With the development and refinement of mass spectrometry methods for identifying protein-protein interactions, and the ease with which they can now be applied, there is a growing need for applications that can simplify the challenges associated with managing, analyzing and visualizing the results obtained. Typically, interaction proteomics experiments will employ some kind of affinity purification for a protein of interest (bait) and generate a quantitative view of its recovered potential interaction partners (preys). While there are several established tools for scoring protein interaction datasets (e.g. 1, 2), the development of toolkits for easily and consistently visualizing scored data has received less attention. Generally scored datasets are loaded into network tools such as Cytoscape3, although these node-link visualizations do not easily convey changes in abundance or confidence scores associated with different bait purifications, or effectively communicate quantitative differences between baits or preys. We therefore sought to develop visualization tools to summarize and display scored quantitative protein interaction data in intuitive ways that provide seamless integration with existing analysis pipelines. Here we present a suite of visualization tools called ProHits-viz (prohits-viz.lunenfeld.ca) that run directly in the browser with no setup required. ProHits-viz explicitly supports data input from two widely used analysis pipelines: SAINT and the CRAPome1, 4, while also supporting a generic tabular input format for those using other tools. Supported input formats feature automated selection of bait, prey, prey abundance and score columns to use for analysis (Fig. 1a), along with suggested parameter defaults that can, however, be easily changed during input as desired by the user (Supplementary Fig. 1).
Figure 1.
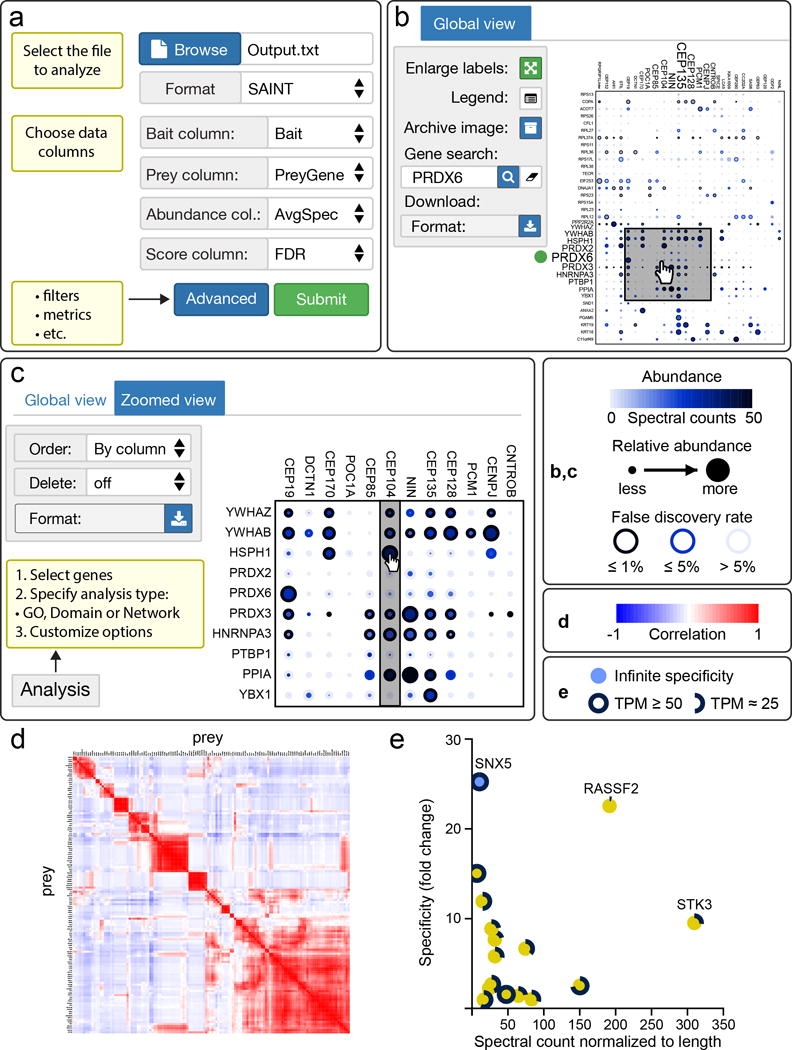
ProHits-viz at a glance. (a) Input and parameter selection overview for the analytical tools at ProHits-viz (also see Supplementary Fig. 1; a test data set is available as Supplementary Table 1). (b) Global dot plot view in the ProHitz-viz interactive viewer; a subset of the options available are highlighted. When necessary, axis labels can be enlarged relative to the cursor position (fish-eye zooming). A region of interest can be selected for zooming using the cursor. (c) Zoomed view of the selected dot plot region from b highlighting features that enable reorganization of the figure for publication or further analysis. After selecting the ordering/deletion group, the user can move/delete rows by clicking and dragging the desired row/column. (d) Global heat map resulting from prey-prey correlation analysis. (e) Example output from the specificity tool for a specific bait (STK4) across a dataset. The abundance of the corresponding prey mRNAs in the cell line used for affinity purification is displayed as the edge length (TPM: transcripts per million). Here, specificity scores from our fold change enrichment metric are shown, but CompPASS2 scoring is also available. Abundance values were normalized to protein length (x-axis). Specific legends for each of the panels are displayed at right.
