Abstract
Transcriptional silencing is a conserved process used by embryonic germ cells to repress somatic fate and maintain totipotency and immortality. In Drosophila, this transcriptional silencing is mediated by polar granule component (pgc). Here, we show that in the adult ovary, pgc is required for timely germline stem cell (GSC) differentiation. Pgc is expressed transiently in the immediate GSC daughter (pre-cystoblast), where it mediates a pulse of transcriptional silencing. This transcriptional silencing mediated by pgc indirectly promotes the accumulation of Cyclin B (CycB) and cell cycle progression into late-G2 phase, when the differentiation factor bag of marbles (bam) is expressed. Pgc mediated accumulation of CycB is also required for heterochromatin deposition, which protects the germ line genome against selfish DNA elements. Our results suggest that transient transcriptional silencing in the pre-cystoblast “re-programs” it away from self-renewal and toward the gamete differentiation program.
Keywords: Transcriptional silencing, Cell cycle, Differentiation, Germ line, Stem cells, Heterochromatin
1. Introduction
Germ cells provide continuity between generations for all sexually reproducing organisms. They do this by repressing somatic fate, controlling differentiation of gametes, and protecting the genome (Cinalli et al., 2008; Seydoux and Braun, 2006). During embryogenesis, germ cells use the conserved process of transcriptional silencing to counter signaling pathways from the soma and avoid a somatic fate (Nakamura and Seydoux, 2008). In Drosophila embryogenesis, this transcriptional silencing is mediated by polar granule component (pgc), which blocks the Positive Transcription Elongation Factor (P-TEFb) complex consisting of Cyclin T (CycT) and Cyclin dependent kinase 9 (Cdk9), from acting on RNA Polymerase II to promote transcriptional elongation (Hanyu-Nakamura et al., 2008; Martinho et al., 2004). Loss of pgc results in transcription of somatic genes and changes to the epigenetic landscape, leading to loss of germ cell fate (de Las Heras et al., 2009; Martinho et al., 2004). Thus, pgc is essential to preserve germ cell fate during embryogenesis.
In Drosophila adults, germline stem cells (GSCs) differentiate into egg and sperm. In the ovary, GSCs are attached to a niche formed by somatic cells (Fig. 1A) (Xie et al., 2008; Xie and Spradling, 1998). In general, the GSC divides asymmetrically (Chen and McKearin, 2003; Jin et al., 2008). One daughter maintains close contact with the somatic niche and remains as a stem cell while the other daughter, the cystoblast (CB), loses contact with the niche and will differentiate into a germ line cyst. In the process of GSC division, several stages of CB maturation can be distinguished (Gilboa et al., 2003; McKearin and Ohlstein, 1995). During the early stage, called the ‘pre-CB’ stage, prominent heterochromatin marks appear that persist throughout oogenesis; these have been linked to the repression of mobile element activity in differentiating germ cells (Rangan et al., 2011). At a later stage, called the ‘CB stage’, the differentiation factor bag of marbles (bam) is expressed. Bam protein is both necessary for germ line cyst differentiation and sufficient to drive GSCs into differentiation (McKearin and Spradling, 1990; Ohlstein and McKearin, 1997). In bam mutants, pre-CB-like cells undergo additional divisions leading to the accumulation of undifferentiated germ cell tumors. The Bam expressing CB divides synchronously four times with incomplete cytokinesis, creating a 16-cell germline cyst (Fig. 1A). One of the cyst cells becomes the oocyte while the others form nurse cells that support the developing oocyte. The events that lead to heterochromatin formation and expression of the differentiation factor Bam during CB maturation are not well understood.
Fig. 1. Pgc is expressed during G2 phase in the differentiating GSC daughter.
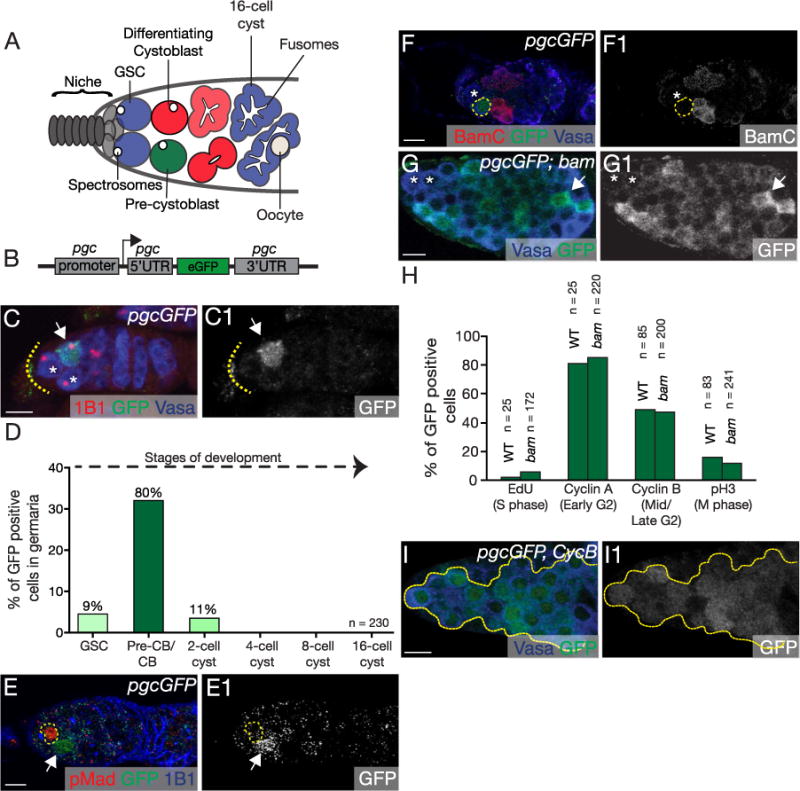
(A) A schematic of the Drosophila female germarium. Stem cells (blue) are attached to the somatic niche (grey). The stem cells divide asymmetrically to renew and to give rise to the pre-cystoblast (pre-CB) (green). The pre-CB expresses Bam and is referred to as the cystoblast (CB) (red). The CB undergoes four incomplete rounds of divisions to give rise to a 16-cell cyst. The undifferentiated cells are marked by structures called spectrosomes while the differentiating cysts are marked by structures called fusomes. (B) The Pgc reporter (pgcGFP) was created by replacing the coding region of pgc with eGFP, leaving the pgc promoter, 5′ UTR and 3′ UTR intact. (C, C1) Germarium of pgcGFP transgenic female stained for 1B1 (red), GFP (green) and Vasa (blue). Pgc is expressed in a single cell of the germarium (white arrow), usually in the cell that is one-cell diameter away from the somatic niche (dotted line). Cells closest to the somatic niche are the germline stem cells (GSC) marked with white asterisks. (D) Quantification of cells expressing Pgc in the germaria (n=230 germaria). 24% of the germaria show pgcGFP expression and 80% of those were one cell diameter away from the niche. Later stages showed no prominent Pgc expression. (E, E1) Germarium of pgcGFP flies stained with pMAD (red), GFP (green) and 1B1 (blue). Pgc expressing cells are not positive for GSC specific marker, pMAD. GSC is marked with a yellow circle. GFP channel is shown in E1. (F, F1) Germarium of pgcGFP flies stained with differentiation marker BamC (red), GFP (green) and Vasa (blue). Pgc expressing cell (yellow circle) is not positive for Bam. BamC channel is shown in F1. White asterisk represents a GSC. (G, G1) pgcGFP; bam mutant germarium stained with GFP (green) and Vasa (blue). 23% of the CB in the bam tumor showed high levels of Pgc expression (white arrow) (n=974 cells, 12 tumors). White asterisk represents GSCs. (H) Quantification of CBs positive for both Pgc and cell cycle markers. Pgc expression correlated mostly with G2 phase markers, CycA (81% in pgcGFP, n=25 germaria and 85% in pgcGFP; bam n=220 cells) and CycB (49% in pgcGFP, n=85 germaria and 47% in pgcGFP; bam, n=200 cells). For representative images see Fig. S1B–I2. (I, I1) Germarium of CycB mutant carrying Pgc reporter stained with Vasa (blue) and GFP (green) show 70% of undifferentiated cells expressing Pgc (n=136 cells, 5 tumors). GFP channel is shown in I1. Scale: 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A number of repressive factors have been identified that either favor GSC self-renewal or promote differentiation to a cyst (Slaidina and Lehmann, 2014; Spradling et al., 2011). The somatic niche is the source of Decapentaplegic (Dpp) ligand that signals to the GSC via Thickveins (TKV) and Saxophone (Sax) receptors expressed in the GSCs (Twombly et al., 1996; Xie and Spradling, 1998). In response to Dpp signaling from the somatic niche, a transcriptional modulator Mothers-against-dpp is phosphorylated (pMad) and then translocates to the nucleus to silence transcription of the differentiation factor, bam. These signaling events promote GSC self-renewal (Kai and Spradling, 2003). GSCs also express high levels of the RNA-binding proteins Nanos and Pumilio, which repress translation of mRNAs that promote GSC differentiation (Forbes and Lehmann, 1998; Gilboa and Lehmann, 2004; Wang and Lin, 2004). After GSC division, TKV/Sax receptors are marked for turnover by the Fused/Smurf complex in the pre-CB to promote Bam expression (Casanueva and Ferguson, 2004; Xia et al., 2010). Bam then downregulates Nanos, promoting expression of differentiation mRNAs (Li et al., 2009). Thus, protein turnover and translational control play critical roles in mediating the switch between GSC self-renewal and differentiation.
