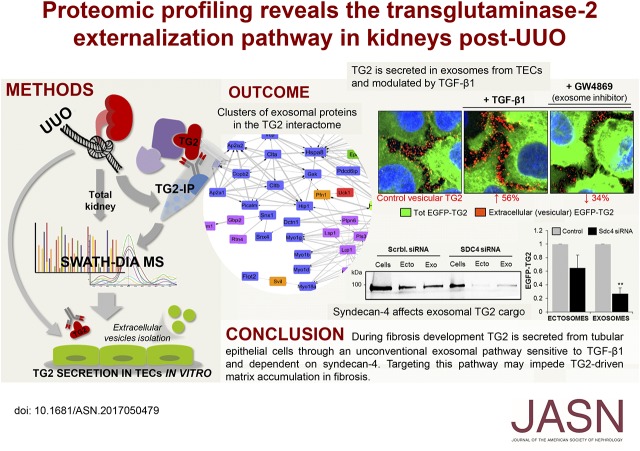
Abstract
Increased export of transglutaminase-2 (TG2) by tubular epithelial cells (TECs) into the surrounding interstitium modifies the extracellular homeostatic balance, leading to fibrotic membrane expansion. Although silencing of extracellular TG2 ameliorates progressive kidney scarring in animal models of CKD, the pathway through which TG2 is secreted from TECs and contributes to disease progression has not been elucidated. In this study, we developed a global proteomic approach to identify binding partners of TG2 responsible for TG2 externalization in kidneys subjected to unilateral ureteric obstruction (UUO) using TG2 knockout kidneys as negative controls. We report a robust and unbiased analysis of the membrane interactome of TG2 in fibrotic kidneys relative to the entire proteome after UUO, detected by SWATH mass spectrometry. The data have been deposited to the ProteomeXchange with identifier PXD008173. Clusters of exosomal proteins in the TG2 interactome supported the hypothesis that TG2 is secreted by extracellular membrane vesicles during fibrosis progression. In established TEC lines, we found TG2 in vesicles of both endosomal (exosomes) and plasma membrane origin (microvesicles/ectosomes), and TGF-β1 stimulated TG2 secretion. Knockout of syndecan-4 (SDC4) greatly impaired TG2 exosomal secretion. TG2 coprecipitated with SDC4 from exosome lysate but not ectosome lysate. Ex vivo, EGFP-tagged TG2 accumulated in globular elements (blebs) protruding/retracting from the plasma membrane of primary cortical TECs, and SDC4 knockout impaired bleb formation, affecting TG2 release. Through this combined in vivo and in vitro approach, we have dissected the pathway through which TG2 is secreted from TECs in CKD.
Keywords: chronic kidney disease, fibrosis, UUO, extracellular vesicles, transglutaminase, interactome
Fibrotic remodeling is the primary pathologic process associated with progressive CKD leading to end stage renal disease.1,2 Increased synthesis and especially, externalization of protein crosslinking enzyme transglutaminase-2 (TG2) by cortical tubular epithelial cells (TECs) into the surrounding interstitium are significant features of progressive kidney scarring.3–5 TG2 is the most widespread member of a family of Ca2+-dependent enzymes, and it is able to catalyze an acyl transfer reaction between peptide-bound glutamine residues and peptide-bound lysine residues, leading to the post-translational modification of proteins through the formation of intra- or intermolecular Nε(γ-glutamyl)lysine bonds. After it is outside the cell, TG2 accelerates the deposition of available extracellular matrix (ECM) substrates and confers ECM resistance to proteases.4 Furthermore, TG2 enhances TGF-β activation,6–8 the prototypical profibrotic cytokine in CKD.9 Inhibition of extracellular TG2 activity10,11 or knockout (KO) of the heparan sulfate (HS) proteoglycan syndecan-4 (SDC4),12 which is responsible for TG2 extracellular trafficking through an undiscovered mechanism,13 ameliorates kidney fibrosis in experimental models of CKD.
Despite a link between externalization of TG2 and tubulointerstitial fibrosis that is well established,10 the pathway through which TG2 is secreted from TECs, a major source of TG2 in CKD,4 has not been fully elucidated, because TG2 lacks the leader peptide for the classic endoplasmic reticulum-Golgi secretory pathway. Preliminary work on TG2 release by TECs has suggested that an intact fibronectin (FN)-binding N-terminal β-sandwich domain is crucial for TG2 secretion but that the extracellular trafficking of TG2 is independent of FN.14 Alternative secretory pathways have been proposed,15 either investigated in transfected NIH3T3 fibroblasts involving TG2 loading into recycling endosomes at the perinuclear recycling compartment, small Rab11 GTPase activity, and binding to endosomal phosphoinositides16 or in transfected HEK293 cells involving opening of a purinergic P2X7 receptor-dependent membrane pore.17 Furthermore, we know that the association of TG2 with the HS proteoglycan SDC4 favors the enzyme secretion in mouse dermal fibroblasts13 and in vivo.12
With the aim of unraveling the mechanism of TG2 secretion in CKD, in this study, we report a comprehensive and unbiased analysis of the membrane interactome of TG2 in kidneys subjected to unilateral ureteric obstruction (UUO). An enrichment of extracellular vesicle (EV) proteins was identified in TG2 membrane complexes. This formed the hypothesis of an EV-dependent secretion for TG2. This hypothesis was tested in established TEC lines and primary TECs. Our findings suggest, for the first time, a pathway through which TG2 is secreted from TECs and reveal the involvement of SDC4 in CKD pathogenesis.
Results
Quantitative Proteomic Approach for the Analysis of TG2 Interactome in UUO Kidneys
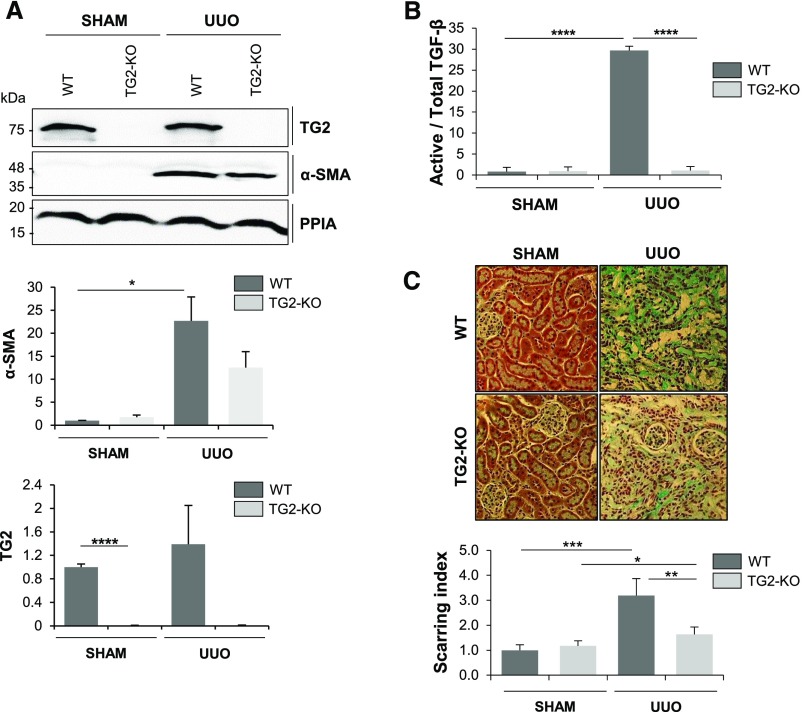
Induction of renal fibrosis was performed in wild type (WT) and TG2-KO animals. TG2−/− and TG2+/+ inbred C57BL/6J mice were subjected to UUO of the left kidney or a sham operation, and kidneys were harvested at 21 days postsurgery. WT UUO kidneys were positive to α-smooth muscle actin (Figure 1A), displayed a significantly higher level of active TGF-β (mink lung epithelial cell assay) (Figure 1B), and showed increased collagen deposition in the interstitium and periglomerulus (Figure 1C). Notably, TG2-null UUO kidneys had a lower level of α-smooth muscle actin (Figure 1A) and active TGF-β (Figure 1B). Moreover, TG2-null kidneys displayed a reduced level of collagen deposition as confirmed by image analysis (Figure 1C). These data reveal that TG2 expression is important in fibrosis development post-UUO, because the disease develops more slowly if TG2 is absent from the extracellular environment.
Figure 1.
A UUO model in WT and TG2 KO kidneys shows that absence of TG2 reduces the development of experimental fibrosis. TG2 KO and WT mice were subjected to UUO or a sham operation. (A) Western blot analyses for the expression of TG2, α-SMA, and cyclophilin-A (PPIA; loading control) in the obstructed kidneys (21 days). Data represent mean band intensities normalized to PPIA relative to the WT sham-operated control (equalized to one) ±SEM; n=3 kidney lysates per group. (B) Active and total TGF-β quantified via the mink lung epithelial cell bioassay. Data represent the ratio of active TGF-β to total TGF-β expressed relative to the WT sham-operated control (equalized to one) ±SEM; n=3 kidney lysates per group. (C) Representative micrographs of Masson's trichrome staining; scarring index was determined as the ratio of collagen staining (light blue) over cytoplasmic staining (pink). Data represent mean values relative to the WT sham (equalized to one) ±SEM; n=24 nonoverlapping fields per treatment covering tubular and glomerular areas. Magnification, ×20. *P<0.05; **P<0.01; ***P<0.01; ****P<0.001.
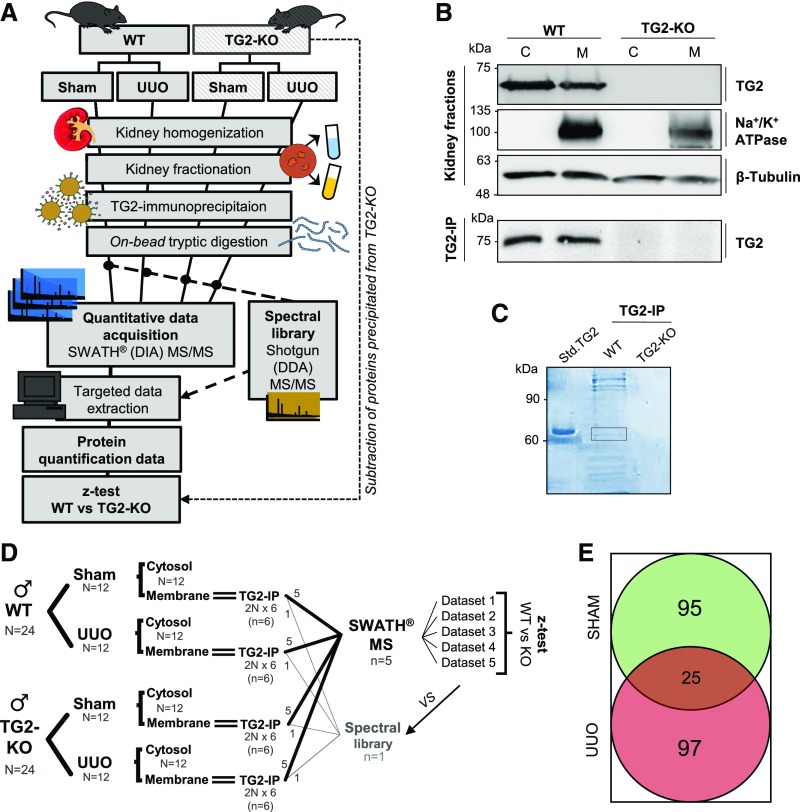
To identify protein partners of TG2 in the murine UUO model of CKD (the “TG2 interactome”), we combined TG2 immunoprecipitation (IP) from whole-kidney membrane preparations of WT and TG2-null kidneys, sham and UUO, with quantitative proteomics by sequential window acquisition of all theoretical fragment ion spectra (SWATH) mass spectrometry (MS).18 The TG2 IP proteome from the TG2 KO was subtracted from the respective TG2 IP WT proteome to reveal only the TG2-dependent interactions. An outline of this original approach is shown in Figure 2A. Cytosol and membrane fractions from WT and TG2 KO kidneys were validated by detection of Na+/K+ ATPase, which was solely displayed by the membrane lysate, and β-tubulin, which was present in both fractions (Figure 2B). TG2-associated complexes were isolated by IP using magnetic beads coated with anti-TG2 antibody. Western blotting confirmed the presence of TG2 in WT fractions (Figure 2B); additionally, SDS-PAGE followed by Coomassie blue staining revealed the presence of TG2-associated proteins in the membrane lysate of WT kidneys immunoprecipitated with anti-TG2 antibody and a very low level of proteins in TG2-null kidneys processed in the same way (Figure 2C). Proteins recruited by the anti-TG2 antibody beads were analyzed by quantitative proteomics. We used SWATH acquisition MS18 to resolve proteomes at the highest possible sensitivity, reproducibility, and proteome coverage. To avoid bias from individual donors and achieve generalizable results, we performed five independent IP experiments, each on the basis of lysates generated from two animal donors, and a sixth IP to build the spectral library as outlined in Figure 2D; we used only male mice to avoid any sex/hormonal bias. Differences between WT and TG2-null precipitated proteins (representing the background) were established by a paired sample z test after analysis by SWATH processing (Figure 2, A and D, Supplemental Material). In this way, we could distinguish TG2-dependent protein recruitment from nonspecific background. However, not all of the TG2-associated proteins were expected to be directly bound to TG2; some molecules could be indirect partners of TG2, bound to its direct interactors. The complete list of 217 proteins directly or indirectly associated with TG2 in UUO (122 proteins) or sham (120 proteins) kidney membranes, which were characterized by either a plasma membrane (PM) or ECM compartmentalization or by a location in biologic fluids, is shown in Tables 1 and 2, respectively (P≤0.05; identified in at least four of five experiments; n≥4). Original processed data with statistical analysis are provided as Supplemental Appendix 1. There was little overlap (11.5%) between cell matrix proteins associated with TG2 in the normal (sham) and fibrotic kidneys (UUO) (Figure 2E). This is not surprising, because TG2 becomes activated in disease both in terms of export4,8 and in terms of enzymatic activity.19,20 There was a considerable cluster of proteins uniquely part of the UUO TG2 interactome (44.7%) or exclusively associated with TG2 in the normal kidneys (43.8%) (Figure 2E). Membrane proteins previously reported to be exclusively located in nucleus and mitochondrial membranes were manually selected from the SWATH MS dataset according to the subcellular localization database “COMPARTMENTS” and UniProtKB (Supplemental Figure 1). Ribosomal proteins and Igs found in the membrane preparation are shown in Supplemental Table 1.
Figure 2.
A comparative proteomic approach for the analysis of TG2 interactome in kidney fibrotic membranes. (A) Workflow of the strategy for the isolation of TG2-interacting proteins. TG2 was immunoprecipitated from kidney membrane fractions obtained from WT and TG2-null kidneys (used as negative controls). TG2-associated proteins were isolated by IP using a monoclonal anti-TG2 antibody (IA12) crosslinked to magnetic beads. The TG2 coimmunoprecipitates (TG2 IP) were proteolytically digested directly on beads and analyzed via SWATH acquisition MS. Five TG2 IP samples per treatment were analyzed by data-independent acquisition (DIA) using 34 static SWATH windows of m/z 15. A spectral library, produced by shotgun/data-dependent acquisition (DDA) MS on a pool of all samples, was used for extracting the SWATH quantitative data. Differences between WT and TG2 KO precipitated proteins (representing the background) were identified by a paired sample z test, leading to the TG2 interactome. (B) Kidney fractions (membrane [M] and cytosolic [C]) were validated by Western blotting for enrichment of membrane marker (Na+/K+ ATPase) or tubulin. TG2 was detected in Western blots of the fractions before and after TG2 IP. (C) Coomassie staining of TG2 immunoprecipitates separated by SDS-PAGE. The rectangular area identifies the precipitated TG2. Std. TG2, purified guinea pig liver TG2. (D) Sample size and animal numbers (N) used in the study; n indicates independent experiments. (E) Number of proteins identified as specifically associated with TG2 in UUO and sham-operated kidney membranes by z test (P≤0.05; n≥4) after exclusion of nuclear, mitochondrial, and ribosomal proteins.
