Abstract
Enveloped viruses enter cells by using their fusion proteins to merge the virus lipid envelope and the cell membrane. While crystal structures of the water-soluble ectodomains of many viral fusion proteins have been determined, the structure and assembly of the C-terminal transmembrane domain (TMD) remains poorly understood. Here we use solid-state NMR to determine the backbone conformation and oligomeric structure of the TMD of the parainfluenza virus 5 fusion protein. 13C chemical shifts indicate that the central leucine-rich segment of the TMD is α-helical in POPC/cholesterol membranes and POPE membranes, while the Ile- and Val-rich termini shift to the β-strand conformation in the POPE membrane. Importantly, lipid mixing assays indicate that the TMD is more fusogenic in the POPE membrane than in the POPC/cholesterol membrane, indicating that the β-strand conformation is important for fusion by inducing membrane curvature. Incorporation of para-fluorinated Phe at three positions of the α-helical core allowed us to measure interhelical distances using 19F spin diffusion NMR. The data indicate that, at peptide:lipid molar ratios of ~1:15, the TMD forms a trimeric helical bundle with inter-helical distances of 8.2–8.4 Å for L493F and L504F and 10.5 Å for L500F. These data provide high-resolution evidence of trimer formation of a viral fusion protein TMD in phospholipid bilayers, and indicate that the parainfluenza virus 5 fusion protein TMD harbors two functions: the central α-helical core is the trimerization unit of the protein, while the two termini are responsible for inducing membrane curvature by transitioning to a β-sheet conformation.
Keywords: magic-angle-spinning NMR, trimer formation, conformational plasticity, spin diffusion
Introduction
Enveloped viruses enter cells by fusing the virus lipid envelope with the cell membrane using trimeric membrane proteins of the virus. Class I viral fusion proteins such as hemagglutinin of the influenza virus and gp41 of the HIV virus contain two hydrophobic domains, an N-terminal fusion peptide (FP) domain and a C-terminal transmembrane domain (TMD). These are linked by a water-soluble ectodomain of variable length that contains two coiled-coil segments [1]. Receptor binding or low pH triggers the protein to adopt a fusogenic form [2], which unfolds to expose the hydrophobic FP and insert it into the cell membrane, while the TMD remains anchored in the virus envelope. This extended intermediate [3,4] continues to refold to bend into a helical hairpin, in doing so pulling the virus envelope and the cell membrane together. In the postfusion state, the protein is characterized by a trimeric helical hairpin between the two coiled coils, hence forming a six-helix bundle. Further membrane merger and fusion-pore formation require destabilization of each lipid bilayer, which is likely accomplished by the FP and the TMD, but the detailed mechanism of this membrane disruption remains largely opaque. The ectodomain of a number of viral fusion proteins has been captured in high-resolution crystal structures [5–9]; however, the structures of the hydrophobic FP and TMD are not yet well understood. The secondary structure and the membrane-bound topology of the HIV and influenza FPs have been extensively investigated using NMR [10–18], while the C-terminal TMD is much less studied. Given the profound conformational changes of the ectodomain along the fusion pathway, the membrane-bound FP and TMD are expected to undergo similar structural transformations [4,8,19,20]. Finally, the oligomeric structures of the TMD and the FP in the prefusion state, hemifusion intermediates, and the postfusion state have been rarely determined with high resolution.
The fusion protein F of the parainfluenza virus 5 (PIV5), which is responsible for infant respiratory diseases such as croup cough, is a class I fusion protein and is activated when hemagglutinin neuraminidase binds to cell surface receptors [1]. Crystal structures of the prefusion and postfusion states of the F ectodomain have been determined [4,19]. However, the TMD was replaced by GCNt in the prefusion crystal construct and was removed in the postfusion construct. Thus, the structural role of the TMD in PIV5 virus–cell fusion is not known. Biochemical and biophysical data have increasingly pointed to the importance of viral fusion protein TMDs for function and for protein stability. For example, mutations at L486A and I488A of the PIV5 TMD abolished both lipid mixing and content mixing [21]. Removal of the PIV5 TMD reduced the trimerization efficiency of the protein, necessitating the addition of the trimeric coiled coil, GCNt, to the C-terminus of the ectodomain to stabilize the trimer [4]. Sedimentation equilibrium data of detergent-bound TMDs of the fusion proteins of three paramyxoviruses, Hendra, PIV5, and human metapneumovirus, indicate that these TMDs exist in a monomer–trimer equilibrium [22,23]. Lipid mixing assays showed that the influenza hemagglutinin TMD is essential for the transition from the hemifusion state to the postfusion state [24,25]. Mutations of the HIV gp41 TMD destabilized the conformation and reduced the fusion activity of the protein [26–28]. SDS-PAGE analysis of a gp41 TMD construct showed that mutation of a GxxxG motif together with truncation of the cytoplasmic domain disrupted trimer formation [29]. Consistently, ectodomain constructs of gp41 without the TMD were found to dissociate into monomers in the presence of detergents [30,31].
Recently, we investigated the backbone conformation and lipid interactions of the PIV5 TMD in phospholipid bilayers. Using a peptide containing six 13C, 15N-labeled residues, we found that the secondary structure and curvature-inducing ability of the TMD depend sensitively on the membrane composition [18,32]. The peptide is predominantly α-helical in phosphatidylcholine-rich lamellar bilayers, but adopts increasing β-strand conformation at the two termini in negative-curvature phosphatidylethanolamine (PE) membranes. Furthermore, the β-strand-rich conformation dehydrates the PE membrane and induces a bicontinuous cubic phase with negative Gaussian curvature [33], which are essential for hemifusion intermediates and postfusion pores [34,35]. These results suggest that the β-strand conformation is the fusion-active structure. However, it is not known whether the strand–helix–strand conformation is oligomerized to affect membrane fusion. In the current study, we employ 19F spin diffusion NMR to determine the oligomeric structure of the TMD in two lipid membranes, POPC/cholesterol and POPE, which promote the α-helical and β-sheet conformation, respectively. 19F–19F distance measurements show that the central α-helical segment of the peptide is trimeric in both lipid membranes, suggesting that this domain may act as the trimerization core of the protein. We also incorporated additional 13C, 15N-labeled residues into the TMD and measured their chemical shifts, thus determining the backbone (φ, ψ) torsion angles of the majority of this domain. These data further define the membrane-dependent secondary structure of the TMD, indicating that the fusion function is located at the β-sheet-rich N- and C-termini, while the trimerization function is sequestered in the central α-helical core of the TMD.
Results
The backbone conformation of the TMD is membrane dependent
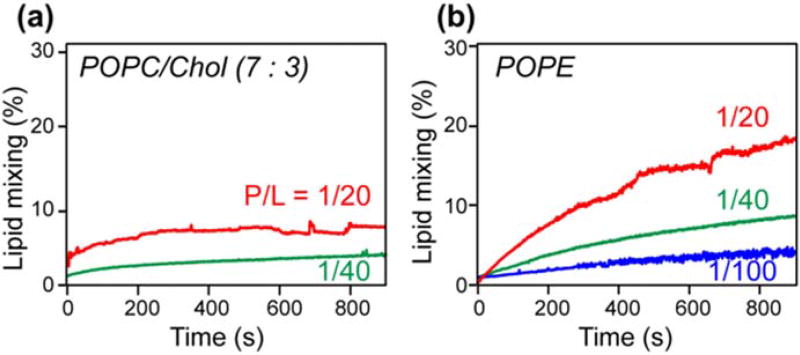
To investigate whether the PIV5 TMD has fusion activity in the absence of the rest of the protein, we carried out a lipid mixing assay that measures fluorescence dequenching induced by the peptide. POPC/cholesterol of 150 µM (7:3 mol ratio) and POPE large unilamellar vesicles were prepared. Ten percent of the vesicles contain 2 mol% of the fluorescent lipid NBD-PE and 2 mol% of the quenching lipid Rh-PE. These were diluted with 90% unlabeled vesicles. If the addition of the TMD causes lipid mixing, then the fluorescent NBD-PE and Rh-PE lipids will be separated into different vesicles, thus causing an increase of the fluorescence intensity. The degree of lipid mixing is reported as the ratio of the fluorescence intensity relative to the maximum intensity obtained when Triton X-100 was added to disrupt the vesicles and cause complete lipid mixing. Figure 1 shows that the PIV5 TMD caused 10% mixing of the POPC/cholesterol vesicles in 15 min when the peptide/lipid molar ratio was 1:20, while lower peptide concentrations reduced the extent of mixing. In comparison, the TMD caused ~20% mixing of the POPE vesicles in the same time period, indicating that the peptide has higher fusion activity in a membrane in which it has larger β-sheet content [18,32]. The increasing fusion activity with increasing peptide/lipid molar ratios suggests that TMD oligomerization promotes membrane fusion.
