Abstract
Ganodermataceae is a remarkable group of polypore fungi, mainly characterized by particular double-walled basidiospores with a coloured endosporium ornamented with columns or crests, and a hyaline smooth exosporium. In order to establish an integrative morphological and molecular phylogenetic approach to clarify relationship of Neotropical Amauroderma s.lat. within the Ganodermataceae family, morphological analyses, including scanning electron microscopy, as well as a molecular phylogenetic approach based on one (ITS) and four loci (ITS-5.8S, LSU, TEF-1α and RPB1), were carried out. Ultrastructural analyses raised up a new character for Ganodermataceae systematics, i.e., the presence of perforation in the exosporium with holes that are connected with hollow columns of the endosporium. This character is considered as a synapomorphy in Foraminispora, a new genus proposed here to accommodate Porothelium rugosum (≡ Amauroderma sprucei). Furtadoa is proposed to accommodate species with monomitic context: F. biseptata, F. brasiliensis and F. corneri. Molecular phylogenetic analyses confirm that both genera grouped as strongly supported distinct lineages out of the Amauroderma s.str. clade.
Keywords: Amauroderma, Ganoderma, polyporales, systematics, ultrastructure
INTRODUCTION
Ganodermataceae is mainly characterized by pileate basidiomata, sessile to stipitate, hyphal system dimitic, with arboriform and skeleto-binding hyphae and double-walled basidiospores with a coloured endosporium ornamented with columns and crests, and a hyaline smooth exosporium. The family has a cosmopolitan distribution with about 220 species, as saprotrophs in dead wood, associated with roots of living and dead trees, and also as parasites/pathogens, causing white rot in woody tissues (Moncalvo & Ryvarden 1997, Ryvarden 2004).
Taxonomy of the family was almost exclusively based on morphological characteristics, such as appearance of pilear surface (i.e., dull or laccate), disposition of the hyphae in the pilear surface (i.e., anamixoderm, characoderm, cortex, hymeniderm, trichoderm) and basidiospore characters (shape and ornamentation pattern including some ultrastructural approaches). Despite extensive studies at generic and infrageneric levels (Furtado 1962, 1965, 1981, Steyaert 1972, 1980, Ryvarden & Johansen 1980, Corner 1983, Gottlieb & Wright 1999a, b, Ryvarden 2004, Torres-Torres & Guzmán-Dávalos 2012), only five genera are currently widely accepted, i.e., Amauroderma, Ganoderma, Haddowia, Humphreya and Tomophagus (Moncalvo et al. 1995, Moncalvo & Ryvarden 1997, Ryvarden 2004, Kirk et al. 2008, Tham et al. 2012). Ganoderma is characterized by ellipsoid to ovoid basidiospores, with a truncate apex and an endosporium with columnar ornamentations. Tomophagus also has basidiospores with a truncate apex; however, it is characterized by a pale and soft floccose context where chlamydospores are produced. Humphreya has basidiospores with truncate apex and the endosporium ornamented by typical longitudinal ridges. Amauroderma and Haddowia have basidiospores without truncate apex, differing mainly due to the ornamentation pattern of the endosporium, i.e., columnar to semi-reticulate in Amauroderma and with longitudinal ridges in Haddowia (Furtado 1981, Steyaert 1972, Ryvarden 2004, Tham et al. 2012).
In this current classification into five genera, several taxa are considered ‘deviating elements’ either by their microscopical characters (basidiospore shape and ornamentation or hyphal system), macroscopical characters (as stipe presence or context colour and consistence) or a combination of these features. In particular, regarding neotropical Amauroderma species there are taxa which not fit within the phylogenetic delimitation of Amauroderma s.str. senso Costa-Rezende et al. (2016), such as Amauroderma sprucei which distinguishes within the genus by its whitish context with hyaline dextrinoid skeletal hyphae and a vivid orange pore surface in most of the specimens (Decock & Herrera-Figueroa 2006). There are also monomitic or nearly so species within Amauroderma, as A. trichodermatum and A. brasiliense (Robledo et al. 2015), as well as species with basidiospores with reticulate endosporium (A. deviatum) (Ryvarden 2004).
Based on phylogenetic evidence it has been shown that Amauroderma is polyphyletic, with Amauroderma s.str. forming a monophyletic clade and some Amauroderma species defined in its broad morphological sense grouped out of Amauroderma s.str. (Gomes-Silva et al. 2015, Costa-Rezende et al. 2016). Although several molecular phylogenetic studies have been published on Ganoderma and Amauroderma, no synthesis of molecular data has been presented with a phylogenetic overview in context of Ganodermataceae.
Regarding the ‘deviating elements’ in Neotropical Amauroderma and the scarce phylogenetic evidence around Ganodermataceae, the aim of our work was to develop an integrative morphological and molecular phylogenetic approach to clarify the relationship of Neotropical Amauroderma s.lat. within the Ganodermataceae family.
MATERIAL AND METHODS
Specimens and morphological studies
The studied specimens are deposited in FLOR, HUEFS and CORD herbaria. Herbarium acronyms follow Thiers (continuously updated, http://sweetgum.nybg.org/science/ih/). Microscopic examinations and measurements were done using Melzer’s reagent, Cotton blue and/or 3–5 % KOH as mounting media. For the study of the hyphal system, sections of the basidiomata were incubated in hot (40 °C) 3 % NaOH solution, then dissected under a stereomicroscope and finally examined at 3 % NaOH solution at room temperature (Decock et al. 2013). Basidiospore-walls designations follow the concept of Furtado (1962). Melzer’s reagent was used to check dextrinoid and amyloid reactions. In order to determine the size range of pores, hyphae and basidiospores, 5 % of the measurements at each end of the range are given in parentheses, when relevant, and forty basidiospores were measured.
For ultrastructural observations, both basidiospores with and without exospore were observed. In the first case, fragments of tubes were placed on stubs, then metalized with gold and observed at SEM. To observe the ornamentation in detail, we removed the outer layer of basidiospores according to Crespo & Robledo (2016). Fragments of tubes were placed on chromic acid (H2CrO4) crystal, covered by enough water drops to dissolve the crystals, and stored around 20 minutes. Then, this solution and dissepiment fragments were filtered (0.45 μm filter) by vacuum, adding water to remove acid. The filter was dried at room temperature and finally scraped with a blade in a stub with a drop of 70 % alcohol, metalized with gold and observed at SEM. The analyses were performed in Scanning Electronic Microscope (SEM) Zeiss LEO 1450VP of the Laboratorio de Microscopía Electrónica y Microanalisis (LABMEM) of the Universidad Nacional de San Luis, Argentina and JEOL JSM-6390LV.
DNA extraction and sequencing
DNA was extracted from dried basidiomata following the protocol of Doyle & Doyle (1987) modified by Góes-Neto et al. (2005). Primer pairs ITS8-F/ITS6-R (Dentinger et al. 2010) and LR0R/LR7 (Vilgalys & Hester 1990) were used to amplify the internal transcribed spacer (ITS) and large subunit (LSU) rDNA regions, respectively. Primer pairs RPB1-Af/RPB1-Cr (Matheny et al. 2002) and EF1-983F/EF1- 2212R (Rehner & Buckley 2005) were used to amplify the protein-coding genes RNA polymerase II largest subunit (RPB1) and translation elongation factor-1α (TEF-1α), respectively. Sanger Sequencing was performed with BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, California, USA) following manufacturer procedures. The same oligos were used as forward and reverse sequencing primers for the ITS, RPB1 and TEF-1α. For LSU the primer LR7 was replaced by the LR5. The sequencing was performed at LAMOL (Universidade Estadual de Feira de Santana) and FIOCRUZ-MG (Brazil), as part of the FungiBrBol project.
Phylogenetic analyses
Chromatograms were manually edited using Geneious v. 6.1.8 (http://www.geneious.com). The sequences generated in this work were combined with ITS, LSU, RPB1 and TEF-1α sequences of Ganodermataceae and outgroups (Perenniporia medulla-panis, Perenniporiella chaquenia and P. pendula) retrieved from GenBank (NCBI). Five datasets were constructed: one of them (ITS) is composed by the majority of the phylogenetic species of Ganodermataceae; the others (ITS, LSU, RPB1 and TEF-1α) are composed of sequences from vouchers belonging to the main putative phylogenetic lineages of the Ganodermataceae family which have available sequences of at least two of the molecular markers mentioned above (except for G. subresinosum and A. brasiliense which were included even having only ITS sequences), in order to perform a multiloci phylogenetic analyses. The newly generated sequences and additional sequences downloaded from GenBank are listed in the Table 1.
Table 1.
