Abstract
Shewanella oneidensis produces an extensive electron transfer network that results in metabolic flexibility. A large number of c-type cytochromes are expressed by S. oneidensis and these function as the fundamental electron transport chain proteins. Although several S. oneidensis cytochromes have been well-characterized, little is known about how their expression is regulated. In this study, we investigate the role of the ferric uptake regulator (Fur) and the sRNA RyhB in regulation. Our results demonstrate that loss of Fur leads to diminished growth and an apparent decrease in heme-containing proteins. Remarkably, deleting the Fur-repressed ryhB gene almost completely reverses these physiological changes, indicating that the phenotypes resulting from loss of Fur are (at least partially) dependent on RyhB. RNA sequencing identified a number of possible RyhB repressed genes. A large fraction of these encode c-type cytochromes, among them two of the most abundant periplasmic cytochromes CctA (also known as STC) and ScyA. We show that RyhB destabilizes the mRNA of four of its target genes, cctA, scyA, omp35, and nrfA and this requires the presence of the RNA chaperone Hfq. Iron limitation decreases the expression of the RyhB target genes cctA and scyA and this regulation relies on the presence of both Fur and RyhB. Overall, this study suggests that controlling cytochrome expression is of importance to maintain iron homeostasis and that sRNAs molecules are important players in the regulation of fundamental processes in S. oneidensis MR-1.
Keywords: Shewanella oneidensis, cytochromes, RyhB, sRNA, Fur
Introduction
The gammaproteobacterium Shewanella oneidensis is a facultative anaerobe that is remarkable for its metabolic versatility. In the absence of dissolved oxygen, it can utilize a large number of soluble compounds as terminal electron acceptors, and is furthermore capable of transferring electrons to reduce insoluble metal oxides in the extracellular environment (for review, see Hau and Gralnick, 2007). This remarkable respiratory capacity has been attributed to the large number of c-type cytochromes encoded in the genome of S. oneidensis MR-1 and serving as fundamental electron transport chain proteins (Meyer et al., 2004). A total of 42 possible c-type cytochrome genes were identified, but one of these is a pseudogene (Meyer et al., 2004). Some, but not all, of the remaining 41 c-type cytochromes have been characterized and their function determined, with most scrutiny given to the proteins involved in electron transfer to ferric iron.
Reduction of insoluble iron compounds occurs at the bacterial surface and requires electron transfer from the quinone pool in the cytoplasmic membrane to the terminal reductases located in the outer membrane. CymA is a tetraheme cytochrome attached to the periplasmic side of the cytoplasmic membrane. It delivers electrons to multiple partners in the periplasmic space and is required for the reduction of Fe(III), DMSO, fumarate, nitrate, and nitrite (Myers and Myers, 1997, 2000; Schwalb et al., 2003). The most abundant periplasmic c-type cytochromes are the small tetraheme CctA (also called STC), the fumarate reductase FccA, and the small monoheme ScyA (Tsapin et al., 2001; Meyer et al., 2004), all of which have been shown to be involved in direct electron transfer with CymA (Schwalb et al., 2003; Schuetz et al., 2009; Firer-Sherwood et al., 2011; Schütz et al., 2011; Fonseca et al., 2013). In addition to CymA, reduction of Fe(III) oxides by S. oneidensis relies on the proteins MtrA, MtrB, MtrC, and OmcA (for review, see Richter et al., 2012; Shi et al., 2012). MtrA is a periplasmic decaheme c-type cytochrome that also associates with the outer membrane β-barrel protein MtrB and the outer membrane decaheme c-type cytochrome MtrC. OmcA is another decaheme c-type cytochrome associated with the outer surface that interacts with MtrC. MtrA reportedly transfers electrons across the outer membrane to the terminal reductases, MtrC and OmcA, through the porin formed by MtrB. MtrA can accept electrons directly from CymA or presumably indirectly via other periplasmic cytochromes, such as CctA and FccA (Schwalb et al., 2003; Schuetz et al., 2009; Firer-Sherwood et al., 2011; Fonseca et al., 2013; Sturm et al., 2015).
The presence of individual anaerobic electron acceptors seems, for the most part, not to regulate expression of specific c-type cytochromes (Beliaev et al., 2005) (with the exception of the nitrite reductase NrfA), and a core set of periplasmic cytochromes are expressed regardless of electron acceptor (Sturm et al., 2015). Instead, a number of c-type cytochromes respond to oxygen availability (Jin et al., 2013; Barchinger et al., 2016), and iron availability (Yang et al., 2009).
Iron is an essential nutrient for most bacteria as a protein cofactor in pathways such as respiration, metabolism and genome repair (for review, see Andrews et al., 2003; Frawley and Fang, 2014). It is the most common redox active metal in proteins and is found in heme and iron-sulfur groups. To ensure sufficient intracellular iron for protein cofactors, bacteria employ a number of iron scavenging and iron uptake systems. However, maintaining iron homeostasis is of crucial importance because free iron is potentially damaging to the bacterial cell due to radicals formed by Fenton chemistry. As a result, iron uptake and iron storage are tightly regulated. Many bacteria use the transcription factor Fur (ferric uptake regulator) as the major regulator of iron homeostasis. Fur binds Fe2+ and typically acts directly as a repressor of transcription, by binding to DNA recognition sequences when the iron concentration is sufficiently high. In S. oneidensis, deletion of fur results in a pale color of the colonies formed by the Δfur strain compared to the reddish color of the wild-type S. oneidensis colonies, suggesting less heme is present in the Δfur strain (Yang et al., 2008).
Under iron limiting conditions, Fur-dependent repression is relieved and Fur-repressed genes are expressed, including the small RNA RyhB in E. coli and other species (Massé and Gottesman, 2002; Oglesby-Sherrouse and Murphy, 2013). In the studied bacterial genera (Escherichia and Vibrio), one of the functions of Fur and RyhB is to control global iron usage (Massé et al., 2005; Mey et al., 2005).
Small RNAs (sRNAs) are regulators of many cellular processes and play important roles in adaptive changes. They are frequently transcribed after exposure to stress and act as post-transcriptional regulators. They generally act by base pairing in trans with sequences in target mRNAs, either blocking or freeing access to the ribosome binding site. Base pairing between a sRNA and mRNA commonly affect mRNA stability by promoting degradation by RNases or alternatively preventing RNase cleavage (Nitzan et al., 2017). RyhB has been extensively studied in E. coli where it most commonly negatively regulates genes by base-pairing with their mRNA and affecting translation and/or transcript stability, but where it can also affect expression positively (Oglesby-Sherrouse and Murphy, 2013).
Here, we present a study of the role of Fur and RyhB in S. oneidensis physiology and evidence their impact on growth and production of heme-containing proteins. We identify a number of genes as potential targets of RyhB regulation and demonstrate that RyhB affects the stability of specific c-type cytochrome mRNA transcripts. The findings imply that iron levels control production of cytochrome expression in S. oneidensis through Fur and RyhB.
Materials and methods
Bacterial strains and culture conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table 1. S. oneidensis MR-1 wild-type and mutants were routinely grown in LB medium at 30°C and supplemented with 5 μg ml−1 tetracycline and/or 50 μg ml−1 kanamycin when indicated. All experiments were performed with oxygen as the electron acceptor and bacterial cultures were grown in Erlenmeyer flasks with shaking.
E. coli strains DH5α or DH5αλpir, used for cloning purposes, were grown in LB medium at 37°C supplemented with 5 μg ml−1 tetracycline or 50 μg ml−1 kanamycin when indicated. Diaminopilimic acid (DAP) was added to LB medium for growth of strain WM3064, which was used for conjugation of plasmids into S. oneidensis. The DAP dependence of WM3064 was utilized as counter-selection tool after conjugation.
Construction of Δfur, ΔryhB, ΔfurΔryhB, and Δhfq mutants of S. oneidensis MR-1
The yeast recombineering vector pMQ150 (Shanks et al., 2009) was digested with NheI and NotI, treated with Klenow enzyme and ligated. The resulting suicide vector, pMQS, was used as a backbone plasmid for targeted gene deletions in S. oneidensis MR-1. Plasmids for construction of markerless gene deletions were constructed by amplifying regions of 500 to 600 bp flanking the individual genes (using primers Dgene_5F+Dgene_5R and Dgene_3F+Dgene_3R, respectively). All primers used in this study are listed in Supplementary Table 2. Deletions in fur and hfq were in-frame and left the sequences encoding the first four amino acids and the last four or two amino acids, respectively, of the two proteins. Crossover PCR was performed to join the up- and down-stream regions followed by cloning into pMQS. pMQS-derivatives containing inserts for deletion of fur, ryhB, or hfq were introduced in S. oneidensis MR-1 by conjugation from E. coli strain WM3064. Colonies with single crossover plasmid insertions were selected on LB agar plates containing kanamycin, purified once on agar plates with kanamycin and resistant colonies were subsequently grown overnight in LB (containing no NaCl) without antibiotic. Double crossover mutants were selected on LB agar plates (containing no NaCl) supplemented with 10% sucrose. Sucrose resistant and kanamycin sensitive colonies were checked by colony PCR for gene deletion using primers flanking the deleted region (gene_FO + gene_RO). Selected clones were purified, genomic DNA isolated, and the region containing the deleted gene was amplified by PCR and the deletion verified by Sanger sequencing. The double mutant, ΔfurΔryhB, was made by deleting ryhB in the Δfur mutant.
Construction of pKM033 and pKM033-ryhB
Plasmid pKM001 was constructed by digesting pBAD-fccA-His (Schuetz et al., 2009) with EcoRV and ligating the vector backbone, thereby deleting almost the entire fccA gene and part of araC. The E. coli lacI gene was amplified by PCR (using primers ELacI_F+ELacI_R) from plasmid pET28b(+) and cloned into the EcoRV site of pKM001 after digestion with SmaI. An artificial promoter containing a Lac operator between the −35 and −10 motifs was created by annealing two oligonucleotides (PlacO_f and PlacO_r), followed by insertion into the HindIII site, thereby creating pKM033. The sequence from −1 to −6 relative to transcription start in this construct is a BamHI restriction site, allowing cloning of genes resulting in transcription initiating at their native transcription start site. In addition, a KpnI site is located at position +1 to +6. The ryhB gene was amplified by PCR (using primers ryhB_F_(+1) and ryhB_R_Stop) and the PCR product inserted into BamHI/HindIII digested pKM033 after digestion with the same enzymes. The transcription start site of ryhB (from Shao et al., 2014) follows the BamHI site at position +1 in plasmid KM033.
