Abstract
Species of Colletotrichum are considered important plant pathogens, saprobes, and endophytes on a wide range of plant hosts. Several species are well-known on citrus, either as agents of pre- or post-harvest infections, such as anthracnose, postbloom fruit drop, tear stain and stem-end rot on fruit, or as wither-tip of twigs. In this study we explored the occurrence, diversity and pathogenicity of Colletotrichum spp. associated with Citrus and allied genera in European orchards, nurseries and gardens. Surveys were carried out during 2015 and 2016 in Greece, Italy, Malta, Portugal and Spain. A total of 174 Colletotrichum strains were isolated from symptomatic leaves, fruits, petals and twigs. A multi-locus phylogeny was established based on seven genomic loci (ITS, GAPDH, ACT, CAL, CHS-1, HIS3 and TUB2), and the morphological characters of the isolates determined. Preliminary pathogenicity tests were performed on orange fruits with representative isolates. Colletotrichum strains were identified as members of three major species complexes. Colletotrichum gloeosporioides s.str. and two novel species (C. helleniense and C. hystricis) were identified in the C. gloeosporioides species complex. Colletotrichum karstii, C. novae-zelandiae and two novel species (C. catinaense and C. limonicola) in the C. boninense species complex, and C. acutatum s.str. was also isolated as member of C. acutatum species complex. Colletotrichum gloeosporioides and C. karstii were the predominant species of Colletotrichum isolated. This study represents the first report of C. acutatum on citrus in Europe, and the first detection of C. novae-zelandiae from outside New Zealand. Pathogenicity tests revealed C. gloeosporioides s.str. to be the most virulent species on fruits. The present study improves our understanding of species associated with several disease symptoms on citrus fruits and plants, and provides useful information for effective disease management.
Keywords: Anthracnose, Citrus, multi-locus sequence typing, pathogenicity
INTRODUCTION
Colletotrichum is one of the most important genera of plant pathogenic fungi, responsible for several diseases in many crops worldwide (Sutton 1992, Cannon et al. 2000, 2012, Cai et al. 2009, Udayanga et al. 2013). Colletotrichum spp. were recently included in the list of the 10 most important plant pathogenic fungi in the world, based on perceived scientific and economic importance (Dean et al. 2012). Agricultural production losses caused by Colletotrichum spp. involve important staple food crops grown in developing countries throughout the tropics and subtropics (Dean et al. 2012). Colletotrichum species can infect more than 30 plant genera (Perfect et al. 1999, Farr et al. 2006, Damm et al. 2012a, b, Farr & Rossman 2017), causing anthracnose disease and postharvest decay on a wide range of tropical, subtropical and temperate fruits, grasses, vegetable crops and ornamental plants (Bailey & Jeger 1992, Bernstein et al. 1995, Freeman & Shabi 1996, Crouch et al. 2009, Lima et al. 2011, Damm et al. 2012a, b, Anderson et al. 2013, Crous et al. 2016b, Guarnaccia et al. 2016, De Silva et al. 2017). Moreover, many Colletotrichum species are latent plant pathogens, endophytes, epiphytes or saprobes, switching to a pathogenic lifestyle when host plants are subjected to stress conditions, or placed in postharvest storage (Crous et al. 2016a). Appressoria that develop from germinating spores, start plant infection by penetration of the cuticle (Deising et al. 2000) and occasionally also of the epidermal cells via fungal hyphae (Bailey & Jeger 1992).
The taxonomy of Colletotrichum species has recently been reviewed in several impactful studies (Cannon et al. 2008, Cai et al. 2009, Damm et al. 2009, 2012a, b, 2013, 2014, Weir et al. 2012, Liu et al. 2014, 2015, 2016). Before the molecular era, morphological characters such as size and shape of conidia and appressoria, presence or absence of setae, aspect, colour and growth rate of the colonies, formed the basis to study and compare the taxonomy of Colletotrichum species (Von Arx 1957, Sutton 1980, 1992). Modern studies demonstrated that these characters are not reliable for species level identification due to their variability under changing environmental conditions (Cai et al. 2009, Liu et al. 2016).
Following adoption of the use of multi-gene phylogenetic analysis, the polyphasic protocols for studying the genus Colletotrichum significantly changed the classification and species concepts in Colletotrichum (Cannon et al. 2012, Damm et al. 2012a, b, 2013, 2014, Weir et al. 2012). Several systematic studies of nearly all acknowledged species have led to the identification of 11 Colletotrichum species complexes, and more than 20 singleton species (Cannon et al. 2012, Liu et al. 2014, 2016, Marin-Felix et al. 2017). In plant pathology the most important species are members of the C. gloeosporioides (Cannon et al. 2008, Phoulivong et al. 2010, Weir et al. 2012), C. acutatum (Marcelino et al. 2008, Shivas & Tan 2009, Damm et al. 2012a, Baroncelli et al. 2015), C. boninense (Moriwaki et al. 2003, Yang et al. 2009, Damm et al. 2012b) and C. truncatum (Damm et al. 2009, Cannon et al. 2012) complexes. The use of multi-locus phylogenetic analyses revealed many cases, in which certain Colletotrichum spp. that were historically considered to be causal agents of economically important plant disease, were then revealed to be different species, such as C. alienum which seems to be the most important species in Proteaceae cultivation (Liu et al. 2013), and not C. gloeosporioides s.str. as previously assumed (Lubbe et al. 2004).
The citrus industry is one of the most important fruit industries worldwide. The Mediterranean countries are second only to China for fruit production, and are the largest fruit exporter after South Africa (FAO 2016). Therefore, the study and knowledge of all the pathogens affecting this crop is imperative. The use of a polyphasic approach in the past revealed many cryptic and new Colletotrichum species associated with citrus, belonging to four species complexes, namely: the C. boninense species complex (C. boninense, C. citricola, C. constrictum, C. karstii and C. novae-zelandiae) (Damm et al. 2012b, Huang et al. 2013); the C. acutatum species complex (C. abscissum, C. acutatum, C. citri, C. godetiae, C. johnstonii, C. limetticola and C. simmondsii) (Damm et al. 2012a, Huang et al. 2013, Crous et al. 2015); the C. truncatum species complex (C. truncatum) (Damm et al. 2009) and the C. gloeosporioides species complex (C. fructicola, C. gloeosporioides, C. kahawae subsp. ciggaro and C. siamense) (Weir et al. 2012, Huang et al. 2013, Perrone et al. 2016, Liu et al. 2016). Further Colletotrichum species such as C. brevisporum and C. tropicicola have been reported in association with citrus (Huang et al. 2013).
Several major diseases of citrus are caused internationally by Colletotrichum species (Timmer et al. 2000, Lima et al. 2011). According to several reports published before the main Colletotrichum revisions (Damm et al. 2009, 2012a, b, 2013, 2014, Weir et al. 2012), C. gloeosporioides and C. abscissum (previously known as C. acutatum) are the causal agents of postbloom fruit drop (PFD) in Brazil (Peres et al. 2008, Lima et al. 2011, Crous et al. 2015) and Bermuda (McGovern et al. 2012), causing petal necrosis, abscission of developing fruit and the formation of persistent calyces of various citrus species. A recent extensive investigation in citrus orchards of São Paulo state (Brazil), revealed only C. abscissum and C. gloeosporioides s.str. associated with PFD disease (Silva et al. 2016). Key lime anthracnose (KLA), a disease complex relating to leaves, flowers and fruits of Key lime, was initially reported to be caused by C. acutatum (Brown et al. 1996, Peres et al. 2008, MacKenzie et al. 2009), but later classified as C. limetticola (Damm et al. 2012a). Colletotrichum gloeosporioides was previously thought to be the only Colletotrichum species causing post-harvest anthracnose (Brown 1975, Sutton 1980, Freeman & Shabi 1996), but recent works showed that several species of Colletotrichum are associated with fruit decay worldwide (Peng et al. 2012, Damm et al. 2012a, b, Weir et al. 2012). Huang et al. (2013) demonstrated the ability of C. fructicola and C. truncatum to cause anthracnose on citrus fruits. Moreover, C. gloeosporioides s.lat. was also reported to cause pre-harvest symptoms such as wither-tip on twigs, tear-stain (Klotz 1961, Benyahia et al. 2003) and stem-end rot on fruit (Kaur et al. 2007).
Recently, various infections caused by Colletotrichum spp. strongly compromised citrus production in different Mediterranean countries: heavy pre-harvest anthracnose symptoms appeared on orange fruits and lesions on leaves of mandarins in Italy (Aiello et al. 2015, Perrone et al. 2016), twig wither-tip symptoms were observed on cultivated orange trees in Tunisia (Rhaiem & Taylor 2016), and severe anthracnose symptoms on unripe and ripe lemon fruits were recorded in Portugal (Ramos et al. 2016). In these studies, Colletotrichum species belonging to the C. acutatum species complex were never found associated with citrus. However, C. acutatum s.lat. was reported in Mediterranean countries causing diseases on several hosts such as Fragaria × ananassa (Garrido et al. 2008), Arbutus unedo (Polizzi et al. 2011) and Olea europaea (Talhinhas et al. 2011). Because of the commercial yield losses in citrus orchards caused by Colletotrichum infections, the recent findings and the changes in the species concepts, new surveys are required to study the Colletotrichum species diversity related to citrus and their occurrence and association with foliar and fruit diseases.
The current study aimed to investigate the major citrus production areas in Europe by large-scale sampling, and to identify isolates via morphology and multi-locus phylogeny based on modern taxonomic concepts. In 2015 and 2016 several surveys were conducted in commercial nurseries, citrus orchards, gardens, backyards and plant collections to determine the occurrence of Colletotrichum spp. associated with Citrus and allied genera (Atlantia, Fortunella, Microcitrus, Murraya, Poncirus). In particular the objectives of the present study were:
i) to conduct extensive surveys for sampling fresh plant materials;
ii) to cultivate as many Colletotrichum isolates as possible;
iii) to subject those isolates to DNA sequence analyses combined with morphological characterisation;
iv) to compare the obtained results with the data from other phylogenetic studies on the genus; and
v) to evaluate the pathogenicity of Colletotrichum species to citrus fruit.
MATERIALS AND METHODS
Sampling and isolation
During 2015 and 2016 several surveys were conducted in many of the main citrus-producing regions of Europe. Fruits and leaves with lesions and typical anthracnose symptoms and twigs showing wither-tip were collected from more than 70 sites in Andalusia, Valencia, Balearic Islands (Spain), Apulia, Calabria, Sicily, Eoalian Islands (Italy), Algarve (Portugal), Missolonghi, Nafplio, Arta, Crete (Greece) and Malta and Gozo (Malta). Investigated species of Citrus and allied genera such as Atlantia, Fortunella, Microcitrus, Murraya and Poncirus (Rutaceae) included Australasian lime, citranges, citrons, kumquat, mandarins, oranges, pummelo, grapefruit, limes, lemons and ornamental brushes.
Fungal isolates were obtained following two procedures. In the first, leaf, fruit and twig fragments (5 × 5 mm) were surface-sterilised in a sodium hypochlorite solution (10 %) for 20 s, followed by 70 % ethanol for 30 s, and rinsed three times in sterilised water. The fragments were dried in sterilised tissue paper, placed on malt extract agar (MEA; Crous et al. 2009) amended with 100 μg/mL penicillin and 100 μg/mL streptomycin (MEA-PS) and incubated at 25 °C until characteristic Colletotrichum colonies were observed. In the second procedure, plant material was incubated in moist chambers at room temperature (25 °C ± 3 °C) for up to 10 d and inspected daily for fungal sporulation. Sporulating conidiomata obtained through both procedures were collected and crushed in a drop of sterile water and then spread over the surface of MEA-PS plates. After 24 h germinating spores were individually transferred onto MEA plates. The isolates used in this study are maintained in the culture collection of the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands, and in the working collection of Pedro Crous (CPC), housed at the Westerdijk Institute.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega Corporation, WI, USA) following the manufacturer’s instructions. Partial regions of seven loci were amplified. The primers ITS5 and ITS4 (White et al. 1990) were used to amplify the internal transcribed spacer region (ITS) of the nuclear ribosomal RNA operon, including the 3’ end of the 18S rRNA, the first internal transcribed spacer region, the 5.8S rRNA gene; the second internal transcribed spacer region and the 5’ end of the 28S rRNA gene. The primers CL1 and CL2 (O’Donnell et al. 2000) were used to amplify part of the calmodulin (CAL) gene. The partial glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was amplified using primers GDF1 + GDR1 (Guerber et al. 2003). The primers ACT-512F and ACT-783R (Carbone & Kohn 1999) were used to amplify part of the actin gene (ACT). The partial beta-tubulin (TUB2) gene was amplified with primers T1 (Glass & Donaldson 1995) and Bt-2b (O’Donnell & Cigelnik 1997). The primers CHS-79F and CHS-345R (Carbone & Kohn 1999) were used to amplify part of the chitin synthase 1 (CHS-1) gene. The partial histone3 (HIS3) gene was amplified with primers CYLH3F and CYLH3R (Crous et al. 2004b).