Fig. 1.
PIV5 TMD induces mixing of POPC/cholesterol vesicles (a) and POPE vesicles (b). The peptide causes stronger lipid mixing in the POPE membrane than in the POPC/cholesterol membrane. Moreover, the extent of lipid mixing increases with the peptide/lipid molar ratio.
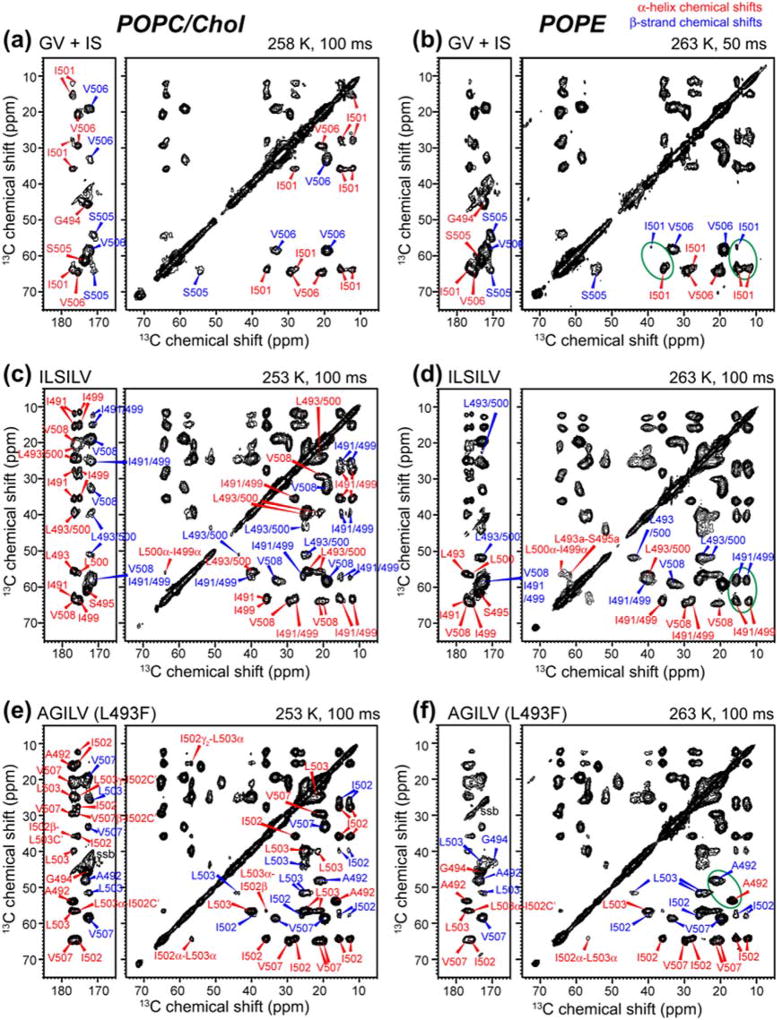
The previous result of membrane-dependent conformation of the TMD was based on the measurement of the chemical shifts of six labeled residues distributed throughout the peptide [32]. To obtain more complete information about the back-bone conformation, we have now labeled a total of 18 residues (Table 1) and measured their chemical shifts using 2D 13C–13C (Fig. 2) and 15N–13C correlation experiments. At moderately low temperature (253–263 K) that suppresses peptide motion, both α-helical and β-sheet signals were observed. In the POPC/cholesterol (7:3) membrane, the TMD exhibits stronger α-helix signals than β-strand signals (Fig. 2a, c, e). Residues in the center of the sequence such as S495 and I501 show exclusively α-helical intensities, while N-terminal residues such as A492 show dominant (~90%) α-helical signals. In comparison, C-terminal residues such as S505 and V506 have significant β-strand intensities that coexist with the α-helical signals. Based on the relative intensities of the cross peaks at α-helical and β-sheet chemical shifts, the helical content of residues from S505 to V508 is 35–79% in the POPC/cholesterol membrane (Table S1). Averaged overall residues, the α-helicity of the TMD in the POPC/cholesterol membrane is 79%. In the POPE membrane, the TMD conformational equilibrium shifts significantly toward the β-strand structure (Fig. 2b, d, f). For example, residues N-terminal to L493 show much higher β-sheet signals than α-helical peaks; similarly, residues V506 to V508 show stronger β-sheet intensities than α-helical intensities.
Table 1.
13C, 15N-labeled (red underlined) and fluorinated (blue bolded) residues in the PIV5F TMD (485–510) peptides used in this study
| Samples | Labeled residues | POPC/ Chol |
POPE | |
|---|---|---|---|---|
| 1 | IAGLSV: I488, A490, G497, L498, S505, V506 |
|
x | x |
| 2 | 1:1 mixed GV and IS G494, V506 and I501, S505 |
|
x | x |
| 3 | ILSILV: I491, L493, S495, I499, L500, V508 |
|
x | x |
| 4 | AGILV (L493F): A492, 4- 19F-L493F, G494, I502, L503, V507 |
|
x | x |
| 5 | 1:1 mixed ILSILV and AGILV (L493F) |
|
- | x |
| 6 | Diluted ILSILV | ILSILV : Unlabeled peptide = 1: 2 | x | - |
| 7 | L504F |
|
x | - |
| 8 | L500F |
|
x | - |
Sample 1 (IAGLSV) was used in a previously reported study [32], while samples 2–8 are produced specifically for this study.
Fig. 2.
2D 13C–13C DARR correlation spectra of the TMD in the POPC/cholesterol (left) and POPE (right) membranes. (a, b) Mixed GV and IS-labeled TMD. (c, d) ILSILV-labeled TMD. (e, f) AGILV-labeled TMD. Red and blue assignments denote α-helical and β-strand chemical shifts. The peptide shows a mixture of α-helix and β-strand chemical shifts in the membrane.
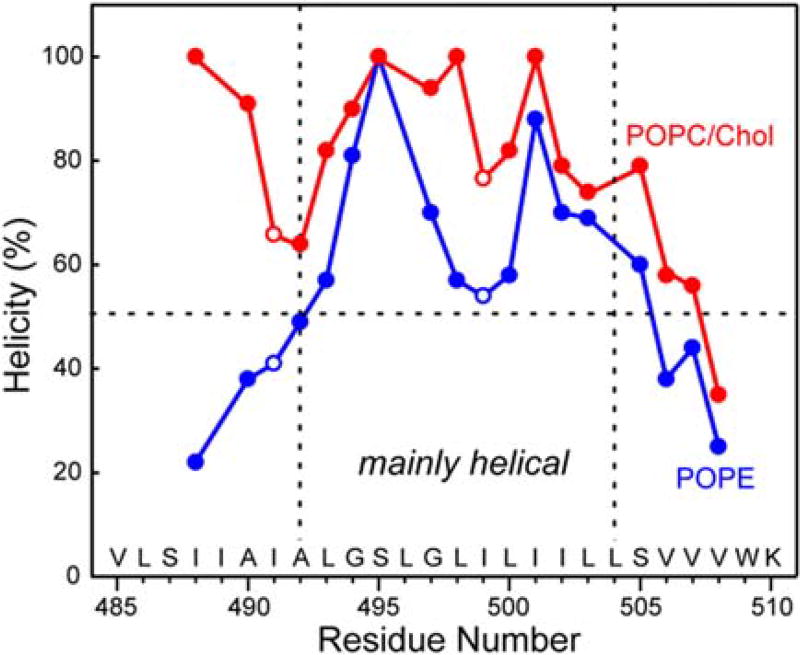
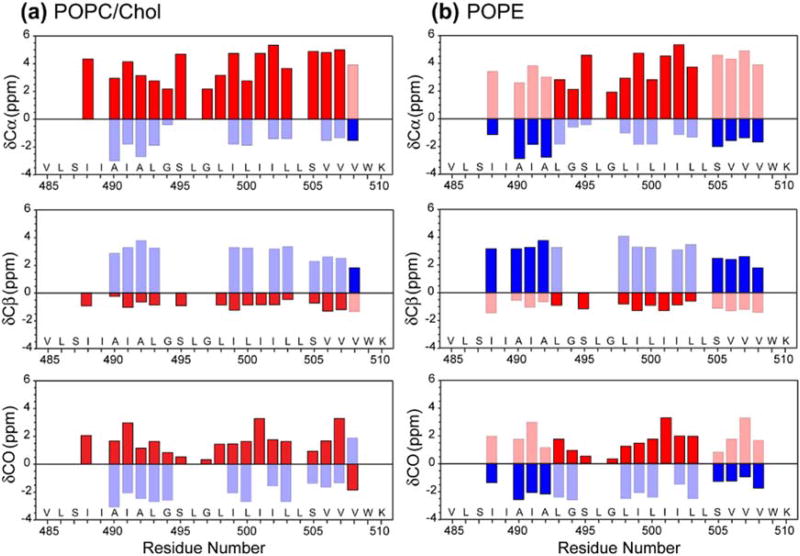
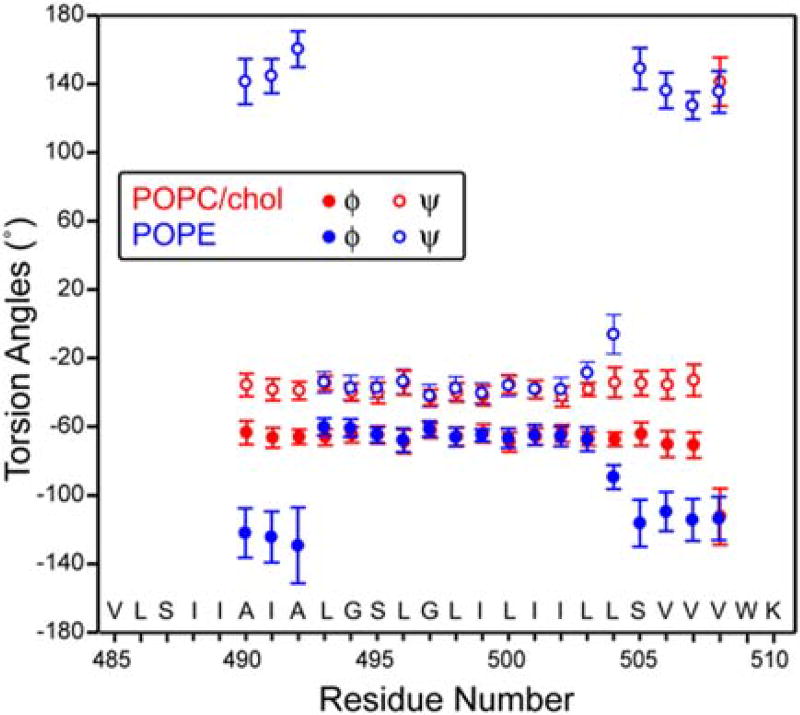
Figure 3 plots the residue-specific α-helicity obtained from the cross peak intensities. As can be seen, the TMD is predominantly helical in the POPC/cholesterol membrane except for the C-terminal end of the peptide (V508), while in the POPE membrane, residues N-terminal to L493 and C-terminal to S505 show less than 50% helicity (Table S1). Thus, the central 12-residue segment of the peptide from L493 to L504 is the α-helical core, while the two termini are sensitive to the membrane spontaneous curvature and convert to a predominantly β-strand conformation in negative-curvature PE membranes. Based on the measured Cα, Cβ and CO chemical shifts (Table S2), we calculated the secondary structure-dependent chemical shifts for the major and minor conformers (Fig. 4). In the POPC/cholesterol membrane, the major conformer is a continuous α-helix, with positive Cα and CO secondary shifts and negative Cβ secondary shifts, while in the POPE membrane, the major conformer is a central helix sandwiched by two β-strands at the termini [32]. The (φ, ψ) torsion angles predicted from these chemical shifts using TALOS-N [36] (Fig. 5) show the same trend for the major conformer: in the POPC/cholesterol membrane, the TMD is predominantly α-helical, while in the POPE membrane, the major conformer has a strand–helix–strand motif.
