Abstract
Trypanosomes are protistan parasites that diverged early in evolution from most eukaryotes. Their streamlined genomes are packed with arrays of tandemly linked genes that are transcribed polycistronically by RNA polymerase (pol) II. Individual mRNAs are processed from pre-mRNA by spliced leader (SL) trans splicing and polyadenylation. While there is no strong evidence that general transcription factors are needed for transcription initiation at these gene arrays, a RNA pol II transcription pre-initiation complex (PIC) is formed on promoters of SLRNA genes, which encode the small nuclear SL RNA, the SL donor in trans splicing. The factors that form the PIC are extremely divergent orthologues of the small nuclear RNA-activating complex, TBP, TFIIA, TFIIB, TFIIH, TFIIE and Mediator. Here, we functionally characterized a heterodimeric complex of unannotated, nuclear proteins that interacts with RNA pol II and is essential for PIC formation, SL RNA synthesis in vivo, SLRNA transcription in vitro, and parasite viability. These functional attributes suggest that the factor represents TFIIF although the amino acid sequences are too divergent to firmly make this conclusion. This work strongly indicates that early-diverged trypanosomes have orthologues of each and every general transcription factor, requiring them for the synthesis of SL RNA.
INTRODUCTION
In eukaryotes, RNA pol II core promoters direct the formation of the transcription pre-initiation complex (PIC) which comprises the general transcription factors TFIID, TFIIA, TFIIB, TFIIE, TFIIF, TFIIH and the polymerase. In addition, the large Mediator complex has an important role in PIC formation and stability (1). In a first step, core promoters of protein coding genes are recognized and bound by TFIID which consists of the TATA-binding protein (TBP) and several TBP-associated factors whereas promoters of small RNA genes are contacted by a different DNA binding factor, namely the small nuclear RNA-activating protein complex or SNAPc (2,3). Overall, PIC formation is highly conserved and the structures of the core PIC in the yeast Saccharomyces cerevisiae and in humans are nearly identical (4).
The protist Trypanosoma brucei is a lethal parasite of humans in sub-Saharan Africa and a member of the phylogenetic family Trypanosomatidae which harbors other important parasites such as Trypanosoma cruzi and Leishmania spp. (5). Trypanosomatids diverged very early in evolution from most eukaryotes and they express their genes in an unusual manner. Their protein coding genes are arranged in long dense tandem arrays which are transcribed polycistronically by RNA polymerase (pol) II, and individual mRNA molecules are processed from pre-mRNA by spliced leader (SL) trans splicing and polyadenylation (6,7). In trans splicing, the capped 5/ portion of the small nuclear SL RNA, the SL or mini-exon, is fused to the 5/ end of every mRNA. Since SL RNA is consumed in the process, trypanosome viability crucially depends on strong and continuous SL RNA production. T. brucei harbors approximately 100 tandem SLRNA gene copies on chromosome 9 whose expression ensures sustained gene expression in the parasite. While trypanosome genes of small cytoplasmic and nuclear RNAs are transcribed by RNA pol III (8,9), SLRNAs are transcribed by RNA pol II (10). However, in contrast to protein coding gene arrays, SLRNAs are transcribed monocistronically. Each gene copy has its own promoter that recruits RNA pol II to a distinct transcription initiation site. Conversely, transcription of protein coding gene arrays appears to start at multiple positions over broad regions either in divergent strand switch regions in which gene arrays are arranged head-to-head or in non-divergent regions upstream of an array (11,12).
Trypanosomes form a PIC at the SLRNA promoter (13,14). Identification of the general transcription factors, however, proved to be difficult because their amino acid sequences are extremely divergent from those of their eukaryotic counterparts - genome annotation merely identified TBP [published and previously referred to as TBP-related factor 4 (15)] and the two TFIIH helicases Xeroderma pigmentosum B (XPB) and XPD (16). Identification of other factors and subunits relied predominantly on biochemical and genetic methods. Consistent with SLRNA encoding a small nuclear RNA, the first transcription factor purified and characterized was a trimeric SNAPc that formed a larger complex with TBP, the small subunit of TFIIA (TFIIA2), and a sixth protein, originally termed TFIIA1, whose orthologous status remains uncertain (17,18). This breakthrough was followed by the identification of trypanosome TFIIB (19,20) and the biochemical characterization of TFIIH. Unlike human and yeast TFIIH, which consists of a core of seven subunits and a trimeric cyclin-dependent kinase 7 (CDK7; in yeast Kin28) complex, trypanosome TFIIH comprised a full core complex but was not associated with a kinase (21–23). Instead, the complex was stably associated with two other proteins which likely represent the heterodimeric TFIIE (23). Finally, a TFIIH-associated complex of nine subunits was discovered that exhibited no motif or sequence conservation that could reveal its identity. However, molecular structure analysis of the purified complex by electron microscopy (EM) and its functional role in stabilizing the PIC, identified the factor as the trypanosome Mediator complex MED-T which structurally resembles the head module of the much larger Mediator complex of other eukaryotes (24). As shown by gene knockdowns, in vitro transcription assays and chromatin precipitation (ChIP) analyses, these factors assemble at the SLRNA promoter and are indispensable for SLRNA transcription and trypanosome viability. It is not known whether any of these factors is required for transcription of protein coding genes.
The missing general transcription factor in trypanosomes is TFIIF. Eukaryotic TFIIF is a heterodimer (mammals) or trimer (yeast) that binds tightly to RNA pol II which is recruited to the PIC as a RNA pol II-TFIIF complex by TFIIB (25,26). Two distinct roles in transcription initiation have been associated with TFIIF. Firstly, TFIIF is critical for the formation of a stable PIC, in particular for recruitment, retention and positioning of TFIIB, and may function in initial transcriptional steps such as open complex formation, determination of the correct transcription initiation site and promoter escape of RNA pol II (27–30). Secondly, by recruiting and stimulating the phosphatase FCP1, yeast TFIIF is involved in dephosphorylation of the CTD of the largest RNA pol II subunit RPB1 during the transcription cycle (31). Conserved from yeast to humans this CTD harbors 26–52 repeats of the heptad motif YSPTSPS. Phosphorylation and dephosphorylation of this motif regulate the transcription cycle (32,33). The CTD in trypanosomes does not contain the heptad or other repetitive motifs and a CDK7 ortholog seems to be missing in these organisms (34). On the other hand, the CTD was shown to be essential for RNA pol II transcription in vivo (35,36), 17 phospho-sites were identified within the CTD (37), and evidence obtained in the related organism Trypanosoma cruzi suggested that RNA pol II association with chromatin requires the CTD to be phosphorylated (38).
Here, tandem affinity purification of T. brucei RNA pol II revealed for the first time all twelve subunits (RPB1–12), several proteins that are known to associate with the enzyme as well as four proteins of unknown function. We show that two of the latter form a heterodimeric complex which localizes to the nucleus, occupies the SLRNA promoter, and is essential for trypanosome viability and SLRNA transcription. Moreover, depletion of this complex significantly decreased the presence of TFIIB at the SLRNA promoter, indicating that the complex is important for stable PIC formation. While all these attributes are in accordance with the complex being trypanosome TFIIF, the amino acid sequences have no resemblance to TFIIF subunit sequences of other eukaryotes. Accordingly, we termed the complex tentatively TFIIF-like or TFL. Since we additionally provide structure modeling evidence that the trypanosome TFIIH subunit TSP2 is potentially the homolog of TFIIEα, this report strongly indicates that trypanosomatids possess a full set of RNA pol II general transcription factors.
MATERIALS AND METHODS
DNA
pRPB9-PTP-NEO was generated by inserting 383 bp of the C-terminal RPB9 coding region (position 17 to position 399 relative to the translation initiation codon) into the pC-PTP-NEO tagging construct (39), using ApaI and NotI restriction sites (please note that the PTP tag consists of a protein C epitope [ProtC], a tobacco etch virus [TEV] protease site, and a tandem protein A domain [ProtA]). By introducing the silent T174G mutation, a unique restriction site for AflII was engineered into the cloned RPB9 sequence. pTFL1-PTP-NEO and pTFL2-PTP-NEO were obtained correspondingly, introducing 460 bp of TFL1 (position 1101–1560) and 1002 bp of TFL2 (position 241 to position 1242) coding region into pC-PTP-NEO, respectively. For TFL1 gene silencing 330 bp of the 3/ UTR sequence (position 1564 to position 1893) were integrated in a stem-loop arrangement into the pT7-stl vector (40) to obtain plasmid T7-TFL1-stl. pT7-TFL2-stl for TFL2 silencing was obtained accordingly, integrating the coding region from nucleotide position 363 to position 863. For the expression of N-terminal glutathione-S-transferase (GST)-TFL2 fusion protein in Escherichia coli, the complete coding region of TFL2 was cloned into pGEX-6p-1 (GE Healthcare) using the vector's BamHI and EcoRI restriction sites to generate plasmid GST-TFL2. pGPEET-trm and pSLins19, the template constructs for in vitro transcription assays, have been described in detail previously (41,42).
Trypanosomes
Culturing and stable transfection of procyclic T. brucei brucei strain 427 wild-type cells and the genetically modified cell line 29–13 (43) were carried out as described previously (44). Transfected cells were cloned by limiting dilution. Cell line TbRPB9ee was generated by targeted insertion of AflII-linearized pRPB9-PTP-NEO into one RPB9 allele and by knocking out the remaining wild-type allele with a PCR product of the hygromycin phosphotransferase coding sequence surrounded by 100 bp of RPB9 5/ and 3/ gene flanks. Cell line TbT2PTPee was generated accordingly with SmaI-linearized pTFL2-PTP-NEO and a corresponding hygromycin resistance-promoting PCR product. Procyclic cells that express TFL1-PTP were generated by transfecting BbS1-linearized pTFL1-PTP-Neo. For conditional TFL1 and TFL2 silencing experiments, pT7-TFL1-stl and pT7-TFL2-stl were linearized with SacII and EcoRV, respectively, transfected into 29–13 cells and targeted for integration into the transcriptionally silent spacer region of the rRNA gene locus. Transfected cells were selected with phleomycin. The concentrations of selecting antibiotics in medium was 40 μg/ml of G418, 40 μg/ml of hygromycin and 2.5 μg/ml of phleomycin. For each transfection, correct DNA integration was confirmed by PCR of genomic DNA with at least one oligonucleotide hybridizing outside of the transfected nucleotide sequence. Conditional gene silencing experiments were performed by incubating trypanosomes with medium containing 2 μg/ml of doxycycline. Culture growth was monitored by a Z1 Coulter Particle Counter (Beckman Coulter), and cells were diluted daily to a density of 2 × 106 cells/ml.
