Abstract
Ceramides are central intermediates of sphingolipid metabolism with dual roles as mediators of cellular stress signaling and mitochondrial apoptosis. How ceramides exert their cytotoxic effects is unclear and their poor solubility in water hampers a search for specific protein interaction partners. Here, we report the application of a photoactivatable and clickable ceramide analog, pacCer, to identify ceramide binding proteins and unravel the structural basis by which these proteins recognize ceramide. Besides capturing ceramide transfer protein (CERT) from a complex proteome, our approach yielded CERT-related steroidogenic acute regulatory protein D7 (StarD7) as novel ceramide binding protein. Previous work revealed that StarD7 is required for efficient mitochondrial import of phosphatidylcholine (PC) and serves a critical role in mitochondrial function and morphology. Combining site-directed mutagenesis and photoaffinity labeling experiments, we demonstrate that the steroidogenic acute regulatory transfer domain of StarD7 harbors a common binding site for PC and ceramide. While StarD7 lacks robust ceramide transfer activity in vitro, we find that its ability to shuttle PC between model membranes is specifically affected by ceramides. Besides demonstrating the suitability of pacCer as a tool to hunt for ceramide binding proteins, our data point at StarD7 as a candidate effector protein by which ceramides may exert part of their mitochondria-mediated cytotoxic effects.
Keywords: click chemistry, lipid transfer protein, mitochondria, phosphatidylcholine, photoaffinity labeling, ceramide transfer protein, steroidogenic acute regulatory protein D7
Sphingolipids are abundant components of eukaryotic membranes that participate in a wide array of cellular processes by modulating vital physical membrane properties and as signaling molecules in responses to physiological cues and stresses (1–4). Notably ceramides, the central intermediates of sphingolipid metabolism, receive considerable attention as key mediators of anti-proliferative cellular responses including apoptosis, autophagy, cell cycle arrest, and senescence (5–7). Various stress stimuli, such as TNFα, ionizing radiation, and chemotherapeutic drugs, trigger ceramide accumulation through activation of sphingomyelinases, stimulation of de novo ceramide synthesis, or both (8–10). Interventions that suppress ceramide accumulation render cells resistant to these stress-inducing agents, while their biological effects can be partially mimicked by addition of exogenous ceramides (11).
Despite numerous reports on cellular processes controlled by ceramides, the mechanisms by which ceramides exert their signaling functions are unclear. It has been suggested that ceramides can generate or stabilize lipid microdomains that function as platforms for the recruitment of other signaling molecules (12, 13). Ceramides can also form stable channels in planar membranes, and it has been suggested that the pore-forming activity of ceramides promotes the cytosolic release of cytochrome c and other apoptogenic proteins during the execution phase of mitochondrial apoptosis (14, 15). An alternative mechanism of ceramide signaling is through direct interaction with target proteins. However, only a few specific ceramide binding proteins have been described to date. These include the ceramide transfer protein CERT (16), the kinase suppressor of Ras (17), protein kinase c-Raf (18), cathepsin D (19), and protein phosphatase 2A (PP2A) inhibitor, SET (20). Identification of additional ceramide binding proteins is desirable, as this would likely lead to further mechanistic insights into ceramide-mediated signaling pathways and expand opportunities for exploiting their therapeutic potential.
Several proteome-wide methods have been developed to detect specific lipid-protein interactions, which include the application of protein microarrays in a screen for novel phosphoinositide binding proteins (21). In an inverted setup, lipid strips have been used to obtain lipid-binding fingerprints for a large number of proteins with predicted lipid binding domains (22). Column-based affinity purification strategies with lipids immobilized onto magnetic beads have also been utilized (23, 24). However, a major drawback of screens using lipids immobilized on solid supports is that such lipids are not presented in their natural state. Moreover, interactions where the lipid has to enter a deep hydrophobic binding pocket within the protein are likely to be missed. In recent years, bifunctional lipid analogs have emerged as promising new tools to circumvent some of these disadvantages, enabling global profiling of lipid-protein interactions in living cells (25–28). Bifunctional lipids possess a small diazirine group to allow photo-cross-linking with their protein interaction partners and a terminal alkyne or clickable group for functionalization. Biotinylation of cross-linked lipid-protein complexes enables their affinity purification and identification, while their subcellular location can be visualized by click reaction with a fluorophore (29). A recent study combined the advantages of bifunctional and coumarin caged lipids to facilitate identification of protein binding partners of the signaling lipids, sphingosine and diacylglycerol (DAG) (30).
In the present study, we report the synthesis and application of a bifunctional ceramide analog, pacCer, to search for novel ceramide binding proteins. Besides proteins involved in DNA damage response pathways, protein ubiquitination, membrane trafficking, and signal transduction, our approach yielded CERT and the CERT-related phosphatidylcholine (PC) transfer protein, steroidogenic acute regulatory protein D7 (StarD7). Using molecular modeling in combination with photoaffinity labeling and lipid transfer assays, we demonstrate that StarD7 harbors a lipid-binding pocket with dual specificity for ceramide and PC, and pinpoint structural determinants of lipid recognition. As StarD7 is required for normal respiratory activity and cristae structure of mitochondria (31, 32), its ability to bind ceramides may be relevant to the mechanism by which ceramides mediate their cytotoxic effects.
MATERIALS AND METHODS
Reagents and antibodies
The 1-palmitoyl-2-oleoyl-sn-glycerol (DAG, 16:0/18:1; catalog number 800815), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), POPC, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-lactosyl (lactosyl-PE), C18-ceramide (d18:1/18:0; catalog number 860518), and C16:0-ceramide (d18:1/16:0; catalog number 860516) were obtained from Avanti Polar Lipids. D-erythro-sphingosine was obtained from Enzo Biochem. Alexa Fluor647-N3 and biotin-N3 were from Thermo Fischer Scientific. Other fine chemicals were from Sigma-Aldrich. The antibodies used were: rabbit polyclonal anti-StarD7 (1:1,000; catalog number 15689-1-AP, Proteintech), mouse monoclonal anti-PARP-1 (1:1,000; catalog number sc8007, Santa Cruz), mouse monoclonal anti-mitochondrial surface protein p60 (1:1,000; catalog number MAB1273, Millipore), affinity-purified rabbit polyclonal anti-SM synthase-related protein (SMSr) antibody [1:1,000; (33)], mouse monoclonal anti-biotin antibody conjugated to horseradish peroxidase (1:1,000; catalog number 200-032-211, Jackson ImmunoResearch), goat anti-mouse and goat anti-rabbit IgG conjugated to horseradish peroxidase (1:1,000; catalog numbers 31430 and 31460, respectively, Thermo Fischer Scientific).
Synthesis of bifunctional lipid analogs
A 15 carbon-long FA containing a photo-activatable diazerine and clickable alkyne group, pacFA, was synthesized in three steps from commercially available educts, as described in (25) (see the supplemental information for further details). Next, pacFA was coupled to D-erythro-sphingosine using a combination of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) and hydroxybenzotriazole as condensing reagents, yielding the photo-activatable and clickable C15-ceramide analog, pacCer (85% overall yield). pacPC was synthesized starting from 1-oleoyl-2-hydroxy-sn-glycero-3-phosphocholine (Avanti Polar Lipids) and pacFA under the action of N,N-dicyclohexylcarbodiimide and 4-dimethylaminopyridine (DMAP) with satisfactory yield (39%). pacDAG was synthesized in three steps starting from 1-oleoyl-sn-glycerol (Santa Cruz Biotechnology). First, the primary HO-group was protected with the triphenylmethyl protecting group (trityl-chloride/pyridine; 92% overall yield). The glycerol obtained was coupled with the pacFA using EDCI/DMAP activation (58% overall yield). The final deprotection step was achieved using trifluoroacetic acid to generate pacDAG (28% overall yield). pacPE was synthesized in three steps starting from 1-oleoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine (Avanti Polar Lipids). First, the amino-group was protected with the tert-butoxycarbonyl protecting group (di-tert-butyl dicarbonate/triethylamine; 98% overall yield). The ethanolamine obtained was coupled with pacFA using EDCI/DMAP activation in a good yield (52%). The final deprotection step was achieved with trifluoroacetic acid to generate pacPE (35% overall yield). pacSM was synthesized starting from sphingosylphosphorylcholine (lyso-SM d18:1; Avanti Polar Lipids) and pacFA under the action of EDCI/hydroxybenzotriazole (78% overall yield). pacGlcCer was synthesized from 1-β-D-glucosylsphingosine (Matreya) and pacFA in the presence of triphenylphosphine and dithiopyridine, essentially as described by (34) (62% overall yield). The synthesis of C1-deoxy-pacCer, C3-deoxy-pacCer, and C3-deoxy-N-methyl-pacCer is described in the supplemental information. All synthetic compounds were purified by thin-layer chromatography to a high degree (purity >98%) and their structures were confirmed by 1H and 13C NMR and ESI MS.
DNA constructs
Bacterial maltose binding protein (MBP) expression construct pMAL-c5X was obtained from New England Biolabs. A DNA insert encoding the steroidogenic acute regulatory transfer (START) domain of human CERT was amplified from cDNA (kindly provided by K. Hanada) and cloned into the BamHI and NotI restriction sites of bacterial expression vector pET24a(+). DNA inserts encoding full-length human StarD7 (StarD7 isoform-I) and StarD7 lacking the N-terminal mitochondrial targeting sequence [StarD7 isoform-II; (35)] were PCR amplified from IMAGE clone 3842611 and cloned into the SalI and XhoI restriction sites of pET24a(+). DNA inserts encoding full-length human StarD2 and StarD10 were PCR amplified from IMAGE clones 4575824 and 4301295, respectively, and cloned into the EcoRI and XhoI restriction sites of pET24a(+). Single amino acid substitutions were introduced by site-directed mutagenesis according to the QuickChangeII™ manual (Agilent Technologies) with modifications. All expression constructs were verified by DNA sequencing.
Production of recombinant protein
Escherichia coli BL21(DH3) pLysS cells transformed with the expression construct were grown in LB medium supplemented with 0.1 mM isopropyl-D-thiogalactoside for 2 h at 30°C. MBP was purified from cell lysates in-batch using amylose resin (New England Biolabs) according to the manufacturer’s instructions. Poly-His-tagged proteins were purified by Ni2+-NTA affinity (Qiagen) using an in-batch protocol, eluted in 50 mM Tris/HCl (pH 7,4), 300 mM NaCl, 300 mM imidazole, 2.5 mM β-mercaptoethanol, and protease inhibitor cocktail (150 nM aprotinin, 1 μM leupeptin, 1.5 μM pepstatin, 7.5 μM antipain, and 1 mM benzamidine), supplemented with 10% glycerol (volume), aliquoted, and stored at −80°C until further use. Protein concentrations were determined by SDS-PAGE and Coomassie staining using BSA as reference protein. StarD7 isoform-I was expressed and purified as described in (25), and used in the experiments shown in Fig. 3. All other experiments were performed with StarD7 isoform-II.