The PCR amplification mixtures and cycling conditions for all seven loci were followed as described by Damm et al. (2012b). The PCR products were sequenced in both directions using the BigDye® Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems Life Technologies, Carlsbad, CA, USA), after which amplicons were purified through Sephadex G-50 Fine columns (GE Healthcare, Freiburg, Germany) in MultiScreen HV plates (Millipore, Billerica, MA). Purified sequence reactions were analysed on an Applied Biosystems 3730xl DNA Analyzer (Life Technologies, Carlsbad, CA, USA). The DNA sequences generated were analysed and consensus sequences were computed using the program SeqMan Pro (DNASTAR, Madison, WI, USA).
Phylogenetic analyses
Novel sequences generated in this study were blasted against the NCBIs GenBank nucleotide database to determine the closest relatives for a taxonomic framework of the studied isolates. Alignments of different gene regions, including sequences obtained from this study and sequences downloaded from GenBank, were initially performed by using the MAFFT v. 7 online server (http://mafft.cbrc.jp/alignment/server/index.html) (Katoh & Standley 2013), and then manually adjusted in MEGA v. 6.06 (Tamura et al. 2013). To establish the identity of isolates at species level, phylogenetic analyses were conducted first individually for each locus (data not shown) and then as concatenated analyses of seven loci. Two separate analyses were conducted for the C. boninense species complex and the remainder of the Colletotrichum spp. included in this study. Additional reference sequences were selected based on recent studies on Colletotrichum species (Damm et al. 2012a, b, Weir et al. 2012, Huang et al. 2013). Phylogenetic analyses were based on Maximum Parsimony (MP) for all the individual loci and on both MP and Bayesian Inference (BI) for the multilocus analyses. For BI, the best evolutionary model for each partition was determined using MrModeltest v. 2.3 (Nylander 2004) and incorporated into the analyses. MrBayes v. 3.2.5 (Ronquist et al. 2012) was used to generate phylogenetic trees under optimal criteria per partition. The Markov Chain Monte Carlo (MCMC) analysis used four chains and started from a random tree topology. The heating parameter was set to 0.2 and trees were sampled every 1 000 generations. Analyses stopped once the average standard deviation of split frequencies was below 0.01. The MP analyses were done using PAUP (Swofford 2003). Phylogenetic relationships were estimated by heuristic searches with 100 random addition sequences. Tree bisection-reconnection was used, with the branch swapping option set on ‘best trees’ only with all characters weighted equally and alignment gaps treated as fifth state. Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC) were calculated for parsimony and the bootstrap analyses (Hillis & Bull 1993) were based on 1 000 replications. Sequences generated in this study were deposited in GenBank (Table 1) and alignments and phylogenetic trees in TreeBASE (www.treebase.org).
Table 1.
Collection details and GenBank accession numbers of isolates included in this study.
| Species | Culture no.1 | Host | Locality | Associated symptoms | GenBank no.2 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | ACT | CAL | CHS-1 | TUB2 | HIS3 | |||||
| Colletotrichum abscissum | COAD 1876 | Citrus sinensis | Brazil | – | KP843124 | KP843127 | KP843139 | – | KP843130 | KP843133 | KP843136 |
| COAD 1877 | Citrus sinensis | Brazil | – | KP843126 | KP843129 | KP843141 | – | KP843132 | KP843135 | KP843138 | |
| C. acutatum | CBS 112759 | Hakea sericea | South Africa | – | JQ948391 | JQ948722 | JQ949712 | – | JQ949052 | JQ950042 | JQ949382 |
| CBS 112996 | Carica papaya | Australia | – | JQ005776 | JQ948677 | JQ005839 | – | JQ005797 | JQ005860 | JQ005818 | |
| CBS 129952 | Olea europaea | Portugal | – | JQ948364 | JQ948695 | JQ949685 | – | JQ949025 | JQ950015 | JQ949355 | |
| CBS 142407 = CPC 27005 | Citrus sinensis | Italy, Messina | Leaf lesion | KY856397 | KY856221 | KY855968 | – | KY856133 | KY856479 | KY856303 | |
| CPC 26987 | Citrus limon | Italy, Messina | Leaf lesion | KY856398 | KY856222 | KY855969 | – | KY856134 | KY856480 | KY856304 | |
| C. alienum | ICMP 12071 | Malus domestica | New Zealand | – | JX010251 | JX010028 | JX009572 | JX009654 | JX009882 | JX010411 | – |
| C. annellatum | CBS 129826 | Hevea brasiliensis | Colombia | – | JQ005222 | JQ005309 | JQ005570 | JQ005743 | JQ005396 | JQ005656 | JQ005483 |
| C. asianum | CBS 130418 | Coffea arabica | Thailand | – | FJ972612 | JX010053 | JX009584 | FJ917506 | JX009867 | JX010406 | KY856305 |
| C. boninense | CBS 123755 | Crinum asiaticum ‘Sinicum’ | Japan | – | JQ005153 | JQ005240 | JQ005501 | JQ005674 | JQ005327 | JQ005588 | JQ005414 |
| GZAAS5.09505 | Citrus medica | China | – | JQ247622 | JQ247598 | JQ247646 | – | – | JQ247634 | – | |
| C. brevisporum | GZAAS5.09545 | Citrus medica | China | – | JQ247623 | JQ247599 | JQ247647 | JQ247589 | – | JQ247635 | – |
| C. camelliae | ICMP 10643 | Camellia × williamsii | UK | – | JX010224 | JX009908 | JX009540 | JX009630 | JX009891 | JX010436 | – |
| C. catinaense | CBS 142416 = CPC 28019 | Citrus sinensis | Portugal, Mesquita | Fruit tear stain | KY856399 | KY856223 | KY855970 | KY856052 | KY856135 | KY856481 | KY856306 |
| CBS 142417 = CPC 27978 | Citrus reticulata | Italy, Catania | Leaf lesion | KY856400 | KY856224 | KY855971 | KY856053 | KY856136 | KY856482 | KY856307 | |
| CPC 28149 | Citrus aurantiifolia | Italy, Catania | Twigs wither-tip | KY856401 | KY856225 | KY855972 | KY856054 | KY856137 | KY856483 | KY856308 | |
| C. citri | CBS 134233 | Citrus aurantiifolia | China | – | KC293581 | KC293741 | KY855973 | KC293701 | KY856138 | KC293661 | KY856309 |
| CBS 134234 | Citrus aurantiifolia | China | – | KC293582 | KC293742 | KY855974 | KC293702 | KY856139 | KC293662 | KY856310 | |
| C. citricola | CBS 134228 | Citrus unchiu | China | – | KC293576 | KC293736 | KC293616 | KC293696 | KY856140 | KC293656 | KY856311 |
| CBS 134229 | Citrus unchiu | China | – | KC293577 | KC293737 | KC293617 | KC293697 | KY856141 | KC293657 | KY856312 | |
| C. constrictum | CBS 128504 | Citrus limon | New Zealand | – | JQ005238 | JQ005325 | JQ005586 | JQ005759 | JQ005412 | JQ005672 | KY856313 |
| C. fructicola | CBS 238.49 | Ficus edulis | Germany | – | JX010181 | JX009923 | JX009495 | JX009671 | JX009839 | JX010400 | KY856314 |
| CBS 125397 | Tetragastris panamensis | Panama | – | JX010173 | JX010032 | JX009581 | JX009674 | JX009874 | JX010409 | KY856315 | |
| ICMP 18581 | Coffea arabica | Thailand | – | JX010165 | JX010033 | FJ907426 | FJ917508 | JX009866 | JX010405 | – | |
| C. gloeosporioides | CBS 112999 | Citrus sinensis | Italy | – | JX010152 | JX010056 | JX009531 | JX009731 | JX009818 | JX010445 | KY856316 |
| CBS 142408 = CPC 28059 | Citrus sinensis ‘Lanelate’ | Spain, Moncada | Petal lesions | KY856402 | KY856226 | KY855975 | KY856055 | KY856142 | KY856484 | KY856317 | |
| CPC 26172 | Citrus sinensis ‘Tarocco Tapi’ | Italy, Catania | Twigs wither-tip | KY856403 | KY856227 | KY855976 | KY856056 | KY856143 | KY856485 | KY856318 | |
| CPC 26178 | Citrus sinensis ‘Tarocco Tapi’ | Italy, Catania | Leaf lesion | KY856404 | KY856228 | KY855977 | KY856057 | KY856144 | KY856486 | KY856319 | |
| CPC 26371 | Citrus sinensis ‘Valencia’ | Italy, Catania | Twigs wither-tip | KY856405 | KY856229 | KY855978 | KY856058 | KY856145 | KY856487 | KY856320 | |
| CPC 26373 | Citrus limon | Italy, Catania | Twigs wither-tip | KY856406 | KY856230 | KY855979 | KY856059 | KY856146 | KY856488 | KY856321 | |
| CPC 26376 | Citrus paradisi | Italy, Catania | Twigs wither-tip | KY856407 | KY856231 | KY855980 | KY856060 | KY856147 | KY856489 | KY856322 | |
| CPC 26381 | Citrus limon | Italy, Catania | Twigs wither-tip | KY856408 | KY856232 | KY855981 | KY856061 | KY856148 | KY856490 | KY856323 | |
| CPC 26479 | Citrus sinensis | Italy, Enna | Fruit lesion | KY856409 | KY856233 | KY855982 | KY856062 | KY856149 | KY856491 | KY856324 | |
| CPC 26486 | Citrus sinensis | Italy, Enna | Fruit lesion | KY856410 | KY856234 | KY855983 | KY856063 | KY856150 | KY856492 | KY856325 | |
| CPC 26488 | Citrus sinensis | Italy, Catania | Fruit lesion | KY856411 | KY856235 | KY855984 | KY856064 | KY856151 | KY856493 | KY856326 | |
| CPC 26515 | Citrus medica | Italy, Catania | Leaf lesion | KY856412 | KY856236 | KY855985 | KY856065 | KY856152 | KY856494 | KY856327 | |
| CPC 26803 | Citrus sinensis ‘Tarocco Meli’ | Italy, Catania | Twigs wither-tip | KY856413 | KY856237 | KY855986 | KY856066 | KY856153 | KY856495 | KY856328 | |
| CPC 26809 | Citrus limon | Spain, Malaga | Leaf lesion | KY856414 | KY856238 | KY855987 | KY856067 | KY856154 | KY856496 | KY856329 | |
| CPC 26823 | Citrus paradisi | Spain, Malaga | Leaf lesion | KY856415 | KY856239 | KY855988 | KY856068 | KY856155 | KY856497 | KY856330 | |
| CPC 26937 | Citrus paradisi | Spain, Malaga | Twigs wither-tip | KY856416 | KY856240 | KY855989 | KY856069 | KY856156 | KY856498 | KY856331 | |
| CPC 26957 | Citrus reticulata ‘Nova’ | Greece, Nafplio | Leaf lesion | KY856417 | KY856241 | KY855990 | KY856070 | KY856157 | KY856499 | KY856332 | |
| CPC 26965 | Citrus sinensis | Italy, Vibo Valentia | Fruit lesion | KY856418 | KY856242 | KY855991 | KY856071 | KY856158 | KY856500 | KY856333 | |
| CPC 26975 | Citrus paradisi | Italy, Vibo Valentia | Twigs wither-tip | KY856419 | KY856243 | KY855992 | KY856072 | KY856159 | KY856501 | KY856334 | |
| CPC 26985 | Citrus reticulata ‘Nova’ | Italy, Vibo Valentia | Leaf lesion | KY856420 | KY856244 | KY855993 | KY856073 | KY856160 | KY856502 | KY856335 | |
| CPC 27019 | Citrus limon | Italy, Cosenza | Twigs wither-tip | KY856421 | KY856245 | KY855994 | KY856074 | KY856161 | KY856503 | KY856336 | |
| CPC 27021 | Fortunella margarita | Italy, Vibo Valentia | Twigs wither-tip | KY856422 | KY856246 | KY855995 | KY856075 | KY856162 | KY856504 | KY856337 | |
| CPC 27088 | Citrus reticulata | Greece, Missolonghi | Leaf lesion | KY856423 | KY856247 | KY855996 | KY856076 | KY856163 | KY856505 | KY856338 | |
| CPC 27127 | Citrus maxima | Greece, Missolonghi | Twigs wither-tip | KY856424 | KY856248 | KY855997 | KY856077 | KY856164 | KY856506 | KY856339 | |
| CPC 27129 | Citrus bergamia | Greece, Missolonghi | Fruit lesion | KY856425 | KY856249 | KY855998 | KY856078 | KY856165 | KY856507 | KY856340 | |
| CPC 27839 | Citrus sinensis | Italy, Catania | Leaf lesion | KY856426 | KY856250 | KY855999 | KY856079 | KY856166 | KY856508 | KY856341 | |
| CPC 27841 | Citrus sinensis | Italy, Catania | Leaf lesion | KY856427 | KY856251 | KY856000 | KY856080 | KY856167 | KY856509 | KY856342 | |
| CPC 27905 | Citrus limon | Malta, Gozo | Twigs wither-tip | KY856428 | KY856252 | KY856001 | KY856081 | KY856168 | KY856510 | KY856343 | |
| CPC 27923 | Citrus sinensis | Malta, Gozo | Leaf litter | KY856429 | KY856253 | KY856002 | KY856082 | KY856169 | KY856511 | KY856344 | |
| CPC 27939 | Citrus limon | Portugal, Faro | Leaf lesion | KY856430 | KY856254 | KY856003 | KY856083 | KY856170 | KY856512 | KY856345 | |
