Abstract
The human cellular reverse transcriptase, telomerase, is very tightly regulated in large long-lived species. Telomerase is expressed during early human fetal development, is turned off in most adult tissues, and then becomes reactivated in almost all human cancers. However, the exact mechanism regulating these switches in expression are not known. We recently described a phenomenon where genes are regulated by telomere length dependent loops (telomere position effects over long distances; TPE-OLD). The hTERT gene is ~1.2 Mb from the human chromosome 5p end. We observed that when telomeres are long hTERT gene expression is repressed and a probe next to the 5p telomere and the hTERT locus are spatially co-localized. When telomeres are short at least one of the hTERT alleles is spatially separated from the telomere, developing more active histone marks and changes in DNA methylation in the hTERT promoter region. These findings have implications for how cells turn off telomerase when telomeres are long during fetal development and how cancer cells reactivate telomerase in cells that have short telomeres. In addition to TPE-OLD, in proliferating stem cells such as activated T-lymphocytes, telomerase can be reversibly activated and silenced by telomere looping. In telomerase positive cancer cells that are induced to differentiate and downregulate telomerase, telomere looping correlates with silencing of the hTERT gene. These studies and others support a role of telomeres in regulating gene expression via telomere looping that may involve interactions with internal telomeric sequences (ITS). In addition to telomere looping, TPE-OLD may be one mechanism of how cells time changes in physiology without initiating a DNA damage response.
Keywords: telomere looping, telomerase, TRF2, TERRA, TERT, chromatin
1. Introduction
Telomere and Telomerase
Telomeres are long tracks of noncoding repetitive (TTAGGGn) sequences at the ends of chromosomes (Figure 1). During early human development telomerase is expressed until telomeres reach a specific length and then telomerase is silenced. Due to the end-replication problem (Watson, 1972) and oxidative damage (von Zglincki et al, 2005) telomeres then progressively shorten with cell division throughout life in vitro (Harley et al, 1990) and in vivo (Hastie et al, 1990). Progressive telomere shortening generally correlates with in vitro cell divisions (Harley et al, 1990) and in vivo with increased age (Lindsey et al, 1991). There are telomere maintenance spectrum disorders (also termed telomeropathies) that also correlate with the onset of diseases such as dyskeratosis congenita and familial idiopathic pulmonary fibrosis (Holohan et al, 2014), but it remains to be demonstrated that the shortened telomeres directly cause age-related diseases. In cancer cells telomeres are maintained by activating or upregulating telomerase, a cellular ribonucleoprotein enzyme complex that is expressed during early human fetal development (Wright et al, 1996), turned off in most adult tissues, and becomes reactivated in almost all human cancers (Kim et al, 1994) (Figure 2).
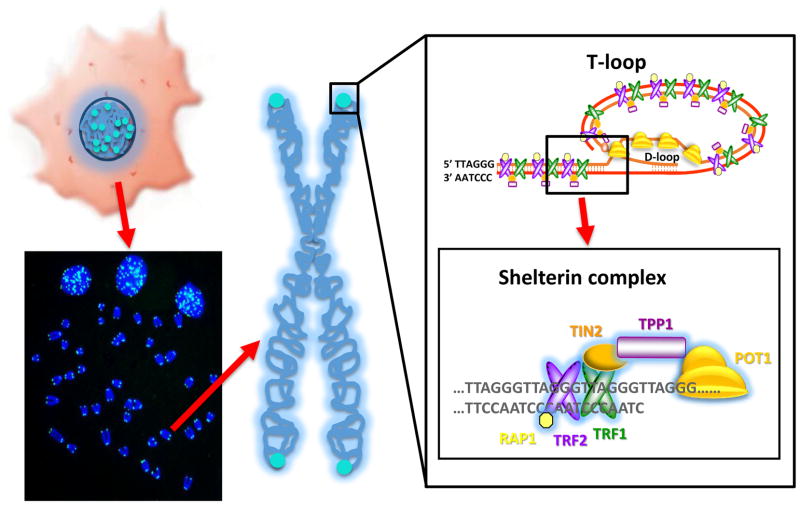
Figure 1.
Telomeres are repetitive DNA sequences at the ends of linear chromosomes. In a normal human cells, there are 46 chromosomes and 92 ends. In total the telomere ends only account for about 1/6000th of the total DNA in a cell. Telomeres can be visualized by using a labeled probe to the TTAGGG repeats and they appear as small fluorescent puncta (in blue) in interphase cells and as signals (blue) at the ends of chromosomes in metaphase cells (left, bottom). An enlargement of the telomere shows the very ends of telomeric DNA consists of double and single stranded TTAGGG repeats ending in a single stranded G-rich overhang. This overhang folds back and forms a T-loop structure (right, top) to hide the ends of the telomeres from being recognized as DNA double strand breaks. In addition, there are a series of proteins that interact directly and indirectly with the telomeric repeats termed the shelterin complex (right, bottom) that also protect the telomeres from DNA repair mechanisms, as well as to regulate telomerase activity access to the telomeres.
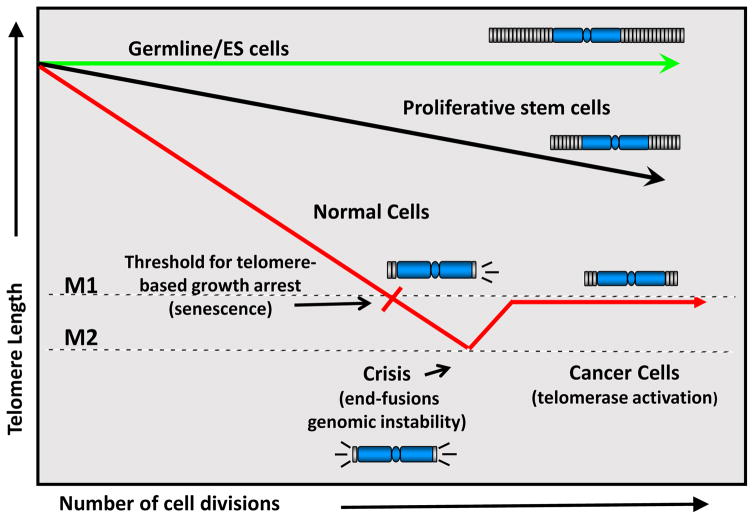
Figure 2.