Table 1.
Specific TG2 partner proteins in the UUO kidney (membrane fraction)
| Identification | Name | n | P Value | U/S |
|---|---|---|---|---|
| DESP | Desmoplakin | 5 | 1.22E−13 | U |
| HSP7C | Heat shock cognate 71-kD protein | 5 | 7.67E−10 | U |
| CALM | Calmodulin | 5 | 1.83E−08 | U/S |
| CAZA2 | F-actin–capping protein subunit-α2 | 5 | 1.97E−08 | U |
| GPX1 | Glutathione peroxidase 1 | 5 | 4.52E−07 | U |
| ADH1 | Alcohol dehydrogenase 1 | 5 | 9.40E−07 | U/S |
| POSTN | Periostin | 4 | 1.03E−06 | U |
| MYO1D | Unconventional myosin-Id | 5 | 3.52E−06 | U |
| GLRX1 | Glutaredoxin-1 | 4 | 5.61E−06 | U |
| GLRX3 | Glutaredoxin-3 | 5 | 1.40E−05 | U |
| PROF1 | Profilin-1 | 5 | 1.89E−05 | U |
| TGM2 | Transglutaminase 2 | 5 | 3.31E−05 | U/S |
| HSPB1 | Heat shock protein-β1 | 5 | 3.68E−05 | U |
| VATH | V-type proton ATPase subunit H | 5 | 5.98E−05 | U |
| ADDG | γ-Adducin | 5 | 6.66E−05 | U |
| AT2A2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 5 | 8.58E−05 | U |
| SVIL | Supervillin | 4 | 1.01E−04 | U |
| COR1C | Coronin-1C | 5 | 1.38E−04 | U |
| IQGA1 | Ras GTPase-activating–like protein IQGAP1 | 5 | 1.50E−04 | U/S |
| TCPE | T-complex protein 1 | 5 | 3.17E−04 | U |
| UBP5 | Ubiquitin C-terminal hydrolase 5 | 5 | 4.59E−04 | U |
| FLOT2 | Flotillin-2 | 4 | 5.38E−04 | U |
| NAMPT | Nicotinamide phosphoribosyltransferase | 5 | 6.56E−04 | U/S |
| PCKGC | Phosphoenolpyruvate carboxykinase | 5 | 8.93E−04 | U/S |
| RAB1A | Ras-related protein Rab-1A | 5 | 9.16E−04 | U |
| SDC4 | Syndecan-4 | 5 | 9.34E−04 | U |
| LDHA | l-lactate dehydrogenase A chain | 4 | 1.04E−03 | U |
| MYO1G | Unconventional myosin-Ig | 5 | 1.21E−03 | U |
| K1C20 | Keratin, type I cytoskeletal 20 | 4 | 1.35E−03 | U |
| COEA1 | Collagen-α1(XIV) chain | 5 | 1.64E−03 | U |
| COCA1 | Collagen-α1(XII) chain | 4 | 1.68E−03 | U |
| HIP1 | Huntingtin-interacting protein 1 | 4 | 1.86E−03 | U |
| LIMS1 | LIM and senescent cell antigen-like–containing domain protein 1 | 5 | 2.47E−03 | U/S |
| SAR1B | GTP binding protein SAR1b | 5 | 2.79E−03 | U |
| SPTA1 | Spectrin α-chain, erythrocytic 1 | 5 | 2.92E−03 | U/S |
| FLNA | Filamin-A | 5 | 2.98E−03 | U |
| ANK3 | Ankyrin-3 | 5 | 3.03E−03 | U |
| PSD11 | 26S proteasome non-ATPase regulatory subunit 11 | 5 | 3.09E−03 | U |
| PGBM | Basement membrane–specific heparan sulfate proteoglycan core protein (Perlecan) | 5 | 3.16E−03 | U |
| LSP1 | Lymphocyte-specific protein 1 | 5 | 3.53E−03 | U |
| GELS | Gelsolin | 5 | 4.61E−03 | U |
| YKT6 | Synaptobrevin homolog YKT6 | 5 | 4.72E−03 | U |
| PTN6 | Tyrosine-protein phosphatase nonreceptor type 6 | 4 | 4.84E−03 | U/S |
| FLNB | Filamin-B | 5 | 5.03E−03 | U |
| TLN2 | Talin-2 | 5 | 5.28E−03 | U |
| PGK1 | Phosphoglycerate kinase 1 | 4 | 5.39E−03 | U/S |
| PICAL | Phosphatidylinositol binding clathrin assembly protein | 4 | 5.65E−03 | U/S |
| F120A | Constitutive coactivator of PPARγ-like protein 1 | 5 | 6.27E−03 | U/S |
| PRDX2 | Peroxiredoxin-2 | 4 | 6.87E−03 | U/S |
| KCC2D | Calcium/calmodulin-dependent protein kinase type II subunit-δ | 5 | 7.10E−03 | U |
| RTN4 | Reticulon-4 | 5 | 7.57E−03 | U |
| SERA | D-3-phosphoglycerate dehydrogenase | 5 | 7.78E−03 | U/S |
| KC1A | Casein kinase I isoform-α | 5 | 8.06E−03 | U |
| DCTN1 | Dynactin subunit 1 | 5 | 8.52E−03 | U |
| ADDA | α-Adducin | 5 | 8.61E−03 | U |
| PGS2 | Decorin | 5 | 1.09E−02 | U |
| IF4G3 | Eukaryotic translation initiation factor 4-γ3 | 5 | 1.11E−02 | U/S |
| RHG18 | Rho GTPase-activating protein 18 | 4 | 1.11E−02 | U |
| CAPZB | F-actin–capping protein subunit-β | 5 | 1.11E−02 | U |
| MVP | Major vault protein | 5 | 1.14E−02 | U |
| TERA | Transitional endoplasmic reticulum ATPase | 5 | 1.21E−02 | U/S |
| ACTB | Actin, cytoplasmic 1 | 5 | 1.24E−02 | U |
| PGS1 | Biglycan | 5 | 1.30E−02 | U/S |
| K1C14 | Keratin, type I cytoskeletal 14 | 5 | 1.33E−02 | U |
| PLSL | Plastin-2 | 5 | 1.34E−02 | U |
| AP2A2 | AP-2 complex subunit-α2 | 5 | 1.52E−02 | U |
| FINC | Fibronectin | 5 | 1.57E−02 | U/S |
| CLCB | Clathrin light-chain b | 5 | 1.65E−02 | U |
| SNX4 | Sorting nexin-4 | 5 | 1.67E−02 | U |
| SPTB1 | Spectrin β-chain, erythrocytic | 5 | 1.73E−02 | U |
| ZO1 | Tight junction protein ZO-1 | 4 | 1.79E−02 | U |
| DREB | Drebrin | 5 | 1.83E−02 | U |
| CAN1 | Calpain1 | 5 | 1.84E−02 | U |
| PDLI5 | PDZ and LIM domain protein 5 | 5 | 1.85E−02 | U |
| PUR6 | Multifunctional protein ADE2 | 5 | 1.88E−02 | U |
| MY18A | Unconventional myosin-XVIIIa | 5 | 1.93E−02 | U |
| CLCA | Clathrin light-chain A | 5 | 2.07E−02 | U |
| IRGM1 | Immunity-related GTPase family M protein 1 | 5 | 2.09E−02 | U |
| NEB2 | Neurabin-2 | 5 | 2.11E−02 | U |
| K2C6B | Keratin, type II cytoskeletal 6B | 5 | 2.14E−02 | U |
| AP2A1 | AP-2 complex subunit-α1 | 5 | 2.15E−02 | U |
| LYPA1 | Acyl-protein thioesterase 1 | 5 | 2.18E−02 | U |
| PDC6I | Programmed cell death 6–interacting protein | 5 | 2.38E−02 | U/S |
| AP2B1 | AP-2 complex subunit-β | 5 | 2.43E−02 | U |
| K1C19 | Keratin, type I cytoskeletal 19 | 5 | 2.58E−02 | U |
| GRP78 | 78-kD Glucose-regulated protein | 5 | 2.59E−02 | U |
| ARK72 | Aflatoxin B1 aldehyde reductase member 2 | 5 | 2.61E−02 | U |
| SNTB2 | β2-Syntrophin | 5 | 2.63E−02 | U |
| MYO1B | Unconventional myosin-Ib | 5 | 2.67E−02 | U |
| K6PP | ATP-dependent 6-phosphofructokinase, platelet type | 5 | 2.67E−02 | U |
| C1QB | Complement C1q subcomponent subunit B | 5 | 2.71E−02 | U |
| F213A | Redox-regulatory protein FAM213A | 5 | 2.79E−02 | U/S |
| HS90A | Heat shock protein HSP 90α | 5 | 2.85E−02 | U |
| GAK | Cyclin-G–associated kinase | 5 | 2.86E−02 | U |
| SPTN1 | Spectrin α-chain, nonerythrocytic 1 | 5 | 3.08E−02 | U/S |
| UCK1 | Uridine-cytidine kinase 1 | 5 | 3.12E−02 | U |
| ECHP | Peroxisomal bifunctional enzyme | 5 | 3.15E−02 | U/S |
| ES8L2 | EGF receptor kinase substrate 8–like protein 2 | 4 | 3.29E−02 | U |
| MOES | Moesin | 5 | 3.33E−02 | U |
| PNCB | Nicotinate phosphoribosyltransferase | 5 | 3.34E−02 | U |
| MYH10 | Myosin-10 | 5 | 3.59E−02 | U |
| RBGPR | Rab3 GTPase-activating protein noncatalytic subunit | 5 | 3.65E−02 | U |
| VIME | Vimentin | 5 | 3.66E−02 | U |
| SERPH | Serpin 1H | 5 | 3.81E−02 | U |
| RPN1 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | 4 | 3.89E−02 | U |
| LAMB2 | Laminin subunit-β2 | 4 | 3.92E−02 | U/S |
| GSTT1 | Glutathione S-transferase-θ1 | 5 | 3.95E−02 | U |
| TCPQ | T-complex protein 1 | 5 | 3.96E−02 | U |
| TCPZ | T-complex protein 1 | 5 | 4.23E−02 | U/S |
| IRAK4 | IL-1 receptor-associated kinase 4 | 5 | 4.24E−02 | U |
| C1TC | C-1–tetrahydrofolate synthase | 5 | 4.28E−02 | U |
| ARC1B | Actin-related protein 2/3 complex subunit 1B | 5 | 4.28E−02 | U |
| SCFD1 | Sec1 family domain-containing protein 1 | 5 | 4.30E−02 | U/S |
| COPB2 | Coatomer subunit-β | 5 | 4.42E−02 | U |
| FA49B | Protein FAM49B | 5 | 4.62E−02 | U |
| MYH14 | Myosin-14 | 5 | 4.79E−02 | U |
| SNX1 | Sorting nexin-1 | 4 | 4.81E−02 | U |
| PLST | Plastin-3 | 5 | 4.82E−02 | U |
| GBP2 | IFN-induced guanylate binding protein 2 | 5 | 5.00E−02 | U |
| SYEP | Bifunctional glutamate/proline–tRNA ligase | 5 | 5.17E−02 | U |
| TCPA | T-complex protein 1 | 5 | 5.25E−02 | U |
| CLIC1 | Chloride intracellular channel protein 1 | 5 | 5.33E−02 | U |
The association was evaluated by z analysis58 (P≤0.05; n≥4) of n=5 independent experiments, which combined TG2 IP and SWATH MS, using the TG2-null mice as background control (as outlined in Figure 2, A and D). Nuclear and mitochondrial membrane proteins (Supplemental Figure 1) as well as ribosomal proteins (Supplemental Table 1) were manually removed. Proteins are denoted by full name and UniProtKB protein entry name (identification), and they are listed according to the specificity of the interaction with TG2 (P value). U/S, TG2-associated proteins in UUO and sham control membranes; U, TG2-associated proteins uniquely found in UUO membranes.
Table 2.
