Aminoglycosides are important antimicrobials used worldwide for prophylaxis and/or therapy in multiple production animal species. The emergence of new resistance genes jeopardizes current pathogen detection and treatment methods. The risk of resistance gene transfer to other animal and human pathogens is elevated when resistance genes are carried by mobile genetic elements. This study identified a new variant of a spectinomycin/streptomycin resistance gene harbored in a self-transmissible mobile element. The gene was also present in four different bovine pathogen species.
KEYWORDS: Histophilus somni, Mannheimia haemolytica, Pasteurella multocida, aminoglycoside-modifying enzymes, aminoglycosides, antibiotic resistance, bovine respiratory disease, cosmid library, integrative and conjugative element, spectinomycin
ABSTRACT
A novel variant of the AAD(3″) class of aminoglycoside-modifying enzymes was discovered in fatal bovine respiratory disease-associated pathogens Pasteurella multocida and Histophilus somni. The aadA31 gene encodes a spectinomycin/streptomycin adenylyltransferase and was located in a variant of the integrative and conjugative element ICEMh1, a mobile genetic element transmissible among members of the family Pasteurellaceae. The gene was also detected in Mannheimia haemolytica from a case of porcine pneumonia and in Moraxella bovoculi from a case of keratoconjunctivitis.
IMPORTANCE Aminoglycosides are important antimicrobials used worldwide for prophylaxis and/or therapy in multiple production animal species. The emergence of new resistance genes jeopardizes current pathogen detection and treatment methods. The risk of resistance gene transfer to other animal and human pathogens is elevated when resistance genes are carried by mobile genetic elements. This study identified a new variant of a spectinomycin/streptomycin resistance gene harbored in a self-transmissible mobile element. The gene was also present in four different bovine pathogen species.
OBSERVATION
In the last decade, multidrug-resistant (MDR) bacterial isolates of the bovine respiratory disease (BRD) complex have emerged that harbor mobile genetic elements (MGEs) containing genes that confer resistance to many veterinary antimicrobials (1). Widespread resistance in BRD bacterial pathogens—primarily the gammaproteobacterial Pasteurellaceae family members Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni (2)—would be devastating to the beef and dairy industries (3). Discovered in the 1940s, aminoglycosides are broad-spectrum antimicrobials; veterinary formulations may include spectinomycin, dihydrostreptomycin, gentamicin, hygromycin B, and neomycin (4, 5). Collectively, they are the fifth-most-used drug class, accounting for 2%, by weight (344,120 kg), of the veterinary antimicrobials sold (5). Notwithstanding off-label usage, spectinomycin has been withdrawn from therapeutic BRD usage in Canada but continues to be used in the United States and is approved in both countries for in-feed prophylaxis for chickens and swine, often in combination with lincomycin (FDA Center for Veterinary Medicine, Health Canada Drug Product Database). Aminoglycoside resistance may involve 16S rRNA target modification or methylation, efflux, decreased permeability, and enzymatic inactivation by aminoglycoside-modifying enzymes (AMEs) (4, 6). The AMEs include N-acetyltransferases, O-phosphotransferases, and O-adenylyltransferases (AADs/ANTs). Inactivation of spectinomycin is commonly via adenylylation, and AAD(3″) class enzymes confer resistance to both spectinomycin and streptomycin, despite structural dissimilarities between the two drugs. The AAD(3″) enzymes adenylylate streptomycin at the 3″-OH position of the streptomycin glucosamine ring and adenylylate spectinomycin at the 9-OH position of the spectinomycin actinamine ring (4, 6, 7). The favored gene nomenclature is aadA, and versions/alleles are numbered (8).
We functionally identified the mechanism of spectinomycin resistance in two pathogens from diseased lung tissue from separate fatal cases of BRD in Alberta, Canada, isolated and identified to the species level as previously described (9). Unable to detect known AMEs via PCR, we constructed genomic DNA cosmid libraries to functionally screen for the spectinomycin antimicrobial resistance gene (ARG) and to examine the ARG context. Briefly, DNA was purified from P. multocida strain PM13 and H. somni strain HS31 via phenol-chloroform extraction (9). Two large (~35-kb)-insert libraries were constructed in Escherichia coli EPI300-T1R in accordance with the instructions for the pCC1FOS CopyControl fosmid library production kit (Lucigen), with modification (10). Transformants were plated onto Luria-Bertani (LB) agar containing chloramphenicol (12.5 µg/ml), and harvested colonies (~14,000/library) were pooled. The number of clones ensuring a 99% probability that a given sequence of a typical P. multocida genome (~2.3 Mb) will be contained within a 35-kb insert cosmid library was determined to be ~300.3 colonies (11).
