ABSTRACT
Skeletal muscle is primarily composed of large myofibers containing thousands of post-mitotic nuclei distributed throughout a common cytoplasm. Protein production and localization in specialized myofiber regions is crucial for muscle function. Myonuclei differ in transcriptional activity and protein accumulation, but how these differences among nuclei sharing a cytoplasm are achieved is unknown. Regulated nuclear import of proteins is one potential mechanism for regulating transcription spatially and temporally in individual myonuclei. The best-characterized nuclear localization signal (NLS) in proteins is the classical NLS (cNLS), but many other NLS motifs exist. We examined cNLS and non-cNLS reporter protein import using multinucleated muscle cells generated in vitro, revealing that cNLS and non-cNLS nuclear import differs among nuclei in the same cell. Investigation of cNLS nuclear import rates in isolated myofibers ex vivo confirmed differences in nuclear import rates among myonuclei. Analyzing nuclear import throughout myogenesis revealed that cNLS and non-cNLS import varies during differentiation. Taken together, our results suggest that both spatial and temporal regulation of nuclear import pathways are important in muscle cell differentiation and protein regionalization in myofibers.
KEY WORDS: Myofiber, Myotube, Myonuclei, cNLS
Summary: Skeletal muscle nuclei sharing a common cytoplasm differ from one another in nuclear import. These differences in nuclear import could help establish regional specialization in large multinucleated muscle cells.
INTRODUCTION
Skeletal muscle is crucial for survival and quality of life as it is required for respiration, ingestion and locomotion. The primary cell type in skeletal muscle is the myofiber, which is a very large contractile cell containing thousands of post-mitotic nuclei. Myofibers are initially formed by fusion of differentiated precursor cells during embryogenesis. Additional muscle precursor cells fuse as the muscle grows and matures. In the event of muscle injury, resident stem cells proliferate, differentiate and fuse with myofibers, leading to regeneration in the area of injury. The process of differentiation and fusion has been modeled in vitro and results in the formation of multinucleated muscle cells called myotubes. How gene expression is coordinated among multiple nuclei in myofibers to produce proteins necessary to maintain muscle function remains an open question.
A single myofiber can extend the entire length of a muscle and has three distinct regions: the myotendinous junction (MTJ) on each end of the myofiber, which anchors the myofiber to the tendon; the body of the myofiber, which is primarily responsible for contraction; and the neuromuscular junction (NMJ), where a motor neuron synapses onto the myofiber. Each of these regions requires certain proteins to function properly. Although they exist in a common cytoplasm, nuclei in the NMJ and the MTJ specialized regions differ from other nuclei in the myofiber. Nuclei close to the MTJ are more tightly packed than elsewhere in the myofiber (Bruusgaard et al., 2003; Rosser and Bandman, 2003). When a muscle is stressed by stretching, nuclei at the MTJ increase production of myosin heavy chain (Dix and Eisenberg, 1990), one of the major components of the contractile sarcomere. The three to eight nuclei at the NMJ are also tightly clustered and are larger and rounder than other nuclei in the myofiber (Couteaux and Pecot-Dechavassine, 1973). In addition to morphological differences, some proteins are selectively associated with NMJ nuclei, such as Syne-1, a component of the LINC complex, which connects the nuclear lamina to the cytoskeleton (Apel et al., 2000). Additionally, transcripts such as N-CAM, 43k-rapsyn, S-laminin (Moscoso et al., 1995), acetylcholine esterase (Jasmin et al., 1993) and acetylcholine receptor subunits α (Fontaine and Changeux, 1989) and ε (Brenner et al., 1990) are produced exclusively or preferentially by NMJ nuclei, whereas transcripts for actin and myosin (some of the most highly expressed proteins in myofibers) are seldom produced by NMJ nuclei (Moscoso et al., 1995). Although these differences between nuclei in specialized regions and other nuclei have been well described, it is not known how these nuclei are distinguished from other nuclei within the syncytia.
Although the differences between specialized and non-specialized nuclei are well established, nuclei in non-specialized regions also differ from one another in protein accumulation and gene expression. Differences among nuclei in single muscle cells have been detected both in vivo and in vitro. Some, but not all, nuclei in a single multinucleated muscle cell accumulate NFATc1 (Abbott et al., 1998), NFAT5 (O'Connor et al., 2007), myogenin (Ferri et al., 2009; Ishido et al., 2004), MyoD (Ishido et al., 2004; Yamamoto et al., 2008), Myo18B (Salamon et al., 2003) or myostatin (McPherron et al., 1997; Artaza et al., 2002). Additionally, within a single myofiber, some nuclei accumulate EndoG, an apoptosis-associated endonuclease, and undergo DNA fragmentation in response to muscle atrophy whereas others do not (Dupont-Versteegden et al., 2006). Similarly, some but not all nuclei within a muscle cell produce α skeletal actin, troponin 1 slow (Newlands et al., 1998) or myostatin (Artaza et al., 2002). This compartmentalization of gene expression probably facilitates regional production of proteins. Despite the functional significance of this nonequivalence among myonuclei in multinucleated muscle cells both in vivo and in vitro, the molecular mechanisms governing differential protein localization and regulation of gene expression are unknown.
One mechanism that could dictate accumulation of proteins in some nuclei, but not others, in multinucleated muscle cells is regulated nuclear import. The nucleus is compartmentalized from the cytoplasm by the nuclear envelope, which contains nuclear pore complexes (NPC) that mediate movement of molecules between the nuclear and cytoplasmic compartments. Small molecules and ions freely diffuse through NPCs whereas proteins larger than 40 kDa must be bound by nuclear transport receptors to traverse the NPC. Nuclear transport receptors bind to a nuclear localization signal (NLS) within a cargo protein and mediate movement of the cargo through the NPC. Once in the nucleus, a small GTPase (RanGTP) binds to the nuclear transport receptor and triggers a conformational change that releases the cargo. The nuclear transport receptor bound by RanGTP is recycled to the cytoplasm where RanGTP is converted to RanGDP, releasing the nuclear transport receptor. The nuclear transport receptor is then free to import a new cargo protein. The directionality of nuclear transport is maintained by localizing the Ran guanine nuclear exchange factor RCC1 to the nucleus and the Ran GTPase activating proteins to the cytoplasm (Tran et al., 2014; Chook and Süel, 2011). Nuclear transport maintains privileged access of proteins to the nuclear compartment, thus protecting the genome and facilitating gene regulation.
Several distinct NLS sequences within cargo proteins have been characterized. The most common NLS is the classical nuclear localization signal (cNLS). Proteins containing a cNLS are imported to the nucleus by the nuclear transport receptor heterodimer comprised of importin α and importin β. Importin α recognizes and binds to the cNLS in the cargo protein and acts as an adapter for importin β. Importin β mediates translocation of the complex through the NPC. Although the cNLS is the most common NLS, many proteins contain non-classical NLSs (non-cNLSs), whose import is mediated by direct binding to a nuclear transport receptor from the importin β superfamily. These nuclear transport receptors do not require an importin α adapter but directly bind the non-cNLS in the cargo protein and facilitate movement through the NPC. Apart from the PY-NLS, which is recognized and imported by the nuclear transport receptor transportin, few of the non-cNLSs have been extensively characterized or shown to have a conserved motif or bind directly to specific, non-redundant nuclear transport receptors. Understanding of these less-characterized non-cNLS import pathways is currently based on studies of individual protein cargos. The diversity of NLSs could enable specialization of the nuclear transport system for classes of cargo proteins.
Multinucleated muscle cells present an intriguing biological system; many nuclei all share the same common cytoplasm but nonetheless behave differently. We investigated nuclear import as a potential mechanism for achieving differences in nuclear activity. Here, we present our findings of differences in nuclear import among myonuclei within the same skeletal muscle cell. Using an in vitro import assay, we show that nuclei in myotubes in vitro differ in the activity of classical and non-classical nuclear import pathways. The activities of these nuclear import pathways also vary at different stages of myogenesis. We further identified differences in the rates of nuclear transport for nuclei in myofibers ex vivo by using fluorescence recovery after photobleaching (FRAP). These findings highlight the remarkable variability of nuclei within the same muscle cell, exposed to the same cytoplasmic factors, and identify a potential mechanism for achieving diversity in protein accumulation and nuclear activity among myonuclei within a single muscle cell.
RESULTS
Nuclear import varies among nuclei within single cultured multinucleated myotubes
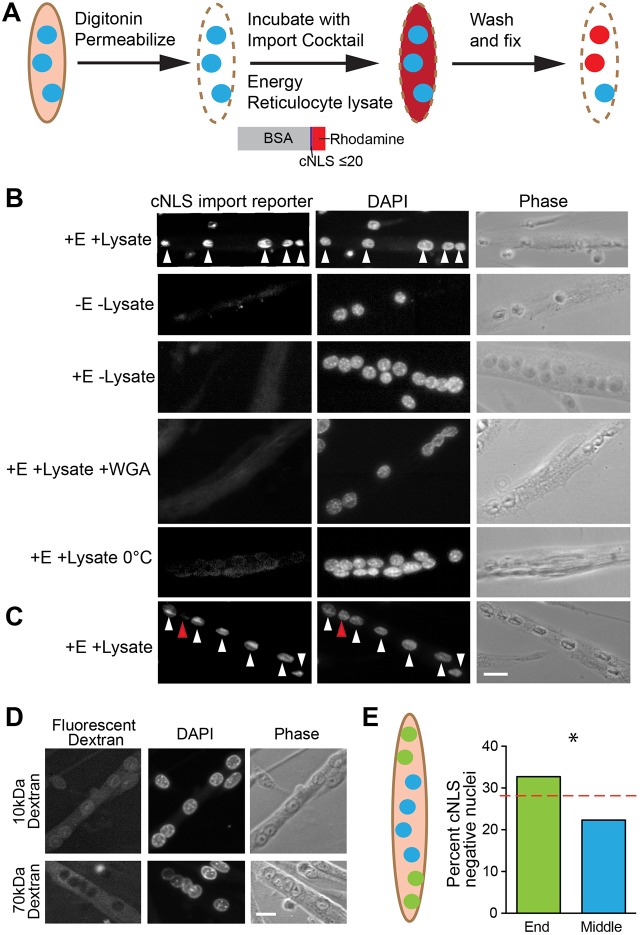
To analyze nuclear import in multinucleated myotubes, we adapted an established in vitro nuclear import assay (Moore and Schwoebel, 2001). Primary mouse myotubes were generated in vitro by inducing differentiation and fusion of precursor myoblasts. In this adaptation of the assay, illustrated in Fig. 1A, the myotube plasma membrane was permeabilized with digitonin, which selectively extracts cholesterol and leaves the nuclear envelope intact. The cytoplasm was washed out and the permeabilized cells were incubated with an import cocktail containing reticulocyte lysate to provide soluble nuclear transport factors, an energy regenerating system and a fluorescent nuclear import reporter containing multiple copies of a cNLS. The reporter was approximately 78 kDa, which is above the NPC size exclusion limit (Lénárt et al., 2003), making this cargo dependent on facilitated nuclear import to enter the nucleus. After the incubation, excess reporter was removed by washing and the cells were fixed and examined by fluorescence microscopy. Because the cell cytoplasm had been removed, this assay specifically assessed nuclear-intrinsic differences in the import of the fluorescent nuclear import reporter.
Fig. 1.