RNA sequencing
Triplicate cultures of S. oneidensis MR-1 and mutants were independently grown in LB medium to OD600nm ~0.7. For ΔryhB containing either pKM033 or pKM033-ryhB, triplicate independent cultures were grown in LB medium to OD600nm ~0.2 at which point 0.5 mM IPTG was added and growth continued for 60 min. Cells were harvested and total RNA isolated using Trizol reagent (Invitrogen) followed by purification of the aqueous phase on a RNeasy column (Qiagen). RNA was treated with DNase I for 45 min at 37°C and again purified on an RNeasy column. The integrity of the RNA was assessed on a fragment analyzer (Advanced Analytical Technologies, Ankeny, Iowa, USA). RNA sequencing libraries were constructed after rRNA depletion (with the Illumina Ribo-Zero rRNA removal kit) using the Truseq stranded mRNA library prep kit (without polyA selection using the strand-specific kit (Illumina, San Diego, CA, USA)). Library construction and RNA sequencing was performed at the Lausanne Genomic Technologies Facility at the University of Lausanne, Switzerland.
Analysis of RNAseq data
RNA sequencing reads were aligned with TopHat2 v.2.0.11 (Kim et al., 2013) (parameters: –library-type fr-firststrand -p 6 -g 2 -G). Reads were counted using featureCounts (Liao et al., 2014) (parameters: -t CDS -g gene_name -s 2), included in the SourceForge Subread v.1.5.1 package (Liao et al., 2013). Reference genome and annotation files were downloaded from NCBI RefSeq database (NCBI reference: chromosome: NC_004347.2; megaplasmid: NC_004349.1.).
Once the matrix of read counts was created, only those genes having more than 10 counts per million mapped reads (CPM) in at least 3 of all the samples were kept. Normalization and conversion to logCPM, was performed using the edgeR package v.3.14.0 (Robinson et al., 2010), and the limma-trend approach from the Limma package (Ritchie et al., 2015), in order to complete the differential expression analysis. Both edgeR and Limma are R Bioconductor packages (Huber et al., 2015).
Only genes which adj.p.value < 0.05 and logFC > |1| were considered differentially expressed. Genes expressed at lower level in Δfur relative to wild-type according to these criteria are shown in Table 1.
Table 1.
RNAseq data of genes expressed at lower level in Δfur mutant relative to wild-type S. oneidensis MR-1a.
| Δfur vs. WT | ΔfurΔryhB vs. Δfur | ΔfurΔryhB vs. WT | RyhB overexpressionb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Annotation | log2R | adj.P.Val | log2R | adj.P.Val | log2R | adj.P.Val | log2R | adj.P.Val | Groupc |
| SO_0002 | Glutathione uptake transporter | −1.02 | 4.9E-06 | 0.80 | 1.39E-04 | −0.22 | 2.41E-01 | −0.67 | 7.17E-04 | Ia |
| SO_0264 | Periplasmic monoheme cytochrome c5 ScyA | −2.76 | 1.7E-10 | 2.71 | 2.16E-10 | −0.04 | 8.18E-01 | −3.57 | 5.32E-12 | Ia |
| SO_0608 | Ubiquinol-cytochrome c reductase iron-sulfur subunit Peta | −1.04 | 4.0E-07 | 0.91 | 5.55E-06 | −0.12 | 4.21E-01 | −1.29 | 1.26E-07 | Ia |
| SO_0609 | Ubiquinol-cytochrome c reductase cytochrome b subunit Petb | −1.11 | 1.9E-07 | 0.99 | 2.46E-06 | −0.12 | 4.11E-01 | −1.40 | 5.47E-08 | Ia |
| SO_0610 | Ubiquinol-cytochrome c reductase cytochrome c1 subunit Petc | −1.01 | 3.0E-08 | 0.88 | 6.01E-07 | −0.13 | 2.44E-01 | −1.23 | 1.30E-08 | Ia |
| SO_1236 | Purine transporter | −1.23 | 4.7E-06 | 1.08 | 6.43E-05 | −0.16 | 5.09E-01 | −0.97 | 1.67E-04 | Ia |
| SO_1718 | Protein of unknown function | −1.18 | 8.2E-05 | 0.92 | 1.57E-03 | −0.26 | 3.86E-01 | −1.09 | 6.65E-04 | Ia |
| SO_2373 | Drug:H+ antiporter | −1.17 | 3.3E-06 | 0.91 | 1.18E-04 | −0.26 | 2.01E-01 | −0.80 | 3.74E-04 | Ia |
| SO_2727 | Periplasmic tetraheme cytochrome c CctA | −1.97 | 1.1E-08 | 1.88 | 8.31E-08 | −0.09 | 6.94E-01 | −2.19 | 1.30E-08 | Ia |
| SO_2797 | Glutaredoxin | −1.05 | 1.3E-03 | 0.71 | 2.46E-02 | −0.34 | 3.68E-01 | −0.75 | 3.04E-02 | Ia |
| SO_2881 | Superoxide dismutase SodB | −1.42 | 1.1E-07 | 1.27 | 1.69E-06 | −0.15 | 4.09E-01 | −2.22 | 5.82E-09 | Ia |
| SO_3195 | Proton-dependent oligopeptide transporter | −1.07 | 5.6E-07 | 0.93 | 9.75E-06 | −0.15 | 3.50E-01 | −1.16 | 8.58E-07 | Ia |
| SO_3420 | Monoheme cytochrome c' | −1.22 | 1.2E-06 | 1.26 | 3.48E-06 | 0.04 | 8.63E-01 | −2.09 | 1.34E-08 | Ia |
| SO_3920 | Periplasmic [Fe-Fe] hydrogenase large subunit HydA | −1.79 | 2.7E-06 | 1.46 | 6.89E-05 | −0.33 | 2.77E-01 | −0.87 | 4.52E-03 | Ia |
| SO_3980 | Ammonia-forming nitrite reductase NrfA | −1.95 | 1.6E-05 | 1.43 | 7.04E-04 | −0.53 | 1.98E-01 | −1.53 | 5.69E-04 | Ia |
| SO_4047 | SoxA-like diheme cytochrome c | −1.11 | 6.5E-06 | 1.51 | 1.59E-06 | 0.40 | 5.19E-02 | −1.63 | 5.14E-07 | Ia |
| SO_4340 | Putative transport protein | −1.47 | 5.9E-06 | 0.96 | 6.61E-04 | −0.50 | 6.09E-02 | −0.59 | 2.19E-02 | Ia |
| SO_4506 | Iron-sulfur cluster-binding protein | −1.20 | 2.1E-05 | 1.11 | 1.51E-04 | −0.10 | 7.50E-01 | −0.67 | 8.00E-03 | Ia |
| SO_4507 | Formate dehydrogenase chaperone FdhT | −1.08 | 1.1E-05 | 0.96 | 1.17E-04 | −0.12 | 6.01E-01 | −0.83 | 4.71E-04 | Ia |
| SO_4509 | Formate dehydrogenase molybdopterin-binding subunit FdhA | −1.30 | 3.4E-05 | 1.17 | 2.90E-04 | −0.13 | 6.86E-01 | −0.68 | 1.54E-02 | Ia |
| SO_4510 | Formate dehydrogenase FeS subunit FdhB | −1.05 | 2.6E-06 | 0.97 | 2.27E-05 | −0.08 | 6.75E-01 | −0.70 | 3.74E-04 | Ia |
| SO_4591 | Membrane anchored tetraheme cytochrome c CymA | −1.75 | 2.3E-05 | 1.40 | 5.00E-04 | −0.35 | 3.68E-01 | −0.90 | 1.32E-02 | Ia |
| SO_0403 | Predicted outer membrane protein | −1.39 | 2.5E-03 | 1.50 | 3.36E-03 | 0.11 | 8.77E-01 | −0.61 | 2.30E-01 | Ib |
| SO_0404 | Zinc dependent metalloprotease domain lipoprotein | −1.33 | 5.6E-05 | 1.51 | 7.44E-05 | 0.18 | 6.07E-01 | −0.54 | 6.49E-02 | Ib |
| SO_0435 | Uroporphyrinogen decarboxylase HemE | −1.20 | 4.0E-05 | 0.92 | 1.04E-03 | −0.28 | 3.15E-01 | −0.27 | 3.00E-01 | Ib |
| SO_0827 | Lactate permease | −1.81 | 8.9E-08 | 1.51 | 2.43E-06 | −0.30 | 1.68E-01 | −0.34 | 9.19E-02 | Ib |
| SO_1072 | N-acetylglucosamine-binding protein A GbpA | −2.24 | 3.1E-06 | 1.77 | 9.80E-05 | −0.47 | 2.30E-01 | −0.25 | 5.53E-01 | Ib |
| SO_1192 | Predicted extracytoplasmic protein | −1.11 | 1.6E-04 | 1.06 | 7.01E-04 | −0.06 | 8.81E-01 | 0.09 | 8.09E-01 | Ib |
| SO_2806 | Phospahte starvation inducible E-like protein | −1.27 | 9.3E-06 | 0.81 | 1.12E-03 | −0.46 | 5.93E-02 | −0.30 | 1.98E-01 | Ib |
| SO_2824 | Pseudogene | −2.92 | 5.9E-04 | 2.86 | 1.66E-03 | −0.06 | 9.57E-01 | 1.46 | 8.81E-02 | Ib |
| SO_2905 | O-methyltransferase | −1.23 | 3.4E-04 | 0.86 | 8.67E-03 | −0.37 | 3.20E-01 | −0.35 | 3.26E-01 | Ib |
| SO_3284 | Protein YbgT | −1.24 | 4.2E-05 | 0.79 | 3.57E-03 | −0.45 | 1.20E-01 | −0.19 | 5.38E-01 | Ib |
| SO_3285 | Cytochrome d ubiquinol oxidase subunit II CydB | −1.24 | 3.6E-05 | 0.80 | 2.83E-03 | −0.43 | 1.24E-01 | −0.20 | 4.82E-01 | Ib |
| SO_3286 | Cytochrome d ubiquinol oxidase subunit I CydA | −1.27 | 7.0E-05 | 0.90 | 2.61E-03 | −0.37 | 2.38E-01 | −0.21 | 5.16E-01 | Ib |
| SO_3705 | 5-Methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | −1.79 | 5.6E-07 | 1.39 | 2.45E-05 | −0.40 | 1.23E-01 | 0.09 | 7.67E-01 | Ib |
| SO_3706 | Nucleoside:proton symporter NupX | −1.90 | 2.8E-07 | 1.50 | 1.16E-05 | −0.41 | 1.11E-01 | 0.23 | 3.74E-01 | Ib |
| SO_3874 | Transcriptional regulator LysR family | −1.17 | 4.7E-04 | 0.67 | 2.92E-02 | −0.50 | 1.68E-01 | −0.09 | 8.57E-01 | Ib |
| SO_4085 | Chitinase ChiA | −1.05 | 5.6E-05 | 0.66 | 4.97E-03 | −0.40 | 1.16E-01 | −0.30 | 2.13E-01 | Ib |
