Abstract
The order Phyllachorales (Pezizomycotina, Ascomycota) is a group of biotrophic, obligate plant parasitic fungi with a tropical distribution and high host specificity. Traditionally two families are recognised within this order: Phyllachoraceae and Phaeochoraceae, based mostly on morphological and host characteristics. Currently, the position of the order within the class Sordariomycetes is inconclusive, as well as the monophyly of the order, and its internal phylogenetic structure. Here we present a phylogeny of the order Phyllachorales based on sequence data of 29 species with a broad host range resulting from a wide geographical sampling. We inferred Maximum Likelihood and Bayesian phylogenies from data of five DNA regions: nrLSU rDNA, nrSSU rDNA, ITS rDNA, and the protein coding genes RPB2, and TEF1. We found that the order Phyllachorales is monophyletic and related to members of the subclass Sordariomycetidae within Sordariomycetes. Within the order, members of the family Phaeochoraceae form a monophyletic group, and the family Phyllachoraceae is split into two lineages. Maximum Likelihood ancestral state reconstructions indicate that the ancestor of Phyllachorales had a monocotyledonous host plant, immersed perithecia, and a black stroma. Alternative states of these characters evolved multiple times independently within the order. Based on our results we redefine the family Phyllachoraceae and propose the new family Telimenaceae with Telimena erythrinae as type species, resulting in three families in the order. Species of Telimena spp. occur in several monocotyledonous and eudicotyledonous host plants except Poaceae, and generally have enlarged black pseudostroma around the perithecia, a character not present in species of Phyllachoraceae.
Keywords: ancestral state reconstruction, plant parasitic, tar spot fungi, Telimenaceae
INTRODUCTION
Phyllachorales is an order of biotrophic, obligate plant parasitic fungi in the class Sordariomycetes, i.e., inoperculate pyrenomycetes. About 1 226 species are currently accepted in the order (Kirk et al. 2008), although 160 000 species have been estimated to occur worldwide (Cannon 1997). Phyllachorales are highly diverse in the tropics, relatively common in disturbed and natural vegetation, and likewise found in open and forested areas (Piepenbring et al. 2011).
Species of Phyllachorales are leaf- or steam-inhabiting microfungi with shiny black stromata, which gave them the common name ‘tropical tar spot fungi’. They are morphologically characterised by: deep black stromata of various shapes (except in species of Polystigma which have brightly coloured stromata); pseudostroma inside the host tissue and usually beneath an epidermal clypeus; perithecia usually strongly melanised that may be superficial, erumpent or immersed in the host tissue (Fig. 1a–f); thin-walled paraphyses which frequently deliquesce; unitunicate asci of cylindrical to clavate shape, with an ascus crown and an inconspicuous apical ring not staining blue in iodine; and globose to filiform ascospores, which in most species are hyaline and 1-celled, with only a few genera including species with brown or septate ascospores (Parbery 1967, Cannon 1991, 1997). Although it has been difficult to connect asexual and sexual morphs in the order due to their obligately parasitic condition, some species of Phyllachorales have been linked to the asexual genus Linochora (Von Höhnel 1910), which may be inconspicuous and spermatial in function (Parbery & Langdon 1963, Parbery 1996).
Fig. 1.
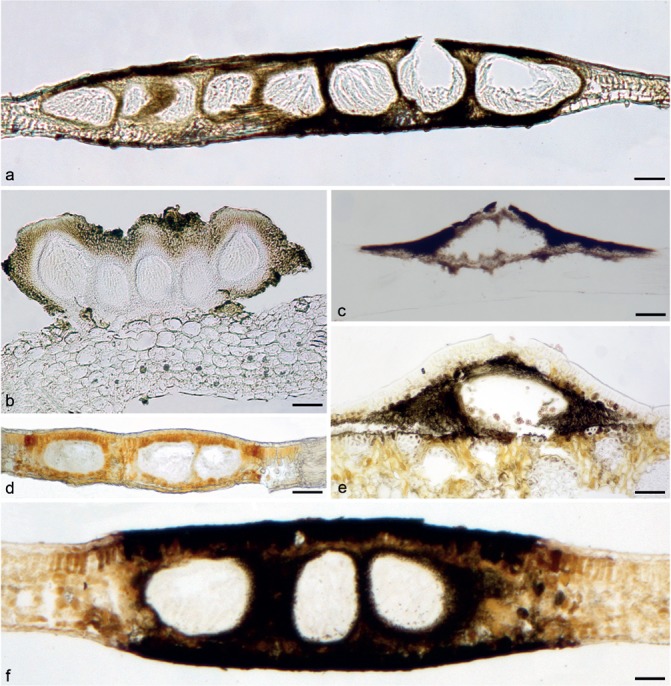
Perithecia of species of Phyllachorales in different positions in the mesophyll of the leaves. a. Phyllachora graminis (isotype CUP3536) with immersed perithecia and few pseudostroma; b. Coccodiella miconiae (ppMP1342) with superficial perithecia; c. Camarotella costaricensis (MM-21) with erumpent perithecium; d. Polystigma pusillum (MM-113) with brightly coloured stroma; e. Serenomyces phoenicis (F59049) with subcuticular perithecium and without a clypeus; f. Telimena bicincta (epitype MM-133) with immersed perithecia and strongly developed pseudostroma. — Scale bars = 100 μm.
Due to their biotrophic nutrition mode, high host specificity is assumed and species concepts are based partly on the identity and systematic position of the corresponding host plants. Hence to identify species of Phyllachorales, it is necessary to identify the host plant. Traditionally, new species have been described on the basis of new host records at generic level. However, examination of species of Phyllachorales on the host families Poaceae and Fabaceae demonstrated that tropical tar spot species are not restricted to a single host genus, but may occur on species belonging to a group of closely related genera (Parbery 1978, Cannon 1991, 1997). Species delimitation based mainly on host identity may therefore lead to over-splitting of species (Cannon 1997).
Phyllachorales are associated with diverse host plants, and most of the species are linked to angiosperms, with a few exceptions including the lichenicolous Lichenochora species, the marine algicolous genus Phycomelaina, and some species on ferns and gymnosperms. Within the angiosperms, the following families are preferentially parasitized: Arecaceae, Fabaceae, Lauraceae, Melastomataceae, Moraceae, Myrtaceae, and Poaceae (Cannon 1997). Monographs for phyllachoraceous species are available for Arecaceae (Hyde & Cannon 1999), Fabaceae (Cannon 1991), and Poaceae (Orton 1944, Parbery 1967). Additional host families studied include Asclepiadaceae (Pearce et al. 1999), Erythroxylaceae (Cannon & Evans 1999), Proteaceae (Pearce et al. 2001), and Rosaceae (Cannon 1996).
The genus Phyllachora was introduced on a herbarium label in Fuckels exsiccate series ‘Fungi Rhenani’ with a single species, P. agrostis (Fuckel 1867 in Cannon 1991), currently accepted as Scirrhia agrostis in Dothideales (Eriksson 1967). Later the genus Phyllachora was lectotypified with Phyllachora graminis as generic type (Clements & Shear 1931), and the genus name in the sense of Fuckel (1870) was conserved to allow continued use in its currently accepted circumscription. The order Phyllachorales was formally described by Barr (1983). However, throughout the history of the group, several authors have placed phyllachoraceous fungi into various families and orders, stressing different morphological and ecological characteristics: Diaporthales (Cannon 1988), Dothideales (Saccardo 1876, Theissen & Sydow 1915), Polystigmatales or Polystigmataceae (Von Arx & Müller 1954, Eriksson 1982, Hawksworth et al. 1983), Sphaeriales (Nannfeldt 1932, Luttrell 1951, Müller & Von Arx 1962, 1973), and Xylariales (Barr 1983). For a detailed description of the taxonomical history of the order see Cannon (1991) and Pearce & Hyde (2006).
The order Phyllachorales comprises the families Phyllachoraceae and Phaeochoraceae. The family Phyllachoraceae was erected by Theissen & Sydow (1915) and is by far the largest family within the order with almost 1 200 described species (Kirk et al. 2008). The number of genera varies between 51 (Kirk et al. 2008) and 73 (www.indexfungorum.org). Many of these genera, however, have less than ten species and 27 are monotypic. Phyllachora is the largest genus with 994 species; and Coccodiella, Lichenochora, Ophiodothella, Polystigma, and Trabutia are also large genera with 22, 40, 36, 24, and 35 species, respectively (www.indexfungorum.org). The family Phaeochoraceae is a small group with 19 accepted species in the genera Cocoicola, Phaeochora, Phaeochoropsis, and Serenomyces and is known to occur only associated with species of Arecaceae (Hyde et al. 1997). Phaeochoraceae were provisionally assigned to Phyllachorales since the family was erected (Hyde et al. 1997), mainly due to their unusual stromatic characteristics. However, no molecular studies are available to clarify the family’s phylogenetic placement.
Molecular phylogenetic studies including members of the Phyllachorales are infrequent, mainly because it is difficult to obtain cultures of these biotrophic fungi. The existing molecular studies indicate, without support and with a limited sampling, that the order might be related to Sordariales or Boliniales (Winka & Eriksson 2000, Wanderlei-Silva et al. 2003, Inderbitzin et al. 2004, Trampe 2010). A recent large-scale phylogenetic study confirms the position of Phyllachorales in the subclass Sordariomycetidae with high support (Maharachchikumbura et al. 2015). However, in this phylogeny based on four loci, Phyllachorales are represented only by three taxa and the single locus nrSSU.
The phylogeny within the order Phyllachorales is still almost unresolved and its monophyly has not yet been resolved. It is known that the Glomerella/Colletotrichum complex that was placed within Phyllachorales in the past, belongs to the Glomerellales (Wanderlei-Silva et al. 2003, Réblová et al. 2011) fungi. Some studies suggested that Phyllachorales are a polyphyletic assemblage, since several taxa had to be excluded from this order: Ophiodothella and Sphaerodothis were transferred to Xylariales and Hypocreales, respectively (Wanderlei-Silva et al. 2003); Plectosphaera eucalypti to Xylariales (Summerell et al. 2006), and Polystigma amygdalinum to subclass Xylariomycetidae (Habibi et al. 2015). These studies also suggested that among the studied taxa, Phyllachora and Coccodiella are the only genera forming a monophyletic clade, closely related to Sordariales, and considered as true Phyllachorales. Due to the limited availability of molecular phylogenetic data, information is lacking concerning the evolution of morphological traits and the co-evolution trails with the hosts.
The aims of this study are:
1 to confirm the phylogenetic position of Phyllachorales within Sordariomycetidae;
2 to determine the monophyly of Phyllachorales;
3 to define monophyletic clades within the order for the delimitation of families; and
4 to reconstruct the evolution of morphological and ecological characteristics to assess their value as systematic criteria.
To achieve these objectives, we inferred the first comprehensive multilocus phylogeny for the order Phyllachorales.
MATERIALS AND METHODS
Taxon sampling
Fresh specimens representing the two recognised families of Phyllachorales were collected mainly in Costa Rica during 2012–2015 and Western Panama during 2007–2015 (Trampe 2010). Additional specimens were collected from Benin, Ecuador, Germany, Thailand, and the USA. A total of 48 collections of tropical tar spot fungi, representing 29 species and six genera were sequenced. Specimens collected in the context of the present study were deposited in the following herbaria: FR, M, UCHI, USJ, and HUTPL.
Extraction, amplification, and sequencing of DNA
DNA was isolated directly from hymenia of fresh, recently collected material or from dry specimens except for the species belonging to family Phaeochoraceae, which were available as cultures previously isolated by Elliott & Des Jardin (2014). To extract non-melanised cells with high quality DNA, for each stroma, the clypeus was cut off in half to exposed the hymenia of 1–10 perithecia (diam c. 0.2–0.5 mm) that were removed and placed into 1.5 mL sterilised microtubes containing Cetyltrimethyl ammonium bromide (2 % CTAB). The isolation of genomic DNA from fresh material was performed with 600 μL of extraction buffer (2 % CTAB; 100 mM Tris-HCl, pH 8; 1.4 M NaCl, and 20 mM EDTA) and the DNA was extracted using phenol-chloroform : isoamyl alcohol (24 : 1). For dry material the E.Z.N.A® Forensic DNA Extraction Kit (VWR-Omega, USA) was used following the manufacturer’s instructions with a few modifications. The material was homogenized for 5–10 min using a Retsch Mixer Mill MM301 with STL buffer and 2.5 mm Zirconia beads. Isolated DNA was resuspended in sterile water and stored at −20 °C. DNA concentration was checked by electrophoresis in 0.8 % agarose and by the spectrophotometer NanoDrop 2000c (Thermo Fisher Scientific, USA). Several attempts were made to extract DNA from older herbarium specimens (type specimens) but they were unsuccessful. For that reason, the macro- and micro-morphology of extracted phyllachoraceous specimens were rigorously compared with the respective type specimen when it was possible.