| CPC 27941 | Citrus sinensis | Portugal, Silves | Twigs wither-tip | KY856431 | KY856255 | KY856004 | KY856084 | KY856171 | KY856513 | KY856346 | |
| CPC 27971 | Citrus sinensis ‘Valencia’ | Portugal, Mesquita | Fruit lesion | KY856432 | KY856256 | KY856005 | KY856085 | KY856172 | KY856514 | KY856347 | |
| CPC 27991 | Citrus sinensis ‘Valencia’ | Portugal, Mesquita | Fruit tear stain | KY856433 | KY856257 | KY856006 | KY856086 | KY856173 | KY856515 | KY856348 | |
| CPC 28001 | Citrus paradisi | Portugal, Faro | Leaf lesion | KY856434 | KY856258 | KY856007 | KY856087 | KY856174 | KY856516 | KY856349 | |
| CPC 28021 | Citrus sinensis | Portugal, Mesquita | Twigs wither-tip | KY856435 | KY856259 | KY856008 | KY856088 | KY856175 | KY856517 | KY856350 | |
| CPC 28023 | Citrus limon | Portugal, Monchique | Leaf lesion | KY856436 | KY856260 | KY856009 | KY856089 | KY856176 | KY856518 | KY856351 | |
| CPC 28029 | Citrus sinensis | Portugal, Silves | Twigs wither-tip | KY856437 | KY856261 | KY856010 | KY856090 | KY856177 | KY856519 | KY856352 | |
| CPC 28052 | Citrus reticulata | Spain, Algemesi | Twigs wither-tip | KY856438 | KY856262 | KY856011 | KY856091 | KY856178 | KY856520 | KY856353 | |
| CPC 28056 | Citrus sinensis ‘Lanelate’ | Spain, Moncada | Petal lesions | KY856439 | KY856263 | KY856012 | KY856092 | KY856179 | KY856521 | KY856354 | |
| CPC 28061 | Citrus sinensis | Spain, Castellò | Leaf lesion | KY856440 | KY856264 | KY856013 | KY856093 | KY856180 | KY856522 | KY856355 | |
| CPC 28063 | Citrus sinensis | Spain, Castellò | Leaf lesion | KY856441 | KY856265 | KY856014 | KY856094 | KY856181 | KY856523 | KY856356 | |
| CPC 28155 | Citrus floridana | Italy, Catania | Fruit lesion | KY856442 | KY856266 | KY856015 | KY856095 | KY856182 | KY856524 | KY856357 | |
| CPC 28159 | Citrus digitata | Italy, Catania | Leaf lesion | KY856443 | KY856267 | KY856016 | KY856096 | KY856183 | KY856525 | KY856358 | |
| CPC 28196 | Atlantia citroides | Spain, Soller | Leaf lesion | KY856444 | KY856268 | KY856017 | KY856097 | KY856184 | KY856526 | KY856359 | |
| CPC 28197 | Microcitrus australasica | Spain, Soller | Twigs wither-tip | KY856445 | KY856269 | KY856018 | KY856098 | KY856185 | KY856527 | KY856360 | |
| ICMP 12938 | Citrus sinensis | New Zealand | – | JX010147 | JX009935 | JX009560 | JX009732 | JX009746 | – | – | |
| ICMP 18695 | Citrus sp. | USA | – | JX010153 | JX009979 | JX009494 | JX009735 | JX009779 | – | – | |
| ICMP 18730 | Citrus sp. | New Zealand | – | JX010157 | JX009981 | JX009548 | JX009737 | JX009861 | – | – | |
| ICMP 18738 | Carya illinoinensis | Australia | – | JX010151 | JX009976 | JX009542 | JX009730 | JX009797 | – | – | |
| C. godetiae | CBS 133.44 | Clarkia hybrida | Denmark | – | JQ948402 | JQ948733 | JQ949723 | – | JQ949063 | JQ950053 | JQ949393 |
| C. helleniense | CBS 142418 = CPC 26844 | Poncirus trifoliata | Greece, Arta | Twigs wither-tip | KY856446 | KY856270 | KY856019 | KY856099 | KY856186 | KY856528 | KY856361 |
| CBS 142419 = CPC 27107 | Citrus reticulata | Greece, Arta | Fruit lesion | KY856447 | KY856271 | KY856020 | KY856100 | KY856187 | KY856529 | KY856362 | |
| CPC 26845 | Poncirus trifoliata | Greece, Arta | Twigs wither-tip | KY856448 | KY856272 | KY856021 | KY856101 | KY856188 | KY856530 | KY856363 | |
| CPC 27108 | Citrus reticulata | Greece, Arta | Fruit lesion | KY856449 | KY856273 | KY856022 | KY856102 | KY856189 | KY856531 | KY856364 | |
| C. hystricis | CBS 142411 = CPC 28153 | Citrus hystrix | Italy, Catania | Leaf lesion | KY856450 | KY856274 | KY856023 | KY856103 | KY856190 | KY856532 | KY856365 |
| CBS 142412 = CPC 28154 | Citrus hystrix | Italy, Catania | Leaf lesion | KY856451 | KY856275 | KY856024 | KY856104 | KY856191 | KY856533 | KY856366 | |
| C. johnstonii | CBS 128532 | Citrus sp. | New Zealand | – | JQ948443 | JQ948774 | JQ949764 | – | JQ949104 | JQ950094 | JQ949434 |
| C. kahawae subsp. kahawae | ICMP 17816 | Coffea arabica | Kenya | – | JX010231 | JX010012 | JX009452 | JX009642 | JX009813 | JX010444 | – |
| C. kahawae subsp. ciggaro | ICMP 18539 | Olea europaea | Australia | – | JX010230 | JX009966 | JX009523 | JX009635 | JX009800 | JX010434 | – |
| C. karstii | CBS 126532 | Citrus sp. | South Africa | – | JQ005209 | JQ005296 | JQ005557 | JQ005730 | JQ005383 | JQ005643 | JQ005470 |
| CBS 127597 | Diospyros australis | Australia | – | JQ005204 | JQ005291 | JQ005552 | JQ005725 | JQ005378 | JQ005638 | JQ005465 | |
| CBS 128551 | Citrus sp. | New Zealand | – | JQ005208 | JQ005295 | JQ005556 | JQ005729 | JQ005382 | JQ005642 | JQ005469 | |
| CBS 129829 | Gossypium hirsutum | Germany | – | JQ005189 | JQ005276 | JQ005537 | JQ005710 | JQ005363 | JQ005623 | JQ005450 | |
| CBS 129833 | Musa sp. | Mexico | – | JQ005175 | JQ005262 | JQ005523 | JQ005696 | JQ005349 | JQ005609 | JQ005436 | |
| CBS 134226 | Citrus limon | China | – | KC293570 | KC293730 | KC293610 | KC293690 | KY856192 | KC293650 | KY856367 | |
| CBS 142415 = CPC 26379 | Fortunella margarita | Italy, Catania | Fruit tear stain | KY856452 | KY856276 | KY856025 | KY856105 | KY856193 | KY856534 | KY856368 | |
| CPC 26375 | Citrus paradisi | Italy, Catania | Twigs wither-tip | KY856453 | KY856277 | KY856026 | KY856106 | KY856194 | KY856535 | KY856369 | |
| CPC 27023 | Citrus sinensis | Italy, Cosenza | Leaf lesion | KY856454 | KY856278 | KY856027 | KY856107 | KY856195 | KY856536 | KY856370 | |
| CPC 27035 | Citrus paradisi | Spain, Almeria | Leaf lesion | KY856455 | KY856279 | KY856028 | KY856108 | KY856196 | KY856537 | KY856371 | |
| CPC 27063 | Fortunella margarita | Italy, Vibo Valentia | Leaf lesion | KY856456 | KY856280 | KY856029 | KY856109 | KY856197 | KY856538 | KY856372 | |
| CPC 27065 | Citrus sinensis | Spain, Almeria | Leaf lesion | KY856457 | KY856281 | KY856030 | KY856110 | KY856198 | KY856539 | KY856373 | |
| CPC 27077 | Citrus reticulata ‘Nova’ | Spain, Almeria | Twigs wither-tip | KY856458 | KY856282 | KY856031 | KY856111 | KY856199 | KY856540 | KY856374 | |
| CPC 27817 | Citrus paradisi | Italy, Catania | Twigs wither-tip | KY856459 | KY856283 | KY856032 | KY856112 | KY856200 | KY856541 | KY856375 | |
| CPC 27845 | Citrus sinensis | Italy, Catania | Twigs wither-tip | KY856460 | KY856284 | KY856033 | KY856113 | KY856201 | KY856542 | KY856376 | |
| CPC 27853 | Citrus sinensis | Italy, Catania | Fruit lesion | KY856461 | KY856285 | KY856034 | KY856114 | KY856202 | KY856543 | KY856377 | |
| CPC 27979 | Citrus reticulata | Italy, Catania | Leaf lesion | KY856462 | KY856286 | KY856035 | KY856115 | KY856203 | KY856544 | KY856378 | |
| CPC 27989 | Citrus sinensis | Portugal, Mesquita | Twigs wither-tip | KY856463 | KY856287 | KY856036 | KY856116 | KY856204 | KY856545 | KY856379 | |
| CPC 27999 | Citrus limon | Portugal, Faro | Twigs wither-tip | KY856464 | KY856288 | KY856037 | KY856117 | KY856205 | KY856546 | KY856380 | |
| CPC 28065 | Citrus limon | Spain, Castellò | Leaf lesion | KY856465 | KY856289 | KY856038 | KY856118 | KY856206 | KY856547 | KY856381 | |
| CPC 28142 | Citrus limon | Italy, Catania | Fruit lesion | KY856466 | KY856290 | KY856039 | KY856119 | KY856207 | KY856548 | KY856382 | |
| CPC 31139 | Citrus sinensis | Italy, Catania | Leaf lesion | KY856467 | KY856291 | KY856040 | KY856120 | KY856208 | KY856549 | KY856383 | |
| CPC 31143 | Citrus sinensis | Malta, Zurrieq | Twigs wither-tip | KY856468 | KY856292 | KY856041 | KY856121 | KY856209 | KY856550 | KY856384 | |
| CPC 31144 | Citrus sinensis | Malta, Zurrieq | Twigs wither-tip | KY856469 | KY856293 | KY856042 | KY856122 | KY856210 | KY856551 | KY856385 | |
| CPC 31196 | Murraya paniculata | Italy, Catania | Leaf lesion | KY856470 | KY856294 | KY856043 | KY856123 | KY856211 | KY856552 | KY856386 | |
| C. limetticola | CBS 114.14 | Citrus aurantifolia | USA, Florida | – | JQ948193 | JQ948523 | JQ949514 | – | JQ948854 | JQ949844 | JQ949184 |
| C. limonicola | CBS 142409 = CPC 27861 | Citrus limon | Malta, Gozo | Leaf lesion | KY856471 | KY856295 | KY856044 | KY856124 | KY856212 | KY856553 | KY856387 |
| CBS 142410 = CPC 31141 | Citrus limon | Malta, Gozo | Leaf lesion | KY856472 | KY856296 | KY856045 | KY856125 | KY856213 | KY856554 | KY856388 | |
| CPC 27862 | Citrus limon | Malta, Gozo | Leaf lesion | KY856473 | KY856297 | KY856046 | KY856126 | KY856214 | KY856555 | KY856389 | |
| C. musae | CBS 116870 | Musa sp. | USA | – | JX010146 | JX010050 | JX009433 | JX009742 | JX009896 | HQ596280 | – |
| C. novae-zelandiae | CBS 128505 | Capsicum annuum | New Zealand | – | JQ005228 | JQ005315 | JQ005576 | JQ005749 | JQ005402 | JQ005662 | JQ005489 |
| CBS 130240 | Citrus medica | New Zealand | – | JQ005229 | JQ005316 | JQ005577 | JQ005750 | JQ005403 | JQ005663 | JQ005490 | |
| CBS 142413 = CPC 26949 | Citrus paradisi | Greece, Missolonghi | Leaf lesion | KY856474 | KY856298 | KY856047 | KY856127 | KY856215 | KY856556 | KY856390 | |
| CBS 142414 = CPC 27888 | Citrus sinensis | Malta, Gozo | Twigs wither-tip | KY856475 | KY856299 | KY856048 | KY856128 | KY856216 | KY856557 | KY856391 | |
| CPC 27864 | Citrus limon | Malta, Gozo | Twigs wither-tip | KY856476 | KY856300 | KY856049 | KY856129 | KY856217 | KY856558 | KY856392 | |
| CPC 27890 | Citrus sinensis | Malta, Gozo | Twigs wither-tip | KY856477 | KY856301 | KY856050 | KY856130 | KY856218 | KY856559 | KY856393 | |
| CPC 27957 | Citrus limon | Malta, Gozo | Leaf lesion | KY856478 | KY856302 | KY856051 | KY856131 | KY856219 | KY856560 | KY856394 | |
| C. siamense | GZAAS5.09506 | Murraya sp. | China | – | JQ247633 | JQ247609 | JQ247657 | JQ247596 | – | JQ247644 | – |
| C. simmondsii | CBS 122122 | Carica papaya | Australia | – | JQ948276 | JQ948606 | JQ949597 | – | JQ948937 | JQ949927 | JQ949267 |
| GZAAS5.09510 | Murraya sp. | China | – | JQ247631 | JQ247607 | JQ247655 | JQ247595 | – | JQ247643 | – | |
| C. ti | ICMP 4832 | Cordyline sp. | New Zealand | – | JX010269 | JX009952 | JX009520 | JX009649 | JX009898 | JX010442 | – |
| C. tropicale | CBS 124949 | Theobroma cacao | Panama | – | JX010264 | JX010007 | JX009489 | JX009719 | JX009870 | JX010407 | KY856395 |
| C. tropicicola | BCC 38877 | Citrus maxima | Thailand | – | JN050240 | JN050229 | JN050218 | – | – | JN050246 | – |
| C. truncatum | CBS 151.35 | Phaseolus lunatus | USA | – | GU227862 | GU228254 | GU227960 | KY856132 | GU228352 | GU228156 | GU228058 |
| CBS 134232 | Citrus limon | China | – | KC293580 | KC293740 | KC293620 | KC293700 | KY856220 | KC293660 | KY856396 | |
| Moniolochaetes infuscans | CBS 869.96 | Ipomoea batatas | South Africa | – | JQ005780 | JX546612 | JQ005843 | – | JQ005801 | JQ005864 | JQ005822 |
1 BCC: Culture Collection, National Center for Genetic Engineering and Biotechnology (BIOTEC), Khlong Luang, Pathumthani, Thailand; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; COAD: Coleção Octávio Almeida Drummond, Viçosa, Brazil; CPC: Culture collection of P.W. Crous, housed at the Westerdijk Institute; GZAAS: Guizhou Academy of Agricultural Science herbarium, Guizhou Province, China; ICMP: International Collection of Microorganisms from Plants, Landcare Research, Auckland, New Zealand. Ex-type and ex-epitype cultures are indicated in bold.