Cell cycle control is a critical aspect in the decision between GSC maintenance versus differentiation. Drosophila GSCs have a short G1 and a long G2 phase (Hsu et. al, 2008). The new GSC daughter and the CB remain connected throughout most of the cell cycle, and complete abscission between the cells is not completed until G2 phase (de Cuevas and Spradling, 1998; Mathieu and Huynh, 2017). Cyclin B (CycB) is a late-G2 phase regulator that controls the G2-M phase transition but also plays additional roles in GSC development (Chau et al., 2012; Wang and Lin, 2005). Loss of CycB leads to GSC self-renewal and maintenance defects (Wang and Lin, 2005). CycB is also required for the abscission of the CB from the GSCs, and for CB proliferation (Chau et al., 2012; Mathieu et al., 2013). Thus, CycB plays a pivotal role in the early germ line to ensure GSC self-renewal, maintenance, and proliferation of the GSC daughter. While cell cycle regulators, such as CycB, are themselves transcriptionally and translationally regulated (Kronja and Orr-Weaver, 2011), it is not known how the levels of CycB are modulated during the course of GSC differentiation.
Here, we report that an active silencing of transcription promotes differentiation of the CB. We find that the transcriptional repressor Pgc is transiently expressed in the pre-CB prior to the expression of the differentiation factor Bam. Pgc expression mediates a pulse of transcriptional silencing that is required for timely CB differentiation. This transcriptional silencing by Pgc indirectly promotes accumulation of CycB to modulate the cell cycle state of the pre-CB as it transitions into the CB. Additionally, Pgc promotes the formation of genome-protecting heterochromatic marks in the pre-CB via regulation of the cell cycle. Our results suggest that during the GSC to CB transition, Pgc mediated transcriptional silencing has a role in reprogramming self-renewal programs, such as cell cycle, to permit efficient differentiation. We propose that, in addition to mechanisms such as protein turnover and translational control (Casanueva and Ferguson, 2004; Li et al., 2009; Xia et al., 2010), transient transcriptional silencing can provide a mechanism to clear the stem cell program for another, the differentiation program, within one cell division.
2. Results
2.1. The transcriptional repressor Pgc is expressed in the GSC daughter
pgc encodes a small 71 amino acid peptide that is expressed in early primordial germ cells of the Drosophila embryo (Hanyu-Nakamura et al., 2008). We generated a pgc reporter by fusing the pgc promoter and its regulatory 5′ and 3′ untranslated region (UTR) elements to eGFP (pgcGFP) to observe the expression pattern of Pgc (Fig. 1B). This reporter recapitulated the expression of endogenous Pgc during embryogenesis, where pgcGFP was translated only in early germ cells and not in the germ plasm (Hanyu-Nakamura et al., 2008; Rangan et al., 2009) (Fig. S1A–A1). This pgcGFP reporter was also expressed transiently during the early stages of GSC development (Fig. 1C–C1). To determine the identity of the cell expressing Pgc, we used antibodies against Vasa to mark the germ line and antibodies against the Adducin-like protein Hts (1B1) to distinguish between the GSCs and their progeny (de Cuevas et al., 1996; Lasko and Ashburner, 1990; Zaccai and Lipshitz, 1996). 1B1 stains a round, membranous structure called the spectrosome in both the GSC and in its daughters, the pre-CB and CB (Fig. 1A) (de Cuevas and Spradling, 1998; Lasko and Ashburner, 1990). As the CB divides to develop into a sixteen cell germline cyst, the spectrosome grows into a branched structure, called the fusome, that connects the cells within cysts (Fig. 1A) (de Cuevas and Spradling, 1998). We found that Pgc is primarily expressed in the spectrosome-containing germ cell that was not in direct association with the stem cell niche (Fig. 1C–D). This suggests that Pgc is expressed in the undifferentiated GSC daughter cell.
To test our hypothesis that Pgc is not expressed in the GSC, but rather in the GSC daughter, we co-stained pgcGFP expressing germaria with antibodies against the GSC marker, pMad, and the CB differentiation marker Bam (Kai and Spradling, 2003; McKearin and Ohlstein, 1995). GSCs robustly express pMad, but not Bam. During the pre-CB stage pMad is still weakly expressed, but Bam is not expressed, while differentiating CBs do not express pMad, but express Bam strongly. We detected GFP expression only in cells with low pMad expression (Fig. 1E–E1) (n=10 germaria) and no Bam expression (Fig. 1F–F1) (n=18 germaria), suggesting that Pgc is expressed in the pre-CB stage. If Pgc was expressed prior to Bam, we would expect that Pgc expression be independent of Bam. In bam mutants, GSC daughter cells accumulate that cannot differentiate but continue to divide resulting in tumors enriched for pre-CB markers (Gilboa et al., 2003; McKearin and Ohlstein, 1995). We found that Pgc was expressed in a subset of cells that accumulate in bam mutant tumor (23%, n=974 cells from 12 tumors) (Fig. 1G–G1). Collectively our data show that Pgc is expressed prior to and independent of the expression of the differentiation factor Bam.
2.2. Pgc is expressed in a cell cycle dependent manner in the GSC daughter
The pre-CB and the CB stage describe developmental time points within the same differentiating cell. Our data suggest that Pgc is expressed during the pre-CB stage, however, in bam mutants not all undifferentiated cells express Pgc. This observation led us to ask if Pgc expression was cell cycle regulated. To correlate stages of the cell cycle with Pgc expression, we used the following cell cycle markers: a pulse of the nucleotide analogue 5-Ethynyl deoxyUridine (EdU) to identify S phase, and antibodies against CycA, CycB and phosphorylated histone 3 (pH3) to mark the early-G2 phase, late-G2 phase, and M phase, respectively (Fig. 1H) (Hsu and Drummond-Barbosa, 2011). pgcGFP expression was correlated with particular cell cycle markers in both wild-type cells and in pre-CB-enriched pgcGFP; bam tumors (Fig. 1H, S1B–I2). The majority of Pgc expressing cells expressed the G2 phase markers CycA and CycB. The early-G2 marker CycA showed the strongest correlation (Fig. 1H, S1F–G2) followed by the late-G2 phase marker CycB (Fig. 1H, S1H–I2). CycA is primarily present in the cytoplasm (Ohlmeyer and Schupbach, 2003). However, we observed CycA as puncta in the germ line. Our staining is consistent with CycA staining previously observed in the Drosophila testes (Yuan et al., 2012). The puncta are observed with both, methanol and paraformaldehyde fixations, and are specific, as they are absent or significantly reduced in CycA mutants (Fig. S1J–O1). To further explore the cell cycle-dependent regulation of Pgc, we analyzed its distribution in CycB mutants, where the cell cycle is arrested at the end of G2 phase, before the G2 to M phase transition. While CycB mutants lose some GSCs due to under-proliferation, as previously reported (Wang and Lin, 2005), we also observed an accumulation of undifferentiated cells. These undifferentiated cells in CycB mutants can enter S phase as they incorporate EdU (Fig. S1P–P1) but are arrested in early G2 phase as they express CycA (Fig. S1Q–Q1). 70% of these undifferentiated cells expressed Pgc (n=136 cells, 5 germaria) (Fig. 1I–I1). Taken together, we conclude that Pgc is expressed in the GSC daughter during the G2 phase of the cell cycle prior to the expression of differentiation factor Bam.
The expression of Pgc in G2 phase is intriguing, as previous studies have shown that the cellular connection between the GSC and CB is only severed during the G2 phase (de Cuevas and Spradling, 1998; Hsu and Drummond-Barbosa, 2011; Hsu et al., 2008; Mathieu and Huynh, 2017). These studies showed that spectrosome morphology during GSC division is a sensitive marker to follow the separation of the GSC from its daughter. During GSC division, cytokinesis is delayed and the spectrosome continues to bridge the new GSC and the CB. Then at G2, the connection between the two cells is severed (de Cuevas and Spradling, 1998; Hsu and Drummond-Barbosa, 2011). To determine how Pgc expression is linked to the separation of the CB from the GSC, we tracked spectrosome morphology of the CB along with Pgc expression and found that Pgc was expressed in the “round” stage (Fig. S2A–C2). Consistent with our cell cycle analysis, we conclude that Pgc is expressed during G2 phase of the cell cycle, after the cytoplasmic connection between the GSC and CB has been severed.