Specific TG2 partner proteins in the sham control kidney (membrane fraction)
| Identification | Name | n | P Value | U/S |
|---|---|---|---|---|
| PRS4 | 26S protease regulatory subunit 4 | 5 | 5.22E−09 | S |
| PSME1 | Proteasome activator complex subunit 1 | 5 | 8.91E−07 | S |
| NEDD4 | E3 ubiquitin-protein ligase NEDD4 | 5 | 3.03E−05 | S |
| K2C1B | Keratin, type II cytoskeletal 1b | 4 | 8.40E−05 | S |
| UN45A | Protein unc-45 homolog A | 5 | 1.04E−04 | S |
| TGM2 | Transglutaminase 2 | 5 | 1.21E−04 | U/S |
| PRUNE | Protein prune homolog | 5 | 1.31E−04 | S |
| NIBL1 | Niban-like protein 1 | 5 | 1.41E−04 | S |
| PLEC | Plectin | 5 | 1.80E−04 | S |
| IF4G3 | Eukaryotic translation initiation factor 4-γ3 | 5 | 1.82E−04 | U/S |
| SERA | D-3–phosphoglycerate dehydrogenase | 5 | 1.85E−04 | U/S |
| TERA | Transitional endoplasmic reticulum ATPase | 5 | 2.49E−04 | U/S |
| PH4H | Phenylalanine-4-hydroxylase | 5 | 2.67E−04 | S |
| TOM1 | Target of Myb protein 1 | 4 | 3.37E−04 | S |
| PTN6 | Tyrosine-protein phosphatase nonreceptor type 6 | 4 | 4.04E−04 | U/S |
| ACY3 | N-acyl-aromatic-l-amino acid amidohydrolase | 4 | 5.54E−04 | S |
| TCPZ | T-complex protein 1 | 5 | 6.52E−04 | U/S |
| DEST | Destrin | 5 | 8.51E−04 | S |
| UB2D3 | Ubiquitin-conjugating enzyme E2 D3 | 5 | 9.25E−04 | S |
| KAD1 | Adenylate kinase isoenzyme 1 | 5 | 9.92E−04 | S |
| G3P | Glyceraldehyde-3-phosphate dehydrogenase | 5 | 1.05E−03 | S |
| USP9× | Probable ubiquitin C-terminal hydrolase FAF-X | 5 | 1.27E−03 | S |
| PDIA1 | Protein disulfide-isomerase | 4 | 1.40E−03 | S |
| CASP3 | Caspase-3 | 4 | 1.42E−03 | S |
| UBP24 | Ubiquitin C-terminal hydrolase 24 | 4 | 1.43E−03 | S |
| MEP1A | Meprin A subunit-α | 5 | 1.68E−03 | S |
| ARF6 | ADP-ribosylation factor 6 | 5 | 1.88E−03 | S |
| SPTA1 | Spectrin α-chain, erythrocytic 1 | 5 | 1.97E−03 | U/S |
| TM55B | Type 1 phosphatidylinositol 4,5-bisphosphate 4-phosphatase | 4 | 3.22E−03 | S |
| KCRB | Creatine kinase B type | 5 | 4.11E−03 | S |
| RHEB | GTP binding protein Rheb | 5 | 4.12E−03 | S |
| SCFD1 | Sec1 family domain-containing protein 1 | 5 | 4.24E−03 | U/S |
| ST1A1 | Sulfotransferase 1A1 | 5 | 4.51E−03 | S |
| ANXA2 | Annexin A2 | 5 | 5.38E−03 | S |
| CALM | Calmodulin | 5 | 5.39E−03 | U/S |
| F120A | Constitutive coactivator of PPARγ-like protein 1 | 5 | 5.77E−03 | U/S |
| OLFM4 | Olfactomedin-4 | 5 | 5.80E−03 | S |
| IST1 | IST1 homolog | 5 | 6.01E−03 | S |
| PRDX2 | Peroxiredoxin-2 | 4 | 6.07E−03 | U/S |
| PP1A | Serine/threonine-protein phosphatase PP1-α catalytic subunit | 4 | 6.08E−03 | S |
| NLTP | Nonspecific lipid transfer protein | 5 | 6.29E−03 | S |
| PPBT | Alkaline phosphatase, tissue-nonspecific isozyme | 5 | 6.39E−03 | S |
| PDC6I | Programmed cell death 6–interacting protein | 5 | 6.41E−03 | U/S |
| ENOA | α-Enolase | 4 | 6.90E−03 | S |
| FBLN1 | Fibulin-1 | 5 | 7.33E−03 | S |
| LIMS1 | LIM and senescent cell antigen-like–containing domain protein 1 | 5 | 7.55E−03 | U/S |
| TCPG | T-complex protein 1 subunit-γ | 5 | 7.61E−03 | S |
| GGA1 | ADP-ribosylation factor binding protein GGA1 | 5 | 8.30E−03 | S |
| IRF3 | IFN regulatory factor 3 | 4 | 8.39E−03 | S |
| TBB5 | Tubulin β5-chain | 5 | 8.92E−03 | S |
| FARP1 | FERM, RhoGEF, and pleckstrin domain-containing protein 1 | 5 | 9.63E−03 | S |
| DJB11 | DnaJ homolog subfamily B member 11 | 4 | 1.03E−02 | S |
| ARF5 | ADP-ribosylation factor 5 | 5 | 1.03E−02 | S |
| ARPC5 | Actin-related protein 2/3 complex subunit 5 | 4 | 1.06E−02 | S |
| ARHGC | Rho guanine nucleotide exchange factor 12 | 5 | 1.10E−02 | S |
| ARL2 | ADP-ribosylation factor–like protein 2 | 5 | 1.14E−02 | S |
| AT1B1 | Sodium/potassium-transporting ATPase subunit-β1 | 5 | 1.21E−02 | S |
| MGST3 | Microsomal glutathione S-transferase 3 | 5 | 1.21E−02 | S |
| NSF | Vesicle-fusing ATPase | 5 | 1.28E−02 | S |
| PGK1 | Phosphoglycerate kinase 1 | 4 | 1.38E−02 | U/S |
| ARL1 | ADP-ribosylation factor–like protein 1 | 5 | 1.42E−02 | S |
| AT1A1 | Sodium/potassium-transporting ATPase subunit-α1 | 5 | 1.43E−02 | S |
| CUL5 | Cullin-5 | 5 | 1.43E−02 | S |
| VILI | Villin-1 | 5 | 1.51E−02 | S |
| S12A3 | Solute carrier family 12 member 3 | 5 | 1.55E−02 | S |
| KS6A3 | Ribosomal protein S6 kinase-α3 | 5 | 1.58E−02 | S |
| TBA4A | Tubulin α4A-chain | 5 | 1.62E−02 | S |
| VATA | V-type proton ATPase catalytic subunit A | 5 | 1.70E−02 | S |
| SYWC | Tryptophan–tRNA ligase | 5 | 1.75E−02 | S |
| CAH9 | Carbonic anhydrase 9 | 4 | 1.76E−02 | S |
| RAC2 | Ras-related C3 botulinum toxin substrate 2 | 5 | 1.85E−02 | S |
| HBA | Hemoglobin subunit-α | 5 | 1.88E−02 | S |
| RAB10 | Ras-related protein Rab-10 | 5 | 1.98E−02 | S |
| PCKGC | Phosphoenolpyruvate carboxykinase | 5 | 2.08E−02 | U/S |
| NRK1 | Nicotinamide riboside kinase 1 | 5 | 2.11E−02 | S |
| VPS35 | Vacuolar protein sorting–associated protein 35 | 5 | 2.25E−02 | S |
| SNX3 | Sorting nexin-3 | 5 | 2.27E−02 | S |
| ACSA | Acetyl-CoA synthetase | 5 | 2.29E−02 | S |
| DNJA2 | DnaJ homolog subfamily A member 2 | 5 | 2.29E−02 | S |
| ST1D1 | Sulfotransferase 1 family member D1 | 5 | 2.40E−02 | S |
| ANFY1 | Rabankyrin-5 | 5 | 2.42E−02 | S |
| GLCTK | Glycerate kinase | 5 | 2.42E−02 | S |
| ARC1A | Actin-related protein 2/3 complex subunit 1A | 5 | 2.42E−02 | S |
| PDIA6 | Protein disulfide-isomerase A6 | 5 | 2.53E−02 | S |
| VATB2 | V-type proton ATPase subunit B | 5 | 2.59E−02 | S |
| GCYA3 | Guanylate cyclase–soluble subunit-α3 | 5 | 2.67E−02 | S |
| LYPA2 | Acyl-protein thioesterase 2 | 5 | 2.68E−02 | S |
| RUFY3 | Protein RUFY3 | 4 | 2.72E−02 | S |
| PICAL | Phosphatidylinositol binding clathrin assembly protein | 4 | 2.73E−02 | U/S |
| TBC9B | TBC1 domain family member 9B | 5 | 2.78E−02 | S |
| IQGA1 | Ras GTPase-activating–like protein IQGAP1 | 5 | 2.99E−02 | U/S |
| RCN1 | Reticulocalbin-1 | 4 | 3.03E−02 | S |
| ARP3 | Actin-related protein 3 | 5 | 3.04E−02 | S |
| PGS1 | Biglycan | 5 | 3.18E−02 | U/S |
| NAMPT | Nicotinamide phosphoribosyltransferase | 5 | 3.20E−02 | U/S |
| ECHP | Peroxisomal bifunctional enzyme | 5 | 3.21E−02 | U/S |
| HS90B | Heat shock protein HSP 90β | 5 | 3.38E−02 | S |
| ITM2B | Integral membrane protein 2B | 5 | 3.60E−02 | S |
| MP2K2 | Dual specificity mitogen-activated protein kinase kinase 2 | 5 | 3.62E−02 | S |
| DHRS4 | Dehydrogenase/reductase SDR family member 4 | 4 | 3.62E−02 | S |
| TBA1A | Tubulin α1A-chain | 5 | 3.66E−02 | S |
| IF4A1 | Eukaryotic initiation factor 4A-I | 5 | 3.68E−02 | S |
| OXSR1 | Serine/threonine-protein kinase OSR1 | 5 | 3.69E−02 | S |
| F213A | Redox-regulatory protein FAM213A | 5 | 3.76E−02 | U/S |
| CLIC4 | Chloride intracellular channel protein 4 | 5 | 3.82E−02 | S |
| VDAC1 | Voltage-dependent anion-selective channel protein 1 | 5 | 3.87E−02 | S |
| CAN2 | Calpain-2 catalytic subunit | 5 | 3.95E−02 | S |
| NDRG3 | Protein NDRG3 | 5 | 4.06E−02 | S |
| CLH1 | Clathrin heavy-chain 1 | 5 | 4.11E−02 | S |
| LAMB2 | Laminin subunit-β2 | 4 | 4.21E−02 | U/S |
| FINC | Fibronectin | 5 | 4.27E−02 | U/S |
| TS101 | Tumor susceptibility gene 101 protein | 4 | 4.31E−02 | S |
| DPYL3 | Dihydropyrimidinase-related protein 3 | 5 | 4.39E−02 | S |
| ISG15 | Ubiquitin-like protein ISG15 | 5 | 4.53E−02 | S |
| DRG2 | Developmentally regulated GTP binding protein 2 | 5 | 4.71E−02 | S |
| RN213 | E3 ubiquitin-protein ligase RNF213 | 4 | 4.73E−02 | S |
| BAX | Apoptosis regulator BAX | 5 | 4.73E−02 | S |
| ADH1 | Alcohol dehydrogenase 1 | 5 | 4.74E−02 | U/S |
| SPTN1 | Spectrin α-chain, nonerythrocytic 1 | 5 | 5.25E−02 | U/S |
| EF1A1 | Elongation factor 1-α1 | 5 | 5.34E−02 | S |
The association was evaluated as described in Table 1. U/S, TG2-asociated proteins in UUO and sham-operated membranes; S, TG2-associated proteins uniquely found in sham-operated membranes.
TG2 Interaction Networks Reveal a Predominance of Vesicular Trafficking and Actin Dynamics Proteins in the UUO Kidney
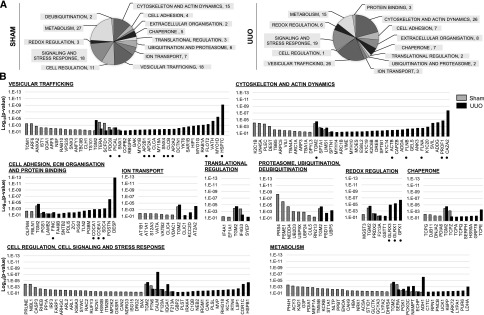
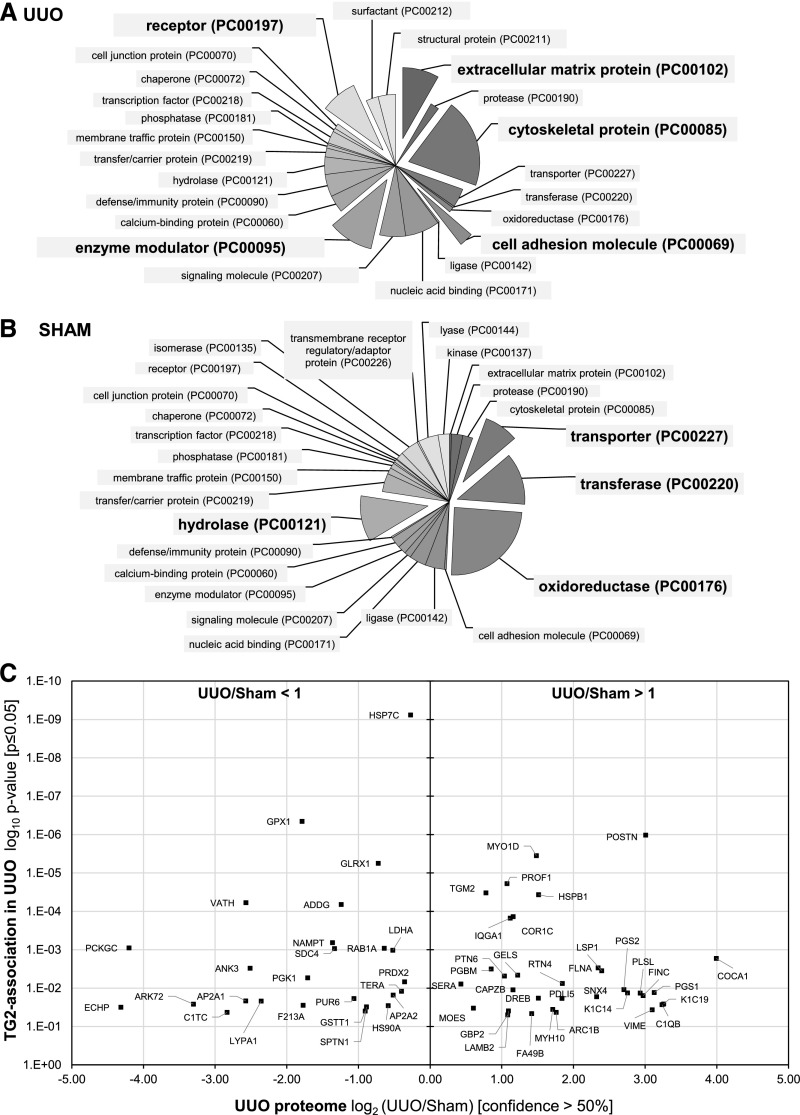
Analysis of functions obtained by UniprotKB database (Mus musculus) revealed enrichment of TG2 interactions with proteins involved in vesicular trafficking and cytoskeletal actin dynamics in the UUO kidney (Figure 3A). Canonical TG2 partner proteins were present (e.g., in cell adhesion, FN, SDC4, and collagens [COCA1 and COEA1]; in cytoskeleton, actin and filamin [FLNA and FLNB] [Figure 3B]), but there were also unexpected absences, such as the integrin family. This may be because of the transitory nature of integrin complexes, which can be only effectively captured by stabilizing the interaction with a chemical crosslinker.21 Desmoplakin (involved in interepithelial cell adhesion), F-actin capping protein (CAZA2), and profilin-1 (PROF1; involved in actin cytoskeleton remodeling) were new highly significant partners of TG2 in UUO. Proteins that had never been associated or connected with TG2, such as a series of vesicular trafficking proteins like clathrin and adaptor proteins (CLCA, CLCB, AP2A1, AP2A2, and AP2B1), sorting nexins (SNX1 and SNX4), Alix (PDC6I), flotillin-2 (FLOT2), and heat shock cognate 71-kD protein (HSP7C), formed a large part of the TG2 UUO interactome (Figure 3B). Moreover, redox regulation proteins, such as glutathione peroxidase-1, glutaredoxin-1 (GLRX1), and GLRX3, were significant TG2 partners in the UUO (Figure 3B). There was a definite difference in the composition of the TG2 partners between the UUO and normal (sham) status, and proteins associated with TG2 uniquely in the UUO state were likely to influence its trafficking, activity, and regulation in fibrosis.
Figure 3.
Functional distribution of TG2-associated proteins in kidney membranes reveals a difference in TG2 partners between the UUO and normal (sham) status. TG2-associated proteins were clustered depending on their functions in M. musculus investigated by protein identification search on UniProtKB database. (A) Pie charts display the distribution of the different functions of the TG2-associated proteins in UUO or sham controls, with numbers next to each function name denoting how many proteins fall in the various functional categories. (B) Column charts list proteins in the order of significance of their association with TG2 (log10 P value) and are grouped according to their function in UUO and sham controls (histogram bars). Dots under protein names denote proteins described in Results.
