Abstract
Cancer results from a breakdown of normal gene expression control, so the study of gene regulation is critical to cancer research. To gain insight into the transcriptional and epigenetic factors regulating abnormal gene expression patterns in cancers, we developed the Cistrome Cancer web resource (http://cistrome.org/CistromeCancer/). We conducted the systematic integration and modeling of over ten thousand tumor molecular profiles from The Cancer Genome Atlas (TCGA) with over twenty-three thousand ChIP-seq and chromatin accessibility profiles from our Cistrome collection. The results include reconstruction of functional enhancer profiles, “super-enhancer” target genes, as well as predictions of active transcription factors (TF) and their target genes for each TCGA cancer type. Cistrome Cancer reveals novel insights from integrative analyses combining chromatin profiles with tumor molecular profiles, and will be a useful resource to the cancer gene regulation community.
INTRODUCTION
Gene expression misregulation plays a critical role in tumorigenesis and progression (1), so cancer-specific transcription factor (TF) and cis-element activities of gene expression are essential for understanding the molecular mechanisms of cancer. The Cancer Genome Atlas (TCGA) consortium has generated mutation, copy number variation, DNA methylation, transcriptome profiling as well as patient survival data for over ten thousand primary tumor in over 30 cancer types (2). However, no ChIP-seq data characterizing TF binding locations, has been produced from TCGA due to the technical difficulty of ChIP-seq with limited cell numbers in primary tumor samples. Nevertheless, tens of thousands of ChIP-seq datasets are available in the public domain, generated in a variety of cell line models and primary tissues, by large consortia like the Encyclopedia of DNA Elements (ENCODE) (3) and Roadmap Epigenomics (4), as well as by individual laboratories world-wide.
To study gene regulation in cancer, we designed comprehensive modeling approaches to integrate these publicly available chromatin profiling data with TCGA data, and developed the Cistrome Cancer web resource (http://cistrome.org/CistromeCancer/) to report the data integration results. We have previously developed the Cistrome Data Browser (DB) (5). It contains over twenty three thousand processed and quality controlled (6) ChIP-seq and chromatin accessibility (DNase-seq and ATAC-seq) profiles from human and mouse genomes from sources including Gene Expression Omnibus (GEO), ENCODE, and Roadmap Epigenomics (Fig. 1a). We have also developed Model-based Analysis of Regulation of Gene Expression (MARGE) (7), a computational method for predicting cis-regulatory (functional enhancer) profiles to interpret differential expression gene sets by leveraging a compendium of H3K27ac ChIP-seq datasets from human or mouse genomes. We integrated ChIP-seq and chromatin accessibility data from Cistrome DB with TCGA profiles to impute functional enhancer profiles, “super-enhancer” target genes, and active TF target genes for each TCGA cancer type. The results of our integrative modeling are available for browsing and download. A video demonstration can be found in Video 1 as well as on the website homepage.
Figure 1. Cistrome Cancer.
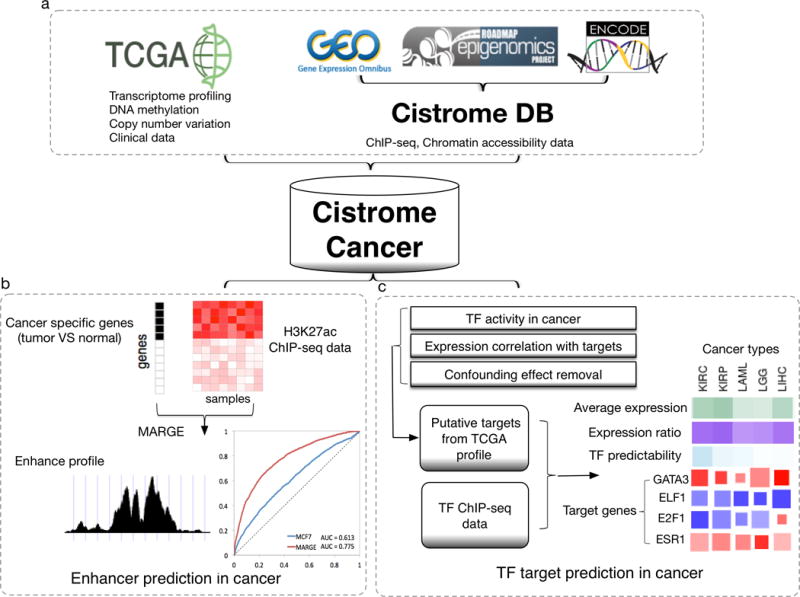
a. Data sources: Cancer molecular profiling data, including transcriptomic profiles, DNA methylation profiles, copy number variation, and clinical survival information, were collected from TCGA. ChIP-seq and chromatin accessibility profiling data downloaded from GEO, ENCODE and Roadmap Epigenomics were curated, processed, and stored in the Cistrome DB.
b. Enhancer prediction in cancer. For each cancer type, cancer specific genes (marked in black) were identified as upregulated in tumor compared to normal samples; MARGE was used to calculate gene regulatory potential scores (heatmap) from a compendium of H3K27ac ChIP-seq profiles and to predict enhancer profiles from selected relevant H3K27ac samples. Multiple H3K27ac profiles used in combination can better model breast cancer specific genes than H3K27ac from a single breast cancer cell line MCF7.
