± 0.04 and n H = 2.06 ± 0.27 for LIN) showing no significant shift in the activation curve in the presence of LIN. (D) Whole‐cell currents recorded from rat ASIC3 channels held at pH 7.8 in response to different pH 5.5 stimulus, with (red) or without (black) 15 s pre‐incubation of LIN at different concentrations. Data are mean ± SEM (n = 5).
LIN‐induced recovery of proton‐activated hASIC3 currents
We propose that the transient potentiation of proton‐activated currents is a result of LIN‐induced recovery of hASIC3 channels from desensitization.
To test this idea, we used the protocol illustrated in Figure 7A, where the pH stimulus was applied in the presence of LIN after cells were pre‐incubated with LIN at pH 7.3. Maintaining cells at pH 7.3 without LIN completely eliminated the transient component of the proton‐activated hASIC3 current, apparently due to desensitization (black traces in Figure 7A). In contrast, LIN pre‐incubation restored this transient current (Figure 7A, red traces). Moreover, the amplitude of this restored transient current sometimes exceeded the amplitude of the control current recorded in response to pH reduction from 7.8 to pH of the corresponding acidic stimulus.
Figure 7.
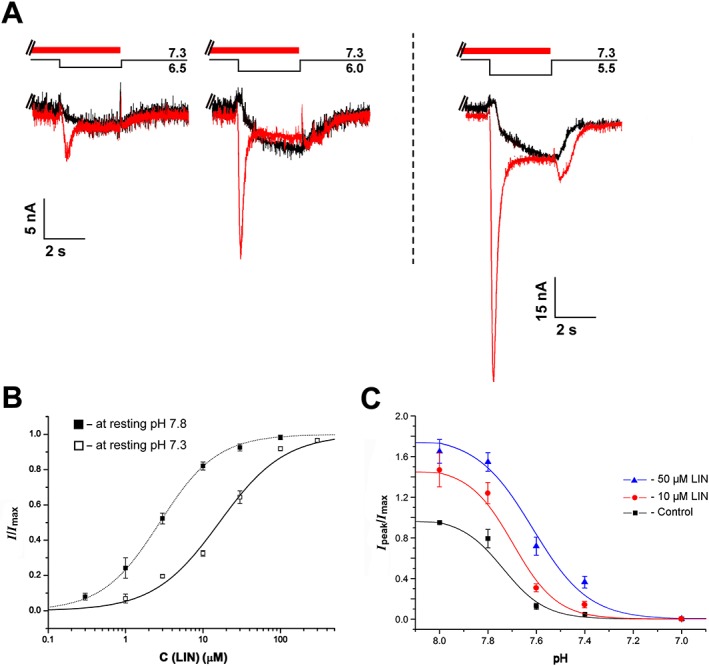
LIN‐induced recovery of the transient hASIC3 current. (A) Whole‐cell currents recorded from human ASIC3 channels held at pH 7.3 in response to different pH stimuli in the presence (red) or absence (black) of 0.1 mM LIN pre‐incubated for 15 s before the pH stimulus. (B) Dose–response curves for potentiation of hASIC3 currents by LIN at the holding pH 7.3 and 7.8. For both pHs, the pH of the activating stimulus was 5.5. The maximum amplitude (I max) was calculated for each cell by individual fitting, and the data were normalized to it, then the normalized data were averaged and fitted by the logistic equation. Correspondingly, solid and dotted lines through the points fit with the logistic equation. Data are mean ± SEM (n = 6). (C) pH dependence of hASIC3 steady‐state desensitization (SSD) in the absence and presence of 10 and 50 μM LIN. hASIC3 held at various conditioning pHs was activated by pH 5.5 stimulus. The peak current amplitudes are plotted as a function of pH and fitted (solid lines) with the logistic equation [pHSSD50 = 7.73 ± 0.03 (n H = 6.90 ± 0.74) for control vs. pHSSD50 = 7.70 ± 0.03 (n H = 6.81 ± 2.82) for 10 μM LIN and pHSSD50 = 7.62 ± 0.04 (n H = 3.1 ± 1.4) for 50 μM LIN] showing no significant shift in the SSD curve in the presence of LIN. Data are mean ± SEM (n = 6).