Germline and embryonic stem (ES) cells retain full or almost full telomere length due to expression of telomerase activity. Pluripotent proliferative stem cells have regulated telomerase activity and thus they lose telomeres throughout life but at a reduced rate. Most somatic cells do not express telomerase activity and thus lose telomere length with each division at a faster rate compared to stem cells until the cells uncap a few of their telomeres and undergo a growth arrest called replicative senescence (or M1, Mortality Stage 1). In the absence of cell cycle checkpoints (e.g. p53/pRB pathway), cells bypass senescence until they reach crisis (or M2, Mortality Stage 2). In crisis, telomeres are so short that chromosome end fusions occur and there is increased genomic instability (probably due to chromosomal, breakage-fusion- bridge cycles). A rare cell that escapes crisis almost universally does so by reactivating telomerase and this cell can now become a cancer cell with limitless potential to divide. When telomerase is activated in crisis, most cancer cells have short telomeres and just the right amount of telomerase to maintain the shortest telomeres. However, in certain types of cancer such as malignant melanoma some invasive tumors have very long telomeres but lack telomerase or an ALT mechanism to maintain the telomeres (as illustrated in Shay, 2017). In this case, it is possible that when cells escape from crisis, there is a large amount of telomerase produced, telomeres grow substantially, and due to telomere looping the TERT locus is again silenced. These tumors can be aggressive since they have very long telomeres but they would still progressively shorten their telomeres with each cell division (Shay, 2017).
Upon somatic cell differentiation, telomerase is downregulated in normal stem cells and in cancer cells (Holt et al, 1996). However, the exact mechanism(s) regulating these switches in expression are not known. Human TERT, the catalytic reverse transcriptase component of telomerase, is tightly regulated by transcriptional, epigenetic and post-transcriptional regulatory mechanisms (Shay, 2016). In cancer, TERT promoter mutations are emerging as one of the most common non-coding mutations in cancer (Huang et al, 2013), and altered pre-mRNA splicing of TERT is another important mechanism of telomerase regulation (Wong et al, 2014). While telomeres are rate-limiting for in vitro cell division of normal human cells (Shay et al, 1991, Wright and Shay, 1992a), introduction of hTERT into most cells is sufficient to elongate telomeres permitting greatly extended lifespan in vitro (Bodnar et al, 1998). These experiments (Bodnar et al, 1998) directly demonstrate a cause and effect relationship between telomere shortening and cellular senescence. Overall, telomerase is carefully regulated in humans and other large long-lived mammals where anti-cancer protection mechanisms must have evolved.
Telomeres are fully maintained by telomerase in human embryonic stem (ES) cells (Figure 2) and at least partially maintained by telomerase in transit amplifying proliferative somatic stem cells in the adult (Shay, 2016) (Figure 2). Telomerase is a multi-component enzyme complex that adds telomeric repeats to the ends of chromosomes (Greider and Blackburn, 1985). Telomeres are important in protecting linear chromosome ends from being recognized as DNA double-stranded breaks by forming special telomeric structures called T-loops (Griffith et al, 1999) (Figure 1). In addition, telomeres recruit shelterin complexes (de Lange et al, 2005) for maintaining genome integrity (Figure 1). Thus, the regulation of telomere length and the spatio-temporal expression of telomerase are tightly regulated in human cells. Regulated telomerase expression in normal human replicating stem-like cells is believed to be important in the maintenance of tissue functions by slowing down or counteracting telomere losses in highly proliferative cells, such as in the bone marrow, intestine and skin.
Telomere Position Effect (TPE)
In Saccharomyces cerevisiae, telomere position effect (TPE) results in the silencing of genes near telomeres by a mechanism that depends both on telomere length and on the distance to the gene (Gottschling et al, 1990, Kueng et al, 2013). TPE provides a mechanism to alter cell and tissue function in telomerase silent tissues with increasing cellular age (Wright and Shay, 1992b) without necessarily initiating a DNA damage response. The existence of TPE in mammalian cells (Baur et al, 2001) raises the possibility that some changes in gene expression could be “programmed” by the progressive shortening of telomeres with ongoing cell divisions, leading to incremental altered patterns of gene expression that might affect both cell and organ function. For TPE to communicate to neighboring genes, it has previously been thought that the telomere and gene needed to be physically close so as telomeres shortened heterochromatin changes would progressively occur (Figure 3). We term this classic TPE for telomere length dependent position effects in genes close (in the subtelomeric region) to a telomere. In classic TPE, the degree of repression declines with distance from the telomere. We previously reported ISG15, a gene ~1Mb from the chromosome 1p telomere, was regulated by what we thought of as classic TPE in vitro and in vivo (Lou et al, 2009). However, we later discovered that there were genes between the telomere and ISG15 that were not regulated by telomere length. Recently, we have identified a series of additional genes regulated by telomere length over long distances (Robin et al, 2014 and 2015, Kim et al, 2016). To distinguish this phenomenon from classic TPE (Figure 3) we refer to it as telomere looping or TPE-OLD (telomere position effect over long distances) (Figure 3). Using a combination of 3D FISH and protein expression levels using cells with long and short telomeres, we have demonstrated (Figure 3) that ISG15 is silenced in young cells and young individuals with long telomeres and is progressively expressed in old cells and older individuals with short telomeres (Lou et al, 2009). Importantly, we demonstrated that re-elongating telomeres in old cells resulted in silencing of ISG15 demonstrating that this regulation was not due to DNA damage signaling from a too short telomere (Lou et al, 2009). Thus, TPE-OLD could affect gene regulation at remote sites far from a telomere many megabases distal to a telomere.