ProHits-viz consists of four data analysis and image generation tools, complemented by interactive viewers. The analysis tools can: 1) cluster baits and preys for dot plot images, 2) perform correlation analysis, 3) calculate prey specificity scores and 4) generate detailed bait-bait comparisons. Dot plots are a simple way of summarizing protein interaction data5, displaying raw prey abundance, relative prey abundance across baits and prey confidence (Fig. 1b, c). The correlation analysis tool performs both bait and prey correlation and visualizes the results as clustered heat maps (Fig. 1d). Correlation of prey-prey profiles is particularly valuable for large datasets and offers an alternative prey-centric means of visualizing data that aids in identifying preys that may co-localize, belong to the same complex or even be direct interactors. The specificity tool is useful for medium to large datasets and calculates prey specificity scores for each prey-bait pair (Fig. 1e). The specificity scores are helpful in identifying preys preferentially associated with one or a small number of baits amongst a number of related baits. Available scores include a crude fold enrichment calculation as well as re-implementation of scores from CompPASS2. Finally, the bait-bait comparison tool takes information on two baits, as selected by the user, and plots their significant preys in a way that allows for comparison in a compact scatter plot (Supplementary Protocol, p. 19). Results from each analytical tool at ProHits-viz can be passed directly to interactive visualization interfaces or downloaded as a folder that contains images in PDF format. Details about each tool and usage options can be found in the Supplementary Protocol and Supplementary Methods.
An important feature at ProHits-viz is the ability to view images interactively in the browser. The interactive dot plot/heat map viewer enables visualization of the results from the dot plot and correlation tools. For large images, axes labels will follow the page as it scrolls, small labels can be selectively enlarged using fisheye zooming and regions of interest can be zoomed for closer inspection (Fig. 1b, c). Zooming enables deletion or reordering of particular rows and columns (Fig. 1c; Supplementary Protocol, p. 31), as well as GO, domain and network analysis of the selected plot region. The scatter plot viewer enables exploration of the images generated by the specificity and bait-bait comparison tools, supporting zooming and selective node labeling. All images can be exported directly from the interactive viewers in SVG format.
In summary, ProHits-viz provides an easy-to-use, interactive user interface for visualization, analysis and generation of publication quality figures for interaction data. This toolkit will continue to expand in the future thanks to user suggestions and the introduction of new technologies and experimental protocols.
Supplementary Material
Acknowledgments
We are grateful to all members of the Gingras lab for feedback on ProHits-viz, in particular Ji-Young Youn and Amber L. Couzens for comments during manuscript preparation. We would also like to thank Dattatreya Mellacheruvu for help supporting CRAPome output. We acknowledge funding from the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-088 and OGI-097 to A.-C.G. and L.P.), the Canadian Institutes of Health Research (Foundation grant FDN143301 to A.-C.G.); the US National Institutes of Health (to A.I.N. and A.-C.G.; 5R01GM94231); and the Singapore Ministry of Education (to H.C.; MOE T2-2-084). A.-C.G. is the Canada Research Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics.
Abbreviations
- CRAPome
Contaminant Repository for Affinity Purification
- SAINT
Significance Analysis of INTeractomes
- CompPASS
Comparative Proteomics Analysis Software Suite
- SVG
Scalable Vector Graphics
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
J.D.R.K. and A.-C.G. conceived the project. J.D.R.K. designed and implemented the software. H.C. contributed to tool development. G.D.G. provided input on the centrosome dataset (L.P. supervised G.D.G.). B.R., A.I.N. and H.C. contributed to feature design. A.-C.G. supervised the project. J.D.R.K. and A.-C.G. wrote the manuscript with input from all authors.
References
- 1.Choi H, et al. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shannon P, et al. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellacheruvu D, et al. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight JD, et al. Proteomics. 2015;15:1432–1436. doi: 10.1002/pmic.201400429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