2 ITS: internal transcribed spacers 1 and 2 together with 5.8S nrDNA; GAPDH: partial glyceraldehyde-3-phosphate dehydrogenase gene; ACT: partial actin gene; CAL; partial calmodulin gene; CHS-1: partial chitin synthase 1 gene; TUB2: partial beta-tubulin gene; HIS3: histone3. Sequences generated in this study are indicated in italics.
Morphology
Agar plugs (6-mm-diam) were taken from the edge of actively growing cultures on PDA and transferred to the centre of 9-cm-diam Petri dishes containing PDA and synthetic nutrient-poor agar medium (SNA; Nirenberg 1976) as described in recent studies (Huang et al. 2013, Diao et al. 2017). Cultures were incubated at 25 °C with a 12/12 h fluorescent light/dark cycle for 10 d. Colony characters and pigment production on PDA and SNA were noted after 10 d. Colony colours were rated according to Rayner (1970). Cultures were examined periodically for the development of ascomata, conidiomata and setae. Colony diameters were measured after 7 and 10 d. The morphological characteristics were examined by mounting fungal structures in clear lactic acid and 30 measurements at × 1 000 magnification were determined for each isolate using a Zeiss Axioscope 2 microscope with interference contrast (DIC) optics. Descriptions and illustrations of taxonomic novelties were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004a).
Pathogenicity
Fruits of two sweet orange (Citrus sinensis) clones (‘Tarocco Scirè’ and ‘Tarocco Nucellare’) were collected in Sicily during the veraison stage and used for artificial inoculation. A subset of 13 isolates representing the Colletotrichum species isolated from specimens collected in Europe (Table 2) were inoculated following the adapted wound/drop method (Cai et al. 2009, Aiello et al. 2015, Cristóbal-Martínez et al. 2017). Eight fruits for each isolate/clone combination were inoculated. Fruits were washed and surface-disinfected by immersion in 70 % ethanol for 10 min, and rinsed twice in sterilised water. Six inoculation points per fruit were labelled with a dot made with a permanent marker and were injured using a sterile pipette tip (wounds of 2 mm diam). A spore suspension (1.0 × 105 conidia/mL) was obtained from cultures grown on PDA for 15 d at 27 °C, and 10 μL were injected into each inoculation point. Control fruits were inoculated with sterile water. The inoculated oranges were placed in plastic containers, covered with plastic bags and incubated in a growth chamber with 100 % relative humidity at 25 °C under a lighting rig providing a 12 h photoperiod. Symptom development was evaluated 10 d after inoculation and the percentage of infected inoculation points was calculated per each isolate/clone combination. This percentage value was calculated by the formula [(%) = (infected inoculation points / inoculated inoculation points) × 100 %].
Table 2.
Pathogenicity testing of Colletotrichum species: percentage of infected inoculation points of citrus fruits.
| Species | Isolates | Infected inoculation points (%) |
|
|---|---|---|---|
| Tarocco ‘Scirè’ | Tarocco ‘Nucellare’ | ||
| Colletotrichum acutatum | CBS 142407 = CPC 27005 | 0 | 0 |
| C. catinaense | CBS 142417 = CPC 27978 | 12.5 | 4.1 |
| C. catinaense | CBS 142416 = CPC 28019 | 18.75 | 6.2 |
| C. gloeosporioides | CBS 142408 = CPC 28059 | 87.5 | 83.3 |
| C. helleniense | CBS 142418 = CPC 26844 | 14.6 | 8.3 |
| C. helleniense | CBS 142419 = CPC 27107 | 31.2 | 16.6 |
| C. hystricis | CBS 142411 = CPC 28153 | 20.8 | 8.3 |
| C. hystricis | CBS 142412 = CPC 28154 | 16.6 | 10.4 |
| C. karstii | CBS 142415 = CPC 26379 | 8.3 | 6.2 |
| C. limonicola | CBS 142409 = CPC 27861 | 25 | 8.3 |
| C. limonicola | CBS 142410 = CPC 31141 | 16.6 | 12.5 |
| C. novae-zelandiae | CBS 142413 = CPC 26949 | 20.8 | 16.6 |
| C. novae-zelandiae | CBS 142414 = CPC 27888 | 10.4 | 4.1 |
The inoculated fungi were re-isolated from the obtained lesions and the identity of the re-isolated fungi confirmed by sequencing the loci ACT and GAPDH, thus fulfilling Koch’s postulates.
RESULTS
Sampling and isolation
Symptoms of anthracnose caused by Colletotrichum spp. were frequently observed on several Citrus species in all countries investigated. The leaves presented necrotic, more or less circular spots. These lesions appeared with a pale brown to purple margin and produced the fruiting bodies of the fungus (Fig. 1a–b). Different symptoms appeared on fruits. Irregular and sunken lesions, of variable size, from purple-brown to black with acervuli (Fig. 1d–g), were observed. Further, fruits showed tear stain (Fig. 1h), as superficial, reddish brown streaks or bands (down) along the fruit. Moreover, a dark-brown to black rot, with a well-defined margin at the stem-end was occasionally detected (Fig. 1i). Mummified fruits were occasionally observed in association with affected tips (Fig. 1c). Twigs showed typical dieback and wither-tip (Fig. 1k). Under high moisture conditions, pink masses of spores appeared sporulating in acervuli on dead twigs. A total of 174 monosporic isolates resembling those of the genus Colletotrichum were collected. The Colletotrichum isolates were recovered from 17 species of Citrus at 44 different sites in multiple locations of Greece, Italy, Malta, Spain and Portugal. Among them, 67 isolates were obtained from leaves, 72 were associated with twigs, 28 from fruits and seven were isolated from petals. Based on initial ITS and GAPDH sequencing, 82 representative isolates were selected (Table 1) for phylogenetic analysis and further taxonomic study.
Fig. 1.

Symptoms on citrus tissues with associated Colletotrichum spp. a–b. Anthracnose symptoms on leaves of naturally infected: a. Citrus bergamia and b. Fortunella margarita; c. mummified fruit of Citrus limon; d–g. various symptoms on fruits: d. diverse lesions and e–f. sunken lesions on orange and g. on mandarin; h. tear stain on grapefruit; i. stem-end rot on orange; j. typical anthracnose on fallen orange fruits; k. wither-tip of Citrus sinensis tree.
Phylogenetic analyses
The 14 MP trees derived from the single gene sequence alignments (ITS, GAPDH, ACT, CAL, CHS-1, HIS3 and TUB2) for both the C. boninense species complex and the remainder of the Colletotrichum spp., produced topologically similar trees, and confirmed that 30 isolates recovered in this study belong to the C. boninense species complex, 50 to C. gloeosporioides species complex and two to C. acutatum species complex. The combined species phylogeny of the C. boninense species complex consisted of 45 sequences, including the outgroup sequences of C. acutatum (culture CBS 112996). All the species belonging to the C. gloeosporioides and C. acutatum species complexes were included in a combined phylogeny consisting of 86 sequences, including the outgroup sequences of Moniolochaetes infuscans (CBS 896.96). A total of 3 149 characters (ITS: 1–549, GAPDH: 556–863, ACT: 870–1166, CAL: 1173–1946, CHS-1: 1953–2237, TUB2: 2244–2754, HIS3: 2761–3149) were included in both phylogenetic analyses. For the phylogeny of the C. boninense species complex, 411 characters were parsimony-informative, 454 were variable and parsimony-uninformative and 2 248 characters were constant. A maximum of 1 000 equally most parsimonious trees were saved (Tree length = 1 236, CI = 0.871, RI = 0.947 and RC = 0.825). Regarding the C. gloeosporioides and C. acutatum species complexes, 1 171 characters were parsimony-informative, 319 were variable and parsimony-uninformative and 1 623 characters were constant. A maximum of 1 000 equally most parsimonious trees were saved (Tree length = 3 238, CI = 0.736, RI = 0.921 and RC = 0.678). Bootstrap support values from the parsimony analysis were plotted on the Bayesian phylogenies presented in Fig. 2, 3. For both of the Bayesian analyses, MrModeltest suggested that all partitions should be analysed with dirichlet state frequency distributions, except for the CHS-1 partition, which was analysed with a fixed state frequency distribution. The following models were recommended by MrModeltest and used: GTR+I+G for ITS, CAL and HIS3, HKY+I+G for GAPDH and TUB2, HKY+G for ACT and SYM+I+G for CHS-1. In the Bayesian analysis of the C. boninense species complex, the ITS partition had 68 unique site patterns, the GAPDH partition 147, the ACT partition 108, the CAL partition 144, the CHS-1 partition 51, the TUB2 partition 146, the HIS3 partition 72 and the analysis ran for 2 260 000 generations, resulting in 4 522 trees of which 3 392 trees were used to calculate the posterior probabilities. Regarding the C. gloeosporioides and C. acutatum species complex, the ITS partition had 167 unique site patterns, the GAPDH partition 247, the ACT partition 183, the CAL partition 304, the CHS-1 partition 81, the TUB2 partition 257, the HIS3 partition 123 and the analysis ran for 4 890 000 generations, resulting in 9 782 trees of which 7 338 trees were used to calculate the posterior probabilities.
Fig. 2.
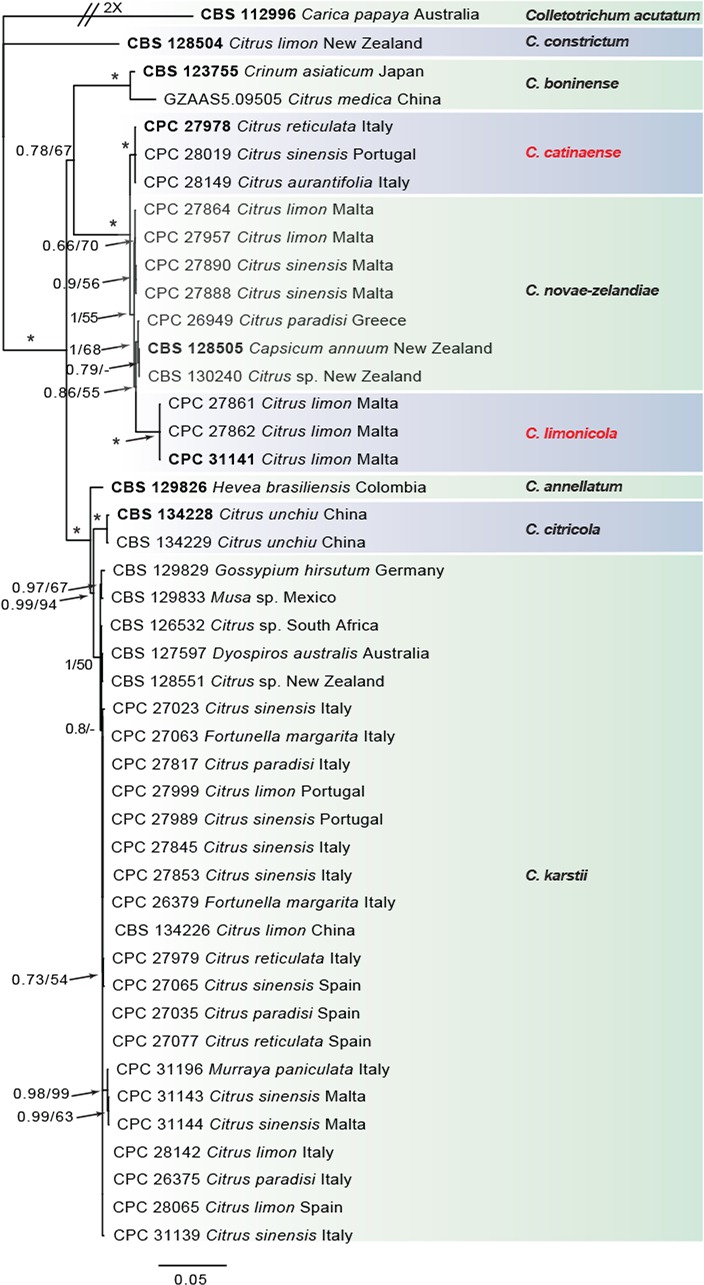
Consensus phylogram of 4 522 trees resulting from a Bayesian analysis of the combined ITS, CAL, GAPDH, ACT, TUB2, CHS-1 and HIS3 sequence alignments of the Colletotrichum boninense species complex. Bootstrap support values and Bayesian posterior probability values are indicated at the nodes. The asterisk symbol (*) represents full support (1/100). Substrate and country of origin, where known, are listed next to the strain numbers. In red the novel species. The tree was rooted to Colletotrichum acutatum (CBS 112996).