2.3. Pgc facilitates CB differentiation
Given that Pgc is expressed prior to and independent of the differentiation factor Bam, we asked if Pgc is required for differentiation. We assayed for differentiation defects by examining the expression of the GSC marker, pMad, and the differentiation marker, Bam in both wild-type and pgc mutants (Kai and Spradling, 2003; McKearin and Ohlstein, 1995). We found that pgc mutants accumulated undifferentiated, pre-CB-like cells that were removed from the niche but did not express Bam or pMad when compared to wild-type (Fig. 2A–B, D). This phenotype is rescued by providing two copies of the genomic region containing pgc (Fig. 2A–D). The accumulation of pre-CB-like cells in pgc mutants could be due to either Pgc expression in the pre-CB indirectly promoting GSC proliferation or due to Pgc directly affecting Bam expression in the CB. An effect on GSC proliferation seemed unlikely, as the rate of GSC divisions in pgc mutants (6%; n=80 germaria) as compared to wild-type (7%; n=80 germaria) was not significantly altered (Fig. S3A–B). Instead, pgc mutants showed an accumulation of cells that do not express the reporter for Bam transcription (Fig. S3C–E), suggesting that Pgc promotes differentiation by regulating Bam expression. Because pgc mutant germ cells can differentiate, pgc is not essential for differentiation but rather its expression could serve as a regulatory step in transition from a GSC fate to a more differentiated fate.
Fig. 2. Pgc promotes cystoblast differentiation.
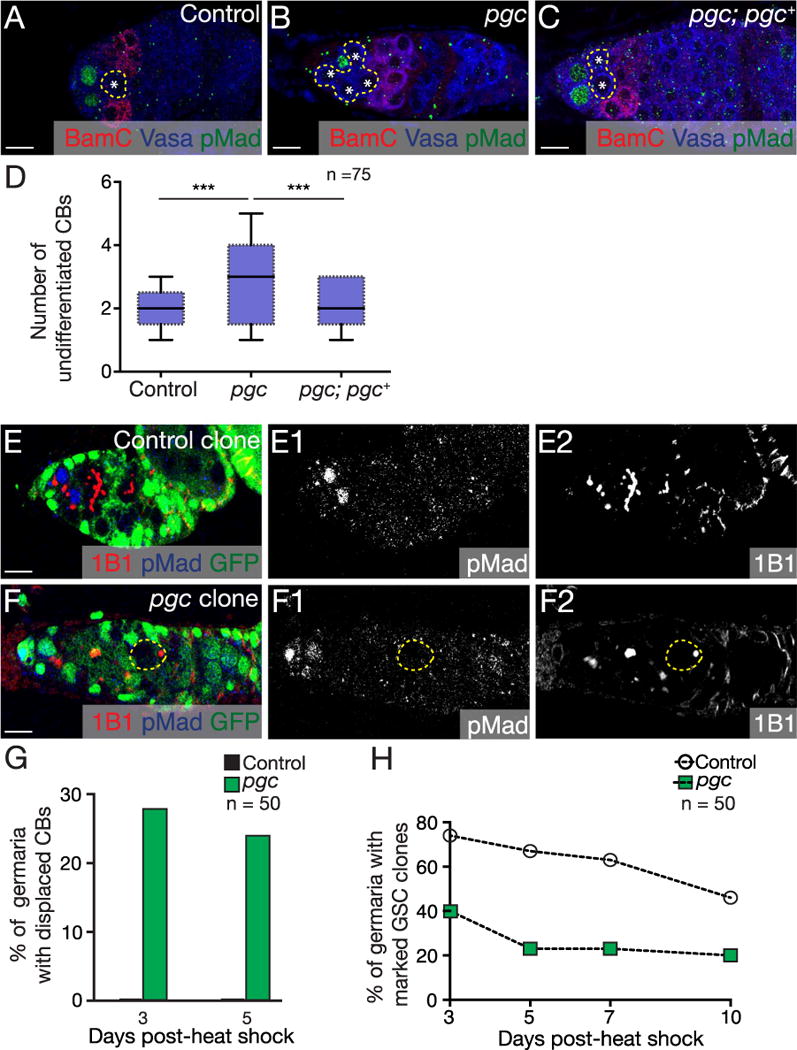
(A–C) Germaria of a control, pgc mutant and pgc; pgc+ ovaries stained with BamC (red), Vasa (blue) and pMad (green). pgc mutants accumulate supernumerary undifferentiated cells. This phenotype is rescued by providing two copies of genomic pgc+ transgene. (D) Quantification of number of undifferentiated CBs in the germaria of WT, pgc mutants and pgc; pgc+ (2.4 ± 0.7 in control, 4.4 ± 1.1 in pgc and 2.7 ± 0.7 in pgc; pgc+, n=75 germaria). Horizontal lines in a box-and-whisker plots represent maximum, third-quartile, median, first quartile and minimum. Paired t-test was performed. *** p < 0.0001. Germaria of (E) control clone and (F) pgc clones stained with 1B1 (red), pMad (blue) and GFP (green). When compared to the control clone 3 days post heat shock, pgc clone germaria show a pMad negative, single undifferentiated cell displaced far away from the niche. pMad and 1B1 channels are shown in E1, F1 and E2, F2 respectively. (G) Quantification of the percentages of single undifferentiated cells found away from the niche 3 and 5 days post heat shock (n=50). (H) Graphical representation of percentage of marked GSC clones present after 3, 5, 7 and 10 days post heat shock (n=50). For representative images see Fig. S3F–K. Scale: 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further determine the role of pgc specifically in the germ line and in the context of wild-type tissue, we generated pgc mutant clones using the Flippase/Flippase Recognition Target (FLP/FRT) recombination system (Song and Xie, 2002). In wild-type control clones, spectrosome-containing undifferentiated cells, such as GSCs and CBs, are only found in close vicinity to the somatic niche but in pgc mutant clones we observed single, undifferentiated cells that were pMad negative located far from the niche (Fig. 2E–G). Although, pgc is not expressed in the GSCs, in addition to the displaced single cell, we also observed that there were a fewer number of marked pgc mutant clone GSCs compared to wild-type marked GSC clones, 3 and 5 days post-heat shock (Fig. 2H, S3F–K). Stem cell clones of differentiation genes such as bam and bcgn, though they do not play a role in GSCs, seem to play a role in stem cell competition in the niche (Jin et al., 2008). Our results suggest that pgc mutant GSCs could be competed out by wild-type GSCs early after clone induction and pgc mutant CBs struggle to differentiate in the context of heterozygous tissue. These results suggest that Pgc acts in germ cells to promote robust CB differentiation.
Continuous egg production requires timely CB differentiation. To test whether the differentiation defects in pgc mutants have an adverse effect on fecundity, we carried out an egg-laying test to compare pgc mutant to control. We found that pgc mutant flies consistently laid fewer eggs than wild-type over 21 days (~19% less), supporting the conclusion that the delay we see in CB differentiation has a direct effect on egg production (Fig. S3L). Thus, pgc is required for proper CB differentiation and fecundity in the ovary.
2.4. Pgc modifies CB cell cycle by regulating Cyclin B to promote differentiation
Because Bam is the major instructor of GSC differentiation and its expression follows that of Pgc, we asked whether Pgc regulates Bam expression. As pgc mutants are capable of differentiation, it is less likely that Pgc promotes differentiation by increasing the levels of Bam expression needed for differentiation. Alternatively, it is possible that Pgc controls the timing of Bam expression. We therefore tested if loss of pgc caused a delay in the expression of Bam due to changes in the cell cycle. We analyzed cell cycle progression in pgc mutants using markers for individual phases of the cell cycle (Fig. S4A–H). To circumvent the problem that CBs are connected to GSCs until the early-G2 phase and progression into M phase results in cyst differentiation, we compared cell cycle phases in bam mutants with those of pgc; bam double mutants. Both these genotypes enrich for the same pre-CB cell type that either express Pgc or lack pgc respectively. This allowed us to evaluate the cell cycle of pre-CBs individually. We observed a higher proportion of cells in S and early-G2 phase in pgc; bam double mutants compared to bam mutants (Fig. 3A, S4A–D). Conversely, pgc; bam double mutants had fewer cells positive for both the late-G2 phase marker, CycB, and M phase marker, pH3, compared to bam mutants (Fig. 3A, S4E–H). This suggested that pgc mutants have defects in entering and progressing towards the late-G2 phase of the cell cycle. To determine if pgc single mutants are also accumulating cells in early-G2 phase, we stained for early-G2 marker CycA, and found that the majority of the single undifferentiated cells in pgc mutants were CycA positive (55% in bamGFP compared to 70% in pgc; bamGFP germaria, n=45) (Fig. S4I–J1). This indicates that pgc mutants are accumulating cells in the early-G2 phase.
