Abstract
E1784K is the most common mixed syndrome SCN5a mutation underpinning both Brugada syndrome type 1 (BrS1) and Long-QT syndrome type 3 (LQT3). The charge reversal mutant enhances the late sodium current (INa) passed by the cardiac voltage-gated sodium channel (NaV1.5), delaying cardiac repolarization. Exercise-induced triggers, like elevated temperature and cytosolic calcium, exacerbate E1784K late INa. In this study, we tested the effects of Ranolazine, the late INa blocker, on voltage-dependent and kinetic properties of E1784K at elevated temperature and cytosolic calcium. We used whole-cell patch clamp to measure INa from wild type and E1784K channels expressed in HEK293 cells. At elevated temperature, Ranolazine attenuated gain-of-function in E1784K by decreasing late INa, hyperpolarizing steady-state fast inactivation, and increasing use-dependent inactivation. Both elevated temperature and cytosolic calcium hampered the capacity of Ranolazine to suppress E1784K late INa. In-silico action potential (AP) simulations were done using a modified O’Hara Rudy (ORd) cardiac model. Simulations showed that Ranolazine failed to shorten AP duration, an effect augmented at febrile temperatures. The drug-channel interaction is clearly affected by external triggers, as reported previously with ischemia. Determining drug efficacy under various physiological states in SCN5a cohorts is crucial for accurate management of arrhythmias.
Introduction
The alpha subunit of the cardiac voltage-gated sodium channel, NaV1.5, is encoded by the SCN5a gene. Mutations in this gene usually cause long-QT syndrome type 3 (LQT3), Brugada syndrome type 1 (BrS1), or both (mixed syndromes)1–5. These clinical conditions are elicited by expression of gating dysfunctions in NaV1.56–10. Gain- and loss-of-function mutations can modify the inward sodium current (INa). Gain-of-function (GoF) in NaV1.5 arises from loss in channel fast inactivation, thereby increasing the non-inactivating, late INa, underlying LQT37,11–16. Loss-of-function (LoF) mainly arises from decreased peak INa resulting in BrS11,6,17–20. Interestingly, both GoF and LoF defects can occur simultaneously in a number of mutants6,12,17,21–24.
A guanine to an adenine substitution at position 5349 in SCN5a expresses the charge reversal mutant, E1784K, in the NaV1.5 C-terminal domain (CTD)25,26. E1784K is the most common mixed syndrome mutant, particularly prevalent in the Okinawa Islands in Japan, where carriers mainly express diagnostic LQT327. Clinical studies reveal differential phenotypic expressivity in E1784K cohorts12,22,28,29.
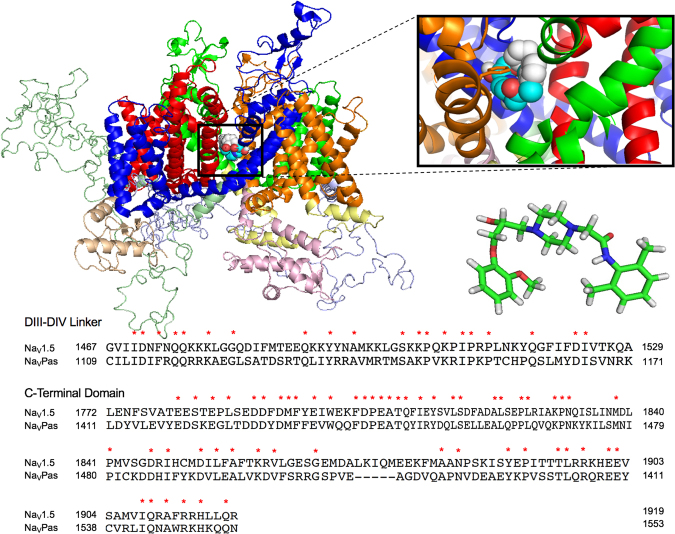
E1784 is located directly upstream of the acidic globular EF-like hand domain (α1–α4). The residue contributes to the electrostatic interactions formed between the acidic domain and the downstream basic IQ domain (α6, Fig. 1 compares WT to E1784K structure)30–34. The proximal CTD, in which E1784K resides, has the largest effects on kinetics and steady-state inactivation30,33,35,36. The charge reversal mutant, E1784K, is thought to disturb the integrity of CTD, causing the α6 to become more mobile (Fig. 1)37. A disturbance to α6 integrity has been correlated with elevations in late INa and enhanced slow inactivation16,35,38–40, which are key biophysical attributes in E1784K12,22,28.
Figure 1.
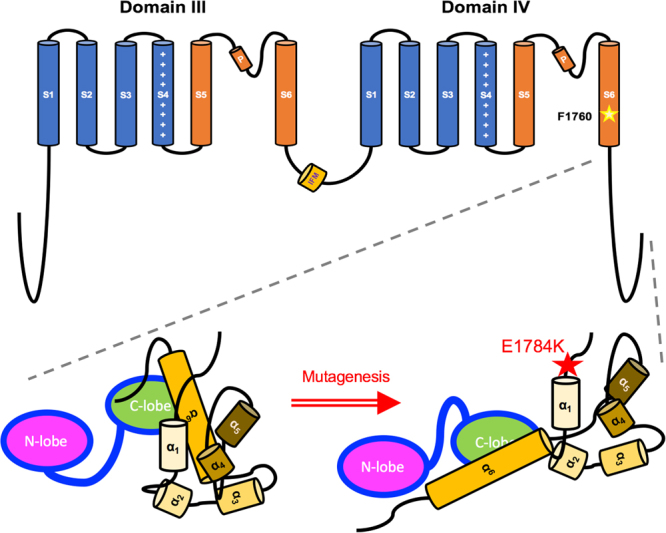
Sodium Channel Schematic Diagram. Domain III and Domain IV, along with their inter-linker and the CTD, regulate channel function and calcium sensitivity. DIII-DIV linker contains the fast inactivation “particle” (IFM motif). DIV-S6 contains the putative drug-binding residue, F1760, which is key for Ranolazine binding. The enlarged CTD contains six α-helices which aggregate to form an intact domain for calmodulin binding. Calmodulin (CaM) binds to the IQ-domain (α6) under low cytosolic calcium conditions (structure adapted from Chagot et al., 2009 and Gabelli et al., 2014). E1784K may rearrange CTD structure affecting Calmodulin interaction with other channel sites, such as the DIII-DIV linker.
Recent studies characterized the effects of exercise-induced triggers on E1784K. These triggers include acidosis, elevated temperatures, and cytosolic calcium. Acidosis and elevated temperatures augment late INa and decrease peak INa in E1784K41–43. Use-dependence in E1784K is reduced with high stimulation frequencies at elevated temperatures41. Compared to other mutants, E1784K tends to hamper the native potency of cytosolic calcium to block late INa in NaV1.537,44. Elevated cytosolic calcium augments channel availability in E1784K by depolarizing the voltage-dependence of slow inactivation37. Dynamic in silico action potential (AP) simulations in cardiac cells show E1784K-induced alternans at sinus rhythm and with tachycarida37,41.
We hypothesize that Ranolazine, which preferentially blocks late INa, is suitable for ameliorating the thermal and calcium-induced defects in E1784K. Although prescribed as an anti-anginal drug for diastolic dysfunction treatment45–47, Ranolazine has anti-arrhythmic efficacy proven to be useful in treating SCN5a inherited conditions48–51. Ranolazine efficacy is enhanced with SCN5a mutations or channel triggers, such as acidosis, which augment late INa48,49,52–54.We predicted that the channel mutation-trigger interaction may alter drug efficacy. Our goal is to study the effects of Ranolazine on E1784K under conditions of elevated temperature and cytosolic calcium levels.
Results
Ranolazine binds to NaV1.5 inner vestibule
The NaV1.5 homology model based on NaVPas (NaV1.5-NaVPas) is shown in Fig. 2. The side view of the channel shows the four domains and their putative voltage and pore-forming segments (including the p-helices, extracellular and intracellular linkers). NaVPas shares about 32% sequence identity with NaV1.5. The aligned sequences for DIII-DIV linker and CTD are shown in Fig. 2. Ranolazine was auto-docked against NaV1.5-NaVPas using AutoDock4. The highest affinity (−7.7 kcal/mol) binding mode is enlarged in Fig. 2. The compound formed polar and Van der Waals interaction with various residues located in all four domains: S401, V405, C896, N927, F1418, S1458, L1462, N1463, I1466, F1760, V1764, I1768. The aromatic residue, F1760, is outlined in Fig. 2 as it is a key putative binding site for many anti-arrhythmics, local anesthetics, and anticonvulsants52,55. F1760 orientation with respect to Ranolazine supports its critical role in drug binding.
Figure 2.
Ranolazine docked to NaV1.5-NaVPas. The side view of NaV1.5-NaVPas homology model is shown docked to Ranolazine. The enlarged inset shows the cartoon structure of the drug binding to the central domains of the channel. The aromatic F1760 residue is outlined. Below the inset is a 3D-structure of Ranolazine (Nitrogen is blue, Oxygen is red, Carbon is green, and Hydrogen is grey). Conserved residues in DIII-DIV linker and CTD between NaV1.5 and NaVPas are indicated by a red asterisk.
Ranolazine does not affect conductance
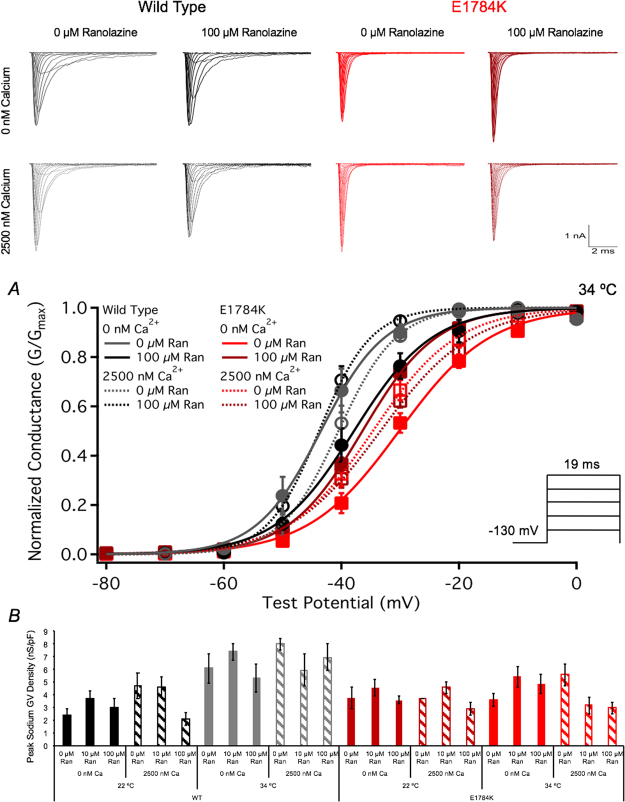
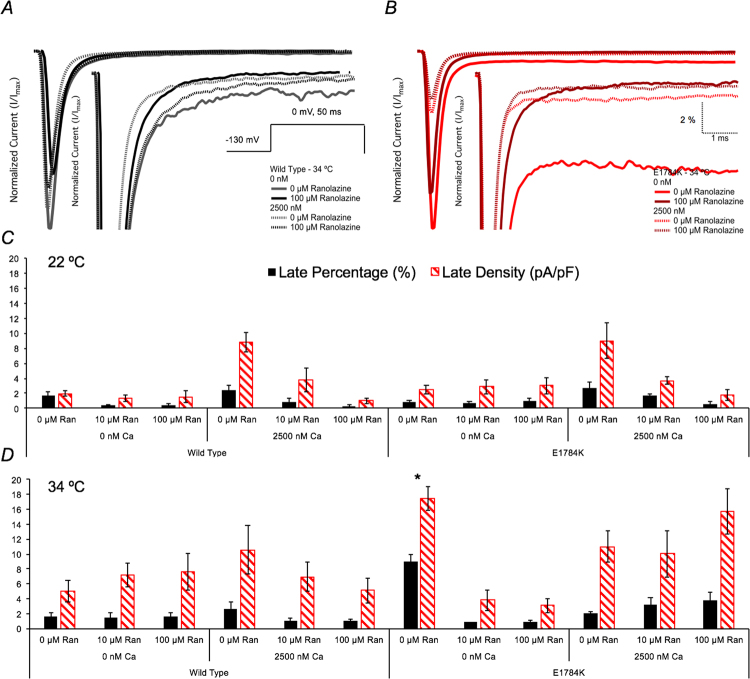
Raw current traces in Fig. 3 show the effects of 0 µM and 100 µM Ranolazine on WT and E178K at 0 nM and 2500 nM cytosolic calcium (only 34 °C shown). E1784K reduced (p < 0.0001) the peak current and conductance density compared to WT. Elevated temperature (34 °C) increased (p < 0.0001) peak current and conductance density in WT but not in E1784K. Ranolazine had no effect (p > 0.05) on peak current or conductance density (Table 1 and Fig. 3B).
Figure 3.
Ranolazine effects on channel conductance. Top traces show raw INa traces recorded at 34 °C. Panel A shows normalized conductance plotted against the test potential (pulse protocol shown in inset) at 34 °C. Panel C shows the peak conductance density bar graph versus all conditions at both 22 °C and 34 °C.
Table 1.