Five partial nuclear gene regions (three ribosomal loci and two protein-coding genes) were amplified and sequenced: one fragment of the large subunit nuclear ribosomal DNA (nrLSU) with primers NL1 and NL2 (O’Donnell 1993), one fragment of the small subunit nuclear ribosomal DNA (nrSSU) with primers NS1 and NS4 (White et al. 1990), the complete internal transcribed spacer region of ribosomal DNA (ITS1-5.8S-ITS2) with primers ITS5 and ITS4 (White et al. 1990), one fragment of the second largest subunit of RNA polymerase II (RPB2) with primers fRPB2-5f and fRPB2-7cr (Liu et al. 1999), and one fragment of the translation elongation factor 1 (TEF1) with primers EF1-983f (Carbone & Kohn 1999) and EF1-2218r (Rehner & Buckley 2005). PCR reactions were performed on a PEQSTAR 2X GRADIENT Thermal Cycler (PEQLAB, Erlangen, Germany) using VWR Taq DNA polymerase (VWR-Omega, USA).
The reactions followed this protocol: Each 50 μL PCR mixture included 10 μL of 5 × buffer, 3 μL (25 mM) of magnesium chloride (MgCl2), 0.8 μL (20 mM) of dNTP mix, 1 μL (10 mM) of each primer, 0.4 μL (5U/μL) of Taq Polymerase, 1 μL of template DNA, and 37.8 μL of sterile distilled water. For RPB2 and TEF1 4 μL of each primer and 4 μL of template DNA were used, and the amount of water decreased accordingly. Conditions of the PCR for nrLSU, nrSSU, and ITS were as follows: DNA denaturation 94 °C for 4 min; 35 cycles of DNA denaturation 94 °C for 30 s, primer annealing 55 °C for 30 s and TAQ extension 72 °C for 45 s, and a final TAQ extension 72 °C for 5 min, followed by storage at 8 °C. Thermal cycling parameters of the RPB2 and TEF1 genes were performed as described by Liu et al. (1999) and Rehner & Buckley (2005), respectively.
PCR-products were checked on 1.5 % agarose electrophoresis gels stained with ethidium bromide. Amplified PCR products were purified with the Cycle Pure Kit (VWR-Omega, USA). The sequencing in both directions was performed with the same PCR primers with an Applied Biosystems 3730 DNA Analyzer in the BiK-F Laboratory Centre, Frankfurt am Main, Germany.
Sequence alignment and model determination
Alignments for each gene were made by MAFFT v. 7.164b (Katoh & Standley 2013) using the L-INS-i algorithm, and adjusted manually using Mega v. 6.06 (Tamura et al. 2013). The program Gblocks v. 0.91b (Talavera & Castresana 2007) was used to remove poorly aligned positions and divergent regions from the DNA alignment using the parameters for a less stringent selection.
Among the acquired sequences, often multiple sequences were recovered from a single stroma for a given genetic locus. To distinguish DNA of contaminants from phyllachoraceous DNA, all sequences were subjected to BLAST searches to verify the identity, and preliminary phylogenetic analyses were also performed including common plant parasitic fungi in Pezizomycotina.
To test the level of congruence among loci, the Congruence Among Distance Matrices test (CADM, Legendre & Lapointe 2004) was performed using patristic distance matrices to test the null hypothesis of complete incongruence among loci. This analysis has been shown to have an accurate type-I error rate (Campbell et al. 2011).
We assembled two datasets for phylogenetic analyses: a three-locus concatenated alignment (nrSSU, nrLSU, RPB2) including 88 specimens representing the three recognized subclasses of the Sordariomycetes following the classification of Zhang et al. (2006). The purpose of this analysis was to infer the position of Phyllachorales within Sordariomycetes, and to test the monophyly of the order. Sequences of further representative species of orders in Sordariomycetes were downloaded from GenBank (Table 1) mostly from studies published by Spatafora et al. (2006) and Zhang et al. (2006). Leotia lubrica was selected as outgroup taxon based on Zhang et al. (2006), and missing data were shown as gaps.
Table 1.
Sequences downloaded from GenBank (in alphabetical order) used in this study.
| Species | Order | Source | GenBank Accession Numbers |
Reference | ||
|---|---|---|---|---|---|---|
| nrLSU | nrSSU | RPB2 | ||||
| Aniptodera chesapeakensis | Microascales | ATCC 32818 | U46882 | U46870 | DQ470896 | Spatafora et al. (2006) |
| Balansia henningsiana | Hypocreales | AEG96-27a | AY489715 | AY489683 | DQ522413 | Spatafora et al. (2006) |
| Bionectria ochroleuca | Hypocreales | AFTOL-ID 187 | DQ862027 | DQ862044 | DQ862013 | Zhang et al. (2006) |
| Bombardia bombarda | Sordariales | SMH 3391 | DQ470970 | DQ471021 | DQ470923 | Spatafora et al. (2006) |
| Buergenerula spartinae | Magnaporthales | ATCC 22848 | DQ341492 | DQ341471 | – | Thongkantha et al. (2009) |
| Calosphaeria pulchella | Calosphaeriales | CBS 115999 | AY761075 | AY761071 | GU180661 | Réblová & Seifert (2004) |
| Camarops amorpha | Boliniales | SMH1450 | AY780054 | – | AY780156 | Miller & Huhndorf (2005) |
| Camarops microspora | Boliniales | CBS 649.92 | AY083821 | DQ471036 | DQ470937 | Spatafora et al. (2006) |
| Camarops tubulina | Boliniales | SMH4614 | AY346266 | – | AY780157 | Miller & Huhndorf (2005) |
| Camarops ustulinoides | Boliniales | DEH 2164 | DQ470941 | DQ470989 | DQ470882 | Spatafora et al. (2006) |
| Cercophora coprophila | Sordariales | SMH3794 | AY780058 | – | AY780162 | Miller & Huhndorf (2005) |
| Chaetosphaerella phaeostroma | Coronophorales | SMH4257 | AY695264 | – | FJ968940 | Huhndorf et al. (2004) |
| Chrysoporthe cubensis | Diaporthales | CBS 101281 | AF408338 | DQ862047 | DQ862016 | Zhang et al. (2006) |
| Cordyceps cardinalis | Hypocreales | OSC 93610 | AY184963 | AY184974 | EF469106 | Sung et al. (2007) |
| Cryphonectria parasitica | Diaporthales | ATCC 38755 | NG027589 | DQ862048 | DQ862017 | Zhang et al. (2006) |
| Cryptosporella hypodermia | Diaporthales | CBS 171.69 | DQ862028 | DQ862049 | DQ862018 | Zhang et al. (2006) |
| Diaporthe eres | Diaporthales | CBS 109767 | AF408350 | DQ471015 | DQ470919 | Spatafora et al. (2006) |
| Diatrype disciformis | Xylariales | CBS 197.49 | DQ470964 | DQ471012 | DQ470915 | Zhang et al. (2006) |
| Eutypa lata | Xylariales | CBS 208.87 | DQ836903 | DQ836896 | DQ836889 | Zhang et al. (2006) |
| Falcocladium multivesiculatum | Falcocladiales | CBS 120386 | JF831932 | JF831928 | – | Jones et al. (2014) |
| Falcocladium sphaeropedunculatum | Falcocladiales | CBS 111292 | JF831933 | JF831929 | – | Jones et al. (2014) |
| Gelasinospora tetrasperma | Sordariales | CBS 178.33 | DQ470980 | DQ471032 | DQ470932 | Spatafora et al. (2006) |
| Glomerella cingulata | Glomerellales | CBS 114054 | AF543786 | AF543762 | DQ522441 | Farr et al. (2006) |
| Gnomonia gnomon | Diaporthales | CBS 199.53 | AF408361 | DQ471019 | DQ470922 | Spatafora et al. (2006) |
| Graphium penicillioides | Microascales | CBS 506.86 | AF027384 | DQ471038 | DQ470938 | Spatafora et al. (2006) |
| Halosphaeria appendiculata | Microascales | CBS 197.60 | U46885 | U46872 | – | Zhang et al. (2006) |
| Hypocrea lutea | Hypocreales | ATCC 208838 | AF543791 | AF543768 | DQ522446 | Spatafora et al. (2007) |
| Kylindria peruamazonensis | Glomerellales | CBS 838.91 | GU180638 | GU180609 | GU180656 | Réblová et al. (2011) |
| Lasiosphaeria ovina | Sordariales | CBS958.72 | AY587946 | AY083799 | AY600286 | Miller & Huhndorf (2004) |
| Melanospora tiffanii | Melanosporales | ATCC15515 | AY015630 | AY015619 | AY015637 | Zhang & Blackwell (2002) |
| Melanospora zamiae | Melanosporales | ATCC 12340 | AY046579 | AY046578 | AY046580 | Zhang & Blackwell (2002) |
| Microascus trigonosporus | Microascales | CBS 218.31 | DQ470958 | DQ471006 | DQ470908 | Spatafora et al. (2006) |
| Monilochaetes infuscans | Glomerellales | CBS 869.96 | GU180639 | GU180620 | GU180657 | O’Connell et al. (2012) |
| Ophioceras dolichostomum | Magnaporthales | CBS 114926 | JX134689 | JX134663 | – | Luo & Zhang (2013) |
| Ophioceras leptosporum | Magnaporthales | CBS 894.70 | JX134690 | JX134664 | – | Luo & Zhang (2013) |
| Ophiocordyceps irangiensis | Hypocreales | OSC 128578 | DQ518770 | DQ522556 | DQ522445 | Spatafora et al. (2007) |
| Ophiodothella vaccinii | Phyllachorales | ATCC 36333 | – | U78777 | – | Wanderlei-Silva et al. (2003) |
| Ophiostoma piliferum | Ophiostomatales | CBS 158.74 | DQ470955 | DQ471003 | DQ470905 | Spatafora et al. (2006) |
| Ophiostoma stenoceras | Ophiostomatales | CBS 139.51 | DQ836904 | DQ836897 | DQ836891 | Zhang et al. (2006) |
| Papulosa amerospora | Cordanales | JK 5547F | DQ470950 | DQ470998 | DQ470901 | Spatafora et al. (2006) |
| Polystigma amygdalinum | Phyllachorales | EA-1 | KM111540 | KM111539 | – | Habibi et al. (2015) |
| Pseudohalonectria lignicola | Magnaporthales | M95 | JX134691 | JX134665 | – | Luo & Zhang (2013) |
| Roumegueriella rufula | Hypocreales | GJS 91-164 | EF469082 | EF469129 | EF469116 | Sung et al. (2007) |
| Sordaria fimicola | Sordariales | CBS 15.5973 | AY545728 | AY545724 | AY780194 | Zhang et al. (2006) |
| Sordaria macrospora | Sordariales | AFTOL-ID 393 | AY346301 | AY641007 | AY641074 | Huhndorf et al. (2004) |
| Sphaerodothis acrocomiae | Phyllachorales | – | – | U76340 | – | Wanderlei-Silva et al. (2003) |
| Sphaerostilbella berkeleyana | Hypocreales | CBS 102308 | U00756 | AF543770 | DQ522465 | Spatafora et al. (2007) |
| Togninia vibratilis | Togniniales | CBS 117115 | DQ649065 | – | HQ878611 | Réblová & Mostert (2007) |
| Tolypocladium capitatum | Hypocreales | OSC 71233 | AY489721 | AY489689 | DQ522421 | Spatafora et al. (2007) |
| Tolypocladium japonicum | Hypocreales | OSC 110991 | DQ518761 | DQ522547 | DQ522428 | Spatafora et al. (2007) |
| Valsa ambiens | Diaporthales | AR 3516 | AF362564 | DQ862056 | DQ862025 | Zhang et al. (2006) |
| Verticillium dahliae | Glomerellales | ATCC 16535 | DQ470945 | AY489705 | DQ522468 | Spatafora et al. (2006) |
| Xylaria acuta | Xylariales | ATCC 56487 | AY544676 | AY544719 | DQ247797 | Zhang et al. (2006) |
The second dataset is a four-locus concatenated alignment (nrSSU, ITS, RPB2, TEF1) including 51 specimens representing members of the order Phyllochorales. The alignment contained mostly sequences generated in this study plus all sequences of the Phyllachorales available from GenBank. The purpose of this analysis was to infer the phylogenetic relationships within Phyllachorales. Camarops ustulinoides and Camarops microspora (Boliniales) were used as outgroup taxa, because some of our previous analyses (unpubl. data) have shown Boliniales as the sister group of Phyllachorales. The taxa of Phyllachorales used in both analyses are listed in Table 2 together with their location, host plant, and GenBank accession numbers. The alignments were deposited in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S19724).
Table 2.