Fig. 3. Pgc promotes cell cycle progression.
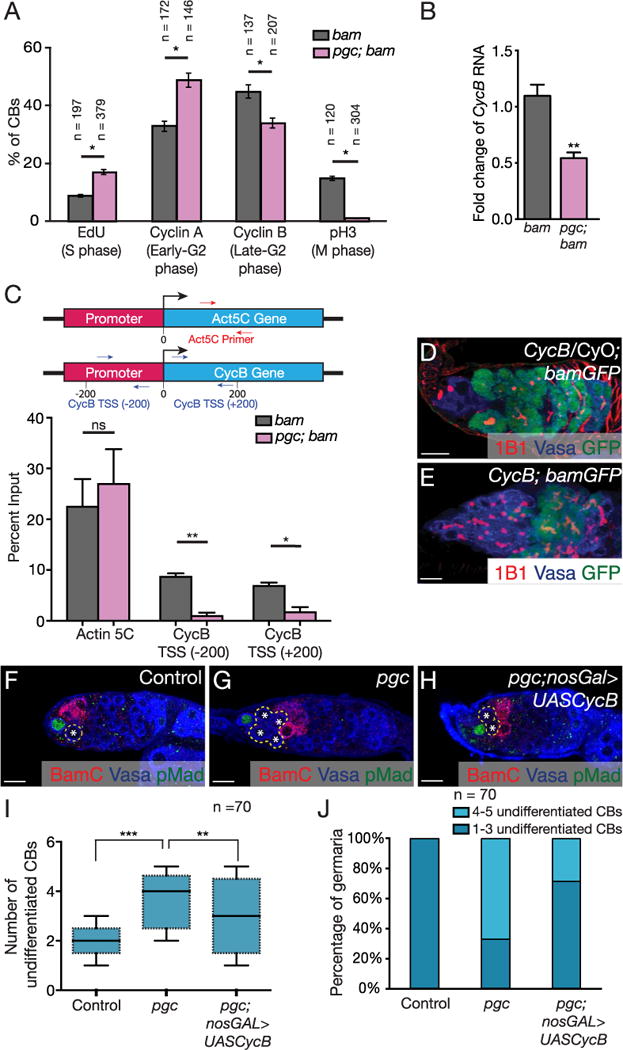
(A) Quantification of CBs positive for different cell cycle markers in both bam and pgc; bam mutants. An increase in S phase cells in pgc; bam mutants was observed (9% in bam, n=197 cells and 17% in pgc; bam, n=379 cells, p < 0.05). pgc; bam mutants had a higher number of cells positive for early-G2 phase marker, CycA (33% in bam, n=172 cells and 49% in pgc; bam, n=146 cells, p < 0.05). pgc; bam mutants also showed fewer cells positive for the late-G2 phase marker, CycB (45% in bam, n=137 cells and 34% in pgc; bam, n=207 cells, p < 0.05), and even lower M phase positive cells (15% in bam, n=120 cells and 1% in pgc; bam, n=304 cells, p < 0.05). For representative images see Fig. S4A–H. Two-way ANOVA test was performed. * p < 0.05. (B) qPCR analysis shows CycB mRNA is down regulated in pgc; bam tumors when compared to bam tumors. Paired t-test was performed. **p < 0.001. (C) qPCR analysis on positive control Act5c intron1 fragment and CycB transcription start site (−200) and (+200) after Pol II ChIP experiments in bam and pgc; bam mutants. CycB transcription in pgc; bam mutant shows a decrease as compared to bam mutant, whereas transcription of Act5c does not change. (D) Germarium of CycB/ CyO; bamGFP flies stained with 1B1 (red), Vasa (blue) and GFP (green) show timely expression of Bam. (E) Germarium of CycB; bamGFP flies stained with 1B1 (red), Vasa (blue) and GFP (green) show a significant accumulation of undifferentiated cells with delayed Bam expression. (F–H) Germaria of a control, pgc mutant and pgc; nosGal4 > UASCycB ovaries stained with BamC (red), Vasa (blue) and pMad (green). pgc mutants accumulate undifferentiated cells which is partially rescued by overexpressing CycB in the germ line. (I) Quantification of number of undifferentiated CBs in the germaria of WT, pgc mutants and pgc; nosGal4 > UASCycB (2.1 ± 0.5 in control, 3.9 ± 1.0 in pgc and 3.0 ± 0.9 in pgc; UASCycB n=70 germaria). Horizontal lines in a box-and-whisker plots represent maximum, third-quartile, median, first quartile and minimum. Paired t-test was performed. *** p < 0.0001, ** p < 0.001 (J) Stacked graph representing the distribution of pre-CB numbers observed in WT, pgc mutant and pgc; nosGal4 > UASCycB germaria. Scale: 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The observation that the G2 phase is altered in pgc mutants indicated CycB as a downstream target of pgc regulation. CycB is required for the progression from G2 to M phase and its expression peaks prior to expression of the differentiation factor Bam (Fig. S4K–K1). We asked if CycB transcription is affected in pgc mutants by measuring the levels of CycB transcript. We compared CycB transcript levels in bam mutants with pgc; bam double mutants and found that CycB transcript levels are indeed reduced (Fig. 3B). To determine if the changes in transcript levels of CycB were due to differential RNA Pol II occupancy on their promoters, we carried out ChIP-PCR on the G2 regulator, CycB. We found that, compared to bam mutants, pgc; bam mutants displayed lower Pol II occupancy on the CycB promoter compared to a control gene, actin5C (Fig. 3C). Taken together, our data suggest that pgc is required for the timely progression into late-G2 by regulating CycB mRNA levels transcriptionally by repressing yet undetermined factors.
If pgc regulates Bam expression by modulating CycB expression and cell cycle progression, lowering CycB levels independent of pgc should mimic the pgc mutant phenotype. CycB mutants were previously reported to lose GSCs (Fig. S5A–B) (Wang and Lin, 2005). In addition to this, we observed that 35% of cycB mutant germaria accumulate more than 5 single, spectrosome containing cells (Fig S5C–E). To determine the identity of these cells, we assayed for bam transcription in CycB mutants using the transcriptional reporter for Bam, and the GSC marker, Daughters-against-dpp fused to a LacZ reporter (Dad-lacZ). We found that CycB mutants accumulated both Dad-lacZ and BamGFP-negative undifferentiated cells (Fig. 3D–E, S5F–H). Thus, reduction of CycB causes an accumulation of pre-CBs similar to that observed in pgc mutants. If pgc was indeed regulating CycB expression to promote timely differentiation, we hypothesized that over-expression of CycB could rescue the delay in differentiation observed in pgc mutants. Therefore, we overexpressed CycB in the germ line of pgc mutants and assayed for the number of undifferentiated CBs by staining for pMad and BamC. We found that over-expression of CycB significantly reduced the number of accumulating pre-CBs in pgc mutants (Fig. 3F–J). However, this rescue was only partial (Fig. 3I–J). Our data suggests that Pgc modulates CycB to promote pre-CB differentiation.
2.5. Pgc causes transcriptional silencing
During embryogenesis, Pgc acts as a global transcriptional repressor. To test if Pgc expression affects overall transcription in the pre-CB, we assayed for active transcription by incubating ovaries with a pulse of 5-Ethynyl Uridine (EU), which incorporates into nascent RNAs (Navarro-Costa et al., 2016). Pre-CBs that expressed Pgc showed reduced EU incorporation, as compared to neighboring GSCs and differentiating cysts in both wild-type and in pgcGFP positive bam mutant cells. (Fig. 4A–B3). This phase of transcriptional repression is concurrent with CycA (Fig. S6A–A3) and occurs prior to CycB and Bam expression (Fig. S6B–B3). To test if loss of pgc would result in higher number of transcriptionally active pre-CBs, we assayed for EU incorporation in the pre-CBs of bam and pgc; bam mutants. We observed an increase in transcriptionally active pre-CBs in pgc; bam mutants when compared to bam mutants (Fig. 4C–E). Taken together, our data suggests that Pgc expressing pre-CBs undergo a phase of transcriptional quiescence prior to differentiation.
Fig. 4. Pgc causes transcriptional silencing in the stem cell daughter via the P-TEFb complex.