Species, vouchers and accession numbers of the specimens used in phylogenetic analyses.
| Genbank acession numbers |
|||||
|---|---|---|---|---|---|
| Species name | Voucher | ITS | LSU | RPB1 | TEF-1α |
| Amauroderma aurantiacum | FLOR52205 | KR816510 | KU315205 | – | – |
| DHCR540 (HUEFS) | MF409961 | MF409953 | MF436687 | – | |
| URM78847 | JX310840 | – | – | – | |
| A. calcigenum | FLOR52315 | KR816514 | – | – | – |
| A. calcitum | FLOR50931/DHCR538 (HUEFS) | KR816528 | KU315207 | MF436690 | – |
| FLOR52230 | KR816529 | – | – | – | |
| A. elegantissimum | URM82789 | JX310844 | KT006617 | – | – |
| URM82787 | JX310843 | KT006616 | – | – | |
| A. exile | URM82794 | JX310845 | – | – | – |
| A. floriformum | URM83250 | JX310846 | – | – | – |
| A. intermedium | GAS910 (HUEFS) | MF409959 | – | MF436685 | – |
| FLOR52248 | KR816527 | KU315209 | – | – | |
| A. omphalodes | DHCR499/501 (HUEFS) | MF409956 | MF409951 | MF436682 | MF421238 |
| DHCR500 (HUEFS) | MF409957 | MF409952 | MF436683 | MF421239 | |
| A. partitum | URM83039 | JX310853 | – | – | – |
| URM82882 | JX310852 | – | – | – | |
| A. perplexum | CUI6496 | KJ531650 | KU220001 | – | – |
| WEI5562 | KJ531652 | – | – | – | |
| DAI10811 | KJ531651 | KU220002 | – | – | |
| A. aff. praetervisum | FLOR52249 | KR816511 | – | – | – |
| A. praetervisum | REC18707 | JX310855 | – | – | – |
| URM84230 | KC348461 | – | – | – | |
| GOMES SILVA 909 | JX310856 | – | – | – | |
| A. pseudoboletum | FLOR52318 | KR816516 | – | – | – |
| A. rude | CANB643174 | KU315197 | – | – | – |
| CANB795782 | KU315198 | – | – | – | |
| CANB359451 | KU315199 | – | – | – | |
| A. rugosum | CUI9012 | KJ531665 | KU220011 | – | KU572503 |
| ZHOU547 | KJ531675 | – | – | – | |
| CUI9011 | KJ531664 | KU220010 | – | KU572504 | |
| A. schomburgkii | DHCR504 (HUEFS) | MF409958 | – | MF436684 | – |
| FLOR52177 | KR816522 | KU315215 | – | – | |
| URM83228 | JX310848 | – | – | – | |
| A. sp. | INPA249751 | KR816525 | – | – | – |
| A. subresinosum | WEI5569 | KJ531649 | – | – | – |
| THP48 | FJ154784 | – | – | – | |
| THP16 | FJ154782 | – | – | – | |
| A. yunnanense | CUI7974 | KJ531653 | KU220013 | – | – |
| DAI13021 | KJ531654 | – | – | – | |
| YUAN2253 | KJ531655 | – | – | – | |
| Furtadoa brasiliensis | URM83578 | JX310841 | – | – | – |
| TBG58 | JX982569 | – | – | – | |
| F. biseptata | FLOR50932 | KU315196 | KU315206 | – | – |
| Foraminisporus sprucei | FLOR52191 | KU315200 | KU315216 | – | – |
| FLOR52184 | KU315201 | – | – | – | |
| FLOR52195 | KU315202 | – | – | – | |
| DHCR512 (HUEFS) | MF409960 | – | MF436686 | MF421240 | |
| DHCR554 (HUEFS) | MF409962 | MF409954 | MF436688 | – | |
| DHCR560 (HUEFS) | MF409963 | MF409955 | MF436689 | MF421241 | |
| Ganoderma adspersum | R1212 | AJ006685 | – | – | – |
| GATO00 | AM906057 | – | – | – | |
| GAD3 | JN222418 | – | – | – | |
| G. annulare | KCTC16803 | JQ520160 | – | – | – |
| G. applanatum | KM120830 | AY884178 | – | – | – |
| GA165 | DQ425009 | – | – | – | |
| GA117 | DQ424996 | – | – | – | |
| ATCC44053 | JQ520161 | – | – | – | |
| WEI5787 | KF495001 | KF495011 | KF494978 | – | |
| Dai 12483 | KF494999 | KF495009 | – | KF494977 | |
| G. aridicola | DAI 12588 | KU572491 | – | – | KU572502 |
| G. cf. australe | K621 | JN596327 | – | – | – |
| G561 | JN596326 | – | – | – | |
| G. australe | DHCR411 (HUEFS) | MF436675 | MF436672 | MF436680 | MF436677 |
| DHCR417 (HUEFS) | MF436676 | MF436673 | MF436681 | MF436678 | |
| GDGM25745 | JX195205 | – | – | – | |
| HMAS86596 | AY884180 | – | – | – | |
| G. australe cplx | FLOR52289 | KU315203 | KU315217 | – | – |
| G. austroafricanum | CMW41454 | KM507324 | – | – | – |
| G. boninense | WD2085 | KJ143906 | – | KJ143945 | KJ143925 |
| WD2028 | KJ143905 | – | KJ143944 | KJ143924 | |
| G. carnosum | KM109415 | AY884175 | – | – | – |
| GCR1 | JN222419 | – | – | – | |
| G. chalceum | URM80457 | JX310812 | – | – | – |
| G. coffeatum | FLOR50933 | KU315204 | – | – | – |
| G. cupreum | GANOTK7 | JN105702 | – | – | – |
| GANOTK4 | JN105701 | – | – | – | |
| KR61 | FJ655470 | – | – | – | |
| KL161 | FJ655466 | – | – | – | |
| G. curtisii | CBS100132 | JQ520164 | – | KJ143947 | KJ143927 |
| CBS100131 | JQ781848 | – | KJ143946 | KJ143926 | |
| G. enigmaticum | DAI 15970 | KU572486 | – | – | KU572496 |
| DAI 15971 | KU572487 | – | – | KU572497 | |
| G. flexipes | WEI5494 | JN383979 | – | – | – |
| WEI5491 | JQ781850 | – | – | – | |
| G. fornicatum | TN231 | FJ655476 | – | – | – |
| KL231 | FJ655471 | – | – | – | |
| G. fulvellum | XSD08051 | FJ478088 | – | – | – |
| G. gibbosum | XSD34 | EU273513 | – | – | – |
| KUT0805 | AB733121 | – | – | – | |
| G1 | JN596331 | – | – | – | |
| G. hoehnelianum | DAI12096 | JN383980 | – | – | – |
| GDGM25735 | JX195203 | – | – | – | |
| G. japonicum | AS5.69 | AY593864 | – | – | – |
| AS5.69 | AY593865 | – | – | – | |
| G. leucocontextum | DAI 15601 | KU572485 | – | – | KU572495 |
| GDGM44490 | KM396272 | – | – | – | |
| G. lingzhi | DAI12574 | KJ143908 | – | JX029985 | JX029977 |
| DAI12426 | JQ781870 | – | – | – | |
| CUI9166 | KJ143907 | – | JX029982 | JX029974 | |
| G. lipsiense | NOR5311432 | EF060005 | – | – | – |
| FIN131R610 | EF060004 | – | – | – | |
| G. lobatum | JV 1212/10J | KF605676 | – | – | KU572501 |
| G. lucidum | BEOFB 432 | KX371595 | – | – | KX371598 |
| BEOFB 431 | KX371594 | – | – | KX371597 | |
| K175217 | KJ143911 | – | KJ143950 | KJ143929 | |
| CUI9207 | KJ143910 | – | KJ143949 | KJ143928 | |
| GL16 | HM053438 | – | – | – | |
| GL14 | HM053436 | – | – | – | |
| GL951 | KC311371 | – | – | – | |
| G. martinicense | LIPSWMart0844 | KF963257 | – | – | – |
| LIPSWMart0855 | KF963256 | – | – | – | |
| G. mastoporum | PM21 | JQ409361 | – | – | – |
| G. meredithae | ASI7140 | JQ5201911 | – | – | – |
| ATCC64492 | JQ520190 | – | – | – | |
| G. multipileum | DAI9447 | KJ143914 | – | KJ143953 | KJ143932 |
| CWN04670 | KJ143913 | – | KJ143952 | KJ143931 | |
| DAI9447 | KF494997 | – | – | – | |
| G. multiplicatum | DAI12320 | KU572490 | – | – | KU572500 |
| DAI13710 | KU572489 | – | – | KU572499 | |
| URM83346 | JX310823 | – | – | – | |
| G. orbiforme | URM83334 | JX310814 | – | – | – |
| URM83336 | JX310816 | – | – | – | |
| G. oregonense | CBS266.88 | JQ781876 | – | KJ143955 | – |
| CBS265.88 | JQ781875 | – | KJ143954 | KJ143933 | |
| G. parvulum | URM83345 | JX310820 | – | – | – |
| URM80765 | JX310822 | – | – | – | |
| G. perzonatum | SP445985 | KJ792745 | – | – | – |
| SP4459871 | KJ792747 | – | – | – | |
| G. pfeifferi | KM120818 | AY884185 | – | – | – |
| GPF1 | JN222420 | – | – | – | |
| G. philippii | E7098 | AJ536662.2 | – | – | – |
| E7092 | AJ608710 | – | – | – | |
| G. pudoferreum | CATASGp008 | FJ392284 | – | – | – |
| G. pseudoferreum | CATASGp005 | FJ392281 | – | – | – |
| G. ramosissimum | XSD08032 | EU918700 | – | – | – |
| XSD08085 | FJ478127 | – | – | – | |
| G. resinaceum | CBS 194.76 | X78737/X78758 | – | KJ143956 | KJ143934 |
| IUM3651 | JQ520204 | – | – | – | |
| ASI7143 | JQ520203 | – | – | – | |
| BR4150 | KJ143915 | – | KJ143915 | – | |
| G. sessile | JV1209/9 | KF605629 | – | KJ143958 | KJ143936 |
| JV1209/27 | KF605630 | – | KJ143959 | KJ143937 | |
| G. sichuanense | CGMCC55331 | JN197284 | – | – | – |
| HMAS1301281 | JF915404 | – | – | – | |
| G. sinense | XZGC1 | HQ235633 | – | – | – |
| GDGM25829 | KC415760 | – | – | – | |
| WEI5327 | KF494998 | KF495008 | – | KF494976 | |
| G. sp. | PALCOSTPBP10 | KJ792084 | – | – | – |
| PALCOSTPBP09 | KJ792083 | – | – | – | |
| GD026 (HUEFS) | MF436674 | MF436671 | MF436679 | – | |
| G. aff. steyaertanum | C17274 | EU239388 | – | – | – |
| G. steyaertanum | MEL2382783 | KP012964 | – | – | – |
| G. stipitatum | THC16 | KC884264 | – | – | – |
| G. subamboinense | GSUB1371 | DQ425006 | – | – | – |
| GSUB1361 | DQ425005 | – | – | – | |
| G. tornatum | URM82776 | JQ514110 | – | – | – |
| TBG01AM2009 | JQ514108 | – | – | – | |
| G. tropicum | YUAN3490 | JQ781880 | – | – | – |
| DAI9724 | JQ781879 | – | – | – | |
| G. tsugae | DAI3937 | JQ781853 | – | – | – |
| AFTOL ID 771 | DQ206985 | AY684163 | – | DQ059048 | |
| DAI12760 | KJ143920 | – | KJ143961 | KJ143940 | |
| G. tsunodae | GR3631 | FJ154773 | – | – | – |
| WD2034 | AB588989 | AB368069 | – | – | |
| G. tuberculosum | LIPSWMart0845 | KF963258 | – | – | – |
| LIPRCMart1075 | KF963255 | – | – | – | |
| G. weberianum | GANOTK16 | JN105704 | – | – | – |
| GANOTK06 | JN105703 | – | – | – | |
| GW11 | GU726935 | – | – | – | |
| GW10 | GU726934 | – | – | – | |
| TN21 | FJ491988 | – | – | – | |
| TN15 | FJ491986 | – | – | – | |
| G. zonatum | FL03 | KJ143922 | – | – | KJ143942 |
| FL02 | KJ143921 | – | KJ143962 | KJ143941 | |
| Perenniporia medulla-panis | MUCL43250 | NR119717 | – | – | – |
| Perenniporiella chaquenia | MUCL49758 | NR111365 | FJ393857 | – | HM467602 |
| P. pendula | MUCL47129 | FJ411082 | FJ393854 | – | HM467600 |
| Tomophagus cattienensis | CT119 | JN184398 | – | – | – |
| CT99 | JN184397 | – | – | – | |
| T. colossus | TC02 | KJ143923 | – | KJ143963 | KJ143943 |
| URM80450 | JX310825 | JX310839 | – | – | |
| URM83330 | JQ618247 | JX310811 | – | – | |
The datasets were aligned using MAFFT v. 7 (Katoh & Standley 2013), under the G-INS-i criteria. Then, they were manually inspected using MEGA v. 6 (Tamura et al. 2013). Both ITS datasets were subdivided into three data partitions, ITS1, 5.8S and ITS2, while RPB1 and TEF-1α were subdivided in introns, and 1st, 2nd and 3rd codon positions.