Taxa of Phyllachorales used in this study. Sequences in bold were isolated/sequenced in the present study.
| Species | Locality | Voucher | Host | Host family | GenBank Accession Numbers |
||||
|---|---|---|---|---|---|---|---|---|---|
| nrLSU | nrSSU | ITS | RPB2 | TEF1 | |||||
| Camarotella costaricensis | Panama | MM-149 | Acrocomia aculeata | Arecaceae | KX430484 | KX451863 | KX451913 | KX451954 | KX451982 |
| Panama | MM-21 | Acrocomia aculeata | Arecaceae | KX430490 | KX451851 | KX451900 | KX451963 | KX451988 | |
| Camarotella sp. | Panama | MM-27 | Unknown | Arecaceae | KX430492 | KX451852 | KX451901 | – | – |
| Coccodiella melastomatum | Venezuela | CMU78543 | Miconia sp. | Melastomataceae | – | U78543 | – | – | – |
| Coccodiella miconiae | Panama | ppMP1342 | Miconia sp. | Melastomataceae | KX430506 | KX451871 | – | – | – |
| Coccodiella miconiicola | Panama | TH571 | Ossaea micrantha | Melastomataceae | KX430512 | KX451880 | – | – | – |
| Coccodiella sp. | Ecuador | MM-165 | Unknown | Melastomataceae | KX430488 | KX451865 | KX451917 | KX451957 | KX451986 |
| Coccodiella toledoi | Venezuela | Unknown | Miconia sp. | Melastomataceae | – | U78544 | – | – | – |
| Cocoicola californica | USA | F59034 | Washingtonia robusta | Arecaceae | KX430468 | KX451866 | KX451918 | KX451958 | KX451995 |
| USA | F59038 | Washingtonia robusta | Arecaceae | KX430469 | KX451867 | KX451919 | KX451959 | KX451996 | |
| Phyllachora graminis | Unknown | Unknown | Unknown | Poaceae | – | – | AF257111 | – | – |
| Canada | DAOM240981 | Unknown | Poaceae | – | – | HQ317550 | – | – | |
| Germany | RoKi3084 | Arrhenatherum elatius | Poaceae | – | KX451872 | – | – | – | |
| Germany | MM-166 | Hordelymus europaeus | Poaceae | – | KX451869 | KX451920 | KX451962 | KX452001 | |
| Panama | TH544 | Dichanthelium viscidellum | Poaceae | KX430508 | KX451873 | – | – | – | |
| Sweden | UME31349 | Unknown | Poaceae | – | – | AF064051 | – | – | |
| Phyllachora maydis | USA | BPI893231 | Zea mays | Poaceae | – | – | KU184459 | – | – |
| Phyllachora sp. 1 | Thailand | MM-130 | Unknown | Poaceae | – | KX451883 | – | KX451949 | KX451976 |
| Phyllachora sp. 2 | Thailand | MM-128 | Bamboo | Poaceae | – | KX451859 | KX451908 | KX451964 | KX451973 |
| Phyllachora sp. 2 | Thailand | MM-129 | Bamboo | Poaceae | – | KX451860 | KX451909 | KX451948 | KX451974 |
| Phyllachora sp. 3 | Costa Rica | MM-135 | Chusquea longifolia | Poaceae | – | KX451885 | – | KX451951 | KX451978 |
| Phyllachora sp. 3 | Costa Rica | MM-78 | Chusquea sp. | Poaceae | – | KX451853 | – | KX451942 | KX451990 |
| Phyllachora sp. 3 | Costa Rica | MM-98 | Chusquea longifolia | Poaceae | KX430502 | KX451856 | – | KX451945 | KX451994 |
| Phyllachora sp. 3 | Costa Rica | MM-134 | Chusquea longifolia | Poaceae | KX430479 | KX451884 | – | KX451968 | – |
| Phyllachora sp. 3 | Ecuador | SO-07 | Chusquea sp. | Poaceae | – | KX451890 | – | – | KX452009 |
| Phyllachora sp. 4 | Benin | RMB1061 | Panicum maximum | Poaceae | – | KX451870 | KX451921 | – | KX452002 |
| Polystigma pusillum | Costa Rica | MM-113 | Andira inermis | Fabaceae | KX430474 | KX451858 | KX451907 | KX451947 | KX451972 |
| Costa Rica | MM-147 | Andira inermis | Fabaceae | – | KX451862 | – | – | KX451981 | |
| Panama | MM-19 | Andira inermis | Fabaceae | KX430489 | KX451850 | KX451899 | KX451941 | KX451987 | |
| Polystigma sp. | Ecuador | MM-163 | Paspalum sp. | Poaceae | KX430487 | KX451864 | KX451916 | – | KX451985 |
| Serenomyces phoenicis | USA | PLM314 | Phoenix canariensis | Arecaceae | – | KX451868 | KX451928 | KX451960 | KX451997 |
| USA | PLM315 | Phoenix canariensis | Arecaceae | KX430505 | KX451886 | – | KX451961 | KX451998 | |
| Telimena aequatoriensis | Ecuador | SO-05 | Monnina hirta | Polygalaceae | – | KX451889 | – | – | KX452008 |
| Telimena bicincta | Costa Rica | MM-133 | Picramnia antidesma | Picramniaceae | KX430478 | KX451861 | KX451910 | KX451950 | KX451977 |
| Costa Rica | MM-108 | Picramnia antidesma | Picramniaceae | – | KX451857 | KX451906 | KX451946 | KX451971 | |
| Telimena canafistulae | Panama | MM-13 | Cassiafistula | Fabaceae | KX430477 | KX451849 | KX451898 | – | KX451975 |
| Telimena engleri | Ecuador | MM-153 | Anthurium sp. | Araceae | – | KX451888 | KX451914 | KX451955 | KX451983 |
| Ecuador | MM-159 | Anthurium sp. | Araceae | – | – | KX451915 | KX451956 | KX451984 | |
| Panama | TH551 | Anthurium concinnatum | Araceae | KX430511 | KX451875 | KX451895 | KX451939 | KX451969 | |
| Ecuador | SO-09 | Anthurium cf. triphyllum | Araceae | – | – | KX451934 | – | KX452010 | |
| Telimena leeae | Panama | TH549 | Cissus trianae | Vitaceae | KX430509 | KX451874 | – | – | – |
| Telimena picramniae | Panama | MM-05 | Picramnia sp. | Picramniaceae | KX430470 | KX451848 | KX451896 | KX451940 | KX451970 |
| Telimena sp. 1 | Panama | MM-143 | Eugenia acapulcensis | Myrtaceae | – | KX451887 | KX451911 | KX451952 | KX451979 |
| Telimena sp. 1 | Panama | MM-144 | Eugenia acapulcensis | Myrtaceae | – | – | KX451912 | KX451953 | KX451980 |
| Telimena sp. 2 | Costa Rica | MM-92 | Eugenia sp. | Myrtaceae | KX430501 | KX451855 | KX451905 | KX451944 | KX451993 |
| Telimena sp. 3 | Costa Rica | MM-88 | Symplocos panamensis | Symplocaceae | KX430499 | KX451854 | KX451904 | KX451943 | KX451991 |
| Telimena sp. 4 | Costa Rica | MM-47 | Rinorea sp. | Violaceae | – | – | KX451902 | – | KX451989 |
| Telimena sp. 5 | Ecuador | SO-14 | Wettinia sp. | Arecaceae | – | KX451892 | KX451936 | – | – |
| Telimena sp. 5 | Ecuador | SO-21 | Wettinia sp. | Arecaceae | – | KX451893 | KX451937 | – | KX452012 |
| Telimena sp. 5 | Ecuador | SO-22 | Unknown | Arecaceae | – | KX451894 | KX451938 | – | KX452013 |
| Telimena ulei | Ecuador | SO-12 | Dioscorea meridensis | Dioscoreaceae | – | KX451891 | KX451935 | – | KX452011 |
| Panama | TH574 | Dioscorea urophylla | Dioscoreaceae | – | KX451877 | – | – | – | |
| Telimena zanthoxylicola | Panama | TH550 | Zanthoxylum scheryi | Rutiaceae | KX430510 | KX451879 | – | – | – |
PartitionFinder v. 1.1.1 (Lanfear et al. 2012) following Akaike Information Criterion (AIC) was used to select the best-fit model of evolution for each gene fragment separately for Bayesian and Maximum Likelihood (ML) analyses. Data were partitioned by gene and by codon position in the case of the protein-coding sequences. For the first dataset, a GTR+G model was applied to nrLSU, TrN+G model to nrSSU, and TIM+I+G to RPB2. For the second dataset, a TrNef+G model was applied to ITS, TrN+G model to nrSSU, SYM+G model to TEF1, and TVMef+G to RPB2.
Phylogenetic tree inference
Phylogenetic analyses for each dataset were conducted applying Maximum Likelihood (ML) and Bayesian methods. The ML analyses were performed in RAxML (Stamatakis 2014) implemented in raxmlGUI v. 0.9b2 (Silvestro & Michalak 2012). One thousand non-parametric bootstrap iterations were used with the available models of generalized time reversible (GTRGAMMA model) and a discrete gamma distribution (Stamatakis et al. 2008). Bayesian analyses were performed with the program MrBayes v. 3.2.6 (Ronquist et al. 2012) on XSEDE (Miller et al. 2010) in the CIPRES Science Gateway web portal (http://www.phylo.org/sub_sections/portal/). Two parallel runs with eight chains of Metropolis-coupled Markov chain Monte Carlo iterations were performed. Analyses were run for 100 million generations, with trees sampled every 1 000th generation. Burn-ins were determined by checking the likelihood trace plots in Tracer v. 1.6 (Rambaut et al. 2014) and subsequently discarded. Tracer and the online version of AWTY (Nylander et al. 2008) were used to test convergence; no indication of lack of convergence was detected. Bayesian posterior probabilities (BPP) ≥ 95 % and Bootstrap values (BS) ≥ 70 % were considered to be significant.
Ancestral state reconstruction of morphological and ecological characteristics
An ancestral state reconstruction of four morphological and ecological characteristics used to delimit genera in Phyllachorales was performed with the Likelihood Ancestral States method of Mesquite v. 2.74 with an asymmetrical two-parameter model for binary data and a Mk1 model for multistate data (Maddison & Maddison 2015). The likelihood decision threshold value was set to 2. The Bayesian consensus tree based on the four-locus dataset was used for this reconstruction. The analysis was restricted to members of the Phyllachorales. The characteristics considered were: monocotyledonous or eudicotyledonous host plant, position of the perithecia in the leaves (completely immersed, erumpent, subcuticular, or superficial), the presence or absence of clypeus, and the colour of the stroma (black or brightly coloured). Other characteristics such as the family of the host plant, the presence or absence of ascospore septa, ascospore colour (hyaline or brown), and the anamorphic state also were considered, but they did not yield conclusive results because they lacked variation or data were missing for numerous taxa. Observations were taken from the respective specimen and from published literature. The outgroups were coded as uncertain. The character matrix used for this analysis is provided in Appendix 1.
RESULTS
Sequences and alignments produced in this study
We generated a total of 156 sequences from 27 species of Phyllachorales: 23 sequences of nrLSU, 40 of nrSSU, 31 of ITS, 26 of RPB2, and 36 of TEF1.
Congruence among loci
For the three-locus dataset, CADM results showed no significant incongruence among loci, thus allowing concatenation of the three loci. The null hypothesis of complete incongruence among loci was rejected (W = 0.75; p < 0.0001). For the four-locus dataset, the null hypothesis of complete incongruence among loci was also rejected (W = 0.48; p < 0.0001), and the four loci were concatenated. Initially, we had planned to compile a five-locus dataset, also including nrLSU, however, the null hypothesis of complete incongruence among loci was accepted for nrLSU (W = 0.37; p > 0.05), and thus we did not consider nrLSU in the concatenated dataset.
Phyllachorales within Sordariomycetes
The separately aligned datasets for each marker consisted of 76 sequences/790 base pairs for nrLSU, 77/908 for nrSSU, and 61/939 for RPB2. The three-locus dataset consisted of 88 specimens representing 72 species in 16 orders of Sordariomycetes. The final alignment was 2 637 base pairs in length. No conflicts were detected among the phylogenies produced by ML and Bayesian analyses; therefore we present only the ML tree for this dataset (Fig. 2). Support values for nodes were consistently higher in Bayesian analyses than in ML analyses.
Fig. 2.
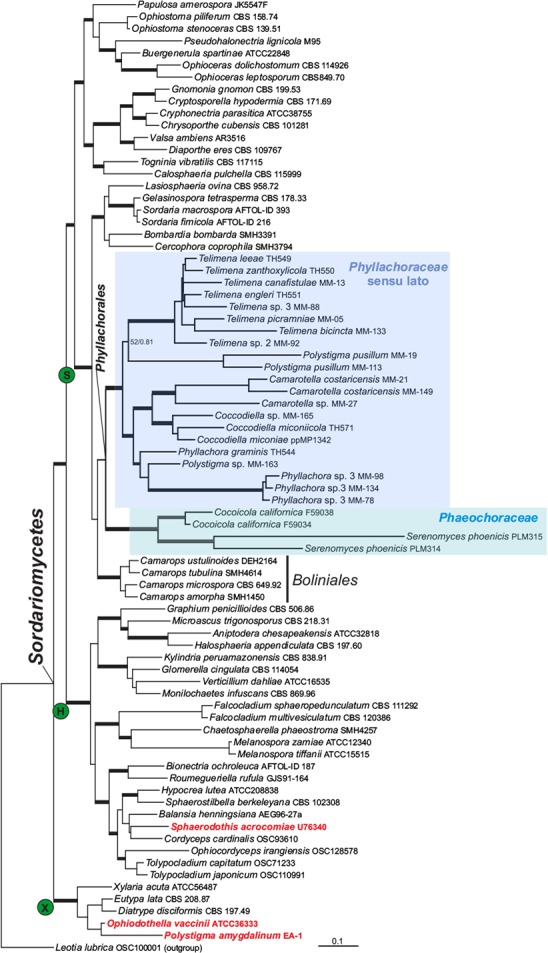
Phylogenetic position of Phyllachorales within Sordariomycetes. This is a Maximum Likelihood phylogeny based on three nuclear markers (nrLSU, nrSSU, RPB2). Support values are ML bootstrap values based on 1 000 replicates, and posterior probabilities from a Bayesian analysis. Nodes receiving ML bp > 70 %, or Bayesian PP > 0.94 are considered as strongly supported and are indicated by thickened branches; see TreeBASE files for individual support. Phyllachoraceous taxa which fall outside the order are indicated in red. Abbreviations: S = subclass Sordariomycetidae; H = subclass Hypocreomycetidae; X = subclass Xylariomycetidae.