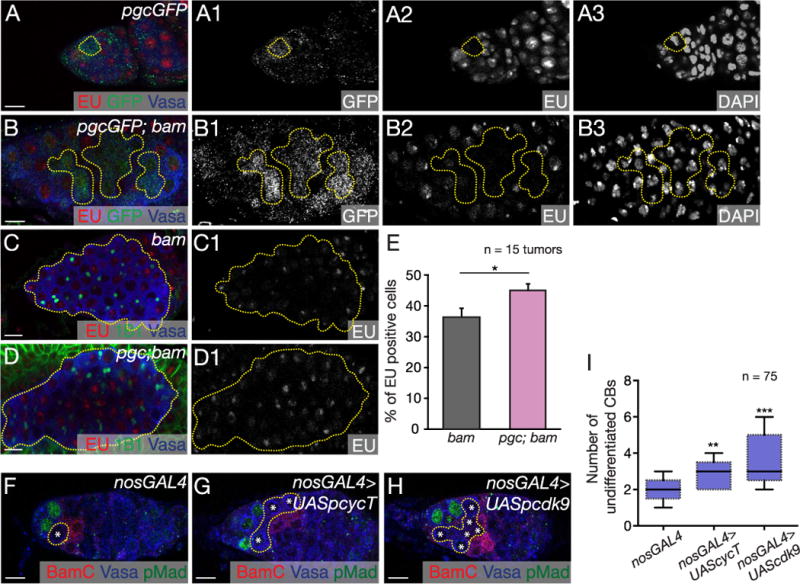
(A–A3) pgcGFP germarium pulsed with EU (red) and stained with GFP (green) and Vasa (blue) showed no transcription in Pgc expressing cell (yellow circle). GFP, EU and DAPI channels are shown in A1–A3 respectively. (B–B3) pgcGFP; bam tumors pulsed with EU (red) and stained for GFP (green) and Vasa (blue) show 64% of Pgc expressing CBs are transcriptionally quiescent (n=20 tumors). GFP, EU and DAPI channels are shown in B1–B3 respectively. (C–D1) bam and pgc; bam tumors pulsed with EU (red) and stained with 1B1 (green) and Vasa (blue). EU channels are shown in C1 and D1 respectively. (E) Quantification of cells positive for EU in bam and pgc; bam tumors. pgc; bam mutants had 45% of cells positive for EU incorporation compared to 36% of cells in bam mutant (n=15 tumors). Error bars are represented by standard error. Paired t-test was performed. * p < 0.05. Germaria of (F) nosGal4, (G) nosGal4 > UASp cyclinT and (H) nosGal4 > UASp cdk9 flies stained with pMad (green), Vasa (blue) and BamC (red). (I) Quantification of undifferentiated CBs in nosGal4 (1.9 ± 0.6), nosGal4 > UASp cyclinT (2.9 ± 0.7) and nosGal4 > UASp cdk9 (3.4 ± 1) germaria. nosGal4 > UASp cyclinT and nosGal4 > UASp cdk9 germaria showed accumulation of undifferentiated cells when compared to nosGal4 control (n=75 germaria). Horizontal lines in a box-and-whisker plots represent maximum, third-quartile, median, first quartile and minimum. Paired t-test was performed. ** p < 0.001, ***p < 0.0001. Scale: 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
During embryogenesis, Pgc mediates transcriptional silencing in the germ cells by inhibiting the P-TEFb complex that consists of CycT and Cdk9 (Hanyu-Nakamura et al., 2008; Martinho et al., 2004; Seydoux and Dunn, 1997). The P-TEFb complex mediates the switch of RNA Pol II from the initiation to the elongation phase (Saunders et al., 2006). Loss of pgc, or overexpression of components of the P-TEFb complex, results in spurious transcription in embryonic germ cells (Hanyu-Nakamura et al., 2008; Martinho et al., 2004). To test if Pgc also acted as a suppressor of transcription via inhibition of the P-TEFb complex during oogenesis, we overexpressed two components of the P-TEFb complex, Cdk9 and CycT, individually in the germ line during oogenesis. We found that overexpression of either Cdk9 or CycT resulted in accumulation of undifferentiated cells that were both pMad and Bam negative (Fig. 4F–I). This phenotype mimics the pgc mutant phenotype suggesting that Pgc promotes CB differentiation by inhibition of the P-TEFb complex. Alternatively, we also tried to rescue pgc mutant phenotype by depleting Cdk9 and CycT in the germ line of pgc mutants. However, P-TEFb is required for transcription elongation of all RNA Pol II dependent genes, and its depletion in the germ line results in germ line death (Fig. S6C–D). Therefore, rescuing the pgc mutant phenotype by depleting Cdk9 and CycT during oogenesis was not feasible. Altogether, our data suggest that a Pgc-induced period of transcriptional silencing causes downregulation of yet unknown GSC transcriptional regulators during the pre-CB stage. This transition promotes cell cycle regulation via CycB and Bam expression leading to differentiation.
2.6. Pgc controls increase in heterochromatin marks during differentiation
The cell cycle is intimately linked to epigenetic changes, as DNA replication presents a window of opportunity to change epigenetic states during differentiation (Probst et al., 2009). We previously showed that heterochromatin marks increase dramatically during the pre-CB stage compared to GSCs (Rangan et al., 2011). An important role of heterochromatin in the germ line is to protect the integrity of the genome. This is, in part, achieved by heterochromatin regulating the production of piRNAs and controlling transposon activity (Mohn et al., 2014; Rangan et al., 2011; Zhang et al., 2014). To determine whether Pgc is required to regulate the level of heterochromatin formation in the pre-CB, we co-stained for Pgc (pgcGFP) and the heterochromatin marker Histone 4 lysine 20 methyl 3 (H4K20me3). We found that pre-CBs expressing Pgc showed prominent heterochromatin marks, suggesting that the increase in heterochromatin and transcriptional silencing occur at the same phase of the CB cell cycle (Fig. 5A–A1). Next, to determine if loss of pgc during oogenesis resulted in changes in heterochromatin formation, we stained pgc; bam mutant germaria for heterochromatin and found that heterochromatin marks were reduced in the majority of pre-CBs, as compared to those in bam tumors (Fig. 5B–C1, E) (Rangan et al., 2011). We next asked if Pgc regulates heterochromatin formation directly or indirectly through its regulation of the cell cycle, via CycB. To test this, we assayed heterochromatin formation in CycB mutants and found decreased heterochromatic marks only in the germ line but not in the soma (Fig. 5D–E). We conclude that Pgc modulates the cell cycle of the CB daughter to allow heterochromatin formation during mid- to late-G2 phase. In embryonic germ cells, loss of pgc also results in acquisition of active chromatin marks (Martinho et al., 2004). We therefore assayed for active chromatin marks using antibodies to H3K4me3 in the germarium. We observed that H3K4me3 mark was not altered in pgc; bam compared to bam mutants (Fig. S7A–C). Thus, we conclude that during oogenesis pgc does not affect active epigenetic marks, but regulates genome-protecting heterochromatic marks.
Fig. 5. Pgc regulates heterochromatin formation via cell cycle during differentiation.
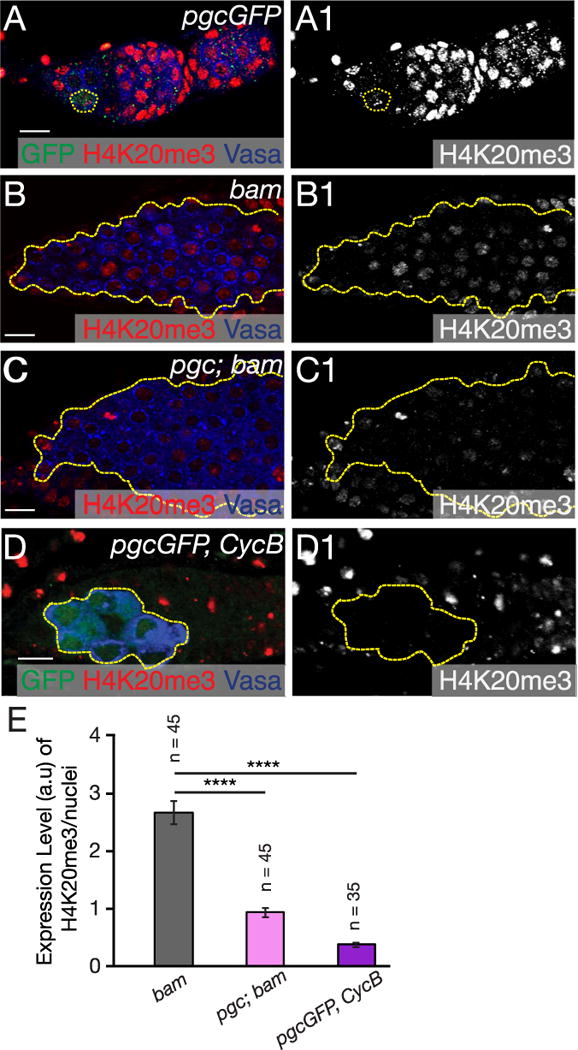
(A) Germarium of pgcGFP stained with GFP (green), H4K20me3 (red) and Vasa (blue) show Pgc expression concurrent with increasing heterochromatin in the CB. (B) bam mutant germarium stained with H4K20me3 (red) and Vasa (blue) show majority of the CBs with increased heterochromatin. (C) pgc; bam mutant germarium stained with Vasa (blue) and H4K20me3 (red) show a large fraction of cells with no heterochromatin formation. (D) pgcGFP, CycB germarium stained with GFP (green), H4K20me3 (red) and Vasa (blue) show Pgc expressing CBs do not form heterochromatin in the germ line in absence of CycB. (E) A graph depicting quantification of arbitrary units (a.u) of H4K20me3 expression levels in bam (n=45 cells), pgc; bam (n=45 cells) and pgcGFP, CycB (n=35 cells) germaria. Compared to bam mutant cells, H4K20me3 marks are significantly reduced in pgc; bam and pgcGFP, CycB mutant cells. Student t-test was performed. ****p < 0.00001 Scale: 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
Our studies identify the G2 phase of the cell cycle as a regulated stage in the transition from a stem cell to a more differentiated fate. Transient transcriptional silencing, mediated by the transcriptional repressor Pgc, is important for this transition. The physical properties of the Drosophila germarium allowed us to follow this transition with exquisite spatial and temporal resolution. We find that a Pgc reporter is expressed in the G2 phase of the GSC daughter after it loses contact with the GSC. At this stage of the cell cycle, which has been referred to as the pre-CB stage, the GSC daughter has reduced expression of typical GSC markers, but has yet to turn on expression of the differentiation marker Bam (Gilboa et al., 2003; McKearin and Ohlstein, 1995). It is likely that our pgc reporter accurately accounts for the onset of expression, but due to the stability of GFP, may overestimate the duration of Pgc expression. The expression of Pgc in the pre-CB at its G2 phase is reminiscent of Pgc expression in embryonic germ cells, where it is expressed in post replicative germ cells that have recently lost contact with the soma and have arrested in G2 phase (Cinalli and Lehmann, 2013; Rangan et al., 2009; Su et al., 1998).