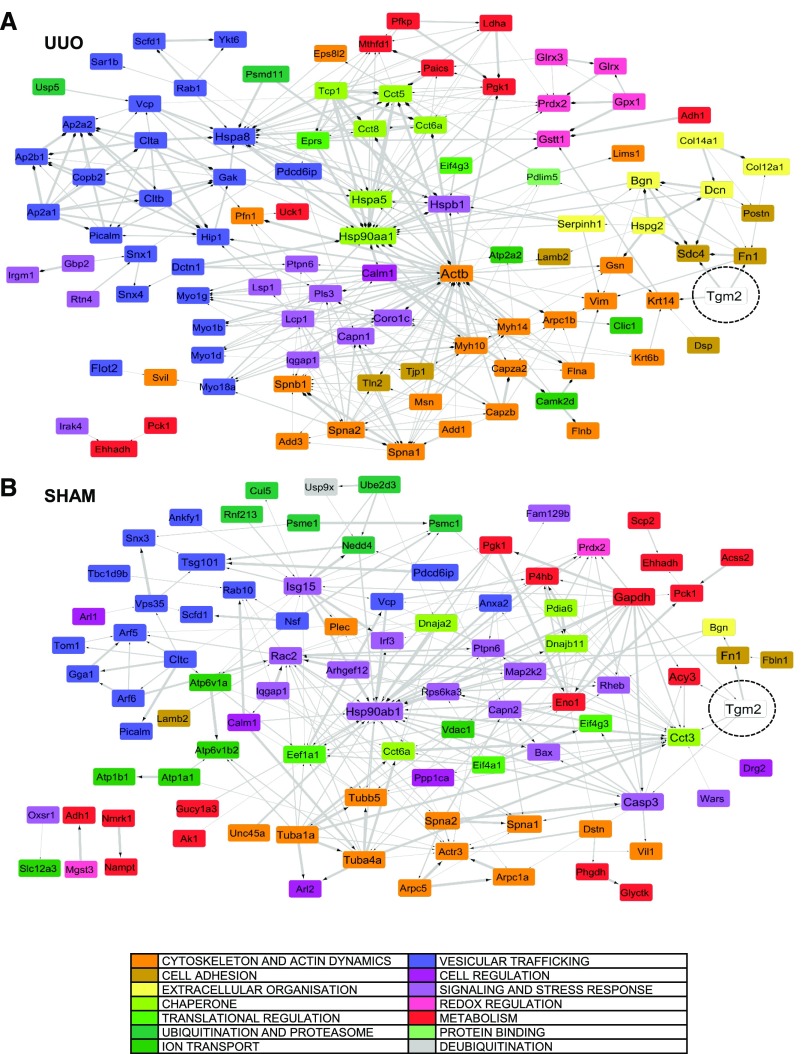
Network analysis of the TG2 interactomes built using the STRING v10 bioinformatics tool (on the basis of known and predicted protein-protein interactions) showed a clear cluster of proteins associated with membrane vesicles (Figure 4A, light blue), redox regulation (Figure 4A, pink), and cytoskeletal dynamics (Figure 4A, orange) in the UUO kidney. In contrast, in the TG2 interactome of the normal kidney (Figure 4B), there were changes in the topology of interconnections among the same functionally related protein groups, which were less represented, but denser clusters of metabolic proteins were evident (Figure 4B). Therefore, TG2 clearly becomes more interactive with vesicular trafficking proteins in the UUO as well as proteins linked to actin dynamics and ECM events.
Figure 4.
The interactome of TG2 in UUO and sham control kidney reveals increased interactivity of TG2 with proteins related to vesicular trafficking, cytoskeleton, and ECM dynamics in the UUO model. The protein interaction network built from TG2-associated proteins in (A) UUO and (B) sham-operated kidney membranes was mapped against the M. musculus reference database using String V10.0 (http://string-db.org). Candidates were selected using both known and predicted protein interactions and a threshold confidence level of 0.4 (default). Networks were imported in Cytoscape and color coded according to the main functional nodes. The thickness of the lines is proportional to the confidence of the interactions.
Matching of the TG2 Interactome with the UUO Proteome
To exclude any concentration-dependent association with TG2, we evaluated if the proteins of the TG2 UUO interactome were also over-represented in the UUO proteome or associated with TG2 without being upregulated in the UUO. To this aim, we profiled the UUO and sham-operated kidney proteomes by SWATH MS (Supplemental Tables 2 and 3). This was necessary, because current UUO proteomes are of limited coverage.22 Of the 2106 proteins quantified in the UUO proteome, 195 were significantly upregulated in kidneys post-UUO (Supplemental Table 2), and 458 were downregulated (Supplemental Table 3) (confidence >80%). Original processed data are provided as Supplemental Appendix 2.
TG2 signal was increased but only less than twice the physiologic level (1.71-fold) (Supplemental Table 2, TGM2), consistent with prior observations that UUO leads to an increase in TG2 secretion rather than expression.12 Protein class annotation terms were associated with ECM components, cytoskeletal organization, and cell adhesion, forming significant clusters of the UUO proteome (P≤0.05) (Figure 5A, Supplemental Table 4A).
Figure 5.
Interrogation of the UUO proteome with TG2-interacting proteins uncovers essential partners of TG2 in UUO kidney membranes. The proteomes of the UUO kidneys and sham-operated control kidneys were resolved by quantitative SWATH acquisition proteomics. (A and B) Functional distribution of proteins with signal (A) increased (n=195) or (B) decreased (n=458) on UUO compared with sham control (confidence ≥80%) was performed in PANTHER using the relative annotation terms ontologies. Each pie chart sector represents the percentage of the proteome belonging to the specific PANTHER class over the total number of class hits. (C) Interrogation of the UUO proteome (confidence ≥50%) with the TG2-interacting proteins identified in UUO kidney membranes (P≤0.05) and reported in Table 1. The 122 proteins specifically associated with TG2 in UUO kidney membranes were plotted on the ordinate axis according to the significance (P value) of TG2 association and the abscissa according to their expression in the UUO kidney proteome compared with sham control proteome: log2(UUO/sham). A positive value on the abscissa indicates the increased signal of a given TG2-associated protein partner on UUO compared with the sham-operated condition (UUO/sham >1), and a negative value indicates its decreased signal on UUO (UUO/sham <1).
Among the proteins most significantly associated with TG2 in the UUO kidney (P≤0.001 in Table 1), which were also intensified in the UUO proteome (Figure 5C, right panel), there were periostin (a marker of progressive kidney injury in animal models of CKD),23 myosin-1D, PROF1, CAZA-2, heat shock protein β1, FN, laminin, collagen (COCA1), and actin nucleation complex ARC1B. A number of these TG2 partners sat in pathways over-represented in the UUO model (“ECM-receptor interaction” and “regulation of the actin cytoskeleton”) (Supplemental Figure 2), suggesting that they act in synergy with TG2 in fibrosis progression. However, a significant level of TG2 partners, such as HSP7C, SDC4, and the antioxidant proteins glutathione peroxidase-1 and GLRX1 (Figure 5C, left panel), was either decreased or was not significantly over-represented in the UUO proteome, raising the possibility that they could be essential partners of TG2, some of which were potentially implicated in the externalization pathway of TG2 post-UUO.
TG2 Association with Exosomal Proteins Post-UUO
The TG2 interactome has revealed that TG2 primarily interacts with a number of proteins associated with vesicular trafficking and EVs, especially in the UUO condition. When the TG2 partners identified in the UUO model (Table 1) were matched with proteins collected in the EVs (exosome) database ExoCarta (and its compendium Vesiclepedia), almost all proteins associated with TG2, either directly or indirectly, were nominated in the database (Table 3). These included the prominent exosome marker Alix (PDC6I), HSP7C, PROF1, FLOT2, and heat shock protein HSP90-α, all among the top 50 proteins reported in exosomes. Other significant exosomal TG2 partners less recurrent in the exosome database were PRDX2, RAB1A, and SDC4 (Table 3). These findings support the hypothesis that TG2 may be trafficked and externalized via EVs, either of endosomal origin (exosomes) or shed from the PM (ectosomes).
Table 3.
Matching of TG2 partner proteins to exosomal proteins
| Identification | TG2-Associated Candidate Names in UUO | Found in Exosomes | |
|---|---|---|---|
| Homo sapiens | Rattus norvegicus | ||
| DESP | Desmoplakin | Match | |
| HSP7Ca | Heat shock cognate 71-kD protein | Match | Match |
| CALM | Calmodulin | Match | Match |
| CAZA2 | F-actin–capping protein subunit-α2 | Match | Match |
| GPX1 | Glutathione peroxidase 1 | Match | Match |
| ADH1 | Alcohol dehydrogenase 1 | Match | Match |
| POSTN | Periostin | Match | Match |
| MYO1D | Unconventional myosin-Id | Match | |
| GLRX1 | Glutaredoxin-1 | Match | |
| GLRX3 | Glutaredoxin-3 | Match | Match |
| PROF1a | Profilin-1 | Match | Match |
| TGM2 | Transglutaminase 2 | Match | Match |
| HSPB1 | Heat shock protein-β1 | Match | Match |
| VATH | V-type proton ATPase subunit H | Match | Match |
| ADDG | γ-Adducin | ||
| AT2A2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | Match | Match |
| SVIL | Supervillin | ||
| COR1C | Coronin-1C | Match | Match |
| IQGA1 | Ras GTPase-activating–like protein IQGAP1 | Match | Match |
| TCPE | T-complex protein 1 | ||
| UBP5 | Ubiquitin C-terminal hydrolase 5 | ||
| FLOT2a | Flotillin-2 | Match | Match |
| NAMPT | Nicotinamide phosphoribosyltransferase | Match | Match |
| PCKGC | Phosphoenolpyruvate carboxykinase | Match | Match |
| RAB1A | Ras-related protein Rab-1A | Match | Match |
| SDC4 | Syndecan-4 | Match | Match |
| LDHAa | l-lactate dehydrogenase A-chain | Match | Match |
| MYO1G | Unconventional myosin-Ig | Match | |
| K1C20 | Keratin, type I cytoskeletal 20 | Match | |
| COEA1 | Collagen-α1(XIV) chain | Match | |
| COCA1 | Collagen-α1(XII) chain | Match | |
| HIP1 | Huntingtin-interacting protein 1 | Match | |
| LIMS1 | LIM and senescent cell antigen-like–containing domain protein 1 | Match | Match |
| SAR1B | GTP binding protein SAR1b | Match | |
| SPTA1 | Spectrin α-chain, erythrocytic 1 | ||
| FLNA | Filamin-A | Match | Match |
| ANK3 | Ankyrin-3 | Match | |
| PSD11 | 26S proteasome non-ATPase regulatory subunit 11 | Match | |
| PGBM | Basement membrane–specific heparan sulfate proteoglycan core protein (Perlecan) | ||
| LSP1 | Lymphocyte-specific protein 1 | Match | |
| GELS | Gelsolin | Match | Match |
| YKT6 | Synaptobrevin homolog YKT6 | Match | |
| PTN6 | Tyrosine-protein phosphatase nonreceptor type 6 | Match | |
| FLNB | Filamin-B | Match | Match |
| TLN2 | Talin-2 | Match | Match |
| PGK1a | Phosphoglycerate kinase 1 | Match | Match |
| PICAL | Phosphatidylinositol binding clathrin assembly protein | Match | Match |
| F120A | Constitutive coactivator of PPARγ-like protein 1 | Match | |
| PRDX2 | Peroxiredoxin-2 | Match | Match |
| KCC2D | Calcium/calmodulin-dependent protein kinase type II subunit-δ | Match | |
| RTN4 | Reticulon-4 | Match | Match |
| SERA | D-3-phosphoglycerate dehydrogenase | Match | |
| KC1A | Casein kinase I isoform-α | Match | |
| DCTN1 | Dynactin subunit 1 | Match | Match |
| ADDA | α-Adducin | Match | |
| PGS2 | Decorin | Match | |
| IF4G3 | Eukaryotic translation initiation factor 4-γ3 | Match | |
| RHG18 | Rho GTPase-activating protein 18 | Match | |
| CAPZB | F-actin–capping protein subunit-β | Match | Match |
| MVP | Major vault protein | Match | Match |
| TERAa | Transitional endoplasmic reticulum ATPase | Match | |
| ACTBa | Actin, cytoplasmic 1 | Match | Match |
| PGS1 | Biglycan | Match | |
| K1C14 | Keratin, type I cytoskeletal 14 | Match | |
| PLSL | Plastin-2 | Match | Match |
| AP2A2 | AP-2 complex subunit-α2 | Match | |
| FINC | Fibronectin | Match | |
| CLCB | Clathrin light-chain b | Match | |
| SNX4 | Sorting nexin-4 | Match | |
| SPTB1 | Spectrin β-chain, erythrocytic | Match | |
| ZO1 | Tight junction protein ZO-1 | Match | |
| DREB | Drebrin | Match | |
| CAN1 | Calpain1 | Match | |
| PDLI5 | PDZ and LIM domain protein 5 | Match | |
| PUR6 | Multifunctional protein ADE2 | Match | Match |
| MY18A | Unconventional myosin-XVIIIa | Match | |
| CLCA | Clathrin light-chain A | Match | Match |
| IRGM1 | Immunity-related GTPase family M protein 1 | ||
| NEB2 | Neurabin-2 | ||
| K2C6B | Keratin, type II cytoskeletal 6B | Match | |
| AP2A1 | AP-2 complex subunit-α1 | Match | |
| LYPA1 | Acyl-protein thioesterase 1 | Match | Match |
| PDC6Ia | Programmed cell death 6–interacting protein | Match | Match |
| AP2B1 | AP-2 complex subunit-β | Match | Match |
| K1C19 | Keratin, type I cytoskeletal 19 | Match | |
| GRP78 | 78-kD Glucose-regulated protein | Match | Match |
| ARK72 | Aflatoxin B1 aldehyde reductase member 2 | Match | Match |
| SNTB2 | β2-Syntrophin | Match | |
| MYO1B | Unconventional myosin-Ib | Match | |
| K6PP | ATP-dependent 6-phosphofructokinase, platelet type | ||
| C1QB | Complement C1q subcomponent subunit B | Match | Match |
| F213A | Redox-regulatory protein FAM213A | Match | |
| HS90Aa | Heat shock protein HSP 90α | Match | Match |
| GAK | Cyclin-G–associated kinase | Match | |
| SPTN1 | Spectrin α-chain, nonerythrocytic 1 | Match | |
| UCK1 | Uridine-cytidine kinase 1 | ||
| ECHP | Peroxisomal bifunctional enzyme | Match | Match |
| ES8L2 | EGF receptor kinase substrate 8–like protein 2 | Match | |
| MOESa | Moesin | Match | Match |
| PNCB | Nicotinate phosphoribosyltransferase | Match | Match |
| MYH10 | Myosin-10 | Match | Match |
| RBGPR | Rab3 GTPase-activating protein noncatalytic subunit | Match | |
| VIME | Vimentin | Match | Match |
| SERPH | Serpin 1H | Match | Match |
| RPN1 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | Match | |
| LAMB2 | Laminin subunit-β2 | Match | Match |
| GSTT1 | Glutathione S-transferase-θ1 | Match | |
| TCPQ | T-complex protein 1 | ||
| TCPZ | T-complex protein 1 | Match | |
| IRAK4 | IL-1 receptor–associated kinase 4 | ||
| C1TC | C-1-tetrahydrofolate synthase | Match | Match |
| ARC1B | Actin-related protein 2/3 complex subunit 1B | Match | Match |
| SCFD1 | Sec1 family domain–containing protein 1 | Match | |
| COPB2 | Coatomer subunit-β | Match | Match |
| FA49B | Protein FAM49B | Match | Match |
| MYH14 | Myosin-14 | Match | |
| SNX1 | Sorting nexin-1 | Match | |
| PLST | Plastin-3 | Match | |
| GBP2 | IFN-induced guanylate binding protein 2 | Match | |
| SYEP | Bifunctional glutamate/proline–tRNA ligase | Match | Match |
| TCPAa | T-complex protein 1 | Match | Match |
| CLIC1a | Chloride intracellular channel prot 1 | Match | Match |
TG2 Secretion via Exosomes and Ectosomes in Established Epithelial Cell Lines
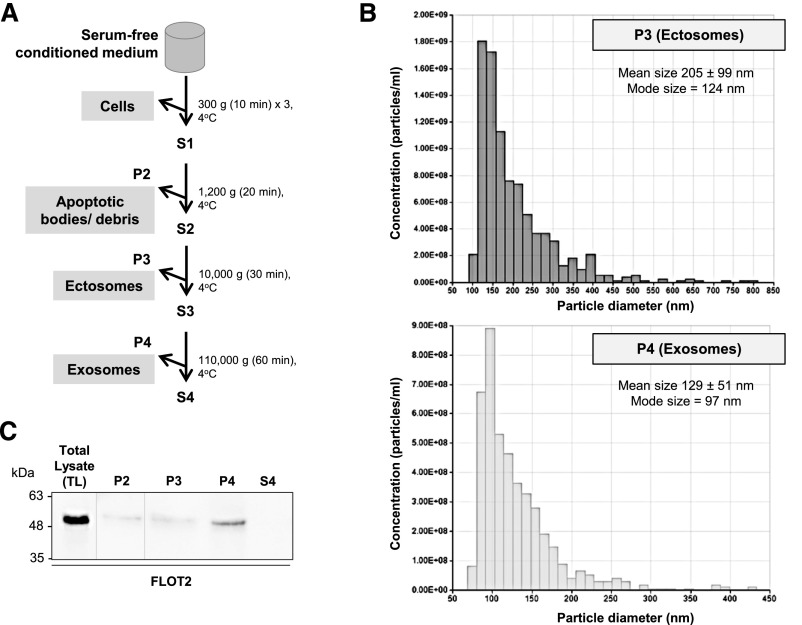
Previous reports have shown that TG2 is mainly secreted by TECs, although renal fibroblasts and mesangial cells are less investigated sources.4,5 To test the hypotheses that TG2 is externalized in the UUO in EVs and that this pathway is increased in the pathologic state, we used an established rat renal TEC line (NRK52E) stably transfected with EGFP-tagged TG2 to facilitate tracking of TG2. Exosomes and ectosomes were isolated from the supernatant of cells according to standard protocols24,25 (Figure 6A). After removal of cells and cellular debris, ectosomes were recovered by differential centrifugation in the P3 fraction (2.97×107 particles per 1 ml medium) and smaller exosomes were recovered in the P4 fraction (5.89×107 particles per 1 ml medium) as revealed by tunable resistive pulse sensing analysis with a qNano instrument (Figure 6B). The EV-free medium was collected (S4), and all nonvesicular proteins were recovered by TCA precipitation. The fractions were subjected to Western blot analysis using antibodies toward the EV marker FLOT2 (Figure 6C).