The phylogenies inferred from individual genes (data not shown) and the three-locus phylogeny (Fig. 2) showed the Sordariomycetes as a robust monophyletic clade comprising three well-supported subclasses, Hypocreomycetidae, Xylariomycetidae, and Sordariomycetidae, with Phyllachorales grouping within Sordariomycetidae. The Phyllachorales appeared as a monophyletic, but moderately to weakly supported clade (0.94/77), including taxa of the two families Phaeochoraceae and Phyllachoraceae with the Boliniales as a sister group (0.97/74).
Three phyllachoraceous taxa fell outside the Phyllachorales clade. Polystigma amygdalinum and Ophiodothella vaccinii were located with weak support within the Xylariomycetidae. The single sequence representing Sphaerodothis acrocomiae formed a weakly supported clade together with taxa of Hypocreales (Fig. 2). This sequence may stem from a hypocrealen hyperparasite. It was not possible, however, to test possible incongruence regarding these taxa since they are represented by sequences downloaded from GenBank and two of them were represented by only one of the markers (nrSSU).
Phylogenetic relationships within the Phyllachorales
The four-locus dataset included 53 sequences representing 29 species of Phyllachorales. The dataset was supplemented with additional sequences from GenBank (two SSU and four ITS sequences). The alignment consisted of 2 728 total characters.
The topology of the tree identified by Bayesian analysis was almost identical to the one obtained by the ML analyses, therefore we present the Bayesian tree for this dataset (Fig. 3). In the Bayesian tree, the 51 sequences of Phyllachorales clustered into one major clade with high support (100/1.0). Within the Phyllachorales, three clades (I–III) can be identified.
Fig. 3.
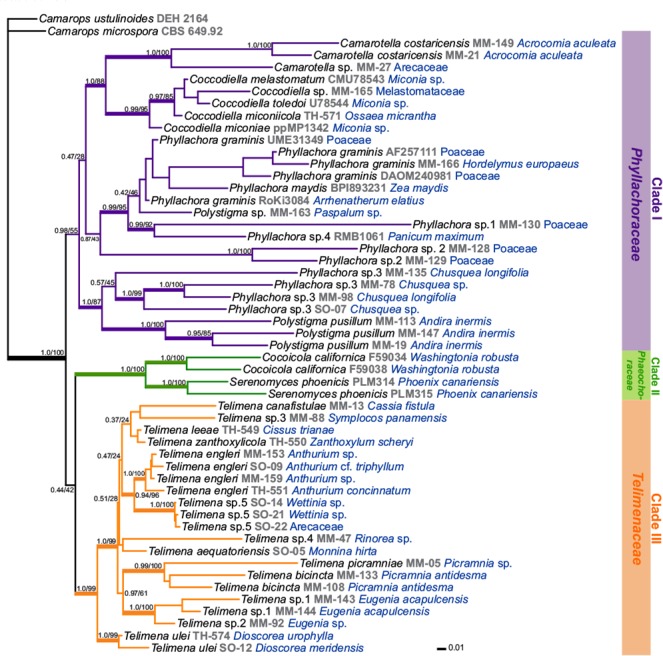
Phylogenetic relationships within the order Phyllachorales. This is a Bayesian analyses based on four nuclear markers (nrSSU, ITS, RPB2, TEF1). Support values are posterior probabilities from a Bayesian analysis and ML bootstrap values based on 1 000 replicates. Nodes receiving Bayesian PP > 0.94, or ML bp > 70 % are considered as strongly supported and are indicated by thickened branches. Hosts are indicated in blue text.
Clade I, which received weak support in the ML analysis (0.98/55), was divided into three subclades: the well-supported subclade one containing members of the genera Camarotella on Arecaceae and Coccodiella on Melastomataceae; subclade two containing the type species Phyllachora graminis, other species of Phyllachora, and one species of Polystigma, all of them growing on Poaceae; and subclade three including other graminicolous species of Phyllachora on Chusquea spp. and Polystigma pusillum on Fabaceae.
Clade II (1.0/100) is a monophyletic, strongly supported group restricted to species growing on Arecaceae, i.e., members of the genera Cocoicola and Serenomyces in the family Phaeochoraceae.
Clade III is also strongly supported (1.0/99) and included species of tar spot fungi with immersed perithecia and infecting species belonging to many different plant families, but not Poaceae. Two subclades can be distinguished, one containing members growing on several monocotyledonous and eudicotyledonous host families and a second group with species growing on Dioscoreaceae. The internal relationships within Clade III were mostly unresolved, with low posterior probability and bootstrap values. Clades II and III may be sister groups but this relationship was not strongly supported (0.44/42).
Our results indicated that the family Phyllachoraceae and the genus Phyllachora are polyphyletic being represented in two distinct clades, called Clades I and III here. The genus Polystigma is polyphyletic, with the three species treated in this study included in three groups, one in Xylariomycetidae and the other two within two different lineages within Clade I.
There were several examples of different species from the same host species, genus, or family forming supported clades, for example two different species growing on Picramnia spp. (Picramniaceae), several species on Eugenia spp. (Myrtaceae) or Chusquea spp. (Poaceae), and Coccodiella spp. on Melastomataceae.
Ancestral state reconstruction of ecological and morphological characteristics
The evolution of one ecological and three morphological characteristics was reconstructed by employing the Bayesian tree sampling of the four-locus dataset (Fig. 4a–d). The analysis of the host relationships suggested that the ancestor of Phyllachorales was growing most likely on a monocotyledonous plant (Proportional Likelihood (PL) 0.9966, Fig. 4a). The ancestor of each clade most probably was also growing on a monocotyledonous plant (Clade I, PL = 0.9981; Clade II, PL = 0.9960; Clade III, PL = 0.9685). The position of the perithecia in the leaves varies from immersed in the mesophyll to superficial (Fig. 1a–f). This reconstruction suggested that for species of Phyllachorales the ancestral state were perithecia completely immersed in the mesophyll (PL = 0.9988, Fig. 4b), while erumpent or superficial perithecia apparently evolved at least once each within Clade I. The presence of subcuticular perithecia was supported as the ancestral state in Clade II (PL = 0.9928). The ancestor of Phyllachorales was predicted to have had a clypeus (PL = 0.9989) while the lack of a clypeus was predicted as the ancestral state (PL = 0.9933) for species within Clade II (Fig. 4c). A black colour of stromata was well supported as the ancestral state (PL = 0.9999) for Phyllachorales, and bright coloured stromata apparently evolved at least twice in Clade I (Fig. 4d).
Fig. 4.
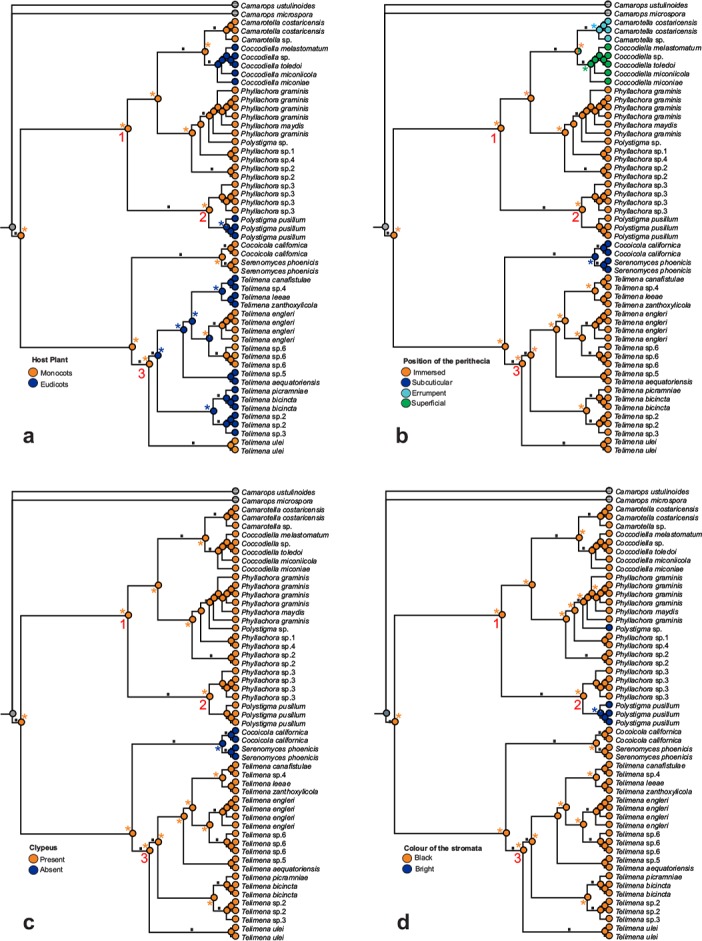
Ancestral character state reconstruction in the Phyllachorales based on the Bayesian tree. All analyses are based on maximum likelihood reconstruction with asymmetrical two-parameter (a, c, d) or Mk1 (b) models. The characteristics considered were: a. Host plant (possible states are monocotyledonous or eudicotyledonous host plant); b. position of the perithecia in the leaves (immersed, erumpent, subcuticular, or superficial); c. presence or absence of clypeus; d. colour of the stroma (black or brightly coloured). Relative likelihood probabilities for each character state are represented with a pie chart at the nodes. Squares denote supported nodes for which posterior probabilities and bootstrap values are presented in Fig. 3. Coloured asterisks near pies indicate that the corresponding state is judged best according to the threshold.
TAXONOMY
Based on the phylogenetic relationships revealed by this study, as well as the ecological and morphological characteristics of the species grouped in the observed clades within the Phyllachorales, three families were recognised: the Phaeochoraceae were accepted as previously described (Hyde et al. 1997), the Phyllachoraceae and Phyllachora need to be emended, while the Telimenaceae are newly described here. The genus Telimena is emended to accommodate established species of Phyllachora that belong to the family Telimenaceae.
Phaeochoraceae K.D. Hyde et al.
Species of Phaeochoraceae present black stromata, usually significantly raising the substratum surface, perithecia immersed, clustered forming a single cavity, embedded in pseudostromata and not covered by a clypeus as in other species of Phyllachorales. Ascospores are typically thick-walled, olivaceous to brownish, aseptate and usually with a delicate striate ornamentation. They are biotrophic or saprotrophic on palms.
For a more detailed description of this family see Hyde et al. (1997).
Genera included in this family: Cocoicola, Phaeochora, Phaeochoropsis, Serenomyces.
Phyllachoraceae Theiss. & P. Syd., Ann. Mycol. 13, 3/4: 168. 1915. emend. Mardones, Trampe & M. Piepenbr.
Stroma of various shapes, covered by a cuticular or epidermal shiny black clypeus, sometimes bright coloured, development around the ostioles of perithecia. Ascomata perithecioid, amphigenous, epiphyllous or hyphophyllous, uni- to multiloculate, sometimes confluent, frequently surrounded by a bright yellow to reddish discolouration zone, and when superficial with an hypostroma anchoring the ascomata with the host tissue. Pseudostroma sparse or absent. Perithecia superficial, erumpent or immersed in the host tissue, pyriform, globose, lenticular, or deformed by vascular bundles, with a periphysate ostiole, with a hyaline to pigmented peridium composed of textura intricata. Paraphyses hyaline, thin-walled, slightly longer than the asci, septate, often dissolving during maturation. Asci unitunicate, clavate or cylindrical, usually 8-spored, rarely 4-spored, apical ring normally not turning blue in iodine reagent (J-), sometimes with an ascus crown. Ascospores usually hyaline, rarely pale brown, globose to filiform, mostly cylindrical, thin and smooth-walled, mostly aseptate, sometimes surrounded by gel. Structures supposed to be spermogonia infrequently found, pycnidial, spermatiogenous cells cylindrical, tapering towards the tip, proliferating percurrently, producing filiform, hyaline, aseptate scolecospores that are probably spermatial in function.
Mostly biotrophic, growing mainly on members of Poaceae, but also associated with other families.
Generic type of the family. Phyllachora Nitschke ex Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 216. 1870 (1869–1870).
Phyllachora Nitschke ex Fuckel., Jahrb. Nassauischen Vereins Naturk. 23–24: 216 (1870) emend. Mardones, Trampe & M. Piepenbr.
Etymology. Name probably referring to the leaf habitat (gr. phyllas: leaf, chora: location, position).
Type species. Phyllachora graminis (Pers.) Nitschke. In Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 216. 1870 (1869–1870).
Infection spot variable in outline, often roundish, black, shiny. Clypeus mostly epidermal. Pseudostroma absent or sparse. Perithecia immersed in the host tissue. Periphyses present. Paraphyses filiform, septate, hyaline, often deliquescent. Asci cylindrical to clavate, with or without apical ring that does not stain blue in iodine, mostly 8-spored. Ascospores mostly hyaline, aseptate, smooth, mostly without gelatinous sheaths. Spermogonia acervulate or pycnidial, variable in shape, often associated with ascomata. Spermatiogenous cells cylindrical, tapering toward the apex, proliferation percurrent. Spermatia filiform, curved, hyaline.