Peak INa Current and Conductance Density.
| Condition | Peak INa Density (pA/pF) | N | Peak GV Density (nS/pF) | N |
|---|---|---|---|---|
| WT - 22 °C - 0 nM Ca2+ - 0 µM Ran | 148.97 ± 35.10 | 7 | 2348.13 ± 584.87 | 7 |
| WT - 22 °C - 0 nM Ca2+ - 10 µM Ran | 241.31 ± 41.80 | 5 | 3708.29 ± 580.79 | 5 |
| WT - 22 °C - 0 nM Ca2+ - 100 µM Ran | 184.16 ± 40.16 | 5 | 3030.10 ± 640.76 | 5 |
| WT - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 302.91 ± 59.07 | 9 | 4714.99 ± 971.64 | 9 |
| WT - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 250.88 ± 45.34 | 7 | 4599.97 ± 788.18 | 7 |
| WT - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 128.74 ± 34.69 | 9 | 2088.79 ± 517.47 | 9 |
| WT - 34 °C - 0 nM Ca2+ - 0 µM Ran | 354.74 ± 83.12 | 7 | 6067.75 ± 1197.96 | 6 |
| WT - 34 °C - 0 nM Ca2+ - 10 µM Ran | 472.24 ± 55.47 | 5 | 7370.72 ± 654.58 | 7 |
| WT - 34 °C - 0 nM Ca2+ - 100 µM Ran | 369.56 ± 81.13 | 8 | 5321.49 ± 1074.25 | 8 |
| WT - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 535.63 ± 70.44 | 6 | 7979.56 ± 433.11 | 5 |
| WT - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 447.46 ± 106.69 | 6 | 5948.51 ± 1253.7 | 6 |
| WT - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 599.72 ± 113.81 | 6 | 6935.36 ± 1048.34 | 5 |
| EK - 22 °C - 0 nM Ca2+ - 0 µM Ran | 214.56 ± 50.18 | 7 | 3762.09 ± 835.36 | 7 |
| EK - 22 °C - 0 nM Ca2+ - 10 µM Ran | 265.65 ± 51.36 | 8 | 4534.15 ± 651.10 | 10 |
| EK - 22 °C - 0 nM Ca2+ - 100 µM Ran | 240.86 ± 24.43 | 7 | 3576.84 ± 301.37 | 7 |
| EK - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 170.41 ± 31.44 | 5 | 3660.43 ± 1006.25 | 6 |
| EK - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 250.36 ± 33.96 | 6 | 4562.87 ± 453.93 | 8 |
| EK - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 183.02 ± 44.18 | 5 | 2886.72 ± 498.80 | 5 |
| EK - 34 °C - 0 nM Ca2+ - 0 µM Ran | 257.99 ± 34.07 | 8 | 3613.63 ± 524.71 | 9 |
| EK - 34 °C - 0 nM Ca2+ - 10 µM Ran | 322.26 ± 47.45 | 6 | 5422.63 ± 833.24 | 8 |
| EK - 34 °C - 0 nM Ca2+ - 100 µM Ran | 278.25 ± 67.75 | 8 | 4834.84 ± 766.20 | 7 |
| EK - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 354.25 ± 64.03 | 8 | 5587.74 ± 831.96 | 7 |
| EK - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 227.98 ± 41.82 | 5 | 3154.90 ± 685.41 | 5 |
| EK - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 212.41 ± 28.73 | 9 | 2959.51 ± 471.73 | 9 |
Figure 3A shows normalized conductance plotted against the test potential at 34 °C. E1784K (p < 0.0001) and elevated temperature (p = 0.0003) depolarized the conductance midpoint (GV-V1/2). The conductance slope (GV-z) was reduced (p < 0.0001) in E1784K compared to WT and increased (p < 0.0001) when temperature was elevated in both channel variants. The interaction between channel variant and temperature had no effect on GV-V1/2 and GV-z. Normalized conductance was unchanged in all conditions with Ranolazine (Table 2).
Table 2.
Activation and Steady-State Fast Inactivation.
| Condition | GV-V1/2 (mV) | GV-z | N | SSFI-V1/2 (mV) | SSFI-z | N |
|---|---|---|---|---|---|---|
| WT - 22 °C - 0 nM Ca2+ - 0 µM Ran | −40.45 ± 1.24 | 3.76 ± 0.33 | 7 | −88.62 ± 1.99 | −3.93 ± 0.26 | 7 |
| WT - 22 °C - 0 nM Ca2+ - 10 µM Ran | −45.59 ± 1.09 | 5.37 ± 0.29 | 6 | −86.51 ± 1.36 | −3.95 ± 0.16 | 7 |
| WT - 22 °C - 0 nM Ca2+ - 100 µM Ran | −44.13 ± 3.07 | 4.84 ± 0.56 | 5 | −88.45 ± 3.18 | −2.87 ± 0.11*2 | 6 |
| WT - 22 °C - 2500 nM Ca2+ - 0 µM Ran | −42.22 ± 1.16 | 4.08 ± 0.25 | 9 | −91.72 ± 1.72 | −3.68 ± 0.15 | 9 |
| WT - 22 °C - 2500 nM Ca2+ - 10 µM Ran | −44.40 ± 2.22 | 3.87 ± 0.28 | 7 | −91.44 ± 1.63 | −3.93 ± 0.18 | 7 |
| WT - 22 °C - 2500 nM Ca2+ - 100 µM Ran | −44.76 ± 2.09 | 4.38 ± 0.36 | 9 | −90.94 ± 1.64 | −2.20 ± 0.13*2 | 7 |
| WT - 34 °C - 0 nM Ca2+ - 0 µM Ran | −43.44 ± 2.06 | 6.00 ± 0.45 | 6 | −80.31 ± 1.17 | −4.72 ± 0.18 | 6 |
| WT - 34 °C - 0 nM Ca2+ - 10 µM Ran | −37.93 ± 1.97 | 4.95 ± 0.28 | 7 | −82.51 ± 2.26 | −4.00 ± 0.14 | 8 |
| WT - 34 °C - 0 nM Ca2+ - 100 µM Ran | −37.58 ± 2.03 | 4.43 ± 0.33 | 7 | −87.30 ± 2.73 | −2.82 ± 0.16*2 | 7 |
| WT - 34 °C - 2500 nM Ca2+ - 0 µM Ran | −40.62 ± 1.44 | 5.96 ± 0.35 | 6 | −80.25 ± 2.30 | −4.24 ± 0.08 | 7 |
| WT - 34 °C - 2500 nM Ca2+ - 10 µM Ran | −38.03 ± 3.69 | 5.15 ± 0.40 | 6 | −83.18 ± 3.95 | −4.00 ± 0.13 | 6 |
| WT - 34 °C - 2500 nM Ca2+ - 100 µM Ran | −43.69 ± 0.98 | 6.30 ± 0.48 | 6 | −90.43 ± 2.80 | −3.18 ± 0.2*2 | 6 |
| EK - 22 °C - 0 nM Ca2+ - 0 µM Ran | −35.13 ± 2.70 | 2.99 ± 0.27 | 8 | −100.46 ± 1.51 | −2.97 ± 0.08 | 8 |
| EK - 22 °C - 0 nM Ca2+ - 10 µM Ran | −35.83 ± 1.65 | 3.14 ± 0.27 | 10 | −99.37 ± 1.61 | −2.88 ± 0.08 | 10 |
| EK - 22 °C - 0 nM Ca2+ - 100 µM Ran | −36.16 ± 1.87 | 3.95 ± 0.17 | 7 | −103.83 ± 2.42 | −1.77 ± 0.06*2 | 8 |
| EK - 22 °C - 2500 nM Ca2+ - 0 µM Ran | −39.33 ± 2.02 | 3.09 ± 0.16 | 5 | −101.30 ± 3.14 | −3.07 ± 0.21 | 6 |
| EK - 22 °C - 2500 nM Ca2+ - 10 µM Ran | −34.88 ± 1.57 | 2.83 ± 0.15 | 8 | −100.76 ± 1.83 | −3.11 ± 0.13 | 8 |
| EK - 22 °C - 2500 nM Ca2+ - 100 µM Ran | −33.72 ± 1.52 | 3.02 ± 0.35 | 5 | −106.45 ± 2.29 | −2.12 ± 0.08*2 | 5 |
| EK - 34 °C - 0 nM Ca2+ - 0 µM Ran | −30.22 ± 1.27 | 3.68 ± 0.28 | 9 | −91.02 ± 2.79 | −3.18 ± 0.14 | 9 |
| EK - 34 °C - 0 nM Ca2+ - 10 µM Ran | −36.80 ± 1.20 | 4.31 ± 0.38 | 9 | −94.20 ± 3.73 | −3.14 ± 0.13 | 9 |
| EK - 34 °C - 0 nM Ca2+ - 100 µM Ran | −35.53 ± 1.18 | 4.11 ± 0.22 | 8 | −105.48 ± 1.64*1 | −2.07 ± 0.07*2 | 8 |
| EK - 34 °C - 2500 nM Ca2+ - 0 µM Ran | −34.74 ± 1.48 | 4.06 ± 0.41 | 8 | −96.78 ± 4.70 | −3.26 ± 0.12 | 8 |
| EK - 34 °C - 2500 nM Ca2+ - 10 µM Ran | −31.25 ± 2.04 | 3.92 ± 0.35 | 7 | −95.01 ± 3.23 | −2.94 ± 0.15 | 7 |
| EK - 34 °C - 2500 nM Ca2+ - 100 µM Ran | −32.33 ± 1.64 | 3.39 ± 0.15 | 9 | −105.73 ± 2.44 | −2.00 ± 0.06*2 | 10 |
*1p < 0.01 vs 0 µM and 10 µM Ran of same condition.
*2p < 0.05 vs 0 µM and 10 µM Ran of same condition.
E1784K availability is decreased in Ranolazine
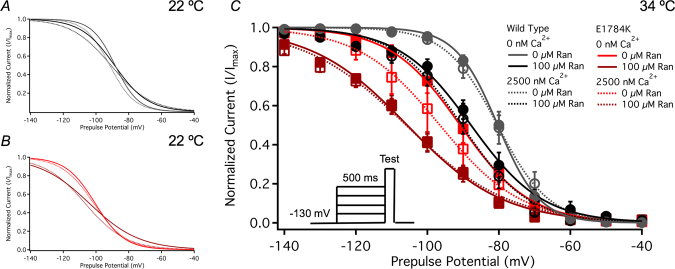
Normalized current is plotted against membrane potential in Fig. 4. E1784K hyperpolarized (p < 0.0001) the SSFI midpoint (SSFI-V1/2) compared to WT. Elevated temperature depolarized (p < 0.0001) SSFI-V1/2 in both WT and E1784K. At 34 °C and 0 nM cytosolic calcium, SSFI-V1/2 in E1784K was hyperpolarized (p < 0.0001) in 100 µM Ranolazine compared to WT (Fig. 4C and Table 2). This effect was not significant at 2500 nM cytosolic calcium. Analogous to the shifts on GV-z, SSFI-z was decreased in E1784K and increased with elevated temperature (p < 0.0001). The slope was reduced in all conditions when Ranolazine was increased from 10 µM to 100 µM (p < 0.05, Table 2).
Figure 4.
Ranolazine effects on steady-state fast inactivation. Panels A,B (22 °C) and C (34 °C) shows steady-state fast inactivation as normalized current plotted against the prepulse potential (pulse protocol shown in C inset).
Fast inactivation onset kinetics are not altered with Ranolazine
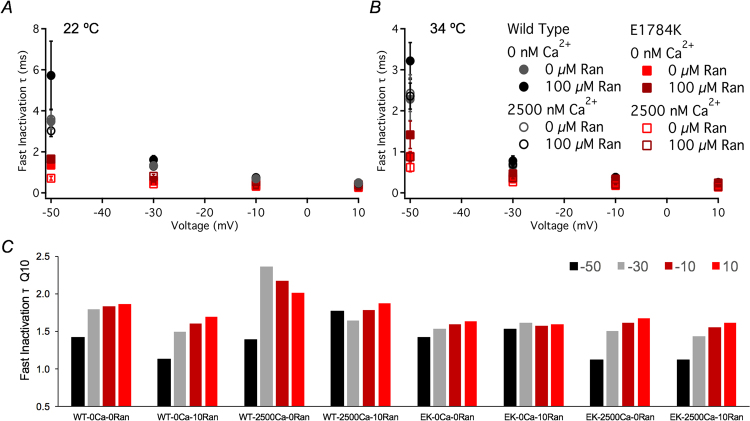
Fast inactivation onset kinetics at depolarized potentials (>−50 mV) were measured from τon of the mono-exponential fits. E1784K fast inactivation onset kinetics were accelerated regardless of temperature (Fig. 5A,B). Onset kinetics were accelerated (decreased τon) with elevated temperature in WT compared to E1784K (p < 0.01). WT and E1784K onset kinetics were decelerated (increased τon, p < 0.05) in Ranolazine as a function of voltage and cytosolic calcium at 22 °C (values reported in Table 3). These drug effects on τon were not significant at elevated temperature.
Figure 5.
Ranolazine effects on fast inactivation onset time constants. Panels A,B show the single-exponential time constants plotted against voltage. The pulse protocol is identical to that used to measure channel conductance (refer to Methods). Panel C includes Q10 coefficient values for all conditions between −50 mV to +10 mV. Cytosolic calcium seems to modulate Ranolazine effects on WT Q10, as elevated cytosolic calcium heightened thermosensitivity. Cytosolic calcium made −50 mV fast inactivation onset in E1784K less thermosensitive, consistent with decoupling between CTD and Domain III-IV linker mechanism, explained in the discussion.
Table 3.