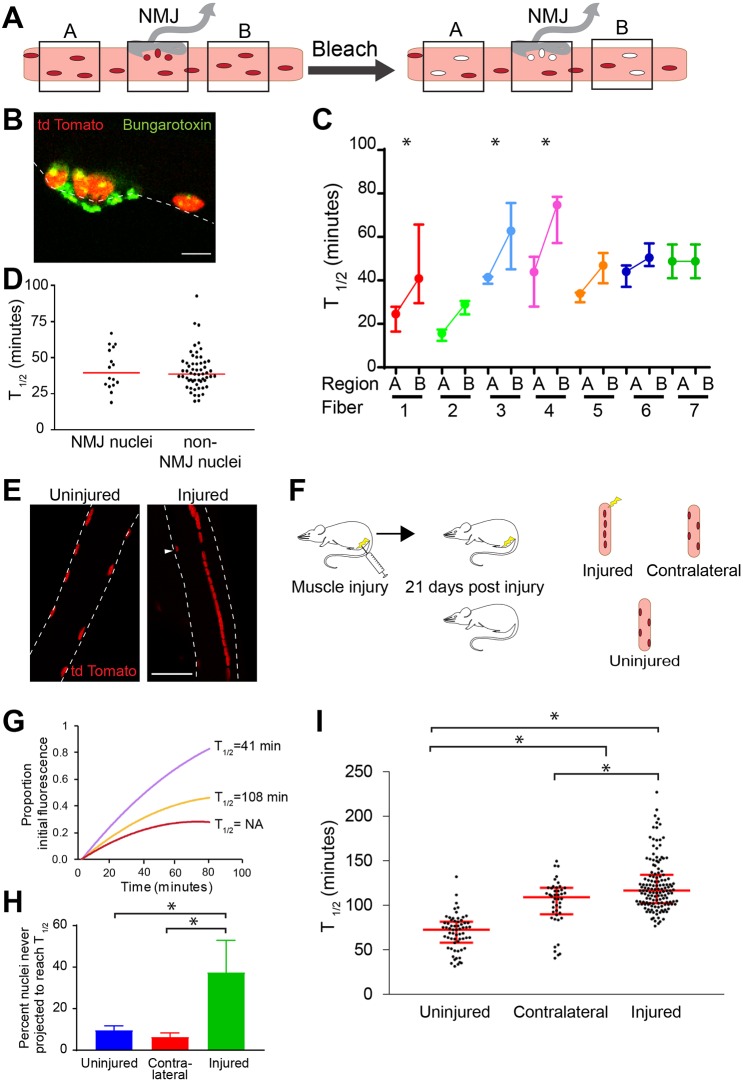
Some, but not all, nuclei within myotubes import a fluorescent nuclear import reporter. (A) To perform the in vitro import assay, the plasma membrane of myotubes was permeabilized using digitonin and the cytoplasm washed out. Cells were incubated with reticulocyte lysate, an energy regenerating cocktail and a fluorescent nuclear import reporter composed of BSA conjugated to many SV40 T-antigen cNLSs. After washing and fixation, nuclei were DAPI stained and imaged. (B) cNLS-positive nuclei (white arrowheads) were only detected when permeabilized cells were incubated with both the energy regenerating system (E) and reticulocyte lysate (lysate) in addition to the import reporter. Nuclear import was blocked by addition of wheat germ agglutinin (WGA) or by low temperature. (C) Within single myotubes, both cNLS-positive (white arrowheads) and cNLS-negative (red arrowhead) nuclei were present. (D) All myotube nuclei were permeable to 10 kD fluorescent dextran but appropriately excluded 70 kD dextran, indicating that the nuclear envelopes were not damaged by permeabilization and that cNLS-import negative nuclei do not have blocked NPCs. (E) The percentages of cNLS-negative nuclei in the middle or on the ends of a myotube were compared with the percentage expected assuming random distribution of positive and negative nuclei in a myotube (red dotted line). The two nuclei on either end of myotubes were cNLS-negative more frequently than predicted by random distribution of cNLS-positive and cNLS-negative nuclei (chi squared *P<0.001, n=450 myotubes from three independent cell isolates). Scale bars: 20 µm.
When all components of the import cocktail were present, nuclear import was readily detectable (Fig. 1B). However, if reticulocyte lysate or the energy regenerating system was omitted, nuclear import did not occur. These results are consistent with the requirement for nuclear transport factors and energy in the form of RanGTP for nuclear import (Adam et al., 1990; Newmeyer et al., 1986). Furthermore, nuclear import was abolished if the cells were treated with wheat germ agglutinin (WGA), which binds to glycosylated nucleoporin (Nup) residues (Hanover et al., 1987) and thereby blocks the NPC (Finlay et al., 1987). Nuclear import was also blocked when cells were incubated with the complete import cocktail at 0°C, consistent with the temperature dependence of nuclear import (Adam, 2016; Adam et al., 1990). Together, these results indicate that import of the fluorescent nuclear import reporter detected by this assay is mediated by active transport proceeding through the NPC.
To analyze whether nuclear import varies among myonuclei within a single myotube, fluorescence microscopy was used to score nuclear fluorescence manually following the in vitro nuclear import assay. Nuclei within myotubes containing 4–17 nuclei were scored either as positive for the nuclear import reporter (cNLS-positive) or negative for the nuclear import reporter (cNLS-negative). Within single multinucleated myotubes, we observed that not all the nuclei were cNLS-positive (Fig. 1C); 71% of myotubes contained at least one cNLS-negative nucleus. Of the 1422 nuclei analyzed in 227 myotubes with both cNLS-positive and cNLS-negative nuclei, 69% of nuclei were cNLS-positive. The number of nuclei per myofiber did not affect the percentage of cNLS-positive nuclei (Fig. S1). To examine whether cNLS-negative nuclei resulted from completely blocked NPCs, we incubated digitonin-permeabilized myotubes with fluorescent dextrans. The 70 kDa dextran is above the size exclusion limit for NPCs and is excluded from intact nuclei; the 10 kDa dextran is below this size cut off and can diffuse freely through NPCs (Adam, 2016). We found that 94% of nuclei appropriately excluded the 70 kDa dextran and 100% of nuclei were permeable to the small 10 kDa dextran (Fig. 1D). These results indicate that nuclei were not damaged by digitonin permeabilization and that cNLS-negative nuclei within myotubes do not result from blocked NPCs.
Having established that this assay reliably evaluates nuclear import and that differences among nuclei in myotubes are the result of intrinsic nuclear characteristics, we investigated whether there was a connection between the position of a nucleus within a myotube and cNLS import. We defined the two nuclei at each end of a myotube as end nuclei and the other nuclei as middle nuclei (Fig. 1E). Our results indicate that a cNLS-negative nucleus can be located in any position within a myotube and suggest that nuclear position in a myotube could contribute to differences in nuclear import (Fig. 1E).
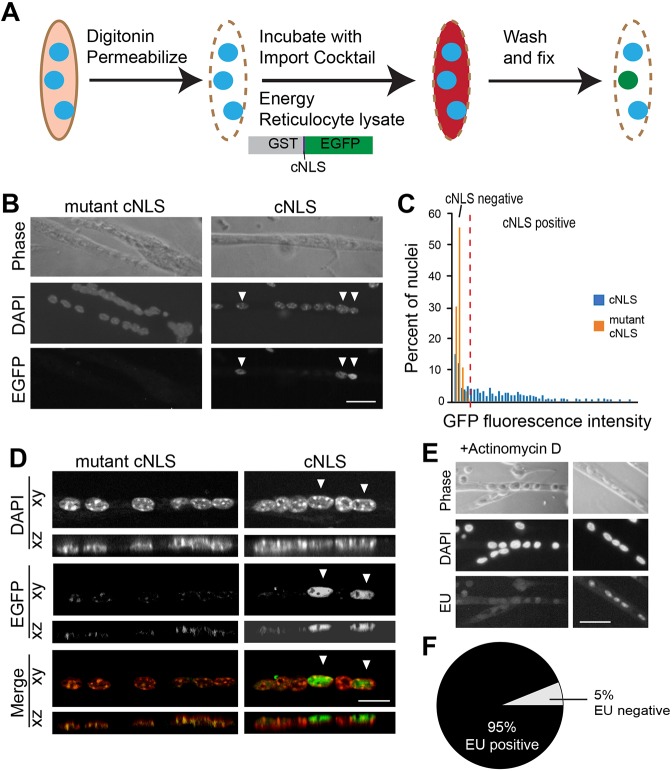
The fluorescent import reporter used in the initial experiments contained up to 20 cNLS peptides conjugated to a single bovine serum albumin (BSA) protein (Fig. 1). This high concentration of cNLS peptides allowed us to examine the nuclear import system at saturation. However, endogenous proteins typically contain a single encoded NLS. To model nuclear import better, we used a more biologically relevant reporter containing glutathione S-transferase (GST), a single cNLS and enhanced green fluorescent protein (EGFP) (GST–cNLS–EGFP). At 61 kDa, this reporter is larger than the NPC diffusion limit and so relies on facilitated, nuclear transport receptor-mediated import to enter the nucleus. A fluorescent reporter with a mutant cNLS, which is not bound by importin α and consequently does not accumulate in the nucleus or on the nuclear envelope (Butel et al., 1969; Rapp et al., 1969), was used as a control. The import assay was performed in myotubes using the reporter with a single cNLS sequence, as schematized in Fig. 2A. Like the reporter with the multiple cNLS peptides conjugated to BSA, some but not all nuclei were cNLS-positive (Fig. 2B). Nuclei within myotubes containing more than six nuclei were visualized by fluorescence microscopy and the intensity of integrated EGFP measured by an automated image analysis pipeline. Background fluorescence was established by the peak fluorescence intensity of nuclei incubated with the mutant cNLS reporter. Nuclei within myotubes incubated with the cNLS reporter with fluorescence higher than background were defined as cNLS-positive; those with fluorescence less than or equal to the background level were defined as cNLS-negative (Fig. 2C). As expected, performing the in vitro import assay with the new reporter containing a single cNLS yielded fewer cNLS-positive nuclei (25%; n=5 independent experiments with 150 nuclei per experiment) compared with the reporter containing up to 20 cNLS peptides (Fig. 1; 69% cNLS-positive). To check whether the fluorescent reporter might be concentrated in some areas of the nucleus, we examined multiple planes of focus through nuclei. Analysis by confocal fluorescence microscopy confirmed that on no plane of focus were cNLS-negative nuclei more fluorescent than those incubated with the mutant cNLS reporter (Fig. 2D). Thus, the encoded cNLS and mutant cNLS results provide confidence that active nuclear import can be tracked in these multinucleated cells. Detection of differences in nuclear import among nuclei using two distinct reporter cargos additionally lends confidence that the differences represent actual variations in nuclear import activity among these nuclei.
Fig. 2.
Nuclear import depends on the NLS. (A) The in vitro import assay was performed on myotubes using purified recombinant protein composed of GST, a single cNLS and EGFP. (B) Some, but not all, nuclei were cNLS-positive (arrowheads), similar to those observed with the reporter containing multiple cNLS in Fig. 1. A reporter protein containing a mutant cNLS was not imported into nuclei. Scale bar: 30 μm. (C) A nucleus was determined to be cNLS-positive if its fluorescence exceeded the background established by the mutant cNLS control protein. (D) cNLS-positive (arrowheads) and cNLS-negative nuclei were examined using confocal microscopy. cNLS-negative nuclei remained negative regardless of the plane of focus. Scale bar: 15 μm. (E) To examine the general transcriptional activity of nuclei within myotubes, RNA production was examined by incorporation of EU, a uridine analog. Actinomycin treatment effectively prevented EU incorporation. Scale bar: 30 μm. (F) Almost all nuclei (95% of 680 examined) were EU-positive, indicating that they were transcriptionally active. The high percentage of EU-positive nuclei compared with the relatively low percentage of cNLS-positive nuclei indicates that cNLS-negative nuclei are transcriptionally active.
