Short abstract
Organs-on-Chips (OoCs) are poised to reshape dramatically the study of biology by replicating in vivo the function of individual and coupled human organs. Such microphysiological systems (MPS) have already recreated complex physiological responses necessary to simulate human organ function not evident in two-dimensional in vitro biological experiments. OoC researchers hope to streamline pharmaceutical development, accelerate toxicology studies, limit animal testing, and provide new insights beyond the capability of current biological models. However, to develop a physiologically accurate Human-on-a-Chip, i.e., an MPS homunculus that functions as an interconnected, whole-body, model organ system, one must couple individual OoCs with proper fluidic and metabolic scaling. This will enable the study of the effects of organ-organ interactions on the metabolism of drugs and toxins. Critical to these efforts will be the recapitulation of the complex physiological signals that regulate the endocrine, metabolic, and digestive systems. To date, with the exception of research focused on reproductive organs on chips, most OoC research ignores homuncular endocrine regulation, in particular the circadian rhythms that modulate the function of all organ systems. We outline the importance of cyclic endocrine regulation and the role that it may play in the development of MPS homunculi for the pharmacology, toxicology, and systems biology communities. Moreover, we discuss the critical end-organ hormone interactions that are most relevant for a typical coupled-OoC system, and the possible research applications of a missing endocrine system MicroFormulator (MES-µF) that could impose biological rhythms on in vitro models. By linking OoCs together through chemical messenger systems, advanced physiological phenomena relevant to pharmacokinetics and pharmacodynamics studies can be replicated. The concept of a MES-µF could be applied to other standard cell-culture systems such as well plates, thereby extending the concept of circadian hormonal regulation to much of in vitro biology.
Impact statement
Historically, cyclic endocrine modulation has been largely ignored within in vitro cell culture, in part because cultured cells typically have their media changed every day or two, precluding hourly adjustment of hormone concentrations to simulate circadian rhythms. As the Organ-on-Chip (OoC) community strives for greater physiological realism, the contribution of hormonal oscillations toward regulation of organ systems has been examined only in the context of reproductive organs, and circadian variation of the breadth of other hormones on most organs remains unaddressed. We illustrate the importance of cyclic endocrine modulation and the role that it plays within individual organ systems. The study of cyclic endocrine modulation within OoC systems will help advance OoC research to the point where it can reliably replicate in vitro key regulatory components of human physiology. This will help translate OoC work into pharmaceutical applications and connect the OoC community with the greater pharmacology and physiology communities.
Keywords: organs-on-chips, diurnal rhythms, microformulator, pharmacology, toxicology, endocrine
Introduction
In recent years, Organ-on-a-Chip (OoC) systems have been able to replicate a variety of complex physiological processes and provide novel insights into previously uncharacterized phenomena.1–16 Recent studies published in this journal have developed functioning tissue for every major organ system of the body, including the blood-brain barrier, liver, kidney, lung, reproductive system, skin, and cardiopulmonary system, and even disease-specific models.17–27 Researchers have long known, however, that to harness the true power of OoCs these systems must one day be combined together to create a fully functioning microphysiological system (MPS), which when implemented on a single microfluidic chip has been described as a body-on-a-chip or human-on-a-chip,28,29 and more generally can be termed a homunculus.7 By linking OoC systems together to create an MPS homunculus, researchers may finally investigate in vitro interactions between drugs, diseases, and human organs in unparalleled detail.
MPS homunculi may provide detailed information regarding drug pharmacokinetics and pharmacodynamics beyond the current capabilities of either in vitro or in vivo models.30 Critical to these efforts will be the recreation of the complex physiological signals that regulate the endocrine, metabolic, and digestive systems. Understanding the interplay between these three major systems is crucial for OoC development and may contribute additional value in the study of pharmacokinetic and pharmacodynamic (PKPD) models in drug development.29,31–35 However, it is well beyond the capabilities of current in vitro drug or toxicology studies to simulate endogenous circadian rhythms because the cultured cells used in classical biology-on-plastic typically have their media changed every day or two, precluding the simulation of endogenous circadian rhythms through hormonal oscillations.12 Hence there should be utility in MPS homunculi that contain not only the desired end organ and the other organs whose metabolic activities are known to affect that target, but also the full endocrine system whose signals will affect the interconnected systems.
To explain the complexity of the endocrine regulatory system and its circadian modulation,36 and to propose an alternative to creating the full system in vitro, this review first presents a compendium from the literature describing the cyclic, hormonal modulation of the liver, neurovascular unit, intestine, kidney, skeletal muscle, adipose tissue, and heart – all organs that are the focus of intense investigation by the OoC community. We then suggest that it may be possible to utilize a device under development at Vanderbilt, termed a Missing Endocrine System MicroFormulator (MES-µF), to replicate this modulation. The device would obviate the need to create in vitro multiple, interlinked endocrine organs and glands, and instead draws from hormone reservoirs to formulate the combination of hormones needed during a particular time interval, for example each hour of the day.
For the purpose of this review, a MES-µF can be thought of as simply a self-contained system of pumps and valves that can add, on demand, the requisite time-dependent concentrations of hormones to the perfusate of one or more organs-on-chips.4 Time-division multiplexing may be used to support the delivery of multiple hormones at the same time. The concept is to use the MES-µF to recreate the empirically derived temporal profile of hormonal secretions of the endocrine organs immediately upstream of the target organ. These secretions, controlled by a separate master control unit, would thereby preclude the need to recreate in vitro the full host of human endocrine organs. We emphasize at the outset that we are presenting only a first-pass, literature-based review of how circadian hormonal rhythms might be incorporated into individual organ-on-chip and coupled human-on-chip models. Undoubtedly, we will have failed to include the favorite hormone of particular readers, and in anticipation of these omissions emphasize that we view this paper as a conceptual demonstration that provides a foundational bibliography and, in the Supplementary Materials, the tools that allow expansion of the analysis for even greater physiological realism.
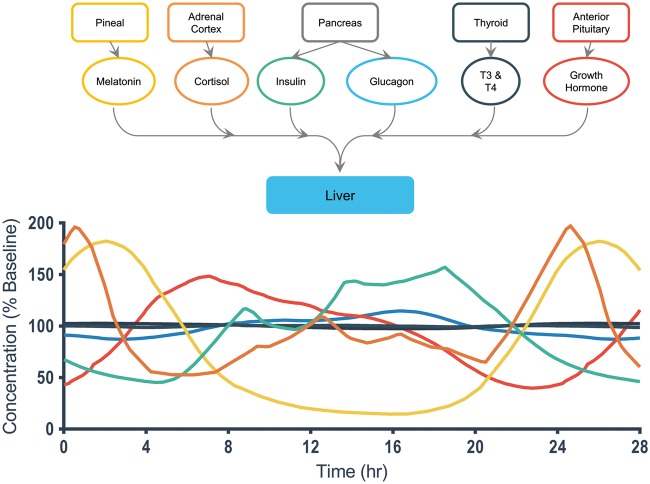
The liver serves as a prime example of the importance of time-dependent physiological signals, illustrated in Fig. 1.37 The hormones insulin, cortisol, growth hormone, and melatonin connect and feed into the liver from upstream endocrine organs such as the pancreas and adrenal cortex.38,39 These hormones help drive liver function and stimulate drug metabolism and hepatic blood flow.37 More important, these hormones regulate liver function in a time-dependent manner. Over the course of the 28 hours plotted, we see clear yet different 24-hour oscillatory cycles of multiple liver-regulating hormones, where the hormone concentrations fluctuate around a baseline concentration level, illustrating that their relative functional role changes over time.38–48 Liver metabolic functions are well known to exhibit circadian oscillations.37,49 Even more complicated is the cyclic variation of transcriptional regulation in the liver, which arises not only from central circadian regulation, but also hepatic clocks and environmental factors.50–52 Other examples can be found in the denervation of kidney53 and skeletal muscle,54 which causes organ-specific circadian genes to exhibit major changes in expression level, phase, and amplitude, while the core clock genes exhibit only minor differences. Dysregulation of circadian clocks has been hypothesized to drive metabolic comorbidities associated with psychiatric disorders55 and increase the risk of cardiometabolic disease.56
Figure 1.
Hormones that play important roles in driving liver physiology over a 24-hour cycle. Multiple hormones contribute to the generation of time-dependent fluctuations in liver physiology.38–44 The top image illustrates the upstream endocrine organs that produce the most important hormones relevant to liver function. All of the upstream endocrine organs interact with the liver through hormone chemical messengers that change in concentration throughout the circadian cycle. The concentrations over time for the important target hormones were constructed by selecting data from the literature, smoothing the data using an end-capped approach to limit loss, and plotting each hormone over a 28-hour cycle to illustrate day-to-day changes over their 24-hour cycle. The hormones are color-coded to correspond with the graph below that depicts their relative concentration change, as a percentage of their normal baseline level, over the course of the day. Cortisol, melatonin, and growth hormone exhibit the most significant daily changes with three-fold or larger peak-to-valley changes in concentration over their cycle.
Other than an elegant demonstration of hormonal oscillations in coupled female reproductive organs-on-chips,21,57–59 to date there has been no meaningful effort to study the impact of the breadth of oscillatory hormonal rhythms in generic OoC systems. To realize their full potential, OoC systems must capture a greater fraction of human physiological function, including the time-dependent fluctuations in hormone, nutrient, drug, and metabolite levels. As the field moves towards coupled organ systems, i.e., MPS homunculi, it is important to remember that humans sleep, eat, eliminate wastes, and exercise with distinct circadian rhythms that are subject to and can influence cyclic hormonal control.
For the context of this review, a circadian rhythm is a biological rhythm that oscillates over a period of 24 hours, synchronizes to an internal or external stimulus, for example a genetic clock, light, the environment, or social practices, and has a periodicity that is consistent across a range of physiological temperatures.36,56,60 We use the term “circadian” in the most general sense to cover the full extent of approximately 24-hour physiological rhythmicity. Diurnal rhythms, focused on day/night differences, are a specific subset of the circadian ones that are entrained by light and thus exhibit modulation via the sleep-wake cycle. Some hormones exhibit diurnal rhythms, e.g., insulin, but some do not, so in this review we simply consider all rhythms discussed to be circadian. One of the most well-studied examples of circadian rhythms is that of blood glucose levels and plasma insulin levels in response to food intake.36 For this review, which is based upon literature reports of experimental measurements on humans, we do not separate circadian rhythms that are driven by clock proteins from those that are driven by diurnal behavior (with our apologies to the circadian rhythm community). Although beyond the scope of this review, one must note that some clock proteins are central to the hypothalamic-pituitary-adrenal axis and act on peripheral organs by hormonal or autonomic control, while peripheral organs have localized clock proteins that may or may not be phase-locked with the central clocks.50,56,61,62 As we will discuss later, it should be possible to simulate behaviors such as daily exercise in an MPS homunculus, and hence it is up to the user of our analytical approach to determine which rhythms to incorporate explicitly through centralized cyclic hormones and which will occur spontaneously due to the intrinsic clocks that are endogenous to each organ.
Given that many research groups now culture their organs-on-chips for up to 28 days, it would be interesting to ascertain whether individual or connected organ chips in vitro are themselves exhibiting circadian variations in gene expression, as has been verified rigorously for twelve organs in the mouse,52 and the extent to which these oscillations are driven by intrinsic local clocks or by humoral or autonomic factors driven by other, upstream organs.
It is well understood that physiological systems exist within a state of homeostatic flux. Numerous physiological parameters have been observed to oscillate across the 24-hour circadian scale, and these include plasma concentrations of hormones,36 neurotransmitters,63 glucose,64 plasma proteins, second messengers (e.g., cortisol, insulin, atrial natriuretic hormone, noradrenaline, cAMP, melatonin), blood pressure,65 blood flow, the renin-angiotensin-aldosterone system,66 liver metabolism,67 and the first-pass effect.68,69 In fact, many physiological systems exhibit a temporal rhythmicity, several of which are relevant to the study of pharmacokinetics and pharmacodynamics (Table 1). Hence it is important to think of homeostasis not as fixed values of a large number of physiological variables but as an attractor in a high-dimensioned phase space about which these variables orbit.70–75
Table 1.