The best-fit model of nucleotide evolution to the datasets was selected by AIC (Akaike Information Criterion) using jModelTest2 v. 1.6 (Guindon & Gascuel 2003, Darriba et al. 2012). For the phylogenetic reconstruction two datasets were analyzed, the ITS dataset and the multiloci dataset (ITS+LSU+RPB1+TEF-1α). Bayesian Inference (BI) and Maximum Likelihood (ML) phylogenetic analyses were applied to the datasets. BI was performed using MrBayes 3.1.2 (Ronquist & Huelsenbeck 2003) with two independent runs, each one beginning from random trees with four simultaneous independent chains, performing 1 × 107 replications, sampling one tree every 1 × 103th generation. The first 2.5 × 106 sampled trees were discarded as burn-in and checked by the convergence criterion (frequencies of average standard deviation of split < 0.01), while the remaining ones were used to reconstruct a 50 % majority-rule consensus tree and calculate Bayesian posterior probabilities (BPP) of the clades. ML searches were conducted with RAxML-HPC v. 8.2.3 (Stamatakis 2014), available in the CIPRES science gateway (Miller et al. 2010; http://www.phylo.org/). The analysis first involved 100 ML searches, each one starting from one randomized stepwise addition parsimony tree, under a GTRGAMMA model, with all other parameters estimated by the software. Only the best scored likelihood tree from all the searches was kept to access the reliability of the nodes. Multiparametric bootstrapping replicates under the same model are computed, allowing the program to halt bootstrapping automatically by the autoMRE option. An additional alignment partition file to force RAxML software to search for a separate evolution model for each partition was used.
A node was considered to be strongly supported if it showed a BPP ≥ 0.95 and/or BS ≥ 70 %. The final alignment and the retrieved topologies were deposited in TreeBASE (http://www.treebase.org), under accession ID: 20193 (http://purl.org/phylo/treebase/phylows/study/TB2:S20193).
RESULTS
Molecular Phylogeny
The final ITS dataset (Fig. 1) included sequences from 157 fungal specimens, with 659 characters, of which 320 were constant and 267 parsimony informative. The combined (ITS+LSU+RPB1+TEF-1α) dataset (Fig. 2) included sequences from 68 fungal specimens, with 3 489 characters, of which 2 415 were constant and 813 parsimony informative. The evolutionary models selected for ITS dataset were TIM2+G (ITS1), TIM1ef+I+G (5.8S) and HKY+I+G (ITS2). For the multiloci dataset the selected models were TVM+I+G (ITS1), K80+I (5.8S), TPM3+G (ITS2), TIM2+I+G (LSU), HKY+G (RPB1 introns), TRN+I (RPB1 1st codon), HKI+I (2nd codon), TIM2+G (3rd codon), TPM3u+I+G (TEF-1α introns), GTR+I (TEF-1α 1st codon), TVM+I+G (TEF-1α 2nd codon) and TIM2+G (TEF-1α 3rd codon).
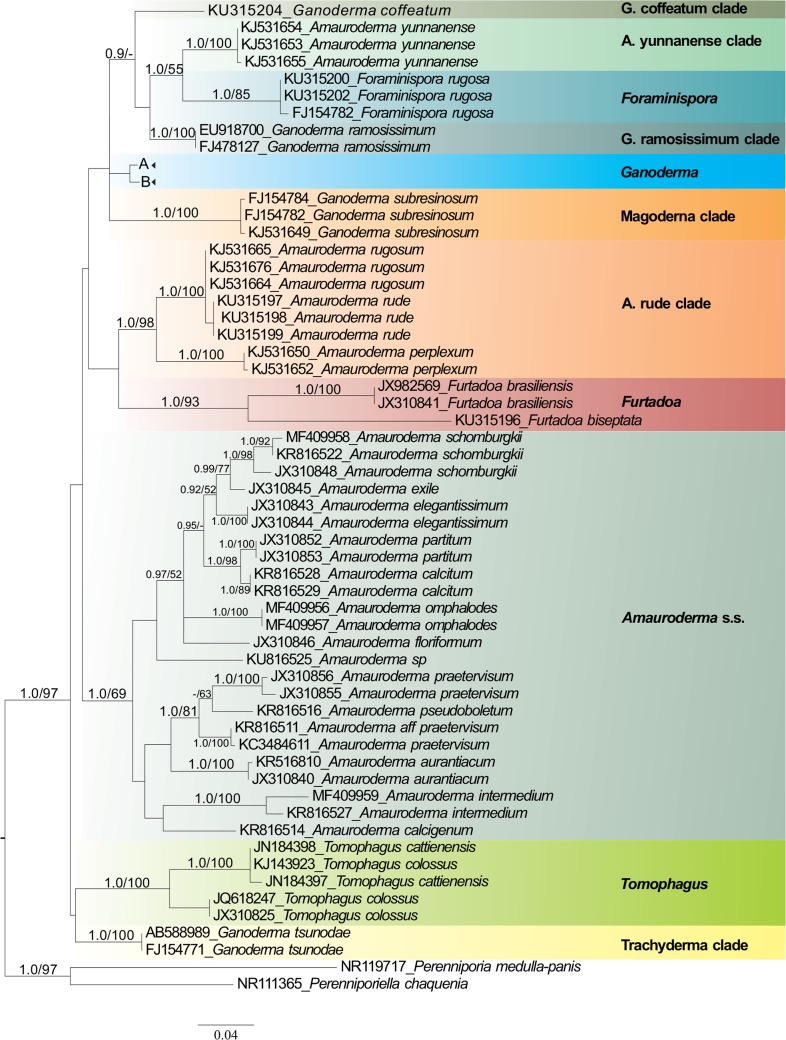
Fig. 1.
Maximum likelihood (ML) tree of Ganodermataceae based on dataset of ITS sequences. Bayesian posterior probability above 0.7 and Bootstrap values above 50 % are shown.
