Abstract
Triatomines are hematophagous arthropod vectors of Trypanosoma cruzi, the causative agent of Chagas Disease. Panstrongylus lignarius, also known as Panstrongylus herreri, is considered one of the most versatile triatomines because it can parasitize different hosts, it is found in different habitats and countries, it has sylvatic, peridomestic and domestic behavior and it is a very important vector of Chagas disease, especially in Peru. Molecules produced and secreted by salivary glands and fat body are considered of important adaptational value for triatomines because, among other functions, they subvert the host haemostatic, inflammatory and immune systems and detoxify or protect them against environmental aggressors. In this context, the elucidation of the molecules produced by these tissues is highly valuable to understanding the ability of this species to adapt and transmit pathogens. Here, we use high-throughput sequencing techniques to assemble and describe the coding sequences resulting from the transcriptome of the fat body and salivary glands of P. lignarius. The final assembly of both transcriptomes together resulted in a total of 11,507 coding sequences (CDS), which were mapped from a total of 164,676,091 reads. The CDS were subdivided according to their 10 folds overexpression on salivary glands (513 CDS) or fat body (2073 CDS). Among the families of proteins found in the salivary glands, lipocalins were the most abundant. Other ubiquitous families of proteins present in other sialomes were also present in P. lignarius, including serine protease inhibitors, apyrase and antigen-5. The unique transcriptome of fat body showed proteins related to the metabolic function of this organ. Remarkably, nearly 20% of all reads mapped to transcripts coded by Triatoma virus. The data presented in this study improve the understanding on triatomines’ salivary glands and fat body function and reveal important molecules used in the interplay between vectors and vertebrate hosts.
Author summary
This work describes an in-depth salivary and fat body transcriptome from the triatomine P. lignarius, the main vector of Chagas disease in Peru. Following “de novo” assembly of over 160 million reads, 11 thousand coding sequences were obtained, 9,013 of which were submitted to GenBank. Those over expressed in the salivary glands or fat bodies were identified, leading to a detailed insight of the specificities of each organ. This work will serve future scientists not only for its increased knowledge on triatomine proteins, but also for its public protein sequences that will serve scientists as a target database in mass spectrometry experiments aiming at the identification of P. lignarius salivary pharmacological peptides or immunological targets of vector exposure.
Introduction
Panstrongylus lignarius, also known as Panstrongylus herreri (WALKER, 1873) [1], is a triatomine species found in tropical and subtropical forests of South American countries including Peru, Ecuador, Colombia, Guyana, Suriname, Venezuela and Brazil [2, 3]. This species presents sylvatic behavior in the Amazon basin [4, 5] and peridomestic and domestic behavior in Peru [6]. Concerning its medical importance, this species is strongly synanthropic and is considered the major vector of Chagas disease in Peru [6, 7]. Among the triatomines of the genus Panstrongylus, the species P. lignarius is notable because, among other characteristics, it is capable of parasitizing different species of animals including marsupials, rabbits, spiny rats, anteaters, bats, chickens, toucans and pigeons [3]. In the Amazon region located in Peru, a considerable amount of the triatomines of this species are naturally infected with Trypanosoma cruzi (62.4%), and among those with identified food sources, 18.2% have been fed with human blood [8].
Among the mechanisms related to triatomine’s adaptation, it has been suggested that their saliva, which is inoculated during hematophagy, is crucial for the parasitism process and pathogen transmission. Indeed, the saliva of hematophagous arthropods, including triatomines, has inhibitory molecules of different defense mechanisms including platelet aggregation, inflammation, vasoconstriction, blood coagulation, and immune responses, which has been demonstrated to facilitate hematophagy and transmission of disease-causing agents [9].
In addition to saliva, molecules produced by the fat body from hematophagous arthropods have a substantial role in the detoxification of heme from blood, in developmental regulation and in the production of antimicrobial peptides and immunity [10–12]. Beyond these functions, the fat body is a multifunctional organ that has a pivotal role in nutrient and energy storage, in the synthesis of biomolecules and the whole metabolism [13]. It acts as a storage of energetic sources, important for the metamorphosis, egg maturation, reproduction and to survive long starvation periods. The fat body synthesizes and releases peptides, carbohydrates and lipids according to the metabolic needs and hormonal regulation [14].
It has been demonstrated through proteomic studies that triatomines of the P. lignarius species present a large number of bioactive molecules, but these molecules have a high interspecific functional biodiversity when compared to the molecules of the species Triatoma lecticularia and Rhodnius prolixus [15]. It has also been recently described that salivary molecules of P. lignarius, when compared to saliva of triatomines of the genus Triatoma, Meccus and Rhodnius have a remarkable ability to modulate dendritic cells and facilitate their invasion by T. cruzi [16].
The isolation and characterization of bioactive molecules in different tissues of blood-feeding insects has grown significantly in recent years and this scenario is mainly due to high-throughput sequencing techniques associated with bioinformatic tools. Different databases searches reveal genomes and sialomes of hematophagous arthropods such as ticks, mosquitoes and triatomines [17–24]. Here, we use high-throughput sequencing techniques to assemble and describe the coding sequences derived from a transcriptome of salivary glands and fat body of P. lignarius.
Material and methods
Ethics statement
The experiments were approved by the Institutional Animal Care and Use Committee—CEUA (protocol numbers 220 and 320).
Insects
P. lignarius was obtained from the insectary of the Universidade Federal do Triângulo Mineiro, Uberaba, Minas Gerais, Brazil. The colonies were maintained in cylindrical recipients and fed weekly on chickens. The experiments were approved by the Institutional Animal Care and Use Committee—CEUA (protocol numbers 220 and 320). Fed adults, including 7 female and 7 male insects were used to collect salivary glands (SG) and fat body (FB). One couple was dissected every other day for 14 days. The SG and FB were stored in 200 μl and 400 μl of RNA later (Qiagen, Valencia, CA) respectively, at 4°C for 48 hours and then maintained at -80°C until the day of shipping. The samples from the 14 days were pooled together and used for qRT-PCR or sent lyophilized to NIH Intramural Sequencing Center (5625 Fishers Lane—Rockville, MD 20852).
Sequencing
All procedures, including RNA extraction, libraries construction and sequencing were performed as previously described [23], with modifications. Briefly, RNA from each sample was collected using the Micro FastTrack-mRNA isolation kit (Invitrogen, Grand Island, NY) according to the manufacturer’s protocol. Following the isolation, total RNA integrity was checked using the BioAnalyser instrument (Agilent Technologies, Santa Clara, CA). The construction of mRNA libraries and sequencing were done at the NIH Intramural Sequencing Center. The fragments of cDNAs were made using a Covaris E210 (Covaris, Woburn, MA) and the libraries of SG and FB were constructed separately using the TruSeq RNA sample prep kit, v. 2 (Illumina Inc., San Diego, CA). Both libraries were amplified using eight cycles to minimize the risk of over-amplification. The sequencing of SG and FB were performed on a HiSeq 2000 (Illumina) with v. 3 flow cells and sequencing reagents. A paired-end protocol was used.
Bioinformatics
Raw data were processed using RTA 1.12.4.2 and CASAVA 1.8.2. The reads were trimmed of low quality regions, and only those with an average Illumina quality of 20 or more were used. Afterwards, they were assembled using ABySS software (Genome Sciences Centre, Vancouver, BC, Canada) [25, 26]. SOAPdenovo-Trans assembler [27] was also used because the ABySS may misassemble highly expressed transcripts. Assemblies were then joined using BLAST and Cap3 assembler [28]. All coding sequences (CDS) from SG and FB were selected based on similarities with known proteins or containing signal peptide using an automated pipeline [29]. The CDS and their respective protein sequences were placed in a hyperlinked Excel spreadsheet [30]. Software from the Center for Biological Sequence Analysis (Technical University of Denmark, Lyngby, Denmark) were used to predict Signal peptide, transmembrane domains, furin cleavage sites, and mucin-type glycosylation [29, 31–33]. Blastn [34] was used to map the reads into contigs with a word size of 25. The resulting contigs and RPKM values were also mapped to the Excel spreadsheet available as supplemental S1 and S2 Spreadsheets. Differential expression of the reads mapping to contigs between the two libraries were done using the X2 test. Relative expression of the transcripts of each separate transcriptome was evaluated using the “expression index”, which is the number of reads to a particular CDS divided by the largest number of reads mapped for a single CDS. The automated annotation of the proteins was based in the matches to various databases, including Gene Ontology, Pfam, Swissprot, KOG, SMART, Refseq-invertebrates and sequences containing Hemiptera[organism] protein sequences obtained from GenBank. The manual annotation was performed as detailed in [28].
Phylogenetic analysis
Evolutionary analyses were conducted in MEGA6 [35]. The evolutionary history of selected protein sequences was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the figure branches [36]. The trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [37] and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The sequences are shown with the first 3 letters of the genus name followed by the first 3 of the species name followed by their GenBank accession code.
qRT-PCR
Tissue expression of randomly chosen sixty genes were evaluated by RT-PCR. Briefly, RNA of salivary glands and fat body were extracted using a RNA SV Total RNA Isolation System (Promega, USA) according to the manufacturer’s recommendations. The cDNA was prepared using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) according to the manufacturer’s recommendations and samples were then frozen at −20°C until analysis. Gene expression was evaluated using a Sybr Green Master Mix (Roche, EUA) and specific primers (forward and reverse) as described in S1 Spreadsheet. Ultrapure DNA/RNA-free water was used as negative control. Relative gene expression was determined by ΔΔCT comparative method using PhSigP-51408_FR4_55–276 as reference gene (similar expression in salivary gland and fat body).
