ABSTRACT
Recent large-scale RNA sequencing efforts have revealed the extensive diversity of mRNA molecules produced from most eukaryotic coding genes, which arises from the usage of alternative, cryptic or non-canonical splicing and intronic polyadenylation sites. The prevailing view regarding the tremendous diversity of coding gene transcripts is that mRNA processing is a flexible and more-or-less noisy process leading to a diversity of proteins on which natural selection can act depending on protein-mediated cellular functions. However, this concept raises two main questions. First, do alternative mRNA processing pathways have a role other than generating mRNA and protein diversity? Second, is the cellular function of mRNA variants restricted to the biogenesis of functional protein isoforms? Here, I propose that the co-transcriptional use of alternative mRNA processing sites allows first, the resolution of co-transcriptional biophysical constraints that may otherwise result in DNA instability, and second, increases the diversity of cellular functions of mRNAs in a manner that is not restricted to protein synthesis.
KEYWORDS: Transcription, RNA metabolism, Genomic instability
Introduction
Most eukaryotic coding genes produce different transcripts through alternative mRNA processing pathways resulting from the co-transcriptional usage of alternative or cryptic splicing and intronic polyadenylation sites (i.e., alternative RNA processing sites).1,2 The flexibility of using alternative mRNA processing sites increases mRNA and potentially protein diversity. However, it is tempting to speculate that some mRNAs are ‘error‘ or ‘junk’ and are the result of ‘noisy’ (inaccurate) biological processes when they lack canonical mRNA characteristics and may not give rise to proteins, are weakly expressed or rapidly degraded, or do not seem to have cellular functions.3–6 The prevailing view is that mRNA processing is a flexible and more-or-less noisy process leading to a diversity of proteins on which natural selection can act depending on the protein-mediated cellular functions. In this view, some mRNA processing sites are positively or negatively selected during evolution depending on the cellular functions of the resulting protein isoforms, while other mRNA processing sites would be neutral and result in the biogenesis of tolerated “junk” mRNAs. This view raises two main questions. First, do alternative mRNA processing pathways have a role other than generating mRNA and protein diversity? Second, is the cellular function of mRNA variants restricted to the biogenesis of functional protein isoforms?
In the first part of this manuscript, I propose that the diversity of co-transcriptional mRNA processing pathways themselves plays an important role in sustaining the gene expression flow and in protecting DNA from transcription-mediated damage independently of the functions of the generated gene products. If the act of co-transcriptional mRNA processing is per se playing an important role independently of the cellular function of the resulting gene products, does this means that some mRNA variants are merely by-products without any cellular functions? In the second part, I propose that the cellular functions of alternatively processed pre-mRNAs that are independent of their capacity to give rise stable functional proteins have been overlooked and that mRNAs are much more than passive intermediates of gene-to-protein information transfer. In conclusion, it will be proposed that the co-transcriptional use of alternative mRNA processing sites alleviates transcription-dependent DNA damage and reduces hazardous mutational processes (e.g., transcriptional-mediated DNA instability) within coding genes. Therefore, the co-transcriptional use of alternative mRNA processing sites reduces genetic variability and it simultaneously increases the functional diversity of the RNAs produced from coding genes.
Alternative co-transcriptional mRNA processing pathways and DNA stability
Co-transcriptional biophysical constraints and DNA damage
The synthesis of mRNA molecules creates biophysical constraints that can challenge the DNA and cellular integrity. First, transcription induces negative and positive DNA supercoilings behind and ahead, respectively, of the transcribing RNA polymerase II (RNAPII)7 (Fig. 1A). Positive DNA supercoilings as well as nucleosomes and DNA-binding proteins can create physical transcription and replication roadblocks that can lead to DNA instability (e.g., DNA breaks).7 Second, transcription can trigger the formation of R-loops that consist of the nascent pre-mRNA re-hybridizing to the DNA template and the displaced single-stranded DNA.8 These R-loops are a major source of DNA instability.8 The inherent capacity of nascent pre-mRNA to interact with a wide array of DNA- and chromatin-associated proteins9,10 may also create constraints to RNAPII. For example, the interaction between the nascent RNA and RNAPII can cause RNAPII pausing.11 Co-transcriptional inter-molecular RNA-RNA interactions can also be formed12 and RNA molecules and their associated proteins are prone to form potentially cellular toxic aggregates.13 Therefore, the synthesis of new mRNA molecules can challenge the DNA and cellular integrity in an RNA-dependent and -independent manner.7 While exosome-mediated co-transcriptional RNA degradation may eliminate some RNA-mediated physical constraints on DNA,14 it will be shown below that co-transcriptional mRNA processing contributes to sustain the gene expression flow and to maintain genomic stability.
Figure 1.
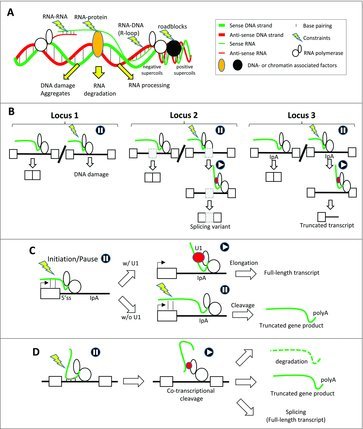
Co-transcriptional constraints trigger co-transcriptional mRNA processing, which resolves the constraints. (A) Transcription creates topological constraints, like positive supercoils that together with nucleosomes and DNA-associated factors (black circle) create transcription and replication roadblocks. The nascent RNA can interact with the DNA template (R-loops), DNA- and chromatin-associated proteins, or with other RNAs. Co-transcriptional physical constraints (yellow lightening) can induce DNA damage (e.g., DNA breaks) and aggregates or trigger RNA degradation or RNA processing. (B) Co-transcriptional physical constraints within an intron (yellow lightening in locus 1) can lead to DNA damage. When physical constraints occur in the vicinity of alternative or cryptic splicing sites in an intron (grey rectangle, locus 2), these sites can be recognized by splicing factors (red circle), which resolves the constraints (e.g., by tethering the nascent RNA on RNAPII CTD). However, the splicing factor recruitment results in the production of a splicing variant. Likewise, the usage of alternative intronic polyadenylation sites (IpA, Locus 3) owing to the recruitment of dedicated factors (ref circle) resolves the constraints and results in the production of a truncated gene product. (C) Co-transcriptional biophysical constraints (lightning bolt) at a gene's 5′-end trigger RNA polymerase pausing. The recruitment of the U1 snRNP to the nascent RNA alleviates biophysical constraints and leads to promoter clearance. In the absence of splicing factor recruitment, a proximal intronic polyadenylation site (IpA) downstream of the 5′ splicing site of the first intron triggers co-transcriptional RNA cleavage and leads to the biogenesis of a truncated gene product. (D) Co-transcriptional biophysical constraints (lightning bolt) trigger co-transcriptional RNA cleavage (e.g., by endoribonucleases) within different kinds of RNA sequences (red dots). Co-transcriptional cleavage of the nascent RNAs tethered to RNAPII though RNA binding proteins (red circles) prevents aggregate and R-loop formation and results in RNA degradation or the production of truncated gene products or alternative splicing variants.