| SO_4138 | Putative periplasmic protein | −1.07 | 1.8E-04 | 0.94 | 1.31E-03 | −0.13 | 7.05E-01 | −0.06 | 8.86E-01 | Ib |
| SO_4157 | Thiosulfate/tetrathionate-responsive two component signal transduction system response regulator Ttr | −1.00 | 2.0E-03 | 1.07 | 3.03E-03 | 0.06 | 8.96E-01 | −0.14 | 7.66E-01 | Ib |
| SO_4274 | Undecaprenol diphosphatase UppP | −1.49 | 3.8E-07 | 1.18 | 1.49E-05 | −0.31 | 1.32E-01 | 0.24 | 2.17E-01 | Ib |
| SO_4513 | Fnr-inducilble formate dehydrogenase molybdopterin-binding subunit FdhA | −1.07 | 2.6E-03 | 1.00 | 8.22E-03 | −0.07 | 8.95E-01 | −0.33 | 4.33E-01 | Ib |
| SO_4623 | Two component signal transduction system response regulator | −1.15 | 2.6E-03 | 1.00 | 1.15E-02 | −0.14 | 7.89E-01 | −0.11 | 8.58E-01 | Ib |
| SO_4625 | Predicted phosphoribosyltransferase ComF family | −1.33 | 1.1E-04 | 1.55 | 1.16E-04 | 0.22 | 5.57E-01 | 0.02 | 9.72E-01 | Ib |
| SO_4719 | ABC-type tungstate uptake system substrate-binding component TupA | −1.38 | 1.8E-05 | 0.87 | 1.99E-03 | −0.51 | 7.74E-02 | 0.01 | 9.81E-01 | Ib |
| SO_4720 | ABC-type tungstate uptake system permease component TupB | −1.11 | 2.5E-05 | 0.87 | 6.39E-04 | −0.24 | 3.21E-01 | −0.10 | 7.30E-01 | Ib |
| SO_1777 | Extracelllular iron oxide respiratory system periplasmic decaheme cytochrome c component MtrA | −2.02 | 1.3E-06 | 0.59 | 3.02E-02 | −1.43 | 1.05E-04 | −0.91 | 4.47E-03 | IIa |
| SO_1778 | Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC | −2.24 | 1.2E-06 | 0.66 | 2.87E-02 | −1.58 | 1.05E-04 | −0.74 | 2.64E-02 | IIa |
| SO_1948 | Glutamate/aspartate:proton symporter GltP | −1.13 | 1.6E-07 | 0.57 | 3.08E-04 | −0.56 | 4.37E-04 | −0.81 | 1.19E-05 | IIa |
| SO_2879 | N-acetylglucosamine/uracil transporter UraA | −1.20 | 1.6E-08 | 0.78 | 5.50E-06 | −0.42 | 1.35E-03 | −0.36 | 4.47E-03 | IIa |
| SO_3099 | Outer membrane long-chain fatty acid receptor FadL family | −1.25 | 2.0E-05 | 0.73 | 3.71E-03 | −0.52 | 4.89E-02 | −0.50 | 4.20E-02 | IIa |
| SO_3896 | outer membrane porin Omp35 | −1.80 | 7.3E-08 | 1.23 | 1.30E-05 | −0.57 | 8.77E-03 | −1.95 | 9.64E-08 | IIa |
| SO_4232 | Long-chain fatty acid transport protein | −1.43 | 5.7E-07 | 0.71 | 8.58E-04 | −0.72 | 1.24E-03 | −1.37 | 2.56E-06 | IIa |
| SO_4666 | Diheme cytochrome c4 CytcB | −1.30 | 6.0E-07 | 0.91 | 6.89E-05 | −0.39 | 3.99E-02 | −1.36 | 1.29E-06 | IIa |
| SO_0141 | Nitrate/nitrite-responsive bifunctional diguanylate cyclase/phosphodiesterase with PAS sensory domain | −1.58 | 9.3E-06 | 0.96 | 1.52E-03 | −0.62 | 4.28E-02 | −0.26 | 4.00E-01 | IIb |
| SO_2384 | Site-specific recombinase phage integrase family | −1.95 | 8.7E-07 | 1.27 | 1.49E-04 | −0.68 | 2.24E-02 | −0.03 | 9.45E-01 | IIb |
| SO_4131 | Nucleoside-specifc outer membrane porin Tsx family | −1.95 | 5.9E-06 | 1.03 | 2.81E-03 | −0.92 | 1.25E-02 | −0.25 | 5.01E-01 | IIb |
| SO_4355 | cAMP-binding regulator | −1.37 | 1.6E-05 | 0.73 | 5.57E-03 | −0.64 | 2.46E-02 | −0.21 | 4.77E-01 | IIb |
| SO_1776 | Extracellular iron oxide respiratory system outer membrane component MtrB | −1.61 | 1.2E-06 | 0.40 | 5.91E-02 | −1.21 | 5.10E-05 | −0.73 | 4.22E-03 | III (IIa) |
| SO_1111 | Bacterioferritin subunit 2 Bfr2 | −2.25 | 3.0E-07 | −0.16 | 5.42E-01 | −2.42 | 2.46E-07 | 1.29 | 2.19E-04 | III |
| SO_1112 | Bacterioferritin subunit 1 Bfr1 | −2.22 | 3.8E-07 | −0.18 | 5.05E-01 | −2.40 | 2.68E-07 | 1.31 | 2.02E-04 | III |
| SO_1779 | Extracelllular iron oxide respiratory system surface decaheme cytochrome c component OmcA | −2.49 | 1.3E-07 | 0.51 | 5.31E-02 | −1.98 | 2.51E-06 | 0.15 | 6.80E-01 | III |
| SO_1977 | Inner membrane of unknown function | −1.25 | 1.3E-05 | 0.18 | 3.91E-01 | −1.06 | 1.92E-04 | 0.49 | 3.77E-02 | III |
| SO_0396 | Quinol:fumarate reductase menaquinol-oxidizing subunit FrdC | −1.38 | 1.2E-02 | 0.92 | 1.11E-01 | −0.46 | 5.26E-01 | −0.09 | 9.35E-01 | other |
All genes that are downregulated at least 2-fold (and with adjusted p < 0.05) in Δfur relative to the wild-type strain are listed in the table. For these genes, expression ratios and adjusted p-values are shown for other strains comparisons. For all strain comparisons, the Log2R (log2 of the expression ratio) with adjusted p < 0.05 are indicated in bold, also when expression ratios are <2-fold.
RyhB overexpression data are from strains ΔryhB/pKM033-ryhB vs. ΔryhB/pKM033.
The genes were divided into groups based on their expression pattern in various strains. Group I genes are significantly (p < 0.05) upregulated in ΔfurΔryhB relative to Δfur and show no significant difference in expression between wild-type and ΔfurΔryhB. Group II genes are differently expressed in ΔfurΔryhB relative to both Δfur (upregulated) and relative to wild-type (downregulated). Group III genes are not differently expressed in ΔfurΔryhB relative to Δfur. Genes in groups Ia and IIa are downregulated after RyhB over-expression.
The computations were performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the SIB Swiss Institute of Bioinformatics.
RNAseq data are available GEO (Gene Expression Omnibus) with accession number GSE104952.
RNA stability assay
S. oneidensis wild-type and mutants (with or without pKM033 or pKM033-ryhB) were grown to mid-exponential phase and transcription was stopped by adding 500 μg ml−1 rifampicin. For experiments with RyhB overexpression, 0.5 mM IPTG was added to the cultures for 30 min before addition of rifampicin. Samples were taken at 0 min (immediately before adding rifampicin), 4 min, 8 min, and 15 min after transcription was terminated. Cultures (2 ml) were centrifuged at maximum speed at room temperature for one min and the bacterial pellet was immediately frozen in dry ice/ethanol.
Iron depletion experiment
Bacterial cultures were grown to mid-exponential phase in LB medium at which time iron chelator (2,2′-dipyridyl) was added at a concentration of 250 μg ml−1, and incubation was continued for 30 min. Samples were taken just before addition of iron chelator (Fe-rich) and at the end of the experiment (Fe-limitation).
qRT-PCR
RNA was isolated from liquid cultures of S. oneidensis wild-type and mutants using Trizol reagent followed by purification of the aqueous phase on RNeasy columns (Qiagen), with either on-column or in-solution DNase digestion (in latter case followed by a second RNeasy purification step). cDNA synthesis was performed according to the protocol provided by with the reverse transcriptase GoScript (Promega) using 250–500 ng total RNA and random hexamers. qPCR of diluted cDNA was performed in 10 μl volume [containing 1xSensiFast SYBR Mix (BioLine) and 200 nM primers] in a Mic qPCR Cycler (Bio Molecular Systems) with a 2-step program (5 min 95°C, followed by 40 cycles of 95°C for 5 s and 60°C for 20 s). Four repetitions of each sample and each primer set were performed, as well as negative controls (no reverse transcription and no template controls). Data were analyzed with the analysis software provided with the Mic qPCR cycler that uses the REST tool for relative expression analysis (Pfaffl et al., 2002). Specifically, quantification cycle Cq values and reaction efficiencies were calculated using the LinRegPCR method and relative quantification of specific genes were determined using two reference genes, rpoB and gyrB. For RNA stability assays, the 16S rRNA gene was used as the reference gene. Each experiment was independently performed twice.