Circadian modulation of parameters that cause physiological changes relevant to pharmacokinetic studies
| System | Parameter | Peak Time with Peak Percent Change Above Baseline | Source |
|---|---|---|---|
| Plasma Concentrations | Glucose | 09.00, 14.00, 19.00 h; 60 min after meal (50%) | (Zimmet et al., 1974)64 |
| Hormones and Chemical Messengers | Cortisol 07.00-08.00 h (95%) | (Gamble et al., 2014)36 | |
| Melatonin 02.00-03.00 h (150%) | |||
| Growth Hormone 01.00-02.00 h (340%) | (Nussey, and Whitehead, 2001)38 | ||
| TSH 24.00-01.00 h (27%) | (Russell et al., 2009)41 | ||
| FT3 24.00-01.00 h (4%) | |||
| FT4 07.00-08.00 h (6%) | |||
| Testosterone 07.00-08.00 h (20%) | (Bremner et al., 1983)47 | ||
| PTH 02.00-03.00 h (17%) | (Fuleihan et al., 1997)43 | ||
| PRL 04.00-05.00 h (80%) | (Miyatake et al., 1980)44 | ||
| Progesterone 07.00-08.00 h (50%) | (Bungum et al., 2013)76 | ||
| Estradiol 02.00,12.00, 16.00 h (10%) | |||
| Norepinephrine 15.00-16.00 h (60%) | (Linsell et al., 1985)42 | ||
| Epinephrine 16.00- 17.00 h (60-80%) | |||
| Leptin 24.00-01.00 h (15%) | (Wardlaw et al., 2014)77 | ||
| Insulin 09.00, 14.00, 19.00 h; (150%) | (Yu et al., 2011)39 | ||
| Glucagon 18.00 h (30%) | |||
| ANP 04.00 h (50%) | (Portaluppi et al., 1990)46 | ||
| Aldosterone 06.00-07.00 h (60%) | (Hurwitz et al., 2004)45 | ||
| Liver | Drug metabolism | Drug-dependent | (Gachon and Firsov, 2011)67 |
| Hepatic blood flow | 08.00 h | (Lemmer and Nold, 1991)65 | |
| Kidney | Glomerular filtration | 16.00-17.00 h (33%) | (Koopman et al., 1989)66 |
| Renal blood flow | 19.00-20.00 h (34%) | (Wuerzner et al., 2014)78 | |
| Gastrointestinal (GI) | Gastric emptying time | Morning | (Goo et al., 1987)68 |
| GI motility | 08.00 h and after meal | (Rao et al., 2001)69 |
Not only do endocrine factors oscillate over the 24-hour circadian period, but so too does the responsiveness of target tissues – the hormones have differing time-courses of their actions, and each organ-on-chip should have an appropriate physiological response to the hormone depending upon whether the hormone invokes direct action on receptors, transporters, or enzymes, etc., or adjusts transcriptional regulation, which is much slower. We have focused this paper on a literature-based review of the timing of hormonal rhythms in the regulation of a selected set of organs, which we term End Organs, but it is important to recognize that as a result of global and local influences, the gene expression levels in each of these organs also fluctuates with differing phases.52 For example, in the mouse the liver demonstrates the largest number of genes with statistically significant circadian fluctuations (3,186), and the hypothalamus the least (642).52
In addition to studies of the physiology of hormonal regulation, OoC systems may provide an unparalleled opportunity to study in vitro detailed, organ-specific, pharmacokinetic drug-organ-hormone interactions. Most OoC systems currently under development operate under static conditions, however; that is, they maintain static levels of glucose, insulin, and other factors through continuous, single-pass perfusion,79 although a small number of systems have been shown to operate under long-term recirculation but without any cyclic circadian modulation of media components.28,29,59,80–84 In contrast, typical biology-on-plastic utilizes daily feedings, between which there are significant decreases in nutrients and increases in waste products.12 As a result, today’s OoC systems are in some ways limited in their applicability to the study of drug-organ interactions because they are missing a major component of cyclic hormonal organ regulation.85
By incorporating oscillatory endocrine regulation, OoC systems could surpass the ability of current animal studies to model advanced pharmacokinetic and pharmacodynamic effects of drug-organ-hormone interactions in humans. One consequence of studying animal models is that they have been shown to be incapable of fully accounting for the broad range of physiological phenomena operating within a human.79,86–89 For example, insulin secretion displays a diurnal rhythm in humans which peaks at 1700 h and has a trough at 0400 h.36 Such a rhythm is not observed in mouse models, due to a distinctly different diet and feeding schedule.56 The ovulatory cycle of a rat repeats every four days.90 Similar issues arise with in vitro Liberation, Absorption, Distribution, Metabolism, and Elimination (LADME) studies. It may be worthwhile to examine as early as possible in the drug development process the role of circadian modulation of factors that affect LADME, as summarized in Table 2. Hence OoC systems offer the possibility of recreating in vitro biological phenomena that heretofore have remained unstudied. OoC systems may provide the ability to access, measure, and control in vitro the dynamics of key human physiological variables which could not be studied due to practical and ethical limitations in human experiments. In addition, the opportunity for OoC researchers to elucidate complex physiological interactions between organ systems and their associated circadian rhythms may hold promise in accelerating the adoption and physiological relevance of OoC systems for therapeutic testing.20,29,31
Table 2.
Liberation, Absorption, Distribution, Metabolism, Excretion (LADME) parameters affected by the circadian clock (Adapted from Baraldo, 200891)
| Action | Modulated Physiological Activity |
|---|---|
| Liberation | Time-specified (immediate, controlled, etc.) release of a drug |
| Absorption | Perfusion, gastric pH change, acid secretion, motility, gastric emptying, sleep-wake rhythm |
| Distribution | Blood distribution, peripheral resistance, blood cell changes, serum protein levels |
| Metabolism | First-pass effect, enzyme activity, protein binding |
| Elimination | Renal perfusion, glomerular filtration, urine excretion, urine pH, electrolytes |
Important metabolic enzymes and pharmacodynamic processes tied to drug functionalization, inhibition, activation, conjugation, and excretion have already been associated with circadian changes.67,91–95 Drugs have variable pharmacokinetic and pharmacodynamic properties depending on the time of day,31 and 54 of the top 100 drugs have targets that are subject to circadian modulation.52 As an example in the liver, acetaminophen toxicity is controlled by both the central and hepatic clocks, but the contribution of the latter is more significant, suggesting that decoupling of the central and hepatic clocks in shift workers and international travelers could have adverse consequences.50 As a result of circadian effects in drug metabolism, dosages for drugs are established based on the time of day (Fig. 2), in recognition that the efficacy of a drug may differ by a factor of 2 to 10 over the course of a 24-hour interval.92 Current therapeutic regimens have ideal dosage times corresponding to the timing of the circadian clock. The optimal time of dosage for a compound is distinctly human and not well suited for study in mice because they exhibit variable activity cycles not correlated with human physiology. Consequently, there is a major shortcoming presented by animal models when comparing time of dosage and drug kinetics – a disadvantage avoided by OoC systems when attempting to examine the complex pharmacokinetic and pharmacodynamics involved in drug metabolism.
Figure 2.
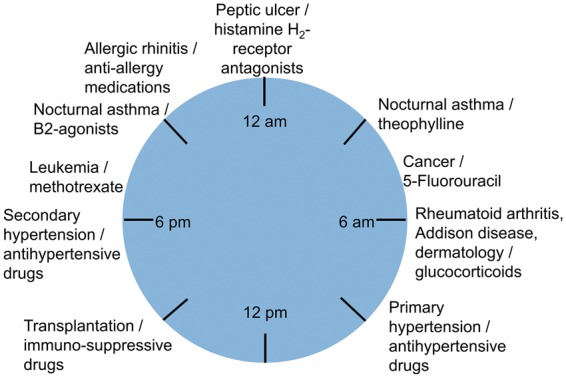
Time-dependent administration of pharmaceuticals whose responses are linked to the physiological clock. Different classes of drugs have optimal times when they should be administered based on their pharmacokinetic and pharmacodynamic properties. These time-dependent dosages are designed to correlate with changes in physiology that are, in turn, driven by changes in the circadian clock, indicating that the physiological clock is an important mechanism that must be included in the study of drug release and administration. (Adapted from Baraldo, 200891)
We recommend the recreation of such circadian oscillatory networks in OoC systems as a meaningful contribution towards the adoption of OoC micro-homunculi in the drug development process. Investigations of drug-OoC interactions may better inform results correlated with information utilized by toxicologists and pharmacologists, including, but not limited to, plasma half-life, area under the curve, bioavailability, volume of distribution, total clearance, and steady-state concentration,34,96–99 concepts that are already being applied to microphysiological systems.29,33,35,100–109
End organ interaction mapping provides insights into the connections between organ systems
To assess how the complete endocrine system affects our selected target organs, we constructed an End Organ Interaction Map to evaluate the role of chemical messengers in different organ systems (Fig. 3). The map, created from a review of the relevant literature36,38,39,41–48,76,77,110–124 and plotted using yED,125–128 provides a detailed view of where a chemical messenger is derived, the type of hormone, what tissues it interacts with, and whether the organ is receiving or secreting that hormone. Primary literature studies on human subjects from the clinical chemistry, toxicology, and metabolism fields were used as the basis for mapping interactions. Experimental evidence from at least one source, and oftentimes multiple sources, was used to generate a linkage, and the accompanying sources are listed as numbers along each linkage. The map serves as a tool that can be used to determine the most important hormones of interest to an organ system, and which endocrine organs are directly responsible for the modulation of a particular target organ. As an example of external modulation that may have a diurnal component, the pink kite for glucose is in the middle right of the figure while the kite in the upper left of the figure shows where alcohol, with a socially driven diurnal variation and circadian modulation of metabolism,129–133 should be introduced into the regulatory network. It is well known that the activity of alcohol dehydrogenase, the enzyme critical to alcohol metabolism, is itself modulated by an intrinsic circadian rhythm.122,134 A high-resolution, plottable version of this map is found in Supplement S1; see also Supplement S2, which is a version of the map that can be viewed interactively using yED (http://www.yworks.com/products/yed). Each ED link contains the reference numbers of the articles used to establish that link, listed in Supplement S3.
Figure 3.
An end-organ connectome highlighting the interactions between chemical messengers. A high-level view of the chemical interactions between organ systems provides insights into the complexity driving organ physiology. The most important end-organ hormone interactions were selected from the literature and depicted using an integrated network based in yED. An interactive version of this map and a high-resolution plottable one are in the Supplementary Materials (Supplements S2 and S1, respectively).36,38, 39, 41–48, 76,77, 110–124 Key: Organ: square = tissue, light blue = target tissue; Signals: ellipse = chemical messenger (salmon = peptide hormone, green = amino acid hormone, purple = steroid hormone, pink = other), kite = nutrient; Regulation: arrow = upregulation, bar = downregulation, solid line = organ receiving the hormone, dotted line = organ releasing the hormone.
Table 3 (and the expanded version in Supplement S4) presents details of the hormones we have identified as the most important for general OoC studies. We selected hormones based on their prevalence throughout the literature, their oscillatory nature, and their role in the organs that are being targeted by the MPS community. Some hormones, such as a cortisol, have wide-reaching effects across numerous organ systems and play an important part in regular tissue function. We evaluate the effects of different hormones on critical organ systems in further detail below. This analysis provides a detailed picture of the relevant hormones for an individual OoC system so that researchers may rapidly evaluate candidate hormones for inclusion in their studies.
Table 3.