Open-State Fast Inactivation Time Constant.
| Condition | −50 mV τ (ms) | −30 mV τ (ms) | −10 mV τ (ms) | +10 mV τ (ms) | N |
|---|---|---|---|---|---|
| WT - 22 °C - 0 nM Ca2+ - 0 µM Ran | 3.47 ± 0.22 | 1.32 ± 0.11 | 0.69 ± 0.07 | 0.48 ± 0.04 | 7 |
| WT - 22 °C - 0 nM Ca2+ - 10 µM Ran | 3.28 ± 0.45 | 1.13 ± 0.06 | 0.61 ± 0.04 | 0.44 ± 0.03 | 5 |
| WT - 22 °C - 0 nM Ca2+ - 100 µM Ran | 5.72 ± 1.67*1 | 1.61 ± 0.20*2 | 0.74 ± 0.06 | 0.46 ± 0.03 | 5 |
| WT - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 3.59 ± 0.49 | 1.34 ± 0.12 | 0.69 ± 0.03 | 0.47 ± 0.01 | 8 |
| WT - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 3.03 ± 0.13 | 1.09 ± 0.05 | 0.58 ± 0.01 | 0.42 ± 0.01 | 6 |
| WT - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 3.02 ± 0.28 | 1.30 ± 0.09 | 0.71 ± 0.05 | 0.49 ± 0.04 | 9 |
| WT - 34 °C - 0 nM Ca2+ - 0 µM Ran | 2.28 ± 0.52 | 0.66 ± 0.07 | 0.34 ± 0.02 | 0.23 ± 0.01 | 5 |
| WT - 34 °C - 0 nM Ca2+ - 10 µM Ran | 2.82 ± 0.46 | 0.70 ± 0.08 | 0.35 ± 0.04 | 0.23 ± 0.02 | 7 |
| WT - 34 °C - 0 nM Ca2+ - 100 µM Ran | 3.22 ± 0.45 | 0.78 ± 0.12 | 0.38 ± 0.06 | 0.22 ± 0.02 | 5 |
| WT - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 2.43 ± 0.45 | 0.48 ± 0.04 | 0.27 ± 0.01 | 0.20 ± 0.01 | 6 |
| WT - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 1.53 ± 0.24 | 0.60 ± 0.10 | 0.29 ± 0.03 | 0.20 ± 0.02 | 4 |
| WT - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 2.36 ± 0.32 | 0.69 ± 0.08 | 0.38 ± 0.04 | 0.25 ± 0.02 | 6 |
| EK - 22 °C - 0 nM Ca2+ - 0 µM Ran | 1.35 ± 0.14 | 0.57 ± 0.03 | 0.40 ± 0.02 | 0.33 ± 0.02 | 7 |
| EK - 22 °C - 0 nM Ca2+ - 10 µM Ran | 1.46 ± 0.17 | 0.54 ± 0.03 | 0.36 ± 0.02 | 0.30 ± 0.02 | 10 |
| EK–22 °C - 0 nM Ca2+ - 100 µM Ran | 1.65 ± 0.16 | 0.61 ± 0.03 | 0.37 ± 0.01 | 0.29 ± 0.01 | 6 |
| EK - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 0.71 ± 0.09 | 0.44 ± 0.02 | 0.32 ± 0.01 | 0.26 ± 0.01 | 6 |
| EK - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 1.23 ± 0.09 | 0.54 ± 0.03 | 0.37 ± 0.01 | 0.30 ± 0.01 | 7 |
| EK - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 1.65 ± 0.13 | 0.81 ± 0.09 | 0.54 ± 0.06*3 | 0.40 ± 0.03*4 | 5 |
| EK - 34 °C - 0 nM Ca2+ - 0 µM Ran | 0.89 ± 0.12 | 0.34 ± 0.02 | 0.23 ± 0.02 | 0.18 ± 0.01 | 9 |
| EK - 34 °C - 0 nM Ca2+ - 10 µM Ran | 0.88 ± 0.11 | 0.30 ± 0.02 | 0.21 ± 0.01 | 0.17 ± 0.01 | 9 |
| EK - 34 °C - 0 nM Ca2+ - 100 µM Ran | 1.41 ± 0.34 | 0.47 ± 0.09 | 0.34 ± 0.07 | 0.24 ± 0.05 | 8 |
| EK - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 0.62 ± 0.11 | 0.27 ± 0.02 | 0.18 ± 0.01 | 0.14 ± 0.01 | 9 |
| EK - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 1.08 ± 0.07 | 0.35 ± 0.05 | 0.22 ± 0.02 | 0.17 ± 0.01 | 6 |
| EK - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 0.88 ± 0.11 | 0.37 ± 0.02 | 0.21 ± 0.01 | 0.16 ± 0.01 | 8 |
*1p < 0.05 vs 0 µM and 10 µM Ran of same condition.
*2p < 0.05 vs 10 µM Ran of same condition.
*3p < 0.01 vs 0 µM Ran of same condition.
*4p < 0 ± 0.05 vs 0 µM Ran of same condition.
Figure 5C shows the Q10 values at 0 µM and 10 µM Ranolazine for all conditions. We observed high variability in the temperature coefficient at −50 mV compared to other voltages. At −50 mV, both Ranolazine and cytosolic calcium mutually affect thermosensitivity in WT: Q10 decreased at 0 nM cytosolic calcium and increased at 2500 nM cytosolic calcium in 10 µM Ranolazine. At more depolarized voltages than −50 mV, subtle alterations occurred in Q10 (Fig. 5C). E1784K Q10 was not sensitive to Ranolazine. However, E1784K thermosensitivity was dampened in cytosolic calcium at −50 mV compared to other voltages and to WT.
Ranolazine does not suppress thermosensitive late INa in E1784K with elevated cytosolic calcium
Representative normalized late INa current traces are shown in Fig. 6A,B at 0 µM and 100 µM Ranolazine (only 34 °C shown). Late INa percent and density are shown in Fig. 6C,D as bar graphs. Late INa percent and density in E1784K increased (p < 0.01) by 11-fold and 7-fold, respectively, with elevated temperature at 0 nM cytosolic calcium (Fig. 6D and Table 4). This increase in late INa was almost fully attenuated in 10 µM Ranolazine. Late INa percent decreased in elevated cytosolic calcium (p < 0.01) but there was no effect on late INa density in E1784K. Late INa percent and density in E1784K were not suppressed with Ranolazine at 2500 nM cytosolic calcium (Fig. 6D and Table 4).
Figure 6.
Ranolazine effects on late INa. Panels A,B show normalized current traces, with emphasis on the non-inactivating, late INa, at 34 °C. The normalized late INa in E1784K is enhanced drastically by elevated temperature. Cytosolic calcium suppresses the E1784K normalized late INa (addressed in discussions). Ranolazine suppresses late INa only at 0 nM compared to 2500 nM cytosolic calcium. Panels C,D show the late INa percent and density bar graphs for all conditions at 22 °C and 34 °C, respectively.
Table 4.
Late INa Current Density and Percentage.
| Condition | Late INa Density (pA/pF) | N | Late Percent (%) | N |
|---|---|---|---|---|
| WT - 22 °C - 0 nM Ca2+ - 0 µM Ran | 1.95 ± 0.34 | 9 | 1.71 ± 0.54 | 9 |
| WT - 22 °C - 0 nM Ca2+ - 10 µM Ran | 1.35 ± 0.44 | 8 | 0.35 ± 0.12 | 8 |
| WT - 22 °C - 0 nM Ca2+ - 100 µM Ran | 1.55 ± 0.79 | 6 | 0.45 ± 0.20 | 6 |
| WT - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 8.83 ± 1.33 | 8 | 2.41 ± 0.72 | 6 |
| WT - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 3.78 ± 1.64 | 8 | 0.80 ± 0.48 | 7 |
| WT - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 1.01 ± 0.38 | 7 | 0.30 ± 0.12 | 6 |
| WT - 34 °C - 0 nM Ca2+ - 0 µM Ran | 5.04 ± 1.42 | 7 | 1.57 ± 0.52 | 9 |
| WT - 34 °C - 0 nM Ca2+ - 10 µM Ran | 7.21 ± 1.61 | 8 | 1.54 ± 0.54 | 8 |
| WT - 34 °C - 0 nM Ca2+ - 100 µM Ran | 7.66 ± 2.47 | 9 | 1.67 ± 0.42 | 9 |
| WT - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 10.57 ± 3.2 | 7 | 2.71 ± 0.90 | 9 |
| WT - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 6.95 ± 1.97 | 8 | 1.09 ± 0.26 | 8 |
| WT - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 5.11 ± 1.67 | 4 | 0.98 ± 0.25 | 5 |
| EK - 22 °C - 0 nM Ca2+ - 0 µM Ran | 2.51 ± 0.63 | 9 | 0.85 ± 0.20 | 9 |
| EK - 22 °C - 0 nM Ca2+ - 10 µM Ran | 2.86 ± 0.88 | 9 | 0.68 ± 0.21 | 8 |
| EK - 22 °C - 0 nM Ca2+ - 100 µM Ran | 3.01 ± 1.08 | 10 | 1.02 ± 0.39 | 8 |
| EK - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 9.02 ± 2.31 | 5 | 2.64 ± 0.81 | 4 |
| EK - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 3.68 ± 0.51 | 9 | 1.65 ± 0.30 | 9 |
| EK - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 1.78 ± 0.65 | 8 | 0.61 ± 0.30 | 6 |
| EK - 34 °C - 0 nM Ca2+ - 0 µM Ran | 17.47 ± 1.54*1 | 6 | 8.97 ± 1.02*1 | 6 |
| EK - 34 °C - 0 nM Ca2+ - 10 µM Ran | 3.84 ± 1.39 | 4 | 0.86 ± 0.04 | 4 |
| EK - 34 °C - 0 nM Ca2+ - 100 µM Ran | 3.09 ± 0.96 | 6 | 0.89 ± 0.22 | 6 |
| EK - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 11.01 ± 2.12 | 7 | 2.00 ± 0.34 | 9 |
| EK - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 10.05 ± 3.08 | 5 | 3.28 ± 0.93 | 6 |
| EK - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 15.73 ± 6.24 | 5 | 3.76 ± 1.10 | 4 |
*1p < 0.01 vs 10 µM and 100 µM Ran of same condition.
Ranolazine does not enhance UDI in E1784K with elevated cytosolic calcium
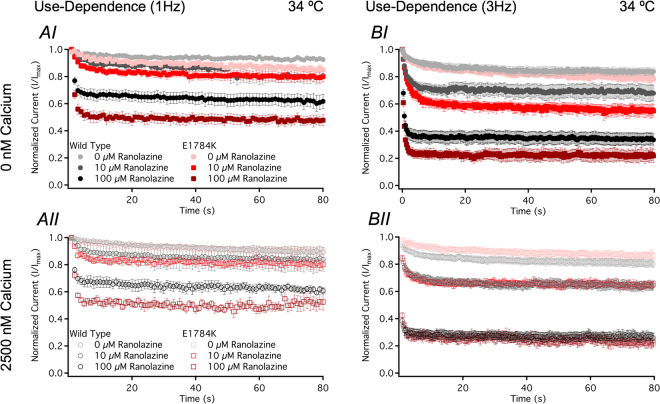
Sustained or repetitive depolarizations induce slow inactivation in NaV1.5, which was indirectly measured by the use-dependent inactivation (UDI) protocols described in the methods. We were unable to measure fast inactivation recovery kinetics in E1784K which may be decelerated by Ranolazine and contribute to UDI54. Use-dependence was measured at 1 Hz and 3 Hz, mimicking resting heart rate (60 bpm) and tachycardia (180 bpm), respectively. Normalized current plotted against time for UDI measured at 1 Hz and 3 Hz are shown in Fig. 7 (only 34 °C shown).
Figure 7.
Ranolazine effects on use-dependence. Panels AI-II and Panels BI-II show normalized current versus time measuring UDI at 1 Hz and 3 Hz, respectively. Insets are excluded for visual clarity (refer to methods for pulse protocols). All three drug concentrations are included in the figures to show 10 µM Ranolazine effect on UDI (3 Hz) in E1784K at 0 nM compared to 2500 nM.
UDI plateau (y0) was greater (p = 0.0430) at elevated temperature at both 1 Hz and 3 Hz, but the shift was larger in E1784K at 1 Hz (Table 5). y0 decreased to different levels in Ranolazine (reported in Table 5). At high UDI frequencies, E1784K y0 decreased in Ranolazine at 34 °C compared to WT (Fig. 7 shows only 34 °C, Table 5). Our statistical results suggest that the drug effects on UDI (3 Hz) in E1784K are limited in elevated cytosolic calcium (Table 5).
Table 5.
Use-Dependence (1 Hz and 3 Hz).
| Condition | 1 Hz–y0 | 1 Hz–τ1 (s) | 1 Hz–τ2 (s) | N | 3 Hz–y0 | 3 Hz–τ1 (s) | 3 Hz–τ2 (s) | N |
|---|---|---|---|---|---|---|---|---|
| WT - 22 °C - 0 nM Ca2+ - 0 µM Ran | 0.84 ± 0.03 | 3.13 ± 0.88 | 47.88 ± 9.44 | 6 | 0.63 ± 0.04 | 2.03 ± 0.41 | 40.53 ± 5.42 | 7 |
| WT - 22 °C - 0 nM Ca2+ - 10 µM Ran | 0.77 ± 0.01 | 2.96 ± 0.52 | 18.55 ± 4.50 | 7 | 0.52 ± 0.02 | 3.39 ± 0.62 | 23.00 ± 7.54 | 7 |
| WT - 22 °C - 0 nM Ca2+ - 100 µM Ran | 0.44 ± 0.02*1 | 2.19 ± 0.44 | 9.70 ± 3.31 | 5 | 0.15 ± 0.02*1 | 0.92 ± 0.18 | 7.02 ± 4.44 | 4 |
| WT - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 0.81 ± 0.02 | 8.22 ± 2.84 | 114.32 ± 22.42 | 7 | 0.66 ± 0.03 | 1.83 ± 0.23 | 36.4 ± 7.98 | 7 |
| WT - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 0.78 ± 0.01 | 6.93 ± 0.94 | 31.93 ± 10.85 | 6 | 0.52 ± 0.02 | 1.19 ± 0.19 | 8.57 ± 0.42 | 5 |
| WT - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 0.35 ± 0.03*1 | 1.67 ± 0.12 | 34.32 ± 22.87 | 7 | 0.20 ± 0.03*1 | 0.69 ± 0.32 | 14.56 ± 13.09 | 3 |
| WT - 34 °C - 0 nM Ca2+ - 0 µM Ran | 0.92 ± 0.01 | 4.77 ± 3.14 | 39.42 ± 20.36 | 4 | 0.84 ± 0.03 | 2.75 ± 1.00 | 17.73 ± 9.63 | 5 |
| WT - 34 °C - 0 nM Ca2+ - 10 µM Ran | 0.76 ± 0.05 | 5.41 ± 3.35 | 62.60 ± 20.25 | 5 | 0.65 ± 0.06 | 0.98 ± 0.10 | 16.65 ± 6.29 | 6 |
| WT - 34 °C - 0 nM Ca2+ - 100 µM Ran | 0.60 ± 0.06*2 | 0.66 ± 0.10 | 43.51 ± 18.93 | 8 | 0.33 ± 0.05*1 | 0.39 ± 0.08 | 13.99 ± 8.03 | 8 |
| WT - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 0.88 ± 0.03 | 30.10 ± 6.41*3 | 59.96 ± 37.08 | 4 | 0.79 ± 0.04 | 1.46 ± 0.17 | 38.85 ± 12.19 | 8 |
| WT - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 0.82 ± 0.04 | 2.83 ± 1.02 | 67.05 ± 23.05 | 5 | 0.60 ± 0.03*4 | 1.02 ± 0.16 | 17.28 ± 6.89 | 7 |
| WT - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 0.64 ± 0.02 | 0.44 ± 0.16 | 8.44 ± 2.77 | 5 | 0.27 ± 0.03*1 | 0.41 ± 0.06 | 17.80 ± 9.98 | 5 |
| EK - 22 °C - 0 nM Ca2+ - 0 µM Ran | 0.69 ± 0.06 | 3.37 ± 0.64 | 69.20 ± 25.09 | 6 | 0.79 ± 0.01 | 3.08 ± 1.61 | 32.65 ± 20.62 | 4 |
| EK - 22 °C - 0 nM Ca2+ - 10 µM Ran | 0.57 ± 0.05 | 3.25 ± 0.63 | 64.71 ± 37.46 | 7 | 0.47 ± 0.02*4 | 2.40 ± 0.37 | 16.28 ± 3.57 | 6 |
| EK - 22 °C - 0 nM Ca2+ - 100 µM Ran | 0.27 ± 0.05*1 | 1.87 ± 0.25 | 74.43 ± 28.56 | 8 | 0.14 ± 0.03*1 | 1.00 ± 0.34 | 1.29 ± 0.19 | 4 |
| EK - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 0.73 ± 0.02 | 1.99 ± 1.02 | 31.37 ± 6.74 | 4 | 0.69 ± 0.06 | 3.59 ± 1.84 | 8.20 ± 3.73 | 4 |
| EK - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 0.59 ± 0.06 | 7.17 ± 1.35 | 75.52 ± 20.26 | 5 | 0.47 ± 0.04 | 3.38 ± 0.43 | 13.21 ± 1.51 | 5 |
| EK - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 0.24 ± 0.04*1 | 2.68 ± 1.13 | 70.86 ± 29.29 | 3 | 0.20 ± 0.06*2 | 0.82 ± 0.29 | 1.94 ± 0.31 | 3 |
| EK - 34 °C - 0 nM Ca2+ - 0 µM Ran | 0.82 ± 0.04 | 6.76 ± 1.43 | 68.66 ± 25.39 | 8 | 0.77 ± 0.03 | 2.66 ± 0.45 | 49.11 ± 13.27 | 9 |
| EK - 34 °C - 0 nM Ca2+ - 10 µM Ran | 0.77 ± 0.06 | 2.40 ± 0.55 | 78.22 ± 35.32 | 7 | 0.55 ± 0.03*4 | 1.63 ± 0.32 | 27.16 ± 15.43 | 5 |
| EK - 34 °C - 0 nM Ca2+ - 100 µM Ran | 0.47 ± 0.04*1 | 0.77 ± 0.07 | 19.07 ± 9.49 | 6 | 0.21 ± 0.05*1 | 0.40 ± 0.04 | 24.08 ± 18.78 | 5 |
| EK - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 0.87 ± 0.03 | 15.05 ± 3.35*3 | 65.60 ± 19.19 | 5 | 0.74 ± 0.08 | 2.50 ± 0.65 | 30.82 ± 2.43 | 6 |
| EK - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 0.77 ± 0.04 | 1.77 ± 0.48 | 48.78 ± 22.76 | 5 | 0.63 ± 0.03 | 1.13 ± 0.12 | 6.41 ± 3.13 | 6 |
| EK - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 0.46 ± 0.05*1 | 1.21 ± 0.20 | 38.24 ± 19.24 | 5 | 0.26 ± 0.03*1 | 0.54 ± 0.14 | 6.89 ± 4.62 | 5 |
*1p < 0.0001 vs 0 µM and 10 µM Ran of same condition.