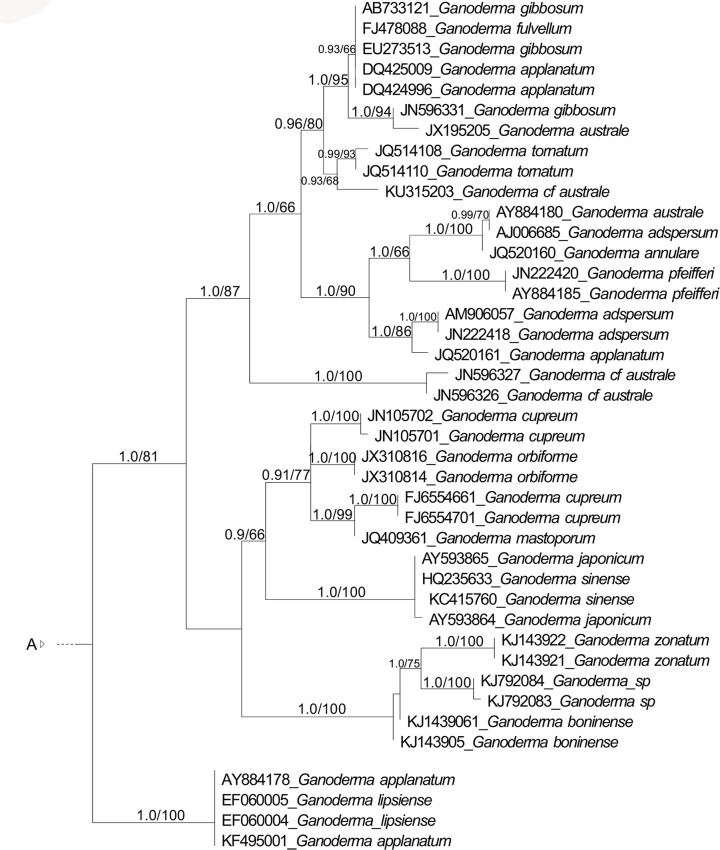
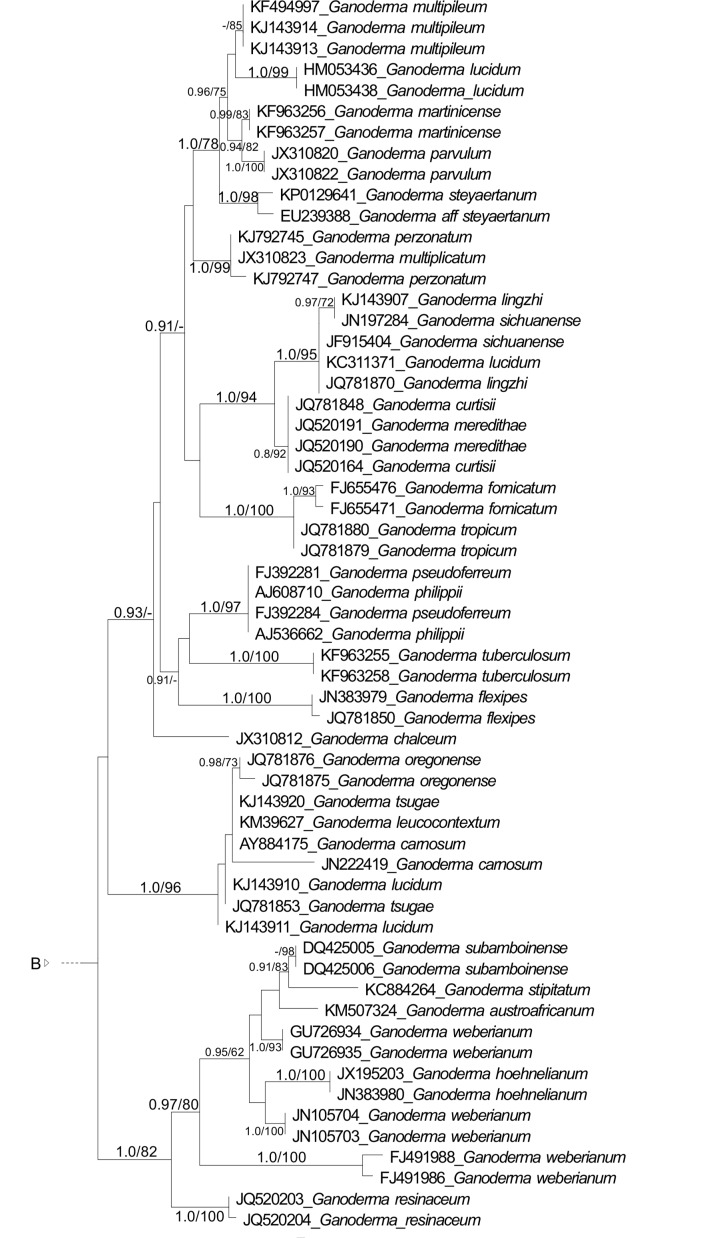
Fig. 2.
Maximum likelihood (ML) tree of Ganodermataceae based on concatenated ITS, LSU, RPB1, TEF-1α sequence data. Bayesian posterior probability above 0.7 and Bootstrap values above 50 % are shown.
Eleven major lineages were recovered in ITS analyses. Two of them corresponded to the new genera proposed here, i.e., Furtadoa (1.0 BPP, 95 % BS) and Foraminispora (1.0 BPP, 100 % BS). Three distinct lineages were composed of species currently classified in the genus Amauroderma, here named the Amauroderma s.str. (1.0 BPP, 63 % BS), ‘Amauroderma rude’ clade (1.0 BPP) and ‘Amauroderma yunannense’ clade (1.0 BPP, 99 % BS), which clustered as the sister clade of Foraminispora (0.98 BPP). Four distinct lineages were composed of species currently classified in the genus Ganoderma, which are Ganoderma, ‘Ganoderma coffeatum’ clade, ‘Ganoderma ramosissimum’ clade (1.0 BPP, 100 % BS) , ‘Magoderna’ clade (1.0 BPP, 100 % BS) and ‘Trachyderma’ clade (1.0 BPP, 100 % BS). Finally, Tomophagus (1.0 BPP, 100 % BS) represented an independent lineage composed of two species.
The multiloci dataset recovered nine main clades, which consists of the clades in the ITS dataset, with exception to ‘Ganoderma coffeatum’ clade and ‘Ganoderma ramosissimum’ clade which were not included in the analyses. The clades are Amauroderma s.str. (1.0 BPP, 89 % BS), Ganoderma (1.0 BPP, 90 % BS), ‘Magoderna’ clade (1.0 BPP, 100 % BS), ‘Trachyderma’ clade (1.0 BPP, 100 % BS), Tomophagus (1.0 BPP, 100 % BS), ‘Amauroderma rude’ clade (1.0 BPP, 96 % BS), ‘Amauroderma yunannense’ clade (1.0 BPP, 99 % BS), and the new genera proposed here, Furtadoa (1.0 BPP, 92 % BS) and Foraminispora (1.0 BPP, 100 % BS). ‘Amauroderma yunnanense’ clade clustered as the sister clade of Foraminispora (1.0 BPP, 96 % BS) and this assemblage as a sister clade of Ganoderma (0.98 BPP, 52 % BS).
Taxonomy
Foraminispora Robledo, Costa-Rezende & Drechsler-Santos, gen. nov. — MycoBank MB819015
Etymology. Referring to the basidiospores with hollow endosporic projections which are continuous until the exospore wall. Foramen means hole, while spora means spore in Latin.
Typification. Porothelium rugosum Berk., Hooker’s J. Bot. Kew Gard. Misc. 8: 237. 1856.
Diagnosis — Similar to Amauroderma, differing by the spores with endosporic ornamentation as hollow columns, which are continuous until the exospore wall.
Basidiomata annual, stipe pleuropodal to pseudomesopodal, pileus circular to spathulate. Pilear surface glabrous, greyish brown to dark brown, concentrically zonate with thin blackish bands, radially rugose. Context white, homogenous, in section with a shiny black cuticle. Tubes slightly darker than context. Pore surface whitish to vivid orange. Pores regular, circular to angular. Dissepiments thick, entire. Stipe cylindrical, pale to dark brown, finely tomentose, solid to hollow, context homogeneous, whitish, in section with a shiny dark cuticle. Hyphal system dimitic, generative hyphae clamped, arboriform and skeleto-binding hyphae almost hyaline, dextrinoid. Cystidia and cystidioles absent. Basidia clavate, with four sterigmata. Basidiospores subglobose, hyaline to pale brown, double walled, with conspicuous ornamentation as endosporic projections column-like, some of them with a hole, that persists up to the exospore, IKI-.
Ecology & Distribution — Specimens growing on the ground or on decayed angiosperm wood in Brazil, Venezuela, French Guiana, Costa Rica and Cuba (Decock & Herrera-Figueroa 2006).
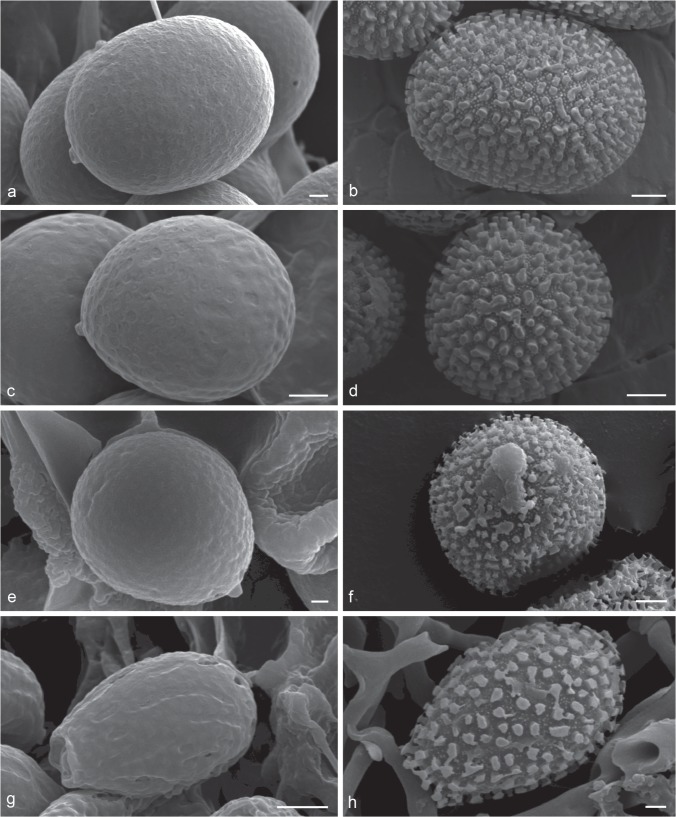
Notes — The new genus is characterized by stipitate basidiomata, dull pilear surface, whitish context, a dimitic hyphal system, skeleto-binding hyphae with lateral and apical branches and arboriform skeletal hyphae, both dextrinoid, and globose to subglobose, hyaline to pale brown spores, with conspicuous endosporic projections. Under SEM, it is possible to observe that some of the columnar endosporic projections are hollow and these holes persist until the exospore wall (Fig. 3). This feature is unique within Ganodermataceae, thus, it is considered as an exclusive feature for this genus.
Fig. 3.
Basidiospores of Foraminispora rugosa. a–b. Optical microscopy (KOH and Cotton blue, respectively). — c–f. SEM micrographs. c. General view showing holes in exospore; d. general view of endospore showing hollow columns; e–f. detail in connection between the hollow columns and exospore holes. — Scale bars: a–b = 10 μm; c = 2 μm; d–f = 1 μm.
The genus clearly fits into Ganodermataceae circumscription, due to its hyphal system with clamped generative and arboriform skeletal hyphae, as well as the double-walled basidiospores, with the inner layer ornamented. Both macro- and microscopic features of Foraminispora are shared with the genus Amauroderma, i.e., stipitate and annual basidiomata, a dimitic hyphal system and non-truncate basidiospores (Furtado 1962, 1981, Ryvarden & Johansen 1980, Corner 1983, Ryvarden 2004). However, an ultrastructural examination of some species of Amauroderma (A. calcigenum, A. pseudoboletus and A. schomburgkii) led us to conclude that the perforated column is absent in this genus (Fig. 4a–f).
