ABSTRACT
The 7SK small nuclear RNA is a multifunctional transcriptional regulatory RNA that controls the nuclear activity of the positive transcription elongation factor b (P-TEFb), specifically targets P-TEFb to the promoter regions of selected protein-coding genes and promotes transcription of RNA polymerase II-specific spliceosomal small nuclear RNA genes.
KEYWORDS: 7SK snRNA, 7SK snRNP, P-TEFb, RNA polymerase II pausing, transcription regulation
Introduction
Human 7SK RNA is an abundant 331-nucleotide-long, evolutionarily conserved small nuclear RNA (snRNA) that, in the form of 7SK small nuclear RNP (snRNP), regulates RNA polymerase II (RNAPII) transcription through controlling the activity of a major transcription elongation factor, P-TEFb. The 7SK snRNA is an RNAPIII transcript that undergoes post-transcriptional modifications. The gamma phosphate of the 5′-terminal triphosphate of the nascent 7SK RNA is methylated by the methyl phosphate capping enzyme (MePCE) and the U250 residue is converted into pseudouridine.1 The mature 7SK snRNA folds into 4 major hairpin domains which provide docking sites for 7SK snRNP core proteins and several other transiently interacting 7SK snRNP proteins. The classical 7SK structure has been proposed to undergo conformational rearrangements to support transient association with various 7SK snRNP proteins.2-4
The 7SK snRNA is incorporated into the functionally active 7SK core snRNP that also contains the capping enzyme MePCE and the La-related protein 7 (Larp7).3,5 While MePCE binds to the 5′-terminal region of the mature 7SK snRNA, Larp7 binds to its 3′-terminal hairpin and the following oligouridylate tail. Both MePCE and Larp7 are fundamental to the accumulation and stability of 7SK snRNA. Besides binding to 7SK, MePCE and Larp7 also interact with each other to form the metabolically stable 7SK core snRNP.
The 7SK core snRNP provides a scaffold for dynamic assembly of structurally and functionally distinct larger 7SK snRNP particles. The most abundant and most extensively characterized 7SK particle contains the 7SK core snRNP, a hetero- or homodimer of HEXIM1/2 and the RNAPII transcription elongation factor, P-TEFb. P-TEFb is a protein kinase composed of cyclin-dependent kinase 9 (Cdk9) and its regulatory cyclin partner CycT1 or T2. The 7SK/HEXIM/P-TEFb snRNP functions as a negative transcriptional regulatory RNP in which the kinase activity of P-TEFb is inhibited.6 In response to various transcriptional stimuli, however, active P-TEFb can be rapidly mobilized from the transcriptionally inactive 7SK/HEXIM/P-TEFb. According to a recently emerging view, instead of simply serving as a nucleoplasmic P-TEFb reservoir, the 7SK/HEXIM/P-TEFb snRNP can directly target inactive P-TEFb to the promoter regions of protein-coding genes to promote “on site” P-TEFb mobilization.5
Recently, 2 novel, functionally distinct 7SK-containing transcriptional regulatory complexes have been identified.7,8 No evidence supports the presence of P-TEFb in these new complexes, suggesting that 7SK can also regulate RNAPII transcription through P-TEFb-independent mechanisms. Here, we discuss the recent advances on the well-established P-TEFb-dependent and the newly reported P-TEFb-independent mechanisms of transcriptional regulation by 7SK snRNP.