Having determined that some nuclei were cNLS-negative, we hypothesized that these nuclei could be dormant or simply inactive; therefore, we examined transcription as a marker of general nuclear activity. To analyze transcription in myotubes, cultures were incubated with alkyne-modified 5-ethynyl uridine (EU), a uracil analog that is incorporated into newly synthesized RNA. Nuclei were subsequently visualized using chemoselective ligation of Alexa Fluor 594 azide with EU (Fig. 2E). Surprisingly, 95% of 680 nuclei in four independent experiments were EU-positive, indicating that almost all nuclei in myotubes are transcriptionally active (Fig. 2F), even though 75% of nuclei are cNLS-negative for the GST–cNLS–EGFP reporter. Together, these results indicate that differences in cNLS nuclear import among nuclei within single myotubes are not the result of using a saturated NLS reporter or an artifact of the plane of focus. Furthermore, these results demonstrate that cNLS-negative nuclei are still transcriptionally active.
Independence of nuclear import pathways in cultured myotubes
Having determined that some nuclei within individual myotubes are cNLS-negative but almost all nuclei are transcriptionally active, the question naturally arose as to whether these nuclei can import proteins through other non-classical NLS (non-cNLS)-dependent nuclear import pathways. To address this question, fluorescent reporters for specific non-classical nuclear import pathways were required. Many proteins have poorly defined NLSs and are imported by uncharacterized or redundant nuclear transport factors. We selected NLS sequences from proteins with a well-defined NLS that had been characterized as directly bound and imported by a single specific nuclear transport receptor. We identified three such proteins for analysis, hnRNPA1, Nup53 and PTHrP (Table 1), and cloned each of the three non-cNLSs into the GST–NLS–EGFP reporter used above and into a GST–NLS–mCherry reporter of comparable size (61 kDa and 57 kDa, respectively). We produced both EGFP and mCherry fluorescent reporter proteins for the cNLS, mutant cNLS and each non-cNLS. Each reporter differed from the others only in which NLS was included, allowing comparison of different import pathways.
Table 1.
NLS sequences and associated nuclear transport receptors

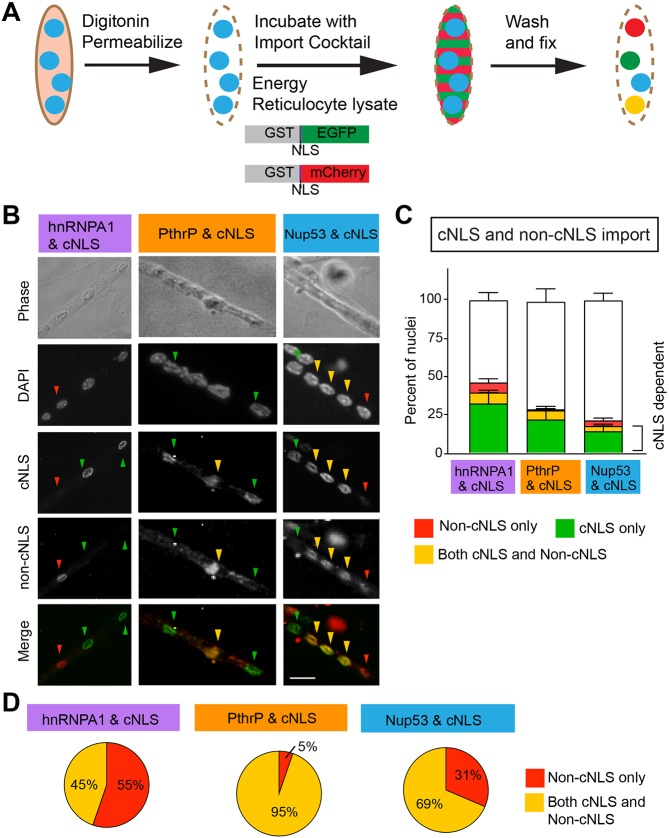
Using these reporters, we performed the nuclear import assay making systematic pairwise comparisons between the cNLS reporter and one of the non-cNLS reporters (Fig. 3A). In each comparison, the assay was always performed in duplicate with the reciprocal fluorophores. For example, in a single experiment, one coverslip was incubated with equal concentrations of GST–cNLS–EGFP and GST–hnRNPA1 NLS–mCherry while another coverslip was incubated with equal concentrations of GST–hnRNPA1 NLS–EGFP and GST–cNLS–mCherry. A third coverslip was incubated with equal concentrations of the control reporters GST–mutant cNLS–EGFP and GST–mutant cNLS–mCherry. For each pairwise comparison, some nuclei were import-positive for one, both or neither pathway (Fig. 3B). Nuclei were identified by fluorescence microscopy, and the fluorescence intensities of the EGFP and mCherry channels were measured by an automated image analysis pipeline. The percentages of nuclei that were cNLS-positive only, both cNLS- and non-cNLS-positive, non-cNLS-positive only or positive for neither NLS were calculated for each reporter (Fig. 3C). For each set of reporters, the highest percentage of nuclei was negative for both reporters; the next highest percentage of nuclei was cNLS-positive. In each pairwise comparison, the percentage of cNLS-positive nuclei was significantly higher than the percentage of non-cNLS-positive nuclei. For example, comparing hnRNPA1-NLS and cNLS, 39% of nuclei were cNLS-positive and 14% were positive for hnRNPA1-NLS. These data demonstrate that individual nuclei can be positive for one, multiple or no import pathways and that the cNLS import pathway is the predominant import pathway in myotubes.
Fig. 3.
A single nucleus with a myotube can import through multiple import pathways, a single import pathway, or no detectable import pathway. (A) Purified recombinant protein composed of GST fused to an NLS (cNLS, hnRNPA1 NLS, PthrP NLS, Nup53 NLS or mutant cNLS) followed by a fluorescent protein (either EGFP or mCherry) was expressed and isolated. The in vitro import assay was performed on myotubes using a GST–NLS–EGFP reporter and a distinct GST–NLS–mCherry reporter to compare the relative import of the two selected NLSs. Pairwise comparisons were made both with cNLS in green and non-cNLS in red and with cNLS in red and non-cNLS in green. (B) For each pairwise comparison, some nuclei were import-positive for both reporters (yellow arrowheads), only one reporter (red and green arrowheads) or none. Scale bar: 10 µm. (C) The proportions of nuclei in each pair of NLSs examined that were positive for the cNLS only, both the cNLS and non-cNLS, the non-cNLS only or neither NLS reporter were quantified. The percentage of nuclei that were negative for both cNLS and non-cNLS reporters differed significantly between the cNLS-hnRNP pairing and the other two cNLS–non-cNLS pairings. Data are mean±s.e.m.; comparisons by ANOVA with Tukey correction for multiple comparisons. For hnRNP, n=4 independent experiments were carried out; for PthrP and Nup53, n=5 independent experiments, each analyzing 200–300 nuclei. (D) For each pair of NLSs examined, the proportions of nuclei importing the non-cNLS only or both the cNLS and non-cNLS were compared. The findings differed significantly from the percentage of nuclei predicted to be positive for the non-cNLS reporter or positive for both reporters by random distribution of active NLS pathways (P<0.0001 by chi squared analysis; hnRNP n=4 independent experiments, PthrP and Nup53 n=5 independent experiments, each analyzing 200–300 nuclei).
To examine the degree of independence between the cNLS and non-cNLS reporters in individual nuclei, we compared the percentage of nuclei that were only non-cNLS-positive with those that were both non-cNLS- and cNLS-positive. We found that the degree of overlap with the cNLS reporter varied for each non-cNLS reporter (Fig. 3D). The hnRNPA1 NLS and cNLS reporters overlapped the least, as only 45% of hnRNPA1 NLS-positive nuclei were positive for both reporters. For the Nup53-NLS and cNLS reporter pairing, 69% of Nup53 NLS-positive nuclei were also cNLS-positive. In contrast, 95% of the PTHrP NLS-positive nuclei were positive for both the PTHrP NLS and cNLS reporters. The high degree of overlap with the PTHrP NLS and cNLS reporters compared with the other non-cNLS reporters is not surprising because both these NLSs rely on importin β; the cNLS additionally requires importin α (Kalderon et al., 1984) whereas the PTHrP-NLS is independent of importin α (Cingolani et al., 2002; Lam et al., 2001). Together, these data support the conclusion that the cNLS import pathway is the predominant mode of nuclear import for myonuclei and that each import pathway can be differentially regulated in individual nuclei.
Variation in cNLS import among myofiber nuclei ex vivo
Although cultured myotubes are large compared with other cell types in vitro, relative to myofibers in vivo, they are quite small and lack the extensive subcellular specialization found in myofibers. To examine whether nuclear import is also variable among nuclei in myofibers (as shown for cultured myotubes), we performed FRAP on isolated myofibers to examine nuclear import ex vivo.
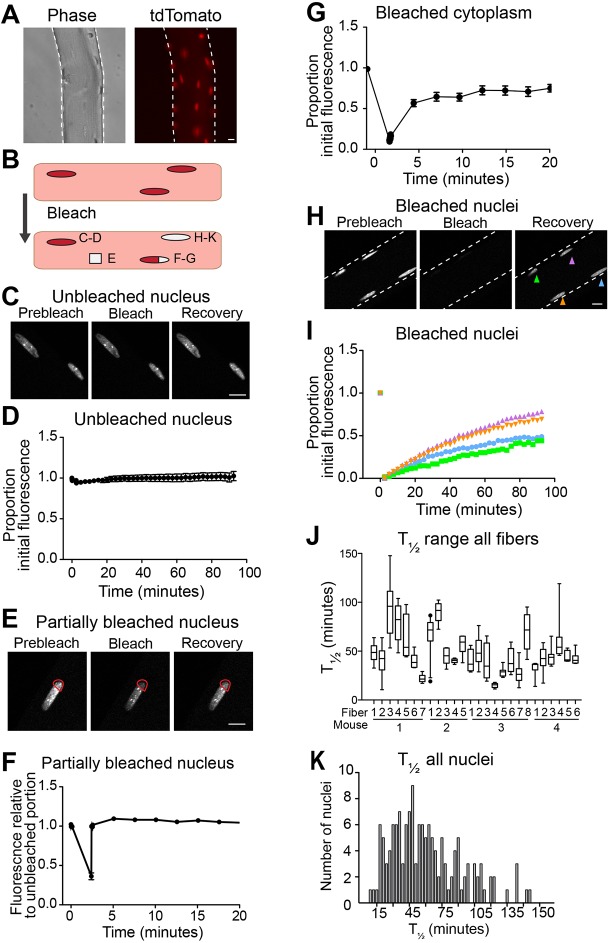
Myofibers were isolated from the gastrocnemius muscles of mice constitutively expressing cNLS–tdTomato (Prigge et al., 2013). These myofibers exhibit strong nuclear fluorescence with very little fluorescence in the cytoplasm (Fig. 4A). To determine whether we could monitor nuclear transport by FRAP, we measured fluorescence over time of unbleached nuclei, partially bleached nuclei, bleached cytoplasm and fully bleached nuclei (Fig. 4B). Unbleached nuclei maintained their fluorescence over the course of the 94-min assay with only minor decreases in fluorescence over time (Fig. 4C,D). This result demonstrates the photostability of cNLS–tdTomato. When a portion of a nucleus was bleached, the region rapidly recovered to the level of the unbleached area, with a time to half recovery (T1/2) of 0.5±0.02 s (mean±s.d.) (Fig. 4E,F). A section of bleached cytoplasm also quickly recovered fluorescence, with T1/2=5.4±0.14 s (mean±s.d.) (Fig. 4G). The substantially longer T1/2 of bleached cytoplasm relative to nucleoplasm could reflect reduced mobility in the cytoplasm resulting from tightly packed, highly ordered myofibrils. These two controls demonstrate that the experiment can detect bleaching and recovery and that the diffusion rates of cNL–tdTomato within either the nucleus or the cytoplasm of a myofiber are rapid and consistent. However, when a whole nucleus was bleached, it recovered fluorescence much more slowly than a partially bleached nucleus, with a rate best modeled by a quadratic function (Fig. 4H,I). This dramatically slower rate, in conjunction with the 95 kDa size of the tdTomato molecule (above the NPC diffusion limit), supports the validity of the FRAP assay for measuring nuclear import rates.