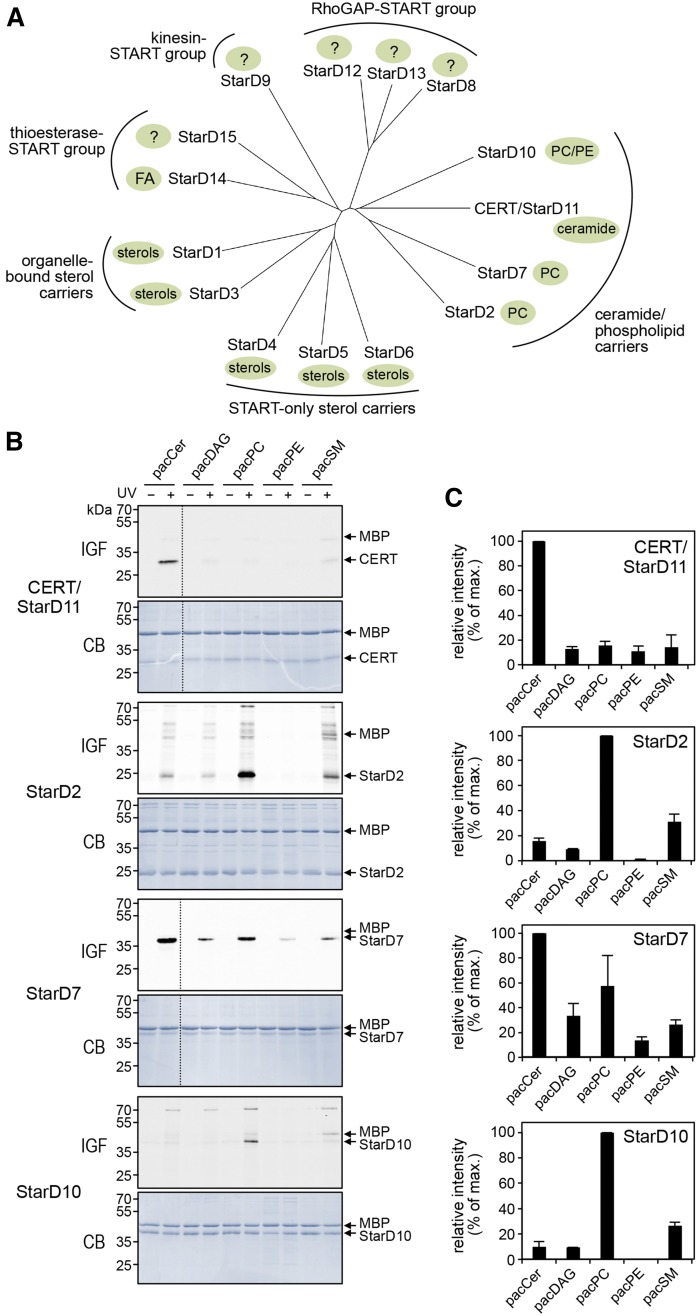
Fig. 3.
Lipid specificity profiling of CERT-related StarD proteins. A: Phylogenetic tree of the human START domain-containing family of lipid transfer proteins, grouped by their known lipid ligands and additional functional domains. Note that the closest relatives of CERT (StarD11) are the PC-carrier proteins, StarD2, StarD7, and StarD10. The phylogenetic tree was produced with ClustalW and Phylodendron using protein sequences of START domains predicted by PROSITE in UniProt accession numbers: StarD1 (P49675), StarD2 (Q9UKL6-1), StarD3 (Q14849), StarD4 (Q96DR4), StarD5 (Q9NSY2), StarD6 (P59095), StarD7 (Q9NQZ5), StarD8 (Q92502), StarD9 (Q9P2P6), StarD10 (Q9Y365), CERT (Q9Y5P4), StarD12 (Q96QB1), StarD13 (Q9Y3M8), StarD14 (Q8WXI4-1), and StarD15 (Q8WYK0). B: START domains of CERT, StarD2, StarD7, and StarD10 were produced in E. coli, purified, mixed with MBP, incubated with liposomes containing 1 mol% of pacCer, pacDAG, pacPC, pacPE, or pacSM, and processed as in Fig. 2B. C: Quantitative analysis of photoaffinity labeling of START domains by pacLipids as in B. Maximum labeling intensity was set at 100%. Data are shown as the mean ± error range of two independent experiments. IGF, in-gel-fluorescence; CB, Coomassie staining.
Cell culture, RNAi, and generation of StarD7−/− cells
Human cervical carcinoma HeLa (ATCC-CCL2), Chinese hamster ovary (CHO)-K1 (ATCC-CCL-61), and mouse melanoma GM95 cells (kindly provided by Hein Sprong, University of Utrecht, The Netherlands) were grown in Dulbecco’s Modified Eagle’s medium supplemented with 4.5 g/l glucose, 10% FCS, and GlutaMAX™ (Invitrogen) at 37°C with 5% CO2. To knock out StarD7 in HeLa cells, we obtained a mix of three different CRISPR/Cas9 plasmids and the corresponding HDR plasmids from Santa Cruz (sc-405820). The StarD7-specific gRNA sequences were: A/sense, 5′-ATCCAACTAACACAGTAGCG-3′ B/sense, 5′-GCTCACCTCGGTACTGGTAA-3′ and C/sense, 5′-ACCCACCTTTACCAGTACCG-3′. HeLa cells were cotransfected with both plasmid mixes using Effectene (Qiagen) and grown for 48 h without selection. Next, the cells were grown for 2 weeks under selection pressure with 2 μg/ml puromycin. Individual drug-resistant clones were picked and analyzed for StarD7 expression by immunoblot analysis using anti-StarD7 antibody. Two independent StarD7−/− cell lines, StarD7-KO#1 and StarD7-KO#2, were used for subsequent RNAi experiments. To this end, cells were transfected with siRNA (Qiagen) using Oligofectamine reagent (Invitrogen) as described previously (36). The siRNA target sequences used were: nonsilencing RNA (siNS) (nonsense), 5′-AAUUCUCCGAACGUGUCACGU-3′, and siSMSr, 5′-CAAGAAGCUGGAAUUUCUUGC-3′. Both adherent and nonadherent cells were harvested 72 h posttransfection, washed twice in ice-cold 0.25 M sucrose, and homogenized in ice-cold IM buffer [5 mM HEPES-KOH (pH 7.0), 250 mM mannitol, and 0.5 mM EGTA] supplemented with 0.1 mM phenylmethanesulfonyl fluoride and protease inhibitor cocktail (150 nM aprotinin, 1 μM leupeptin, 1.5 μM pepstatin, 7.5 μM antipain, and 1 mM benzamidine). To this end, cells were flushed through a Balch homogenizer 20–30 times using a 2 ml syringe. Cell homogenates were centrifuged twice at 600 g maximum for 5 min at 4°C to remove nuclei. The protein concentration of postnuclear supernatants was determined by Bradford assay (Bio-Rad). Postnuclear supernatants were normalized for total protein content prior to immunoblot analysis.
Preparation of liposomes
Liposomes used in the photoaffinity experiments with cytosolic fractions were prepared in PBS (1.4 M NaCl, 27 mM KCl, 18 mM KH2PO4, and 126 mM Na2HPO4) from a mixture of egg-PC and pacLipid (95/5 mol%). Liposomes used in the photoaffinity experiments with purified recombinant proteins were prepared from a defined lipid mixture (DOPC/DOPE/pacLipid, 80/20/1 mol%) in CHCl3/methanol (9/1, v/v). For competition assays, 0.5 or 0.25 mol% pacCer was used and C16:0 ceramide was added in 10- to 40-fold molar excess at the expense of DOPC and DOPE, keeping the DOPC/DOPE ratio constant. In brief, 10 μmol of total lipid were dried in a Rotavap and the resulting lipid film was resuspended in 1 ml buffer L [50 mM Tris-HCl (pH 7.4) and 50 mM NaCl] by vigorous vortexing and sonication, yielding a 10 mM lipid suspension. Liposomes with an average diameter of ∼100 nm were obtained by sequential extrusion of the lipid suspension through 0.4, 0.2, and 0.1 micron track-etched polycarbonate membranes (Whatman-Nuclepore) using a mini-extruder (Avanti Polar Lipids). Acceptor liposomes used in lipid transfer assays were prepared in buffer L using a mixture of DOPG and DOPE (80/20 mol%). Donor liposomes were prepared using a mixture of DOPG, DOPE, lactosyl-PE, and either C16:0-ceramide or DOPC (65/16/10/10 mol%). Donor liposomes used in competition assays were prepared using a mixture of DOPG, DOPE, lactosyl-PE, DOPC, and C16:0-ceramide or DAG (69/17/10/5/10 mol%). All liposomes were stored under N2 at 4°C and used within 2–3 days after preparation.
Photoaffinity labeling of cytosolic fractions
Five 15 cm dishes each of GM95 and HeLa cells and two 15 cm dishes of CHO cells were resuspended in 2 ml of ice-cold lysis buffer [50 mM Tris (pH 6.8), 1 mM EDTA, 0.3 M sucrose, 1 mM PMSF, and 1× protease inhibitor cocktail). Cells were homogenized by passing them through a 26Gx1″ (0.45 × 25 mm) needle using a 1 ml syringe. The suspension was centrifuged at 100,000 g for 1 h at 4°C to remove cell debris, nuclei, and membranes. The protein concentration in the obtained cytosol was determined by Bradford assay and adjusted to 1.2 mg/ml with PBS. Fifty-five microliters of cytosol were mixed with 55 μl of liposome suspension containing 5 mol% of pacLipid and incubated for 30 min at room temperature with gentle shaking. Subsequently, the samples were placed on ice and irradiated for 60 s using a 1,000 W mercury lamp equipped with a dichroic mirror and a 345 nm bandpass filter (Newport) at 30 cm distance. Protein was recovered by chloroform-methanol precipitation and the air-dried protein pellet was dissolved in 20 μl of 1% SDS in PBS with vigorous shaking for 10 min at 70°C. Click reactions were performed by adding 8 μl of a freshly prepared “click” mix {40 μl of 25 mM Tris(2-carboxyethyl)phosphine hydrochloride, 40 μl of 2.5 mM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine, 40 μl of 25 mM CuSO4, and 40 μl of 25 mM biotin-N3} per sample followed by incubation at room temperature for 2 h while shaking. After addition of 0.25 vol of 5× sample buffer [0.3 M Tris/HCl (pH 6.8), 10% SDS, 50% glycerol, 0.025% bromphenol blue, and 10% β-mercaptoethanol], samples were boiled for 5 min at 95°C and subjected to SDS-PAGE and immunoblotting or Coomassie staining.
For the identification of ceramide binding proteins, a cytosolic fraction was prepared from twenty 15 cm dishes of GM95 cells and diluted to a protein concentration of 1.4 mg/ml in PBS. Four hundred and fifty microliters of cytosol were mixed with 450 μl of a liposome suspension containing 5 mol% of pacCer or pacGlcCer and incubated for 30 min at room temperature with gentle shaking. Samples were split into four portions of 200 μl, UV irradiated, and subjected to chloroform-methanol precipitation as above. Protein pellets were combined, resuspended in 400 μl 1% SDS in PBS, and solubilized for 10 min at 70°C. Click reactions were performed by adding 80 μl of freshly prepared click mix containing biotin-N3, as above. Samples were split in two portions of 240 μl and subjected to chloroform-methanol precipitation twice. Protein pellets were resuspended in 200 μl of 1% SDS in PBS, solubilized by vigorous shaking for 10 min at 70°C, and then diluted 5-fold in PBS. After centrifugation at 20,000 g for 1 min at room temperature, supernatants were collected and combined. Supernatant (1,600 μl) was mixed with 50 μl of a 50% slurry of NeutrAvidin beads (NeutrAvidin™ agarose resin; Thermo Scientific) equilibrated in 0.2% SDS in PBS and incubated at room temperature with rotation. The beads were collected by centrifugation (100 g, 1 min, room temperature) and washed three times with 1 ml 0.1% SDS in PBS and three times with 1 ml of PBS. To elute bound proteins, the beads were incubated in SDS-PAGE sample buffer for 5 min at 95°C. The eluates were analyzed by SDS-PAGE and Coomassie staining. Lanes of interest were cut into 10 equal sections, cut from the gel, and protein in each section was subjected to trypsin digestion and peptides were analyzed by LC-MS/MS using an LTQ-Orbitrap mass spectrometer (Thermo Scientific) connected to an Agilent 1200 series nano LC system, as described in (25). The following criteria were applied to select for pacCer-modified cytosolic proteins: i) no spectral counts in the controls (no pacLipid/±UV, pacCer/−UV) and three or more spectral counts in the pacCer photoaffinity-labeled samples (pacCer/+UV) for experiment #1 or experiment #2; ii) a 5-fold enrichment in spectral counts for pacCer photoaffinity-labeled samples (pacCer/+UV) in comparison with the controls for experiment #1 or #2; and iii) a spectral count ratio for pacCer/+UV over pacGlcCer/+UV samples of one or more for experiment #1 or experiment #2. High-confidence ceramide binding proteins were selected based on a spectral count ratio for pacCer/+UV over pacGlcCer/+UV samples of two or more for both experiment #1 and experiment #2.
