Abstract
Survival biomass production and crop yield are heavily constrained by a wide range of environmental stresses. Several phytohormones among which abscisic acid (ABA), ethylene and salicylic acid (SA) are known to mediate plant responses to these stresses. By contrast, the role of the plant hormone auxin in stress responses remains so far poorly studied. Auxin controls many aspects of plant growth and development, and Auxin Response Factors play a key role in the transcriptional activation or repression of auxin-responsive genes through direct binding to their promoters. As a mean to gain more insight on auxin involvement in a set of biotic and abiotic stress responses in tomato, the present study uncovers the expression pattern of SlARF genes in tomato plants subjected to biotic and abiotic stresses. In silico mining of the RNAseq data available through the public TomExpress web platform, identified several SlARFs as responsive to various pathogen infections induced by bacteria and viruses. Accordingly, sequence analysis revealed that 5’ regulatory regions of these SlARFs are enriched in biotic and abiotic stress-responsive cis-elements. Moreover, quantitative qPCR expression analysis revealed that many SlARFs were differentially expressed in tomato leaves and roots under salt, drought and flooding stress conditions. Further pointing to the putative role of SlARFs in stress responses, quantitative qPCR expression studies identified some miRNA precursors as potentially involved in the regulation of their SlARF target genes in roots exposed to salt and drought stresses. These data suggest an active regulation of SlARFs at the post-transcriptional level under stress conditions. Based on the substantial change in the transcript accumulation of several SlARF genes, the data presented in this work strongly support the involvement of auxin in stress responses thus enabling to identify a set of candidate SlARFs as potential mediators of biotic and abiotic stress responses.
Introduction
Tomato (Solanum lycopersicum) is one of the major crops with significant economic and scientific interests and as in the case of other crop plants, environmental stresses negatively affect tomato growth, productivity and quality [1]. Developing new tomato cultivars with enhanced tolerance to abiotic and biotic stress would have a significant impact on global food production in many regions. Plant responses to environmental stresses are extremely complex and involve changes at the transcriptome, cellular and physiological levels in order to prevent damage and ensure survival [2]. Better understanding of the molecular mechanisms by which plants adapt to constantly changing environmental conditions is a topic of prime interest in the context of global climate warming. However, our knowledge of the actors and pathways underlying plant tolerance or susceptibility to environmental stresses remains scarce. Several phytohormones are known for their role in the response to abiotic and biotic stresses [1]. Abiotic stress responses are largely controlled by the hormone ABA while defense against different biotic constraints is specified by antagonism between the salicylic acid (SA) and jasmonic acid (JA) or ethylene signaling pathways [3,4]. This creates a complex network of interacting pathways and hormones cross-talk at different levels [5].
Even though auxin is known to control a wide range of plant developmental processes, strikingly, its putative role in modulating plant growth under stress responses remains poorly understood. Auxin is involved in different aspects of plant development throughout the plant life cycle, including apical dominance, tropic responses, vascular development, organ patterning, flower and fruit development [6–12]. Significant progress has been made towards understanding the mechanisms by which this hormone impacts plant growth and development. By contrast, our knowledge about auxin implication in biotic and abiotic stress responses remains quite limited [13]. Recent studies including expression profiling suggested that auxin might act as a regulator of plant responses to environmental stresses [11,14–18].
Auxin regulates the cell-specific transcription of auxin response genes via three types of transcriptional regulators, Auxin/Indole-Acetic Acid (Aux/IAAs), Auxin Response Factors (ARFs) and TOPLESS proteins (TPS) [19–21]. In Arabidopsis, 23 ARFs were identified based on the presence of a conserved N-terminal DNA-binding domain (DBD), a variable central transcriptional regulatory region which can function as activator or a repressor domain, and a carboxy-terminal dimerization domain (CTD) that contributes to the formation of either ARF/ARF homo- and hetero-dimers or ARF/Aux/IAA hetero-dimers [21–23]. The DBD enables ARFs to specifically bind the conserved Auxin Response Element (AuxRE, 5’ TGTCTC 3’) present in the promoters of various Auxin-regulated genes and in this way to control gene transcription associated with plant responses to auxin [23–26].
ARF proteins orchestrate several biological and physiological processes such as embryogenesis, leaf expansion and senescence, lateral root development and fruit development by regulating the expression of auxin response genes [27–30]. The ARF gene family has been identified and well characterized in many crop species, such as Arabidopsis (Arabidopsis thaliana) [31], Maize (Zea mays) [32], Rice (Oryza sativa) [11,15], Poplar (Populus trichocarpa) [33], Tomato (Solanum lycopersicum) [21,34,35], Chinese cabbage (Brassica rapa) [36], Sorgho (Sorghum bicolor) [18], and Banana (Musa acuminata) [37]. In recent years, small RNA, known as miRNA, have been shown to substantially contribute to the regulation of plant development, physiology and stress responses [38]. To date, 872 miRNAs, belonging to 42 families, have been identified in 71 plant species by genetic screening, direct cloning, computational strategies and EST analysis [38,39]. Noteworthy, among the first miRNAs to be identified and characterized, miR160 has sequence complementarities with ARF10, ARF16 and ARF17 [21,40] and miR167 potentially regulates ARF6A, ARF6B and ARF8A [21,41]. Therefore, ARF expression in tomato is likely to be regulated at the post-transcriptional level by a large family of miRNAs that are non-coding RNAs playing a critical role in regulating auxin-dependent gene expression at the post-transcriptional level [42,43].
To address the putative involvement of tomato SlARF genes in plant responses to stresses we performed in the present study in silico analyses of the 5’ regulatory regions of using PLACE web plateform (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en) [44] and established their expression profiles under biotic stress conditions using TomExpress web platform [45]. We then, assessed the transcript accumulation of SlARF genes under salt, drought and flooding stresses using a qPCR experimental approach. We also examined the expression of miRNA precursor genes to check whether their expression is regulated under stress conditions and whether they are involved in post-transcriptional regulation of their target ARF genes under these conditions. Overall, the study provides guidance on the implication of auxin signaling pathway in plant responses to biotic and abiotic stresses and defines new targets towards engineering tomato plants better adapted to adverse environmental conditions.
Materials and methods
Plant materials
Tomato (Solanum lycopersicum cv Micro-Tom) plants of Wild type and transgenic DR5::GUS, pARF8A::GUS and pARF10::GUS lines generated in house (GBF laboratory) were used in this study.
Histochemical analysis of GUS expression
Transgenic plants expressing pARF8A::GUS and pARF10A::GUS were generated by A. tumefaciens-mediated transformation according to Wang et al. (2005) [46]. For that, PCR was performed on the genomic DNA of tomato ‘Micro-Tom’ (10 ng.ml–1) using specific primers. The corresponding amplified fragment was cloned into the pMDC162 vector containing the GUS reporter gene using Gateway technology (Invitrogen). The cloned SlARF promoter was sequenced from both sides using vector primers in order to see whether the end of the promoter is matching with the beginning of the reporter gene. Sequence results were carried out using the Vector NTI (Invitrogen) and ContigExpress software by referring to ARF promoter sequences.