7SK is a master regulator of nuclear P-TEFb activity
Since P-TEFb is a major regulator of RNAPII transcription, controlling its nuclear activity is of particular importance. P-TEFb controls the conversion of transcriptionally engaged but promoter proximally paused RNAPII into elongation-competent polymerase.5,6 Shortly after transcription initiation, the early elongating RNAPII is arrested by binding of negative transcription elongation factors, NELF and DSIF. Through phosphorylation of NELF, DSIF and the C-terminal domain (CTD) of RNAPIl at serine 2, P-TEFb allows RNAPII elongation to resume. During the past decade, P-TEFb-mediated release of paused RNAPII has been recognized as a major pervasive checkpoint of RNAPII transcription that controls expression of most RNAPII-transcribed genes with special importance in controlling stimulus-induced and developmentally-regulated genes. The nuclear activity of P-TEFb is regulated mainly by the 7SK snRNP and HEXIM1/2. Mechanistically, a hetero- or homo-dimer of HEXIM1/2 binds to conserved motifs of the long 5′-terminal hairpin of 7SK snRNA. Docking of 7SK triggers a conformational change that unveils the otherwise inaccessible P-TEFb binding surface of HEXIM proteins. The 7SK-associated “activated” HEXIM proteins bind to both subunits of P-TEFb and mask the catalytic site of Cdk9.3,6,9
The active and inactive forms of P-TEFb coexist in the nucleus (Fig. 1). While the active pool of P-TEFb seems to be sufficient to support basal transcription, P-TEFb rapidly mobilized from the inactive 7SK/HEXIM/P-TEFb reservoir can reinforce the increased transcriptional demand of the cell. In response to cellular stress or transcriptional blockade, the 7SK/HEXIM/P-TEFb RNP rapidly disassembles and releases ready-to-use P-TEFb, whereas the free 7SK core snRNP associates with a set of abundant heterogeneous ribonucleoproteins (hnRNPs) A1, A2/B1, R, Q and RNA helicase A.10,11 The hnRNP proteins presumably ‘capture’ the newly released 7SK snRNP to prevent its re-association with HEXIM and P-TEFb. Recent RNA structural analysis revealed that stress-dependent remodeling of the 7SK snRNP is accompanied by structural rearrangements of the 7SK snRNA.4 In the 7SK/HEXIM/P-TEFb snRNP, the 5′-terminal region of 7SK snRNA folds into the classical extended hairpin structure that supports HEXIM and P-TEFb binding. Upon 7SK/HEXIM/P-TEFb disassembly, 7SK adopts a closed conformation in which the 5′- and 3′-terminal domains are folded close to each other. RNA helicase A may promote the structural rearrangements of 7SK and the associated hnRNP proteins may stabilize the closed conformation of 7SK snRNA that prevents HEXIM and P-TEFb binding.11
Figure 1.
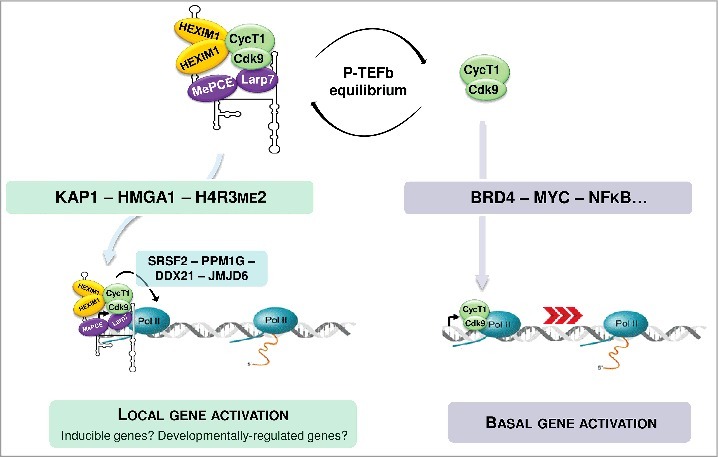
Both active and inactive forms of P-TEFb are recruited to their target gene promoters through association with specific transcription factors.
