Abstract
Background
Circular RNAs (circRNAs) are a class of novel RNAs with important biological functions, and aberrant expression of circRNAs has been implicated in human diseases. However, the feasibility of using blood circRNAs as disease biomarkers is largely unknown.
Methods
We explored the potential of using human peripheral blood mononuclear cell (PBMC) circRNAs as marker molecules to diagnose active pulmonary tuberculosis (TB).
Findings
First, we demonstrated that circRNAs are widely expressed in human PBMCs and that many are abundant enough to be detected. Second, we found that the magnitude of PBMC circRNAs in TB patients was higher than that in the paired healthy controls. Compared with host linear transcripts, the circRNAs within several pathways are disproportionately upregulated in active TB patients, including “Cytokine-cytokine receptor interaction”, “Chemokine signaling pathway”, “Neurotrophin signaling pathway”, and “Bacterial invasion of epithelial cells”. Based on the differentially expressed circRNAs within these pathways, we developed a PBMC circRNA-based molecular signature differentiating active TB patients from healthy controls. We validated the classification power of the PBMC circRNA signature in an independent cohort with the area under the receiver operating characteristic curve (AUC) at 0.946.
Interpretation
Our results suggest that PBMC circRNAs are potentially reliable marker molecules in TB diagnosis.
Keywords: Circular RNA, Tuberculosis, Biomarker, PBMC
Highlights
-
•
Circular RNAs (circRNAs) are abundant in human peripheral blood mononuclear cells (PBMCs).
-
•
CircRNAs are dysregulated in PBMCs from patients with active tuberculosis (TB) compared with healthy controls.
-
•
A PBMC circRNA-based molecular signature can differentiate active TB patients from healthy controls.
Circular RNAs (circRNAs) are emerging as novel regulatory RNAs that have been found in various tissues and cell types. Notably, circRNAs are enriched and stable in whole blood, platelets, and exosomes, which makes circRNAs in human peripheral blood good candidates for diagnostic or prognostic biomarkers of human diseases. Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis. Some novel diagnostic approaches, particularly a rapid, low-cost, and non-sputum-based tests, are essential to identify patients with active TB. Here, we propose that circRNA expression in human peripheral blood mononuclear cells potentially delivers useful information in TB diagnosis.
1. Introduction
Circular RNAs (circRNAs) are a class of novel RNAs that are expressed across the eukaryotic tree of life (Jeck and Sharpless, 2014, Wang et al., 2014). CircRNAs can be formed by back-splicing, which is a non-canonical process in RNA splicing (Jeck and Sharpless, 2014). Although little is known regarding the exact mechanisms of circRNA biogenesis, some factors have been shown to regulate this process, such as flank intronic sequences (Liang and Wilusz, 2014, Vicens and Westhof, 2014, Zhang et al., 2014) and RNA-binding proteins (Barrett et al., 2015, Qu et al., 2015). CircRNAs have been revealed to perform several important functions, including microRNA (miRNA) sponges (Hansen et al., 2013, Memczak et al., 2013), alternative RNA splicing regulation (Conn et al., 2017), and transcriptional regulation of the parental gene (Li et al., 2015b). Recent studies have also suggested that some circRNAs can be translated in a cap-independent translation manner and function in myogenesis (Legnini et al., 2017, Pamudurti et al., 2017, Yang et al., 2017). Furthermore, circRNAs have been found to be differentially expressed across human tissues and cell types (Guo et al., 2014, Salzman et al., 2013) and extraordinarily enriched in the mammalian brain (Rybak-Wolf et al., 2015, You et al., 2015). Although circRNAs are generally expressed at low levels (Chen, 2016, Guo et al., 2014), some circRNAs are expressed more abundantly than their linear counterparts (Salzman et al., 2013). Dynamic circRNA profiles are related to neurogenesis (Rybak-Wolf et al., 2015), mouse brain development (You et al., 2015), and human epithelial-mesenchymal transition (Conn et al., 2015). Aberrant expression of many circRNAs has been implicated in several human diseases, including cancers (Meng et al., 2017), neurodegenerative diseases (Kumar et al., 2017), and some hematological malignancies (Bonizzato et al., 2016). Notably, circRNAs are enriched and stable in whole blood (Memczak et al., 2015), platelets (Alhasan et al., 2016), and exosomes (Li et al., 2015a). These features make circRNAs in human peripheral blood good candidates for diagnostic or prognostic biomarkers of human diseases.
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis (Pai et al., 2016). Although TB incidence, prevalence, and mortality have decreased since 2000, the World Health Organization (WHO) estimated that there were 1.5 million TB-associated deaths and 10.5 million new TB cases worldwide in 2015 (World Health Organization, 2016). In particular, China has the third largest number of TB cases in the world, which is approximately 10% of the world total (World Health Organization, 2016). To achieve better TB management, some novel diagnostic methods, especially a rapid, low-cost, and non-sputum-based test, are required to screen active TB patients at the primary-care level (World Health Organization, 2016). Recently, several studies have developed blood transcriptomic signatures to distinguish active pulmonary TB patients from other pulmonary disease cohorts and healthy controls (Bloom et al., 2013, Qian et al., 2016, Roe et al., 2016, Sweeney et al., 2016) and predicted the risk of developing active TB (Zak et al., 2016). However, the expression of mRNA transcripts in peripheral blood can be obscured by blood collection procedures, which may slow down the process of their clinical applications (Dvinge et al., 2014). As an alternative to blood transcriptional signatures, Liu et al. also developed a method of quantifying the circulating Mycobacterium tuberculosis antigen peptides for active TB diagnosis and treatment monitoring (Liu et al., 2017). Compared with blood transcriptional signatures, the amount of circulating bacterial antigens in blood could be too low to be detected at the early stage of disease (Abbosh et al., 2017). Therefore, new markers in the peripheral blood are needed to facilitate the early diagnosis and treatment of active pulmonary TB.