*2p < 0.0001 vs 0 µM Ran of same condition.
*3p < 0.0001 vs 10 µM and 100 µM Ran of same condition.
*4p < 0.0001 vs 0 µM and 100 µM Ran of same condition.
UDI onset kinetics were accelerated in E1784K and elevated temperature (p < 0.05), measured by τ1, at 1 Hz and 3 Hz. Onset kinetics decelerated in elevated cytosolic calcium (p < 0.0001) at 1 Hz predominately in WT compared to E1784K. WT and E1784K τ1 was decreased (p < 0.0001) in Ranolazine at 2500 nM cytosolic calcium and 34 °C (Table 5). τ1 at 3 Hz was unaffected in Ranolazine. τ2 was unaffected at 1 Hz and 3 Hz in all experimental conditions.
E1784K-induced alternans is exacerbated with Ranolazine
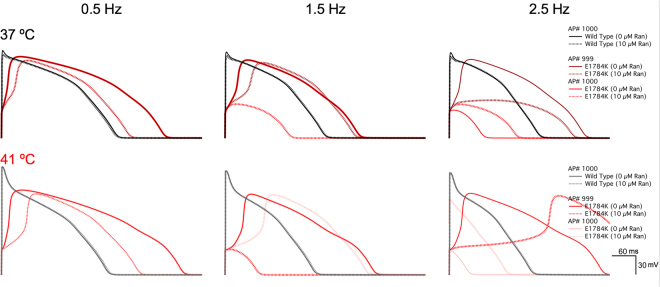
Biophysical data were extrapolated to physiological (37 °C) and febrile (41 °C) temperatures using a Q10 coefficient (Equation 6). Extrapolations were independently supported using Arrhenius relationships (Equation 7 and Table 6). Action potential (AP) traces are shown in Fig. 8 for simulations conducted in endocardial cells at three frequencies: 0.5 Hz (bradycardia), 1.5 Hz (sinus rhythm), and 2.5 Hz (tachycardia). At febrile temperature, simulated APs show accelerated depolarizations and repolarizations in WT compared to E1784K.
Table 6.
Values at 37 °C and 41 °C based on Q10 and Arrhenius Calculations.
| Condition |
37 °C
(Q 10 ) |
41 °C
(Q 10 ) |
37 °C
(Arrhenius) |
41 °C
(Arrhenius) |
|---|---|---|---|---|
| Peak GV Density (nS/pF) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 16179.18 | 55592.62 | 8124.68 | 10881.05 |
| WT - 0 nM Ca2+ - 10 µM Ran | 22678.58 | 99329.99 | 9107.70 | 11251.36 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 27586.50 | 131656.89 | 9377.01 | 11024.82 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 15043.82 | 48489.81 | 6439.00 | 6969.03 |
| EK - 0 nM Ca2+ - 0 µM Ran | 3575.20 | 3527.30 | 3569.09 | 3525.15 |
| EK - 0 nM Ca2+ - 10 µM Ran | 12400.15 | 35482.80 | 5731.14 | 6055.55 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 14940.82 | 51538.73 | 6366.24 | 7251.15 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 2860.49 | 2527.87 | 2816.21 | 2513.95 |
| Peak I Na Density (pA/pF) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 446.63 | 597.28 | 463.00 | 604.67 |
| WT - 0 nM Ca2+ - 10 µM Ran | 564.39 | 706.75 | 580.56 | 713.78 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 623.14 | 754.26 | 637.95 | 760.26 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 521.76 | 633.36 | 534.72 | 638.92 |
| EK - 0 nM Ca2+ - 0 µM Ran | 270.93 | 288.18 | 272.97 | 288.90 |
| EK - 0 nM Ca2+ - 10 µM Ran | 339.21 | 361.89 | 341.86 | 362.80 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 430.22 | 549.75 | 443.92 | 556.03 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 222.38 | 215.51 | 221.51 | 215.22 |
| Late I Na Density (pA/pF) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 6.48 | 8.89 | 6.74 | 9.01 |
| WT - 0 nM Ca2+ - 10 µM Ran | 11.26 | 19.76 | 12.09 | 20.26 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 11.08 | 11.77 | 11.17 | 11.80 |
| WT–2500 nM Ca2+ - 10 µM Ran | 8.17 | 10.02 | 8.38 | 10.11 |
| EK - 0 nM Ca2+ - 0 µM Ran | 29.25 | 56.02 | 31.75 | 57.68 |
| EK - 0 nM Ca2+ - 10 µM Ran | 4.15 | 4.58 | 4.20 | 4.60 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 11.61 | 12.41 | 11.71 | 12.45 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 13.12 | 18.37 | 13.69 | 18.65 |
| Late Percent (%) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 1.54 | 1.50 | 1.53 | 1.49 |
| WT - 0 nM Ca2+ - 10 µM Ran | 2.27 | 3.71 | 2.41 | 3.79 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 2.80 | 2.91 | 2.81 | 2.92 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 1.18 | 1.30 | 1.19 | 1.31 |
| EK - 0 nM Ca2+ - 0 µM Ran | 11.64 | 16.17 | 12.13 | 16.41 |
| EKv0 nM Ca2+ - 10 µM Ran | 0.92 | 1.00 | 0.93 | 1.00 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 1.86 | 1.69 | 1.84 | 1.69 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 3.94 | 4.96 | 4.05 | 5.01 |
| GV-V 1/2 (mV) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | −44.27 | −45.34 | −44.40 | −45.39 |
| WT - 0 nM Ca2+ - 10 µM Ran | −36.12 | −33.96 | −35.84 | −33.87 |
| WT - 2500 nM Ca2+ - 0 µM Ran | −40.20 | −39.68 | −40.13 | −39.66 |
| WT - 2500 nM Ca2+ - 10 µM Ran | −36.50 | −34.66 | −36.26 | −34.58 |
| EK - 0 nM Ca2+ - 0 µM Ran | −29.04 | −27.61 | −28.86 | −27.56 |
| EK - 0 nM Ca2+ - 10 µM Ran | −37.06 | −37.39 | −37.10 | −37.40 |
| EK - 2500 nM Ca2+ - 0 µM Ran | −33.62 | −32.25 | −33.44 | −32.19 |
| EK - 2500 nM Ca2+ - 10 µM Ran | −30.35 | −29.25 | −30.21 | −29.21 |
| GV-z | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 6.79 | 7.93 | 6.92 | 7.99 |
| WT - 0 nM Ca2+ - 10 µM Ran | 4.84 | 4.71 | 4.82 | 4.71 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 6.59 | 7.49 | 6.70 | 7.53 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 5.55 | 6.11 | 5.62 | 6.13 |
| EK - 0 nM Ca2+ - 0 µM Ran | 3.89 | 4.17 | 3.93 | 4.19 |
| EK - 0 nM Ca2+ - 10 µM Ran | 4.68 | 5.21 | 4.75 | 5.23 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 4.36 | 4.78 | 4.41 | 4.80 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 4.28 | 4.78 | 4.34 | 4.80 |
| SSFI-V 1/2 (mV) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | −78.24 | −75.70 | −77.91 | −75.59 |
| WT - 0 nM Ca2+ - 10 µM Ran | −81.49 | −80.21 | −81.32 | −80.15 |
| WT - 2500 nM Ca2+ - 0 µM Ran | −77.46 | −74.07 | −77.02 | −73.92 |
| WT - 2500 nM Ca2+ - 10 µM Ran | −81.11 | −78.58 | −80.79 | −78.47 |
| EK - 0 nM Ca2+ - 0 µM Ran | −88.67 | −85.79 | −88.30 | −85.67 |
| EK - 0 nM Ca2+–10 µM Ran | −92.87 | −91.23 | −92.66 | −91.15 |
| EK - 2500 nM Ca2+ - 0 µM Ran | −95.62 | −94.16 | −95.43 | −94.10 |
| EK - 2500 nM Ca2+ - 10 µM Ran | −93.54 | −91.72 | −93.31 | −91.64 |
| SSFI-z | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | −4.95 | −5.26 | −4.99 | −5.27 |
| WT - 0 nM Ca2+ - 10 µM Ran | −4.02 | −4.04 | −4.02 | −4.04 |
| WT - 2500 nM Ca2+ - 0 µM Ran | −4.41 | −4.63 | −4.44 | −4.63 |
| WT - 2500 nM Ca2+ - 10 µM Ran | −4.02 | −4.05 | −4.03 | −4.05 |
| EK - 0 nM Ca2+ - 0 µM Ran | −3.24 | −3.31 | −3.25 | −3.32 |
| EK - 0 nM Ca2+ - 10 µM Ran | −3.21 | −3.30 | −3.22 | −3.31 |
| EK - 2500 nM Ca2+ - 0 µM Ran | −3.31 | −3.38 | −3.32 | −3.38 |
| EK - 2500 nM Ca2+ - 10 µM Ran | −2.90 | −2.85 | −2.89 | −2.84 |
| −50mV FI τ (ms) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 2.04 | 1.77 | 2.00 | 1.76 |
| WT - 0 nM Ca2+ - 10 µM Ran | 2.71 | 2.58 | 2.69 | 2.57 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 2.19 | 1.92 | 2.15 | 1.91 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 1.27 | 1.01 | 1.24 | 1.00 |
| EK - 0 nM Ca2+ - 0 µM Ran | 0.80 | 0.69 | 0.78 | 0.69 |
| EK - 0 nM Ca2+ - 10 µM Ran | 0.77 | 0.65 | 0.75 | 0.64 |
| EK - 2500 nM Ca2+ - - 0 µM Ran | 0.60 | 0.57 | 0.59 | 0.57 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 1.04 | 0.99 | 1.03 | 0.99 |
| −30mV FI τ (ms) | ||||
| WT–0 nM Ca2+ - 0 µM Ran | 0.54 | 0.43 | 0.53 | 0.43 |
| WT - 0 nM Ca2+–10 µM Ran | 0.61 | 0.52 | 0.60 | 0.52 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 0.36 | 0.26 | 0.35 | 0.25 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 0.51 | 0.42 | 0.50 | 0.42 |
| EK - 0 nM Ca2+ - 0 µM Ran | 0.30 | 0.25 | 0.29 | 0.25 |
| EK - 0 nM Ca2+ - 10 µM Ran | 0.26 | 0.21 | 0.25 | 0.21 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 0.23 | 0.20 | 0.23 | 0.20 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 0.31 | 0.27 | 0.31 | 0.27 |
| −10mV FI τ (ms) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 0.28 | 0.22 | 0.27 | 0.21 |
| WT - 0 nM Ca2+ - 10 µM Ran | 0.30 | 0.25 | 0.29 | 0.25 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 0.21 | 0.16 | 0.20 | 0.15 |
| WT–2500 nM Ca2+ - 10 µM Ran | 0.24 | 0.19 | 0.23 | 0.19 |
| EK - 0 nM Ca2+ - 0 µM Ran | 0.20 | 0.16 | 0.19 | 0.16 |
| EK - 0 nM Ca2+ - 10 µM Ran | 0.18 | 0.15 | 0.18 | 0.15 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 0.15 | 0.13 | 0.15 | 0.13 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 0.19 | 0.16 | 0.18 | 0.16 |
| +10 mV FI τ (ms) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 0.19 | 0.14 | 0.18 | 0.14 |
| WT - 0 nM Ca2+ - 10 µM Ran | 0.20 | 0.16 | 0.19 | 0.16 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 0.16 | 0.12 | 0.16 | 0.12 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 0.16 | 0.13 | 0.16 | 0.12 |
| EK - 0 nM Ca2+ - 0 µM Ran | 0.16 | 0.13 | 0.15 | 0.13 |
| EK - 0 nM Ca2+ - 10 µM Ran | 0.15 | 0.12 | 0.14 | 0.12 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 0.12 | 0.10 | 0.12 | 0.10 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 0.15 | 0.12 | 0.14 | 0.12 |
| 1 Hz-y0 | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 0.94 | 0.97 | 0.94 | 0.97 |
| WT - 0 nM Ca2+ - 10 µM Ran | 0.76 | 0.76 | 0.76 | 0.76 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 0.90 | 0.93 | 0.90 | 0.93 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 0.83 | 0.85 | 0.84 | 0.85 |
| EK - 0 nM Ca2+ - 0 µM Ran | 0.86 | 0.92 | 0.87 | 0.92 |
| EK - 0 nM Ca2+ - 10 µM Ran | 0.83 | 0.91 | 0.84 | 0.92 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 0.92 | 0.97 | 0.92 | 0.97 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 0.83 | 0.90 | 0.84 | 0.91 |
| 1 Hz-τ1 (s) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 5.34 | 6.15 | 5.43 | 6.18 |
| WT - 0 nM Ca2+ - 10 µM Ran | 6.34 | 7.76 | 6.51 | 7.83 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 42.49 | 65.65 | 44.88 | 66.93 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 2.23 | 1.65 | 2.15 | 1.63 |
| EK - 0 nM Ca2+ - 0 µM Ran | 8.14 | 10.28 | 8.38 | 10.38 |
| EK - 0 nM Ca2+ - 10 µM Ran | 2.22 | 2.01 | 2.19 | 2.00 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 25.75 | 50.72 | 28.04 | 52.25 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 1.22 | 0.77 | 1.15 | 0.75 |
| 1 Hz-τ2 (s) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 37.43 | 35.07 | 37.13 | 34.97 |
| WT - 0 nM Ca2+ - 10 µM Ran | 86.46 | 129.96 | 91.00 | 132.31 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 50.52 | 40.70 | 49.16 | 40.30 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 81.65 | 104.69 | 84.25 | 105.85 |
| EK - 0 nM Ca2+ - 0 µM Ran | 68.52 | 68.34 | 68.50 | 68.33 |
| EK - 0 nM Ca2+ - 10 µM Ran | 82.26 | 87.65 | 82.91 | 87.89 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 79.80 | 102.18 | 82.29 | 103.26 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 43.43 | 37.52 | 42.64 | 37.27 |