Fitting the amplitude of the LIN‐induced transient current with the logistic equation (EC50 = 16.2 ± 2.2 μM, n H = 1.03 ± 0.13, n = 6) showed a fivefold reduction in the LIN apparent affinity at pH 7.8 (Figure 7B). Thus, LIN can induce the recovery of hASIC3 channels from desensitization, and an increase in proton concentration during LIN pre‐incubation resulted in weakening of its potentiating effect.
The effect of LIN on proton dependence of hASIC3 desensitization
The proton dependence of desensitization has been extensively studied in rat ASIC3 channels, with the pH50 values reported in the pH range of 6.9 to 7.1 (Waldmann et al., 1997a; Gründer and Pusch, 2015). In contrast, such data have not been available for human ASIC3 channels. Our results show that the conditioning pH value, which causes 50% desensitization of the transient current (pHSSD50), is 7.73 ± 0.03 (Figure 7C, black curve). As illustrated in Figure 7C, LIN displayed a significant potentiating effect at pH 8.0, when desensitization is negligible. Additionally, at 50 μM concentration, LIN caused a rightward shift of the desensitization curve pHSSD50 = 7.62 ± 0.04. This shift was accompanied by a decrease in the value of the Hill coefficient: n H = 6.9 ± 0.7 for the control versus n H = 3.1 ± 1.4 for 50 μM LIN.
This result suggests that the mechanism of LIN‐induced current potentiation is more complicated than just a recovery of hASIC3 channels from desensitization.
Discussion
ASICs were discovered in 1980 (Krishtal and Pidoplichko, 1980) and cloned and characterized in 1997 (Waldmann et al., 1997b). Although they have attracted the attention of many laboratories around the world, there is no consensus about whether protons are the only or the most effective activators of ASICs (Askwith et al., 2000; Krishtal, 2015). Even though other channel activators, such as GMQ and agmatine, have been identified, there is still an open question regarding the availability of proton‐independent activators. For example, GMQ (Yu et al., 2010) did not activate rat ASIC3 at pH 7.8, thus acting as a proton‐dependent gating modulator at pH 7.4 (Alijevic and Kellenberger, 2012; Gründer and Pusch, 2015). Similar results were obtained for PcTx1, which promoted ASIC1b opening under slightly acidic conditions (Chen et al., 2006).
Here, we present the activation of ASIC3 channels by the bisbenzylisoquinoline alkaloid LIN. Bisbenzylisoquinoline alkaloids are structural dimers of 1‐benzylisoquinolines, a family of plant secondary metabolites derived from pharmacologically active tyrosine. For example, (+)‐tubocurarine, one of the prominent bisbenzylisoquinoline alkaloids, is the principal neuromuscular blocking agent found in curare. Tetrandrine and dauricine are calcium channel blockers with antihypertensive and anti‐arrhythmic actions (Qian, 2002). First isolated from the plant Triclisia sacleuxii, LIN was characterized as an AChE inhibitor with micromolar affinity (Murebwayire et al., 2009; Liu et al., 2012).
Our results show that LIN represents the first proton‐independent activator of ASIC3, eliciting slowly developing, non‐desensitizing currents at basic pH. Previously, it was postulated that ASIC activation required electrostatic interaction of charged ligands with charged binding sites on the channel (Jasti et al., 2007; Paukert et al., 2008; Yu et al., 2010; Tikhonova et al., 2014). Contrary to this postulate, the LIN molecule is neutral, i.e., it lacks positive or negative charge at pH varying from 1 to 8 (Supporting Information Figure S2). Accordingly, LIN is most likely to interact allosterically with ASIC3 channels. LIN activates ASICs at high pH, when the concentration of protons is too low to elicit the proton‐activated currents. In contrast to GMQ, when the pH was shifted from neutral to alkaline, the activating effect of LIN not only did not decrease, but, on the contrary, increased (Figure 2F). It follows that proton binding is not necessary for ASIC3 activation. Thus, LIN was shown to be a truly proton‐independent activator.