RNA analysis
To compare RNA amounts between uninduced and gene-silenced trypanosomes, total RNA was prepared from 108 cells using the Trizol reagent (ThermoFisher Scientific) and reverse-transcribed with SuperScript II (ThermoFisher Scientific) and random hexamer or Oligo (dT)20 primers both according to the manufacturer's protocol. cDNA sequences were then subjected to semi-quantitative and quantitative PCR (qPCR). For each semi-quantitative PCR, the number of cycles for the linear amplification range was determined empirically. qPCR reactions were carried out with the SsoFast EvaGreen Supermix (Biorad) on a CFX96 cycler (Biorad). Suitability of each oligonucleotide pair was determined by standard agarose gel electrophoresis, melting curve analysis, and a standard curve obtained from serial dilutions of DNA samples whose coefficient of determination (R2) needed to be in the range of 0.98–1.0. Relative abundances of SL RNA and U2 snRNA were assayed by primer extension of total RNA using 5/-P32-endlabeled oligonucleotides SL_PE and U2f. Sequences of oligonucleotides that were used in qPCR and primer extension are listed in Supplementary Table S1.
Newly synthesized RNA was labeled in lysolecithin-permeablized, procyclic cells (45), by incubating these cells with transcription cocktail (20 mM HEPES–KOH, pH 7.7, 20 mM potassium l-glutamate, 3 mM MgCl2, 1 mM dithiothreitol (DTT), 25 mM creatine phosphate, 0.6 mg/ml creatine kinase, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM CTP, 1 mM GTP, 2 mM ATP, 5 μM UTP and 100 μCi of [α-32P]UTP [3000Ci/mol]) for 8 min at 28°C. Total RNA from these reactions was separated electrophoretically in 6% acrylamide/50% urea gels, and labeled RNA visualized through autoradiography.
Tandem affinity purification and sedimentation analysis
Preparation of extract and tandem affinity purification of PTP-tagged RPB9 and TFL1 was carried out exactly as described previously (39). Purified proteins were separated on denaturing SDS-10 to 20% polyacrylamide gradient gels (Biorad) and visualized by staining with either SYPRO Ruby (Biorad) or Gelcode Coomassie blue (Thermo Fisher Scientific) according to manufacturers’ specifications. For protein identification, individual protein bands were excised from the gel or, for a complete analysis of an eluate, the whole gel lane was divided up into seven pieces. Proteins were digested with trypsin, eluted from gel pieces and subjected to liquid chromatography–tandem mass spectrometry (LC/MS/MS) at the Keck Biotechnology Resource Laboratory of Yale University.
For sedimentation analysis, a complete final eluate of a standard PTP tandem affinity purification was vacuum-concentrated from a volume of 1.8 ml to a volume of ∼0.6 ml, dialyzed overnight against E-80 buffer (150 mM sucrose, 20 mM HEPES–KOH, pH7.7, 20 mM potassium l-glutamate, 3 mM MgCl2, 80 mM KCl, 0.2 mM EDTA, 0.5 mM EGTA, 0.1% Tween20) at 4°C and further concentrated to a volume of 0.2 ml in an Amicon Ultra 0.5 ml centrifugal filter device (Millipore Sigma). The eluate was then loaded onto a linear 3.8 ml 10–40% sucrose gradient in E-80 buffer and sedimented by ultra-centrifugation at 41,000 rpm in a Beckman SW55Ti rotor for 19 h at 4°C. The gradient was fractionated in twenty 0.2 ml aliquots from top to bottom. Proteins from 0.1 ml of each fraction were collected by hydrophobic Strataclean resin (Stratagene) and analyzed by denaturing SDS-PAGE while the remaining material was used for co-immunoprecipitation (IP) and in in vitro transcription assays (see below).
Antibodies, immunoblotting and co-IPs
To raise an anti-TFL2 immune serum in rats, we transformed the BL21 strain of Escherichia coli with pGST-TFL2 and purified recombinant TFL2-GST from BL21 extract by glutathione affinity chromatography (GE Healthcare) following the manufacturer's protocol. The recombinant protein was then used to immunize Sprague-Dawley rats as described previously (20) and according to an animal use protocol that was approved by the UConn Health Institutional Animal Care and Use Committee (Public Health Service [PHS] assurance number A3471-01) and in accordance with the PHS Policy for the Humane Care and Use of Vertebrate Animals and the Guide for the Care and Use of Laboratory Animals.
For immunoblot analyses, proteins were separated by SDS-PAGE, electroblotted onto PVDF membrane and detected with the following antibodies. PTP-tagged proteins were recognized by the ProtA-specific Peroxidase Anti-Peroxidase (PAP) reagent (ThermoFisher Scientific) or by the mouse anti-ProtC monoclonal antibody HPC4 (Roche). Trypanosome TFL2 was detected with rat anti-TFL2 immune serum (this study), RPB1 with an equivalent immune serum (46), and α tubulin with a commercially available mouse monoclonal anti-[human] α tubulin antibody (Millipore Sigma). Primary antibodies/sera were diluted 1:2,000 except for the anti-α tubulin antibody which was diluted 1:200,000. As secondary antibodies we used peroxidase-conjugated goat anti-mouse IgG antibody (Millipore Sigma) at 1:6,000 dilution and goat anti-rat IgG antibody (ThermoFisher Scientific) at 1:8,000 dilution. Blots were developed using the BM chemiluminescence blotting substrate (Roche) according to the manufacturer's specifications.
To precipitate TFL1-PTP from trypanosome extract, 25 μl of pelleted IgG beads (GE Healthcare), which bind to ProtA of the PTP tag, were equilibrated in TET150 buffer (150 mM NaCl, 20 mM Tris–HCl, pH 8.0, 3 mM MgCl2, 0.1% Tween20), mixed into extract and incubated on ice for 30 min. After washing the beads six times with TET150, proteins were eluted by incubating the beads for 30 min at 28°C in TET150 buffer containing 0.5 mM EDTA, 0.1 mM DTT, and 20 units of AcTEV protease (ThermoFisher Scientific). IP of untagged TFL2 from fraction 9 of the sucrose gradient was carried out basically as described above with antibodies from the anti-TFL2 immune serum immobilized on protein G Sepharose (GE Healthcare). Precipitated proteins were eluted twice with 0.2 mM glycine–HCl, pH2.0, immediately followed by pH neutralization with one tenth volume of 1 M Tris–HCl, pH 8.0.
In vitro transcription
Preparation of transcriptionally active extracts and in vitro transcription reactions were essentially performed as described previously except that the volume of extract was reduced (44). Briefly, trypanosomes were broken with a glass douncer and nuclear proteins extracted into the soluble fraction by adding 1/10 volume of extraction buffer (150 sucrose, 20 mM HEPES–KOH, pH 7.7, 1500 mM KCl, 3 mM MgCl2) to the suspension. After reducing the KCl concentration to ∼100 mM by dilution, and concentrating the extract about fourfold with a 10K centrifugal filter (Sigma Millipore), extracts were obtained with a protein concentration of ∼25 μg/μl. Transcription reactions were carried out in a volume of 40 μl for 1 h at 27°C and contained 6 μl of extract, 20 mM HEPES–KOH, pH 7.7, 20 mM potassium l-glutamate, 20 mM KCl, 3 mM MgCl2, 0.5 mM of each nucleoside triphosphate, 20 mM creatine phosphate, 0.48 mg/ml creatine kinase, 2.5% polyethylene glycol, 0.2 mM EDTA, 0.5 mM EGTA, 4 mM DTT, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 12.5 μg/ml empty vector DNA, 20 μg/ml GPEET-trm template and 7.5 μg/ml SLins19 template. In reactions with immune serum, 6 μl of extract were pre-incubated with 1 μl of serum for 30 min on ice before reactions were started. TFL2-PTP was depleted or mock-depleted from extract by IgG or protein G affinity chromatography in which 600 μl of extract was mixed with 50 μl of pelleted, E buffer-equilibrated beads. After rotating the mixtures for 1 h at 4°C, extracts were separated from beads by centrifugation and directly used in transcription reactions. SLins19 transcription was reconstituted in reactions with TFL2-PTP-depleted extract by adding 2 or 4 μl of sucrose gradient fraction 9. Transcription reactions were generally stopped by adding the Trizol reagent to prepare total RNA. SLins19 and GPEET-trm transcripts carry unique oligonucleotide tags and were analyzed in these RNA preparations by primer extension of 5′-32P-endlabeled, tag-complementary oligonucleotides SL_Tag and Tag_PE, respectively.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as described previously (21,22). Briefly, 108 procyclic trypanosomes were fixed in 1% formaldehyde for 20 min and chromatin of these cells was fragmented with a Bioruptor sonicator (Diagenode) to an average DNA length of ∼500 bp by 25 sonication cycles (30 s on/30 s off) at high settings. TFL2-PTP-crosslinked chromatin was precipitated with a ChIP-grade, rabbit polyclonal anti-ProtA antibody (Sigma) that was immobilized on protein A beads whereas chromatin fragments containing untagged TFIIB were precipitated with rat anti-TFIIB immune serum (20) immobilized on protein G beads. In anti-TFL2-PTP ChIP assays we carried out negative control precipitations with a comparable nonspecific rabbit immune serum. For these assays we calculated the percentage of precipitated DNA relative to input DNA, standardizing the data by subtraction of the percent IP of the negative control. For anti-TFIIB ChIP assays, we omitted the negative control precipitation and, instead, used PicoGreen (ThermoFisher Scientific) to quantify the concentrations of DNA prepared from input and precipitated chromatin samples, enabling us to quantify by qPCR the fold enrichment of SLRNA promoter DNA in precipitated DNA relative to that of the α tubulin coding region which served as a negative control in qPCR.
Microscopy
The cellular localization of TFL1-PTP and TFL2-PTP was determined by indirect fluorescence microscopy using a rabbit polyclonal anti-ProtA antibody (Sigma) at 1:40,000 dilution followed with the Alexa 594 conjugated secondary antibody (ThermoFisher Scientific) at 1:400 dilution as previously published (47). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) at a final concentration of 2 μg/ml. Images were acquired on laser scanning confocal microscope (LSM780, Carl Zeiss) with a 63× 1.4 numerical aperture and oil immersion objective using ZEN software and processed with ZEN lite software (Carl Zeiss).