| 3 Hz-y0 | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 0.90 | 0.99 | 0.91 | 1.00 |
| WT - 0 nM Ca2+ - 10 µM Ran | 0.69 | 0.75 | 0.70 | 0.75 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 0.83 | 0.88 | 0.84 | 0.88 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 0.63 | 0.65 | 0.63 | 0.66 |
| EK - 0 nM Ca2+ - 0 µM Ran | 0.79 | 0.82 | 0.79 | 0.83 |
| EK - 0 nM Ca2+ - 10 µM Ran | 0.57 | 0.60 | 0.57 | 0.60 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 0.76 | 0.78 | 0.76 | 0.78 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 0.68 | 0.76 | 0.69 | 0.76 |
| 3 Hz-τ1 (s) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 2.97 | 3.29 | 3.01 | 3.30 |
| WT - 0 nM Ca2+ - 10 µM Ran | 0.70 | 0.46 | 0.67 | 0.46 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 1.38 | 1.28 | 1.36 | 1.27 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 0.98 | 0.93 | 0.97 | 0.92 |
| EK - 0 nM Ca2+ - 0 µM Ran | 3.07 | 3.69 | 3.14 | 3.72 |
| EK - 0 nM Ca2+ - 10 µM Ran | 1.47 | 1.29 | 1.45 | 1.28 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 2.27 | 2.01 | 2.24 | 2.00 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 0.84 | 0.58 | 0.81 | 0.57 |
| 3 Hz-τ2 (s) | ||||
| WT - 0 nM Ca2+ - 0 µM Ran | 14.24 | 10.80 | 13.75 | 10.66 |
| WT - 0 nM Ca2+ - 10 µM Ran | 15.28 | 13.72 | 15.08 | 13.65 |
| WT - 2500 nM Ca2+ - 0 µM Ran | 39.52 | 40.39 | 39.63 | 40.43 |
| WT - 2500 nM Ca2+ - 10 µM Ran | 20.81 | 26.32 | 21.43 | 26.60 |
| EK - 0 nM Ca2+ - 0 µM Ran | 54.26 | 61.55 | 55.14 | 61.90 |
| EK - 0 nM Ca2+ - 10 µM Ran | 31.12 | 36.95 | 31.81 | 37.24 |
| EK - 2500 nM Ca2+ - 0 µM Ran | 43.81 | 68.26 | 46.41 | 69.75 |
| EK - 2500 nM Ca2+ - 10 µM Ran | 5.29 | 4.15 | 5.13 | 4.11 |
Figure 8.
Endocardial action potential simulations. AP simulations are plotted against time (inset is shown in bottom right corner) at 37 °C and 41 °C. The last two AP beats were plotted in E1784K to show alternans-induction. Simulations only included therapeutic concentrations of Ranolazine (10 µM).
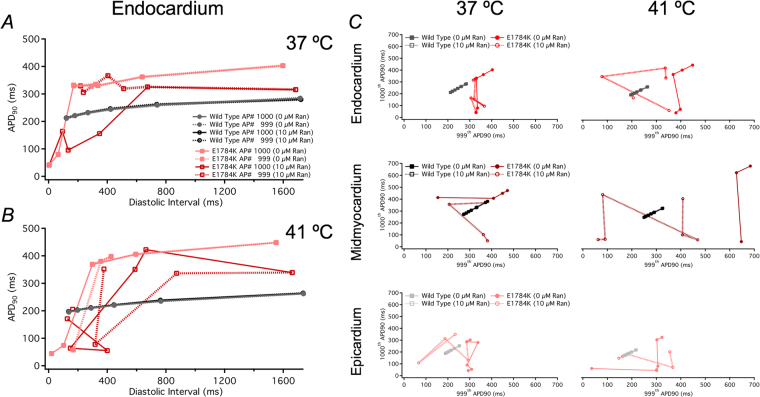
Electrical restitution curves (ERCs) at 90% repolarization were constructed from plotting APD90 against the diastolic interval as shown in Fig. 9A,B in endocardial cells. The last two beats were included in the ERCs to exemplify bifurcation and alternans-induction at critical diastolic intervals. The APD90 for WT follows a similar trend to previously published ERCs, typifying a relatively stable APD rate dependence56–58. E1784K has a higher APD90 in all cardiac cells, especially the mid-myocardium, compared to WT (not shown in Fig. 9). At 37 °C, the mutant causes bifurcation in APD90, mainly in epicardial cells, indicative of alternans (not shown); however, endocardial cells also experience alternans at febrile temperature in addition to epicardial cells (Fig. 9B). Upon drug perfusion, bifurcations were observed at higher diastolic intervals in E1784K (Fig. 9). The drug-induced bifurcations in ERCs were augmented at febrile temperature in all cardiac cells.
Figure 9.
Cardiac electrical restitution properties. Panel A,B shows the endocardial ERC curves at 37 °C and 41 °C. Panels C shows plots of the last two AP beats to determine alternans-induction in the three myocardial cells at 37 °C and 41 °C.
A linear relationship is established between the last two AP beats at each frequency step (shown in Fig. 9C), with no alternans. Divergence from linearity is indicative of alternans occurrence. At both 37 °C and 41 °C, WT cells had no alternans, even upon 10 µM Ranolazine perfusion, showing linearity with a slope = 1 (Fig. 9C). In drug-free conditions, E1784K had a linear relationship at high BCLs (low frequencies), but deviated from linearity beginning at intermediate BCLs; distortion in linearity is observed at lower frequencies in epicardial cells (Fig. 9C). This relationship in E1784K is augmented with febrile temperature. The prolonged APD90 in E1784K were shortened with Ranolazine at very low frequencies, and alternans were quickly induced even during bradycardia, an effect exacerbated by febrile temperature (Fig. 9C).
Discussion
Our goal was to determine whether Ranolazine reduced channel dysfunction in E1784K under the triggering conditions of elevated temperature and cytosolic calcium. Ranolazine did not attenuate gain-of-function in E1784K when temperature and cytosolic calcium were elevated. Ranolazine has minimal effects on conductance in NaV1.548,49. The drug follows the modulated receptor hypothesis, targeting the open/inactivated states at depolarized potentials, thereby suppressing late INa59. Physiological events, such as acidaemia, enhance Ranolazine antiarrhythmic effect by augmenting late INa, thus providing the drug with a larger open-state channel substrate to target48,60,61. In addition to physiological modulators, SCN5a mutations often alter voltage-dependence of the channel, which modify drug effects on NaV1.5. To date, Ranolazine has been screened against only ∆KPQ50,52, Y1767C49, R1623Q51, and D1790G62. Our study is the first to show the combined external triggers and SCN5a mutation effects on Ranolazine.
Ranolazine efficacy was enhanced at elevated temperature. Similar to acidosis effects, elevated temperature increases the late open probability in E1784K48,53. Late INa percent and density increased by 11- and 7-fold, respectively, with elevated temperature. We previously reported a 3.54-fold increase in late INa percent when temperature was elevated from 22 °C to 34 °C41; however, we used CHOK1 cells to study E1784K thermosensitivity. The temperature coefficient (Q10) partly depends on lipid-channel interactions in the membrane, which differ between heterologous expression systems63,64. The HEK293 lipid bilayer is less viscous than CHOK1 cells as observed in our whole-cell recordings, which may justify the heightened Q10 in the late INa measurements. At elevated stimulation frequencies and temperature, therapeutic Ranolazine decreased channel availability by increasing channel use-dependence in E1784K.
Although Ranolazine efficacy appears to be increased by temperature, its efficacy appears to be dampened by the combination of both elevated temperature and cytosolic calcium. At 34 °C, E1784K late INa percent was depressed with elevated cytosolic calcium, but there was no effect on late INa density. These opposing changes in late INa percent and density may be attributed to the increased peak INa density with elevated cytosolic calcium at 34 °C. Although not significant, the shift contributes to late INa percent calculation (Table 1). Late INa in E1784K was not attenuated by Ranolazine at elevated temperature and cytosolic calcium.
Drug Binding in Mutant-Trigger Context
Ranolazine action on NaV1.5 is commonly associated with the modulated receptor hypothesis65. Sokolov et al. argued that the drug follows a modified form of binding, as their pharmacological analysis suggested drug-binding at hyperpolarized voltages48. Ranolazine exhibits partial preference to the closed state in NaV1.5, as suggested by the hyperpolarizing shift in SSFI observed with Ranolazine. The shift mainly decreases channel availability near resting potential in cardiomyocytes. Other classic antiarrhythmics, anticonvulsants, and local anesthetics have similar effects, yet follow the modulated receptor hypothesis59,66. Sokolov et al. also report that the drug-induced block in late INa occurs by a slow-mode recovery in slow inactivation, which is exacerbated at low pH48. This effect was mildly observed in this study. However, it is difficult to correlate the drug effects in E1784K UDI to late INa, as the former had minor but significant shifts.
Ranolazine shares a very similar structure with the class 1b antiarrhythmic drug lidocaine, which, like other sodium blockers, preferentially binds to F1760 and, to a lesser extent, Y1767 in DIVS649,52. Ranolazine has high lipophilicity and can only bind to its receptor sites by traversing the phospholipid membrane and entering the central cavity through the inner vestibule. However, lateral pores, known as fenestrations, are alternative routes for large compounds like Ranolazine to access the central cavity. The fenestrations in the NaVPas-NaV1.5 model were unavailable for drug binding in auto-docking due to their constricted sizes67. It would be interesting to determine whether E1784K alters fenestration size in NaV1.5, modifying drug entry via the fenestrations.
The interaction between Ranolazine and extracellular channel regions is unknown, but is unlikely due to its lipophilicity; a crystal structure of the channel/drug interaction would elucidate the drug-induced modifications in gating. The newly discovered aryl sulfonamide antagonists preferentially stabilize NaV1.7 DIVS4 activation thereby stabilizing the fast inactivated state and suppressing late INa68. Ranolazine may be structurally modified to include other moieties, like anionic aryl sulfonamides, for further optimizing its selectivity for targeting late versus peak INa.
E1784K-induced Structural Rearrangements in NaV1.5 and its Impact on Ranolazine
We speculate E1784K affects fast inactivation via two possible mechanisms in NaV1.5, thereby altering drug-channel interactions. Figure 1 shows the channel structures discussed, as follows:
E1784K hyperpolarizes the voltage-dependence of SSFI, thus stabilizing the interaction between the channel and the fast inactivation particle7,11,12,22,28. Fast inactivation onset is correlated with DIVS4 activation, whereas channel recovery is rate limited by charge immobilization of DIVS469,70. The charge reversal mutant, E1784K, may enhance the transition of DIVS4 between closed and open states, as suggested by the Peters-Ruben model43. We postulate that this effect may be due to an electrostatic repulsion between the CTD mutant and conserved positive residues in DIVS4, given their close proximity67. This repulsion could make the DIVS4 in E1784K more mobile, which might explain the accelerated fast inactivation onset and recovery kinetics22,37,41,43. Fast inactivation kinetics in E1784K are not enhanced by temperature, so it does not seem justified to attribute the thermosensitive late INa in E1784K to increased recovery kinetics. Rather a rearrangement may occur in DIVS4, conforming the voltage sensor to a state in which conductance in E1784K is higher43.