Fig. 4.
cNLS nuclear import varies among nuclei within single myofibers. (A) Myofibers isolated from gastrocnemius muscles of mice constitutively expressing nuclear-targeted cNLS–tdTomato exhibit strong nuclear fluorescence and minimal cytoplasmic fluorescence. (B) Schematic representing the cellular parameters that were studied in FRAP experiments: an unbleached nucleus, a partially bleached nucleus, a bleached portion of cytoplasm and a fully bleached nucleus. (C) Representative images of unbleached nuclei at different times points. (D) Unbleached nuclei consistently maintained fluorescence throughout the entire period of imaging (n=33, mean±s.e.m.). (E) Representative partially bleached nucleus prior to bleaching, immediately after bleaching and after recovery. Red outlines the bleached area. (F) Bleached sections of partially bleached nuclei rapidly recover to baseline fluorescence (n=11, mean±s.e.m.). (G) Regions of bleached cytoplasm rapidly recover to their initial fluorescence (n=19, mean±s.e.m.). (H) Representative bleached nuclei prior to bleaching, immediately after bleaching and after recovery. (I) The time to half recovery (T1/2) of four bleached nuclei within a single myofiber differ from one another, as calculated by quadratic regression. (J) The median and range of T1/2 of nuclei greatly vary among myofibers, indicating differences in the rate of cNLS import among nuclei, both within a myofiber and among myofibers (4–8 nuclei measured per myofiber). (K) The histogram of T1/2 of bleached nuclei reveals a continuous broad population with median T1/2 of 45 min (n=128). Scale bars: 10 µm.
To assess whether nuclei within the same myofiber differed in cNLS nuclear import rates, we performed FRAP on several nuclei in a single myofiber. When multiple nuclei within a single myofiber were bleached, we observed a clear variation in the rate of fluorescence recovery among nuclei, with T1/2 ranging from 15 to 146 min (schematic in Fig. 4I). To determine whether this variability in T1/2 was observed in other myofibers, we examined 26 myofibers isolated from four different mice. We found that the T1/2 of nuclei varied within any given myofiber, with both the median T1/2 and degree of T1/2 variability differing among myofibers (Fig. 4J). Hypothesizing that distinct classes of nuclei could exist within myofibers (i.e. those importing rapidly and those importing more slowly), we compared the T1/2 of all 128 nuclei examined and found that the rates of import reflected a single broad population with median T1/2 of 45 min and not distinct subpopulations (Fig. 4K).
Given the variability in ranges of nuclear import rates among myofibers and the correlation detected between nuclear import and nuclear position within a myotube (Fig. 3C), we compared nuclear import rates between nuclei in two different non-NMJ regions of the same myofiber. We also compared the import rates of populations of nuclei located at the NMJ and in non-NMJ regions of the same fibers (Fig. 5A). The NMJ is a well-characterized, easily identifiable, specialized region of the myofiber containing a cluster of acetylcholine receptors (AChRs) (Couteaux and Pecot-Dechavassine, 1973). We identified the NMJ on myofibers by fluorescent bungarotoxin staining of AChRs (Fig. 5B). By comparing nuclei from two non-NMJ regions in a single myofiber (four to nine nuclei in each region), we determined that the median and range of T1/2 could differ significantly between the two non-NMJ regions of the same fiber (Fig. 5C). Comparing the population of nuclei directly beneath the NMJ with paired nuclei in non-NMJ regions of the myofiber revealed no difference between the NMJ nuclei and non-NMJ nuclei (Fig. 5D). These results indicate that, although NMJ nuclei differ from other nuclei in the myofiber transcriptionally (Fontaine and Changeux, 1989; Moscoso et al., 1995), they do not differ in cNLS nuclear import rates; however, nuclei in two non-NMJ regions can differ significantly in nuclear import rates.
Fig. 5.
Rates of cNLS nuclear import can vary between regions of a single myofiber. (A) FRAP experiments were performed on nuclei within two different myofiber regions (A or B) not near the NMJ and nuclei directly under the NMJ using myofibers isolated from gastrocnemius muscles of mice constitutively expressing nuclear-targeted cNLS–tdTomato. (B) The NMJ was identified by FITC-conjugated bungarotoxin staining of acetylcholine receptors. (C) Nuclei were bleached, imaged and the T1/2 calculated as in Fig. 4. The T1/2 of 4–8 nuclei in disparate non-NMJ regions (A and B in Fig. 5A) of a myofiber can vary. Data are median±interquartile range. (D) T1/2 of NMJ (n=15) and non-NMJ (n=58) nuclei were compared. The red line represents the median. (E) In uninjured myofibers, nuclei are peripherally located. After injury, nuclei are centrally located in regenerated myofibers. Arrow indicates a non-central nucleus in a regenerated myofiber. (F) Schematic representing the injury experiment: one gastrocnemius muscle was injured by barium chloride injection. After 21 days, the injured muscle, the contralateral gastrocnemius and a gastrocnemius of an uninjured littermate were collected. (G) The fluorescence recovery of three bleached nuclei is represented by quadratic regression as in Fig. 4I. The recovery of some nuclei plateaus before reaching T1/2. (H) The percentage of nuclei that plateau before recovering to T1/2 was calculated for nuclei in myofibers from uninjured (8%; n=69 from three mice), contralateral (4%; n=52 from three mice) and injured (38%; n=304 from three mice) muscle. Data represent mean±s.d. (I) T1/2 values for nuclei in myofibers isolated from uninjured, contralateral and injured muscles were calculated. The median T1/2 of nuclei from contralateral (109 min) and injured myofibers (123 min) were significantly higher than for uninjured control myofibers (65 min). T1/2 values for contralateral and injured myofibers also differed significantly. Uninjured n=63, contralateral n=44, injured n=143. Red lines represent the median and quartile range. *P<0.001, comparisons by ANOVA with Tukey correction for multiple comparisons. Scale bars: 10 µm (B), 50 µm (E).
We next assessed whether nuclear import rates change in response to physiological conditions. Skeletal muscle injury has a well-defined physiological response and a well-characterized nuclear phenotype. After injury, skeletal muscle regenerates and many of the nuclei incorporated during regeneration are localized in the center of the myofiber rather than at the periphery (Fig. 5E). To determine whether central nuclei differ from non-central nuclei with respect to nuclear import rates, we compared T1/2 of nuclei in myofibers isolated from gastrocnemius muscles 21 days post injury, contralateral gastrocnemius muscles and gastrocnemius muscles isolated from uninjured mice (schematic in Fig. 5F). When fluorescence recovery was modeled by a quadratic equation, the recovery of some nuclei plateaued before half recovery was reached (Fig. 5G, red curve). Consequently, these nuclei never reach T1/2. Our data indicate that 8% of nuclei from uninjured muscles and 4% of nuclei from contralateral muscles never reach T1/2. In contrast, the fluorescent recovery of 38% of nuclei in injured myofibers plateaued before reaching T1/2 (Fig. 5H). The nuclei in injured myofibers for which a T1/2 could be calculated recovered significantly more slowly (median T1/2=123 min) than the nuclei in myofibers isolated from uninjured mice (median T1/2=62 min). Surprisingly, the nuclei in myofibers isolated from the contralateral leg also recovered significantly more slowly (median T1/2=109 min) than nuclei from uninjured mice (Fig. 5I). These findings demonstrate that nuclear import is responsive to muscle injury in injured myofibers. Furthermore, nuclear import is affected in myofibers distant from the site of injury. Together, our findings suggest both local and systemic modulation of reduced nuclear import rates following injury.
Nuclear import varies with muscle differentiation
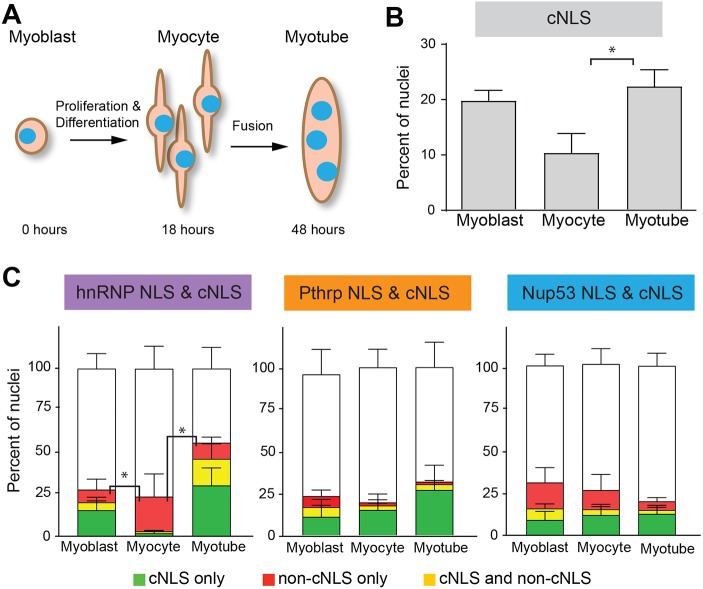
The nuclei in newly regenerated myofibers are derived from muscle stem cells that have recently differentiated and fused. Given the decreased rate of nuclear import in newly regenerated myofibers, we examined whether global changes in nuclear import occur during differentiation in an in vitro model of myogenesis (Bondesen et al., 2004). During in vitro myogenesis, mononucleated myoblasts cease proliferating and differentiate to become myocytes. Subsequently, myocytes fuse to form multinucleated myotubes. We differentiated pure cultures of primary mouse myoblasts in vitro and performed the in vitro import assay using the fluorescent cNLS reporter (Fig. 2) at myoblast (0 h), myocyte (18 h) and myotube (48 h) stages of differentiation (Fig. 6A). We found that the proportion of nuclei that were cNLS-positive varied across the stages of differentiation. The highest proportion of nuclei was cNLS-positive at the myoblast (20%) and myotube (23%) stages, whereas only 9% of myocyte nuclei were cNLS-positive (Fig. 6B).
Fig. 6.
Nuclear import varies during myogenesis in vitro. (A) Pure cultures of primary myoblasts were differentiated to form myocytes or myotubes and the in vitro nuclear import assay was performed. (B) The percentage of cNLS-positive nuclei was high in myoblasts (20%), dropped dramatically in myocytes (9%) and then rose again to pre-differentiation levels in myotubes (23%). (C) The in vitro import assay was performed using cNLS and non-cNLS import reporters to compare the relative import of the cNLS and non-cNLSs at different stages of myogenesis. For each pair of NLS reporters examined, the proportions of nuclei importing the non-cNLS only or both the cNLS and non-cNLS were compared, as in Fig. 3C. The percentage of hnRNP NLS import differed significantly across differentiation. The hnRNP reporter displayed relatively low import in the myoblast and myotube stages but relatively high import in the myocyte stage. All data are mean±s.e.m.; *P<0.05, comparisons by ANOVA with Tukey correction for multiple comparisons; n=3 independent experiments, each with ∼200 nuclei per differentiation stage.
Curious about the low percentage of cNLS-positive myocyte nuclei, we investigated whether the frequency of import for non-cNLS pathways also changed with myogenesis. We used pairwise comparisons of cNLS and non-cNLS reporters, as shown in Fig. 3. In the PTHrP NLS and cNLS as well as the Nup53 and cNLS reporter pairings, the percentage of nuclei positive for only the non-cNLS reporter did not change significantly across differentiation. In contrast, the percentage of nuclei positive for only hnRNPA1 NLS was significantly higher at the myocyte stage (21%) than at either the myoblast (8%) or myotube (10%) stages. This preferential import of the hnRNPA1 reporter occurred to the exclusion of the cNLS reporter. Together, these results show that differentiation state affects nuclear import.