GUS assays were conducted on DR5::GUS, pARF8A::GUS and pARF10::GUS tomato lines. After being surface sterilized, seeds were cultivated in Petri dishes containing half strength Murashige & Skoog medium for 7 days in a growth chamber at 25°C with 16h light/ 8h dark cycle. One week plants were then grown hydroponically for two weeks in BD (BROUGHTON & DILLWORTH) liquid medium [47]. Three week-old plants were subjected to salt and drought treatment. Salt stress was performed by adding 250 mM of NaCl to the culture medium. After 24 hours of the salt stress application, plants were incubated in GUS solution. Drought stress was conducted by adding 15% of PEG 20000 to the liquid culture solution. Plants were collected after 5 days of stress application and were incubated in the GUS solution. For each stress condition, control plants were cultivated in BD liquid medium for the same period.
GUS staining was performed overnight at 37°C in 3 mM XGluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide (Duchefa Biochemie, Haarlem, The Netherlands), 0.1% (v/v) Triton X-100 Sigma, Steinhaim, Germany, 8 mM β-mercaptoethanol and 50 mM Na2HPO4/NaH2PO4 (pH 7.2), then followed by a destaining in EtOH.
Plant growth and stress conditions
Wild type tomato seeds were sterilized for 10 min in 50% sodium hypochlorite, rinsed four times with sterile distilled water and sown in pots containing peat. Then they were incubated in a culture room with 16h light/ 8h dark photoperiod and 25± 2°C temperature. After 3 weeks, plants were subjected to salt, drought and flooding stresses. Salt stress was performed by watering daily the plants with 250mM of NaCl solution. Control plants were daily watered with distilled water. Leaves and roots samples were harvested after 2 and 24 hours of salt stress application. Drought stress was performed on three week-old plants by water holding for 48 hours and for 5 days. Watering continued normally throughout for control plants. Leave and root samples were collected after drought stress application. For flooding stress, three week-old plants were flooded with deionized water which was maintained at the level of cotyledonary node throughout the experiment. After 48 hours, the leaves and roots were harvested. Control plants were daily watered. Three biological replicates were done for each condition and three independent biological replicates were done for each experiment.
RNA extraction
Total RNA was extracted from leaves and roots samples by using the Plant RNeasy extraction kit (RNeasy Plant Mini Kit, Qiagen, Valencia, CA, USA). To remove any residual genomic DNA, the RNA was treated with an RNase-Free DNase according to the manufacturer’s instruction (Ambion® DNA-freeTMDNase). The concentration of RNA was accurately quantified by spectrophotometric measurement and 1μg of total RNA was separated on 2% agarose gel to monitor its integrity. DNase-treated RNA (2μg) was then reverse-transcribed in a total volume of 20μl using the Omniscript Reverse Transcription Kit (Qiagen).
Real time PCR
The real-time quantification of cDNA corresponding to 2μg of total RNA was performed in the ABI PRISM 7900HT sequence detection system (Applied biosystems) using the QuantiTech SYBR Green RT-PCR kit (Qiagen). The Gene-specific primers used are listed in S1 Table. The reaction mixture (10μl) contained 2μg of total RNA, 1,2 μM of each primer and appropriate amounts of enzymes and fluorescent dyes as recommended by the manufacturer. Actin gene was used as reference. Real-Time PCR conditions were as follow: 50°C for 2 min, 95°C for 10 min, then 40 cycles of 95°C for 15 s and 60°C for 1 min, and finally one cycle at 95°C for 15 s and 60°C for 15 s. Three independent biological replicates and three technical replicates of each sample were used for real-time PCR analysis. For each data point, the CT value was the average of CT values obtained from the three biological replicates and three technical replicates.
Three stress related genes were used as markers in these experiments. CI7 gene was used in both leaves and roots for salt and drought stresses [48,49]. For flooding, ACO1 gene was used as a marker in leaves and PDC gene in roots [50].
Tomato ARF gene expressions under biotic stress conditions
The expression pattern of 22 SlARF genes was analyzed by RNAseq technology in several studies and the results presented here were extracted from the TOMEXPRESS database (published online on October 2014) (S2 Table). TOMEXPRESS database includes 16 RNA-seq projects and 124 unique global conditions [45].
Analysis of Cis-acting regulatory elements
Using database associated search tools, 2 kb of 5′ regulatory region of each ARF transcription factor gene in tomato was scanned for the presence of putative cis-acting regulatory elements and motifs registered in Plant PLACE database (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en) (consulted in March 22nd, 2015) [44]. The cis-acting elements analyzed are listed in S3 Table.
Statistical analysis
Data presented in this work are expressed as arithmetic means +/- SD of replicate plants within an experiment. The data shown are representative of a total of three independent biological replicates. The results were statistically analyzed using Student’s t test; Statistica soflware version 10 (Statsoft, Tulsa, USA). A p value of <0,05 was considered statistically significant. In all the figures presented, error bars indicate standard deviation.
Results
Analysis of SlARF promoter genes to search for stress response cis-acting regulatory elements
PLACE signal scan analysis revealed the presence of several cis-regulatory elements that are putatively associated with plant response to stress in the 5’ regulatory region (2kb upstream region from the translation start codon) of SlARFs (S1 Fig). The names of the identified cis-acting elements and their predicted functions are listed in S3 Table. Some of these elements (CCATBOX1, WBOXNTERF3, RAV1AAT, MYCCONSENSUSAT, GT1GMSCAM4, MYB2A; MYBCORE and MYBCONCENSUSAT) are common to the majority of the analyzed SlARFs. A high number of these conserved motifs is identified for SlARF1, SlARF2A, SlARF4, SlARF6B, SlARF9A, SlARF16A and SlARF16B whereas a low number are found for SlARF10A and SlARF10B genes.
Abiotic stress associated cis-acting regulatory elements were found for all SlARFs (S1 Fig). MYBCORE cis-acting element related to drought stress response [51] was detected in most of the 22 SlARFs except for SlARF5, SlARF10A and SlARF24 while the MYB1AT [52] motif was only located in 13 of the 22 SlARFs. Noteworthy, MYCONSENSUSAT motif [53] was found in all the 22 SlARFs genes with varying occurrence. MYCATERD1 motif [52] was only found in SlARF1, SlARF6A, SlARF7B, SlARF8A, SlARF8B, SlARF9B, SlARF16A and SlARF24 while the presence of the other MYC binding site “MYCATRD22” [51] was specific to SlARF4, SlARF5, SlARF7A, SlARF7B, SlARF8B, SlARF16A and SlARF16B. Screening for motifs involved in temperature variation showed that with the exception of SlARF8A and SlARF18, all other SlARFs promoter regions were enriched with the two cis-acting elements RAV1AAT and CCAATBOX1 boxes [54,55]. Cis-acting element associated with biotic stress response such as WBOXNTCHN48, WBOXNTERF3 and GT1GMSCAM4 were located in most SlARF promoters with differences in their occurrence. The WBOXNTERF3 motif, involved in wounding [56], was found in the SlARFs gene promoters except for SlARF3, SlARF7A and SlARF9B while the elicitor binding motif WBOXNTCHN48 [57] was only detected in SlARF2A, SlARF5 and SlARF6B. Finally, the GT1GMSCAM4 motif [58] was present in all the SlARFs promoter regions.