Results and discussion
General description of salivary gland and fat body transcriptome
The final assembly of both transcriptomes generated a total of 11,507 CDS, which were mapped from a total of 164,676,091 reads. The 11,507 CDS were subdivided per their putative function (Table 1 and S1 Spreadsheet) as Housekeeping, Secreted, Viral, Transposons and Unknown. The housekeeping (H) class had 5,460 CDS, corresponding to 47% of the total. The putative secreted (S) had 2,943 CDS, or 25% of all CDS. Remarkably, 20% of the reads mapped to transcripts coding for putative viral proteins, particularly to Triatoma virus proteins. Transposable elements (TE) accounted to only 3% of the CDS and 0.82% of the reads. Approximately 7% of the reads, corresponding to 2,734 CDS, were not classified and were placed in the unknown (U) class (Table 1).
Table 1. General classification of all coding sequences (CDS) from the combined salivary glands and fat body transcriptome of P. lignarius.
| Class | Number of CDS | % of total | Number of Reads | % of total |
|---|---|---|---|---|
| Housekeeping | 5,460 | 47.45 | 80,227,208 | 48.72 |
| Secreted | 2,943 | 25.58 | 38,247,116 | 23.23 |
| Viral | 12 | 0.10 | 33,602,364 | 20.41 |
| Transposons | 358 | 3.11 | 1,343,408 | 0.82 |
| Unknown | 2,734 | 23.76 | 11,255,995 | 6.84 |
| Total | 11,507 | 100 | 164,676,091 | 100 |
Housekeeping (H) genes of the combined transcriptome
The 5,460 CDS classified as housekeeping genes were characterized in 21 subgroups depending of their putative functions (S1 Spreadsheet and Table 2). In these subgroups, the category “signal transduction” had the highest expression presenting 11% of the mapped reads from class H, followed by “storage” and “protein synthesis machinery”.
Table 2. Classification of all coding sequences (CDS) with putative housekeeping function extracted from the transcriptome of P.lignarius.
| Class | Number of Contigs | % of total | Number of Reads | % of total |
|---|---|---|---|---|
| Signal transduction | 1,009 | 18.48 | 9,118,215 | 11.37 |
| Storage | 18 | 0.33 | 7,216,235 | 8.99 |
| Protein synthesis machinery | 280 | 5.13 | 6,379,967 | 7.95 |
| Nuclear regulation | 315 | 5.77 | 6,211,075 | 7.74 |
| Transcription machinery | 595 | 10.90 | 6,207,164 | 7.74 |
| Transporters/storage | 458 | 8.39 | 5,503,602 | 6.86 |
| Protein modification machinery | 311 | 5.70 | 4,996,742 | 6.23 |
| Cytoskeletal | 329 | 6.03 | 4,788,334 | 5.97 |
| Metabolism, lipid | 282 | 5.16 | 4,584,580 | 5.71 |
| Extracellular matrix/cell adhesion | 199 | 3.64 | 3,512,502 | 4.38 |
| Proteasome machinery | 242 | 4.43 | 3,112,153 | 3.88 |
| Metabolism, energy | 190 | 3.48 | 3,011,966 | 3.75 |
| Metabolism, amino acid | 116 | 2.12 | 2,696,206 | 3.36 |
| Metabolism, carbohydrate | 191 | 3.50 | 2,537,516 | 3.16 |
| Oxidant metabolism/detoxification | 144 | 2.64 | 2,378,367 | 2.96 |
| Protein export machinery | 331 | 6.06 | 2,360,314 | 2.94 |
| Immunity | 105 | 1.92 | 2,124,090 | 2.65 |
| Metabolism, nucleotide | 103 | 1.89 | 1,199,974 | 1.50 |
| Metabolism, intermediate | 60 | 1.10 | 978,596 | 1.22 |
| Transcription factor | 148 | 2.71 | 946,838 | 1.18 |
| Nuclear export | 34 | 0.62 | 362,772 | 0.45 |
| Total | 5,460 | 100 | 80,227,208 | 100 |
Of the 11,507 CDS of the combined transcriptome, 513 were found 10x or more expressed in the salivary glands, 2,073 were found similarly overexpressed in the fat bodies, and 8,921 CDS were not particularly overexpressed in either organ (Fig 1). We will proceed analyzing these enriched subsets, as they represent what is possibly specific for each tissue type.
Fig 1. Venn diagram of SG and FB transcriptomes of P. lignarius.
SG10x, 10-fold overexpressed transcripts in the salivary gland compared to fat body; FB10x, 10-fold overexpressed transcripts in the fat body compared to the salivary gland; and, Not Enriched, transcripts expressed in both SG and FB with less than a 10-fold variation.
The enriched salivary gland transcriptome of Panstrongylus lignarius
A total of 513 CDS appeared at least 10 X overexpressed in salivary glands (now referred to SG enriched transcriptome) when compared to the fat body transcriptome (S2 Spreadsheet and Table 3). The majority of these were associated with secreted products as expected for the SG.
Table 3. Classification of coding sequences (CDS) that are at least 10 x overexpressed in the salivary glands transcriptome as compared to the fat body transcriptome.
| SG overexpressed 10 x | Average FPKM SG | N | % of total | Average FPKM FB | Ratio FPKM |
|---|---|---|---|---|---|
| Secreted | 2,005.45 | 328 | 64.13 | 6.66 | 301.07 |
| Housekeeping | 221.60 | 136 | 26.51 | 4.28 | 51.74 |
| Unknown | 830.20 | 44 | 8.58 | 3.95 | 210.38 |
| Transposons | 6.37 | 4 | 0.78 | 0.31 | 20.58 |
| Total or average | 513 | 100 | 145.94 |
The SG enriched transcriptome contains 136 transcripts attributed to the housekeeping class, as further detailed in Table 4.
Table 4. Classification of coding sequences (CDS) 10X overexpressed in the salivary transcriptome with putative housekeeping function.
| Subclass | No. of CDS | No. of reads | % Total |
|---|---|---|---|
| Detoxification | 17 | 673,883 | 34.84 |
| Cytoskeletal | 4 | 267,216 | 13.84 |
| Lipid metabolism | 18 | 203,530 | 10.54 |
| Protein synthesis machinery | 4 | 185,847 | 9.62 |
| Unknown conserved | 15 | 143,472 | 7.42 |
| Transporters | 33 | 140,626 | 7.28 |
| Signal transduction | 24 | 89,506 | 4.63 |
| Glycosyl transferase | 1 | 76,195 | 3.95 |
| 15-hydroxyprostaglandin dehydrogenase | 4 | 65,330 | 3.38 |
| Transcription machinery | 6 | 52,714 | 2 2.72 |
| Nuclear Regulation | 2 | 23,056 | 1.19 |
| Salivary ubiquitin | 1 | 3,581 | 0.18 |
| Transcription Factors | 3 | 2,156 | 0.11 |
| Protein export and modification | 2 | 2,114 | 0.11 |
| Intermediate metabolism | 1 | 1,181 | 0.06 |
| Energy metabolism | 1 | 644 | 0.03 |
| Total | 136 | 1,931,051 | 100 |
Transcripts belonging to the detoxification class are the most abundant of the SG overexpressed CDS of the housekeeping class. Of these 17 transcripts, 12 are members of the cytochrome P450 family. Additionally, 4 CDS matches 15-hydroxyprostaglandin dehydrogenase, a similar finding in previous triatomine transcriptomes of Triatoma [18, 38, 39] and Panstrongylus megistus [40]. In a previous review, it was stated that this combination of transcripts suggested a role of triatomine salivary glands in the manufacture of eicosanoids [9]. However, a search for prostaglandins in the saliva of triatomines was negative [16]. Since these enzymes are associated with prostaglandin catabolism, it is here speculated that prostaglandins may function as salivary secretagogues and that the enzyme is associated with agonist detoxification. Alternatively, non-prostaglandin eicosanoids may be produced by the SG.
Transcripts coding for putative secreted proteins in Panstrongylus lignarius salivary glands
Lipocalins and the small molecule binding proteins with a JH binding motif comprised over 40% of the secreted transcripts that are overexpressed in the SG transcriptome (Table 5).
Table 5. Classification of coding sequences with putative secretory function extracted from sialotranscriptome of P. lignarius.
| Subclass | No. of CDS | No. of reads | % Total |
|---|---|---|---|
| Lipocalins | 88 | 6,280,411 | 29.09 |
| JH binding protein | 7 | 2,629,387 | 12.18 |
| Kazal-type peptides | 18 | 2,343,412 | 10.85 |
| Glycine rich proteins | 17 | 1,951,574 | 9.04 |
| Salivary proteases | 33 | 1,829,373 | 8.47 |
| Other secreted proteins | 94 | 1,580,689 | 7.32 |
| Conserved insect family 15 | 4 | 1,349,810 | 6.25 |
| Conserved insect family 12 | 17 | 1,000,854 | 4.64 |
| Immunity related | 8 | 983,020 | 4.55 |
| Inositol-145-triphosphate 5-phosphatase | 4 | 414,855 | 1.92 |
| Amylase/Maltase | 4 | 302,186 | 1.40 |
| Antigen-5 | 3 | 245,873 | 1.14 |
| Hemiptera specific family 225 | 5 | 171,874 | 0.80 |
| Mucin | 3 | 155,075 | 0.72 |
| Serpin | 2 | 86,723 | 0.40 |
| Apyrase | 2 | 83,292 | 0.39 |
| Hemiptera specific family 210 | 10 | 51,196 | 0.24 |
| Transferrin | 1 | 35,869 | 0.17 |
| Endonuclease | 1 | 35,862 | 0.17 |
| Ribonuclease | 1 | 28,532 | 0.13 |
| Lipase | 3 | 12,443 | 0.06 |
| Phosphatase | 1 | 11,308 | 0.05 |
| Odorant binding protein | 2 | 5,412 | 0.03 |
| Total | 328 | 21,589,030 | 100.00 |
Lipocalins are widely distributed in vertebrates, invertebrates, plants and bacteria [41, 42] and it is one of the main classes of proteins on the salivary glands of ticks and triatomines [43, 44]. Lipocalins possess a conserved three-dimensional structure and include an extensive group of extracellular proteins that generally bind to small hydrophobic proteins, extracellular ligands and other proteins. Triatomine lipocalins were shown to have vasodilator, anticoagulant and antiplatelet activities [45]. Some of these functions, such as the anticlotting activities of triabin [46] or nitrophorin 2 [47, 48], are exerted by interactions of the lipocalin with a clotting cascade protein, while other functions relate to their strong binding to agonists of hemostasis or inflammation, namely their kratagonist function (from the Greek “kratos” = to seize) [49]. Recently, a salivary lipocalin of Rhodnius prolixus was shown to antagonize cysteinyl leukotrienes [50].
