Suppose you are a physician with a patient whose complaint could arise from multiple diseases. To attain a specific diagnosis, you might ask yourself a series of yes/no questions depending on observed features describing the patient, such as clinical test results and reported symptoms. As some questions rule out certain diagnoses early on, each answer determines which question you ask next. With about a dozen features and extensive medical knowledge, you could create a simple flow chart to connect and order these questions. If you had observations of thousands of features instead, you would probably want to automate. Machine learning methods can learn which questions to ask about these features to classify the entity they describe. Even when we lack prior knowledge, a classifier can tell us which features are most important and how they relate to, or interact with, each other. Identifying interactions with large numbers of features poses a special challenge. In PNAS, Basu et al. (1) address this problem with a new classifier based on the widely used random forest technique. The new method, an iterative random forest algorithm (iRF), increases the robustness of random forest classifiers and provides a valuable new way to identify important feature interactions.
Random forests came into the spotlight in 2001 after their description by Breiman (2). He was largely influenced by previous work, especially the similar “randomized trees” method of Amit and Geman (3), as well as Ho’s “random decision forests” (4). Random forests have since proven useful in many fields due to their high predictive accuracy (5, 6). In biology and medicine, random forests have successfully tackled a range of problems, including predicting drug response in cancer cell lines (7), identifying DNA-binding proteins (8), and localizing cancer to particular tissues from a liquid biopsy (9). Random forests have also recognized speech (10, 11) and handwritten digits (12) with high accuracy.
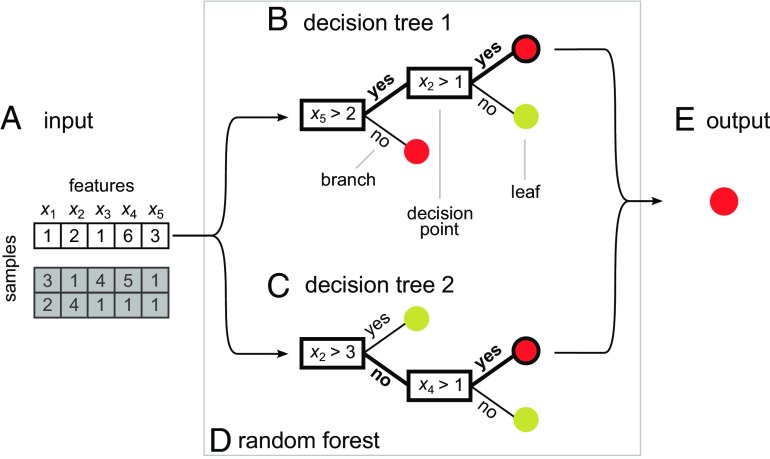
Like their real-world counterparts, random forests consist of trees. Specifically, random forests are ensembles of decision trees. Morgan and Sonquist (13) proposed the decision tree methodology in 1963, formalizing an intuitive approach to simplifying the analysis of multiple features during prediction tasks. We use the decision tree on an input dataset made up of a collection of samples, each described by features (Fig. 1A). Each sample represents an entity, such as a protein, that we want to assign to a class, such as “binds DNA” or “does not bind DNA” (8). The decision tree classifies samples through a forking path of decision points (Fig. 1B). Each decision point has a rule determining which branch to take. As we move down the tree, we stop at each decision point to apply its rule to one of the sample’s features. Eventually, we arrive at the end of the branch, or leaf. The leaf has a class label, and we conclude our path through the tree by assigning the sample to that class.
Fig. 1.
Individual decision trees vote for class outcome in a toy example random forest. (A) This input dataset characterizes three samples, in which five features (x1, x2, x3, x4, and x5) describe each sample. (B) A decision tree consists of branches that fork at decision points. Each decision point has a rule that assigns a sample to one branch or another depending on a feature value. The branches terminate in leaves belonging to either the red class or the yellow class. This decision tree classifies sample 1 to the red class. (C) Another decision tree, with different rules at each decision point. This tree also classifies sample 1 to the red class. (D) A random forest combines votes from its constituent decision trees, leading to a final class prediction. (E) The final output prediction is again the red class.
While we can easily use a decision tree’s rules to classify a sample, where do those rules come from? We can construct them using training data in which a known class accompanies each sample’s features. Our goal is to create a tree that can later predict classes correctly from the features alone. There are a variety of algorithms to train decision trees (14–16), but we will describe one of the simplest methods (17). This method minimizes the heterogeneity, or impurity, of the classes of training data assigned to each branch. First, we identify the rule that will split the training data into two branches with the least class impurity, and establish a decision point with this rule. We then further subdivide the resulting branches by creating new rules in the same way. We continue splitting until we can find no rule that further reduces class impurity. This training process generates a trained decision tree made up of multiple decision points, with each possible path through the tree terminating in a class label.
Despite ease of interpretation, decision trees often perform poorly on their own (18). We can improve accuracy by instead using an ensemble of decision trees (Fig. 1 B and C), combining votes from each (Fig. 1D). A random forest is such an ensemble, where we select the best feature for splitting at each node from a random subset of the available features (5, 18). This random selection causes the individual decision trees of a random forest to emphasize different features. The resulting diversity of trees can capture more complex feature patterns than a single decision tree and reduces the chance of overfitting to training data. In this way, the random forest improves predictive accuracy.
