Abstract
The aim of this study, comprising 88 suicide attempters, was to identify hypothalamic-pituitary-adrenal (HPA) -axis coupled CpG-sites showing methylation shifts linked to severity of the suicide attempt. Candidate methylation loci were further investigated as risk loci for a general psychiatric risk score in two cohorts of adolescents (cohort 1 and 2). The genome-wide methylation pattern was measured in whole blood using the Illumina Infinium Methylation EPIC BeadChip. Subjects were stratified into high-risk and low-risk groups based on the severity of the suicidal behavior. We included CpG sites located within 2000 basepairs away from transcriptional start site of the following HPA-axis coupled genes: corticotropin releasing hormone (CRH), corticotropin releasing hormone binding protein (CRHBP), corticotropin releasing hormone receptor 1 (CRHR1), corticotropin releasing hormone receptor 2 (CRHR2), FK506-binding protein 51 (FKBP5) and the glucocorticoid receptor (NR3C1). The methylation state of two corticotropin releasing hormone (CRH)-associated CpG sites were significantly hypomethylated in the high-risk group of suicide attempters (n = 31) (cg19035496 and cg23409074) (p < 0.001). Adolescent cohort 1 and 2 consisted of 129 and 93 subjects, respectively, and were stratified by the in silico generated DAWBA measurements of a general psychiatric risk score into high-risk group (>~50% risk) or controls. In adolescent cohort 2, cg19035496 was hypermethylated in subjects with a high general psychiatric risk score. Our results show epigenetic changes in the CRH gene related to severity of suicide attempt in adults and a general psychiatric risk score in adolescents.
Keywords: Suicide, Suicide attempt, HPA axis, CRH gene, Epigenetics, Methylation, Adolescent depression
Highlights
-
•
Two CRH-associated CpG sites were significantly hypomethylated in the high-risk group of suicide attempters.
-
•
In adolescent cohort, cg19035496 was hypermethylated in subjects with a high general psychiatric risk score.
-
•
Epigenetic modulatory effects on the HPA axis dysregulation are associated with psychiatric illness and suicidal behavior.
In this study, comprising 88 suicide attempters, we aimed to identify epigenetic changes in stress system linked to severity of the suicide attempt. In the next step, we investigated if the same epigenetic changes could be detected in adolescents with high risk for psychiatric illness. The methylation pattern was measured in blood and subjects were stratified into high-risk and low-risk groups based on the severity of the suicidal behavior. One corticotropin releasing hormone (CRH)-a key regulator of stress system-associated CpG site showed less methylation in the high-risk group and was hypermethylated in adolescents with a high general psychiatric risk score. Epigenetic changes in the CRH gene were related to severity of suicide attempt in adults and a general psychiatric risk score in adolescents.
1. Introduction
Suicide is a major preventable public health concern and suicide attempt is associated with a considerable reduction in life expectancy compared to general population (Jokinen et al., 2017b). Suicidal behaviors have multifactorial aetiology including both distal risk factors, e.g. early life adversity (ELA), which through epigenetic modifications may alter the development and lead to dysregulation of emotional and behavioral traits, and proximal risk factors, like serious life events and substance abuse (Turecki, 2014). Suicide is a complex, heterogeneous phenotype, where the severity of suicidal behavior has been proposed to include both the high intent to die and the choice of lethal suicide attempt method (Silverman et al., 2007). Both high intent to die (Stefansson et al., 2010) and a choice of a violent suicide attempt method (Stenbacka and Jokinen, 2015) evince a higher risk of completed suicide in follow-up studies of suicide attempters and serious suicide attempts may have distinct neurobiological underpinnings.
The cortisol stress response is one of the most promising candidate suicide endophenotypes (Mann et al., 2009). Dysregulation in form of hyperactivity of the hypothalamic–pituitary–adrenal (HPA) axis measured with the dexamethasone test (DST) (non-suppression) has been associated with increased risk of suicide in patients with mood disorders in prospective studies (Coryell and Schlesser, 2001), for review (Mann et al., 2006), especially in patients with a clinical history of attempted suicide (Jokinen et al., 2007). Further, higher cortisol levels in cerebrospinal fluid (CSF) and plasma have been reported in suicide attempters compared to healthy volunteers (Chatzittofis et al., 2013) as well as increased corticotropin-releasing hormone (CRH) levels in the CSF in depressed suicide victims (Arato et al., 1989). Suicide vulnerability may be due to epigenetic alterations in molecular pathways involved in HPA axis function. DNA methylation changes at the NR3C1 gene, encoding the glucocorticoid receptor, are higher than normal in the hippocampus of suicide victims exposed to early life adversity (McGowan et al., 2009). Moreover, spindle and kinetochore associated complex subunit 2 (SKA2), which modulates cortisol suppression, has been proposed as a novel genetic and epigenetic target involved in the pathophysiology of suicidal behaviors (Guintivano et al., 2014, Pandey et al., 2016). Corticotropin releasing hormone binding protein (CRHBP), FK506-binding protein 51 (FKBP5) and glucocorticoid receptor gene (NR3C1) promoters were significantly hypermethylated in depressed patients with largest effects in patients with severe suicidal ideation (Roy et al., 2017). Even genetic studies point to significant associations between FKBP5 and CRHR1 genes and a high rate of attempted suicide (De la Cruz-Cano, 2017) and some genetic studies have reported that CRHBP and FKBP5 genes interact with childhood trauma to increase the risk for suicidal behavior (Roy et al., 2012).
The aim of this study was to detect HPA-axis coupled CpG-sites, which showed alterations at the epigenetic profile in relationship with severity of suicidal behavior. Since significant proportion of psychopathological changes has early onset in childhood and adolescence (Teicher and Samson, 2013), the candidate methylation loci were further investigated and confirmed as risk loci for a general psychiatric risk score in independent population-based cohorts of adolescents.
2. Materials and Methods
2.1. Ethics
The study protocols were approved by the Regional Ethical Review Board in Stockholm (Dnrs: 00-194, 2015/1454-32) and the participants gave their written informed consent to the study. Concerning adolescent cohorts and expression data set, the studies were approved by the Regional Ethical Review Board in Uppsala, and all participants gave their written informed consent.
2.2. Patients
Patients (age of 18 years or older) followed-up clinically after a recent suicide attempt at the Suicide Prevention Clinic at the Karolinska University Hospital were invited to participate in the research study with focus on neurobiological and psychological risk factors for suicidal behavior. Individuals with schizophrenia spectrum psychosis, intravenous drug abuse, mental retardation or dementia were excluded from the study. A suicide attempt was defined as a self-destructive act with some degree of intent to die.
During the study period (between years 2000–2005), a total of 258 patients (89 men and 169 women) from the catchment area came into contact with the Suicide Prevention Clinic after attempted suicide. Among them, 61 patients met the exclusion criteria above, 50 patients declined to participate in the study. Due to other reasons, e.g. initial refusal to have a clinical follow-up, or moving to another part of the country, 47 patients were not proposed to participate in the study A total of 100 patients with a recent suicide attempt (33 men and 67 women) participated in the study, yet only 88 of them had DNA samples.
2.3. Cohorts of Adolescents
In the next step, we made use of two previously published cohorts of adolescents termed adolescent cohort 1 and adolescent cohort 2, respectively. For adolescent cohort 1, a total of 129 adolescent volunteers aged 14 to 16 years, were randomly selected from public school in Uppsala County between November 2012 and January 2013. The study had the aim to investigate risk factors for psychiatric illness. Adolescent cohort 2 included 93 adolescents aged 14–17, which were recruited between 2013 and 2014. More details on the samples and preprocessing of the methylation specimens have been previously published (Ciuculete et al., 2017).
2.4. Assessments of Severity of Suicidal Behavior
Patients were stratified into two groups (high-risk or low-risk) based on the severity of the suicide attempt using following criteria: violent suicide attempt method or a high score in the Freeman scale or information of later completed suicide. The classification of violent and non-violent suicide attempts was performed using a dichotomization with relevance concerning biological differences. Self-poisoning and cuts in one wrist were considered as non-violent suicide attempts, while all other, i.e. attempted drowning, shooting, gassing or hanging were considered to be violent (Traskman et al., 1981).