Selected major hormones and major actions. The most important hormones with details on their interactions. The complete list of hormones in Fig. 3 is provided in the Supplementary Materials (Table S1). (Adapted from Tietz Textbook of Clinical Chemistry Chapter 29, 201240)
| Endocrine Organ and Hormone | Chemical Nature of Hormone | Major Sites of Action | Principal Actions |
|---|---|---|---|
| Anterior Pituitary Lobe | |||
| Growth hormone (GH) or somatotropin | Peptide (191aa) | Liver | Production of IGF-1 (promoting growth) |
| Liver and peripheral tissues | Anti-insulin and anabolic effects | ||
| Posterior Pituitary Lobe | |||
| Vasopressin or ADH | Peptide (9aa) | ArteriolesRenal tubules | Elevation of blood pressure; water reabsorption |
| Pineal Gland | |||
| Melatonin | Indoleamine | Hypothalamus | Suppression of gonadotropin and GH secretion; induction of sleep |
| Thyroid Gland | |||
| Thyroxine (T4) and triiodothyronine (T3) | Iodoamino acids | General body tissue | Stimulation of oxygen consumption and metabolic rate of tissue |
| Parathyroid Gland | |||
| Parathyroid hormone (PTH) or parathyrin | Peptide (84aa) | Kidney | Increased calcium reabsorption, inhibited phosphate reabsorption; increased production of 1,25- dihydroxycholecalciferol |
| Skeleton | Increased bone resorption | ||
| Adrenal Cortex | |||
| Aldosterone | Steroid | Kidney | Salt and water balance |
| Cortisol | Steroid | Many | Metabolism of carbohydrates, proteins, and fats; anti-inflammatory effects; others |
| Adrenal Medulla | |||
| Norepinephrine and epinephrine | Aromatic amines | Sympathetic receptors | Stimulation of sympathetic nervous system |
| Epinephrine | Liver and muscle, adipose tissue | Glycogenolysis Lipolysis | |
| Ovary | |||
| Estrogens | Phenolic steroids | Female accessory sex organs | Development of secondary sex characteristics |
| Bone | Control of skeletal maturation, etc. | ||
| Testis | |||
| Testosterone | Steroid | Male accessory sex organs | Development of secondary sex characteristics, maturation, and normal function |
| Pancreas | |||
| Glucagon | Peptide (29aa) | Liver | Glycogenolysis |
| Insulin | Peptide g | Liver, fat, muscle | Regulation of carbohydrate metabolism; lipogenesis |
| Gastrointestinal Tract | |||
| Glucagon-like peptide-1 | Peptide (30-31aa) | Gastrointestinal tract | Increase insulin and decrease glucagon secretion; inhibit gastric emptying |
| Kidney | |||
| Erythropoietin | Peptide (165aa) | Bone marrow | Stimulation of red cell formation |
| Renin-angiotensin-aldosterone system | Peptides (renin, 297aa; Ang I, 10aa; Ang II, 8aa) | Renin (from kidney) catalyzes hydrolysis of angiotensinogen (from liver, 485aa) to ang I in the intravascular space | Ang II increase of blood pressure and stimulation of secretion of aldosterone (see adrenal) |
| Liver | |||
| IGF-1, formerly called somatomedin | Peptide (70aa) | Most cells | Stimulation of cellular and linear growth |
| IGF-2 | Peptide (67aa) | Most cells | Insulin-like activity |
| Heart | |||
| Atrial natriuretic peptide (ANP, Atriopeptin) | Peptide with an intrachain disulfide bond (28aa) | Vascular, renal, and adrenal tissues | Regulation of blood volume and blood pressure |
| B-type natriuretic peptide (BNP) | Peptide with an intrachain disulfide bond (32aa) | Vascular, renal, and adrenal tissues | Regulation of blood volume and blood pressure |
| Adipose Tissue | |||
| Adiponectin | Peptide oligomers of 30 kDa subunits | MuscleLiver | Increase of fatty acid oxidation Suppression of glucose formation |
| Leptin | Peptide (167aa) | Hypothalamus | Inhibition of appetite, stimulation of metabolism |
| Multiple Cell Types | |||
| Estrogens | See above | See above | See above |
| Growth factors (e.g., epidermal growth factor, fibroblast growth factor, transforming growth factor family, platelet-derived growth factor, nerve growth factors) | Peptides | Many | Stimulation of cellular growth |
The data that directed our hormone analysis were drawn from published experimental studies on circadian hormonal oscillations in human subjects.38–48 Each hormone was evaluated over the course of a 24-hour circadian cycle, and the raw data from each study were extracted using a MATLAB script. The raw data were then extended on both sides of a 24-hour time interval so that the data could be smoothed using a locally weighted scatterplot smoothing (LOWESS) function. The smoothed data over a 28-hour time interval are presented in Figs. 4–9 as well as Supplement S5, and the raw, experimental data from the literature are available for download and analysis from Supplement S6.
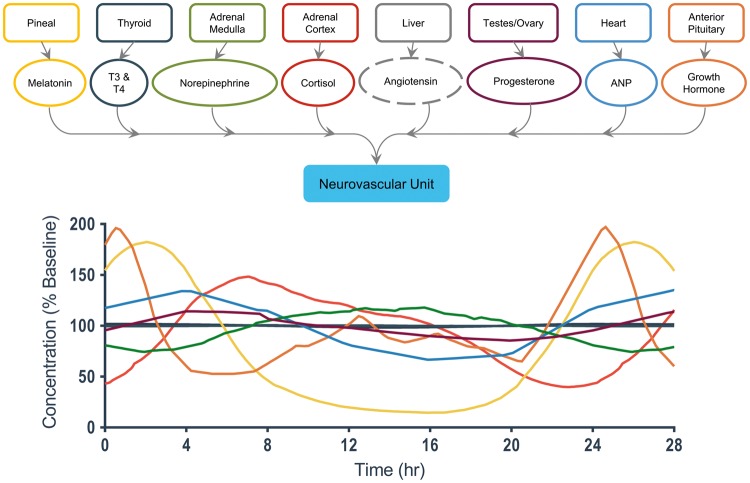
Figure 4.
A detailed view of end-organ interactions for the neurovascular unit.38, 40–42, 44,46, 48 The hormones are color-coded to correspond with the graph below that depicts their relative concentration change, as a percentage of their normal baseline level, over the course of the day. Hormones that did not have significant literature support for a circadian oscillation are given a dashed line to denote a constant concentration.
Figure 5.
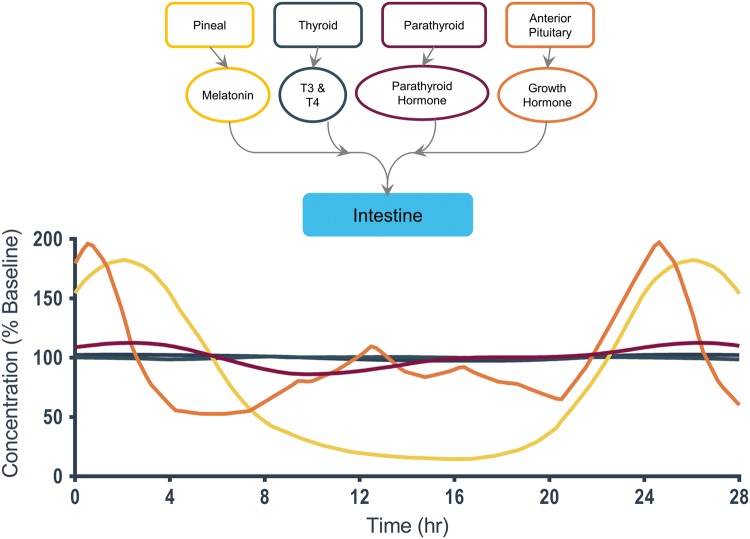
A detailed view of end-organ interactions for the intestine.38,40,43
Figure 6.
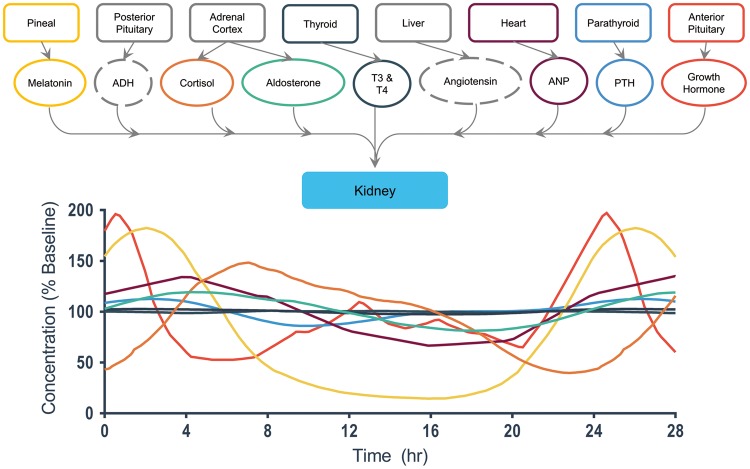
A detailed view of end-organ interactions for the kidney.38,40,41,43–46
Figure 7.
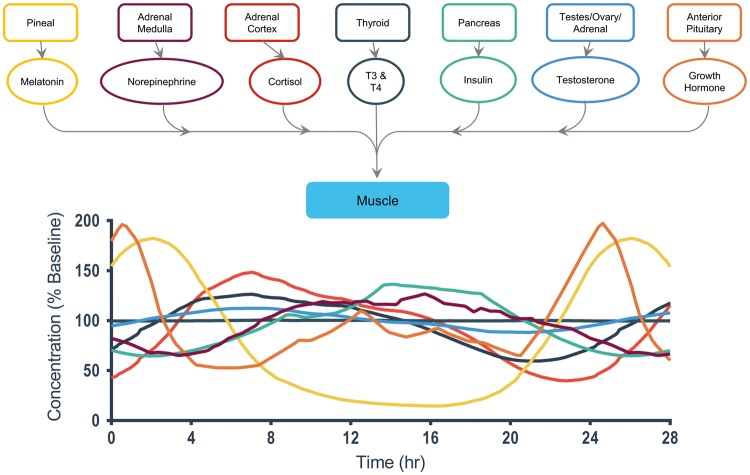
A detailed view of end-organ interactions for skeletal muscle.38, 40–42, 44,47
Figure 8.
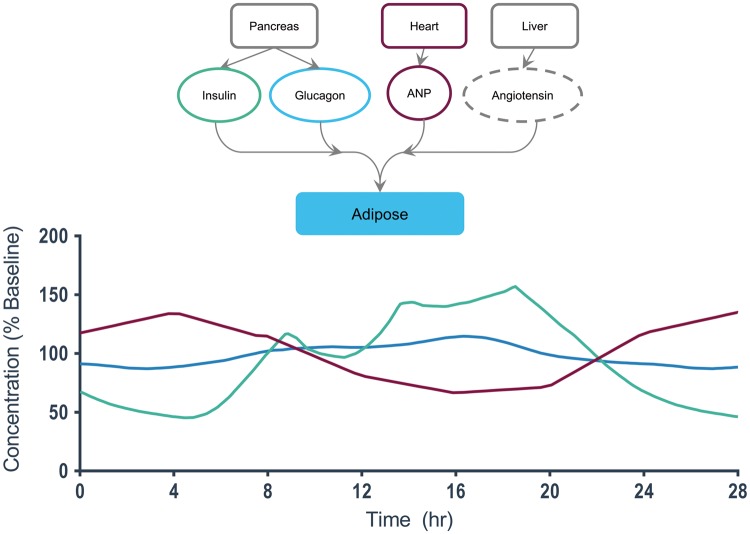
A detailed view of end-organ interactions for adipose tissue.38–40, 46
Figure 9.