Creation of a lacZ reporter system
To create a translational reporter system, the lacZ gene beginning at codon 9 was amplified (primers lacZ_9F+lacZ_R) from genomic DNA of E. coli strain MG1655 and inserted into HindIII-PstI digested pME6031 (Heeb et al., 2000), creating plasmid pKM002. The KpnI site in the MCS region was then inactivated by digestion with KpnI and treatment with Klenow DNA polymerase followed by ligation, generating plasmid pKM202. The artificial promoter containing a Lac operator between the −35 and −10 motifs (created by annealing the two oligonucleotides PlacO_f and PlacO_r) was inserted into HindIII digested plasmid pKM202 thereby forming pKM232. The 5′ untranslated region (5′UTR) and the beginning of selected genes were amplified by PCR and cloned as KpnI-HindIII fragments to construct translational fusions with lacZ. The transcription start site used was based on the data by Shao et al. (2014) and was located immediately after the KpnI site in all constructs.
β-galactosidase assay
Bacteria (ΔryhB or Δhfq) containing pKM232-derivatives and pKM033 or pKM033-ryhB were grown in LB medium supplemented with kanamycin and tetracycline. Over-night cultures were diluted to OD600nm ~0.025 and incubated for 90 min at which time expression was induced by addition of 0.5 mM IPTG and cultures were grown an additional 90 min. One hundred microliters of bacterial culture was lyzed in 1 ml Z-bufffer (0.1M KCl, 1 mM MgSO4, 60 mM Na2HPO4, 40 mM NaH2PO4; pH 7) with 0.27% β-mercaptoethanol, 50 μl chloroform and 25 μl 0.1% SDS. After equilibration at 30°C, 200 μl o-nitrophenyl-β-D-galactopyranoside (ONPG, 4 mg/ml) was added and reactions were stopped when a yellow color developed by adding 500 μl 1M Na2CO3. Each strain was grown in duplicate with triplicate samples taken from each and the experiment was independently repeated twice. Specific activities were calculated as Miller units [1,000*(OD420)/(time*volume*OD600)] and shown as the mean ± SD of all samples.
Results
Fur and RyhB control growth and expression of heme-containing proteins
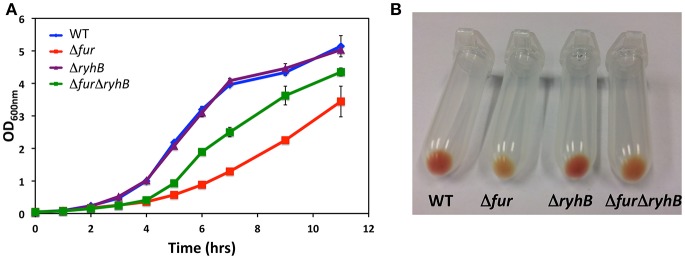
A transposon screen in S. oneidensis identified a mutant with an insertion in the gene locus encoding the ferric uptake regulator Fur, resulting in decreased expression of the outer membrane multiheme c-type cytochrome MtrC (K. L. Meibom and R. Bernier-Latmani, unpublished data). Deletion of the fur gene in wild-type S. oneidensis showed that loss of Fur affected growth of S. oneidensis in liquid culture and the mutant formed colonies of smaller size than the wild-type strain (Figure 1A and Supplementary Figure 1A), in accordance with an earlier report (Yang et al., 2008). Complementation with a plasmid encoding fur restored colony size (Supplementary Figure 1B). It is well-known from studies in other bacterial species that even though Fur is generally a repressor, it can control expression of target genes positively through the sRNA RyhB (Oglesby-Sherrouse and Murphy, 2013). S. oneidensis MR-1 encodes a RyhB homolog between genes SO_4716 and SO_4717 and the gene appear to encode a ~127 nt transcript based on the previously identified transcription start site (Shao et al., 2014) and prediction of a rho-independent terminator using the tool ARNold (Naville et al., 2011; Supplementary Figures 2A,B). To study the role of RyhB in S. oneidensis, we deleted the gene in both the wild-type strain and in the Δfur mutant strain. Northern blot analysis confirmed the expression of RyhB and the approximate size of the RNA (Supplementary Figure 2C). Deleting ryhB had no obvious phenotype in the wild-type strain, whereas deletion of ryhB in the Δfur strain almost reversed the observed growth defect of the Δfur strain (Figure 1A and Supplementary Figure 1A). Also, overproduction of RyhB from a plasmid had a negative effect on growth of the wild-type and ΔryhB strains (Supplementary Figure 1C), indicating that the increased expression of RyhB in the Δfur strain is responsible for this phenomenon. Another phenotype is the pale color of the colonies formed by the Δfur strain compared to the reddish color of the wild-type S. oneidensis colonies. The reddish color is attributed to the high level of cytochromes in S. oneidensis. We observed the same phenotype in our mutant strain in liquid culture but it was more readily noticeable in bacterial pellets (Figure 1B). Again, the deletion of ryhB in the Δfur mutant nearly reverted the phenotype to wild-type appearance (Figure 1B). Furthermore, heme-staining of cell lysates after polyacrylamide gel electrophoresis showed that the Δfur mutant contains a lower level of heme-containing proteins and this can be nearly completely reversed by deleting ryhB (Supplementary Figure 1D). Taken together, these results suggest that the Fur protein, through the sRNA RyhB, controls expression of proteins with importance for growth and controls the production of heme-containing proteins.
Figure 1.
Deletion of ryhB nearly abolishes the phenotypes of a fur mutant. (A) Growth of S. oneidensis wild-type, Δfur, ΔryhB, and ΔfurΔryhB strains in LB medium at 30°C with shaking. (B) Bacterial pellets of stationary phase cultures of S. oneidensis wild-type and mutants.
Identification of potential RyhB repressed genes
In order to identify targets of RyhB regulation in S. oneidensis, we used RNA sequencing (RNAseq) to assess the transcriptomes in various strains. We grew the wild-type, Δfur, and ΔfurΔryhB strains in LB medium and isolated total RNA from three independent cultures of each. We hypothesized that Fur acts indirectly as a positive regulator via RyhB. According to the presumed regulatory mechanism, genes negatively regulated by RyhB should exhibit decreased expression in the Δfur mutant, where RyhB levels are high relative to the wild-type strain. Conversely, we would expect an increased expression of a target gene in the ΔfurΔryhB mutant compared to the Δfur mutant. The RNA sequencing confirmed that Fur acts as a repressor of ryhB expression as the RyhB level increases in the Δfur mutant, as well as a repressor of the expression level of a number of other genes (Supplementary Table 3). Analysis of the RNAseq data showed that 64 genes were expressed at a lower level in the Δfur mutant relative to the wild-type strain (minimum 2-fold difference and p < 0.05; Table 1).
Of the 64 genes we identified as expressed at a lower level in the Δfur mutant relative to wild-type, 46 genes were expressed at a higher level in the ΔfurΔryhB mutant relative to the Δfur mutant (p < 0.05), but were not differently expressed in the ΔfurΔryhB mutant relative to the wild-type strain (Table 1, group I). This result indicates that RyhB regulates these 46 genes and is responsible for the entire decrease in expression seen in the Δfur mutant relative to the wild-type. A second subset of the 64 Fur-activated genes, consisting of 12 genes, was expressed at a significantly different level (p < 0.05) in the ΔfurΔryhB mutant relative to both the Δfur mutant and to the wild-type strain (Table 1, group II). Specifically, these genes were expressed at a higher level in the ΔfurΔryhB mutant compared to the Δfur mutant and at a lower level relative to the wild-type. This expression pattern suggests that RyhB regulates these genes, but that other regulators could contribute as well. Finally, 6 genes did not differ in expression level in the ΔfurΔryhB mutant compared to the Δfur mutant (Table 1, group III and other), suggesting that Fur, but not RyhB regulates these genes.
To obtain a more direct measure of the targets of RyhB regulation, we performed RNAseq of total RNA isolated from a ΔryhB mutant containing either a plasmid with an inducible ryhB gene or an empty plasmid. This allowed us to divide the 64 genes expressed at lower levels in the Δfur strain into further subgroups. Of the 46 genes in group I, we found that 22 were expressed at a lower level when RyhB was overexpressed (Table 1, group Ia), strongly indicating that these genes are directly controlled by RyhB. Conversely, 24 genes belonging to group I did not change expression after overexpression of RyhB (group Ib). Of the group II genes, 8 out of 12 genes exhibited decreased expression when RyhB was overexpressed (group IIa), pointing to a direct control of these by RyhB.
Thus, genes from both group Ia and group IIa show a direct response to RyhB overexpression, indicating that they are regulated by Fur via RyhB. But, group Ia likely is regulated by Fur through RyhB exclusively, while group IIa is regulated by Fur through RyhB but also likely through another Fur-dependent or iron-dependent regulator.
We observe that groups Ia and IIa are comprised of many genes encoding iron-containing proteins such as cytochromes and proteins with iron-sulfur clusters. In fact, genes encoding iron-containing proteins represent about 50% of the RyhB-regulated genes (Table 1). Our data show that RyhB influences the expression of 10 of the 41 possible functional c-type cytochromes: scyA, cctA, SO_3420, nrfA, cymA, petC, mtrA, mtrC, SO_4047, and SO_4666. Membrane proteins and transporters, including Omp35 (SO_3896), account for most of the remaining RyhB regulated genes, in both group Ia and IIa.
It is also noteworthy, that one of the first genes known to be regulated by RyhB in E. coli, sodB encoding superoxide dismutase (Massé and Gottesman, 2002), is also regulated by this sRNA in S. oneidensis as was suggested by an earlier study (Yang et al., 2010).