Photoaffinity labeling of recombinant protein
Recombinant proteins were diluted in 50 mM Tris-HCl (pH 7.4) and 300 mM NaCl to a final concentration of 0.1 μg/μl. Twenty-five microliters of this solution were combined with 25 μl of 0.1 μg/μl MBP, which served as internal negative control. The sample was then mixed with 50 μl of a liposome suspension containing 1 mol% pacLipid and incubated for 30 min at 37°C with gentle shaking. For UV cross-linking, samples were transferred into a precooled 96-well plate on ice and irradiated for 90 s, as described above. The samples were transferred to Eppendorf tubes and subjected to chloroform-methanol precipitation after addition of 20 μg soybean trypsin inhibitor as carrier to aid protein recovery. The protein pellet was dissolved in 100 μl of 1% SDS in PBS with vigorous shaking for 10 min at 37°C. Twenty microliters of each sample were used for click reaction for 1 h at 37°C, as described above, except that biotin-N3 was replaced by 80 μM Alexa Fluor647-N3. After addition of 0.25 vol of 5× sample buffer, samples were boiled for 5 min at 95°C, subjected to SDS-PAGE, and then analyzed by in-gel fluorescence and Coomassie staining. To this end, run gels were fixed in 40% ethanol and 10% acetic acid for 30 min and then washed in water for 1 h at room temperature. Gels were scanned with a Typhoon FLA 9500 (GE Healthcare) with a 635 nm laser and LPR filter. Fluorescence intensities were quantified using ImageQuant TL. Next, gels were stained with Coomassie and the amount of recombinant protein was determined by measuring the staining intensity using Image Lab 5.2 software. Fluorescence intensities of recombinant protein were corrected for background fluorescence in the same lane and then divided by the total amount of protein. The specific fluorescence intensity of UV-irradiated protein was determined after subtraction of fluorescence intensity of non-UV-irradiated protein.
Modeling of StarD7 lipid-binding pocket and docking studies
A database search of SWISS-MODEL (37–40) using the primary sequence of human StarD7 START (35) retrieved human StarD2 START [Protein Data Bank (PDB) 1LN1 (41)] as the closest related protein sequence (29.19% identity). The search also yielded CERT START [PDB 2E3R (42)] as a more distant related protein (16.40% identity). As both StarD2 and StarD7 are known as PC transfer proteins, the crystal structure, 1LN1, was chosen as a starting point. Three separate runs using SWISS-MODEL standard parameters were carried out, the resultant homology models were subsequently tested for anomalies by their Ramachandran plots. The best three models, one from each run, were superimposed and energy minimized using MMFF94x force field to yield the final model that was used as a reference. The homology model was tested by docking PC (16:1, 16:1) into the binding pocket using SWISS-DOCK (43, 44). The interaction maps revealed high similarity to the identified residues in the orthologous StarD2 protein. C16:0-ceramide was then docked into the refined model, as described above. Note that a significant difference in volume of the ligand-binding pockets between StarD7 (∼2,298 Å3 1LN1) and CERT (∼1,650 Å3, 2E3P and 2E3R; CHIMERA, http://www.cgl.ucsf.edu/chimera/docs/morerefs.html) resulted in different binding modes of ceramide. Only those modes that showed a mixture of hydrophilic and hydrophobic interaction were considered for this study. Protein structures were displayed using Pymol software.
Lipid transfer assay
For lipid transfer assays, 14 pmol of CERT START or 70 pmol of StarD7 isoform-II protein in 50 μl of transfer buffer [50 mM Tris-HCl (pH 7.4) and 300 mM NaCl) were mixed with 40 μl of 10 mM acceptor liposomes on ice. The reaction was started by addition of 10 μl donor liposomes followed by incubation for 20 min at 37°C while shaking mildly. To stop the reaction, 20 μl of agglutinin (2.5 mg/ml in transfer buffer) were added and samples were incubated on ice for 15 min. For control samples, the protein was mixed with acceptor liposomes as above, but kept on ice, and the donor liposomes were added just after addition of agglutinin. Donor liposomes were removed by centrifugation at 16,000 g for 5 min at 4°C, and 100 μl of the supernatant were transferred to a fresh Eppendorf tube. Either C18-ceramide (d18:1/18:0; Avanti, 860518) or DOPC were added in appropriate amounts as internal standard. Lipids were extracted by addition of 320 μl CHCl3:methanol (1:2) and 340 μl water with subsequent centrifugation at 16,000 g for 5 min at room temperature. The organic phase was collected in a fresh Eppendorf tube and the samples were subjected to a second round of extraction by addition of 100 μl CHCl3 followed by the same treatment as above. The organic phase was dried down in a centrifugal evaporator and lipids were dissolved in 10 μl of CHCl3. For MS analysis, 1 μl of superDHB matrix (50 mg/ml in 30% acetonitrile and 0.1% TFA) was spotted on a stainless steel MALDI plate and dried before spotting 1 μl per lipid sample. The spots were analyzed in a Bruker UltrafleXtreme™ MALDI-TOF/TOF instrument in positive ion mode. Peptide calibration standard II (Brucker) was used for calibration.
RESULTS
A search for cytosolic ceramide binding proteins using bifunctional ceramide analog pacCer yields CERT
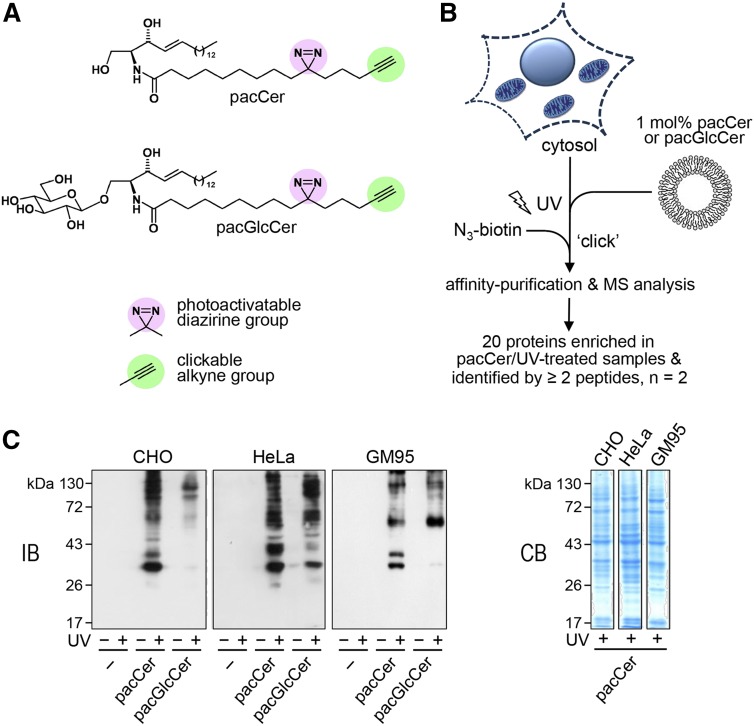
As part of our ongoing efforts to identify novel effector proteins involved in ceramide-mediated stress signaling and apoptosis, we synthesized a bifunctional analog of ceramide containing a photoactivatable diazirine and clickable alkyne group in its N-linked 15-carbon-long acyl chain (pacCer; Fig. 1A). UV irradiation of the diazirine group generates a highly reactive pacCer intermediate that can form a covalent linkage with proteins in its direct vicinity. Click chemistry is then used to react the alkyne group with a fluorophore or biotin as reporter molecule to allow visualization or affinity purification of photoaffinity labeled proteins, respectively [Fig. 1B (29)]. To test whether pacCer can be used to capture ceramide binding proteins, cytosolic fractions prepared from CHO, human cervix carcinoma HeLa, and mouse melanoma GM95 cells were incubated with liposomes containing 5 mol% of pacCer and subjected to UV cross-linking. Photoaffinity labeling experiments with cytosol and liposomes containing 5 mol% of a bifunctional analog of glucosylceramide (pacGlcCer; Fig. 1A) served as control. Following a click reaction with biotin-N3, photoaffinity-labeled cytosolic proteins were visualized by immunoblotting using an anti-biotin antibody. As shown in Fig. 1C, a subset of cytosolic proteins from each cell line could be UV-cross-linked with both lipid probes. However, some cytosolic proteins appeared to have a higher affinity for pacCer than pacGlcCer and vice versa.
Fig. 1.
Screen for ceramide binding proteins using bifunctional lipid analogs. A: Bifunctional analogs of ceramide (pacCer) and glycosylceramide (pacGlcCer) containing a photoactivatable diazirine group (pink) and clickable alkyne group (green). B: General outline of a screen for soluble ceramide binding proteins. Cytosolic fractions from mammalian cells are incubated with liposomes containing 5 mol% of pacCer or pacGlcCer and then subjected to UV irradiation. Click chemistry is then used to label the alkyne group in the pacLipid with biotin-N3 or Alexa Fluor647-N3 (not shown), allowing the respective identification or visualization of cross-linked protein-lipid complexes. C: Cytosolic fractions from CHO, HeLa, and GM95 cells were incubated with liposomes containing pacCer, pacGlcCer, or no pacLipid (−) for 30 min at room temperature, subjected to UV irradiation and then clickreacted with biotin-N3. Samples were analyzed by immunoblotting using an anti-biotin antibody (IB) or by Coomassie staining (CB).
To identify soluble proteins that preferentially bind ceramide, the cytosolic fraction derived from GM95 cells was photoaffinity labeled with pacCer or pacGlcCer and subjected to a click reaction with biotin-N3. Next, biotinylated proteins were affinity-purified using NeutrAvidin beads, separated by SDS-PAGE, subjected to in-gel tryptic digestion, and identified by MS. Two independent experiments yielded 67 proteins, in total, that were either exclusively present or enriched in affinity-purified fractions of pacCer-incubated and UV-irradiated cytosol (supplemental Table S1). Nearly 30% of these proteins (20 out of 67) were marked as high-confidence ceramide binding proteins based on a spectral count ratio for pacCer/+UV over pacGlcCer/+UV samples of two or more in both experiments (Table 1). These proteins participate in a wide range of cellular processes, including membrane trafficking (sorting nexins), signal transduction (Unc-119 proteins), DNA damage response pathways (DNA damage binding protein-1, DnaJ homolog DnaJc7), and protein ubiquitination (COP9 signalosome subunit). A possible involvement of these proteins in ceramide-dependent cellular processes is currently under investigation and will be discussed elsewhere. Reassuringly, the list of high-confidence ceramide binding proteins also included CERT, a cytosolic protein responsible for nonvesicular delivery of newly synthesized ceramides from the endoplasmic reticulum (ER) to the site of SM production in the trans-Golgi (16). The latter finding qualifies pacCer as a suitable tool to capture ceramide binding proteins from a complex proteome.
TABLE 1.