Co-transcriptional biophysical constraints trigger co-transcriptional alternative mRNA processing pathways
While transcription is coupled to translation in prokaryotic cells, transcription is coupled to mRNA processing, packaging and nuclear export in eukaryotic cells (Box). Consequently, co-transcriptional constraints, that trigger RNAPII pausing, impact on several steps of the gene expression process. For example, biophysical constraints at gene 5′-end may play an important role in the complex interplay between RNAPII promoter release, transcription elongation, mRNA capping, and 5′ splice site recognition of the first intron.15 Indeed, the GC-rich 5′-end of nascent mRNAs can interact with both the complementary DNA strand and with the RNA exit channel of RNAPII, which collectively could contribute to RNAPII pausing, increasing the time window for the recruitment of factors involved in 5′-end RNA processing11,16 (Box). In addition, biophysical constraints, such as transcriptional roadblocks created by nucleosomes and chromatin compactness impact on the recognition of splice sites during transcription elongation.17,18 One prevailing model is that impediments during transcription elongation slow down RNAPII and this increases the time window for the recognition of splicing sites or regulatory sequences by RNA processing factors.18 Finally, transcription termination and subsequent 3′-end RNA processing (i.e., cleavage and polyadenylation) are the consequence of biophysical constraints resulting from transcriptional roadblocks, R-loop formation and/or the interaction between the nascent mRNA and RNAPII.16,19 In this context, it is interesting to note that massive variations of the selection process of splicing and 3′-end mRNA processing sites can be induced by inhibition of DNA topoisomerases that are involved in resolving DNA topological constraints, suggesting a role in DNA topological constraints on RNA processing site selection.20,21
In conclusion, co-transcriptional constraints occurring when canonic or cryptic splicing and polyadenylation sites emerge from RNAPII impact on the utilization of these sites. Since different RNAPII molecules that are simultaneously producing transcripts from a given locus may encounter different constraints from one another, different mRNA processing sites can be selected during transcription, leading to the biogenesis of a diversity of mRNAs from one locus. While coupling of transcription and mRNA processing explains why co-transcriptional constraints can trigger different mRNA processing pathways, it will be shown below that co-transcriptional mRNA processing pathways can in turn resolve the initiating constraints.
Co-transcriptional use of alternative RNA processing sites to resolve co-transcriptional biophysical constraints
There is now compelling evidence of a widespread role in genome stability for mRNA processing factors involved in splicing, 3′-end mRNA processing, mRNA packaging and/or export.8,22–27 For example, inactivation of splicing or depletion of human splicing factors like SRSF1, induces R-loop formation, DNA rearrangements and genome instability.28,29 Depletion of core splicing factors like the SNRPA1 (U2 snRNP) and DDX23 (PRP28 from the U5 snRNP) or over-expression of mutated U2AF1 also triggers R-loop formation and genome instability.30–32 R-loops can equally be induced by drug-mediated inhibition of splicing and displacement from DNA of the assembled co-transcriptional spliceosome from lesion-arrested RNAPII.31–33 Similar observations were made in yeast.34,35
The protective effects on DNA of splicing factors may result from their ability to coat the nascent RNAs, to tether the nascent RNAs to RNAPII, and/or by reducing the complementarity between intron-containing DNA and nascent RNA after splicing.8,24–27,36,37 RNA biding protein-mediated RNA folding may also play an important role as nascent RNA folding mitigates transcription associated-mutagenesis.38 In this context, it is interesting to underline that an increasing number of RNA helicases that can contribute to RNA folding and modify or resolve RNA-DNA and RNA-protein interactions play a major role in driving nascent RNAs toward different RNA metabolism pathways as well as in genomic stability.32,39-41 The splicing process may also obviate potentially genotoxic co-transcriptional biophysical constraints by cleaving the nascent RNA, as the splicing process involves two consecutive RNA cleavages, followed by a ligation step.42 The first RNA cleavage occurs when the 5′ donor splicing site attacks the intronic branch site. Remarkably, large-scale sequencing approaches have recently indicated that usage of alternative branch-point sites within a given intron is more widespread than previously though.43,44 The same is true for recursive splicing, which allows the removal of large introns by iterative used of 5′ splice sites in intron. Recursive splicing can be viewed as the removal of “virtual” zero-length exons.43–45 This means that the use of different splicing sites can give rise to the same final gene product. Collectively, these observations support a model in which splicing-mediated cleavage of nascent RNAs does not necessarily affect the nature of the gene products and could “just” be a way to make the nascent RNAs going through RNA processing pathways.