Telimenaceae Mardones, Trampe & M. Piepenbr., fam. nov. — MycoBank MB818222
Stroma of various shapes, covered by a cuticular or epidermal shiny blackened clypeus, which may have limited development around the ostiole or extensively above the ascomata and in some cases below the ascomata. Ascomata perithecioid, amphigenous, epiphyllous or hyphophyllous, uni- to multiloculate, sometimes confluent, frequently surrounded by a bright yellow to reddish discolouration zone. Pseudostroma strongly developed, interfusing and conspicuously expanding into the host tissue. Perithecia subcuticular, epidermal, subepidermal or immersed in the host tissue, pyriform, globose, lenticular, or deformed by vascular bundles, with a periphysate ostiole, with a hyaline to pigmented peridium composed of textura intricata. Paraphyses hyaline, thin-walled, slightly longer than the asci, septate, often dissolving during maturation. Asci unitunicate, clavate or cylindrical, usually 8-spored, rarely 4-spored, apical ring normally not turning blue in iodine reagent (J-). Ascospores usually hyaline, rarely pale brown, globose to filiform, mostly cylindrical, thin and smooth-walled, aseptate to 3-septate, sometimes surrounded by gel. Spermogonia infrequently found, pycnidial, spermatiogenous cells cylindrical, tapering towards the tip, proliferating percurrently, developing filiform, hyaline, aseptate scolecospores, probably spermatial in function.
Mostly biotrophic, growing on several monocotyledoneous and dicotyledonous families, except Poaceae.
Type genus. Telimena Racib., Parasit. Alg. Pilze Java’s (Jakarta) 1: 18. 1900.
Telimena Racib., Parasit. Alg. Pilze Java’s (Jakarta) 1: 18. 1900. emend. Mardones, Trampe & M. Piepenbr.
Etymology. The name of the genus refers to the name of a Polish hero in literary works of A. Mickievicz.
Type species. Telimena erythrinae Racib., Parasit. Alg. Pilze Java’s (Jakarta) 1: 18. 1900. Java, Merapi, on Erythrina variegata L. (as E. lithosperma Miq.), s.d., Raciborski s.n. (type IMI302320!).
Infection spots dark, on living or dead leaves. Ascomata solitary to aggregated, subcuticular, epidermal, subepidermal or immersed in the host tissue, amphigenous, epiphyllous or hypophyllous, clypeate, uni- to multilocular, ostiolate. Clypeus subcuticular or epidermal. Pseudostroma strongly developed. Hamathecium with periphyses in the ostiole and evanescent paraphyses. Asci unitunicate, cylindrical to broadly ellipsoidal, with iodine-negative apical ring, 8-spored. Ascospores hyaline to pale brown when mature, globose to filiform, straight to curved, smooth, 0–3-septate.
Additional specimens examined. Telimena bicincta. Costa Rica, San Jose, on Picramnia antidesma, 10 Mar. 1890, A. Tonduz 2183 (type BR-76016-65). Telimena ecastophylli (as ‘T. caudata’). Ecuador, on Pterocarpus amazonum (as P. ulei), 24 Feb. 1938, H. Sydow s.n. (IMI307885); on Pterocarpus sp., 22 Feb. 1938, H. Sydow s.n. (IMI346458). Venezuela, Puerto La Cruz, El Limón, Pterocarpus rohrii, 22 Jan. 1928, H. Sydow s.n. (IMI18828). Telimena graminella. Philippines, Luzon, on Paspalum sp., Sept. 1913, M. Ramos in Flora of the Philippines 8224 (type IMI18829). Telimena haraeana (as ‘T. arundinariae’). Japan, Nagato Prov., Shimonoseki, on Pleioblastus simonii, 5 May 1955, Katumoto s.n. (IMI63381). Telimena rhoina (as ‘Homostegia rhoina’). USA, California, San Diego, on Rhus integrifolia, Mar. 1895, K. Brandegee No. 15 (type NY00830409).
Notes — The genus Telimena was originally described by Raciborski (1900) for phyllachora-like species with 3-septate ascospores. Currently, 14 species have been described within this genus. Its type species, T. erythrinae, is a parasite of the dicotyledonous plant host Erythrina variegata (Fabaceae). In the past, Telimena have been related with genera Telimenopsis (currently a synonym), Telimenella and Telimenochora (Petrak 1931, Müller 1975, Barr 1977). Morphological characteristics of the ascopores, i.e., shape, number of septa and position of the septa, have been used to separate these genera from each other. For a detailed description of the former and their main differences see Sivanesan (1987).
Examination of specimens of Telimena spp. (cited above, including type material) showed that stromata of Telimena spp. are similar to those of Phyllachora spp. with aseptate ascospores. Photos of sections of ascomata of Ph. graminis and T. bicincta are provided for comparison (Fig 1a, f). These sections show immersed perithecia in the mesophyll of the leaf, surrounded by pseudostroma. The stromatic development in species of Telimena seems rather variable, like in Phyllachora spp., with reduced stromatic development in species occurring in grasses and abundant pseudostroma in the remaining species. Our observations show that ascospores of Phyllachora spp. sometimes are septate when mature, and we repeatedly observed aseptate ascospores as well as ascospores with one, two or three septa in the same specimen.
The following taxa are combined into Telimena based on their phylogenetic position as shown by data presented herein. In addition, we propose a recently collected specimen of Telimena bicincta as epitype.
Telimena aequatoriensis (Theiss. & Syd.) Mardones, Trampe & M. Piepenbr., comb. nov. — MycoBank MB818223
Basionym. Phyllachora aequatoriensis Theiss. & Syd., Ann. Mycol. 13, 5/6: 521. 1915.
Synonym. Phyllachora dendritica Rehm, Hedwigia 31: 305. 1892. Ecuador, Quito, Río Machangara, on Monnina sp., 10 Apr. 1892, G.V. Lagerheim in Rehm 1072 Ex. Herb. Sydow (type S F9301).
Telimena bicincta (E. Bommer & M. Rousseau) Theiss. & Syd., Ann. Mycol. 13, 5/6: 601. 1915
Basionym. Montagnella bicincta E. Bommer & M. Rousseau, Bull. Soc. Roy. Bot. Belgique 35: 163. 1896. Costa Rica, San Jose, on Picramnia antidesma, 10 Mar. 1890, A. Tonduz 2183 (holotype BR-76016-65!).
Epitype (MycoBank MBT375061, designated here): Costa Rica, San José, San Pedro Montes de Oca, Campus Universidad de Costa Rica, N9°56′17" W84°2′59″, on Picramnia antidesma, 19 Jan. 2015, Mardones MM-133 (epitype USJ 108929; isoepitype M).
Telimena canafistulae (F. Stevens & Dalbey) Mardones, Trampe & M. Piepenbr., comb. nov. — MycoBank MB818224
Basionym. Phyllachora canafistulae F. Stevens & Dalbey, Bot. Gaz. 68: 55. 1919. Puerto Rico, Mayaguez, on Cassia fistula, 14 June 1915, F.L. Stevens 7022 (holotype ILL00011456!; isotypes BPI 636604, BPI 636617, BPI 844649!, K, MAPR, NY 00986162; fide Cannon 1991).
Synonym. Phyllachora azuanensis Petr. & Cif., Ann. Mycol. 30, 3/4: 235. 1932. Dominican Republic, Azua, on Barbieria pinnata, 25 Ago. 1929, E.L. Ekman 3565 in Herb. Ciferri (holotype BPI 636400 n.v.; isotypes NY 00986150, S F49803 n.v.).
Telimena engleri (Speg.) Mardones, Trampe & M. Piepenbr., comb. nov. — MycoBank MB818225
Basionym. Phyllachora engleri Speg., Anales Soc. Ci. Argent. 19, 2: 96. 1885. Paraguay, Barrancas de San Antonio, on Spathicarpa lanceolata, Jan. 1882, B. Balansa 3746 (holotype LPS130!).
Synonyms. Botryosphaeria anthuriicola Massee, Bull. Misc. Inform. Kew: 185. 1899. Costa Rica, Cartago, on Anthurium gracile, s.d., Donnell Smith 6813 (type K(M) 190501 n.v.; isotype BPI 797076 n.v.).
Dothidella bifrons Starbäck, Bih. Kongl. Svenska Vetensk.-Akad. Handl., Afd. 3 25, no. 1: 46. 1899. Paraguay, Concepcion, on Araceae, 17 Sept. 1893, G.A. Malme s.n. (holotype S F9201 n.v.).
Phyllachora anthurii (E. Bommer & M. Rousseau) Speg., Bol. Acad. Nac. Ci. Córdoba 23, 3-4: 567. 1919. (1918).
Dothidea anthurii E. Bommer & M. Rousseau, Bull. Soc. Roy. Bot. Belgique 35: 163. 1896.
Phyllachora dioscoreae Rehm, Hedwigia 36, 6: 370. 1897. Brazil, Brasilien, on leaves of Dioscoreaceae, s.d., E. Ule 217 in Herb. Berol. Ex Herb. Rehm (syntype S F218511 n.v.).
Phyllachora engleri f. anthurii Speg., Anales Soc. Ci. Argent. 26, 1: 37. 1888. Paraguay, on Anthurium sp., Sept. 1883, B. Balansa 4082-4106 (type LPS 130!).
Phyllachora engleri var. anthurii (Speg.) Pat., Bull. Herb. Boissier 3: 71. 1895.
Phyllachora philodendri Pat. (as ‘philodendronis’), Bull. Soc. Mycol. France 8, 3: 134. 1892. Ecuador, on Philodendron sp., Jan. 1892, Lagerheim s.n. (type FH n.v.).
Phyllachora phylloplaca Chardón, Mycologia 32, 2: 197. 1940. Brazil, Vicosa, on Diclidanthera laurifolia, 22 Apr. 1933, Muller 491 (type CUP-MG-000491 n.v.).
Additional specimens examined. Dothidea phylloplaca (as ‘phylloplacus’). Guyana, prope Cayennam, on unidentified leaves, s.d., Leprieur 1152 (type PC 96772!). Phyllachora ipirangae. Brazil, Sao Paulo, Ipiranga, Villa Marianna, on Eugenia sp., 23 Aug. 1906, A. Usteri s.n. (type S F8918!). Sphaeria phylloplaca (as ‘phylloplacus’). Surinam, on unidentified leaves, 1827, Weigelt s.n. (isotypes HBG 6597, PC 96768).
Notes — Currently, P. engleri is treated as a synonym of Phyllachora phylloplaca in Index Fungorum. Montagne (1855) published Dothidea phylloplaca and gave Sphaeria phylloplaca as synonym. Later, Saccardo (1883) cited Montagne’s description of D. phylloplaca recombining the species to P. phylloplaca. Theissen & Sydow (1915) cited Sphaeria phylloplaca and P. ipirangae, as synonyms of P. phylloplaca on Eugenia sp. (Myrtaceae), and considered as distinct from P. engleri. However, this species is currently accepted as illegitimate (superfluous name). Re-examination of the type material of D. phylloplaca and S. phylloplaca confirmed that both specimens are growing on dicotyledonous plant hosts. Furthermore, D. phylloplaca is not accepted as a synonym of S. phylloplaca as described by Montagne (1855), because leaf material differs significantly in habitus and texture.
Telimena leeae (Koord.) Mardones, Trampe & M. Piepenbr., comb. nov. — MycoBank MB818226
Basionym. Phyllachora leeae Koord., Verh. Kon. Akad. Wetensch., Afd. Natuurk., sect. 2, 13, 4: 182. 1907. Java, Gombong, on Leea rubra, 18 Mar. 1905, Kooders s.n. (holotype S F49896!).
Telimena picramniae (Syd. & P. Syd.) Mardones, Trampe & M. Piepenbr., comb. nov. — MycoBank MB818227
Basionym. Dothidella picramniae Syd. & P. Syd., Ann. Mycol. 11, 3: 266. 1913. Costa Rica, San Jose, on Picramnia bonplandiana (as P. antidesma), 10 Nov. 1912, Ad. Tonduz s.n. (isotype CUP Syd. F.exot.ex.0134 n.v.).
Synonyms. Endodothella picramniae (Syd. & P. Syd.) Syd., in Theissen & Sydow, Ann. Mycol. 13, 5/6: 590. 1915.
Phyllachora picramniae (Syd. & P. Syd.) Petr., Ann. Mycol. 38, 2/4: 259. 1940.
Phyllachora picramniae F. Stevens, Illinois Biol. Monogr. (Urbana) 11, 2: 190. 1927. Costa Rica, Aserri, on Picramnia bonplandiana (as P. antidesma), 26 June 1923, F.L. Stevens 119 (holotype ILL00005686!; isotypes BPI 639002!, CUP-014698, MICH 14837; paratypes CUP-014697, CUP-014699, ILL00005686, ILL00005687).
Telimena ulei (G. Winter) Mardones, Trampe & M. Piepenbr., comb. nov. — MycoBank MB818228
Basionym. Phyllachora ulei G. Winter, Grevillea 15, no. 75: 90. 1887. Brazil, Sao Francisco, on unknown plant, Aug. 1884, Ule 143 (holotype S F8887!).
Telimena zanthoxylicola (Seaver) Mardones, Trampe & M. Piepenbr., comb. nov. — MycoBank MB818229
Basionym. Phyllachora zanthoxylicola Seaver, Mycologia 20, 4: 225. 1928. Jamaica, on Zanthoxylum insularis, s.d., E.G. Britton 443 (holotype NY 01089448!).