In this study, we investigated the potential of using circRNAs in peripheral blood mononuclear cells (PBMCs) as biomarkers for pulmonary TB diagnosis. First, we characterized circRNA expression profiles in human PBMCs using ribosomal RNA-depleted (rRNA-depleted) RNA-seq and circRNA expression microarrays. We compared the circRNA repertoire in PBMCs with that in whole blood and other blood components, such as platelets and red blood cells. Under the assumption that the function of a given circRNA could be associated with the known function of its parental gene, we performed a pathway-based analysis to identify the PBMC circRNAs that are dysregulated in TB patients. Compared with host linear transcripts, the circRNAs within several Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) physiological pathways were observed to be disproportionately upregulated in active TB patients. Based on these differentially expressed circRNAs, we developed a PBMC circRNA-based molecular signature that can differentiate active TB patients from healthy controls. Finally, we validated the performance of our circRNA signature in an independent cohort using quantitative real-time PCR (qRT-PCR). Our results suggest the feasibility of using PBMC circRNAs as molecular markers to diagnose active TB. Fig. 1 provides an overview of the experimental design.
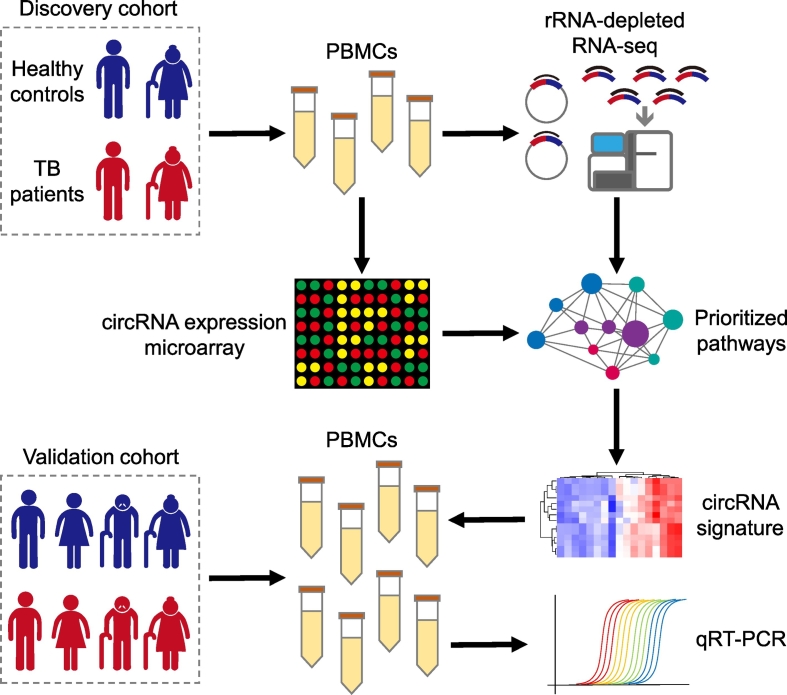
Fig. 1.
The experimental scheme of the study. The discovery cohort was composed of two TB patients and two age- and gender-matched healthy controls: one young male case-control pair and one senior female case-control pair. PBMC circRNA expression in the discovery cohort was profiled by both rRNA-depleted RNA-seq and a circRNA expression microarray. We aggregated circRNAs into pathway-level mechanisms using KEGG pathway definitions. Pathways with significant circRNA dysregulation were prioritized by performing a paired Wilcoxon signed-rank test between paired control and TB samples. A molecular signature was developed based on the dysregulated circRNAs within the prioritized pathways. This circRNA signature was further validated by qRT-PCR in a validation cohort with 11 healthy controls and 10 TB patients.
2. Materials and Methods
2.1. Human Subjects
This study was approved by the Ethics Committee of Bengbu Medical College, with written informed consent obtained from all subjects. This was conformed to standards indicated by the Declaration of Helsinki. The diagnosis of active pulmonary TB was based on established international guidelines (Lewinsohn et al., 2017). Subjects with other concurrent infectious diseases were excluded. All the subjects investigated were of Chinese Han descent. The discovery cohort included two subjects with active pulmonary TB and two age- and gender-matched healthy controls: one young male case-control pair and one senior female case-control pair (Supplementary Table S1). The validation cohort consisted of 10 TB patients and 11 healthy controls, with no significant difference in age and gender between the cases and controls (Supplementary Table S2). The TB patients in the discovery and validation cohorts were recruited from the First Hospital of Huainan City and the Infectious Disease Hospital of Bengbu City, respectively. The healthy controls were recruited from Bengbu Medical College. Several clinical TB status indicators of each individual in the validation cohort, including the number of cavities, diameter of the largest cavities, and sputum smear grade were obtained.
2.2. Cell Purification, RNA Isolation and circRNA Expression Profiling
PBMCs were collected from all subjects in the discovery and validation cohorts. To profile PBMC circRNA expression in the discovery cohort, we constructed ribosomal RNA (rRNA)-depleted RNA-seq libraries using PBMC total RNA from each person in the discovery cohort. The expression of PBMC circRNAs in the discovery cohort was also profiled using the CapitalBio Technology Human CircRNA Array v2 (CapitalBio Technology, Beijing, China). The experimental details of cell purification, RNA isolation, RNA-seq library preparation and sequencing, and circRNA microarray expression profiling were available in Supplementary Methods. Raw sequencing reads were available at the NCBI Short Read Archive (SRA) (Kodama et al., 2012) under the accession number SRP115429, and circRNA microarray data were deposited at the NCBI Gene Expression Omnibus (GEO) (Edgar et al., 2002) under the accession number GSE103188.
2.3. RNA-seq Data of Whole Blood and Other Blood Components
To compare the circRNA repertoire in PBMCs with that in whole blood and other blood components, we collected several RNA-seq datasets from public databases, which included six samples of human whole blood (Memczak et al., 2015), three samples of human blood platelets (Kissopoulou et al., 2013), and one sample of human red blood cells (RBCs) (Alhasan et al., 2016) (Supplementary Table S3). All these samples were sequenced using rRNA-depleted RNA-seq libraries in their original studies (Alhasan et al., 2016, Kissopoulou et al., 2013, Memczak et al., 2015). We downloaded the raw RNA-seq reads of these samples from SRA (Kodama et al., 2012).