Spectinomycin resistance screening was performed by separately inoculating 5 ml of LB broth with the pooled cosmid libraries, which were grown for ~2 h at 37°C with shaking at 200 rpm. Cultures were standardized to an optical density at 600 nm of 0.1, and a 10-fold dilution series was plated on LB agar with or without spectinomycin (50 µg/ml; Millipore Sigma). Empty vector EPI300 was used as a comparator for false-positive enumeration, but no false-positive colonies were observed after 24 h of growth at 37°C. For each library, 12 spectinomycin-resistant cosmids were extracted (EZ-10 Plasmid DNA Miniprep kit; BioBasic) and analyzed for uniqueness by EcoRI/BamHI restriction digestion (New England Biolabs) (not shown). DNA from three unique cosmids from both HS31 and PM13 libraries was prepared for Illumina MiSeq PE250 sequencing (Génome Québec). Reads (~175,000/cosmid) and the Illumina adaptor were trimmed with Trimmomatic 0.36 (criteria: phred33, LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36; Reads surviving = >98%) (12), assembled with SPAdes 3.10.1 (13), and annotated with PROKKA (14) by using default settings. The pCC1FOS sequence (GenBank accession no. EU140751.1) was manually subtracted from resultant contigs in Geneious 8.1.9. The smallest cosmid insert was 28,221 bp, and the largest was 38,394 bp. The three contigs/cosmid inserts obtained for each strain were assembled to produce finalized sequences 48,228 and 30,236 bp in length for HS31 and PM13, respectively.
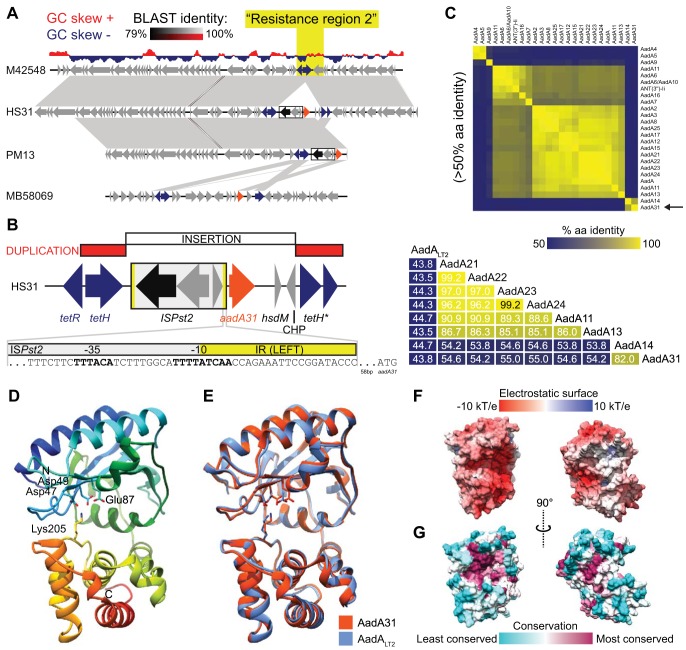
The HS31 and PM13 sequences shared 87.9% pairwise identity (MUSCLE alignment) and shared synteny with the integrative and conjugative element (ICE) MGE of M. haemolytica strain M42548 (GenBank accession no. CP005383) (Fig. 1A), which was isolated from a BRD case in Pennsylvania (15). Annotation identified a 789-bp open reading frame designated adenylyltransferase (ANT1), which was further identified as a streptomycin-3″-adenylyltransferase via a conserved-domain BLAST search. BLAST searches against the NCBI nonredundant (nr) database showed that the gene identified had been detected yet had not been characterized. We designated the gene aadA31—to the best of our knowledge, the lowest unassigned number.
FIG 1.