Candidate ceramide-binding proteins
| Number | Accession Number | Name | MM (kDa) | Control | pacCer #1 | pacGlcCer #1 | pacCer #2 | pacGlcCer #2 | |||||
| −UV | +UV | −UV | +UV | −UV | +UV | −UV | +UV | −UV | +UV | ||||
| 1 | IPI00321734 | Lactoylglutathione lyase Glo1 | 21 | 0 | 0 | 0 | 12 | 0 | 2 | 0 | 5 | 0 | 1 |
| 2 | IPI00230084 | Aldehyde dehydrogenase-7a1 | 59 | 0 | 0 | 0 | 14 | 0 | 0 | 1 | 9 | 2 | 1 |
| 3 | IPI00622364 | Sorting nexin-1 | 59 | 0 | 0 | 0 | 18 | 2 | 8 | 1 | 8 | 4 | 3 |
| 4 | IPI00109212 | Sorting nexin-2 | 58 | 0 | 0 | 0 | 16 | 0 | 4 | 0 | 10 | 2 | 0 |
| 5 | IPI00131315 | Unc-119 homolog A | 27 | 0 | 0 | 0 | 10 | 0 | 1 | 0 | 6 | 0 | 1 |
| 6 | IPI00225371 | Unc-119 homolog B | 28 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 5 | 0 | 0 |
| 7 | IPI00111167 | CERT | 71 | 0 | 0 | 0 | 5 | 0 | 2 | 0 | 3 | 0 | 1 |
| 8 | IPI00348414 | Ado, 2-aminoethanethiol dioxygenase | 28 | 0 | 0 | 0 | 10 | 1 | 0 | 0 | 6 | 0 | 0 |
| 9 | IPI00421223 | Tropomyosin α-4 chain | 28 | 0 | 0 | 0 | 5 | 0 | 2 | 0 | 6 | 1 | 1 |
| 10 | IPI00131870 | COP9 signalosome subunit 3 | 48 | 0 | 0 | 0 | 8 | 3 | 2 | 0 | 3 | 1 | 1 |
| 11 | IPI00134334 | Prostaglandin reductase-2 | 38 | 0 | 0 | 0 | 9 | 1 | 2 | 0 | 4 | 1 | 1 |
| 12 | IPI00314510 | Aspartoacylase-2 | 35 | 0 | 0 | 0 | 5 | 0 | 1 | 0 | 4 | 0 | 0 |
| 13 | IPI00331707 | Hydroxymethylglutaryl-CoA synthase | 58 | 0 | 0 | 0 | 12 | 2 | 4 | 0 | 2 | 1 | 0 |
| 14 | IPI00310658 | Aldo-ketoreductase-1c13 | 37 | 0 | 0 | 0 | 7 | 0 | 1 | 0 | 4 | 1 | 0 |
| 15 | IPI00111959 | CTP synthase-1 | 67 | 0 | 0 | 0 | 5 | 0 | 2 | 0 | 3 | 1 | 0 |
| 16 | IPI00114818 | Sec14-like protein-2 | 46 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | 0 |
| 17 | IPI00316740 | DNA damage binding protein-1 | 127 | 0 | 0 | 0 | 16 | 0 | 8 | 0 | 3 | 1 | 0 |
| 18 | IPI00223861 | Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | 55 | 0 | 0 | 0 | 6 | 0 | 1 | 0 | 3 | 0 | 0 |
| 19 | IPI00128880 | Importin-4 | 119 | 0 | 1 | 0 | 18 | 1 | 8 | 1 | 6 | 3 | 3 |
| 20 | IPI00331385 | DnaJ homolog DnaJc7 | 65 | 0 | 0 | 0 | 10 | 0 | 3 | 0 | 2 | 2 | 0 |
MM, molecular mass.
Probing the lipid-binding pocket of CERT using bifunctional lipid analogs
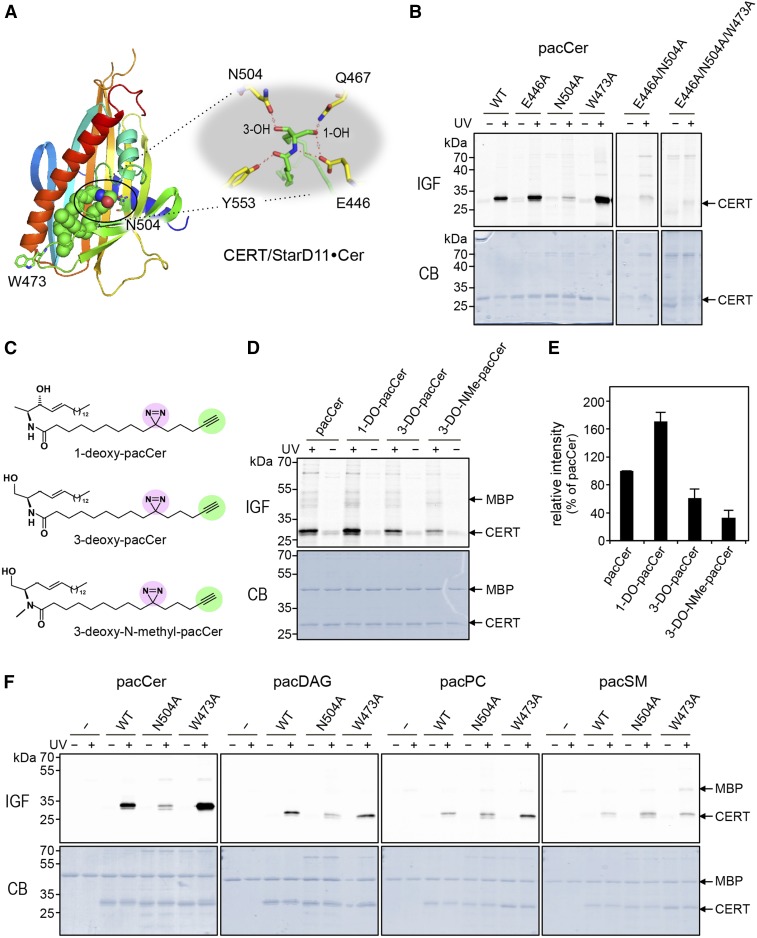
CERT, also referred to as StarD11, is a member of the START domain containing proteins (45). Members of this family are defined by the presence of a conserved ∼210 amino acid sequence that folds into an α/β helix-grip structure forming a hydrophobic cavity for ligand binding. A crystal structure of the START domain of CERT with one ceramide molecule buried in its amphiphilic cavity has been solved at 1.40 Å resolution (42). At the far end of the cavity, the amide and hydroxyl groups of ceramide form a hydrogen bond network with specific amino acid residues that play key roles in ceramide recognition. For instance, the carboxy-group of Glu446 forms two hydrogen bonds with the amide-nitrogen and C1-hydroxyl group of ceramide. In addition, the oxygen atom of Asn504 forms a hydrogen bond with the C3-hydroxyl group of ceramide (Fig. 2A). This led us to examine the impact of alanine substitutions of these residues on pacCer labeling of CERT START produced in E. coli. Contrary to recombinant MBP, CERT START could be readily UV-cross-linked with pacCer (Fig. 2B, F). Substitution of Glu446 with Ala had no obvious impact on pacCer labeling of the protein. In contrast, substitution of Asn504 for Ala virtually abolished photoaffinity labeling, also in combination with the E446A substitution (Fig. 2B, F). These results are largely consistent with the previous observation that the ceramide extraction activity of recombinant CERT START is severely compromised by an N504A substitution and only partially affected by an E446A substitution (42). Interestingly, Ala substitution of a Trp residue exposed on the protein’s exterior near the entry site of the amphiphilic cavity, i.e., Trp473, resulted in a marked increase in pacCer labeling. However, pacCer labeling of CERT STARTW473A was abolished upon substitution of Glu446 and Asn504 for Ala (Fig. 2B).
Fig. 2.
Probing the lipid-binding pocket of CERT using bifunctional lipid analogs. A: Structure of the START domain of CERT in complex with ceramide (PDB 2E3O). The START domain is displayed as a rainbow-colored ribbon representation and the ceramide molecule as spheres, with green, red, and blue color representing C, O, and N atoms, respectively. The small inset highlights residues involved in coordinating the ceramide molecule in the lipid-binding pocket. B: WT and point-mutants of CERT START were produced in E. coli, purified, and then incubated with liposomes containing 1 mol% of pacCer for 30 min at 37°C. Samples were either UV-irradiated (+) or kept in the dark (−), clickreacted with Alexa Fluor647-N3, and then analyzed by in-gel-fluorescence (IGF) and Coomassie staining (CB). C: Bifunctional analogs of C1-deoxy-ceramide (1-DO-pacCer), C3-deoxy-ceramide (3-DO-pacCer), and C3-deoxy-N-methyl-ceramide (3-DO-NMe-pacCer). D: WT CERT START was mixed with maltose-binding protein (MBP), incubated with liposomes containing 1 mol% of pacCer, 1-DO-pacCer, 3-DO-pacCer, or 3-DO-NMe-pacCer, and processed as in B. E: Quantitative analysis of photoaffinity labeling of CERT START by deoxy derivatives of pacCer relative to control (pacCer). The labeling intensity of CERT START by pacCer was set at 100%. Data are shown as the mean ± SD of three independent experiments. F: WT and point-mutants of CERT START were mixed with MBP, incubated with liposomes containing 1 mol% of pacCer, pacDAG, pacPC, or pacSM, and processed as in B.
Together, the above data indicate that photoaffinity labeling of CERT START with pacCer occurs within the protein’s lipid-binding pocket and that hydrogen bonding between Asn504 and the C3-hydroxyl of pacCer is particularly critical for stabilizing the protein-lipid interaction. Indeed, removal of the C3-hydroxyl group strongly reduced the ability of pacCer to label CERT START, whereas removal of the C1-hydroxyl group actually enhanced photoaffinity labeling (Fig. 2C–E). Furthermore, UV-cross-linking experiments with bifunctional analogs of DAG (pacDAG), phosphatidylethanolamine (pacPE), phosphotidylcholine (pacPC), and SM (pacSM) revealed that photoaffinity labeling of CERT START with pacCer is specific (Fig. 2F). Altogether, our findings demonstrate the suitability of pacCer as a chemical probe to trace ceramide binding proteins and map their lipid-binding pocket.
Lipid specificity profiling of CERT-related proteins indicates that StarD7 binds ceramide
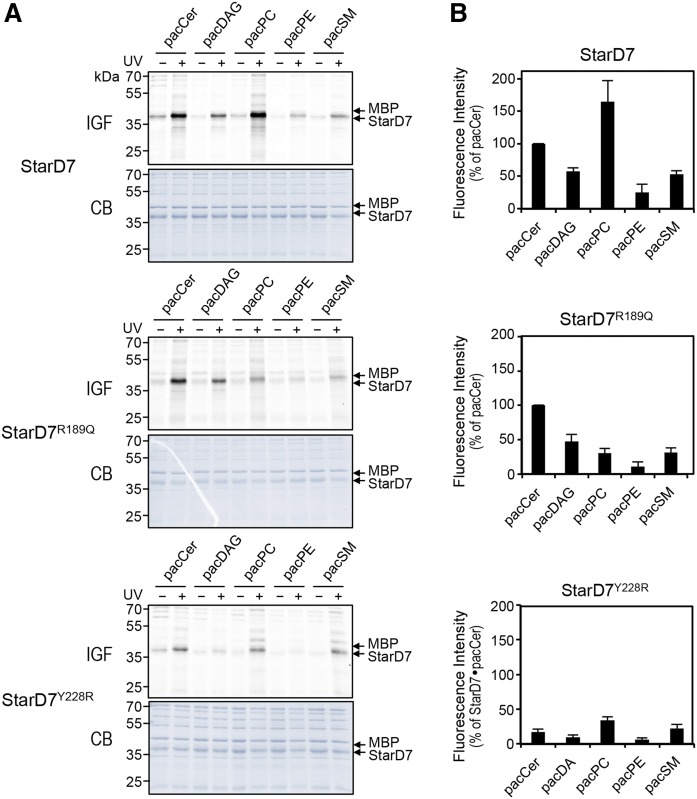
The human genome encodes 15 START domain-containing proteins that can be categorized into six subfamilies based on sequence homology within the START domain (45). According to this phylogenetic classification, proteins in the same clade mainly show specificity toward a common lipid ligand (Fig. 3A). As the lipid ligands of StarD9 and members of the Rho-GAP START group (i.e., StarD8, StarD12, and StarD13) are not known, we first determined whether any of these putative lipid carriers bind ceramides. To this end, we produced their START domains in E. coli and carried out pacCer labeling experiments on the purified recombinant proteins in the presence of MBP as internal control. This revealed that, contrary to CERT START, none of these proteins displayed any notable affinity for ceramide (data not shown). We then asked whether any of the START domains of the CERT-related phospholipid carrier proteins (i.e., StarD2, StarD7, and StarD10) bind ceramide in addition to their phospholipid ligand. As expected, START domains of the PC transfer proteins, StarD2 [PCTP (46)] and StarD10 (47) preferentially labeled with pacPC over all other bifunctional lipid analogs tested (Fig. 3B, C). These domains could be labeled only modestly with pacSM and lacked any pronounced affinity for pacCer, pacDAG, or pacPE, even though StarD10 has been shown to possess dual specificity lipid transfer activity for PC and PE in vitro (47). Strikingly, the START domain of StarD7, which was so far thought to only bind PC (35), consistently displayed a very strong photoaffinity labeling signal with pacCer (Fig. 3B, C). Photoaffinity labeling of StarD7 START with pacPC was also pronounced, while pacDAG, pacSM, and pacPE each gave rise to substantially lower labeling intensities. As PC and ceramide are structurally very different lipids, we next asked whether their interaction with StarD7 involves a shared lipid-binding pocket.