2.4. Identification and Expression Quantification of circRNAs From RNA-seq Data
The raw RNA-seq reads were filtered by removing adaptor sequences, contamination, and low-quality reads. To assure the sequencing performance and library quality, we used RNA-SeQC (DeLuca et al., 2012) to assess the data quality of each RNA-seq dataset. For each sample, we identified all putative circRNAs using CIRI (Gao et al., 2015) with default parameter settings. After circRNA identification, we quantified the expression values of all identified circular transcripts and known linear transcripts in the Ensembl (Cunningham et al., 2015) human gene annotation (release 87) using our recent developed tool, Sailfish-cir (Li et al., 2017), with default settings. For each host gene, the transcripts per million (TPM) values of both linear and circular transcripts were recorded. These circRNA identification and quantification procedures were performed for both our PBMC and the public (the other blood component) RNA-seq raw data. The normalized TPM values of circRNAs were used to explore the global differences of circRNA repertoire among human blood components.
2.5. Inferring Differentially Expressed Pathways and circRNAs
To discover PBMC circRNA signature in differentiating TB patients from healthy controls, we identified differentially expressed pathways and circRNAs from human PBMC RNA-seq and microarray data, respectively. First, we aggregated circRNAs into pathway-level mechanisms using KEGG annotations. A paired Wilcoxon signed-rank test was performed to compare the paired circRNA expression profiles restricted to a given KEGG pathway between paired control and TB samples (Gardeux et al., 2014). The pathways with an adjusted P-value < 0.05 after Benjamini and Hochberg correction were deemed differentially expressed. Next, the edgeR (Robinson et al., 2010) and SAM (Tusher et al., 2001) algorithms were used to identify the differentially expressed circRNAs. For the RNA-seq data, only the circRNAs with a reads-per-million value > 5 in at least two samples were included. We further excluded the circRNAs with average junction read counts < 5. For the microarray data, only the circRNAs without absent calls were retained. We limited our analysis to the circRNAs with unique circBase (Glazar et al., 2014) annotation and removed the circRNAs on chromosomes X and Y to avoid the potential confounding sex factor. The circRNAs with a false discovery rate (FDR) < 5% were deemed differentially expressed.
2.6. qRT-PCR Validation
We validated the performance of the dysregulated PBMC circRNAs in differentiating active pulmonary TB patients from healthy controls in an independent validation cohort. qRT-PCRs were performed to quantify the expression of the prioritized PBMC circRNAs. We designed divergent primers for the selected circRNAs using CircInteractome (Dudekula et al., 2016) (Supplementary Table S4). qRT-PCRs were performed on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Massachusetts, USA) using a SYBR Green Real Master Mix with Rox (Tiangen, Beijing, China). The housekeeping gene, β-actin, was used as an internal control. The comparative CT (2− ΔΔCT) method was used to obtain the fold change of circRNA expression levels, and Student's t-test was used to test its statistical significance.
2.7. CircRNA-based TB Index
We applied a scoring method used in our previous study to assign each human subject a TB index (Qian et al., 2016): . Here, I is the circRNA-based TB index; n is the number of circRNAs; ei denotes the expression level of circRNA i; and μi and τi are the mean and standard deviation of the expression of circRNA i across all samples, respectively. A higher TB index implies a higher likelihood of active TB.
3. Results
3.1. Landscape of circRNA Expression in Human PBMCs
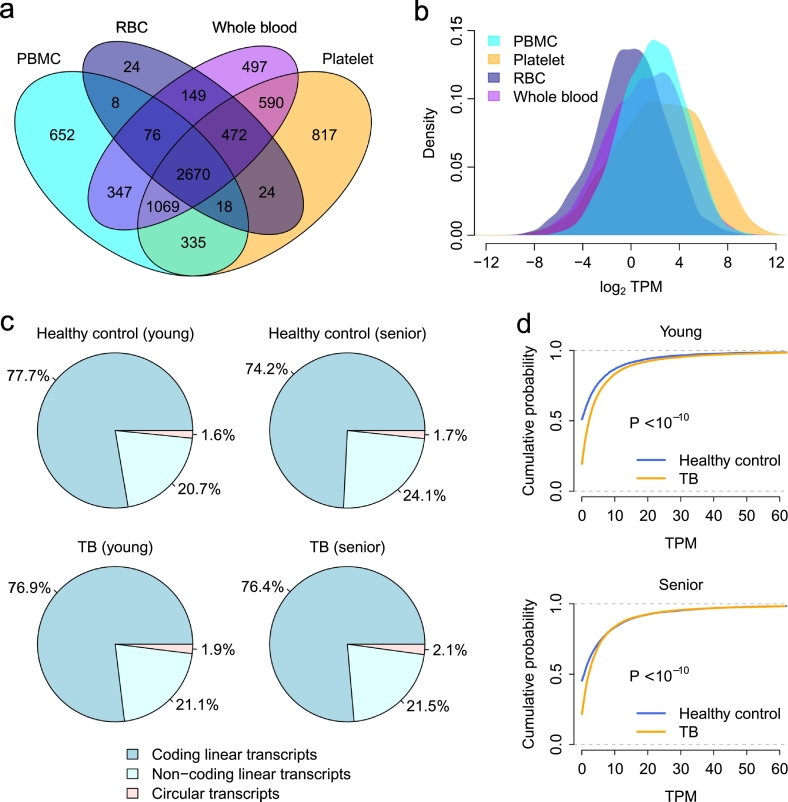
Previous studies have suggested that circRNAs are enriched in human peripheral whole blood (Memczak et al., 2015), platelets (Alhasan et al., 2016), and exosomes (Li et al., 2015a), and circRNAs in whole blood and blood components could be used as biomarker molecules. To explore whether PBMC circRNAs are potential molecular markers as well, we first investigated the landscape of expressed circular transcripts and their expression pattern in human PBMCs. We sequenced four rRNA-depleted RNA-seq libraries of human PBMC samples in the discovery cohort (Supplementary Table S3). The expression of both mRNAs and circRNAs was simultaneously quantified by Sailfish-cir (Li et al., 2017). In total, 5175 parental genes were found to express circular transcripts in these four human PBMC samples, among which the circular transcripts of 131 parental genes were highly expressed with an average TPM > 100.