Genomic context, sequence identity, and structural homology modeling of the aadA31 gene detected in P. multocida PM13 and H. somni HS31. (A) Genomic BLAST comparison of the PM13 and HS31 cosmid sequences, highlighting the insertion containing aadA31. The aadA31 gene is absent from M. haemolytica M42548 but resides with the ISPst2 element in a variant region syntenous with ICEMh1. The aadA31 gene is also present in the genome of M. bovoculi MB58069. (B) Gene context schematic of the insertion, depicting the novel insertion sequence from HS31 and the tetR-tetH duplication (truncated tetH* is represented as two arrows; truncated tetR is not shown). An expanded view of the ISPst2 left terminal inverted repeat (IR) showing the putative promoter for aadA31 (−35 and −10 boxes in bold) is also presented. ISPst2 is inversely oriented such that the left inverted repeat is 58 bp from the aadA31 start codon. (C) Predicted amino acid (aa) sequence pairwise identity matrix of aadA homologues (only those with >50% amino acid sequence identity in the CARD database). Shown at the bottom are the percent amino acid sequence identity values of selected AadA enzymes. (D) Predicted RobettaCM (21) ribbon model of the overall structure of AadA31. The N terminus is blue, and the C terminus is red. Conserved active site residues (20) are represented by sticks. (E) Superposition of the structures of AadA31 (orange) and AadALT2 (light blue) (F) Electrostatic surface potential of AadA31 (showing orientations 90° apart). The highest electropositive and electronegative surfaces are blue and red, respectively. (G) Surface conservation plotted by ConSurf (22). The highest degree of conservation is magenta, and the lowest is cyan. All of the images are in the same orientation, except where indicated. Electrostatic surface potential was calculated with the APBS plugin and visualized in Chimera UCSF (23).
Two instances of aadA31 with 100% nucleotide identity were detected by a BLAST search of the nr database (i) in the draft genome sequence of M. haemolytica MhSwine2000, an isolate from the pneumonic lung of a juvenile pig in an Iowa grower house (M. J. Hauglund et al., USDA, unpublished data; GenBank accession no. ATTA00000000.1), and (ii) in the whole-genome sequence of Moraxella bovoculi strain Mb58069 (GenBank accession no. CP011374), an isolate from a case of infectious bovine keratoconjunctivitis in a yearling crossbred steer in Nebraska (16).
In PM13 and HS31, aadA31 was adjacent to a 2,985-bp three-gene insertion (IS) element with 97% nucleotide sequence identity to ISPst2, a widespread IS element identified in Pseudomonas stutzeri strain OX1 (17) (Fig. 1B). Discrimination of the element’s imperfect terminal inverted repeats (5′ GGGTATMCGGAWTTWMTGGTTGAT 3′) indicated that aadA31 was harbored downstream from ISPst2. Although the 58-bp sequence upstream of the aadA31 start codon is conserved among PM13, HS31, and Mb58069, it does not appear to contain a promoter. Instead, putative −10 (TTTTATCAA) and −35 (TTTACA) boxes are partially located in the ISPst2 terminal repeat. Downstream from aadA31 are genes for a putative type I restriction enzyme (hsdM; truncated) and a conserved hypothetical protein (CHP). Together, this sequence is a variation of syntenous resistance region 2 of ICEMh1 (15). Interestingly, these features reside within an insertion in tetR, which appears to have duplicated part of tetR and the tetH sequence, resulting in two copies that are either conventional or truncated (Fig. 1B). This suggests that resistance region 2 of ICEMh1 has a history of recombination.
We also tested for aadA31 in a larger in-house collection of BRD agents isolated from feedlot cattle that died of BRD. Standard PCR amplification was performed in 20-μl reaction mixtures (HotStarTaq Plus master mix; Qiagen) containing 1 μl of heat-lysed bacteria in TE (10 mM Tris-HCl, 1 mM EDTA) and 0.5 μM oligonucleotides (ANT1_F, 5′ ATGACTACTAAACTAGATACCATAT 3′; ANT1_R, 5′ CTATTGCAGCTTCGTCGTC 3′; Eurofins Genomics). For positive controls, PCR was also performed with standard 16S oligonucleotides 27F and 1492R (18). The aadA31 gene was detected in 40/42 P. multocida isolates and 5/5 H. somni isolates.