Tomato ARF responsiveness to biotic stress
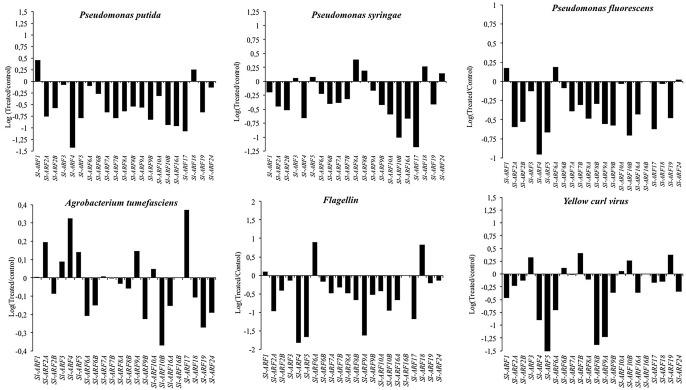
We mined the public tomato RNA-Seq web platform TomExpress [59] to examine the expression profile of tomato SlARF genes upon biotic stress. Among the expression data available in the TomExpress platform, we focused on those related to flagellin, bacteria (Pseudomonas putida, Pseudomonas fluorescens, Pseudomonas syringae DC3000 and Agrobacterium tumefaciens) and Yellow curl leave virus YCV infection (Fig 1). When tomato leaves were exposed six hours to Pseudomonas strains (Pseudomonas putida, Pseudomonas fluorescens or Pseudomonas syringae DC3000), most SlARF genes were down-regulated.
Fig 1. SlARFs gene expression in tomato plant leaves exposed to various biotic stresses: Flagellin, Pseudomonas putida, Pseudomonas syringae, Pseudomonas fluorescens, Agrobacterium tumefaciens and Yellow curl virus.
All the data presented here were extracted from TOMPEXPRESS database (http://gbf.toulouse.inra.fr/tomexpress).
Flagellin is considered as the main building unit of the bacterial motility organ and constitutes an important microbial pattern for virulence through its use by plants to recognize bacterial pathogens. When tomato leaves are exposed six hours to flagellin [60], the expression of almost all members of the SlARF family is repressed except for SlARF6A and SlARF18 that showed a twofold induction.
In response to P. putida, the expression of SlARFs was also repressed except for SlARF1 and SlARF18. Likewise, upon P. syringae infection the expression of the overwhelming majority of SlARF genes was downregulated although SlARF8A was induced. P. fluorescens infection also resulted in down-regulation of most ARF genes, regulation that is highly marked for SlARF4. Noteworthy, none of the 22 SlARFs showed a significant change in their expression at the transcription level upon Agrobacterium tumefaciens infection.
Tomato yellow leaf curl virus (TYLCV) designates a complex of geminiviruses infecting tomato cultures worldwide. This virus is transmitted by a single insect species, the whitefly Bemisia tabaci. In response to TYLCV, the expression of SlARF4, SlARF5, SlARF6A, SlARF8B and SlARF9A genes was downregulated in leaf tissues whereas the expression of the remaining SlARFs did not exhibit significant change. Overall, down-regulation seems to be clearly the main trend for SlARF genes upon bacterial infection.
Tomato ARF responsiveness upon abiotic stress
Distribution of auxin response activated upon abiotic stress
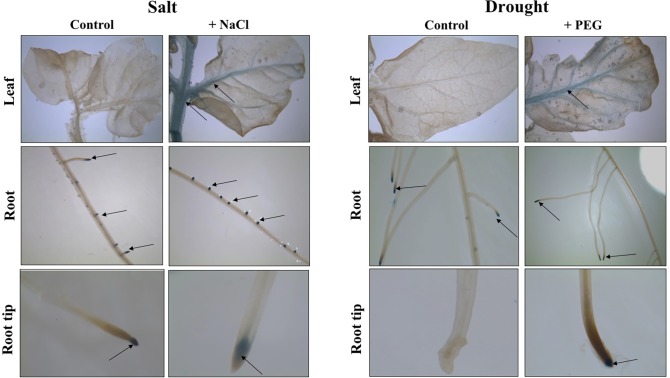
The extent of changes in auxin distribution and auxin signaling upon application of salt and drought stresses, were assessed using tomato lines expressing the GUS reported gene driven by the DR5 auxin-responsive promoter [61]. DR5::GUS tomato transgenic lines were subjected to salt and drought stress by adding 250 mM of NaCl and 15% PEG20000 to the nutrient solution, respectively (Fig 2). In leaf organs, GUS activity appears after 24 hours of salt stress and is localized in leaf veins and petioles while it is barely detectable in untreated plants. In salt stressed roots, the distribution of GUS expression is observed in lateral root primordia and in primary root tip. The control plants showed also a blue coloration in primary and lateral root tips. After 5 days of PEG treatment, a slight increase in GUS activity was detected in leaf veins and primary and lateral root tips while it scarcely appears in lateral root primordia of the untreated plants.
Fig 2. GUS activity in DR5::GUS tomato lines in salt or drought stress conditions.
Salt and drought stresses were performed on three week-old tomato plants by adding 250 mM of NaCl or 15% PEG 20000 to the nutrient solution. Black arrows show the location of the GUS activity in the different tissues analyzed.
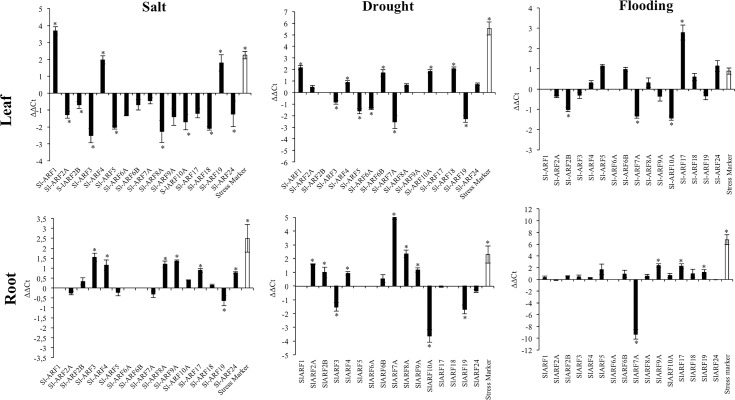
Expression profiling of SlARF genes in response to salt stress
We analyzed the expression profiles of SlARF genes in tomato leaves and roots after 24 hours of salt treatment. The expression of Cold Inducible 7 gene (CI7), a known marker for salt stress [48], was significantly induced in both leaves and roots as expected, thus validating the efficiency of the applied stress in our condition. The data reveal that almost all SlARF genes were downregulated in leaves except SlARF1, SlARF4 and SlARF19 whose expression was significantly induced (Fig 3). Among the down-regulated genes, SlARF3, SlARF5, SlARF8A and SlARF18 were highly repressed. In roots, 9 SlARF genes were up regulated among which SlARF3, SlARF4, SlARF8A and SlARF9A showed high induction (Fig 3). The expression of SlARF2A, SlARF5 and SlARF7A were slightly repressed though not statistically significant while the expression of SlARF19 was significantly repressed.
Fig 3. SlARFs gene expression under salt, drought and flooding conditions.
Values are mean ± SD of three biological replicates. White bars represent the expression of stress marker: CI7 for salt and drought stresses (Leaves & Roots), ACO1 in flooded leaves and PDC1 in flooded roots. Stars (*) indicate the statistical significance (p<0,05) using Student’s t-test.