We also analyzed the rRNA-depleted RNA-seq data of human whole blood, platelets, and RBCs (Supplementary Table S3). The PBMC circRNA repertoire was then compared with the circRNAs identified in whole blood, platelets, and RBCs. We observed that the number of parental genes with circular transcripts was comparable among PBMCs, platelets, and whole blood (Fig. 2a). There were 3739 host genes commonly expressing circular transcripts in PBMCs, platelets, and whole blood (Fig. 2a). The circRNA expression level in PBMCs was significantly higher than that in whole blood and RBCs (t-test: P < 10− 10) but lower than that in platelets (t-test: P < 10− 10) (Fig. 2b). In comparison, human RBCs had the least number of identified circRNAs (Fig. 2a) and the lowest level of circRNA expression (Fig. 2b). These results suggest that circRNAs are widely expressed in human PBMCs. Furthermore, the expression of a substantial number of circular transcripts was high enough to be detected, which ensures that circRNAs in PBMCs, like those in whole blood and platelets, meet the essential requirements as molecular markers.
Fig. 2.
Landscape of PBMC circRNA expression. (a) A Venn diagram of the parental genes with circRNA transcripts among PBMCs, platelets, RBCs, and whole blood. (b) Distribution of log2-transformed TPM values of circular transcripts in PBMCs, platelets, RBCs, and whole blood. (c) The fraction of RNA-seq reads from coding linear, non-coding linear, and circular transcripts. (d) The cumulative distribution of TPM values of circular transcripts. A significantly increased circRNA TPM was observed in the TB patients compared with the healthy controls. The P-values were computed by the Kolmogorov-Smirnov test.
3.2. Pathway-based Analysis of PBMC circRNA Dysregulation in TB Using RNA-seq
Next, we investigated the difference in PBMC circRNA expression between TB patients and healthy controls in the discovery cohort. We observed that the RNA-seq reads fraction of circRNA transcripts in the PBMC transcriptome was consistently increased in both young and senior TB patients compared with healthy controls (Fig. 2c). The circRNA TPM of the TB patients was significantly higher than that of the healthy controls (Kolmogorov-Smirnov test: P < 10− 10) (Fig. 2d).
Given the broad differences in PBMC circRNA expression between TB patients and healthy controls, we performed a pathway-based paired-sample analysis to identify the dysregulated biological processes by pooling circRNAs into host gene pathways. In this analysis, we assumed that the function of a given circRNA could be associated with the known function of its parental gene, as circRNAs can potentially cis-regulate their corresponding host genes (Chen, 2016, Li et al., 2015b, Veno et al., 2015). Briefly, a paired Wilcoxon signed-rank test was applied to compare the paired circRNA expression profiles restricted to a given KEGG pathway between healthy controls and TB patients for the young and senior pairs, respectively (Gardeux et al., 2014). Unlike conventional transcriptomic approaches that have been developed to uncover molecular signatures across a large population of human subjects, this approach enables discovery of dysregulated pathways unique to individual case-control pairs and highlights both the individuality and commonality of a clinical trait (Gardeux et al., 2014).
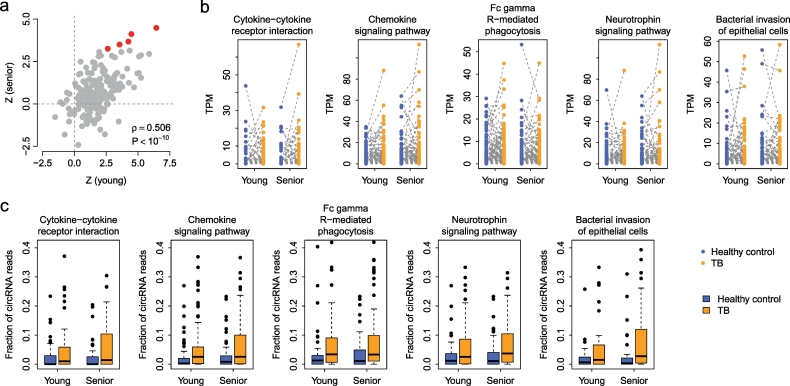
In total, five KEGG pathways were found to be commonly upregulated (adjusted P < 0.05) regarding circRNA expression in both young and senior TB patients (Fig. 3a), including “Cytokine-cytokine receptor interaction”, “Chemokine signaling pathway”, “Fc gamma R-mediated phagocytosis”, “Neurotrophin signaling pathway”, and “Bacterial invasion of epithelial cells” (Fig. 3b and Supplementary Table S5). The Z-score of the Wilcoxon test was significantly and positively correlated between the young and senior TB control pairs (Spearman's rank correlation test: ρ = 0.506 and P < 10− 10) (Fig. 3a), which reflects the commonality in circRNA dysregulation patterns between young and senior TB patients.
Fig. 3.
The KEGG pathways enriched by upregulated circRNAs. A paired Wilcoxon test was applied to identify the KEGG pathways enriched by dysregulated circRNAs in young and senior TB patients. (a) The correlation in Z-scores (computed by a paired Wilcoxon test) between young and senior case-control pairs. Each dot represents one KEGG pathway. The red points stand for the pathways with circular transcripts commonly upregulated in both young and senior TB patients. (b) Paired comparison of circRNA expression in the five prioritized KEGG pathways. (c) Comparison of the fraction of circRNA reads in the five pathways between healthy controls and TB patients.