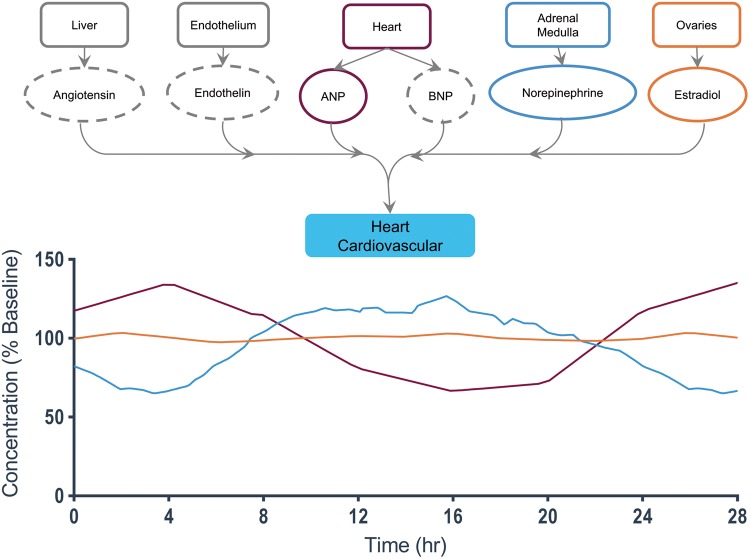
A detailed view of end-organ interactions for the heart/cardiovasculature.38,40, 42,46
Neurovascular unit
The neurovascular unit (NVU) on-a-chip (Fig. 4) recreates the minimal functional unit of the cerebrovascular system, in that it comprises the blood-brain barrier (BBB) and functioning central nervous system (CNS) neurons incorporated into an extracellular matrix, and hence is of critical importance to the study of the function of both the BBB and the CNS in pharmacology, toxicology, and brain injury.135,136 The NVU has eight regulating hormones that feed into the system over a 24-hour cycle. The hormones with the largest daily change include melatonin, cortisol, and growth hormone. Melatonin exhibits greater than a three-fold daily concentration change throughout the period of the day, falling over the first 8 hours of the daily cycle and then peaking at 175% of the baseline concentration after midnight. (As we will see later, of the seven targeted end organs examined, all except adipose tissue and heart/cardiovasculature appear to be affected by melatonin, which has broad-ranging actions and a role in a variety of diseases.137) Growth hormone presents a sharp decrease in concentration over the first 4 hours of wakefulness before steadily climbing back up to a peak value of 200% baseline.138 Cortisol levels have a gradual increase from 0-8 hours up to 150% baseline concentration, followed by a decrease from 8-22 hours down to 50% baseline before the cycle begins again.139 While angiotensin affects the neurovascular unit, we have not found reports of any significant circadian variation in angiotensin concentration, and hence the ellipse for that hormone is drawn as a dashed line (and a MES-µF could simply deliver that hormone at a chosen, constant concentration). The importance of any one of these time-varying hormones on NVU function should depend upon exactly which function is being evaluated, but this will be difficult to ascertain in vitro without the capability to modulate independently each hormone during a long-term experiment. An obvious question would be to ask how circadian hormonal rhythms might affect the observed signaling and metabolomic responses of the barrier functions of an NVU to inflammatory cytokines.136
Intestine
The intestine (Fig. 5) has fewer regulating hormones than the neurovascular unit. Parathyroid hormone exhibits a gradual but distinct oscillation around 110% to 85% baseline concentration.140 In vivo, the function of the intestine will also be modulated by the daily rhythms of eating and clearance, which are in turn affected not only by intrinsic hormonal circadian rhythms, but also social customs and environmental variables. It will be interesting to observe the changes that an intestine-on-a-chip undergoes upon cyclic feeding.
Kidney
The kidney (Fig. 6) has the most regulating hormones and demonstrates the most significant amplitude variation during the daily cycle. Aldosterone exhibits a gradual increase over the first 4 hours of wakefulness to 125% baseline, before decreasing over a period of 20 hours to a level of 85% baseline concentration.141 Neither antidiuretic hormone (ADH) nor angiotensin levels are observed to have daily oscillations based on current literature.
Muscle
The muscle system (Fig. 7) has several regulating hormones, including insulin, testosterone, and norepinephrine. Insulin levels increase over a 14-hour period up to 130% baseline and then gradually fall back down to nighttime levels of 60% baseline. Testosterone levels fluctuate in a symmetric fashion, rising to 120% baseline and then falling to 80% baseline across the 14-hour mark. Were one to include a muscle-on-a-chip in an MPS homunculus, it would be worthwhile to examine the effect on the entire system of increased daytime physical activity and even intense periods of regular exercise. This in turn could affect the consumption of nutrients that might otherwise be available to the rest of the homunculus, the production of a variety of systemic hormones, the withdrawal of fatty acids from adipose tissue, and the levels of lactic acid and other metabolites in the circulating perfusate. Imagine the opportunities offered by the ability to take one’s MPS homunculus out for a morning or evening jog, or even simulate other intense physical activities.
Adipose tissue
Adipose tissue (Fig. 8) has relatively few hormonal interactions, but they are significant. Insulin and A-natriuretic protein (ANP) display the most profound changes. Insulin levels rise periodically after each of three meals from approximately 60% baseline to over 150% baseline at hour 19. ANP levels gradually rise and fall from 120% baseline to 60% baseline.142,143 It would be important to determine from the literature what fraction of the reported circadian changes in insulin, for example, is due to intrinsic circadian control versus entrainment by external feeding schedules. In a long-term experiment, a MES-µF could deliver increased glucose to simulate regular feeding intervals, and when an obese MPS homunculus visited an endocrinologist treating its diabetes, the MES-µF could deliver a glucose-challenge test.
Heart and cardiovasculature
The heart (Fig. 9) has six regulating hormones, but many do not exhibit measurable oscillations over a daily period (and hence are plotted as dashed ellipses). The most important time-varying hormones include norepinephrine and ANP, which see an average two-fold change over the course of the day.142,143
Future work
The purpose of this review is to provide a framework upon which further investigations of OoC physiological regulation may begin. It is unlikely that the map in Fig. 3 is complete, and the hormones shown in Figs. 1 and 4–9 are intended to be representative of the most important hormones that affect the organs we are considering, not an exhaustive list of regulatory hormones for each organ. Obviously, specialists in the physiology, pathology, or toxicology of any of these organs may wish to add hormones of interest to their respective areas of study. The central issue we raise for OoC studies is the need to identify those hormones whose circadian rhythms produce significant changes in the function of the organ or organs under test, and then apply those changes to the MPS homunculus. For those hormones whose modulation has no observed effect, it may be sufficient to include that hormone in the base media without modulation. The need for modulation may have to be determined experimentally. It will be important to confirm that each of the organ-chips expresses appropriate levels of each hormone receptor and demonstrates the expected cellular response to that hormone, and also has the appropriate rhythmic modulation of the affected physiological processes. It will be interesting, and straightforward, to examine non-invasively the fluctuations in an organ’s metabolomic and signaling profile that might arise from external hormonal modulation, possibly by ion mobility-mass spectrometry136,144,145 or other high-throughput analytical techniques. Tracking fluctuations in gene expression will be possible if the cells that comprise the organ-chip tissues include, for example, selected optical reporter genes, but tracking of large sets of genes52 is not yet possible without destroying the set of organ-chips, which at present cost much more than a mouse.
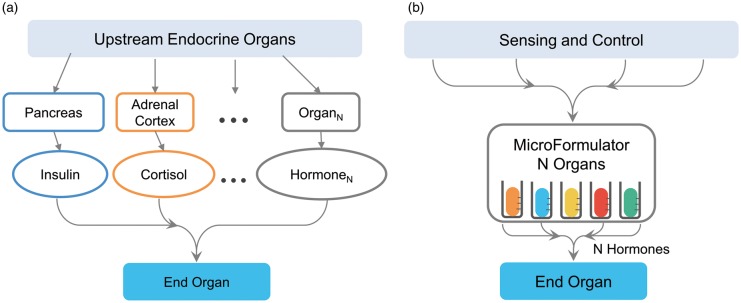
Total recreation of the physiological timing cues relevant to a whole-body MPS homunculus may never be achieved (and one might argue should never be attempted), but to neglect their role is to severely limit the potential application and validity of OoC systems. In place of replicating the endocrine organs that control a specific target organ, which might require up to nine separate endocrine organs for some target organs, we propose that the MES-µF could consolidate hormonal sensing and control into a centralized unit and account for the contribution of any endocrine organs that were missing from the multi-organ MPS homunculus under development. The centralized unit would be connected to each organ within a homunculus to provide time-dependent dosages of hormones to a specific organ, thereby replacing the natural endocrine system, shown in Fig. 10A, with the simulated one in Fig. 10B. The MES-µF has the distinct advantage that it would interface with all the organs within an MPS homunculus and control their circadian rhythms, and allow the investigator to interrogate the homunculus and test hypotheses regarding hormonal regulation and organ-organ and organ-drug-organ interactions. Obviously, it could be useful to have closed-loop hormonal sensing and control, which might in turn be driven by a computational model of hormonal regulation.146 It would remain to be seen the extent to which this control system might be able to simulate the high-level control exerted by the hypothalamus61 and pituitary62 on the intermediate endocrine organs removed from direct regulation of a target organ.
Figure 10.
A missing endocrine system MicroFormulator (MES-µF) that could help recapitulate human endocrine regulation in vitro for microphysiological homunculi without the need to implement multiple, interlinked endocrine organs. A) Standard biology uses multiple endocrine organs (Organ1 to OrganN) to release numerous hormones (Hormone1 to HormoneN) that can engage with a targeted end organ. Instead of recreating the entire network of endocrine organs within an MPS homunculus, the MES-µF (B) combines the function of multiple endocrine organs into a single unit capable of recreating N endocrine organs by the time-dependent delivery of N hormones. The endocrine MicroFormulator contains a variety of vessels that each contain an important chemical messenger for the target organ. The MES-µF would either operate based upon an internally programmed clock or could sense the homeostatic state of the MPS homunculus in its entirety or simply that of a single target organ and adjust the hormone concentrations in a fashion similar to the feedback loops present in normal human biological endocrine modulation.
Once circadian-regulated organs-on-chips are coupled together to create an MPS homunculus with appropriate scaling of circulating fluid volumes,5 each of the “end” organs could affect that organ as well as others in the system, possibly to the extent of recreating closed-loop physiological control. This would constitute an even higher level of physiological regulation than envisioned at the outset of the MPS initiatives. At this point, the concept of “end organ” would become irrelevant.
We have already discussed the difficulty in recreating autonomic and immune modulation of an MPS homunculus. Obviously, a separate microformulator could feed the gastrointestinal system. There are other distributed regulatory systems in the body, and some might be addressable with a microformulator; for example, a microformulator could provide time-varying levels of carrier proteins in the circulating blood surrogate were the organs that produced them in vivo not included in the MPS homunculus. There is a practical limit to the detail that an MPS homunculus should attempt to capture, as discussed in Watson et al.12
The MES-µF could be entrained onto a daily rhythm cycle that helps replicate key parameters of drug administration and absorption. The ability to specifically and accurately evaluate drug delivery and absorption into end-organ tissues has not yet been fully refined and may provide critical insights for development of new pharmaceuticals. Furthermore, different organ systems could have their drug delivery entrained at different time intervals to simulate dose responses to differing time-of-day administration protocols. For example, a researcher interested in the excretion of a lead compound could program the MES-µF to simulate ingestion at different times of day and measure excretion as a function of time. Furthermore, if the excretion pattern is disrupted by the new compound, the MES-µF could introduce different compounds and individually investigate the chemical messengers at work.
The MES-µF could greatly expand the physiological processes recreated by MPS homunculi and advance their utility and adoption into the pharmacology space. Furthermore, the challenges associated with recreating the complex physiological signals that regulate organ function present new opportunities for research. Principal among these are the technologies required for fluidic control and analytic sensing.4,146
The many hormones, metabolites, and nutrients that are transported throughout the body in plasma may affect the design of suitable blood surrogates for OoC systems.4,107 The surrogate must not only serve as the cell culture media for the different tissues of the MPS homunculus, but also as a transporter of critical regulatory messengers, including hormones, and hence the volume of media in the perfusion system could affect the concentration of metabolites generated by the target organs.5 The role of organ-specific endothelial cells and their transporter proteins at the interface between the common perfusion media and the stromal cells of each organ may mitigate the need for universal media. The presence or absence of an endothelial barrier and/or transport proteins may have to be accounted for when specifying the concentration of hormone delivered by the MES-µF.