Expression profiling of SlARF genes in response to drought stress
The responsiveness of SlARF members under water deficit condition was investigated by qPCR after five days of drought stress application. The drought marker gene C17, unsurprisingly, showed the same pattern (high induction) in leaves and roots as previously reported by Kirch et al., (1997) [48]. In leaves, solely, SlARF1, SlARF4, SlARF6B, SlARF10A and SlARF18 showed significant increase in expression, while the expression of SlARF3, SlARF5, SlARF6A, SlARF7A and SlARF19 was significantly repressed (Fig 3).
In roots, the expression of SlARF2A, SlARF2B, SlARF4, SlARF7A, SlARF8A and SlARF9A was significantly increased upon drought stress while SlARF3, SlARF10A, SlARF19 displayed significant down-regulation. Of particular note, SlARF7A with 32 fold induction displayed the most dramatic up-regulation compared to CI7 control gene which showed only four times increase in the stressed roots (Fig 3).
Expression profiling of SlARF genes in response to flooding stress
After 48 hours of flooding, 3 SlARFs (SlARF2B, SlARF7A and SlARF9A) were significantly downregulated and 4 were upregulated (SlARF5, SlARF6B, SlARF17 and SlARF24) in leaves (Fig 3). The highest changes were observed within SlARF7 and SlARF17 whose expression was three times more repressed or ten times more induced respectively. As validation of the flooding stress, the expression of the amino-cyclopropane -1- carboxylate oxidase (ACO1) and pyruvate decarboxylase 1 (PDC1) flooding marker genes was substantially increased in both leaves and roots as reported by Nie et al., (2002) [50] and Gharbi et al., (2007) [62], respectively. In root parts, SlARF7A was strongly repressed (512 times) (Fig 3).
Investigating the expression of SlARFs gene family in salt, drought and flooding conditions have shown that several members of these transcriptional factors are regulated. Among the candidate genes identified, SlARF8A and SlARF10A were significantly regulated in salt and water stresses. Moreover, for these two genes, we were able to generate transgenic lines expressing promoter-GUS construct (pARF8A::GUS or pARF10As::GUS). All this prompt us to check SlARF8A and SlARF10A genes expressions in planta in response to salt and drought stresses and also to examine a possible involvement of related miRNA in their regulation.
Spatio-temporal analysis of SlARF8A and SlARF10A expression under salt and drought stress conditions
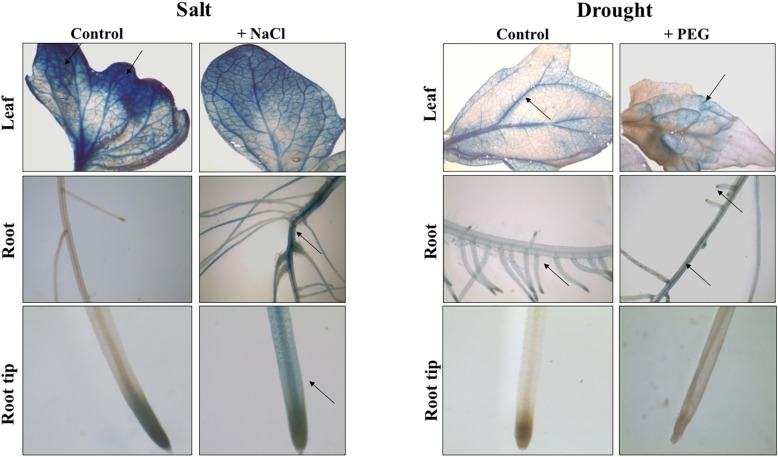
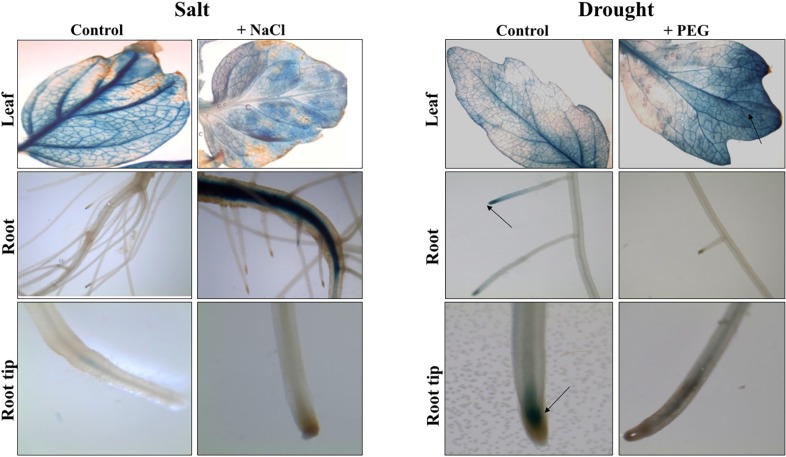
Since SlARF8A and SlARF10A were significantly regulated by salt and drought stresses, we sought to examine their spatiotemporal expression in planta. Tomato transgenic lines harboring pARF8A::GUS or pARF10A::GUS constructs were therefore generated and GUS expression was analyzed under stress conditions. GUS staining performed on three week-old plants expressing pARF8A::GUS fusion construct and exposed to 24 hours salt stress reveals a strong expression in leaves and lateral roots compared to untreated plants showing GUS expression only in leaves top and leaves veins (Fig 4). After 5 days of drought stress, GUS activity was detected in the primary root and in the middle part of leaves while the control plants showed a uniform blue coloration in leaf veins, primary root and lateral root tips (Fig 4).
Fig 4. GUS activity in pARF8A::GUS tomato lines in salt or drought stress conditions.
Salt and drought stresses were performed on three week-old tomato plants by adding 250 mM of NaCl or 15% PEG 20000 to the nutrient solution. Black arrows show the location of the GUS activity in the different tissues analyzed.
The GUS activity in pARF10A::GUS transgenic plants remained similar to the control plants after 24 hours on salt stress application in leaf tissues while it becomes intense in stressed roots. The control showed a uniform blue coloration all over the leaf. Five days exposure to drought restricted the GUS expression to the apical part of the leaf while the control plants displayed a blue coloration in the whole leaf and in the primary and lateral root tips (Fig 5).
Fig 5. GUS activity in pARF10A::GUS tomato lines in salt or drought stress conditions.
Salt and drought stresses were performed on three week-old tomato plants by adding 250 mM of NaCl or 15% PEG 20000 to the nutrient solution. Black arrows show the location of the GUS activity in the different tissues analyzed.
SlARF specific miRNAs responsiveness under abiotic stress conditions
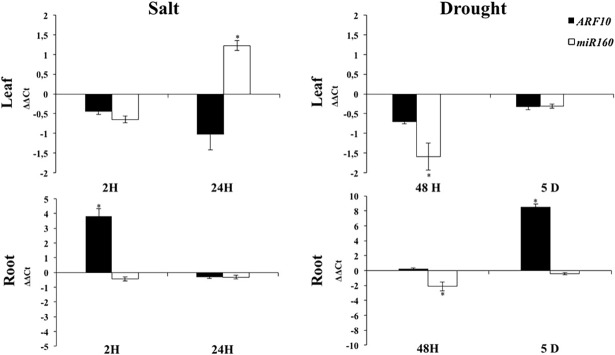
miRNAs are small non-coding RNA molecules that negatively regulate their target genes at the post-transcriptional level [38,42]. ARF family members are known to be subject to this type of regulation. SlARF8 and SlARF6 are known to be targeted by miR167 and SlARF10 is specifically regulated by miR160 [41,63]. Since the data described above (Fig 3) indicated that the expression of SlARF8A and SlARF10A was altered by both salt and drought stress, we checked whether the modification in the expression of these SlARF was correlated with the expression of their specific miRNA regulators. qPCR analysis showed that the expression pattern of miR160 and its target gene SlARF10A changed significantly in response to salt stress (Fig 6). In leaves, miR160 was two times induced after 24 hours of salt treatment while ARF10A gene was concomitantly downregulated. In roots, SlARF10A was up-regulated during the first two hours of salt treatment while the expression of miR160 remained unchanged. Tomato miR160 was significantly repressed in leaves after 48 hours of drought stress whereas the expression of ARF10A showed a high induction in 5 days drought stressed roots while the expression of miR160 remained similar to the control.