To understand whether the alteration of circRNA abundance in the five prioritized pathways is simply a function of the host gene dysregulation, we computed the fraction of circRNA reads over the total read count (reads from both circular and linear transcripts) for each host gene. We found that the fraction of circRNA reads was significantly higher in TB patients than in healthy controls in all five pathways (paired Wilcoxon test: P < 0.05) (Fig. 3c), which suggests that circRNA expression is somewhat independent of host linear transcripts and, at least to a certain extent, the circRNA dysregulation in TB PBMCs is not a secondary consequence of the host gene expression.
3.3. Technical Validation of the Prioritized Pathways Using a circRNA Microarray
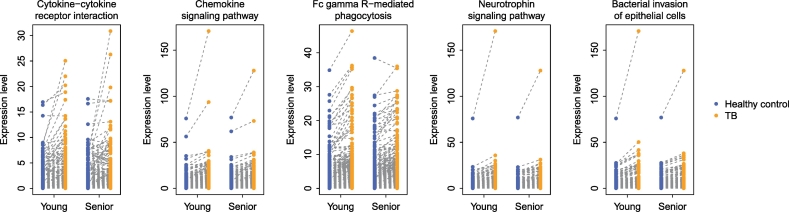
To verify that the above findings were not caused by potential technical bias introduced by RNA-seq procedures (Lahens et al., 2014, Zheng et al., 2011), we further performed circRNA microarray analysis of the same PBMC RNA samples treated by RNase R (Jeck et al., 2013). The CapitalBio Technology Human CircRNA Array v2 (4 × 180K) was used to quantify circRNA expression. As indicated in Fig. 4, the microarray data perfectly mirrored our findings from the RNA-seq analysis. The five prioritized pathways were consistently upregulated with respect to circRNA expression (paired Wilcoxon test: P < 0.05) (Fig. 4). These results confirmed that the dysregulated pathways we prioritized from TB PBMC circRNAs were independent of the circRNA detection and quantification methods.
Fig. 4.
Technical validation of the five prioritized KEGG pathways using a circRNA expression microarray. Total RNA was treated with RNase R to remove linear transcripts and then subject to microarray hybridization.
3.4. Classification Power of PBMC circRNA Expression in TB
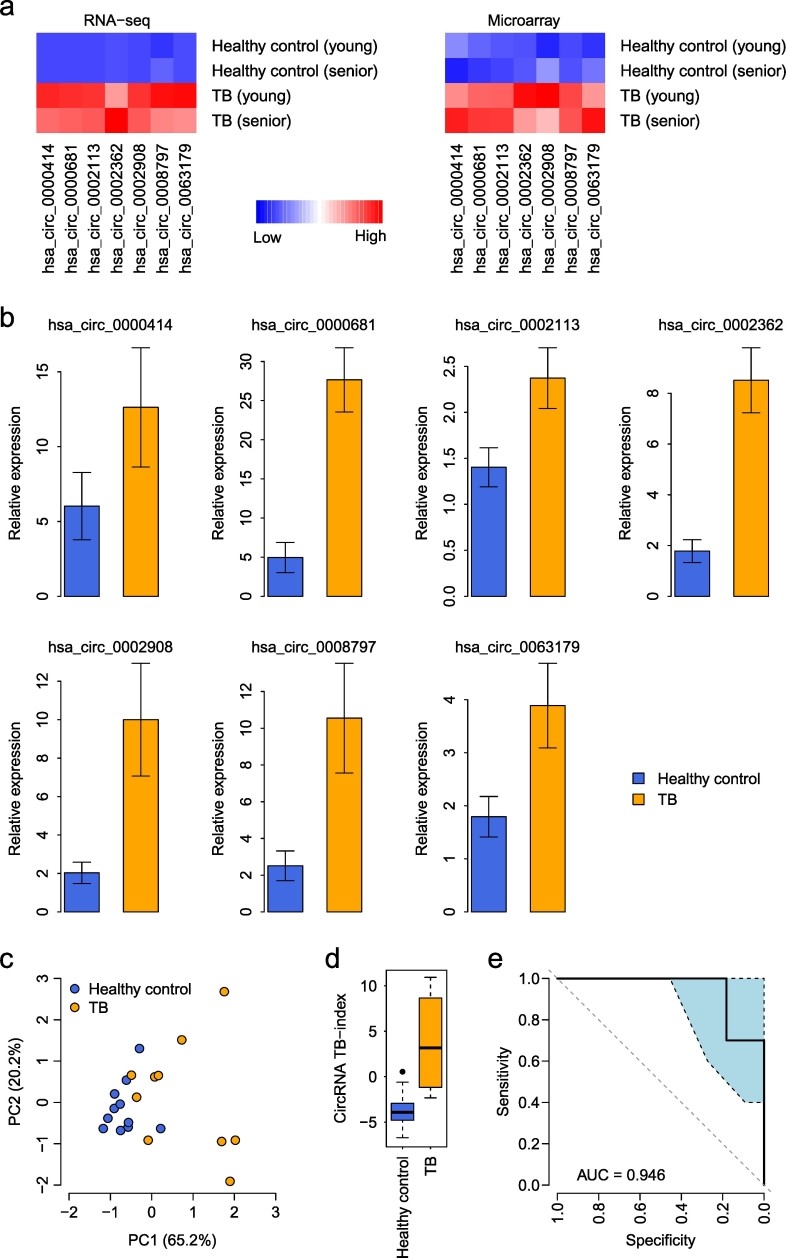
Finally, we explored the predictive power of PBMC circRNA expression in differentiating TB patients from a healthy population. We further explored the RNA-seq and microarray data to compare the expression of individual circRNAs within the five prioritized pathways between TB patients and healthy controls. In total, seven circRNAs with unique circBase annotations were identified to be commonly upregulated in TB PBMCs in both the RNA-seq and microarray data (see Methods for details), which was designated the 7-circRNA signature (Table 1 and Fig. 5a).
Table 1.