The non‐desensitizing, LIN‐induced, proton‐independent currents have onset kinetics, similar to those of the proton‐induced sustained currents. The significant difference between the EC50 and τ off values for human and rat ASIC3 suggests that LIN has higher affinity to human channels. A higher value of I max also indicates a greater efficacy of opening of human channels by LIN.
Apart from proton‐independent ASIC activation, LIN also potentiated a transient component of ASIC responses to protons. This potentiation was species‐specific, being observed for hASIC3 but not for rASIC3 (Figure 6B). LIN therefore represents the first ASIC3 positive modulator that displays a pronounced species specificity. In contrast to other ASIC activators (GMQ and LPC) (Alijevic and Kellenberger, 2012; Marra et al., 2016), LIN did not shift the pH‐activation curve, implying that LIN potentiation is not a result of increased channel affinity to protons in the presence of LIN (Figure 6С). In our experiments, hASIC3‐mediated currents were completely desensitized at pH 7.3. However, pre‐incubation of hASIC3 channels with micromolar concentrations of LIN restored the transient current component. For both effects, the n H ~1 value suggests that both potentiation and recovery of ASIC from desensitization result from LIN binding to a single site. As the potentiating effect of LIN was observed at pH 8.0, when proton‐induced desensitization is negligible (Figure 7С), we assume that LIN exerts its potentiating effect both in protonated and non‐protonated states of hASIC3 channels.
Because pH reduction caused a rightward shift of the dose–response curve (Figure 7B) and LIN caused a rightward shift of the desensitization curve (Figure 7C), LIN appears to compete with protons for binding to the same sites. Therefore, it is very likely that proton binding sites responsible for desensitization and the LIN binding site overlap. On the other hand, as activation of transient current by protons was observed in the presence of LIN (Figures 6A and 7A), we propose that the proton sites responsible for activation of the transient current component does not overlap with the binding site of LIN. An ‘acidic pocket’ at the intersubunit interfaces that contains several proton sites (Jasti et al., 2007; Baconguis and Gouaux, 2012; Baconguis et al., 2014) may be one of the possible candidates for LIN binding. Mutagenesis combined with single‐channel recordings will be necessary to understand ASIC regulation by LIN in more detail.
The effects of LIN in recovering and potentiating the transient ASIC currents may be especially interesting for its clinical application as they can be useful for normalizing the dynamic processes of pain sensitivity, disturbed by pathological conditions. Significant differences in the activating and, more importantly, potentiating effects of LIN on ASICs should be taken into account when evaluating the clinical potential of this alkaloid. The strongly expressed species‐selective action of LIN makes it a valuable tool for determining the molecular basis of differences in the pharmacology of rat and human ASIC3 channels.
Author contributions
D.I.O., S.G.K., Y.A.A. and S.A.K. designed the experiment; D.I.O. performed the isolation of the ASIC3 ligand; M.A.D., V.S.K. and R.G.E. performed the structure elucidation; S.G.K., D.I.O. and A.I.S. performed the electrophysiological study and analysed the data; and D.I.O., S.G.K, Y.A.A., A.I.S. and S.A.K. wrote the manuscript.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Figure S1 Purification of LIN. A, The first separation stage of the acetic extract of L. nobilis leaves on a reverse‐phase column LunaC18 (10x250mm) in 0.1% TFA with a flow rate 5 ml/min using a linear gradient of acetonitrile concentration. B, Final purification on a column VydacC18 (4.6x250mm) in 0.1% TFA with a flow rate of 1 ml/min using a linear gradient of acetonitrile concentration. Fractions containing the active component are marked by an arrow.