Diverse stimuli and signaling cascades, including the PI3K/Akt and PP1α/PP2B pathways, have been shown to induce 7SK/HEXIM/P-TEFb dissociation through addition or erasure of specific post-translational modifications on the components of the 7SK/HEXIM/P-TEFb snRNP.5 Whether the released P-TEFb stimulates transcription of different sets of genes depending on the activated cellular pathway remains unknown. Once released from 7SK/HEXIM/P-TEFb, the free P-TEFb has to find its way to the target genes. A number of transcription factors and co-activators, including NF-κB, c-Myc, CIITA, p53, the bromodomain protein 4 (Brd4), the super elongation complexes (SECs) and the human immunodeficiency virus (HIV) transactivator Tat can directly bind and target P-TEFb to paused RNAPII.5,6 Brd4 seems to be the major nuclear binding partner of active P-TEFb that directs it to the promoters of many primary response genes through interacting with acetylated chromatin.12 Releasing of P-TEFb from the 7SK/HEXIM/P-TEFb snRNP concomitantly augments Brd4/P-TEFb level, suggesting that Brd4 directly captures 7SK-evicted P-TEFb for transcription stimulation.13 More recently, P-TEFb has been identified as a constitutive component of SECs which are composed of the ELL, AFF and ENL/AF9 proteins.14 SECs have been proposed to deliver and tether P-TEFb to promoters of protein-coding genes through association with Mediator and Integrator complexes.15-17 It remains to be elucidated how P-TEFb is distributed among Brd4, SECs and other transcription factors under normal and stress conditions. Moreover, recent studies demonstrated that the 7SK-associated inactive form of P-TEFb can also be targeted to genes, adding a new layer to this intricate regulatory network.
The 7SK/HEXIM/P-TEFb snRNP is targeted to the chromatin
The nuclear 7SK snRNP, mostly in the form of 7SK/HEXIM/P-TEFb, accumulates mainly in the soluble fraction of the nucleoplasm. Cell fractionation experiments, however, also detected a significant fraction of 7SK snRNA being associated with chromatin.18 This raised the possibility that the 7SK/HEXIM/P-TEFb negative transcriptional regulatory snRNP may directly interact with P-TEFb-dependent genes. The first support to this idea was provided by detection of HEXIM1, Cdk9 and Larp7 on the HIV long-terminal repeat (LTR) promoter.19 It was proposed that tethering 7SK/HEXIM/P-TEFb to the HIV LTR promoter can promote RNAPII transcription activation through “on site and on time” release of active P-TEFb. Chromatin-proximal P-TEFb activation could rapidly increase P-TEFb concentration at the promoter regions of target genes to facilitate P-TEFb capturing by gene-specific transcription factors. Next, dissociation of chromatin-bound 7SK/HEXIM/P-TEFb in response to specific cellular stimuli was proposed to induce rapid transcriptional activation of primary response genes which are heavily loaded with promoter proximally paused RNAPII.20 More recently, genome-wide studies suggested that chromatin-tethering of 7SK-repressed P-TEFb is a common mechanism controlling transcription elongation on most protein-coding genes.21,22 Chromatin immunoprecipitation and deep sequencing (ChIP-seq) experiments detected Larp7, Cdk9 and HEXIM1 co-enrichments at the promoter regions of more than 15,000 human genes.22 Likewise, chromatin isolation by RNA purification followed by deep sequencing (ChIRP-seq) analysis detected 7SK snRNA occupancy on the transcribed regions of a large set of human genes, largely resembling RNAPII distribution.8 Widespread 7SK/HEXIM/P-TEFb anchoring to chromatin was proposed to be mediated by the Kruppel-associated box-interacting protein KAP1 that has been shown to facilitate both P-TEFb recruitment and RNAPII elongation on early-response genes upon stimulation. Moreover, KAP1 shows significant co-occupancy with components of the 7SK snRNP on most protein-coding genes controlled by RNAPII pausing.22 Other mechanisms targeting 7SK/HEXIM/P-TEFb