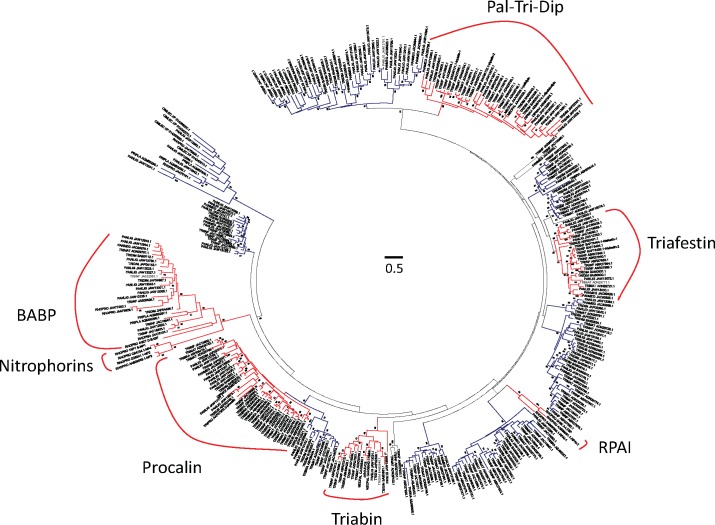
The assembly of the P. lignarius transcriptome revealed 78 contigs coding for full length lipocalins, all at least 10 times overexpressed in the SG when compared to the FB, and averaging over 3,000 fold overexpression. The protein sequences of these 78 gene products were aligned with 252 other triatomine lipocalins, producing a dendrogram where at least 15 clades with strong bootstrap support are observed (Fig 2). Eight P. lignarius sequences are found within the Pal-Tri-Dip clade which includes the platelet aggregation inhibitors pallidipin [51, 52], triplatin [53, 54] and dipetalodipin [55] from the Triatoma and Dipetalogaster genera. Triplatin and dipetalodipin were shown to be kratagonists of eicosanoids, possibly the same mechanism occurring in pallidipin. Triafestins are inhibitors of the activation of the kinin system [56] found in T. infestans. The two characterized sequences are within a clade with strong bootstrap support. Four P. lignarius sequences are found within this clade. The clade containing Rhodnius platelet aggregation inhibitor (RPAI) [57, 58], as well the Rhodnius prolixus leukotriene binding protein (LBP) [50] is Rhodnius specific, thus not containing any Panstrongylus sequences. It, however, has a sister clade, with low bootstrap support, containing several Triatoma and Panstrongylus sequences, including two from P. lignarius. The clade named triabin contains the anti-thrombin inhibitor from T. infestans [46, 59] and several other sequences, including two from P. lignarius. A large clade contains the salivary antigen procalin [60], of unknown physiological function. The BABP clade, which is sister to the uniquely Rhodnius nitrophorin clade, has the Rhodnius prolixus Biogenic Amine Binding Protein [61, 62], a protein having anti-platelet and vasodilatory activities. Ten P. lignarius sequences populate this clade. This analysis may help to design experiments with recombinant triatomine proteins aiming at determining their functions.
Fig 2. Phylogram of the lipocalin family proteins from P. lignarius and their best matches.
The optimal tree with the sum of branch length = 123.81570372 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The bar at the center of the graph indicates a value of 0.5. The analysis involved 330 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 423 positions in the final dataset. The values near the branches represent the percentage of bootstrap support. Values below 50% are not shown. For more details, see Material and Methods.
Apyrases are enzymes that hydrolyze ATP and ADP to AMP and orthophosphate. These are abundant in the saliva of blood feeding arthropods presumably because they destroy these agonists of platelet aggregation and inflammation [63]. In mosquitoes [64] and triatomine bugs [65], but not in Rhodnius, salivary apyrases belong to the 5’-nucleotidase family, while in Cimex [66] and sand flies [67], and probably in Rhodnius [68], they belong to the Cimex family of apyrases. In P. lignarius, two apyrase-like proteins of the 5’-nucleotidase family are highly expressed in the SG with a total of 83,000 reads (Table 5). A third member of the family was additionally identified, but it is not particularly enriched in either FB or SG transcriptomes. This third member has a glycophosphatidylinositol (GPI) anchor as predicted by the big-PI Predictor site [69], while the two overexpressed proteins do not, indicating they are secreted and not membrane bound. This is in accordance with the postulated evolution of secreted salivary apyrases which included a step of gene duplication of an ancestral, membrane-bound product, plus loss of the GPI anchor [64]. The phylogram of the three P. lignarius members of the apyrase/5’-nucleotidase family together with their best matches by Blastp to GenBank proteins displays robust clades for various insect orders of families, the P. lignarius proteins each sharing a robust clade with other triatomine proteins, indicative of their long evolutionary history (Fig 3). Tellingly is the absence of Rhodnius proteins, supporting the monophyletic status of this genus with relation to its evolution to blood feeding [70].
Fig 3. Phylogram of the apyrase/5’-nucleotidase family of proteins from P. lignarius and their best matches.
The optimal tree with the sum of branch length = 15.40595903 is shown. The values near the branches represent the percentage of bootstrap support. Values below 75% are not shown. The analysis involved 41 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 689 positions in the final dataset. For more details, see Material and Methods.
The inositol phosphate 5-phosphatase (IPP) enzymes were predicted in 4 CDS. Previously, the Rhodnius homolog was shown to act both on soluble inositol phosphatase and the substrate phosphoinositide [71]. These phospholipids are involved in several cellular processes related to signal transduction, secretion and cytoskeletal structure. Although IPP was found to be produced by Rhodnius prolixus salivary glands in 2006 [71], it still has an unknown function in this organism. These studies suggested that IPP ejected in saliva act by decreasing the concentration of PI (4,5) P2 e PI(3,4,5) P3, which are present in the plasma membrane of cells and platelets, causing changes in the cytoskeletal architecture [72]; however, how they could enter the cell to perform this function is still a puzzle.
Among other enzymes overexpressed in the salivary glands, we address a cathepsin D, which is normally a housekeeping enzyme associated with lysosomes, but is over 1,000 fold overexpressed in the SG, and has a signal peptide indicative of secretion, indicating this protein may have a role in blood meal acquisition by interfering with host hemostasis. Serine proteases were also found, these having been regularly detected in triatomine sialomes [9]. The function of these proteases in feeding, however, is unknown, but could be related to fibrin hydrolysis, as occur with salivary serine proteases from a tabanid [73].
Somewhat surprising is the finding of amylases and maltase overexpressed in the SG. These enzymes are usually found in mosquito sialomes, and are associated with the sugar feeding mode of these organism [63]. Is it possible that P. lignarius feeds on plants? Recently it was shown that R. prolixus feeds on plants [74] and perhaps this behavior is more widespread.