Synonym. Telimenopsis fagarae Speer, Trans. Brit. Mycol. Soc. 75, 3: 504. 1981 (1980). Ecuador, Galapagos Islands, Insula Santa Cruz, on Zanthoxylum fagara, 16 Oct. 1976, Gard & For.Nobis, s.n. (holotype IMI 245878 n.v., fide Speer 1980).
DISCUSSION
Phyllachorales within Sordariomycetes
Our findings support the placement of Phyllachorales within the subclass Sordariomycetidae in the class Sordariomycetes, as suggested by previous molecular studies (Winka & Eriksson 2000, Wanderlei-Silva et al. 2003). The extended taxon sampling and the use of three markers (nrLSU, nrSSU, and RPB2) allow us to strongly corroborate these findings.
This study confirms that the Phyllachorales and Boliniales are closely related orders, disproving that the sister group of Phyllachorales may be the Diaporthales (Cannon 1988). Although the order Boliniales mostly comprises saprotrophic fungi, species of both orders have perithecia immersed in stromata and unitunicate asci with an inamyloid apical ring (Untereiner et al. 2013). Also, in some species of Boliniales, the black stromata are described as clypeate (Sivanesan 1975, Réblová 1997).
The Phyllachorales are supported as monophyletic based on the three-gene tree with moderate support. This is the first analysis that demonstrates the monophyly of the order including members of both families currently accepted in the order. The reason for the lack of a strong support for the monophyly of the order seems to be the sister group relationship among the three major clades. The three-locus dataset showed a close relationship between Clades I and III, and a separate Clade II while the four-locus dataset showed Clade II to be more closely related to Clade III. Species excluded from the Phyllachorales are Polystigma amygdalinum, Ophiodothella vaccinii, and Sphaerodothis acrocomiae. The reasons for the previous inclusion of these species in Phyllachorales were their biotrophic condition, immersed perithecia, and presence of stromatic tissue. Other molecular studies also supported these exclusions (Wanderlei-Silva et al. 2003, Habibi et al. 2015).
Phylogenetic relationships within the Phyllachorales: clade-based assessment
Our current study presents the up to now largest analysis of the Phyllachorales, with four gene regions from 29 species, yielding the most reliable phylogenetic analyses of Phyllachorales so far. Based on this analysis, three distinct monophyletic clades can be distinguished within the Phyllachorales.
Clade I includes the type species of the order, P. graminis, together with other tar spot fungi on Poaceae having immersed stromata, Po. pusillum on Fabaceae, Camarotella spp. on palms, and Coccodiella spp. on Melastomataceae. Species on Poaceae are grouped in two subclades, one containing P. graminis, P. maydis, one species of Polystigma sp., and two species on bamboo from Thailand. The other subclade contains Phyllachora spp. growing on Chusquea spp. In our study, the ML analysis provided no support for this clade. However, due to the limited taxon sampling included in this phylogeny, it seems too early to further subdivide this clade. A better-sampled phylogenetic study of these species as well as more species on Chusquea spp., and other grasses are needed to resolve the systematics of these species. Within the clade, sequences of P. graminis show a high level of variation, so apparently P. graminis is an assemblage of cryptic species. The position of Po. pusillum remains uncertain. In the three-locus dataset, sequences of Po. pusillum formed a separate clade outside Clade III without support (0.81/52). The same occurred with the analysis restricted to the nrLSU marker. This situation was the main cause of incongruent results that did not allow us to concatenate the five-locus dataset.
The other subclade in Clade I comprises Camarotella spp. with strongly erumpent and flattened stromata and Coccodiella spp. with superficial stromata. These two genera seem to be monophyletic and closely related. Hyde & Cannon (1999) suggested that the species of Oxodeora and Coccostromopsis, also with typically erumpent stromata, are probably closely related to Coccodiella and Camarotella. However, no molecular data is available so far to corroborate this relationship. These results indicate that, surprisingly, tar spot fungi on Poaceae with immersed perithecia are more closely related to Camarotella spp. and Coccodiella spp. with perithecia in erumpent stromata, than to species with rather similar immersed perithecia growing on other monocotyledonous and dicotyledonous host plants in Clade III.
Polystigma spp., which are leaf parasites with brightly coloured stromata, also are polyphyletic in Clade I (Cannon 1991). Three species of Polystigma were included in our analyses: Po. amygdalinum on Prunus dulcis, Po. pusillum on Andira inermis and Polystigma sp. on Paspalum sp. Several authors already suggested that the genus Polystigma is polymorphic, containing at least five well-defined assemblages of species (Cannon 1996, 1997, Pearce & Hyde 2006, Habibi et al. 2015). According to Cannon (1996) and his examination of the type species Polystigma rubrus on Prunus domestica, members of Polystigma should be restricted to species growing on Euro-Asiatic species of Rosaceae (Prunus spp.). However, the species of this group included in our analyses, Po. amygdalinum, was not grouped among phyllachoraceous fungi but with Trichosphaeriales and Xylariales in the Xylariomycetidae (Fig. 2), as previously reported by Habibi et al. (2015). This exclusion from Phyllachorales is morphologically supported by the presence of sympodial proliferation of conidia rather than percurrent proliferation typical in species of Phyllachorales, and also by the accumulation of starch in the stromata of Polystigma spp., which is unusual in species of Phyllachorales. The other two species included in our analyses, Po. pusillum and Polystigma sp., were both placed in Clade I but not as closely related species. Polystigma pusillum has been related to the genus Physalospora (Hyponectriaceae) mainly due to the fact that microscopic features of the two genera are largely similar (Cannon 1991). As we mentioned before, our results confirm its placement within Phyllachorales, although its phylogenetic placement within the order is still uncertain. Another Polystigma sp. on Poaceae was grouped together with species of P. graminis. These results suggest that brightly coloured stromata evolved several times in the Phyllachorales. There are several Phyllachora spp. and Stigmatula spp. with poorly developed blackened tissue, so it is possible that species with brightly coloured stromata might be species of Phyllachora with reduced melanin pigmentation (Cannon 1996).
Clade II includes species growing on Arecaceae, which are characterised by the lack of a clypeus and a more developed pseudostroma, as well as, by saccate evanescent asci and ascospores with appendages or weak striations (Hyde et al. 1997). Species of two genera were included in the analyses, Serenomyces and Cocoicola, which formed two different subclades. Serenomyces spp. can be distinguished by the presence of individual ascomata with distinct necks, while Cocoicola spp. present multi-ostiolate ascomata without necks (Hyde & Cannon 1999).
Clade III includes species of Phyllachorales growing on plants of numerous families of eudicots and monocots, except the family Poaceae. All the species belonging to this clade have immersed perithecia and hyaline ascospores, mostly without septa. Only one species, Telimena bicincta on Picramnia spp., shows 3-septate ascospores but it is closely related to other species with aseptate ascospores. The number of septa in the ascospores has been used to separate Telimena spp. from species of other genera, but our results show that the presence or absence of septa in the spores is not always systematically informative in the present context.
Taxonomic implications
According to the current classification, Clades I and III include members of the accepted family Phyllachoraceae and Clade II corresponds to the family Phaeochoraceae. The two traditionally accepted families in Phyllachorales can be distinguished morphologically by the lack of a clypeus and a more developed pseudostroma in species of Phaeochoraceae, which is in contrast to mostly immersed perithecia beneath a clypeus typical for species of Phyllachoraceae; as well as by the presence of olivaceous to brownish ascospores with striate ornamentation in Phaeochoraceae instead of smooth and hyaline ascospores typical of Phyllachoraceae.
Species of Phyllachora in the traditional sense are present in Clades I and III, so the genus needs to be redefined. Therefore, we revise the taxonomy of Phyllachora and the Phyllachoraceae to be consistent with the multi-gene phylogeny and the host relationships. The type species, P. graminis, forms part of a strongly supported clade of grass-associated species. This group is well studied (Orton 1944, Parbery 1967), morphologically homogenous, and should be considered Phyllachora s.str. The family Phyllachoraceae s.str. is emended and includes Phyllachora spp. that possess immersed perithecia and occur on Poaceae, and the erumpent perithecia genera Camarotella and Coccodiella. The suggested placement of Po. pusillum within Phyllachoraceae could not be confirmed.
Further Phyllachora species on other monocotyledonous and eudicotyledonous hosts form the strongly supported Clade III. The emendation of the family Phyllachoraceae and the genus Phyllachora require the recognition of a new family and at least one genus to accommodate the species clustering in Clade III. We propose to transfer these species to the genus Telimena (Raciborski 1900), based on the phylogenetic position of T. bicincta, which is the oldest name available for species included in Clade III, and based on the type species of Telimena, T. erythrinae, which does not occur on a species of Poaceae but on Erythrina variegata (Fabaceae). Telimena was described for species with Phyllachora type stromata and 3-septate ascospores, instead of aseptate ascospores in typical Phyllachora spp. As mentioned above, molecular results indicate that the number of septa of the ascospore is not a reliable character to delimit genera in the present context; therefore we include species with septate as well as aseptate ascospores in the same genus. The examination of the type specimen of T. erythrinae and specimens of other Telimena spp. showed that apart from the septation of the ascospores, stromatic characteristics of Telimena spp. are similar to those of species of Phyllachora not growing on Poaceae, as has been pointed out by several authors (Raciborski 1900, Von Höhnel 1911, Müller 1975, Barr 1977, Sivanesan 1987). The fact that Telimena comprises species with ascomata similar to those of Phyllachora spp. means that the generic concept of Telimena can be easily emended to include species considered Phyllachora spp. up to now. We assume that further molecular sequence data of the remaining Phyllachora spp. not occurring in Poaceae will not belong to Phyllachora s.str. but to Clade III due to their eudicotylenous host. We propose the new family Telimenaceae for taxa belonging to Clade III.
Species that are proposed here as new combinations into the genus Telimena correspond to species collected and sequenced by us. All of them have been compared with the corresponding type specimen and exhibited the same morphological characteristics. As no molecular sequence data could be obtained from the type material and most sequenced specimens were not collected at type localities, we mostly refrained from epitypification following recommendations by Hyde & Zhang (2008) as well as Zhang et al. (2013). Therefore, we decided to designate an epitype only for T. bicincta, which was obtained from the same location and host species as the type of this species.
Our findings strongly support the separation of Clades I and III from Clade II based on morphological, molecular, and host data, but it is difficult to identify morphological synapomorphies to separate Clade I from Clade III. Nevertheless, a careful re-examination of morphological characteristics of species classified in the families Phyllachoraceae s.str. (Clade I) and Telimenaceae (Clade III) revealed characteristics that allow distinguishing species of the two families morphologically: Stromata of tar spot fungi classified in the new family Telimenaceae are located either in subcuticular, epidermal, subepidermal, or immersed position, whereas stromata of species in Phyllachoraceae are either completely immersed in host tissue in the case of Phyllachora spp. on grasses or erumpent to superficial in species of the other genera. In Phyllachora spp. on grasses examined by Parbery & Langdon (1964) the pseudostroma was absent; they did not document any stromatic tissue apart from the clypeus. On the contrary, in Telimena spp. on dicotyledonous plants, pseudostroma often is strongly developed, interfuses between adjacent stromata, and conspicuously expands into the host tissue.
Evolution of parasite-host relationship
Members of the order Phyllachorales mainly infect monocotyledonous or dicotyledonous plant hosts. Our analyses show that the ancestor of Phyllachorales may have grown on monocotyledonous hosts. All three families recognized in this study include monocotyledonous plants as hosts. These results strongly suggest intimate relationships between Phyllachorales and monocotyledonous plants in the early evolution of the order. In general, long-term evolutionary dynamics or coevolution between hosts and their symbionts (parasitic or mutualistic relationships) operates in parallel by co-speciation or through speciation by host shifts. Co-speciation usually involves the speciation of a symbiont at the same time as another species, while host-shift speciation can occur when the symbiont moves to a new host on which the symbiont’s immediate ancestor did not occur, and gives rise to new host-symbiont combinations (De Vienne et al. 2013).
Based on the host expansion strategy, two groups can be distinguished in the Phyllachorales, the family Phaeochoraceae which apparently did not expand its host range outside Arecaceae, and the families Telimenaceae and Phyllachoraceae, which expanded their host ranges to rather distant hosts. The facts that the phylogeny of Phyllachorales is not consistent with the phylogeny of their host plants, that a high number of distant host families are infected by phyllachoraceous fungi, and that several terminal groups of Phyllachorales species concentrate on hosts belonging to the same family, suggest that several host-shift speciation events followed by co-speciation might explain host-parasite patterns in Phyllachoraceae and Telimenaceae. We speculate that phyllachoraceous fungi first infected monocots, radiated on monocotyledonous hosts, and later expanded their host range to other dicotyledonous plant families by host jumps. For instance, in Clade I (Phyllachoraceae), Phyllachora species are restricted to grasses but the family also includes the genera Camarotella and Coccodiella with species growing on other hosts. In the genus Coccodiella, C. arundinariae (not included in our analyses) is the only species which occurs on the monocotyledonous family Poaceae, specifically on bamboos in Far East Asia, but most of the species of the genus occur on the dicotyledonous family Melastomataceae. We hypothesize that the ancestor of Coccodiella on Poaceae probably infected a melastomataceous plant and expanded its host range within this family.