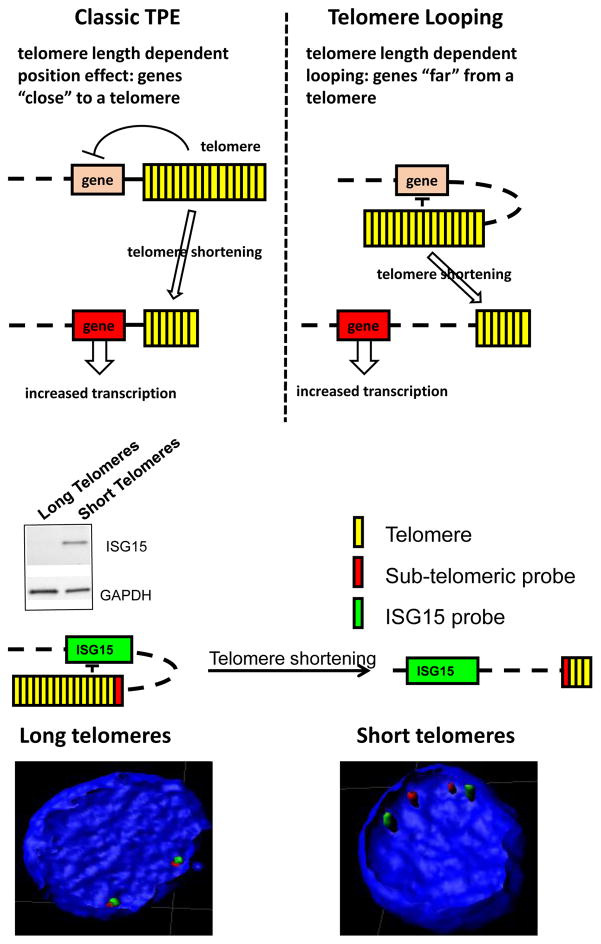
Figure 3.
Comparison of classic telomere position effects (TPE) to telomere looping. TPE (top). In the budding yeast and in mammalian cells TPE is primarily due to deacetylation of subtelomeric nucleosomes. In humans, DUX4, C1S and SORBS2, ISG15, and hTERT expression, some of which are located up to 10 Mb from telomeres can also depend on TPE but by a slightly different mechanism. This novel mechanism involves physical association of the telomere through looping to chromatin (often internal telomere sequences) near these genes and is called TPE-OLD (over long distances). TPE-OLD genes are generally repressed in cells with long telomeres and become expressed in cells with shorter telomeres. Importantly, there are genes between the telomeres and the TPE-OLD regulated genes that are not regulated by telomere length. One example is illustrated for the ISG15 gene (bottom). In young cells with long telomeres ISG15 is repressed but in older cells with short telomere ISG15 is expressed. Increasing telomeres length by introduction of hTERT into old cells with short telomere repressed ISG15 expression but only after substantial telomere re-elongation. In addition to protein expression changes, a subtelomeric probe next to the canonical TTAGGG repeats at the chromosome end are adjacent to the ISG15 locus in young cells with long telomeres while separated in old cells with short telomeres.
3D Genome Organization
How chromosome looping regulates gene expression over long distances in normal development and aging as well as in diseases such as cancer remains a central question in biology. Models have been presented to suggest that the spatial organization of the 3D genome is a key mechanism to control gene expression via regulatory elements that are only at the earliest stages of being understood (Dekker et al, 2013). While some of the details are starting to emerge about long-range chromosomal loops that bring regulatory elements in close proximity with genes they control (Dekker and Mirny, 2016), much remains to be discovered. Using a variety of techniques including methods to detect chromatin interactions, such as chromosome conformation capture (3C) and Hi-C (that combines 3C with deep sequencing), it has been suggested that chromosome looping occurs in discrete blocks or chromosome territories called topologically associating domains (TADs) where most looping interactions occur locally (Dekker et, 2013). The cohesin ring-shaped protein complex and the co-localization of cohesins with CTCF (CCCTC DNA binding factor) boundary proteins may be an almost universal mechanism of limiting looping extrusion (Canela et al, 2017). However, this does not explain how cohesins are recruited to specific sites or how specific genes are regulated in a tissue-restricted manner. In addition, the inner nuclear lamina associates with large chromatin domains that appear to establish interphase chromosome regions that may be important in regulating the silencing of large blocks of genes.
Similar to chromosome looping, telomeres (~1/6000th of the genome) can also make looping structures adjacent to internal loci and regulate gene expression (Robin et al, 2014, 2015; Kim et al, 2016). This mechanism may partially explain the relationship between progressive shortening of telomeres and changes in gene expression. Chromatin modifications in the hTERT genomic locus region may also be associated with CTCF-binding sites (Renaud et al, 2007) since knockdown of CTCF can lead to changes in telomere looping in the hTERT locus (Kim et al, 2016). Some of these telomere loops may be associated with the underlying nuclear membrane through interactions with lamin A/C which directly interacts with a shelterin protein, TRF2 both at the nuclear lamina and throughout the nucleoplasm (Woods et al, 2014, 2015). In other cases telomeres may interact with interstitial genomic sites in a cell-type specific manner that are termed interstitial telomere loops (ITLs) but this may or may not be related to progressive telomere shortening (Woods et al, 2014, 2015). The concept that progressive telomere shortening with increased age can regulate changes in gene expression at genomic sites far removed from a telomere is a novel mechanism that we have termed TPE-OLD (telomere position effect over long distances). We show that stable chromosomal loop formation occurs during development and is altered by progressive telomere shortening. In addition, we show that dynamic telomere loops may engage telomeres with distal chromosome regions to regulate gene expression in proliferating stem-like cells in a temporal manner (see results). The regulation of transit amplifying telomerase positive cells with signals to differentiate and silence telomerase may also be regulated in part by telomere looping. This may have evolved to limit the amount of expressed telomerase in long-lived and large mammals to prevent the early onset of cancer. In addition, in diseases associated with premature aging such as progeria, changes in telomere loops coincides with telomere changes (Woods et al, 2014)
Regulation of the rate limiting telomerase catalytic hTERT protein
Recently, we demonstrated that expression of the human TERT gene (only 1.2 Mb from the end of chromosome 5p) is in part regulated by the length of the telomere (Kim et al, 2016). We demonstrated that normal young human cells with long telomeres have a repressed hTERT epigenetic states (chromatin and DNA methylation) but the epigenetic state is altered when telomeres become short. The change in epigenetic status correlates with altered expression of hTERT and genes nearby hTERT, including the CLPTM1L gene, indicating a change in chromatin structure near the hTERT locus. Further, we identified a telomere loop by droplet digital chromatin conformation capture (dd3C) to a region near hTERT in cells with long telomeres that is disengaged when telomeres shorten. This region is ~100 kb from hTERT and contains a series of interstitial TTAGGG repeats that may recruit TRF2 or TERRA to form a stable interaction in cells with long telomeres. Finally, using siRNA experiments and ChIP-qPCR analysis we obtained support for a role of the TRF2 protein, and TERRA, in the telomere looping maintenance mechanism through interactions with interstitial TTAGGG repeats adjacent to hTERT (Kim et al, 2016).