DISCUSSION
Our study reveals that nuclear import varies among nuclei in the same multinucleated muscle cell. Nuclei within in vitro generated myotubes differ from one another in accumulation of fluorescent reporter proteins, which indicates differential usage of the cNLS import pathway and non-cNLS import pathways. Additionally, rates of cNLS import vary among nuclei in isolated myofibers ex vivo. Variation in nuclear import correlated with nuclear position within the cell and progression through myogenesis. These differences in nuclear import among muscle nuclei could play a role in differentiation and in establishing regional specialization in mature myofibers.
We determined that nuclei within a single myotube differ in cNLS nuclear import. Furthermore, examination of non-cNLS import pathways revealed that, like the cNLS pathway, some but not all nuclei were positive for the non-cNLS pathways. Individual nuclei could be positive for one, both or neither examined import reporter. The percentage of overlap between cNLS and non-cNLS reporters indicated that individual import pathways can be differentially regulated. Because these experiments were performed on permeabilized cells from which the cytoplasm had been removed, these findings reflect intrinsic differences among myonuclei and not changes in soluble transport factors.
In addition to examining nuclear import pathways, we used FRAP to investigate cNLS nuclear import rates in isolated myofibers ex vivo. Nuclei within individual myofibers differed in the rate of nuclear import. Although individual nuclei could differ greatly in import rate, the import rates of all nuclei examined formed a single broad population. The import rates of nuclei located at the NMJ did not differ significantly from nuclei elsewhere in a myofiber. Thus, although nuclei associated with the NMJ are distinct from other nuclei both in protein content (Apel et al., 2000) and transcript production (Moscoso et al., 1995; Brenner et al., 1990; Jasmin et al., 1993; Fontaine and Changeux, 1989), these differences are not caused by detectable variation in cNLS nuclear import rates. The specialization of NMJ nuclei could be achieved through selective activation of non-cNLS import pathways in NMJ nuclei or through subcellular enrichment of NMJ targeted cargo in the cytoplasm surrounding NMJ nuclei.
When the median rate of nuclear import was compared between two morphologically non-specialized regions of a single myofiber, we found that the median rate and range could vary significantly between different regions of the same myofiber. A study describing variability in expression of both housekeeping and muscle-specific genes among nuclei noted that, although nuclei producing housekeeping gene transcripts were randomly distributed throughout a myofiber, nuclei producing muscle-specific transcripts clustered together (Newlands et al., 1998). This selective distribution of nuclei producing muscle-specific transcripts prompted the authors to suggest the existence of domains in a myofiber that could be defined functionally rather than morphologically. Our finding that the median and range of nuclear import rates can vary significantly between two morphologically non-specialized myofiber regions supports this model.
To investigate whether nuclear transport was altered by physiological stimuli, we examined cNLS nuclear import rates in response to skeletal muscle injury. Following injury, muscle-resident stem cells called satellite cells proliferate, differentiate and fuse to repair multinucleated myofibers (Ratnayake and Currie, 2017). Myonuclei in regenerated myofibers localize in the center of the myofiber rather than at the periphery and remain centralized for months after the injury has healed (Wada et al., 2008). We found that the T1/2 of nuclei in regenerated myofibers was significantly slower than in control nuclei. A central nucleus completely surrounded by dense myofibrils could have reduced access to diffusible proteins, resulting in apparent decreased transport rates. However, the T1/2 of nuclei in myofibers isolated from the contralateral limb, in which the nuclei remain peripheral, was also significantly slower than for uninjured control myofibers. These observations indicate that the decreased nuclear import rate in response to injury is not solely the result of decreased diffusion. The increased T1/2 in myonuclei in the contralateral limb suggests that systemic factors can modulate nuclear import in response to injury. Previous studies have demonstrated that circulating hepatocyte growth factor activator (HGFA), which is activated upon injury (Rodgers et al., 2017), induces a state of readiness in satellite cells distant from the location of the injury (Rodgers et al., 2014). A circulating factor could similarly mediate changes in nuclear transport in response to tissue injury.
In support of the biological relevance of altered nuclear import, we found that nuclear import is dynamic across differentiation. A study that examined steady-state nucleocytoplasmic localization of fluorescent cNLS reporters in differentiation of the C2C12 murine muscle cell line reported decreased reporter steady-state nuclear localization in myotubes compared with myoblasts. This difference in steady state localization was attributed to changes in nuclear export rather than nuclear import rates (Asally et al., 2011). However, the technical details of the approaches used to examine nuclear transport differed greatly between the previous study and the current work. The assays employed here directly examine import in the absence of export and thus represent differences in nuclear import.
Our conclusion that nuclear import varies with differentiation is based on experiments conducted in permeabilized cells, indicating that inherent differences between nuclei are responsible for the nuclear import variability we detected. Similarly, a comparison of steady-state localization of a fluorescent nuclear import protein and nuclear export reporter protein in myotubes and myoblasts treated with polyethylene glycol to induce fusion found that multinucleated myoblasts were more like mononucleated myoblasts than like multinucleated myotubes (Asally et al., 2011). This finding indicates that differentiation of a nucleus has a greater impact on nuclear transport than simply existing in a multinucleated environment. Our observed changes in nuclear import with differentiation could result in part from the stress of serum starvation, as various kinds of cellular stress induce changes in nuclear transport (Kodiha et al., 2009; Regot et al., 2013; Yasuda et al., 2006; Kodiha et al., 2008). Whether resulting from a stress response or regulated differentiation-related changes, the alterations in nuclear import are most likely mediated by changes to the NPC.
Variation in nuclear import pathway usage or the rate of nuclear import could represent a mechanism for achieving diversity among myonuclei in a single cell. Nuclear transport is affected by NPC composition, Nup post-translational modification, the RanGTP gradient between the nucleus and the cytoplasm, the levels and modification state of nuclear transport receptors and the levels and modification state of cargo protein (Sekimoto and Yoneda, 2012; Hung and Link, 2011). The in vitro import assay specifically examines nuclear intrinsic variables, which include the RanGTP gradient among nuclei, variable NPC composition and post-translational Nup modification. Thus, although additional variables probably contribute to regulating nucleocytoplasmic transport in vivo, the differences in nuclear import that we identified among myonuclei are rooted in intrinsic nuclear properties.
Differences in NPC composition among myonuclei could drive differential import pathway usage. Differences in the stoichiometry of various Nups within the NPC can affect nuclear import (Crampton et al., 2009; Gustin and Sarnow, 2002, 2001). Although the core NPC proteins are incorporated into the nuclear envelope as it re-forms after mitosis and experience minimal exchange over the cell lifetime (Savas et al., 2012), some peripheral Nups dynamically associate with the NPC (Toyama et al., 2013). Nups can specifically interact with chromatin proteins, enzymes or nuclear transport receptors. For example, specific Nups are required for SMAD nuclear transport (Xu et al., 2007) but not cNLS import (Chen and Xu, 2010), whereas other Nups play a more crucial role in cNLS import (Sabri et al., 2007). Myonuclear NPC composition changes during differentiation, with additional Nup358 present at each pore (Asally et al., 2011) and incorporation of Nup210 into the NPC (D'Angelo et al., 2012). Although the increased level of these Nups is necessary for proper differentiation, their depletion does not alter nucleocytoplasmic transport (Asally et al., 2011; D'Angelo et al., 2012). Indeed, the effect of Nup210 on differentiation is independent of incorporation into the NPC (Gomez-Cavazos and Hetzer, 2015). Our own comparison of FG Nups, Nup358, RanGAP1 and Kapβ levels in myonuclei in myotubes by immunofluorescence did not reveal obvious differences among nuclei (Fig. S2A,B) and we could not conclusively detect a correlation between intensity of Nup staining and nuclear import (Fig. S2C,D). Although challenging to detect, dynamic changes in NPC composition could contribute to the differences in nuclear import.
Post-translational modification of Nups could also contribute to differences in nuclear transport among myonuclei. Both phosphorylation and O-linked β-N-acetylglucosamine (O-GlcNAc) glycosylation of Nups are dynamic transient modifications that change in response to cellular physiology (Kodiha et al., 2009; Regot et al., 2013; Crampton et al., 2009). Increased O-GlcNAc modification of Nups can decrease association of nuclear export factors with the NPC, decreasing nuclear export (Crampton et al., 2009). Interestingly, in C2C12 cells, differentiation from myoblasts to myotubes is accompanied by a global decrease in O-GlcNAc modification, and pharmacological inhibition of O-GlcNAc removal blocks myoblast fusion and decreases expression of myogenic regulatory factors (Ogawa et al., 2012). Similarly, changes in Nup phosphorylation affect nuclear transport. Dynamic phosphorylation of specific Nups leads to decreased binding of importin β and decreased nuclear import (Kosako et al., 2009). Many NPC proteins undergo differentiation-dependent phosphorylation, with a general trend of reduced phosphorylation as differentiation progresses (Rigbolt et al., 2011). Thus, transient Nup post-translational modifications could result in nucleus-to-nucleus differences in nuclear import within a single muscle cell.
Identifying the biochemical mechanisms responsible for the differences in nuclear import among nuclei is technically challenging. Standard biochemical assays require large amounts of purified cNLS-positive nuclei separated from cNLS-negative nuclei. Furthermore, many Nups have additional nuclear functions unrelated to their role at the NPC (Gomez-Cavazos and Hetzer, 2015; Buchwalter et al., 2014; Morchoisne-Bolhy et al., 2015). Thus, Nup protein levels in a nucleus might not reflect the amount of a Nup at the nuclear pore. NPC heterogeneity within a single nucleus would also complicate analysis. A myonucleus has approximately 360,000 NPCs (Asally et al., 2011). Therefore, even if an individual nucleus could be biochemically analyzed, the result would represent the average of a large population that could vary within a single nucleus (Kinoshita et al., 2012). Post-translational modifications would be difficult to identify for many of the same reasons as changes in NPC stoichiometry. Differences in modification would be additionally challenging to study because many of the modifications that impact nuclear transport occur in hydrophobic regions. These regions are highly repetitive and share homology between Nups, which reduces confidence in detecting the modifications and in assigning detected modifications to a single protein. Additionally, each NPC contains 16–32 copies of each Nup (Hetzer and Wente, 2009) and individual copies of the same Nup might not all be modified in the same way. This level of NPC complexity decreases the probability of detecting the modification using an approach that examines a population of nuclei. As additional techniques are developed (Cutler et al., 2017; Jankowska et al., 2016; Krishnaswami et al., 2016; Porpiglia et al., 2017), identifying the biochemical mechanisms regulating differences in nuclear import among nuclei sharing a common cytoplasm will become increasingly feasible.
Understanding how individual nuclei within a single multinucleated muscle cell behave differently has broader implications than skeletal muscle biology. Although most eukaryotic cells have a single nucleus, some specialized cells are multinucleated. Ranging from single-celled organisms, to fungi, to differentiated mammalian cells, cells with multiple nuclei present an intriguing biological system. Not only must gene expression be regulated in multiple nuclei in a coordinated fashion to achieve the necessary cellular protein levels, but also nuclei within these multinucleated cells differ from one another in many cases. Tetrahymena, a binucleated single-celled organism, contains a germline micronucleus and a non-germline macronucleus that are structurally and functionally distinct (Orias et al., 2011). The NPCs of the two Tetrahymena nuclei differ in composition and, when Nups specific to one nucleus are targeted to the other nucleus, cargo proteins are inappropriately localized to the wrong nucleus (Iwamoto et al., 2009). Multinucleated fungi grow by mitosis uncoupled from cytokinesis and, in contrast to the Drosophila embryo, nuclei in some species divide independently of neighboring nuclei (Gladfelter et al., 2006; Anderson et al., 2013). Mammalian osteoclasts contain approximately 20 nuclei, which differ from one another in chromatin structure and markers of transcriptional activity (Youn et al., 2010). The nuclei of the syncytial trophoblast in placental mammals differ from one another in chromatin arrangement and transcriptional activity (Ellery et al., 2009). These examples of multinucleated cells from several different phyla highlight the unique biology of multinucleated cells, which allows nuclei exposed to the same cytoplasm to behave differently. The mechanisms governing variation in myonuclei could be more generally applicable to these other cell types and could represent conserved mechanisms for coordinating activity among nuclei in a syncytium.