Figure 6.
A differential centrifugation approach allows the isolation of EVs subpopulations from NRK52E conditioned medium. (A) Flowchart of the differential centrifugation steps for the isolation of EVs. P1–P4 indicate pellets, and S1–S4 indicate supernatants. (B) Analysis of microparticle size distribution in fractions P3 and P4 was performed by nanoparticle tracking analysis with qNano (Izon). Ectosomes were recovered from the P3 fraction, and exosomes were recovered from the P4 fraction. Representative particle size distributions of an ectosome-enriched 10,000-g pellet (dark gray) and an exosome-enriched 110,000-g pellet (light gray). (C) Expression of FLOT2 was measured by Western blotting in equal amounts from the differential centrifugation fractions obtained from the conditioned medium of NRK52E cells cultured in serum-free conditions.
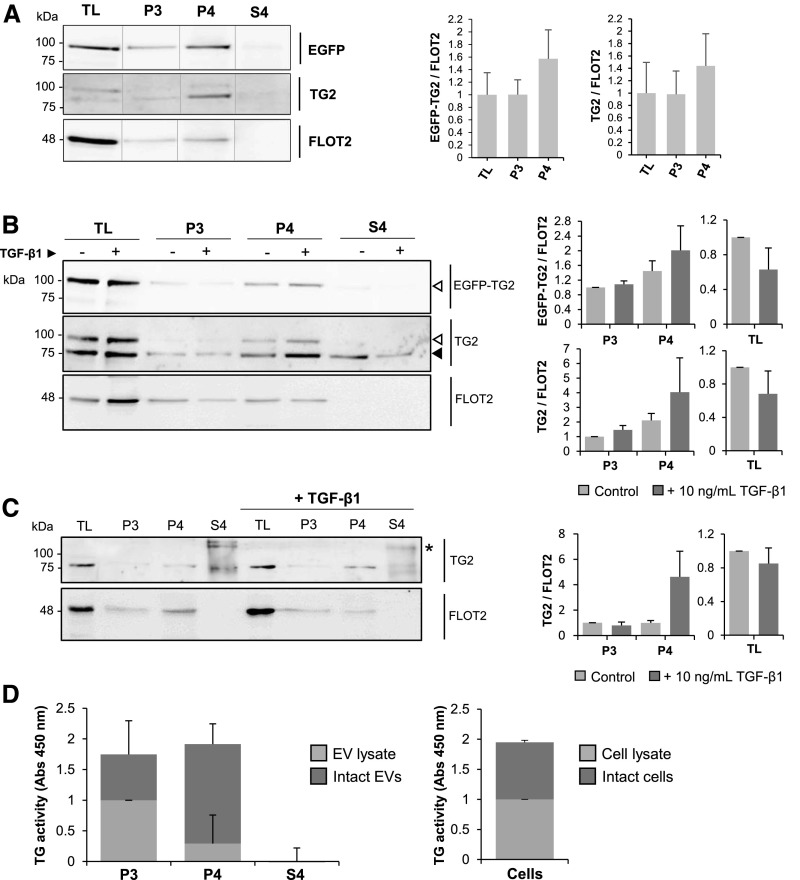
To test for the presence of TG2 in the EVs fractions, equal amounts of proteins (20 μg) extracted from ectosomes (P3) and exosomes (P4) and those from the vesicles-free medium (S4; after TCA protein precipitation) were immunoprobed using antibodies against TG2 and the EGFP tag. Both endogenous (75 kD) and EGFP-tagged TG2 (100 kD) were clearly present in the exosomes (P4) and in lower amounts in the ectosome (P3) fraction (Figure 7A). Densitometric analysis of TG2 relative to FLOT2 revealed a trend of enrichment of TG2 in the exosome fraction (P4) (Figure 7A). Cells were also stimulated by recombinant TGF-β1 (10 ng/ml) to simulate conditions of fibrogenesis in vitro. TGF-β1 did not change the level of TG2 in the total cell lysate (TL) but caused a trend of increase in the exosome-associated TG2 (Figure 7B). The cytokine affected the expression of FLOT2 in TL and EVs but without reaching significance (Figure 7B). When the same experiment was conducted in the nontransfected NRK52E cells, TG2 was present prevalently in exosomes (Figure 7C). Treatment with TGF-β1 caused a trend of increase in TG2 in the exosomal fraction, which became enriched in TG2 relative to FLOT2 expression. As an additional analysis, we isolated exosomes and ectosomes from an established renal fibroblast cell line (NRK49F). TG2 was present in the ectosomal and exosomal fractions, and the band was increased by TGF-β1 (Supplemental Figure 3). The EV-free medium (S4) was also collected from both the EGFP-TG2 NRK52E clone and the WT NRK52E, and all proteins were recovered by TCA precipitation. A trace of endogenous TG2 was occasionally present freely in the culture medium (S4) of NRK52E-expressing EGFP-TG2 on TCA precipitation (Figure 7, A and B), and TG2 immunoreactivity was seen in high molecular weight polymers in the S4 fraction of WT NRK52E (Figure 7C). Our data suggest that the free TG2 may be transient and unstable and that it either self-polymerizes or becomes incorporated in circulating proteins (Figure 7C, asterisk); moreover, it is not increased by TGF-β1 (Figure 7, B and C).
Figure 7.
TG2 is present in vesicles of endosomal and plasma membrane origin in established TEC lines. (A and B) NRK52E-expressing EGFP-TG2 and (C) WT NRK52E cells were grown in serum-free medium for 36 hours (A) without and (B and C) with supplementation of 10 ng/ml TGF-β1. After incubation, culture medium was collected, and vesicular fractions (P3 and P4) were separated by serial centrifugation as shown in Figure 6. Proteins from vesicles-deprived medium were concentrated by TCA precipitation (S4). Fractions and cell lysate (TL) were immunoprobed with primary antibodies toward the EGFP tag, TG2, or FLOT2. Intensity of immunoreactive bands was quantified by densitometric analysis and normalized to FLOT2 expression. Data represent mean values of three independent experiments ±SEM. Black arrowhead, endogenous TG2; white arrowheads, EGFP-TG2. *TG2 polymers. (D) EVs obtained from NRK52E EGFP-TG2 conditioned medium were either lysed in a mild lysis buffer to obtain a total lysate or resuspended as intact vesicles in sample buffer (from a tissue transglutaminase picoassay kit). The transamidating activity of TG2 was measured in either the whole EVs or the EV lysate by a sensitive “picoassay.” As a control, TG2 activity was also measured in TL and whole cells. Data represent mean values of three independent experiments ±SEM.
To gain insights into the location of TG2 within EVs, we devised an experiment aimed at measuring the catalytic activity of TG2 of whole EVs and lysed EVs (providing the total EVs homogenate) isolated from NRK52E cells. As a control, we also measured TG2 activity in lysed cell homogenates and whole cells. In the ectosomes (P3), TG2 activity was similar in the intact vesicles and vesicles lysed in a buffer dissolving cytosolic proteins; in the exosomes (P4), there was a higher proportion of TG2 detected in the intact particles compared with the EV lysate, suggesting that TG2 is enriched on the surface of exosomes but without reaching significance (P=0.14). No TG2 activity was detected in the EV-free medium, and levels of TG2 were similar on the surface of NRK52E cells and in the cell lysate (Figure 7D).
Collectively, these results show that TG2 is clearly present in EVs and suggest that TGF-β1 directs TG2 toward exosome secretion.
Inhibition of Neutral Sphingomyelinase Affects Exosome Release of TG2
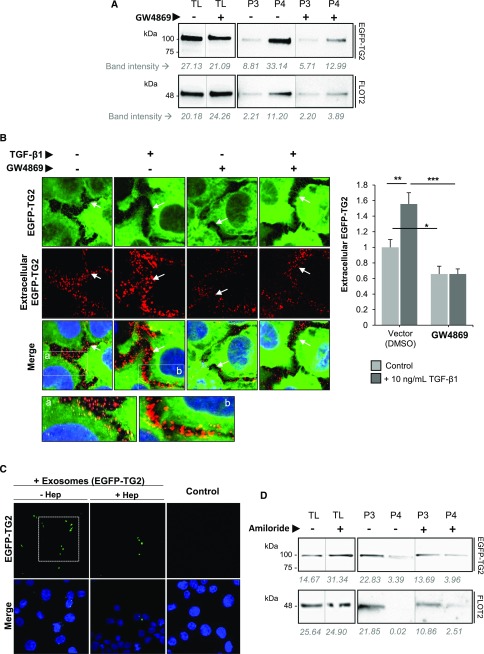
In several mammalian cell types, the biogenesis of exosome secretion depends on sphingomyelinase for the production of ceramide of which exosomes are rich.26 We, therefore, investigated whether the sphingomyelinase inhibitor GW4869, which blocks neutral sphingomyelinase (N-SMase), affected exosome release of TG2. After treatment of NRK52E-expressing EGFP-TG2 with GW4869 (10 μM) for 16 hours,27 FLOT2 was decreased in the exosomal fraction (P4) (Figure 8A), consistent with a reduction of total exosome release, and this was accompanied by a marked reduction of TG2 in the same fraction (Figure 8A). The ectosome fraction (P3) did not seem to be sensitive to inhibition of N-SMase as previously described in glial cells,24 and GW4869 produced no difference in the ectosome release of TG2 (Figure 8A). Vesicular export of TG2 was visualized by immunofluorescence staining using high-resolution confocal microscopy in EGFP-TG2–expressing NRK52E cells. Extracellular TG2 was detected by an anti-EGFP antibody in nonpermeabilized cells (denoted by the red fluorescence), whereas the green fluorescence represents the total EGFP-TG2. Minimum washing of the monolayer before fixation to prevent exosome loss revealed an intense punctate pattern of extracellular EGFP-TG2 staining, which was increased in TGF-β1–stimulated cells. GW4869 visibly inhibited extracellular EGFP-TG2 staining, suggesting that it inhibits TG2 export (Figure 8B). EGFP fluorescence visualization of exosomes isolated from NRK52E-expressing EGFP-TG2 and incubated with WT NRK52E revealed a similarly punctuated staining, which was retained in the WT NRK52E monolayer after several washes, suggesting that this consists of EVs containing EGFP-TG2 (Figure 8C, −Hep). Pretreatment of NRK52E with heparitinase to remove the HS chains before addition of EGFP-TG2–containing exosomes reduced the exosome uptake by 49% (P<0.01) (Figure 8C, +Hep), suggesting a role for cell surface HS in the capturing of EVs.
Figure 8.
EV release of TG2 is reduced by inhibition of exosome and ectosome synthesis. (A) Exponentially growing NRK52E-expressing EGFP-TG2 cells were cultured in serum-free medium supplemented with GW4869 (10 μM; +) or DMSO (vector; −) for 16 hours. TL, ectosome (P3), and exosome (P4) fractions were blotted with anti-GFP (EGFP) and anti-FLOT2 antibodies. Band intensities per area measured by densitometric analysis are shown underneath the blots. (B) NRK52E EGFP-TG2 cells were grown in an eight-well chamber with and without 10 ng/ml TGF-β1 and treated with GW4869 as described in A. Extracellular EGFP-TG2 was detected in cells fixed by paraformaldehyde (3% [wt/vol]) but not permeabilized using a rabbit polyclonal anti-GFP antibody followed by a goat anti-rabbit Alexa Fluor 568 antibody (red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). The green fluorescence denotes total EGFP-TG2. Representative confocal microscopy sections (10 μm) are presented, and they were acquired by a Leica TCS confocal microscope. White arrows identify the cell perimeter. Extracellular EGFP-TG2 was quantified by ImageJ intensity analysis on at least eight nonoverlapping images per treatment and is presented as mean relative intensity of red (extracellular EGFP-TG2) over green (total EGFP-TG2) ±SEM expressed relative to the control cells without GW4869 (equalized to one). *P<0.05; **P<0.01; ***P<0.01. (C) Exosomes from NRK52E EGFP-TG2 cells were isolated and incubated on a monolayer of WT NRK52E cells for 3 hours. Cells were washed three times in PBS and fixed in paraformaldehyde, but they were not permeabilized. EGFP fluorescence (green) was clearly visible in particles with estimated diameter of <0.5 μm. Nuclei were stained with 4′,6-diamidino-2-phenylindole. Pretreatment of the NRK52E cell monolayer with heparitinase (+Hep) to digest cells surface HS chains led to a decrease in the uptaken EGFP-TG2–bearing exosomes. (D) NRK52E-expressing EGFP-TG2 cells were exposed to 1 μM amiloride for 10 minutes in serum-free medium. TL, ectosome (P3), and exosome (P4) fractions were blotted with anti-GFP and anti-FLOT2 antibodies. Band intensities per area were measured as described in A.