Fig 6. SlARF10A and miR160 expression under salt and drought stress conditions.
Values are mean ± SD of three biological replicates. Stars (*) indicate the statistical significance (p<0,05) using Student’s t-test.
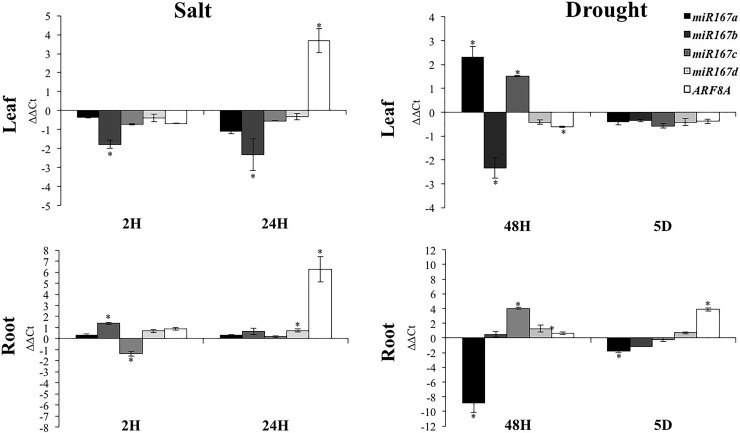
The expression of miR167 precursors and their target gene SlARF8A were modified by salt and drought stresses (Fig 7). In salt stressed leaves, SlARF8A was highly induced up to 16 times after 24 hours of salt treatment while the expression miR167b was in a concurring of way highly repressed. In roots, SlARF8A was highly up-regulated after 24 hours of salt treatment while no strong changes occurred in the expression of miR167 precursors. After 48 hours of drought stress exposure, SlARF8A gene expression in leaves was not strongly affected while miR167a and miR167c genes were highly induced when miR167b was repressed. In roots, miR167a was highly repressed when miR167c was induced while SlARF8A expression did not change. Five days of drought stress exposure resulted in an over-expression of ARF8A gene and down expression of miR167a in roots.
Fig 7. SlARF8A and miR167s (miR167a, miR167b, miR167c, miR167d) expression under salt and drought stress conditions.
Values are mean ± SD of three biological replicates. Stars (*) indicate the statistical significance (p<0,05) using Student’s t-test.
Discussion
Plants are constantly exposed to various abiotic and biotic stresses in their immediate environment, limiting considerably their growth rate and productivity. Plants have evolved complex signaling pathways in response to various stimuli such as salt, drought, cold, wounding, or pathogen invasion in order to minimize damages while conserving valuable resources for growth and reproduction. These responses were found to be mediated by plant growth regulators [64]. Several phytohormones such as abscisic acid, ethylene, salicylic acid and jasmonate were identified as key regulators in various stress responses. Auxin is implicated in many plant developmental processes and recent studies suggest its involvement in stress [65,66]. It has been reported that the endogenous auxin level decreased substantially upon drought stress conditions but increased in response to cold and heat stresses in Rice and during pathogen infection in Arabidopsis thaliana [67–69]. The expression of Aux⁄IAA and ARF genes is altered during cold acclimation in Arabidopsis thaliana and during salt, water stresses and biotic stresses in Oryza sativa [16,70,71]. In this work, we investigated the implication of some actors of auxin signalling pathway in tomato responses to biotic and abiotic stresses.
Auxin signaling pathway is altered by pathogen infection in tomato
The auxin response has emerged recently as an active actor in plant defense against pathogens [72]. Auxin coordinates plant development essentially through two transcriptional regulators Aux/IAA and ARFs [24]. In tomato, 22 SlARFs were previously isolated and characterized [21]. In silico promoter analysis of SlARFs using Plant PLACE database have revealed that at least one copy of the pathogenesis induced element (GT1GMSCAM4) was found in SlARFs 5’ regulatory regions. Furthermore, the expression of almost the entire SlARF gene family was altered in response to some of the pathogen tested based on the results related to biotic stresses that are available in TOMEXPRESS platform [59]. This suggests that auxin might be involved in biotic stress responses through ARFs. Other previous studies have underlined the involvement of auxin responsive genes in biotic stress responses. Auxin responsive genes are downregulated in Arabidopsis thaliana upon Botrytis cinerea infection [73]. In cotton, gene expression profiling in response to infection with Fusarium oxysporum f. sp vasinfectum also revealed differential expression of auxin responsive genes [74]. Studies conducted on rice had revealed that only two OsARFs were responsive to Magnaporthe grisea (ascomycete fungus) and Striga hermonthica (obligate root hemiparasite) infection [71]. The present results revealed that the expression of the SlARF gene family was altered by pathogen infections. Our findings suggest that the regulation of auxin pathway might be an important aspect of the defense response through the regulation of the expression of ARF gene family in tomato.
Auxin accumulation is modified within high salinity and water deficit in tomato
Auxin and its gradients are found to be closely associated to some morphological changes observed in plants exposed to environmental stresses. These changes are likely associated with lateral root formation and axillary branching and represent one of the most specific responses to abiotic stresses [75,76]. In tomato, we showed that auxin distribution is modified by the stress application. Auxin was accumulated only in lateral root primordia and tips of DR5::GUS tomato transgenic plants exposed to salt and drought stress conditions. In support to these results, Zolla et al., (2009) [77] reported that salt stress stimulates the development of a larger number of lateral root primodia in Arabidopsis. This stimulation was associated with auxin accumulation in the lateral root primordia [78–80].