Fig. 3.
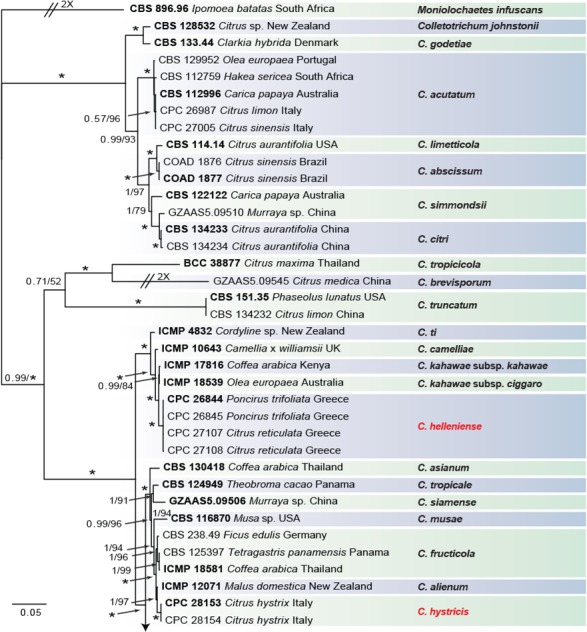
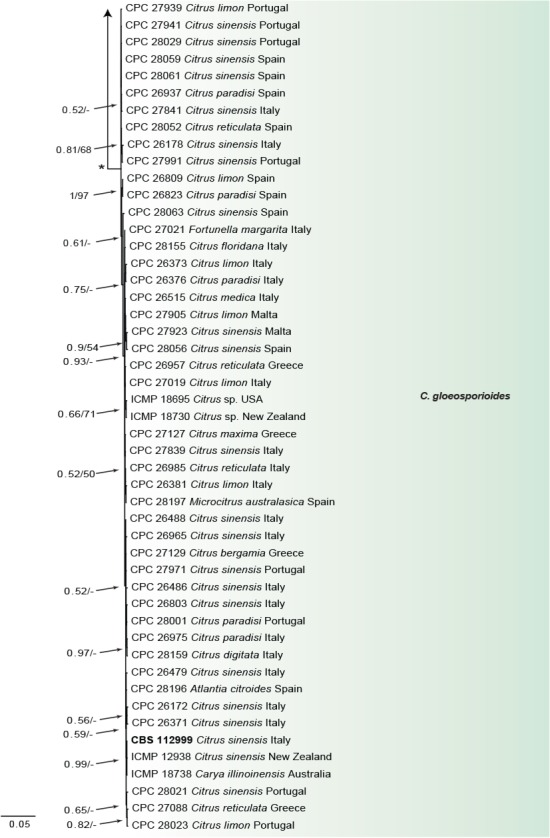
Consensus phylogram of 9 782 trees resulting from a Bayesian analysis of the combined ITS, CAL, GAPDH, ACT, TUB2, CHS-1 and HIS3 sequence alignments of Colletotrichum acutatum and C. gloeosporioides species complex. Bootstrap support values and Bayesian posterior probability values are indicated at the nodes. The asterisk symbol (*) represents full support (1/100). Substrate and country of origin, where known, are indicated next to the strain numbers. The tree was rooted to Moniolochaetes infuscans (CBS 896.96).
In the C. boninense species complex analysis 19 Citrus isolates clustered with six reference strains of C. karstii, whilst five isolates clustered with the ex-type of C. novae-zelandiae. Moreover, three isolates were identified as C. catinaense and a further three as C. limonicola, forming two highly supported subclades (1.00/100) which are embedded in the same clade with C. novae-zelandiae. In the other analyses two isolates clustered with the ex-type strain of C. acutatum s.str. and 44 isolates with the ex-type strain and other reference strains of C. gloeosporioides s.str. Furthermore, two isolates were identified as C. hystricis (closely related to C. alienum) and four as C. helleniense (close to C. kahawae subspecies) within the C. gloeosporioides species complex.
The individual alignments and trees of the seven single genes in both analyses, were compared as well with respect to their performance in species recognition. In the C. boninense species complex analysis, TUB2 differentiated all the taxa. Moreover, the single loci CAL and GAPDH, clearly separated C. catinaense and C. limonicola, respectively. In the other analyses, all the Colletotrichum species collected from citrus in this study differed in GAPDH sequences. Furthermore, C. helleniense was separated also by CAL and TUB2, whilst ACT and CHS-1 distinguished C. hystricis.
Taxonomy
Morphological observations, supported by phylogenetic inference, were used to identify four known species (C. gloeosporioides, C. novae-zelandiae, C. karstii and C. acutatum) and to describe four novel species. Culture characteristics were assessed, and the colour of upper and lower surfaces of Petri dishes determined as shown in Fig. 4, 5, 6, 7. Hyphal appressoria were abundantly observed on the reverse side of colonies growing on SNA plates. Based on the results of both the phylogenetic and morphological analyses, the four distinct novel species are described below.
Fig. 4.
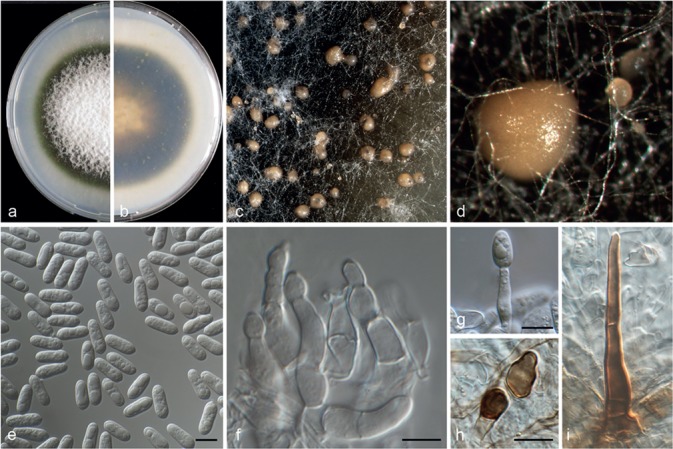
Colletotrichum catinaense (CBS 142417). a–b. Colonies on PDA above and below; c–d. conidiomata; e. conidia; f–g. conidiophores; h. appressoria; i. seta (a–g, i from PDA; h from SNA). — Scale bars = 10 μm.
Fig. 5.
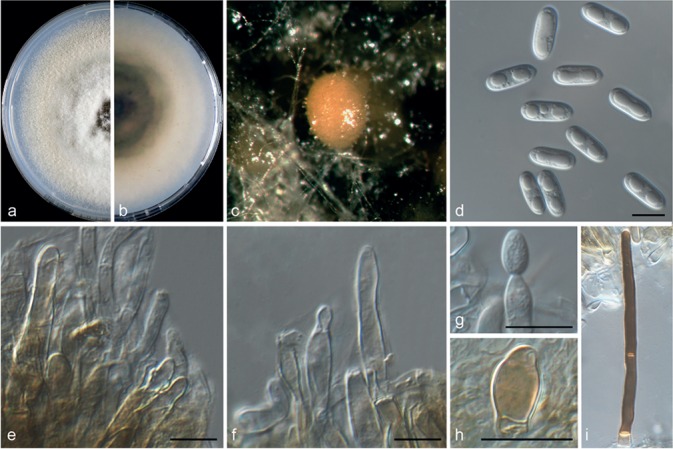
Colletotrichum helleniense (CBS 142418). a–b. Colonies on PDA above and below; c. conidiomata; d. conidia; e–g. conidiophores; h. appressoria; i. seta (a–f, i from PDA; g–h from SNA). — Scale bars = 10 μm.
Fig. 6.
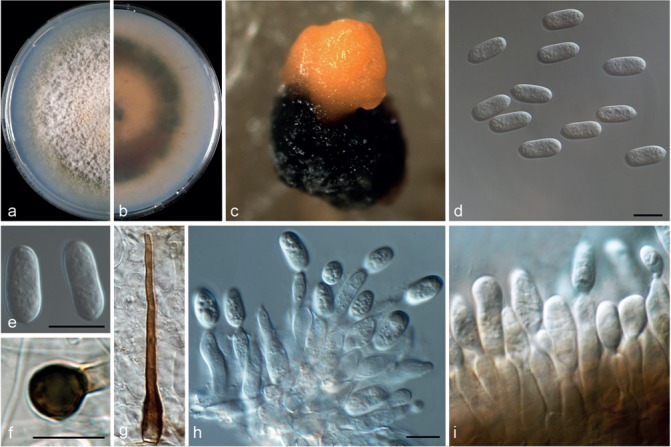
Colletotrichum hystricis (CBS 142411). a–b. Colonies on PDA above and below; c. conidiomata; d. conidiophores; e–f. conidia; g–h. appressoria; i. seta (a–g, i from PDA; h from SNA). — Scale bars = 10 μm.
Fig. 7.
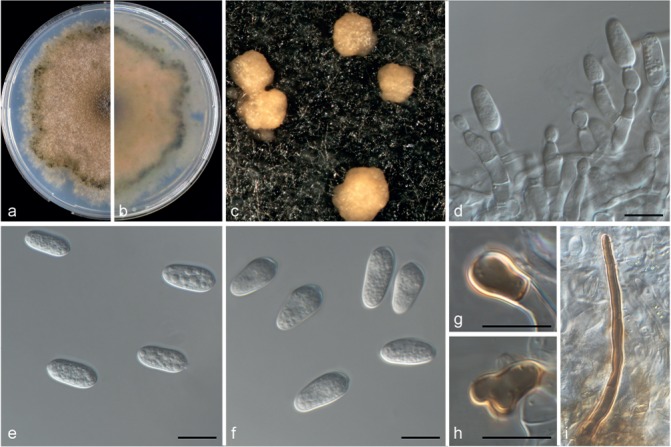
Colletotrichum limonicola (CBS 142410). a–b. Colonies on PDA above and below; c. conidiomata; d–e. conidia; f. appressoria; g. seta; h–i. conidiophores (a–e, g–i from PDA; f from SNA). — Scale bars = 10 μm.
Colletotrichum catinaense Guarnaccia & Crous, sp. nov. — MycoBank MB820247; Fig. 4
Etymology. Named after the city where the first strain was collected, Catania (ancient Latin name, Catina).
Asexual morph on SNA. Vegetative hyphae hyaline, septate, branched, 1–9 μm diam. Conidiomata, chlamydospores and setae absent. Conidiophores hyaline, smooth-walled, septate, branched, to 40 μm long, formed from hyphae. Conidiogenous cells hyaline, smooth-walled, cylindrical to inflated, 5–18 × 4–5 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, rounded apex and base, contents granular and guttulate, 11.5–15 × 4–5.5 μm, mean ± SD = 13.5 ± 0.9 × 4.8 ± 0.5 μm, L/W ratio = 2.7. Appressoria medium to dark brown, roundish with an undulate margin, single, 3.5–6 × 3–5.5 μm, mean ± SD = 4.8 ± 0.9 × 4.2 ± 0.5 μm, L/W ratio = 1.2.
Asexual morph on PDA. Conidiomata acervular, conidiophores and setae formed on a cushion of pale brown, thick-walled, angular cells, 3.5–7 μm diam. Setae brown, smooth, 2–3-septate, 50–120 μm long, base conical or inflated, dark brown, tip rounded. Conidiophores hyaline, smooth-walled, septate and branched, to 40 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical, 5–16 × 4–5 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, rounded apex and base, contents granular and guttulate, 13–16 × 4.5–6 μm, mean ± SD = 14.3 ± 1 × 5.5 ± 0.5 μm, L/W ratio = 2.6.
Culture characteristics — Colonies on SNA flat with entire margin, hyaline, 35–37 mm diam in 7 d (49–52 mm in 10 d). Colonies on PDA flat with entire margin, buff honey in the centre to green olivaceous at the margin, partly covered with floccose white aerial mycelium and with black conidiomata. Conidia present in orange to pale brown mass. Reverse buff, pale luteous to isabelline, dark green in the margin, 66–68 mm diam in 7 d (75–76 mm diam in 10 d).
Materials examined. Italy, Mineo, Catania, from leaf lesion of Citrus reticulata (mandarin), 23 Sept. 2015, V. Guarnaccia (CBS H-23024 holotype, culture ex-type CBS 142417 = CPC 27978). – Portugal, Mesquita, from fruit tear-stain of Citrus sinensis (orange), 7 Oct. 2015, V. Guarnaccia (culture CBS 142416 = CPC 28019).