Based on these observations, an interesting possibility is that potentially genotoxic co-transcriptional constraints can trigger the use of alternative or cryptic splicing sites and branch-points embedded within a given intron during transcription, which in turn, solve the co-transcriptional constraints occurring during the transcription of this intron. In other words, co-transcriptional usage of alternative splicing sites could protect DNA from transcription- and RNA-mediated genotoxicity. As depicted on Fig. 1B, intronic co-transcriptional constraints may result in DNA damage in the absence of mRNA processing sites (locus 1), while their presence and use may prevent DNA damage and lead to the biogenesis of the same or an alternative transcript (locus 2).
Because of the coupling between splicing and 3′-end RNA processing, splicing inhibition may also disturb the removal of the nascent RNA from chromatin (see Box). In this context, it has been shown that impairing mRNA 3′-end processing and/or transcription termination trigger R-loop-mediated genome instability.46–50 Along the same line, alteration of factors involved in the coupling between transcription and mRNA export or between mRNA processing, packaging and export also results in R-loop-mediated genome instability. This has been shown for several components of the THO, TREX and TREX-2 complexes that couple transcription and mRNA export and those mutations affect transcription elongation, mRNA export and induce R-loop formation and transcription-associated hyper-recombination.34,51–55 The DNA instability that is mediated by depletion of the factors that couple transcription and mRNA export may result from defects in mRNA packaging, increases of the time residency of the neo-synthetized RNA in the vicinity of the DNA template, or increases in the nuclear mRNA half-life.8,24–27 Interestingly, alteration of factors involved in nuclear RNA decay also induces R-loop formation and DNA instability.56–58 Based on these observations, the many alternative intronic cryptic polyadenylation sites that have been uncovered through massive RNA sequencing1 could contribute to solve potentially genotoxic co-transcriptional constraints in the same way as alternative splicing sites (Fig. 1B, “locus 3”). In this context, it is important to highlight the discovery of the strong interplay between the spliceosomal U1 snRNP that recognizes 5′ splice sites and the selection of intronic polyadenylation sites within first introns.59–61 Since the binding of the U1 snRNP on the nascent RNA has been shown to inhibit the usage of downstream intronic polyadenylation site, an interesting possibility is that the presence of polyadenylation sites in the first introns may overcome promoter-proximal constraints by removing the cleaved nascent transcript from chromatin in the case that the U1 snRNP has not been recruited on a nascent pre-mRNA59–61 (Fig. 1C). Supporting this possibility, UV-induced DNA damage has been observed to decrease the level of U1 snRNA and lead to the activation of intronic alternative cleavage and polyadenylation sites at gene 5′-ends.62 It would be interesting to look whether co-transcriptional RNA cleavage could occur through a diversity of processes mediated by different kind of endoribonucleases or ribozymes and could actually be a widespread mechanism protecting DNA from damage by triggering either RNA degradation, polyadenylation or splicing1,14,37,63 (Fig. 1D).
In conclusion, since co-transcriptional mRNA processing and packaging protect DNA from transcription- and RNA-mediated DNA damages and allows the neo-synthetized transcript to dissociate from chromatin, the use of cryptic RNA processing sites may play a major role in DNA homeostasis. The co-transcriptional use of alternative mRNA processing sites would be a “safeguard” mechanism ensuring genomic stability by i) being triggered by potentially genotoxic co-transcriptional biophysical constraints and ii) allowing the resolution of these constraints. Indeed, the co-transcriptional use of alternative or cryptic RNA-processing sites that result from co-transcriptional constraints would in turn ensure the dynamic movement of RNAPII and processing, packaging and chromatin release of the neo-synthetized RNA molecule, which collectively would decrease the probability of aggregate formation and/or spurious interaction between the nascent RNA and the DNA or DNA-associated proteins. It is also interesting to note that many proteins involved in mRNA processing are directly involved in DNA repair.64 Therefore, RNA processing sites emerging from RNAPII could contribute to DNA stability by locally recruiting mRNA processing factors, preventing RNA-mediated toxic effects and by contributing to DNA repair. If the co-transcriptional use of alternative mRNA processing sites alleviates transcription-dependent DNA damage, this means that the diversity of mRNAs generated by a genome may not be the results of inaccurate or error-prone biological processes but could rather reflects molecular mechanisms that protect DNA from damage. This model certainly does not exclude the possibly that cellular context-dependent regulatory processes preferentially direct nascent RNAs through one specific processing pathway and lead to the biogenesis of functional protein isoforms.
If the act of co-transcriptional mRNA processing per se has a role in maintaining genomic stability independently of the function of the generated gene product, does this mean that some mRNA variants generated by alternative processing pathways are merely “by-products” with no cellular function? The following section will address this question by challenging the notion of by-products and by reviewing examples from the literature of mRNAs with non-conventional functions (i.e., independent of the production of stable proteins).
Non-conventional roles of pre-mRNAs, introns and mRNAs
Noncoding-related functions of RNAs produced from coding genes
Increasing evidence indicates that transcriptional products can feedback on transcription and chromatin regulation (Fig. 2A). Indeed, pre-mRNAs, mRNA processing “by-products”, intronic circular RNAs, and the so-called ‘extra-coding’ RNAs have been shown to be involved in chromatin and transcription regulation in cis.10,65–67 The structured 3′-UTR of the HIC mRNA interacts with and activates the P-TEFb transcription elongation factor complex by displacing the 7SK noncoding RNA from it.68 Some mRNA processing by-products, like the intronic lariat-derived sno-lncRNAs may also play a role in RNA processing regulation.69
Figure 2.
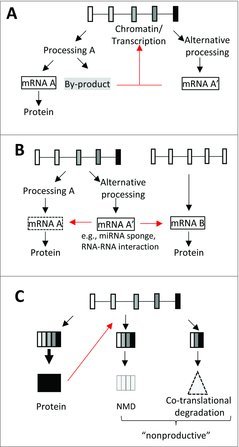
Non-conventional functions of mRNAs. (A) By-products or products of alternative processing pathways (mRNA A’) play a role in the regulation (red arrow) of the loci from which they originate. (B) mRNAs resulting from alternative RNA processing pathways (mRNA A’) can regulate the metabolism of the mRNA originating from the same gene (mRNA A) or those of a different gene (mRNA B) by acting as miRNA sponges or through mRNA-mRNA interaction. (C) Coding genes can give rise to “productive” translatable mRNAs or to “non-productive” mRNAs that are more-or-less rapidly degraded for example by the NMD pathway. The protein resulting from the productive processing pathway can cause feedback inhibition by stimulating the unproductive processing pathway (red arrow). Alternative mRNA processing pathways may also lead to the production of proteins that are co-translationally degraded.