The Freeman scale consists of two parts, Reversibility and Interruption Probability, and showed very good discriminating validity, when it was applied to a large sample of suicide attempt and suicide death cases (Freeman et al., 1974). These two scales capture important aspects of the suicide intent. Interrater reliability of reversibility of method rating was 0.97 and 0.80 for interruption probability (Freeman et al., 1974). The Reversibility scale takes into account the quantity and type of drug taken and the degree of self-injury inflicted. A high score indicates low reversibility of the suicide attempt method, i.e. a more serious suicide attempt with high risk of death, and includes shooting or hanging, while a low score indicates that the method is reversible and suicide death less likely. The second part of the Freeman scales measures the probability of interruption by others, where high scores indicate that interruption is very unlikely. Both categories are coded on a 1–5 graded scale and the range of scores on total Freeman scale is between 2 and 10 (Freeman et al., 1974). A cut-off > 6 was applied to define a serious suicide attempt.
All patients were linked to national Cause of Death register by their own unique identification number. Four patients completed suicide before January 2011. Three patients had died by hanging and one patient with substance intoxication.
2.5. Phenotype Assessment in the Cohorts of Adolescents
The risk for psychiatric diseases was assessed by performing the Development and Well-Being Assessment (DAWBA) web-based interview designed for individuals in the age range 5–17 years to generate Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) and International Classification of Diseases (ICD)-10-based psychiatric diagnoses. DAWBA consists of two versions of individual standardized questionnaires, administrated to adolescents and their parents. Computer generated diagnostic predictions were used in the present study. The average of the ‘probability bands’ was computer-assisted generated referring to several diagnoses such as anxiety disorders, depression, post-traumatic stress disorder, autism, separation anxiety disorder and obsessive compulsive disorder. DAWBA diagnoses are given in the range of < 0.1% to over 70% probability that the individual could experience one of the mentioned DSM-IV or ICD-10-based diagnosis.
2.6. Blood Sample Collection and Methylation Profiling
Blood samples from non-fasting participants were collected in the morning according to standard procedures. Genomic DNA was extracted from 88 samples using the phenol-chloroform method (Sambrook et al., 1989). Subsequently, the EZ DNA Methylation – GoldTM kit (ZymoResearch, USA) was used for bisulfite conversion. Bisulfite converted DNA was thereafter hybridized to the Illumina Infinium Methylation EPIC BeadChip, representing the methylation state of over 850K CpG sites. The array was imaged using the Illumina iScan system (Illumina, San Diego, CA, USA) and the methylation state percent of each CpG site was quantified for the entire study group.
2.7. Data Processing
Preprocessing of the methylation data was performed by background correction, adjustment of probe type differences, removal of batch effects and probe exclusion. Subsequently, the global DNA methylation pattern was adjusted for white blood cell-type heterogeneity. Principal component analysis (PCA) was used to identify sample outliers in the methylation data. Methylation preprocessing steps were performed using the minfi (Aryee et al., 2014), watermelon (Schalkwyk, 2013), sva (Leek et al., 2012), ChAMP (Morris et al., 2013) and FactoMineR (Lê et al., 2008) packages of the Bioconductor project operable in R software, version 3.3.0.
2.8. Background Correction, Adjustment of Type I and Type II Probes, Removal of Batch Effects and Probe Exclusion
Methylation idat files were first loaded into the R environment using the ‘read.metharray.exp’ function of the minfi package (v 1.18.2). Thereafter, the efficient NOOB method was selected to correct for background artifacts (Triche Jr. et al., 2013). Probes on the Illumina Methylation EPIC BeadChip array come in two different designs which differ in dynamic range and distribution of the DNA methylation pattern. We used the Beta Mixture Quantile Dilation (BMIQ) function of the wateRmelon package to adjust the methylation data for these probe type differences (Teschendorff et al., 2012). In addition, the use of different analysis plates could result in undesired batch effects and we used the ‘ComBat’ function of the sva package to correct for this potential bias (Johnson et al., 2007). A total of 2483 probes were also filtered out as 75% or more of the samples exhibited a detection p-value > 0.01. Moreover, methylation levels of CpG sites annotated to known SNP loci could be affected by single nucleotide polymorphisms (SNP:s) (Chen et al., 2013) and probes located on sex chromosomes have been shown to be more difficult to accurately normalize (Fortin et al., 2014). 197,719 CpG sites were thus subsequently excluded as they were located on sex determining chromosomes or covering known SNP loci, according to the Illumina annotation. In addition, probes with SNPs at target CpG or within probe and cross-reactive probes were excluded based on Chen et al.'s annotation (n = 22, 577) (Chen et al., 2013).
2.9. Correction for White Blood Cell Type Heterogeneity
DNA methylation measured in whole blood is composed of different cell populations. Rask-Andersen et al. showed that changes in leukocyte fractions could introduce significant variability in the DNA methylation pattern, an effect that could bias downstream analyses. It is thus important to adjust the global DNA methylation pattern for white blood cell type heterogeneity (Rask-Andersen et al., 2016). We implemented a minfi-based statistical procedure of the Houseman algorithm (Houseman et al., 2012) to adjusted the global DNA methylation data for white blood cell type heterogeneity, which uses raw intensity files to calculate the relative proportion of CD4 + and CD8 + T cells, monocytes (Mono), granulocytes (Gran), B cells (Bcell) and natural killer (NK) cells.
2.10. Criteria of Sample Exclusion
To investigate the global DNA methylation pattern regarding potential sample outliers, the ‘PCA’ function of the FactoMineR package was used (31). A total of 31,493 probes were further studied and included in the covariance matrix based on a threshold of 0.2 and a 95% reference range, as performed by Voisin et al. (2015). The first principal component explained 15.34% of the total variance and successively studied vectors did not add significantly to the total variance. Outliers were identified by visual inspection of the graphical display of the first principal component, where no samples have been excluded from further analysis.
2.11. CpG Site Annotation and Selection of HPA-Axis Coupled Probes
More than 90% of the probes on the Illumina 450K methylation Beadchip are also present on the Illumina EPIC BeadChip (Pidsley et al., 2016). Therefore we used the expanded annotation produced by Price et al., originally designed for the 450K array, to define, for each CpG site, the distance to the closest transcriptional start site (TSS) and the associated gene (Price et al., 2013). As such, only CpG-sites present on both the EPIC array and Illumina 450K methylation beadchip were considered for further analysis. In addition, we only considered CpG sites located within 2000 base pairs (bp) up and downstream of the TSS. Wagner et al. demonstrated that DNA methylation and gene expression are closely related within this region (Wagner et al., 2014).
Based on the hypothesis that HPA-axis is associated with severity of suicidal behavior, we considered the following HPA-axis coupled genes: Corticotropin releasing hormone (CRH), corticotropin releasing hormone binding protein (CRHBP), corticotropin releasing hormone receptor 1 and 2 (CRHR1), (CRHR2), FK506-binding protein 51 (FKBP5) and the glucocorticoid receptor (NR3C1). After the preprocessing steps outlined above, all CpG sites annotated to any of the aforementioned genes were included in the study, resulting in 72 CpG sites investigated in the subsequent analysis.
2.12. Characterization of the Expression Data Set
Eleven healthy male volunteers aged between 18 and 40 years were recruited from the region of Uppsala, Sweden, between 2013 and 2014. Blood analyses were performed before and after a meal intake. For the purpose of this study, only the non-fasting blood samples were further studied to match the prandial state. The Illumina Infinium 450K BeadChip was used to measuere the genome-wide DNA methylation pattern. RNA microarray expression was measured and analyzed using the Affymetrix GeneChip Human Gene 2.1 ST array (Li and Wong, 2001). More details on the sample and preprocessing of the methylation and RNA specimens have been previously published (Rask-Andersen et al., 2016).