Auxin response factors are potential key actors in abiotic stress responses in tomato
In silico, SlARFs promoter analysis has shown that SlARF regulatory regions were enriched with stress associated motifs. Drought and salt related cis-acting elements were found in all the 22 SlARF promoter regions. SlARF1, SlARF4, SlARF8A, SlARF19 and SlARF24 regulatory regions were more enriched in the salt induced element (GT1GMSCAM4). The promoter regions of SlARF3 and SlARF9A although present respectively one copy of the GT1GMSCAM4 salt induced element. The MYC (MYCATERD1, MYCATRD22 and MYCCONSENSUSAT) were found in several copies SlARF1, SlARF4, SlARF6A, SlARF7A, SlARF8A, SlARF9A and SlARF18 regulatory regions while the MYB motifs (MYBCORE, MYB1AT, MYB2AT and MYB2CONSENSUSAT) was highly present in SlARF2A, SlARF2B, SlARF4, SlARF6B SlARF9A and SlARF18 promoter regions. All these finding suggest that the functions of these transcriptional factors may be associated with environmental stresses response. Moreover, investigating the expression of Auxin Response Factor family under abiotic stress conditions showed that most of the tomato ARFs were responsive and some of them were significantly regulated. Among the regulated genes, SlARF1, SlARF4, SlARF8A, SlARF19 and SlARF24 showed a significant upregulation in response to salt stress while SlARF1, SlARF2A, SlARF2B SlARF4, SlARF6A, SlARF6B, SlARF7A, SlARF8A, SlARF9A and SlARF18 were substantially induced in drought stress conditions. In rice, several OsARFs were shown to be involved in salt and drought stress responses. Jain and Khurana (2009) [11] had reported that both OsARF11 and OsARF15 genes had shown differential expression in salt stress conditions and suggested that they are involved in rice response to stress. Zhou et al., (2007) [81] had shown the involvement of nine ARFs genes in response to water deficit (OsARF2, OsARF4, OsARF10, OsARF14, OsARF16, OsARF18, OsARF19, OsARF22 and OsARF23). In Sorghum bicolor, Wang et al., (2010) [17] reported that SbARF10, SbARF16, and SbARF21 genes were significantly induced in leaves and roots tissues exposed to drought conditions. However, among the 50 ARF isolated and identified in Soybean (Glycine max), only GmARF33 and GmARF50 were responsive (induced) to water deficit and were suggested as excellent candidates for drought stress responses in this plant [18].
The expression of some SlARFs showed different pattern in leaf and root under the same treatment. This difference might be associated with the function of these SlARFs. Some ARF functions are more related to the underground part more than to the aerial part. This is the case for SlARF7A, whose expression was significantly repressed or induced respectively in shoot and root parts under drought treatment. This gene seems to act in the underground part of the plant. This assumption is supported by Okushima et al., (2007) [82] and Goh et al., (2012) [83] who had shown that ARF7 is involved in the control of lateral root formation in Arabidopsis.
Functional studies have shown that ARF genes are involved in the control of plant growth and development. Actually, Okushima et al., (2007) [82] and had reported that AtARF7 acts synergistically with AtARF19 in the control of lateral root formation and hypocotyl gravitotropism. Otherwise, Solanum lycopersicum ARF2 has been proposed as a key regulator of apical hook formation while, in Arabidopsis thaliana, the same gene seems to act as a positive activator of flowering senescence and abscission and as a repressor of cell growth in the presence or absence of light and differential hypocotyl growth [84,85]. All these results suggest that auxin, through ARF gene family, activates stress specific auxin response genes in order to mitigate the negative effects of abiotic stresses.
ARF8A and ARF10 are good indicators for salt and drought response in tomato
Our data have shown that SlARF gene family expression is modified by salinity and water deficit. Among these genes, SlARF8A and SlARF10A expressions are altered by the abiotic stress conditions. Moreover, their regulatory regions contain several copies of salt and/or drought induced elements. Investigating their expression in planta had revealed that their expression is clearly modified by salt and drought stress conditions. We found that SlARF8A promoter is especially expressed in lateral roots after 24 hours of exposure to both stresses. Previous studies confirmed the involvement of ARF8 in abiotic stress response. Tian et al., (2004) [86] have suggested that AtARF8 is stably expressed in lateral roots of Arabidopsis thaliana under light conditions and appears to control hypocotyl elongation.
Histochemical analysis of pARF10A::GUS tomato lines in salt stress conditions revealed that ARF10A gene is particularly more expressed in primary and lateral roots. The SlARF10A regulatory region presents four different salt/drought related motifs, which suggests that this gene might be implicated in root growth and development under stress conditions. Wang et al., 2005 [87] have found that Arabidopsis thaliana ARF10A is implicated in root formation and architecture by restricting cell division and promoting cell differentiation in the distal region which might suggest its implication in root development under stress conditions.
miRNAs contribute to ARF gene regulation under stress conditions
miRNA are post transcriptional regulators of a large number of target genes by guiding target mRNAs for degradation or by repressing translation. Zhao and Srivastava (2007) [88] reported that some miRNAs are regulated and could be involved in cell responses to abiotic stresses such as salinity, cold and dehydration. Some miRNAs, including miR160 and miR167, known to regulate the levels of transcription factor transcripts and protein abundance showed altered expression profiles in salt and drought conditions. Our results show that the downregulation of miR167 precursors (miR167a, miR167b, miR167c and miR167d) was negatively correlated with the accumulation of SlARF8A transcripts in leaves after 24 hours of salt exposure. This finding was also observed in roots after 5 days of drought treatment. Knowing that ARF8A is implicated in control and development of vegetative and floral organs in Dicots, the downregulation of miR167 gene precursors might enhance the auxin response and thus enhance shoot and leaf development. In cassava (Manihot esculenta), Xia et al., (2014) [89] showed that miR167 expression was modified in response to extreme temperature and could induce the cleavage of its target gene, ARF8, due to the presence of the miR167 cleavage sites on it. The expression of miR160 was also negatively correlated with the expression of its target gene SlARF10A in roots exposed to drought stress. This finding suggests that miR160 might be implicated in the post-transcriptional regulation of the expression of ARF10A gene under stress conditions.
Conclusion
Auxin plays crucial roles in various aspects of plant growth and development. This phytohormone acts on the transcriptional regulation of target genes, mainly through Auxin Response Factors. The current study provide several clues on the potential involvement of many ARF genes as mediators of the auxin action in biotic and abiotic stress responses in tomato. The 5’ regulatory regions analysis of the SlARFs genes indicates the presence of several biotic and abiotic stress-responsive cis-elements. Moreover, transcriptome analysis reveals the responsiveness of SlARF genes to a wide range of biotic and abiotic stresses. Additionally, SlARF8A and SlARF10A specific miRNA were involved in the regulation of these genes suggesting the importance of post-transcriptional regulation in auxin signaling pathway and plant response to abiotic stresses. Taking together, the data presented in this work brings new elements and open new ways to explore the molecular mechanisms associated with stress tolerance in tomato. This provides new insights into tomato selection and breeding for environmental stress-tolerant.
Supporting information
(PDF)
The data presented in this table were extracted from TOMEXPRESS database [44].
(PDF)
The 2 Kb of 5’ regulatory region were analyzed using the Place software.
(PDF)
The consensus sequences corresponding to the various putative cis-elements are described in S2 Table. Positions are with respect to the first base of translation start site.