Homology modeling
Homology models of the C-terminal domain of TFL1 (residues 464–520; Tb927.11.1040) and the N-terminal domain of TSP2 (residues 108–227; Tb11.01.5700) were generated using the PHYRE 2 protein structure prediction server (48). The amino-acid sequences of these domains underwent secondary structure prediction; the outputs were aligned and scored against the PHYRE database to produce a tertiary homology model ensemble. From this ensemble, the top scoring PHYRE homology model was used as a representative domain structure for both proteins. For TFL1, the highest confidence model (95% of residues modeled at > 72% confidence) was based on the NMR structure of the DNA-binding domain of human RAP30 (49) (protein data bank identifier [PDB ID]: 2BBY) and the crystal structure of the C-terminal domain of human RAP74 (50) (PDB ID: 1I27). For TSP2, the highest confidence model (75% of residues modeled at >68% confidence) was based on the high-resolution cryo-EM structure of the yeast TFIIEα subunit (26) (PDB ID: 5YFW).
RESULTS
Re-visiting RNA pol II tandem affinity purification (TAP)
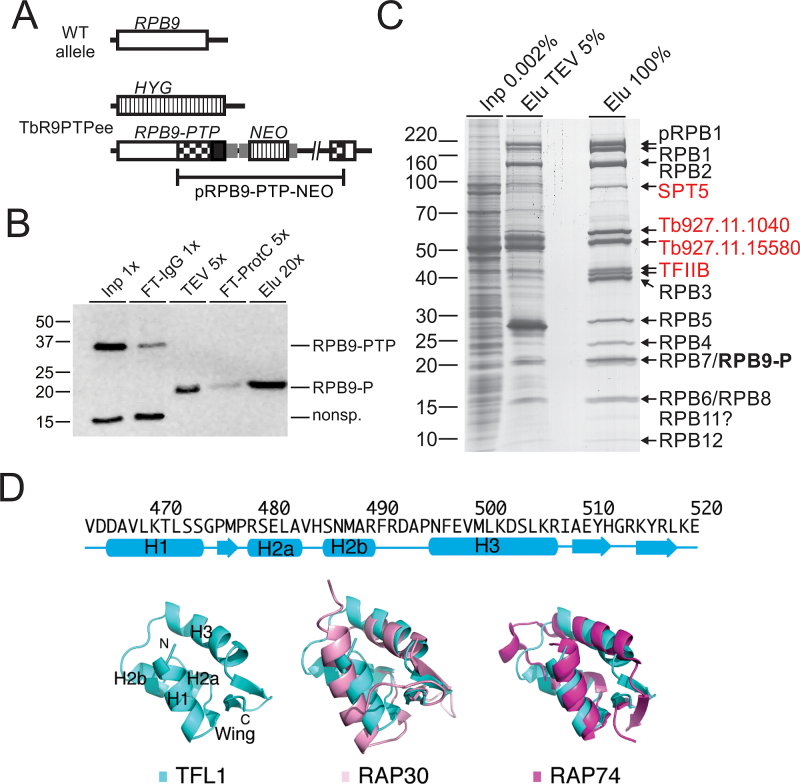
Conserved from yeast to humans, RNA pol II consists of subunits RPB1–12 and is known to be associated with TFIIB, TFIIF and several transcription elongation factors such as SPT5, SPT6, TFIIS, and FACT (facilitates chromatin transcription). In previous purifications of RNA pol II from T. brucei and the related organism Leishmania major, the original TAP tag (51) was fused to the RNA pol II-specific subunits RPB4 (52), RPB9 (N-terminal tag) (53), or RPB2 (54). In these studies, TAP revealed most but not all of the RNA pol II subunits as well as several additional proteins among which only the FACT subunit SPT16 was a known transcription factor. Since we had modified the TAP procedure based on the composite PTP tag (39), we revisited RNA pol II purification in the hope to identify new transcription-relevant proteins. For the purification we fused the PTP tag C-terminally to RPB9 (RPB9-PTP) and generated the clonal procyclic cell line TbR9PTPee, which exclusively expressed RPB9-PTP and no untagged RPB9, by two consecutive transfections. In a first step, we integrated plasmid RPB9-PTP-NEO into one RPB9 allele, fusing the PTP sequence to the RPB9 coding sequence, and, in a second transfection of a PCR amplification product that comprised the hygromycin phosphotransferase (HYG) coding region surrounded by RPB9 gene flanks, we eliminated the remaining wild-type allele (Figure 1A). Since RPB9 is an essential gene in T. brucei (53) and TbR9PTPee cells exhibited only a mild proliferation defect (Supplementary Figure S1), we concluded that C-terminally tagged RPB9 was functional.
Figure 1.
Tandem affinity purification of RNA pol II via RPB9-PTP. (A) Schematic depiction (not to scale) of a wild-type RPB9 allele (open box) and the two modified RPB9 alleles in TbR9PTPee trypanosomes. In this cell line, one RPB9 allele was knocked out by a hygromycin phosphotransferase gene (HYG, striped box) and integration of plasmid RPB9-PTP-NEO into the second allele fused the PTP sequence (black box) to the RPB9 coding region and added a neomycin phosphotransferase gene (NEO; striped box). Smaller gray boxes indicate gene flanks for RNA processing signals and checkered boxes depict RPB9 sequences encoded in the plasmid. (B) Immunoblot monitoring of RPB9-PTP tandem affinity purification, detecting the tagged protein with the monoclonal anti-ProtC antibody HPC4 in extract (Inp), flowthrough of IgG affinity chromatography (FT-IgG), TEV protease eluate, flowthrough of anti-ProtC immunoaffinity chromatography (FT-ProtC) and the final eluate (Elu). x-Values indicate relative amounts analyzed (Please note that TEV protease, cleaving off ProtA, reduces RPB9-PTP to RPB9-P, which corresponds to molecular mass reduction of ∼20 kDa). Marker sizes in kDa are indicated on the left. (C) RPB9-PTP co-purified proteins (Elu) and, for comparison, proteins of extract (Inp) and the TEV eluate were separated on a 10–20% SDS polyacrylamide gradient gel and stained with Coomassie blue. Individual protein bands were excised and proteins identified by LC/MS/MS as indicated on the right. (D) Predicted secondary-structure elements and homology model for the C-terminal winged-helix DNA-binding motif of TFL1. Top, secondary-structure assignments predicted by PHYRE 2 are shown as cyan cylinders (α helices) and arrows (β strands). Bottom, ribbon diagram of top scoring PHYRE homology model of the C-terminal domain of TFL1 (left), and supposition of TFL1 (cyan) with the C-terminal WH DNA-binding domain of human RAP30 (PDB ID: 2BBY) (pink; middle) and RAP74 (PDB ID: 1I27) (magenta; right).
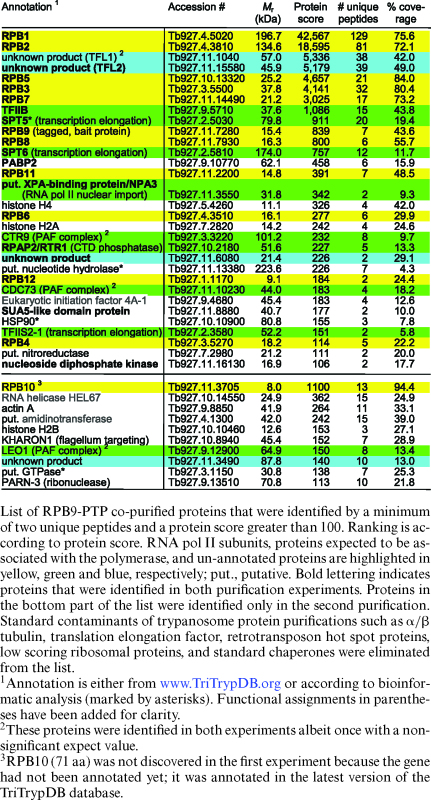
Immunoblot monitoring of the purification showed that IgG affinity chromatography (IgG binds to ProtA), TEV protease cleavage and anti-ProtC immunoaffinity chromatography of RPB9-PTP was efficient (Figure 1B). Analysis of the final eluate by SDS-PAGE revealed more distinct protein bands than in previous RNA pol II purifications. In particular, two prominent bands of ∼55 kDa and ∼60 kDa were not seen before. To identify the proteins, individual bands were excised, trypsin-digested and analyzed by LC/MS/MS (Figure 1C). In addition, we analyzed the complete eluate of a comparable purification in the same way (Table 1). In a later repeat of the experiment, the protein banding pattern was largely the same, except that the strength of the 55/60 kDa bands could not be reproduced (Supplementary Figure S2). This difference was most likely due to variation of the salt concentration in extracts (see below). Nonetheless, for the first time, all twelve subunits were biochemically identified as part of an isolated RNA pol II complex. Since trypanosomes possess two RPB10 paralogs, detection of RPB10 in the eluate confirmed that this paralog and not RPB10z (Tb927.3.1250) is a subunit of RNA pol II (55). Among the known general transcription factors, TFIIB consistently co-purified with the polymerase with high protein scores, raising the possibility that, in addition to its known essential role in SLRNA transcription, it is important for gene array transcription as well.
Table 1. Mass spectrometric identification of RPB9-PTP co-purified proteins.
|
Besides RNA pol II subunits and TFIIB we identified known and newly annotated transcription elongation factors. The minor band of ∼100 kDa contained the elongation factor SPT5, which is conserved among eubacteria, archaea and eukaryotes and which, by binding to the polymerase, increases its processivity by closing the DNA binding channel (56). The previously characterized TFIIS2–1 (57) and the histone chaperone SPT6 are also proteins which likely contribute to transcription elongation. We also identified all three trypanosome subunits of the conserved PAF (polymerase associated factor) complex (58) which in yeast and humans associates with RNA pol II, facilitates transcription elongation through chromatin, and interacts with transcription-relevant proteins (59). Interestingly, in trypanosomes, interaction of the PAF complex with RNA pol II could not be established previously which suggested a transcription-independent role of the factor in the expression of many genes (58). However, detection of all three subunits as RPB9-PTP-copurifying proteins (Table 1) strongly indicated that the trypanosome PAF complex does have a transcriptional role as its counterpart in other eukaryotes.
The proteins of the 55 and 60 kDa bands appear to be TFIIF proteins
The proteins of the 60 kDa and 55 kDa bands were identified as products of genes Tb927.11.1040 and Tb927.11.15580, respectively. While the proteins are well conserved among trypanosomatid organisms (Supplementary Figure S3), they have no sequence similarity to proteins of other eukaryotes. However, the facts that both proteins co-purified in seemingly stoichiometric amounts (Figure 1C), that, in other eukaryotes, TFIIB binds to RNA pol II that is associated with TFIIF and TFIIB was readily detectable in our RPB9-PTP purifications, and that extreme sequence divergence is a hallmark of all trypanosome RNA pol II general transcription factors (14), made us speculate that these two proteins form the heterodimeric TFIIF complex of trypanosomes. We therefore tentatively named these proteins TFL1 (Tb927.11.1040) and TFL2 (Tb927.11.15580).