In summary, we found that both cNLS and non-cNLS nuclear import activities vary among nuclei in a multinucleated muscle cell. Such variation in nuclear import could be a mechanism for achieving differences in transcript production among myonuclei, leading to functional specialization in myofibers. The biochemical and signaling mechanisms that modulate nuclear import and target NPCs in individual myonuclei in a common cytoplasm remain unclear and the focus of future studies. Future studies could also include defining whether physiological changes such as aging or disease spatially or temporally alter nuclear import or the ratio between cNLS and non-cNLS nuclear import in myofibers. A better understanding of the biochemical and signaling mechanisms driving differences in nuclear import among nuclei in a skeletal muscle cell can also provide further insight into the biology of other mammalian and non-mammalian syncytial cells.
MATERIALS AND METHODS
Mice
Wild-type C57BL6 mice were obtained from Charles River laboratories (Willmington, MA). nTnG mice containing an allele for nuclear targeted tdTomato and EGFP reporter proteins [B6N.129S6-Gt(ROSA)26Sortm1(CAG-tdTomato*,-EGFP*)Ees/J] (Prigge et al., 2013) were purchased from Jackson Laboratory (Bar Harbor, ME).
All experiments were performed using tissues from 3-month-old male C57BL6 mice or 6-month-old female mice homozygous for the nTnG transgene. Experiments were performed in accordance with approved guidelines and ethical approval from Emory University's Institutional Animal Care and Use Committee and in compliance with the National Institutes of Health.
In vitro myogenesis
Primary myoblasts were isolated from hindlimb muscles of 3-month-old male C57BL/6 mice as described previously, with the exception of the Percoll gradient (Bondesen et al., 2004). Myoblasts were cultured on dishes coated with bovine collagen I (Gibco, Gaithersburg, MD) in growth medium [Ham's F10, 20% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin and 5 ng/ml fibroblast growth factor] at 37°C and 5% CO2. For import assays, differentiation was induced by plating 2×105 myoblasts on 18 mm diameter coverslips coated with elastin, collagen, laminin (ECL; Gibco) and culturing in differentiation medium [Dulbecco's modified Eagle's medium, 10 mg/l insulin, 5.5 mg/l transferrin and 6.7 µg/l selenium (Gibco), 100 U/ml penicillin, 100 µg/ml streptomycin] for 18 h for myocytes or 48 h for myotubes. To analyze nuclear import in myoblasts, myoblasts were similarly plated on ECL-coated coverslips and cultured in growth medium for 18–24 h.
Fluorescent dextran
Myotubes were differentiated on glass coverslips as described above. Coverslips were placed on ice for 5 min and the medium was gently aspirated. The cells were washed once with transport buffer (20 mM HEPES, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM DTT) and then permeabilized on ice with 25 µM digitonin in transport buffer for 5 min. Permeabilized myotubes were stained with DAPI to identify nuclei and then incubated with 10 kDa or 70 kDa Texas Red-conjugated dextran for 15 min at room temperature (Sigma-Aldrich, St Louis, MO). The 10 kDa dextran enters all nuclei without blocked NPCs. The 70 kDa dextran is excluded from nuclei with intact nuclear envelopes and NPCs (D'Angelo et al., 2009). Cells were imaged after incubation with the fluorescent dextran.
In vitro nuclear import assay
The in vitro nuclear import assay was modified from previously established protocols (Moore and Schwoebel, 2001; Adam, 2016). Myoblasts, myocytes and myotubes were generated as described above. Coverslips were placed on ice for 5 min and the medium was gently aspirated. The cells were washed once with transport buffer and then permeabilized on ice with 25 µM digitonin in transport buffer for 5 min. After permeabilization, cells were gently washed with transport buffer. Cells were than incubated at 30°C for 15 min in either transport buffer or transport buffer containing 5.6 mM WGA (Vector Laboratories, Burlingame, CA). Coverslips were then placed inverted on 100 µl import cocktail (2% BSA), 20% rabbit reticulocyte lysate (Promega, Madison, WI), 4 mM import cargo, 0.1 mM GTP, 7.6 mM creatine phosphate, 15 U/ml creatine phosphokinase, 1 mM ATP, 20 mM HEPES, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA and 2 mM DTT. Fluorescent import reporters were either purchased (Sigma) or generated in the laboratory by fast protein liquid chromatography (FPLC) purification of recombinant proteins. Coverslips were incubated at 30°C in a humidified chamber for 30 min. After incubation, coverslips were washed three times with 1% BSA in transport buffer. Cells were then fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences, Hatfield, PA) and 0.1% glutaraldehyde on ice for 15 min. After fixation, cells were stained with DAPI to identify nuclei.
Labeling of newly synthesized RNA
The Click-iT RNA imaging kit (Molecular Probes, Eugene, OR) was used to label newly synthesized RNA. Primary mouse myotubes were generated by 48 h of differentiation on coverslips. Control coverslips were pretreated with 5 µg/ml actinomycin D (Molecular Probes) for 1 h at 37°C. All coverslips were then treated with 2 mM EU in differentiation medium for 30 min. Cells were fixed with 3.7% formaldehyde for 15 min on ice and further processed per the manufacturer's instructions.
Plasmids used
The pGEX4TH-SV40NLS-EGFP plasmid (Shumaker et al., 2005) (gift from Steven Adam, Northwestern University, IL) was used to generate the cNLS–EGFP reporter protein. The mutant cNLS control reporter protein was generated by site-directed mutagenesis. The GST–cNLS–mCherry construct was cloned by ligation following cleavage of the vector backbone and insert by KpnI and NotI. The insert was produced by PCR amplification of the mCherry gene using the following primers: forward, 5′-CGACTCTAGAGGATCCCCGGGTACCGGTCGCCACCATGGTGAGCAAGGGCGAGGAGGAT-3′; reverse, 5′-ATCGTCAGTCAGTCACGATGCGGCCGCTTTACTTGTACAGCTCGTCCATGCCGCC-3′. The GST mutant cNLS–EFGP construct and both PTHrP NLS constructs were cloned by the quick change method using the following primers: for the mutant cNLS forward primer, 5′-CGAAACGTAAAGTGGAAGATGGTCCGCCGCAT-3′; reverse primer, 5′-TTTTCGGCGGACCACCTTCGAGTCGACCCGGG-3′. Primers for the PTHrP NLS constructs were forward, 5′-GCAACCCCTGAAAACCCCCGGTAAGAAAAAAAAAGGAAAGCCTCATGCCTGCAGGTCGACTCTAGAG-3′; reverse, 5′-TCTTTGTAGGTTTCCACTTTATTAGTTTCCTGCGTCAGGTAGCGTTCGAGTCGACCCGGGAATTCCGG-3′. For Nup53 NLS and hnRNPA1 NLS constructs, gBlocks were purchased from Integrated DNA Technologies and ligated into the vector backbone after cleavage with EcoRI and XbaI. All constructs were sequence verified.
Recombinant protein expression and purification
BL21(DE3) competent Escherichia coli were transformed and grown in selective Luria–Bertani broth to 0.5 optical density. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 30°C for 6 h. Pelleted bacteria were resuspended in lysis buffer (1 mM phenylmethylsulfonyl fluoride (PMSF), 20 mM Tris pH 8.0, 100 mM NaCl, 10% glycerol, 1 µM ZnCl2) and lysed by French press. The lysate was passed through a 2 µm filter. Recombinant proteins were purified over a 5 ml GSTrap FF column (GE Healthcare Life Sciences, Pittsburgh, PA), using a P-500 pump (Pharmacia Biotech, Pittsburgh, PA) and a Controller LCC-501 Plus (Pharmacia Biotech). Purified protein was eluted in a gradient of lysis buffer and lysis buffer containing 10 mM glutathione. Fractions were collected using FRAC-100 (Pharmacia Biotech). Proteins were dialyzed against dialysis buffer (20 mM Tris pH 8.5,150 mM NaCl, 2 mM magnesium acetate, 2 µM ZnCl2, 2% glycerol) using a 10 kD membrane (Spectrum Laboratories Inc, Rockleigh, NJ), tested for purity by gel electrophoresis and Coomassie staining and then concentrated using a Vivaspin 30,000 Da molecular weight cut-off column (Sartorius Stedim Biotech, Göttingen, Germany). A representative Coomassie stained gel of purified proteins showed that the reporter proteins were close to the same molecular weight (Fig. S3). The molecular weights of the purified proteins were cNLS–EGFP 61.2 kDa, cNLS–mCherry 57.3 kDa, mutant cNLS–EGFP 61.2 kDa, mutant cNLS–EGFP 61.2 kDa, hnRNPA1 NLS–EGFP 64.0 kDa, hnRNPA1 NLS–mCherry 60.1 kDa, Nup53 NLS–EGFP 65.2 kDa, Nup53 NLS–mCherry 61.3 kDa, PThrP NLS–EGFP 63.0 kDa and PThrP NLS–mCherry 59.1 kDa.
Myofiber isolation
Gastrocnemius muscles from 6-month-old female C57BL/6 mice were dissected, enzymatically digested and further processed as described previously (Pichavant and Pavlath, 2014). Briefly, muscles were gently dissected and cut twice longitudinally in order to increase the surface area of the muscle in contact with the enzyme. Muscles were incubated for 80 min in a benchtop Enviro-genie (Scientific Industries, Inc., Bohemia, NY) rocking at 26 rpm at 37°C in digestion medium (Dulbecco's modified Eagle's medium, 25 mM HEPES, 400 U/ml collagenase type I; Worthington Biochemical, Lakewood, NJ). All subsequent steps were carried out at room temperature. The samples were washed three times to remove debris and transferred to a Petri dish. Single myofibers were picked using a Pasteur pipette and placed in eight-well chamber coverslips (Ibidi, Martinsried, Germany) precoated with 4% Matrigel (BD Biosciences, San Jose, CA) containing imaging medium (Dulbecco's modified Eagle's medium, 20% FBS, 5 mM EGTA, 1 mM MgCl2). Chamber coverslips were centrifuged at 1100×g for 20 min to adhere the myofibers. Myofibers were incubated at 37°C and 5% CO2 for 1 h prior to imaging.
Bungarotoxin staining
Isolated myofibers were incubated with 2 µg/ml fluorescein-conjugated α-bungarotoxin (Molecular Probes) for 15 min at 37°C. Myofibers were then washed three times with PBS and centrifuged into eight-well coverslips as described above.
Barium chloride muscle injury
Mice were anesthetized with intraperitoneal injection of a solution containing 80 mg/kg ketamine HCl and 5 mg/kg xylazine. For analgesia, mice were injected subcutaneously with 0.1 mg/kg buprenorphine before and after muscle injury. Injury was induced in the gastrocnemius muscles of anesthetized mice by injection of 40 μl of 1.2% BaCl2 (Griffin et al., 2009) (Sigma-Aldrich). Muscles were collected 21 days post injury.