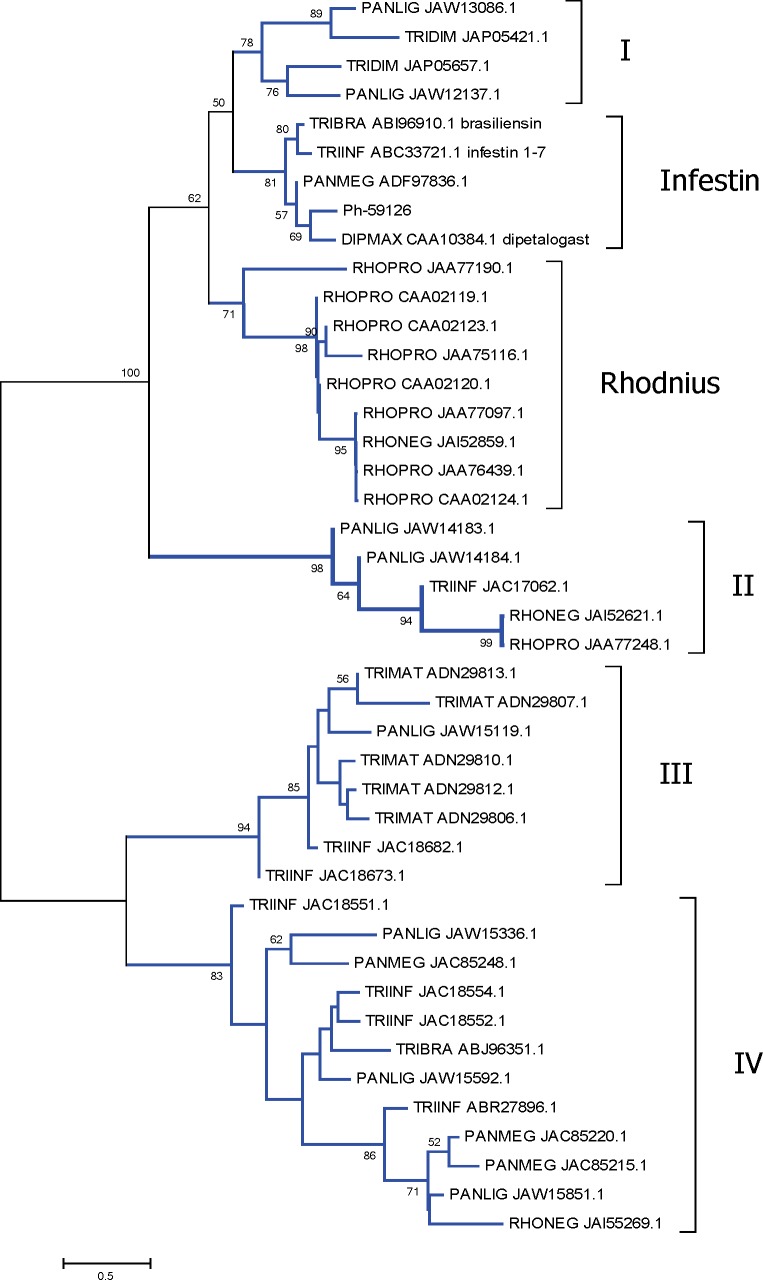
Serine-proteases inhibitory proteins were found in the salivary gland transcriptome. These may play a role in inhibiting the coagulation cascade or the activation of the complement system. These proteins are subdivided according to their domain, such as the Kazal domain and serpins [75]. The Kazal family was the second most abundant in P. lignarius SG, corresponding to ~ 10% of the total reads. They consist in molecules with single or multiple domains with a shared conserved motif and a distinct pattern of cysteine distribution. Several proteins from the Kazal family were already described in vertebrates and invertebrates, including triatomines. Kazal domain containing peptides are typical inhibitors of serine proteases. Indeed, the two Kazal domain protein Rhodnin, isolated from the crop of R. prolixus, was shown to inhibit thrombin [76]; similarly, dipetalogastin was isolated from D. maximus [77] and brasiliensin from T. brasiliensis [78] guts. Infestins, with up to seven Kazal domains, were isolated from T. infestans midgut [79–82] and shown to inhibit thrombin, neutrophil elastase and Factor XIIa. The salivary gland transcriptome of P. lignarius discloses a seven-Kazal domain containing peptide that is the most expressed member of this peptide class, encoded by Ph-59126. On the other hand, a KGD-containing Kazal peptide in tabanids named vasotab was shown to have vasodilatory activity, in addition to anti-platelet activity [83]. Phylogenetic analysis of P. lignarius Kazal-domain containing peptides aligned with other related triatomine proteins indicates the complexity of this family (Fig 4). A clade containing the triatomine intestinal serine protease inhibitors (named Infestin in Fig 4) contains the seven-Kazal peptide from P. lignarius mentioned above, indicating that P. lignarius may have co-opted this peptide family for salivary expression. Four other robust clades, plus one Rhodnius-specific clade indicate the diversity of these peptides in triatomines. Clade IV includes JAW15592.1, JAW15851.1 and JAW15336.1 which have weak similarity to vasotab; however, the KGD domain of vasotab associated with anti-platelet function is not found in P. lignarius.
Fig 4. Phylogram of the Kazal-domain containing family of proteins from P. lignarius and their best matches.
The optimal tree with the sum of branch length = 14.99541793 is shown. The values near the branches represent the percentage of bootstrap support. Values below 50% are not shown. The analysis involved 43 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 969 positions in the final dataset. For more details, see Material and Methods. Robust clades are named I-IV (where no member has been functionally characterized); Infestin and Rhodnius indicate clades containing previously functionally characterized proteins.
The serpin domain containing proteins, or serpin-like, are also found in the saliva of arthropods and they affect hemostasis, including platelet adhesion, fibrinolysis and coagulation, facilitating the blood feeding. Two salivary enriched serpins were found in the P. lignarius transcriptome.
The Antigen-5 protein family is a group of proteins belonging to the cysteine-rich secretory proteins (CRISP) superfamily which have been identified through sialotranscriptomes in the saliva of different hematophagous insects, such as mosquitoes [84, 85], phlebotomines [86–88], and triatomines [17, 18, 21, 22, 24, 89–91]. Although it is frequently found in the saliva of hematophagous arthropods, its function is mostly unknown and it was described to be part of the toxin repertoire of snake venom [92]. An antigen-5 protein from Dipetalogaster maxima was shown to inhibit platelet aggregation by low doses of collagen and to have superoxide dismutase activity [93]. The current transcriptome of P. lignarius identified three members of this family that are highly expressed in the SG.
Immunity-related proteins and peptides are broadly found in several organisms from mammals to plants and hematophagous arthropods. In the saliva of these insects and ticks, those peptides, such as lysozyme and defensins may help in controlling the microbial growth in the ingested blood and, in the vertebrate host, they may prevent microbial infections at the biting place [94]. Pathogen pattern recognition proteins such as lectins were also found in this group; these proteins may play a role in modulating host immunity.
Only two transcripts coding for members of the Odorant/Pheromone-Binding Family (OBP) were found in the enriched SG transcriptome, corroborating the findings from other studies with Rhodnius, Triatoma, Panstrongylus and Cimex, where in Cimex they were more abundant. The properties of these proteins in blood-feeding is unknown [9]. We also identified 7 CDS putatively coding for Juvenile hormone binding proteins (JH binding proteins) with very long sequences and representing ~12% of the total reads. These proteins were already identified in the SG of Rhodnius neglectus [91], Anopheles culicifacies [95], Anopheles gambiae [96] and Aedes aegypti [97], however, no single CDS coding for JH binding proteins was found in the sialotranscriptome of P. megistus [24]. JH in adult insects is responsible for controlling the reproductive maturation and inducing vitellogenesis, as JH and vitellogenin are negatively correlated. Additionally, JH carrier and odorant binding proteins have a pivotal function in the regulation of feeding behavior of hematophagous arthropods [98]. Despite these more conventional functions, it is possible that these gene products may have been recruited to a salivary function to act as kratagonists of lipidic mediators of hemostasis.
The sialome of P. lignarius revealed at least two protein family expansions that appear exclusive of Hemiptera, and three protein family expansions that appear to be exclusive of insects. Some of the transcripts are highly expressed, such as Ph-55919, with a RPKM = 14,784. Their functions are unknown.
Fat body
There are few transcripts analyses of fat body published so far, such as Bactocera dorsalis [12], Melipona scutellaris [99], and Aedes aegypti [100], and none are related to triatomines.
The evaluation of the entire transcriptome from the fat body of P. lignarius (Table 6) showed the most prevalent subclass was classified as secreted proteins with ~20.5% of the total reads, which agrees with one of the main functions of this tissue, the synthesis of peptides with distinct functions, many to be destined to the hemolymph compartment. Following the secreted proteins, the next subclass with high FB expression was storage with ~8% of the reads, also associated with secreted hemolymph proteins. On the 10x overexpressed subset (Table 7), the most prevalent classes of molecules were also related to secreted proteins and storage, followed by proteins with unknown function and cytoskeletal proteins.
Table 6. Classification of all coding sequences (CDS) from the fat body transcriptome of P. lignarius.
| Subclass | No. of CDS | No. of reads | % Total |
|---|---|---|---|
| Secreted protein | 2,664 | 16,900,616 | 20.40 |
| Storage | 17 | 6,882,176 | 8.23 |
| Signal transduction | 985 | 5,648,555 | 6.82 |
| Transcription machinery | 590 | 4,808,076 | 5.80 |
| Viral product | 11 | 4,200,886 | 5.07 |
| Protein synthesis machinery | 276 | 4,173,641 | 5.04 |
| Transporters and channels | 425 | 3,879,540 | 4.68 |
| Unknown product | 1,559 | 3,429,273 | 4.14 |
| Cytoskeletal proteins | 325 | 3,399,663 | 4.10 |
| Unknown conserved | 881 | 3,356,864 | 4.05 |
| Nuclear regulation | 313 | 2,975,729 | 3.60 |
| Protein modification | 295 | 2,844,094 | 3.43 |
| Proteasome machinery | 241 | 2,378,803 | 2.87 |
| Lipid metabolism | 260 | 2,260,869 | 2.73 |
| Amino acid metabolism | 115 | 2,250,741 | 2.71 |
| Energy metabolism | 188 | 2,075,093 | 2.50 |
| Carbohydrate metabolism | 185 | 1,698,819 | 2.05 |
| Protein export | 329 | 1,392,878 | 1.68 |
| Extracellular matrix | 182 | 1,298,620 | 1.56 |
| Unkown conserved membrane protein | 235 | 1,029,230 | 1.24 |
| Transposable element | 353 | 1,020,635 | 1.23 |
| Nucleotide metabolism | 102 | 979,483 | 1.18 |
| Immunity | 97 | 922,667 | 1.11 |
| Intermediary metabolism | 59 | 789,404 | 0.95 |
| Detoxification | 72 | 762,671 | 0.92 |
| Transcription factor | 145 | 677,034 | 0.81 |
| Oxidant metabolism/Detoxification | 56 | 496,585 | 0.60 |
| Nuclear export | 34 | 287,566 | 0.34 |
| Total | 10,994 | 82,820,211 | 100 |
Table 7. Coding sequences (CDS) 10X overexpressed in fat body compared to salivary glands from transcriptome of P. lignarius.