2.13. The Genomic Context Analysis
The genomic context of the identified CpG sites (cg19035496, cg23409074) was investigated using the WashU Epigenome Browser, 37/hg19 version. Moreover, potential gene interactions and transcription factor binding sites were derived from chromatin analysis by paired-end tag sequencing (ChiP-seq) libraries from the ENCODE consortium (The E.P.C., 2012). Two different cell lines were analyzed for long-range interactions, i.e. erythrocytic leukaemia cells (K562) and breast cancer (MCF-7), targeting the transcription factors RNA polymerase II, estrogen receptor alpha (ERalpha) and CCCTC-binding factor (CTCF).
2.14. Comparing the Methylation Levels in Blood and Brain Regions
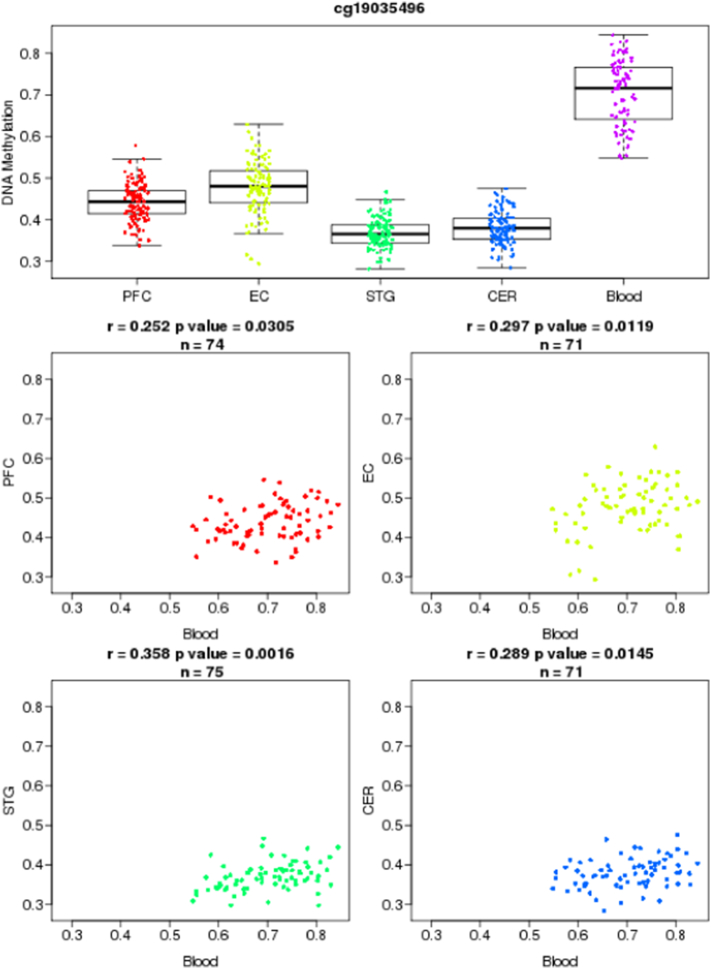
To assess whether the methylation levels that we observed in blood might be present in brain, we made use of the recently developed Blood Brain DNA Methylation Comparison Tool (http://epigenetics.iop.kcl.ac.uk/bloodbrain/). Using this tool, we were able to investigate the correlation coefficients between DNA methylation levels in whole blood and four brain regions. Whole blood and brain samples were collected using the Infinium HumanMethylation450 BeadChip, in a study described in more details elsewhere (Hannon et al., 2015). The methylation levels between blood and four different brain regions, i.e. prefrontal cortex (PFC), entorhinal cortex (EC), superior temporal gyrus (STG) and cerebellum (CER), were correlated from 71 to 75 matched samples. Pearson's correlations having p-values < 0.05 were considered statistically significant.
2.15. Statistical Analyses
All statistical analyses were performed using R statistics, version 3.3.0.
2.15.1. Data Analysis
Skewness and kurtosis of the distribution of continuous variables were evaluated with the Shapiro-Wilk's test. Both age and BMI of participants were normally distributed. The t-test was subsequently used to investigate group differences in continuous variables between the high- and low-risk groups in an unadjusted manner. Chi-squared tests computing p-values by Monte Carlo simulation were used to detect differences in categorical variables.
2.15.2. Determination of Covariates
We considered the following variables as potential confounders on the association analysis between DNA methylation and suicide risk group, i.e. age, sex, BMI, depression, occurrence of borderline personality disorder, other personality disorder, alcohol dependence, substance dependence, occurrence of completed suicide and the KIVS subscales. To avoid overfitting by including too many covariates, we only included parameters where there were group differences between the high- and low-risk groups with a p-value < 0.1, evaluated through t-test and chi-squared tests. As such, gender (p = 6.29E − 03), occurrence of other (non-borderline) personality disorder (p = 6.60E − 02), alcohol dependence (p = 9.35E − 02) and occurrence of completed suicide (p = 1.25E − 02) were included as co-variates.
2.15.3. Main Analysis
Associations between DNA methylation patterns and severe suicidal behavior were tested through linear models using the ‘limma’ package for R, applying an empirical Bayes method based on a moderated t-statistic (Smyth, 2004). We assumed a linear model where the M values of each CpG site were used as a quantitative dependent trait against suicide risk group, where, sex, occurrence of other (non-borderline) personality disorder, alcohol dependence and occurrence of completed suicide were included as covariates. Confirmatory analysis of candidate CpG loci were then performed by binomial logistic regression models of a binary outcome variable of suicide attempt severity (high vs low) to methylation M-values and adjusting for the same co-variates.
All analyses of the 72 tested CpG sites on the candidate genes were accounted for multiple testing using the Bonferroni method. A Bonferroni-adjusted p-value < 0.05 was considered significant.
2.15.4. Adolescent Cohort Analysis
The adolescent cohorts were stratified by the in silico generated DAWBA measurements of a general psychiatric risk score into high-risk group (>~50% risk) or controls. We investigated candidate CpG-sites by binomial logistic regression models, contrasting methylation M-values in high-risk subjects and controls, and adjusting for age, sex and BMI, where age and gender were treated as factor variables.
2.15.5. Investigation of Methylation and Expression Correlations in an Independent Cohort
We investigated candidate CpG sites with regard to their modulatory effect on transcriptional expression of the respective gene in the expression data set. Methylation M-values were correlated with normalized gene expression data using robust linear regressions from the ‘lmRob’ function of the “robust” package for R (Hampel et al., 1986).
3. Results
3.1. Behavior of the Clinical Outcome Variables
The mean age of the patients was 34 years (SD = 12.4; range 18—67). Eighty-six percent of the suicide attempters fulfilled criteria for at least one actual Axis I psychiatric diagnosis. Most of them (71%) had mood disorders, 5% adjustment disorder, 6% anxiety disorders and 4% alcohol abuse. Concerning psychiatric comorbidity, 25% of the patients had a comorbid anxiety disorder, 12% had a comorbid substance-related disorder (mainly alcohol dependence) and 4% had a comorbid eating disorder (bulimia nervosa). A personality disorder was prevalent among 28% of the patients.
Using above mentioned criteria, 31 patients (35%) were classified as a high-risk/severe phenotype. The high-risk group (n = 31) included significantly more males (p < 0.01). There were no between-group differences in BMI, occurrence of borderline or any other personality disorder, alcohol dependence or substance abuse. Patients from both groups scored equally on the Karolinska Interpersonal Violence Subscales measuring expressed violent behavior during childhood and adulthood as well as exposure to violent behavior during childhood and adulthood (Table 1).
Table 1.