Mature coding-gene transcripts produced from alternative processing pathways can also have nuclear non-coding functions. For example, the ASCC3 coding gene produces a long coding mRNA and a short non-coding mRNA that plays a role in transcription recovery after UV-induced DNA damage.70 Likewise, splicing variants produced from two coding genes, namely SRAP and LXR-b, act as non-coding RNA transcriptional co-activators.71,72 An mRNA containing trinucleotide repeats produced from the FMR1 gene has been shown to form an RNA:dsDNA triplex and repress the promoter it originates from.73
Some mRNAs also play regulatory noncoding functions in the cytoplasm (Fig. 2B). In one example, an alternative mRNA produced from the UBE3 coding gene acts as a noncoding competing endogenous RNA (ceRNA) that titrates miR-134, which impacts the miR-134-dependent neuronal regulatory circuit.74,75 While it has been shown that Natural Antisense Transcripts (NATs) bind to and regulate the expression level of the corresponding sense coding gene products, recent reports indicate that mRNA-mRNA interactions also regulate gene expression level. For example, the 5′ UTR of the Insulin Receptor Substrate 1 (IRS1) mRNA can interact with the retinoblastoma (Rb) mRNA and repress its translation.76 In yeast, interactions between the 3′-UTRs of different mRNAs can result in mRNA degradation through what is known as the ‘no-go’ decay pathway.77 Since a number of mRNA-mRNA pairs have recently been identified,78 it can be anticipated that mRNA-mRNA interaction has an important general role in gene expression regulation.
mRNA molecules can also achieve noncoding functions through mRNA-protein interactions. For example, p53 mRNA interacts with and regulates the nuclear trafficking and protein activity of the MDM2 protein.79 Several mRNAs regulate the activity of the RNA-activated protein kinase, PKR, which plays a role in translation regulation.80 Coding RNAs are known to interact with numerous RNA-binding proteins (e.g. hnRNPs) and this was presumed to be in order just to package and process the RNA. However the functions of RNA:protein interactions may be multi-faced as a surprisingly large number of proteins that are not related to known RNA-binding proteins have also been recently found to interact with mRNAs.81 Some of these mRNA-interacting proteins play a role in cellular metabolism, cytoskeleton organization or signaling pathways.81 Although these unconventional mRNA-binding proteins may play a role in mRNA metabolism, mRNAs may actually also regulate the RNA-binding proteins' enzymatic activity or cellular function.81,82 Alternatively, these mRNA-protein interactions may play a role in the co-translational assembly of specific complexes. For example, a long 3′ UTR of the CD47 mRNA has been shown to interact with the SET protein. SET then binds to the neo-synthetized CD47 protein and aids its translocation to the plasma membrane.83
There are also numerous examples of mRNAs, in various species, that play a structural role in cytoskeleton organization. For example, the Oskar mRNA serves as a scaffold for the assembly of cytoplasmic complexes in Xenopus oocytes.84 Increasing evidence indicates that mRNAs play a driving role in establishing membrane-less subcellular compartments, including granules and droplet organelles.85,86 These compartments are not only involved in mRNA metabolism (e.g., mRNA storage and decay) but they are also hubs for several signaling pathways, and they may create an intracellular molecular environment conducive to specific biological processes.85
Several observations support the possibility that some mRNAs give rise to functional mRNA-derived small RNAs. First, an increasing number of endoribonucleases have been recently characterized87 and the recent discovery of cytoplasmic capping and polyadenylation activities raises the possibility that mRNAs cleaved by endoribonucleases can be re-processed in the cytoplasm.88,89 Supporting this notion, all mature noncoding RNAs (e.g., tRNA, rRNAs, miRNAs, snoRNAs) can be further cleaved to generate functional derived RNAs.90 In addition, mRNA fragments, corresponding in particular to 3′ UTRs, have been identified.90,91 Finally, the production of functional small RNAs from cleaved mRNAs is well established in prokaryotes92 and it has been recently shown in yeast that an mRNA-derived noncoding RNA regulates the ribosome.93 The frontier between noncoding and coding RNAs is less clear-cut that previously thought since some lncRNAs contain productive small ORFs.94
Role of alternative mRNA processing pathways in mRNA and protein homeostasis
Gene expression level is partly regulated by alternative RNA processing pathways that can lead to the biogenesis of more or less unstable and unproductive mRNAs. Two mechanisms, namely intron retention and nonsense mediated decay (NMD), play major roles in protein homeostasis by allowing a switch from the biogenesis of “productive” (i.e., translated) to “unproductive” mRNAs.95,96 For example, a large number of splicing regulators regulate the splicing of their own gene by modulating intron retention or the inclusion of exons containing pre-mature stop codons. The result of this auto-regulatory mechanism is usually that splicing factors limit their own concentration by causing their own alternative mRNAs to be degraded by NMD96 (Fig. 2C). Alternative mRNA processing pathways can also adapt the production rate of specific proteins. For example, a recent class of ‘detained introns’ has been shown to remain un-excised during transcription, unlike most introns.97 Instead, the regulated nuclear post-transcriptional removal of these detained introns is triggered by cellular stresses.97 The regulation of intron removal may actually play a more widespread role in protein homeostasis under various physiological conditions than previously anticipated.98 Although this is still a matter of debate, cytoplasmic splicing may also play a role in regulated post-transcriptional intron removal.99
There is also debate around what proportion of coding genes' transcript variants is translated.100–103 While ribosome profiling at the RNA levels suggests that many coding mRNA variants are translated, proteomic analyses have revealed so far that coding genes may give rise to mostly one major principal protein isoform, as the other predicted alternative protein isoforms were not detected across a large number of analyzed samples.100 Even though technical issues (e.g., coverage currently achieved by transcriptome and proteome technologies) may explain the discrepancy between conclusions drawn from proteomic and transcriptomic analyses, another possible explanation is that some mRNA splicing variants do give rise to stable proteins. Some protein isoforms could be highly unstable or might mis-fold and therefore could be co-translationally degraded. Supporting this possibility, many alternative exons code for intrinsically disordered regions that are aggregate prone and that can mis-fold.104–106 Isoform-specific co-translational protein degradation could work together with the NMD pathway to maintain protein homeostasis (Fig. 2C).