The closest relative of AadA31 is AadA14 (82% amino acid sequence identity) (Fig. 1C), which was first identified as plasmid borne in a bovine P. multocida isolate from Belgium (7). The next closest relative of both AadA31 and AadA14 is AadA23 from Salmonella enterica serovar Agona, which has 55.0 and 54.6% amino acid sequence identity, respectively (19). To provide further evidence that AadA31 is an AadA enzyme, we performed structural homology comparisons against the structure of AadALT2 from S. enterica serovar Typhimurium LT2 (PDB code 4cs6) (20) with RobettaCM (21). AadA31 shares 43.8% amino acid sequence identity with AadALT2. Least-squares superposition indicated that the predicted structure of AadA31 was consistent with the structure of AadALT2 (root mean square deviation of 0.42 Å over 252 residues). Consurf (22) was used to calculate that the positioning of residues implicated in ligand binding and catalysis (20) was conserved within the active site, a highly electronegative cleft (Fig. 1D to G) (23).
To confirm that aadA31 confers spectinomycin resistance and to assess resistance to other aminoglycosides, promoterless aadA31 was cloned into E. coli under the control of the lac promoter of high-copy-number blue-white screening vector pGEM-T (Promega). Taq-based PCR amplification was performed as before with purified HS31 cosmid DNA in lieu of heat-lysed extractions. The amplicon was purified (Zymo DNA Clean and Concentrator kit) and ligated overnight at 16°C to pGEM-T in a standard 15-μl T4 DNA ligase reaction mixture (New England Biolabs). Ligated plasmids were transformed into chemically competent E. coli DH5α (Invitrogen) and plated on LB agar supplemented with isopropyl-β-d-thiogalactopyranoside (IPTG) at 0.5 mM, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 20 μg/ml, and ampicillin at 100 µg/ml (Fisher Scientific). Following overnight growth at 37°C, a white insert-containing colony was selected for susceptibility testing via the microtiter broth dilution method for MIC determination (24). The drug concentration range was 512 to 0.25 µg/ml, tested in triplicate in 96-well microtiter plates (Greiner CELLSTAR) with Mueller-Hinton broth (BD Difco). The MIC was the lowest concentration inhibiting visually detectable growth. Compared to the empty-vector control, the pGEM-aadA31-carrying strain exhibited resistance to spectinomycin (MIC, >512 µg/ml) and streptomycin (MIC, 256 µg/ml) but not to kanamycin, amikacin, or gentamicin (each MIC, 0.5 µg/ml). To demonstrate horizontal transfer of the ICEMh1 variant and to assess the spectinomycin/streptomycin MIC for aadA31 under native promoter control in E. coli, P. multocida strain PM13 was mated with recipient rifampin-resistant E. coli DH5α as previously described (15). Transconjugants were selected on LB agar supplemented with rifampin (25 µg/ml; Millipore Sigma) and spectinomycin (50 µg/ml). Transconjugants were confirmed by PCR for aadA31 as described above and with the UAL754 and UAR900 oligonucleotides for the E. coli-specific uidA gene (25). The spectinomycin and streptomycin MICs for a PM13-E. coli transconjugant versus empty-vector DH5α were >512 and 256 µg/ml versus 4 and 2 µg/ml, respectively. This was consistent with high-level (7) resistance activity of AadA class AMEs (6).
In conclusion, we identified a new aadA gene variant via functional screening of genomic cosmid-based libraries. Our investigation provides a striking example of the spread of ARGs; in this case, emerging in multiple bacterial species associated with bovine and swine respiratory disease and bovine keratoconjunctivitis. The results of this study also hint at the ongoing recombination of ARGs into and from MGEs such as ICEMh1, a finding consistent with another published study (7) that also showed that MDR members of the family Pasteurellaceae can carry ARGs whose sequences have diverged considerably from those of homologues in other bacteria. These findings also highlight the interconnectedness of ARG transmission in MDR production animal and zoonotic pathogens and suggest that antimicrobial usage elsewhere in the production spectrum may impact the development and dissemination of resistant pathogens in distant animals.
Accession number(s).
The aadA31, PM13, and HS31 cosmid DNA sequences obtained in this study have been deposited in GenBank under accession numbers MG520668, MG520669, and MG520670, respectively.
ACKNOWLEDGMENTS
We thank Wendi Smart, Ruth Barbieri, and Cheyenne Conrad for technical assistance.
A.C. is supported by an NSERC postdoctoral fellowship, and the program is supported by funding from the Alberta Livestock and Meat Agency (ALMA) and the Beef Cattle Research Council (BCRC)—Agriculture and Agri-Food Canada beef cluster. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
C.K., A.C., R.Z., and T.A.M. conceived and designed the experiments. A.C., R.H., and C.K. performed the experiments. A.C., R.Z., and R.J.G. analyzed the data. A.C. and R.J.G. wrote the paper. R.Z., R.H., C.K., and T.A.M. edited the paper. T.A.M. obtained funding.