to various subsets of RNAPII-transcribed genes may also exist. The CTIP2 transcriptional repressor can directly interact with both HEXIM1 and 7SK snRNA to position 7SK/HEXIM/P-TEFb on a specific set of human promoters through forming a direct interaction with HMGA1.23 Finally, the 7SK snRNA has been reported to directly bind to a specific chromatin signature, the repressive H4R3me2 mark, at a class of enhancers also occupied by Brd4 and JMJD6.24 This interaction could tether the 7SK/HEXIM/P-TEFb snRNP to specific enhancers and allow local P-TEFb activation upon removal of the H4R3me2 mark by the JMJD6 demethylase. On the HIV LTR promoter and primary response genes, P-TEFb release from 7SK/HEXIM/P-TEFb is triggered by recruitment of the PPM1G phosphatase that disassembles the 7SK/HEXIM/P-TEFb snRNP through dephosphorylation of the Cdk9 T-loop at Thr186. In this case, release of RNAPII pausing would require de novo re-phosphorylation of the Cdk9 T-loop, since this phosphorylation is absolutely required for Cdk9 kinase activity. RNA binding proteins have been also implicated in chromatin-associated P-TEFb activation. SRSF2, a serine/arginine-rich splicing factor, was found to be associated with 7SK/HEXIM/P-TEFb on several target promoters.21 Emergence of SRSF2 binding sites on the nascent RNA transcripts captures SRSF2 and promotes 7SK/HEXIM/P-TEFb dissociation to trigger RNAPII pause release. The DDX21 DEAD-box RNA helicase has been also proposed to stimulate RNAPII transcription through promoting ‘on site’ disassembly of chromatin-associated 7SK/HEXIM/P-TEFb through remodeling of the 7SK secondary structure by its helicase activity.25
The above studies have described multiple mechanisms to position and disassemble the 7SK/HEXIM/P-TEFb snRNP directly on the chromatin encompassing protein genes. Now, a major question of the field is how widespread these regulatory mechanisms are? A great fraction of P-TEFb exists in its active free form in the nucleoplasm, and some transcription factors, including Brd4 and c-myc, can directly recruit and use active P-TEFb to stimulate global transcription elongation by RNAPII (Fig. 1). It is unclear why other transcription activators cannot directly capture and utilize transcriptionally active nucleoplasmic P-TEFb. While coordinated ‘on site’ P-TEFb activation can represent an apparent advantage for orchestrated activation of stimulus-inducible and developmentally-regulated genes, the benefit of such P-TEFb activation strategy is less obvious for ubiquitously-expressed genes. Nevertheless, if 7SK/HEXIM/P-TEFb delivers P-TEFb to a great fraction of genomic promoters, the loss of 7SK snRNP should lead to important transcriptional defects on RNAPII-transcribed genes. However, depletion of 7SK snRNP by RNA interference had only a limited impact on the expression of a subset of P-TEFb-dependent genes.7 Another genome-wide study found that 7SK depletion in mouse embryonic stem cells alters expression of less than 500 genes which are mainly upregulated, pointing to a gene-specific repressor function of 7SK.26 Further investigations are therefore required to determine whether anchoring 7SK/HEXIM/P-TEFb to chromatin is a pre-requisite for basal transcription by RNAPII or it regulates only specific cohorts of genes.
7SK snRNA regulates RNAPII transcription in a P-TEFb-independent manner
In addition to protein-coding genes, RNAPII also transcribes genes encoding small nuclear (sn) and small nucleolar (sno) RNAs. The RNAPII-specific snRNA and snoRNA genes possess specific structural and functional features distinct from protein-coding genes.27 They display specific promoter and 3′ end processing elements which are recognized by snRNA gene-specific factors. After transcription initiation on sn/snoRNA genes, RNAPII elongation is not interrupted by promoter-proximal pausing and does not require P-TEFb activity.28 We have recently determined the genome-wide chromatin occupancy of the 7SK snRNP protein Larp7 in human HeLa cells.7 We detected marked Larp7 enrichments on RNAPII-transcribed sn/snoRNA genes. Other components of the 7SK core snRNP, MePCE and 7SK snRNA, were also detected on U2 snRNA genes, but not HEXIM1. We found that the 7SK snRNP interacts with an sn/snoRNA gene-specific transcription factor, the Little Elongation Complex (LEC) composed of Ice1, Ice2, ELL and ZC3H8.29 LEC promotes RNAPII recruitment to the promoter of sn/snoRNA genes, and stimulates sn/snoRNA transcription through traveling with the elongating polymerase.29,30