Fig. 4.
Scanning Electron Micrograph of basidiospores of Amauroderma s.str. and Ganoderma. — a–b. Amauroderma calcigenum (CORD Robledo 394). a. General view showing exospore without holes; b. general view of endospore showing solid columns and smaller secondary ornamentation. — c–d. Amauroderma pseudoboletus (CORD Robledo 1441). c. General view showing exospore without holes; d. general view of endospore showing solid columns and smaller secondary ornamentation. — e–f. Amauroderma schomburgkii (CORD Robledo 909). e. General view showing exospore without holes; f. general view of endospore showing solid columns and smaller secondary ornamentation. — g–h. Ganoderma australe (CORD Robledo 3181). g. General view showing exospore without holes; h. general view of endospore showing solid columns and smaller secondary ornamentation. — Scale bars: a, c, e, h = 1 μm; b, d, f = 2 μm; g = 3 μm.
Ganoderma also presents species with pale context and double-walled spores with endosporic ornamentation (Ryvarden & Johansen 1980, Corner 1983, Ryvarden 2004, Torres-Torres & Guzmán-Dávalos 2012); however, the absence of the hollow columns (G. australe; Fig. 4g–h) and the truncate apex of basidiospores clearly distinguish this genus from Foraminispora. Ganoderma also has holes in the exospore of some species (G. lucidum, G. pfeifferi, G. valesiacum). Nevertheless, the holes are formed among the columns (Pegler & Young 1973). Haddowia and Humphreya also present species with pale context and double-walled spores with endosporic ornamentation; however, the ornamentation is formed by ridges. Tomophagus mainly differs from Foraminispora by its laccate and soft pileus and truncate basidiospores (Murrill 1905, Steyaert 1972, Ryvarden 2004, Tham et al. 2011). Since only Foraminispora rugosa is known to bear this feature, its whitish context and the vivid orange pore surface seem to be remarkable features of this genus in its current circumscription.
Foraminispora rugosa (Berk.) Costa-Rezende, Drechsler-Santos & Robledo, comb. nov. — MycoBank MB819019; Fig. 3
= Polyporus dubiopansus Lloyd, Lloyd Myco. Writ. 3: 125. 1921.
≡ Porothelium rugosum Berk., Hooker’s J. Bot. Kew Gard. Misc. 8: 237. 1856.
≡ Ganoderma sprucei Pat., Bull. Soc. Mycol. France 10: 75. 1894.
≡ Amauroderma sprucei (Pat.) Torrend, Brotéria, Sér. Bot. 18: 121. 1920
≡ Amauroderma dubiopansum (Lloyd) Ryvarden, Neotropical Polypores, Syn. Fungorum 19: 52. 2004.
Description — Decock & Herrera-Figueroa (2006) as Amauroderma sprucei.
Specimens examined. Brazil, Amazonas, Panure, Spruce 44, isotype herb. BPI 237203; Mato Grosso, Chapada dos Guimarães, Parque Nacional da Chapada dos Guimarães, Sítio Vale do Rio Claro, 7 Jan. 2013, D.H. Costa-Rezende 113, FLOR52191; ibid., 7 Jan. 2013, D.H. Costa-Rezende 114, FLOR 52184; ibid., 7 Jan. 2013, D.H. Costa-Rezende 115, FLOR 52192; ibid., 12 Jan. 2014, L. Pereira-Silva 21, FLOR52190; ibid., 12 Jan. 2014, L. Pereira-Silva 22, FLOR 52189; ibid., 12 Jan. 2014, L. Pereira-Silva 58, FLOR52186; ibid., 12 Jan. 2014, L. Pereira-Silva 77, FLOR52187; ibid., 12 Jan. 2014, L. Pereira-Silva 79, FLOR52185. – Argentina, Jujuy, Depto Ledesma, Parque Nacional Calilegua, Abra de Cañas, S23°40′38.2″ O64°53′46.3″, alt. 1730 m above sea level, 21 May 2007, Robledo 1507, CORD.
Notes — The dull concentric zonate pilear surface, the whitish context, the ochraceous to vivid orange pore surface, the small pores (5–7(–8) pores/mm), a crust with a short trichoderm in the pilear surface, the strongly dextrinoid skeletal hyphae and the predominantly subglobose basidiospores ((7–)8–10 × 7–9 μm), with conspicuous hollow columnar ornamentation are characteristic of this species. The species was described with a di-trimitic hyphal system, with generative and vegetative hyphae in all portions of basidioma, and the trama of tubes as dimitic with arboriform skeletal hyphae (Decock & Herrera-Figueroa 2006). In our observations, the hyphal system is considered dimitic. In the context, we have observed clamped generative hyphae, intercalary skeleto-biding hyphae, with long lateral and apical, thin branches, and skeletal hyphae (up to 7 μm diam), tortuous, with few apical ramifications. The trama of the tubes is composed of clamped generative, arboriform skeletals, and thick-walled skeleto-binding hyphae, formed by a main stalk and very short lateral branches, with or without two thin apical branches.
When Porothelium rugosum was combined in Ganoderma the epithet ‘rugosum’ was already occupied by Ganoderma rugosum, then the nome novum Ganoderma sprucei was proposed. The same happened when Torrend combined P. rugosum in Amauroderma, because the epithet ‘rugosum’ was occupied as well (Amauroderma rugosum). Torrend therefore continued to use ‘sprucei’, the earliest epithet available in Amauroderma. Considering the combination of Porothelium rugosum in Foraminispora the epithet is available.
Furtadoa Costa-Rezende, Robledo & Drechsler-Santos, gen. nov. — MycoBank MB819014
Etymology. Named in honour of Dr. João Salvador Furtado, due to his contribution to the taxonomy of Ganodermataceae.
Typification. Furtadoa biseptata gen. & sp. nov.
Diagnosis — Similar to Amauroderma, differing by presenting a monomitic context.
Basidiomata annual, stipe pleuropodal to pseudomesopodal, soft when fresh, light and fragile when dried, pileus circular to almost flabelliform or funnel-shaped. Pilear surface dull, glabrous, greyish brown, azonate. Context white to pale brown, homogenous. Tubes slightly darker than context. Pore surface pale brown. Pores angular, sometimes radially elongated. Dissepiments thin, entire to lacerate. Stipe yellowish brown, finely tomentose, solid to hollow, context homogeneous, pale brown. Hyphal system dimitic. Context composed of clamped to simple-septate generative hyphae, thin to slightly thick-walled, some distinctly wider, with a swollen apex. Trama of tubes composed of clamped generative and arboriform skeletal hyphae. Cystidia and cystidioles not seen. Basidia clavate, with four sterigmata. Basidiospores subglobose to ellipsoid, hyaline, double walled, with ornamentation as endosporic projections column-like, IKI-.
Ecology & Distribution — Specimens growing on the ground or on decayed angiosperm wood from Brazil, Guyana and Venezuela (Ryvarden 2004, Coelho et al. 2007, Gomes-Silva et al. 2015, as Amauroderma brasiliense).
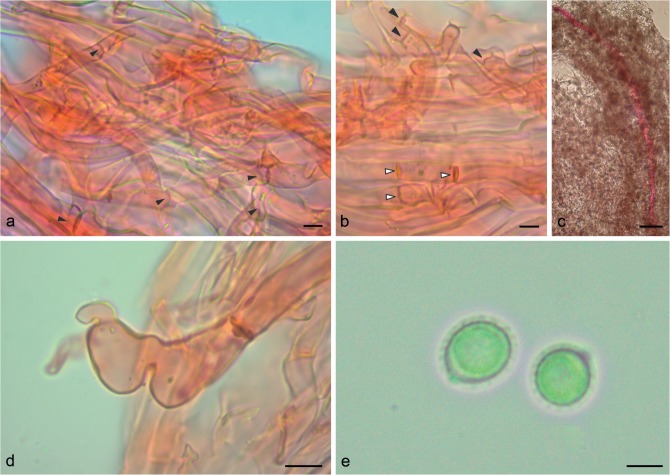
Notes — This new genus is characterized by a stipitate basidiomata, soft when fresh, dull pilear surface, pale context, a dimitic hyphal system, with a monomitic context, composed of both clamped and simple-septate generative hyphae (Fig. 5), thin to slightly thick-walled and dimitic trama of tubes, composed of clamped generative hyphae and arboriform skeletal hyphae and double-walled, ornamented basidiospores.
Fig. 5.
Micromorphology of Furtadoa biseptata. a–b. General view of monomitic hyphal system from context. a. Arrows indicates clamp connections; b. black arrows indicate clamp connections, white arrows indicate simple septate hyphae; c. general view of gloeoporus-like hyphae from context; d. detail in gloeoporus-like hyphae from context; e. basidiospores. — Scale bars: a–b, e = 5 μm; c = 50 μm; d = 10 μm.
Considering the double-walled basidiospores with the inner layer ornamented, the genus fits into Ganodermataceae circumscription. Both macro- and microscopic features of Furtadoa are shared with the genus Amauroderma, i.e., stipitate and annual basidiomata, presence of arboriform skeletal hyphae in the trama of tubes and double-walled, non-truncate basidiospores (Furtado 1962, 1981, Ryvarden & Johansen 1980, Corner 1983, Ryvarden 2004). However, the monomitic context with simple-septate generative hyphae is exclusive of this new genus in the context of the family. Regarding the other accepted genera in Ganodermataceae, besides the difference in the hyphal system, Ganoderma, Humphreya and Tomophagus have truncate basidiospores, and Haddowia has basidiospores with mainly longitudinal ridges (Steyaert 1972, Ryvarden 2004, Tham et al. 2012).
Furtadoa biseptata Costa-Rezende, Drechsler-Santos & Reck, sp. nov. — MycoBank MB819016; Fig. 5
Etymology. The species epithet refers to the two different septa in the generative hyphae that compose the context of the species.