In addition to appropriate perfusion media and hormone concentrations, each MPS homunculus will require chemical sensors capable of quantifying with adequate fidelity the temporal variation in the concentration of the intrinsic and externally controlled chemical messengers in near real time, since delays between sampling and the reporting of the concentrations could adversely affect the stability of the controlled system.4,146–149 It may be useful to have sensors that operate on both the interstitial fluid of each end organ and the common circulating media.84,135,136 The sensors will enable the study of both open- and closed-loop operation of the MPS homunculus, so that their results may be compared to both whole-body function and isolated end-organ responses. Computational in silico models may be employed to further enhance our understanding of independent organ and system-level behaviors.29,33,107 Computer-based simulations and tests may fine tune control of physical flow rates or better predict hormone regulation and production. Furthermore, these models may better predict critical pharmacokinetic phenomena and inform experimental design, as has been demonstrated by Yu et al.33 in a micro-perfused liver/immune MPS. The integrated experimental/computational paradigm put forth by Yu illustrates the power of a self-regulating MES-µF model which can both test and inform new experimental designs.150–152
Just as biological rhythms have shown increasing importance in drug pharmacology and toxicology, so too should they play an active role in OoC investigations. Without the physiological regulatory cues that are inherent to native biology, we simply have a clock with no hands. While the different physiological systems may work independently in their own harmony, it is only when we combine the systems and produce a working synchrony that we truly begin to realize the power of the interconnected whole-body Human-on-a-Chip. The MES-µF may provide the first mechanism for studying the complex physiological signals that regulate the endocrine, metabolic, and digestive systems in real-time at the functional level of the end organ. To date, this feat has yet to be fully investigated through in vitro or in vivo models. Thus, many new discoveries remain on the horizon for the interconnected and programmed Human-on-a-Chip. The most rewarding applications are those that will enable new insights beyond current in vitro biology-on-plastic.12 These advances will ultimately enable OoC systems to fulfill their potential and unite the pharmacology, toxicology, and systems biology communities.
Our proposed development of a novel Missing Endocrine System MicroFormulator will allow researchers to replicate the time- and dose-dependent variations in circadian hormonal oscillations. We introduced the concept of the missing-organ microformulator in 2013,4 and now apply the concept specifically to the endocrine system. The MES-µF, to be implemented in the near future by a system of computer-controlled microfluidic pumps and valves153–157 that supports time-division multiplexing of multiple reagents, will certainly accelerate our exploration of the numerous physiological processes linked to LADME, which are of primary importance to pharmacology and toxicology. Our MES-µF will enable the study of time- and dose-dependent delivery of hormones and drug compounds specific to the intended MPS homunculus. It will allow OoC researchers to study ultradian rhythms that are more frequent than circadian,158–161 or slower, as with the female reproductive system.21,59 The MES-µF might allow researchers to examine whether the "gender" of a single homunculus could be altered by adjusting the delivery of gender-specific combinations of time-dependent steroidal sex hormones while maintaining the existing X and Y chromosome presentation of the cells used to construct the homunculus. It would be interesting to determine whether this could be used to address the sex bias in research.162 Could this approach be used to create a transsexual homunculus?
The capabilities of the MES-µF could play an important role in allowing OoC systems to recapitulate the physiology of a particular individual. Custom-tailoring of individual pharmaceutical regimes is a hallmark of personalized medicine, and the MES-µF would enable, for example, the simulation in a personalized MPS homunculus of a patient’s abnormal circadian hormonal rhythms. Just as insulin pumps are changing diabetic care, a cyclic pump for circadian hormones might prove useful in the care of some patients, but today it would be difficult to conduct the requisite experiments on humans. MPS homunculi driven by a MES-µF could enhance the adoption of personalized medicine within clinical research and thereby drive better outcomes in therapeutic development.
The circadian MES-µF might enable in vitro homuncular studies of circadian contributions to a number of physiological and pathophysiological processes, including metabolism, diabetes and obesity,56,159,163–165 aging,166 exercise,161 sleep deprivation,167 and psychiatric disorders.55 The ability to simulate specific organ-organ hormonal interactions will enable an unprecedented level of understanding on the given effects a new therapeutic may have on an organ-specific basis, since it will be possible to isolate and test specific interactions between organ, receptor, and a pharmacologic agent. It is our hope that the recapitulation of the oscillatory endocrine cycle and time-dose-dependent delivery of drug will advance the study of OoC systems and prepare them for even wider adoption by the greater pharmacology and toxicology community. Central to the success of current OoC systems have been the collaborative efforts of engineers, biologists, physicists, pharmacologists, physicians, and toxicologists; the MES-µF will simplify the introduction of endocrinologists to this community so that their contributions can help accelerate the future adoption of OoC systems. That said, there is every reason to apply the concept of a MES-µF to other in vitro cell culture systems, including flasks, Petri dishes, well-plates, or transwell assays – the only issue is the requirement that one must add and remove sufficient culture media and provide adequate mixing to ensure that all of the cells being cultured together are in fact exposed to the same temporal hormonal profile. This can be addressed by providing pumps, valves, and fluid distribution systems that can support the necessary flow rates.
Without a doubt, there are regulatory processes that will be hard to replicate within an MPS homunculus, most notably those associated with the autonomic nervous and immune systems. How might one incorporate a vagal nerve to regulate heart, lung, kidney, liver, and digestive tract organs-on-chips, and could one account for the hypothalamic control of the circadian rhythms that affect sympathetic and parasympathetic activity and hence adrenal medullary and pancreatic secretions?61 Although it may be difficult to incorporate the adaptive immune system into an MPS, innate immune cells present in a variety of organs would respond to circadian immune modulators.168 While it will be important to include endocrine function and other components in MPS homuncular models so as to increase their physiological realism, it is critical to recognize that the goal of the larger organ-on-chip exercise is to study a specific physiological process. It is a futile exercise to recreate a complete, in vitro microhuman when only a minimal set of features is required to recapitulate the particular physiological process or system under study and allow its examination without the complication of other organs that may be difficult to both understand and control. The MES-µF should prove to be a valuable addition to the constructionist closure of the hermeneutic circle, as we continue to understand both the individual “parts” identified by recent advances in reductionist biology and the function of the whole organism as revealed by more than two millennia of physiological studies.7,8 We conclude by predicting that the combination of analyses such as ours and devices functionally equivalent to the forthcoming MES-µF will enable OoC homunculi to play a significant role in elucidating how endogenous hormonal rhythms affect organ-organ and organ-drug-organ interactions.
Note
To open the second supplemental file ‘Endocrine Hormone Organ Connectome Map’, download the yED software from http://www.yworks.com/products/yed
Author Contributions
All authors curated research articles, synthesized novel insights, and outlined key applications relevant to the OoC community. JPW led the conception and design of the study. KJC acquired data, analyzed and interpreted findings, and drafted figures. OMA collected the articles, references, and background information necessary to produce the end-organ connectome and oscillatory hormone graphs. KJC and JPW drafted the article. All authors critically revised and edited the manuscript.
Supplementary Material
Acknowledgements
We thank David Wasserman and Jake Hughey for their insights and critical review of the manuscript. Additional thanks are due Allison Price and Don Berry for their editorial and bibliographic assistance, as well as Sean Bedingfield, Alex Boyer, Mike Jacobs, Reid McCallister, Jason Miller, Oscar Ortega, Stephen Petty-Valenzuela, Emily Schafer, Ian Setliff, and LaNell Williams, from Dr. Wikswo’s 2016 class in Automated Biology: Sensors, Controls, Scaling and Topology, for their lively participation in discussions regarding the MES-µF and hormonal regulation.
Declaration of Conflicting Interests
John Wikswo has one awarded patent and four pending patent applications on microformulator technologies. Pump and valve patents have been licensed to KIYATEC Inc. The development of the microformulator at Vanderbilt has been funded by AstraZeneca and CFD Research Corporation, but there have been no discussions or experiments regarding hormonal modulation. A license agreement for the MicroFormulator is under negotiation with a manufacturer. Neither of the other authors has a conflict to disclose.
Funding
This work has been supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH) under Awards 5UH3TR000491, 5UH3TR000503, and 5UH3TR000504, and Contract No. HHSN271201600009C; and the Environmental Protection Agency (EPA) Assistance Agreement No. 83573601. The views expressed in this paper are solely those of the authors and do not necessarily reflect those of the funding agencies. The EPA does not endorse any products or commercial services mentioned in this publication.
References
- 1.Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: Organs-on-chips. Lab Chip. 2012; 12:2156–64 [DOI] [PubMed] [Google Scholar]
- 2.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Organs-on-a-chip: A focus on compartmentalized microdevices. Ann Biomed Eng. 2012; 40:1211–27 [DOI] [PubMed] [Google Scholar]
- 3.Moraes C, Labuz JM, Leung BM, Inoue M, Chun TH, Takayama S. On being the right size: scaling effects in designing a human-on-a-chip. Integr Biol. 2013; 5:1149–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikswo JP, Block III FE, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, Samson PC, Schaffer DK, Seale KT, Sherrod SD. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng. 2013; 60:682–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wikswo J, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip. 2013; 13:3496–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014; 32:760–72 [DOI] [PubMed] [Google Scholar]
- 7.Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med. 2014; 239:1061–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wikswo JP, Porter AP. Biology coming full circle: Joining the whole and the parts. Exp Biol Med. 2015; 240:3–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell. 2016; 164:1105–9 [DOI] [PubMed] [Google Scholar]