Fig. 3.
Residue-specific α-helicity of the TMD in POPC/cholesterol and POPE membranes, obtained from the relative intensities of the cross peaks in 2D 13C–13C correlation spectra. The peptide has much lower helicity at the N- and C-termini than at the central segment in the POPE membrane. In the POPC/cholesterol membrane, the peptide is mostly helical except for the C-terminal end.
Fig. 4.
13C secondary chemical shifts of the TMD in the POPC/cholesterol (a) and POPE membrane (b). α-Helical secondary shifts are shown in red, while β-strand secondary shifts are shown in blue. Shaded bars denote the minor conformation. The TMD exhibits mainly α-helical structure in the POPC/cholesterol membrane but predominant β-strand conformation at the N- and C-terminal regions in the POPE membrane.
Fig. 5.
Backbone (φ, ψ) torsion angles of the TMD in the POPC/cholesterol (red) and POPE (blue) membrane, predicted by TALOS-N from the measured 13C and 15N chemical shifts. Only the torsion angles of the major conformer are shown.
Mobility and depth of insertion of the TMD in the membrane
To probe whether the TMD is mobile in the membrane, we measured 13C–1H dipolar couplings using the 2D DIPSHIFT experiment [37]. At 303 K where the POPE membrane is in the liquid-crystalline phase, most residues show significant dipolar dephasing (Fig. S1), indicating large 13C–1H couplings. Best-fit simulations of the Cα–Hα dephasing curves indicate order parameters of 0.82–0.95 for the α-helical conformation and 0.86–0.92 for the β-sheet conformation. Thus, both conformations are immobilized in the liquid-crystalline membrane, with only small-amplitude local motions, suggesting that the peptide is oligomerized in the membrane. 2D lipid–protein 1H–13C correlation spectra of POPE-bound TMD show clear cross peaks between the lipid chain CH2 and the peptide 13C signals for both the helical and sheet conformations after 100-ms 1H spin diffusion (Fig. S2), indicating that the TMD is well inserted into the membrane. Taken together, these data indicate that the TMD spans the lipid bilayer as an immobilized oligomer in both POPC/cholesterol and POPE membranes.
Oligomeric structure of the TMD in lipid membranes
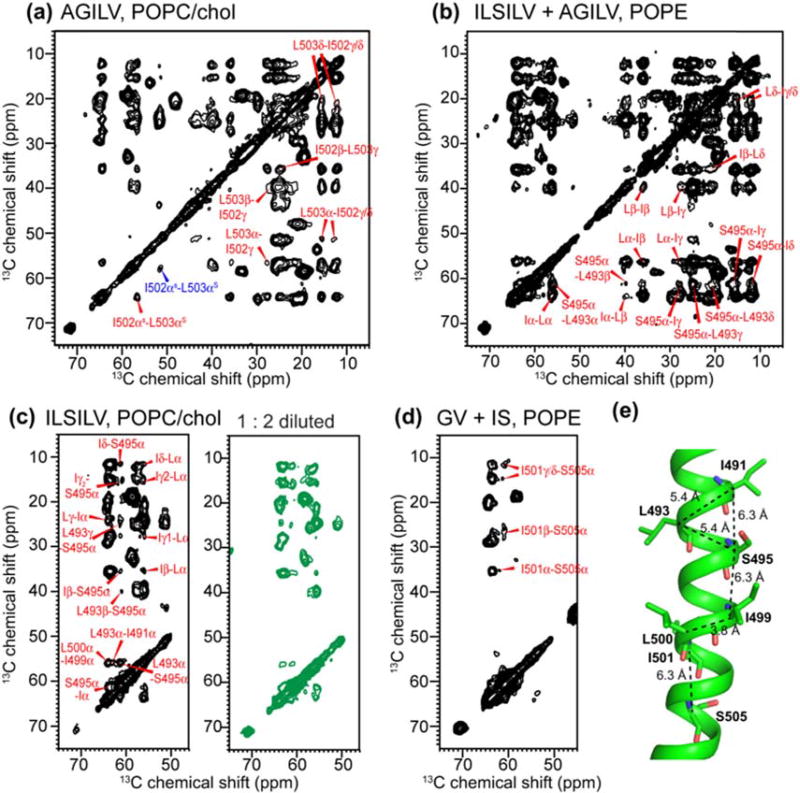
To determine the intermolecular assembly of the TMD, we first conducted 1H-driven 13C spin diffusion (PDSD) 2D 13C–13C correlation experiments using long mixing times of 0.5 and 1.0 s, which detect internuclear distances up to ~8 Å [38,39]. For AGILV and ILSILV-labeled peptides, multiple inter-residue cross peaks such as sequential I50–-L503 and I499– L500 cross peaks and non-sequential I491–L493, L493–S495, and S495–I491 or S495–I499 cross peaks are observed (Fig. 6). To investigate if any of the cross peaks are intermolecular in origin, we measured a 1:1 mixture of the ILSILV and AGILV-labeled peptide. No A492 cross peaks with either Ile or Leu were observed, which would provide unambiguous evidence of intermolecular contacts since A492 is too far from I502 and L503 in the same AGILV-labeled peptide but can be potentially close to I491 and L493 in the ILSILV-labeled peptide. S495 cross peaks with Ile chemical shifts are detected in this mixture. These could result from intramolecular contacts of S495 with I491 and/or I499, or from intermolecular contacts with I502 in AGILV. To distinguish these two possibilities, we mixed the ILSILV-labeled peptide at a 1:2 M ratio with unlabeled peptide. In this 33% diluted sample, all inter-residue cross peaks seen in the undiluted sample remained with comparable intensities (Fig. 6c), indicating that all inter-residue cross peaks are intramolecular in origin. Finally, 1:1 mixing of a G494, V506-labeled peptide with an I501, S505-labeled peptide showed only intramolecular I501–S505 cross peaks (Fig. 6d) and no intermolecular S505-V506 cross peaks. Thus, 13C spin diffusion did not detect any intermolecular cross peaks. To estimate the internuclear distances for the observed intramolecular cross peaks, we generated an α-helical monomer using the chemical-shift constrained (φ, ψ) torsion angles for residues A490 to V508 (Fig. 6e). The Cα–Cα cross peaks seen in the 2D PDSD spectra correspond to intra-helical distances of 5.0–6.3 Å, which are within 13C spin diffusion reach [39], further supporting the assignment of the observed cross peaks to the same helix.