Table S1 NMR chemical shifts (ppm), multiplicities (Hz) and NMR HMBC/ROESY/COSY contacts in LIN. Conditions: D2O, pH 3.0, 14 °C, Bruker Avance III 600 MHz. Atom numbering follows Fig 1B. a)Heavy atom number of corresponding NMR cross‐peak outline HMBC contacts (italic), ROESY contacts (bold) and COSY ones (underlined). b) Not applicable.
Figure S2 The 1H NMR spectra of LIN at different pH values (D2O, 14 °C, 600 MHz). Absence of any observable chemical shift changes in 1H NMR and 2D 13C–HSQC/13C–HMBC NMR spectra (data not shown) proves absence of ionogenic groups with pKa in range 1.8 in LIN.
Figure S3 LIN application on uninjected oocytes. Whole‐cell current traces recorded from oocytes held at pH 8.5, 7.8 and 7.3 in response to corresponding bath solution (left panel) and to 1 mM LIN (right panel).
Figure S4 LIN application on rat ASIC1a and rat ASIC2a channels. (A) Whole‐cell current traces recorded from rat ASIC1a channels held at pH 7.8 in response to pH 5.5 stimulus (left panel) and to 1 mM LIN (right panel). The mean of LIN‐induced current was 0.55 ± 0.13% of pH 5.5‐induced current, n = 6. Data are mean ± SEM. (B) Whole‐cell traces recorded from rat ASIC2a channels held at pH 7.8 in response to pH 5.5 stimulus (left panel) and to 1 mM LIN (right panel). The mean of LIN‐induced current was 0 ± 0% of pH 5.5‐induced current, n = 5.
Acknowledgements
This article is dedicated to the memory of Professor Eugene V. Grishin, whose guidance made this work possible. We are grateful to Sylvie Diochot (Institutde Pharmacologie Moleculaire et Cellulaire, Valbonne, France) for PCi plasmids containing cDNA of rat ASIC1a, ASIC2a and ASIC3. Experiments were partly carried out using the equipment provided by the IBCH сore facility (CKP IBCH, supported by the Russian Ministry of Education and Science, grant RFMEFI62117X0018). This study was supported, in part, by the Russian Foundation for Basic Research (grant no. 15‐04‐04666), the ‘Molecular and Cell Biology’ programme of the Presidium of the Russian Academy of Sciences ‘Molecular and Cell Biology’. A.I.S. was supported by the National Institutes of Health (grants R01 NS083660 and R01 CA206573), the Pew Charitable Trusts (Pew Scholar Award in Biomedical Sciences) and the Irma T. Hirschl Trust (Irma T. Hirschl Career Scientist Award).
Osmakov, D. I. , Koshelev, S. G. , Andreev, Y. A. , Dubinnyi, M. A. , Kublitski, V. S. , Efremov, R. G. , Sobolevsky, A. I. , and Kozlov, S. A. (2018) Proton‐independent activation of acid‐sensing ion channel 3 by an alkaloid, lindoldhamine, from Laurus nobilis . British Journal of Pharmacology, 175: 924–937. doi: 10.1111/bph.14134.
Contributor Information
Alexander I Sobolevsky, Email: as4005@cumc.columbia.edu.
Sergey A Kozlov, Email: serg@ibch.ru.