The lipid-binding pocket of StarD7 has dual specificity for PC and ceramide
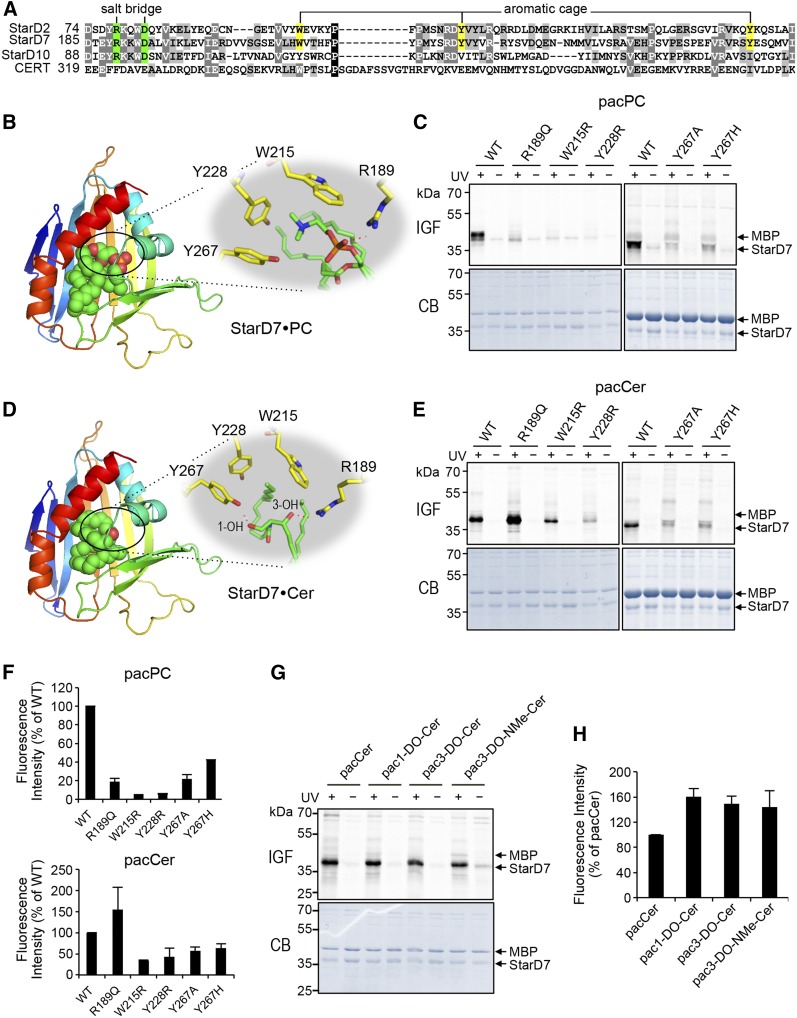
As there is no detailed structural information on the START domain of StarD7, we created a 3D homology model based on the crystal structures of the closely-related START domains of CERT [PDB 2E3R (42)] and PC transfer protein StarD2 [PDB 1LN1 (41)]. StarD7 START shares 17% and 28% sequence identity with the START domains of CERT and StarD2, respectively, whereas the latter two domains share 22% sequence identity. Next, the structure of C16:0-ceramide or di-palmitoleoyl-PC was docked to the StarD7 START lipid-binding pocket (see the Materials and Methods for further details). In the PC-bound model, a conserved Arg residue (Arg189) makes direct contact to the lipid’s phosphoryl group, probably by forming a salt bridge with the help of Asp193 (Fig. 4A, B), in line with what has been reported for PC when bound to StarD2 (41). Moreover, the quaternary amine of PC interacts with Trp215 and Tyr228, which likely form part of an aromatic cage similar to the one described in the START domain of StarD2 (Fig. 4A, B). In StarD10, the Arg-Asp pair is conserved and embedded in a consensus sequence (YRKKWD) that is also present in StarD2 and StarD7, but absent in CERT (Fig. 4A). Contrary to this, the residues that form the aromatic cage are not conserved in StarD10, which might explain why pacPC labeling of this protein is overall much less efficient than for StarD2 and StarD7 (Fig. 3B).
Fig. 4.
The lipid-binding pocket of StarD7 has dual specificity for PC and ceramide. A: Sequence alignment of lipid-binding regions in the START domains of StarD2, StarD7, StarD10, and CERT. The residues predicted to form a salt bridge involved in coordinating the phosphoryl-group of PC are marked in green. The residues predicted to form an aromatic cage around the quaternary amine of PC are marked in yellow. B: Model of the START domain of StarD7 in complex with PC. The START domain is displayed in a rainbow-colored ribbon representation and the PC molecule as spheres with green, red, orange, and blue color representing C, O, P, and N atoms, respectively. The small inset highlights residues involved in coordinating the PC molecule in the lipid-binding pocket. See the text for further details. C: WT and point-mutants of StarD7 were produced in E. coli, purified, mixed with MBP, incubated with liposomes containing 1 mol% pacPC, and processed as in Fig. 2B. D: Model of the START domain of StarD7 in complex with ceramide with color code as in B. See the text for further details. E: WT and point-mutants of StarD7 were mixed with MBP, incubated with liposomes containing 1 mol% pacer, and processed as in Fig. 2B. F: Quantitative analysis of photoaffinity labeling of StarD7 by pacLipids as in C and E. The labeling intensity of WT-StarD7 was set at 100%. Data are shown as the mean ± error range of two independent experiments. G: WT-StarD7 and MBP were mixed, incubated with liposomes containing 1 mol% of pacCer, 1-DO-pacCer, 3-DO-pacCer, or 3-DO-NMe-pacCer, and processed as in Fig. 2B. H: Quantitative analysis of photoaffinity labeling of StarD7 by pacLipids as in G. The labeling intensity of WT-StarD7 by pacCer was set at 100%. Data are shown as the mean ± SD of three independent experiments. IGF, in-gel-fluorescence; CB, Coomassie staining.
To validate our model of the StarD7 PC binding site, we next substituted Arg189, Trp215, and Tyr228 with other residues using site-directed mutagenesis and then analyzed the impact of each substitution on PC binding by photoaffinity labeling of the mutant proteins with pacPC. Substitution of Arg189 with Gln (R189Q) caused a major (∼80%) reduction in pacPC labeling, indicating that Arg189 is a critical determinant of PC binding by StarD7 (Fig. 4C, F). Substitution of Trp215 or Tyr228 with Arg (W215R, Y228R) in each case essentially abolished pacPC labeling. The marked impact of the latter substitutions is likely due to the introduction of a positive charge close to the quaternary amine within the PC head group, which would strongly interfere with its binding. In our initial model, Tyr267 did not show up directly as being part of the PC binding pocket in StarD7, but was nevertheless assumed to be part of an aromatic cage around the quaternary amine of PC, analogous to how StarD2 binds PC (41). To verify this assumption, we tried a slightly different approach by substituting Tyr267 on the one hand for a neutral residue, i.e., Ala, and on the other hand for another aromatic, but charged, residue, i.e., His. Substitution of Tyr276 with Ala (Y267A) reduced the intensity of pacPC labeling by 80% as compared with WT, while the Y267H substitution was slightly less effective (Fig. 4C, F). Together, these data indicate that Trp215, Tyr228, and Tyr276 in StarD7 likely form an aromatic cage that coordinates the quaternary amine of the PC head group, whereas Arg189 stabilizes PC binding by forming a salt bridge with the lipid’s phosphoryl group.
In the ceramide-bound model, Arg189 is predicted to also contribute to ceramide binding by forming a hydrogen bond with the lipid’s C3-hydroxyl group (Fig. 4D). However, substitution of Arg189 with Gln (R189Q), if anything, enhanced photoaffinity labeling of StarD7 START with pacCer (Fig. 4E, F), while having either no or a slightly negative impact on labeling with pacDAG, pacPE, and pacSM (Fig. 5). We reasoned that the C3-hydroxyl group of ceramide might also form a hydrogen bond with a glutamine at position 189. Instead, we found that the C3-hydroxyl group is fully dispensable for ceramide binding, as StarD7 START could be equally well labeled with pacCer, C3-deoxy-pacCer, or C3-deoxy-N-methyl-pacCer (Fig. 4G, H). This indicates that Arg189, a critical determinant of PC binding, does not significantly contribute to ceramide binding. In contrast, substitution of any of the residues that form the aromatic cage (i.e., Trp215, Tyr228, and Tyr267) clearly reduced pacCer labeling, with the introduction of a positive charge (i.e., W215R, Y228R) having the strongest impact (Fig. 4E, F). Substitution of Tyr228 for Arg also reduced pacDAG labeling (Fig. 5), suggesting that StarD7 START can also accommodate DAG. However, the efficiency of pacCer labeling consistently exceeded that of pacDAG. From this we conclude that StarD7 has a significantly higher affinity for ceramide than for DAG. Even though Tyr267 was predicted to participate in hydrogen bonding with the C1-hydroxyl group of ceramide (Fig. 4D), removal of this group did not reduce ceramide binding, as judged from our photoaffinity labeling experiments (Fig. 4G, H).
Fig. 5.
Lipid specificity profiling of StarD7 mutants deficient in PC and ceramide binding. A: WT-StarD7, PC-binding deficient mutant StarD7R189Q, and PC/ceramide-binding deficient mutant StarD7Y228R were mixed with MBP, incubated with liposomes containing 1 mol% pacCer, pacDAG, pacPC, pacPE, or pacSM, and processed as in Fig. 2B. B: Quantitative analysis of photoaffinity labeling of StarD7 by pacLipids as in A. Labeling intensity of WT-StarD7 by pacCer was set at 100%. Data are shown as the mean ± SD of three independent experiments. IGF, in-gel-fluorescence; CB, Coomassie staining.
Collectively, our data indicate that the START domain of StarD7 provides a common binding pocket for PC and ceramide, which includes residues that form an aromatic cage involved in coordinating the quaternary amine of PC. Exactly how these residues contribute to ceramide binding remains to be established, but based on our results, it seems unlikely that hydrogen bonding of the ceramide head group makes a major contribution to the interaction. We also uncovered differences regarding the involvement of the conserved Arg residue in the PC binding pocket, which proved to be dispensable for ceramide binding.