Notes — Colletotrichum catinaense was isolated from several hosts in Italy and Portugal. The isolation of this species from multiple combinations of organ/host demonstrates its ability to colonise different citrus tissues. This species is phylogenetically close to but clearly differentiated from C. novaezelandiae in CAL and TUB sequences. Colletotrichum novae-zelandiae formed a separate lineage/cluster in all single-gene phylogenies (Damm et al. 2012b) before this study. Based on multi-locus phylogenetic analyses performed in this study (Fig. 2), C. catinaense together with C. limonicola (described below) are new species belonging to the same clade of C. novaezelandiae within the C. boninense species complex. This species is morphologically indistinguishable from the other two species of the same clade.
Colletotrichum helleniense Guarnaccia & Crous, sp. nov. — MycoBank MB820249; Fig. 5
Etymology. Named after the country where it was collected, Greece (ancient name, Hellas).
Asexual morph on SNA. Vegetative hyphae hyaline, septate, branched, 1–8 μm diam. Conidiomata, chlamydospores and setae absent. Conidiophores formed from hyphae, hyaline, smooth to finely verruculose, septate, branched, to 50 μm long. Conidiogenous cells are hyaline, smooth-walled, cylindrical to inflated, 5–15 × 4–5 μm. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, rounded apex and base, contents granular or guttulate, 11–14.5 × 4–5.5 μm, mean ± SD = 12.2 ± 0.7 × 4.7 ± 0.5 μm, L/W ratio = 2.6. Appressoria medium to dark brown, roundish or irregular in shape, single or in small groups, 5–10 × 7–15 μm.
Asexual morph on PDA. Conidiomata acervular, conidiophores and setae formed on a cushion of pale brown, thick-walled, angular cells, 3.5–7 μm diam. Setae brown, smooth, 2-septate, 55–90 μm long, base conical or inflated, dark brown, tip rounded. Conidiophores hyaline, smooth to undulate walled, septate and branched, to 35 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical, 5–15 × 4–5.5 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, rounded apex and base, contents granular or guttulate, 11.5–14.5 × 4–5.5 μm, mean ± SD = 12.7 ± 0.8 × 4.7 ± 0.5 μm, L/W ratio = 2.7.
Culture characteristics — Colonies on SNA flat with entire margin, hyaline, 40–46 mm diam in 7 d (54–59 mm diam in 10 d). Colonies on PDA with entire margin, green to grey in the centre and white to pale buff in the margin, entirely covered with floccose to dense, white to grey aerial mycelium and with black conidiomata. Conidia present in pinkish orange mass. Reverse grey to buff, pale luteous, 59–62 mm diam in 7 d (72–75 mm diam in 10 d).
Materials examined. Greece, Arta, from wither-tip twigs of Poncirus trifoliata (citrumelo), 20 May 2015, V. Guarnaccia (CBS H-23025 holotype, culture ex-type CBS 142418 = CPC 26844); from fruit lesions of C. reticulata (mandarin), 20 May 2015, V. Guarnaccia (culture CBS 142419 = CPC 27107).
Notes — Colletotrichum helleniense was isolated from Citrus reticulata fruit lesions and from Poncirus trifoliata wither-tip twigs in Greece. Poncirus is an allied genus of Citrus, in the Rutaceae, containing species mostly used as rootstock for citrus. These results show the ability of C. helleniense to colonise tissues of different genera within the Rutaceae. This species is phylogenetically close to but clearly differentiated from C. kahawae based on GAPDH, CAL and TUB2. Two subspecies of C. kahawae were described in the past; C. kahawae subsp. kahawae and C. kahawae subsp. ciggaro (Weir et al. 2012). Recently, the legitimacy of this distinction has been supported by Batista et al. (2016), who accepted the two subspp. as two cryptic species. Colletotrichum helleniense is clearly separate from both C. kahawae subspecies and from further species such as C. aotearoa, C. clidemiae, C. cordylinicola, C. jiangxiense, C. psidii, C. rhexiae (data not shown) belonging to the same clade (Diao et al. 2017, Weir et al. 2012). Therefore, C. helleniense represents a new species in the C. kahawae clade, belonging to the C. gloeosporioides species complex.
Colletotrichum hystricis Guarnaccia & Crous, sp. nov. –– MycoBank MB820252; Fig. 6
Etymology. In reference to its occurrence on Citrus hystrix.
Asexual morph on SNA. Vegetative hyphae hyaline, septate, 1–7 μm diam. Conidiomata, chlamydospores and setae absent. Conidiophores formed from hyphae, hyaline, smooth-walled, septate, branched, to 40 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical, 5–10 × 4–5 μm. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical to obovoidal, rounded apex and base, contents granular, 13–15 × 4–5.5 μm, mean ± SD = 14 ± 1.3 × 4.8 ± 0.5 μm, L/W ratio = 2.8. Appressoria dark brown, globose to irregular shape, single, with irregular lobes, 3.5–8 × 3–5.5 μm, mean ± SD = 6 ± 0.9 × 4.2 ± 0.5 μm, L/W ratio = 1.4.
Asexual morph on PDA. Conidiomata acervular, conidiophores and setae formed on a cushion of pale brown, thick-walled, angular cells, 3.5–7 μm diam. Setae brown, smooth, 2–3-septate, curved, 50–100 μm long, base conical, dark brown, tip rounded. Conidiophores hyaline, smooth-walled, septate, branched, to 50 μm long. Conidiogenous cells hyaline, smooth-walled to undulate, cylindrical, 5–20 × 3–5 μm. Conidia hyaline, smooth-walled, cylindrical to obovoidal, aseptate, rounded apex and base, contents granular, 14–16 × 4.5–6 μm, mean ± SD = 13.8 ± 1 × 5.1 ± 0.4 μm, L/W ratio = 2.7.
Culture characteristics — Colonies on SNA flat with entire margin, hyaline, 60–61 mm diam in 7 d (72–75 mm diam in 10 d). Colonies on PDA flat with entire margin, buff honey to pinkish, green to grey in the margin, entirely covered with white aerial mycelium and with black conidiomata. Conidia present in orange mass. Reverse buff, pale luteous to dark green, 69–71 mm diam in 7 d (80–82 mm diam in 10 d).
Materials examined. Italy, Mascali, Catania, from leaf lesion of Citrus hystrix, 30 Jan. 2016, V. Guarnaccia (CBS H-23026 holotype, culture ex-type CBS 142411 = CPC 28153); ibid., (culture CBS 142412 = CPC 28154).
Notes — Colletotrichum hystricis was isolated from Citrus hystrix leaf lesions in Sicily, Italy. This species differs from closely related species in GAPDH, ACT and CHS-1 sequences. Colletotrichum hystricis is similar to C. alienum and other species such as C. aenigma, C. conoides and C. nupharicola (Weir et al. 2012, Diao et al. 2017) but represents a distinct taxon, supported also by morphological differences. Colletotrichum hystricis differs from C. alienum in having obovoidal conidia (also on SNA) and a slower growth rate.
Colletotrichum limonicola Guarnaccia & Crous, sp. nov. — MycoBank MB820254; Fig. 7
Etymology. In reference to its occurrence on Citrus limon.
Asexual morph on SNA. Vegetative hyphae hyaline, septate, branched, 1–10 μm diam. Conidiomata, chlamydospores and setae absent. Conidiophores formed from hyphae, hyaline, smooth-walled, septate, branched, to 50 μm long. Conidiogenous cells are hyaline, smooth-walled, cylindrical to inflated, 5–20 × 4–5 μm. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, rounded apex and base, contents granular, 9–15 × 4–6 μm, mean ± SD = 12.2 ± 1.3 × 6 ± 0.5 μm, L/W ratio = 2.5. Appressoria medium to dark brown, roundish, single, 3–6 × 3–5.5 μm, mean ± SD = 4.5 ± 0.9 × 4.2 ± 0.5 μm, L/W ratio = 1.1.
Asexual morph on PDA. Conidiomata acervular, conidiophores and setae formed on a cushion of pale brown, thick-walled, angular cells, 3.5–7 μm diam. Setae brown, smooth, 2–3-septate, 45–100 μm long, base conical or inflated, dark brown, tip rounded. Conidiophores hyaline, smooth walled, septate and branched, to 50 μm long. Conidiogenous cells hyaline, smooth-walled, cylindrical, 7–16 × 4–5.5 μm. Conidia hyaline, smooth-walled, aseptate, cylindrical, rounded apex and base, contents granular, 9.5–15.5 × 4–6 μm, mean ± SD = 12.7 ± 1.3 × 5 ± 0.5 μm, L/W ratio = 2.5.
Culture characteristics — Colonies on SNA flat with entire margin, hyaline, 44–46 mm diam in 7 d (58–60 mm diam in 10 d). Colonies on PDA flat with entire margin, buff honey in the centre to green olivaceous in the margin, entirely covered with floccose white aerial mycelium and with black conidiomata. Conidia present in orange to pale brown mass. Reverse buff, pale luteous to dark green, 64–66 mm diam in 7 d (75–76 mm diam in 10 d).
Materials examined. Malta, Gozo, from wither-tip twigs of Citrus limon (lemon), 11 July 2016, V. Guarnaccia (CBS H-23027 holotype, culture ex-type CBS 142410 = CPC 31141); from leaf lesions of C. limon, 22 Sept. 2015, V. Guarnaccia (culture CBS 142409 = CPC 27861).
Notes — Colletotrichum limonicola was isolated from leaf lesions and twigs with wither-tip symptoms on Citrus limon in Malta. This species is phylogenetically close to but clearly differentiated from C. novae-zelandiae based on GAPDH and TUB. Colletotrichum limonicola and C. catinaense (described above) are new species belonging to the same clade of C. novaezelandiae in the C. boninense species complex.
Pathogenicity
All tested isolates except that of C. acutatum were pathogenic to most of the detached orange fruits (Table 2). Both Citrus sinensis clones tested developed typical brown lesions around the fruit wounds after 10 d (Fig. 8). Colletotrichum gloeosporioides and C. karstii, respectively, showed the highest and the weakest aggressiveness among the eight species inoculated. Clone ‘Tarocco Scirè’ was more susceptible. The inoculated Colletotrichum isolates were re-isolated from the symptomatic tissues, fulfilling Koch’s postulates. No symptoms developed on the negative controls.
Fig. 8.

Pathogenicity test of selected Colletotrichum isolates on Citrus sinensis fruits after 10 d. Fruits inoculated with: a–d. C. gloeosporioides (CBS 142408); e. C. catinaense (CBS 142417); f. C. limonicola (CBS 142410); g. C. novae-zelandiae (CBS 142414); h. C. hystricis (CBS 142411); i. C. helleniense (CBS 142418); j. C. karstii (CBS 142415).
DISCUSSION
Recent studies of phylogenetic analyses in the genus Colletotrichum revealed species to cluster in 11 major clades, as well as a number of small clusters and isolated species (Cannon et al. 2012, Marin-Felix et al. 2017). Four of these major clades represent important species complexes (C. acutatum, C. boninense, C. gloeosporioides and C. truncatum) (Damm et al. 2009, 2012a, b, Weir et al. 2012), which include very important plant pathogenic species. In these studies, a large number of taxa were differentiated and described. The recent revision and epitypification of the main Colletotrichum species complexes (Damm et al. 2009, 2012a, b, 2013, 2014, Weir et al. 2012, Liu et al. 2014), as well as several studies that focussed on citrus diseases (Peng et al. 2012, Huang et al. 2013, Aiello et al. 2015, Perrone et al. 2016), facilitated the description of several new species on Citrus and allied genera in this study (Table 3).
Table 3.