References
- Akopian AN, Chen CC, Ding Y, Cesare P, Wood JN (2000). A new member of the acid‐sensing ion channel family. Neuroreport 11: 2217–2222. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Peters JA, Kelly E, Marrion N, Faccenda E, Harding SD et al (2017). The Concise Guide to PHARMACOLOGY 2017/18: Ligand‐gated ion channels. Br J Pharmacol 174 (Suppl 1): S130–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alijevic O, Kellenberger S (2012). Subtype‐specific modulation of acid‐sensing ion channel (ASIC) function by 2‐guanidine‐4‐methylquinazoline. J Biol Chem 287: 36059–36070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez de la Rosa D, Krueger SR, Kolar A, Shao D, Fitzsimonds RM, Canessa CM (2003). Distribution, subcellular localization and ontogeny of ASIC1 in the mammalian central nervous system. J Physiol 546: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith CC, Cheng C, Ikuma M, Benson C, Price MP, Welsh MJ (2000). Neuropeptide FF and FMRFamide potentiate acid‐evoked currents from sensory neurons and proton‐gated DEG/ENaC channels. Neuron 26: 133–141. [DOI] [PubMed] [Google Scholar]
- Askwith CC, Wemmie JA, Price MP, Rokhlina T, Welsh MJ (2004). Acid‐sensing ion channel 2 (ASIC2) modulates ASIC1 H+‐activated currents in hippocampal neurons. J Biol Chem 279: 18296–18305. [DOI] [PubMed] [Google Scholar]
- Babinski K, Lê KT, Séguéla P (1999). Molecular cloning and regional distribution of a human proton receptor subunit with biphasic functional properties. J Neurochem 72: 51–57. [DOI] [PubMed] [Google Scholar]
- Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E (2014). X‐ray structure of acid‐sensing ion channel 1–snake toxin complex reveals open state of a Na+‐selective channel. Cell 156: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis I, Gouaux E (2012). Structural plasticity and dynamic selectivity of acid‐sensing ion channel–spider toxin complexes. Nature 489: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson CJ, Xie J, Wemmie JA, Price MP, Henss JM, Welsh MJ et al (2002). Heteromultimers of DEG/ENaC subunits form H+‐gated channels in mouse sensory neurons. Proc Natl Acad Sci U S A 99: 2338–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen CJ, Chesler AT, Sharif‐naeini R, Medzihradszky KF, Zhou S, King D et al (2011). A heteromeric Texas coral snake toxin targets acid‐sensing ion channels to produce pain. Nature 479: 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, England S, Akopian AN, Wood JN (1998). A sensory neuron‐specific, proton‐gated ion channel. Proc Natl Acad Sci U S A 95: 10240–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kalbacher H, Gründer S (2005). The tarantula toxin psalmotoxin 1 inhibits acid‐sensing ion channel (ASIC) 1a by increasing its apparent H+ affinity. J Gen Physiol 126: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kalbacher H, Gründer S (2006). Interaction of acid‐sensing ion channel (ASIC) 1 with the tarantula toxin psalmotoxin 1 is state dependent. J Gen Physiol 127: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Bond RA, Spina D, Ahluwalia A, Alexander S, Giembycz MA et al (2015). Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol 172 (14): 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval E, Noël J, Gasull X, Delaunay A, Alloui A, Friend V et al (2011). Acid‐sensing ion channels in postoperative pain. J Neurosci 31: 6059–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval E, Noël J, Lay N, Alloui A, Diochot S, Friend V et al (2008). ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J 27: 3047–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diochot S, Baron A, Rash LD, Deval E, Escoubas P, Scarzello S et al (2004). A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid‐sensitive channel in sensory neurons. EMBO J 23: 1516–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese MA, Craner MJ, Etzensperger R, Vergo S, Wemmie JA, Welsh MJ et al (2007). Acid‐sensing ion channel‐1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nat Med 13: 1483–1489. [DOI] [PubMed] [Google Scholar]