Evolution of morphological characteristics
The position of the perithecia varies from completely immersed in the mesophyll of the leaf as in Phyllachora s.lat. spp. to completely superficial as in Coccodiella spp. (Fig. 1a–f). Several other genera have been erected based on this characteristic, i.e., Camarotella spp. with erumpent perithecia, Trabutia spp. with subcuticular perithecia or Catacauma spp. with perithecia inserted between the clypeus and the epidermis. However, the location of the perithecia in the leaf has been suggested to be greatly influenced by the consistency of the host tissue (Parbery & Langdon 1964, Cannon 1991), and therefore not very reliable as a taxonomical criterion. The reconstruction presented here suggests that the ancestral state for Phyllachorales was completely immersed perithecia, which apparently was lost in the family Phaeochoraceae and evolved to erumpent or superficial perithecia in some members of Phyllachoraceae. In the genus Telimena the position of perithecia varies from immersed to subepidermal. Based on our results, when the perithecia are superficial or erumpent, as in the genera Coccodiella and Camarotella, this characteristic is reliable to define genera, while subepidermal and subcuticular positions of perithecia might be dependent on the texture and anatomy of the host tissue.
Our data also indicate that phyllachoraceous fungi growing on palms form a distinctive group, probably due to the very particular anatomical characteristics of palms. In these fungi, the expansion of the stromata and the shape of the perithecia are affected by leaves with closely spaced, strongly lignified, parallel vascular bundles, typical of Arecaceae. Our phylogeny shows that palm-inhabiting species have evolved independently at least three times within Phyllachorales, as Arecaceae is the only host family occurring in all three fungal families. According to Hyde & Cannon (1999), there are three different types of stromata in phyllachoraceous fungi on palms, one elongated and erumpent as in Camarotella spp. (Clade I, Phyllachoraceae), another one inserted between the outermost layers of the host tissue, as in family Phaeochoraceae (Clade II), and a few species having immersed perithecia, as in Telimenaceae (Clade III), which are confined to species of palms with less lignified leaf tissue. Our results do not confirm that the ancestor of Phyllachorales was growing on a palm. Therefore, more research is needed to elucidate the role of palm-inhabiting species in the evolutionary history of Phyllachorales.
Most species within Phyllachorales produce black stromata caused by dense fungal cells with melanin deposits, the only exception being Polystigma spp. Our analyses suggest that the presence of black stromata is the ancestral state in Phyllachorales, and that brightly coloured stromata evolved at least twice in the order.
The clypeus is a shield of black fungal cells located above the perithecia, which is restricted to the epidermal cells of the host and is thought primarily to protect the developing tissues from damage caused by UV radiation or to absorb heat from the sun promoting growth (Durrell & Shields 1960, Sherwood 1981). Species of Phyllachorales share the presence of a clypeus as a synapomorphy, which apparently was lost only once in the Phaeochoraceae, thus, the clypeus is thought to represent an evolutionarily stable characteristic in the order.
Conclusions and future work
This study demonstrates the monophyly of Phyllachorales and its placement in Sordariomycetidae with Boliniales as a sister group. Although several genera, which possibly do not belong to Phyllachorales, were not represented in our dataset, it is clear that there is a core group representing this order. The phylogenetic relationships within the order are partially elucidated. The placement of Phaeochoraceae within Phyllachorales is confirmed, however, more sampling in the three families is necessary to better assess their internal relationships. Our data also supports the split of the family Phyllachoraceae and the genus Phyllachora, and the establishment of the additional family Telimenaceae and the genus Telimena. With monophyly demonstrated, efforts should be made to find characteristics that help to distinguish between families. In this study, apart from the molecular and ecological distinctions, few morphological synapomorphies were detected to differentiate the families Telimenaceae and Phyllachoraceae. Potentially informative characteristics that should be evaluated are the ascus wall and the morphology of the ascus apex. Additional morphological and ultrastructural work by electron microscopy will contribute to our understanding of the asci. Other valuable characteristics might be the asexual/spermatogonial states of Phyllachorales and the morphology of the haustoria. Also, additional molecular markers are necessary for a profound phylogenetic study of some specific clades. Although our study contains a comprehensive dataset, it is still not possible to clearly circumscribe the family Phyllachoraceae and to elucidate the monophyly of the genera within Phyllachorales. DNA should be generated for species of Phyllachora on grasses and for several other genera that have been erected historically based on single characteristics of ascospores, i.e., Apiosphaeria, Ophiodothella, Sphaerodothis, Stigmochora, Telimenochora. Obtaining fresh specimens of these fungi to place them within a molecular framework should be an important objective of future studies, to prove the validity of the characteristics used to delimit them. Further molecular analyses including sequences of more species, from new locations and host plants, will contribute to an understanding of the evolution of host ranges. We predict that this re-evaluation will produce the reassessment of several genera.
The results of ancestral state reconstruction analyses rely on the phylogeny of the present analysis that includes only a small fraction of species known for the corresponding systematic relationships. Therefore, the ancestral state reconstructions are not certain and the true number of state changes is probably underestimated. A broader sampling will probably reveal further state changes specially regarding subsequent host jumps and also concerning the position of the perithecia in the leaves in species belonging to Phyllachoraceae and Telimenaceae.
Acknowledgments
We thank Orlando Cáceres, Julieta Carranza, Darío Cruz, Tina Hofmann, and Carlos Rojas for their help during the fieldwork, José Macia-Vicente for fruitful discussion on an early state of the manuscript on molecular phylogeny, Ralph Mangelsdorff for valuable advice on nomenclature and Federico Albertazzi (CIBCM, UCR) for facilities for DNA extraction in Costa Rica. Special thanks to Ralph Mangelsdorff, Carlos Morales, Rafael Rincón, and Héctor Zumbado for their help with the identification of host plants. We would like to thank Roland Kirschner, Tina Hofmann, and Ralph Mangelsdorff for providing samples for this work. We also thank the Autoridad Nacional del Ambiente (ANAM) in Panama, MINAE (SINAC and CONAGEBIO) in Costa Rica, and Ministerio de Ambiente in Ecuador for collecting and export permits. We thank the curators of the Herbaria BPI, ILL, IMI, LPS, M, PC, S, and USJ for the loan of specimens. We thank the Deutsche Forschungsgemeinschaft (DFG) for financial support of the project Plant Parasitic Microfungi of Panama (ppMP). Melissa Mardones received an ALECOSTA PhD scholarship from DAAD and OAICE (University of Costa Rica).
Appendix 1.
Character matrix used for ancestral state reconstruction analyses of members of Phyllachorales.
| Species | Voucher | Hosta | Perithecial positionb | Clypeusc | Stromata colourd | Morphological observations based on |
|---|---|---|---|---|---|---|
| Camarotella costaricensis | MM-149 | 0 | 1 | 0 | 0 | specimen |
| MM-21 | 0 | 1 | 0 | 0 | specimen | |
| Camarotella sp. | MM-27 | 0 | 1 | 0 | 0 | specimen |
| Coccodiella melastomatum | CMU78543 | 1 | 2 | 0 | 0 | Léveillé 1845 (as Sphaeria melastomatum) |
| Coccodiella miconiae | ppMP1342 | 1 | 2 | 0 | 0 | specimen |
| Coccodiella miconiicola | TH571 | 1 | 2 | 0 | 0 | specimen |
| Coccodiella sp. | MM-165 | 1 | 2 | 0 | 0 | specimen |
| Coccodiella toledoi | Unknown | 1 | 2 | 0 | 0 | Chardón & Toro 1934 (as Bagnisiopsis toledoi) |
| Cocoicola californica | F59034 | 0 | 3 | 1 | 0 | specimen |
| F59038 | 0 | 3 | 1 | 0 | specimen | |
| Phyllachora graminis | AF257111 | 0 | 0 | 0 | 0 | Parbery 1967 |
| DAOM240981 | 0 | 0 | 0 | 0 | Parbery 1967 | |
| RoKi3084 | 0 | 0 | 0 | 0 | specimen | |
| MM-166 | 0 | 0 | 0 | 0 | specimen | |
| UME31349 | 0 | 0 | 0 | 0 | Parbery 1967 | |
| Phyllachora maydis | BPI893231 | 0 | 0 | 0 | 0 | Parbery 1967 |
| Phyllachora sp. 1 | MM-130 | 0 | 0 | 0 | 0 | specimen |
| Phyllachora sp. 2 | MM-128 | 0 | 0 | 0 | 0 | specimen |
| Phyllachora sp. 2 | MM-129 | 0 | 0 | 0 | 0 | specimen |
| Phyllachora sp. 3 | MM-135 | 0 | 0 | 0 | 0 | specimen |
| Phyllachora sp. 3 | MM-78 | 0 | 0 | 0 | 0 | specimen |
| Phyllachora sp. 3 | MM-98 | 0 | 0 | 0 | 0 | specimen |
| Phyllachora sp. 3 | SO-07 | 0 | 0 | 0 | 0 | specimen |
| Phyllachora sp. 4 | RMB1061 | 0 | 0 | 0 | 0 | specimen |
| Polystigma pusillum | MM-113 | 1 | 0 | 0 | 1 | specimen |
| MM-147 | 1 | 0 | 0 | 1 | specimen | |
| MM-19 | 1 | 0 | 0 | 1 | specimen | |
| Polystigma sp. | MM-163 | 0 | 0 | 0 | 1 | specimen |
| Serenomyces phoenicis | PLM314 | 0 | 3 | 1 | 0 | specimen |
| PLM315 | 0 | 3 | 1 | 0 | specimen | |
| Telimena aequatoriensis | SO-05 | 1 | 0 | 0 | 0 | specimen |
| Telimena bicincta | MM-133 | 1 | 0 | 0 | 0 | specimen |
| MM-108 | 1 | 0 | 0 | 0 | specimen | |
| Telimena canafistulae | MM-13 | 1 | 0 | 0 | 0 | specimen |
| Telimena engleri | MM-153 | 0 | 0 | 0 | 0 | specimen |
| MM-159 | 0 | 0 | 0 | 0 | specimen | |
| TH551 | 0 | 0 | 0 | 0 | specimen | |
| SO-09 | 0 | 0 | 0 | 0 | specimen | |
| Telimena leeae | TH549 | 1 | 0 | 0 | 0 | specimen |
| Telimena picramniae | MM-05 | 1 | 0 | 0 | 0 | specimen |
| Telimena sp. 1 | MM-57 | 1 | 0 | 0 | 0 | specimen |
| Telimena sp. 2 | MM-143 | 1 | 0 | 0 | 0 | specimen |
| Telimena sp. 2 | MM-144 | 1 | 0 | 0 | 0 | specimen |
| Telimena sp. 3 | MM-92 | 1 | 0 | 0 | 0 | specimen |
| Telimena sp. 4 | MM-88 | 1 | 0 | 0 | 0 | specimen |
| Telimena sp. 5 | MM-47 | 1 | 0 | 0 | 0 | specimen |
| Telimena sp. 6 | SO-14 | 0 | 0 | 0 | 0 | specimen |
| Telimena sp. 6 | SO-21 | 0 | 0 | 0 | 0 | specimen |
| Telimena sp. 6 | SO-22 | 0 | 0 | 0 | 0 | specimen |
| Telimena ulei | SO-12 | 0 | 0 | 0 | 0 | specimen |
| TH574 | 0 | 0 | 0 | 0 | specimen | |
| Telimena zanthoxylicola | TH550 | 1 | 0 | 0 | 0 | specimen |
a 0 = monocotyledonous host plant; 1 = dicotyledonous host plant.
b 0 = immersed in the mesophyl of the leaf; 1 = errumpent; 2 = superficial; 3 = subcuticular.
c 0 = present; 1 = absent.
d 0 = black; 1 = bright coloured.