| Subclass | No. of CDS | No. of reads | % Total |
|---|---|---|---|
| Secreted protein | 611 | 11,982,880 | 32.26 |
| Storage | 9 | 6,803,966 | 18.32 |
| Unknown product | 387 | 2,081,905 | 5.60 |
| Cytoskeletal proteins | 112 | 1,794,794 | 4.83 |
| Transcription machinery | 45 | 1,696,142 | 4.57 |
| Transporters and channels | 89 | 1,609,127 | 4.33 |
| Signal Transduction | 168 | 1,242,131 | 3.34 |
| Amino acid metabolism | 35 | 1,065,122 | 2.87 |
| Unknown conserved | 156 | 1,000,574 | 2.69 |
| Carbohydrate metabolism | 30 | 826,799 | 2.23 |
| Energy metabolism | 33 | 799,257 | 2.15 |
| Proteasome machinery | 29 | 717,203 | 1.93 |
| Extracellular matrix | 52 | 681,489 | 1.83 |
| Nucleotide metabolism | 11 | 677,461 | 1.82 |
| Protein modification | 65 | 561,469 | 1.51 |
| Transposable element | 18 | 494,232 | 1.33 |
| Detoxification | 9 | 490,735 | 1.32 |
| Unkown conserved membrane protein | 31 | 459,387 | 1.24 |
| Lipid metabolism | 43 | 450,227 | 1.21 |
| Protein synthesis machinery | 16 | 390,148 | 1.05 |
| Immunity | 31 | 346,241 | 0.93 |
| Nuclear regulation | 22 | 283,827 | 0.76 |
| Intermediary metabolism | 6 | 230,441 | 0.62 |
| Oxidant metabolism/Detoxification | 14 | 195,373 | 0.53 |
| Protein export | 32 | 136,685 | 0.37 |
| Transcription factor | 17 | 73,442 | 0.20 |
| Nuclear export | 2 | 53,368 | 0.14 |
| Total | 2,073 | 37,144,425 | 100 |
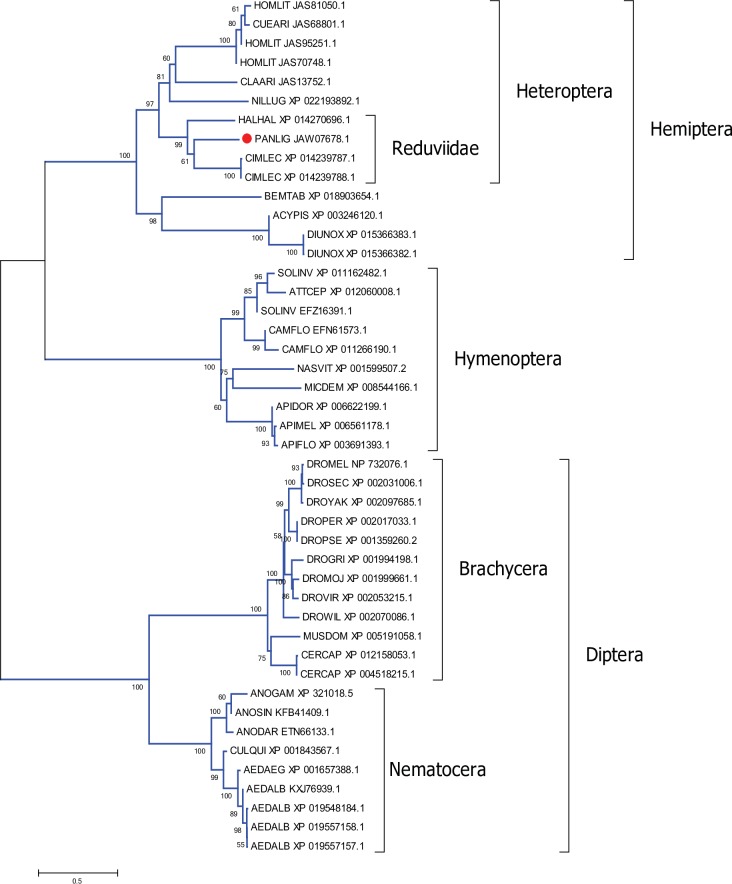
Vitellogenin is an important protein precursor of vitelline, used in insect oocytes’ formation and maturation, and it is produced exclusively in the fat body of insects and then processed and secreted in hemolymph [101]. The transcriptome of P. lignarius allowed for the disclosure of JAW07678.1, a 1,324 aa long protein with N-terminal Vitellogenin-N and carboxyterminal VWD motifs, typical of insect vitellogenins. Alignment of this protein sequence with its best matches from the NCBI database allows identification of the insect order-specific clades Hemiptera, Hymenoptera and Diptera, including the Diptera sub-orders Brachycera and Nematocera, and the Heteroptera sub-order within the Hemiptera, and within Heteroptera, the Reduvidae family (Fig 5).
Fig 5. Phylogram of the vitellogenin protein of P. lignarius and their best matches.
The optimal tree with the sum of branch length = 11.41384103 is shown. The values near the branches represent the percentage of bootstrap support. Values below 50% are not shown. The analysis involved 45 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1713 positions in the final dataset. For more details, see Material and Methods. The Panstrongylus lignarius sequence is shown with a red marker.
Transferrins are glycoproteins found in different animals, such as mammals, marsupials, fish and in more than 34 species of invertebrates, including R. prolixus [102]. These proteins may function in insect defense mechanisms [103]. In insects, transferrins are synthesized and stored in the fat body for posterior secretion to the hemolymph, where they participate in iron uptake and distribution with ferritin [104]. A transferrin was identified in the fat body transcriptome of P. lignarius. This 656 aa long protein was aligned with its best matches from the NCBI database to produce the phylogram shown in Fig 6, where robust clades of several insect orders are observed.
Fig 6. Phylogram of the transferrin protein of P. lignarius and their best matches.
The optimal tree with the sum of branch length = 10.61177438 is shown. The values near the branches represent the percentage of bootstrap support. Values below 50% are not shown. The analysis involved 51 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1487 positions in the final dataset. For more details, see Material and Methods. The Panstrongylus lignarius sequence is shown with a red marker.
Triatoma virus
Previous sialotranscriptomes from triatomines reported a low expression of diverse viruses. For example, in an Illumina-based sialotranscriptome of Panstrongylus megistus seven transcripts were reported that best matched viral proteins. However, these transcripts were poorly expressed, reaching expression indexes below 0.0025 [40]. Another Illumina-based sialotranscriptome of Triatoma infestans reported 38 transcripts similarly coding for putative viruses, two of which had an expression index between 1 and 2.7, most similar to Deformed winged virus and to Drosophila A virus [39]. Similarly, the P. lignarius transcriptome uncovered 12 transcripts putatively coded by viruses. Remarkably, two transcripts coding for the capsid P1 polyprotein (Genbank AHB63946.1) and the nonstructural protein precursor (Genbank NP_620562.1) of Triatoma virus were very highly expressed, attaining expression indexes of 100 (most expressed transcript) and 18 in the SG. Each transcript accrued more than 9% of the totality of reads, summing up to near 20% of all transcriptome reads deriving from the viral genome (Table 1 and S1 Spreadsheet). The relative expression was high both on the salivary gland as well as in the fat body transcriptomes. Triatoma virus was first discovered infecting T. infestans in Argentina [105], and later shown to infect other Triatoma species as well as Psammolestes coreodes [106]. A survey of laboratory reared insects additionally detected several species harboring the virus, including specimens from the Barbacena insectary from where the P. lignarius used in this work derived [107]. However, P. lignarius was not analyzed in that study, and thus this species can be added to the 15 previously found infected with this virus, which included a single Panstrongylus species (P. guentheri).
Mice inoculation with the virus resulted in a non-infective immune response [108], and it was found that people with Chagas’ disease living in Bolivia, Argentina and Mexico developed a detectable immune response to the virus [109]. The high levels of transcription found in the salivary glands suggest that in addition to the previously proposed route of fecal contamination, humans and rodents could be infected via direct salivary inoculation. From the standpoint of insect to insect propagation, it has been proposed that viral transmission occurs via the fecal-oral route or by cleptohematophagy [107]. It should be added that, to the extent mature viral particles are secreted in the bugs’ saliva, co-feeding bugs could promote transmission between insects, as their feeding mechanism includes frequent reversal of the ingestion pump, possibly to dislodge incipient platelet plugs [110, 111], thus spreading the virus in the skin vasculature from where it could reach co-feeding insects.
Determination of relative tissue expression of randomly selected genes
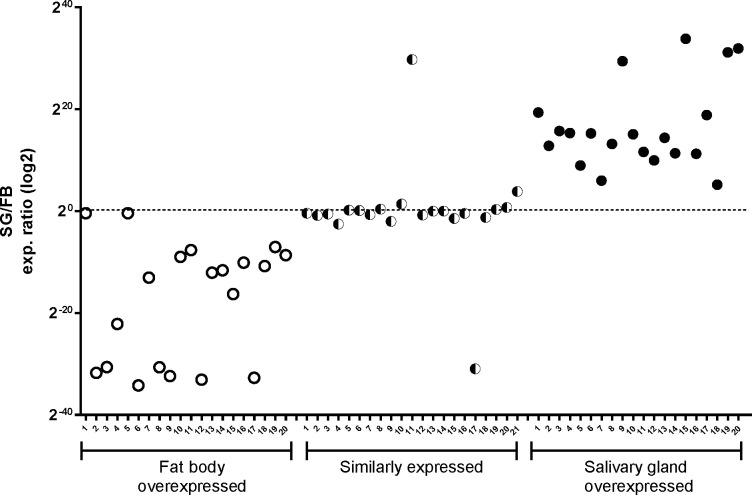
Confirmation of the gene expression of randomly selected genes from the salivary gland and fat body transcriptomes of P. lignarius was performed. We selected twenty 10X overexpressed SG genes, twenty 10X overexpressed FB genes and twenty genes with similar expression in both tissues. As demonstrated in Fig 7, SG or FB overexpression or similar expression between these tissues was confirmed by qRT-PCR for most of the evaluated genes, as predicted by transcriptome analysis.
Fig 7. Validation by qPCR of the differential expression of transcripts between salivary gland and fat body libraries.
Relative gene expression was calculated by ΔΔCT method using PhSigP-51408_FR4_55–276 as reference gene (similar expression in salivary gland and fat body). Scatter plot presenting the SG:FB expression ratio of FB overexpressed, similarly expressed and SG overexpressed genes. The Y axis is the log2 of the observed ratio between SG and FB by qPCR on three groups of arbitrarily selected transcripts that are overexpressed in either tissue, or similarly expressed. S1 Table provides for the transcript names and primer sequences used in this experiment.