In early embryonic germ cells, Pgc causes global transcriptional silencing by inhibiting the recruitment of the P-TEFb complex to chromatin (Hanyu-Nakamura et al., 2008; Martinho et al., 2004). Consistently, we observed a significant decrease in EU incorporation in pre-CBs concurrent with Pgc expression. Furthermore, we found that overexpression of P-TEFb components caused a delay in differentiation similar to that of pgc mutants. These results suggest that, during GSC division, the daughter cell destined for differentiation undergoes a period of transcriptional quiescence. During embryogenesis, Pgc is expressed when germ cells switch from fast cycling syncytial nuclei to a slow cycling state, suggesting that changes to the cell cycle program may be critical to maintain germ cell identity (Hanyu-Nakamura et al., 2008; Rangan et al., 2009). In both embryogenesis and oogenesis, Pgc expression and subsequent transcriptional silencing occurs in G2 right after S phase, where overall transcriptional activity is low and chromatin marks are being reset through the passage of the replication fork (Probst et al., 2009). Therefore, we reason that global transcriptional silencing at this stage is advantageous, as transcriptional activity is already low and epigenetic marks can be easily altered to reset cell fate.
We propose that Pgc mediates the efficient transition toward differentiation by altering the CB cell cycle. Our data suggests that transient Pgc expression during G2 phase leads to up-regulation of CycB. Changes to the cell cycle may be critical to promote GSC differentiation, as these cells are slow cycling, with short G1 and long G2 phase (Hsu and Drummond-Barbosa, 2011). At this point, we do not know how Pgc function leads to increased transcription of CycB and how many genes are targets of pgc in the pre-CB. During differentiation, GSC daughter cells progress into G2 phase and change their epigenetic landscape (Ables and Drummond-Barbosa, 2013; Hsu and Drummond-Barbosa, 2011). It is known from yeast that progression into G2 phase promotes recruitment of epigenetic modifiers that establish heterochromatin and aids in its spreading (Chen et al., 2008). Our finding that progression of the pre-CB into mid-late G2 phase promotes the formation of heterochromatin is consistent with this observation.
B-Lymphocyte-Induced Maturation Protein 1 (BLIMP1) mediates the transcriptional silencing of genes required for somatic differentiation in early embryonic germ cells in the mouse (Hayashi et al., 2007). Like pgc mutants, germ cells are not properly specified in Blimp1 mutants. Blimp1, along with other transcription factors, can reprogram epiblast-like cells into a germ cell fate (Nakaki et al., 2013), demonstrating that transcriptional silencing is an important part of resetting cell fate. Blimp1 function, unlike Pgc, is not restricted to germ cells, and loss of Blimp1 also results in improper differentiation of somatic cells during development. Based on our data, we propose that transient transcriptional silencing mediated by Pgc “clears” the previous stem cell state by temporarily preventing transcription of existing cell fate regulators and allowing the resetting of epigenetic marks. As this “clearing” is not itself instructive to specify cell fate, it could be used to efficiently reprogram somatic cells back to pluripotency and GSCs toward oocyte differentiation.
4. Materials and methods
4.1. Fly Stocks
Drosophila were grown on corn flour and agar media with brewer’s yeast. All strains were grown at 25 °C. pgcGFP, pgcΔ and pgcΔFRT fly lines were generated for this study. liprinγH1 and bamΔ86 flies were acquired from the Treisman Lab (Astigarraga et al., 2010) and McKearin Lab (McKearin and Ohlstein, 1995) respectively. nos-Gal4::VP16, UASCycB (Mathieu et al., 2013) and sco/CyO; MKRS/TM6 was acquired from the Lehmann lab. UASpCycT-nos3′UTR, UASpcdk9-nos3′UTR and Genomic pgc+ transgene was given to us by the Nakamura Lab (Hanyu-Nakamura et al., 2008). CycB2, bamGFP, hsFLP, 42DFRT-GFP, CycA[c03456], CycA[03946] and CycA[c8LR1] are available at Bloomington Drosophila Stock Center, Bloomington, IN (Wang and Lin, 2005). CycTRNAi (v103387) and cdk9RNAi (v103569) lines were acquired from VDRC stock center.
4.2. Immunostaining
Female Drosophila ovaries were dissected in 1X PBS and fixed in 4% paraformaldehyde for 30 min. Then the tissue was permeabilized in 1 mL of PBST (1X PBS, 0.2% Tween and 1% Triton-X). After permeabilization the tissues were blocked in 1 mL of BBT (0.5% BSA in PBST). Then 0.5 mL of primary antibody was added and tissues were kept in 4 °C overnight. For methanol fixation, ovaries were fixed at 80 °C for 10 min in 90% methanol and 3% formaldehyde. The ovaries were then washed three times with PBT (0.3% Triton-X, 0.25% BSA) for ten minutes each wash. After washes, primary antibody was added as described above. Concentration used for each antibody has been detailed below. After overnight incubation, ovaries were washed three times in 1 mL of BBT for 10, 15, 30 min. An additional wash for 30 min was carried on by adding 2% Donkey serum to 1 mL of BBT. After the last wash secondary antibody in 0.5 mL of BBT with 4% Donkey serum was added and incubated for 2 h away from light. Secondary antibodies used in this study have also been listed below. After the 2-h incubation, ovaries were washed in 1 mL of PBST for five times. After the washed one-drop of Vectashield was added and then the tissue was mounted on a glass slide and a coverslip was placed on the slide. Antibodies used in this study are Rb Vasa (1:5000 dilution) and Ch Vasa (1:500 dilution) was generated in our lab. Mo 1B1 (1:20) and Mo Cyclin A (1:10) is from DSHB, Iowa city, IA. Ch GFP (Ab13970) (1:1000), pSmad3 (ab52903) (1:150), Mo H4K20me3 (ab78517) (1:500), H3K4me3 (ab8580) (1:500) were acquired from abcam, Cambridge MA. Rb pH3 (9701) (1:200) is from Cell Signaling, Beverly, MA. Rb Cyclin B (Sc-25764) (1:200) is from Santa Cruz Biotechnology, Dallas, TX. EU (C10330) and EdU (C10338) kits are from Life technologies, Grand Island, NY. Alexa 488 (Molecular Probes), Cy3 and Cy5 (Jackson Labs) conjugated secondary antibodies were used at a concentration of 1:500. All ovary images were taken either with a Carl Zeiss 510 meta or 710 meta confocal microscopes using a 40× oil immersion objective. Embryo images were taken with a Carl Zeiss 510 meta confocal microscope using a 20X dry objective. All scale bars were generated using Zen Blue (Lite) software. Images were processed using ImageJ and Photoshop (Adobe) software.
4.3. EdU and EU pulsing
Fly ovaries were dissected in Grace’s Media. 1 mL of 10 μM solution of EdU (Click-iT® Plus EdU Kits, Life Technologies, Grand Island, NY [Catalog number: C10338]) and 1 mM EU solution (Click-iT® RNA Imaging Kit, Life Technologies, Grand Island, NY [Catalog number: C10330]) was added to each genotype and pulsed for one hour. After pulsing, the tissues were fixed in 5% formaldehyde for 30 min. After fixation, tissues were washed once with 1X PBS and permeabilized in 1%Triton-X PBST for one hour. Next, EdU and EU was detected by adding 500 μl of detection cocktail according to the recommended recipe (Click-iT® Plus EdU Kits, Click-iT® RNA Imaging Kits). After EdU and EU detection, immunostaining was carried out according to the aforementioned protocol.