Type. Brazil, Mato Grosso, Chapada dos Guimarães, Parque Nacional da Chapada dos Guimarães, Sítio Véu da Noiva, on the ground, 26 Mar. 2013, D.H. Costa-Rezende 128, holotype herb. FLOR50932.
Diagnosis — This species differs from F. brasiliensis by its thinner basidiomata, darker context, and the presence of simple-septate generative hyphae in the context.
Basidiomata stipitate, pleuropodal, single; pileus 25–45 mm diam, up to 10 mm thick, almost flattened to slightly convex, soft when fresh, corky when dry; margin incurved and irregular, becoming strongly involute upon dried. Pilear surface greyish brown, azonate, radially finely strigose, wrinkled at the center, glabrous. Context corky, pale brown, homogeneous, 0.3–5 mm thick, thinner near the margin. Tubes slightly darker than context, up to 3 mm long. Pore surface concolorous to context; pores circular, 3–5(–6) per mm, (200–)250–400 μm diam, (mean = 358.2 μm); dissepiment entire, 90–230 μm thick, (mean = 155.9 μm). Stipe solid to hollow, straight to tortuous, up to 50 mm long and 5 mm diam; surface velutinous, longitudinally corrugated, pale brown; context with the same consistency and concolorous with pilear context. Pilear surface composed of generative hyphae, 4–7 μm diam, thin to slightly thick-walled, parallel to the contextual hyphae. Hyphal system mono-dimitic; context composed of two kinds of generative hyphae: one clamped to occasionally simple-septate, 3–7 μm diam, hyaline, thin to slightly thick-walled, straight to tortuous, branched; the second gloeopleurous-like, rarely simple-septate, with long stretches without septa (up to 1 600 μm), 10–15 μm diam, hyaline, thin to slightly thick-walled, straight to tortuous, mostly unbranched, but eventually presenting some lateral short prolongations; trama of tubes composed of clamped generative hyphae, 3–5 μm diam, hyaline, thin walled; and arboriform skeletal hyphae with few apical, 4.5–6 μm diam in main stalk. Basidia subglobose to clavate, 4-sterigmate, 12–15 × 8–10 μm. Basidiospores subglobose to ellipsoid, ((6–)7–10 × (5.5–)6–8(–9) μm), (mean = 7.6 × 6.5 μm), Q = 1.07–1.33 (1.36), (mean-Q = 1.18), hyaline, double-walled with the inner layer finely and regular ornamented, verrucose under SEM, IKI-.
Notes — Furtadoa biseptata presents macro- and micromorphology that resembles Furtadoa brasiliensis, mainly differing by a thinner and darker pileus and by the presence of simple septa (Fig. 5). Furtadoa corneri differs from the new species by the funnel-shaped basidiomata and the thinner pileus, as well as by slightly larger basidiospores (8–10 × 6–8(–9) μm, mean = 8.2 × 7.4). Furtadoa biseptata was collected just once, even with several field expeditions across four years in the type locality, suggesting it to be a rare species.
Furtadoa brasiliensis (Singer) Costa-Rezende, Drechsler-Santos & Robledo, comb. nov. — MycoBank MB819017
≡ Scutiger brasiliensis Singer, Nova Hedwigia, Beih. 77: 22, 1983.
≡ Amauroderma brasiliense (Singer) Ryvarden, Syn. Fungorum 19: 44, 2004 ‘as A. brasilensis’.
Description — Singer et al. (1983) 22, ‘as Scutiger brasiliensis’.
Notes — Since Scutiger brasiliense was proposed, some different interpretations in its morphology have been raised. Scutiger brasiliense was described based on a specimen from Brazilian Amazonia and a specimen from Santa Catarina collected by Rick (Singer et al. 1983), with stipitate basidiomata with a white and soft-flesh context, monomitic hyphal system and inamyloid and ellipsoid to almost subglobose spores (7–9.3 × 6.3–8 μm) as the diagnostic characters. Amauroderma corneri was proposed fifteen years later to accommodate another monomitic species with Amauroderma-like basidiospores, based on a specimen from Atlantic Rain Forest in Brazil (Gulaid & Ryvarden 1998). However, the species was later considered under synonymy of A. brasiliense (Ryvarden 2004, Coelho et al. 2007, Gomes-Silva et al. 2015). In accordance with the morphological differences reported, i.e., A. corneri has a thin and funnel- to fan-shaped pileus, whitish when fresh, turning orange to brown when dried and A. brasiliense presents a thick and permanently pale basidiomata (Gomes-Silva et al. 2015), we preferred to maintain both taxa as independent species.
Furtadoa corneri (Gulaid & Ryvarden) Robledo & Costa-Rezende, comb nov. — MycoBank MB819018
≡ Amauroderma corneri Gulaid & Ryvarden, Mycol. Helv. 10 (1): 28. 1998.
Description — Gulaid & Ryvarden (1998) 28, as ‘A. corneri’.
Specimen examined. Brazil, São Paulo, Reg. Santos, Cananeia, Ilha do Cardoso, L. Ryvarden 24745, holotype herb. SP 213543.
Notes — Furtadoa corneri is characterized by a thin, funnel- to fan-shaped pileus, monomitic context and subglobose to ellipsoid basidiospores (8–10 × 6–8(–9) μm, mean = 8.2 × 7.4), IKI-.
DISCUSSION
Furtadoa, Foraminispora and Amauroderma s.str. within Ganodermataceae
In this work, we presented a molecular phylogenetic overview of the Ganodermataceae based on analyses with a wide dataset composed of the majority of the phylogenetic species with ITS sequences available in GenBank (NCBI) and a multiloci dataset (ITS+LSU+RPB1+TEF-1α) with a narrower sampling. These analyses, combined with morphological analyses evidenced new ultrastructural characters that enable a better understanding of the generic delimitation in the family. Our results agree with the polyphyletic status of Amauroderma previously proposed with morphological and phylogenetic approaches (Steyaert 1972, Gomes-Silva et al. 2015, Costa-Rezende et al. 2016).
A detailed examination of the morphology of some neotropical ‘deviating’ specimens of Amauroderma, previously determined as A. brasiliense and A. sprucei led us to observe some remarkable morphological features. Our phylogenetic analyses showed that those specimens grouped on different separated lineages, distinct from Amauroderma s.str., and, thus, two new genera are proposed to accommodate those species, as well as a new species is proposed. Furtadoa is proposed to accommodate 3 monomitic species (F. biseptata, F. brasiliensis and F. corneri) while Foraminispora was proposed to accommodate A. sprucei.
The monomitic context of F. biseptata (Fig. 5), F. brasiliensis and F. corneri may represent a synapomorphy of Furtadoa. As A. trichodermatum also has a monomitic context, future studies will probably point out that this species should be better placed in Furtadoa, as already suggested by Robledo et al. (2015), who speculated that A. trichodermatum and A. brasiliense could be related. Furtadoa appears as not closely related to Amauroderma s.str. in both analyses (Fig. 1, 2). Furtadoa brasiliensis and F. biseptata (both as A. brasiliense) appeared in a distinct lineage from Amauroderma s.str. in previous studies carried out by Gomes-Silva et al. (2015) and Costa-Rezende et al. (2016), supporting our proposition. Furthermore, hyphal system structure has been considered as a character to support the proposition of new genera among Agaricomycetes, especially polypores, such as in Perenniporiella, Yuchengia, Sanghuangporus, Tropicoporus and Phellinotus (Decock & Ryvarden 2003, Robledo et al. 2009, Zhao et al. 2013, Zhou et al. 2015, Drechsler-Santos et al. 2016).
The new species (F. biseptata) appears in a long branch in the retrieved phylogenetic trees, clustered as the sister clade of F. brasiliensis, which represents that there is a high genetic divergence between the taxa, in spite of their morphological similarity.
Foraminispora has a unique morphological feature among Ganodermataceae, the hollowed columnar endosporic projections of basidiospores, which is continuous until the exospore wall (Fig. 3). The ontogeny of endosporic ornamentation in Ganodermataceae is currently unexplored but it should be investigated in order to contribute to the taxa delimitation, as already observed in other polypore fungi, such as in Perenniporia s.lat. (Decock & Ryvarden 2003). Based both in nrITS and combined phylogenies, Fo. rugosa is not related to the Amauroderma s.str. clade (Fig. 1, 2), as observed by Costa-Rezende et al. (2016, as A. sprucei), corroborating the proposition of the new genus. In both phylogenetic analyses Foraminispora clustered as a sister group of ‘Amauroderma yunannense’ clade, which is composed only of A. yunnanense. This species also presents a homogeneous whitish to pale yellow context, similarly to Fo. rugosa (Li & Yuan 2015). Future studies based on basidiospores ultrastructure may point out that A. yunnanense should be placed in Foraminispora. Despite presenting basidiospores which are subglobose and not truncate, Foraminispora is more related to Ganoderma (Fig. 2; 0.98 BPP, 52 % BS) than to Amauroderma.
The genus Amauroderma, as usually morphologically circumscribed, comprises sessile to stipitate polypores with globose to ellipsoid basidiospores, without a truncate apex, double-walled basidiospores with the inner layer ornamented (rarely smooth, as in A. coltricioides), associated with fallen dead wood or roots of living or dead trees, with a tropical and subtropical distribution (Ryvarden 2004). Besides Furtadoa, Foraminispora and ‘Amauroderma yunannense’ clade, species usually included in Amauroderma clustered in two unrelated clades in both analysis (Fig. 1, 2). One of them is Amauroderma s.str., a taxon comprising neotropical species, which shares a sessile to stipitate basidiomata with a di-trimitic hyphal system, composed of clamped generative hyphae, arboriform to skeleto-binding hyphae (both in context and tubes) and non-truncated, double walled spores with solid columnar to semi-reticulate endosporic ornamentation. The second is the ‘Amauroderma rude’ clade, which is composed of species occurring outside the neotropical region (A. perplexum, A. rude, A. rugosum) and clustered in a distinct lineage from Amauroderma s.str., as also observed by Costa-Rezende et al. (2016). Further studies are needed to clarify the taxonomic status of this group since supposedly there are no morphological differences between these species and those of Amauroderma s.str.