Photolabeling of CERT and StarD7 with pacCer in the presence of excess ceramide
To obtain complementary evidence that photolabeling of the START domains of CERT and StarD7 with pacCer occurs at an authentic ceramide-binding site, we also examined whether pacCer labeling of these protein domains could be competitively inhibited by natural C16:0-ceramide. Using the START domains of WT CERT and the ceramide-binding-deficient mutant, CERTN506A, as references, we determined that liposomes containing 0.5 mol% of pacCer (instead of the usual 1 mol%) in combination with an UV cross-linking time of 90 s gave the highest specific labeling efficiency. As shown in supplemental Fig. S1A, B, lowering the pacCer concentration to 0.25 mol% or shortening the UV cross-linking time to 30 s diminished the specific labeling efficiency so that these conditions were discarded for the competition experiments. Photolabeling of CERT with 0.5 mol% pacCer in the presence of 10- to 40-fold excess of ceramide (i.e., pacCer:Cer, 0.5:5 to 0.5:20 mol%) showed a trend toward inhibition that was not statistically significant (supplemental Fig. S1C, D). Addition of excess ceramide also did not lead to any significant reduction in pacCer labeling of either StarD7 or the PC-binding-deficient mutant, StarD7R189Q, which was included to facilitate ceramide binding (see Fig. 4E, F). While these results are hard to reconcile with our site-directed mutagenesis data and argue against the notion that pacCer labels an authentic ceramide-binding site in CERT and StarD7, it deserves mention that competitive inhibition of ligand photolabeling could also not be demonstrated for known ligand-receptor interactions, such as the GABA receptor and neurosteroids (48) (see also Discussion). As the competition experiments did not yield any conclusive insights, we next set out to examine possible functional implications of a dual binding specificity of StarD7 for ceramide and PC.
Ceramide modulates the PC transfer activity of StarD7
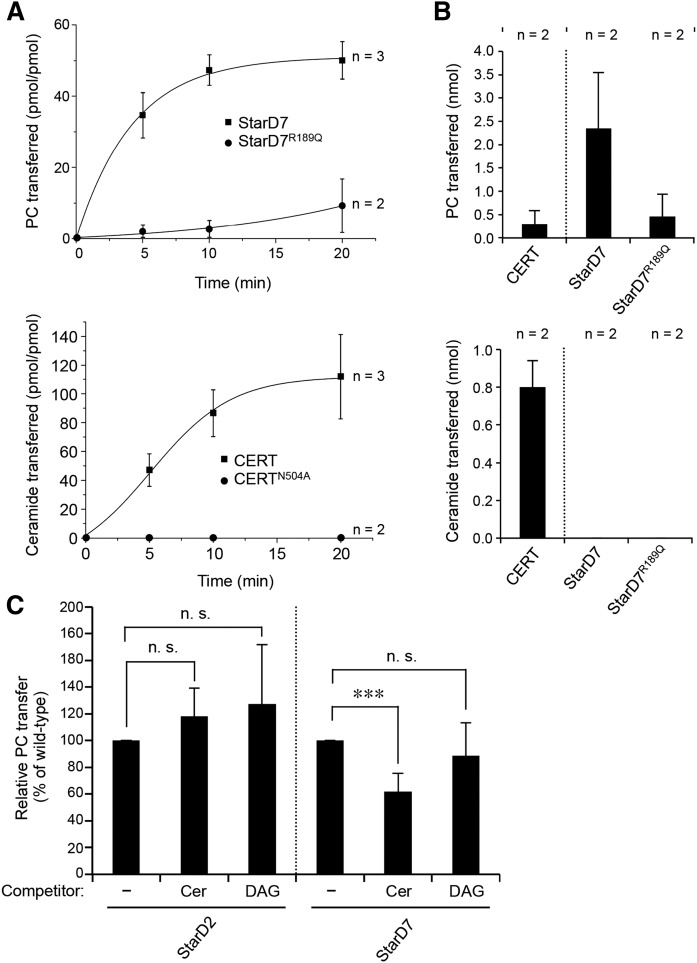
To elucidate the functional relevance of ceramide binding by StarD7, we first asked whether StarD7 acts as a bona fide ceramide transfer protein. Thus, we analyzed the ability of its START domain to catalyze the transfer of ceramide between liposomes using a MALDI-based read out assay. As we wanted to avoid any competing effect of the liposome backbone lipids on PC or ceramide transfer, we chose a mixture of phosphatidylglycerol (PG) and PE for the liposomes in our assay. With this assay at hand, ceramide transfer by the START domain of CERT, as well as PC transfer by the START domain of StarD7, could be readily monitored in a time-dependent manner (Fig. 6A). For the START domain of CERT, the transfer activity was approximately 8 pmol of ceramide per picomole of protein transferred per minute, which is comparable to previously published data (16). As expected, substitution of Asn504 for Ala in the START domain of CERT abolished ceramide transfer activity (Fig. 6A, B). The START domain of StarD7 showed an activity of approximately 7 pmol of PC transferred per picomole of protein per minute (Fig. 6A), a rate that is substantially higher than the one reported by Horibata and Sugimoto (35). This discrepancy is likely due to the fact that, in their assay, Horibata and Sugimoto (35) measured the transfer of radiolabeled PC between liposomes containing bulk amounts of unlabeled PC. In line with our photoaffinity labeling experiments, substitution of Arg189 for Gln in the START domain of StarD7 strongly reduced its PC transfer activity (Fig. 6A, B). In contrast to CERT START, neither WT StarD7 START nor the R189Q mutant displayed any measurable ceramide transfer activity (Fig. 6B).
Fig. 6.
PC transfer activity of StarD7 is negatively influenced by ceramide. A: Time course of PC transfer by StarD7 and StarD7R189Q (top) and of ceramide transfer by CERT START and CERT STARTN504A (bottom). Lipid transfer from donor liposomes containing 10 mol% PC or ceramide was measured as described in the Materials and Methods. Data are shown as the mean ± error range of two or three independent experiments, as indicated. B: Quantitative analysis of the PC (top) and ceramide (bottom) transfer activity of CERT, StarD7, and StarD7R189Q was performed as described in the Materials and Methods. Data are shown as the mean ± error range of two independent experiments. C: Quantitative analysis of the PC transfer activity of StarD2, StarD7, and StarD7R189Q using donor liposomes containing 5 mol% PC in the absence (−) or presense of 10 mol% ceramide (Cer) or DAG. The analysis was performed as described in the Materials and Methods. Data are shown as the mean ± SD of at least three independent experiments. ***P < 0.001; paired t-test.
It should be noted that, owing to its hydrophobicity, the detection limit of ceramide by MALDI is considerably higher than that of PC. Thus, a low ceramide transfer activity may go unnoticed in our assay. Still, the foregoing experiments demonstrate that under conditions where CERT actively transports ceramide, StarD7 does not, even though it displays a robust PC transfer activity under the same conditions. As our site-directed mutagenesis and photoaffinity labeling studies indicate that StarD7 START has dual affinity for PC and ceramide, we wondered whether ceramide binding could influence its PC transfer activity. To test this idea, we next performed PC transfer assays with donor liposomes containing ceramide in 2-fold excess over PC. As shown in Fig. 6C, ceramide addition caused a significant (∼40%) reduction in StarD7-mediated PC transfer. By contrast, including DAG in 2-fold excess over PC in donor liposomes had no obvious impact on PC transfer by StarD7, in line with our finding that StarD7 preferentially labels with pacCer over pacDAG (Figs. 3–5). Moreover, addition of ceramide did not affect PC transfer by StarD2 (Fig. 6C), a StarD7-related protein lacking affinity for pacCer (Fig. 3). Besides providing complementary evidence that StarD7 has dual specificity for PC and ceramide, these results indicate that ceramide modulates the PC transfer activity of StarD7, presumably by acting as a competitive inhibitor of PC binding to the protein’s START domain.
StarD7 is dispensable for ceramide-induced apoptosis in SMSr-depleted cells
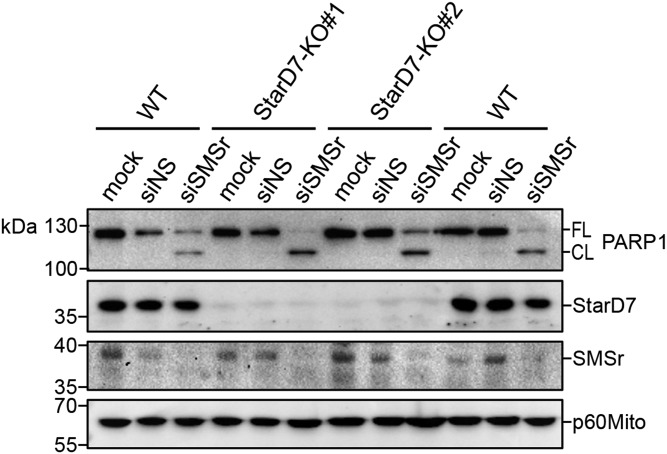
We previously showed that acute inactivation of SM synthase-related protein SMSr/SAMD8 causes a rise in ER ceramides and their mistargeting to mitochondria, triggering a mitochondrial pathway of apoptosis (36). How ER ceramides reach mitochondria in cells lacking functional SMSr is not known. StarD7 has been reported to serve a crucial role in mitochondrial import of PC (31, 35). Given our current finding that StarD7 has dual affinity for PC and ceramide, we wondered whether the protein might participate in the mitochondrial translocation of ER ceramides to commit SMSr-deficient cells to death. To address this, we created two independent HeLa cell-lines in which expression of StarD7 was eliminated using CRISPR/Cas9 technology (StarD7-KO#1 and StarD7-KO#2). WT HeLa cells treated with SMSr-targeting siRNAs (siSMSr) readily underwent apoptosis, as evidenced by proteolytic cleavage of the caspase substrate, poly-ADP ribose polymerase-1 (PARP1; Fig. 7). No PARP1 cleavage was observed when cells were treated with nonsilencing RNA (siNS). Importantly, removal of StarD7 in HeLa cells did not lead to reduced PARP1 cleavage upon siSMSr-treatment. From this we conclude that StarD7 is not a critical component of the mechanism by which ER ceramides can reach mitochondria to induce apoptosis in SMSr-depleted cells.
Fig. 7.
StarD7 is dispensable for apoptosis induction in SMSr-depleted cells. WT, StarD7-KO#1, and StarD7-KO#2 HeLa cells were mock-treated or treated with siNS or siSMSr for 3 days. The cells were lysed and subjected to immunoblot analysis using antibodies against PARP, StarD7, SMSr, and mitochondrial marker, p60Mito. FL, full length; CL, cleaved.
DISCUSSION
Here, we report the successful application of a photoactivatable and clickable ceramide analog, pacCer, to capture ceramide-binding proteins and unravel the structural basis by which these proteins recognize ceramide. This approach yielded CERT-related PC transfer protein, StarD7, as a novel ceramide-binding protein. Previous work revealed that StarD7 is required for efficient PC import by mitochondria and has a critical role in mitochondrial function and morphogenesis (31, 35, 49). Combining site-directed mutagenesis and photoaffinity-labeling experiments, we show that the START domain of StarD7 harbors a common binding site for PC and ceramide. While StarD7 lacks robust ceramide transfer activity in vitro, we observed that its ability to shuttle PC between two membranes is strongly affected by ceramide. Thus, StarD7 qualifies as a candidate effector protein of ceramide, a lipid known for its ability to initiate a variety of mitochondria-mediated cytotoxic effects (50–52).
The suitability of pacCer as a chemical tool to capture ceramide binding proteins from a complex proteome is reflected by the identification of CERT among the photoaffinity-labeled proteins in a cytosolic fraction of mouse melanoma GM95 cells. Interestingly, our primary set of pacCer-labeled proteins also included the 65 kDa regulatory subunit A of protein phosphatase PP2A, a ceramide-inducible serine/threonine phosphatase with potent tumor suppressor activity (53, 54). As this subunit also displayed affinity for pacGlcCer (supplemental Table S1), it did not pass our stringent selection criteria for high-confidence ceramide binding proteins (Table 1). Nevertheless, whether direct binding of ceramide to PP2A subunit A is part of the mechanism by which the phosphatase is activated merits further investigation. High-confidence ceramide binding proteins included proteins involved in membrane trafficking (sorting nexins), signal transduction (Unc-119 proteins), DNA damage response pathways (DNA damage binding protein-1, DnaJ homolog DnaJc7), and protein ubiquitination (COP9 signalosome subunit). Addressing whether the cellular activities of these proteins are responsive to ceramides is subject to ongoing investigations. StarD7 was not among the ceramide binding proteins originally identified by photoaffinity labeling of a cytosolic fraction prepared from GM95 cells. However, lipid specificity profiling of the recombinant protein with our bifunctional lipid probes clearly indicated that StarD7 has dual specificity for ceramide and PC. In this respect, it deserves mention that StarD7 primarily localizes to mitochondria and distributes only to the cytosol when cells are grown to very high density (35). As this was not the case for the GM95 cells used to prepare a cytosolic fraction, StarD7 likely escaped detection in our screen for cytosolic ceramide binding proteins. In addition, it is unlikely that this screen reached saturation.