Characteristics of subjects.
| Attempted suicide (n = 88) |
|||
|---|---|---|---|
| High-risk group | Low-risk group | Statistics (t-test, Kruskal-Wallis, Chisq. test), p-value) | |
| N | 31 | 57 | |
| Age (years) | 35.16 (12.3) | 33.6 (12.2) | ns |
| Men: women, n (%) | 16(51.6): 15(48.4) | 12 (21.1): 45 (78.9) | 6.92E − 03 |
| BMI, mean (SD) | 24.3 (4.6) | 24.9 (4.3) | ns |
| Depression, n (%) | 23 (74.2%) | 37 (68.5) | ns |
| Borderline personality disorder, n (%) | 7 (22.6) | 5 (8.8) | ns |
| Other personality disorder, n (%) | 11 (35.5) | 10 (17.5) | 6.60E − 02 |
| Alcohol dependence, n (%) | 9 (29.0) | 8 (14.0) | 9.35E − 02 |
| Substance dependence, n (%) | 6 (19.4) | 9 (15.8) | ns |
| Completed suicide, n (%) | 4 (12.9%) | 0 (0.0) | 1.25E − 02 |
| KIVS subscale, n (%) | |||
| 1Expressed violent behavior during | |||
| Childhood | 0 (0.0) | 1 (1.8) | ns |
| Adulthood | 6 (19.4) | 4 (7.0) | ns |
| 2Exposure to violent behavior during | |||
| Childhood | 10 (32.3) | 15 (26.3) | ns |
| Adulthood | 15 (48.4) | 19 (33.3) | ns |
Values are shown as mean (SD) unless otherwise specified. p-Values were calculated by means of t-test, Kruskal-Wallis' test or chi-squared test, contrasting values for subjects in the high-risk vs low-risk suicide attempt group. A one-tailed p-value < 0.05 was considered significant. Abbreviations: KIVS, Karolinska Interpersonal Violence Scale; ns, not significant.
Significant findings in bold.
3.2. Investigation of 72 HPA-Axis Coupled Probes Reveals two CRH Associated CpG Sites to Be Significantly Hypomethylated in Patients With Severe Suicidal Behavior
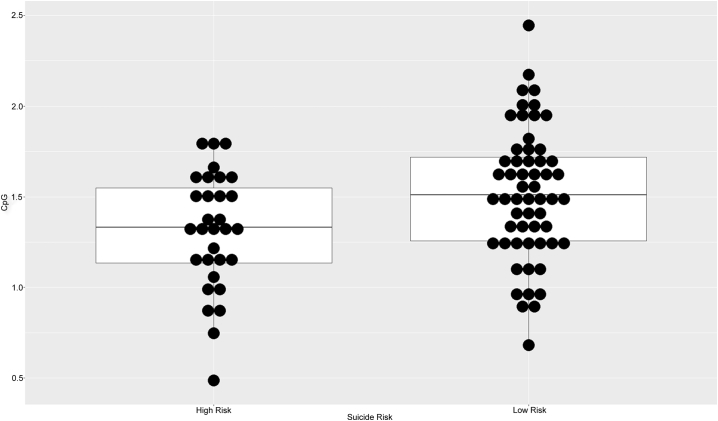
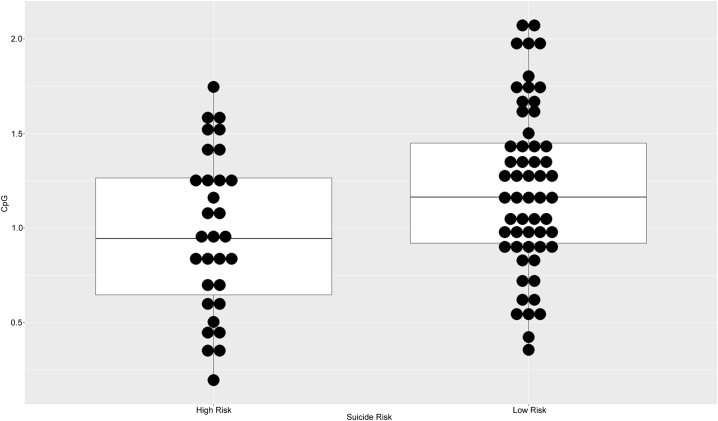
We performed linear regression models including methylation M-values at each CpG site against a binary outcome variable of suicide attempt severity (high vs low), adjusting for sex, alcohol dependence, occurrence of any non-borderline personality disorder and occurrence of completed suicide. Out of 72 tested individual CpG sites, four were nominally significant (p < 0.05), associated with the genes CRH, CRHBP and CRHR1. Using the bonferroni multiple-testing correction, two of them, i.e. cg19035496 (padj. = 0.0498) and cg23409074 (padj. = 0.0165) – located 54 and 48 bp:s upstream of the TSS of CRH gene, – were significantly hypomethylated in the high-risk group (Table 2, Fig. 1, Fig. 2). These results were confirmed by independent binomial logistic regressions, adjusting for the same co-variates, showing significant hypomethylation for both cg19035496 (p = 0.00751) and cg23409074 (p = 0.00275) (Supplementary Table 1).
Table 2.
Suicide attempt severity associated methylation changes in HPA-axis coupled CpG sites.
| Gene | Transcript | Illumina ID | logFC | t | P.Value | adj.P.Val | B |
|---|---|---|---|---|---|---|---|
| CRH | NM_000756 | cg23409074 | − 0.33 | − 3.84 | 2.29E-04 | 1.65E-02 | 0.36 |
| CRH | NM_000756 | cg19035496 | − 0.27 | − 3.52 | 6.92E-04 | 4.98E-02 | − 0.64 |
| CRHBP | AK311052 | cg13777717 | 0.16 | 2.49 | 1.45E-02 | ns | − 3.35 |
| CRHR1 | EU012435 | cg24063856 | − 0.15 | − 2.04 | 4.39E-02 | ns | − 4.31 |
| CRHR1 | BC037967 | cg12577105 | 0.12 | 1.91 | 5.90E-02 | ns | − 4.55 |
| NR3C1 | HQ450644 | cg08845721 | − 0.12 | − 1.90 | 6.12E-02 | ns | − 4.57 |
| CRH | NM_000756 | cg08215831 | − 0.15 | − 1.86 | 6.69E-02 | ns | − 4.65 |
| CRHR2 | EU012442 | cg01972879 | 0.10 | 1.71 | 9.13E-02 | ns | − 4.89 |
| CRHR1 | EU012435 | cg11338426 | 0.07 | 1.54 | 1.28E-01 | ns | − 5.14 |
| CRHR2 | EU012442 | cg04923928 | − 0.10 | − 1.51 | 1.35E-01 | ns | − 5.20 |
Analysis by multiple linear regression models of methylation M-values to a binary outcome variable of suicide attempt severity (high vs low), adjusting for gender, occurrence of completed suicide, alcohol dependence and occurrence of other (non-borderline) personality disorder. 72 individual HPA-axis associated CpG-sites were studied. p-Values were corrected for multiple testing using the bonferroni-method. Abbreviations: B, log-odds that the CpG-site is differentially methylated; logFC, log fold change; ns, not significant; p, p-value; t, moderated t-statistic.
Significant findings in bold.
Fig. 1.
Dot plot of cg19035496 methylation M-values by high risk group suicide attempter cohort.
Fig. 2.
Dot plot of cg23409074 methylation M-values by high risk group suicide attempter cohort.
3.3. Methylation Variation in Adolescents With High Risk of Psychiatric Illness
In the adolescent cohorts, subjects were stratified into high-risk group (>~50% risk) and controls based on the in silico generated DAWBA measurements of a general psychiatric risk score. The outcome of demographic and clinical variables of the adolescent cohort 1 and 2, respectively, is illustrated in Table 3. The first cohort consisted of 129 adolescents, where 16 of them were in the high-risk group and 92 were controls. The subjects were in the majority females and the mean age was 15.3 ± 0.60 years. There were no between-group differences in age, sex or BMI. The second cohort consisted of 93 adolescents, also in the majority female subjects. The mean age was 15.7 ± 0.63 years. Controls had significantly more males (p = 0.019). There were no between-group differences in participants' age or BMI.
Table 3.