4.4. Fluorescence imaging
The tissues were visualized under 10X, 20X and 40X objective lenses. The images were acquired by using a Zeiss LSM-510/710 confocal microscope under 40X objective.
4.5. Analysis of cell cycle
To quantitate the number of Pgc expressing cells in pgcGFP and pgcGFP; bam germaria that were positive for different cell cycle markers, the ovaries from respective genotypes were co-stained for GFP and cell cycle markers. First, all cells positive for GFP was counted. Second, only cells that were positive for both, GFP and cell cycle markers, were counted. Then the percentage of GFP positive cells positive for that marker in that germaria was calculated using: (GFP and cell cycle marker positive cells/GFP positive cells) × 100.
To quantitate the difference in cell cycle in bam and pgc; bam CBs, the ovaries were stained with Vasa and cell cycle markers. To avoid discrepancies in expression levels of different cell cycle markers between genotypes, all images for the same marker was taken under the same confocal settings. In a single z-stack plane of each tumor for both genotypes, the total number of Vasa positive cells were counted. Then, the total number of Vasa and cell cycle marker positive cells were counted. Then the percentage of Vasa positive cells positive for that marker in that germaria was calculated using: (Vasa and cell cycle positive cells/all Vasa positive cells) × 100.
4.6. Arbitrary units (a.u) quantification and analysis
In order to calculate intensities of H4K20me3 and H3K4me3 in bam, pgc; bam and pgcGFP, CycB tumors, Z stack planes were taken under the same confocal settings. Chromatin marks in Vasa positive cells were outlined and the pixel intensity of the highlighted area was analyzed using ImageJ. The ratio between mean and area was calculated for each cell. The average a.u was then calculated and compared between control and experimental samples. Paired student t-test was carried out between samples to generate a P-value.
4.7. Clone induction experiments
hsFLP;42DFRT-GFP flies were crossed to pgcΔFRT flies and were grown at 25 °C. Once adults started eclosing, clones were generated by heat-shocking adults for 3 h in a 37 °C walk-in twice a day for 2 days. The flies were then transferred to a 25 °C incubator and dissected 3 days or 5 days post heat-shock and immuostaining was carried out as described above.
4.8. Generation of pgc deletion
The pgc deletion allele was generated by imprecise excision of P(EPgy2)pgcEY07794. This is a 475 nucleotide deletion of the genomic DNA that completely deletes the pgc ORF. pgc mRNA signal was not detected by in situ hybridization and RT-PCR in the embryo (data not shown).
4.9. Egg laying test
Experiments were performed on pgcΔ/liprinγH1; MKRS/TM6 and Sco/CyO; MKRS/TM6 flies. Several bottles of each genotype were created to ensure adequate numbers of females would hatch on a daily basis. One the day before the experiment, bottles were cleared of flies, allowing for collection of newly hatched females, which would all be approximately the same age (~1 day old) the next day. Ten female flies were then placed in a vial with adequate yeast to last for one day, and supplemented with two wild-type male to ensure proper fertilization and egg laying. Because some flies had still not reached sexual maturity, egg laying on day one was not included in our report; however, every day after this, flies were flipped at the same time into new food vials with yeast. The previous day’s vials were then counted to determine the total number of eggs laid. This number was then divided by the number of female flies to calculate the number of eggs laid per female fly over a period of 21 days. Comparison of means statistical test was carried out to obtain a P-value.
4.10. Cloning of pgc cassette
The pgc cassette contains 2777 bp of genomic sequence upstream of the Pgc translational start site, an N-terminal TC2 tag, restriction enzyme sites (SpeI/XhoI) for in frame cloning, the pgc 3′UTR, and 703 bp of downstream genomic sequence. The construct was generated using two rounds of PCR. The first round generated two overlapping fragments using either primer set A:
-
(5′ GCGGCCGCATAAAAGACTCAAGTTGACCGACATCC-CCTTCC-3′ and 5′-TGGCTCCATACAACATCCTGGGCAGCAGTTGAGGAACATTTTCGGGTCTTCTTGTAGTTCAAAGCT-GCAAG-3′)
Or primer set B:
(5′ GCCCAGGATGTTGTATGGAGCCAACTAGTTGACTCGAGCTGG-ACCTCCCAAAAGCCAACTTATTGTG-3′ and 5′ GCGGCCGCTGTTGTAAACGAATGAGTCTTTATTGTGCACGGG-3′).
The second round used the purified first round PCRs as templates for amplification with the forward primer of set A and the reverse primer of set B generating a 3931 bp product.
The PCR product was cloned using a Topo TA cloning kit (Invitrogen) and sequenced.
The fragment was excised from the Topo TA cloning vector and subcloned into pCasper2 P element transformation vector using NotI. In the resulting construct, eGFP was subcloned using Spe1 and Xho1 restriction enzymes generating the Pgc cassette plasmid. The construct was sent for P-element mediated transformation into white1118 flies (Genetic Services).
4.11. Quantitative real-time PCR (qRT-PCR)
Expression levels of CycB in bam and pgc; bam mutants was analyzed using real-time, quantitative PCR. Reverse transcription was performed with Super Script II (Invitrogen, Catalog Number: 18064-014). The cDNA samples were diluted to 50 ng/μl. CycB specific primer was used (Life Technologies, Catalog Number: DM01817445G1). All real-time PCR reactions were carried out using the ABI 7700 sequence detection system (Applied Biosystems) and the amplifications were done using the iTaq Universal Probes Supermix (Biorad, Catalog Number: 172-5130). The thermal cycling conditions consisted of 50 °C for 2 min, 95 °C for 10 min, 40 cycles at 95 °C for 15 s, and 60 °C for 60 s. The experiments were carried out in technical triplicate and three biological replicates for each data point.
4.12. CHIP-qPCR
Flies were fattened with active dry yeast overnight. 150 flies were dissected in 1 x PBS, 1 mM PMSF, and 1 pill/1 mL protease inhibitors. The samples were crosslinked with 1% formaldehyde for 15 min at room temperature. Glycine was then added for 10 min at room temperature to stop crosslinking (final concentration of 10 mM glycine). The supernatant was removed and 100 μl of FA lysis buffer (50 HEPES, 140 mM NaCl, 1 mM EDTA, 1% Triton-X, 0.1% NaDOC) with 1 mM PMSF, 10 μg/mL leupeptin and 10 μg/mL pepsustatin was added prior to be homogenizing with blue homogenizer (Fisher Cat# 749521-1590). Tissues were incubated for 30 min at 4 °C with gentle mixing. The lysed samples were sonicated (Diagenode Bioruptor 300) at low setting for 20 cycles (30 s ON and 30 s OFF). Sonicated samples were diluted to 1.0 mL with FA lysis buffer, 1 mPMSF, 10 μg/mL leupeptin and 10 μg/mL pepstatin. Meanwhile, 25 μl of Dynobeads Protein A (Life Technologies Cat#10001D) were washed with 600 μl of 1 × PBS mixing for 2 min at room temperature twice. 500 μl of supernatant was discarded, leaving Dynobeads suspended in 100 μl of 1 × PBS. The Dynobeads were pre-cleared with lysed sample without antibodies for 1 h gently mixing at 4 °C. A 100 μl of input sample was put aside for the later qPCR analysis. The beads were discarded. Another set of Dynobeads were incubated with RNA polymerase II antibodies (Covance Cat#mms-126R) with a ratio of 1:100, mixing for 1 h at room temperature. The antibodies were removed after an hour. The pre-cleared sample was then added to the Dynobeads. The mixture was incubated mixing at 4 °C overnight. Supernatant was removed and the Dynobeads were washed with the following buffer for 10 min at 4 °C on each following step: twice with 1 mL of RIPA buffer (1.68 mL of FA Lysis buffer with 315 μl 7x protease inhibitors and 20 μl of 0.5 mM PMSF), twice with 1 mL of RIPA buffer with 500 mM NaCl, twice with 1 mL of LiCl buffer (10 mM Tris-HCl, 250 mM LiCl, 0.5% NP-40, 1 mM EDTA, 0.5% NaDOC), once with 1 mL of TE buffer (10 mM Tris-HCl and 1 mM EDTA). The beads and the input sample were re-suspended in 100 μl Elution buffer (100 μl TE buffer, 10 mM EDTA, and 1% SDS). The reverse crosslinking was performed at 100 °C shaking at 1200 rpm for 10 min. The sample was then centrifuged at 16,000 rpm for 5 min at room temperature, and the supernatant was transferred to a new tube. DNA sample was then purified with Qiagene PCR purification kit (Qiagene Cat#28106) prior to performing quantitative PCR (qPCR) assay. The primers (IDT DNA) were designed approximately 200 bp before and after the transcription start site of, CycB where a high occupancy of RNA polymerase II is expected. Power SYBA Green PCR Master Mix (Applied Biosystem Cat#4367659) was used for the qPCR assay.