Remarkably, our efforts to demonstrate that photolabeling of the START domains of CERT and StarD7 with pacCer occurs at an authentic ceramide-binding site using natural ceramide as competitive inhibitor were unsuccessful. Even addition of ceramide in 40-fold excess over pacCer did not result in any statistically significant reduction in photolabeling. We noticed that other known or predicted ligand-protein interactions also could not be validated by competitive inhibition of ligand photolabeling (48, 55). Possible reasons for why competitive inhibition may be hard to demonstrate in certain cases include the irreversible nature of photolabeling, a gradual depletion of the photoactivatable analog by solvent labeling, and potential differences in binding affinities between the analog and its natural counterpart for the target protein (56). A further complication potentially relevant to our studies is that ceramide forms gel-like domains in synthetic phospholipid bilayers (57), which may reduce its capacity to serve as effective competitive inhibitor in pacCer photolabeling experiments.
As a complementary approach to demonstrate that pacCer retains the physicochemical properties important for recognition by ceramide binding proteins, we took advantage of crystal structures of the START domain of CERT in complex with ceramides of different acyl chain lengths (42). In these structures, Asn504 is predicted to form a hydrogen bond with the C3-hydroxyl of ceramide, which appears critical for efficient CERT-mediated ceramide transport (42). Consistent with this notion, substitution of Asn504 for Ala virtually abolished pacCer labeling of the CERT START domain. Moreover, removal of the C3-hydroxyl group from pacCer strongly reduced its ability to label CERT START. Two other residues, Glu446 and Gln467, form a hydrogen bond network with the amide and C1-hydroxyl groups of ceramide (42). Surprisingly, removal of the C1-hydroxyl group actually enhanced pacCer labeling of CERT START, whereas substitution of Glu446 to Ala did not have any obvious impact. Hence, we anticipate that the multilateral hydrogen bonding, involving the amide and C1-hydroxyl groups, is more adaptable to changes in one of its constituents. Our finding that substitution of Trp473, a residue located in the Ω1 loop near the entry of the amphiphilic cavity, increased pacCer labeling was unexpected, as previous work revealed that the membrane affinity of CERT STARTW473A is reduced and that ceramide extraction by a CERT STARTW473A/W562A mutant is almost completely blocked (42, 58). The Ω1 loop of CERT START is thought to function as a gate that opens the amphiphilic cavity upon interaction of Trp473 and also Trp562 with the target membrane (42, 58). Conceivably, substitution of Trp473 for Ala may lead to enhanced pacCer labeling by causing a constitutive opening of the amphiphilic cavity. However, addressing the precise role of Trp473 in the transfer cycle of CERT will require a more detailed functional analysis.
Our finding that StarD7 has dual specificity for PC and ceramide is striking given the considerable structural differences between the two lipids. A homology model based on crystal structures of the closely related PC transfer protein, StarD2, allowed us to map potential structural determinants of PC binding in the START domain of StarD7 and verify their functional relevance by site-directed mutagenesis and photoaffinity labeling analysis. Our data indicate that Trp215, Tyr228, and Tyr276 in StarD7 START form an aromatic cage that coordinates the quaternary amine of the PC head group and Arg189 promotes PC binding by forming a salt bridge with the lipid’s phosphoryl group, analogous to the PC binding site described in StarD2 (41). Whereas Arg189 is fully dispensable for ceramide binding, substitution of any of the residues forming the aromatic cage caused a significant reduction in pacCer labeling, indicating that PC and ceramide share a common binding site. However, given that StarD2 lacks affinity for ceramide, it is clear that ceramide recognition by StarD7 involves additional structural determinants.
Other examples of lipid transfer proteins with dual lipid specificity include the mammalian oxysterol binding protein, OSBP, and its yeast homolog, Osh4p, which catalyze nonvesicular transport of both sterols and phosphatidylinositol-4-phosphate (PI4P) (59–61). Molecular dynamics simulations revealed that the binding energy of the Osh4p-PI4P pair is mostly mediated by electrostatic interactions between specific polar residues and the lipid head group. For the Osh4p-sterol pair, on the other hand, binding is achieved by van der Waals contacts that are dispersed over the entire lipid molecule, involving numerous Osh4p residues (61, 62). Our data suggest that similar principles may govern the dual specificity of StarD7 for PC and ceramide. Indeed, PC binding was overall more sensitive to single residue substitutions than ceramide binding. Moreover, neither hydroxyl group in ceramide proved essential for its interaction with StarD7. As StarD7 displays only a reduced affinity for DAG, it appears that ceramide recognition relies, at least in part, on the lipid’s sphingoid backbone. Given that its lipid-binding START domain can accommodate both PC and ceramide, it is remarkable that StarD7 does not display strong affinity for SM. However, we found that residues that form the aromatic cage that coordinates the quaternary amine of PC are also critical for ceramide binding. Thus, it is conceivable that SM shows only low affinity for StarD7 because its phosphocholine head group precludes specific binding of the ceramide backbone to the protein’s START domain. Moreover, the dynamic volume of SM likely deviates significantly from that of PC. Indeed, even minor changes in the lipid backbone can have considerable impact on the dynamic volume of phospholipids by altering the P-N head group angle (63). This effect may reduce the affinity of the phosphocholine head group for the lipid-binding pocket of StarD7 in SM, as compared with PC. Further insights into the different binding modes of PC and ceramide will require crystal structures or molecular dynamics simulations of StarD7 bound to these ligands.
As opposed to OSBP and Osh4p, which catalyze nonvesicular transport of both PI4P and sterols, we were unable to detect any StarD7-mediated transfer of ceramides with a protein that showed robust PC transfer activity in vitro. Nevertheless, ceramides caused a significant reduction in StarD7-mediated PC transfer in vitro, whereas PC transfer by StarD2 was unaffected. DAG had no obvious impact on PC transfer by StarD7, in agreement with our finding that StarD7 START displays only a reduced affinity for this lipid. The most straightforward explanation for these findings is that ceramide, contrary to DAG, effectively competes with PC for binding to the START domain of StarD7. As StarD7 has a crucial role in mitochondrial import of PC from the ER (31, 35, 60), we also addressed a potential involvement of StarD7 in the mitochondrial translocation of ER ceramides and subsequent induction of apoptosis in cells depleted of SMSr (36). This revealed that StarD7 is fully dispensable for apoptosis induction in SMSr-depleted cells. While our current findings do not exclude that StarD7 acts as a CERT in vivo, they raise the possibility that StarD7-mediated PC transport is subject to negative regulation by ceramides.
PC is a vital component of mitochondria. As mitochondria lack the enzymes necessary for PC synthesis, they rely on PC import for proper function. Loss of StarD7 causes a significant reduction in mitochondrial PC levels, impaired respiratory activity, overproduction of reactive oxygen species, and disrupted cristae structures (31, 32). StarD7 resides in the outer mitochondrial membrane and is believed to shuttle PC between the ER and outer mitochondrial membrane at ER-mitochondria contact sites (35, 64). As ceramide biosynthesis occurs in mitochondria-associated ER membranes (MAMs) (61, 62) and is upregulated in response to various stress stimuli (8, 9), it is conceivable that the local concentration of ceramides at ER-mitochondria contact sites can rise to a level where it affects StarD7-mediated mitochondrial PC import. Moreover, it is well-known that a rise in mitochondrial ceramide levels has a detrimental impact on mitochondria function and is associated with overproduction of reactive oxygen species and stress-induced mitochondrial apoptosis (50–52, 65–67). In view of our current findings, the notion that ceramides may exert their toxic effects, in part, by acting through StarD7 merits further investigation.
Supplementary Material
Footnotes
Abbreviations:
- CHO
- Chinese hamster ovary
- DAG
- diacylglycerol
- DMAP
- 4-dimethylaminopyridine
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOPG
- 1,2-dioleoyl-sn-glycero-3-phosphoglycerol
- EDCI
- 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- ER
- endoplasmic reticulum
- lactosyl-PE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-lactosyl
- MBP
- maltose binding protein
- PARP1
- poly-ADP ribose polymerase-1
- PC
- phosphatidylcholine
- PDB
- Protein Data Bank
- PE
- phosphatidylethanolamine
- PI4P
- phosphatidylinositol-4-phosphate
- PP2A
- protein phosphatase 2A
- SMSr
- SM synthase-related protein
- StarD7
- steroidogenic acute regulatory protein D7
- START
- steroidogenic acute regulatory transfer
This work was supported by a Marie Curie Intra-European Fellowship (to J.G.M.M.), European Commission Seventh Framework Programme (Marie-Curie ITN “Sphingonet”) Grant 289278, and Deutsche Forschungsgemeinschaft Grant SFB944-P14 (to J.C.M.H.). Additional support was provided by Netherlands Organisation for Scientific Research Grant 184.032.201, through the funding of the large-scale proteomics facility, Proteins@Work, embedded in the Netherlands Proteomics Centre. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Lingwood D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50. [DOI] [PubMed] [Google Scholar]
- 2.Lippincott-Schwartz J., and Phair R. D.. 2010. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu. Rev. Biophys. 39: 559–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hla T., and Dannenberg A. J.. 2012. Sphingolipid signaling in metabolic disorders. Cell Metab. 16: 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wennekes T., van den Berg R. J. B. H. N., Boot R. G., van der Marel G. A., Overkleeft H. S., and Aerts J. M. F. G.. 2009. Glycosphingolipids–nature, function, and pharmacological modulation. Angew. Chem. Int. Ed. Engl. 48: 8848–8869. [DOI] [PubMed] [Google Scholar]
- 5.Hannun Y. A., and Obeid L. M.. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 6.Morad S. A. F., and Cabot M. C.. 2013. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer. 13: 51–65. [DOI] [PubMed] [Google Scholar]
- 7.Galadari S., Rahman A., Pallichankandy S., and Thayyullathil F.. 2015. Tumor suppressive functions of ceramide: evidence and mechanisms. Apoptosis. 20: 689–711. [DOI] [PubMed] [Google Scholar]
- 8.Bose R., Verheij M., Haimovitz-Friedman A., Scotto K., Fuks Z., and Kolesnick R.. 1995. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 82: 405–414. [DOI] [PubMed] [Google Scholar]
- 9.Deng X., Yin X., Allan R., Lu D. D., Maurer C. W., Haimovitz-Friedman A., Fuks Z., Shaham S., and Kolesnick R.. 2008. Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science. 322: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Ruiz C., Colell A., Marí M., Morales A., Calvo M., Enrich C., and Fernández-Checa J. C.. 2003. Defective TNF-alpha-mediated hepatocellular apoptosis and liver damage in acidic sphingomyelinase knockout mice. J. Clin. Invest. 111: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stover T. C., Sharma A., Robertson G. P., and Kester M.. 2005. Systemic delivery of liposomal short-chain ceramide limits solid tumor growth in murine models of breast adenocarcinoma. Clin. Cancer Res. 11: 3465–3474. [DOI] [PubMed] [Google Scholar]
- 12.Grassmé H., Jendrossek V., Bock J., Riehle A., and Gulbins E.. 2002. Ceramide-rich membrane rafts mediate CD40 clustering. J. Immunol. 168: 298–307. [DOI] [PubMed] [Google Scholar]