- 10.Hutson MS, Alexander PG, Allwardt V, Aronoff DM, Bruner-Tran KL, Cliffel DE, Davidson JM, Gough A, Markov DA, McCawley LJ, McKenzie JR, McLean JA, Osteen KG, Pensabene V, Samson PC, Senutovitch NK, Sherrod SD, Shotwell MS, Taylor DL, Tetz LM, Tuan RS, Vernetti LA, Wikswo JP. Organs-on-chips as bridges for predictive toxicology. Appl In Vitro Toxicol. 2016; 2:97–102 [Google Scholar]
- 11.Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: Intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep. 2017; 7:42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson DE, Hunziker R., Wikswo JP. Fitting tissue chips and microphysiological systems into the grand scheme of medicine, biology, and toxicology. Exp Biol Med 2017:Accepted [DOI] [PMC free article] [PubMed]
- 13.Low LA, Tagle DA. Organs-on-chips: Progress, challenges, and future directions. Exp Biol Med 2017: doi: 10.1177/1535370217700523. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 14.Knudsen T, Klieforth B, Slikker W. Programming microphysiological systems for children's health protection. Exp Biol Med 2017: doi: 10.1177/1535370217717697. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 15.Ewart L, Fabre K, Chakilam A, Dragan Y, Duignan D, Eswaraka J, Gan J, Guzzie-Peck P, Otieno M, Jeong C, Keller D, Van Vleet T, Radhakrishna S, de Morais S, Phillips J, Proctor W, Watson D, Will Y, Tagle D, Berridge B. Navigating tissue chips from development to dissemination: A pharmaceutical industry perspective. Exp Biol Med 2017: doi: 10.1177/1535370217715441. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 16.Slikker W., Jr Of human-on-a-chip and humans: Considerations for creating and using microphysiological systems. Exp Biol Med. 2014; 239:1078–9 [DOI] [PubMed] [Google Scholar]
- 17.Nichols JE, Niles JA, Vega SP, Argueta LB, Eastaway A, Cortiella J. Modeling the lung: Design and development of tissue engineered macro- and micro-physiologic lung models for research use. Exp Biol Med. 2014; 239:1135–69 [DOI] [PubMed] [Google Scholar]
- 18.Clark AM, Wheeler SE, Taylor DP, Pillai VC, Young CL, Prantil-Baun R, Nguyen T, Stolz DB, Borenstein JT, Lauffenburger DA, Venkataramanan R, Griffith LG, Wells A. A microphysiological system model of therapy for liver micrometastases. Exp Biol Med. 2014; 239:1170–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander PG, Gottardi R, Lin H, Lozito TP, Tuan RS. Three-dimensional osteogenic and chondrogenic systems to model osteochondral physiology and degenerative joint diseases. Exp Biol Med. 2014; 239:1080–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabre K, Livingston C, Tagle DA. Organs-on-Chips (Microphysiological Systems): Tools to expedite efficacy and toxicity testing in human tissue. Exp Biol Med. 2014; 239:1073–7 [DOI] [PubMed] [Google Scholar]
- 21.Young AN, Moyle-Heyrman G, Kim JJ, Burdette JE. Microphysiologic systems in female reproductive biology. Exp Biol Med 2017: doi: 10.1177/1535370217697386 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 22.Soto-Gutierrez A, Gough A, Vernetti LA, Taylor DL, Monga SP. Pre-clinical and clinical investigations of metabolic zonation in liver diseases: The potential of microphysiology systems. Exp Biol Med 2017: doi: 10.1177/1535370217707731. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 23.Sheehy SP, Grosberg A, Qin P, Behm DJ, Ferrier JP, Eagleson MA, Nesmith AP, Krull D, Falls JG, Campbell PH, McCain ML, Willette RN, Hu E, Parker KK. Toward improved myocardial maturity in an organ-on-chip platform with immature cardiac myocytes. Exp Biol Med 2017: doi: 10.1177/1535370217701006. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 24.Phan DT, Bender RHF, Andrejecsk JW, Sobrino A, Hachey SJ, George SC, Hughes CC. Blood–brain barrier-on-a-chip: Microphysiological systems that capture the complexity of the blood–central nervous system interface. Exp Biol Med 2017: doi: 10.1177/1535370217694100. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 25.Lee-Montiel FT, George SM, Gough AH, Sharma AD, Wu J, DeBiasio R, Vernetti LA, Taylor DL. Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Exp Biol Med 2017: doi: 10.1177/1535370217703978 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 26.Barry C, Schmitz MT, Propson NE, Hou Z, Zhang J, Nguyen BK, Bolin JM, Jiang P, McIntosh BE, Probasco MD, Swanson S, Stewart R, Thomson JA, Schwartz MP, Murphy WL. Uniform neural tissue models produced on synthetic hydrogels using standard culture techniques. Exp Biol Med 2017: doi: 10.1177/1535370217715028 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 27.Vernetti LA, Senutovitch N, Boltz R, DeBiasio R, Shun TY, Gough A, Taylor DL. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med. 2016; 241:101–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuler ML, Esch MB. Body-on-a chip: Using microfluidic systems to predict human responses to drugs. Pure Appl Chem. 2010; 82:1635–45 [Google Scholar]
- 29.Sung JH, Srinivasan B, Esch MB, McLamb WT, Bernabini C, Shuler ML, Hickman JJ. Using physiologically-based-pharmacokinetic-guided "Body-on-a-Chip" systems to predict mammalian response to drug and chemical exposure. Exp Biol Med. 2014; 239:1225–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reardon S. 'Organs-on-chips' go mainstream. Nature. 2015; 523:266. [DOI] [PubMed] [Google Scholar]
- 31.Lemmer B. Chronobiology, drug-delivery, and chronotherapeutics. Adv Drug Del Rev. 2007; 59:825–7 [DOI] [PubMed] [Google Scholar]
- 32.Jones HM, Rowland-Yeo K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacometrics Syst Pharmacol,. 2013; 2:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Cilfone NA, Large EM, Sarkar U, Wishnok JS, Tannenbaum SR, Hughes DJ, Lauffenburger DA, Griffith LG, Stokes CL, Cirit M. Quantitative systems pharmacology approaches applied to microphysiological systems (MPS): Data interpretation and multi-MPS integration. CPT Pharmacometrics Syst Pharmacol,. 2015; 4:585–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obach RS, Baxter JG, Liston TE, Silber BM, Jones BC, Macintyre F, Rance DJ, Wastall P. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J Pharmacol Exp Ther. 1997; 283:46–58 [PubMed] [Google Scholar]
- 35.Sung JH, Shuler ML. In vitro microscale systems for systematic drug toxicity study. Bioprocess Biosystems Eng. 2010; 33:5–19 [DOI] [PubMed] [Google Scholar]
- 36.Gamble KL, Berry R, Frank SJ, Young ME. Circadian clock control of endocrine factors. Nat Rev Endocrinol. 2014; 10:466–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwighaft Z, Reinke H, Asher G. The liver in the eyes of a chronobiologist. J Biol Rhythms. 2016; 31:115–24 [DOI] [PubMed] [Google Scholar]
- 38.Nussey S, Whitehead S. Endocrinology: An integrated approach: Oxford: BIOS Scientific Publishers; 2001. Available from: http://www.ncbi.nlm.nih.gov/books/NBK22/. [PubMed]
- 39.Yu HY, Xia FZ, Lam KSL, Wang Y, Bao YQ, Zhang JL, Gu YJ, Zhou PC, Lu JX, Jia WP, Xu AM. Circadian rhythm of circulating fibroblast growth factor 21 is related to diurnal changes in fatty acids in humans. Clin Chem. 2011; 57:691–700 [DOI] [PubMed] [Google Scholar]
- 40.Kleerekoper M. Hormones In: Burtis CA, Ashwood ER, Bruns DE, editors. Tietz Textbook of Clinical Chemistry and Molecular Diagnostics, 12th ed Elsevier Sanders, 2012, p. 837–49 [Google Scholar]
- 41.Russell W, Harrison RF, Smith N, Darzy K, Shalet S, Weetman AP, Ross RJ. Free triiodothyronine has a distinct circadian rhythm that is delayed but parallels thyrotropin levels. J Clin Endocrinol Metab. 2008; 93:2300–6 [DOI] [PubMed] [Google Scholar]
- 42.Linsell CR, Lightman SL, Mullen PE, Brown MJ, Causon RC. Circadian rhythms of epinephrine and norepinephrine in man. J Clin Endocrinol Metab. 1985; 60:1210–5 [DOI] [PubMed] [Google Scholar]
- 43.Fuleihan GE, Klerman EB, Brown EN, Choe Y, Brown EM, Czeisler CA. The parathyroid hormone circadian rhythm is truly endogenous - A general clinical research center study. J Clin Endocrinol Metab. 1997; 82:281–6 [DOI] [PubMed] [Google Scholar]
- 44.Miyatake A, Morimoto Y, Oishi T, Hanasaki N, Sugita Y, Iijima S, Teshima Y, Hishikawa Y, Yamamura Y. Circadian rhythm of serum testosterone and its relation to sleep: Comparison with the variation in serum luteinizing hormone, prolactin, and cortisol in normal men. J Clin Endocrinol Metab. 1980; 51:1365–71 [DOI] [PubMed] [Google Scholar]
- 45.Hurwitz S, Cohen RJ, Williams GH. Diurnal variation of aldosterone and plasma renin activity: timing relation to melatonin and cortisol and consistency after prolonged bed rest. J Appl Physiol. 2004; 96:1406–14 [DOI] [PubMed] [Google Scholar]
- 46.Portaluppi F, Bagni B, Uberti ED, Montanari L, Cavallini R, Trasforini G, Margutti A, Ferlini M, Zanella M, Parti M. Circadian rhythms of atrial-natriuretic-peptide, renin, aldosterone, cortisol, blood-pressure and heart-rate in normal and hypertensive subjects. J Hypertens. 1990; 8:85–95 [DOI] [PubMed] [Google Scholar]
- 47.Bremner WJ, Vitiello MV, Prinz PN. Loss of circadian rhythmicity in blood testosterone levels with aging in normal men. J Clin Endocrinol Metab. 1983; 56:1278–81 [DOI] [PubMed] [Google Scholar]
- 48.Graham JD, Clarke CL. Physiological action of progesterone in target tissues. Endocr Rev. 1997; 18:502–19 [DOI] [PubMed] [Google Scholar]
- 49.Reinke H, Asher G. Circadian clock control of liver metabolic functions. Gastroenterology. 2016; 150:574–80 [DOI] [PubMed] [Google Scholar]
- 50.Johnson BP, Walisser JA, Liu Y, Shen AL, McDearmon EL, Moran SM, McIntosh BE, Vollrath AL, Schook AC, Takahashi JS, Bradfield CA. Hepatocyte circadian clock controls acetaminophen bioactivation through NADPH-cytochrome P450 oxidoreductase. Proc Natl Acad Sci USA. 2014; 111:18757–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Lin JD. Transcriptional control of circadian metabolic rhythms in the liver. Diabetes Obes Metab. 2015; 17:33–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014; 111:16219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen WD, Yeh JK, Peng MT, Shie SS, Lin SL, Yang CH, Chen TH, Hung KC, Wang CC, Hsieh IC, Wen MS, Wang CY. Circadian CLOCK mediates activation of transforming growth factor-β signaling and renal fibrosis through cyclooxygenase 2. Am J Pathol. 2015; 185:3152–63 [DOI] [PubMed] [Google Scholar]
- 54.Dyar KA, Ciciliot S, Tagliazucchi GM, Pallafacchina G, Tothova J, Argentini C, Agatea L, Abraham R, Ahdesmaki M, Forcato M, Bicciato S, Schiaffino S, Blaauw B. The calcineurin-NFAT pathway controls activity-dependent circadian gene expression in slow skeletal muscle. Mol Metab. 2015; 4:823–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barandas R, Landgraf D, McCarthy MJ, Welsh DK. Circadian clocks as modulators of metabolic comorbidity in psychiatric disorders. Curr Psychiatry Rep. 2015; 17:98. [DOI] [PubMed] [Google Scholar]
- 56.McGinnis GR, Young ME. Circadian regulation of metabolic homeostasis: causes and consequences. Nat Sci Sleep. 2016; 8:163–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laronda MM, Burdette JE, Kim JJ, Woodruff TK. Recreating the female reproductive tract in vitro using iPSC technology in a linked microfluidics environment. Stem Cell Res Ther 2013;4 (Supp.1):Article S13 [DOI] [PMC free article] [PubMed]
- 58.Eddie SL, Kim JJ, Woodruff TK, Burdette JE. Microphysiological modeling of the reproductive tract: a fertile endeavor. Exp Biol Med. 2014; 239:1192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, McKinnon KE, Dokic D, Rashedi AS, Haisenleder DJ, Malpani SS, Arnold-Murray CA, Chen K, Jiang M, Bai L, Nguyen CT, Zhang J, Laronda MM, Hope TJ, Maniar KP, Pavone ME, Avram MJ, Sefton EC, Getsios S, Burdette JE, Kim JJ, Borenstein JT, Woodruff TK. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun. 2017; 8:14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hastings M, O'Neill JS, Maywood ES. Circadian clocks: regulators of endocrine and metabolic rhythms. J Endocrinol. 2007; 195:187–98 [DOI] [PubMed] [Google Scholar]