We also find that when telomeres are long the human (h) TERT gene expression is generally repressed and the telomere and hTERT locus are spatially co-localized. When telomeres are short at least one of the hTERT alleles is spatially separated from the telomere, developing more active histone marks and changes in DNA methylation in the TERT promoter region. These findings have implications for how human cells and tissues turn off telomerase when telomeres are long during human fetal development and how cancer cells reactivate telomerase in cells that have short telomeres (Shay, 2016; Shay and Wright, 2000). In mice the TERT locus is not close to a telomeres and adult mice express telomerase in many tissues. These studies add growing support for how telomeres regulate gene expression in humans via TPE-OLD and provide new insights into how the changes in genome structure during replicative aging result in an increased susceptibility to aged-related diseases and cancer in some cases prior to the initiation of a DNA damage signal.
2. Material and Methods
Purification of CD4+ T lymphocytes from human PBMC
Human peripheral blood monocyte cells (PBMC) were acquired from volunteered individuals in University of Texas Southwestern Medical Center at Dallas under informed consent. All the human samples were handled under the University’s guidelines, and this study was pre-approved by the institutional review board. BD IMag™ Human CD4 T Lymphocyte Enrichment Set- DM (BD biosciences, San Jose, CA, USA) was used to selectively purify CD4+ T lymphocytes by following the manufacturer’s instructions.
Cell culture
Purified CD4+ T lymphocytes were cultivated in RPMI-1640 (Invitrogen, Waltham MA, USA) containing 10% FBS, 10mM HEPES, and 2mM L-glutamine. HL60 cells were cultivated in RPMI-1640 containing 10% FBS. Cells were incubated at 37°C with 5% of CO2. For stimulation, purified CD4+ T lymphocytes were transferred to anti-CD3ε antibody (Fisher, Waltham, MA, USA)-coated dishes for 2 hours at 37°C. Antibody-containing solution was changed with fresh culture medium containing 2μg/ml of anti-CD28 antibody (Fisher, Waltham, MA, USA). Differentiation of HL60 cells was performed by adding 1.5% of DMSO into fresh medium. After an additional 24 to 72 hours of cultivation, cells were harvested for further analyses.
3D-FISH
Slides were washed with phosphate buffered saline (PBS) three times before cell harvest. 5.0 × 105 purified CD4+ T lymphocytes were prepared using a cytospin centrifuge at 2,000 rpm for 3 minutes. Cells were immediately incubated with 0.3X PBS for 3 minutes. Next, cells were fixed with 4% paraformaldehyde/0.3X PBS for 10 minutes at room temperature. For all remaining steps, we followed our previously described protocol for 3D-FISH (Robin et al, 2014).
Gel-based TRAP and droplet digital TRAP (ddTRAP)
Gel-based TRAP was performed as previously described (Mender et al, 2015). ddPCR was performed by following the manufacturer’s instructions (QX200 Droplet Digital PCR System by Bio-Rad). For ddTRAP analysis, harvested cells were lysed in NP-40 lysis buffer containing 1mM Tris-HCl (pH 8.0), 1mM MgCl2, 1mM EDTA, 1% NP40, 0.25mM sodium deoxycholate, 10% glycerol, 0.15M NaCl, 0.1mM AEBSF, and 5mM β-mercaptoethanol. Telomerase substrate extension and PCR amplification were performed as described previously (Ludlow et al, 2014).
3. Results
Telomere looping (TL) in stem cells
There is a lack of mechanistic understanding of telomerase regulation in transit-amplifying stem cells including more differentiated lymphocytes when activated. T lymphocytes play important roles in the immune system, and stimulation of T lymphocytes through the T cell receptor triggers transient proliferation. Cell proliferation is achieved during several days to generate significant numbers of T lymphocytes for immune responses. It is well established that the enzymatic activity of telomerase is elevated during T lymphocyte stimulation (Hiyama et al, 1995; Weng, 1997; Huang et al, 2017) but even with continued stimulation telomerase is reduced significantly even though some cell division continues. The role and mechanism of this transient telomerase activation has not been elucidated. Thus, we tested if mitogenic signaling induces genome structural changes that would correlate with activation of telomerase activity.
We first tested resting T lymphocytes and, as expected, they do not express telomerase activity (Figure 4, top left). By day 3–4 post-stimulation robust telomerase was detected (Figure 4, top left (ddTRAP) and right (gel-based TRAP) and mostly correlated with increased cell proliferation. We also examined transcription of hTERT. Resting CD4+ T lymphocytes express non-catalytic splice variants of hTERT (data not shown). However, we detected increased levels of full length catalytically active hTERT transcripts, in the activated CD4+ T lymphocytes 4 days post-stimulation that correlated well with telomerase activity (data not shown).