c. Transcription factor (TF) target prediction in cancer. For each TF in each cancer, TF activity and expression correlation, corrected for confounding effects from DNA methylation, CNV and tumor purity, were used to identify putative target genes. TF ChIP-seq data best matching the putative target genes were integrated with expression correlation to make the final prediction. The result is presented in a heatmap, in which the top rows represent the TF average expression level, percent of tumors expressing the TF above baseline, and TF ChIP-seq predictability, in green, purple, and cyan, respectively. Predicted target genes in each cancer type were listed in columns, where the color and size of each square represents the expression correlation and ChIP-seq regulatory potential score percentile.
METHODS AND RESULTS
To integrate the orthogonal data contained in TCGA and Cistrome DB, TCGA RNA-seq profiles were re-clustered into 29 reannotated cancer types (Figs. S1, S2, and Supplementary Table 1). Cistrome Cancer has two main functional modules: enhancer and target prediction (Fig. 1b), and transcription factor (TF) target prediction (Fig. 1c). Details for the two modules are described below as well as in the Supplementary Material.
Differentially expressed genes in cancers can be driven by unknown TFs bound to distal enhancers, so genome-wide cis-regulatory profiles imputed from public H3K27ac ChIP-seq can be useful for understanding cancer-specific gene expression regulation. To this end, we first identified up-regulated genes by comparing the RNA-seq data of tumor over normal samples for each of the 15 cancer types that have over 15 normal samples. For each H3K27ac ChIP-seq profile in the Cistrome DB, the regulatory potential, which is defined as the ChIP-seq signal weighted by the distance to the transcription start site, was calculated for each gene indicating the level of gene expression reflected from H3K27ac. As a quantitative gene-centric approach, the regulatory potential defined in MARGE is more informative to identify target genes of “super-enhancers” (8). We applied the logistic regression function in MARGE to over 1,200 H3K27ac profiles and retrieved 10 relevant H3K27ac profiles that best model the up-regulated genes in each cancer type. The selected H3K27ac profiles in combination can better model cancer-specific genes than any single H3K27ac ChIP-seq dataset from an individual cancer cell line (Fig. 1b). Next we adopted the semi-supervised learning approach in MARGE to weigh the selected H3K27ac profiles and used the union DNaseI hypersensitive sites (UDHS) ranked by the weighted integration of H3K27ac signal as the predicted profile of the enhancers regulating these genes. The cancer-upregulated genes, predicted cis-regulatory (enhancer) profile, as well as the “super-enhancer” targets quantified by MARGE-integrated regulatory potential for each cancer type can be downloaded for downstream analysis or visualized on genome browsers.
TF targets in a given cancer type can be predicted. If a TF is active in a given cancer type, its expression is correlated with its targets across tumor samples, and its ChIP-seq profiles provide evidence of strong binding, this information can be used to predict potential targets of this TF. In addition, TF regulation of target genes could be continuous from weak to strong in a context specific manner, rather than a strict binary mode of regulation. Therefore, we chose a loose cutoff and provide users detailed information on TF expression, TF and target gene expression correlation, and TF binding evidence, so users interested in specific TF could set stricter cutoffs for in depth study in specific cancer type. We consider a TF to be active in a cancer type if a sufficient percentage of tumors express the TF above a TF-dependent baseline (Fig. S3a). We identified putative targets of an active TF as those genes that are correlated with the TF in the tumor samples to a significantly higher level than random gene pairs in the same cancer type (Fig. S3b). We then examined all the ChIP-seq datasets of this TF and used logistic regression to select a small subset of ChIP-seq datasets whose regulatory potentials best model the targets identified in the correlation analysis. In addition, we used the likelihood ratio test to ensure that the selected TF ChIP-seq profiles have better signal than the best matching chromatin input for the putative targets. All together, we predicted target genes for 575 TFs and made them available on the Cistrome Cancer website, with an example of androgen receptor (AR) targets shown in Fig. S4. For each TF, users can see the TF expression reads per kilobase per million, percentage of tumors expressing the TF above baseline, and ChIP-seq regression likelihood ratio for each cancer type. In the Cistrome Cancer web interface, each putative target gene for each TF in each cancer is represented by a square, where the color and size indicate supporting evidence from gene expression correlation and ChIP-seq binding, respectively (Fig. 1c).
We demonstrate the utility of Cistrome Cancer through analyses of selected TFs. We found that FOXM1 is consistently overexpressed in most cancer types (Fig. S5a), and that luminal breast cancer patients with high FOXM1 expression have poor clinical outcomes (P = 0.018, Fig. S5b). Comparing FOXM1 target genes with targets of other TFs identified in Cistrome Cancer, we found that target genes of MYBL2, EZH2, E2F1, E2F2, E2F8, CBX3, TTF2, BRCA1, NCAPG, SSRP1 and LIN9 to have the largest overlap with those of FOXM1 (Fig. S5c). Analysis of ChIP-seq data for these TFs reveals a high degree of binding overlap between FOXM1, E2F1 and MYBL2 (Fig. S5d), suggesting that these three factors form a regulatory module in cancer. These Cistrome Cancer results are consistent with previous studies of FOXM1 showing its elevated expression and role in cancer related biological processes including cell proliferation, cell cycle progression and DNA damage repair (9, 10). The target genes of FOXM1 inferred from Cistrome Cancer, including cell cycle regulators Cyclin B1 and CENP-A, have also been reported as FOXM1 targets in many cancer types (11).