E1784K alters the structure of CTD by disrupting the native hydrophobic and electrostatic interactions holding the EF-like hand domain (α1–α4) tight with the IQ motif (α6)31. Calcium sensitivity is imparted in NaV1.5 via CaM, which binds to the IQ motif (α6) via its C-lobe or N-lobe depending on cytosolic calcium levels71–73. During diastole or systole, CaM is calcified to different extents at its N-lobe74. Calcified CaM has a lower affinity for the IQ motif and binds, via its C-lobe, to DIII-DIV linker, forming a tripartite complex. This interaction is thought to prevent the DIII-DIV linker fast inactivation particle from occluding the pore, increasing channel availability near resting potential71,73. With depolarized potentials, the CaM C-lobe stabilizes the fast inactivation particle, suppressing late INa, as in ∆KPQ and other mixed syndrome mutants37,40,44. Some studies refute the tripartite complex formation and favor a CaV1.2-like regulation of inactivation in NaV1.574,75. In those studies, the calcium-calmodulin complex is localized to CTD76,77. The NaVPas structure showed intermolecular interactions between DIII-DIV linker, CTD, and DIVS467. Motoike et al. reported CaM-independent interactions between the inter-linkers in NaV1.536, suggesting that CaM acts as an auxiliary channel modifier during a calcium signal75. Calcium regulation in NaV1.5 is mediated by CaM since the dual EF-like hand domains in CTD do not bind calcium74,75. In light of these structural models, we speculate that E1784K decouples both the calcium-dependent and calcium–independent interactions between the DIII-DIV linker and the CTD. Thus, E1784K inhibits calcium-dependent facilitation in NaV1.5. We propose that the decoupling in CTD caused by E1784K may create a high entropy, unstable structure. Upon a calcium signal, the calcified calmodulin has reduced affinity for the IQ motif, thus augmenting CTD entropy73.
Both mechanisms (1) and (2) may occur simultaneously in E1784K. The calcium effects in NaV1.5 are localized to CTD. No reports have shown direct interaction between calcium-calmodulin and DIVS4, so if mechanism (2) occurs, it may be via an indirect effect on DIVS4.
In light of the discussed structural insights, we speculate that Ranolazine can easily access the inner vestibule with non-calcified calmodulin, since the molecule binds tightly to the IQ motif. Ranolazine efficacy, however, is hampered by cytosolic calcium, suggesting an interaction between the drug and the channel at CTD. The high entropy CTD in calcified calmodulin seems to physically hinder Ranolazine from entering into the inner vestibule.
Physiological and Medical Implications
Elevated temperature and cytosolic calcium are two of many other physiological triggers that occur during exercise and are common to other pathophysiological states, such as myocardial ischemia or infarction, and heart failure78,79. The majority patients with SCN5a mutations show ameliorated LQT3 phenotype during exercise80. Functional studies have correlated this to a stimulation frequency or calcium-induced reduction in late INa44. However, it is clear from our study, focusing on E1784K, that the SCN5a mutant response to triggers can be unique37,41. Thus, it is necessary to study antiarrhythmics in SCN5a cohorts during different physiological states as the mutant-trigger effect may determine drug efficacy.
Our AP simulations clearly show pro-arrhythmic effects of Ranolazine, which are exacerbated by febrile temperatures. Electrical restitution curves clearly show a critical diastolic interval at which alternans are triggered. Our AP simulations provide evidence of Ranolazine’s arrhythmogenicity, as it does not shorten APD90 in E1784K at high heart rates. At low heart rates and at body core temperature, the drug shortens APD90 in cardiac cells. However, with normal and elevated heart rates, the drug induces alternans, an effect exacerbated at febrile temperature. The critical diastolic intervals at which alternans are caused by the drug appear earlier (at higher BCLs) at febrile temperature.
E1784K induces alternans with higher prevalence in epicardial cells at low heart rates. This result coincides with the phase 2 re-entry phenomenon constituting the repolarization hypothesis in BrS178. The high IKto density, especially in the right epicardium, results in complete action potential failure81. E1784K channels are less available for activation due to the hyperpolarized SSFI-V1/2. This seems to be the main mechanism behind the decrease in AP upstroke velocity in cardiac cells, especially the epicardial cells, despite the mutant and triggers-exacerbated increased late INa. Thus, E1784K expresses both gain- and loss-of-function at the electrical level in cardiac cells. However, this expressivity is finely tuned by channel switches, like temperature and cytosolic calcium.
Our previous and current data suggest exercise, and its accompanying physiological triggers, differentially affect mixed syndrome mutations, especially E1784K22,37,41,42. The action of different antiarrhythmics appear to differ depending on physiological state.
Conclusions
Appropriate management of cardiac arrhythmias in SCN5a patients requires careful investigation of antiarrhythmic drug efficacy under various physiological states. Our results suggest that Ranolazine may increase the susceptibility for arrhythmia development in E1784K carriers at sinus rhythm and tachycardia. The risk is augmented under febrile conditions. Although exercise is commonly associated with high heart rates, other pathophysiological states share common triggers, as in heart failure or myocardial ischemia and infarction. Other antiarrhythmics should also be screened against E1784K and other channel mutants under various physiological conditions.
Methods
Homology Modelling and Auto-Docking
Homology modeling was performed using the Swiss-Model server (https://swissmodel.expasy.org)82. The newly cryo-EM solved American cockroach voltage-gated sodium channel (NaVPas) structure (3.8-Å resolution) was used as a template against the NaV1.5 sequence. Modeling was done according to the protocol established by Bordoli et al.82. Sequence alignment was performed using Uniprot Align (http://www.uniprot.org/align/) for SCN5A_HUMAN (NaV1.5) and SCNA1_PERAM (NaVPas).
Ranolazine was virtually docked using AutoDock4 against the NaV1.5 homology model built on NaVPas (NaV1.5-NaVPas)83. PyMOL-pdb viewer was used for optimization and visualization of the auto-docking results.
Ethical approval
The research was approved by Biohazards review 251–2012 issued by the office of the Environmental Health and Safety at Simon Fraser University, Burnaby, BC, Canada.
Cell Culture
HEK293 cells were grown at pH 7.4 in a DMEM (1x) nutrient medium (Life Technologies, NY, USA), supplemented with 10% FBS and maintained in a humidified environment at 37 °C with 5% CO2. The α subunits (WT or E1784K) were co-transfected with the β1 subunit and green fluorescent protein, eGFP (1.50 µg: 0.75 µg: 1.50 µg, respectively). The cDNA mixture was then allowed to incubate with the HEK293 cells before plating on coverslips. The HEK293 cells were selected for this study since they contain a relatively elevated [CaM]free level compared to other cell lines, thereby controlling for calcium-calmodulin effects on NaV1.584.
Electrophysiology
Whole-cell patch clamp recordings were performed in extracellular solution containing (mM): 96 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, and 10 HEPES (pH 7.4). Solutions were titrated with CsOH to pH 7.4. Pipettes were fabricated with a P-1000 puller using borosilicate glass (Sutter Instruments, CA, USA), dipped in dental wax to reduce capacitance, then thermally polished to a resistance of 1.0–1.5 MΩ. Low resistance electrodes were used to minimize series resistance between pipette and intracellular solution resulting in typical access resistances of 3.5 MΩ or less, thereby minimizing voltage measurement error. Pipettes were filled with intracellular solution. For minimal cytosolic calcium levels, pipettes contained (mM): 130 CsF, 9.6 NaCl, 10 HEPES, and 10 EGTA titrated to pH 7.4. The intracellular pipette solution was manipulated to mimic peak systolic cytosolic calcium85,86. To do so, we calculated, using the Ca-EGTA Calculator v1.3, the amount of CaCl2 (in mM) added to bring cytosolic calcium to 2500 nM at both 22 °C and 34 °C: 9.53 and 9.60, respectively.
All recordings were made using an EPC-9 patch-clamp amplifier (HEKA Elektronik, Lambrecht, Germany) digitized at 20 kHz using an ITC-16 interface (HEKA Elektronik, Lambrecht, Germany). Data were acquired and low-pass-filtered (5 kHz) using PatchMaster/FitMaster software (HEKA Elektronik, Lambrecht, Germany) running on an Apple iMac (Apple Computer, Cupertino, CA). Leak subtraction was performed online using a P/4 procedure. Bath solution temperature was controlled using a Peltier device driven by a TC-10 Temperature Controller (Dagan, Minneapolis, MN). Bath temperature was maintained at 22 °C or 34 °C. Experiments were not performed at physiological temperatures because of the inherent instability of cells at temperatures above 34 °C. We extrapolated data to physiological temperatures using a Q10 relationship, which was supported with Arrhenius calculations, (described below). After a giga ohm seal resistance was achieved, the whole-cell configuration was attained. The holding potential between protocols was −110 mV. We recorded INa from cells that expressed currents no greater than −5 nA. The average voltage error calculated for all cells used in this study (n = 250) is 6.06 mV ± 0.40 mV obtained (Table 7). There are no differences between the voltage-errors in the different conditions (p > 0.05).
Table 7.
Voltage Error.
| Condition | Voltage Error (mV) | N |
|---|---|---|
| WT - 22 °C - 0 nM Ca2+ - 0 µM Ran | 3.52 ± 0.97 | 9 |
| WT - 22 °C - 0 nM Ca2+ - 10 µM Ran | 7.83 ± 1.47 | 8 |
| WT - 22 °C - 0 nM Ca2+ - 100 µM Ran | 5.70 ± 2.05 | 6 |
| WT - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 4.89 ± 1.29 | 9 |
| WT - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 6.54 ± 1.30 | 11 |
| WT - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 3.12 ± 0.88 | 10 |
| WT - 34 °C - 0 nM Ca2+ - 0 µM Ran | 8.35 ± 1.56 | 11 |
| WT - 34 °C - 0 nM Ca2+ - 10 µM Ran | 9.08 ± 1.32 | 9 |
| WT - 34 °C - 0 nM Ca2+ - 100 µM Ran | 7.18 ± 1.16 | 19 |
| WT - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 8.64 ± 0.98 | 7 |
| WT - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 8.53 ± 1.79 | 9 |
| WT - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 8.14 ± 1.57 | 6 |
| EK - 22 °C - 0 nM Ca2+ - 0 µM Ran | 4.41 ± 0.65 | 19 |
| EK - 22 °C - 0 nM Ca2+ - 10 µM Ran | 5.99 ± 1.22 | 10 |
| EK - 22 °C - 0 nM Ca2+ - 100 µM Ran | 4.28 ± 0.72 | 14 |
| EK - 22 °C - 2500 nM Ca2+ - 0 µM Ran | 3.01 ± 0.71 | 6 |
| EK - 22 °C - 2500 nM Ca2+ - 10 µM Ran | 4.49 ± 1.04 | 9 |
| EK - 22 °C - 2500 nM Ca2+ - 100 µM Ran | 4.63 ± 1.21 | 10 |
| EK–34 °C - 0 nM Ca2+ - 0 µM Ran | 6.50 ± 1.09 | 19 |
| EK - 34 °C - 0 nM Ca2+ - 10 µM Ran | 6.74 ± 1.73 | 10 |
| EK - 34 °C - 0 nM Ca2+ - 100 µM Ran | 5.52 ± 0.98 | 12 |
| EK - 34 °C - 2500 nM Ca2+ - 0 µM Ran | 5.63 ± 1.32 | 8 |
| EK - 34 °C - 2500 nM Ca2+ - 10 µM Ran | 8.95 ± 1.97 | 8 |
| EK - 34 °C - 2500 nM Ca2+ - 100 µM Ran | 3.67 ± 0.69 | 11 |
Drug Preparation
Ranolazine was obtained from Gilead Sciences (Foster City, CA) in powder form, diluted to 100 mM stock in 0.1 M HCl, aliquoted at 10 mM and stored at −20 °C. Working concentrations of 10 µM (therapeutic concentration) or 100 μM (non-therapeutic) were freshly prepared in bath solution. pH was readjusted before performing electrophysiological experiments. Due to the large number of experimental conditions and the challenges of maintaining whole-cell recordings at elevated temperature, we performed unmatched pair experiments.
Analysis and Statistics
Analysis and graphing were done using FitMaster software (HEKA Elektronik, Lambrecht, Germany) and Igor Pro (Wavemetrics, Lake Oswego, OR, USA) with statistical information derived using JMP statistical software. Statistical significance was accepted at p < 0.05 using a four-factor completely randomized design (CRD) ANOVA test followed by a post-hoc Tukey test. Our statistical model was a full factorial in which all the factors were allowed to interact together yielding multiple effect tests: Ranolazine, Channel Variant, Ranolazine × Channel Variant, Temperature, Ranolazine × Temperature, Channel Variant × Temperature, Ranolazine × Channel Variant × Temperature, Calcium, Ranolazine × Calcium, Channel Variant × Calcium, Ranolazine × Channel Variant × Calcium, Temperature × Calcium, Ranolazine × Temperature × Calcium, Channel Variant × Temperature × Calcium, Ranolazine × Channel Variant × Temperature × Calcium. All values reported in the results sections are given as means ± standard error of means.
Voltage Protocols
Current Density
We measured current density from the ratio of current amplitude to the cell membrane capacitance (pA/pF).
Conductance Density
Channel conductance was calculated from peak INa using Ohm’s law at 0 mV.
| 1 |
where GNa is sodium channel conductance, INa is peak sodium current in response to the command potential V = 0 mV, and Erev is the reversal potential. We measured conductance density from the ratio of conductance to the cell membrane capacitance (nS/pF).
Activation (GV)
To determine the voltage dependence of activation, we measured the peak current amplitude at test pulse potentials ranging from −100 mV to +80 mV in increments of +10 mV for 19 ms. Prior to the test pulse, channels were allowed to recover from fast inactivation at −130 mV for 197 ms. Channel conductance was calculated from peak INa using Formula (1). Calculated values for conductance were normalized to the maximal conductance and fit with the Boltzmann function:
| 2 |
where G/G max is the normalized conductance amplitude, Vm is the command potential, z is the apparent valence, e0 is the elementary charge, V1/2 is the midpoint voltage, k is the Boltzmann constant, and T is temperature in °K.
Steady-State Fast Inactivation (SSFI)
The voltage-dependence of SSFI was measured by preconditioning the channels to a hyperpolarizing potential of −130 mV and then eliciting prepulses from −130 or −150 to +10 mV in increments of 10 mV for 500 ms. Channel availability was assessed during a test pulse to 0 mV. Normalized current amplitude as a function of voltage was fit using the Boltzmann function:
| 3 |
where I/Imax is the normalized current amplitude, z is apparent valence, e0 is the elementary charge, Vm is the prepulse potential, V1/2 is the midpoint voltage of SSFI, k is the Boltzmann constant, and T is temperature in °K.