Fig. 6.
2D 13C–13C PDSD spectra of the TMD with long mixing times of 0.5 and 1.0 s, measured at 253–263 K. Only inter-residue cross peaks are assigned. (a) AGILV (L493F)-labeled TMD in the POPC/cholesterol membrane. (b) 1:1 mixture of AGILV (L493F) and ILSILV-labeled TMD in the POPE membrane. All inter-residue cross peaks are intramolecular ones. (c) Undiluted and 1:2 diluted (green) ILSILV-labeled TMD in the POPC/cholesterol membrane. The same inter-residue cross peaks were observed, indicating that all cross peaks are intramolecular. (d) Mixed GV and IS-labeled TMD in the POPE membrane, showing I501–S505 cross peaks. (e) Intramolecular Cα–Cα distances of the TMD generated using the measured (φ, ψ) torsion angles.
Since the absence of intermolecular 13C–13C cross peaks may be due to the inability of 13C spin diffusion to measure distances longer than ~8 Å rather than the absence of TMD oligomerization, we next turned to 19F spin diffusion to investigate intermolecular assembly. The larger 19F gyromagnetic ratio compared to 13C allows 19F spin diffusion to measure distances to the 10- to 15-Å range, using the Centerband-Only Detection of Exchange (CODEX) technique [40–44]. The CODEX technique detects the oligomeric number of molecular assemblies through the equilibrium intensity of a stimulated echo: an oligomeric number of n gives an equilibrium echo intensity of 1/n [45]. Inter-fluorine distances can be quantified based on the decay rate to equilibrium using an exchange matrix formalism. We mutated the TMD at three positions: L493F, L500F, and L504F, where the Phe residues are para-fluorinated. These three residues were chosen based on cysteine disulfide crosslinking data [21], which showed that L493C and L504C have high disulfide crosslinking fractions, while L500C has vanishing crosslinking. 2D 13C–13C correlation spectra of AGILV (L493F)-labeled TMD give the same chemical shifts and intensity distributions as the non-fluorinated peptide, indicating that the Leu-to-Phe mutation and Phe fluorination do not perturb the TMD structure.
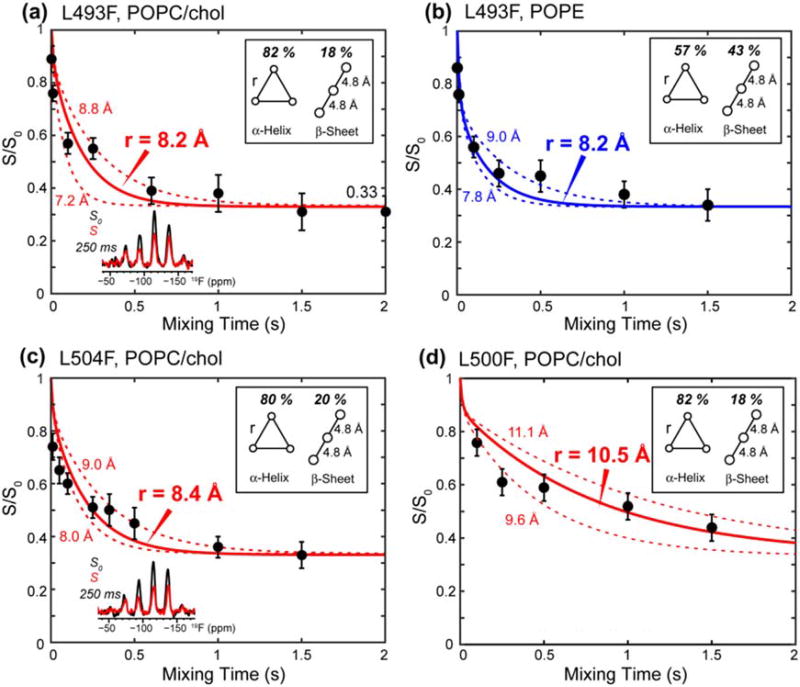
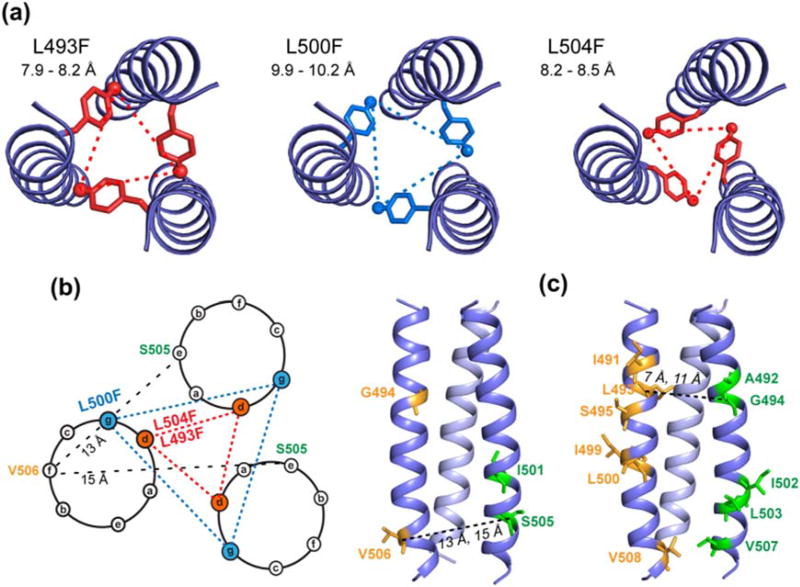
The 19F CODEX experiment was carried out on both POPC/cholesterol-bound TMD and POPE-bound TMD. Figure 7a, b shows that the CODEX intensities, S/S0, of L493F decayed to 0.33 by 1.5 s in both membranes, thus directly proving that the TMD is trimerized. The CODEX data exhibit a small fraction of an initial fast decay together with a major fraction of a slower decay. Since L493 is 82% α-helical in the POPC/cholesterol membrane and 57% helical in the POPE membrane (Table S1), we attribute this distance distribution to the coexistence of the helix and sheet conformations. Thus, we fit the measured CODEX intensities using a weighted average of two decay curves: the minor β-strand conformer is assumed to have a 19F–19F distance of 4.8 Å, which is the backbone separation of a parallel in-register β-sheet [46], while the distance for the major α-helical conformer is varied to fit the slow-decaying component. The resulting best-fit simulations yielded a 19F–19F distance of 8.2 Å at L493F in both POPC/cholesterol and POPE membranes, indicating that the α-helical core of the TMD has similar trimer structures in the two membranes. The L504F-labeled TMD shows a similar CODEX decay rate as L493F and is best fit to an interhelical 19F–19F distance of 8.4 Å in the POPC/cholesterol membrane (Fig. 7c). The similar interhelical distances for these two well-separated residues in the peptide suggest a uniform trimer structure for the α-helical domain. In comparison, L500F shows much slower CODEX decay, with an S/S0 value of ~0.50 at 1.5-s mixing (Fig. 7d), indicating that the interhelical distance is much longer at this residue, in good qualitative agreement with the vanishing crosslinking at this residue. Best-fit simulations gave an interhelical distance of 10.5 Å for the slow component at L500F.
Fig. 7.
19F CODEX data of membrane-bound TMD. (a) L493F-TMD in the POPC/cholesterol membrane. Representative S0 and S spectra are shown. (b) L493F-TMD in the POPE membrane. (c) L504F-TMD in the POPC/cholesterol membrane. (d) L500F-TMD in the POPC/cholesterol membrane. The data were acquired at 230 K under 8 kHz MAS. The percentages of α-helix and β-strand are taken from Table S1, and intermolecular 19F–19F distances in the β-sheet are fixed to 4.8 Å in the simulation. The CODEX intensities of L493F and L504F equilibrate to 0.33, indicating that the TMD is trimerized. The L500F CODEX decay is slower, indicating longer interhelical distances.