To summarize, deciphering what fraction of the mRNAs that are generated through alternative mRNA processing pathways is biologically relevant and how much is just “noise”, junk or functionless cannot rely only on the potential production of functional proteins. The capacity of mRNAs to give rise to proteins may be just one of their many cellular functions.
Conclusion
A paradigm shift is needed in the way we think about coding gene expression. First, the diversity of alternative co-transcriptional mRNA processing pathways is functionally important in terms of DNA homeostasis as well as to regulate the expression of coding genes, independently of any resulting protein function. Second, any processed mRNA might function either by feeding back on the locus and pathway it originates from or by participating in various cellular processes other than protein synthesis. Coding genes are likely producing both coding mRNAs and functional non-coding RNAs and even coding mRNAs seem to be able to work as do non-coding RNAs. Only by considering these complexities can we hope to integrate the current tsunami of RNA sequencing data into a coherent biological framework. The terms ‘error’, ‘noise’, ‘junk’ and ‘quality control’ should be used with caution when addressing the diversity of coding-gene products. These terms often imply a kind of function-related hierarchy among coding gene products and put too much emphasis on protein-related functions. As recently proposed for pre-mRNAs,10 mRNAs are much more than passive intermediates used to transfer information from genes to proteins.
In conclusion, the use of alternative mRNA processing sites during transcription alleviates transcription-dependent DNA damage and therefore decreases the instability of transcribed protein-coding loci. The resulting mRNAs are not the results of inaccurate biological processes but rather of mechanisms that protect DNA from damage. Nor are they functionless “by-products” as they can feedback on chromatin and transcription regulation or can be used in a diversity of cellular processes other than to translation. Therefore, the co-transcriptional use of alternative mRNA processing sites not only increases genomic stability by reducing hazardous mutational processes (e.g., transcriptional-mediated DNA instability), therefore reducing genetic variability but it simultaneously increases the functional diversity of the RNAs produced from coding genes.
Box: Coupling of nuclear steps of the gene expression process
The nascent RNA emerging from RNAPII is coated by RNA-binding proteins (RBPs) that are involved in the mRNA processing steps (capping, splicing, and 3′-end mRNA processing), which mostly occur during transcription.15,107,108 The proximity between the nascent RNA and RBPs is in part mediated by the carboxy-terminal domain of RNAPII, and constraints affecting RNAPII dynamics impact on the different mRNA processing steps. For example, chromatin compaction can affect splicing and polyadenylation site selection.15,18 In addition, to be coupled to transcription, the mRNA processing steps are coupled to one another. Capping is coupled to splicing and 3′-end mRNA processing, and splicing is coupled to 3′-end mRNA processing.15,107 Transcription, capping, splicing and 3′-end mRNA processing are also tightly connected to mRNA packaging and mRNP biogenesis.15,107–109 For example, the splicing process contributes to the recruitment of exon junction complexes on nascent RNAs.110 Finally, co-transcriptional mRNA processing is tightly connected to removal of the neo-synthetized RNAs from chromatin, and both transcription and mRNA processing are coupled to subsequent mRNA export.15,107,108,110 Interestingly, pre-mRNA, mRNA processing by-products, as well as factors involved in RNA processing can all feedback on chromatin, transcription initiation and elongation.10 For example, the spliceosome or splicing factors can promote transcription initiation, promoter release or transcription elongation.111–116 An emerging picture is that the extensive network of coupling described above provides a molecular framework “ensuring” that the nascent and neo-synthetized RNAs are constantly directed toward specific molecular interactions.39 This broad vision helps explain why the presence of introns in eukaryotic genes enhances gene expression at multiple levels (from transcription to translation).117–119 It is important to underline that the global enhancing effect of RNA processing (e.g., splicing) on gene expression, does not exclude that recognition of RNA processing sites may create local RNAPII pauses. For example, a local decrease in RNAPII velocity, mediated by the splicing process in the vicinity of exons, may be required for proper processing, which next would resume transcription elongation and improve downstream steps.107,120–122 In general, coupling between the gene expression steps may not only increase efficiency but also protect from hazards. Indeed, by directing the nascent and neo-synthetized RNAs toward specific molecular interactions, these extensive inter-connections prevent the formation of RNA-dependent aggregates and potentially toxic spurious interactions, like RNA-DNA hybrids.