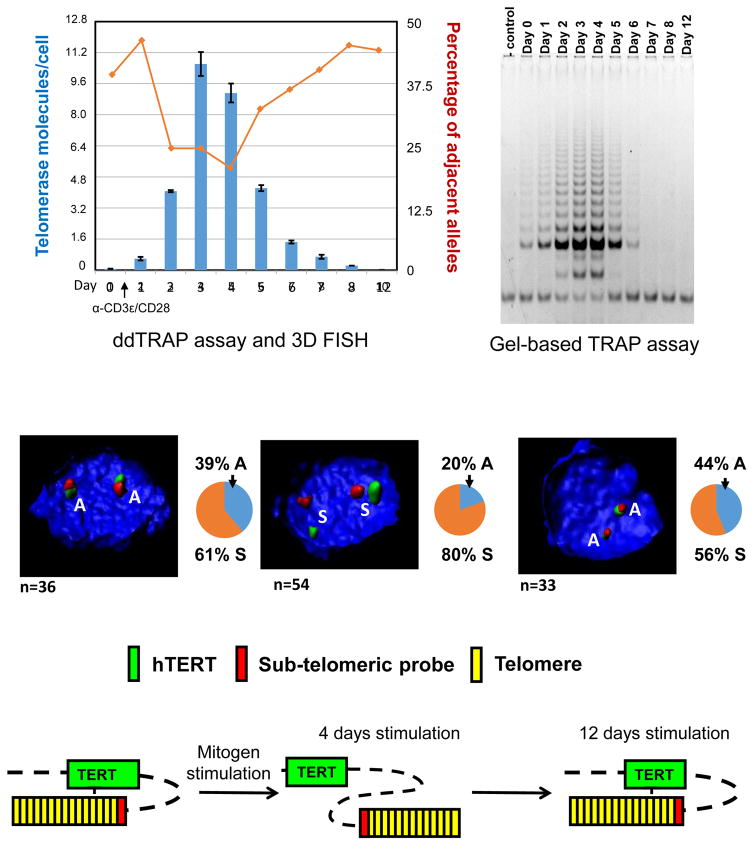
Figure 4.
Changes in the number of telomerase molecules per cell (blue bars) and percentage of adjacent alleles during CD4+ T lymphocytes stimulation (red line) plotted on the same graph. The red line shows changes in the percentage of adjacent alleles between the hTERT locus and a subtelomere 5p probe at various time points after CD4+ T lymphocytes stimulation. The blue bars represent the number of active telomerase molecules per cell during CD4+ T lymphocytes stimulation, which is determined by ddTRAP. Comparison of ddTRAP to the gel based TRAP (top, right) shows telomerase activity is the highest at 3–4 days after mitogenic stimulation in primary human CD4+ T lymphocytes that decreases over the next week of continuous stimulation. DNA probes against hTERT locus (green) and subtelomeric 5p region (red) are depicted to show physical proximity using 3D-FISH analyses. Representative images show the proximity of two loci in response to mitogenic signaling. Control primary human CD4+ T lymphocytes shows more adjacent (A) alleles while CD3ε/CD28-activated T lymphocyte shows more separated (S) alleles when telomerase activity is at peak levels. However, when cells slow-down in proliferation, even in the presence of continued mitogen stimulation, the number of adjacent alleles increased as telomerase decreases. DNA was counterstained with DAPI. One model of telomere looping (TL) is depicted (bottom).
We next examined if the genome configuration changed during CD4+ T lymphocyte proliferation by three-dimensional (3D) fluorescence in situ hybridization (3D-FISH). Anti-CD3ε and CD28 antibodies were used to activate primary human T lymphocytes. At various time points we performed 3D-FISH to analyze genomic distances between the hTERT locus and the 5p subtelomeric region in resting T-cells, at day 4 after stimulation, and 12 days after stimulation (Figure 4). Human resting CD4+ T lymphocytes showed ~39% of allele pairs between the hTERT locus and the 5p subtelomere were adjacent (A). However, 4 days after stimulation the percent of adjacent allele pairs was decreased to 20% while separated (S) alleles increased to 80% (Figure 4, middle), followed by rapid return to the resting level by day 12 even with continued mitogen stimulation (Figure 4, middle, right side).
These results can be interpreted to imply that the genomic environment at the hTERT locus has two different stages in terms of telomere looping. We term this telomere looping (TL) to help explain transient telomerase expression in T cells (and perhaps other transit amplifying stem like cells such as in the skin and the GI track), since this type of telomerase regulation does not appear to be influenced by telomere length (Figure 4, bottom). Taken together, the results show that telomere looping near the hTERT locus occurs regardless of telomere length changes. We have recently shown using standard TRF analyses that there is no detectable telomere length changes during 10 days of mitogen stimulation of T-cells (Huang et al, 2017). The looping change is induced by mitogenic signals and returns back to a resting stage even in the presence of additional mitogen stimulation. We also showed that there were no significant changes in nuclear volume to explain the changes in distances between the subtelomeric probe on chromosome 5p and the TERT locus about 1.2 Mb away. Since it has been demonstrated that some telomeres can associate via lamin A/C and the nuclear lamina (inner nuclear envelop membrane) via TRF2 interactions in stable heterochromatic clusters, it is possible that in resting T cells the TERT locus is physically associated with the nuclear lamina. However, when T-cells are stimulated this may signal chromatin modifications permitting the TERT locus to separate from heterochromatin clusters next to the nuclear envelope and increase TERT mRNA production.
Telomerase regulation during telomerase positive cancer cell-induced differentiation
Telomerase is known to be cell cycle regulated (Holt et al, 1996). However, the mechanisms silencing telomerase in quiescent or differentiated cancer cells is not well understood. There are E-boxes in the telomerase proximal promoter so a cell cycle early response regulated gene, such as c-Myc, could be involved. We used the promyelocytic human leukemia cell line, HL60 to study myeloid cell proliferation and differentiation (Birnie, 1988). The HL60 cells differentiate into neutrophil-like cells with various stimuli such as DMSO (Xu et al, 1999).