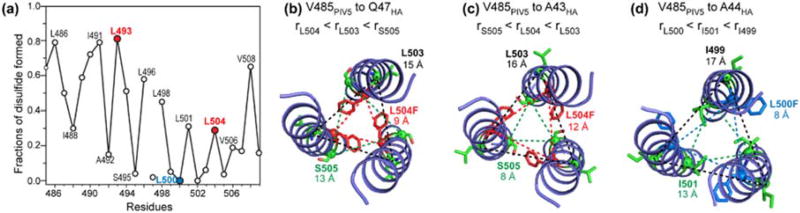
These sparse but long-range interhelical distances strongly constrain the trimer structure. We built a trimer model using the crystal structure of low-pH hemagglutinin (residues 47–71) (PDB code: 1HTM) [47] as the starting template. We replaced the hemagglutinin backbone by the PIV5 TMD residues, and modified the (φ, ψ) torsion angles to conform with the chemical-shift constrained torsion angles (Fig. 5). To determine the alignment between the PIV5 TMD sequence and the hemagglutinin sequence, we satisfied both the measured interhelical 19F–19F distances and the cysteine crosslinking data. The heptad positions of the residues directly dictate the residues' distances from the helix–helix interface and hence the cross-linking efficiencies. The crosslinking data, reproduced in Fig. 8a, show local maxima at L493 and L504 but local minimum at L500, in good agreement with the 19F CODEX data. Three sequence alignments, with V485 placed at hemagglutinin residues A43, A44, and Q47, resulted in possible agreement with the measured 19F–19F distances at residues 493, 500, and 504. However, matching V485 with A43 and A44 contradicted the cysteine crosslinking data (Fig. 8). For example, alignment of V485 with A43 (Fig. 8c) produced shorter interhelical Cα–Cα distances at S505 than at L504, which contradict the local maximum cross-linking for L504. Similarly, alignment of V485 with A44 (Fig. 8d) of hemagglutinin dictates longer distances for I499 and I501 than for L500, which contradict the fact that L500 has vanishing crosslinking, while L501 has significant crosslinking. Thus, the relative crosslinking fractions between sequential residues provide strong constraints to the rotation angle of the helix relative to the trimer interface. In addition, aligning V485 with A43 and A44 could agree with the 19F–19F distances only by using sterically unfavorable Phe χ1 torsion angles of 120° and 30°, which contradict the dominant trans χ1 rotamer of Phe in α-helices [48]. The only alignment that agrees with both the crosslinking data and the 19F–19F distances while using the dominant trans rotamer is when V485 matches residue Q47, which puts L493F and L504F at the “d” position of the heptad repeat and L500F at the “g” position. The interhelical distances in this proposed trimer model were 7.9–8.2 Å for L493F, 8.2–8.5 Å for L504F, and 9.9–10.2 Å for L500F, in good agreement with the measured interhelical 19F–19F distances (Fig. 9a). The Phe χ1 torsion angles in this trimer structure are −149° for L493F, −158° for L500F, and 142° for L504F, which are close to the trans conformation. The helical wheel diagram of the trimer illustrates that L493F and L504F sidechains point to the helix–helix interface, consistent with the disulfide crosslinking data (Fig. 9b) [21]. Residues L486, A490, L493, I501, L504, and V508, which have local maximum in the disulfide crosslinking [21], lie at the helix–helix interface. The side views of the trimer model (Fig. 9c) also illustrate the reason for the lack of 13C–13C cross peaks for the two mixed labeled samples (Fig. 6): the labeled residues are located either away from the helix–helix interface or at significantly different heights along the helix axis.
Fig. 8.
Interhelical 19F–19F distances and disulfide crosslinking data rule out several trimer structural models. (a) Fractions of disulfide bond formation reproduced from Ref. [21]. The three Leu residues that were replaced by 4-19F-Phe in this study are colored. (b) Proposed trimer structure model, obtained by aligning PIV5 V485 with hemagglutinin Q47. The interhelical Cα–Cα distances for L504 are shorter than the distances for the neighboring L503 and S505, consistent with the crosslinking data. (c) Alternative trimer model obtained by aligning PIV5 V485 with hemagglutinin A43. The interhelical Cα–Cα distance of S505 is shorter than that of L504, which is inconsistent with the disulfide crosslinking data. (d) Alternative trimer model obtained by aligning V485 of PIV5 with A44 of hemagglutinin. L500 gives shorter interhelical Cα–Cα distances than the neighboring I499 and I501, which is inconsistent with the crosslinking data.
Fig. 9.
Proposed trimer structure model of the PIV5 TMD. (a) Top view of the trimer structure, showing L493F, L500F, and L504F interhelical 19F–19F distances. These agree with the experimentally measured 19F–19F distances. (b) Helical wheel diagram for the trimeric TMD, with L493F and L504F at the d position of the heptad repeat, forming close interhelical contacts, while L500F lies at the g position, giving longer interhelical distances. (c) Side views of the trimer structure, showing the locations of 13C, 15N-labeled residues in the two mixed labeled samples (Table 1). Representative interhelical distances between S505 and V506, and between G494 and L495, are much longer than can be measured using 13C spin diffusion NMR. This is further illustrated in the helical wheel diagram for the S505–V506 pair.
Discussion
The present study provides more complete information about the membrane-dependent backbone conformation of the TMD of the PIV5 fusion protein and moreover determines the oligomeric structure of this domain. To our knowledge, this is the first time that the trimerization and interhelical packing of a viral fusion protein TMD has been determined in lipid bilayers. The 13C chemical shifts for POPC/cholesterol and POPE membrane-bound TMD (Fig. 2) confirm and extend the previous finding that the TMD has significant conformational plasticity, induced by the lipid membrane. The two termini adopt significant β-strand conformations in the negative-curvature POPE membrane, while the central segment of the peptide, spanning residues 493–504, is largely helical in both membranes [32]. Importantly, the peptide has higher lipid mixing activities in POPE membranes, indicating that the β-sheet-rich conformation is correlated with membrane fusion. As proposed before, the terminal β-sheet conformation may facilitate membrane fusion by differential perturbation of the lipid headgroup packing of the two leaflets and by dehydrating the membrane surface [49,50].
The higher β-strand content in the two termini of the TMD (Table S1) most likely results from the β-branched Val and Ile in these regions. The amino acid sequence of the PIV5 TMD shows an interesting separation of an Ile/Val-rich domain and a Leu-rich domain. For the N-terminal eight residues and C-terminal six residues, which show less than 50% helicity in the POPE membrane (Fig. 3), seven residues are β-branched Val and Ile, representing 50% of these regions. In contrast, in the central 12 residues of the peptide that exhibits high α-helical content, six residues are Leu while only three are Ile. β-Branched Ile and Val residues are known to promote β-sheet conformations [51,52] and have been implicated in the fusogenicity of other fusion proteins. For example, SNARE proteins have a large number of Ile and Val residues in their TMDs and their fusion activities decrease with increasing stability of the α-helical structure [53]. The vesicular stomatitis virus G protein shows lower fusion activity upon mutation of Ile to Leu or Ala [54]. De novo designed LV peptides with varying ratios of Leu and Val residues exhibit higher fusion activity as more Val residues are used [55–57]. Moreover, fusogenicity increased further when the Val residues were concentrated to the termini of the peptide sequence [58], in excellent agreement with our conclusion that β-strand conformation at the termini of the TMD promotes membrane fusion.
While viral fusion proteins have trimeric ectodomains in both prefusion and postfusion states, the oligomeric structure of the membrane-bound TMD, as well as the oligomeric structure of the N-terminal FP after its insertion into the target membrane, has been rarely studied. Solution NMR data of a gp41 construct spanning residues 677–716, which include part of the MPER, the TMD, and part of the cytoplasmic domain, showed a three-chain coiled coil in DMPC/DHPC bicelles [29]. This trimer is stabilized by a GxxxG motif at residues G690 to G694, as shown by SDS-PAGE analysis of a G690 mutant [29]. The trimer may be further stabilized by inter-chain hydrogen bonds between the three transmembrane arginine residues, as shown by molecular dynamics simulations [59] and mutagenesis data [27]. These data suggest that the gp41 TMD may be an autonomous trimerization domain for the full-length protein. Indeed, NMR relaxation data of gp41 ectodomain constructs without the membrane-interacting FP, TMD, and MPER showed a lack of interhelical interactions in the presence of DPC micelles, indicating that the protein dissociates into monomers that interact instead with detergents [30]. For the TMD of the PIV5 fusion protein, functional evidence for oligomerization was obtained from cysteine scanning mutagenesis, which found approximately helical periodicity in the fractions of disulfide bond formation [21]. Sedimentation equilibrium data for three paramyxovirus fusion proteins' TMDs in isolation as well as when appended to staphylococcal nuclease showed that a monomer–trimer equilibrium gives the best fit to the data, indicating these TMDs self-associate into trimers in detergents [23]. Despite these biochemical and biophysical measurements, atomic-resolution information about the oligomeric structure of the TMD had not been available.