- 61.Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci. 2001; 2:521–6 [DOI] [PubMed] [Google Scholar]
- 62.Gloe T, Sohn HY, Meininger GA, Pohl U. Shear stress-induced release of basic fibroblast growth factor from endothelial cells is mediated by matrix interaction via integrin alpha(v)beta(3). J Biol Chem. 2002; 277:23453–8 [DOI] [PubMed] [Google Scholar]
- 63.Matsumura T, Nakagawa H, Suzuki K, Ninomiya C, Ishiwata T. Influence of circadian disruption on neurotransmitter levels, physiological indexes, and behaviour in rats. Chronobiol Int. 2015; 32:1449–57 [DOI] [PubMed] [Google Scholar]
- 64.Zimmet PZ, Wall JR, Rome R, Stimmler L, Jarrett RJ. Diurnal variation in glucose tolerance: Associated changes in plasma insulin, growth hormone, and non-esterified fatty acids. Br Med J. 1974; 1:485–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lemmer B, Nold G. Circadian changes in estimated hepatic blood flow in healthy subjects. Br J Clin Pharmacol. 1991; 32:627–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koopman MG, Koomen GCM, Krediet RT, Demoor EAM, Hoek FJ, Arisz L. Circadian rhythm of glomerular filtration rate in normal individuals. Clin Sci. 1989; 77:105–11 [DOI] [PubMed] [Google Scholar]
- 67.Gachon F, Firsov D. The role of circadian timing system on drug metabolism and detoxification. Expert Opin Drug Metab Toxicol. 2011; 7:147–58 [DOI] [PubMed] [Google Scholar]
- 68.Goo RH, Moore JG, Greenberg E, Alazraki NP. Circadian variation in gastric-emptying of meals in humans. Gastroenterology. 1987; 93:515–8 [DOI] [PubMed] [Google Scholar]
- 69.Rao SSC, Sadeghi P, Beaty J, Kavlock R, Ackerson K. Ambulatory 24-h colonic manometry in healthy humans. Am J Physiol Gastrointest Liver Physiol. 2001; 280:G629–G39 [DOI] [PubMed] [Google Scholar]
- 70.Huang S, Kauffman SA. Complex Gene Regulatory Networks - from Structure to Biological Observables: Cell Fate Determination In: Meyers RA, editor. Encyclopedia of Complexity and Systems Science, Springer New York, 2009, p. 1180–213 [Google Scholar]
- 71.de Bivort B, Huang S, Bar-Yam Y. Dynamics of cellular level function and regulation derived from murine expression array data. Proc Natl Acad Sci. 2004; 101:17687–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wikswo J. Challenges in characterizing and controlling complex cellular systems. Bull Am Phys Soc. 2011; 56:Abstract: X38.00002 [Google Scholar]
- 73.Hughey JJ, Hastie T, Butte AJ. ZeitZeiger: supervised learning for high-dimensional data from an oscillatory system. Nucleic Acids Res. 2016; 44:e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughey JJ. Machine learning identifies a compact gene set for monitoring the circadian clock in human blood. Genome Med. 2017; 9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hughey JJ. Monitoring the circadian clock in human blood using personalized machine learning. bioRxiv preprint first posted online Jul. 31, 2016; doi: http://dx.doi.org/10.1101/066126
- 76.Bungum L, Franssohn F, Bungum M, Humaidan P, Giwercman A. The circadian variation in Anti-Müllerian hormone in patients with polycystic ovary syndrome differs significantly from normally ovulating women. PLoS One. 2013; 8:e68223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wardlaw SL, Burant CF, Klein S, Meece K, White A, Kasten T, Lucey BP, Bateman RJ. Continuous 24-hour leptin, proopiomelanocortin, and amino acid measurements in human cerebrospinal fluid: Correlations with plasma leptin, soluble leptin receptor, and amino acid levels. J Clin Endocrinol Metab. 2014; 99:2540–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wuerzner G, Firsov D, Bonny O. Circadian glomerular function: from physiology to molecular and therapeutical aspects. Nephrol Dial Transplant. 2014; 29:1475–80 [DOI] [PubMed] [Google Scholar]
- 79.Eckel-Mahan K, Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol Rev. 2013; 93:107–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang YI, Oleaga C, Long CJ, Esch MB, McAleer CW, Miller PG, Hickman JJ, Shuler ML. Self-contained, low-cost Body-on-a-Chip systems for drug development. Exp Biol Med 2017: doi: 10.1177/1535370217694101 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 81.Materne EM, Maschmeyer I, Lorenz AK, Horland R, Schimek KMS, Busek M, Sonntag F, Lauster R, Marx U. The multi-organ chip - a microfluidic platform for long-term multi-tissue coculture. J Vis Exp. 2015; 98:Article e52526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hubner J, Lindner M, Drewell C, Bauer S, Thomas A, Sambo NS, Sonntag F, Lauster R, Marx U. A. four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015; 15:2688–99 [DOI] [PubMed] [Google Scholar]
- 83.Wagner I, Materne EM, Brincker S, Sussbier U, Fradrich C, Busek M, Sonntag F, Sakharov DA, Trushkin EV, Tonevitsky AG, Lauster R, Marx U. A. dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip. 2013; 13:3538–47 [DOI] [PubMed] [Google Scholar]
- 84.Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, Massa S, Riahi R, Chae S, Hu N, Avci H, Zhang W, Silvestri A, Nezhad AS, Manbohi A, De, Ferrari F, Polini A, Calzone G, Shaikh N, Alerasool P, Budina E, Kang J, Bhise N, Ribas J, Pourmand A, Skardal A, Shupe T, Bishop CE, Dokmeci MR, Atala A, Khademhosseini A. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci USA. 2017; 114:E2293–E302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Musiek ES, FitzGerald GA. Molecular Clocks in Pharmacology In: Kramer A, Merrow M, editors. Circadian Clocks. Handbook of Experimental Pharmacology Berlin, Heidelberg: Springer Berlin Heidelberg, 2013, p. 243–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marx U, Andersson TB, Bahinski A, Beilmann M, Beken S, Cassee FR, Cirit M, Daneshian M, Fitzpatrick S, Frey O, Gaertner C, Giese C, Griffith L, Hartung T, Heringa MB, Hoeng J, de Jong WH, Kojima H, Kuehnl J, Leist M, Luch A, Maschmeyer I, Sakharov D, Sips A, Steger-Hartmann T, Tagle DA, Tonevitsky A, Tralau T, Tsyb S, van de Stolpe A, Vandebriel R, Vulto P, Wang JF, Wiest J, Rodenburg M, Roth A. Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing. ALTEX 2016; 33:272–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van Deun K, Smith P, Berger B, Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000; 32:56–67 [DOI] [PubMed] [Google Scholar]
- 88.Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009; 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Basketter DA, Clewell H, Kimber I, Rossi A, Blaauboer B, Burrier R, Daneshian M, Eskes C, Goldberg A, Hasiwa N, Hoffmann S, Jaworska J, Knudsen TB, Landsiedel R, Leist M, Locke P, Maxwell G, McKim J, McVey EA, Ouedraogo G, Patlewicz G, Pelkonen O, Roggen E, Rovida C, Ruhdel I, Schwarz M, Schepky A, Schoeters G, Skinner N, Trentz K, Turner M, Vanparys P, Yager J, Zurlo J, Hartung T. t4 Report: A roadmap for the development of alternative (non-animal) methods for systemic toxicity testing. ALTEX 2012; 29:5–15 [DOI] [PubMed] [Google Scholar]
- 90.Gruenberg ML, Steger RW, Peluso JJ. Follicular development, steroidogenesis and ovulation within ovaries exposed in vitro to hormone levels which mimic those of the rat estrous cycle. Biol Reprod. 1983; 29:1265–75 [DOI] [PubMed] [Google Scholar]
- 91.Baraldo M. The influence of circadian rhythms on the kinetics of drugs in humans. Expert Opin Drug Metab Toxicol. 2008; 4:175–92 [DOI] [PubMed] [Google Scholar]
- 92.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007; 47:593–628 [DOI] [PubMed] [Google Scholar]
- 93.Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. In: Annu Rev Pharmacol Toxicol. 502010, p. 377-421 [DOI] [PubMed]
- 94.Lemmer B. Circadian rhythms and drug delivery. J Control Release. 1991; 16:63–74 [Google Scholar]
- 95.Lemmer B. Chronopharmacology and controlled drug release. Expert Opin Drug Deliv. 2005; 2:667–81 [DOI] [PubMed] [Google Scholar]
- 96.Segui A, Lebaron JP, Leverge R. Biomedical engineering approach of pharmacokinetic problems: Computer-aided design in pharmacokinetics and bioprocessing. IEE Proc-D. 1986; 133:217–23 [Google Scholar]
- 97.Meibohm B, Derendorf H. Basic concepts of pharmacokinetic/pharmacodynamic (PK/PD) modelling. Int J Clin Pharmacol Ther. 1997; 35:401–13 [PubMed] [Google Scholar]
- 98.Derendorf H, Meibohm B. Modeling of pharmacokinetic/pharmacodynamic (PK/PD) relationships: Concepts and perspectives. Pharm Res. 1999; 16:176–85 [DOI] [PubMed] [Google Scholar]
- 99.Salmon M. Practical Pharmacology for the Pharmaceutical Sciences. Chichester, West Sussex: John Wiley & Sons Inc, 2014, p. 197 [Google Scholar]
- 100.Viravaidya K, Sin A, Shuler ML. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog. 2004; 20:316–23 [DOI] [PubMed] [Google Scholar]
- 101.Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog. 2004; 20:338–45 [DOI] [PubMed] [Google Scholar]
- 102.Chao P, Maguire T, Novik E, Cheng KC, Yarmush ML. Evaluation of a microfluidic based cell culture platform with primary human hepatocytes for the prediction of hepatic clearance in human. Biochem Pharmacol. 2009; 78:625–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010; 10:446–55 [DOI] [PubMed] [Google Scholar]
- 104.Sung JH, Esch MB, Shuler ML. Integration of in silico and in vitro platforms for pharmacokinetic-pharmacodynamic modeling. Expert Opin Drug Metab Toxicol. 2010; 6:1063–81 [DOI] [PubMed] [Google Scholar]
- 105.Lee JB, Sung JH. Organ-on-a-chip technology and microfluidic whole-body models for pharmacokinetic drug toxicity screening. Biotechnol J. 2013; 8:1258–66 [DOI] [PubMed] [Google Scholar]
- 106.Vivares A, Salle-Lefort S, Arabeyre-Fabre C, Ngo R, Penarier G, Bremond M, Moliner P, Gallas JF, Fabre G, Klieber S. Morphological behaviour and metabolic capacity of cryopreserved human primary hepatocytes cultivated in a perfused multiwell device. Xenobiotica. 2014; 45:29–44 [DOI] [PubMed] [Google Scholar]
- 107.Abaci HE, Shuler ML. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr Biol. 2015; 7:383–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee H, Kim DS, Ha SG, Choi I, Lee JM, Sung JH. A. pumpless multi-organ-on-a-chip (MOC) combined with a pharmacokinetic-pharmacodynamic (PK-PD) model. Biotechnol Bioeng. 2016; 114:432–43 [DOI] [PubMed] [Google Scholar]
- 109.Long TJ, Cosgrove PA, Dunn RT, ll, Stolz DB, Hamadeh HK, Afshari C, McBride H, Griffith LG. Modeling therapeutic antibody-small molecule drug-drug interactions using a 3D perfusable human liver co-culture platform. Drug Metab Disposition. 2016; 44:1940–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raven PH, Johnson GB. The Endocrine System. In: Biology, 6th ed. Boston MA: McGraw-Hill, 2002, p. 1125-46
- 111.Dockray GJ. Clinical endocrinology and metabolism. Gastrin. Best Pract Res Clin Endocrinol Metab. 2004; 18:555–68 [DOI] [PubMed] [Google Scholar]
- 112.Kim W, Egan JM. The role of incretins in glucose homeostasis and diabetes treatment. Pharmacol Rev. 2008; 60:470–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dupre J, Ross SA, Watson D, Brown JC. Stimulation of insulin secretion by gastric inhibitory polypeptide in man. J Clin Endocrinol Metab. 1973; 37:826–8 [DOI] [PubMed] [Google Scholar]
- 114.Gregory SJ, Kaiser UB. Regulation of gonadotropins by inhibin and activin. Semin Reprod Med. 2004; 22:253–67 [DOI] [PubMed] [Google Scholar]