It has been reported that telomerase activity in the HL60 model is down-regulated upon various differentiation cues and that the decline of telomerase activity is closely associated with down-regulation of transcription of hTERT (Sharma, et al, 1995). In the present studies we also demonstrated that proliferating HL60 cells express robust telomerase activity (Figure 5, left). However, upon differentiation in the presence of DMSO (Figure 5, left) we observed rapid decreases in hTERT transcription and telomerase activity. Based on the T-lymphocyte experiments we next tested if telomere looping changes occurred upon differentiation of HL60 cells to regulate transcription of hTERT.
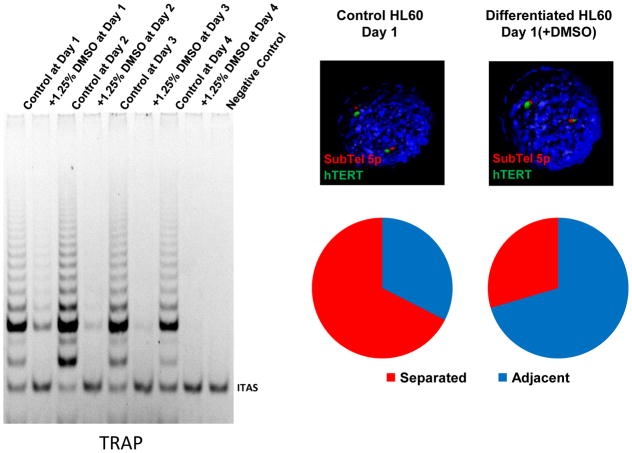
Figure 5.
HL60 human promyelocytic cancer cells are telomerase positive but upon terminal differentiation with DMSO treatment telomerase becomes rapidly silenced. As seen in the gel based TRAP assay within 24 hours of DMSO treatment almost all telomerase activity is gone. HL60 cells also show telomere looping changes between the 5p subtelomere probe (red) and the hTERT locus (green) during DMSO induced differentiation. Representative pictures show mostly separated alleles in control HL60 cells (open configuration) and mostly adjacent alleles in differentiated HL60 cells (closed configuration). The hTERT locus and the subtelomere 5p probe were stained with green and red fluorescent dyes respectively. Blue and red on the pie charts (right, bottom) represent adjacent and separated alleles respectively on control and DMSO-treated cells. 3D-FISH was performed to analyze the proximity between the hTERT locus and the subtelomere 5p probe in control and differentiated HL60 cells. Each allele pair was divided into adjacent or separated. Control HL60 cells showed 67% of separated alleles between the hTERT locus and the subtelomeric 5p probe while differentiated cells show only 30% separated alleles. Volume of the cells did not change upon differentiation.
We analyzed the proximity between the hTERT locus and subtelomere 5p locus by performing 3D-FISH to measure the distance between two loci. The images obtained sorted into two different forms; adjacent alleles (average 0.4μm apart or less) and separated alleles (average 0.85μm apart or greater). Both control and DMSO-treated HL60 cells had the same genomic distance between two loci in each adjacent and separated allele (Fig. 5), so that changes in genomic configuration assessed by 3D-FISH did not result from global nuclear shrinkage or enlargement. Figure 5 (right side) shows proliferating HL60 cells have 67% separated alleles which implies active transcription of hTERT based on our previous studies (Robin et al, 2014). However, differentiation of HL60 cells with treatment of DMSO significantly shifted the proximity between hTERT locus and subtelomere 5p region. For example, 24 hours after DMSO treatment a complete reversal occurred such that there were 70% adjacent alleles while control proliferating HL60 cells showed 33% of allele pairs adjacent.
4. Discussion
TPE-OLD is a new and potentially important epigenetic mechanism during human aging, and in some but not all cases, its role may be to suppress expression of the target locus when the cell is young and the cells have long telomeres (Kim et al, 2016; Robin et al, 2014, 2015). In the current studies we showed that telomere looping (TL) mechanism is also important in lymphocytes activation and cancer cell differentiation that is unrelated to the length of the telomere. The results imply that the change in telomere looping is not only a passive outcome of telomere attrition during aging, but also an active mechanism that participates in the regulation of target genes during various physiological responses including proliferation and differentiation. Both TPE-OLD and TL are not mutually exclusive and may be important regulatory mechanisms since the transcriptome can be changed without initiating a DNA damage response such as when telomeres become critically shortened.
Nuclear Lamina and Internal Telomere Sequences (ITS)
The nuclear lamina has many functions, including chromatin organization, and there is emerging evidence for a link between lamin A/C, a component of the nuclear lamina, and telomere stability (Wood et al, 2014, 2015; Burla, 2016). TRF2, a shelterin protein involved in telomere protection, has been reported to interact with lamin A/C both at the nuclear lamina but also throughout the nucleoplasm (Wood et al, 2014). In addition, there appears to be a functional interaction between lamin A/C and TRF2 at tracks of interstitial telomere sequences (ITS) suggesting that a molecular interaction between TRF2 and lamin A/C could facilitate telomere associations with heterochromatin looping over long distances (Wood et al, 2015). To test for this interaction, co-immunoprecipitation analysis with endogenous TRF2 and lamin A/C was undertaken (Wood et al, 2014). Immunoprecipitation of TRF2 revealed pull down of a GFP-tagged lamin A/C (Wood et al, 2014). The lamin A/C interaction with telomeres and possibly the TERT locus could provide one explanation for telomere looping in T cell activation. For example, upon lymphocyte stimulation TRF2 may dissociate from ITS near the TERT locus resulting in TERT transcriptional upregulation but after a period of time and multiple rounds of cell proliferation, as cell proliferation decreases, TRF2 may re-associate with ITS near the TERT locus again silencing telomerase. In addition, TRF2 is found at promoters of some genes (Simonet, et al, 2011; El Mai et al, 2014; Martinez and Blasco, 2011) and a loss of TRF2 association is observed in cells with shortened telomeres (Kim et al, 2016). These results parallel our work, demonstrating that TRF2 may be necessary to facilitate long-range telomere looping (Kim et al, 2016). Further analysis is necessary to determine whether TRF2 has a direct role in the regulation of gene expression at these promoters or if its role is indirect through the regulation of telomere looping perhaps mediated by TERRA/TRF2 interactions (Deng et al, 2009; Maicher et al, 2012). Previous investigations have reported other shelterin associated proteins such as RAP1 upon DNA damage responses at telomeres may relocate and contribute to global chromatin-mediated gene expression changes and may be related to senescence in yeast with a deficiency in telomerase (Platt et al, 2013). One possible model is that with progressive telomere shortening Rap1 and perhaps TRF2 may be liberated from the telomeres and relocate to new target loci near ITS. Since ITS often co-localize with fragile chromosome sites and may be hotspots for deletions, breakage, recombination, rearrangements, fusions, and amplifications (Bozan, 2017), re-localization of shelterin proteins in cells with short telomeres may be another mechanism to reduce genomic instability (Bosco and de Lange, 2012) as an additional anti-cancer protection mechanism.