The 7-circRNA signature.
| circBase ID | Genomic position | Strand | Length | Gene | Associated KEGG pathway |
|---|---|---|---|---|---|
| hsa_circ_0000414 | chr12:66597490-66605377 | + | 455 | IRAK3 | Neurotrophin signaling pathway |
| hsa_circ_0000681 | chr16:23999828-24046868 | + | 324 | PRKCB | Chemokine signaling pathway; Fc gamma R-mediated phagocytosis |
| hsa_circ_0002113 | chr21:34793786-34805178 | + | 673 | IFNGR2 | Cytokine-cytokine receptor interaction |
| hsa_circ_0002362 | chr2:9458652-9468040 | + | 341 | ASAP2 | Fc gamma R-mediated phagocytosis |
| hsa_circ_0002908 | chr8:141840570-141874498 | − | 286 | PTK2 | Chemokine signaling pathway; Bacterial invasion of epithelial cells |
| hsa_circ_0008797 | chr3:119582265-119624699 | − | 420 | GSK3B | Chemokine signaling pathway; Neurotrophin signaling pathway |
| hsa_circ_0063179 | chr22:37328806-37330036 | + | 303 | CSF2RB | Cytokine-cytokine receptor interaction |
Fig. 5.
The 7-circRNA signature. (a) Expression heatmap of the circRNAs within the 7-circRNA signature in the discovery cohort. The expression measured by both RNA-seq and circRNA microarray is displayed. (b) Comparison of the expression levels of the 7-circRNA signature between the healthy controls and TB patients in the validation cohort. The error bars represent the standard error of the mean. (c) Principal component analysis of the 7-circRNA signature. PC1: the first principal component; PC2: the second principal component. (d) Comparison of the circRNA-based TB index between the healthy controls and TB patients in the validation cohort. (e) The ROC curve of the TB index in distinguishing TB patients from healthy controls. The light blue area indicates the confidence interval of the ROC curve.
The 7-circRNA signature was then assessed in the validation cohort with 11 healthy controls and 10 TB patients. qRT-PCR was used to quantify the expression of each individual circRNA. Elevated expression was observed for all the prioritized circRNAs (t-test: P < 0.05), except for hsa_circ_0000414 (t-test: P = 0.171) (Fig. 5b). Principal component analysis indicated that the 7-circRNA signature differentiated TB patients from healthy controls in the validation cohorts (Fig. 5c). To systematically evaluate the classification power of the 7-circRNA signature, a circRNA-based TB index was assigned to each subject (see Methods for details). The TB index is a linear combination of the 7-circRNA expression values, and a higher TB index implies a higher likelihood of TB. We found that the 7-circRNA-based TB index was significantly higher in TB patients than in healthy controls (t-test: P = 0.001) (Fig. 5d). The area under the receiver operating characteristic (ROC) curve (AUC) was 0.946 (Fig. 5e). We further tested whether the predictive power of the circRNA-based TB index was dependent on standard TB clinical indicators, including number of cavities, diameter of the largest cavities, and sputum smear grade. Multivariate logistic regression indicated that the 7-circRNA-based TB index was the most significant predictor of TB status (P = 0.038) (Supplementary Table S6). This suggests that circRNA expression information adds independent value to TB diagnosis. We also investigated the correlation between the 7-circRNA-based TB index and the above clinical indicators. However, no significant correlation was observed (Supplementary Table S7), which may be partly due to the limited sample size in the validation cohort.
4. Discussion
In this study, we explored the potential of using human PBMC circRNAs as the molecular markers to diagnose active pulmonary TB. Due to the remarkable stability of circRNAs (Enuka et al., 2016), several groups have already investigated circRNA expression in easily accessible human body fluids to determine the potential of circRNAs as biomarkers (Alhasan et al., 2016, Bahn et al., 2015, Li et al., 2015a, Memczak et al., 2015). For example, Memczak et al. reported the detection of thousands of circRNAs and high expression of hundreds of circRNAs in human peripheral whole blood (Memczak et al., 2015). Some comprehensive analysis further revealed stable and enriched circRNAs in some human blood components, including erythrocytes (Alhasan et al., 2016), platelets (Alhasan et al., 2016), and exosomes (Li et al., 2015a). As an alternative to using human peripheral blood, Bahn et al. identified > 400 circRNAs in cell-free saliva samples (Bahn et al., 2015). Unlike the above studies, we investigated the circRNA repertoire in a different blood component, human PBMCs. PBMCs contain any peripheral blood cell with a round nucleus, including monocytes and lymphocytes. In our results, we showed wide and abundant circRNA expression in human PBMCs, which is comparable to the circRNA landscapes in human whole blood, RBCs, and platelets (Fig. 2A and B). This result complements previous observations by Alhasan et al. (2016), who found that circular transcripts are enriched in human platelets, with a 17- to 188-fold change relative to nucleated tissues, which is a common signature of anucleate cells (Alhasan et al., 2016). Our study suggests that, similar to anucleate cells, the magnitude of PBMC circRNA expression may also be higher than that of nucleated tissues and other nucleate cells. This is potentially an exciting feature of human PBMCs, as it implies that circRNAs in human PBMCs may be used as liquid biomarker molecules for human diseases.