Global distribution of Colletotrichum species occurring in Citrus hosts and allied genera.
| Species complex | Species | Host | Organ | Geographical distribution | Reference(s) |
|---|---|---|---|---|---|
| C. acutatum | C. abscissum | Citrus sinensis | Flower | Brazil, USA | Crous et al. (2015), |
| Bragança et al. (2016) | |||||
| C. acutatum | Citrus limon | Leaf | Italy | This study | |
| Citrus sinensis | Leaf | ||||
| C. citri | Citrus aurantiifolia | Twig | China | Huang et al. (2013) | |
| C. godetiae | Citrus aurantium | Fruit | Unknown | Damm et al. (2012a) | |
| C. johnstonii | Citrus sp. | Fruit | New Zealand | Damm et al. (2012a) | |
| C. limetticola | Citrus aurantiifolia | Twig | Cuba, USA | Clausen (1912), | |
| Damm et al. (2012a) | |||||
| C. simmondsii | Citrus reticulata | Fruit | China | Peng et al. (2012), | |
| Phoulivong et al. (2012) | |||||
| Murraya sp. | Leaf | ||||
| C. boninense | C. boninense | Citrus medica | Leaf | China | Peng et al. (2012) |
| C. catinaense | Citrus aurantiifolia | Twig | Italy, Malta, Portugal | This study | |
| Citrus reticulata | Leaf | ||||
| Citrus sinensis | Fruit | ||||
| C. citricola | Citrus unchiu | Leaf | China | Huang et al. (2013) | |
| C. constrictum | Citrus limon | Fruit | New Zealand | Damm et al. (2012b) | |
| C. karstii | Citrus grandis | Leaf, twig | China, Europe, New Zealand, South Africa | Damm et al. (2012b), Peng et al. (2012), Huang et al. (2013), This study | |
| Citrus limon | Fruit, leaf, twig | ||||
| Citrus paradisi | Twig | ||||
| Citrus reticulata | Leaf, twig | ||||
| Citrus sinensis | Fruit, leaf, twig | ||||
| Fortunella margarita | Fruit | ||||
| Murraya paniculata | Leaf | ||||
| C. limonicola | Citrus limon | Leaf | Malta | This study | |
| C. novae-zelandiae | Citrus medica | Fruit | Greece, Malta, New Zealand | Damm et al. (2012b), This study | |
| Citrus limon | Leaf, twig | ||||
| Citrus paradisi | Leaf | ||||
| Citrus sinensis | Twig | ||||
| C. gloeosporioides | C. fructicola | Citrus reticulata | Leaf | China | Huang et al. (2013) |
| Fortunella margarita | Branch | ||||
| C. gloeosporioides | Atlantia citroides | Leaf | Brazil, China, Ethiopia, Ghana, Greece, Italy, Malta, Portugal, Spain, New Zealand, Tunisia, USA | Lima et al. (2011), Weir et al. (2012), Huang et al. (2013), Honger et al. (2016), Moges et al. (2016), Rhaeim & Taylor (2016), This study | |
| Citrus bergamia | Fruit | ||||
| Citrus digitata | Leaf | ||||
| Citrus floridana | Fruit | ||||
| Citrus grandis | Leaf | ||||
| Citrus limon | Fruit, leaf, twig | ||||
| Citrus maxima | Twig | ||||
| Citrus medica | Leaf | ||||
| Citrus paradisi | Leaf, twig | ||||
| Citrus reticulata | Fruit, leaf, twig | ||||
| Citrus sinensis | Flower, fruit, leaf, twig | ||||
| Citrus unchiu | Branch, leaf | ||||
| Fortunella margarita | Twig | ||||
| Microcitrus australasica | Twig | ||||
| C. helleniense | Citrus reticulata | Fruit | Greece | This study | |
| Poncirus trifoliata | Twig | ||||
| C. hystricis | Citrus hystrix | Leaf | Italy | This study | |
| C. kahawae subsp. ciggaro | Citrus reticulata | Leaf | Italy | Perrone et al. (2016) | |
| C. siamense | Murraya sp. | Leaf | China | Liu et al. (2016) | |
| C. truncatum | C. truncatum | Citrus flamea | Twig | China | Huang et al. (2013) |
| Citrus limon | Leaf | ||||
| – | C. brevisporum | Citrus medica | Leaf | China | Peng et al. (2012) |
| – | C. tropicicola | Citrus maxima | Leaf | Thailand | Liu et al. (2014) |
Colletotrichum spp. are frequently associated with several citrus diseases worldwide (Timmer et al. 2000), such as PFD on sweet orange, KLA on lime and wither-tip, leaf spot, pre- and post-harvest anthracnose on different hosts (Brown et al. 1996, Timmer et al. 2000, Peres et al. 2008, Lima et al. 2011, McGovern et al. 2012). Before the multi-gene analysis era, C. acutatum was identified as the only species responsible for PFD (Peres et al. 2008) and KLA (Brown et al. 1996). Similarly, C. gloeosporioides was reported as the only Colletotrichum species to cause citrus fruit anthracnose (Brown 1975, Timmer et al. 2000). During the last decade a polyphasic approach was used in several Colletotrichum studies, revealing new species involved with citrus diseases, such as C. abscissum and C. gloeosporioides associated with PFD (Lima et al. 2011, Crous et al. 2015, Silva et al. 2016).
During the last years Colletotrichum spp. affected several commercial orchards in the main citrus producing areas of Mediterranean, causing a broad variety of symptoms and, consequently, losses of marketable fruits (Aiello et al. 2015, Ramos et al. 2016, Rhaiem & Taylor 2016). Therefore, the need for a large-scale investigation of Colletotrichum spp. associated with citrus infections in Europe was needed. This study provides the first molecular characterisation of Colletotrichum diversity related to citrus production in Europe, combined with morphological characterisation.
We performed single gene and multilocus DNA sequence analyses combining seven loci (ITS, CAL, GAPDH, ACT, TUB2, CHS-1 and HIS3) commonly used in previous phylogenetic studies of the C. gloeosporioides, C. acutatum and C. boninense species complexes (Damm et al. 2012a, b, Weir et al. 2012, Bragança et al. 2016). These species complexes incorporate several taxa (Damm et al. 2012a, b, Weir et al. 2012). However, only the closest taxa to the eight Colletotrichum species recovered in this study, were selected based on BLAST searches of NCBIs GenBank nucleotide database and included in the analyses. The final phylogenetic trees clearly distinguished each of these eight species.
We surveyed several citrus orchards, plant nurseries, private gardens and collections in five Mediterranean European countries. We further investigated host plant members of Citrus-allied genera, also economically important as ornamental (Atlantia, Murraya) or rootstock plants (Poncirus), and also for fruit production (Fortunella, Microcitrus). We obtained 174 Colletotrichum single spore strains from symptomatic tissues. Based on multi-locus data we found species allocated in three species complexes. Colletotrichum gloeosporioides in the C. gloeosporioides species complex, and C. karstii in the C. boninense species complex were the predominant species. However, C. gloeosporioides was found in all the countries investigated, whereas C. karstii was not isolated from samples collected in Greece. Moreover, C. acutatum s.str., part of the C. acutatum species complex, was recovered only on the Aeolian Islands (Italy), a volcanic archipelago to the north of Sicily. Colletotrichum novae-zelandiae was recovered in association with leaf spot on grapefruit in Greece and with twig cankers in orange and lemon trees in Malta. In addition, four new species were detected and described. Colletotrichum catinaense was associated with multiple symptoms on different hosts in Italy and Portugal. Colletotrichum helleniense was isolated from Citrus reticulata fruit anthracnose and from leaf lesions on Poncirus trifoliata in Greece. Colletotrichum hystricis was associated with leaf lesions of young plants of Citrus hystrix cultivated in a greenhouse located on Sicily and C. limonicola was recovered on Malta from leaf lesions on lemon plants.
Pathogenicity of all the species isolated from citrus samples collected in Europe was preliminarily tested on two clones of Citrus sinensis. Representative isolates were selected and artificially inoculated on orange fruits of clones ‘Tarocco Scirè’ and ‘Tarocco Nucellare’ (Rapisarda & Russo 2003). All of the Colletotrichum species tested, except C. acutatum, developed lesions on fruits. These results demonstrated a cross-infection potential between multiple species on fruits of two clones of species as already reported by a previous study on Colletotrichum (Freeman et al. 1998). However, our pathogenicity experiments were conducted under extreme conditions commonly applied in artificial inoculations, and it remains to be seen how easily the symptoms development will happen under natural conditions.
The pathogenicity test performed in this study confirmed that C. acutatum is not able to cause symptoms on citrus fruits. However, the establishment of the PFD disease caused by C. acutatum in Europe should be a focus in future surveys. Colletotrichum gloeosporioides was the most aggressive species, causing typical brown lesions that involved the skin and the albedo tissues. Although C. karstii showed the lowest aggressiveness, the pathogenicity test demonstrated its ability to cause lesions on fruits, which was also true for the remaining species, C. catinaense, C. helleniense, C. hystricis, C. limonicola and C. novae-zelandiae. The clone ‘Tarocco Scirè’ appeared more susceptible than ‘Tarocco Nucellare’ as Aiello et al. (2015) recently demonstrated for C. gloeosporioides and C. karstii.
Colletotrichum acutatum s.lat. is a common pathogen of several crops, including citrus, worldwide (Damm et al. 2012a). In Europe it has been detected on different hosts such as strawberry (Garrido et al. 2008), strawberry tree (Polizzi et al. 2011), olives (Moral et al. 2008), but never on citrus. Furthermore, C. novae-zelandiae was previously recovered from grapefruit and chili in New Zealand (Damm et al. 2012b). Thus, this study represents the first report of C. acutatum associated with citrus in Europe and the first detection of C. novae-zelandiae outside of New Zealand. Colletotrichum karstii, a member of the C. boninense species complex, has been reported on many host plants with a wide geographical distribution (Damm et al. 2012b). This species has been reported on citrus in South Africa, New Zealand and China (Damm et al. 2012b, Peng et al. 2012, Huang et al. 2013) as well as in Europe, where it was reported as citrus pathogen in Italy and Portugal (Aiello et al. 2015, Ramos et al. 2016). In Europe, C. karstii has been detected also on other hosts such as tropical fruits, cotton and lupine plants (Damm et al. 2012b, Ismail et al. 2015). Colletotrichum gloeosporioides was largely dominant in our investigation, in agreement with recent global results (Lima et al. 2011, Huang et al. 2013, Aiello et al. 2015, Honger et al. 2016, Ramos et al. 2016, Rhaiem & Taylor 2016). Colletotrichum gloeosporioides was isolated from all the citrus organs sampled (leaves, flowers, fruit and twigs), and proved to be the most aggressive Colletotrichum species. This species is reported as pathogen of the main cultivated citrus species worldwide (Huang et al. 2013) and to our knowledge the present study represents the first report of C. gloeosporioides associated with citrus flower disease in Europe, previously reported in Brazil (Lima et al. 2011). Colletotrichum catinaense and C. limonicola represent new species in the C. novae-zelandiae clade within the C. boninense species complex. Colletotrichum catinaense was recovered associated with infections of diverse Citrus species, whereas C. limonicola has been isolated only from lemon leaf lesions. Thus, more surveys are needed to investigate distribution and host specificity of this new species. Colletotrichum helleniense was isolated from Citrus reticulata and from Poncirus trifoliata, a member of the Rutaceae family largely cultivated in nurseries as citrus rootstock due to its economically useful traits, including cold temperature and poor soil tolerance, and Citrus Tristeza Virus resistance (Garnsey & Barrett 1987). This report shows the ability of C. helleniense to colonise tissues of different genera within the Rutaceae. Recently, Batista et al. (2016) supported the distinc-tion of two C. kahawae subspp. as two cryptic species. Colletotrichum kahawae subsp. ciggaro, one of these subspecies, has also recently been recorded by Perrone et al. (2016) as a pathogen of mandarin (Citrus reticulata). However, C. helle-niense is phylogenetically close to both C. kahawae subspecies, but clearly differentiated based on multi-locus phylogenetic analyses. As such it thus represents a new species in the C. kahawae clade in the C. gloeosporioides species complex. Colletotrichum hystricis was isolated from lesions on leaves of Citrus hystrix. This Citrus species is commonly cultivated, has a pleasant smell, and is referred to as medicinal lime (Yaacob & Subhadrabandhu 1995). The fruit is not appreciated, but is economically important for the extraction of essential oil used for cooking and cosmetics (Allen 1967). Colletotrichum hystricis is close to but clearly differentiated from C. alienum, which is commonly associated with cultivated fruits (Weir et al. 2012). In the present study it is described as a distinct taxon, supported also by morphological differences such as having obovoidal conidia and a slower growth rate in culture. Moreover, C. alienum is characterised by the development of perithecia in culture, whereas the two strains of C. hystricis did not produce perithecia on artificial media in this study.
The present study provides the first overview of Colletotrichum diversity associated with several disease symptoms on citrus fruits and plants in Europe, and provides useful information for pathogenicity evaluation and effective disease control. Preliminary inoculations also demonstrated the ability of all the Colletotrichum spp. found in Europe to cause infections on orange fruits. Further studies are thus required to resolve the host range and pathogenicity of the Colletotrichum species reported on other Citrus spp. and different plant organs.
Acknowledgments
We are grateful to Arien van Iperen (cultures), Marjan Vermaas (photo plates) and Mieke Starink-Willemse (DNA isolation, amplification and sequencing) for their technical assistance. V.G. would like to thank Antonino Azzaro, Dimitrios Dimou, Amilcar Duarte, Pietro Formica, Anna Guglielmo, Ioannis Livieratos, Leonardo Velasco, Antonio Vicent and Anthony Zammit for the kind support with specimen collection at several sites.