As a second example, we found that STAT4 is significantly overexpressed in kidney renal clear cell carcinoma (KIRC) relative to normal kidney (Fig. S6a) and high STAT4 expression is associated with poor survival (Fig. S6b). STAT4 ChIP-seq target genes have overall higher expression in KIRC (Fig. S6c) and, consistent with known immune related functions of STAT4 (12), target genes are enriched in immune related functions such as T cell activation, leukocyte activation, and immune response. Like STAT4, IRF4 is known to have immune cell specific activity (13). However, IRF4 and its target genes are down-regulated in colon and rectal adenocarcinomas (COAD-READ) (Figs. S6d, S6e, S6f), and higher IRF4 expression is associated with better prognosis (Fig. S6e). We used TIMER (14), a systematic computational approach to analyze tumor immune infiltrations, to estimate the abundance of tumor infiltrating lymphocytes, and found CD8 T cell levels to be higher in KIRC tumors and lower in COAD-READ tumors as compared to their respective normal tissues (Fig. S6g). Interestingly, CD8 T cell abundance is positively correlated with both STAT4 in KIRC (Fig. S6h) and IRF4 in COAD-READ (Fig. S6i). This suggests that the transcriptional activity of STAT4 in KIRC and IRF4 in COAD-READ tumors might reflect the level of infiltrating immune cells instead of regulation in the tumor cells themselves.
DISCUSSION
A few caveats regarding Cistrome Cancer target gene predictions are worth noting. First, Cistrome Cancer determines relevant ChIP-seq datasets using a regression approach independent from cell type annotations. This allows TF binding information to be borrowed across cell types, but may not be accurate in cases where data is absent from closely related cancer types. Users should pay attention to the likelihood ratio test statistics to assess the correspondence between gene expression and TF binding. Second, expression correlation between a TF and another gene does not prove direct TF regulation of the gene, and Cistrome Cancer might miss direct gene targets due to insufficient expression correlation with the TF. Third, as observed in the STAT4 and IRF4 examples, gene expression patterns observed across TCGA samples may reflect differences in subpopulations represented within the overall population instead of gene expression misregulation in cancer. Fourth, Cistrome Cancer TF target predictions are limited to those TFs with ChIP-seq data. In some cancer types there may be active TFs that are not represented by suitable publicly available ChIP-seq data. In evaluating Cistrome Cancer predictions users should take other available information into account rather than relying on any measure in isolation.
In summary, Cistrome Cancer is a web resource that integrates cancer genomics data from TCGA with chromatin profiling data from Cistrome DB to enable cancer researchers to explore regulatory links between TFs and cancer transcriptomes. Exploratory and interactive data visualization can be carried out using the Cistrome Cancer web browser, and regulatory predictions can be downloaded for further analysis. Cistrome Cancer will be a valuable resource for experimental and computational cancer biologists alike.
Supplementary Material
Acknowledgments
Financial support: This work was supported by grants from the US National Institutes of Health [U01CA180980 and R01GM099409] and National Natural Science Foundation of China [31329003].
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
References
- 1.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–51. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu H, Fang X. A brief review on the Human Encyclopedia of DNA Elements (ENCODE) project. Genomics, proteomics & bioinformatics. 2013;11:135–41. doi: 10.1016/j.gpb.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature biotechnology. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mei S, Qin Q, Wu Q, Sun H, Zheng R, Zang C, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic acids research. 2017;45:D658–D62. doi: 10.1093/nar/gkw983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin Q, Mei S, Wu Q, Sun H, Li L, Taing L, et al. ChiLin: a comprehensive ChIP-seq and DNase-seq quality control and analysis pipeline. BMC bioinformatics. 2016;17:404. doi: 10.1186/s12859-016-1274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Zang C, Xiao T, Fan J, Mei S, Qin Q, et al. Modeling cis-regulation with a compendium of genome-wide histone H3K27ac profiles. Genome research. 2016;26:1417–29. doi: 10.1101/gr.201574.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer research. 2011;71:4329–33. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochimica et biophysica acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Molecular and cellular biology. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan MH. STAT4: a critical regulator of inflammation in vivo. Immunologic research. 2005;31:231–42. doi: 10.1385/IR:31:3:231. [DOI] [PubMed] [Google Scholar]
- 13.Biswas PS, Bhagat G, Pernis AB. IRF4 and its regulators: evolving insights into the pathogenesis of inflammatory arthritis? Immunological reviews. 2010;233:79–96. doi: 10.1111/j.0105-2896.2009.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome biology. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