Since multiple sequence alignments of TFIIF subunits from model organisms and TFL1 and TFL2 from several trypanosomatids did not reveal convincing sequence similarities (data not shown), we employed homology modeling to explore the possibility that TFL1 and TFL2 are structurally related to TFIIF subunits. Using the PHYRE 2 protein folding recognition server we were able to generate a homology model for TFL1. Intriguingly, the highest confidence model generated (95% of residues modeled at >72% confidence) is located in the extreme C-terminal region of the protein (residues 464–520) and unequivocally reminiscent of the classic ‘winged’ helix (WH) domain of helix-turn-helix DNA-binding proteins (Figure 1D). This model adopts a compact, α/β mixed fold consisting of four α-helices and an antiparallel three-stranded β sheet that forms the ‘wing’. Mammalian TFIIF is an α/β hetero-oligomer of RAP74 and RAP30 subunits, both of which contain a highly-conserved C-terminal WH domain despite limited sequence similarity (49,50). Correspondingly, the homology model of the WH domain of TFL1 can be superimposed onto those of RAP74 and RAP30 with a root mean square deviation of 3.6 and 2.3 Å, respectively, for the 70 core Cα atoms, although TFL1 and these two proteins share only ∼23% sequence identity. The WH domains of RAP74 and RAP30 have been reported to participate in DNA binding (60–62). Interestingly, the calculated electrostatic potential on the surface of the WH domain of TFL1 flanking the α-helix H3 is largely positive, suggesting that this putative recognition helix can interact with the major groove of DNA, in a mode similar to that of RAP30, but significantly different from RAP74 (Supplementary Figure S4). Thus, the homology model of the WH domain of TFL1 suggested that TFL1 appears to be functionally more related to RAP30 than to RAP74.
TFL1 and TFL2 are essential for parasite viability and SL RNA synthesis in vivo
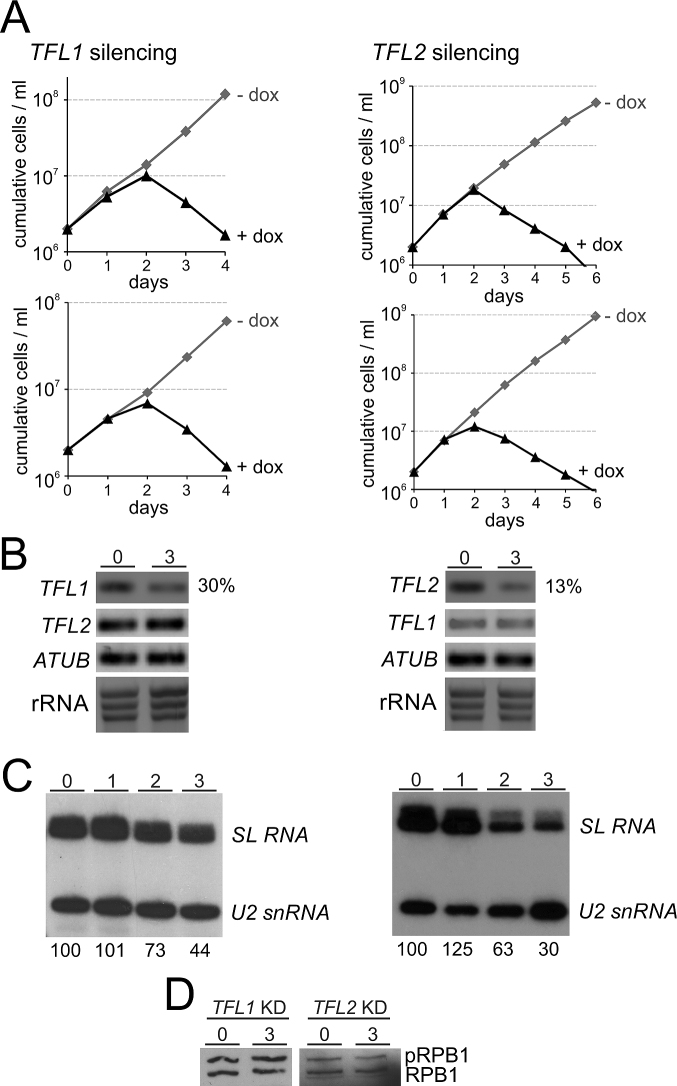
If TFL1 and TFL2 are indeed orthologues of TFIIF subunits, they should be important for SLRNA gene transcription since TFIIF is a critical factor for PIC formation (63). To test this, we generated clonal cell lines for conditional TFL1 and TFL2 silencing. For each gene we generated a construct in which a doxycycline-inducible T7 promoter drives the expression of a hairpin RNA that, via the RNA interference pathway, targets the mRNA of the respective gene. Each construct was integrated into the transcriptionally silent rRNA gene spacer region of procyclic 29–13 trypanosomes which constitutively express T7 RNA polymerase and the tetracycline repressor (43). For each gene knockdown, we generated two independently derived clonal cell lines. Addition of doxycycline to these cells invariably stopped culture growth after the second day of induction and, subsequently, led to a strong reduction of the cell number (Figure 2A), indicating that both genes are essential for procyclic trypanosome viability in culture. We carried out RNA analysis for one of the clonal cell lines for each gene. Semi-quantitative reverse transcription (RT)–PCR analysis of total RNA prepared from uninduced cells and from cells in which TFL1 or TFL2 was silenced for three days showed a specific decline of the target mRNA whereas the abundances of the non-targeted partner mRNA, the α tubulin (ATUB) mRNA or rRNA remained unaffected (Figure 2B). RT-quantitative (q)PCR revealed that, relative to ATUB mRNA, 30% of TFL1 mRNA and 13% of TFL2 mRNA remained three days after inducing the gene knockdowns.
Figure 2.
Silencing of TFL1 or TFL2 was lethal and reduced SL RNA abundance. (A) Cumulative culture growth curves were obtained for TFL1 and TFL2 silencing in the absence and presence of doxycycline (dox), the gene knockdown-inducing small molecule. For each knockdown, two independently derived cell lines were investigated. (B) Analysis of total RNA from one cell line for each knockdown in uninduced state (0) and after 3 days of doxycycline induction. TFL1 and TFL2 and, as a control, α tubulin (ATUB) mRNAs were analyzed by reverse transcription (RT) of oligo-dT and semi-quantitative PCR. To confirm equal RNA content, rRNA was visualized by ethidium bromide staining after agarose gel electrophoresis. Numbers on the right were derived from RT-qPCR assays and specify the percentage of the targeted mRNA that remained relative to ATUB RNA in gene-silenced cells. (C) Relative abundances of SL RNA and U2 snRNA in total RNA of uninduced and 1, 2, and 3 days TFL-silenced cells were determined by primer extension assays, using a SL RNA- and a U2 snRNA-specific primer in the same reactions. Numbers on the bottom were determined by densitometry and specify the strength of the SL RNA signal normalized by the U2 snRNA signal for each lane. The value in uninduced cells was arbitrarily set to 100. The numbers represent the average of two independent experiments. (D) Lysates of uninduced cells and of cells in which either TFL1 or TFL2 was silenced (KD) for 3 days were immunoblotted, detecting phosphorylated (pRPB1) and unphosphorylated RPB1.
To determine the effect of gene silencing on steady-state SL RNA abundance, we carried out a primer extension assay with two oligonucleotides that are complementary to SL RNA and U2 snRNA. Detection of the latter served as a control since U2 snRNA in trypanosomes is synthesized by RNA pol III. Relative to U2 snRNA, SL RNA abundance was reduced to 73% and 63% (day 2) and to 44% and 30% (day 3) of the uninduced level in TFL1- and TFL2-silenced trypanosomes, respectively (Figure 2C). These specific declines of SL RNA were in agreement with potential roles of TFL1 and TFL2 in SLRNA transcription.
We also wanted to know whether the knockdown of either gene would affect the phosphorylation status of RPB1. If TFL1 and TFL2 were TFIIF subunits, it was possible that depleting either protein would prevent the recruitment of an FCP1 phosphatase to RNA pol II and result in an increase in phosphorylated RPB1. In SDS-PAGE, trypanosome RPB1 is typically present in two major bands (see Figure 1C) of which the slower migrating band is phosphorylated RPB1 and the faster migrating band is unphosphorylated RPB1 (46). Using an anti-CTD antibody we found that silencing of either TFL1 or TFL2 did not detectably affect RPB1 phosphorylation, suggesting that the two proteins are not involved in recruiting FCP1 phosphatase to RNA pol II (Figure 2D).
If the TFL1 and TFL2 genes indeed encode factors for SLRNA transcription, their silencing should affect SL RNA synthesis. To test this, we employed a permeable cell system in which uptake and incorporation of radiolabeled UTP into newly synthesized RNA is efficient (45). We permeabilized uninduced cells and cells in which TFL1 or TFL2 were silenced for 2 or 3 days and radiolabeled newly synthesized RNA for eight minutes which is well within the 15–20 min time span of linear RNA synthesis this system supports (45). In this system, the signal of newly synthesized SL RNA is strong enough that it can be visualized as a prominent band of ∼140 nt after separation of total RNA by denaturing PAGE (Figure 3). In addition, tRNA signals can be seen around the 76 nt mark whereas signals of pre-mRNA and pre-rRNA are apparent on top of the gel. Knocking down either gene resulted in a decline of SL RNA relative to the tRNA signal. Consistent with the more efficient knockdown, reduction of newly synthesized SL RNA was strongest when TFL2 was silenced. After two days, the SL RNA signal was reduced to 57% and, after 3 days, only 25% of SL RNA synthesis seen in uninduced cells remained. These numbers are in close agreement with those observed upon silencing the genes of the basal transcription factors TFIIB (20), TFIIH (21), and Mediator (24). Although the SL RNA signal was only reduced by 42% on day 3 of TFL1 silencing, this result did support the notion that both proteins are important for SLRNA transcription.
Figure 3.