- Gao J, Duan B, Wang D‐G, Deng X‐H, Zhang G‐Y, Xu L et al (2005). Coupling between NMDA receptor and acid‐sensing ion channel contributes to ischemic neuronal death. Neuron 48: 635–646. [DOI] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E (2009). Pore architecture and ion sites in acid sensing ion channels and P2X receptors. Nature 460: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründer S, Pusch M (2015). Biophysical properties of acid‐sensing ion channels (ASICs). Neuropharmacology 94: 9–18. [DOI] [PubMed] [Google Scholar]
- Guha KP, Mukherjee B, Mukherjee R (1979). Bisbenzylisoquinoline alkaloids – a review. J Nat Prod 42: 1–84. [DOI] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselager M, Timmermann DB, Ahring PK (2004). pH dependency and desensitization kinetics of heterologously expressed combinations of acid‐sensing ion channel subunits. J Biol Chem 279: 11006–11015. [DOI] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E (2007). Structure of acid‐sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449: 316–323. [DOI] [PubMed] [Google Scholar]
- Kellenberger S, Schild L (2002). Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev 82: 735–767. [DOI] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG (2010). Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishtal O (2015). Receptor for protons: first observations on acid sensing ion channels. Neuropharmacology 94: 4–8. [DOI] [PubMed] [Google Scholar]
- Krishtal OA, Pidoplichko VI (1980). A receptor for protons in the nerve cell membrane. Neuroscience 5: 2325–2327. [DOI] [PubMed] [Google Scholar]
- Li W‐G, Yu Y, Huang C, Cao H, Xu T‐L (2011). Nonproton ligand sensing domain is required for paradoxical stimulation of acid‐sensing ion channel 3 (ASIC3) channels by amiloride. J Biol Chem 286: 42635–42646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y‐W, Min M‐Y, Lin C‐C, Chen W‐N, Wu W‐L, Yu H‐M et al (2008). Identification and characterization of a subset of mouse sensory neurons that express acid‐sensing ion channel 3. Neuroscience 151: 544–557. [DOI] [PubMed] [Google Scholar]
- Lingueglia E, de Weille JR, Bassilana F, Heurteaux C, Sakai H, Waldmann R et al (1997). A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J Biol Chem 272: 29778–29783. [DOI] [PubMed] [Google Scholar]
- Liu Q, Mao X, Zeng F, Jin S, Yang X (2012). Effect of daurisoline on hERG channel electrophysiological function and protein expression. J Nat Prod 75: 1539–1545. [DOI] [PubMed] [Google Scholar]
- Lu S‐T, Chen I‐S (1976). Structure of a new bisbenzylisoquinoline alkaloid, lindoldhamine. Heterocycles 4: 1073. [Google Scholar]
- Marra S, Ferru‐Clément R, Breuil V, Delaunay A, Christin M, Friend V et al (2016). Non‐acidic activation of pain‐related acid‐sensing ion channel 3 by lipids. EMBO J 35: 414–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JC, Lilley E (2015). Implementing guidelines on reporting research using animals (ARRIVE etc.): new requirements for publication in BJP. Br J Pharmacol 172: 3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murebwayire S, Ingkaninan K, Changwijit K, Frédérich M, Duez P (2009). Triclisia sacleuxii (Pierre) Diels (Menispermaceae), a potential source of acetylcholinesterase inhibitors. J Pharm Pharmacol 61: 103–107. [DOI] [PubMed] [Google Scholar]
- Osmakov DI, Andreev YA, Kozlov SA (2014). Acid‐sensing ion channels and their modulators. Biochem 79: 1528–1545. [DOI] [PubMed] [Google Scholar]
- Paukert M, Chen X, Polleichtner G, Schindelin H, Gründer S (2008). Candidate amino acids involved in H+ gating of acid‐sensing ion channel 1a. J Biol Chem 283: 572–581. [DOI] [PubMed] [Google Scholar]
- Qian J‐Q (2002). Cardiovascular pharmacological effects of bisbenzylisoquinoline alkaloid derivatives. Acta Pharmacol Sin 23: 1086–1092. [PubMed] [Google Scholar]