The 7SK/LEC complex is devoid of P-TEFb, indicating that it is structurally and functionally distinct from the canonical 7SK/HEXIM/P-TEFb transcriptional regulatory snRNP (Fig. 2). Depletion of 7SK snRNP decreased both LEC and RNAPII occupancy on snRNA genes and reduced sn/snoRNA synthesis, but had no significant effect on the expression of protein-coding genes including P-TEFb-dependent ones. Interestingly, depletion of the LEC scaffold protein Ice1 resulted in similar transcriptional defects, since it also compromised RNAPII recruitment to and transcription of sn/snoRNA genes.7,30 One intriguing possibility is that on the analogy of directing 7SK/HEXIM/P-TEFb assembly by HEXIM bound to 7SK, binding of Ice1 to the 7SK snRNA is a prerequisite for 7SK/LEC assembly, although Ice1 contains no obvious RNA-binding domain. The mechanism targeting the 7SK/LEC snRNP to sn/snoRNA genes remains elusive. Most likely, the pre-initiation complex formed on the RNAPII-specific sn/snoRNA gene promoters specifically interacts with 7SK/LEC. Lending support to this idea, the 7SK snRNP preferentially interacts with RNAPII hyperphosphorylated at Ser5 and Ser7 that is a hallmark of RNAPII engaged in snRNA gene transcription. For instance, the snRNA-specific transcription factor SNAPc could be a possible candidate for directing 7SK/LEC recruitment to sn/snoRNA genes. The detected interaction between the LEC and MED26 suggests that the Mediator Complex might also participate in LEC recruitment, although MED26 is not specific to snRNA gene promoters.31
Figure 2.
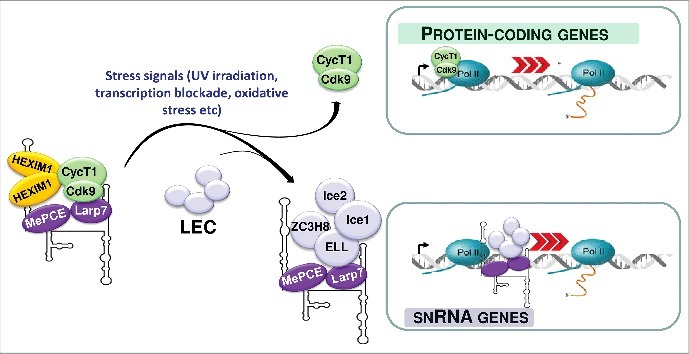
The 7SK/HEXIM/P-TEFb snRNP controls transcription of both protein-coding and RNAPII-specific spliceosomal snRNA genes. In response to transcriptional stress, P-TEFb and 7SK core snRNP are released from the 7SK/HEXIM/P-TEFb snRNP. While P-TEFb stimulates transcription of protein-coding genes through releasing promoter-proximal paused RNAPII, a fraction of the free 7SK core snRNP associates with the Little Elongation Complex (LEC) to facilitate snRNA genes transcription.
We observed strong Larp7 accumulation along the entire sn/snoRNA genes that likely reflects stable 7SK/LEC association with the transcribing RNAPII. Supporting this notion, highly similar distribution profiles were obtained for Larp7, RNAPII and LEC on sn/snoRNA genes. In contrast to RNAPII-specific sn/snoRNA genes, we failed to detect significant Larp7 association with most protein-coding genes,7 suggesting that 7SK/HEXIM/P-TEFb association with protein-coding genes is highly transient and that fast P-TEFb activation on the chromatin may result in quick 7SK snRNP release.
Importantly, 7SK association with LEC is largely enhanced under stress conditions, such as transcriptional blockade or UV irradiation that are known to trigger P-TEFb eviction from 7SK/HEXIM/P-TEFb.7 Stress-induced dissociation of 7SK/HEXIM/P-TEFb might transiently increase the cellular level of free 7SK snRNP available for 7SK/LEC formation. Thus, 7SK/HEXIM/P-TEFb dissociation can simultaneously augment accumulation of active P-TEFb and 7SK/LEC snRNP to synchronously boost transcription of protein-coding and snRNA genes, respectively (Fig. 2). This means that the 7SK/HEXIM/P-TEFb snRNP could be considered as a bifunctional transcriptional regulatory RNP that can coordinate pre-mRNA transcription with spliceosomal snRNA expression.