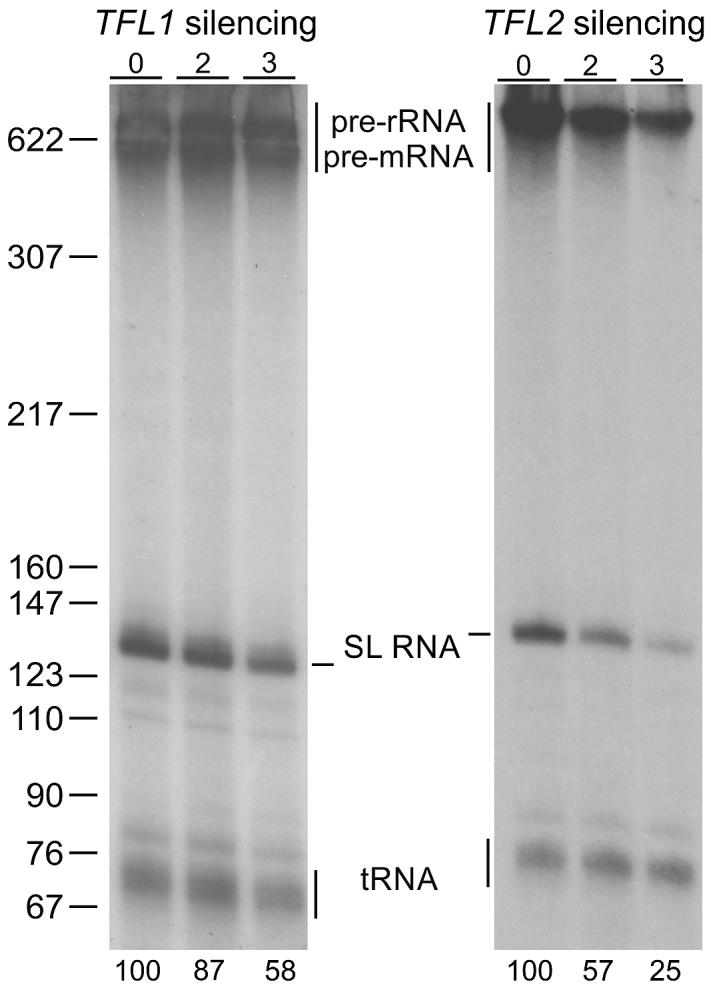
TFL1 and TFL2 silencing affects SL RNA synthesis. Newly synthesized RNA was labeled with [32P-α]UTP in permeabilized procyclic trypanosomes, which were uninduced or gene-silenced for 2 or 3 days, extracted, separated on a 50% urea-6% polyacrylamide gel, and visualized by autoradiography. Numbers on the bottom refer to relative SL RNA signal strengths normalized by the corresponding tRNA signals as determined by densitometry. The numbers represent the average of two independent experiments. On the left, pBR322-MspI marker sizes are indicated.
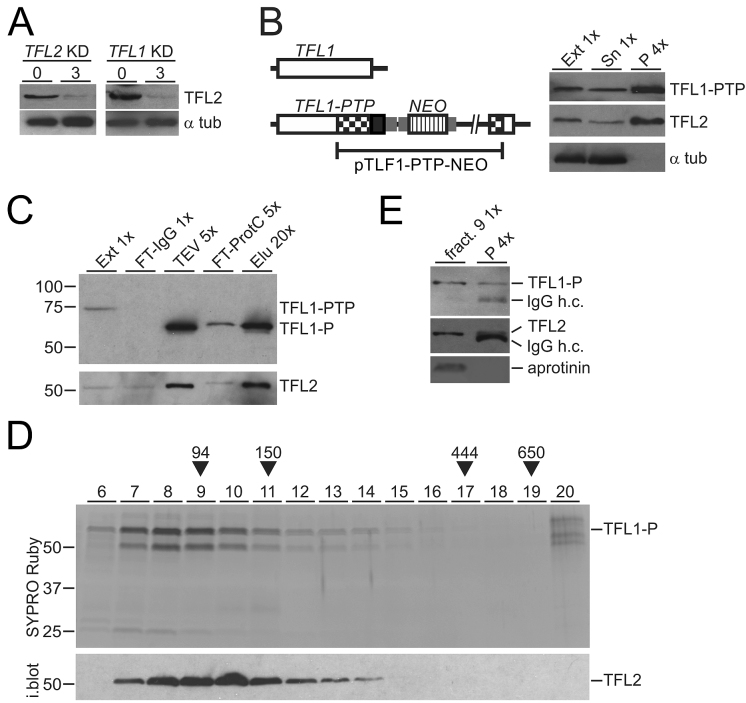
TFL1 and TFL2 form a heterodimeric complex
Another prediction for TFL1 and 2 being TFIIF subunits is that they form a complex and directly interact with each other. As a tool to investigate such a complex we generated rat immune serum against full-length recombinant TFL2 that was N-terminally tagged with GST and expressed in and purified from E. coli (Supplementary Figure S5). With this immune serum at hand, we first confirmed that TFL2 and RNA pol II interact with each other by reciprocal co-IP (Supplementary Figure S6A). Moreover, we could show that TFL2 co-precipitation with RPB9-PTP is salt sensitive and largely disrupted in extracts that contain ∼300 mM KCl (Supplementary Figure S6B). This result strongly indicated that variation of the salt concentration in extracts used for RPB9-PTP tandem affinity purification accounted for the different amounts of TFL co-purification we observed (compare Figure 1C with Supplementary Figure S2B), and it suggested that the purification shown in Figure 1C reflects the true interaction of RNA pol II and TFL in cells. Also, the salt sensitivity of the RNA pol II-TFL interaction is consistent with dissociation of mammalian TFIIF subunits from RNA pol II in a buffer containing 200 mM NaCl (64) and may be the reason why the two TFL proteins were not detected in previous RNA pol II characterizations. First evidence that the two proteins form a complex came from determining TFL2 abundance in TFL-silenced cells. As expected, the TFL2 protein level was strongly reduced in the TFL2 knockdown, but it was similarly affected when the partner gene TFL1 was silenced (Figure 4A), suggesting that TFL2 is only stable when assembled with TFL1 in a complex. Next, by targeted integration of plasmid TFL1-PTP-NEO, we generated a clonal procyclic cell line, that expressed TFL1 with a C-terminal PTP tag (TFL1-PTP), and conducted a co-precipitation experiment. IgG beads, which bind the tandem ProtA domain of the PTP tag, effectively precipitated TFL1-PTP and untagged TFL2 but not α tubulin, demonstrating interaction of TFL1 and TFL2 in extract (Figure 4B). To isolate a TFL1-TFL2 complex, we tandem affinity-purified TFL1-PTP from extract. Immunoblot monitoring of the procedure showed efficient purification of TFL1-P (please note that TFL1-PTP is reduced to TFL1-P in the TEV protease step of the purification) and co-purification of TFL2 (Figure 4C). Subsequent sedimentation of the final eluate in a linear sucrose gradient revealed co-sedimentation of two proteins with sizes that corresponded to TFL1-P and TFL2 with a peak in fractions 8–10. Immunoblotting of the gel confirmed that the lower band was TFL2 (Figure 4D). Furthermore, mass spectrometric analysis identified the minor protein band of ∼27 kDa, seen in fractions 7 and 8, as a degradation product of TFL1 and no other protein was detected. A heterodimeric TFL1-P/TFL2 complex has a calculated molecular mass of 106 kDa which, according to our sedimentation markers, should sediment around fraction 9. Since immunoprecipitation of TFL2 from this fraction, using the polyclonal immune serum, clearly co-precipitated TFL1-P (Figure 4E), we concluded that both TFL proteins directly interact with each other, forming a heterodimeric TFL complex.
Figure 4.
TFL1 and TFL2 form a complex. (A) Lysates of uninduced cells and of cells in which TFL2 or TFL1 was knocked down (KD) for 3 days were immunoblotted and probed with anti-TFL2 immune serum and, as a loading control, with an anti-α tubulin (α tub) antibody. (B) Schematic on the left shows integration of plasmid TFL1-PTP-NEO into the TFL1 locus which fused the PTP tag sequence to the 3′ end of the TFL1 coding region. The immunoblot on the right shows TFL1-PTP, TFL2 and α tubulin in extract (Ext), supernatant (Sn) and precipitate of a TFL1-PTP pull-down with IgG beads. x-Values specify relative amounts loaded. (C) Immunoblot monitoring of TFL1-PTP tandem affinity purification with the HPC4 antibody analogous to RPB9-PTP purification of Figure 1B. Detection of TFL2 on the same blot showed efficient co-purification of this protein. (D) The final eluate of a TFL1-PTP tandem affinity purification was sedimented through a 10–40% linear sucrose gradient by ultracentrifugation, and the gradient fractionated into 20 aliquots from top to bottom. Proteins from fractions 6–20 were separated by SDS-PAGE and stained with SYPRO Ruby. As sedimentation markers with known molecular masses, Taq DNA polymerase (94 kDa), mouse IgG (150 kDa, 6.6S), apoferritin (444 kDa, 17S), and thyroglobulin (650 kDa, 19S) were co-analyzed. After gel destaining, proteins were immunoblotted and probed with anti-TFL2 immune serum (i. blot.). (E) TFL2 was immunoprecipitated from fraction 9 of the sucrose gradient and detected with anti-TFL2 immune serum. Coprecipitation of TFL1-P was analyzed on the same blot with anti-ProtC antibody. Coomassie blue-stained aprotinin which was added to fraction 9 prior to precipitation served as a negative control. In both immunoblot panels, co-eluted IgG heavy chain (IgG h.c.) was detected.
In solution, human TFIIF appears to form a heterotetramer with two RAP74 and two RAP30 subunits (65). A corresponding TFL complex would have a molecular mass of ∼210 kDa and sediment around fraction 13. The complex is clearly detectable in fraction 13 but there is no sedimentation peak in this region (Figure 4E), suggesting that, after tandem affinity purification, the majority of TFL is in the form of a heterodimer and only a minor amount of the purified complex may be in the form of a heterotetramer.