To attenuate the formation of ectosomes, we used the inhibitor of Na+/H+ exchanges amiloride, which interferes with the activity of the efflux pumps expressed on acidic vacuoles and the exocytosis of micropinosomes.28–31 Ectosomes (P3) formed in a greater proportion than exosomes (P4) in a short time of incubation (10 minutes) as previously shown,32 and they were revealed by the faint FLOT2 staining in the exosome fraction (Figure 8D). Amiloride incubation lowered FLOT2 in the ectosomes (P3), being that amiloride is an inhibitor of MV formation,33 and consistent with this, there was a reduction in TG2 in ectosomes (Figure 8D).
Collectively, these results indicate that TG2 is transported into exosomes and ectosomes and that this pathway is stimulated by profibrogenic TGF-β1 in exosomes.
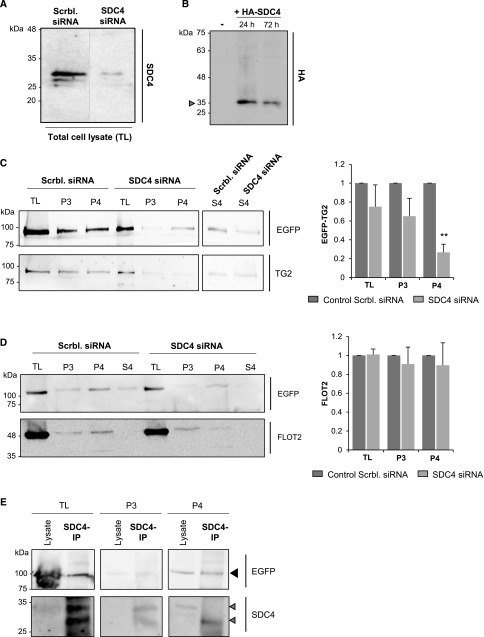
Recruitment of TG2 in Exosomes Depends on SDC4
Another protein that we found specifically present in TG2 UUO interactome is the cell surface HS proteoglycan SDC4 (Table 1). Syndecans have been reported to bind cargo with HS chains, leading to their clustering and recruitment of syntenin-1 and Alix for membrane budding with exosome formation.34 Therefore, we hypothesized that TG2 may be recruited as an HS binding cargo and targeted to exosomes by SDC4. To test the role of SDC4 in EV-associated TG2 secretion, NRK52E-expressing EGFP-TG2 was transiently transfected with either rat SDC4-targeting siRNA or scrambled control siRNA. SDC4 reduction by SDC4-targeting siRNA (SDC4 KO) was confirmed by Western blotting of the TL (Figure 9A). As a control for the proteoglycan immunodetection (which is notoriously difficult), NRK52E transiently transfected with a hemagglutinin (HA)-tagged SDC4 construct revealed a similar band in the 30-kD range when probed with anti-HA antibody (Figure 9B). Culture medium was collected from the SDC4 KO cells and EV fractions separated by serial centrifugation. Proteins and complexes from EV-deprived medium were precipitated by TCA as described before. As shown in Figure 9C, KO of SDC4 by siRNA did not change TG2 expression in TL, but it greatly reduced TG2 in exosome (P4) fractions, affecting the vesicular secretion of TG2. SDC4 KO also reduced the low level of free TG2 found in the EV-free conditioned medium (S4) (Figure 9C). As shown by quantifications (Figure 9D), KO of SDC4 by siRNA did not change expression of FLOT in ectosome (P3) and exosome (P4) fractions. IP of SDC4 from exosomes and ectosome lysates revealed SDC4 immunoreactivity in the two fractions, but SDC4 clearly pulled down TG2 only from exosomes (Figure 9E), suggesting a direct interaction of TG2 and SDC4 within EVs only in exosomes. Given the affinity of TG2 for the HS chains of SDC413,35 and their coassociation within exosomes shown here, our data suggest that TG2 is recruited as an HS binding cargo and targeted to exosomes by SDC4.
Figure 9.
Recruitment of TG2 in exosomes depends on SDC4. (A) EGFP-TG2–expressing NRK52E cells were treated with RNAi-targeting SDC4 (SDC4 KO) or nontargeting RNAi (Scrbl. siRNA; used as a control). TLs were analyzed by Western blotting testing for SDC4. (B) As a blotting control for SDC4, NRK52E cells were transiently transfected with HA-tagged SDC4 cDNA, and cell lysates were blotted for HA. A mock transfection (−) was performed as a control. The arrow points at HA-tagged SDC4 with a molecular mass between 25 and 35 kD. (C and D) Lysates of ectosome (P3) and exosome (P4) fractions isolated from the conditioned medium of SDC4 KO cells obtained as described in A and TLs were tested for (C and D) EGFP, (C) TG2, and (D) FLOT2. Intensity of immunoreactive bands was quantified by densitometric analysis; graphs represent mean values ±SEM of (C) four (EGFP) or (D) three (FLOT2) independent experiments expressed relative to the scrambled nontargeting siRNA control (equalized to one). **P<0.01. (E) SDC4 was immunoprecipitated from vesicular fractions and TLs with a rabbit polyclonal anti-SDC4 antibody. SDC4 immunoprecipitates were analyzed by Western blotting for the presence of EGFP-TG2 (black arrowhead) and SDC4 (gray arrowheads).
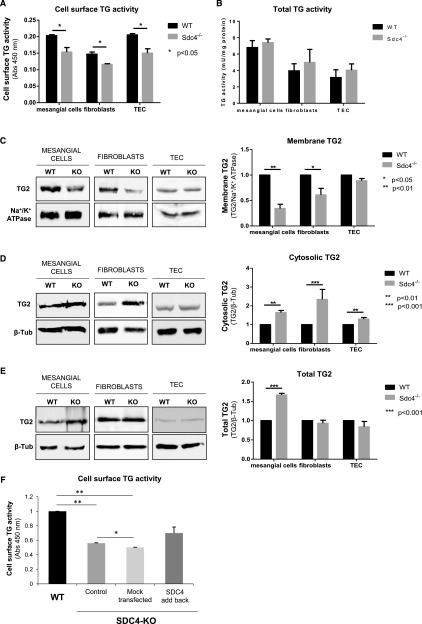
Vesicular Trafficking of TG2 Ex Vivo Depends on SDC4
To examine TG2 release and investigate the role played by SDC4 further, primary cortical TECs, fibroblasts, and mesangial cells were isolated from WT (SDC4+/+) and SDC4 KO (SDC4−/−) mice,36 and the level of extracellular TG2 was measured in live cells by a well established enzymatic assay.37 SDC4 deletion (SDC4−/−) resulted in lower TG2 externalization in all of the three primary renal cell types (Figure 10A), with no changes in the total level of TG2 activity between SDC4+/+ and SDC4−/− genotypes (Figure 10B). TECs displayed the highest level of extracellular TG2 compared with the total TG2 activity (Figure 10, A and B). The decreased TG2 externalization in SDC4−/− cells (Figure 10A) was accompanied by a lower level of membrane TG2 as measured by Western blotting of membrane fractions (Figure 10C) and an increased level of cytosolic TG2 (Figure 10D), suggesting TG2 retention. Total TG2 expression was not lowered by SDC4 KO when revealed by Western blotting of TLs (Figure 10E). The increment in total TG2 protein in SDC4−/− mesangial cells, compared with SDC4+/+ cells, may be a compensatory event of TG2 overproduction consequent to the defect in TG2 secretion (Figure 10E). Therefore, SDC4 KO impaired TG2 membrane distribution and externalization, with an increased retention of intracellular TG2 in the three primary renal cell types. To visualize TG2 trafficking in live cells, the primary WT and SDC4-null TECs were transiently transfected with an EGFP-tagged TG2 cDNA and visualized by live confocal microscopy (Supplemental Movie 1). EGFP-TG2 was recruited in globular elements continuously protruding and retracting from the PM and moving along the PM at the periphery of the cell. Budding of TG2-storing vesicles from the PM was evident. However, EGFP-TG2 was less dynamic and appeared confined in the cytosol in the SDC4-null TECs, which also had less budding activity than the WT TECs. When SDC4 cDNA was transfected back to SDC4-null TECs, EGFP-TG2 vesicular blebbing was largely resumed, and budding was reconstituted to WT levels (quantifications of extracellular TG2 activity are in Figure 10F, Supplemental Movie 1). These data show for the first time a dependence of TG2 vesicular trafficking on SDC4 and also suggest that SDC4 could be implicated in general EV dynamics of TECs involving cargo proteins with affinity for HS, like TG2.
Figure 10.
SDC4 KO leads to decreased TG2 membrane distribution and externalization ex vivo. Kidney glomeruli and tubules were isolated from WT and SDC KO (SDC4−/−) mice. Primary mesangial cells, fibroblasts, and TECs were grown from glomeruli or tubules using selective growth medium. (A) Extracellular TG activity was measured by a well established assay in live cells (4×104 cells per well of a 96-well plate) cultured on FN in the presence of the TG substrate biotin-cadaverine for 1 hour. Data represent mean Abs (450 nM) ±SD of three independent experiments undertaken in triplicates. (B) Total TG activity was similarly assayed but using whole cell lysate (60 μg per well) and specific activities obtained from a standard curve of purified TG2. Data represent mean TG activity ±SEM of three independent experiments undertaken in triplicates. (C–E) TG2 was probed by Western blot using a mouse monoclonal anti-TG2 antibody (IA12) and quantified by densitometric analysis on (C) cell membrane extracts, (D) cytosolic proteins, and (E) TLs. In each experiment, either Na+/K+ ATPase or β-tubulin was used as the loading control. Data represent mean intensities normalized for the loading control ±SEM; n=3 per group. (F) Extracellular TG activity was measured as described in A in WT and SDC4 KO (SDC4−/−) primary TECs. Where indicated, SDC4 KO cells (SDC4−/−) were transfected back with pcDNA3-hSdc4 plasmid or subjected to mock transfection (with transfection efficiency of approximately 30%). Data represent mean Abs (450 nM) ±SEM; n=3 independent experiments undertaken in triplicates. *P<0.05; **P<0.01; ***P<0.01.
TG2 Positive Exosomes in Urine Samples of Patients with CKD
To verify whether TG2 is released via exosomes in human CKD, we isolated EVs from pooled urine samples collected from a small cohort of patients with CKD (n=10) with stages 3 and 4 primary kidney disease and from healthy controls (n=5). Immunoblotting of equal amounts of samples (Supplemental Figure 4) revealed the presence of TG2 exclusively in the exosomes (P4) fractions, with an intensification of the band in the exosomes from the patients with CKD. There was no TG2 band in the ectosomes (P3) or the EV-free urine. FLOT was used as an EV marker, and it also intensified in the exosomes of patients with CKD. We previously reported the increased presence of TG2 in urines of patients with CKD by sandwich ELISA.38 Data in this study show that TG2 is conveyed by EVs in urines, validating our finding in human CKD.
Discussion
This is the first unbiased global analysis of TG2-interacting proteins in a model of CKD (the murine UUO type). TG2-associated proteins were identified using an original targeted proteomic strategy by combining TG2 IP from whole-kidney membrane preparations of WT kidneys with negative control IP from TG2-null kidneys from inbred mice. Furthermore, TG2 complexes have been used to interrogate the UUO proteome. Proteomes were resolved by SWATH data-independent acquisition MS.18 The proposed strategy could be adapted for precision-targeted proteomics in other systems.
Bioinformatic analysis of the TG2-associated proteins in the UUO model revealed enrichment of proteins involved in vesicular transport, and among them, a striking number were exosome-associated proteins. Because the TG2 partners involved in vesicular trafficking were not increased in the UUO proteome compared with the healthy proteome, we deduce that their function in TG2 trafficking is caused by their augmented interaction with TG2 on UUO, excluding the possibility of a concentration-dependent association. However, almost all proteins regulating ECM receptor organization and actin dynamics that we found significantly upregulated in the UUO proteome were also TG2-interacting proteins in the UUO. Collectively, these findings suggest the existence of a pathway of TG2 secretion during fibrosis progression driven by vesicular trafficking followed by the interaction of secreted TG2 with a protein network responsible for ECM dynamics, leading to fibrotic remodeling and expansion. The diminished scarring index and lower level of active TGF-β in the TG2 KO kidneys that we have reported here align well with the molecular connections that link TG2 to the fibrotic kidney proteome generated by this study, confirming the role of TG2 in the extracellular milieu in fibrosis progression.