References
- 1.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. PMID:27677860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blencowe BJ. An exon-centric perspective. Biochem Cell Biol. 2012;90:603–12. doi: 10.1139/o2012-019. PMID:22779775 [DOI] [PubMed] [Google Scholar]
- 3.Wang M, Zhang P, Shu Y, Yuan F, Zhang Y, Zhou Y, Jiang M, Zhu Y, Hu L, Kong X. Alternative splicing at GYNNGY 5′ splice sites: More noise, less regulation. Nucleic Acids Res. 2014;42:13969–80. doi: 10.1093/nar/gku1253. PMID:25428370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. PMID:21151575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–86. doi: 10.1093/nar/gkp471. PMID:19546110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–7. doi: 10.1242/jcs.181008. PMID:26787741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaillard H, Aguilera A. Transcription as a threat to genome Integrity. Annu Rev Biochem. 2016;85:291–317. doi: 10.1146/annurev-biochem-060815-014908. PMID:27023844 [DOI] [PubMed] [Google Scholar]
- 8.Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst). 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. PMID:24746923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D GH, Kelley DR, Tenen D, Bernstein B, Rinn JL. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. doi: 10.1186/s13059-016-0878-3. PMID:26883116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skalska L, Beltran-Nebot M, Ule J, Jenner RG. Regulatory feedback from nascent RNA to chromatin and transcription. Nat Rev Mol Cell Biol. 2017;18:331–7. doi: 10.1038/nrm.2017.12. PMID:28270684 [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Landick R. A Two-way street: Regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem Sci. 2016;41:293–310. doi: 10.1016/j.tibs.2015.12.009. PMID:26822487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–99. doi: 10.1016/j.cell.2014.08.018. PMID:25259926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sicot G, Gomes-Pereira M. RNA toxicity in human disease and animal models: From the uncovering of a new mechanism to the development of promising therapies. Biochim Biophys Acta. 2013;1832:1390–409. doi: 10.1016/j.bbadis.2013.03.002. PMID:23500957 [DOI] [PubMed] [Google Scholar]
- 14.Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol. 2016;17:227–39. doi: 10.1038/nrm.2015.15. PMID:26726035 [DOI] [PubMed] [Google Scholar]
- 15.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–75. doi: 10.1038/nrg3662. PMID:24514444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–600. doi: 10.1101/gr.158436.113. PMID:23868195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimeno-Gonzalez S, Payán-Bravo L, Muñoz-Cabello AM, Guijo M, Gutierrez G, Prado F, Reyes JC. Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing. Proc Natl Acad Sci USA. 2015;112:14840–5. doi: 10.1073/pnas.1506760112. PMID:26578803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–98. doi: 10.1146/annurev-biochem-060614-034242. PMID:26034889 [DOI] [PubMed] [Google Scholar]
- 19.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352:aad9926. doi: 10.1126/science.aad9926. PMID:27284201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutertre M, Chakrama FZ, Combe E, Desmet FO, Mortada H, Polay Espinoza M, Gratadou L, Auboeuf D. A recently evolved class of alternative 3′-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nat Commun. 2014;5:3395. doi: 10.1038/ncomms4395. PMID:24577238 [DOI] [PubMed] [Google Scholar]
- 21.Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17:1358–66. doi: 10.1038/nsmb.1912. PMID:20972445 [DOI] [PubMed] [Google Scholar]
- 22.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–88. doi: 10.1016/j.molcel.2011.10.017. PMID:22195970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al.. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–39. doi: 10.1016/j.molcel.2009.06.021. PMID:19647519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos-Pereira JM, Aguilera A. R loops: New modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–97. doi: 10.1038/nrg3961. PMID:26370899 [DOI] [PubMed] [Google Scholar]
- 25.Montecucco A, Biamonti G. Pre-mRNA processing factors meet the DNA damage response. Front Genet. 2013;4:102. doi: 10.3389/fgene.2013.00102. PMID:23761808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naro C, Bielli P, Pagliarini V, Sette C. The interplay between DNA damage response and RNA processing: The unexpected role of splicing factors as gatekeepers of genome stability. Front Genet. 2015;6:142. doi: 10.3389/fgene.2015.00142. PMID:25926848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–96. doi: 10.1101/gad.242990.114. PMID:24990962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Niu T, Manley JL. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007;13:2108–15. doi: 10.1261/rna.734407. PMID:17959926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–78. doi: 10.1016/j.cell.2005.06.008. PMID:16096057 [DOI] [PubMed] [Google Scholar]
- 30.Tanikawa M, Sanjiv K, Helleday T, Herr P, Mortusewicz O. The spliceosome U2 snRNP factors promote genome stability through distinct mechanisms; transcription of repair factors and R-loop processing. Oncogenesis. 2016;5:e280. doi: 10.1038/oncsis.2016.70. PMID:27991914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol Cell. 2017;65:832–47 e4. doi: 10.1016/j.molcel.2017.01.029. PMID:28257700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sridhara SC, Carvalho S, Grosso AR, Gallego-Paez LM, Carmo-Fonseca M, de Almeida SF. Transcription dynamics prevent RNA-Mediated genomic instability through SRPK2-Dependent DDX23 phosphorylation. Cell Rep. 2017;18:334–43. doi: 10.1016/j.celrep.2016.12.050. PMID:28076779 [DOI] [PubMed] [Google Scholar]
- 33.Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IJcken WF, Grosveld FG, Medema RH, Hoeijmakers JH, et al.. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015;523:53–8. doi: 10.1038/nature14512. PMID:26106861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos-Pereira JM, Herrero AB, García-Rubio ML, Marín A, Moreno S, Aguilera A. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 2013;27:2445–58. doi: 10.1101/gad.229880.113. PMID:24240235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aronica L, Kasparek T, Ruchman D, Marquez Y, Cipak L, Cipakova I, Anrather D, Mikolaskova B, Radtke M, Sarkar S, et al.. The spliceosome-associated protein Nrl1 suppresses homologous recombination-dependent R-loop formation in fission yeast. Nucleic Acids Res. 2016;44:1703–17. doi: 10.1093/nar/gkv1473. PMID:26682798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollander D, Naftelberg S, Lev-Maor G, Kornblihtt AR, Ast G. How are short exons flanked by long introns defined and committed to splicing? Trends Genet. 2016;32:596–606. doi: 10.1016/j.tig.2016.07.003. PMID:27507607 [DOI] [PubMed] [Google Scholar]