REFERENCES
- Barr M. 1977. Magnaporthe, Telimenella, and Hyponectria (Physosporellaceae). Mycologia 69: 952–966. [Google Scholar]
- Barr M. 1983. The ascomycete connection. Mycologia 75: 1–13. [Google Scholar]
- Campbell V, Legendre P, Lapointe F. 2011. The performance of the Congruence Among Distance Matrices (CADM) test in phylogenetic analysis. BMC Evolutionary Biology 11: 64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon PF. 1988. Proposal to merge the Phyllachorales with the Diaporthales, with a new family structure. Systema Ascomycetum 7: 23–43. [Google Scholar]
- Cannon PF. 1991. Revision of Phyllachora and some similar genera on the host family Leguminosae. Mycological Papers 163: 1–302. [Google Scholar]
- Cannon PF. 1996. Systematics and diversity of the Phyllachoraceae associated with Rosaceae, with a monograph of Polystigma. Mycological Research 100: 1409–1427. [Google Scholar]
- Cannon PF. 1997. Diversity of the Phyllachoraceae with special reference to the tropics. In: Hyde K. (ed), Biodiversity of Tropical Microfungi: 255–278. Hong Kong University Press, Hong Kong. [Google Scholar]
- Cannon PF, Evans HC. 1999. Biotrophic species of Phyllachoraceae associated with the angiosperm family Erythroxylaceae. Mycological Research 103: 577–590. [Google Scholar]
- Carbone I, Kohn L. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Chardón CE, Toro RA. 1934. Mycological explorations of Venezuela. Monograph Univ Puerto Rico, Series B 2: 1–353. [Google Scholar]
- Clements F, Shear C. 1931. The genera of fungi. 2nd ed Wilson Co., New York. [Google Scholar]
- De Vienne DM, Refregier G, Lopez-Villavicencio M, et al. 2013. Cospeciation vs host-shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytologist 198: 347–385. [DOI] [PubMed] [Google Scholar]
- Durrell ALW, Shields LM. 1960. Fungi isolated in culture from soils of the Nevada Test Site. Mycologia 52: 636–641. [Google Scholar]
- Elliott ML, Des Jardin EA. 2014. Serenomyces associated with palms in southeastern USA: isolation, culture storage and genetic variation. Mycologia 106: 698–707. [DOI] [PubMed] [Google Scholar]
- Eriksson OE. 1967. On graminicolous pyrenomycetes from Fennoscandia 3. Amerosporous and didymosporous species. Archiv für Botanik ser. 2: 441–466. [Google Scholar]
- Eriksson OE. 1982. Outline of the ascomycetes - 1982. Mycotaxon 15: 203–248. [Google Scholar]
- Farr DF, Aime MC, Rossman AY, et al. 2006. Species of Colletotrichum on Agavaceae. Mycological Research 110: 1395–1408. [DOI] [PubMed] [Google Scholar]
- Fuckel L. 1870. Symbolae mycologicae. Beiträge zur Kenntniss der Rheinischen Pilze. Jahrbücher des Nassauischen Vereins für Naturkunde 23–24: 1–459. [Google Scholar]
- Habibi A, Banihashemi Z, Mostowfizadeh-Ghalamfarsa R. 2015. Phylogenetic analysis of Polystigma and its relationship to Phyllachorales. Phytopathologia Mediterranea 54: 45–54. [Google Scholar]
- Hawksworth D, Sutton B, Ainsworth G. 1983. Ainsworth & Bisby’s dictionary of the fungi. 7th ed CMI Kew, Surrey, UK. [Google Scholar]
- Huhndorf SM, Miller AN, Fernàndez FA. 2004. Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96: 368–387. [PubMed] [Google Scholar]
- Hyde KD, Cannon PF. 1999. Fungi causing tar spots on palmae. Mycological Papers 175: 1–114. [Google Scholar]
- Hyde KD, Cannon PF, Barr ME. 1997. Phaeochoraceae, a new ascomycete family from palms. Systema Ascomycetum 15: 117–120. [Google Scholar]
- Hyde KD, Zhang Y. 2008. Epitypification: Should we epitypify? Journal of Zhejiang University Science B 9: 842–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderbitzin P, Lim SR, Volkmann-Kohlmeyer B, et al. 2004. The phylogenetic position of Spathulospora based on DNA sequences from dried herbarium material. Mycological Research 108: 737–748. [DOI] [PubMed] [Google Scholar]
- Jones EBG, Suetrong S, Cheng WH, et al. 2014. An additional fungal lineage in the Hypocreomycetidae (Falcocladium species) and the taxonomic re-evaluation of Chaetosphaeria chaetosa and Swampomyces species, based on morphology, ecology and phylogeny. Cryptogamie, Mycologie 35: 119–138. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, et al. 2008. Dictionary of fungi. 10 ed CAB International, UK. [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, et al. 2012. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Legendre P, Lapointe F. 2004. Assessing congruence among distance matrices: single-malt scotch whiskies revisited. Australian & New Zealand Journal of Statistics 46: 615–629. [Google Scholar]
- Léveillé JH. 1845. Champignons exotiques. Annales des Sciences Naturelles Botanique 3: 38–71. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among Ascomycetes: Evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Luo J, Zhang N. 2013. Magnaporthiopsis, a new genus in Magnaporthaceae (Ascomycota). Mycologia 105: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Luttrell E. 1951. Taxonomy of the pyrenomycetes. University of Missouri Studies 24: 1–120. [Google Scholar]
- Maddison W, Maddison D. 2015. Mesquite: a modular system for evolutionary analysis. Version 3.04. http://mesquiteproject.org.
- Maharachchikumbura SSN, Hyde KD, Jones EBG, et al. 2015. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity 72: 199–301. [Google Scholar]
- Miller AN, Huhndorf SM. 2004. Using phylogenetic species recognition to delimit species boundaries within Lasiosphaeria. Mycologia 96: 1106–1127. [PubMed] [Google Scholar]
- Miller AN, Huhndorf SM. 2005. Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Molecular Phylogenetics and Evolution 35: 60–75. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), IEEE: 1–8. [Google Scholar]
- Montagne JPFC. 1855. Cryptogamia Guyanensis seu plantarum cellularium in Guyana gallica annis 1835–49 a cl. Leprieur collectarum enumeratio universalis. Pyrenomycetes. Annales des Sciences Naturelles Botanique, Serie 4, 3: 98–135. [Google Scholar]
- Müller E. 1975. Über die Gattung Telimena Raciborski (Ascomycetes). Sydowia 27: 74–77. [Google Scholar]
- Müller E, Von Arx J. 1962. Die Gattungen der didymosporen Pyrenomyceten. Beiträge zur Kryptogamenflora der Schweiz 11: 1–922. [Google Scholar]
- Müller E, Von Arx J. 1973. Pyrenomycetes: Meliolales, Coronophorales, Sphaeriales. In: Ainsworth G, Sparrow F, Sussman A. (eds), The fungi: an advanced treatise 4A: 87–132. Academic Press, New York. [Google Scholar]
- Nannfeldt J. 1932. Studien über die Morphologie und Systematik der nichtlichenisierten inoperculaten Discomyceten. Nova Acta Regiae Societatis Scientiarum Upsaliensis 8: 1–368. [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, et al. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583. [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Thon MR, Hacquard S, et al. 2012. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics 44: 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K. 1993. Fusarium and its near relatives. In: Reynolds D, Taylor J. (eds), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics: 225–233. CAB International, London. [Google Scholar]
- Orton CR. 1944. Graminicolous species of Phyllachora in North America. Mycologia 36: 18–53. [Google Scholar]
- Parbery DG. 1967. Studies on graminicolous species of Phyllachora Nke in Fckl. V. A taxonomic monograph. Australian Journal of Botany 15: 271–375. [Google Scholar]
- Parbery DG. 1978. Phyllachora, Linochora and hyperparasites. Taxonomy of Fungi 1: 263–277. [Google Scholar]
- Parbery DG. 1996. Spermatial states of fungi are andromorphs. Mycological Research 100: 1400. [Google Scholar]
- Parbery DG, Langdon RFN. 1963. Studies on graminicolous species of Phyllachora FCKL. III. The relationship of certain scolecospores to species of Phyllachora. Australian Journal of Botany 11: 141–151. [Google Scholar]
- Parbery DG, Langdon RFN. 1964. Studies on graminicolous species of Phyllachora FCKL. IV. Evaluation of the criteria of species. Australian Journal of Botany 12: 265–281. [Google Scholar]
- Pearce CA, Hyde KD. 2006. Phyllachoraceae of Australia. Fungal Diversity Press, Hong Kong. [Google Scholar]
- Pearce CA, Reddell P, Hyde KD. 1999. A revision of Phyllachora (Ascomycotina) on hosts in the angiosperm family Asclepiadaceae, including P. gloriana sp. nov. on Tylophora benthamii from Australia. Fungal Diversity 3: 123–138. [Google Scholar]
- Pearce CA, Reddell P, Hyde KD. 2001. Revision of the Phyllachoraceae (Ascomycota) on hosts in the angiosperm family Proteaceae. Australian Systematic Botany 14: 283–328. [Google Scholar]
- Petrak F. 1931. Mykologische Notizen IX. Annales Mycologici 29: 339–397. [Google Scholar]
- Piepenbring M, Hofmann T, Kirschner R, et al. 2011. Diversity patterns of neotropical plant parasitic microfungi. Ecotropica 17: 27–40. [Google Scholar]
- Raciborski M. 1900. Parasitische Algen und Pilze Java’s 1: 1–39. [Google Scholar]
- Rambaut A, Suchard M, Xie D, et al. 2014. Tracer v1.6. Available from http://beast.bio.ed.ac.uk/Tracer.
- Réblová M. 1997. Fungal diversity in the Czech Republic. New species of Apiorhynchostoma, Capronia, Ceratosphaeria and Lasiosphaeria. Sydowia 50: 229–251. [Google Scholar]
- Réblová M, Gams W, Seifert K. 2011. Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Studies in Mycology 68: 163–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Mostert L. 2007. Romellia is congeneric with Togninia, and description of Conidiotheca gen. nov. for one species of this genus with polysporous asci. Mycological Research 111: 299–307. [DOI] [PubMed] [Google Scholar]
- Réblová M, Seifert KA. 2004. Cryptadelphia (Trichosphaeriales), a new genus for holomorphs with Brachysporium anamorphs and clarification of the taxonomic status of Wallrothiella. Mycologia 96: 343–367. [DOI] [PubMed] [Google Scholar]
- Rehner S, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo PA. 1876. Fungi Veneti novi vel critici. Series V. Nuovo Giornale Botanico Italiano 8: 162–211. [Google Scholar]
- Saccardo PA. 1883. Sylloge Fungorum 2. Padua, Italy. [Google Scholar]
- Sherwood MA. 1981. Convergent evolution in discomycetes from bark and wood. Botanical Journal of the Linnean Society 82: 15–34. [Google Scholar]
- Silvestro D, Michalak I. 2012. RaxmlGUI: A graphical front-end for RAxML. Organisms Diversity and Evolution 12: 335–337. [Google Scholar]
- Sivanesan A. 1975. New ascomycetes and some revisions. Transactions of the British Mycological Society 65: 19–27. [Google Scholar]
- Sivanesan A. 1987. Telimena, Telimenopsis, and a new ascomycete genus Telimenochora of the Phyllachorales. Transactions of the British Mycological Society 88: 473–477. [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, et al. 2006. A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Sung JM, et al. 2007. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Molecular Ecology 16: 1701–1711. [DOI] [PubMed] [Google Scholar]
- Speer EO. 1980. Telimenopsis fagarae sp. nov., a new parasitic fungus from the Galapagos Islands. Transactions of the British Mycological Society 75: 504–506. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Summerell B, Groenewald JZ, Carnegie AJ, et al. 2006. Eucalyptus microfungi known from culture. 2. Alysidiella, Fusculina and Phlogicylindrium genera nova, with notes on some other poorly known taxa. Fungal Diversity 23: 323–350. [Google Scholar]
- Sung GH, Hywel-Jones NL, Sung JM, et al. 2007. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57: 5–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology 56: 564–577. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen F, Sydow H. 1915. Die Dothideales. Kritisch-systematische Originaluntersuchungen. Annales Mycologici 13, 3–4: 147–746. [Google Scholar]
- Thongkantha S, Jeewon R, Vijaykrishna D, et al. 2009. Molecular phylogeny of Magnaporthaceae (Sordariomycetes) with a new species, Ophioceras chiangdaoense from Dracaena loureiroi in Thailand. Fungal Diversity 34: 157–173. [Google Scholar]
- Trampe T. 2010. Neotropical tar spot fungi: exploration of Phyllachorales in Panama. PhD thesis, Biowissenschaften, Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany. [Google Scholar]
- Untereiner WA, Bogale M, Carter A, et al. 2013. Molecular phylogeny of Boliniales (Sordariomycetes) with an assessment of the systematics of Apiorhynchostoma, Endoxyla and Pseudovalsaria. Mycologia 105: 564–588. [DOI] [PubMed] [Google Scholar]
- Von Arx JA, Müller E. 1954. Die Gattungen der amerosporen Pyrenomyceten. Beiträge zur Kryptogamenflora der Schweiz 11: 1–434. [Google Scholar]
- Von Höhnel F. 1910. Fragmente zur Mykologie: XII. Mitteilung (Nr. 574 bis 641). Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften in Wien Mathematisch-naturwissenschaftliche Klasse, Abt. 1, 119: 877–958. [Google Scholar]
- Von Höhnel F. 1911. Fragmente zur Mykologie, Nr. 709: Über Telimena erythrinae Rac. Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften in Wien Mathematisch-naturwissenschaftliche Klasse 1, Abt. 120: 453–454. [Google Scholar]
- Wanderlei-Silva D, Ramalho Neto E, Hanlin R. 2003. Molecular systematics of the Phyllachorales (Ascomycota, Fungi) based on 18S ribosomal DNA sequences. Brazilian Archives of Biology and Technology 46: 315–322. [Google Scholar]
- White T, Bruns S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, New York. [Google Scholar]
- Winka K, Eriksson OE. 2000. Papulosa amerospora accommodated in a new family (Papulosaceae, Sordariomycetes, Ascomycota) inferred from morphological and molecular data. Mycoscience 41: 97–103. [Google Scholar]
- Zhang K, Zhang N, Cai L. 2013. Typification and phylogenetic study of Phyllosticta ampelicida and P. vaccinii. Mycologia 105: 1030–1042. [DOI] [PubMed] [Google Scholar]
- Zhang N, Blackwell M. 2002. Molecular phylogeny of Melanospora and similar pyrenomycetous fungi. Mycological Research 106: 148–155. [Google Scholar]
- Zhang N, Castlebury LA, Miller AN, et al. 2006. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98: 1076–1087. [DOI] [PubMed] [Google Scholar]