The current 19F CODEX data for residues 493, 500, and 504 provide definitive proof that the TMD alone, without the rest of the PIV5 fusion protein, is trimeric in lipid bilayers. This is true for both the POPC/cholesterol membrane and the POPE membrane, where the TMD has different helical contents. The interhelical 19F–19F distances are shorter for L493F and L504F (8.2 and 8.4 Å) than for L500F (10.5 Å) (Fig. 7), in excellent agreement with cysteine crosslinking data [21]. The similar distance between L493F and L504F is also consistent with the heptad repeat motif that puts residues 493 and 504 on the same face of the helix. The fact that similar intermolecular distances were found for L493F in the POPC/cholesterol and POPE membranes suggests that the interhelical packing is largely independent of the membrane composition. Since all three fluorinated residues lie in the central helical core of the peptide, the similar distances in the two membranes suggest that this central 12-residue segment is the trimerization core of the TMD and by inference may also be the trimerization core of the full-length protein. The separation of functionality is further supported by the fact that among the six Leu residues (L493, L496, L498, L500, L503, and L504) in the central 12-residue segment, two Leus (L493 and L504) lie at the d position of the heptad repeat (Fig. 9b), reminiscent of the leucine zipper motif [60]. In comparison, the Val- and Ile-rich N- and C-termini of the TMD, with its strong β-sheet propensity in negative-curvature PE membranes, may be chiefly responsible for inducing membrane curvature.
The 19F CODEX spin diffusion technique [40–42] is ideal for determining the intermolecular interface in homo-oligomeric helical bundles [61–65] as well as β-sheet assemblies [66]. Not only is 19F–19F dipolar coupling much stronger than 13C–13C or 13C–15N dipolar coupling and thus sensitive to longer distances [45], but also 19F CODEX probes the same residue in different protomers of the oligomeric complex. At the peptide/lipid mass ratio of 1:4 used in our samples, average inter-trimer distances are expected to be much longer (~20 Å) than intra-trimer distances, thus inter-trimer contacts should have little impact on the CODEX decays, as confirmed by dephasing to ~0.33 and not lower. Compared to 19F spin diffusion, the mixed 13C-labeling approach is much less effective for measuring intermolecular distances in α-helical bundles because sequentially labeled residues are located at positions shifted by ~100° in the helical wheel and hence have long intermolecular distances. If different residues facing the helix–helix interface are chosen, for example, at positions d and a of the heptad repeat, then these residues are shifted along the helix axis, and thus may lie outside the distance reach of 13C spin diffusion. This non-ideal placement of differentially labeled residues explains why the two mixed 13C-labeled peptides (Table 1) did not show detectable intermolecular 13C–13C cross peaks, although the peptide is trimerized in the lipid membrane.
In conclusion, the data presented here show that the central Leu-rich α-helical segment of the PIV5 fusion protein TMD is the trimerization core of the peptide, while the Ile- and Val-rich termini of the peptide adopt membrane-dependent secondary structure, with the β-strand conformation in negative-curvature PE membranes responsible for inducing membrane curvature and promoting membrane fusion. The α-helical trimer structure is the same in POPC/cholesterol membrane and in POPE membranes, suggesting that the central segment of the TMD may act as a stable α-helical trimer core during the fusion process.
Materials and Methods
Peptide synthesis and membrane sample preparation
The TMDs of the PIV5 F protein spanning residues 485–510 (VLSIIAIALGSLGLILIILL-SVVVWK) were synthesized using Fmoc chemistry. Six 13C, 15N-labeled and 19F-incorporated peptides were synthe-sized for this study: GV, IS, ILSILV, AGILV (L493F), L504F, and L500F-TMD (Table 1). Together with a previously synthesized IAGLSV-labeled peptide [32], we labeled 18 out of 26 residues of this domain. AGILV (L493F), L500F, and L504F samples were synthesized for the 19F CODEX experiment. Most peptides were synthesized on a NovaPEG Rink Amide resin (Novabiochem) using a Symphony Multiple Peptide synthesizer (Protein Technologies) or a CEM Liberty microwave synthesizer using Fmoc chemistry. All labeled residues were double coupled, while the unlabeled residues were triple coupled (4 eq. excess each time). The L500F peptide was synthesized on an H-Rink amide ChemMatrix resin using a custom-designed flow-based synthesizer [67]. All residues were single coupled (10 eq. excess), while six Leu, six Ile residues, and the L500F mutation site were double coupled (4 × excess each time). The crude peptide was purified to N95% by preparative HPLC using a Vydac diphenyl column (1″ × 12″). The peptide was eluted from the column with a linear gradient starting with water:acetonitrile (8:2, v/v) and mixing with acetonitrile: isopropanol (2:1, v/v) to a final concentration of 100% with 0.1 mM HCl as an ion-pairing agent. The peptide mass was confirmed by MALDI-TOF or electrospray mass spectrometry.
The peptide was reconstituted into POPC/cholesterol (7:3) and POPE membranes at a peptide:lipid mole ratio of 1:15 or 1:13.5, which correspond to a mass ratio of 1:4.2 or 1:3.8 (Table 1). The peptides and lipids were codissolved in TFE/chloroform solution and the solvents were removed under nitrogen gas, then the samples were completely dried under vacuum overnight. The dried samples were resuspended in pH 7.5 Hepes buffer (10 mM Hepes–NaOH, 1 mM EDTA, 0.1 mM NaN3), freeze-thawed seven times between liquid nitrogen temperature and 30 °C to make homogeneous vesicles, then dialyzed for a day. The proteoliposomes were spun down at 40,000 rpm using a Beckman SW60Ti rotor at 4 °C for 4 h to obtain membrane pellets. The hydration of membrane pellets was adjusted ~40 wt% in desiccator, then transferred to magic-angle-spinning (MAS) NMR rotors.
Lipid mixing assays
POPC/cholesterol (7:3) and POPE membranes were used to measure peptide-induced lipid mixing. Lipids were dissolved in chloroform and dried under nitrogen gas. The dried lipids were resuspended in 10 mM Hepes buffer (pH 7.5), freeze-thawed 10–12 times between liquid nitrogen temperature and ~35 °C, then extruded 15–20 times through 100-nm membranes to produce homogeneous large unilamellar vesicles. For fluorescently labeled vesicles, 2 mol% of the fluorescent lipid NBD-PE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl)) and 2 mol% of the quenching lipid Rh-PE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(Lissamine rhodamine B sulfonyl)) were added to the lipid mixture. Unlabeled and labeled vesicles were mixed at a 9:1 molar ratio, and the total lipid concentration was 150 µM. A 3-mM TMD stock solution in TFE was prepared and 1–5 µl of this stock solution was added to 2 ml of lipid vesicle solutions to the desired peptide:lipid mole ratio, which ranges from 1:100 to 1:20. The final TFE concentrations in the lipid vesicle solutions were 0.05–0.25%. Peptide-induced lipid fusion dilutes the fluorescently labeled lipids and increases the distances between NBD-PE and Rh-PE, thus increasing the fluorescence intensity. A HORIBA Fluoromax-P fluorimeter was used to measure fluorescence at an excitation wavelength of 465 nm and an emission wavelength of 530 nm. Each measurement was carried out at 21 °C under continuous stirring in 2 ml of large unilamellar vesicle with a time incrementof1 s. Twenty microliters of 10% Triton X-100 was added to the 2-ml solution to measure the maximum fluorescence, Fmax. We measured the initial fluorescence before (F0) and after the addition of the peptide (Ff). The percent of lipid fusion was calculated as % mixing = [(Ft − F0)/(Fmax − F0)] × 100.
Solid-state NMR experiments
All 13C, 15N, and 1H 2D correlation experiments were measured on Bruker 800-MHz (18.8 T) and 900-MHz spectrometers using 3.2-mm MAS probes. 13C chemical shifts were referenced to the adamantane CH2 signal at 38.48 ppm or the Met Cε peak in the tripeptide N-formyl-Met-Leu-Phe-OH (f-MLF) at 14.0 ppm on the TMS scale. 15N chemical shifts were referenced to the Met 15N peak in f-MLF at 128.0 ppm on the liquid ammonia scale. Typical radiofrequency (rf) field strengths were 63–83 kHz for 1H, 50 kHz for 13C, and 36 kHz for 15N. 2D 13C–13C correlation spectra were measured using spin diffusion mixing times of 50–100 ms for DARR [68] and 0.5–1.0 s for PDSD. 2D 15N–13C correlation spectra were measured using a REDOR sequence with 833-µs mixing time to transfer the 15N and 13C coherence [69]. 2D 13C–1H DIPSHIFT experiments were conducted under 7 kHz MAS at 303 K [37], using the FSLG sequence for 1H–1H homonuclear decoupling [70]. To determine the depth of insertion of the TMD, a 1H–13C 2D correlation experiment with a 1H spin diffusion times of 25 and 100 ms was conducted at 303 K [71]. The 1H chemical shift was measured without homonuclear decoupling; thus, the 1H dimension only exhibits the signals of the mobile lipids and water.