Recently, the 7SK snRNA has been also detected at active enhancers and super-enhancers that are genomic regions encompassing clustered enhancer elements controlling gene expression networks.8 At super-enhancers, neither HEXIM1 nor P-TEFb were detected, supporting the idea that the 7SK snRNP can impact RNAPII transcription independently from P-TEFb regulation. The 7SK snRNA was shown to repress RNAPII-mediated transcription of enhancer RNAs (eRNAs) through facilitating recruitment of the BAF chromatin-remodeling complex, a known repressor of eRNA transcription. 7SK/BAF-promoted reshaping of chromatin structure at super-enhancers would restrict convergent transcription and prevent DNA damage. Of note, while 7SK/HEXIM/P-TEFb incorporates a large fraction of cellular 7SK snRNP, the 7SK/LEC and 7SK/BAF snRNPs are much less abundant in human cells. With development of sensitive RNA-based purification techniques such as ChIRP-MS, one can predict that additional low-abundance 7SK-containing complexes will be uncovered in the future.
Conclusion
Distinct 7SK core snRNP-containing particles associate with the chromatin and regulate multiple aspects of nuclear RNAPII transcription. The 7SK/BAF axis controls production of eRNAs, the 7SK/HEXIM/P-TEFb snRNP regulates RNAPII pause release and/or elongation on protein-coding genes and finally, the 7SK/LEC complex promotes transcription of sn/snoRNA genes. Moreover, recent findings suggest that 7SK also participates in proper transcription termination on a subset of genes.26 Therefore, the 7SK snRNP plays a complex role in orchestration of global RNAPII progression along the nuclear genome. Future efforts will be required to decipher the precise mechanisms by which 7SK is able to fine-tune RNAPII transcriptional activity.
Funding Statement
Our work was supported by grants from la Fondation ARC pour la recherche sur le cancer (to SE), l'Agence Nationale de la Recherche (to T.K.).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are indebted to our laboratory members P. Vitali and B. Jády for helpful discussions.
References
- [1].Zhao Y, Karijolich J, Glaunsinger B, Zhou Q. Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV-1 transcription and escape from latency. EMBO Rep 2016; 17:1441-51; PMID:27558685; http://doi.org/10.15252/embr.201642682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol 2009; 26:2821-30; PMID:19734296; http://doi.org/10.1093/molbev/msp198 [DOI] [PubMed] [Google Scholar]
- [3].Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev 2012; 3:92-103; PMID:21853533; http://doi.org/10.1002/wrna.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brogie JE, Price DH. Reconstitution of a functional 7SK snRNP. Nucleic Acids Res 2017; 45:6864-6880; PMID:28431135; http://doi.org/10.1093/nar/gkx262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Quaresma AJ, Bugai A, Barboric M. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res 2016; 44:7527-39; PMID:27369380; http://doi.org/10.1093/nar/gkw585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhou Q, Li T, Price DH. RNA Polymerase II Elongation Control. Annu Rev Biochem 2012; 81:119-43; PMID:22404626; http://doi.org/10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Egloff S, Vitali P, Tellier M, Raffel R, Murphy S, Kiss T. The 7SK snRNP associates with the little elongation complex to promote snRNA gene transcription. EMBO J 2017; 36:934-948; PMID:28254838; http://doi.org/10.15252/embj.201695740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Flynn RA, Do BT, Rubin AJ, Calo E, Lee B, Kuchelmeister H, Rale M, Chu C, Kool ET, Wysocka J, et al.. 7SK-BAF axis controls pervasive transcription at enhancers. Nat Struct Mol Biol 2016; 23:231-8; http://doi.org/10.1038/nsmb.3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kobbi L, Demey-Thomas E, Braye F, Proux F, Kolesnikova O, Vinh J, Poterszman A, Bensaude O. An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb. Proceedings of the National Academy of Sciences of the United States of America 2016; 113:12721-12726; PMID:27791144; http://doi.org/10.1073/pnas.1612331113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol 2007; 27:6996-7006; PMID:17709395; http://doi.org/10.1128/MCB.00975-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J 2007; 26:3570-80; PMID:17611602; http://doi.org/10.1038/sj.emboj.7601783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 2005; 19:523-34; PMID:16109376; http://doi.org/10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- [13].Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular Cell 2005; 19:535-45; PMID:16109377; http://doi.org/10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- [14].Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 2011; 25:661-72; http://doi.org/10.1101/gad.2015411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gardini A, Baillat D, Cesaroni M, Hu D, Marinis JM, Wagner EJ, Lazar MA, Shilatifard A, Shiekhattar R. Integrator regulates transcriptional initiation and pause release following activation. Mol Cell 2014; 56:128-39; PMID:25201415; http://doi.org/10.1016/j.molcel.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lu X, Zhu X, Li Y, Liu M, Yu B, Wang Y, Rao M, Yang H, Zhou K, Wang Y, et al.. Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res 2016; 44:6853-67; PMID:27353326; http://doi.org/10.1093/nar/gkw571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al.. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 2011; 146:92-104; PMID:21729782; http://doi.org/10.1016/j.cell.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res 2010; 20:899-907; PMID:20404130; http://doi.org/10.1101/gr.103473.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 2010; 17:815-21; http://doi.org/10.1038/nsmb.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McNamara RP, McCann JL, Gudipaty SA, D'Orso I. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep 2013; 5:1256-68; PMID:24316072; http://doi.org/10.1016/j.celrep.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR Proteins Collaborate with 7SK and Promoter-Associated Nascent RNA to Release Paused Polymerase. Cell 2013; 153:855-68; PMID:23663783; http://doi.org/10.1016/j.cell.2013.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].McNamara RP, Reeder JE, McMillan EA, Bacon CW, McCann JL, D'Orso I. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol Cell 2016; 61:39-53; PMID:26725010; http://doi.org/10.1016/j.molcel.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Eilebrecht S, Le Douce V, Riclet R, Targat B, Hallay H, Van Driessche B, Schwartz C, Robette G, Van Lint C, Rohr O, et al.. HMGA1 recruits CTIP2-repressed P-TEFb to the HIV-1 and cellular target promoters. Nucleic Acids Res 2014; 42:4962-71; PMID:24623795; http://doi.org/10.1093/nar/gku168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 2013; 155:1581-95; PMID:24360279; http://doi.org/10.1016/j.cell.2013.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 2015; 518:249-53; PMID:25470060; http://doi.org/10.1038/nature13923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Castelo-Branco G, Amaral PP, Engstrom PG, Robson SC, Marques SC, Bertone P, Kouzarides T. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol 2013; 14:R98; PMID:24044525; http://doi.org/10.1186/gb-2013-14-9-r98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Egloff S, O'Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem Soc Trans 2008; 36:590-4; PMID:18631122; http://doi.org/10.1042/BST0360590 [DOI] [PubMed] [Google Scholar]
- [28].Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J 2005; 24:4154-65; PMID:16308568; http://doi.org/10.1038/sj.emboj.7600876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, et al.. The little elongation complex regulates small nuclear RNA transcription. Mol Cell 2011; 44:954-65; PMID:22195968; http://doi.org/10.1016/j.molcel.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu D, Smith ER, Garruss AS, Mohaghegh N, Varberg JM, Lin C, Jackson J, Gao X, Saraf A, Florens L, et al.. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol Cell 2013; 51:493-505; PMID:23932780; http://doi.org/10.1016/j.molcel.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Takahashi H, Takigawa I, Watanabe M, Anwar D, Shibata M, Tomomori-Sato C, Sato S, Ranjan A, Seidel CW, Tsukiyama T, et al.. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat Commun 2015; 6:5941; PMID:25575120; http://doi.org/10.1038/ncomms6941 [DOI] [PMC free article] [PubMed] [Google Scholar]