Fast Inactivation Onset
Time constants for open-state fast inactivation were derived by fitting a single exponential function to the decay of current obtained from the activation protocol.
| 4 |
where I is current amplitude, Iss is the plateau amplitude, α is the amplitude at time 0 for time constant τ, and t is time.
Late INa Current
Late INa was measured between 40–50 ms during a 50 ms depolarizing pulse to 0 mV from a holding potential of −130 mV. An average of 10 pulses was used to increase the signal-to-noise ratio.
Use-Dependent Inactivation (UDI, 1 Hz and 3 Hz)
Channels accumulated into a use-dependent inactivated state during either a series of 300 380 ms depolarizing pulses to 0 mV followed by a 615 ms–110 mV recovery pulse at a frequency 1 Hz, or 500 220 ms depolarizing pulses to 0 mV followed by a 110 ms–110 mV recovery pulse at a frequency 3 Hz. Normalized current amplitude as a function of time was fit with a double exponential.
| 5 |
where I is current amplitude, Iss is the plateau amplitude, α1 and α2 are the amplitudes at time 0 for time constants τ1 and τ2, and t is time.
Q10 Coefficients
The temperature coefficient for kinetic and thermodynamic parameters plotted as a function temperature was calculated in Igor:
| 6 |
where R is the rate and T is temperature (1 and 2 are the two temperatures measured). Rate was calculated by the inverse of the τ value. Q10 fits for steady-state midpoints and slopes were calculated by replacing the RX with V1/2 and z values. Fits for y0 were calculated based of the 1/y0 to yield optimal Q10 values. The fit was extrapolated to physiological (37 °C) and febrile (41 °C) temperatures.
Arrhenius Calculations
The Arrhenius linear relationship for the natural exponent of kinetic or thermodynamic parameters as a function of inverse temperature was calculated in Igor:
| 7 |
where k is the rate constant, steady-state midpoint, or slope, A is the pre-exponential factor, Ea is the activation energy, R is the universal gas constant, and T is temperature in °K.
Myocardial Action Potential (AP) Modeling
Simulations
Action potentials were simulated using a modified version of the O’Hara Rudy (ORd) model at 37 °C and 41 °C programmed in Matlab87. The sodium data was extrapolated to physiological and febrile temperatures Q10 values for WT and E1784K at 0 µM and 10 µM Ranolazine. The maximal GNa density was 150 mS/µF in all conditions simulated. We modified the gating INa parameters data in accordance with our biophysical data for the various conditions. The GV and SSFI midpoints and slopes were extrapolated to 37 °C and 41 °C and normalized to the original ORd parameters. The phosphorylated steady-state fast inactivation midpoints in all channel variants were equally hyperpolarized by 6.2 mV. Late INa density was normalized to the original ORd value and multiplied by the percentage of late to peak INa calculated above.
To model the calcium-dependence of our late INa data, we fit the biophysical parameters extrapolated to 37 °C and 41 °C with a Hill equation:
| 8 |
where Z is the biophysical parameter of interest, Y0 is the minimum value, YM is the maximum value, X1/2 is the midpoint of the curve, X is the intracellular cytosolic calcium, b is the rate.
Subspace calcium was not accounted for due to the lack of experimental data. Thus, the modified ORd model is a dynamic simulation of the calcium-induced shifts which are observed with increasing intracellular calcium levels as a function of pacing frequency, comprising the positive staircase phenomenon88,89.
Simulations at febrile temperature (41 °C) included modifications to the major ionic currents, IKto90, ICaL91,92, IKr93, and IKs94, in the ORd model based on previously published Q10 values.
Simulations were run on endocardial, midmyocardial, and epicardial ventricular myocytes using a 0.5 ms stimulus pulse with an amplitude of −80 µA/µF. The stimulus protocol was designed to step up the frequency gradually from 0.5 Hz to 2.5 Hz, with a 1000 beats per frequency step to ensure attainment of steady-state.
Analysis
Analysis of APs only included those that fully recovered and were restored to baseline. Action potential duration (APD) was measured at 90% of repolarization by multiplying the resting membrane potential (RMP) value, prior to the current stimulus pulse, by 0.9. The APD90 of the final two beats in the frequency step were plotted versus the diastolic interval (DI = BCL − APD90), where BCL is the basic cycle length, creating electrical restitution curves.
Data Availability
All data generated or analysed during this study are included in this published article.
Acknowledgements
We thank Dr. Elizabeth M. Cherry for helping in A.P. simulation analysis. We are also grateful to both Mr. Colin Peters and Mr. Mohammed-Reza Ghovanloo for their valuable input and support. P.R. and M.A. receive funding from Simon Fraser University, the Natural Sciences and Engineering Research Council of Canada, and the Canadian Foundation for Innovation.
Author Contributions
M.A. and co-author collected, assembled, analyzed, and interpreted the data, designed the experiments, and drafted the manuscript. M.R. collected, assembled, and analyzed data. P.C.R. conceived the experiments and revised the manuscript critically for important intellectual content. All authors approved the final version of the manuscript and qualify for authorship.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bezzina C, et al. A Single Na+ Channel Mutation Causing Both Long-QT and Brugada Syndromes. Circ. Res. 1999;85:1206–1213. doi: 10.1161/01.RES.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 2.Rivolta I. Inherited Brugada and Long QT-3 Syndrome Mutations of a Single Residue of the Cardiac Sodium Channel Confer Distinct Channel and Clinical Phenotypes. J. Biol. Chem. 2001;276:30623–30630. doi: 10.1074/jbc.M104471200. [DOI] [PubMed] [Google Scholar]
- 3.Kapplinger JD, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm Off. J. Heart Rhythm Soc. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antzelevitch C, Brugada P, Brugada J, Brugada R. Brugada syndrome: from cell to bedside. Curr. Probl. Cardiol. 2005;30:9–54. doi: 10.1016/j.cpcardiol.2004.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu W, Antzelevitch C. Differential effects of beta-adrenergic agonists and antagonists in LQT1, LQT2 and LQT3 models of the long QT syndrome. J. Am. Coll. Cardiol. 2000;35:778–786. doi: 10.1016/S0735-1097(99)00582-3. [DOI] [PubMed] [Google Scholar]
- 6.Baroudi G, Chahine M. Biophysical phenotypes of SCN5A mutations causing long QT and Brugada syndromes. FEBS Lett. 2000;487:224–228. doi: 10.1016/S0014-5793(00)02360-7. [DOI] [PubMed] [Google Scholar]
- 7.Dumaine R, et al. Multiple mechanisms of Na+ channel–linked long-QT syndrome. Circ. Res. 1996;78:916–924. doi: 10.1161/01.RES.78.5.916. [DOI] [PubMed] [Google Scholar]
- 8.Sun, A. Y. et al. The S1103Y cardiac sodium channel variant is associated with ICD events in African Americans with heart failure and reduced ejection fraction. Circ. Cardiovasc. Genet. CIRCGENETICS–110 (2011). [DOI] [PMC free article] [PubMed]
- 9.Baroudi G, Acharfi S, Larouche C, Chahine M. Expression and intracellular localization of an SCN5A double mutant R1232W/T1620M implicated in Brugada syndrome. Circ. Res. 2002;90:E11–16. doi: 10.1161/hh0102.102977. [DOI] [PubMed] [Google Scholar]
- 10.Makita N, Mochizuki N, Tsutsui H. Absence of a trafficking defect in R1232W/T1620M, a double SCN5A mutant responsible for Brugada syndrome. Circ. J. Off. J. Jpn. Circ. Soc. 2008;72:1018–1019. doi: 10.1253/circj.72.1018. [DOI] [PubMed] [Google Scholar]
- 11.Chandra R, Starmer CF, Grant AO. Multiple effects of KPQ deletion mutation on gating of human cardiac Na+ channels expressed in mammalian cells. Am. J. Physiol. 1998;274:H1643–1654. doi: 10.1152/ajpheart.1998.274.5.H1643. [DOI] [PubMed] [Google Scholar]
- 12.Wei J, et al. Congenital Long-QT Syndrome Caused by a Novel Mutation in a Conserved Acidic Domain of the Cardiac Na+ Channel. Circulation. 1999;99:3165–3171. doi: 10.1161/01.CIR.99.24.3165. [DOI] [PubMed] [Google Scholar]
- 13.Groenewegen WA, et al. A novel LQT3 mutation implicates the human cardiac sodium channel domain IVS6 in inactivation kinetics. Cardiovasc. Res. 2003;57:1072–1078. doi: 10.1016/S0008-6363(02)00838-6. [DOI] [PubMed] [Google Scholar]
- 14.Moreau A, et al. Sodium overload due to a persistent current that attenuates the arrhythmogenic potential of a novel LQT3 mutation. Front. Pharmacol. 2013;4:126. doi: 10.3389/fphar.2013.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon HW, et al. Long QT syndrome and dilated cardiomyopathy with SCN5A p.R1193Q polymorphism: cardioverter-defibrillator implantation at 27 months. Pacing Clin. Electrophysiol. PACE. 2012;35:e243–246. doi: 10.1111/j.1540-8159.2012.03409.x. [DOI] [PubMed] [Google Scholar]
- 16.Bankston JR, et al. A novel LQT-3 mutation disrupts an inactivation gate complex with distinct rate-dependent phenotypic consequences. Channels Austin Tex. 2007;1:273–280. doi: 10.4161/chan.4956. [DOI] [PubMed] [Google Scholar]
- 17.Veldkamp MW, et al. UltraRapid Communication. Circ Res. 2000;86:e91–e97. doi: 10.1161/01.RES.86.9.e91. [DOI] [PubMed] [Google Scholar]
- 18.Mok N-S, et al. A newly characterized SCN5A mutation underlying Brugada syndrome unmasked by hyperthermia. J. Cardiovasc. Electrophysiol. 2003;14:407–411. doi: 10.1046/j.1540-8167.2003.02379.x. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, H. W. et al. R1193Q of SCN5A, a Brugada and long QT mutation, is a common polymorphism in Han Chinese. J. Med. Genet. 42, e7; author replye8 (2005). [DOI] [PMC free article] [PubMed]
- 20.Wang DW, Makita N, Kitabatake A, Balser JR, George AL. Enhanced Na+ channel intermediate inactivation in Brugada syndrome. Circ. Res. 2000;87:e37–e43. doi: 10.1161/01.RES.87.8.e37. [DOI] [PubMed] [Google Scholar]
- 21.Postema PG, et al. Founder mutations in the Netherlands: SCN5a 1795insD, the first described arrhythmia overlap syndrome and one of the largest and best characterised families worldwide. Neth. Heart J. Mon. J. Neth. Soc. Cardiol. Neth. Heart Found. 2009;17:422–428. doi: 10.1007/BF03086296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makita, N. et al. The E1784K mutation in SCN5A is associated with mixed clinical phenotype of type 3 long QT syndrome. J. Clin. Invest. 10.1172/JCI34057 (2008). [DOI] [PMC free article] [PubMed]
- 23.Huang H, Zhao J, Barrane F-Z, Champagne J, Chahine M. Nav1.5/R1193Q polymorphism is associated with both long QT and Brugada syndromes. Can. J. Cardiol. 2006;22:309–313. doi: 10.1016/S0828-282X(06)70915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, J. et al. Cardiac sodium channel mutation associated with epinephrine-induced QT prolongation and sinus node dysfunction. Heart Rhythm Off. J. Heart Rhythm Soc. 10.1016/j.hrthm.2015.08.021 (2015). [DOI] [PubMed]
- 25.Splawski I, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.CIR.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 26.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, et al. High prevalence of the SCN5A E1784K mutation in school children with long QT syndrome living on the Okinawa islands. Circ. J. Off. J. Jpn. Circ. Soc. 2014;78:1974–1979. doi: 10.1253/circj.cj-13-1516. [DOI] [PubMed] [Google Scholar]
- 28.Deschênes I, et al. Electrophysiological characterization of SCN5A mutations causing long QT (E1784K) and Brugada (R1512W and R1432G) syndromes. Cardiovasc. Res. 2000;46:55–65. doi: 10.1016/S0008-6363(00)00006-7. [DOI] [PubMed] [Google Scholar]
- 29.Sumitomo N. E1784K mutation in SCN5A and overlap syndrome. Circ. J. Off. J. Jpn. Circ. Soc. 2014;78:1839–1840. doi: 10.1253/circj.cj-14-0564. [DOI] [PubMed] [Google Scholar]
- 30.Mantegazza M, Yu FH, Catterall WA, Scheuer T. Role of the C-terminal domain in inactivation of brain and cardiac sodium channels. Proc. Natl. Acad. Sci. USA. 2001;98:15348–15353. doi: 10.1073/pnas.211563298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chagot B, Potet F, Balser JR, Chazin WJ. Solution NMR Structure of the C-terminal EF-hand Domain of Human Cardiac Sodium Channel NaV1.5. J. Biol. Chem. 2009;284:6436–6445. doi: 10.1074/jbc.M807747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cormier JW, Rivolta I, Tateyama M, Yang A-S, Kass RS. Secondary Structure of the Human Cardiac Na+ Channel C Terminus: Evidence For a Role of Helical Structures In Modulation of Channel Inactivation. J. Biol. Chem. 2002;277:9233–9241. doi: 10.1074/jbc.M110204200. [DOI] [PubMed] [Google Scholar]
- 33.Glaaser IW, Bankston JR, Liu H, Tateyama M, Kass RS. A carboxyl-terminal hydrophobic interface is critical to sodium channel function. Relevance to inherited disorders. J. Biol. Chem. 2006;281:24015–24023. doi: 10.1074/jbc.M605473200. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, et al. Calmodulin Mediates Ca2+ Sensitivity of Sodium Channels. J. Biol. Chem. 2004;279:45004–45012. doi: 10.1074/jbc.M407286200. [DOI] [PubMed] [Google Scholar]
- 35.Glaaser IW, et al. Perturbation of sodium channel structure by an inherited Long QT Syndrome mutation. Nat. Commun. 2012;3:706. doi: 10.1038/ncomms1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motoike HK. The Na+ Channel Inactivation Gate Is a Molecular Complex: A Novel Role of the COOH-terminal Domain. J. Gen. Physiol. 2004;123:155–165. doi: 10.1085/jgp.200308929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelsayed, M. et al. Differential calcium sensitivity in NaV1.5 mixed syndrome mutants: Calcium and mixed syndrome mutants. J. Physiol. 10.1113/JP274536 (2017). [DOI] [PMC free article] [PubMed]