In addition, both TRF1 and TRF2 have been reported to bind to ITS repeats (Simonet et al, 2011). Using chromatin immunoprecipitation and DNA sequencing, not only did TRF1/2 bind to ITS but also to non-ITS sites such as centromeric and pericentromeric satellite DNA (Simonet et al, 2011). Interestingly, TRF1/2 non-ITS binding sites were frequently located within or less than 100kb from coding sequences (genic) regions. This is consistent with long-range interactions between telomeric repeats, shelterin proteins and a telomere looping mechanism such as TPE-OLD or telomere looping (TL) that may influence the chromatin landscape and in turn the transcriptome. In the case of the hTERT gene, there are a cluster of ITS ~100kb from the TERT promoter but this is on a linear map. However, chromosome/genomic folding at this region can easily be modeled to position the TERT promoter close to the ITS region (Kim et al, 2016). While we are still in the early stages in understanding long-range chromosome organization, there are some mechanisms that could be proposed for future testing. For example, TERRA (telomeric repeat-containing RNA) and TRF2 have been shown to interact and facilitate heterochromatin formation (Cusanelli, 2015; Deng et al, 2009) and may be important in some human diseases (Maicher, et al, 2012). In addition, TERRA and TRF2 bind to intramolecular G-quadruplex structures (Martadinata et al, 2011) and longer stretches of ITS may form secondary structures such as G-quadruplexes that may promote recombination events at these fragile sites. While TERRA and TRF2 interact with telomere canonical ends, it is also possible that TERRA/TRF2 interactions may also bind to ITS repeats which are known G-quadruplex structures as well as non-ITS G-quadruplexes such as in the CDKN1A (p21) and the c-MYC promoter. Finally, telomere length may determine TERRA and R-loop regulation in a cell cycle manner (Graf et al, 2017).
TRF2 protein binding to some interstitial sites suggests these sites may be involved in regulating gene transcription (Yang et al. 2011). ITSs, however, do not shorten during DNA replication or believed to contribute to senescent cell changes. Estimates of the average percent of ITS within a species is from 15–45% and there is also intra-variation between individuals within a species (Bolzan, 2017). Combining chromatin immunoprecipitation with high-throughput sequencing to map chromosomal sites of TRF1 and TRF2 binding, it was shown that these shelterin proteins can bind to extra telomeric regions associated with ITS sites (Simonet et al, 2011). Thus, one model that is consistent with the TPE-OLD results are that as telomeres shorten shelterin proteins such as TRF2 may be released from the true telomeric repeats at the end of the chromosomes and bind to ITS sites to modify cellular transcription. Taken together, we suggest that both progressive telomere shortening induced changes in telomere looping (TPE-OLD) and telomere looping (TL) that may also be associated in some instances with changes in telomere length are both closely associated with changes in chromatin modifications and genomic configurations at the hTERT and other loci. A more thorough understanding of this epigenetic and functional regulation of gene expression during age associated and transient telomerase activation will enable us to better elucidate the mechanisms involved in the in vivo regulation of telomerase.
Highlights.
Telomere shortening in normal cells alters telomere looping over long distances
Telomeres interacts with internal telomeric sequences (ITS) via TRF2
Telomere looping regulates gene expression at distal genomic sites
The human TERT gene regulates itself by telomere looping and telomere length
Activated lymphocytes regulate telomerase expression via telomere looping
Acknowledgments
Acknowledgements/Funding: This work was supported by AG01228 from the National Institute on Aging (JWS), NCI SPORE P50CA70907 (JWS), the Harold Simmons NCI Designated Comprehensive Cancer Center Support Grant (CA142543), the CPRIT Training Grant RP140110, and the Southland Financial Corporation Distinguished Chair in Geriatric Research (JWS. and WEW). This work was performed in laboratories constructed with support from NIH grant C06 RR30414.