Comments on Ganoderma, Tomophagus and unresolved taxa
Tomophagus was proposed to accommodate Polyporus colossus due to its light weight basidiomata and thick, soft spongy context, differing from Ganoderma. The genus was recovered as monophyletic both in the nrITS and combined analysis in the present study, as also observed in earlier studies (Moncalvo et al. 1995, Hong & Jung 2004, Tham et al. 2012, De Lima Júnior et al. 2014). Our results sustain the independency of Tomophagus against its synonymy under Ganoderma.
The Trachyderma clade is composed only of G. tsunodae, which is the type of Trachyderma, a genus that was mainly characterized by a fleshy succulent context when growing, differing from Ganoderma (Imazeki 1939, 1952). Unfortunately, according to the International Code of Nomenclature for algae, fungi, and plants the name Trachyderma is not valid since the name was first given to a lichenized Ascomycota. Therefore, further studies are needed to point out if the taxon is congeneric to Tomophagus, or represent a genus that should be properly proposed.
Except for G. coffeatum, G. ramosissimum G. subresinosum and G. tsunodae (treated above), all the Ganoderma species clustered in an homogeneous clade (Fig. 1, 2) mainly characterized by presenting a coriaceous to wood basidiomata and truncate spores with column-like endosporic projections (Fig. 4g–h), which in future studies could be attributed to Ganoderma s.str. The recovered topologies (Fig. 1, 2) does not corroborate the distinction between the genera Ganoderma and Elfvingia, even at subgeneric level (G. subg. Ganoderma and G. subg. Elfvingia) since none of these groups with dull and laccate species, respectively, were monophyletic, contrary to previous results, in which the laccate and the dull species appeared as two distinct clades (Moncalvo et al. 1995, Hong & Jung 2004).
Ganoderma subresinosum (Magoderna clade) was recovered in our topologies in a distinct lineage from Amauroderma s.str. and Ganoderma, as also observed by Gomes-Silva et al. (2015, as A. subresinosum) and Costa-Rezende et al. (2016, as A. subresinosum). Steyaert (1972) proposed the genera Haddowia, Humphreya and Magoderna, the last one typified by M. subresinosus, and contains two other species (M. infundibuliforme and M. vansteenisii), and was proposed to accommodate species with dimidiate to pleuropodal basidiomata, anticlinal hyphae (hymenioderm) in the pilear surface and ovoid-ellipsoid to spherical basidiospores without a truncate apex. Although the genus has been considered as synonym of Amauroderma (Furtado 1981) or Ganoderma (http://www.indexfungorum.org/names/Names.asp), according to our topology and the morphological circumscription of Steyaert (1972), Magoderna might be accepted at generic level.
Steyaert (1972) proposed the genus Humphreya to accommodate A. lloidii, P. coffeatus and H. endertii due to their hyphal disposition (peri- or pantoclinal) and basidiospore ornamentation (reticulate or disjointed cristae). Decock & Herrera-Figueroa (2007) reported that G. coffeatum has typical basidiospores with endosporic ornamentation as predominantly longitudinal ridges and with a known distribution in South and Central America. These authors refuted Steyaert’s combination since the vicinity of G. coffeatum and H. lloydii is uncertain. In our work, G. coffeatum clustered in an independent clade from the typical Ganoderma species (Fig. 1). In this way, the Steyaert’s concept of Humphreya may represent a genus independent of Ganoderma, but, since we have no other sequences from Humphreya, we consider that its position at genus level is still uncertain.
ANNOTATED KEY TO GENERA, PHYLOGENETIC CLADES AND GROUPS OF GANODERMATACEAE
This key includes accepted genera in the strict sense and phylogenetic groups as defined in the multigene phylogenetic analyses of this work. Species not included in our analysis that does not fits with any of the defined groups of the key are included in s.lat. genera concepts.
1. Endosporium with simple ornamentation, composed of single columns, occasionally 2–3 columns fused forming short isolated crests . . . . . . . . . . . . . . . 2
1. Endosporium with complex ornamentation, longitudinal or transversal crests, or a reticulated pattern. . . . . . . . . . . . . . . 11
2. Basidiospores truncate . . . . . . . . . . . . . . . 3
2. Basidiospores non truncate. . . . . . . . . . . . . . . 5
3. Vegetative hyphae brown to pale brown, context hard and fibrous, dark brown, brown to pale brown . . . . . . . . . . . . . . . Ganoderma1
3. Vegetative hyphae hyaline to pale yellowish, context soft, white, creamy white, to very pale brown. . . . . . . . . . . . . . . 4
4. Chlamydospores scattered in the context and trama, globose, reddish brown in KOH, basidiospores > 20 μm long. . . . . . . . . . . . . . . Tomophagus2
4. Chlamydospores absent, basidiospores < 20 μm long. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Trachyderma clade3
5. Hyphal system monomitic . . . . . . . . . . . . . . . 6
5. Hyphal system dimitic brown, dark to pale . . . . . . . . . . . . . . . 7
6. Pilear surface glabrous. . . . . . . . . . . . . . . Furtadoa4
6. Pilear surface hirsute strigose. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Amauroderma trichodermatum5
7. Context whitish, spores subglobose. . . . . . . . . . . . . . . 8
7. Context brown to pale, vegetative hyphae brown to pale, IKI-, spores subglobose to ellipsoid or ovoid . . . . . . . . . . . . . . . 9
8. Vegetative hyphae hyaline and dextrinoid. . . . . . . . . . . . . . . Foraminispora6
8. Vegetative hyphae pale yellow, IKI- . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Amauroderma yunnanense clade7
9. Neotropical species. . . . . . . . . . . . . . . Amauroderma s.str.8
9. Paleotropical species. . . . . . . . . . . . . . . 10
10. Basidiomata with whitish context and laccate pilear surface, basidiospores ovoid. . . . . . . . . . . . . . . Magoderna clade9
10. Basidiomata with pale brown context and upper surface dull, basidiospores typically ellipsoid to subglobose or globose. . . . . . . . . . . . . . . Amauroderma rude clade10
11. Endosporium with double longitudinal crests, partly connected by short transverse walls. . . . . . . . . . . . . . . Haddowia
11. Endosporium with crests or ridges ordered in a reticulated, longitudinal, transversal or ‘honey-comb’ pattern ornamentation. . . . . . . . . . . . . . . 12
12. Basidiospore truncate . . . . . . . . . . . . . . . Humphreya11
12. Basidiospore not truncate . . . . . . . . . . . . . . . Amauroderma deviatum12
Acknowledgments
The authors acknowledge the staff of the Parque Nacional da Chapada dos Guimarães for support in the field expeditions; Luciana Pereira-Silva for specimen collections; herbaria mentioned for the loan of reference material; Connie Baak for advising in the final manuscript editing; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for providing PhD and PDSE scholarships to DHCR; Fiocruz and LAMOL for performing the molecular sequencing; PPGBot UEFS, PPGFAP and BrBOL for partial financing of the research. GR acknowledges the assistance of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Córdoba for the support facilities used in this work. Financial support was provided by FONCYT (PICT-2015-0830) to G. Robledo. Authors kindly acknowledge Idea Wild for their support with technical equipment; and L. Caeiro (CPA CONICET-UNC) and D. Franchi for their technical support. This study is part of the project Fungos poliporóides (Agaricomycetes) do PARNA Chapada dos Guimarães, Mato Grosso–Políporos PNCG-MT.
Footnotes
1 Ganoderma includes traditional dull and shiny complexes/groups: Ganoderma australe/aplanatum complex, Ganoderma lucidum complex, Ganoderma resinaceum complex and others.
2 Tomophagus is so far represented by 2 species: T. collosus, the type species, and T. catienensis. Tomophagus collosus was suggested to be congeneric with G. tsunodae (Hattori & Ryvarden 1994). Although our analyses suggest a relationship between these species, whether the taxa are congeneric or not remains unclear.
3 Trachyderma clade is so far represented by Ganoderma tsunodae. Imazeki (1939, 1952) proposed Trachyderma as a new genus for this species. However, the generic name is illegitimate as a homonym of Trachyderma Norm. 1853 as pointed out by Ryvarden (1991).
4 Furtadoa is distinct from Amauroderma s.str. by presenting a monomitic hyphal system in context and a dimitic trama of tubes.
5 Amauroderma s.lat. species. The hyphal system structure and the pale colour of the context suggest a relationship with Furtadoa (Robledo et al. 2015).
6 Foraminispora rugosa is so far the only representative of Foraminispora, being characterized by a whitish context, dextrinoid vegetative hyphae and subglobose spores with conspicuous ornamentation as endosporic projections column-like, some of them with a hole, that persists up to the exospore.
7 Amauroderma s.lat. species. According to our phylogenetic analyses this species is related to Foraminisporus and further ultrastructural examination of basidiospores could prove that the taxa belongs to this genus.
8 Amauroderma s.str. is typified by A. schomburkii and as defined phylogenetically is so far restricted to the neotropical region. Morphologically the genus is characterized by stipitate basidiomata with a di-trimitic hyphal system, composed of clamped generative hyphae, arboriform to skeletobinding hyphae (both in context and tubes) and non-truncated, globose to ellipsoid spores with solid columnar to semi-reticulate endosporic ornamentation. The sessile species of Amauroderma were not included in phylogenetic analyses so far, so the inclusion of them in Amauroderma s.str. remains uncertain.
9 Magoderna is composed by M. subresinosus (type), M. infundibuliforme and M. vansteenisii, and was proposed to accommodate species with dimidiate to pleuropodal basidiomata, anticlinal hyphae (hymenioderm) in the pilear surface and ovoid-ellipsoid to globose basidiospores without a truncate apex (Steyaert 1972).
10 Amauroderma s.lat. species. Amauroderma perplexum, A. rude and A. rugosum presents typical morphology of Amauroderma s.str.; however, they are restricted to Paleotropics (Furtado 1981, Corner 1983). Further morphological and phylogenetic studies might corroborate the clade as a new genus.