- 13.Thorsell A-G., Lee W. H., Persson C., Siponen M. I., Nilsson M., Busam R. D., Kotenyova T., Schüler H., and Lehtiö L.. 2011. Comparative structural analysis of lipid binding START domains. PLoS One. 6: e19521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siskind L. J., Kolesnick R. N., and Colombini M.. 2002. Ceramide channels increase the permeability of the mitochondrial outer membrane to small proteins. J. Biol. Chem. 277: 26796–26803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombini M. 2017. Ceramide channels and mitochondrial outer membrane permeability. J. Bioenerg. Biomembr. 49: 57–64. [DOI] [PubMed] [Google Scholar]
- 16.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., and Nishijima M.. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426: 803–809. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Yao B., Delikat S., Bayoumy S., Lin X. H., Basu S., McGinley M., Chan-Hui P. Y., Lichenstein H., and Kolesnick R.. 1997. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 89: 63–72. [DOI] [PubMed] [Google Scholar]
- 18.Huwiler A., Brunner J., Hummel R., Vervoordeldonk M., Stabel S., van den Bosch H., and Pfeilschifter J.. 1996. Ceramide-binding and activation defines protein kinase c-Raf as a ceramide-activated protein kinase. Proc. Natl. Acad. Sci. USA. 93: 6959–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich M., Wickel M., Schneider-Brachert W., Sandberg C., Gahr J., Schwandner R., Weber T., Saftig P., Peters C., Brunner J., et al. 1999. Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J. 18: 5252–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay A., Saddoughi S. A., Song P., Sultan I., Ponnusamy S., Senkal C. E., Snook C. F., Arnold H. K., Sears R. C., Hannun Y. A., et al. 2009. Direct interaction between the inhibitor 2 and ceramide via sphingolipid-protein binding is involved in the regulation of protein phosphatase 2A activity and signaling. FASEB J. 23: 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. 2001. Global analysis of protein activities using proteome chips. Science. 293: 2101–2105. [DOI] [PubMed] [Google Scholar]
- 22.Gallego O., Betts M. J., Gvozdenovic-Jeremic J., Maeda K., Matetzki C., Aguilar-Gurrieri C., Beltran-Alvarez P., Bonn S., Fernández-Tornero C., Jensen L. J., et al. 2010. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 6: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kota V., Szulc Z. M., and Hama H.. 2012. Identification of C(6)-ceramide-interacting proteins in D6P2T Schwannoma cells. Proteomics. 12: 2179–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bidlingmaier S., Ha K., Lee N-K., Su Y., and Liu B.. 2016. Proteome-wide identification of novel ceramide-binding proteins by yeast surface cDNA display and deep sequencing. Mol. Cell. Proteomics. 15: 1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haberkant P., Raijmakers R., Wildwater M., Sachsenheimer T., Brügger B., Maeda K., Houweling M., Gavin A-C., Schultz C., van Meer G., et al. 2013. In vivo profiling and visualization of cellular protein-lipid interactions using bifunctional fatty acids. Angew. Chem. Int. Ed. Engl. 52: 4033–4038. [DOI] [PubMed] [Google Scholar]
- 26.Hulce J. J., Cognetta A. B., Niphakis M. J., Tully S. E., and Cravatt B. F.. 2013. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods. 10: 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niphakis M. J., Lum K. M., Cognetta A. B., Correia B. E., Ichu T-A., Olucha J., Brown S. J., Kundu S., Piscitelli F., Rosen H., et al. 2015. A global map of lipid-binding proteins and their ligandability in cells. Cell. 161: 1668–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haberkant P., Stein F., Höglinger D., Gerl M. J., Brügger B., Van Veldhoven P. P., Krijgsveld J., Gavin A-C., and Schultz C.. 2016. Bifunctional sphingosine for cell-based analysis of protein-sphingolipid interactions. ACS Chem. Biol. 11: 222–230. [DOI] [PubMed] [Google Scholar]
- 29.Haberkant P., and Holthuis J. C. M.. 2014. Fat & fabulous: bifunctional lipids in the spotlight. Biochim. Biophys. Acta. 1841: 1022–1030. [DOI] [PubMed] [Google Scholar]
- 30.Höglinger D., Nadler A., Haberkant P., Kirkpatrick J., Schifferer M., Stein F., Hauke S., Porter F. D., and Schultz C.. 2017. Trifunctional lipid probes for comprehensive studies of single lipid species in living cells. Proc. Natl. Acad. Sci. USA. 114: 1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horibata Y., Ando H., Zhang P., Vergnes L., Aoyama C., Itoh M., Reue K., and Sugimoto H.. 2016. StarD7 protein deficiency adversely affects the phosphatidylcholine composition, respiratory activity, and cristae structure of mitochondria. J. Biol. Chem. 291: 24880–24891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L., Na C-L., Luo S., Wu D., Hogan S., Huang T., and Weaver T. E.. 2017. The phosphatidylcholine transfer protein Stard7 is required for mitochondrial and epithelial cell homeostasis. Sci. Rep. 7: 46416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bickert A., Ginkel C., Kol M., vom Dorp K., Jastrow H., Degen J., Jacobs R. L., Vance D. E., Winterhager E., Jiang X-C., et al. 2015. Functional characterization of enzymes catalyzing ceramide phosphoethanolamine biosynthesis in mice. J. Lipid Res. 56: 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishimoto Y., and Costello C.. 1975. Rearrangement of 3-ketoceramide. Chem. Phys. Lipids. 15: 27–32. [DOI] [PubMed] [Google Scholar]
- 35.Horibata Y., and Sugimoto H.. 2010. StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. J. Biol. Chem. 285: 7358–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tafesse F. G., Vacaru A. M., Bosma E. F., Hermansson M., Jain A., Hilderink A., Somerharju P., and Holthuis J. C. M.. 2014. Sphingomyelin synthase-related protein SMSr is a suppressor of ceramide-induced mitochondrial apoptosis. J. Cell Sci. 127: 445–454. [DOI] [PubMed] [Google Scholar]
- 37.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., et al. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42: W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiefer F., Arnold K., Kunzli M., Bordoli L., and Schwede T.. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37: D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex N., Peitsch M. C., and Schwede T.. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 30(Suppl 1): S162–S173. [DOI] [PubMed] [Google Scholar]
- 40.Arnold K., Bordoli L., Kopp J., and Schwede T.. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 41.Roderick S. L., Chan W. W., Agate D. S., Olsen L. R., Vetting M. W., Rajashankar K. R., and Cohen D. E.. 2002. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 9: 507–511. [DOI] [PubMed] [Google Scholar]
- 42.Kudo N., Kumagai K., Tomishige N., Yamaji T., Wakatsuki S., Nishijima M., Hanada K., and Kato R.. 2008. Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc. Natl. Acad. Sci. USA. 105: 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grosdidier A., Zoete V., and Michielin O.. 2011. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 32: 2149–2159. [DOI] [PubMed] [Google Scholar]
- 44.Grosdidier A., Zoete V., and Michielin O.. 2011. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39: W270–W277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alpy F., and Tomasetto C.. 2005. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 118: 2791–2801. [DOI] [PubMed] [Google Scholar]
- 46.Wirtz K. W. A. 1991. Phospholipid transfer proteins. Annu. Rev. Biochem. 60: 73–99. [DOI] [PubMed] [Google Scholar]
- 47.Olayioye M. A., Vehring S., Muller P., Herrmann A., Schiller J., Thiele C., Lindeman G. J., Visvader J. E., and Pomorski T.. 2005. StarD10, a START domain protein overexpressed in breast cancer, functions as a phospholipid transfer protein. J. Biol. Chem. 280: 27436–27442. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z-W., Manion B., Townsend R. R., Reichert D. E., Covey D. F., Steinbach J. H., Sieghart W., Fuchs K., and Evers A. S.. 2012. Neurosteroid analog photolabeling of a site in the third transmembrane domain of the β3 subunit of the GABA(A) receptor. Mol. Pharmacol. 82: 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flores-Martín J., Reyna L., Ridano M. E., Panzetta-Dutari G. M., and Genti-Raimondi S.. 2016. Suppression of StarD7 promotes endoplasmic reticulum stress and induces ROS production. Free Radic. Biol. Med. 99: 286–295. [DOI] [PubMed] [Google Scholar]
- 50.García-Ruiz C., Colell A., Marí M., Morales A., and Fernández-Checa J. C.. 1997. Direct effect of ceramide on the mitochondrial electron transport chain leads to generation of reactive oxygen species. Role of mitochondrial glutathione. J. Biol. Chem. 272: 11369–11377. [DOI] [PubMed] [Google Scholar]
- 51.Lee H., Rotolo J. A., Mesicek J., Penate-Medina T., Rimner A., Liao W-C., Yin X., Ragupathi G., Ehleiter D., Gulbins E., et al. 2011. Mitochondrial ceramide-rich macrodomains functionalize Bax upon irradiation. PLoS One. 6: e19783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain A., Beutel O., Ebell K., Korneev S., and Holthuis J. C. M.. 2017. Diverting CERT-mediated ceramide transport to mitochondria triggers Bax-dependent apoptosis. J. Cell Sci. 130: 360–371. [DOI] [PubMed] [Google Scholar]
- 53.Dobrowsky R. T., Kamibayashi C., Mumby M. C., and Hannun Y. A.. 1993. Ceramide activates heterotrimeric protein phosphatase 2A. J. Biol. Chem. 268: 15523–15530. [PubMed] [Google Scholar]
- 54.Oaks J., and Ogretmen B.. 2015. Regulation of PP2A by sphingolipid metabolism and signaling. Front. Oncol. 4: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Budelier M. M., Cheng W. W., Bergdoll L., Chen Z-W., Janetka J. W., Abramson J., Krishnan K., Mydock-McGrane L., Covey D. F., Whitelegge J. P., et al. 2017. Photoaffinity labeling with cholesterol analogues precisely maps a cholesterol-binding site in voltage-dependent anion channel-1. J. Biol. Chem. 292: 9294–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woll K. A., Dailey W. P., Brannigan G., and Eckenhoff R. G.. 2016. Shedding light on anesthetic mechanisms: application of photoaffinity ligands. Anesth. Analg. 123: 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staneva G., Momchilova A., Wolf C., Quinn P. J., and Koumanov K.. 2009. Membrane microdomains: role of ceramides in the maintenance of their structure and functions. Biophys. Biophys. Acta. 1788: 666–675. [DOI] [PubMed] [Google Scholar]
- 58.Kudo N., Kumagai K., Matsubara R., Kobayashi S., Hanada K., Wakatsuki S., and Kato R.. 2010. Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer. J. Mol. Biol. 396: 245–251. [DOI] [PubMed] [Google Scholar]
- 59.de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., and Drin G.. 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195: 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B.. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 155: 830–843. [DOI] [PubMed] [Google Scholar]
- 61.Moser von Filseck J., Vanni S., Mesmin B., Antonny B., and Drin G.. 2015. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat. Commun. 6: 6671. [DOI] [PubMed] [Google Scholar]
- 62.Singh R. P., Brooks B. R., and Klauda J. B.. 2009. Binding and release of cholesterol in the Osh4 protein of yeast. Proteins. 75: 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contreras F-X., Ernst A. M., Haberkant P., Björkholm P., Lindahl E., Gönen B., Tischer C., Elofsson A., von Heijne G., Thiele C., et al. 2012. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 481: 525–529. [DOI] [PubMed] [Google Scholar]
- 64.Horibata Y., Ando H., Satou M., Shimizu H., Mitsuhashi S., Shimizu Y., Itoh M., and Sugimoto H.. 2017. Identification of the N-terminal transmembrane domain of StarD7 and its importance for mitochondrial outer membrane localization and phosphatidylcholine transfer. Sci. Rep. 7: 8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bionda C., Portoukalian J., Schmitt D., Rodiguez-Lafrasse C., and Ardail D.. 2004. Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 382: 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chipuk J. E., McStay G. P., Bharti A., Kuwana T., Clarke C. J., Siskind L. J., Obeid L. M., and Green D. R.. 2012. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 148: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Birbes H., El Bawab S., Hannun Y. A., and Obeid L. M.. 2001. Selective hydrolysis of a mitochondrial pool of sphingomyelin induces apoptosis. FASEB J. 15: 2669–2679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.