Footnotes
Author Contributions: WK conducted the experimental results. JWS wrote the original draft. WK edited. Both authors provided substantial contributions, design, acquisition of data, or data analysis and interpretation. Both authors gave final approval of the submitted version. Conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baur JA, Zou Y, Shay JW, et al. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- Birnie GD. The HL60 cell line: a model system for studying human myeloid cell differentiation. Br J Cancer Suppl. 1988;9:41–45. [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Bolzan AD. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability, and evolution. Mutation Res. 2017;773:51–65. doi: 10.1016/j.mrrev.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Bosco N, de Lange T. A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma. 2012;121(5):465–474. doi: 10.1007/s00412-012-0377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burla R, LaTorre M, Saggio I. Mammalian telomeres and their partnership with lamins. Nucleus. 2016;7(2):187–202. doi: 10.1080/19491034.2016.1179409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S. Genome organization drives chromosome fragility. Cell. 2017;170:507–521. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet. 2015;6:143–149. doi: 10.3389/fgene.2015.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14(6):390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1120. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, et al. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35(4):403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mai M, Wagner KD, Michiels JF, et al. The telomeric protein TRF2 regulates angiogenesis by binding and activating the PDGFRβ promoter. Cell Rep. 2014;9(3):1047–1060. doi: 10.1016/j.celrep.2014.09.038. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, et al. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Graf M, Bonetti D, Lockhart A, et al. Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell. 2017;170:72–85. doi: 10.1016/j.cell.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher BA, Greider CW. Telomeres shorten during aging of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Dempster M, Dunlop MG, et al. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- Huang E, Tedone E, O’Hara R, et al. The maintenance of telomere length in CD28+ T cells during lymphocyte stimulation. Scientific Reports. 2017:7. doi: 10.1038/s41598-017-05174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama K, Hirai Y, Kyoizumi S, et al. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J Immunol. 1995;155(8):3711–3715. [PubMed] [Google Scholar]
- Holohan B, Wright WE, Shay JW. Impaired telomere maintenance spectrum disorders. J Cell Biology. 2014;205(3):289–299. doi: 10.1083/jcb.201401012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SE, Wright WE, Shay JW. Regulation of telomerase activity in immortal cell lines Mol. Cell Biol. 1996;16:2932–2939. doi: 10.1128/mcb.16.6.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kim W, Ludlow A, Min J, et al. Regulation of the human telomerase gene TERT by telomere position effect-over long distances (TPE-OLD): Implications for aging and cancer. PLoS Biol. 2016;14(12):e2000016. doi: 10.1371/journal.pbio.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Oppikofer M, Gasser SM. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- Lindsey J, McGill NI, Lindsey LA, et al. In vivo loss of telomeric repeats with age in humans. Mut Res. 1991;256:45–48. doi: 10.1016/0921-8734(91)90032-7. [DOI] [PubMed] [Google Scholar]
- Lou Z, Jun W, Riethman H, et al. Endogenous genes near telomeres regulated by telomere length in human cells. Aging. 2009;1:608–621. doi: 10.18632/aging.100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow AT, Robin JD, Sayed M, et al. Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic Acids Res. 2014;42(13):e104. doi: 10.1093/nar/gku439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicher A, Kastner L, Luke B. Telomeres and disease - enter TERRA. RNA Biology. 2012;9(6):843–849. doi: 10.4161/rna.20330. [DOI] [PubMed] [Google Scholar]
- Martadinata H, Heddit B, Lim KW, et al. Structure of long human telomeric RNA (TERRA): G-quadruplexe formed by four and eight UUAGGG repeats are stable building blocks. Biochemistry. 2011;50(29):6455–6461. doi: 10.1021/bi200569f. [DOI] [PubMed] [Google Scholar]
- Martínez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- Mender I, Shay JW. Telomerase Repeated Amplification Protocol (TRAP) Bio-protocol. 2015 Nov 20;5(22):e1657. doi: 10.21769/bioprotoc.1657. pii: e1657. http://www.bioprotocol.org/e1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt JM, Ryvkin O, Wanat JJ, et al. Rap1 relocalization contributes to the chromatin mediated gene expression profile and pace of cell senescence. Genes Dev. 2013;27:1406–1420. doi: 10.1101/gad.218776.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S, Loukinov D, Abdullaev Z, et al. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35:1245–1256. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin JD, Ludlow AT, Chen M, et al. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014;28(22):2464–2476. doi: 10.1101/gad.251041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin JD, Ludlow AT, Batten K, et al. SORBS2 transcription is activated by telomere position effect-over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 2015;25(12):1781–1790. doi: 10.1101/gr.190660.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma HW, Sokoloski JA, Perez JR, et al. Differentiation of immortal cells inhibits telomerase activity. Proc Natl Acad Sci U S A. 1995;92(26):12343–12346. doi: 10.1073/pnas.92.26.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE, Werbin H. Defining the molecular mechanism of human cell immortalization. Biochim Biophys Acta. 1991;1072:1–7. doi: 10.1016/0304-419x(91)90003-4. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Implications of mapping the human telomerase genes (hTERT) as the most distal gene on chromosome 5p. Neoplasia. 2000;2:195–196. doi: 10.1038/sj.neo.7900093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6(6):584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW. New insights into melanoma development (perspective) Science. 2017;357:1358–1359. doi: 10.1126/science.aao6963. [DOI] [PubMed] [Google Scholar]
- Simonet T, Laurew-Emanuelle Z, Philippe C, et al. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Research. 2011;21:1028–1038. doi: 10.1038/cr.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Zglinicki T, Saretzki G, Ladhoff J, et al. Human cell senescence as a DNA damage responses. Mechan Ageing Devel. 2005;26(1):111–117. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Weng NP, Palmer LD, Levine BL, et al. Tales of tails: regulation of telomere length and telomerase activity during lymphocyte development, differentiation, activation, and aging. Immunol Rev. 1997;160:43–54. doi: 10.1111/j.1600-065x.1997.tb01026.x. [DOI] [PubMed] [Google Scholar]
- Wood AM, Laster K, Rice EI, et al. A beginning of the end: new insights into the functional organization of telomeres. Nucleus. 2015;3:172–178. doi: 10.1080/19491034.2015.1048407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Rendtlew Danielsen JM, Lucas CA, et al. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nature Com. 2014;5:5467. doi: 10.1038/ncomms6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MS, Wright WE, Shay JW. Alternative splicing regulation of telomerase: a new paradigm? Trends in Genetics. 2014;10:430–438. doi: 10.1016/j.tig.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp Gerontol. 1992a;27:383–389. doi: 10.1016/0531-5565(92)90069-c. [DOI] [PubMed] [Google Scholar]
- Wright WE, Shay JW. Telomere positional effects and the regulation of cellular senescence. Trends Genet. 1992b;8:193–197. doi: 10.1016/0168-9525(92)90232-s. [DOI] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, et al. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Xu D, Gruber A, Bjorkholm M, et al. Suppression of telomerase reverse transcriptase (hTERT) expression in differentiated HL-60 cells: regulatory mechanisms. Br J Cancer. 1999;80(8):1156–1161. doi: 10.1038/sj.bjc.6690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, et al. Human telomeric proteins occupy selective interstitial sites. Cell Research. 2011;21:1013–1021. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]