(PDF)
Acknowledgments
The authors are grateful to L. Lemonnier, D. Saint-Martin and O. Berseille for genetic transformation and culture of tomato plants. This research was supported by the “Laboratoire d'Excellence” (LABEX) entitled TULIP (ANR-10-LABX-41) and benefited from networking activities within the European COST Action FA1106, from the ANR TomEpiSet (ANR-16-CE20-0014) project, the TomGEM H2020 project, by Hubert-Curien partnership program Volubilis (MA/12/280-27105PL) and CMCU program (08G0906) granted to Dr Habib Khoudi.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the “Laboratoire d'Excellence” (LABEX) entitled TULIP (ANR-10-LABX-41) and benefited from networking activities within the European COST Action FA1106, from the ANR TomEpiSet (ANR-16-CE20-0014) project, the TomGEM H2020 project, by Hubert-Curien partnership program Volubilis (MA/12/280-27105PL) and CMCU program (08G0906) granted to Dr Habib Khoudi. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Popko J, Hänsch R, Mendel R, Polle A, Teichmann T. The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol. 2010;12: 242–258. doi: 10.1111/j.1438-8677.2009.00305.x [DOI] [PubMed] [Google Scholar]
- 2.Atkinson NJ, Urwin PE. The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot. 2012;63: 3523–3543. doi: 10.1093/jxb/ers100 [DOI] [PubMed] [Google Scholar]
- 3.Tuteja N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal Behav. 2007;2: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorantes-Acosta AE, Sánchez-Hernández CV, Arteaga-Vazquez MA. Biotic stress in plants: life lessons from your parents and grandparents. Front Genet. 2012;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuteja N, Sopory SK. Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal Behav. 2008;3: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leyser H, Pickett FB, Dharmasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR‐AC1 promoter. Plant J. 1996;10: 403–413. [DOI] [PubMed] [Google Scholar]
- 7.Reinhardt D, Pesce E-R, Stieger P, Mandel T, Baltensperger K, Bennett M, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426: 255–260. doi: 10.1038/nature02081 [DOI] [PubMed] [Google Scholar]
- 8.Mattsson J, Ckurshumova W, Berleth T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 2003;131: 1327–1339. doi: 10.1104/pp.013623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci U S A. 2006;103: 236–241. doi: 10.1073/pnas.0507127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, Zhao Y. A role for auxin in flower development. J Integr Plant Biol. 2007;49: 99–104. [Google Scholar]
- 11.Jain M, Khurana JP. Transcript profiling reveals diverse roles of auxin‐responsive genes during reproductive development and abiotic stress in rice. Febs J. 2009;276: 3148–3162. doi: 10.1111/j.1742-4658.2009.07033.x [DOI] [PubMed] [Google Scholar]
- 12.Krizek BA. Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family. J Exp Bot. 2011;62: 3311–3319. doi: 10.1093/jxb/err127 [DOI] [PubMed] [Google Scholar]
- 13.Kazan K. Auxin and the integration of environmental signals into plant root development. Ann Bot. 2013;112: 1655–1665. doi: 10.1093/aob/mct229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong YH, Chang H-S, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129: 661–677. doi: 10.1104/pp.002857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, et al. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene. 2007;394: 13–24. doi: 10.1016/j.gene.2007.01.006 [DOI] [PubMed] [Google Scholar]
- 16.Song Y, Wang L, Xiong L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta. 2009;229: 577–591. doi: 10.1007/s00425-008-0853-7 [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Bai Y, Shen C, Wu Y, Zhang S, Jiang D, et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics. 2010;10: 533–546. doi: 10.1007/s10142-010-0174-3 [DOI] [PubMed] [Google Scholar]
- 18.Van Ha C, Le DT, Nishiyama R, Watanabe Y, Sulieman S, Tran UT, et al. The Auxin Response Factor Transcription Factor Family in Soybean: Genome-Wide Identification and Expression Analyses During Development and Water Stress. DNA Res Int J Rapid Publ Rep Genes Genomes. 2013;20: 511–524. doi: 10.1093/dnares/dst027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319: 1384–1386. doi: 10.1126/science.1151461 [DOI] [PubMed] [Google Scholar]
- 20.Causier B, Lloyd J, Stevens L, Davies B. TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal Behav. 2012;7: 325–328. doi: 10.4161/psb.19283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zouine M, Fu Y, Chateigner-Boutin A-L, Mila I, Frasse P, Wang H, et al. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PloS One. 2014;9: e84203 doi: 10.1371/journal.pone.0084203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci. 1999;96: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19: 309–319. [DOI] [PubMed] [Google Scholar]
- 24.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10: 453–460. doi: 10.1016/j.pbi.2007.08.014 [DOI] [PubMed] [Google Scholar]
- 25.Chandler JW. Auxin response factors. Plant Cell Environ. 2016;39: 1014–1028. doi: 10.1111/pce.12662 [DOI] [PubMed] [Google Scholar]
- 26.Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 2007;17: 1784–1790. doi: 10.1016/j.cub.2007.09.025 [DOI] [PubMed] [Google Scholar]
- 27.Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin‐induced lateral root formation. Plant J. 2005;43: 118–130. doi: 10.1111/j.1365-313X.2005.02432.x [DOI] [PubMed] [Google Scholar]
- 28.De Jong M, Mariani C, Vriezen WH. The role of auxin and gibberellin in tomato fruit set. J Exp Bot. 2009; erp094. [DOI] [PubMed] [Google Scholar]
- 29.Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, et al. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot. 2010;61: 1419–1430. doi: 10.1093/jxb/erq010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sagar M, Chervin C, Mila I, Hao Y, Roustan J-P, Benichou M, et al. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 2013;161: 1362–1374. doi: 10.1104/pp.113.213843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liscum E, Reed J. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49: 387–400. [PubMed] [Google Scholar]
- 32.Xing H, Pudake RN, Guo G, Xing G, Hu Z, Zhang Y, et al. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics. 2011;12: 178 doi: 10.1186/1471-2164-12-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalluri UC, DiFazio SP, Brunner AM, Tuskan GA. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007;7: 59 doi: 10.1186/1471-2229-7-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, et al. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 2011;30: 2059 doi: 10.1007/s00299-011-1113-z [DOI] [PubMed] [Google Scholar]
- 35.Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485: 635 doi: 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mun J-H, Yu H-J, Shin JY, Oh M, Hwang H-J, Chung H. Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol Genet Genomics. 2012;287: 765–784. doi: 10.1007/s00438-012-0718-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu W, Zuo J, Hou X, Yan Y, Wei Y, Liu J, et al. The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci. 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunkar R, Zhu J-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16: 2001–2019. doi: 10.1105/tpc.104.022830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Jia W, Yang J, Ismail AM. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006;97: 111–119. [Google Scholar]
- 40.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17: 1360–1375. doi: 10.1105/tpc.105.031716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu N, Wu S, Van Houten J, Wang Y, Ding B, Fei Z, et al. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J Exp Bot. 2014;65: 2507–2520. doi: 10.1093/jxb/eru141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. cell. 2004;116: 281–297. [DOI] [PubMed] [Google Scholar]
- 43.Curaba J, Singh MB, Bhalla PL. miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot. 2014;65: 1425–1438. doi: 10.1093/jxb/eru002 [DOI] [PubMed] [Google Scholar]
- 44.SOGO [Internet]. [cited 6 Dec 2017]. Available: https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en