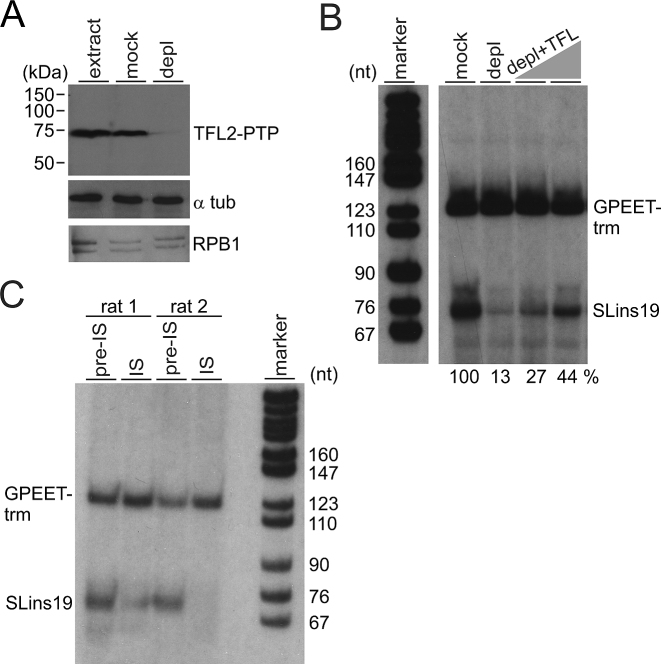
TFL is important for SLRNA transcription in vitro
To provide further evidence for TFL’s role in SLRNA transcription, we employed an in vitro system which supports correctly initiated transcription of an oligonucleotide-tagged SLRNA promoter template (SLins19) and of the similarly tagged control template GPEET-trm that recruits RNA pol I (41,44). First, employing the same strategy as for TbR9PTPee cells outlined in Figure 1A, we generated a procyclic cell line that exclusively expressed TFL2-PTP and no untagged TFL2. Anti-TFL2 immunoblotting of extract from these TbT2PTPee cells confirmed replacement of endogenous untagged TFL2 by a band that was ∼20 kDa larger due to the PTP tag (Supplementary Figure S5). Next we prepared transcriptionally active extract from these cells and depleted TFL2-PTP by IgG affinity chromatography or, in parallel, mock-treated the extract with protein G beads (Figure 5A). Detection of RPB1 in these extracts indicated that RNA pol II was not detectably co-depleted by TFL2-PTP removal. Carrying out co-transcription assays of SLins19 and GPEET-trm templates in mock-treated and TFL2-PTP-depleted extracts, we saw strong and specific reduction of the SLins19 transcription signal (Figure 5B). This signal could be partially reconstituted in a dose-dependent manner by adding back TFL from fraction 8 of the linear sucrose gradient shown in Figure 4D (Figure 5B). In a separate approach, we analyzed whether anti-TFL2 antibodies, by binding to TFL2, had an inhibitory effect on SLins19 transcription. Since we raised the anti-TFL2 immune serum in two rats we pre-incubated transcription extract with [pre-]immune serum from each rat before carrying out transcription reactions. As shown in Figure 5C, adding pre-immune serum from either rat to extract resulted in clear SLins19 and GPEET-trm transcriptional signals. As predicted, the SLins19 signal, but not the GPEET-trm signal, was strongly reduced or abolished when the corresponding immune serum derived from rat 1 or 2 was added. Together, these results demonstrated TFL’s important function in SLRNA transcription.
Figure 5.
TFL is essential for SLRNA transcription in vitro. (A) Immunoblot of extract from TbT2PTPee trypanosomes that was untreated (extract), mock-treated or TFL2-PTP-depleted (depl), detecting TFL2-PTP, α tubulin (α tub) and RNA pol II subunit RPB1 with specific antibodies. (B) Cotranscription of the SLRNA promoter template SLins19 and the RNA pol I-transcribed template GPEET-trm in mock-treated and TFL2-PTP-depleted extract. The latter reaction was reconstituted with 2 and 4 μl of sucrose gradient fraction 8 shown in Figure 4D. GPEET-trm and SLins19 RNA were detected by primer extension reactions using radiolabeled oligonucleotides that specifically hybridized to these RNAs. Extension products were separated on 50% urea/6% polyacrylamide gels and visualized by autoradiography. The correct length of the extension products verified correctly initiated transcription. Numbers at the bottom show the relative SLins19 signal strengths normalized by those of the GPEET-trm signals as determined by densitometry. The signal in mock-treated extract was arbitrarily set to 100. The pBR322-MspI marker lane on the left was derived from the same gel. (C) Cotranscription reactions were carried out in extract that was either mixed with pre-immune serum or anti-TFL2 immune serum derived from two different rats.
TFL1-PTP and TFL2-PTP are localized in the nucleus and TFL2-PTP occupies the SLRNA promoter
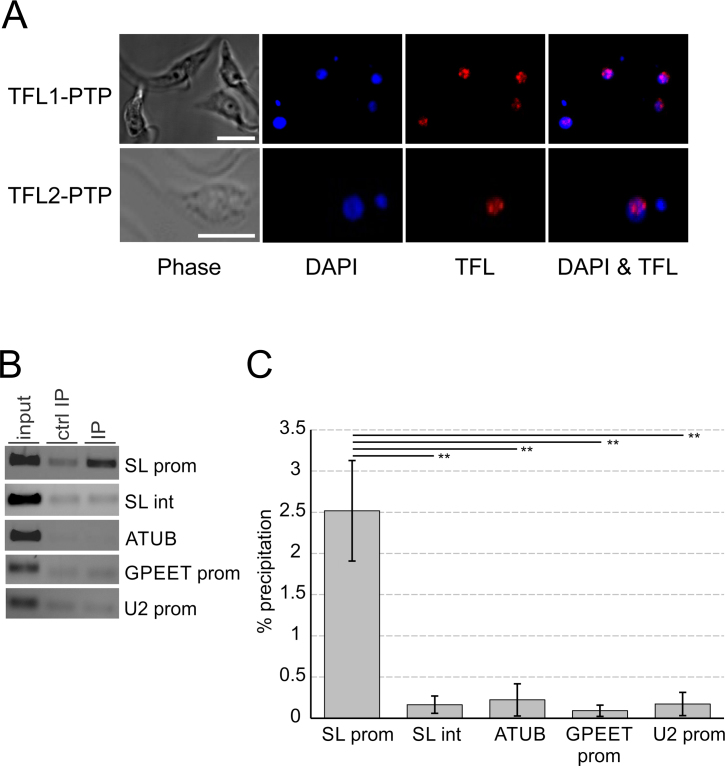
To determine the cellular localization of TFL1 and TFL2, we performed indirect anti-ProtA immunofluorescence microscopy with cells that expressed either TFL1-PTP or TFL2-PTP. Trypanosomes have two stainable DNA bodies, namely the nucleus and the kinetoplast which consists of catenated DNA of the trypanosome's single mitochondrion. Trypanosomes in G1 phase of the cell cycle possess one kinetoplast and one nucleus (1K1N), whereas trypanosomes at the end of S-phase and postmitotic trypanosomes are 2K1N and 2K2N cells, respectively (66). Independent of the cell cycle stage, both TFL subunits were concentrated in the nucleus (Figure 6A, Supplementary Figure S7). In most images, the TFL proteins were localized in one or two major spots around the nucleolus, a nuclear area of less stain, with diffuse staining around these spots. The spots are reminiscent of the so-called SL ribonucleoprotein (RNP) factory in which SLRNAs are transcribed and SL RNPs are assembled (7). Thus, this result suggested that TFL is predominantly localized at SLRNAs, the only genes in trypanosomes known to form a RNA pol II PIC.
Figure 6.
TFL is localized in the nucleus and occupies the SLRNA promoter in vivo. (A) Localization of TFL1-PTP or TFL2-PTP (red) in procyclic trypanosomes. DNA was stained with DAPI (blue) showing nuclei and smaller kinetoplasts. The nucleolus can be recognized within a nucleus as a spherical structure of low DNA density. White bars in left panels represent 5 μm. (B) Anti-TFL2-PTP ChIP assay using a polyclonal anti-ProtA antibody (IP) and, in a control precipitation, a comparable nonspecific immune serum (ctrl IP). DNA from total chromatin (input) and precipitated DNA was analyzed by semi-quantitative PCR, amplifying the SLRNA promoter (SL prom), part of the SLRNA intergenic region (SL int), a fragment of the α tubulin coding region (ATUB), the GPEET procyclin promoter (GPEET prom), and the U2 snRNA gene promoter (U2 prom). (C) Corresponding qPCR analysis from three independent experiments. For each region amplified the percent precipitation was calculated and the values corrected by the corresponding values of the control precipitations. The difference in TFL2-PTP occupancy between the SLRNA promoter and the other amplified regions was statistically analyzed by paired two-tailed student's t-tests.
To test this directly, we performed chromatin immunoprecipitation (ChIP) analysis of TFL2-PTP using TbT2PTPee cells, because PTP-tagged proteins can be effectively precipitated by a commercially available, ChIP-grade rabbit anti-ProtA antibody [see e.g. (24)]. Anti-TFL2-PTP ChIP revealed efficient precipitation of the SLRNA promoter region when compared to a negative control precipitation with a non-specific rabbit immune serum whereas the silent SLRNA intergenic region, the α tubulin coding region, the RNA pol III-recruiting U2 snRNA promoter and the RNA pol I-specific GPEET procyclin promoter did not reveal detectable precipitation over the control in these assays (Figure 6B). qPCR analysis of three independent experiments confirmed this result, revealing statistical significance for TFL2-PTP occupancy of the SLRNA promoter (Figure 6C). Hence, TFL appeared to be part of the SLRNA PIC.
TFL2 is important for PIC formation at the SLRNA promoter
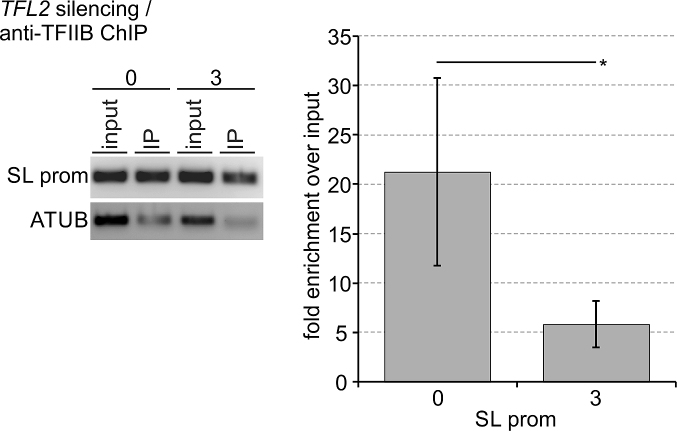
Eukaryotic TFIIF has several distinct functions in PIC assembly and stability, being particularly important for recruitment and/or retention of TFIIB (25,63). Our own studies of trypanosome PIC formation had shown that Mediator depletion caused a loss of TFIIB occupancy of the SLRNA promoter, indicating that stable integration of trypanosome TFIIB into the PIC depends on other factors as well (24). Thus, if TFL did represent the trypanosome TFIIF, we expected TFL depletion to reduce TFIIB occupancy of the SLRNA promoter. To test this, we conducted anti-TFIIB ChIP in uninduced cells and in procylic cells in which TFL2 was silenced for 3 days, using a rat anti-T. brucei TFIIB immune serum that previously allowed us to detect significant TFIIB occupancy changes of the SLRNA promoter in Mediator depletions (24). Conducting four independent experiments, this immune serum enriched the SLRNA promoter on average 21.2-fold over input in uninduced cells (Figure 7). This value dropped significantly to 5.8-fold when TFL2 was silenced for 3 days, strongly indicating that PIC formation became impaired in TFL-depleted cells and that TFL is indispensable for TFIIB recruitment to and/or retention at the SLRNA promoter. Thus, TFL is apparently required for PIC formation in trypanosomes as is true for TFIIF in other eukaryotes.