19F CODEX spectra were measured on a 400-MHz spectrometer using a 4-mm MAS probe. The samples were spun at 8 kHz MAS and kept at 228–233 K [40] to ensure that only distance-dependent spin diffusion effects were measured while slow reorientational motions were frozen. 19F chemical shifts were referenced to the 19F signal of Teflon at −122 ppm. The 4-19F-Phe chemical shift was −115 ppm and the line widths were about 5.0 ppm, consistent with previously measured membrane peptides [61,63,65]. Typical rf field strengths were 71.4 kHz for 1H decoupling and 62.5 kHz for the 19F pulses. The total 19F CSA recoupling period was 0.50 or 0.25 ms, corresponding to 4 or 2 rotor periods of π pulses spaced half a rotor period apart (Table S3). To account for 19F T1 relaxation during the mixing time, a control experiment (S0) and a dephased experiment (S) were conducted for each mixing time, tmix [40]. The dephased experiment places the variable tmix period between the two CSA recoupling periods and a short tz period of 10 µs after the second CSA recoupling period, while the control experiment puts the 10-µs z-filter between the two CSA recoupling periods followed by the tmix period. The normalized intensities (S/S0) as a function of the tmix time, which ranged from 1 ms to 2.0 s, reflect pure spin diffusion, free of T1 relaxation effects.
5-19F-Trp was used to optimize the CODEX experiments. The 19F 180° pulse length was optimized by minimizing the S/S0 value at 50-ms mixing. The ideal equilibrium value for 5- 19F-Trp is 0.5 since there are two inequivalent molecules in the asymmetric unit cell, with an intermolecular distance of 4.62 Å (Fig. S3a). The CSA of 5- 19F-Trp is 54 ppm or 20 kHz at the magnetic field of 9.4 Tesla, similar to the CSA of 4- 19F-Phe, which is ~58 ppm. The normalization of the S intensity by S0 also removes potential pulse imperfections during the CSA recoupling periods such as the finite 19F pulse lengths, large 19F CSAs, and residual 1H–19F dipolar coupling. Even when pulse imperfections are explicitly considered, SIMPSON simulations [72] using realistic combinations of 19F CSAs and rf field strengths show that with CSA recoupling periods of 0.25 and 0.50 ms, the recoupled 19F CSA interaction causes complete dephasing of the 19F magnetization (Fig. S3b), indicating that the mixing time-dependent CODEX decay is sensitive to spin exchange between sites with small orientation differences.
19F CODEX simulation for distance extraction
19F CODEX curves were fit using a custom-written MATLAB program [42,61] that employs an exchange matrix formalism to obtain the internuclear distances in the three-spin system. The 3 × 3 exchange matrix contains rate constants, which are proportional to the square of the 19F–19F dipolar couplings and the overlap integral, F(0). The overlap integral depends on the 19F CSA and the MAS frequency, and has been previously calibrated on 5-19F-Trp to be 37 µs under 8 kHz MAS (Fig. S3a).
Structural modeling
We built an α-helical monomer for the TMD using the (φ, ψ) torsion angles obtained from 13C and 15N chemical shifts of the peptide in POPC/cholesterol membrane [36]. To generate a three-helix bundle, we aligned three copies of the α-helical monomers to residues 47–71 of the low-pH influenza hemagglutinin structure (PDB: 1HTM, 2.5 A [47]). Residues L493, L500, and L504 in the PIV5 TMD sequence were changed to 4-19F-Phe and their χ1 torsion angles were set to ~180°, since the trans conformation is the most populated rotamer of Phe in α-helices. The alignment of PIV5 residue V485 with the hemagglutinin residue Q47 resulted in a structural model that agrees with both the experimentally measured interhelical 19F–19F distances and the cysteine crosslinking data [21]. In the final structural model, the (χ1, χ2) angles of L493F, L500F, and L504F were (−149°, 80°), (−158°, 80°), and (142°, 80°), respectively.
Supplementary Material
Acknowledgments
The authors would like to thank Prof. Barbara Imperiali and Dr. Jean-Marie Swiecicki for help with lipid mixing assay. This work was supported by National Institutes of Health grant GM066976 to M.H.
Abbreviations
- TMD
transmembrane domain
- PIV5
parainfluenza virus 5
- FP
fusion peptide
- CODEX
Centerband-Only Detection of Exchange
- MAS
magic angle spinning
- GO
Gene ontology
- DRIP
DNA:RNA immuno-precipitation
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2018.01.002.
References
- 1.Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 3.Kim YH, Donald JE, Grigoryan G, Leser GP, Fadeev A, Lamb RA, et al. Capture and imaging of prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin HS, Wen XL, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:287–288. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison SC. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 10.Qiang W, Sun Y, Weliky DP. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sackett K, Nethercott MJ, Zheng Z, Weliky DP. Solid-state NMR spectroscopy of the HIV gp41 membrane fusion protein supports intermolecular antiparallel β sheet fusion peptide structure in the final six-helix bundle state. J. Mol. Biol. 2014;426:1077–1094. doi: 10.1016/j.jmb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Weliky DP. 13C– 13C correlation spectroscopy membrane-associated influenza virus fusion peptide strongly supports a helix-turn-helix motif and two turn conformations. J. Am. Chem. Soc. 2009;131:13228–13229. doi: 10.1021/ja905198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 14.Lai AL, Moorthy AE, Li Y, Tamm LK. Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. J. Mol. Biol. 2012;418:3–15. doi: 10.1016/j.jmb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamm LK, Lai AL, Li Y. Combined NMR and EPR spectroscopy to determine structures of viral fusion domains in membranes. Biochim. Biophys. Acta. 1768;2007:3052–3060. doi: 10.1016/j.bbamem.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao H, Hong M. Membrane-dependent conformation, dynamics, and lipid interactions of the fusion peptide of the paramyxovirus PIV5 from solid-state NMR. J. Mol. Biol. 2013;425:563–576. doi: 10.1016/j.jmb.2012.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao HW, Hong M. Conformation and lipid interaction of the fusion peptide of the paramyxovirus PIV5 in anionic and negative-curvature membranes from solid-state NMR. J. Am. Chem. Soc. 2014;136:2611–2624. doi: 10.1021/ja4121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin HS, Paterson RG, Wen XL, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pancera M, Zhou TQ, Druz A, Georgiev IS, Soto C, Gorman J, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455-+. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bissonnette MLZ, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J. Mol. Biol. 2009;386:14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith EC, Culler MR, Hellman LM, Fried MG, Creamer TP, Dutch RE. Beyond anchoring: the expanding role of the Hendra virus fusion protein transmembrane domain in protein folding, stability, and function. J. Virol. 2012;86:3003–3013. doi: 10.1128/JVI.05762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith EC, Smith SE, Carter JR, Webb SR, Gibson KM, Hellman LM, et al. Trimeric transmembrane domain interactions in paramyxovirus fusion proteins. Roles in protein folding, stability, and function. J. Biol. Chem. 2013;288:35726–35735. doi: 10.1074/jbc.M113.514554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 25.Armstrong RT, Kushnir AS, White JM. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang L, Hunter E. Residues in the membrane-spanning domain core modulate conformation and fusogenicity of the HIV-1 envelope glycoprotein. Virology. 2010;404:158–167. doi: 10.1016/j.virol.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owens RJ, Burke C, Rose JK. Mutations in the membrane-spanning domain of the human-immunodeficiency-virus envelope glycoprotein that affect fusion activity. J. Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyauchi K, Komano J, Yokomaku Y, Sugiura W, Yamamoto N, Matsuda Z. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J. Virol. 2005;79:4720–4729. doi: 10.1128/JVI.79.8.4720-4729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dev J, Park D, Fu QS, Chen J, Ha HJ, Ghantous F, et al. Structural basis for membrane anchoring of HIV-envelope spike. Science. 2016;353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roche J, Louis JM, Aniana A, Ghirlando R, Bax A. Complete dissociation of the HIV-1 gp41 ectodomain and membrane proximal regions upon phospholipid binding. J. Biomol. NMR. 2015;61:235–248. doi: 10.1007/s10858-015-9900-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roche J, Louis JM, Grishaev A, Ying J, Bax A. Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3425–3430. doi: 10.1073/pnas.1401397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao HW, Lee MW, Waring AJ, Wong GCL, Hong M. Viral fusion protein transmembrane domain adopts beta-strand structure to facilitate membrane topological changes for virus–cell fusion. Proc. Natl. Acad. Sci. U. S. A. 2015;112:10926–10931. doi: 10.1073/pnas.1501430112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Yao H, Hong M. Distinguishing bicontinuous lipid cubic phases from isotropic membrane morphologies using 31P solid-state NMR spectroscopy. J. Phys. Chem. 2015;119:4993–5001. doi: 10.1021/acs.jpcb.5b01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GC. Influenza virus a M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J. Am. Chem. Soc. 2013;135:13710–13719. doi: 10.1021/ja400146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt N, Mishra A, Lai GH, Wong GC. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 36.Shen Y, Bax A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 2015;1260:17–32. doi: 10.1007/978-1-4939-2239-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong M, Gross JD, Rienstra CM, Griffin RG, Kumashiro KK, Schmidt-Rohr K. Coupling amplification in 2D MAS NMR and its application to torsion angle determination in peptides. J. Magn. Reson. 1997;129:85–92. doi: 10.1006/jmre.1997.1242. [DOI] [PubMed] [Google Scholar]
- 38.Wang T, Park YB, Caporini MA, Rosay M, Zhong L, Cosgrove D, et al. Sensitivity-enhanced solid-state NMR detection of expansin's target in plant cell walls. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16444–16449. doi: 10.1073/pnas.1316290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T, Williams JK, Schmidt-Rohr K, Hong M. Relaxation-compensated difference spin diffusion NMR for detecting 13C– 13C long-range correlations in proteins and polysaccharides. J. Biomol. NMR. 2015;61:97–107. doi: 10.1007/s10858-014-9889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.deAzevedo ER, Hu WG, Bonagamba TJ, Schmidt-Rohr K. Centerband-only detection of exchange: efficient analysis of dynamics in solids by NMR. J. Am. Chem. Soc. 1999;121:8411–8412. [Google Scholar]
- 41.Buffy JJ, Waring AJ, Hong M. Determination of peptide oligomerization in lipid membranes with magic-angle spinning spin diffusion NMR. J. Am. Chem. Soc. 2005;127:4477–4483. doi: 10.1021/ja043621r. [DOI] [PubMed] [Google Scholar]
- 42.Luo W, Hong M. Determination of the oligomeric number and intermolecular distances of membrane protein assemblies by anisotropic 1H-driven spin diffusion NMR spectroscopy. J. Am. Chem. Soc. 2006;128:7242–7251. doi: 10.1021/ja0603406. [DOI] [PubMed] [Google Scholar]
- 43.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wi S, Sinha N, Hong M. Long range 1H-19F distance measurement in peptides by solid-state NMR. J. Am. Chem. Soc. 2004;126:12754–12755. doi: 10.1021/ja0462732. [DOI] [PubMed] [Google Scholar]
- 45.Hong M, Schmidt-Rohr K. Magic-angle-spinning NMR techniques for measuring long-range distances in biological macromolecules. Acc. Chem. Res. 2013;46:2154–2163. doi: 10.1021/ar300294x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balbach JJ, Petkova AT, Oyler NA, Antzutkin ON, Gordon DJ, Meredith SC, et al. Supramolecular structure in full-length Alzheimer's beta-amyloid fibrils: evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys. J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 48.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 49.Doherty T, Waring AJ, Hong M. Dynamic structure of disulfide-removed linear analogs of tachyplesin-I in the lipid bilayer from solid-state NMR. Biochemistry. 2008;47:1105–1116. doi: 10.1021/bi701390t. [DOI] [PubMed] [Google Scholar]
- 50.Doherty T, Waring AJ, Hong M. Membrane-bound conformation and topology of the antimicrobial peptide tachyplesin-I by solid-state NMR. Biochemistry. 2006;45:13323–13330. doi: 10.1021/bi061424u. [DOI] [PubMed] [Google Scholar]
- 51.Li SC, Deber CM. Glycine and beta-branched residues support and modulate peptide helicity in membrane environments. FEBS Lett. 1992;311:217–220. doi: 10.1016/0014-5793(92)81106-v. [DOI] [PubMed] [Google Scholar]
- 52.Li SC, Deber CM. A measure of helical propensity for amino-acids in membrane environments. Nat. Struct. Biol. 1994;1:368–373. doi: 10.1038/nsb0694-368. [DOI] [PubMed] [Google Scholar]
- 53.Langosch D, Crane JM, Brosig B, Hellwig A, Tamm LK, Reed J. Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity. J. Mol. Biol. 2001;311:709–721. doi: 10.1006/jmbi.2001.4889. [DOI] [PubMed] [Google Scholar]
- 54.Langosch D, Brosig B, Pipkorn R. Peptide mimics of the vesicular stomatitis virus G-protein transmembrane segment drive membrane fusion in vitro. J. Biol. Chem. 2001;276:32016–32021. doi: 10.1074/jbc.M102579200. [DOI] [PubMed] [Google Scholar]
- 55.Hofmann MW, Weise K, Ollesch J, Agrawal P, Stalz H, Stelzer W, et al. De novo design of conformationally flexible transmembrane peptides driving membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14776–14781. doi: 10.1073/pnas.0405175101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poschner BC, Fischer K, Herrmann JR, Hofmann MW, Langosch D. Structural features of fusogenic model transmembrane domains that differentially regulate inner and outer leaflet mixing in membrane fusion. Mol. Membr. Biol. 2010;27:1–10. doi: 10.3109/09687680903362044. [DOI] [PubMed] [Google Scholar]
- 57.Ollesch J, Poschner BC, Nikolaus J, Hofmann MW, Herrmann A, Gerwert K, et al. Secondary structure and distribution of fusogenic LV-peptides in lipid membranes. Eur. Biophys. J. 2008;37:435–445. doi: 10.1007/s00249-007-0233-4. [DOI] [PubMed] [Google Scholar]
- 58.Poschner BC, Quint S, Hofmann MW, Langosch D. Sequence-specific conformational dynamics of model transmembrane domains determines their membrane fusogenic function. J. Mol. Biol. 2009;386:733–741. doi: 10.1016/j.jmb.2008.12.077. [DOI] [PubMed] [Google Scholar]
- 59.Kim JH, Hartley TL, Curran AR, Engelman DM. Molecular dynamics studies of the transmembrane domain of gp41 from HIV-1. BBA-Biomembranes. 1788;2009:1804–1812. doi: 10.1016/j.bbamem.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gurezka R, Laage R, Brosig B, Langosch D. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J. Biol. Chem. 1999;274:9265–9270. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- 61.Luo W, Mani R, Hong M. Sidechain conformation and gating of the M2 transmembrane peptide proton channel of influenza a virus from solid-state NMR. J. Phys. Chem. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 62.Joh NH, Wang T, Bhate MP, Acharya R, Wu Y, Grabe M, et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science. 2014;346:1520–1524. doi: 10.1126/science.1261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams JK, Shcherbakov AA, Wang J, Hong M. Protonation equilibria and pore-opening structure of the dual-histidine influenza B virus M2 transmembrane proton channel from solid-state NMR. J. Biol. Chem. 2017;292:17876–17884. doi: 10.1074/jbc.M117.813998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandala VS, Liao SY, Kwon B, Hong M. Structural basis for asymmetric conductance of the influenza M2 proton channel investigated by solid-state NMR spectroscopy. J. Mol. Biol. 2017;429:2192–2210. doi: 10.1016/j.jmb.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams JK, Zhang Y, Schmidt-Rohr K, Hong M. pH-dependent conformation, dynamics, and aromatic interaction of the gating tryptophan residue of the influenza M2 proton channel from solid-state NMR. Biophys. J. 2013;104:1698–1708. doi: 10.1016/j.bpj.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon MD, Heider PL, Adamo A, Vinogradov AA, Mong SK, Li X, et al. Rapid flow-based peptide synthesis. Chembiochem. 2014;15:713–720. doi: 10.1002/cbic.201300796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takegoshi K, Nakamura S, Terao T. C-13–H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 69.Hong M, Griffin RG. Resonance assignments for solid peptides by dipolar-mediated C-13/N-15 correlation solid-state NMR. J. Am. Chem. Soc. 1998;120:7113–7114. [Google Scholar]
- 70.Bielecki A, Kolbert AC, Levitt MH. Frequency-switched pulse sequences—homonuclear decoupling and dilute spin NMR in solids. Chem. Phys. Lett. 1989;155:341–346. [Google Scholar]
- 71.Huster D, Yao XL, Hong M. Membrane protein topology probed by 1H spin diffusion from lipids using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2002;124:874–883. doi: 10.1021/ja017001r. [DOI] [PubMed] [Google Scholar]
- 72.Bak M, Rasmussen T, Nielsen NC. SIMPSON: a general simulation program for solid-state NMR spectroscopy. J. Magn. Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.