One of the genes assigned to group III (genes not controlled by RyhB), SO_1776 encoding the outer membrane component MtrB of the MtrABC system for metal respiration, was actually downregulated by overexpression of RyhB (1.7-fold, p = 0.004). Indeed, revisiting the data showed that mtrB was expressed at a very slightly increased level in the ΔfurΔryhB mutant compared to the Δfur, but not significantly (p = 0.059). In light of the RyhB overexpression data and that fact the mtrB is located within an operon with mtrC and mtrA, that both belong to group IIa, we believe mtrB is likely to belong to that group of genes as well.
Surprisingly, three group III genes that did not appear to be controlled by RyhB (based on the fact that the expression level did not increase when ryhB was deleted in the Δfur mutant) had elevated expression when RyhB was overexpressed. The genes encode an inner membrane protein of unknown function (SO_1977) and bacterioferritin subunits 1 and 2, an iron-storage complex.
Iron-availability controls expression of periplasmic c-type cytochromes ScyA and CctA
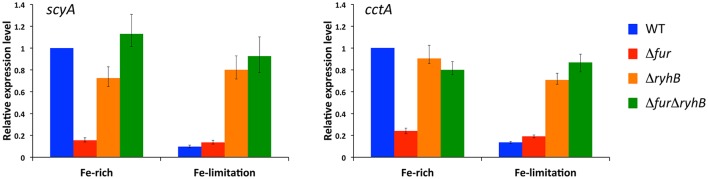
Our RNAseq data suggested that two major periplasmic c-type cytochromes ScyA and CctA are indirectly activated by Fur, through RyhB. Because Fur responds to iron availability, we next assessed whether iron levels influence expression of scyA and cctA in the genetic backgrounds we constructed. Figure 2 (Supplementary Figure 3) show the relative expression values of scyA and cctA in the wild-type, Δfur, ΔryhB, and ΔfurΔryhB strains during growth in LB medium (Fe-rich) and 30 min after addition of the iron chelator 2,2′-dipyridyl (Fe-limitation). Iron-limitation dramatically decreased expression of both scyA and cctA in the wild-type strain, to about 10–20% of the level under iron-rich conditions. In contrast, iron depletion did only very slightly influence expression in the Δfur mutant, where expression was at a low level under both conditions. This suggests that sensing of iron availability through Fur controls expression of these two cytochromes. Deletion of ryhB in the wild-type background had no or a very minor effect on scyA and cctA expression. Their expression level remained relatively high under iron limitation, demonstrating that RyhB is required for the decreased expression under iron limiting conditions. Likewise, expression levels of scyA and cctA in the ΔfurΔryhB mutant were not or only very slightly affected by iron-limitation and stayed at a level comparable to the wild-type strain under iron-rich conditions. Taken together, these data strongly indicate that iron limitation decreases expression of the periplasmic cytochromes ScyA and CctA in a Fur-dependent manner that relies on the presence of RyhB.
Figure 2.
Expression of scyA and cctA is regulated by iron-availability through Fur and RyhB. Transcript levels were assessed by qRT-PCR in S. oneidensis wild-type, Δfur, ΔryhB, and ΔfurΔryhB strains in rich medium (iron-rich) and after 30 min growth with the iron chelator 2,2′-dipyridyl (iron-limitation). Transcript levels are shown relative to wild-type level in rich medium as expression ratio of four technical replicates (error bars, standard error). Results from an independent experiment are shown in Supplementary Figure 3.
RyhB alters the stability of several target transcripts
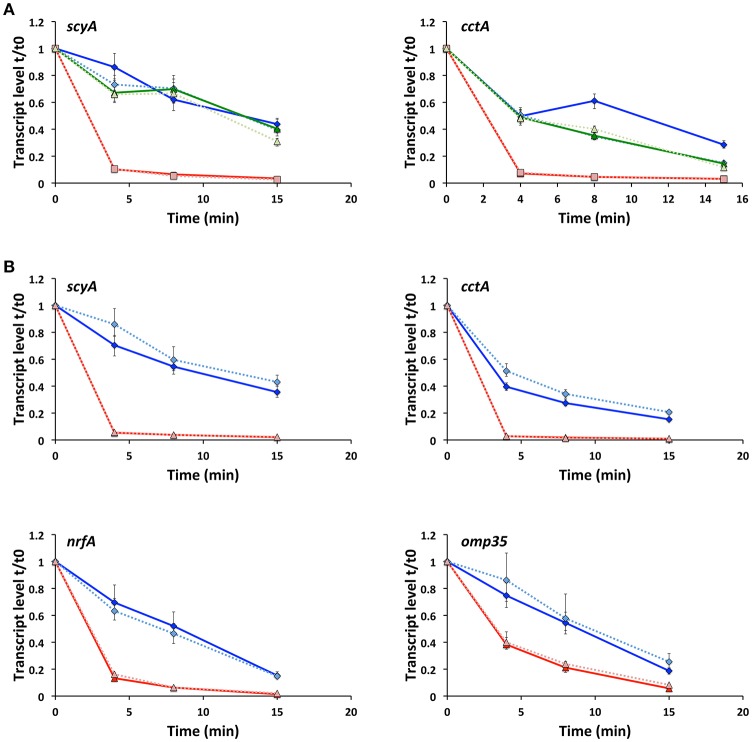
To establish whether RyhB regulates the expression of gene targets in S. oneidensis by affecting the stability of their mRNAs, we determined the transcript levels of scyA and cctA at specific times after transcription was stopped in the wild-type, Δfur and ΔfurΔryhB strains (Figure 3A). We observed that the amount of transcript decreases in all strains over time, but dramatically faster in Δfur than in the wild-type or ΔfurΔryhB strains, showing that RyhB affects transcript stability. When RyhB is overexpressed from a plasmid, it has a similar severe effect on transcript stability (Figure 3B), suggesting that the decreased transcript levels observed in the Δfur strain, is due to mRNA destabilization rather than decreased transcription. We wondered whether this effect would also be observed with other of the RyhB regulated target genes we identified by RNAseq analysis. As can be seen in Figure 3B, both omp35 and nrfA transcript stabilities were significantly altered after RyhB overexpression. In contrast, this was not the case for the RyhB-independent recA gene, verifying that overexpression of RyhB does not generally affect mRNA stability in S. oneidensis (Supplementary Figure 4). The results show that RyhB affect the mRNA stability of its targets ScyA, CctA, NrfA, and Omp35 and suggest that RyhB binding to the target mRNAs promotes degradation by RNases.
Figure 3.
Production of RyhB affects the stability of the target mRNA. (A) Transcript levels of scyA and cctA genes in the wild-type (blue diamonds), Δfur (red squares) and ΔfurΔryhB (green triangles) strains at 0, 4, 8, and 15 min after transcription was stopped. qRT-PCR was used to determine the transcript level relative to time 0, just before addition of rifampicin. Data shown are from two experiments; each with four replicate cDNA samples used in qPCR and shown as expression ratio ± standard error. For all strains, data from experiment 2 are depicted in lighter color and using a dashed line. (B) Transcript levels of scyA, cctA, nrfA, and omp35 genes at 0, 4, 8, and 15 min after transcription was stopped in the ΔryhB mutant containing an empty plasmid (pKM033) (blue diamonds) or a plasmid expressing RyhB (pKM033-ryhB) (red triangles). qRT-PCR was used to determine the transcript level relative to time 0, just before addition of rifampicin. Data are from two experiments; each with four replicate cDNA samples used in qPCR and shown as expression ratio ± standard error. For both strains, data from experiment 2 are depicted in lighter color and using a dashed line.
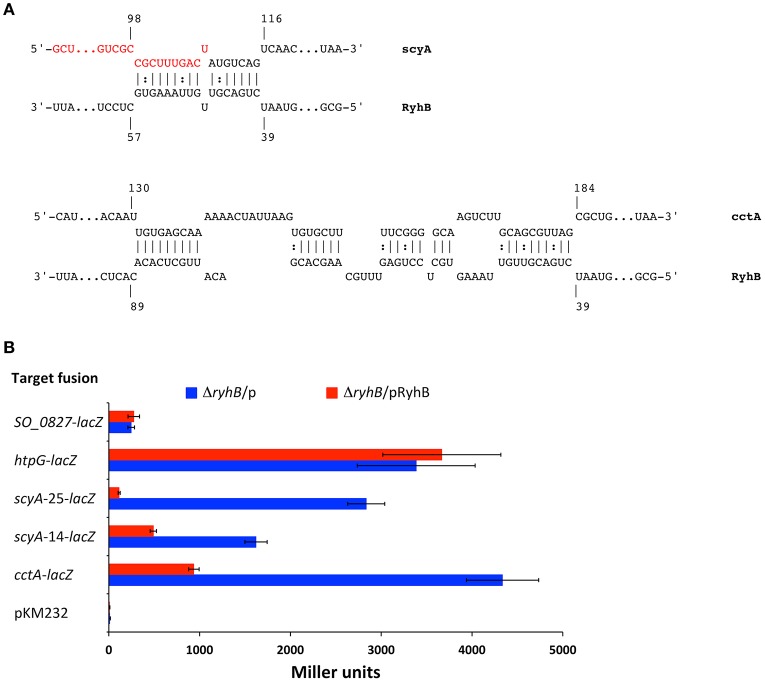
RyhB down-regulation of cytochromes CctA and ScyA is mediated by the 5′-end of target genes
Our data show that RyhB down-regulates expression of the cctA and scyA genes by affecting their mRNA stability. In silico analysis using the IntaRNA software (Busch et al., 2008; Wright et al., 2014; Mann et al., 2017) predicts that RyhB interacts with scyA mRNA sequences at the beginning of the coding region but down-stream from the ribosome binding site and start codon (Figure 4A). RyhB is predicted to interact with a relatively long sequence of the cctA mRNA beginning at the start codon. In order to investigate whether RyhB interacts with the 5′end of these two target genes and to present further evidence for RyhB-dependent regulation of these genes, we established a lacZ-based reporter system in S. oneidensis. The 5′UTR (transcription start sites are designated as +1 and are based on Shao et al., 2014) of cctA and scyA and the beginning of the genes were fused in-frame to the E. coli lacZ missing the first 8 codons. The fusions were expressed on a plasmid from an artificial Plac promoter containing a LacI binding site between the −10 and −35 sequences. At the same time, RyhB is expressed from the same promoter on a second plasmid, which also contains the lacI gene from E. coli. The cctA-lacZ fusion comprises the first 26 codons of cctA, which contains the entire sequence predicted to interact with RyhB. We made two different fusions of scyA with lacZ, one containing the first 25 codons of scyA, which include the entire sequence predicted to interact with RyhB (scyA-25-lacZ) and a second encompassing only part of the interaction region (scyA-14-lacZ) (Figure 4A). When the fusions were expressed in the ΔryhB strain in combination with the plasmid expressing RyhB, the activity of the fusion protein was lower than when RyhB was not expressed (Figure 4B). Even though the activity of both scyA fusions were lower in the presence of RyhB, the fusion containing the entire region predicted to interact with RyhB was decreased to a significantly higher extent (25- vs. 3.3-fold decrease). This indicates that the predicted region is involved in RyhB-dependent regulation and that the entire sequence is required for high-level regulation. No RyhB-dependent decrease in the activity of two control fusions was observed (SO_0827-lacZ and htpG-lacZ) ruling out a general effect of overproducing RyhB on LacZ activity. Thus, the reporter fusion data show that the 5′ ends of the cctA and scyA mRNAs are sufficient to ensure down-regulation of expression by RyhB and provide further evidence of RyhB-dependent down-regulation of the two cytochromes.