In addition to high predictive performance, random forest classifiers can reveal feature importance (5), telling us how much each feature contributes to class prediction. It is here where the new method of Basu et al. (1) delivers its most important advance. By weighting features according to feature importance, the authors grow more relevant trees to uncover complex interactions. To do this, they iteratively refine a random forest, leading to iRF. First, they begin with a weighted random forest, one in which each feature has equal weight, indicating an equal probability of being chosen. In the initial round, the weighted random forest behaves in the same way as Breiman’s original random forest (2). Second, they repeatedly train weighted random forests, using the feature importance from one iteration as the weights in the next. Third, they use the final weights to generate several weighted random forests, each trained on a random selection of samples. This is a bootstrap selection, meaning each sample can appear more than once. Fourth, Basu et al. (1) use the random intersection trees algorithm (19) to find subsets of features that often co-occur. Fifth, they assess the extracted interactions with a stability score averaged over all bootstrap selections. The stability score describes the fraction of times a recovered interaction occurs, with stable interactions having scores greater than 0.5. A higher stability score means it is less likely that random chance alone caused identification of the interaction.
To demonstrate iRF’s efficacy, Basu et al. (1) apply it to several genomic problems, detecting multiway interactions between chromatin-interacting proteins, both known and novel. This moves beyond popular techniques that focus on pairwise interactions. For example, they use iRF to predict genomic enhancers in Drosophila melanogaster from quantitative signal of transcription factor and histone modification presence within
In addition to high predictive performance, random forest classifiers can reveal feature importance, telling us how much each feature contributes to class prediction. It is here where the new method of Basu et al. delivers its most important advance.
each genomic region. They identify 20 pairwise transcription factor interactions, of which 16 are consistent with previously reported physical interactions. They also identify novel third-order interactions involving the early regulatory factor Zelda. This provides an intriguing path to further investigating Zelda, a link to the past reports of codependency with other factors that drive enhancer activity.
Of course, iRF provides a flexible method, whose utility extends past genomics to any classification and feature selection task. In simulations, iRF successfully detects up to order-8 interactions. At the same time, iRF maintains predictive performance similar to conventional random forests. To improve further, one might explore ways to combine iRF with other ensemble methods. As Basu et al. (1) mention, AdaBoost (20) focuses on the least reliable parts of decision trees and could complement iRF’s focus on the most reliable parts. Building on iRF in this way will prove easier due to the installable R package the authors provide, making their methodology accessible to users and extenders alike. iRF holds much promise as a new and effective way of detecting interactions in a variety of settings, and its use will help us ensure no branch or leaf is ever left unturned.
Acknowledgments
This work was supported by Natural Sciences and Engineering Research Council of Canada Grant RGPIN-2015-03948 (to M.M.H.) and a Canada Graduate Scholarship-Master’s (to D.D.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 1943.
References
- 1.Basu S, Kumbier K, Brown JB, Yu B. Iterative random forests to discover predictive and stable high-order interactions. Proc Natl Acad Sci USA. 2018 doi: 10.1073/pnas.1711236115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 3.Amit Y, Geman D. Shape quantization and recognition with randomized trees. Neural Comput. 1997;9:1545–1588. [Google Scholar]
- 4.Ho TK. 1995. Random decision forests. Proceedings of the Third International Conference on Document Analysis and Recognition (IEEE Computer Society, Los Alamitos, CA), Vol 1, pp 278–282.
- 5.Touw WG, et al. Data mining in the life sciences with Random Forest: a walk in the park or lost in the jungle? Brief Bioinform. 2013;14:315–326. doi: 10.1093/bib/bbs034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verikas A, Gelzinis A, Bacauskiene M. Mining data with random forests: a survey and results of new tests. Pattern Recognit. 2011;44:330–349. [Google Scholar]
- 7.Riddick G, et al. Predicting in vitro drug sensitivity using Random Forests. Bioinformatics. 2011;27:220–224. doi: 10.1093/bioinformatics/btq628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nimrod G, Szilágyi A, Leslie C, Ben-Tal N. Identification of DNA-binding proteins using structural, electrostatic and evolutionary features. J Mol Biol. 2009;387:1040–1053. doi: 10.1016/j.jmb.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JD, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. January 18, 2018 doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue J, Zhao Y. Random forests of phonetic decision trees for acoustic modeling in conversational speech recognition. IEEE Trans Audio Speech Lang Process. 2008;16:519–528. [Google Scholar]
- 11.Su Y, Jelinek F, Khudanpur S. INTERSPEECH 2007. International Speech Communication Association; Baixas, France: 2007. Large-scale random forest language models for speech recognition; pp. 598–601. [Google Scholar]
- 12.Bernard S, Adam S, Heutte L. Ninth International Conference on Document Analysis and Recognition. IEEE Computer Society; Los Alamitos, CA: 2007. Using random forests for handwritten digit recognition; pp. 1043–1047. [Google Scholar]
- 13.Morgan JN, Sonquist JA. Problems in the analysis of survey data, and a proposal. J Am Stat Assoc. 1963;58:415–434. [Google Scholar]
- 14.Morgan JN, Messenger RC. 1973. THAID: A Sequential Analysis Program for the Analysis of Nominal Scale Dependent Variables (Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, MI)
- 15.Meisel WS, Michalopoulos DA. A partitioning algorithm with application in pattern classification and the optimization of decision trees. IEEE Trans Comput. 1973;C-22:93–103. [Google Scholar]
- 16.Gordon L, Olshen RA. Asymptotically efficient solutions to the classification problem. Ann Stat. 1978;6:515–533. [Google Scholar]
- 17.Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and Regression Trees. CRC Press; Boca Raton, FL: 1984. [Google Scholar]
- 18.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning: With Applications in R. Springer; New York: 2014. [Google Scholar]
- 19.Shah RD, Meinshausen N. Random intersection trees. J Mach Learn Res. 2014;15:629–654. [Google Scholar]
- 20.Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci. 1997;55:119–139. [Google Scholar]