Compared with previous blood circRNA studies (Alhasan et al., 2016, Li et al., 2015a, Memczak et al., 2015), we pushed several steps forward in investigating the potential of using blood circRNAs as diagnostic markers of human diseases. In addition to the exploration of the circRNA repertoire in human PBMCs, we investigated several key questions in biomarker development, including the differential expression of circRNAs between cases and controls and the development and validation of a circRNA signature. We showed that the magnitude of PBMC circRNAs in TB patients was significantly higher than that of the paired healthy controls (Fig. 2C and D). A KEGG pathway-based paired-sample analysis identified several dysregulated biological processes in human PBMCs, including “Cytokine-cytokine receptor interaction”, “Chemokine signaling pathway”, “Fc gamma R-mediated phagocytosis”, “Neurotrophin signaling pathway”, and “Bacterial invasion of epithelial cells” (Fig. 3b and Supplementary Table S5). Most of these dysregulated pathways are relevant to the occurrence and development of TB. For example, cytokines, chemokines, and their receptors are critical factors in achieving an optimal balance between innate and adaptive immune cells, which can determine the progression or outcome of Mycobacterium tuberculosis infection (Dorhoi and Kaufmann, 2016, Monin and Khader, 2014), while Fc gamma R may play a role in Fc-mediated Ab effector functions in pathogen clearance (Lu et al., 2016). Based on these dysregulated pathways, we prioritized seven differentially expressed PBMC circRNAs in TB patients (Fig. 5a), yielding a 7-circRNA TB molecular signature. We further validated the predictive power of the 7-circRNA signature to differentiate TB patients from healthy controls in an independent cohort (Fig. 5b). Our results strongly suggest that PBMC circRNAs are promising biomarkers to differentiate active TB patients from healthy controls (Fig. 5c–e). However, more thorough studies are needed to test the performance of our 7-circRNA signature as a TB diagnostic biomarker. Due to the limited sample size of the discovery cohort, we used a pathway-based algorithm to control false discovery rate of the prioritized circRNAs. This method potentially overlooks the circRNAs without pathway annotation. In future study, unbiased screening of circRNAs at the whole transcriptomic level in a large cohort will be highly helpful. Moreover, some other diseases with similar clinical phenotypes, such as lung cancers, pneumonias, and pulmonary sarcoidosis, may confound TB diagnosis in clinical application. In future studies, these diseases should be considered to justify the signature's diagnostic specificity when developing TB-specific biomarkers.
Although several studies have developed transcriptomic signatures to diagnose active pulmonary TB (Bloom et al., 2013, Qian et al., 2016, Roe et al., 2016, Sweeney et al., 2016) or predict the risk of developing active TB (Zak et al., 2016), a PBMC-based circRNA signature is still meaningful with practical significance in TB management. In previous studies, mRNA transcripts in peripheral blood were used as marker molecules (Bloom et al., 2013, Qian et al., 2016, Roe et al., 2016, Sweeney et al., 2016). However, mRNA expression-based gene signatures do not always deliver accurate information with reproducible and robust performance, which is especially important when translating the signatures into clinic diagnostics (Byron et al., 2016). This is even more obvious when using peripheral blood as the sample source, as standard blood collection procedures cause rapid changes in the mRNA profiles of hematopoietic cells (Dvinge et al., 2014). Given the higher stability of circRNAs (Enuka et al., 2016), circRNAs are ideal candidate molecules to overcome this shortcoming. Many circRNA biomarkers have been introduced to manage human cancers (Meng et al., 2017). However, most of these circRNA biomarkers were identified from tissue samples. Because tissue samples are impossible to collect in clinical practice of infectious disease management, such as TB, a tissue-based circRNA biomarker is not applicable in TB diagnosis. In a recent study, a peripheral blood circRNA, hsa_circ_0124644, was proposed to diagnose coronary artery disease (Zhao et al., 2017). However, gene expression measurements in human peripheral whole blood are more variable among individuals than several blood components, including PBMCs, lymphoblastoid cell lines, and CD19- and CD20-specific B-cell subsets (Min et al., 2010). Because monocytes (Lastrucci et al., 2015) and lymphocytes (Jasenosky et al., 2015) in human PBMCs are both relevant to TB, a PBMC circRNA signature is able to directly reflect the host responses to infection of Mycobacterium tuberculosis. Compared with circRNAs in exosomes or platelets, the TB signature based on PBMC circRNAs should be much stronger and more easily detected. In addition, PBMC circRNA-based TB index may add additional values to existing clinical tests in TB diagnosis, such as chest X-ray and sputum smear (Supplementary Table S6). Given the advantage of high accuracy, easy sample acquisition and short turnaround time, PBMC circRNA expression signature has the potential to complement current clinical settings in diagnosing active TB and monitoring TB treatment outcomes.
In conclusion, we suggested that PBMC circRNAs are potentially reliable marker molecules to diagnose active TB, which may have translational implications in TB management.
Funding Sources
This work was supported by grants from International Science and Technology Cooperation Project-Key Research and Development Program of Anhui Province (1604b0602026 to ZQ) and from National Natural Science Foundation of China (61571109 to WG). The funders had no role in study design, data collection, data analysis, interpretation and writing of the report.
Conflicts of Interest
The authors declare no competing financial interests.
Author Contributions
WG and TZ conceived this study. ZQ, HL, ML, JS, NL, YZ, XZ, JL, XX, YB, QG, EK, HT, TW, XW, TZ, and WG performed the analysis. ZQ, ZW, TZ, and WG interpreted the results. WG, TZ, ZQ, and HL wrote the manuscript. All authors contributed to the final version of the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.12.007.
Contributor Information
Tong Zhou, Email: tongz@med.unr.edu.