11 Humphreya was proposed by Steyaert (1972) to accommodate species bearing basidiospores with reticulate, honey-comb or cristulate endosporium. Our results showed G. coffeatum as an independent clade, i.e., Ganoderma coffeatum clade. The relationship of H. coffeatum (and G. flaviporum, a species recently recovered from synonym of H. coffeatum) with Humphreya is uncertain, as previously suggested by Decock & Herrera-Figueroa (2007).
12 Amauroderma s.lat. species. Amauroderma deviatum presents broadly ellipsoid up to subglobose or slightly ovoid spores, with well-marked endosporic ridges,reticulated forming a ‘honey-comb’ pattern and secondary, lower ridges forming an irregularly reticulate pattern (Decock & Herrera-Figueroa 2007).
REFERENCES
- Coelho G, Cortez VG, Guerrero RT. 2007. New morphological data on Amauroderma brasiliense (Polyporales, Basidiomycota). Mycotaxon 100: 177–183. [Google Scholar]
- Corner EJH. 1983. Ad Polyporaceas I. Amauroderma and Ganoderma. Beihefte zur Nova Hedwigia, Weinheim. [Google Scholar]
- Costa-Rezende DH, Gugliotta AM, Góes-Neto A, et al. 2016. Amauroderma calcitum sp. nov. and notes on taxonomy and distribution of Amauroderma species (Ganodermataceae). Phytotaxa 244, 2: 101–124. [Google Scholar]
- Crespo EM, Robledo GL. 2016. La microscopía electrónica de barrido revela nuevos caracteres ultraestructurales en las basidiósporas de Amauroderma (Ganodermataceae, Basidiomycota). Acta Microscopica 25 (A). [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9 (8): 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lima Júnior NC, Gibertoni TB, Malosso E. 2014. Delimitation of some neotropical laccate Ganoderma (Ganodermataceae): molecular phylogeny and morphology. Revista de Biologia Tropical 62, 3: 1197–1208. [PubMed] [Google Scholar]
- Decock C, Amalfi M, Robledo G, et al. 2013. Phylloporia nouraguensis, an undescribed species on Myrtaceae from French Guiana. Cryptogamie, Mycologie 34: 15–27. [Google Scholar]
- Decock C, Herrera-Figueroa S. 2006. Neotropical Ganodermataceae (Basidiomycota): Amauroderma sprucei and A. dubiopansum. Criptogamie, Mycologie 27, 1: 3–10. [Google Scholar]
- Decock C, Herrera-Figueroa S. 2007. Studies in Ganodermataceae (Basidiomycota): the concept of Ganoderma coffeatum in the Neotropics and East Asia. Criptogamie, Mycologie 28, 2: 77–89. [Google Scholar]
- Decock C, Ryvarden L. 2003. Perenniporiella gen. nov. segregated from Perenniporia, including a key to neotropical Perenniporia species with pileate basidiomes. Mycological Research 107, 1: 93–103. [DOI] [PubMed] [Google Scholar]
- Dentinger BTM, Margaritescu S, Moncalvo JM. 2010. Rapid and reliable high-throughput methods of DNA extraction for use in barcoding and molecular systematics of mushrooms. Molecular Ecology Resources 10: 628–633. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid isolation procedure for small quantities of fresh tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Drechsler-Santos ER, Robledo GL, Lima-Júnior NC, et al. 2016. Phellinotus, a new neotropical genus in the Hymenochaetaceae (Basidiomycota, Hymenochaetales). Phytotaxa 261, 3: 218–239. doi: http://dx.doi.org/10.11646/phytotaxa.261.3.2. [Google Scholar]
- Furtado JS. 1962. Structure of the spore of the Ganodermoideae Donk. Rickia 1: 227–241. [Google Scholar]
- Furtado JS. 1965. Relation of microstructures to the taxonomy of the Ganodermoideae (Polyporaceae) with special reference to the structure of the cover of the pilear surface. Mycologia 57: 588–611. [Google Scholar]
- Furtado JS. 1981. Taxonomy of Amauroderma (Basidiomycetes, Polyporaceae). Memoirs of the New York Botanical Garden 34: 1–109. [Google Scholar]
- Góes-Neto A, Loguercio-Leite C, Guerrero RT. 2005. DNA extraction from frozen field-collected and dehydrated herbarium fungal basidiomata: performance of SDS and CTAB-based methods. Biotemas 18, 2: 19–32. [Google Scholar]
- Gomes-Silva AC, Lima-Júnior N, Malosso E, et al. 2015. Delimitation of taxa in Amauroderma (Ganodermataceae, Polyporales) based in morphology and molecular phylogeny of Brazilian specimens. Phytotaxa 227, 3: 201–228. [Google Scholar]
- Gottlieb AM, Wright JE. 1999a. Taxonomy of Ganoderma from southern South America: subgenus Ganoderma. Mycological Research 103, 6: 661–673. [Google Scholar]
- Gottlieb AM, Wright JE. 1999b. Taxonomy of Ganoderma from southern South America: subgenus Elfvingia. Mycological Research 103, 10: 1289–1298. [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Gulaid H, Ryvarden L. 1998. Two new species of Amauroderma (Ganodermataceae, Basidiomycetes). Mycologia Helvetica 10, 1: 25–30. [Google Scholar]
- Hattori T, Ryvarden L. 1994. Type studies in the Polyporaceae. 25. Species described from Japan by R. Imazeki & A. Yasuda. Mycotaxon 50: 27–46. [Google Scholar]
- Hong SG, Jung HS. 2004. Phylogenetic analysis of Ganoderma based on nearly complete mitochondrial small-subunit ribosomal DNA sequences. Mycologia 96, 4: 742–755. [DOI] [PubMed] [Google Scholar]
- Imazeki R. 1939. Studies in Ganoderma of Nippon. Bulletin of the Tokyo Science Museum 1: 29–52. [Google Scholar]
- Imazeki R. 1952. A contribution to the fungous flora of Dutch New Guinea. Bulletin of the Government Forest Experimental Station Meguro 57: 87–128. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, et al. 2008. Ainsworth & Bisby’s Dictionary of the Fungi. CABI, The Netherlands. [Google Scholar]
- Li MJ, Yuan HS. 2015. Type studies on Amauroderma species described by J.D. Zhao et al. and the phylogeny of species in China. Mycotaxon 130: 79–89. [Google Scholar]
- Matheny PB, Liu YJ, Ammirati JF, et al. 2002. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany 89: 688–698. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010: 1–8. New Orleans, LA. [Google Scholar]
- Moncalvo J, Hsi-Hua W, Ruey-Shyang H. 1995. Phylogenetic relationships in Ganoderma inferred from the Internal Transcribed Spacers and 25S ribosomal DNA sequences. Mycologia 87, 2: 223–238. [Google Scholar]
- Moncalvo J, Ryvarden L. 1997. A nomenclatural study of the Ganodermataceae Donk. Synopsis Fungorum 11. Fungiflora, Oslo. [Google Scholar]
- Murrill WA. 1905. The Polyporaceae of North America: X. A synopsis of the brown pileate species. Bulletin of the Torrey Botanical Club 32, 7: 366. [Google Scholar]
- Pegler DN, Young TWK. 1973. Basidiospore form in the British species of Ganoderma Karst. Kew Bulletin 28: 351–364. [Google Scholar]
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Robledo GL, Amalfi M, Rajchenberg M, et al. 2009. Perenniporiella chaquenia sp. nov. and further notes on Perenniporiella and its relationships with Perenniporia (Poriales, Basidiomycota). Mycologia 101, 5: 657–673. [DOI] [PubMed] [Google Scholar]
- Robledo GL, Newman DS, Popoff OF, et al. 2015. Amauroderma trichodermatum (Ganodermataceae, Basidiomycota): first record from Bolivia and geographic distribution map, with notes on nomenclature and morphology. Check List 11, 4: 1671. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 12: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ryvarden L. 1991. Genera of Polypores. Nomenclature and taxonomy. Synopsis Fungorum 5. Fungiflora: Oslo, Norway. [Google Scholar]
- Ryvarden L. 2004. Neotropical polypores Part 1. Synopsis Fungorum. Fungiflora, Oslo. [Google Scholar]
- Ryvarden L, Johansen I. 1980. A preliminary polypore flora of East Africa. Fungiflora, Oslo. [Google Scholar]
- Singer R, Araujo I, Ivory MH. 1983. The ectotrophically mycorrhizal fungi of the neotropical lowlands, especially central Amazonia. Nova Hedwigia, Beiheft 77: 22. [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: A tool for phylogenetic analysis and post analysis of large phylogenies. Bioinformatics 30, 9: 1312–1313. doi: https://doi.org/10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert RL. 1972. Species of Ganoderma and related genera mainly of the Bogor and Leiden herbaria. Persoonia 7: 55–118. [Google Scholar]
- Steyaert RL. 1980. Study of some Ganodermas pecies. Bulletin du Jardin Botanique National de Belgique 50: 135–186. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham LX, Hung NLQ, Duong PN, et al. 2012. Tomophagus cattienensis sp. nov., a new Ganodermataceae species from Vietnam: Evidence from morphology and ITS DNA barcodes. Mycological Progress 11: 775–780. [Google Scholar]
- Thiers B. [continuously updated]. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/science/ih/ [accessed 5 Jan. 2017].
- Torres-Torres MG, Guzmán-Dávalos L. 2012. The morphology of Ganoderma species with a laccate surface. Mycotaxon 119: 201–216. [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172, 8: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Cui B, Steffen KT. 2013. Yuchengia, a new polypore genus segregated from Perenniporia (Polyporales) based on morphological and molecular evidence. Nordic Journal of Botany 31: 331–338. [Google Scholar]
- Zhou L, Vlasák J, Decock C, et al. 2015. Global diversity and taxonomy of the Inonotus linteus complex (Hymenochaetales, Basidiomycota): Sanghuangporus gen. nov., Tropicoporus excentrodendri and T. guanacastensis gen. et spp. nov., and 17 new combinations. Fungal Diversity 77, 1: 335–347. [Google Scholar]