Immunocytochemistry
In vitro generated myotubes were fixed with 4% PFA (Electron Microscopy Sciences) for 15 min at room temperature and permeabilized with 0.5% Triton X-100 (Fisher Scientific) for 5 min at room temperature. Cells were then blocked with 1% BSA and 0.1% donkey serum in 0.1% Tween-20 Tris-buffered saline for 30 min at 4°C. Coverslips were incubated overnight with primary antibody: RanGap 1:50 (C-5, sc-28322; Santa Cruz Biotechnology), mAb414 (ab24609, 1:500; Abcam, Cambridge, UK), Kapβ1 (ab2811, 1:10,000; Abcam), Nup 358 (1:20,000; gift from Kehlenbach laboratory). Alexa Fluor 594-conjugated secondary anti-mouse IgG or anti-rabbit IgG antibodies (1:200; Jackson ImmunoResearch Laboratories Inc., West Grove, PA) were used to detect primary antibody signal.
Microscopy and image analysis
For quantitative analysis of in vitro nuclear import assays, images were obtained using an Axioplan microscope (Carl Zeiss MicroImaging, Oberkochen, Germany) with either a 0.3 NA 10× Plan-Neofluar objective (Carl Zeiss MicroImaging) or a 0.8 NA 25× Plan-Neofluar objective (Carl Zeiss MicroImaging) and recorded with a CCD camera (Carl Zeiss MicroImaging) and Scion Image 1.63 (Scion Corporation, Torrance, CA) software. Images were uniformly processed; myonuclei within myotubes were identified; and fluorescence intensity in DAPI, EGFP and mCherry channels were measured using an automated CellProfiler version 2.2.0 (Carpenter et al., 2006) pipeline. The automated processing used DAPI intensity to identify nuclei between 20 and 60 pixel units by adaptive three class Otsu thresholding. Clumped nuclei were distinguished by shape. Mean fluorescence intensity for nuclear import reports or mean edge fluorescence intensity for immunocytochemistry staining was measured.
For analysis of multiple focal planes of myonuclei and live cell imaging of FRAP experiments, images were collected using a Nikon A1R confocal microscope (Nikon, Tokyo, Japan) with a CFI Apo TIRF 60× H objective (Nikon) and were recorded with NIS-Elements Microscope Imaging Software (Nikon). FRAP image sets were processed with Fiji Image J2 (Schindelin et al., 2012, 2015) to correct for register drift and quantification of fluorescence intensity. For live cell imaging of myofibers, coverslips were maintained at 37°C and 5% CO2. Nuclei and portions of the cytoplasm were bleached with a 561 nm laser at 80% power (34 s) and imaged continuously for the first 2 min and every 2.5 min thereafter for 90 min.
Statistical analysis
Sample size was determined based upon the variability observed in pilot experiments. Statistical analysis was performed using MATLAB (MathWorks, Natick, MA) to model second-order quadratic curve fitting and determination of time to half recovery (T1/2). Other statistical tests (t-test, ANOVA and chi squared analyses) were performed using GraphPad Prism version 5 (Graphpad Software, San Diego, CA). A P-value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank the Emory Integrated Cellular Imaging core for assistance with fluorescence recovery after photobleaching (FRAP) and confocal imaging, Steve Adam for the gift of GST–cNLS–EGFP and GST-mutant cNLS–EGFP constructs, the Emory Integrated Genomics Core for assistance with cloning, Ralph Kehlenbach for the kind gift of Nup358 antibody, Yuh MinChook for guidance in selecting nuclear import reporters and Patrick Getz for assistance with statistical modeling.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.A.C., A.H.C., G.K.P.; Methodology: A.A.C., A.H.C., G.K.P.; Validation: A.A.C.; Formal analysis: A.A.C.; Investigation: A.A.C., J.B.J., G.K.P.; Resources: A.A.C., A.H.C., G.K.P.; Data curation: A.A.C.; Writing - original draft: A.A.C.; Writing - review & editing: A.A.C., A.H.C., G.K.P.; Supervision: A.H.C., G.K.P.; Project administration: G.K.P.; Funding acquisition: A.A.C., A.H.C., G.K.P.
Funding
This work was supported by the National Institutes of Health (grants AR062483 to G.K.P., AR067645 to A.A.C., and a training grant T32GM008367). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.207670.supplemental
References
- Abbott K. L., Friday B. B., Thaloor D., Murphy T. J. and Pavlath G. K. (1998). Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell 9, 2905-2916. 10.1091/mbc.9.10.2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam S. A. (2016). Nuclear protein transport in digitonin permeabilized cells. Methods Mol. Biol. 1411, 479-487. 10.1007/978-1-4939-3530-7_29 [DOI] [PubMed] [Google Scholar]
- Adam S. A., Marr R. S. and Gerace L. (1990). Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111, 807-816. 10.1083/jcb.111.3.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. A., Eser U., Korndorf T., Borsuk M. E., Skotheim J. M. and Gladfelter A. S. (2013). Nuclear repulsion enables division autonomy in a single cytoplasm. Curr. Biol. 23, 1999-2010. 10.1016/j.cub.2013.07.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel E. D., Lewis R. M., Grady R. M. and Sanes J. R. (2000). Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275, 31986-31995. 10.1074/jbc.M004775200 [DOI] [PubMed] [Google Scholar]
- Artaza J. N., Bhasin S., Mallidis C., Taylor W., Ma K. and Gonzalez-Cadavid N. F. (2002). Endogenous expression and localization of myostatin and its relation to myosin heavy chain distribution in C2C12 skeletal muscle cells. J. Cell. Physiol. 190, 170-179. 10.1002/jcp.10044 [DOI] [PubMed] [Google Scholar]
- Asally M., Yasuda Y., Oka M., Otsuka S., Yoshimura S. H., Takeyasu K. and Yoneda Y. (2011). Nup358, a nucleoporin, functions as a key determinant of the nuclear pore complex structure remodeling during skeletal myogenesis. FEBS J. 278, 610-621. 10.1111/j.1742-4658.2010.07982.x [DOI] [PubMed] [Google Scholar]
- Bondesen B. A., Mills S. T., Kegley K. M. and Pavlath G. K. (2004). The COX-2 pathway is essential during early stages of skeletal muscle regeneration. Am. J. Physiol. Cell Physiol. 287, C475-C483. 10.1152/ajpcell.00088.2004 [DOI] [PubMed] [Google Scholar]
- Brenner H. R., Witzemann V. and Sakmann B. (1990). Imprinting of acetylcholine receptor messenger RNA accumulation in mammalian neuromuscular synapses. Nature 344, 544-547. 10.1038/344544a0 [DOI] [PubMed] [Google Scholar]
- Bruusgaard J. C., Liestøl K., Ekmark M., Kollstad K. and Gundersen K. (2003). Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. 551, 467-478. 10.1113/jphysiol.2003.045328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A. L., Liang Y. and Hetzer M. W. (2014). Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol. Biol. Cell 25, 2472-2484. 10.1091/mbc.E14-04-0865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butel J. S., Guentzel M. J. and Rapp F. (1969). Variants of defective simian papovavirus 40 (PARA) characterized by cytoplasmic localization of simian papovavirus 40 tumor antigen. J. Virol. 4, 632-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J. et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. and Xu L. (2010). Specific nucleoporin requirement for Smad nuclear translocation. Mol. Cell. Biol. 30, 4022-4034. 10.1128/MCB.00124-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y. M. and Süel K. E. (2011). Nuclear import by Karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593-1606. 10.1016/j.bbamcr.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G., Bednenko J., Gillespie M. T. and Gerace L. (2002). Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell 10, 1345-1353. 10.1016/S1097-2765(02)00727-X [DOI] [PubMed] [Google Scholar]
- Couteaux R. and Pecot-Dechavassine M. (1973). [Ultrastructural and cytochemical data on the mechanism of acetylcholine release in synaptic transmission]]. Arch. Ital. Biol. 111, 231-262. [PubMed] [Google Scholar]
- Crampton N., Kodiha M., Shrivastava S., Umar R. and Stochaj U. (2009). Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol. Biol. Cell 20, 5106-5116. 10.1091/mbc.E09-05-0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler A. A., Dammer E. B., Doung D. M., Seyfried N. T., Corbett A. H. and Pavlath G. K. (2017). Biochemical isolation of myonuclei employed to define changes to the myonuclear proteome that occur with aging. Aging Cell 16, 738-749. 10.1111/acel.12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'angelo M. A., Raices M., Panowski S. H. and Hetzer M. W. (2009). Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284-295. 10.1016/j.cell.2008.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'angelo M. A., Gomez-Cavazos J. S., Mei A., Lackner D. H. and Hetzer M. W. (2012). A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell 22, 446-458. 10.1016/j.devcel.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix D. J. and Eisenberg B. R. (1990). Myosin mRNA accumulation and myofibrillogenesis at the myotendinous junction of stretched muscle fibers. J. Cell Biol. 111, 1885-1894. 10.1083/jcb.111.5.1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont-Versteegden E. E., Strotman B. A., Gurley C. M., Gaddy D., Knox M., Fluckey J. D. and Peterson C. A. (2006). Nuclear translocation of EndoG at the initiation of disuse muscle atrophy and apoptosis is specific to myonuclei. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R1730-R1740. 10.1152/ajpregu.00176.2006 [DOI] [PubMed] [Google Scholar]
- Ellery P. M., Cindrova-Davies T., Jauniaux E., Ferguson-Smith A. C. and Burton G. J. (2009). Evidence for transcriptional activity in the syncytiotrophoblast of the human placenta. Placenta 30, 329-334. 10.1016/j.placenta.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri P., Barbieri E., Burattini S., Guescini M., D'emilio A., Biagiotti L., del Grande P., de Luca A., Stocchi V. and Falcieri E. (2009). Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. J. Cell. Biochem. 108, 1302-1317. 10.1002/jcb.22360 [DOI] [PubMed] [Google Scholar]
- Finlay D. R., Newmeyer D. D., Price T. M. and Forbes D. J. (1987). Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104, 189-200. 10.1083/jcb.104.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine B. and Changeux J. P. (1989). Localization of nicotinic acetylcholine receptor alpha-subunit transcripts during myogenesis and motor endplate development in the chick. J. Cell Biol. 108, 1025-1037. 10.1083/jcb.108.3.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Hungerbuehler A. K. and Philippsen P. (2006). Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172, 347-362. 10.1083/jcb.200507003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cavazos J. S. and Hetzer M. W. (2015). The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis. J. Cell Biol. 208, 671-681. 10.1083/jcb.201410047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin C. A., Kafadar K. A. and Pavlath G. K. (2009). MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev. Cell 17, 649-661. 10.1016/j.devcel.2009.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K. E. and Sarnow P. (2001). Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20, 240-249. 10.1093/emboj/20.1.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K. E. and Sarnow P. (2002). Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 76, 8787-8796. 10.1128/JVI.76.17.8787-8796.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover J. A., Cohen C. K., Willingham M. C. and Park M. K. (1987). O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 262, 9887-9894. [PubMed] [Google Scholar]
- Hetzer M. W. and Wente S. R. (2009). Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell 17, 606-616. 10.1016/j.devcel.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M.-C. and Link W. (2011). Protein localization in disease and therapy. J. Cell Sci. 124, 3381-3392. 10.1242/jcs.089110 [DOI] [PubMed] [Google Scholar]
- Ishido M., Kami K. and Masuhara M. (2004). Localization of Myod, myogenin and cell cycle regulatory factors in hypertrophying rat skeletal muscles. Acta Physiol. Scand. 180, 281-289. 10.1046/j.0001-6772.2003.01238.x [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Mori C., Kojidani T., Bunai F., Hori T., Fukagawa T., Hiraoka Y. and Haraguchi T. (2009). Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr. Biol. 19, 843-847. 10.1016/j.cub.2009.03.055 [DOI] [PubMed] [Google Scholar]