REFERENCES
- 1.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J Antimicrob Chemother 67:84–90. doi: 10.1093/jac/dkr406. [DOI] [PubMed] [Google Scholar]
- 2.DeDonder KD, Apley MD. 2015. A literature review of antimicrobial resistance in pathogens associated with bovine respiratory disease. Anim Health Res Rev 16:125–134. doi: 10.1017/S146625231500016X. [DOI] [PubMed] [Google Scholar]
- 3.Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL. 2006. Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J Anim Sci 84:1999–2008. doi: 10.2527/jas.2006-046. [DOI] [PubMed] [Google Scholar]
- 4.Davies J, Wright GD. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol 5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. 2016. FDA annual summary report on antimicrobials sold or distributed in 2015 for use in food-producing animals. FDA Center for Veterinary Medicine, Rockville, MD: https://www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm534244.htm. [Google Scholar]
- 6.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehrenberg C, Catry B, Haesebrouck F, de Kruif A, Schwarz S. 2005. Novel spectinomycin/streptomycin resistance gene, aadA14, from Pasteurella multocida. Antimicrob Agents Chemother 49:3046–3049. doi: 10.1128/AAC.49.7.3046-3049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White PA, Rawlinson WD. 2001. Current status of the aadA and dfr gene cassette families. J Antimicrob Chemother 47:495–496. doi: 10.1093/jac/47.4.495. [DOI] [PubMed] [Google Scholar]
- 9.Klima CL, Cook SR, Zaheer R, Laing C, Gannon VP, Xu Y, Rasmussen J, Potter A, Hendrick S, Alexander TW, McAllister TA. 2016. Comparative genomic analysis of Mannheimia haemolytica from bovine sources. PLoS One 11:e0149520. doi: 10.1371/journal.pone.0149520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taupp M, Lee S, Hawley A, Yang J, Hallam SJ. 2009. Large insert environmental genomic library production. J Vis Exp pii:1387. doi: 10.3791/1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 12.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 15.Eidam C, Poehlein A, Leimbach A, Michael GB, Kadlec K, Liesegang H, Daniel R, Sweeney MT, Murray RW, Watts JL, Schwarz S. 2015. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J Antimicrob Chemother 70:93–97. doi: 10.1093/jac/dku361. [DOI] [PubMed] [Google Scholar]
- 16.Dickey AM, Loy JD, Bono JL, Smith TP, Apley MD, Lubbers BV, DeDonder KD, Capik SF, Larson RL, White BJ, Blom J, Chitko-McKown CG, Clawson ML. 2016. Large genomic differences between Moraxella bovoculi isolates acquired from the eyes of cattle with infectious bovine keratoconjunctivitis versus the deep nasopharynx of asymptomatic cattle. Vet Res 47:31. doi: 10.1186/s13567-016-0316-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolognese F, Di Lecce C, Galli E, Barbieri P. 1999. Activation and inactivation of Pseudomonas stutzeri methylbenzene catabolism pathways mediated by a transposable element. Appl Environ Microbiol 65:1876–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82:6955–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michael GB, Cardoso M, Schwarz S. 2005. Class 1 integron-associated gene cassettes in Salmonella enterica subsp. enterica serovar Agona isolated from pig carcasses in Brazil. J Antimicrob Chemother 55:776–779. doi: 10.1093/jac/dki081. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Näsvall J, Wu S, Andersson DI, Selmer M. 2015. Structure of AadA from Salmonella enterica: a monomeric aminoglycoside (3′′)(9) adenyltransferase. Acta Crystallogr D Biol Crystallogr 71:2267–2277. doi: 10.1107/S1399004715016429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. 2013. High-resolution comparative modeling with RosettaCM. Structure 21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 25.Maheux AF, Picard FJ, Boissinot M, Bissonnette L, Paradis S, Bergeron MG. 2009. Analytical comparison of nine PCR primer sets designed to detect the presence of Escherichia coli/Shigella in water samples. Water Res 43:3019–3028. doi: 10.1016/j.watres.2009.04.017. [DOI] [PubMed] [Google Scholar]