- 115.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Champon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990; 9:1603–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schumacher M, Guennoun R, Robert FO, Carelli C, Gago N, Ghoumari A, Deniselle MCG, Gonzalez SL, Ibanez C, Labombarda F, Coirini H, Baulieu EE, De Nicola AF. . Local synthesis and dual actions of progesterone in the nervous system: neuroprotection and myelination. Growth Horm IGF Res. 2004; 14:S18–S33 [DOI] [PubMed] [Google Scholar]
- 117.Paul M, Mehr AP, Kreutz R. Physiology of local renin-angiotensin systems. Physiol Rev. 2006; 86:747–803 [DOI] [PubMed] [Google Scholar]
- 118.Takahashi Y, Kipnis DM, Daughada WH. Growth hormone secretion during sleep. J Clin Invest. 1968; 47:2079–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Juul A, Jorgensen JOL, Christiansen JS, Muller J, Skakkeboek NE. Metabolic effects of GH: A rationale for continued GH treatment of GH-deficient adults after cessation of linear growth. Horm Res. 1995; 44:64–72 [DOI] [PubMed] [Google Scholar]
- 120.Ryabinin AE, Bachtell RK, Heinrichs SC, Lee S, Rivier C, Olive MF, Mehmert KK, Camarini R, Kim JA, Koenig HN, Nannini MA, Hodge CW, Roberts AJ, Koob GF. The corticotropin-releasing factor/urocortin system and alcohol. Alcohol Clin Exp Res. 2002; 26:714–22 [PubMed] [Google Scholar]
- 121.Rivier C. Role of hypothalamic corticotropin-releasing factor in mediating alcohol-induced activation of the rat hypothalamic-pituitary-adrenal axis. Front Neuroendocrinol. 2014; 35:221–33 [DOI] [PubMed] [Google Scholar]
- 122.Danel T, Touitou Y. Chronobiology of alcohol: From chronokinetics to alcohol-related alterations of the circadian system. Chronobiol Int. 2004; 21:923–35 [DOI] [PubMed] [Google Scholar]
- 123.Morley JE. Overview of the Endocrine System. In: Merck Manual Professional Version Online 2016. Retrieved from http://www.merckmanuals.com/professional/endocrine-and-metabolic-disorders/principles-of-endocrinology/overview-of-the-endocrine-system
- 124.Dubocovich ML, Markowska M. Functional MT1 and MT2 melatonin receptors in mammals. Endocrine. 2005; 27:101–10 [DOI] [PubMed] [Google Scholar]
- 125.Singh A, Mai D, Kumar A, Steyn AJC. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc Natl Acad Sci USA. 2006; 103:11346–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol. 2006; 24:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.yED graph editor, yFiles software, Tubingen, Germany. https://www.yworks.com/products/yed (accessed 7 June 2017)
- 128.Raza S, Robertson KA, Lacaze PA, Page D, Enright AJ, Ghazal P, Freeman TC. A logic-based diagram of signalling pathways central to macrophage activation. BMC Syst Biol. 2008; 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rosenwasser AM, Fecteau ME, Logan RW. Effects of ethanol intake and ethanol withdrawal on free-running circadian activity rhythms in rats. Physiol Behav. 2005; 84:537–42 [DOI] [PubMed] [Google Scholar]
- 130.Kudo T, Tamagawa T, Shibata S. Effect of chronic ethanol exposure on the liver of Clock-mutant mice. J Circadian Rhythms. 2009; 7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wasielewski JA, Holloway FA. Alcohol's interactions with circadian rhythms – A focus on body temperature. Alcohol Res Health. 2001; 25:94–100 [PMC free article] [PubMed] [Google Scholar]
- 132.Sarkar DK. Circadian genes, the stress axis, and alcoholism. Alcohol Res. 2012; 34:362–6 [PMC free article] [PubMed] [Google Scholar]
- 133.Forsyth CB, Voigt RM, Shaikh M, Tang YM, Cederbaum AI, Turek FW, Keshavarzian A. Role for intestinal CYP2E1 in alcohol-induced circadian gene-mediated intestinal hyperpermeability. Am J Physiol Gastrointest Liver Physiol. 2013; 305:G185–G95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sturtevant RP, Garber SL. Circadian rhythms of alcohol dehydrogenase and MEOS in the rat. Proc Soc Exp Biol Med. 1984; 175:299–303 [DOI] [PubMed] [Google Scholar]
- 135.Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, Britt CM, Hoilett OS, Yang Q, Brewer BM, Samson PC, McCawley LJ, Webb DJ, Li D, Bowman AB, Reiserer RS, Wikswo JP. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics. 2015; 9:Article 054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brown JA, Codreanu SG, Shi M, Sherrod SD, Markov DA, Neely MD, Britt CM, Hoilett OS, Reiserer RS, Samson PC, McCawley LJ, Webb DJ, Bowman AB, McLean JA, Wikswo JP. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. J Neuroinflammation. 2016; 13:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pandi-Perumal SR, Trakht I, Srinivasan V, Spence DW, Maestroni GJM, Zisapel N, Cardinali DP. Physiological effects of melatonin: Role of melatonin receptors and signal transduction pathways. Prog Neurobiol. 2008; 85:335–53 [DOI] [PubMed] [Google Scholar]
- 138.Brandenberger G, Weibel L. The 24-h growth hormone rhythm in men: sleep and circadian influences questioned. J Sleep Res. 2004; 13:251–5 [DOI] [PubMed] [Google Scholar]
- 139.Chung S, Son GH, Kim K. Circadian rhythm of adrenal glucocorticoid: Its regulation and clinical implications. Biochim Biophys Acta. 2011; 1812:581–91 [DOI] [PubMed] [Google Scholar]
- 140.Jubiz W, Canterbury JM, Reiss E, Tyler FH. Circadian rhythm in serum parathyroid hormone concentration in human subjects: correlation with serum calcium, phosphate, albumin, and growth-hormone levels. J Clin Invest. 1972; 51:2040–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Charloux A, Gronfier C, Lonsdorfer-Wolf E, Piquard F, Brandenberger G. Aldosterone release during the sleep-wake cycle in humans. Am J Physiol Endocrinol Metab. 1999; 276:E43–E9 [DOI] [PubMed] [Google Scholar]
- 142.Ehlenz K, Schneider H, Tremmel P, Schmidt P, Kaffarnik H. Circadian rhythm of atrial natriuretic peptide in normal subjects In: Kaufmann W, Wambach G, editors. Endocrinology of the Heart, Berlin, Heidelberg: Springer Berlin Heidelberg, 1989, p. 181–3 [Google Scholar]
- 143.Miyamoto S, Shimokawa H, Sumioki H, Touno A, Nakano H. Circadian rhythm of plasma atrial natriuretic peptide, aldosterone, and blood pressure during the 3rd trimester in normal and preeclamptic pregnancies. Am J Obstet Gynecol. 1988; 158:393–9 [DOI] [PubMed] [Google Scholar]
- 144.Marasco CC, Enders JR, Seale KT, McLean JA, Wikswo JP. Real-time cellular exometabolome analysis with a microfluidic mass spectrometry platform. PLoS One. 2013; 10:Article e0117685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brown JA, Sherrod SD, Goodwin CR, Brewer B, Yang L, Garbett KA, Li D, McLean JA, Wikswo JP, Mirnics K. Metabolic consequences of interleukin-6 challenge in developing neurons and astroglia. J Neuroinflammation. 2014; 11:Article 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.LeDuc PR, Messner WC, Wikswo JP. How do control-based approaches enter into biology? Annu Rev Biomed Eng. 2011; 13:369–96 [DOI] [PubMed] [Google Scholar]
- 147.Wikswo JP, Prokop A, Baudenbacher F, Cliffel D, Csukas B, Velkovsky M. Engineering challenges of BioNEMS: the integration of microfluidics, and micro- and nanodevices, models, and external control for systems biology. IEE Proc Nanobiotechnol. 2006; 153:81–101 [DOI] [PubMed] [Google Scholar]
- 148.Velkovsky M, Snider R, Cliffel DE, Wikswo JP. Modeling the measurements of cellular fluxes in microbioreactor devices using thin enzyme electrodes. J Math Chem. 2011; 49:251–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lima EA, Snider RM, Reiserer RS, McKenzie JR, Kimmel DW, Eklund SE, Wikswo JP, Cliffel DE. Multichamber multipotentiostat system for cellular microphysiometry. Sens Actuators B Chem. 2014; 204:536–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bongard J, Zykov V, Lipson H. Resilient machines through continuous self-modeling. Science. 2006; 314:1118–21 [DOI] [PubMed] [Google Scholar]
- 151.Schmidt M, Lipson H. Distilling free-form natural laws from experimental data. Science. 2009; 324:81–5 [DOI] [PubMed] [Google Scholar]
- 152.Schmidt MD, Vallabhajosyula RR, Jenkins JW, Hood JE, Soni AS, Wikswo JP, Lipson H. Automated refinement and inference of analytical models for metabolic networks. Phys Biol. 2011; 8:055011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Block FE III, Samson PC, Wikswo JP, inventors; Vanderbilt University, Nashville TN, assignee. Normally Closed Microvalve and Applications of the Same. US Patent 9,618,129 B2. Apr. 11, 2017.
- 154.Wikswo JP, Markov DA, Reiserer RS, inventors; Vanderbilt University, Nashville TN, assignee. Multicompartment Layered and Stackable Microfluidic Bioreactors and Applications of Same. International Pub. No. WO 2017/091718 A1. June 1, 2017.
- 155.Wikswo JP, Cliffel DE, Markov DA, McLean JA, McCawley LJ, Samson PC, Reiserer RS, Block FE, McKenzie JR., inventors; Vanderbilt University, Nashville TN, assignee. Integrated Organ-on-Chip System and Applications of the Same. International Pub. No. WO 2013/086505 A1. June 13, 2013.
- 156.Wikswo JP, Markov DA, Samson PC, Block III FE, Schaffer DK, Reiserer RS, inventors; Vanderbilt University, Nashville TN, assignee. Interconnections of Multiple Perfused Engineered Tissue Constructs and Microbioreactors, Multi-Microformulators and Applications of the Same. US Pub. No. 20170081625 A1. Mar. 23, 2017.
- 157.Gould PA, Hoang LT, Scherrer JR, Matloff WJ, Seale KT, Curtis EL, Schaffer DK, Hall DJ, Kole A, Reiserer RS, Tidwell H, Samson PC, Wikswo JP, inventors; Vanderbilt University, Nashville TN, assignee. Peristaltic Micropump and Related Systems and Methods. US Pub. No. 2013/0287613 A1. Oct. 31, 2013.
- 158.Yildiz BO, Suchard MA, Wong ML, McCann SM, Licinio J. Alterations in the dynamics of circulating ghrelin, adiponectin, and leptin in human obesity. Proc Natl Acad Sci USA. 2004; 101:10434–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Simon C, Brandenberger G. Ultradian oscillations of insulin secretion in humans. Diabetes. 2002; 51:S258–S61 [DOI] [PubMed] [Google Scholar]
- 160.Veldhuis JD, Iranmanesh A, Johnson ML, Lizarralde G. 24-hour rhythms in plasma concentrations of adenohypophyseal hormones are generated by distinct amplitude and/or frequency modulation of underlying pituitary secretory bursts. J Clin Endocrinol Metab. 1990; 71:1616–23 [DOI] [PubMed] [Google Scholar]
- 161.Nindl BC, Hymer WC, Deaver DR, Kraemer WJ. Growth hormone pulsatility profile characteristics following acute heavy resistance exercise. J Appl Physiol. 2001; 91:163–72 [DOI] [PubMed] [Google Scholar]
- 162.Yoon DY, Mansukhani NA, Stubbs VC, Helenowski IB, Woodruff TK, Kibbe MR. Sex bias exists in basic science and translational surgical research. Surgery. 2014; 156:508–16 [DOI] [PubMed] [Google Scholar]
- 163.Prodanov L, Jindal R, Bale SS, Hegde M, McCarty WJ, Golberg I, Bhushan A, Yarmush ML, Usta OB. Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol Bioeng. 2016; 113:241–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ribas-Latre A, Eckel-Mahan K. Interdependence of nutrient metabolism and the circadian clock system: Importance for metabolic health. Mol Metab. 2016; 5:133–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Putker M, O'Neill JS. Reciprocal control of the circadian clock and cellular redox state - a critical appraisal. Mol Cells. 2016; 39:6–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.van Coevorden A, Mockel J, Laurent E, Kerkhofs M, L'Hermite-Baleriaux M, Decoster C, Neve P, Van Cauter E. Neuroendocrine rhythms and sleep in aging men. Am J Physiol Endocrinol Metab. 1991; 260:E651–E61 [DOI] [PubMed] [Google Scholar]
- 167.Copinschi G, Challet E. Endocrine rhythms, the sleep-wake cycle, and biological clocks In: Groot LJD, Kretser DMd, Giudice LC, Grossman AB, Melmed S, Potts JT, Weir GC, editors. Endocrinology: Adult and Pediatric, 7th ed Philadelphia: W.B. Saunders, 2016, p. 147–73 [Google Scholar]
- 168.Fang J, Cai C, Wang Q, Lin P, Zhao Z, Cheng F. Systems pharmacology-based discovery of natural products for precision oncology through targeting cancer mutated genes. CPT Pharmacometrics Syst Pharmacol. 2017; 6:177–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.