- Jankowska U., Latosinska A., Skupien-Rabian B., Swiderska B., Dziedzicka-Wasylewska M. and Kedracka-Krok S. (2016). Optimized procedure of extraction, purification and proteomic analysis of nuclear proteins from mouse brain. J. Neurosci. Methods 261, 1-9. 10.1016/j.jneumeth.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Jasmin B. J., Lee R. K. and Rotundo R. L. (1993). Compartmentalization of acetylcholinesterase mRNA and enzyme at the vertebrate neuromuscular junction. Neuron 11, 467-477. 10.1016/0896-6273(93)90151-G [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F. and Smith A. E. (1984). Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33-38. 10.1038/311033a0 [DOI] [PubMed] [Google Scholar]
- Kinoshita Y., Kalir T., Dottino P. and Kohtz D. S. (2012). Nuclear distributions of NUP62 and NUP214 suggest architectural diversity and spatial patterning among nuclear pore complexes. PLoS ONE 7, e36137 10.1371/journal.pone.0036137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J. and Matsuura Y. (2013). Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p. J. Mol. Biol. 425, 1852-1868. 10.1016/j.jmb.2013.02.035 [DOI] [PubMed] [Google Scholar]
- Kodiha M., Tran D., Qian C., Morogan A., Presley J. F., Brown C. M. and Stochaj U. (2008). Oxidative stress mislocalizes and retains transport factor importin-alpha and nucleoporins Nup153 and Nup88 in nuclei where they generate high molecular mass complexes. Biochim. Biophys. Acta 1783, 405-418. 10.1016/j.bbamcr.2007.10.022 [DOI] [PubMed] [Google Scholar]
- Kodiha M., Tran D., Morogan A., Qian C. and Stochaj U. (2009). Dissecting the signaling events that impact classical nuclear import and target nuclear transport factors. PLoS ONE 4, e8420 10.1371/journal.pone.0008420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosako H., Yamaguchi N., Aranami C., Ushiyama M., Kose S., Imamoto N., Taniguchi H., Nishida E. and Hattori S. (2009). Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 16, 1026-1035. 10.1038/nsmb.1656 [DOI] [PubMed] [Google Scholar]
- Krishnaswami S. R., Grindberg R. V., Novotny M., Venepally P., Lacar B., Bhutani K., Linker S. B., Pham S., Erwin J. A., Miller J. A. et al. (2016). Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 11, 499-524. 10.1038/nprot.2016.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. H. C., Hu W., Xiao C.-Y., Gillespie M. T. and Jans D. A. (2001). Molecular dissection of the importin beta1-recognized nuclear targeting signal of parathyroid hormone-related protein. Biochem. Biophys. Res. Commun. 282, 629-634. 10.1006/bbrc.2001.4607 [DOI] [PubMed] [Google Scholar]
- Lanford R. E. and Butel J. S. (1984). Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 37, 801-813. 10.1016/0092-8674(84)90415-X [DOI] [PubMed] [Google Scholar]
- Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z. and Chook Y. M. (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543-558. 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P., Rabut G., Daigle N., Hand A. R., Terasaki M. and Ellenberg J. (2003). Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J. Cell Biol. 160, 1055-1068. 10.1083/jcb.200211076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C. P., Makhnevych T., Marelli M., Aitchison J. D. and Wozniak R. W. (2002). Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159, 267-278. 10.1083/jcb.200203079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M., Lusk C. P., Chan H., Aitchison J. D. and Wozniak R. W. (2001). A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153, 709-724. 10.1083/jcb.153.4.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcpherron A. C., Lawler A. M. and Lee S.-J. (1997). Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83-90. 10.1038/387083a0 [DOI] [PubMed] [Google Scholar]
- Moore M. S. and Schwoebel E. D. (2001). Nuclear import in digitonin-permeabilized cells. Curr. Protoc. Cell Biol. Chapter 11, Unit 11.7 10.1002/0471143030.cb1107s05 [DOI] [PubMed] [Google Scholar]
- Morchoisne-Bolhy S., Geoffroy M.-C., Bouhlel I. B., Alves A., Auduge N., Baudin X., van Bortle K., Powers M. A. and Doye V. (2015). Intranuclear dynamics of the Nup107-160 complex. Mol. Biol. Cell 26, 2343-2356. 10.1091/mbc.E15-02-0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso L. M., Chu G. C., Gautam M., Noakes P. G., Merlie J. P. and Sanes J. R. (1995). Synapse-associated expression of an acetylcholine receptor-inducing protein, ARIA/heregulin, and its putative receptors, ErbB2 and ErbB3, in developing mammalian muscle. Dev. Biol. 172, 158-169. 10.1006/dbio.1995.0012 [DOI] [PubMed] [Google Scholar]
- Newlands S., Levitt L. K., Robinson C. S., Karpf A. B. C., Hodgson V. R. M., Wade R. P. and Hardeman E. C. (1998). Transcription occurs in pulses in muscle fibers. Genes Dev. 12, 2748-2758. 10.1101/gad.12.17.2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer D. D., Lucocq J. M., Burglin T. R. and de Robertis E. M. (1986). Assembly in vitro of nuclei active in nuclear protein transport: ATP is required for nucleoplasmin accumulation. EMBO J. 5, 501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'connor R. S., Mills S. T., Jones K. A., Ho S. N. and Pavlath G. K. (2007). A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 120, 149-159. 10.1242/jcs.03307 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Mizofuchi H., Kobayashi Y., Tsuzuki G., Yamamoto M., Wada S. and Kamemura K. (2012). Terminal differentiation program of skeletal myogenesis is negatively regulated by O-GlcNAc glycosylation. Biochim. Biophys. Acta 1820, 24-32. 10.1016/j.bbagen.2011.10.011 [DOI] [PubMed] [Google Scholar]
- Orias E., Cervantes M. D. and Hamilton E. P. (2011). Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol. 162, 578-586. 10.1016/j.resmic.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichavant C. and Pavlath G. K. (2014). Incidence and severity of myofiber branching with regeneration and aging. Skelet. Muscle 4, 9 10.1186/2044-5040-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V. W., Michael W. M., Nakielny S., Siomi M. C., Wang F. and Dreyfuss G. (1996). A novel receptor-mediated nuclear protein import pathway. Cell 86, 985-994. 10.1016/S0092-8674(00)80173-7 [DOI] [PubMed] [Google Scholar]
- Porpiglia E., Samusik N., van Ho A. T., Cosgrove B. D., Mai T., Davis K. L., Jager A., Nolan G. P., Bendall S. C., Fantl W. J. et al. (2017). High-resolution myogenic lineage mapping by single-cell mass cytometry. Nat. Cell Biol. 19, 558-567. 10.1038/ncb3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge J. R., Wiley J. A., Talago E. A., Young E. M., Johns L. L., Kundert J. A., Sonsteng K. M., Halford W. P., Capecchi M. R. and Schmidt E. E. (2013). Nuclear double-fluorescent reporter for in vivo and ex vivo analyses of biological transitions in mouse nuclei. Mamm. Genome 24, 389-399. 10.1007/s00335-013-9469-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp F., Pauluzzi S. and Butel J. S. (1969). Variation in properties of plaque progeny of PARA (defective simian papovavirus 40)-adenovirus 7. J. Virol. 4, 626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake D. and Currie P. D. (2017). Stem cell dynamics in muscle regeneration: insights from live imaging in different animal models. BioEssays 39 doi:10.1002/bies.201700011 10.1002/bies.201700011 [DOI] [PubMed] [Google Scholar]
- Regot S., de Nadal E., Rodríguez-Navarro S., González-Novo A., Pérez-Fernandez J., Gadal O., Seisenbacher G., Ammerer G. and Posas F. (2013). The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J. Biol. Chem. 288, 17384-17398. 10.1074/jbc.M112.444042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt K. T. G., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V. and Blagoev B. (2011). System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3 10.1126/scisignal.2001570 [DOI] [PubMed] [Google Scholar]
- Rodgers J. T., King K. Y., Brett J. O., Cromie M. J., Charville G. W., Maguire K. K., Brunson C., Mastey N., Liu L., Tsai C.-R. et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G(0) to G(Alert). Nature 510, 393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. T., Schroeder M. D., Ma C. and Rando T. A. (2017). HGFA is an injury-regulated systemic factor that induces the transition of stem cells into G(Alert). Cell Rep. 19, 479-486. 10.1016/j.celrep.2017.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser B. and Bandman E. (2003). Heterogeneity of protein expression within muscle fibers. J. Anim. Sci. 81, E94-E101. [Google Scholar]
- Sabri N., Roth P., Xylourgidis N., Sadeghifar F., Adler J. and Samakovlis C. (2007). Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J. Cell Biol. 178, 557-565. 10.1083/jcb.200612135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamon M., Millino C., Raffaello A., Mongillo M., Sandri C., Bean C., Negrisolo E., Pallavicini A., Valle G., Zaccolo M. et al. (2003). Human MYO18b, a novel unconventional myosin heavy chain expressed in striated muscles moves into the myonuclei upon differentiation. J. Mol. Biol. 326, 137-149. 10.1016/S0022-2836(02)01335-9 [DOI] [PubMed] [Google Scholar]
- Savas J. N., Toyama B. H., Xu T., Yates J. R. and Hetzer M. W. (2012). Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942-942 10.1126/science.1217421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Rueden C. T., Hiner M. C. and Eliceiri K. W. (2015). The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518-529. 10.1002/mrd.22489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T. and Yoneda Y. (2012). Intrinsic and extrinsic negative regulators of nuclear protein transport processes. Genes Cells 17, 525-535. 10.1111/j.1365-2443.2012.01609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D. K., Lopez-Soler R. I., Adam S. A., Herrmann H., Moir R. D., Spann T. P. and Goldman R. D. (2005). Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc. Natl. Acad. Sci. USA 102, 15494-15499. 10.1073/pnas.0507612102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soniat M., Cağatay T. and Chook Y. M. (2016). Recognition elements in the histone H3 and H4 tails for seven different importins. J. Biol. Chem. 291, 21171-21183. 10.1074/jbc.M116.730218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama B. H., Savas J. N., Park S. K., Harris M. S., Ingolia N. T., Yates J. R. III and Hetzer M. W. (2013). Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971-982. 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E. J., King M. C. and Corbett A. H. (2014). Macromolecular transport between the nucleus and the cytoplasm: advances in mechanism and emerging links to disease. Biochim. Biophys. Acta 1843, 2784-2795. 10.1016/j.bbamcr.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K.-I., Katsuta S. and Soya H. (2008). Formation process and fate of the nuclear chain after injury in regenerated myofiber. Anat. Rec. 291, 122-128. 10.1002/ar.20626 [DOI] [PubMed] [Google Scholar]
- Xu L., Yao X., Chen X., Lu P., Zhang B. and Ip Y. T. (2007). Msk is required for nuclear import of TGF-{beta}/BMP-activated Smads. J. Cell Biol. 178, 981-994. 10.1083/jcb.200703106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D. L., Csikasz R. I., Li Y., Sharma G., Hjort K., Karlsson R. and Bengtsson T. (2008). Myotube formation on micro-patterned glass: intracellular organization and protein distribution in C2C12 skeletal muscle cells. J. Histochem. Cytochem. 56, 881-892. 10.1369/jhc.2008.951228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda Y., Miyamoto Y., Saiwaki T. and Yoneda Y. (2006). Mechanism of the stress-induced collapse of the Ran distribution. Exp. Cell Res. 312, 512-520. 10.1016/j.yexcr.2005.11.017 [DOI] [PubMed] [Google Scholar]
- Youn M.-Y., Takada I., Imai Y., Yasuda H. and Kato S. (2010). Transcriptionally active nuclei are selective in mature multinucleated osteoclasts. Genes Cells 15, 1025-1035. 10.1111/j.1365-2443.2010.01441.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.