Conclusions
Different species of insects diverge in relation to the molecular compounds of their saliva, which are determined by their evolutionary history including habitat distribution and food source. Sialome studies of several species have already identified a variety of molecules with potential industrial and/or clinical use for their pharmacological activity and discriminating properties as biomarkers of vector exposure, respectively [112]. The present work contributed to the public disclosure of over 9,000 protein sequences that should contribute to the discovery of new pharmacologically active compounds or new vector-exposure immunological markers while serving as a protein database for mass-spectrometric protein identification studies.
Supporting information
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov)
Data Availability
This project was registered with the National Center for Biotechnology Information (NCBI) under Bioproject PRJNA395235 and BioSamples SAMN07374128 and SAMN07374129. The raw reads are available on the Sequence Read Archive (SRA) under accessions SRR5859113 and SRR5859114. The coding sequences and their protein products (numbering 9,013) were submitted to the Transcriptome Shotgun Assembly (TSA) project which were deposited at DDBJ/EMBL/GenBank under the accession GFTR00000000. The version described in this paper is the first version, GFTR01000000.
Funding Statement
CJFO received grants from Fundação de Amparo à Pesquisa do Estado de Minas Gerais – FAPEMIG grants APQ-02898-14 and RED-00013-14 to CJFO (http://www.fapemig.br/), and JMCR received a National Institute of Allergy and Infectious Diseases Intramural Grant Z01 AI000810-18 (https://www.niaid.nih.gov/about/dir). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marcilla A, Bargues MD, Abad-Franch F, Panzera F, Carcavallo RU, Noireau F, et al. Nuclear rDNA ITS-2 sequences reveal polyphyly of Panstrongylus species (Hemiptera: Reduviidae: Triatominae), vectors of Trypanosoma cruzi. Infect Genet Evol. 2002;1(3):225–35. [DOI] [PubMed] [Google Scholar]
- 2.Paula AM, Castrillon MI, Maciel GBML, Galvão C . Registro de ocorrência de Panstrongylus lignarius (Walker, 1837) (Hemiptera: Reduviidae: Triatominae) no Estado de Mato Grosso, Brasil. Acta Amazonica. 2013;43(1):117–9. [Google Scholar]
- 3.Patterson JS, Barbosa SE, Feliciangeli MD. On the genus Panstrongylus Berg 1879: evolution, ecology and epidemiological significance. Acta Trop. 2009;110(2–3):187–99. doi: 10.1016/j.actatropica.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 4.Teixeira ARL, Monteiro PS, Rebelo JM, Argañaraz HR, Vieira D, Lauria-Pires L, et al. Emerging Chagas Disease: trophic network and cycle of transmission of Trypanosoma cruzi from palm trees in the Amazon. Emerg Infect Dis. 2001;7:100–12. doi: 10.3201/eid0701.700100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guhl F, Aguilera G, Pinto N, Vergara D. Actualización de la distribución geográfica y ecoepidemiología de la fauna de triatominos (Reduviidae: Triatominae) en Colombia. Biomédica. 2007;27:143–62. [PubMed] [Google Scholar]
- 6.Cuba Cuba CA, Abad-Franch F, Roldán Rodríguez J, Vargas Vásquez F, Pollack Velásquez L, Miles MA. The triatomine of Northern Peru, with emphasis on the ecology and infection by trypanosomes of Rhodnius ecuadoriensis (Triatominae). Mem Inst Oswaldo Cruz. 2002;97:175–83. [DOI] [PubMed] [Google Scholar]
- 7.Alroy KA, Huang C, Gilman RH, Quispe-Machaca VR, Marks MA, Ancca-Juarez J, et al. Prevalence and Transmission of Trypanosoma cruzi in People of Rural Communities of the High Jungle of Northern Peru. PLoS Negl Trop Dis. 2015;9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto J, Cáceres AG, Vega S, Martínez R, Náquira C. Fuentes de alimentación de Panstrongylus herreri (HEMIPTERA: TRIATOMINAE) capturados en Utcubamba, Amazonas—Peru. Revista Peruana de Medicina Experimental y Salud Pública. 2008;25(2): 179–84. [Google Scholar]
- 9.Ribeiro JMC, Assumpção TC, Francischetti IMB. An Insight into the Sialomes of Bloodsucking Heteroptera. Psyche: A Journal of Entomology. 2012;2012:1–16. [Google Scholar]
- 10.Attardo GM, Strickler-Dinglasan P, Perkin SAH, Caler E, Bonaldo MF, al. e. Analysis of fat body transcriptome from the adult tsetse fly, Glossina morsitans morsitans. Insect Biochem Mol Biol. 2006;15:411–24. [DOI] [PubMed] [Google Scholar]
- 11.Arrese EL, Soulages JL. Insect fat body: energy, metabolism and regulation. Annu Rev Entomol. 2010;55:2017–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang WJ, Yuan GR, Cong L, Xie YF, Wang JJ. De novo cloning and annotation of genes associated with immunity, detoxification and energy metabolism from the fat body of the oriental fruit fly, Bactrocera dorsalis. PLoS One. 2014;9(4):e94470 doi: 10.1371/journal.pone.0094470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Law JH, Wells MA. Insects as biochemical models. J Biol Chem. 1989;264(28):16335–8. [PubMed] [Google Scholar]
- 14.Gade G. Regulation of intermediary metabolism and water balance of insects by neuropeptides. Annu Rev Entomol. 2004;49:93–113. doi: 10.1146/annurev.ento.49.061802.123354 [DOI] [PubMed] [Google Scholar]
- 15.Montandon CE, Barros E, Vidigal PM, Mendes MT, Anhe AC, de Oliveira Ramos HJ, et al. Comparative proteomic analysis of the saliva of the Rhodnius prolixus, Triatoma lecticularia and Panstrongylus herreri triatomines reveals a high interespecific functional biodiversity. Insect Biochem Mol Biol. 2016;71:83–90. doi: 10.1016/j.ibmb.2016.02.009 [DOI] [PubMed] [Google Scholar]
- 16.Mendes MT, Carvalho-Costa TM, da Silva MV, Anhe AC, Guimaraes RM, da Costa TA, et al. Effect of the saliva from different triatomine species on the biology and immunity of TLR-4 ligand and Trypanosoma cruzi-stimulated dendritic cells. Parasit Vectors. 2016;9(1):634 doi: 10.1186/s13071-016-1890-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assumpcao TC, FIM C., Andersen JF, Schwarz A, Santana JM, Ribeiro JM. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas' disease. Insect Biochem Mol Biol. 2008;38(2):213–32. doi: 10.1016/j.ibmb.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Assumpcao TC, Eaton DP, Pham VM, Francischetti IM, Aoki V, Hans-Filho G, et al. An insight into the sialotranscriptome of Triatoma matogrossensis, a kissing bug associated with fogo selvagem in South America. The American journal of tropical medicine and hygiene. 2012;86(6):1005–14. doi: 10.4269/ajtmh.2012.11-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC genomics. 2009;10:57 doi: 10.1186/1471-2164-10-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribeiro JM, Francischetti IMB. Role of arthropod saliva in blood feeding: Sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812 [DOI] [PubMed] [Google Scholar]
- 21.Ribeiro JM. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2004;34(1):61–79. [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro JM, Assumpcao TC, Pham M, Francischetti IM, Reisenman CE. An Insight Into the Sialotranscriptome of Triatoma rubida (Hemiptera: Heteroptera). J Med Entomol. 2012;49(3):563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribeiro JM, Chagas AC, Pham VM, Lounibos LP, Calvo E. An insight into the sialome of the frog biting fly, Corethrella appendiculata. Insect biochemistry and molecular biology. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeiro JM, Schwarz A, Francischetti IM. A Deep Insight Into the Sialotranscriptome of the Chagas Disease Vector, Panstrongylus megistus (Hemiptera: Heteroptera). Journal of medical entomology. 2015;52(3):351–8. doi: 10.1093/jme/tjv023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, et al. De novo transcriptome assembly with ABySS. Bioinformatics (Oxford, England). 2009;25(21):2872–7. [DOI] [PubMed] [Google Scholar]
- 26.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome research. 2009;19(6):1117–23. doi: 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1(1):18 doi: 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS ONE. 2011;6(12):e28525 doi: 10.1371/journal.pone.0028525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein engineering. 1999;12(1):3–9. [DOI] [PubMed] [Google Scholar]
- 30.Guo Y, Ribeiro JM, Anderson JM, Bour S. dCAS: a desktop application for cDNA sequence annotation. Bioinformatics (Oxford, England). 2009;25(9):1195–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17(1):107–12. doi: 10.1093/protein/gzh013 [DOI] [PubMed] [Google Scholar]
- 32.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 33.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15(2):153–64. doi: 10.1093/glycob/cwh151 [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30(12):2725–9. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution; international journal of organic evolution. 1985;39(4):783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- 37.Zuckerkand E, Pauling L. Evolutionary divergence and convergence in proteins In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. New York: Academic Press; 1965. p. 97–166. [Google Scholar]