4.13. Primers used for ChIP-qPCR
|
| |
| F-cycB-TSS−200 | 5′-CAGACGCTTGGCTATCAC-3′ |
| R-cycB-TSS−200 | 5′-GGACAACAAATTCAACAGCTAA-3′ |
| F-cycB-TSS+200 | 5′-GATGCGATACGTGCGATAA-3′ |
| R-cycB-TSS+200 | 5′-TTTATGGGTAGCACTGTTTCA-3′ |
| Actin5C-F | 5′-GACGTAGGCAGCCGTTT-3′ |
| Actin5C-R | 5′-GTGGCTGGTGAATGTTGAATG-3′ |
|
| |
Supplementary Material
Acknowledgments
We would like to acknowledge members of the Lehmann lab, Morse Lab, Belfort lab, Szaro lab, Larsen lab and Pager Lab for helping with reagents. We would also like to thank Karen Preston and Sridar Chittur for running and analyzing the qPCR experiment for us in the East Campus of University at Albany. Dr. Marlene Belfort, Dr. Daria Siekhaus, Dr. Thomas Hurd and members of the Rangan lab provided useful comments and edits on the drafts. We would also like to thank Dr. Rui Martinho, Dr. Akira Nakamura, Bloomington Stock Center and flybase for stocks. This work was initiated when P.R. was in Ruth Lehmann’s lab at NYUMC.
Funding
P.R is funded by NIH/NIGMS RO1 1119484-1-68857, SUNY Presidential Innovative Award and Pew Biomedical Scholars Program.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2017.11.014.
Footnotes
Competing interests
No competing interests declared.
Author contributions
P.F., S.S., and P.R. designed experiments and interpreted data. P.F., S.S., S.W.D., M.D., M.N. and P.R. performed experiments. P.F. and P.R. wrote the manuscript, which all authors edited and approved.
Summary statement
Transient transcriptional silencing promotes timely germline stem cell differentiation in Drosophila by altering the cell cycle and epigenetic landscape.
References
- Ables ET, Drummond-Barbosa D. Cyclin E controls Drosophila female germline stem cell maintenance independently of its role in proliferation by modulating responsiveness to niche signals. Development. 2013;140:530–540. doi: 10.1242/dev.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astigarraga S, Hofmeyer K, Farajian R, Treisman JE. Three Drosophila liprins interact to control synapse formation. J Neurosci: Off J Soc Neurosci. 2010;30:15358–15368. doi: 10.1523/JNEUROSCI.1862-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva MO, Ferguson EL. Germline stem cell number in the Drosophila ovary is regulated by redundant mechanisms that control Dpp signaling. Development. 2004;131:1881–1890. doi: 10.1242/dev.01076. [DOI] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci USA. 2012;109:9465–9470. doi: 10.1073/pnas.1120473109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol: CB. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Cinalli RM, Lehmann R. A spindle-independent cleavage pathway controls germ cell formation in Drosophila. Nat Cell Biol. 2013;15:839–845. doi: 10.1038/ncb2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinalli RM, Rangan P, Lehmann R. Germ cells are forever. Cell. 2008;132:559–562. doi: 10.1016/j.cell.2008.02.003. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Lee JK, Spradling AC. Alpha-spectrin is required for germline cell division and differentiation in the Drosophila ovary. Development. 1996;122:3959–3968. doi: 10.1242/dev.122.12.3959. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- de Las Heras JM, Martinho RG, Lehmann R, Casanova J. A functional antagonism between the pgc germline repressor and torso in the development of somatic cells. EMBO Rep. 2009;10:1059–1065. doi: 10.1038/embor.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Forbes A, Tazuke SI, Fuller MT, Lehmann R. Germ line stem cell differentiation in Drosophila requires gap junctions and proceeds via an intermediate state. Development. 2003;130:6625–6634. doi: 10.1242/dev.00853. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr Biol: CB. 2004;14:981–986. doi: 10.1016/j.cub.2004.05.049. [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, de Sousa Lopes SM, Surani MA. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- Hsu HJ, Drummond-Barbosa D. Insulin signals control the competence of the Drosophila female germline stem cell niche to respond to Notch ligands. Dev Biol. 2011;350:290–300. doi: 10.1016/j.ydbio.2010.11.032. [DOI] [PubMed] [Google Scholar]
- Hsu HJ, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313:700–712. doi: 10.1016/j.ydbio.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Kirilly D, Weng C, Kawase E, Song X, Smith S, Schwartz J, Xie T. Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell. 2008;2:39–49. doi: 10.1016/j.stem.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T, Spradling A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc Natl Acad Sci USA. 2003;100:4633–4638. doi: 10.1073/pnas.0830856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Orr-Weaver TL. Translational regulation of the cell cycle: when, where, how and why?0. Philos Trans R Soc Lond Ser B, Biol Sci. 2011;366:3638–3652. doi: 10.1098/rstb.2011.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990;4:905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci USA. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol: CB. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Cauvin C, Moch C, Radford SJ, Sampaio P, Perdigoto CN, Schweisguth F, Bardin AJ, Sunkel CE, McKim K, et al. Aurora B and cyclin B have opposite effects on the timing of cytokinesis abscission in Drosophila germ cells and in vertebrate somatic cells. Dev Cell. 2013;26:250–265. doi: 10.1016/j.devcel.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Huynh JR. Monitoring complete and incomplete abscission in the germ line stem cell lineage of Drosophila ovaries. Methods Cell Biol. 2017;137:105–118. doi: 10.1016/bs.mcb.2016.03.033. [DOI] [PubMed] [Google Scholar]
- McKearin D, Ohlstein B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 1995;121:2937–2947. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Seydoux G. Less is more: specification of the germline by transcriptional repression. Development. 2008;135:3817–3827. doi: 10.1242/dev.022434. [DOI] [PubMed] [Google Scholar]
- Navarro-Costa P, McCarthy A, Prudencio P, Greer C, Guilgur LG, Becker JD, Secombe J, Rangan P, Martinho RG. Early programming of the oocyte epigenome temporally controls late prophase I transcription and chromatin remodelling0. Nat Commun. 2016;7:12331. doi: 10.1038/ncomms12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Schupbach T. Encore facilitates SCF-Ubiquitin-proteasome-dependent proteolysis during Drosophila oogenesis. Development. 2003;130:6339–6349. doi: 10.1242/dev.00855. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, McKearin D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development. 1997;124:3651–3662. doi: 10.1242/dev.124.18.3651. [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, Jaime-Bustamante K, Coux RX, Martinho RG, Lehmann R. Temporal and spatial control of germ-plasm RNAs. Curr Biol: CB. 2009;19:72–77. doi: 10.1016/j.cub.2008.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Curr Biol: CB. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Slaidina M, Lehmann R. Translational control in germline stem cell development. J Cell Biol. 2014;207:13–21. doi: 10.1083/jcb.201407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xie T. DE-cadherin-mediated cell adhesion is essential for maintaining somatic stem cells in the Drosophila ovary. Proc Natl Acad Sci USA. 2002;99:14813–14818. doi: 10.1073/pnas.232389399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harb Perspect Biol. 2011;3:a002642. doi: 10.1101/cshperspect.a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Campbell SD, O’Farrell PH. The cell cycle program in germ cells of the Drosophila embryo. Dev Biol. 1998;196:160–170. doi: 10.1006/dbio.1998.8855. [DOI] [PubMed] [Google Scholar]
- Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin H. The division of Drosophila germline stem cells and their precursors requires a specific cyclin. Curr Biol: CB. 2005;15:328–333. doi: 10.1016/j.cub.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Xia L, Jia S, Huang S, Wang H, Zhu Y, Mu Y, Kan L, Zheng W, Wu D, Li X, et al. The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell. 2010;143:978–990. doi: 10.1016/j.cell.2010.11.022. [DOI] [PubMed] [Google Scholar]
- Xie T, Song X, Jin Z, Pan L, Weng C, Chen S, Zhang N. Interactions between stem cells and their niche in the Drosophila ovary. Cold Spring Harb Symp Quant Biol. 2008;73:39–47. doi: 10.1101/sqb.2008.73.014. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Yuan H, Chiang CY, Cheng J, Salzmann V, Yamashita YM. Regulation of cyclin A localization downstream of Par-1 function is critical for the centrosome orientation checkpoint in Drosophila male germline stem cells. Dev Biol. 2012;361:57–67. doi: 10.1016/j.ydbio.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccai M, Lipshitz HD. Differential distributions of two adducin-like protein isoforms in the Drosophila ovary and early embryo. Zygote. 1996;4:159–166. doi: 10.1017/s096719940000304x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