REFERENCES
- Aiello D, Carrieri R, Guarnaccia V, et al. 2015. Characterization and pathogenicity of Colletotrichum gloeosporioides and C. karstii causing preharvest disease on Citrus sinensis in Italy. Journal of Phytopathology 163: 168–177. [Google Scholar]
- Allen BM. 1967. Malayan fruits an introduction to the cultivated species. Singapore, Donald Moore Press Ltd. [Google Scholar]
- Anderson JM, Aitken EAB, Dann EK, et al. 2013. Morphological and molecular diversity of Colletotrichum spp. causing pepper spot and anthracnose of lychee (Litchi chinensis) in Australia. Plant Pathology 62: 279–288. [Google Scholar]
- Bailey JA, Jeger MJ. 1992. Colletotrichum. Biology, pathology and control. CABI, Wallingford, UK. [Google Scholar]
- Baroncelli R, Zapparata A, Sarocco S, et al. 2015. Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum species. PLoS One 10(6): e0129140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista D, Silva DN, Vieira A, et al. 2016. Legitimacy and implications of reducing Colletotrichum kahawae to subspecies in plant pathology. Frontiers in Plant Science 7: 2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyahia H, Jrifi A, Smaili C, et al. 2003. First report of Colletotrichum gloeosporioides causing withertip on twigs and tear stain on fruit of citrus in Morocco. Plant Pathology 52: 798. [Google Scholar]
- Bernstein B, Zehr EI, Dean RA, et al. 1995. Characteristics of Colletotrichum from peach, apple, pecan and other hosts. Plant Disease 79: 478–482. [Google Scholar]
- Bragança CA, Damm U, Baroncelli R, et al. 2016. Species of the Colletotrichum acutatum complex associated with anthracnose diseases of fruit in Brazil. Fungal Biology 120: 547–561. [DOI] [PubMed] [Google Scholar]
- Brown AE, Sreenivasaprasad S, Timmer LW. 1996. Molecular characterization of slow-growing orange and key lime anthracnose strains of Colletotrichum from citrus as C. acutatum. Phytopathology 86: 523–527. [Google Scholar]
- Brown GE. 1975. Factors affecting post-harvest development of Colletotrichum gloeosporioides in citrus fruits. Phytopathology 65: 404–409. [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ, et al. 2009. A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204. [Google Scholar]
- Cannon PF, Bridge PD, Monte E. 2000. Linking the past, present, and future of Colletotrichum systematics. In: Prusky D, Freeman S, Dickman MB. (eds), Colletotrichum: Host specificity, pathology, and host pathogen interaction: 1–20. APS Press, St. Paul, Minnesota, USA. [Google Scholar]
- Cannon PF, Buddie AG, Bridge PD. 2008. The typification of Colletotrichum gloeosporioides. Mycotaxon 104: 189–204. [Google Scholar]
- Cannon PF, Damm U, Johnston PR, et al. 2012. Colletotrichum – current status and future directions. Studies in Mycology 73: 181–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Clausen RE. 1912. A new fungus concerned in whiter tip of varieties of Citrus medica. Phytopathology 2: 217–239. [Google Scholar]
- Cristóbal-Martínez AL, De Jesús Yáñez-Morales M, Solano-Vidal R, et al. 2017. Diversity of Colletotrichum species in coffee (Coffea arabica) plantations in Mexico. European Journal of Plant Pathology 147: 605–614. [Google Scholar]
- Crouch JA, Beirn LA, Cortese LM, et al. 2009. Anthracnose disease of switch-grass caused by the novel fungal species Colletotrichum navitas. Mycological Research 113: 1411–1421. [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. 2004a. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ, Risede JM, et al. 2004b. Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Slippers B, et al. 2016a. Global food and fibre security threatened by current inefficiencies in fungal identification. Philosophical Transactions of the Royal Society B: Biological Sciences 371: 1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. 2009. Fungal biodiversity. CBS Laboratory manual series 1. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands. [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, et al. 2015. Fungal Planet description sheets: 320–370. Persoonia 34: 167–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, et al. 2016b. Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Liu F, et al. 2013. The Colletotrichum orbiculare species complex: Important pathogens of field crops and weeds. Fungal Diversity 61: 29–59. [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012a. The Colletotrichum acutatum species complex. Studies in Mycology 73: 37–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012b. The Colletotrichum boninense species complex. Studies in Mycology 73: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, O’Connell RJ, Groenewald JZ, et al. 2014. The Colletotrichum destructivum species complex – hemibiotrophic pathogens of forage and field crops. Studies in Mycology 79: 49–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Woudenberg JHC, Cannon PF, et al. 2009. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity 39: 45–87. [Google Scholar]
- De Silva DD, Ades PK, Crous PW, et al. 2017. Colletotrichum species associated with chili anthracnose in Australia. Plant Pathology 66: 254–267. [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, et al. 2012. The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deising HB, Werner S, Wernitz M. 2000. The role of fungal appressoria in plant infection. Microbes and Infection 2: 1631–1641. [DOI] [PubMed] [Google Scholar]
- Diao YZ, Zhang C, Liu F, et al. 2017. Colletotrichum species causing anthracnose disease of chili in China. Persoonia 38: 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO - Food and Agricultural Organization of the United Nations, Rome , 2016. Citrus fruits fresh and processed: annual statistics. http://www.fao.org/3/a-i5558e.pdf.
- Farr DF, Aime MC, Rossman AY, et al. 2006. Species of Colletotrichum on Agavaceae. Mycological Research 110: 1395–1408. [DOI] [PubMed] [Google Scholar]
- Farr DF, Rossman AY. 2017. Fungal databases, systematic mycology and microbiology laboratory, ARS, USDA. Retrieved February 26.
- Freeman S, Katan T, Shabi E. 1998. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Disease 82: 596–605. [DOI] [PubMed] [Google Scholar]
- Freeman S, Shabi E. 1996. Cross-infection of subtropical and temperate fruits by Colletotrichum species from various hosts. Physiological and Molecular Plant Pathology 49: 395–404. [Google Scholar]
- Garnsey DJ, Barrett SM. 1987. Identification of citrus tristeza virus resistance in citrus relatives and its potential applications. Phytophylactica 19: 187–192. [Google Scholar]
- Garrido C, Carbú M, Fernández-Acero FJ, et al. 2008. Isolation and pathogenicity of Colletotrichum spp. causing anthracnose of strawberry in south west Spain. European Journal of Plant Pathology 120: 409–415. [Google Scholar]
- Glass NL, Donaldson G. 1995. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Vitale A, Cirvilleri G, et al. 2016. Characterisation and pathogenicity of fungal species associated with branch cankers and stem-end rot of avocado in Italy. European Journal of Plant Pathology 146: 963–976. [Google Scholar]
- Guerber JC, Liu B, Correll JC, et al. 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95: 872–895. [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Honger JO, Offei SK, Oduro KA, et al. 2016. Identification and molecular characterisation of Colletotrichum species from avocado, citrus and pawpaw in Ghana. South African Journal of Plant and Soil 33: 177–185. [Google Scholar]
- Huang F, Chen GQ, Hou X, et al. 2013. Colletotrichum species associated with cultivated citrus in China. Fungal Diversity 61: 61–74. [Google Scholar]
- Ismail AM, Cirvilleri G, Yaseen T, et al. 2015. Characterisation of Colletotrichum species causing anthracnose disease of mango in Italy. Journal of Plant Pathology 97: 167–171. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Rewal HS, Sethi A. 2007. Pre-harvest stem-end rot in citrus cultivars due to Colletotrichum gloeosporioides. European Journal of Horticultural Science 72: 20–25. [Google Scholar]
- Klotz J. 1961. Color handbook of Citrus diseases. University of California, California, USA. [Google Scholar]
- Lima WG, Sposito MB, Amorim L, et al. 2011. Colletotrichum gloeosporioides, a new causal agent of citrus post-bloom fruit drop. European Journal of Plant Pathology 131: 157–165. [Google Scholar]
- Liu F, Cai L, Crous PW, et al. 2014. The Colletotrichum gigasporum species complex. Persoonia 33: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Damm U, Cai L, et al. 2013. Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Diversity 61: 89–105. [Google Scholar]
- Liu F, Wang M, Damm U, et al. 2016. Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evolutionary Biology 16: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Weir BS, Damm U, et al. 2015. Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 35: 63–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbe CM, Denman S, Cannon PF, et al. 2004. Characterization of Colletotrichum species associated with diseases of Proteaceae. Mycologia 96: 1268–1279. [PubMed] [Google Scholar]
- MacKenzie SJ, Peres NA, Barquero MP, et al. 2009. Host range and genetic relatedness of Colletotrichum acutatum isolates from fruit crops and leatherleaf fern in Florida. Phytopathology 99: 620–631. [DOI] [PubMed] [Google Scholar]
- Marcelino J, Giordano R, Gouli S, et al. 2008. Colletotrichum acutatum var. fioriniae (teleomorph: Glomerella acutata var. fioriniae var. nov.) infection of a scale insect. Mycologia 100: 353–374. [DOI] [PubMed] [Google Scholar]
- Marin-Felix Y, Groenewald JZ, Cai L, et al. 2017. Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology, doi: 10.1016/j.simyco.2017.04.002. [DOI] [PMC free article] [PubMed]
- McGovern RJ, Seijo TE, Hendricks K, et al. 2012. New report of Colletotrichum gloeosporioides causing postbloom fruit drop on citrus in Bermuda. Canadian Journal of Plant Pathology 34: 187–194. [Google Scholar]
- Moges AD, Admassu B, Belew D, et al. 2016. Development of microsatellite markers and analysis of genetic diversity and population structure of Colletotrichum gloeosporioides from Ethiopia. PLoS ONE 11(3): e0151257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moral J, Bouhmidi K, Trapero A. 2008. Influence of fruit maturity, cultivar susceptibility, and inoculation method on infection of olive fruit by Colletotrichum acutatum. Plant Disease 92: 1421–1426. [DOI] [PubMed] [Google Scholar]
- Moriwaki J, Sato T, Tsukiboshi T. 2003. Morphological and molecular characterization of Colletotrichum boninense sp. nov. from Japan. Mycoscience 44: 47–53. [Google Scholar]
- Nirenberg HI. 1976. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- Nylander JAA. 2004. MrModeltest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, et al. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- Peng LJ, Yang YL, Hyde KD, et al. 2012. Colletotrichum species on Citrus leaves in Guizhou and Yunnan provinces, China. Cryptogamie Mycologie 33: 267–283. [Google Scholar]
- Peres N, MacKenzie S, Peever T, et al. 2008. Postbloom fruit drop of citrus and key lime anthracnose are caused by distinct phylogenetic lineages of Colletotrichum acutatum. Phytopathology 98: 345–352. [DOI] [PubMed] [Google Scholar]
- Perfect SE, Hughes HB, O’Connell RJ, et al. 1999. Colletotrichum: a model genus for studies on pathology and fungal – plant interactions. Fungal Genetics and Biology 27: 186–198. [DOI] [PubMed] [Google Scholar]
- Perrone G, Magistà D, Ismail AM. 2016. First report of Colletotrichum kahawae subsp. ciggaro on mandarin in Italy. Journal of Plant Pathology 98: 12. [Google Scholar]
- Phoulivong S, Cai L, Parinn N, et al. 2010. A new species of Colletotrichum from Cordyline fruticosa and Eugenia javanica causing anthracnose disease. Mycotaxon 114: 247–257. [Google Scholar]
- Phoulivong S, Mckenzie EHC, Hyde KD. 2012. Cross infection of Colletotrichum species; a case study with tropical fruits. Current Research in Environmental & Applied Mycology 2: 99–111. [Google Scholar]
- Polizzi G, Aiello D, Guarnaccia V, et al. 2011. First report of damping-off on strawberry tree caused by Colletotrichum acutatum and C. simmondsii in Italy. Plant Disease 95: 1588. [DOI] [PubMed] [Google Scholar]
- Ramos AP, Talhinhas P, Sreenivasaprasad S, et al. 2016. Characterization of Colletotrichum gloeosporioides, as the main causal agent of citrus anthracnose, and C. karstii as species preferentially associated with lemon twig dieback in Portugal. Phytoparasitica 44: 549–561. [Google Scholar]
- Rapisarda P, Russo G. 2003. Fruit quality of five ‘Tarocco’ selections grown in Italy. In: Proceedings of the International Society of Citriculture. 9th Congress, 3–7 December 2000 International Society of Citriculture, 2: 1149–1153. Riverside, CA, USA. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, England. [Google Scholar]
- Rhaiem A, Taylor PW. 2016. Colletotrichum gloeosporioides associated with anthracnose symptoms on citrus, a new report for Tunisia. European Journal of Plant Pathology 146: 219–224. [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivas RG, Tan YP. 2009. A taxonomic reassessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov. Fungal Diversity 39: 111–122. [Google Scholar]
- Silva AO, Savi DC, Gomes FB, et al. 2016. Identification of Colletotrichum species associated with postbloom fruit drop in Brazil through GAPDH sequencing analysis and multiplex PCR. European Journal of Plant Pathology 147: 731–748 [Google Scholar]
- Sutton BC. 1980. The coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata: 1–696. Commonwealth Mycological Institute, Kew, Surrey, England. [Google Scholar]
- Sutton BC. 1992. The genus Glomerella and its anamorph Colletotrichum. In: Bailey JA, Jeger MJ. (eds), Colletotrichum: biology, pathology and control: 1–26. CAB International, Wallingford, UK. [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4.0b10. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Talhinhas P, Mota-Capitão C, Martins S, et al. 2011. Epidemiology, histopathology and aetiology of olive anthracnose caused by Colletotrichum acutatum and C. gloeosporioides in Portugal. Plant Pathology 60: 483–495. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer LW, Garnsey SM, Graham JH. 2000. Compendium of Citrus diseases (second edition). The American Phytopathological Society. [Google Scholar]
- Udayanga D, Manamgoda DS, Liu X, et al. 2013. What are the common anthracnose pathogens of tropical fruits? Fungal Diversity 61: 165–179. [Google Scholar]
- Von Arx JA. 1957. Die Arten der Gattung Colletotrichum Cda. Phytopathologische Zeitschrift 29: 413–468. [Google Scholar]
- Weir BS, Johnston PR, Damm U. 2012. The Colletotrichum gloeosporioides species complex. Studies in Mycology 73: 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR Protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California. [Google Scholar]
- Yaacob O, Subhadrabandhu S. 1995. The production of economic fruits in South East Asia: 156. Oxford University Press, Kuala Lumpur. [Google Scholar]
- Yang YL, Liu ZY, Cai L, et al. 2009. Colletotrichum anthracnose of Amaryllidaceae. Fungal Diversity 39: 123–146. [Google Scholar]