Figure 4.
RyhB regulation of ScyA and CctA expression is mediated by sequences in the start of the genes. (A) Predicted interactions between RyhB and scyA and cctA mRNAs. Predictions were made with IntaRNA (Busch et al., 2008; Wright et al., 2014; Mann et al., 2017). Nucleotides are numbered according to their transcription start site (Shao et al., 2014). The transcription start site is located 66 nt upstream from the translational start for scyA. cctA contains two transcription start sites located 85 and 131 nt upstream from the translational start site, respectively (the numbers in the figure refer to the start site 131 nt upstream the translational start, which is used for the cctA-lacZ fusion). Nucleotides marked in red are designating the end of scyA codons in the fusion scyA-14-lacZ. (B) Regulation of target gene fusions by expression of RyhB. β-galatosidase activity was quantified from gene-lacZ fusions harbored on a plasmid in the presence of another plasmid with inducible RyhB production, pKM033-ryhB (pRyhB), or in the presence of an empty plasmid, pKM033 (p). pKM232 contains a promoter-less and truncated lacZ gene missing the first 8 codons. Data are from two independent experiments, each with duplicate cultures, and triplicate samples. Activity is expressed as the mean (Miller units) ± SD.
Hfq is necessary for RyhB-dependent regulation
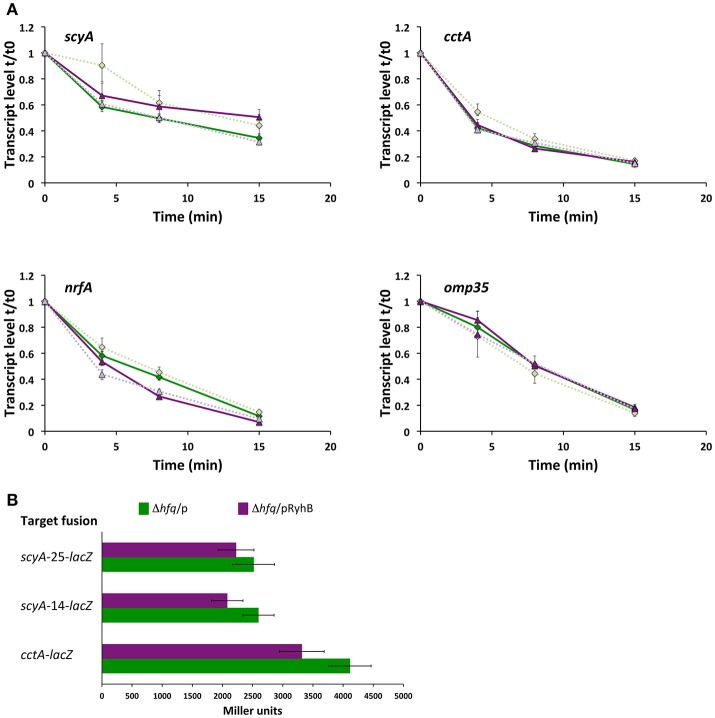
In Gram-negative bacteria, regulation of gene expression by sRNAs often relies on the RNA chaperone Hfq. To determine whether Hfq is required for RyhB function, we constructed a Δhfq mutant strain, which exhibited the small colony and reduced growth phenotype that is well-known for several bacterial species, including S. oneidensis (Dietrich et al., 2009; Meibom et al., 2009; Mégroz et al., 2016). To assess the importance of Hfq for RyhB-dependent destabilization of target mRNAs, we overproduced RyhB in the Δhfq mutant and quantified the mRNA level over time after transcription was stopped. Overproduction of RyhB either did not affect or very slightly affected the stability of the mRNA of the target genes scyA, cctA, nrfA, and omp35 (Figure 5A), which is in stark contrast to the results obtained when Hfq is present (Figure 3B). This finding points to the fact that Hfq is required for the RyhB-induced destabilization of the target mRNAs. The involvement of Hfq was further assessed by measuring the activity of the gene-lacZ fusions in the Δhfq strain (Figure 5B). The activity of the cctA-lacZ and both scyA-lacZ fusions decreased only slightly when RyhB was overproduced in this mutant. From this evidence, we conclude that Hfq is required for optimal function of RyhB during regulation of CctA and ScyA expression.
Figure 5.
Hfq is required for regulation of RyhB targets. (A) Transcript levels of scyA, cctA, nrfA, and omp35 genes at 0, 4, 8, and 15 min after transcription was stopped in a Δhfq mutant containing an empty plasmid (pKM033) (green diamonds) or a plasmid expressing RyhB (pKM033-ryhB) (purple triangles). qRT-PCR was used to determine the transcript level relative to time 0, just before addition of rifampicin. Data are from two experiments; each with four replicate cDNA samples used in qPCR and shown as expression ratio ± standard error. For both strains, data from experiment 2 are depicted in lighter color and using a dashed line. (B) Activity of plasmid encoded target gene fusions in the Δhfq mutant expressing RyhB (from pKM033-RyhB; pRyhB) or containing an empty plasmid (pKM033; p). Activities are shown as the mean (Miller units) ± SD and are the data from two independent experiments, each with duplicate cultures, and triplicate samples.
Discussion
S. oneidensis uses numerous electron acceptors for respiration. Most electron acceptors are reduced in the periplasm or at the bacterial surface, with c-type cytochromes playing a key role. It has been suggested that the periplasmic electron transfer network is dynamic and that simultaneous production of several functional electron transport chains allows S. oneidensis to release electrons quickly to a variety of available electron acceptors (Sturm et al., 2015). The findings presented here strongly indicate that the expression of numerous c-type cytochromes with varying cellular locations and functional roles in response to fluctuating iron availability is regulated by the small RNA RyhB through the master iron regulator Fur.
The presence of a RyhB homolog in S. oneidensis was reported in a previous study that was primarily directed at determining RyhB's potential role in regulating TCA cycle genes (Yang et al., 2010). However, due to unsuccessful attempts to generate a null mutant, a specific role for RyhB was not demonstrated, although it was shown that RyhB expression was increased in a fur mutant and that RyhB overexpression reduced the amount of the sodB transcript, a known RyhB target in E. coli. Our results corroborate the findings that sodB, but not the TCA cycle genes sdhA and acnA, are targets of RyhB (Yang et al., 2010). Also, since we were successful in generating a ryhB mutant in both the wild-type strain and in a Δfur mutant, we were able to characterize this sRNA in S. oneidensis.
RNA sequencing of mutant strains and a RyhB overexpressing strain permitted us to identify putative targets of RyhB regulation. The RNAseq results of the Δfur mutant identified 64 genes downregulated in the mutant relative to wild-type (Table 1). It should be noted that other microarray-based early transcriptomic studies of a Δfur mutant yielded disparate results (Thompson et al., 2002; Wan et al., 2004). The most recent study (Yang et al., 2008) has focused on genes upregulated in the Δfur mutant relative to wild-type. We observed very little overlap between the 64 genes expressed at a lower level in our study, and the 56 expressed a lower level in Δfur in the aforementioned study. This discrepancy is likely due to differing experimental conditions.
Amongst the 64 genes identified above, a sizeable number whose expression level is reduced by the action of RyhB encode proteins that contain iron as a cofactor. In E. coli, RyhB ensures iron conservation in order to guarantee its availability for essential pathways by reducing the expression of iron-binding proteins under iron-limiting conditions (Massé et al., 2005). A recent study found that heme biosynthesis enzymes, cytochrome maturation functions, and heme proteins are one group of iron-containing proteins that appear to mostly escape regulation by RyhB in E. coli (Beauchene et al., 2015). In contrast to E. coli, heme proteins make up the major part of the iron-containing proteins regulated by RyhB in S. oneidensis MR-1. Of the 41 possible functional c-type cytochromes in S. oneidensis, RyhB negatively affected expression of 10: ScyA, PetC, MtrA, MtrC, CctA, SO_3420, NrfA, SO_4047, CymA, and SO_4666. We found that expression of these 10 cytochromes was reduced in a fur mutant (where RyhB levels are elevated) and when RyhB is over-expressed, and conversely that transcript levels increased when ryhB was deleted in the fur mutant. Since RyhB expression is repressed by Fur, its level increases under iron-limiting conditions when less Fur is actively bound to promoter regions. Therefore, expression of RyhB repressed genes is expected to decrease when iron becomes scarce. We confirmed this for scyA and cctA and would assume that other RyhB regulated cytochrome genes respond the same way. In fact, an earlier study of the iron response of S. oneidensis MR-1 found that iron depletion represses expression of a number of genes involved in anaerobic energy metabolism, including cymA, mtrA, mtrC, and omcA (Yang et al., 2009). Our results demonstrate that the previously observed down-regulation of cytochrome gene expression after iron depletion, with the exception of omcA, is likely to be mediated through Fur and RyhB.