- 37.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–59. doi: 10.1016/j.molcel.2006.01.032. PMID:16543153 [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Yang JR, Zhang J. Nascent RNA folding mitigates transcription-associated mutagenesis. Genome Res. 2016;26:50–9. doi: 10.1101/gr.195164.115. PMID:26518484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Biol. 2016;17:426–38. doi: 10.1038/nrm.2016.50. PMID:27251421 [DOI] [PubMed] [Google Scholar]
- 40.Jain A, Bacolla A, Del Mundo IM, Zhao J, Wang G, Vasquez KM. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013;41:10345–57. doi: 10.1093/nar/gkt804. PMID:24049074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodroj D, Recolin B, Serhal K, Martinez S, Tsanov N, Abou Merhi R, Maiorano D. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017;36:1182–98. doi: 10.15252/embj.201695131. PMID:28314779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papasaikas P, Valcarcel J. The Spliceosome: The ultimate RNA chaperone and Sculptor. Trends Biochem Sci. 2016;41:33–45. doi: 10.1016/j.tibs.2015.12.010 . PMID:26682498 [DOI] [PubMed] [Google Scholar]
- 43.Stepankiw N, Raghavan M, Fogarty EA, Grimson A, Pleiss JA. Widespread alternative and aberrant splicing revealed by lariat sequencing. Nucleic Acids Res. 2015;43:8488–501. doi: 10.1093/nar/gkv763. PMID:26261211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly S, Georgomanolis T, Zirkel A, Diermeier S, O'Reilly D, Murphy S, Längst G, Cook PR, Papantonis A. Splicing of many human genes involves sites embedded within introns. Nucleic Acids Res. 2015;43:4721–32. doi: 10.1093/nar/gkv386. PMID:25897131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duff MO, et al.. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature. 2015;521:376–9. doi: 10.1038/nature14475. PMID:25970244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, Boothman DA. XRN2 links transcription termination to DNA damage and replication stress. PLoS Genet. 2016;2:e1006107. doi: 10.1371/journal.pgen.1006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–75. doi: 10.1101/gad.179721.111. PMID:22279048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al.. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–47. doi: 10.1016/j.molcel.2015.01.011. PMID:25699710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. PMID:21700224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaillard H, Aguilera A. Cleavage factor I links transcription termination to DNA damage response and genome integrity maintenance in Saccharomyces cerevisiae. PLoS Genet. 2014;10:e1004203. doi: 10.1371/journal.pgen.1004203. PMID:24603480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. PMID:22144908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–5. doi: 10.1038/nature13374. PMID:24896180 [DOI] [PubMed] [Google Scholar]
- 53.Gavalda S, Santos-Pereira JM, Garcia-Rubio ML, Luna R, Aguilera A. Excess of yra1 RNA-Binding factor causes transcription-Dependent genome instability, replication impairment and telomere shortening. PLoS Genet. 2016;2:e1005966. doi: 10.1371/journal.pgen.1005966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-Aguilera C, Tous C, Gómez-González B, Huertas P, Luna R, Aguilera A. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell. 2008;19:4310–8. doi: 10.1091/mbc.E08-04-0355. PMID:18667528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–35. doi: 10.1093/emboj/cdf335. PMID:12093753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gavalda S, Gallardo M, Luna R, Aguilera A. R-loop mediated transcription-associated recombination in trf4Delta mutants reveals new links between RNA surveillance and genome integrity. PLoS One. 2013;8:e65541. doi: 10.1371/journal.pone.0065541. PMID:23762389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013;2:e00505. doi: 10.7554/eLife.00505. PMID:23795288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al.. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–89. doi: 10.1016/j.cell.2015.04.034. PMID:25957685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–8. doi: 10.1038/nature09479. PMID:20881964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berg MG, et al.. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. PMID:22770214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–3. doi: 10.1038/nature12349. PMID:23792564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Devany E, Park JY, Murphy MR, Zakusilo G, Baquero J, Zhang X, Hoque M, Tian B, Kleiman FE. Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA. Cell Discov. 2016;2:16013. doi: 10.1038/celldisc.2016.13. PMID:27462460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gromak N, Talotti G, Proudfoot NJ, Pagani F. Modulating alternative splicing by cotranscriptional cleavage of nascent intronic RNA. RNA. 2008;14:359–66. doi: 10.1261/rna.615508. PMID:18065715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dutertre M, Lambert S, Carreira A, Amor-Gueret M, Vagner S. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem Sci. 2014;39:141–9. doi: 10.1016/j.tibs.2014.01.003. PMID:24534650 [DOI] [PubMed] [Google Scholar]
- 65.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al.. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. PMID:25664725 [DOI] [PubMed] [Google Scholar]
- 66.Savell KE, Gallus NV, Simon RC, Brown JA, Revanna JS, Osborn MK, Song EY, O'Malley JJ, Stackhouse CT, Norvil A, et al.. Extra-coding RNAs regulate neuronal DNA methylation dynamics. Nat Commun. 2016;7:12091. doi: 10.1038/ncomms12091. PMID:27384705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–70. doi: 10.1038/nsmb.2315. PMID:22659877 [DOI] [PubMed] [Google Scholar]
- 68.Young TM, Tsai M, Tian B, Mathews MB, Pe'ery T. Cellular mRNA activates transcription elongation by displacing 7SK RNA. PLoS One. 2007;2:e1010. doi: 10.1371/journal.pone.0001010. PMID:17925858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–30. doi: 10.1016/j.molcel.2012.07.033. PMID:22959273 [DOI] [PubMed] [Google Scholar]
- 70.Williamson L, Saponaro M, Boeing S, East P, Mitter R, Kantidakis T, Kelly GP, Lobley A, Walker J, Spencer-Dene B, et al.. UV Irradiation Induces a Non-coding RNA that Functionally Opposes the Protein Encoded by the Same Gene. Cell. 2017;168:843–55 e13. doi: 10.1016/j.cell.2017.01.019. PMID:28215706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lanz RB, et al.. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/S0092-8674(00)80711-4. PMID:10199399 [DOI] [PubMed] [Google Scholar]