Wanjun Gu, Email: wanjungu@seu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- Abbosh C., Birkbak N.J., Wilson G.A., Jamal-Hanjani M., Constantin T., Salari R., Le Quesne J., Moore D.A., Veeriah S., Rosenthal R. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhasan A.A., Izuogu O.G., Al-Balool H.H., Steyn J.S., Evans A., Colzani M., Ghevaert C., Mountford J.C., Marenah L., Elliott D.J. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016;127:e1–e11. doi: 10.1182/blood-2015-06-649434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S.P., Wang P.L., Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom C.I., Graham C.M., Berry M.P., Rozakeas F., Redford P.S., Wang Y., Xu Z., Wilkinson K.A., Wilkinson R.J., Kendrick Y. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzato A., Gaffo E., te Kronnie G., Bortoluzzi S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016;6:e483. doi: 10.1038/bcj.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron S.A., Keuren-Jensen K.R., Engelthaler D.M., Carpten J.D., Craig D.W. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet. 2016;17:257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A., Kaufmann S.H. Pathology and immune reactivity: understanding multidimensionality in pulmonary tuberculosis. Semin. Immunopathol. 2016;38:153–166. doi: 10.1007/s00281-015-0531-3. [DOI] [PubMed] [Google Scholar]
- Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H., Ries R.E., Ilagan J.O., Stirewalt D.L., Meshinchi S., Bradley R.K. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16802–16807. doi: 10.1073/pnas.1413374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardeux V., Achour I., Li J., Maienschein-Cline M., Li H., Pesce L., Parinandi G., Bahroos N., Winn R., Foster I. ‘N-of-1-pathways’ unveils personal deregulated mechanisms from a single pair of RNA-Seq samples: towards precision medicine. J. Am. Med. Inform. Assoc. 2014;21:1015–1025. doi: 10.1136/amiajnl-2013-002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Jasenosky L.D., Scriba T.J., Hanekom W.A., Goldfeld A.E. T cells and adaptive immunity to Mycobacterium tuberculosis in humans. Immunol. Rev. 2015;264:74–87. doi: 10.1111/imr.12274. [DOI] [PubMed] [Google Scholar]
- Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissopoulou A., Jonasson J., Lindahl T.L., Osman A. Next generation sequencing analysis of human platelet PolyA + mRNAs and rRNA-depleted total RNA. PLoS One. 2013;8:e81809. doi: 10.1371/journal.pone.0081809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Shumway M., Leinonen R., International Nucleotide Sequence Database, C The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., Shamsuzzama, Haque R., Baghel T., Nazir A. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol. Neurobiol. 2017;54:7224–7234. doi: 10.1007/s12035-016-0213-8. [DOI] [PubMed] [Google Scholar]
- Lahens N.F., Kavakli I.H., Zhang R., Hayer K., Black M.B., Dueck H., Pizarro A., Kim J., Irizarry R., Thomas R.S. IVT-seq reveals extreme bias in RNA sequencing. Genome Biol. 2014;15:R86. doi: 10.1186/gb-2014-15-6-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastrucci C., Bénard A., Balboa L., Pingris K., Souriant S., Poincloux R., Saati T., Rasolofo V., González-Montaner P., Inwentarz S. Tuberculosis is associated with expansion of a motile, permissive and immunomodulatory CD16 + monocyte population via the IL-10/STAT3 axis. Cell Res. 2015;25:1333–1351. doi: 10.1038/cr.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn D.M., Leonard M.K., LoBue P.A., Cohn D.L., Daley C.L., Desmond E., Keane J., Lewinsohn D.A., Loeffler A.M., Mazurek G.H. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention clinical practice guidelines: diagnosis of tuberculosis in adults and children. Clin. Infect. Dis. 2017;64:e1–e33. doi: 10.1093/cid/ciw694. [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Li M., Xie X., Zhou J., Sheng M., Yin X., Ko E.A., Zhou T., Gu W. Quantifying circular RNA expression from RNA-seq data using model-based framework. Bioinformatics. 2017;33:2131–2139. doi: 10.1093/bioinformatics/btx129. [DOI] [PubMed] [Google Scholar]
- Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhao Z., Fan J., Lyon C.J., Wu H.-J., Nedelkov D., Zelazny A.M., Olivier K.N., Cazares L.H., Holland S.M. Quantification of circulating Mycobacterium tuberculosis antigen peptides allows rapid diagnosis of active disease and treatment monitoring. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3969–3974. doi: 10.1073/pnas.1621360114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.L., Chung A.W., Rosebrock T.R., Ghebremichael M., Yu W.H., Grace P.S., Schoen M.K., Tafesse F., Martin C., Leung V. A functional role for antibodies in tuberculosis. Cell. 2016;167:433–443. doi: 10.1016/j.cell.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.L., Barrett A., Watts T., Pettersson F.H., Lockstone H.E., Lindgren C.M., Taylor J.M., Allen M., Zondervan K.T., McCarthy M.I. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics. 2010;11:96. doi: 10.1186/1471-2164-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monin L., Khader S.A. Chemokines in tuberculosis: the good, the bad and the ugly. Semin. Immunol. 2014;26:552–558. doi: 10.1016/j.smim.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai M., Behr M.A., Dowdy D., Dheda K., Divangahi M., Boehme C.C., Ginsberg A., Swaminathan S., Spigelman M., Getahun H. Tuberculosis. Nat. Rev. Dis. Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of CircRNAs. Mol. Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Lv J., Kelly G.T., Wang H., Zhang X., Gu W., Yin X., Wang T., Zhou T. Expression of nuclear factor, erythroid 2-like 2-mediated genes differentiates tuberculosis. Tuberculosis (Edinburgh, Scotland) 2016;99:56–62. doi: 10.1016/j.tube.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H., Circular R.N.A. A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe J.K., Thomas N., Gil E., Best K., Tsaliki E., Morris-Jones S., Stafford S., Simpson N., Witt K.D., Chain B. Blood transcriptomic diagnosis of pulmonary and extrapulmonary tuberculosis. JCI Insight. 2016;1:e87238. doi: 10.1172/jci.insight.87238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:1–16. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T.E., Braviak L., Tato C.M., Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir. Med. 2016;4:213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veno M.T., Hansen T.B., Veno S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q., Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Wang P.L., Bao Y., Yee M.-C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J., Circular R.N.A. Is. Expressed across the eukaryotic tree of life. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . WHO, 2016; 2016. Global T uberculosis Report 2016. [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L., Wang Y. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak D.E., Penn-Nicholson A., Scriba T.J., Thompson E., Suliman S., Amon L.M., Mahomed H., Erasmus M., Whatney W., Hussey G.D. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-O., Wang H.-B., Zhang Y., Lu X., Chen L.-L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L., Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Chung L.M., Zhao H. Bias detection and correction in RNA-sequencing data. BMC Bioinform. 2011;12:290. doi: 10.1186/1471-2105-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material