Characteristics of cases in the PSY-cohort.
| Validation cohort 1 (PSY, n = 129) |
Validation cohort 2 (PSY, n = 93) |
|||||
|---|---|---|---|---|---|---|
| High risk | Low risk | p-Valuea | High risk | Low risk | p-Valuea | |
| N | 16 | 113 | 49 | 44 | ||
| Sex (male) n(%) | 4 (25.0) | 33 (29.2) | ns | 5 (10.2) | 14 (31.8) | 0.019 |
| Age (years) ± SD | 15.25 ± 0.68 | 15.34 ± 0.59 | ns | 15.70 ± 0.64 | 15.77 ± 0.61 | ns |
| BMI (mean) ± SD | 24.68 ± 6.3 | 21.37 ± 2.89 | ns | 22.2 (3.4) | 22.2 (3.1) | ns |
| DAWBA general psychiatric risk score (n (%)) | ||||||
| Level 0 (< 0.1%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Level 1 (~ 0.5%) | 0 (0.0) | 41 (36.3) | 0 (0.0) | 0 (0.0) | ||
| Level 2 (~ 3%) | 0 (0.0) | 51 (45.1) | 0 (0.0) | 0 (0.0) | ||
| Level 3 (~ 15%) | 0 (0.0) | 21 (18.6) | 0 (0.0) | 44 (100.0) | ||
| Level 4 (~ 50%) | 15 (93.8) | 0 (0.0) | 39 (79.6) | 0 (0.0) | ||
| Level 5 (~ 70%) | 1 (6.3) | 0 (0.0) | 10 (20.4) | 0 (0.0) | ||
Abbreviations: BMI, body mass index; DAWBA, Development and Well-Being Assessment. Continuous variables are shown as mean ± s.d. Individuals with a general DAWBA psychiatric risk score below 50% were defined as ‘Low risk’ and included 0 (< 0.1%), 1 (≈ 0.5%), 2 (≈ 3%) and 3 (≈ 15%) level bands of the DAWBA score. Individuals with level bands 4 (≈ 50%) and 5 (> 70%), having a risk higher than 50%, were assigned to the ‘High risk’ category.
Two-tailed analysis tests the difference between the ‘Low risk’ and ‘High risk’ group using the Student's t-test for continuous variables and the χ2-test for categorical variables. Bold value signifies p-values 0.05.
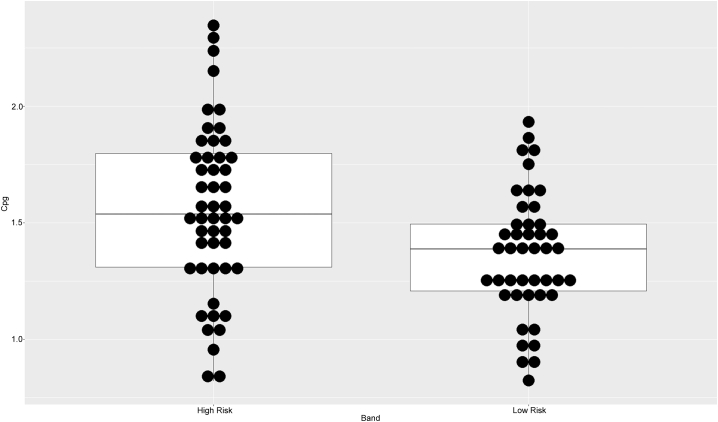
We investigated candidate CpG-sites by binomial logistic regression models, contrasting methylation M-values in high-risk subjects and controls, and adjusting for age, sex and BMI. In adolescent cohort 1, no association was found between DNA methylation of the candidate CpG sites and the general psychiatric risk score. In adolescent cohort 2, cg19035496 was significantly hypermethylated in the high-risk group (p < 0.01) (Table 4, Supplementary Table 2, Fig. 3), while no association was found for cg23409074.
Table 4.
Binomial logistic regressions of candidate CpG site DNA methylation to a general psychiatric risk score.
| Validation cohort 1 (n = 129) |
Validation cohort 2 (n = 93) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CpG | Position (bp) | Chromosome | Dist. TSS | Transcript | Gene | Coef. | p | Coef. | p |
| cg19035496 | 67090792 | 8 | 54 | NM_000756 | CRH | – | ns | 2.17 | 5.94E − 03 |
| cg23409074 | 67090798 | 8 | 48 | NM_000756 | CRH | – | ns | ns | – |
Binomial logistic regressions of DAWBA risk group to CpG-site M-values, age, sex and BMI. Age and gender were treated as factor variables. Shown are the coefficients and p-values for the CpG-site.
Fig. 3.
Boxplot of cg19035496 methylation M-values by PSY general psychiatric risk score group in Adolescent cohort 2.
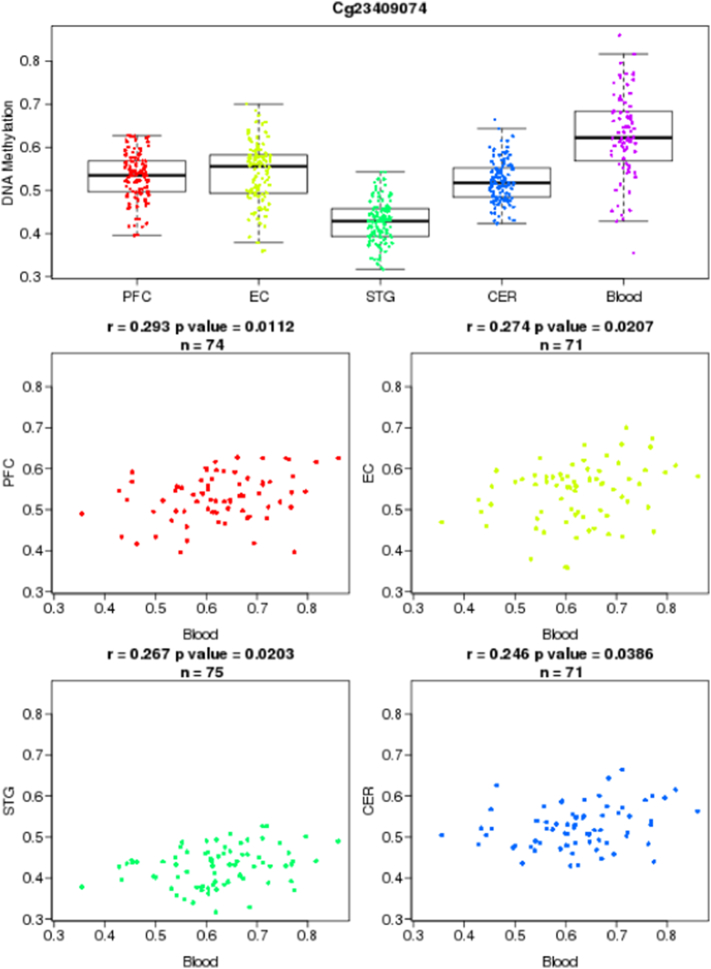
3.4. Methylation Level of cg23409074 is Positively Correlated with Gene Expression of the CRH Gene in an Independent Cohort of 11 Healthy Male Subjects
Methylation M-values of cg23409074 were correlated with normalized expression values of the CRH gene inter-individually, using robust linear regression models. This methylation locus was significantly positively correlated with CRH gene expression in the robust linear regressions (p < 0.05) (Fig. 4), (Table 5).
Fig. 4.
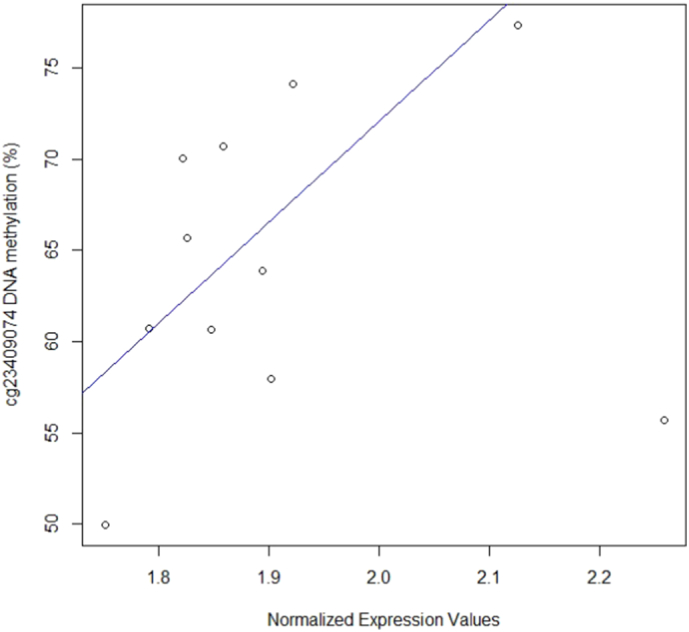
Illustration of (robust) correlation between cg23409074 methylation Beta-values and gene expression levels of the CRH gene.
Table 5.
Methylation/transcription correlations of CpG sites differentially methylated by suicide attempt severity.
| FTO-cohort (n = 11) |
||||
|---|---|---|---|---|
| Robust linear regression | ||||
| Gene | Transcript | Illumina ID | Coef. | p |
| CRH | NM_000756 | cg19035496 | – | ns |
| CRH | NM_000756 | cg23409074 | 3.70 | 2.69E − 02 |
The table lists CpG-sites located within the TSS2000 of HPA-axis genes with significant Suicidality-dependent methylation changes. These methylation probes are investigated for a correlation with transcription in a separate cohort of 11 healthy non-fasting controls. Methylation M-values were correlated with expression values inter-individually, by robust linear regression models. Abbreviations: Coef., regression coefficient; ns, not significant; p, p-value.
3.5. The Genomic Context Analysis
In the genomic context, the identified CpG sites are located close to each other (6 bp apart), within the TSS of CRH. This gene may interact with DNAJC5B, TRIM55, LOC100505676 and RRS1 genes. The genomic region including both CpG sites was a binding site for transcription initiation factor (TFIID) subunit 1, as defined by the ChiP-seq experiments (Supplementary Figure) (The E.P.C., 2012).
3.6. Blood-Brain Correlation of the CRH CpG Site
We showed that methylation variation at the identified cg23409074 and cg19035496 were significantly correlated between blood and four different brain regions. Methylation levels at cg23409074 in blood were positively correlated to cg23409074 methylation levels in all four different brain regions (prefrontal cortex; r = 0.29, p = 0.01; entorhinal cortex; r = 0.27, p = 0.02; superior temporal gyrus; r = 0.26, p = 0.02; cerebellum; r = 0.24, p = 0.03), in linear regression models, (Fig. 5, Fig. 6).
Fig. 5.
Illustration of blood-brain correlations for cg23409074.
Fig. 6.
Illustration of blood-brain correlations for cg19035496.
4. Discussion
In this study, we found that suicide attempters with high risk/severe phenotype had reduced levels of methylation at two methylation loci, i.e. cg19035496 and cg23409074 – located in the promoter region of the CRH gene, compared to suicide attempters exposed to less serious suicide attempts. Further, one of the detected loci cg19035496 was hypermethylated in a cohort of adolescents at high-risk for psychiatric illness. Importantly, methylation shifts at cg23409074 were positively correlated with CRH gene expression in an independent cohort of healthy male subjects. We used genome-wide methylation chips measuring methylation patterns at over 850K CpG sites; however, based on previous literature findings on HPA dysregulation in suicidal behavior, we applied a targeted approach on candidate genes of the HPA axis.
Suicidal behavior was defined as a severe phenotype using established criteria from the literature. Violent attempters may be phenotypically more similar to suicide victims compared to non-violent attempters showing clearly impaired decision-making (Jollant et al., 2005), lower levels of serotonin metabolite 5-hydroxyindolacetic acid (5-HIAA) in CSF (Traskman et al., 1981), more low-grade neuroinflammation (Isung et al., 2014, Lindqvist et al., 2009) and higher quinolinic acid levels than those with non-violent attempts (Erhardt et al., 2013). High suicide intent has been associated with post DST cortisol levels (Lindqvist et al., 2008) and low CSF and plasma oxytocin (Jokinen et al., 2012), reflecting an impaired control of stress. Two studies have reported an association between seriousness of suicide attempts and DST non-suppression either at baseline (Norman et al., 1990, Targum et al., 1983), or during the follow-up (Coryell, 1990). Serious attempt was defined as an attempt with high medical damage (Norman et al., 1990) and necessitating inpatient care (Targum et al., 1983). In the study performed by Coryell et al., DST non-suppression was associated with psychologically serious attempts during the follow-up (Coryell, 1990). In another study, Roy (1992) reported that violent attempters had higher post DST plasma cortisol levels than nonviolent attempters. Further, DST non-suppression predicted future suicide in a long term follow-up study of suicide attempters with mood disorders (Jokinen et al., 2007), pointing that dysregulation of HPA axis can be seen as a candidate endophenotype of high suicide risk. Importantly, suicide attempts with both high suicide intent and violent methods are associated to higher suicide risk compared to suicide attempts with lower intent or non-violent suicide attempt methods (Stefansson et al., 2010, Stenbacka and Jokinen, 2015).
The CRH gene plays a critical role in the regulation of the HPA axis, which is the central system in the stress response. The postmortem studies reported increased numbers of CRH neurons in the paraventricular nucleus of the hypothalamus in depressed suicide victims reflecting excessive activation of the HPA axis (Raadsheer et al., 1994), increased CRH immunoreactivity in monoamine-containing pontine nuclei (Austin et al., 2003), increased CRH in the CSF (Arato et al., 1989), decreased number of CRH receptors in the frontal cortex (Nemeroff et al., 1988) as well as an altered ratio of CRH-R1/R2 in the pituitary glands of suicide victims (Hiroi et al., 2001). Recently, increased expression levels of CRH in young suicide victims was reported (Zhao et al., 2015). Our findings showing reduced levels of methylation at two loci within CRH promoter in serious suicide attempters are in line with the literature conclusions regarding the HPA axis dysregulation and earlier findings concerning CRH in severe suicidal behavior including completed suicide.
Interestingly, one of the detected loci was hypermethylated in adolescents at high risk for psychiatric disorders, where the psychiatric phenotype was assessed by the DAWBA tool. Research supports the theory that a significant proportion of psychopathological changes has early onset in childhood and adolescence (Teicher and Samson, 2013). The early life adversities modulate emotional and cognitive phenotypes through the epigenetic regulation of the HPA, related to increased suicide risk (Turecki et al., 2012, Turecki et al., 2014). In prospective registry studies, adverse prenatal and perinatal events and circumstances, behavioral problems as well as psychiatric illness in childhood have been associated with suicidal behavior in adulthood (Geoffroy et al., 2013). One of the identified differentially methylated CpG loci, in relationship with severe suicidal behavior in adult suicide attempters, showed also methylation variation in adolescents at high risk for psychiatric illness. In adolescents with high risk of psychiatric illness this CpG site was hypermethylated, while in severe suicide attempters we found a hypomethylation. The interpretation of these findings is a challenge since making a direct comparison between adolescent and adult cohorts regarding methylation of genes expressed in the brain is very delicate since the brain development is still in process at the adolescent stage. Most epigenetic markers, including DNA methylation, undergo dynamic and reversible changes during life. DNA methylation occurs for different reasons, but stressful life experiences, are associated with changes in methylation in animal models. In clinical studies, early life adversity is associated with epigenetic changes and is an important confounder to be taken into account. In this study, exposure to early life adversity as well as later stress exposure, measured with the Karolinska Interpersonal Violence Scale, did not differ between the two groups of suicide attempters. This finding of the same differentially methylated CpG loci located in the CRH gene lends further support to the involvement of epigenetic modulatory effect on the HPA axis dysregulation as a shared neurobiological marker of psychiatric illness including suicidal behavior. Further, we have recently reported that the same loci cg23409074 was significantly hypomethylated in men with hypersexual disorder (Jokinen et al., 2017a) indicating shared vulnerability to psychiatric illness associated with epigenetic change in the CRH gene.
The strengths of the study are a representative patient population of suicide attempters with thorough diagnostics of the psychiatric disorders and a careful assessment of severity of suicidal behavior as well as the consideration of possible confounders such as gender, childhood adversity, and comorbidity patterns. Most of the suicide attempters were treated with antidepressants. Medications are potential confounders and epigenetic mechanisms mediate treatment response with potential implications for treatment resistant depression (Bigio et al., 2016).