- 38.Santos A, Ribeiro JM, Lehane MJ, Gontijo NF, Veloso AB, Sant' Anna MR, et al. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae). Insect biochemistry and molecular biology. 2007;37(7):702–12. doi: 10.1016/j.ibmb.2007.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz A, Medrano-Mercado N, Schaub GA, Struchiner CJ, Bargues MD, Levy MZ, et al. An updated insight into the sialotranscriptome of Triatoma infestans: Developmental stage and geographic variations. PLoS neglected tropical diseases. 2014;8(12):e3372 doi: 10.1371/journal.pntd.0003372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribeiro JMC, Schwarz A, Francischetti IMB. A deep insight into the sialotranscriptome of the Chagas disease vector, Panstrongylus megistus (Hemiptera: Heteroptera). J Med Entomol. 2015;52(3):351–8. doi: 10.1093/jme/tjv023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flower DR, North AC, Attwood TK. Structure and sequence relationships in the lipocalins and related proteins. Protein Sci. 1993;2:753–61. doi: 10.1002/pro.5560020507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flower DR. Multiple molecular recognition properties of the lipocalin protein family. J Mol Recogn. 1995;8:185–95. [DOI] [PubMed] [Google Scholar]
- 43.Andersen JF, Gudderra NP, Francischetti IM, Ribeiro JM. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Archives of insect biochemistry and physiology. 2005;58(2):97–105. doi: 10.1002/arch.20032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francischetti IMB, Sá-Nunes A, Mans BJ, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. Frontiers in Biosciences. 2009;14:2051–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersen JF, Gudderra NP, Francischetti IMB, Ribeiro JM. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Archives of insect biochemistry and physiology. 2005;58(2):97–105. doi: 10.1002/arch.20032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuentes-Prior P, Noeske-Jungblut C, Donner P, Schleuning WD, Huber R, Bode W. Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc Natl Acad Sci U S A. 1997;94(22):11845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen JF, Montfort WR. The crystal structure of nitrophorin 2. A trifunctional antihemostatic protein from the saliva of Rhodnius prolixus. The Journal of biological chemistry. 2000;275(39):30496–503. doi: 10.1074/jbc.M002857200 [DOI] [PubMed] [Google Scholar]
- 48.Ribeiro JM, Schneider M, Guimaraes JA. Purification and characterization of prolixin S (nitrophorin 2), the salivary anticoagulant of the blood-sucking bug Rhodnius prolixus. The Biochemical journal. 1995;308 (Pt 1):243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A J.F., R J.M., Feeding SKSoHPEDB. Salivary Kratagonists: Scavengers of Host Physiological Effectors During Blood Feedin Arthropod Vector: Controller of Disease Transmission. London: Elsevier; 2017. p. 51–63. [Google Scholar]
- 50.Jablonka W, Pham V, Nardone G, Gittis A, Silva-Cardoso L, Atella GC, et al. Structure and Ligand-Binding Mechanism of a Cysteinyl Leukotriene-Binding Protein from a Blood-Feeding Disease Vector. ACS Chem Biol. 2016;11(7):1934–44. doi: 10.1021/acschembio.6b00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haendler B, Becker A, N-J C., Krätzschmar J, Donner P, Schleuning WD. Expression of active recombinant pallidipin, a novel platelet aggregation inhibitor, in the periplasm of Escherichia coli. Biochem J. 1995;307:465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noeske-Jungblut C, Krätzschmar J, Haendler B, Alagon A, Possani L, Verhallen P, et al. An inhibitor of collagen-induced platelet aggregation from the saliva of Triatoma pallidipennis. J Biol Chem. 1994;269(7):5050–3. [PubMed] [Google Scholar]
- 53.Ma D, Assumpcao TC, Li Y, Andersen JF, Ribeiro J, Francischetti IM. Triplatin, a platelet aggregation inhibitor from the salivary gland of the triatomine vector of Chagas disease, binds to TXA(2) but does not interact with glycoprotein PVI. Thrombosis and haemostasis. 2012;107(1):111–23. doi: 10.1160/TH11-10-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morita A, Isawa H, Orito Y, Iwanaga S, Chinzei Y, Yuda M. Identification and characterization of a collagen-induced platelet aggregation inhibitor, triplatin, from salivary glands of the assassin bug, Triatoma infestans. The FEBS journal. 2006;273(13):2955–62. doi: 10.1111/j.1742-4658.2006.05306.x [DOI] [PubMed] [Google Scholar]
- 55.Assumpcao TC, Alvarenga PH, Ribeiro JM, Andersen JF, Francischetti IM. Dipetalodipin, a novel multifunctional salivary lipocalin that inhibits platelet aggregation, vasoconstriction, and angiogenesis through unique binding specificity for TXA2, PGF2alpha, and 15(S)-HETE. J Biol Chem. 2010;285(50):39001–12. doi: 10.1074/jbc.M110.152835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isawa H, Orito Y, Jingushi N, Iwanaga S, Morita A, Chinzei Y, et al. Identification and characterization of plasma kallikrein-kinin system inhibitors from salivary glands of the blood-sucking insect Triatoma infestans. The FEBS journal. 2007;274(16):4271–86. doi: 10.1111/j.1742-4658.2007.05958.x [DOI] [PubMed] [Google Scholar]
- 57.Francischetti IM, Andersen JF, Ribeiro JM. Biochemical and functional characterization of recombinant Rhodnius prolixus platelet aggregation inhibitor 1 as a novel lipocalin with high affinity for adenosine diphosphate and other adenine nucleotides. Biochemistry. 2002;41(11):3810–8. [DOI] [PubMed] [Google Scholar]
- 58.Francischetti IM, Ribeiro JM, Champagne D, Andersen J. Purification, cloning, expression, and mechanism of action of a novel platelet aggregation inhibitor from the salivary gland of the blood- sucking bug, Rhodnius prolixus. J Biol Chem. 2000;275(17):12639–50. [DOI] [PubMed] [Google Scholar]
- 59.Noeske-Jungblut C, Haendler B, Donner P, Alagon A, Possani L, Schleuning WD. Triabin, a highly potent exosite inhibitor of thrombin. The Journal of biological chemistry. 1995;270(48):28629–34. [DOI] [PubMed] [Google Scholar]
- 60.Paddock CD, McKerrow JH, Hansell E, Foreman KW, Hsieh I, Marshall N. Identification, cloning, and recombinant expression of procalin, a major triatomine allergen. J Immunol. 2001;167(5):2694–9. [DOI] [PubMed] [Google Scholar]
- 61.Xu X, Chang BW, Mans BJ, Ribeiro JM, Andersen JF. Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi. Acta crystallographica. 2013;69(Pt 1):105–13. doi: 10.1107/S0907444912043326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersen JF, Francischetti IM, Valenzuela JG, Schuck P, Ribeiro JM. Inhibition of hemostasis by a high affinity biogenic amine-binding protein from the saliva of a blood-feeding insect. J Biol Chem. 2003;278(7):4611–7. doi: 10.1074/jbc.M211438200 [DOI] [PubMed] [Google Scholar]
- 63.Ribeiro JMC, Arca B. From sialomes to the sialoverse: An insight into the salivary potion of blood feeding insects. Adv Insect Physiol. 2009;37:59–118. [Google Scholar]
- 64.Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5'-nucleotidase family. Proc Natl Acad Sci U S A. 1995;92(3):694–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faudry E, Lozzi SP, Santana JM, D'Souza-Ault M, Kieffer S, Felix CR, et al. Triatoma infestans apyrases belong to the 5'-nucleotidase family. J Biol Chem. 2004;279(19):19607–13. doi: 10.1074/jbc.M401681200 [DOI] [PubMed] [Google Scholar]
- 66.Valenzuela JG, Charlab R, Galperin MY, Ribeiro JM. Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius. A new type of nucleotide-binding enzyme. J Biol Chem. 1998;273(46):30583–90. [DOI] [PubMed] [Google Scholar]
- 67.Valenzuela JG, Belkaid Y, Rowton E, Ribeiro JM. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J Exp Biol. 2001;204(Pt 2):229–37. [DOI] [PubMed] [Google Scholar]
- 68.Mesquita RD, Vionette-Amaral RJ, Lowenberger C, Rivera-Pomar R, Monteiro FA, Minx P, et al. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(48):14936–41. doi: 10.1073/pnas.1506226112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. Journal of molecular biology. 1999;292(3):741–58. doi: 10.1006/jmbi.1999.3069 [DOI] [PubMed] [Google Scholar]
- 70.Hypsa V, Tietz DF, Zrzavy J, Rego RO, Galvao C, Jurberg J. Phylogeny and biogeography of Triatominae (Hemiptera: Reduviidae): molecular evidence of a New World origin of the Asiatic clade. Molecular phylogenetics and evolution. 2002;23(3):447–57. [DOI] [PubMed] [Google Scholar]
- 71.Andersen JF, Ribeiro JM. A secreted salivary inositol polyphosphate 5-phosphatase from a blood-feeding insect: allosteric activation by soluble phosphoinositides and phosphatidylserine. Biochemistry. 2006;45(17):5450–7. doi: 10.1021/bi052444j [DOI] [PubMed] [Google Scholar]
- 72.Andersen JF, Ribeiro JM. A secreted salivary inositol polyphosphate 5-phosphatase from a bloodfeeding insect: allosteric activation by soluble phosphoinositides and phosphatidylserine. Biochemistry. 2006;45:5450–7. doi: 10.1021/bi052444j [DOI] [PubMed] [Google Scholar]
- 73.Ma D, Wang Y, Yang H, Wu J, An S, Gao L, et al. Anti-thrombosis repertoire of blood-feeding horsefly salivary glands. Mol Cell Proteomics. 2009;8(9):2071–9. doi: 10.1074/mcp.M900186-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diaz-Albiter HM, Ferreira TN, Costa SG, Rivas GB, Gumiel M, Cavalcante DR, et al. Everybody loves sugar: first report of plant feeding in triatomines. Parasit Vectors. 2016;9:114 doi: 10.1186/s13071-016-1401-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30(43):10363–70. [DOI] [PubMed] [Google Scholar]
- 76.Friedrich T, Kroger B, Bialojan S, Lemaire HG, Hoffken HW, Reuschenbach P, et al. A Kazal-type inhibitor with thrombin specificity from Rhodnius prolixus. J Biol Chem. 1993;268(22):16216–22. [PubMed] [Google Scholar]
- 77.Mende K, Petoukhova O, Koulitchkova V, Schaub GA, Lange U, Kaufmann R, et al. Dipetalogastin, a potent thrombin inhibitor from the blood-sucking insect. Dipetalogaster maximus cDNA cloning, expression and characterization. European journal of biochemistry / FEBS. 1999;266(2):583–90. [DOI] [PubMed] [Google Scholar]