- 72.Hashimoto K, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A liver X receptor (LXR)-beta alternative splicing variant (LXRBSV) acts as an RNA co-activator of LXR-beta. Biochem Biophys Res Commun. 2009;390:1260–5. doi: 10.1016/j.bbrc.2009.10.132. PMID:19878653 [DOI] [PubMed] [Google Scholar]
- 73.Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002–5. doi: 10.1126/science.1245831. PMID:24578575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valluy J, Bicker S, Aksoy-Aksel A, Lackinger M, Sumer S, Fiore R, Wüst T, Seffer D, Metge F, Dieterich C, et al.. A coding-independent function of an alternative Ube3a transcript during neuronal development. Nat Neurosci. 2015;18:666–73. doi: 10.1038/nn.3996. PMID:25867122 [DOI] [PubMed] [Google Scholar]
- 75.Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–41. doi: 10.1093/nar/gkq1003. PMID:21149267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagano H, Yamagishi N, Tomida C, Yano C, Aibara K, Kohno S, Abe T, Ohno A, Hirasaka K, Okumura Y, et al.. A novel myogenic function residing in the 5′ non-coding region of Insulin receptor substrate-1 (Irs-1) transcript. BMC Cell Biol. 2015;16:8. doi: 10.1186/s12860-015-0054-8. PMID:25887310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sinturel F, Navickas A, Wery M, Descrimes M, Morillon A, Torchet C, Benard L. Cytoplasmic control of sense-Antisense mRNA pairs. Cell Rep. 2015;12:1853–64. doi: 10.1016/j.celrep.2015.08.016. PMID:26344770 [DOI] [PubMed] [Google Scholar]
- 78.Nguyen TC, Cao X, Yu P, Xiao S, Lu J, Biase FH, Sridhar B, Huang N, Zhang K, Zhong S. Mapping RNA-RNA interactome and RNA structure in vivo by MARIO. Nat Commun. 2016;7:12023. doi: 10.1038/ncomms12023. PMID:27338251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gajjar M, Candeias MM, Malbert-Colas L, Mazars A, Fujita J, Olivares-Illana V, Fåhraeus R. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell. 2012;21:25–35. doi: 10.1016/j.ccr.2011.11.016. PMID:22264786 [DOI] [PubMed] [Google Scholar]
- 80.Nussbaum JM, Gunnery S, Mathews MB. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res. 2002;30:1205–12. doi: 10.1093/nar/30.5.1205. PMID:11861913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Castello A, Hentze MW, Preiss T. Metabolic enzymes enjoying new partnerships as RNA-Binding proteins. Trends Endocrinol Metab. 2015;26:746–57. doi: 10.1016/j.tem.2015.09.012. PMID:26520658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beckmann BM, Castello A, Medenbach J. The expanding universe of ribonucleoproteins: Of novel RNA-binding proteins and unconventional interactions. Pflugers Arch. 2016;468:1029–40. doi: 10.1007/s00424-016-1819-4. PMID:27165283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berkovits BD, Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–7. doi: 10.1038/nature14321. PMID:25896326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–57. doi: 10.1242/dev.01919. PMID:16000384 [DOI] [PubMed] [Google Scholar]
- 85.Nielsen FC, Hansen HT, Christiansen J. RNA assemblages orchestrate complex cellular processes. Bioessays. 2016;38:674–81. doi: 10.1002/bies.201500175. PMID:27172226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? Embo J. 2016;35:1603–12. doi: 10.15252/embj.201593517. PMID:27357569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tomecki R, Dziembowski A. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA. 2010;16:1692–724. doi: 10.1261/rna.2237610. PMID:20675404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schoenberg DR, Maquat LE. Re-capping the message. Trends Biochem Sci. 2009;34:435–42. doi: 10.1016/j.tibs.2009.05.003. PMID:19729311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Norbury CJ. Cytoplasmic RNA: A case of the tail wagging the dog. Nat Rev Mol Cell Biol. 2013;14:643–53. doi: 10.1038/nrm3645. PMID:23989958 [DOI] [PubMed] [Google Scholar]
- 90.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–32. doi: 10.1016/j.tig.2011.06.001. PMID:21741109 [DOI] [PubMed] [Google Scholar]
- 91.Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, Askarian-Amiri ME, Gardiner BB, Goodall GJ, Grimmond SM, et al.. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20:1639–50. doi: 10.1101/gr.112128.110. PMID:21045082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miyakoshi M, Chao Y, Vogel J. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr Opin Microbiol. 2015;24:132–9. doi: 10.1016/j.mib.2015.01.013. PMID:25677420 [DOI] [PubMed] [Google Scholar]
- 93.Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell. 2014;54:147–55. doi: 10.1016/j.molcel.2014.02.024. PMID:24685157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ji Z, Song R, Regev A, Struhl K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015;4:e08890. doi: 10.7554/eLife.08890. PMID:26687005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peccarelli M, Kebaara BW. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot Cell. 2014;13:1126–35. doi: 10.1128/EC.00090-14. PMID:25038084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–9. doi: 10.1038/nature05676. PMID:17361132 [DOI] [PubMed] [Google Scholar]
- 97.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. PMID:25561496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gill J, Park Y, McGinnis JP, Perez-Sanchez C, Blanchette M, Si K. Regulated intron removal integrates motivational state and experience. Cell. 2017;169:836–848 e15. doi: 10.1016/j.cell.2017.05.006. PMID:28525754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buckley PT, Khaladkar M, Kim J, Eberwine J. Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip Rev RNA. 2014;5:223–30. doi: 10.1002/wrna.1203. PMID:24190870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tress ML, Abascal F, Valencia A. Alternative splicing may not be the key to proteome complexity. Trends Biochem Sci. 2017;42:98–110. doi: 10.1016/j.tibs.2016.08.008. PMID:27712956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weatheritt RJ, Sterne-Weiler T, Blencowe BJ. The ribosome-engaged landscape of alternative splicing. Nat Struct Mol Biol. 2016;23:1117–23. doi: 10.1038/nsmb.3317. PMID:27820807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blencowe BJ. The Relationship between Alternative Splicing and Proteomic Complexity. Trends Biochem Sci. 2017;42:407–8. doi: 10.1016/j.tibs.2017.04.001. PMID:28483376 [DOI] [PubMed] [Google Scholar]
- 103.Liu Y, Gonzàlez-Porta M, Santos S, Brazma A, Marioni JC, Aebersold R, Venkitaraman AR, Wickramasinghe VO. Impact of alternative splicing on the human proteome. