Significance
In aging, the brain becomes vulnerable to injury and cognitive function declines, but the mechanisms responsible are unknown. Astrocytes, the most abundant class of glial cells, are vital for the proper function of the central nervous system, and impairment of astrocyte function has been implicated in disease. Here we perform RNA sequencing of astrocytes from different brain regions across the lifespan of the mouse to identify age-related transcriptional changes that could contribute to cognitive decline. We find that aged astrocytes take on a reactive phenotype characteristic of neuroinflammatory reactive astrocytes, and that microglia play a role in inducing astrocyte activation. The aging astrocyte RNA sequencing profiles provide an important new resource for future studies exploring the role of astrocytes in cognitive decline.
Keywords: astrocytes, aging, microglia, RNA sequencing, cognitive decline
Abstract
The decline of cognitive function occurs with aging, but the mechanisms responsible are unknown. Astrocytes instruct the formation, maturation, and elimination of synapses, and impairment of these functions has been implicated in many diseases. These findings raise the question of whether astrocyte dysfunction could contribute to cognitive decline in aging. We used the Bac-Trap method to perform RNA sequencing of astrocytes from different brain regions across the lifespan of the mouse. We found that astrocytes have region-specific transcriptional identities that change with age in a region-dependent manner. We validated our findings using fluorescence in situ hybridization and quantitative PCR. Detailed analysis of the differentially expressed genes in aging revealed that aged astrocytes take on a reactive phenotype of neuroinflammatory A1-like reactive astrocytes. Hippocampal and striatal astrocytes up-regulated a greater number of reactive astrocyte genes compared with cortical astrocytes. Moreover, aged brains formed many more A1 reactive astrocytes in response to the neuroinflammation inducer lipopolysaccharide. We found that the aging-induced up-regulation of reactive astrocyte genes was significantly reduced in mice lacking the microglial-secreted cytokines (IL-1α, TNF, and C1q) known to induce A1 reactive astrocyte formation, indicating that microglia promote astrocyte activation in aging. Since A1 reactive astrocytes lose the ability to carry out their normal functions, produce complement components, and release a toxic factor which kills neurons and oligodendrocytes, the aging-induced up-regulation of reactive genes by astrocytes could contribute to the cognitive decline in vulnerable brain regions in normal aging and contribute to the greater vulnerability of the aged brain to injury.
Astrocytes are the most abundant glial cells and are vital for the proper function of the central nervous system (CNS). They perform many roles, including instructing the formation (1–3) and elimination (4) of neuronal synapses during development. In addition, they provide trophic support to neurons (5), mediate uptake and recycling of neurotransmitters (6), and are involved in maintenance of the blood–brain barrier (7). Given the multitude of vital roles for astrocytes, it is not surprising that impairment of these functions has been implicated in the pathogenesis of many diseases. In particular, astrocyte dysfunction has been linked to age-related neurodegenerative diseases, including Alzheimer’s disease (8–10), dementia (11), and amyotrophic lateral sclerosis (ALS) (12, 13). Several hallmarks of these neurodegenerative diseases, such as cognitive decline and a loss of working memory, also occur during the process of normal aging.
Transcriptomics and epigenetic studies have reported that glial-specific gene expression patterns change significantly with normal aging, while neuron-specific gene expression patterns change relatively little in comparison (14). For instance, microglia, the resident immune cells of brain, up-regulate immune response signaling receptors (15), and neuroprotective signaling pathways (16) in aged mice. Consistent with the observed transcriptional changes, there is evidence that microglia adopt an activated morphology in aged brains (17). The extent to which astrocytes also become activated with age is unknown, although it is reported that the level of glial fibrillary acidic protein (GFAP) and vimentin, two astrocyte-specific genes linked to activation, increase with age in both rodents and humans (18, 19). Regardless, the transcriptional and functional changes occurring in astrocytes in normal aging are not well understood.
The up-regulation of potentially proinflammatory genes by microglia in normal aging raises the question of whether these aging-induced changes in microglia could promote the activation of astrocytes. Recently, we reported that proinflammatory microglia secrete IL-1α, TNF, and C1q, and that these cytokines acting together are necessary and sufficient to activate astrocytes (9). These cytokines induce the formation of a subtype of astrocytes (termed A1 astrocytes) which are strongly neurotoxic and rapidly kill neurons (9). As well as releasing a potent neurotoxin, A1 astrocytes were less able to promote the formation of new synapses, and caused a decrease in the excitatory function of CNS neurons. In addition to changes in the reactive state of astrocytes, it is possible that other transcriptional and functional changes are occurring in astrocytes that contribute to cognitive decline in normal aging.
To investigate the transcriptional changes occurring in astrocytes with aging, we performed RNA sequencing (RNAseq) at five ages across the lifespan of the mouse to determine the aging-induced transcriptional changes occurring in astrocytes. Furthermore, we profiled astrocytes from three functionally distinct brain regions—the hippocampus, striatum and cortex—at these five ages, to investigate brain-region−specific changes in the astrocyte transcriptome in normal aging. Our RNAseq data revealed that astrocytes have distinct region-dependent transcriptional identities and age in a regionally dependent manner. In particular, we found that astrocytes in regions more vulnerable to cognitive decline—the hippocampus and striatum—undergo more dramatic transcriptional changes compared with cortical astrocytes. Differential gene expression analysis revealed that the most prominent class of genes up-regulated by aged astrocytes were reactive astrocyte genes. Interestingly, many of these reactive astrocyte genes are characteristic A1 reactive astrocyte genes that are induced in response to neuroinflammation (9, 20). We found that the aging-induced up-regulation of reactive astrocyte genes was significantly reduced in mice lacking IL-1α, TNF, and C1q, all genes specifically expressed in the CNS by microglia. These data provide evidence that activated microglia play a key role in promoting astrocyte activation not only following acute trauma and chronic neurodevelopmental disease (9) but also in the context of normal brain aging.
Results
Astrocyte RNA Purification and RNAseq Transcriptional Profiling.
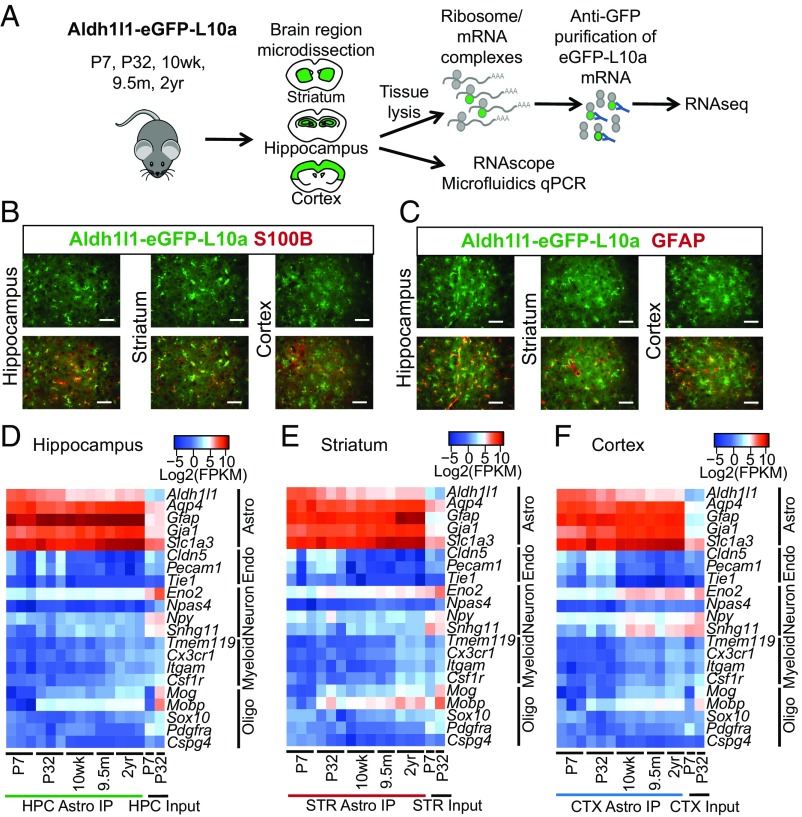
To better understand how normal aging influences the astrocyte phenotype in different brain regions (hippocampus, striatum, and cortex) we generated an RNAseq profiling resource of adolescent [postnatal day 7 (P7)], young adult (P32), mature (10 wk), middle age (9.5 mo), and aged (2 y) astrocytes. A major limitation to sequencing older CNS tissues is the successful purification of intact, viable astrocytes with minimal isolation artifact. To overcome this difficulty, we used the Aldh1l1-eGFP-L10a transgenic mouse line (Fig. 1A), which expresses a fusion of eGFP and a ribosomal protein (L10a) under the control of the astrocyte-specific Aldh1l1 gene. Translating ribosome affinity purification (TRAP) (21, 22) of this fusion protein yields the ribosome and its associated mRNAs, allowing the isolation of transcripts from the eGFP-expressing astrocytes. To first assess the specificity of ALDH1L1-eGFP-L10a protein expression in adult mice (10 wk of age), we performed immunohistochemistry with the known astrocyte markers S100β and GFAP. We found that eGFP colocalized with S100β (Fig. 1B) and/or GFAP (Fig. 1C), but not unlabeled cells, in all three of the brain regions we examined, suggesting highly specific localization of Aldh1l1-eGFP-L10a to only astrocytes.
Fig. 1.
Isolation and validation of the aging astrocyte transcriptome from distinct brain regions. (A) Schematic diagram showing the experimental strategy for isolation of astrocyte RNA by TRAP across the lifespan of the mouse, and validation of differentially expressed aging genes. (B and C) Representative images of astrocytes from the hippocampus, striatum, and cortex in an adult (10 wk old) Aldh1l1-eGFP-L10a mouse showing costaining for (B) S100β and (C) GFAP. (Scale bar, 50 µm.) (D–F) Cell purity heatmaps depicting mean FPKM expression values for cell-type−specific markers of astrocytes, neurons, oligodendrocyte lineage cells (oligo), endothelial cells (endo), and microglia/macrophages (myeloid) in (D) hippocampal, (E) striatal, and (F) cortical astrocyte and input samples. CTX, cortex; HPC, hippocampus; IP, immunoprecipitation; STR, striatum. See also Datasets S1–S4.
We used standard TRAP methods (21, 22) to isolate RNA from the hippocampus, striatum, and cortex at five ages across the lifespan of the mouse (P7, P32, 10 wk, 9.5 mo, and 2 y, in triplicate for each region, except for 9.5 mo of age, which were collected in duplicate for each region). In addition, we sequenced whole-brain input samples for each brain region to verify the astrocyte enrichment of our TRAP samples. We assessed the quality and purity of our RNAseq profiles by mapping quality and the expression of cell-type–specific genes. We mapped >70% of reads for all of our samples. We first determined that these RNAseq profiles were highly enriched for astrocyte-specific genes (Fig. 1 D–F) and depleted of other cell-type−specific genes (Fig. 1 D–F). Despite the high degree of enrichment of astrocyte-specific genes in the majority of samples, we found a small increase in some neuronal and myelin gene contaminants in our adult (10 wk, 9.5 mo, and 2 y) RNAseq samples (Fig. 1 D–F), likely reflecting nonspecific adherence of some mRNAs to beads during purification (23). Since the Aldh1l1-eGFP-L10a protein expression is highly colocalized with known astrocyte markers, the nonspecific pulldown of a small number of eGFP-negative transcripts likely contributes to the low-level sequencing of nonastrocyte transcripts, and highlights the importance of validation.
Comparison of Astrocyte Transcriptomes Throughout Aging.
To investigate astrocyte heterogeneity with both age and brain region, we first mined our RNAseq profiles for the top 50 genes enriched in young P7 cortical astrocytes (24) (Fig. S1A). We found that most of these genes were abundantly expressed in all samples, but the level of expression varied between brain regions with age. Next we compared astrocyte and whole-brain RNA expression profiles [for all genes expressed with fragments per kilobase of transcript per million mapped reads (FPKM) ≥ 5] and found that astrocytes in all brain regions were similar across age (Fig. 2A) compared with the whole brain. Despite astrocyte gene expression being similar across age, unsupervised hierarchical clustering revealed two distinct branches between young (P7 and P32) astrocytes and mature (10 wk, 9.5 mo, and 2 y) astrocytes, indicating that normal aging leads to changes in astrocyte gene expression (Fig. S1B). We found that cortical astrocytes were most similar throughout age. In contrast, both hippocampal and striatal astrocytes segregated into distinct populations with age (Fig. S1B), suggesting that subcortical astrocytes undergo a greater number of aging-induced changes in gene expression.
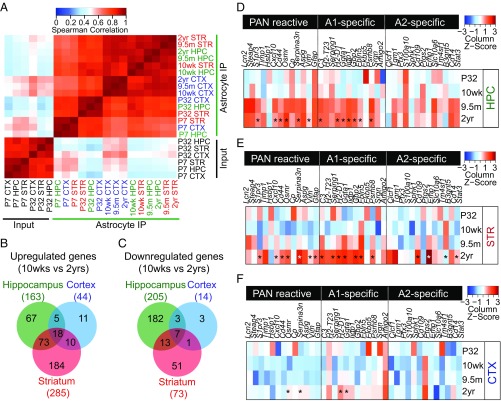
Fig. 2.
Differential gene expression analysis in aging astrocytes from distinct brain regions. (A) Heatmap of Spearman correlation between astrocytes (P7, P32, 10 wk, 9.5 mo, and 2 y) and input samples for three brain regions (hippocampus, striatum, and cortex). Data are mean FPKM values for genes expressed FPKM ≥ 5. (B and C) Venn diagrams showing the number of significantly (P < 0.05) (B) up-regulated and (C) down-regulated genes, determined by edgeR analysis, between mature adult (10 wk) and aged (2 y old) astrocyte samples from the hippocampus, striatum, and cortex. Numbers listed on the outside of the charts represent the total number of up-regulated genes in each brain region [hippocampus (green), striatum (red), and cortex (blue)]. Diagrams are adapted from jvenn (54). (D–F) Heatmaps comparing the mean expression of pan-reactive (genes induced by neuroinflammation or ischemia), A1-specific (genes induced by neuroinflammation), and A2-specfic (genes induced by ischemia) transcripts in TRAP astrocyte RNA samples isolated from the (D) hippocampus, (E) striatum, and (F) cortex of P32, 10-wk-, 9.5-mo-, and 2-y-old mice. Asterisks (*) denote significantly (P < 0.05) increased expression in 2-y-old samples compared with 10 wk by edgeR analysis. See also Figs. S1 and S2 and Datasets S1–S3.
To elucidate the aging-induced gene changes in each brain region, we next conducted edgeR differential gene expression analysis to compare adult (10 wk) and aged (2 y) astrocytes. We considered genes to be differentially expressed if they were up-regulated or down-regulated in at least two out of three replicates, and reached our statistical significance cutoff of P < 0.05. These analyses revealed significant heterogeneity in aging-induced gene expression between brain areas. In particular, a greater number of genes were differentially expressed by striatal and hippocampal astrocytes (358 and 368 genes in the striatum and hippocampus, respectively) compared with cortical astrocytes (58 genes in the cortex) (Fig. 2 B and C). Next, we examined the most up-regulated aging genes in each brain region (Datasets S1–S3). Among the most highly up-regulated genes in all brain regions were a cassette of genes that have been previously identified as reactive astrocyte-associated genes (9, 20). These genes included those involved in the complement (C3 and C4B), cytokine pathway (Cxcl10), antigen presentation (H2-D1 and H2-K1), and peptidase inhibitor (Serpina3n) pathways. Hippocampal and striatal astrocytes up-regulated a larger number of reactive genes (116 and 64 genes in the striatum and hippocampus, respectively) compared with cortical astrocytes (27 genes) (Fig. S2 A–B). We compared the significantly up-regulated genes in our RNAseq profiles with the list of reactive astrocyte genes. These included genes specifically up-regulated by neuroinflammation (A1-specific) or ischemia (A2-specific), or those induced by both neuroinflammation and ischemia (pan-reactive genes) (9, 20). We found that astrocytes from all brain regions up-regulated a larger number of A1 reactive genes (24, 34, and 8 A1 genes were up-regulated by hippocampal, striatal, and cortical astrocytes, respectively) compared with A2 reactive genes (6, 18, and 4 A2 genes were up-regulated by hippocampal, striatal, and cortical astrocytes, respectively) (Fig. 2 D–F and Fig. S2B). In addition to the up-regulation of a larger number of reactive genes, we observed that the fold induction of these genes was greater in hippocampal and striatal astrocytes compared with cortical astrocytes (Fig. 2 D–F and Fig. S2A). Next, to better understand the number of A1 and A2 astrocytes in the aged brain, we carried out in situ hybridization experiments for A1 and A2 genes. In accordance with our RNAseq data, we found that the majority of Slc1a3+ astrocytes express A1 (C3) and/or A2 (Emp1) markers (∼62% of Slc1a3+ astrocytes expressed at least one reactive marker) (Fig. S2 C–F). In particular, we observed that a greater number of Slc1a3+ astrocytes expressed the A1-specific gene C3 (22 ± 3% of astrocytes in the aged striatum), and not the A2-specific gene Emp1 (9 ± 3% of astrocytes in the aged striatum) (Fig. S2 C–F). To our surprise, we also found a substantial number of Slc1a3+ astrocytes expressing detectable levels of both the A1-specific gene C3 and the A2-specific gene Emp1 (31 ± 2% of astrocytes in the aged striatum) (Fig. S2 C–F). Taken together, these data suggest that, at the single-cell level, aged astrocytes can express a combination of A1 and A2 genes, but that a larger number of astrocytes expressing only the A1-specific gene C3 are present in the aged brain, compared with the number of astrocytes expressing only the A2-specific gene Emp1.
To better understand aging-induced gene changes in astrocytes, we performed Ingenuity Pathway Analysis (IPA), for an unbiased analysis of other potentially regulated pathways. These analyses further highlighted that aged astrocytes up-regulate cellular activation pathways and immune response pathways, including acute phase response signaling, complement system, antigen response, and IFN signaling (Fig. S3 A–C). IPA upstream analysis predicted the significant up-regulation of Stat3 in hippocampal (Z score +2.7, P < 0.05) and striatal (Z score +3.3, P < 0.05) astrocytes. This transcription factor is thought to be required for the induction astrocyte gliosis (25, 26). Together, these data provide evidence that normal aging induces astrocyte reactivity, in a region-specific manner—with the hippocampal and striatal astrocytes becoming more reactive than cortical astrocytes.
We next examined the top down-regulated genes with age in all brain regions. Similarly, fewer genes were down-regulated by cortical astrocytes with age compared with hippocampal and striatal astrocytes (Fig. 2C). Among these top down-regulated genes were genes involved in mitochondrial function and energy production, including Ucp2, Cox8b, and Atp5g1, suggesting that aged astrocytes may have dysfunctional mitochondria (Datasets S1–S3). In addition, we found a reduction in antioxidant defense-related genes, including Gpx8 and Atox1 (Datasets S1–S3), suggesting that aged astrocytes may have decreased antioxidant capacity. Taken together, the impaired mitochondrial function and antioxidant capacity of aged astrocytes could contribute to both metabolic and oxidative stress known to occur in the aged brain (27, 28). These findings suggest that aging induces reactivity in astrocytes, which may impact their ability to perform their normal functions in the brain, such as inducing the formation and maturation of synapses (1–3), or eliminating synapses (4).
To assess the ability of aged astrocytes to perform normal physiological functions, we compared the expression of gene families involved in the formation, maturation, and elimination of synapses. The majority of the genes encoding astrocyte-secreted factors known induce the formation and maturation of excitatory synapses (1–3) did not significantly change between adult (10 wk) and aged (2 y) astrocytes (Fig. S3 D–F). To our surprise, we found that Sparcl1 (Hevin) and Sparc, two astrocyte-secreted factors important for the assembly of excitatory synapses (2, 29), were significantly up-regulated by aged astrocytes (Fig. S3 D–F). In addition to the release of synaptogenic secreted factors, astrocytes express phagocytic genes, including Megf10 and Mertk, which are important for the elimination of synapses during development (4).
Comparison of the expression of phagocytic genes revealed that, while a majority of phagocytic genes did not change with age, some phagocytic pathway genes, including Pros1, Mfge8, Megf10, and Lrp1, were significantly up-regulated with age (Fig. S3 G–I). Overall, these data indicate that astrocytes do not down-regulate the expression of genes important for their normal functions at synapses with age, but instead up-regulate reactive genes.
Validation of Top Astrocyte-Specific Aging Genes.
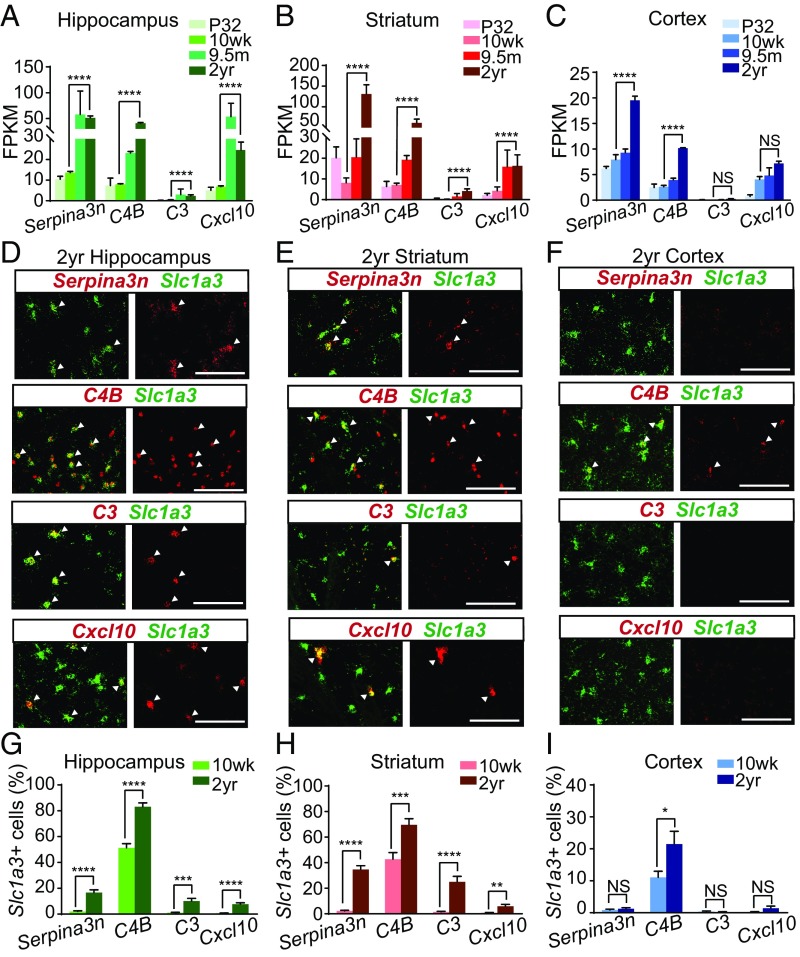
We next sought to validate the increased expression of reactive astrocyte genes in the hippocampus, striatum, and cortex by in situ hybridization. Our RNAseq data indicated that Serpina3n, C4B, C3, and Cxcl10 genes were significantly up-regulated in both hippocampal (Fig. 3A) and striatal astrocytes during aging (Fig. 3B). In contrast, only Serpina3n and C4B were significantly enriched in cortical astrocytes (Fig. 3C). In accordance with our RNAseq data, we found that the number of Slc1a3+ astrocytes expressing detectable levels of Serprina3n, C4B, C3, and Cxcl10 was significantly increased in the hippocampus (Serpina3n+ astrocytes increased from 2 ± 0.5% to 17 ± 2%; C4B+ astrocytes increased from 51 ± 3% to 83 ± 3%; C3+ astrocytes increased from 1 ± 0.3% to 10 ± 2%; and Cxcl10+ astrocytes increased from 0.8 ± 0.2% to 8 ± 1%, at 10 wk and 2 y of age, respectively) (Fig. 3 D and G) and striatum (Serpina3n+ astrocytes increased from 2 ± 0.5% to 35 ± 3%; C4B+ astrocytes increased from 43 ± 5% to 70 ± 5%; C3+ astrocytes increased from 1 ± 0.4% to 25 ± 4%; and Cxcl10+ astrocytes increased from 0.8 ± 0.3% to 6 ± 1%, at 10 wk and 2 y of age, respectively) (Fig. 3 E and H) in aged mice (2 y) compared with adults (10 wk). In contrast, an increase in the number of Slc1a3+ astrocytes expressing C4B mRNA expression could only be detected in aged (2 y) cortical astrocytes (C4B+ astrocytes increased from 11 ± 2% to 21 ± 4%, at 10 wk and 2 y of age, respectively) (Fig. 3 F and I) compared with 10-wk-old astrocytes. Despite quantification of reactive astrocyte transcripts across multiple cortical regions, unlike C4B, we did not detect a significant increase in Serpina3n+ astrocytes by in situ hybridization. It is possible that small subsets of cortical astrocytes up-regulate Serpina3n that we were unable to detect, which could account for the discrepancy between our RNAseq data and in situ hybridization. Collectively, the RNAseq and in situ hybridization data reveal a substantial increase in the expression of reactive astrocyte genes in normal aging, and suggest that two of the brain regions more vulnerable to diseases of aging—the hippocampus and striatum—become more reactive with age compared with the cortex.
Fig. 3.
Validation of aging-induced reactive genes by in situ hybridization. (A–C) Bar plots of RNAseq data showing FPKM expression of four aging-induced genes (Serpina3n, C4B, C3, and Cxcl10) in astrocyte samples across the mouse lifespan in the (A) hippocampus, (B) striatum, and (C) cortex. Error bars depict mean ± SEM. ****P < 0.0001 for values compared between 10-wk- and 2-y-old astrocyte samples by edgeR analysis. (D–F) Representative in situ hybridization images for four aging-induced astrocyte genes (Serpina3n, C4B, C3, and Cxcl10) showing colocalization with the astrocyte marker Slc1a3 in aged mice (2 y) in the (D) hippocampus, (E) striatum, and (F) cortex. (Scale bar, 100 µm.) White arrowheads highlight Slc1a3+ astrocytes expressing reactive genes. (G–I) Bar charts depicting quantification of the number of Slc1a3+ astrocytes expressing detectable levels of Serpina3n, C4B, C3, and Cxcl10 mRNA in the mature adult (10 wk) and aged (2 y old) (G) hippocampus, (H) striatum, and (I) cortex. Error bars depict mean ± SEM. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. NS, nonsignificant; n = 3 animals.
Microglial Influence on the Induction of Aging Astrocyte Gene Expression.
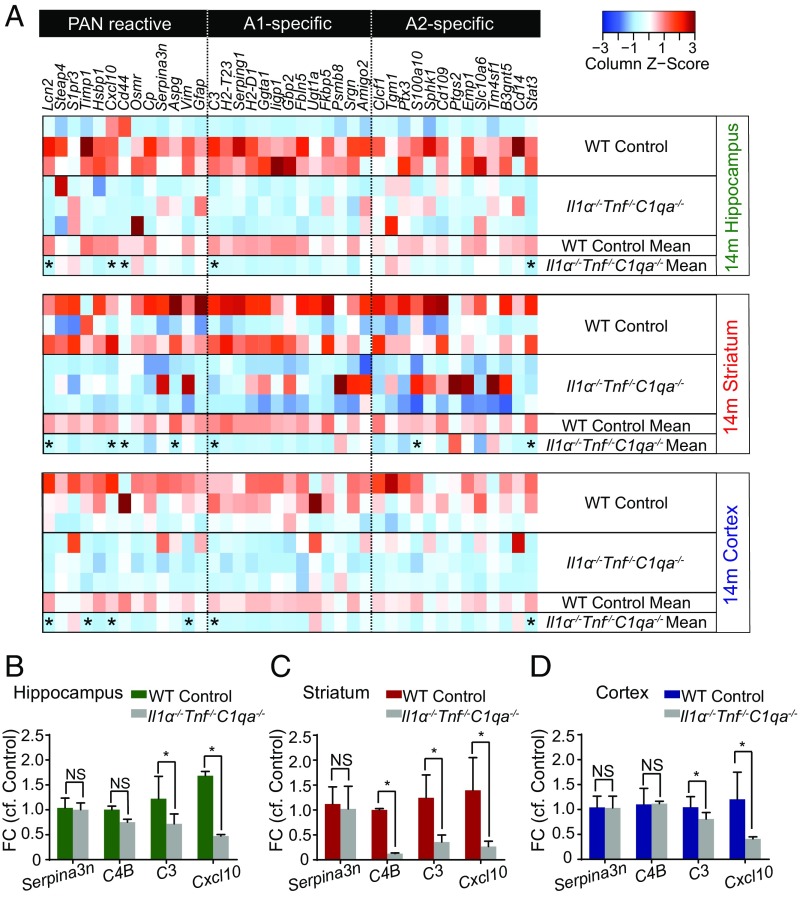
The increased expression of reactive genes in astrocytes with normal aging raises the question of which molecular and cellular pathways induce this activation. Previous work from our laboratory demonstrated that proinflammatory microglia induce the formation A1 reactive astrocytes (9). Another study from our laboratory showed that the C1q protein level, a key inducer of A1 astrocytes, is dramatically up-regulated with normal aging (30). Furthermore, several studies have reported that microglia become more reactive with age (15, 17). Therefore, we next investigated whether aging-induced astrocyte reactivity is reduced in transgenic mice lacking the cytokines known to induce reactive astrocyte gliosis (IL1α−/−;Tnf−/−;C1qa−/− triple knockout mice). We used microfluidics-based qPCR to compare the expression of reactive astrocyte genes in wild-type (WT) and IL1α−/−;Tnf−/−;C1qa−/− aged (14 mo old) mice. We found that a number of pan-reactive genes, including one of the genes most highly up-regulated with age, Cxcl10, were down-regulated in all brain regions examined in IL1α−/−;Tnf−/−;C1qa−/− triple knockout mice compared with age-matched WT mice (Fig. 4). In addition, we observed a significant reduction in the expression of the A1-specific reactive gene complement component C3 in all brain regions of IL1α−/−;Tnf−/−;C1qa−/− knockout mice (Fig. 4). Another highly up-regulated aging gene, complement component C4B, was significantly reduced only in the striatum of IL1α−/−;Tnf−/−;C1qa−/− knockout mice (Fig. 4C). We also noticed that the expression of the A2-specific gene Stat3 was significantly decreased in all three brain regions of IL1α−/−;Tnf−/−;C1qa−/− knockout mice (Fig. 4A). Because these three cytokines are predominantly released by microglia in the CNS in response to neuroinflammation (9), these data suggest that activated microglia promote astrocyte activation in normal aging. Notably, we did not observe a significant decrease in all aging-induced reactive astrocyte genes in IL1α−/−;Tnf−/−;C1qa−/− knockout mice, such as Serpina3n, suggesting that additional signals likely play a role in the induction of astrocyte reactivity with age.
Fig. 4.
Comparison of reactive gene expression in WT and IL-1α, TNF, and C1q knockout mice. (A) Heatmap comparing the expression of pan-reactive (genes induced by neuroinflammation or ischemia), A1-specific (genes induced by neuroinflammation), and A2-specfic (genes induced by ischemia) transcripts in whole brain RNA isolated from 14-mo-old WT and IL-1α, TNF, and C1q mice. Asterisks (*) denote significantly (P < 0.05) reduced expression of transcripts after averaging deltaCT gene Z scores from microfluidics qPCR. (B–D) Bar charts depicting the fold change in expression of four aging-induced (Serpina3n, C4B, C3, and Cxcl10) genes in IL-1α, TNF, and C1q knockout mice compared with WT mice in the (B) hippocampus, (C) striatum, and (D) cortex. Error bars depict mean ± SEM. *P < 0.05; n = 3 animals for each genotype. FC, fold change.
Comparison of LPS-Induced Astrocyte Reactivity Throughout Age.
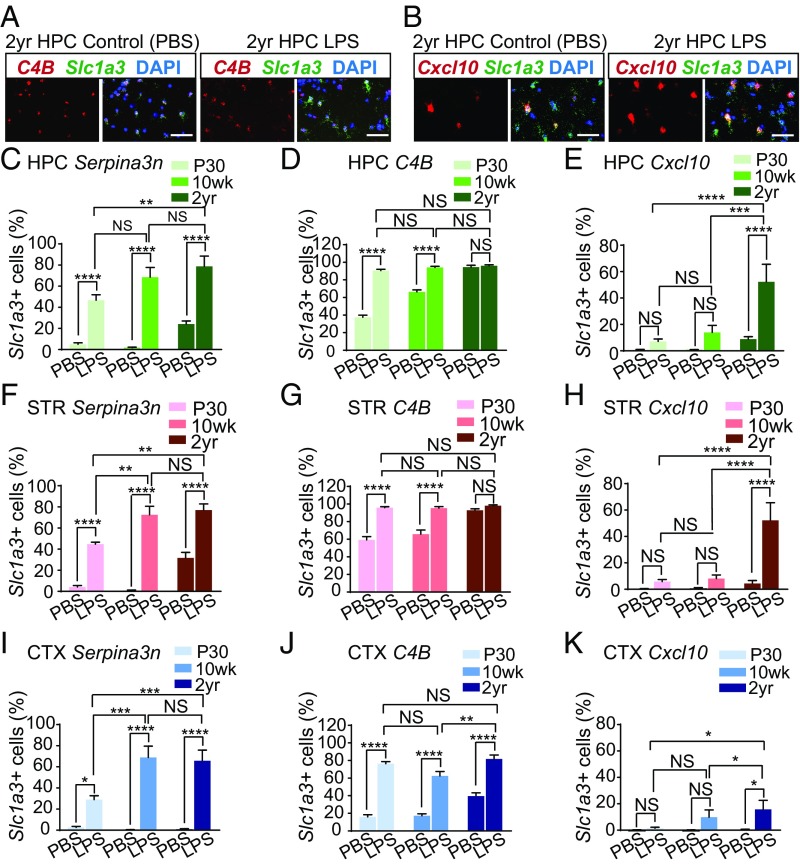
Given that normal aging induces astrocyte reactivity, we wondered if the increased expression of reactive genes could prime astrocytes to become more reactive in response to neuroinflammation. To investigate this we administered 5 mg/kg of lipopolysaccharide (LPS) or vehicle (PBS) by systemic injection to P30, 10-wk-, and 2-y-old Aldh1l1-eGFP-L10a mice, and compared astrocyte reactivity via qPCR and in situ hybridization after 24 h. We observed by qPCR that LPS treatment induces an up-regulation of A1 reactive genes in astrocytes from the hippocampus, striatum, and cortex (Fig. S4). Quantification of the fold induction of reactive genes in P30, 10-wk-, and 2-y-old mice indicated that, despite the up-regulation of many reactive genes in normal aging, the LPS-induced fold change in reactive genes was not significantly different with age (Figs. S4 and S5). Next, using in situ hybridization, we compared the gene expression changes of three of our aging-induced reactive genes (Serpina3n, C4B, and Cxcl10) in response to LPS treatment at P30, 10 wk, and 2 y of age. In accordance with our qPCR data, we observed an increase in the number of Serpina3n, Cxcl10, and C4B Slc1a3+ astrocytes in the hippocampus (Fig. S6A), striatum (Fig. S6B), and cortex (Fig. S6C). Quantification of the number of Slc1a3+ astrocytes expressing detectable levels of reactive genes in response to LPS differed with age (Fig. 5). In particular, we found that the number of astrocytes expressing C4B was not significantly increased after LPS treatment in aged (2 y) animals, since almost all aged astrocytes were C4B+ in control (PBS treated) aged animals (Fig. 5 A, D, G, and J). In contrast, LPS significantly increased the number of Serpina3n+ astrocytes at 2 y compared with younger animals in the hippocampus (47 ± 5% and 79 ± 9% at P30 and 2 y, respectively), striatum (45 ± 2% and 77 ± 6% at P30 and 2 y, respectively), and cortex (29 ± 4% and 66 ± 10% at P30 and 2 y, respectively) (Fig. 5 C, F, and I). Similarly, LPS significantly increased the number of Cxcl10+ astrocytes at 2 y of age, compared with younger animals in the hippocampus (7 ± 2% and 52 ± 13% at P30 and 2 y, respectively), striatum (6 ± 2% and 39 ± 11% at P30 and 2 y, respectively), and cortex (1 ± 1% and 16 ± 7% at P30 and 2 y, respectively) (Fig. 5 B, E, H, and K). Together, these data suggest that aging leads to a significant increase in the number of astrocytes expressing reactive genes in response to neuroinflammation.
Fig. 5.
Comparison of the induction of reactive astrocyte genes in response to LPS treatment with age. (A) Representative images of C4B colocalization with Slc1a3+ astrocytes, by in situ hybridization, in the hippocampus of aged (2 y old) PBS-injected and LPS-treated (5 mg/kg) mice. (Scale bars, 50 µm.) (B) Representative images of Cxcl10 colocalization with Slc1a3+ astrocytes, by in situ hybridization, in the hippocampus of mature adult (10 wk) and aged (2 y old) LPS-treated (5 mg/kg) mice. (Scale bars, 50 µm.) (C–E) Bar graphs comparing the number of hippocampal Slc1a3+ astrocytes expressing reactive astrocyte transcripts [(C) Serpina3n, (D) C4B, and (E) Cxcl10] in PBS-injected and LPS-treated mice at P30, 10 wk, and 2 y of age. (F–H) Bar graphs comparing the number of striatal Slc1a3+ astrocytes expressing reactive astrocyte transcripts [(F) Serpina3n, (G) C4B, and (H) Cxcl10] in PBS-injected and LPS-treated mice at P30, 10 wk, and 2 y of age. (I–K) Bar graphs comparing the number of striatal Slc1a3+ astrocytes expressing reactive astrocyte transcripts [(I) Serpina3n, (J) C4B, and (K) Cxcl10] in PBS-injected and LPS-treated mice at P30, 10 wk, and 2 y of age. Error bars depict mean ± SEM. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; n = 3 animals. See also Figs. S3–S6.
Discussion
New Resource for Understanding the Transcriptional Changes Occurring in Aged Astrocytes.
The mechanisms responsible for the cognitive decline of normal aging are unknown. Although many studies have previously focused on neuronal changes accompanying aging, given the active roles of astrocytes in controlling neuronal function, here we have focused on investigating changes that occur in aging astrocytes. We performed RNAseq at five ages across the lifespan of the mouse to determine the aging-induced transcriptional changes occurring in astrocytes. We also compared aging-astrocyte transcriptomes from three functionally different brain regions (the hippocampus, striatum, and cortex), to investigate brain-region−specific changes in normal aging. Our data show that aged astrocytes significantly up-regulate a cassette of potentially detrimental A1 reactive genes. We found significant heterogeneity in the aging-induced changes in gene expression between brain areas. In the brain regions more vulnerable to neurodegeneration—the hippocampus and striatum—astrocytes up-regulated more reactive genes, compared with the cortex. These RNAseq profiles of astrocytes throughout age are publicly accessible through brainrnaseq.org. This resource provides a new resource that will be helpful for the generation and testing of new hypotheses about the role of astrocytes during normal aging.
A1-Like Reactive Astrocytes Appear with Normal Brain Aging.
Previous studies have used standard astrocyte marker genes such as GFAP and vimentin to assess the development of astrocyte reactivity with aging (18, 19). Here, by analyzing expression of a much larger number of characteristic reactive astrocyte genes, we have found that a surprisingly high percentage of astrocytes take on a partially reactive phenotype with aging. In some brain regions, the majority of astrocytes were affected. Detailed analysis of the differentially expressed genes in aging revealed that aged astrocytes take on a reactive phenotype characteristic of neuroinflammatory A1-like reactive astrocytes. In addition to the up-regulation of many A1 genes, we also observed an up-regulation of characteristic A2 genes in aged astrocytes. This suggests that additional signals may be present in the aged brain, which likely induce the expression of A1 and A2 genes. The increased expression of potentially detrimental A1 genes may exert adverse effects on the neural circuits, since A1 astrocytes lose many normal astrocyte functions such as promoting neuronal survival, produce complement components, and decrease excitatory neuronal function, as well as releasing a toxic factor that kills both neurons and oligodendrocytes (9).
Our study also reveals that the brain regions known to be most vulnerable in neurodegenerative diseases—the hippocampus and striatum—are also the brain regions where astrocytes undergo the more dramatic changes in their transcriptomes, and up-regulate more A1 reactive genes (27, 31). Distinct cortical regions are also vulnerable in neurodegenerative diseases, including Alzheimer’s disease and frontotemporal dementia (32, 33); however, our RNAseq and in situ hybridization experiments indicated that fewer reactive astrocyte genes were up-regulated in cortical regions. Since we did not characterize astrocyte reactivity in detail throughout all cortical regions, it is possible that subsets of astrocytes become more reactive in specific cortical regions associated with cognitive decline; future studies will be required to determine this. Astrocyte heterogeneity has been previously reported in the developing spinal cord (34), and in young adult mice (35, 36), but our study characterizes astrocyte heterogeneity in the aged brain. The heterogeneity in the aging astrocyte transcriptome reported here raises the question of why astrocytes become more reactive in the hippocampus and striatum. Clues from studies of the neurons in these brain regions suggest that they are more sensitive to oxidative stress, metabolic impairments, protein aggregation, mitochondrial instability, and the dysregulation of ion homeostasis (27, 28). Future work will be required to determine whether astrocyte heterogeneity, in the healthy brain or in aging and disease, is intrinsically specified or driven by the local environment.
The finding that astrocytes up-regulate reactive genes in normal aging has important implications for the vulnerability of the aged brain to disease. The aging brain is well documented to be much more vulnerable to injuries and neurodegenerative diseases. Previous studies found neuroinflammation is increased in severity in the aging CNS following peripheral and central immune challenges with LPS (37, 38). Consistent with this, we found that the astrocyte response to LPS-induced neuroinflammation also differed with age. In particular, we found that the number of astrocytes expressing Serpina3n and Cxcl10 in response to LPS treatment was dramatically increased in aged mice, compared with younger mice. This elevation in the number of astrocytes expressing Cxcl10 may lead to the recruitment of a larger number of T lymphocytes (39, 40), exacerbating neuroinflammation after injury.
Furthermore, A1 astrocytes are present in many neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, ALS, and multiple sclerosis (9, 10). The increase in A1-like astrocytes with aging could therefore play a role in enhancing susceptibility of aging brains to neurodegeneration. Indeed, A1 reactive astrocytes are a major source of the classical complement cascade components needed to drive complement-mediated synapse loss, such as C4B and C3. Thus, as many synapses are already highly C1q-coated in the aging brain (30), the appearance of A1 reactive astrocytes with aging would be expected to trigger some synapse loss consistent with the recently reported finding of complement-mediated synapse loss in the aging hippocampus (41).
Do Activated Microglia Induce A1-Like Reactive Astrocytes with Normal Brain Aging?
Previous studies have found that microglia increase in number and become more activated during normal CNS aging (42). Our findings strongly imply that activated microglia formed during aging are responsible for the induction of A1-like reactive astrocytes during normal CNS aging. We found that the aging-induced up-regulation of reactive astrocyte genes was significantly reduced in mice lacking the microglial-secreted cytokines (IL-1α, TNF, and C1q) known to activate astrocytes (9), indicating that microglia play a key role in promoting astrocyte activation in aging. Furthermore, studies have suggested that aged microglia up-regulate their expression of inflammatory cytokines (16, 30, 43). These findings raise the question of why microglia become activated in the aged brain. A recent study found that myelin fragments are released from aging myelin sheaths, and these fragments are cleared by microglia. Myelin fragmentation is significantly increased with age, and aged microglia become overloaded with insoluble lysosomal inclusions, which may contribute to microglial senescence and immune dysfunction in the normal aged brain (44). In addition to the accumulation of myelin fragments, normal aging is also characterized by a buildup of misfolded proteins which form neurofibrillary tangles and amyloid plaques, albeit to a lesser extent than in patients with Alzheimer’s disease (45). The accumulation of myelin and misfolded proteins likely contribute to the impairment of synaptic plasticity reported to occur in aging (31). Indeed, since microglia eliminate synapses during development (46–48), it is possible that impairment of lysosomal degradation pathways reduces the elimination of weak or senescent synapses by microglia, contributing to a reduction in neural circuit plasticity in the aged brain. The induction of astrocyte reactivity in aging may initiate a feedback loop that further promotes neuronal dysfunction and microglial activation, leading to more cognitive decline. Perhaps the replacement of aged microglia with young microglia may promote the removal of fragmented myelin, protein aggregates, and senescent synapses, thereby enhancing synaptic plasticity and improving cognitive function in aging. In addition to the increase of activated microglia in the aged brain, peripheral immune cells are reported to increase in number in the brain (49). Since these cells also release inflammatory cytokines, they could contribute to inducing astrocyte activation in the aged brain and exacerbate cognitive decline.
In summary, our transcriptomic and in situ hybridization approach identified aging-induced transcriptional changes in astrocytes. Furthermore, our data indicate that microglia play a key role in promoting aging-induced astrocyte activation. The aging transcriptomics database reported here provides an important resource for future studies exploring the role astrocytes play in cognitive decline in normal aging. Future studies could investigate the functional role of these aging-induced astrocyte genes on neural circuits and investigate whether these aging-induced changes in astrocytes and microglia drive the cognitive impairments in the aged brain. Finally, by providing evidence for normal aging-induced changes in astrocytes, our data may identify targets for the treatment of cognitive decline in diseases of aging.
Materials and Methods
Contact for Reagent and Resource Sharing.
Individual replicates for all mapped RNAseq data are provided in Dataset S4). Data can also be accessed via this website link: brainrnaseq.org. All raw sequencing data are publicly available at National Center for Biotechnology Information (NCBI) BioProject, https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA417856).
Experimental Model and Subject Details.
All animal procedures were conducted in accordance with the National Institute of Health Stanford University’s Administrative Panel on Laboratory Animal Care. All mice were housed with food and water available ad libitum in a 12-h light/dark environment. Data for experiments were collected from P7, P30, 10-wk-, 9.5-mo-, and 2-y-old mice for RNAseq analysis (data were collected in triplicate for each region, except for the 9.5 mo age, which were collected in duplicate), 10-wk-old mice for immunohistochemistry, and 10-wk- and 2-y-old mice for RNAscope in situ hybridization and qPCR experiments (data were collected in triplicate). Both male and female mice were used in all experiments.
Animals.
WT C57BL/6J mice were from Jackson Laboratories, and aged mice were obtained from the National Institute of Aging. Aldh1l1-eGFP-L10a mice were obtained from D. Rowitch. Triple knockout (Il1α−/−TNF−/−C1qa−/−) animals were from a previous study in our laboratory (9). All lines were maintained by breeding with C57BL/6 mice.
TRAP RNA Isolation.
RNA was collected from hippocampi, striata, and cortex of Aldh1l1-eGFP-L10a mice based on published protocols (21, 22).
RNAseq Library Construction, Sequencing, and Analysis.
RNAseq libraries were made from total RNA isolated from whole-brain input and TRAP-isolated samples. Libraries were sequenced by the Illumina NextSeq sequencer to obtain 75 bp paired-end reads. We used https://usegalaxy.org to run the Tuxedo pipeline. Reads were aligned and mapped to mm9 mouse reference genome (50). We downloaded gene expression (FPKM data) for each sample and merged these into Datasets S1–S4. See Supporting Information for detailed methods.
Differential Expression Analysis.
To calculate fold change differences between samples, we used edgeR (51). To generate raw read counts from TopHat accepted hits output, we used featureCounts (52), an Rsubread package (53). We then shipped the output tables to edgeR for pairwise comparisons between 10-wk- and 2-y-old samples. For each comparison, we filtered out low sequence tags (genes for which two or more sample replicates at each time point had more than 25 counts per million). We merged the output of these comparisons with the FPKM data, displayed in Datasets S1–S3.
Immunohistochemistry.
Mice were anesthetized with a ketamine (100 mg/kg), and perfused with ice-cold PBS followed by ice-cold 4% paraformaldehyde at ∼70% cardiac output. Dissected brains were postfixed overnight in 4% paraformaldehyde at 4 °C, and cryoprotected in 30% sucrose. Brains were embedded in optimal cutting temperature (OCT) compound (Tissue-Tek) and 12- to 14-μm tissue sections were prepared. The following antibodies were used: Rabbit anti-S100beta antibody (ab868, 1:500; Abcam) and Mouse anti-GFAP antibody (G3893, 1:2,000; Sigma Aldrich). Primary antibodies were visualized with appropriate secondary antibodies conjugated with Alexa fluorophores (Invitrogen).
RNAscope in Situ Hybridization.
RNAscope fluorescent in situ hybridization was performed on fresh-frozen tissue. Mice were anesthetized and decapitated, and the brains were rapidly extracted into ice-cold PBS. Brains were embedded in OCT compound (Tissue-Tek) and 12- to 14-μm tissue sections were prepared. Multiplex RNAscope was performed based on manufacturer’s instructions. Probes against the following mRNA were used: Slc1a3, Serpina3n, C4B, C3, Cxcl10, and Emp1 (ACD).
Microfluidics-Based qPCR.
Total RNA was extracted from whole-brain or TRAP-isolated samples, cDNA synthesis was performed, and qPCR reactions were carried out with the BioMark HD Real-Time PCR System (Fluidigm). For protocol and primer sequences see Supporting Information.
Imaging and Image Processing.
Epifluorescence images were acquired with Zeiss Axio Imager M1 and Axiovision with 20× 0.8 NA Plan Apo objective. Images were imported into Image J. Lipofuscin autofluorescence was imaged in the blank green (488 wavelength) image channel and subtracted from red (594 wavelength) and far-red (647 wavelength) channel images. Images were adjusted for brightness and black values.
Statistical Analysis and Power Calculations.
All statistical analyses were done using GraphPad Prism 7.00 software. Most data were analyzed by one-way ANOVA followed by Dunnett’s multiple post hoc test for comparing more than three samples, and two-sample unpaired t tests for comparing two samples with 95% confidence.
Supplementary Material
Acknowledgments
We thank all members of the B.A.B. laboratory, especially S. Sloan, for help with RNAseq library preparation; and M. Bennett and F. C. Bennett for careful review and comments on the manuscript. We thank D. Rowitch (University of California, San Francisco) for the Aldh1l1-eGFP-L10a mice. We thank V. Coates and S. Coates for their generous support. We thank B.A.B. for his wonderful mentorship over the years. This work was supported by the National Institutes of Health (Grant R01 AG048814, to B.A.B.), Glenn Foundation (B.A.B.), Esther B. O’Keeffe Charitable Foundation (B.A.B.), and JPB Foundation (B.A.B.). L.E.C. was supported by a postdoctoral fellowship from European Molecular Biology Organization and a Merck Research Laboratories postdoctoral fellowship (administered by the Life Science Research Foundation). S.A.L. was supported by a postdoctoral fellowship from the Australian National Health and Medical Research Council (GNT1052961), and the Glenn Foundation Glenn Award.
Footnotes
Conflict of interest statement: B.A.B. was a cofounder of Annexon Biosciences Inc., a company working to make new drugs for treatment of neurological diseases.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the BioProject database, https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA417856).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800165115/-/DCSupplemental.
References
- 1.Christopherson KS, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 2.Kucukdereli H, et al. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci USA. 2011;108:E440–E449. doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen NJ, et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung WS, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banker GA. Trophic interactions between astroglial cells and hippocampal neurons in culture. Science. 1980;209:809–810. doi: 10.1126/science.7403847. [DOI] [PubMed] [Google Scholar]
- 6.Rothstein JD, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 7.Bush TG, et al. Leukocyte infiltration, neuronal degeneration, and neurite outgrowth after ablation of scar-forming, reactive astrocytes in adult transgenic mice. Neuron. 1999;23:297–308. doi: 10.1016/s0896-6273(00)80781-3. [DOI] [PubMed] [Google Scholar]
- 8.Sekar S, et al. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging. 2015;36:583–591. doi: 10.1016/j.neurobiolaging.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, et al. Alzheimer’s Disease Neuroimaging Initiative ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549:523–527. doi: 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallmann AL, et al. Astrocyte pathology in a human neural stem cell model of frontotemporal dementia caused by mutant TAU protein. Sci Rep. 2017;7:42991. doi: 10.1038/srep42991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 14.Soreq L, et al. UK Brain Expression Consortium; North American Brain Expression Consortium Major shifts in glial regional identity are a transcriptional hallmark of human brain aging. Cell Rep. 2017;18:557–570. doi: 10.1016/j.celrep.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabert K, et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016;19:504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norden DM, Godbout JP. Review: Microglia of the aged brain: Primed to be activated and resistant to regulation. Neuropathol Appl Neurobiol. 2013;39:19–34. doi: 10.1111/j.1365-2990.2012.01306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols NR, Day JR, Laping NJ, Johnson SA, Finch CE. GFAP mRNA increases with age in rat and human brain. Neurobiol Aging. 1993;14:421–429. doi: 10.1016/0197-4580(93)90100-p. [DOI] [PubMed] [Google Scholar]
- 19.Porchet R, et al. Analysis of glial acidic fibrillary protein in the human entorhinal cortex during aging and in Alzheimer’s disease. Proteomics. 2003;3:1476–1485. doi: 10.1002/pmic.200300456. [DOI] [PubMed] [Google Scholar]
- 20.Zamanian JL, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann JE, et al. STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J Neurosci. 2008;28:7231–7243. doi: 10.1523/JNEUROSCI.1709-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson MA, et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature. 2016;532:195–200. doi: 10.1038/nature17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: From stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 28.Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7:278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh SK, et al. Astrocytes assemble thalamocortical synapses by bridging NRX1α and NL1 via Hevin. Cell. 2016;164:183–196. doi: 10.1016/j.cell.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephan AH, et al. A dramatic increase of C1q protein in the CNS during normal aging. J Neurosci. 2013;33:13460–13474. doi: 10.1523/JNEUROSCI.1333-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burke SN, Barnes CA. Neural plasticity in the ageing brain. Nat Rev Neurosci. 2006;7:30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- 32.Leinonen V, et al. Amyloid and tau proteins in cortical brain biopsy and Alzheimer’s disease. Ann Neurol. 2010;68:446–453. doi: 10.1002/ana.22100. [DOI] [PubMed] [Google Scholar]
- 33.Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet. 2015;386:1672–1682. doi: 10.1016/S0140-6736(15)00461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molofsky AV, et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature. 2014;509:189–194. doi: 10.1038/nature13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chai H, et al. Neural circuit-specialized astrocytes: Transcriptomic, proteomic, morphological, and functional evidence. Neuron. 2017;95:531–549.e9. doi: 10.1016/j.neuron.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morel L, et al. Molecular and functional properties of regional astrocytes in the adult brain. J Neurosci. 2017;37:8706–8717. doi: 10.1523/JNEUROSCI.3956-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Godbout JP, et al. Exaggerated neuroinflammation and sickness behavior in aged mice following activation of the peripheral innate immune system. FASEB J. 2005;19:1329–1331. doi: 10.1096/fj.05-3776fje. [DOI] [PubMed] [Google Scholar]
- 38.Henry CJ, et al. Minocycline attenuates lipopolysaccharide (LPS)-induced neuroinflammation, sickness behavior, and anhedonia. J Neuroinflammation. 2008;5:15. doi: 10.1186/1742-2094-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein RS, et al. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. J Virol. 2005;79:11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hennessy E, Griffin EW, Cunningham C. Astrocytes are primed by chronic neurodegeneration to produce exaggerated chemokine and cell infiltration responses to acute stimulation with the cytokines IL-1β and TNF-α. J Neurosci. 2015;35:8411–8422. doi: 10.1523/JNEUROSCI.2745-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Q, et al. Complement C3-deficient mice fail to display age-related hippocampal decline. J Neurosci. 2015;35:13029–13042. doi: 10.1523/JNEUROSCI.1698-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosher KI, Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer’s disease. Biochem Pharmacol. 2014;88:594–604. doi: 10.1016/j.bcp.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sierra A, Gottfried-Blackmore AC, McEwen BS, Bulloch K. Microglia derived from aging mice exhibit an altered inflammatory profile. Glia. 2007;55:412–424. doi: 10.1002/glia.20468. [DOI] [PubMed] [Google Scholar]
- 44.Safaiyan S, et al. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat Neurosci. 2016;19:995–998. doi: 10.1038/nn.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu L, et al. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 2015;84:927–934. doi: 10.1212/WNL.0000000000001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 47.Schafer DP, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bialas AR, Stevens B. TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci. 2013;16:1773–1782. doi: 10.1038/nn.3560. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Ritzel RM, et al. Age-associated resident memory CD8 T cells in the central nervous system are primed to potentiate inflammation after ischemic brain injury. J Immunol. 2016;196:3318–3330. doi: 10.4049/jimmunol.1502021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Liao Y, Smyth GK, Shi W. The subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: An interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki R, Shimodaira H. 2014 pvclust: Hierarchial clustering with P-values via multiscale bootstrap resampling. Available at cran.r-project.org/web/packages/pvclust/index.html. Accessed October 4, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.