A significant number of the genes in groups I and II were not downregulated after RyhB overexpression (groups Ib and IIb). This suggests that RyhB is not directly regulating these genes. In contrast, deleting ryhB in the Δfur strain increased their expression, suggesting a role for RyhB. We have no conclusive explanation for these apparently contradictory results, but believe it could be a consequence of the effect deleting fur has on the physiology of S. oneidensis. Down-regulation of certain genes in the Δfur mutant relative to the wild-type could be a result of the growth change tied to Fur loss, whereas the increase in expression after the consecutive deletion of ryhB reflects the reversion of growth to wild-type level rather than direct regulation by RyhB.
Three genes that, based on RNAseq data, were not regulated by RyhB (i.e., pertaining to group III) exhibited higher expression upon RyhB overexpression. This result can be attributed to increased intracellular iron concentrations as a result of the decreased production of iron-containing proteins, as observed in E. coli (Jacques et al., 2006). Increased iron concentration would lead to higher activity of Fur as a positive regulator of these genes. We do not know whether Fur directly activates these genes or whether another regulatory molecule, maybe another small RNA, is involved. In agreement with this, we found that some of the Fur-repressed genes (e.g., iron uptake genes that are increased in expression in the Δfur strain) are transcribed at a lower level when RyhB is over-expressed (data not shown) and attribute this to elevated iron levels leading to increased repression by Fur.
We saw, as others before (Yang et al., 2008), that deletion of fur results in several physiological changes, most notably a decreased growth rate and loss of the signature red color of S. oneidensis cultures and colonies. Notably, these phenotypes were almost completely reversed by deletion of ryhB, emphasizing that RyhB controls important cellular processes. The disappearance of the red color can be explained by the significant downregulation of the numerous c-type cytochromes in the fur mutant, which was further supported by visualizing the reduced amount of heme-containing proteins in the Δfur strain. It is also possible that the reduced growth of the Δfur mutant is linked to the decreased production of cytochromes. Two of the most abundant periplasmic c-type cytochromes, ScyA and CctA, as well as the membrane anchored CymA, which functions as the electron donor to several periplasmic electron chains are significantly downregulated in the Δfur mutant. It is known that deletion of cymA results in decreased ability to use the anaerobic electron acceptors Fe(III), Mn (IV), Cr(VI), nitrate, fumarate, and DMSO, but that it does not impact aerobic growth (Myers and Myers, 1997; Schwalb et al., 2003; Gao et al., 2010). The precise role played by ScyA is not known, but it has been identified as a mediator of electron transport between CymA and the diheme cytochrome c5 peroxidase CcpA (Schütz et al., 2011). Whereas a scyA null mutant does not exhibit any major phenotype with respect to growth with a number of electron acceptors (Jin et al., 2013), the gene does appear to have an impact on the fitness of S. oneidensis under some conditions (Deutschbauer et al., 2011; Wetmore et al., 2015).
The genes for the three subunits of the ubiquinol-cytochrome c reductase complex are all expressed at lower level in the fur mutant, and deleting ryhB abolishes this. The three genes encode the Fe-S subunit PetA, the cytochrome b subunit PetB, and the cytochrome c1 subunit PetC. It is possible that the decreased expression of these genes accounts for the growth defect of the fur mutant as both petA and petC mutants exhibit an aerobic growth defect (Gao et al., 2010; Luo et al., 2013).
This study was initiated by searching for regulators controlling expression of the outer membrane decaheme c-type cytochrome MtrC, one of the proteins involved in metal respiration. Our RNA sequencing data strongly indicate that MtrC is controlled by Fur, indirectly by the action of the sRNA RyhB. However, the data also suggest that other regulators contribute, as ryhB deletion does not completely relieve the effect of the fur deletion. Under iron-limiting conditions, when expression of RyhB is increased, mtrC transcript levels decrease, however the mtrC transcript level is also decreased under iron-limitation in a ryhB mutant (data not shown), supporting that other iron-dependent regulators in addition to RyhB control mtrC expression. The identity of this regulator (or regulators) is currently unknown.
In other species, RyhB exerts its function by base-pairing to sequences in target gene mRNAs, which often leads to mRNA degradation (Oglesby-Sherrouse and Murphy, 2013). Hfq is a RNA chaperone that is required for the function of many regulatory sRNAs (Updegrove et al., 2016). Hfq mediates interactions between sRNA molecules and their mRNA targets and stabilizes the sRNA. Since Hfq participates to the function of numerous sRNAs in a single organism, hfq null mutants often exhibit pleiotropic phenotypes. This is also the case for S. oneidensis (Brennan et al., 2013) and specifically it was observed that a hfq mutant synthesizes hemes at a reduced level, although the mechanism remains unknown (Brennan et al., 2014). Our data demonstrate that RyhB requires Hfq for its function in S. oneidensis. Over-production of RyhB strongly destabilizes the mRNA of the examined targets, cctA, scyA, nrfA, and omp35 but this is circumvented in the hfq null mutant. We assume Hfq promotes limited base-pairing between RyhB and the target mRNAs. This is supported by assessing the effect of RyhB over-production on the activity of fusions between lacZ and the mRNA region of scyA and cctA that was proposed in silico to base-pair with RyhB. When Hfq is present, RyhB over-production reduces the activity of the reporter fusions but no decrease was seen when Hfq is absent. As Hfq appears to be required for RyhB function, we would expect the effect on growth observed due to loss of fur that can be alleviated by deleting ryhB, could also be alleviated by deletion of hfq. However, despite numerous attempts, we were unable to construct a double fur hfq null mutant, possibly due the effect the loss of each gene has on the growth.
In summary, we propose a model for regulation of how iron availability controls cytochrome expression through Fur and the sRNA RyhB in S. oneidensis (Figure 6). Under iron-replete conditions, Fur represses of expression of RyhB as well as many proteins involved in iron uptake and c-type cytochromes are expressed. Iron limitation alleviates Fur repression, thus RyhB expression is increased and this reduces production of numerous cytochromes, thereby likely preserving iron for essential proteins. Our data furthermore suggest that some genes, including mtrC, rely not only on RyhB, but also on additional Fur- or iron-dependent molecules(s) for their regulation.
Figure 6.
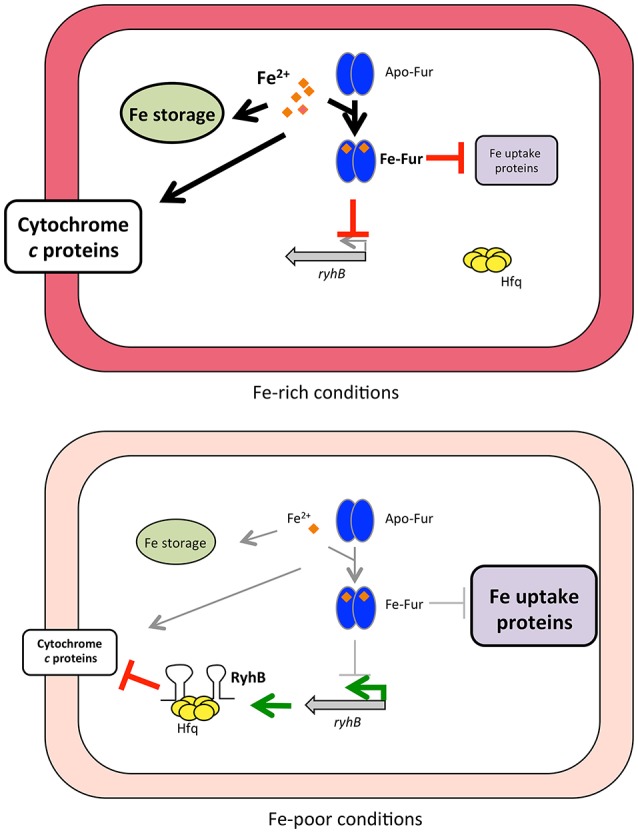
Model of how iron availability controls RyhB regulation of c-type cytochromes. Under iron-rich conditions, Fur associates with Fe2+ (orange diamonds) and binds to Fur-binding sites in the promoter region of target genes, leading to repression of expression (indicated by red blocked lines) of RyhB and other genes (e.g., iron uptake genes). Fur repression is relieved when iron becomes limiting and RyhB is produced (indicated by green arrows), as well as other Fur-repressed genes (including iron uptake genes). RyhB with the aid of the RNA chaperone Hfq represses expression of numerous c-type cytochromes by promoting degradation of the mRNAs. Processes that are favored under either Fe-rich or Fe-poor conditions are shown with thick arrows or thick blocked lines, whereas processes that do not occur (or at diminished rate) are indicated with thin gray arrows or thin gray blocked lines.
Author contributions
KM designed the research, performed the experiments, analyzed, and interpreted the data, and wrote the manuscript. EC analyzed sequencing data and wrote part of the manuscript. RB-L designed the research interpreted results and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the following individuals for providing reagents: Dr. Alain Charbit (INSERM, France) for strain DH5αλpir; Prof. Johannes Gescher (Karlsruhe Institute of Technology, Germany) for plasmids pMQ150 and pBAD-fccA-His; Prof. Haichun Gao (Zhejiang University, China) for plasmid pHGE-Ptac; Dr. Vladimir Sentchilo (University of Lausanne, Switzerland) for plasmid pME3061 and E. coli strain MG1655; Prof. Daad Saffarini (University of Wisconsin-Milwaukee, USA) for strain BG148, and S. Bree Reed (Pacific Northwest National Laboratory, USA) for providing strain WM3064.