- 45.Zouine M, Maza E, Djari A, Lauvernier M, Frasse P, Smouni A, et al. TomExpress, a unified tomato RNA‐Seq platform for visualization of expression data, clustering and correlation networks. Plant J. 2017;92: 727–735. doi: 10.1111/tpj.13711 [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, et al. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell. 2005;17: 2676–2692. doi: 10.1105/tpc.105.033415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broughton W, Dilworth M. Control of leghaemoglobin synthesis in snake beans. Biochem J. 1971;125: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirch H-H, Berkel J van, Glaczinski H, Salamini F, Gebhardt C. Structural organization, expression and promoter activity of a cold-stress-inducible gene of potato (Solanum tuberosum L.). Plant Mol Biol. 33: 897–909. doi: 10.1023/A:1005759925962 [DOI] [PubMed] [Google Scholar]
- 49.Hsieh T-H, Lee J, Charng Y, Chan M-T. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 2002;130: 618–626. doi: 10.1104/pp.006783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nie X, Singh RP, Tai GC. Molecular characterization and expression analysis of 1-aminocyclopropane-1-carboxylate oxidase homologs from potato under abiotic and biotic stresses. Genome. 2002;45: 905–913. [DOI] [PubMed] [Google Scholar]
- 51.Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell. 1997;9: 1859–1868. doi: 10.1105/tpc.9.10.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15: 63–78. doi: 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dang HQ, Tran NQ, Tuteja R, Tuteja N. Promoter of a salinity and cold stress-induced MCM6 DNA helicase from pea. Plant Signal Behav. 2011;6: 1006–1008. doi: 10.4161/psb.6.7.15502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guy C. Molecular responses of plants to cold shock and cold acclimation. J Mol Microbiol Biotechnol. 1999;1: 231–242. [PubMed] [Google Scholar]
- 55.Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol. 2006;61: 897 doi: 10.1007/s11103-006-0057-0 [DOI] [PubMed] [Google Scholar]
- 56.Nishiuchi T, Shinshi H, Suzuki K. Rapid and transient activation of transcription of the ERF3 Gene by Wounding in Tobacco Leaves POSSIBLE INVOLVEMENT OF NtWRKYs AND AUTOREPRESSION. J Biol Chem. 2004;279: 55355–55361. doi: 10.1074/jbc.M409674200 [DOI] [PubMed] [Google Scholar]
- 57.Deb A, Kundu S. Deciphering cis-regulatory element mediated combinatorial regulation in rice under blast infected condition. PloS One. 2015;10: e0137295 doi: 10.1371/journal.pone.0137295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trivedi DK, Ansari MW, Tuteja N. Multiple abiotic stress responsive rice cyclophilin:(OsCYP-25) mediates a wide range of cellular responses. Commun Integr Biol. 2013;6: e25260 doi: 10.4161/cib.25260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.TomExpress Home: visualize and analyse tomato RNA-seq data [Internet]. [cited 28 Mar 2017]. Available: http://gbf.toulouse.inra.fr/tomexpress/www/welcomeTomExpress.php
- 60.Rosli HG, Zheng Y, Pombo MA, Zhong S, Bombarely A, Fei Z, et al. Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 2013;14: R139 doi: 10.1186/gb-2013-14-12-r139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9: 1963–1971. doi: 10.1105/tpc.9.11.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gharbi I, Ricard B, Rolin D, Maucourt M, Andrieu M-H, Bizid E, et al. Effect of hexokinase activity on tomato root metabolism during prolonged hypoxia. Plant Cell Environ. 2007;30: 508–517. doi: 10.1111/j.1365-3040.2007.01640.x [DOI] [PubMed] [Google Scholar]
- 63.Hendelman A, Buxdorf K, Stav R, Kravchik M, Arazi T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol Biol. 2012;78: 561–576. doi: 10.1007/s11103-012-9883-4 [DOI] [PubMed] [Google Scholar]
- 64.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14: 290–295. doi: 10.1016/j.pbi.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 65.Rahman A. Auxin: a regulator of cold stress response. Physiol Plant. 2013;147: 28–35. doi: 10.1111/j.1399-3054.2012.01617.x [DOI] [PubMed] [Google Scholar]
- 66.Liu L, Guo G, Wang Z, Ji H, Mu F, Li X. Auxin in Plant Growth and Stress Responses In: Tran L-SP, Pal S, editors. Phytohormones: A Window to Metabolism, Signaling and Biotechnological Applications. Springer; New York; 2014. pp. 1–35. Available: http://link.springer.com.gate6.inist.fr/chapter/10.1007/978-1-4939-0491-4_1 [Google Scholar]
- 67.Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 2004;39: 790–808. doi: 10.1111/j.1365-313X.2004.02168.x [DOI] [PubMed] [Google Scholar]
- 68.Fu J, Wang S. Insights into auxin signaling in plant–pathogen interactions. Front Plant Sci. 2011;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du H, Liu H, Xiong L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci. 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hannah MA, Heyer AG, Hincha DK. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005;1: e26 doi: 10.1371/journal.pgen.0010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghanashyam C, Jain M. Role of auxin-responsive genes in biotic stress responses. Plant Signal Behav. 2009;4: 846–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312: 436–439. doi: 10.1126/science.aae0382 [DOI] [PubMed] [Google Scholar]
- 73.Llorente F, Muskett P, Sánchez-Vallet A, López G, Ramos B, Sánchez-Rodríguez C, et al. Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol Plant. 2008;1: 496–509. doi: 10.1093/mp/ssn025 [DOI] [PubMed] [Google Scholar]
- 74.Dowd C, Wilson IW, McFadden H. Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp. vasinfectum. Mol Plant Microbe Interact. 2004;17: 654–667. doi: 10.1094/MPMI.2004.17.6.654 [DOI] [PubMed] [Google Scholar]
- 75.Cao Y-R, Chen S-Y, Zhang J-S. Ethylene signaling regulates salt stress response. Plant Signal Behav. 2008;3: 761–763. doi: 10.4161/psb.3.10.5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmad P, Prasad MNV. Abiotic stress responses in plants: metabolism, productivity and sustainability Springer Science & Business Media; 2011. [Google Scholar]
- 77.Zolla G, Heimer YM, Barak S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J Exp Bot. 2009;61: 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G. Shoot‐derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 2002;29: 325–332. [DOI] [PubMed] [Google Scholar]
- 79.López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Pérez-Torres A, Rampey RA, Bartel B, et al. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 2005;137: 681–691. doi: 10.1104/pp.104.049577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He X, Mu R, Cao W, Zhang Z, Zhang J, Chen S. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44: 903–916. doi: 10.1111/j.1365-313X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- 81.Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, et al. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol. 2007;63: 591–608. doi: 10.1007/s11103-006-9111-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19: 118–130. doi: 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA–ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Phil Trans R Soc B. 2012;367: 1461–1468. doi: 10.1098/rstb.2011.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okushima Y, Mitina I, Quach HL, Theologis A. AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 2005;43: 29–46. doi: 10.1111/j.1365-313X.2005.02426.x [DOI] [PubMed] [Google Scholar]
- 85.Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, et al. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot. 2009;60: 1349–1362. doi: 10.1093/jxb/erp009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian C, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, et al. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004;40: 333–343. doi: 10.1111/j.1365-313X.2004.02220.x [DOI] [PubMed] [Google Scholar]
- 87.Wang J-W, Wang L-J, Mao Y-B, Cai W-J, Xue H-W, Chen X-Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17: 2204–2216. doi: 10.1105/tpc.105.033076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32: 189–197. doi: 10.1016/j.tibs.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 89.Xia J, Zeng C, Chen Z, Zhang K, Chen X, Zhou Y, et al. Endogenous small-noncoding RNAs and their roles in chilling response and stress acclimation in Cassava. BMC Genomics. 2014;15: 634 doi: 10.1186/1471-2164-15-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
The data presented in this table were extracted from TOMEXPRESS database [44].
(PDF)
The 2 Kb of 5’ regulatory region were analyzed using the Place software.
(PDF)
The consensus sequences corresponding to the various putative cis-elements are described in S2 Table. Positions are with respect to the first base of translation start site.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.