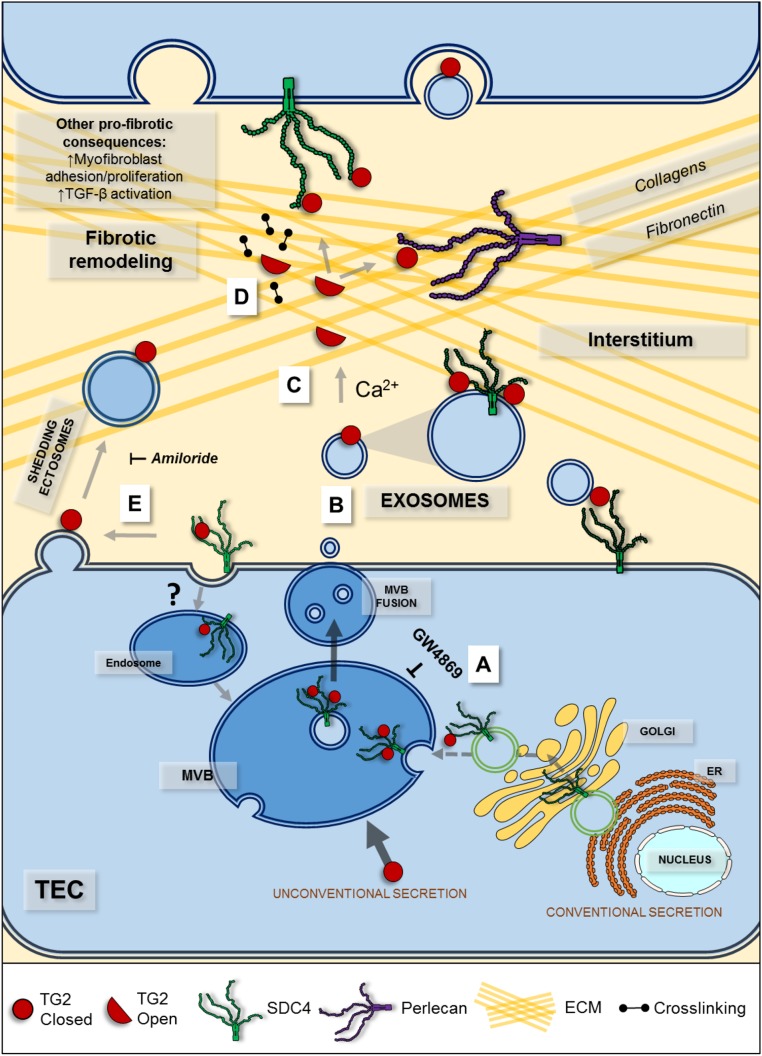
By exploiting the MS data in vitro and ex vivo, we have uncovered a novel pathway for the cellular export of TG2 in TECs. We have shown that TG2 is predominantly secreted in association with exosomes and that TG2-bearing exosomes require the sphingolipid ceramide for their production. Furthermore, TG2 enrichment in exosomes is stimulated by TGF-β1, although on the basis of quantifications of exosomes via FLOT, a well characterized EV marker and partner of TG2 in vivo post-UUO, we cannot rule out a general increase in EV production by TGF-β1. Exosomes form from endosomes by budding into late endosomes (multivesicular bodies), which then fuse with the PM, releasing intraluminal vesicles or exosomes.39 The endosomal sorting complex required for transport (ESCRT) machinery is the main system controlling the sorting of proteins into exosomes.39 Loading of syndecan HS proteoglycans (SDC1 and SDC4) with intracellular cargos able to bind the proteoglycan HS chains leads to syndecan clustering and recruitment of cytosolic adaptor protein syntenin-1, which by direct interaction with Alix, adapts syndecan and syndecan-bound cargoes to the ESCRT at the level multivesicular body formation.34 In recent work, even TG2 itself has been implicated in cargo recruitment to vesicles and is present in exosomes in situations of abnormal proteostasis in an MEF model.40
TG2 is an HS binding protein, and HS affects TG2 trafficking as shown in previous work12,13,35,41; consistent with this observation, SDC4 emerged as a specific partner of TG2 in the UUO kidney in this unbiased analysis. We and others have previously identified SDC4 as a profibrotic partner of TG28,12,13 influencing its secretion but without a clear mechanism. Here, we have for the first time unraveled that the interaction of TG2 with HS SDC4 may play a fundamental role in the targeting of TG2 to exosomes. SDC4 and TG2 were coimmunoprecipitated from exosomal lysates, and SDC4 was required for exosomal secretion of TG2, this being greatly diminished by siRNA targeted to SDC4. Ex vivo, live imaging of primary cortical TECs from SDC4 KO kidney showed reduced vesicular trafficking of TG2 to the cell surface, which was compensated by SDC4 add back. TG2 was retained in the cytosolic fraction not only of SDC4 KO TECs but also, of primary fibroblasts and mesangial cells isolated from SDC4 KO kidneys, suggesting that SDC4 regulates TG2 distribution in all of the main renal cell types. In vivo, SDC4 was a TG2-interacting partner only in the diseased kidney, consistent with the notion that TG2 is secreted during fibrosis progression and only found in low amounts extracellularly in the normal kidney.4,20 Therefore, our data suggest that TG2 would be recruited as an HS binding cargo and targeted to exosomes originating from late endosomes by SDC4. After secreted in the extracellular environment, TG2 meets the high calcium and low trinucleotide phosphates concentrations, which favor the opening of TG2 and the acquisition of a linear conformation on substrate binding,42 that we and others have shown to determine loss of heparin binding.35,41 Consistent with this idea is the finding of a small portion of free soluble TG2 in our cell experiments and the possibility that TG2 and/or EV-bound TG2 may be transferred to HS of other receptor molecules in response to local changes in calcium levels in an autocrine or paracrine fashion (Figure 11). For instance, perlecan is an ECM HS proteoglycan exclusively found in the TG2 UUO interactome, which could further recruit and distribute TG2 in the matrix via the long HS chains. Furthermore, we have shown that exosomal TG2 could be transferred horizontally from the extracellular environment to TECs and that HS chains of proteoglycans are required for the trafficking of TG2-rich exosomes. TG2 was also detected in ectosomes, larger vesicles that bud directly from the PM, although in much lower proportion than in exosomes. There was no clear coprecipitation of SDC4 and TG2 from ectosome lysates (only from exosome lysates), implying that SDC4 has no role or an indirect role in recruiting TG2 to ectosomes.
Figure 11.
The proposed pathway of TG2 cell surface trafficking in TEC during fibrosis progression involves TG2 targeting to exosomes from late endosomes by SDC4. TG2 is secreted unconventionally by exosomes originating from late endosomes as multivesicular bodies (MVBs), which then fuse with the PM, releasing the intraluminal vesicles outside cells. (A) The biogenesis of TG2-bearing exosomes is mediated by ceramide, because the inhibitor of N-SMase GW4869 significantly reduces the presence of TG2 in the exosome fraction. TG2, which has high affinity for HS, is loaded on the surface of the intraluminal vesicle as an HS/SDC4 cargo, and it is exposed on the surface of the formed exosome. (B) After fusion of the outer membrane of MVB with PM, the TG2-bearing exosomes likely accumulate in the ECM by TG2 binding to ECM/cell surface protein partners in an “autocrine” or “paracrine” fashion. (C) After released with exosomes, TG2 could also undergo a conformational change and linearize because of the high Ca2+-to-GTP ratio of the extracellular space, with a lowering or even loss of HS affinity; the free TG2 could bind the HS of other proteoglycans or other ECM partners. (D) In the matrix, TG2 acts as an adhesive protein and matrix crosslinker, promoting fibers accumulation and latent TGF-β1 recruitment. (E) TG2 is also present in ectosomes. It is possible that TG2, after it is secreted via exosomes, is captured by the PM and retained on ectosomes shed by the PM. ER, endoplasmic reticulum.
The role played by EVs in disease is of growing importance, although the field is novel. Exosomes function in intercellular and interorgan communication by transferring proteins, mRNAs, and microRNAs to recipient cells.43 In seminal work, it was shown that hypoxic tubular cells but not normoxic counterparts secreted EVs that induced the expression of TGF-β1, α-SMA, and F-actin in fibroblasts, inducing their subsequent activation.44 Therefore, it is clear that EVs cooperate in spreading and amplifying cell differentiation, leading to sustained fibrosis.43,44 Transfer of exosomal cargo to neighboring cells allows tuning of cell behavior and rapid phenotypic adjustment, including activation of fibroblasts into myofibroblasts.44 Given the location of TG2 outside exosomes and specific interaction of TG2 with cell surface SDC4, it is plausible that, after released, TG2 could assist in the cell-cell trafficking of exosomes, therefore influencing the transfer of pathologic molecules conducive of fibrosis. Furthermore, the topology of TG2 on the outside of EVs would suggest that it may also serve an adhesive function, which is an additional important factor in the chronic healing process underlying fibrosis development. This suggestion is in line with the adhesive role played by TG245 and recent evidence that exosomes influence cell migration by serving as adhesive sites in the matrix.46
TG2 is associated with wound healing and organ fibrosis, but what makes it an attractive target is that the enzyme is only activated in disease on externalization. Therefore, inhibition of TG2 is anticipated not to have an adverse effect in normal cells. We have begun our investigation from an in vivo model of CKD providing a global analysis of the TG2 interactome present at the interface between cells and the ECM niche–surrounding cells and compared this with the CKD model proteome in an unbiased manner. Our approach, which does not rely on chemical crosslinking, has not detected integrin16 or purinergic receptor17 as TG2 secretion partners in fibrotic kidney or attributed TG2 release to PM vesicular shedding as is observed in cancer,47 but it has identified exosome secretion as a key novel mechanism for the cell surface trafficking of fibrogenic TG2 in an SDC4-dependent manner. The presence of TG2 in urinary exosomes from a pool of patients with CKD has confirmed the importance of the finding in human pathology. Our study changes the way that TG2 cell surface trafficking is seen and suggests that therapeutic block of vesicular TG2 could potentially impede TG2-driven matrix accumulation and exosome-mediated transfer of TG2 from cell to cell during fibrosis progression.
Concise Methods
Experimental details are provided in Supplemental Material.
Experimental Models
Experimental UUO48 was performed on fully backcrossed TG2−/− (TG2 KO) mice originally obtained from Gerry Melino49 and WT inbred C57BL/6J mice as described before12 and in Supplemental Material. A total of 32 WT and 32 TG2 KO mice were used, of which 16 per genotype were subjected to either UUO or sham operation. Therefore, four groups of 16 mice were obtained: WT UUO, WT sham, TG2 KO UUO, and TG2 KO sham. The UUO and sham proteome required lysate from one half of a kidney per animal and was carried out in kidneys from four mice per group. Kidneys were halved longitudinally pole to pole through the papilla. Each one half was then cut transversally to provide four identical quarters, with equal cortex and medullary mass in each. The TG2 interactome required two full organs per IP and was carried out in kidneys from 12 male mice per group. Western blotting analysis required one quarter of kidney per animal and was performed using kidneys from three male mice per group as described before.8 Optimal size of biologic replicates for the UUO and sham proteome analysis was determined by a power calculation within Statistica software (Dell), and n=4 individuals per treatment were sufficient to power the experiment above 80%. In keeping with the principle of animal reduction, male and female mice kidneys were used for general proteomics (one male and three female). Linear regression analysis was performed on protein peak areas after SWATH acquisition MS and confirmed that the variability in protein intensities produced by the male mice was not higher than the intrinsic biologic variability detected among the female (P≤0.05; mean standard residuals ≥5). All experimental procedures were carried out under license in accordance with regulations laid down by Her Majesty’s Government, United Kingdom (Animals Scientific Procedures Act ASPA, 1986), and they were approved by the University of Sheffield Animal Ethical Review Committee (ASPA Ethical Review Process) and the Nottingham Trent University Ethical Review Committee (ASPA Ethical Review Process).
Fibrosis Measurement
The level of fibrosis was assessed by Masson trichrome staining as previously described.50
TGF-β Activity
The mink lung epithelial cell bioassay51 was performed as described before.12,52
Isolation of TG2 Membrane Complexes and Data Acquisition by MS
To isolate TG2-associated proteins from kidney fractions, 10% (wt/vol) kidney homogenates were prepared from WT and TG2 KO kidneys, UUO and sham operated, by mechanical homogenization performed on ice using an Ultra Turrax T25 homogenizer (Merck) in homogenization buffer (0.25 M sucrose, 10 mM Tris-HCl, 1 mM MgCl2, and 2 mM EDTA, pH 7.4) containing protease inhibitor cocktail (Sigma-Aldrich). The whole homogenate was centrifuged at 1000×g for 5 minutes at 4°C to remove larger particulates; then, membranes were separated from the cytosolic fraction by centrifugation at 20,000×g at 4°C for 30 minutes as previously described.12 The pellet (crude membrane fraction) was resuspended in IP buffer (25 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% [vol/vol] NP40 detergent solution, and 5% [vol/vol] glycerol, pH 7.4) containing protease inhibitor cocktail and membrane enrichment validated by Western blotting. TG2 with associated proteins was immunoprecipitated from the membrane fraction using the Pierce crosslink magnetic IP kit (Thermo Scientific) by protein A/G magnetic beads to which anti-TG2 antibody (IA12; University of Sheffield) was crosslinked using disuccinimidyl suberate. Incubations of membrane lysates with the antibody-coated beads were performed for 15 hours at 4°C in constant rotation. TG2-associated proteins were subjected to trypsin digestion directly on the beads after washing the beads three times with 50 mM ammonium acetate and incubating for 15 hours with 200 ng/μl of proteomics-grade trypsin (Sigma-Aldrich) in 50 mM ammonium acetate. Peptides were analyzed by MS on removal of the beads using a magnetic stand, by RP-HPLC-ESI-MS/MS using a TripleTOF 5600+ mass spectrometer (SCIEX) in data-dependent acquisition mode for spectral library construction, and in SWATH 2.0 data-independent acquisition mode using static SWATH acquisition windows for quantification as described in Supplemental Material. The MS proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository53 with the dataset identifier PXD008173. To define proteins significantly associated with TG2, a z-test statistical analysis was performed using TG2 IP from TG2 KO kidney membranes as negative controls. Five independent TG2 IP experiments were carried out, each on the basis of a lysate of two kidneys. The obtained interactome has been submitted to the database for CKD-related expression data CKDdb54 (http://www.padb.org/ckddb/) with the dataset identifier ExpGFEVabf79st4na.
The UUO Kidney Proteome
UUO and sham kidney halves were ground in liquid nitrogen, and a 10% (wt/vol) tissue homogenate was prepared in lysis buffer containing 9.5 M urea, 2% (wt/vol) dithiothreitol, 1% (wt/vol) N-octyl-β-glucopyranoside, and protease inhibitors (Sigma-Aldrich) with sonication. Equal amounts of total protein extracts (50 μg) were diluted in 50 mM triethyl ammonium bicarbonate containing a final concentration of 0.02% (wt/vol) ProteaseMAX surfactant (Promega). Proteins were subjected to reduction (5 mM dithiothreitol at 56°C for 20 minutes) and alkylation (15 mM iodoacetamide at room temperature for 15 minutes), and then, they were trypsin digested overnight at 37°C with 0.02 mg/ml MS-grade trypsin (Promega) and 0.01% (wt/vol) ProteaseMAX surfactant in a thermomixer. Samples were vacuum concentrated to dryness and resuspended in 20 μl of 5% (vol/vol) acetonitrile/0.1% (vol/vol) formic acid for MS analysis. SWATH data-independent acquisition MS was performed on the obtained peptides using variable SWATH windows. The MS proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository53 with the dataset identifier PXD008173. Analysis of differentially expressed proteins was performed using the OneOmics cloud processing online platform (SCIEX) as described in Supplemental Material. Proteome data have been submitted to CKDdb54 (http://www.padb.org/ckddb/) with the dataset identifier ExpGFEVyz6gt389dy.
Primary and Immortal Cell Lines
Tubules and glomeruli were isolated from WT and SDC4−/− (SDC4 KO)36 C57BL/6J mice by perfusion of kidneys in situ with BSA-inactivated Dynabeads and subsequent magnetic separation, and they were cultured as previously described and detailed in Supplemental Methods.55 Primary cortical TECs grew out from tubules in selective medium with insulin, transferrin, selenium, and EGF in 0.5% (vol/vol) FBS to cause proliferation of just epithelial cells and not fibroblasts (grown out in medium with 10% FBS); primary mesangial cells were derived from glomeruli in RPMI-1640 containing 20% (vol/vol) FBS. Media composition is detailed in Supplemental Material.
NRK52E rat renal proximal-like TECs and NRK49F renal fibroblasts were obtained from the European Cell Culture Collection and maintained in DMEM supplemented with 5% (vol/vol) or 10% (vol/vol) heat-inactivated FBS, respectively.
Plasmid Constructs and Transfections
The following constructs were prepared: pEGFP-N1-TG2, where TG2 cDNA was fused in frame to the N terminus of EGFP in pEGFP-N1 plasmid (Clontech), and pcDNA3.1(+)-hSdc4, where HA-Sdc4 cDNA (HA downstream of the leader peptide provided by Mark Bass, Sheffield University) was subcloned from pBluescript in pcDNA3.1(+). All constructs were verified by Sanger sequencing. Cell transfection was performed by TransIT (Mirus Bio). NRK52E was stably transfected with pEGFP-N1-TG2 in medium supplemented with 700mg/ml G418, as previously described,56 by using electroporation (Amaxa Nucleofector). SDC4 was knocked down by transient transfection with 100 nM rat SDC4-targeting siRNA or scrambled control siRNA (Dharmacon; Thermo Scientific) for 48 hours using DharmaFECT as transfection reagent.