- 38.Mori M, Konno T, Morii T, Nagayama K, Imoto K. Regulatory interaction of sodium channel IQ-motif with calmodulin C-terminal lobe. Biochem. Biophys. Res. Commun. 2003;307:290–296. doi: 10.1016/S0006-291X(03)01183-5. [DOI] [PubMed] [Google Scholar]
- 39.Yan H, Wang C, Marx SO, Pitt GS. Calmodulin limits pathogenic Na+ channel persistent current. J. Gen. Physiol. 2017;149:277–293. doi: 10.1085/jgp.201611721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Petegem F, Lobo PA, Ahern CA. Seeing the Forest through the Trees: towards a Unified View on Physiological Calcium Regulation of Voltage-Gated Sodium Channels. Biophys. J. 2012;103:2243–2251. doi: 10.1016/j.bpj.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdelsayed, M., Peters, C. H. & Ruben, P. C. Differential thermosensitivity in mixed syndrome cardiac sodium channel mutants. J. Physiol. 10.1113/JP270139 (2015). [DOI] [PMC free article] [PubMed]
- 42.Peters CH, Abdelsayed M, Ruben PC. Triggers for arrhythmogenesis in the Brugada and long QT 3 syndromes. Prog. Biophys. Mol. Biol. 2016;120:77–88. doi: 10.1016/j.pbiomolbio.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Peters CH, Yu A, Zhu W, Silva JR, Ruben PC. Depolarization of the conductance-voltage relationship in the NaV1.5 mutant, E1784K, is due to altered fast inactivation. PLOS ONE. 2017;12:e0184605. doi: 10.1371/journal.pone.0184605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potet, F., Beckermann, T. M., Kunic, J. D. & George, A. L. Intracellular Calcium Attenuates Late Current Conducted by Mutant Human Cardiac SodiumChannels. Circ. Arrhythm. Electrophysiol. 10.1161/CIRCEP.115.002760 (2015). [DOI] [PMC free article] [PubMed]
- 45.Antzelevitch C. Electrophysiological Effects of Ranolazine, a Novel Antianginal Agent With Antiarrhythmic Properties. Circulation. 2004;110:904–910. doi: 10.1161/01.CIR.0000139333.83620.5D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antzelevitch C, et al. Electrophysiologic properties and antiarrhythmic actions of a novel antianginal agent. J. Cardiovasc. Pharmacol. Ther. 2004;9(Suppl 1):S65–83. doi: 10.1177/107424840400900106. [DOI] [PubMed] [Google Scholar]
- 47.Sossalla S, et al. Ranolazine improves diastolic dysfunction in isolated myocardium from failing human hearts—Role of late sodium current and intracellular ion accumulation. J. Mol. Cell. Cardiol. 2008;45:32–43. doi: 10.1016/j.yjmcc.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 48.Sokolov, S., Peters, C. H., Rajamani, S. & Ruben, P. C. Proton-dependent inhibition of the cardiac sodium channel Nav1.5 by ranolazine. Front. Pharmacol. 4, (2013). [DOI] [PMC free article] [PubMed]
- 49.Huang H, Priori SG, Napolitano C, O’Leary ME, Chahine M. Y1767C, a novel SCN5A mutation, induces a persistent Na+ current and potentiates ranolazine inhibition of Nav1.5 channels. AJP Heart Circ. Physiol. 2011;300:H288–H299. doi: 10.1152/ajpheart.00539.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moss AJ, et al. Ranolazine Shortens Repolarization in Patients with Sustained Inward Sodium Current Due to Type-3 Long-QT Syndrome. J. Cardiovasc. Electrophysiol. 2008;19:1289–1293. doi: 10.1111/j.1540-8167.2008.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajamani S, El-Bizri N, Shryock JC, Makielski JC, Belardinelli L. Use-dependent block of cardiac late Na(+) current by ranolazine. Heart Rhythm Off. J. Heart Rhythm Soc. 2009;6:1625–1631. doi: 10.1016/j.hrthm.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fredj S, Sampson KJ, Liu H, Kass RS. Molecular basis of ranolazine block of LQT-3 mutant sodium channels: evidence for site of action. Br. J. Pharmacol. 2006;148:16–24. doi: 10.1038/sj.bjp.0706709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belardinelli L. The mechanism of ranolazine action to reduce ischemia-induced diastolic dysfunction. Eur. Heart J. Suppl. 2006;8:A10–A13. doi: 10.1093/eurheartj/sui091. [DOI] [Google Scholar]
- 54.Peters C, Sokolov S, Rajamani S, Ruben P. Effects of the antianginal drug, ranolazine, on the brain sodium channel NaV1.2 and its modulation by extracellular protons: Effects of ranolazine on Nav1.2 in normal and low extracellular pH. Br. J. Pharmacol. 2013;169:704–716. doi: 10.1111/bph.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lipkind GM, Fozzard HA. Molecular Model of Anticonvulsant Drug Binding to the Voltage-Gated Sodium Channel Inner Pore. Mol. Pharmacol. 2010;78:631–638. doi: 10.1124/mol.110.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nolasco JB, Dahlen RW. A graphic method for the study of alternation in cardiac action potentials. J. Appl. Physiol. 1968;25:191–196. doi: 10.1152/jappl.1968.25.2.191. [DOI] [PubMed] [Google Scholar]
- 57.Franz MR. The Electrical Restitution Curve Revisited:. Steep or Flat Slope-Which is Better? J. Cardiovasc. Electrophysiol. 2003;14:S140–S147. doi: 10.1046/j.1540.8167.90303.x. [DOI] [PubMed] [Google Scholar]
- 58.Jordan PN, Christini DJ. Determining the effects of memory and action potential duration alternans on cardiac restitution using a constant-memory restitution protocol. Physiol. Meas. 2004;25:1013–1024. doi: 10.1088/0967-3334/25/4/018. [DOI] [PubMed] [Google Scholar]
- 59.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc. Natl. Acad. Sci. USA. 1996;93:9270–9275. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vilin, Y. Y., Peters, C. H. & Ruben, P. C. Acidosis Differentially Modulates Inactivation in NaV1.2, NaV1.4, and NaV1.5Channels. Front. Pharmacol. 3, (2012). [DOI] [PMC free article] [PubMed]
- 61.Jones DK, Peters CH, Tolhurst SA, Claydon TW, Ruben PC. Extracellular Proton Modulation of the Cardiac Voltage-Gated Sodium Channel, NaV1.5. Biophys. J. 2011;101:2147–2156. doi: 10.1016/j.bpj.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chorin, E. et al. Ranolazine for Congenital Long-QT Syndrome Type III: Experimental and Long-Term Clinical Data. Circ. Arrhythm. Electrophysiol. 9, (2016). [DOI] [PMC free article] [PubMed]
- 63.Murray KT, Anno T, Bennett PB, Hondeghem LM. Voltage clamp of the cardiac sodium current at 37 degrees C in physiologic solutions. Biophys. J. 1990;57:607. doi: 10.1016/S0006-3495(90)82576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruff RL. Effects of temperature on slow and fast inactivation of rat skeletal muscle Na(+) channels. Am. J. Physiol. 1999;277:C937–947. doi: 10.1152/ajpcell.1999.277.5.C937. [DOI] [PubMed] [Google Scholar]
- 65.Hille B. Local anesthetics: hydrophilic and hydrophobic pathways for the drug-receptor reaction. J. Gen. Physiol. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tikhonov DB, Zhorov BS. Mechanism of sodium channel block by local anesthetics, antiarrhythmics, and anticonvulsants. J. Gen. Physiol. 2017;149:465–481. doi: 10.1085/jgp.201611668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen H, et al. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 2017;355:eaal4326. doi: 10.1126/science.aal4326. [DOI] [PubMed] [Google Scholar]
- 68.Ahuja S, et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 2015;350:aac5464–aac5464. doi: 10.1126/science.aac5464. [DOI] [PubMed] [Google Scholar]
- 69.Capes DL, Goldschen-Ohm MP, Arcisio-Miranda M, Bezanilla F, Chanda B. Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J. Gen. Physiol. 2013;142:101–112. doi: 10.1085/jgp.201310998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kühn FJ, Greeff NG. Movement of voltage sensor S4 in domain 4 is tightly coupled to sodium channel fast inactivation and gating charge immobilization. J. Gen. Physiol. 1999;114:167–183. doi: 10.1085/jgp.114.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shah VN, et al. Calcium-dependent regulation of the voltage-gated sodium channel hH1: intrinsic and extrinsic sensors use a common molecular switch. Proc. Natl. Acad. Sci. USA. 2006;103:3592–3597. doi: 10.1073/pnas.0507397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarhan MF, Van Petegem F, Ahern CA. A Double Tyrosine Motif in the Cardiac Sodium Channel Domain III-IV Linker Couples Calcium-dependent Calmodulin Binding to Inactivation Gating. J. Biol. Chem. 2009;284:33265–33274. doi: 10.1074/jbc.M109.052910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarhan MF, Tung C-C, Van Petegem F, Ahern CA. Crystallographic basis for calcium regulation of sodium channels. Proc. Natl. Acad. Sci. 2012;109:3558–3563. doi: 10.1073/pnas.1114748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ben-Johny M, et al. Conservation of Ca2+/Calmodulin Regulation across Na and Ca2+ Channels. Cell. 2014;157:1657–1670. doi: 10.1016/j.cell.2014.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pitt, G. S. & Lee, S.-Y. Current view on regulation of voltage-gated sodium channels by calcium and auxiliary proteins. Protein Sci. 10.1002/pro.2960 (2016). [DOI] [PMC free article] [PubMed]
- 76.Petegem FV, Chatelain FC, Minor DL. Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain–Ca2 + /calmodulin complex. Nat. Struct. 38 Mol. Biol. 2005;12:1108–1115. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ben-Johny M, Yue DT. Calmodulin regulation (calmodulation) of voltage-gated calcium channels. J. Gen. Physiol. 2014;143:679–692. doi: 10.1085/jgp.201311153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lukas A, Antzelevitch C. Phase 2 reentry as a mechanism of initiation of circus movement reentry in canine epicardium exposed to simulated ischemia. Cardiovasc. Res. 1996;32:593–603. doi: 10.1016/S0008-6363(96)00115-0. [DOI] [PubMed] [Google Scholar]
- 79.Rivera-Fernández, R. et al. Prolonged QT interval in ST-elevation myocardial infarction and mortality: new prognostic scale with QT, Killip and age. J. Cardiovasc. Med. Hagerstown Md10.2459/JCM.0000000000000015 (2014). [DOI] [PubMed]
- 80.Schwartz PJ, et al. Long QT syndrome patients with mutations of the SCN5A and HERG genes have differential responses to Na+ channel blockade and to increases in heart rate. Implications for gene-specific therapy. Circulation. 1995;92:3381–3386. doi: 10.1161/01.CIR.92.12.3381. [DOI] [PubMed] [Google Scholar]
- 81.Di Diego JM, Sun ZQ, Antzelevitch C. I(to) and action potential notch are smaller in left vs. right canine ventricular epicardium. Am. J. Physiol. 1996;271:H548–561. doi: 10.1152/ajpheart.1996.271.2.H548. [DOI] [PubMed] [Google Scholar]
- 82.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2008;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 83.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Black D, Tran Q-K, Persechini A. Monitoring the total available calmodulin concentration in intact cells over the physiological range in free Ca2+ Cell Calcium. 2004;35:415–425. doi: 10.1016/j.ceca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 85.Steenbergen C, Murphy E, Levy L, London RE. Elevation in cytosolic free calcium concentration early in myocardial ischemia in perfused rat heart. Circ. Res. 1987;60:700–707. doi: 10.1161/01.RES.60.5.700. [DOI] [PubMed] [Google Scholar]
- 86.Kirschenlohr HL, Grace AA, Vandenberg JI, Metcalfe JC, Smith GA. Estimation of systolic and diastolic free intracellular Ca2+ by titration of Ca2+ buffering in the ferret heart. Biochem. J. 2000;346(Pt 2):385–391. doi: 10.1042/bj3460385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O’Hara T, Virág L, Varró A, Rudy Y. Simulation of the Undiseased Human Cardiac Ventricular Action Potential: Model Formulation and Experimental Validation. PLoS Comput. Biol. 2011;7:e1002061. doi: 10.1371/journal.pcbi.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kort AA, Capogrossi MC, Lakatta EG. Frequency, amplitude, and propagation velocity of spontaneous Ca++ -dependent contractile waves in intact adult rat cardiac muscle and isolated myocytes. Circ. Res. 1985;57:844–855. doi: 10.1161/01.RES.57.6.844. [DOI] [PubMed] [Google Scholar]
- 89.Song LS, et al. Beta-Adrenergic stimulation synchronizes intracellular Ca(2+) release during excitation-contraction coupling in cardiac myocytes. Circ. Res. 2001;88:794–801. doi: 10.1161/hh0801.090461. [DOI] [PubMed] [Google Scholar]
- 90.Yang F, Zheng J. High temperature sensitivity is intrinsic to voltage-gated potassium channels. eLife. 2014;3:e03255. doi: 10.7554/eLife.03255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.ten Tusscher KHWJ. A model for human ventricular tissue. AJP Heart Circ. Physiol. 2003;286:H1573–H1589. doi: 10.1152/ajpheart.00794.2003. [DOI] [PubMed] [Google Scholar]
- 92.Radzicki D, et al. Temperature-Sensitive Cav1.2 Calcium Channels Support Intrinsic Firing of Pyramidal Neurons and Provide a Target for the Treatment of Febrile Seizures. J. Neurosci. 2013;33:9920–9931. doi: 10.1523/JNEUROSCI.5482-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mauerhöfer M, Bauer CK. Effects of Temperature on Heteromeric Kv11.1a/1b and Kv11.3 Channels. Biophys. J. 2016;111:504–523. doi: 10.1016/j.bpj.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seebohm G, Lerche C, Busch AE, Bachmann A. Dependence of I Ks biophysical properties on the expression system. Pflugers Arch. Eur. J. Physiol. 2001;442:891–895. doi: 10.1007/s004240100608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.