Figure 7.
TFL2 is important for TFIIB recruitment to and retention at the SLRNA promoter. Anti-TFIIB ChIP assays were carried out with chromatin prepared from uninduced cells or cells in which TFL2 was silenced for three days. On the left, semi-quantitative PCR of the SLRNA promoter and part of the tubulin coding region in DNA prepared from total chromatin (input) and precipitate (IP). On the right, corresponding qPCR analysis from four independent experiments in which the TFIIB occupancy of the SLRNA promoter was normalized by that of the ATUB occupancy and the fold enrichment over input was calculated. The difference in TFIIB occupancy between uninduced and TFL2-silenced cells was statistically analyzed by an unpaired, two-tailed Student's t-test assuming equal variance.
DISCUSSION
Does TFL represent the trypanosome TFIIF?
Here we have re-visited isolation and proteomic analysis of T. brucei RNA pol II, identifying for the first time all twelve subunits of the complex. While RPB1 to RPB12 were conserved enough to be unequivocally identified bioinformatically (67), not all of them were shown to be part of the enzyme complex in previous trypanosomatid RNA pol II characterizations (52–54). In addition, several proteins co-purified with the enzyme, most of which were homologs of known RNA pol II-interacting proteins such as transcription elongation factors SPT5, SPT6, TFIIS2-1 and the PAF complex, as well as the CTD phosphatase RPAP2/Rtr1 and TFIIB. Two proteins with unknown function co-purified with RPB9 in seemingly stoichiometric amounts, suggesting that they form a heterodimeric complex and represent the missing general transcription factor TFIIF. According to the known functions of eukaryotic TFIIF, we confirmed that both proteins form a complex which interacts with RNA pol II, that the complex is essential for parasite viability in culture, occupies the SLRNA promoter, is indispensable for SLRNA transcription in vivo and in vitro, and required for stable association of TFIIB with the SLRNA promoter. In addition, we were able to identify a putative WH domain in the C-terminal region of TFL1 that has been identified in both RAP74 and RAP30 orthologues. However, multiple sequence alignments of TFIIF subunits from model organisms and of trypanosomatid TFL proteins did not uncover meaningful sequence similarities. Furthermore, yeast and human TFIIF subunits have been extensively characterized and several distinct functional and structural domains have been described including sigma factor-homologous regions, WH DNA binding domains, dimerization domains and a charged and an acidic region (25,26). Except for the above mentioned putative WH domain in TFL1, we could not recognize these domains in the two TFL proteins. Finally, TFL appears to be not involved in dephosphorylation of the CTD because TFL1 and TFL2 silencing did not lead to an increase in phosphorylated RPB1 and tandem affinity purification of TFL2-PTP did not unequivocally co-purify a FCP1 phosphatase (A.S. & A.G., unpublished data) despite the fact that trypanosomes possess an expanded family of FCP1 phosphatases (68). Hence, if these FCP1 enzymes are involved in CTD dephosphorylation they may be recruited to RNA pol II in a TFL-independent manner. Alternatively, CTD dephosphorylation may be mainly mediated by RPAP2/Rtr1, which consistently co-purified with RNA pol II, whereas FCP1 enzymes may serve RNA pol II-independent functions. This was demonstrated for the FCP1 paralogue TbPIP39 which is targeted to the glycosome, a peroxisome-like organelle for glycolysis, and involved in life cycle regulation (69). Taken together, a firm conclusion of TFL being the trypanosome TFIIF will depend on further studies that functionally characterize the domain arrangement of TFL1 and TFL2 and/or solve the structure of the TFL complex.
The PIC of trypanosomatids
Although the amino acid sequences of trypanosomatid general transcription factors are extremely divergent from those of their eukaryotic counterparts, there is now strong evidence that trypanosomes and their relatives possess a complete set of RNA pol II general transcription factors. TBP and the TFIIH helicases XPB and XPD were sufficiently conserved to be identified in silico (15,16). The sequence of TFIIB was less conserved but functional studies and the crystal structure of its C-terminal domain confirmed the identity of this pivotal transcription factor (17,18,70). The trimeric SNAPc, TFIIA2, and the remaining TFIIH core subunits could only be identified by multiple sequence alignments or by highly conserved sequence motifs once the respective protein complexes were isolated and characterized (17,18,22,23). Two TFIIH subunits, however, did not exhibit any sequence similarity and, therefore, were tentatively termed trypanosomatid-specific proteins 1 and 2 (TSP1 and 2). We have previously argued that these proteins represent trypanosome TFIIE – they are essential for RNA pol II-mediated SLRNA transcription, like TFIIE they interact with TFIIH subunit XPB, and trypanosomatid TSP2s, as all known TFIIEα orthologues, harbor an invariant internal C2C2 zinc-finger motif (23). Here we subjected the TSP sequences to the PHYRE 2 server and received as top homology hits for TSP2 three TFIIEα proteins (PDB Ids: 5FYW, 5IY9, 5FMF). The homology region comprised TSP2’s N-terminal region which, in TFIIEα proteins, is composed of an extended WH domain followed by a Zn ribbon domain (26,71,72). Since the secondary structure prediction of this TSP2 region matched that of the yeast TFIIEα orthologue TFA1, and the TSP2 N-terminal half could be folded into extended WH and Zn ribbon domains (Supplementary Figure S8), we provide further evidence that TSP2 is indeed the orthologue of the TFIIEα. Consequently, characterization of TFL strongly indicates that trypanosomes possess orthologues of each and every RNA pol II general transcription factor including SNAPc and Mediator.
Is PIC formation required beyond SLRNA transcription?
Trypanosomes and related organisms process all pre-mRNA by SL trans splicing, having mRNA 5/ ends determined by SL addition and not, as in other eukaryotes, by transcription initiation. Initiating transcription at concrete sites may therefore not be as important for protein coding gene arrays as for SLRNA genes or protein coding genes in other eukaryotes. Accordingly, RNA-seq of cDNA libraries enriched for 5/-triphosphate RNA ends revealed broad regions of transcription initiation in front of gene arrays (11,12). These transcription initiation regions are enriched in euchromatic histone marks, namely the histone variants H2AZ and H2BV, and the histone modifications H3K4m3, H4K10ac and H3ac (73–75). Since there is evidence in trypanosomes that RNA pol II transcription of reporter genes occurred efficiently in the absence of identifiable core promoters (76,77), it was proposed that a permissive chromatin structure is sufficient for initiating transcription of protein coding gene arrays (12). If so, PIC formation in trypanosomatids may be restricted to SLRNA genes. On the other hand, there are two observations which suggest that general transcription factors do play a role in gene array transcription. ChIP-chip experiments in Leishmania major detected occupancy peaks of the SNAPc subunit SNAP1 (aka SNAP50) and of TBP in transcription initiation regions (75), and TFIIB silencing in procyclic T. brucei reduced synthesis of α tubulin and HSP70 RNA but not that of GPEET procyclin RNA which is mediated by RNA pol I (20). Since these transcription initiation regions apparently function only within their chromosomal context (12) and not in vitro or in episomal plasmids, it will require in vivo experiments that can discriminate between primary effects on gene transcription and secondary effects of SL RNA depletion on mRNA processing to firmly establish whether general transcription factors are required for protein gene transcription in trypanosomes.
CONCLUSION
This report provides strong evidence for an essential heterodimeric TFIIF homolog in trypanosomes and strengthens the hypothesis that the two TFIIH-associated subunits TSP1 and TSP2 represent trypanosome TFIIE. Consequently, early-diverged trypanosomes and their trypanosomatid relatives appear to possess orthologs of each and every RNA pol II general transcription factor including SNAPc and the Mediator complex MED-T. Each one of these factors is extremely divergent in amino acid sequence from their eukaryotic counterparts but essential for SLRNA transcription and trypanosome viability, and, together, they comprise an astonishingly extensive set of 29 proteins so far.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Sung Hee Park from our laboratory who cloned plasmid pRPB9-PTP-NEO and to Christian Tschudi (Yale University) who helped with homology modeling of TFL and critically read our manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Allergy and Infectious Diseases (NIAID) [R01 AI073300 (PI) and R01AI028798 (co-I) to A.G.] and National Institute of General Medical Sciences [R01GM099948 to B.H.] both of the U.S. National Institutes of Health. Funding for open access charge: NIAID.
Conflict of interest statement. None declared.
REFERENCES
- 1. Allen B.L., Taatjes D.J.. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell. Biol. 2015; 16:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henry R.W., Sadowski C.L., Kobayashi R., Hernandez N.. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature. 1995; 374:653–656. [DOI] [PubMed] [Google Scholar]
- 3. Yoon J.B., Murphy S., Bai L., Wang Z., Roeder R.G.. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol. 1995; 15:2019–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hantsche M., Cramer P.. Conserved RNA polymerase II initiation complex structure. Curr. Opin. Struct. Biol. 2017; 47:17–22. [DOI] [PubMed] [Google Scholar]
- 5. Field M.C., Horn D., Fairlamb A.H., Ferguson M.A., Gray D.W., Read K.D., De Rycker M., Torrie L.S., Wyatt P.G., Wyllie S. et al. . Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need. Nat. Rev. Microbiol. 2017; 15:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Günzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified. Eukaryot. Cell. 2010; 9:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011; 6:459–474. [DOI] [PubMed] [Google Scholar]
- 8. Nakaar V., Tschudi C., Ullu E.. An unusual liaison: Small nuclear and cytoplasmic RNA genes team up with tRNA genes in trypanosomatid protozoa. Parasitol. Today. 1995; 11:225–228. [Google Scholar]
- 9. Tschudi C., Ullu E.. Unconventional rules of small nuclear RNA transcription and cap modification in trypanosomatids. Gene Expr. 2002; 10:3–16. [PMC free article] [PubMed] [Google Scholar]
- 10. Gilinger G., Bellofatto V.. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001; 29:1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kolev N.G., Franklin J.B., Carmi S., Shi H., Michaeli S., Tschudi C.. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010; 6:e1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wedel C., Forstner K.U., Derr R., Siegel T.N.. GT-rich promoters can drive RNA pol II transcription and deposition of H2A.Z in African trypanosomes. EMBO J. 2017; 36:2581–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martinez-Calvillo S., Vizuet-de-Rueda J.C., Florencio-Martinez L.E., Manning-Cela R.G., Figueroa-Angulo E.E.. Gene expression in trypanosomatid parasites. J. Biomed. Biotechnol. 2010; 2010:525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Günzl A. Bindereif A. RNA Metabolism in Trypanosomes. 2012; 28, Springer Press; 1–27. [Google Scholar]
- 15. Ruan J.P., Arhin G.K., Ullu E., Tschudi C.. Functional characterization of a Trypanosoma brucei TATA-binding protein-related factor points to a universal regulator of transcription in trypanosomes. Mol. Cell. Biol. 2004; 24:9610–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R. et al. . The genome of the kinetoplastid parasite Leishmania major. Science. 2005; 309:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schimanski B., Nguyen T.N., Günzl A.. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol. Cell. Biol. 2005; 25:7303–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Das A., Zhang Q., Palenchar J.B., Chatterjee B., Cross G.A., Bellofatto V.. Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol. Cell. Biol. 2005; 25:7314–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palenchar J.B., Liu W., Palenchar P.M., Bellofatto V.. A divergent transcription factor TFIIB in trypanosomes is required for RNA polymerase II-dependent spliced leader RNA transcription and cell viability. Eukaryot. Cell. 2006; 5:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schimanski B., Brandenburg J., Nguyen T.N., Caimano M.J., Günzl A.. A TFIIB-like protein is indispensable for spliced leader RNA gene transcription in Trypanosoma brucei. Nucleic Acids Res. 2006; 34:1676–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J.H., Nguyen T.N., Schimanski B., Günzl A.. Spliced leader RNA gene transcription in Trypanosoma brucei requires transcription factor TFIIH. Eukaryot.Cell. 2007; 6:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lecordier L., Devaux S., Uzureau P., Dierick J.F., Walgraffe D., Poelvoorde P., Pays E., Vanhamme L.. Characterization of a TFIIH homologue from Trypanosoma brucei. Mol. Microbiol. 2007; 64:1164–1181. [DOI] [PubMed] [Google Scholar]
- 23. Lee J.H., Jung H.S., Günzl A.. Transcriptionally active TFIIH of the early-diverged eukaryote Trypanosoma brucei harbors two novel core subunits but not a cyclin-activating kinase complex. Nucleic Acids Res. 2009; 37:3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee J.H., Cai G., Panigrahi A.K., Dunham-Ems S., Nguyen T.N., Radolf J.D., Asturias F.J., Günzl A.. A TFIIH-associated mediator head is a basal factor of small nuclear spliced leader RNA gene transcription in early-diverged trypanosomes. Mol. Cell. Biol. 2010; 30:5502–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas M.C., Chiang C.M.. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006; 41:105–178. [DOI] [PubMed] [Google Scholar]
- 26. Plaschka C., Hantsche M., Dienemann C., Burzinski C., Plitzko J., Cramer P.. Transcription initiation complex structures elucidate DNA opening. Nature. 2016; 533:353–358. [DOI] [PubMed] [Google Scholar]
- 27. Yan Q., Moreland R.J., Conaway J.W., Conaway R.C.. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 1999; 274:35668–35675. [DOI] [PubMed] [Google Scholar]
- 28. Cabart P., Ujvari A., Pal M., Luse D.S.. Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:15786–15791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen H.T., Warfield L., Hahn S.. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007; 14:696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fishburn J., Hahn S.. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol. Cell. Biol. 2012; 32:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mayfield J.E., Burkholder N.T., Zhang Y.J.. Dephosphorylating eukaryotic RNA polymerase II. Biochim. Biophys. Acta. 2016; 1864:372–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsin J.P., Manley J.L.. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012; 26:2119–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jeronimo C., Collin P., Robert F.. The RNA Polymerase II CTD: The Increasing Complexity of a Low-Complexity Protein Domain. J. Mol. Biol. 2016; 428:2607–2622. [DOI] [PubMed] [Google Scholar]
- 34. Guo Z., Stiller J.W.. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics. 2004; 5:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Das A., Bellofatto V.. The non-canonical CTD of RNAP-II is essential for productive RNA synthesis in Trypanosoma brucei. PLoS One. 2009; 4:e6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Das A., Banday M., Fisher M.A., Chang Y.J., Rosenfeld J., Bellofatto V.. An essential domain of an early-diverged RNA polymerase II functions to accurately decode a primitive chromatin landscape. Nucleic Acids Res. 2017; 45:7886–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Urbaniak M.D., Martin D.M., Ferguson M.A.. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J. Proteome Res. 2013; 12:2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rocha A.A., Moretti N.S., Schenkman S.. Stress induces changes in the phosphorylation of Trypanosoma cruzi RNA polymerase II, affecting its association with chromatin and RNA processing. Eukaryot. Cell. 2014; 13:855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schimanski B., Nguyen T.N., Günzl A.. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell. 2005; 4:1942–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brandenburg J., Schimanski B., Nogoceke E., Nguyen T.N., Padovan J.C., Chait B.T., Cross G.A., Günzl A.. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 2007; 26:4856–4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laufer G., Günzl A.. In-vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei. Mol. Biochem. Parasitol. 2001; 113:55–65. [DOI] [PubMed] [Google Scholar]
- 42. Günzl A., Ullu E., Dörner M., Fragoso S.P., Hoffmann K.F., Milner J.D., Morita Y., Nguu E.K., Vanacova S., Wünsch S. et al. . Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 1997; 85:67–76. [DOI] [PubMed] [Google Scholar]
- 43. Wirtz E., Leal S., Ochatt C., Cross G.A.M.. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 99:89–101. [DOI] [PubMed] [Google Scholar]
- 44. Laufer G., Schaaf G., Bollgönn S., Günzl A.. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol. 1999; 19:5466–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ullu E., Tschudi C.. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 1990; 18:3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Badjatia N., Ambrósio D.L., Lee J.H., Günzl A.. Trypanosome cdc2-related kinase 9 controls spliced leader RNA cap4 methylation and phosphorylation of RNA polymerase II subunit RPB1. Mol. Cell. Biol. 2013; 33:1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luz Ambrósio D., Lee J.H., Panigrahi A.K., Nguyen T.N., Cicarelli R.M., Günzl A.. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryotic Cell. 2009; 8:990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Groft C.M., Uljon S.N., Wang R., Werner M.H.. Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:9117–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kamada K., De Angelis J., Roeder R.G., Burley S.K.. Crystal structure of the C-terminal domain of the RAP74 subunit of human transcription factor IIF. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:3115–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B.. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999; 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- 52. Das A., Li H., Liu T., Bellofatto V.. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 2006; 150:201–210. [DOI] [PubMed] [Google Scholar]
- 53. Devaux S., Lecordier L., Uzureau P., Walgraffe D., Dierick J.F., Poelvoorde P., Pays E., Vanhamme L.. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol. Biochem. Parasitol. 2006; 148:60–68. [DOI] [PubMed] [Google Scholar]
- 54. Martinez-Calvillo S., Saxena A., Green A., Leland A., Myler P.J.. Characterization of the RNA polymerase II and III complexes in Leishmania major. Int. J. Parasitol. 2007; 37:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nguyen T.N., Schimanski B., Zahn A., Klumpp B., Günzl A.. Purification of an eight subunit RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 2006; 149:27–37. [DOI] [PubMed] [Google Scholar]
- 56. Yakhnin A.V., Babitzke P.. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor. Curr. Opin. Microbiol. 2014; 18:68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uzureau P., Daniels J.P., Walgraffe D., Wickstead B., Pays E., Gull K., Vanhamme L.. Identification and characterization of two trypanosome TFIIS proteins exhibiting particular domain architectures and differential nuclear localizations. Mol. Microbiol. 2008; 69:1121–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ouna B.A., Nyambega B., Manful T., Helbig C., Males M., Fadda A., Clayton C.. Depletion of trypanosome CTR9 leads to gene expression defects. PLoS One. 2012; 7:e34256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jaehning J.A. The Paf1 complex: platform or player in RNA polymerase II transcription. Biochim. Biophys. Acta. 2010; 1799:379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tan S., Garrett K.P., Conaway R.C., Conaway J.W.. Cryptic DNA-binding domain in the C terminus of RNA polymerase II general transcription factor RAP30. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:9808–9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Forget D., Langelier M.F., Therien C., Trinh V., Coulombe B.. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol. Cell. Biol. 2004; 24:1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robert F., Douziech M., Forget D., Egly J.M., Greenblatt J., Burton Z.F., Coulombe B.. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol. Cell. 1998; 2:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Luse D.S. Rethinking the role of TFIIF in transcript initiation by RNA polymerase II. Transcription. 2012; 3:156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sopta M., Carthew R.W., Greenblatt J.. Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 1985; 260:10353–10360. [PubMed] [Google Scholar]
- 65. Flores O., Ha I., Reinberg D.. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J. Biol. Chem. 1990; 265:5629–5634. [PubMed] [Google Scholar]
- 66. Woodward R., Gull K.. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 1990; 95:49–57. [DOI] [PubMed] [Google Scholar]
- 67. Kelly S., Wickstead B., Gull K.. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem. Soc. Trans. 2005; 33:1435–1437. [DOI] [PubMed] [Google Scholar]
- 68. Szöor B. Trypanosomatid protein phosphatases. Mol. Biochem. Parasitol. 2010; 173:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Szöor B., Ruberto I., Burchmore R., Matthews K.R.. A novel phosphatase cascade regulates differentiation in Trypanosoma brucei via a glycosomal signaling pathway. Genes Dev. 2010; 24:1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ibrahim B.S., Kanneganti N., Rieckhof G.E., Das A., Laurents D.V., Palenchar J.B., Bellofatto V., Wah D.A.. Structure of the C-terminal domain of transcription factor IIB from Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13242–13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. He Y., Yan C., Fang J., Inouye C., Tjian R., Ivanov I., Nogales E.. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016; 533:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Murakami K., Tsai K.L., Kalisman N., Bushnell D.A., Asturias F.J., Kornberg R.D.. Structure of an RNA polymerase II preinitiation complex. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Siegel T.N., Hekstra D.R., Kemp L.E., Figueiredo L.M., Lowell J.E., Fenyo D., Wang X., Dewell S., Cross G.A.. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009; 23:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wright J.R., Siegel T.N., Cross G.A.. Histone H3 trimethylated at lysine 4 is enriched at probable transcription start sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 2010; 172:141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thomas S., Green A., Sturm N.R., Campbell D.A., Myler P.J.. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics. 2009; 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Marchetti M.A., Tschudi C., Silva E., Ullu E.. Physical and transcriptional analysis of the Trypanosoma brucei genome reveals a typical eukaryotic arrangement with close interspersionof RNA polymerase II- and III-transcribed genes. Nucleic Acids Res. 1998; 26:3591–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McAndrew M., Graham S., Hartmann C., Clayton C.. Testing promoter activity in the trypanosome genome: isolation of a metacyclic-type VSG promoter, and unexpected insights into RNA polymerase II transcription. Exp. Parasitol. 1998; 90:65–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.