It is important to note that the cross-sectional design of the study prevents to draw any conclusions about causality. Furthermore, the adolescent cohorts consisted of adolescents with high risk scores for psychiatric illness and not only severe suicidal behavior. However, psychiatric illness is one of the most important risk factors for suicidal behavior and HPA axis dysregulation is involved in both mental illness and suicidal behaviors, with often early onset and intertwined trajectories. In addition, while cg23409074 was demonstrated to correlate with gene expression of the CRH gene in healthy controls, it is still not demonstrated to what extent this could reflect modifications occurring in severe suicide attempters and a measure of CRH would have been of value for the study. Whether the whole blood CRH component methylation reflects the effects on the brain is an important question to address. Using a reliable tool to compare methylation between whole blood and brain, the methylation levels at the both identified CRH sites were significantly correlated between blood and four different brain regions providing some support that the differential methylation status observed in whole blood can reflect modifications occurring in certain brain regions. Furthermore, the association analysis of methylation and expression was performed in a relatively small group of healthy volunteers.
In conclusion, our results show epigenetic changes in the CRH gene related to severity of suicide attempt in adults and a general psychiatric risk score in adolescents. Significant blood-brain correlations in methylation suggest these alterations may impact on expressional profile of CRH in brain.
Author Contributions
Design of study (JJ, AEB, MÅ, HBS). Collection of data (JJ, MÅ, AEB, DMC, HBS). Data analysis (AEB, DMC, AD). Drafting of manuscript (JJ, AEB, HBS). (AC, AD) contributed to extensive discussions and critical manuscript reading. All authors contributed to and have approved of the final manuscript. All authors are accountable for all aspects of the work.
Role of Funding Source
Funding for this study was provided by the Swedish Research Council (Marie Åsberg, Jussi Jokinen project numbers: 5454; K2009-61P-21304-04-4; K2009-61X-21305-01-1 and Helgi Schiöth) and the Swedish Brain Research Foundation (Helgi Schiöth) and through a regional agreement between Umeå University and Västerbotten County Council (ALF) (VLL-582221) and by grants provided by the Stockholm County Council (ALF) (SLL-20150269) (Jussi Jokinen).
Acknowledgements
Methylation profiling was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. DNA extraction was performed by Latvian BMC, funded by the Latvian Council of Science European Social Fund and European Regional Development Fund.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.12.018.
Appendix A. Supplementary Data
Supplementary material
References
- Arato M., Banki C.M., Bissette G., Nemeroff C.B. Elevated CSF CRF in suicide victims. Biol. Psychiatry. 1989;25(3):355–359. doi: 10.1016/0006-3223(89)90183-2. [DOI] [PubMed] [Google Scholar]
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M.C., Janosky J.E., Murphy H.A. Increased corticotropin-releasing hormone immunoreactivity in monoamine-containing pontine nuclei of depressed suicide men. Mol. Psychiatry. 2003;8(3):324–332. doi: 10.1038/sj.mp.4001250. [DOI] [PubMed] [Google Scholar]
- Bigio B., Mathé A.A., Sousa V.C., Zelli D., Svenningsson P., McEwen B.S., Nasca C. Epigenetics and energetics in ventral hippocampus mediate rapid antidepressant action: implications for treatment resistance. Proc. Natl. Acad. Sci. U. S. A. 2016;113(28):7906–7911. doi: 10.1073/pnas.1603111113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzittofis A., Nordstrom P., Hellstrom C., Arver S., Asberg M., Jokinen J. CSF 5-HIAA, cortisol and DHEAS levels in suicide attempters. Eur. Neuropsychopharmacol. 2013;23(10):1280–1287. doi: 10.1016/j.euroneuro.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Chen Y.-A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuculete D.M., Bostrom A.E., Voisin S., Philipps H., Titova O.E., Bandstein M. A methylome-wide mQTL analysis reveals associations of methylation sites with GAD1 and HDAC3 SNPs and a general psychiatric risk score. Transl. Psychiatry. 2017;7(1) doi: 10.1038/tp.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coryell W. DST abnormality as a predictor of course in major depression. J. Affect. Disord. 1990;19(3):163–169. doi: 10.1016/0165-0327(90)90086-n. [DOI] [PubMed] [Google Scholar]
- Coryell W., Schlesser M. The dexamethasone suppression test and suicide prediction. Am. J. Psychiatry. 2001;158(5):748–753. doi: 10.1176/appi.ajp.158.5.748. [DOI] [PubMed] [Google Scholar]
- De la Cruz-Cano E. Association between FKBP5 and CRHR1 genes with suicidal behavior: a systematic review. Behav. Brain Res. 2017;15(317):46–61. doi: 10.1016/j.bbr.2016.09.032. [DOI] [PubMed] [Google Scholar]
- Erhardt S., Lim C.K., Linderholm K.R., Janelidze S., Lindqvist D., Samuelsson M. Connecting inflammation with glutamate agonism in suicidality. Neuropsychopharmacology. 2013;38(5):743–752. doi: 10.1038/npp.2012.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.P., Labbe A., Lemire M., Zanke B.W., Hudson T.J., Fertig E.J., Greenwood C.M., Hansen K.D. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman D.J., Wilson K., Thigpen J., McGee P.K. Assessing intention to die in self-injury behaviour. In: Neuringer C., editor. Psychological Assessment of Suicide Risk. Charles Thomas; Springfield: 1974. pp. 18–42. [Google Scholar]
- Geoffroy M.C., Gunnell D., Power C. Prenatal and childhood antecedents of suicide: 50-year follow-up of the 1958 British Birth Cohort Study. Psychol. Med. 2013;29:1–12. doi: 10.1017/S003329171300189X. [DOI] [PubMed] [Google Scholar]
- Guintivano J., Brown T., Newcomer A., Jones M., Cox O., Maher B.S. Identification and replication of a combined epigenetic and genetic biomarker predicting suicide and suicidal behaviors. Am. J. Psychiatry. 2014;171(12):1287–1296. doi: 10.1176/appi.ajp.2014.14010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel F.R., Ronchetti E.M., Rousseeuw P.J., Stahel W.A. John Wiley & Sons; 1986. Robust Statistics: The Approach Based on Influence of Functions. [Google Scholar]
- Hannon E., Lunnon K., Schalkwyk L., Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–1032. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N., Wong M.L., Licinio J., Park C., Young M., Gold P.W. Expression of corticotropin releasing hormone receptors type I and type II mRNA in suicide victims and controls. Mol. Psychiatry. 2001;6(5):540–546. doi: 10.1038/sj.mp.4000908. [DOI] [PubMed] [Google Scholar]
- Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isung J., Aeinehband S., Mobarrez F., Nordstrom P., Runeson B., Asberg M. High interleukin-6 and impulsivity: determining the role of endophenotypes in attempted suicide. Transl. Psychiatry. 2014;4 doi: 10.1038/tp.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jokinen J., Carlborg A., Martensson B., Forslund K., Nordstrom A.L., Nordstrom P. DST non-suppression predicts suicide after attempted suicide. Psychiatry Res. 2007;150(3):297–303. doi: 10.1016/j.psychres.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Jokinen J., Chatzittofis A., Hellstrom C., Nordstrom P., Uvnas-Moberg K., Asberg M. Low CSF oxytocin reflects high intent in suicide attempters. Psychoneuroendocrinology. 2012;37(4):482–490. doi: 10.1016/j.psyneuen.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Jokinen J., Bostrom A.E., Chatzittofis A., Ciuculete D.M., Oberg K.G., Flanagan J.N. Methylation of HPA axis related genes in men with hypersexual disorder. Psychoneuroendocrinology. 2017;80:67–73. doi: 10.1016/j.psyneuen.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Jokinen J., Talbäck M., Feychting M., Ahlbom A., Ljung R. Life expectancy after the first suicide attempt. Acta Psychiatr. Scand. 2017 doi: 10.1111/acps.12842. [DOI] [PubMed] [Google Scholar]