EV Isolation and Characterization
EVs were isolated from conditioned medium of cells by differential centrifugation as described.24,25 Nanoparticle tracking analysis by tunable resistive pulse sensing was performed using qNano (Izon) to examine microparticle size and distribution. In some cases, cells were stimulated by recombinant TGF-β1 (10 ng/ml) to simulate conditions of fibrogenesis in vitro. The EV inhibitor N,N′-Bis[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]-3,3′-p-phenylene-bis-acrylamide dihydrochloride, GW4869 (D1692; Sigma-Aldrich) was used as an inhibitor of N-SMase57 and exosome biogenesis26 at a concentration of 10 μM in serum-free medium for 16 hours. Amiloride hydrochloride (N-Amidino-3,5-diamino-6-chloropyrazinecarboxamide hydrochloride hydrate; A7410; Sigma-Aldrich) was used at a concentration of 1 mM for 10 minutes. For IPs from EV lysates, the Pierce crosslink magnetic IP kit was used followed by Western blotting.
Detection of TG2 in Cell Fractions by Western Blotting
Cells were fractionated into membrane and cytosolic fractions, and TG2 was detected as described before.12,13
Measurements of TG2 Activity at the Cell Surface of Live Cells, in Cell Fractions, and in EVs
TG2 activity associated with the extracellular surface in cells in culture and total TG2 activity in cell homogenates and fractions were assayed by measuring the incorporation of biotin-cadaverine into FN by calcium-dependent transamidation as previously described.13 EVs TG2 activity was measured in both EVs lysed in a sucrose-based buffer (0.32 M sucrose, 5 mM Tris-HCl, and 2 mM EDTA, pH 7.4) with protease inhibitor cocktail and intact EVs kept in suspension in sample buffer (Zedira) using the commercially available tissue transglutaminase picoassay kit (Zedira).
Disclosures
None.
Supplementary Material
Acknowledgments
We are very grateful to Gerry Melino (University of Rome “Tor Vergata”), Eleonora Candi (University of Rome “Tor Vergata”), and Takashi Muramatsu (Aichi Gakuin University) for generously providing transgenic mice. We thank Mark Bass (University of Sheffield) for the HA-Sdc4 construct. We thank Sylvie Ricard-Blum (University of Lyon) for suggesting systems biology resources and Nadia Chuzhanova (Nottingham Trent University) for help with statistics We thank Izhar Burhan (Nottingham Trent University) for invaluable general help and Martina Gabrielli for help with qNano.
The research was supported by Kidney Research UK project grant RP25/2012 (to E.A.M.V., T.S.J., and G.B.) and in part by Wellcome Trust project grant 087163 (to T.S.J. and E.A.M.V.). Work at NTU was also financed by the Research Assessment Exercise-Quality Research (U12) fund (E.A.M.V.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017050479/-/DCSupplemental.
References
- 1.Boor P, Ostendorf T, Floege J: Renal fibrosis: Novel insights into mechanisms and therapeutic targets. Nat Rev Nephrol 6: 643–656, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Webster AC, Nagler EV, Morton RL, Masson P: Chronic kidney disease. Lancet 389: 1238–1252, 2017 [DOI] [PubMed] [Google Scholar]
- 3.Johnson TS, El-Koraie AF, Skill NJ, Baddour NM, El Nahas AM, Njloma M, Adam AG, Griffin M: Tissue transglutaminase and the progression of human renal scarring. J Am Soc Nephrol 14: 2052–2062, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Johnson TS, Skill NJ, El Nahas AM, Oldroyd SD, Thomas GL, Douthwaite JA, Haylor JL, Griffin M: Transglutaminase transcription and antigen translocation in experimental renal scarring. J Am Soc Nephrol 10: 2146–2157, 1999 [DOI] [PubMed] [Google Scholar]
- 5.Verderio EAM, Furini G, Burhan IW, Johnson TS: Transglutaminases: Expression in kidney and relation to kidney fibrosis. In: Transglutaminases, edited by Hitomi K, Kojima S, Fesus L, Tokyo, Japan, Springer, 2015, pp 229–262 [Google Scholar]
- 6.Annes JP, Munger JS, Rifkin DB: Making sense of latent TGFbeta activation. J Cell Sci 116: 217–224, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Shweke N, Boulos N, Jouanneau C, Vandermeersch S, Melino G, Dussaule JC, Chatziantoniou C, Ronco P, Boffa JJ: Tissue transglutaminase contributes to interstitial renal fibrosis by favoring accumulation of fibrillar collagen through TGF-beta activation and cell infiltration. Am J Pathol 173: 631–642, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burhan I, Furini G, Lortat-Jacob H, Atobatele AG, Scarpellini A, Schroeder N, Atkinson J, Maamra M, Nutter FH, Watson P, Vinciguerra M, Johnson TS, Verderio EA: Interplay between transglutaminases and heparan sulphate in progressive renal scarring. Sci Rep 6: 31343, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng XM, Nikolic-Paterson DJ, Lan HY: TGF-β: The master regulator of fibrosis. Nat Rev Nephrol 12: 325–338, 2016 [DOI] [PubMed] [Google Scholar]
- 10.Johnson TS, Fisher M, Haylor JL, Hau Z, Skill NJ, Jones R, Saint R, Coutts I, Vickers ME, El Nahas AM, Griffin M: Transglutaminase inhibition reduces fibrosis and preserves function in experimental chronic kidney disease. J Am Soc Nephrol 18: 3078–3088, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Haylor JL, Hau Z, Jones RA, Vickers ME, Wagner B, Griffin M, Saint RE, Coutts IG, El Nahas AM, Johnson TS: Transglutaminase inhibition ameliorates experimental diabetic nephropathy. Kidney Int 76: 383–394, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Scarpellini A, Huang L, Burhan I, Schroeder N, Funck M, Johnson TS, Verderio EA: Syndecan-4 knockout leads to reduced extracellular transglutaminase-2 and protects against tubulointerstitial fibrosis. J Am Soc Nephrol 25: 1013–1027, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarpellini A, Germack R, Lortat-Jacob H, Muramatsu T, Billett E, Johnson T, Verderio EA: Heparan sulfate proteoglycans are receptors for the cell-surface trafficking and biological activity of transglutaminase-2. J Biol Chem 284: 18411–18423, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou CY, Streets AJ, Watson PF, Huang L, Verderio EA, Johnson TS: A crucial sequence for transglutaminase type 2 extracellular trafficking in renal tubular epithelial cells lies in its N-terminal beta-sandwich domain. J Biol Chem 286: 27825–27835, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckert RL, Kaartinen MT, Nurminskaya M, Belkin AM, Colak G, Johnson GV, Mehta K: Transglutaminase regulation of cell function. Physiol Rev 94: 383–417, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zemskov EA, Mikhailenko I, Hsia RC, Zaritskaya L, Belkin AM: Unconventional secretion of tissue transglutaminase involves phospholipid-dependent delivery into recycling endosomes. PLoS One 6: e19414, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adamczyk M, Griffiths R, Dewitt S, Knäuper V, Aeschlimann D: P2X7 receptor activation regulates rapid unconventional export of transglutaminase-2. J Cell Sci 128: 4615–4628, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R: Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11: O111.016717, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel M, Strnad P, Watts RE, Choi K, Jabri B, Omary MB, Khosla C: Extracellular transglutaminase 2 is catalytically inactive, but is transiently activated upon tissue injury. PLoS One 3: e1861, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skill NJ, Griffin M, El Nahas AM, Sanai T, Haylor JL, Fisher M, Jamie MF, Mould NN, Johnson TS: Increases in renal epsilon-(gamma-glutamyl)-lysine crosslinks result from compartment-specific changes in tissue transglutaminase in early experimental diabetic nephropathy: Pathologic implications. Lab Invest 81: 705–716, 2001 [DOI] [PubMed] [Google Scholar]
- 21.Humphries JD, Byron A, Bass MD, Craig SE, Pinney JW, Knight D, Humphries MJ: Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci Signal 2: ra51, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Q, Yang Y, Wang CL, Hou Y, Chen H: Screening and identification of the differential proteins in kidney with complete unilateral ureteral obstruction. Int J Clin Exp Pathol 8: 2615–2626, 2015 [PMC free article] [PubMed] [Google Scholar]
- 23.Satirapoj B, Tassanasorn S, Charoenpitakchai M, Supasyndh O: Periostin as a tissue and urinary biomarker of renal injury in type 2 diabetes mellitus. PLoS One 10: e0124055, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, Menna E, Saglietti L, Schuchman EH, Furlan R, Clementi E, Matteoli M, Verderio C: Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J 28: 1043–1054, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benussi L, Ciani M, Tonoli E, Morbin M, Palamara L, Albani D, Fusco F, Forloni G, Glionna M, Baco M, Paterlini A, Fostinelli S, Santini B, Galbiati E, Gagni P, Cretich M, Binetti G, Tagliavini F, Prosperi D, Chiari M, Ghidoni R: Loss of exosomes in progranulin-associated frontotemporal dementia. Neurobiol Aging 40: 41–49, 2016 [DOI] [PubMed] [Google Scholar]
- 26.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M: Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319: 1244–1247, 2008 [DOI] [PubMed] [Google Scholar]
- 27.Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ: Exosome release of β-catenin: A novel mechanism that antagonizes Wnt signaling. J Cell Biol 190: 1079–1091, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falcone S, Cocucci E, Podini P, Kirchhausen T, Clementi E, Meldolesi J: Macropinocytosis: Regulated coordination of endocytic and exocytic membrane traffic events. J Cell Sci 119: 4758–4769, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D, De Thonel A, Multhoff G, Hamman A, Martin F, Chauffert B, Solary E, Zitvogel L, Garrido C, Ryffel B, Borg C, Apetoh L, Rébé C, Ghiringhelli F: Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest 120: 457–471, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller ME, Adhikary S, Kolokoltsov AA, Davey RA: Ebolavirus requires acid sphingomyelinase activity and plasma membrane sphingomyelin for infection. J Virol 86: 7473–7483, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savina A, Furlán M, Vidal M, Colombo MI: Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem 278: 20083–20090, 2003 [DOI] [PubMed] [Google Scholar]
- 32.Cocucci E, Meldolesi J: Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol 25: 364–372, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Stelmach H, Rusak T, Tomasiak M: The involvement of the Na(+)/H(+) exchanger in the formation of microvesicles by porcine platelets. Haematologia (Budap) 32: 239–252, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G: Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14: 677–685, 2012 [DOI] [PubMed] [Google Scholar]
- 35.Lortat-Jacob H, Burhan I, Scarpellini A, Thomas A, Imberty A, Vivès RR, Johnson T, Gutierrez A, Verderio EA: Transglutaminase-2 interaction with heparin: Identification of a heparin binding site that regulates cell adhesion to fibronectin-transglutaminase-2 matrix. J Biol Chem 287: 18005–18017, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishiguro K, Kadomatsu K, Kojima T, Muramatsu H, Tsuzuki S, Nakamura E, Kusugami K, Saito H, Muramatsu T: Syndecan-4 deficiency impairs focal adhesion formation only under restricted conditions. J Biol Chem 275: 5249–5252, 2000 [DOI] [PubMed] [Google Scholar]
- 37.Jones RA, Kotsakis P, Johnson TS, Chau DY, Ali S, Melino G, Griffin M: Matrix changes induced by transglutaminase 2 lead to inhibition of angiogenesis and tumor growth. Cell Death Differ 13: 1442–1453, 2006 [DOI] [PubMed] [Google Scholar]
- 38.da Silva Lodge M, El Nahas M, Johnson TS: Urinary transglutaminase 2 as a potential biomarker of chronic kidney disease detection and progression [Abstract]. Lancet 381: S33, 2013 [Google Scholar]
- 39.Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M: Sorting it out: Regulation of exosome loading. Semin Cancer Biol 28: 3–13, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz-Hidalgo L, Altuntas S, Rossin F, D’Eletto M, Marsella C, Farrace MG, Falasca L, Antonioli M, Fimia GM, Piacentini M: Transglutaminase type 2-dependent selective recruitment of proteins into exosomes under stressful cellular conditions. Biochim Biophys Acta 1863: 2084–2092, 2016 [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Collighan RJ, Pytel K, Rathbone DL, Li X, Griffin M: Characterization of heparin-binding site of tissue transglutaminase: Its importance in cell surface targeting, matrix deposition, and cell signaling. J Biol Chem 287: 13063–13083, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinkas DM, Strop P, Brunger AT, Khosla C: Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol 5: e327, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L: Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int 78: 838–848, 2010 [DOI] [PubMed] [Google Scholar]
- 44.Borges FT, Melo SA, Özdemir BC, Kato N, Revuelta I, Miller CA, Gattone VH 2nd, LeBleu VS, Kalluri R: TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J Am Soc Nephrol 24: 385–392, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorand L, Graham RM: Transglutaminases: Crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol 4: 140–156, 2003 [DOI] [PubMed] [Google Scholar]
- 46.Sung BH, Ketova T, Hoshino D, Zijlstra A, Weaver AM: Directional cell movement through tissues is controlled by exosome secretion. Nat Commun 6: 7164, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, Holowka DA, Cerione RA: Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci U S A 108: 4852–4857, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vielhauer V, Anders HJ, Mack M, Cihak J, Strutz F, Stangassinger M, Luckow B, Gröne HJ, Schlöndorff D: Obstructive nephropathy in the mouse: Progressive fibrosis correlates with tubulointerstitial chemokine expression and accumulation of CC chemokine receptor 2- and 5-positive leukocytes. J Am Soc Nephrol 12: 1173–1187, 2001 [DOI] [PubMed] [Google Scholar]
- 49.De Laurenzi V, Melino G: Gene disruption of tissue transglutaminase. Mol Cell Biol 21: 148–155, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang L, Scarpellini A, Funck M, Verderio EA, Johnson TS: Development of a chronic kidney disease model in C57BL/6 mice with relevance to human pathology. Nephron Extra 3: 12–29, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ, Rifkin DB: An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal Biochem 216: 276–284, 1994 [DOI] [PubMed] [Google Scholar]
- 52.Huang L, Haylor JL, Fisher M, Hau Z, El Nahas AM, Griffin M, Johnson TS: Do changes in transglutaminase activity alter latent transforming growth factor beta activation in experimental diabetic nephropathy? Nephrol Dial Transplant 25: 3897–3910, 2010 [DOI] [PubMed] [Google Scholar]
- 53.Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H: The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res 41: D1063–D1069, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandes M, Husi H: Establishment of a integrative multi-omics expression database CKDdb in the context of chronic kidney disease (CKD). Sci Rep 7: 40367, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fisher M, Jones RA, Huang L, Haylor JL, El Nahas M, Griffin M, Johnson TS: Modulation of tissue transglutaminase in tubular epithelial cells alters extracellular matrix levels: A potential mechanism of tissue scarring. Matrix Biol 28: 20–31, 2009 [DOI] [PubMed] [Google Scholar]
- 56.Verderio E, Nicholas B, Gross S, Griffin M: Regulated expression of tissue transglutaminase in Swiss 3T3 fibroblasts: Effects on the processing of fibronectin, cell attachment, and cell death. Exp Cell Res 239: 119–138, 1998 [DOI] [PubMed] [Google Scholar]
- 57.Luberto C, Hassler DF, Signorelli P, Okamoto Y, Sawai H, Boros E, Hazen-Martin DJ, Obeid LM, Hannun YA, Smith GK: Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J Biol Chem 277: 41128–41139, 2002 [DOI] [PubMed] [Google Scholar]
- 58.Cheadle C, Vawter MP, Freed WJ, Becker KG: Analysis of microarray data using Z score transformation. J Mol Diagn 5: 73–81, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathivanan S, Simpson RJ: ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9: 4997–5000, 2009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.