Footnotes
Funding. This work was supported by the Swiss National Science Foundation grant # 310030_156957.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00268/full#supplementary-material
References
- Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- Barchinger S. E., Pirbadian S., Sambles C., Baker C. S., Leung K. M., Burroughs N. J., et al. (2016). Regulation of gene expression in Shewanella oneidensis MR-1 during electron acceptor limitation and bacterial nanowire formation. Appl. Environ. Microbiol. 82, 5428–5443. 10.1128/AEM.01615-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchene N. A., Myers K. S., Chung D., Park D. M., Weisnicht A. M., Keleş S., et al. (2015). Impact of anaerobiosis on expression of the iron-responsive fur and RyhB regulons. MBio 6:e01947-15. 10.1128/mBio.01947-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliaev A. S., Klingeman D. M., Klappenbach J. A., Wu L., Romine M. F., Tiedje J. M., et al. (2005). Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187, 7138–7145. 10.1128/JB.187.20.7138-7145.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. M., Keane M. L., Hunt T. M., Goulet M. T., Mazzucca N. Q., Sexton Z., et al. (2013). Shewanella oneidensis Hfq promotes exponential phase growth, stationary phase culture density, and cell survival. BMC Microbiol. 13:33. 10.1186/1471-2180-13-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. M., Mazzucca N. Q., Mezoian T., Hunt T. M., Keane M. L., Leonard J. N., et al. (2014). Reduced heme levels underlie the exponential growth defect of the Shewanella oneidensis hfq mutant. PLoS ONE 9:e109879. 10.1371/journal.pone.0109879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Richter A. S., Backofen R. (2008). IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinforma. Oxf. Engl. 24, 2849–2856. 10.1093/bioinformatics/btn544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer A., Price M. N., Wetmore K. M., Shao W., Baumohl J. K., Xu Z., et al. (2011). Evidence-based annotation of gene function in Shewanella oneidensis MR-1 using genome-wide fitness profiling across 121 conditions. PLoS Genet. 7:e1002385. 10.1371/journal.pgen.1002385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M., Munke R., Gottschald M., Ziska E., Boettcher J. P., Mollenkopf H., et al. (2009). The effect of hfq on global gene expression and virulence in Neisseria gonorrhoeae. FEBS J. 276, 5507–5520. 10.1111/j.1742-4658.2009.07234.x [DOI] [PubMed] [Google Scholar]
- Firer-Sherwood M. A., Bewley K. D., Mock J.-Y., Elliott S. J. (2011). Tools for resolving complexity in the electron transfer networks of multiheme cytochromes c. Met. Integr. Biometal Sci. 3, 344–348. 10.1039/c0mt00097c [DOI] [PubMed] [Google Scholar]
- Fonseca B. M., Paquete C. M., Neto S. E., Pacheco I., Soares C. M., Louro R. O. (2013). Mind the gap: cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem. J. 449, 101–108. 10.1042/BJ20121467 [DOI] [PubMed] [Google Scholar]
- Frawley E. R., Fang F. C. (2014). The ins and outs of bacterial iron metabolism. Mol. Microbiol. 93, 609–616. 10.1111/mmi.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Barua S., Liang Y., Wu L., Dong Y., Reed S., et al. (2010). Impacts of Shewanella oneidensis c-type cytochromes on aerobic and anaerobic respiration. Microb. Biotechnol. 3, 455–466. 10.1111/j.1751-7915.2010.00181.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau H. H., Gralnick J. A. (2007). Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61, 237–258. 10.1146/annurev.micro.61.080706.093257 [DOI] [PubMed] [Google Scholar]
- Heeb S., Itoh Y., Nishijyo T., Schnider U., Keel C., Wade J., et al. (2000). Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant Microbe Interact. 13, 232–237. 10.1094/MPMI.2000.13.2.232 [DOI] [PubMed] [Google Scholar]
- Huber W., Carey V. J., Gentleman R., Anders S., Carlson M., Carvalho B. S., et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques J. F., Jang S., Prévost K., Desnoyers G., Desmarais M., Imlay J., et al. (2006). RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 62, 1181–1190. 10.1111/j.1365-2958.2006.05439.x [DOI] [PubMed] [Google Scholar]
- Jin M., Jiang Y., Sun L., Yin J., Fu H., Wu G., et al. (2013). Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS ONE 8:e75610. 10.1371/journal.pone.0075610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma. Oxf. Engl. 30, 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Luo Q., Dong Y., Chen H., Gao H. (2013). Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of Tat mutants in Shewanella oneidensis. PLoS ONE 8:e62064. 10.1371/journal.pone.0062064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M., Wright P. R., Backofen R. (2017). IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 45, W435–W439. 10.1093/nar/gkx279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Gottesman S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Vanderpool C. K., Gottesman S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. 10.1128/JB.187.20.6962-6971.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mégroz M., Kleifeld O., Wright A., Powell D., Harrison P., Adler B., et al. (2016). The RNA-binding chaperone Hfq is an important global regulator of gene expression in Pasteurella multocida and plays a crucial role in production of a number of virulence factors, including hyaluronic acid capsule. Infect. Immun. 84, 1361–1370. 10.1128/IAI.00122-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom K. L., Forslund A. L., Kuoppa K., Alkhuder K., Dubail I., Dupuis M., et al. (2009). Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun. 77, 1866–1880. 10.1128/IAI.01496-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey A. R., Craig S. A., Payne S. M. (2005). Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73, 5706–5719. 10.1128/IAI.73.9.5706-5719.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T. E., Tsapin A. I., Vandenberghe I., de Smet L., Frishman D., Nealson K. H., et al. (2004). Identification of 42 possible cytochrome C genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. Omics J. Integr. Biol. 8, 57–77. 10.1089/153623104773547499 [DOI] [PubMed] [Google Scholar]
- Myers C. R., Myers J. M. (1997). Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179, 1143–1152. 10.1128/jb.179.4.1143-1152.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J. M., Myers C. R. (2000). Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J. Bacteriol. 182, 67–75. 10.1128/JB.182.1.67-75.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D. (2011). ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8, 11–13. 10.4161/rna.8.1.13346 [DOI] [PubMed] [Google Scholar]
- Nitzan M., Rehani R., Margalit H. (2017). Integration of Bacterial Small RNAs in Regulatory Networks. Annu. Rev. Biophys. 46, 131–148. 10.1146/annurev-biophys-070816-034058 [DOI] [PubMed] [Google Scholar]
- Oglesby-Sherrouse A. G., Murphy E. R. (2013). Iron-responsive bacterial small RNAs: variations on a theme. Met. Integr. Biometal Sci. 5, 276–286. 10.1039/c3mt20224k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K., Schicklberger M., Gescher J. (2012). Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl. Environ. Microbiol. 78, 913–921. 10.1128/AEM.06803-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz B., Schicklberger M., Kuermann J., Spormann A. M., Gescher J. (2009). Periplasmic electron transfer via the c-type cytochromes MtrA and FccA of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 75, 7789–7796. 10.1128/AEM.01834-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz B., Seidel J., Sturm G., Einsle O., Gescher J. (2011). Investigation of the electron transport chain to and the catalytic activity of the diheme cytochrome c peroxidase CcpA of Shewanella oneidensis. Appl. Environ. Microbiol. 77, 6172–6180. 10.1128/AEM.00606-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb C., Chapman S. K., Reid G. A. (2003). The tetraheme cytochrome CymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochem. Mosc. 42, 9491–9497. 10.1021/bi034456f [DOI] [PubMed] [Google Scholar]
- Shanks R. M., Kadouri D. E., MacEachran D. P., O'Toole G. A. (2009). New yeast recombineering tools for bacteria. Plasmid 62, 88–97. 10.1016/j.plasmid.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Price M. N., Deutschbauer A. M., Romine M. F., Arkin A. P. (2014). Conservation of transcription start sites within genes across a bacterial genus. MBio 5:e01398-14-e01398-14. 10.1128/mBio.01398-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Rosso K. M., Clarke T. A., Richardson D. J., Zachara J. M., Fredrickson J. K. (2012). Molecular underpinnings of Fe(III) oxide reduction by Shewanella Oneidensis MR-1. Front. Microbiol. 3:50. 10.3389/fmicb.2012.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm G., Richter K., Doetsch A., Heide H., Louro R. O., Gescher J. (2015). A dynamic periplasmic electron transfer network enables respiratory flexibility beyond a thermodynamic regulatory regime. ISME J. 9, 1802–1811. 10.1038/ismej.2014.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. K., Beliaev A. S., Giometti C. S., Tollaksen S. L., Khare T., Lies D. P., et al. (2002). Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68, 881–892. 10.1128/AEM.68.2.881-892.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapin A. I., Vandenberghe I., Nealson K. H., Scott J. H., Meyer T. E., Cusanovich M. A., et al. (2001). Identification of a small tetraheme cytochrome c and a flavocytochrome c as two of the principal soluble cytochromes c in Shewanella oneidensis strain MR1. Appl. Environ. Microbiol. 67, 3236–3244. 10.1128/AEM.67.7.3236-3244.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove T. B., Zhang A., Storz G. (2016). Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138. 10.1016/j.mib.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X. F., Verberkmoes N. C., McCue L. A., Stanek D., Connelly H., Hauser L. J., et al. (2004). Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 186, 8385–8400. 10.1128/JB.186.24.8385-8400.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore K. M., Price M. N., Waters R. J., Lamson J. S., He J., Hoover C. A., et al. (2015). Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6, e00306–00315. 10.1128/mBio.00306-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P. R., Georg J., Mann M., Sorescu D. A., Richter A. S., Lott S., et al. (2014). CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 42, W119–W123. 10.1093/nar/gku359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Harris D. P., Luo F., Wu L., Parsons A. B., Palumbo A. V., et al. (2008). Characterization of the Shewanella oneidensis Fur gene: roles in iron and acid tolerance response. BMC Genomics 9(Suppl.1), S11. 10.1186/1471-2164-9-S1-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Harris D. P., Luo F., Xiong W., Joachimiak M., Wu L., et al. (2009). Snapshot of iron response in Shewanella oneidensis by gene network reconstruction. BMC Genomics 10:131. 10.1186/1471-2164-10-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., McCue L. A., Parsons A. B., Feng S., Zhou J. (2010). The tricarboxylic acid cycle in Shewanella oneidensis is independent of Fur and RyhB control. BMC Microbiol. 10:264. 10.1186/1471-2180-10-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.