- Jollant F., Bellivier F., Leboyer M., Astruc B., Torres S., Verdier R. Impaired decision making in suicide attempters. Am. J. Psychiatry. 2005;162(2):304–310. doi: 10.1176/appi.ajp.162.2.304. [DOI] [PubMed] [Google Scholar]
- Lê S., Josse J., Husson F. FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 2008;25(1) [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist D., Traskman-Bendz L., Vang F. Suicidal intent and the HPA-axis characteristics of suicide attempters with major depressive disorder and adjustment disorders. Arch. Suicide Res. 2008;12(3):197–207. doi: 10.1080/13811110802100775. [DOI] [PubMed] [Google Scholar]
- Lindqvist D., Janelidze S., Hagell P., Erhardt S., Samuelsson M., Minthon L. Interleukin-6 is elevated in the cerebrospinal fluid of suicide attempters and related to symptom severity. Biol. Psychiatry. 2009;66(3):287–292. doi: 10.1016/j.biopsych.2009.01.030. [DOI] [PubMed] [Google Scholar]
- Mann J.J., Currier D., Stanley B., Oquendo M.A., Amsel L.V., Ellis S.P. Can biological tests assist prediction of suicide in mood disorders? Int. J. Neuropsychopharmacol. 2006;9(4):465–474. doi: 10.1017/S1461145705005687. [DOI] [PubMed] [Google Scholar]
- Mann J.J., Arango V.A., Avenevoli S., Brent D.A., Champagne F.A., Clayton P. Candidate endophenotypes for genetic studies of suicidal behavior. Biol. Psychiatry. 2009;65(7):556–563. doi: 10.1016/j.biopsych.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan P.O., Sasaki A., D'Alessio A.C., Dymov S., Labonte B., Szyf M. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T.J., Butcher L.M., Feber A., Teschendorff A.E., Chakravarthy A.R., Wojdacz T.K. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2013;30(3):428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff C.B., Owens M.J., Bissette G., Andorn A.C., Stanley M. Reduced corticotropin releasing factor binding sites in the frontal cortex of suicide victims. Arch. Gen. Psychiatry. 1988;45(6):577–579. doi: 10.1001/archpsyc.1988.01800300075009. [DOI] [PubMed] [Google Scholar]
- Norman W.H., Brown W.A., Miller I.W., Keitner G.I., Overholser J.C. The dexamethasone suppression test and completed suicide. Acta Psychiatr. Scand. 1990;81(2):120–125. doi: 10.1111/j.1600-0447.1990.tb06463.x. [DOI] [PubMed] [Google Scholar]
- Pandey G.N., Rizavi H.S., Zhang H., Bhaumik R., Ren X. The expression of the suicide-associated gene SKA2 is decreased in the prefrontal cortex of suicide victims but not of nonsuicidal patients. Int. J. Neuropsychopharmacol. 2016;19(8) doi: 10.1093/ijnp/pyw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P., Van Djik S., Muhlhausler B., Stirzaker C., Clark S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.E., Cotton A.M., Lam L.L., Farré P., Emberly E., Brown C.J. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6(1):4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raadsheer F.C., Hoogendijk W.J., Stam F.C., Tilders F.J., Swaab D.F. Increased numbers of corticotropin-releasing hormone expressing neurons in the hypothalamic paraventricular nucleus of depressed patients. Neuroendocrinology. 1994;60(4):436–444. doi: 10.1159/000126778. [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M., Bringeland N., Nilsson E.K., Bandstein M., Olaya Bucaro M., Vogel H. Postprandial alterations in whole-blood DNA methylation are mediated by changes in white blood cell composition. Am. J. Clin. Nutr. 2016;104(2):518–525. doi: 10.3945/ajcn.115.122366. [DOI] [PubMed] [Google Scholar]
- Roy A. Hypothalamic-pituitary-adrenal axis function and suicidal behavior in depression. Biol. Psychiatry. 1992;32(9):812–816. doi: 10.1016/0006-3223(92)90084-d. [DOI] [PubMed] [Google Scholar]
- Roy A., Hodgkinson C.A., Deluca V., Goldman D., Enoch M.A. Two HPA axis genes, CRHBP and FKBP5, interact with childhood trauma to increase the risk for suicidal behavior. J. Psychiatr. Res. 2012;46(1):72–79. doi: 10.1016/j.jpsychires.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B., Shelton R.C., Dwivedi Y. DNA methylation and expression of stress related genes in PBMC of MDD patients with and without serious suicidal ideation. J. Psychiatr. Res. 2017;89:115–124. doi: 10.1016/j.jpsychires.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Vol. 1,2,3. Cold Spring Harbor Lab Press; 1989. Molecular Cloning A Laboratory Manual Second Edition. [Google Scholar]
- Schalkwyk, L. (2013). wateRmelon: Illumina 450 methylation array normalization and metrics. From https://www.bioconductor.org/packages/release/bioc/html/wateRmelon.html
- Silverman M.M., Berman A.L., Sanddal N.D., O'Carroll P.W., Joiner T.E. Rebuilding the tower of Babel: a revised nomenclature for the study of suicide and suicidal behaviors. Part 2: suicide-related ideations, communications, and behaviors. Suicide Life Threat. Behav. 2007;37(3):264–277. doi: 10.1521/suli.2007.37.3.264. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004:3. doi: 10.2202/1544-6115.1027. (Article 3) [DOI] [PubMed] [Google Scholar]
- Stefansson J., Nordstrom P., Jokinen J. Suicide Intent Scale in the prediction of suicide. J. Affect. Disord. 2010;136(1-2):167–171. doi: 10.1016/j.jad.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Stenbacka M., Jokinen J. Violent and non-violent methods of attempted and completed suicide in Swedish young men: the role of early risk factors. BMC Psychiatry. 2015;15:196. doi: 10.1186/s12888-015-0570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targum S.D., Rosen L., Capodanno A.E. The dexamethasone suppression test in suicidal patients with unipolar depression. Am. J. Psychiatry. 1983;140(7):877–879. doi: 10.1176/ajp.140.7.877. [DOI] [PubMed] [Google Scholar]
- Teicher M.H., Samson J.A. Childhood maltreatment and psychopathology: a case for ecophenotypic variants as clinically and neurobiologically distinct subtypes. Am. J. Psychiatry. 2013;170(10):1114–1133. doi: 10.1176/appi.ajp.2013.12070957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A Beta-Mixture Quantile Normalisation method for correcting probe design bias in Illumina Infinium 450k DNA methylation data. Bioinformatics. 2012;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The E.P.C An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:7414,57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traskman L., Asberg M., Bertilsson L., Sjostrand L. Monoamine metabolites in CSF and suicidal behavior. Arch. Gen. Psychiatry. 1981;38(6):631–636. doi: 10.1001/archpsyc.1981.01780310031002. [DOI] [PubMed] [Google Scholar]
- Triche T.J., Jr., Weisenberger D.J., Van Den Berg D., Laird P.W., Siegmund K.D. Low-level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013;41(7) doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G. The molecular bases of the suicidal brain. Nat. Rev. Neurosci. 2014;15(12):802–816. doi: 10.1038/nrn3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G., Ernst C., Jollant F., Labonte B., Mechawar N. The neurodevelopmental origins of suicidal behavior. Trends Neurosci. 2012;35(1):14–23. doi: 10.1016/j.tins.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Turecki G., Ota V.K., Belangero S.I., Jackowski A., Kaufman J. Early life adversity, genomic plasticity, and psychopathology. Lancet Psychiatry. 2014;1(6):461–466. doi: 10.1016/S2215-0366(14)00022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin S., Almen M.S., Zheleznyakova G.Y., Lundberg L., Zarei S., Castillo S. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:103. doi: 10.1186/s13073-015-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J.R., Busche S., Ge B., Kwan T., Pastinen T., Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15(2):R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Qi X.R., Gao S.F., Lu J., van Wamelen D.J., Kamphuis W., Bao A.M., Swaab D.F. Different stress-related gene expression in depression and suicide. J. Psychiatr. Res. 2015;68:176–185. doi: 10.1016/j.jpsychires.2015.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material