- Sherwood TW, Lee KG, Gormley MG, Askwith CC (2011). Heteromeric acid‐sensing ion channels (ASICs) composed of ASIC2b and ASIC1a display novel channel properties and contribute to acidosis‐induced neuronal death. J Neurosci 31: 9723–9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova TB, Nagaeva EI, Barygin OI, Potapieva NN, Bolshakov KV, Tikhonov DB (2014). Monoamine NMDA receptor channel blockers inhibit and potentiate native and recombinant proton‐gated ion channels. Neuropharmacology 89C: 1–10. [DOI] [PubMed] [Google Scholar]
- Waldmann R, Bassilana F, de Weille J, Champigny G, Heurteaux C, Lazdunski M (1997a). Molecular cloning of a non‐inactivating proton‐gated Na+ channel specific for sensory neurons. J Biol Chem 272: 20975–20978. [DOI] [PubMed] [Google Scholar]
- Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M (1997b). A proton‐gated cation channel involved in acid‐sensing. Nature 386: 173–177. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Taugher RJ, Kreple CJ (2013). Acid‐sensing ion channels in pain and disease. Nat Rev Neurosci 14: 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie JA, Askwith CC, Lamani E, Cassell MD, Freeman JH, Welsh MJ (2003). Acid‐sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J Neurosci 23: 5496–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie JA, Chen J, Askwith CC, Hruska‐Hageman AM, Price MP, Nolan BC et al (2002). The acid‐activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron 34: 463–477. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Price MP, Welsh MJ (2006). Acid‐sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci 29: 578–586. [DOI] [PubMed] [Google Scholar]
- Xiong Z‐G, Zhu X‐M, Chu X‐P, Minami M, Hey J, Wei W‐L et al (2004). Neuroprotection in ischemia: blocking calcium‐permeable acid‐sensing ion channels. Cell 118: 687–698. [DOI] [PubMed] [Google Scholar]
- Yagi J, Wenk HN, Naves LA, McCleskey EW (2006). Sustained currents through ASIC3 ion channels at the modest pH changes that occur during myocardial ischemia. Circ Res 99: 501–509. [DOI] [PubMed] [Google Scholar]
- Yen Y‐T, Tu P‐H, Chen C‐J, Lin Y‐W, Hsieh S‐T, Chen C‐C (2009). Role of acid‐sensing ion channel 3 in sub‐acute‐phase inflammation. Mol Pain 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Z, Li W‐G, Cao H, Feng E‐G, Yu F et al (2010). A nonproton ligand sensor in the acid‐sensing ion channel. Neuron 68: 61–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Purification of LIN. A, The first separation stage of the acetic extract of L. nobilis leaves on a reverse‐phase column LunaC18 (10x250mm) in 0.1% TFA with a flow rate 5 ml/min using a linear gradient of acetonitrile concentration. B, Final purification on a column VydacC18 (4.6x250mm) in 0.1% TFA with a flow rate of 1 ml/min using a linear gradient of acetonitrile concentration. Fractions containing the active component are marked by an arrow.
Table S1 NMR chemical shifts (ppm), multiplicities (Hz) and NMR HMBC/ROESY/COSY contacts in LIN. Conditions: D2O, pH 3.0, 14 °C, Bruker Avance III 600 MHz. Atom numbering follows Fig 1B. a)Heavy atom number of corresponding NMR cross‐peak outline HMBC contacts (italic), ROESY contacts (bold) and COSY ones (underlined). b) Not applicable.
Figure S2 The 1H NMR spectra of LIN at different pH values (D2O, 14 °C, 600 MHz). Absence of any observable chemical shift changes in 1H NMR and 2D 13C–HSQC/13C–HMBC NMR spectra (data not shown) proves absence of ionogenic groups with pKa in range 1.8 in LIN.
Figure S3 LIN application on uninjected oocytes. Whole‐cell current traces recorded from oocytes held at pH 8.5, 7.8 and 7.3 in response to corresponding bath solution (left panel) and to 1 mM LIN (right panel).
Figure S4 LIN application on rat ASIC1a and rat ASIC2a channels. (A) Whole‐cell current traces recorded from rat ASIC1a channels held at pH 7.8 in response to pH 5.5 stimulus (left panel) and to 1 mM LIN (right panel). The mean of LIN‐induced current was 0.55 ± 0.13% of pH 5.5‐induced current, n = 6. Data are mean ± SEM. (B) Whole‐cell traces recorded from rat ASIC2a channels held at pH 7.8 in response to pH 5.5 stimulus (left panel) and to 1 mM LIN (right panel). The mean of LIN‐induced current was 0 ± 0% of pH 5.5‐induced current, n = 5.