- 78.Araujo RN, Campos IT, Tanaka AS, Santos A, Gontijo NF, Lehane MJ, et al. Brasiliensin: A novel intestinal thrombin inhibitor from Triatoma brasiliensis (Hemiptera: Reduviidae) with an important role in blood intake. International journal for parasitology. 2007;37(12):1351–8. doi: 10.1016/j.ijpara.2007.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lovato DV, Nicolau de Campos IT, Amino R, Tanaka AS. The full-length cDNA of anticoagulant protein infestin revealed a novel releasable Kazal domain, a neutrophil elastase inhibitor lacking anticoagulant activity. Biochimie. 2006;88(6):673–81. doi: 10.1016/j.biochi.2005.11.011 [DOI] [PubMed] [Google Scholar]
- 80.Campos IT, Tanaka-Azevedo AM, Tanaka AS. Identification and characterization of a novel factor XIIa inhibitor in the hematophagous insect, Triatoma infestans (Hemiptera: Reduviidae). FEBS letters. 2004;577(3):512–6. doi: 10.1016/j.febslet.2004.10.052 [DOI] [PubMed] [Google Scholar]
- 81.Campos IT, Amino R, Sampaio CA, Auerswald EA, Friedrich T, Lemaire HG, et al. Infestin, a thrombin inhibitor presents in Triatoma infestans midgut, a Chagas' disease vector: gene cloning, expression and characterization of the inhibitor. Insect Biochem Mol Biol. 2002;32(9):991–7. [DOI] [PubMed] [Google Scholar]
- 82.Kolyadko VN, Lushchekina SV, Vuimo TA, Surov SS, Ovsepyan RA, Korneeva VA, et al. New Infestin-4 Mutants with Increased Selectivity against Factor XIIa. PLoS ONE. 2015;10(12):e0144940 doi: 10.1371/journal.pone.0144940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Z, Gao L, Shen C, Rong M, Yan X, Lai R. A potent anti-thrombosis peptide (vasotab TY) from horsefly salivary glands. Int J Biochem Cell Biol. 2014;54:83–8. doi: 10.1016/j.biocel.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 84.Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002;205(Pt 16):2429–51. [DOI] [PubMed] [Google Scholar]
- 85.Arca B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, et al. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect biochemistry and molecular biology. 2007;37(2):107–27. doi: 10.1016/j.ibmb.2006.10.007 [DOI] [PubMed] [Google Scholar]
- 86.Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. J Exp Biol. 2004;207(Pt 21):3717–29. doi: 10.1242/jeb.01185 [DOI] [PubMed] [Google Scholar]
- 87.Anderson JM, Oliveira F, Kamhawi S, Mans BJ, Reynoso D, Seitz AE, et al. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC genomics. 2006;7:52 doi: 10.1186/1471-2164-7-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kato H, Anderson JM, Kamhawi S, Oliveira F, Lawyer PG, Pham VM, et al. High degree of conservancy among secreted salivary gland proteins from two geographically distant Phlebotomus duboscqi sandflies populations (Mali and Kenya). BMC genomics. 2006;7:226 doi: 10.1186/1471-2164-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santos A, Ribeiro JM, Lehane MJ, Gontijo NF, Veloso AB, Sant'Anna MR, et al. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae). Insect biochemistry and molecular biology. 2007;37(7):702–12. doi: 10.1016/j.ibmb.2007.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kato H, Jochim RC, Gomez EA, Sakoda R, Iwata H, Valenzuela JG, et al. A repertoire of the dominant transcripts from the salivary glands of the blood-sucking bug, Triatoma dimidiata, a vector of Chagas disease. Infect Genet Evol. 2010;10(2):184–91. doi: 10.1016/j.meegid.2009.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Santiago PB, Assumpcao TC, de Araujo CN, Bastos IM, Neves D, da Silva IG, et al. A Deep Insight into the Sialome of Rhodnius neglectus, a Vector of Chagas Disease. PLoS neglected tropical diseases. 2016;10(4):e0004581 doi: 10.1371/journal.pntd.0004581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamazaki Y, Morita T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 2004;44(3):227–31. doi: 10.1016/j.toxicon.2004.05.023 [DOI] [PubMed] [Google Scholar]
- 93.Assumpcao TC, Ma D, Schwarz A, Reiter K, Santana JM, Andersen JF, et al. Salivary Antigen-5/CAP family members are Cu2+-dependent antioxidant enzymes that scavenge O2- and inhibit collagen-induced platelet aggregation and neutrophil oxidative burst. The Journal of biological chemistry. 2013;288(20):14341–61. doi: 10.1074/jbc.M113.466995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Raj PA, Dentino AR. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol Lett. 2002;206(1):9–18. [DOI] [PubMed] [Google Scholar]
- 95.Rawal R, Vijay S, Kadian K, Singh J, Pande V, Sharma A. Towards a Proteomic Catalogue and Differential Annotation of Salivary Gland Proteins in Blood Fed Malaria Vector Anopheles culicifacies by Mass Spectrometry. PLoS One. 2016;11(9):e0161870 doi: 10.1371/journal.pone.0161870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Das S, Dimopoulos G. Molecular analysis of photic inhibition of blood-feeding Anopheles gambiae. BMC Physiology. 2008;8(23):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu J, Busche JM, Zhang X. Identification of juvenile hormone target genes in the adult female mosquitoes. Insect Biochem Mol Biol. 2010;40(1):23–9. doi: 10.1016/j.ibmb.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 98.Amsalem E, Malka O, Grozinger C, Hefetz A. Exploring the role of juvenile hormone and vitellogenin in reproduction and social behavior in bumble bees. BMC Evol Biol. 2014;14(1):45 doi: 10.1186/1471-2148-14-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sousa CS, Serrão JE, Bonetti AM, Amaral ImR, Kerr WE, Maranhão AQ, et al. Insights into the Melipona scutellaris (Hymenoptera, Apidae, Meliponini) fat body transcriptome. Genetics and Molecular Biology. 2013;36(2):292–7. doi: 10.1590/S1415-47572013000200022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Price DP, Nagarajan V, Churbanov A, Houde P, Milligan B, Drake LL, et al. The fat body transcriptomes of the yellow fever mosquito Aedes aegypti, pre- and post- blood meal. PLoS One. 2011;6(7):e22573 doi: 10.1371/journal.pone.0022573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Raikhel AS, Dhadialla TS. Accumulation of Yolk Proteins in Insect Oocytes. Annu Rev Entomol 1992;37:217–51. doi: 10.1146/annurev.en.37.010192.001245 [DOI] [PubMed] [Google Scholar]
- 102.Ursic-Bedoya RJ, Lowenberger CA. Rhodnius prolixus: identification of immune-related genes up-regulated in response to pathogens and parasites using suppressive subtractive hybridization. Dev Comp Immunol. 2007;31(2):109–20. doi: 10.1016/j.dci.2006.05.008 [DOI] [PubMed] [Google Scholar]
- 103.Yoshiga T, Hernandez VP, Fallon AM, Law JH. Mosquito transferrin, an acute-phase protein that is up-regulated upon infection. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(23):12337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garrick MD, Garrick LM. Cellular iron transport. Biochim Biophys Acta. 2009;1790(5):309–25. doi: 10.1016/j.bbagen.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 105.Muscio OA, La Torre JL, Scodeller EA. Characterization of Triatoma virus, a picorna-like virus isolated from the triatomine bug Triatoma infestans. The Journal of general virology. 1988;69 (Pt 11):2929–34. [DOI] [PubMed] [Google Scholar]
- 106.Susevich ML, Marti GA, Serena MS, Echeverria MG. New Triatoma virus hosts in wild habitats of Argentina. J Invertebr Pathol. 2012;110(3):405–7. doi: 10.1016/j.jip.2012.03.023 [DOI] [PubMed] [Google Scholar]
- 107.Marti GA, Echeverria MG, Susevich ML, Ceccarelli S, Balsalobre A, Rabinovich JE, et al. Exploration for Triatoma virus (TrV) infection in laboratory-reared triatomines of Latin America: a collaborative study. Int J Trop Insect Sc. 2013;33(4):294–304. [Google Scholar]
- 108.Querido JF, Agirre J, Marti GA, Guerin DM, Silva MS. Inoculation of Triatoma virus (Dicistroviridae: Cripavirus) elicits a non-infective immune response in mice. Parasit Vectors. 2013;6:66 doi: 10.1186/1756-3305-6-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Querido JF, Echeverria MG, Marti GA, Costa RM, Susevich ML, Rabinovich JE, et al. Seroprevalence of Triatoma virus (Dicistroviridae: Cripaviridae) antibodies in Chagas disease patients. Parasit Vectors. 2015;8:29 doi: 10.1186/s13071-015-0632-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Friend WG, Smith JJ. Feeding in Rhodnius prolixus: mouthpart activity and salivation, and their correlation with chanees of electrical resistance. J Insect Physiol. 1971;17(2):233–43. [DOI] [PubMed] [Google Scholar]
- 111.Soares AC, Carvalho-Tavares J, Gontijo Nde F, dos Santos VC, Teixeira MM, Pereira MH. Salivation pattern of Rhodnius prolixus (Reduviidae; Triatominae) in mouse skin. J Insect Physiol. 2006;52(5):468–72. doi: 10.1016/j.jinsphys.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 112.Leitner WW, Wali T, Kincaid R, Costero-Saint Denis A. Arthropod Vectors and Disease Transmission: Translational Aspects. PLoS neglected tropical diseases. 2015;9(11):e0004107 doi: 10.1371/journal.pntd.0004107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
This project was registered with the National Center for Biotechnology Information (NCBI) under Bioproject PRJNA395235 and BioSamples SAMN07374128 and SAMN07374129. The raw reads are available on the Sequence Read Archive (SRA) under accessions SRR5859113 and SRR5859114. The coding sequences and their protein products (numbering 9,013) were submitted to the Transcriptome Shotgun Assembly (TSA) project which were deposited at DDBJ/EMBL/GenBank under the accession GFTR00000000. The version described in this paper is the first version, GFTR01000000.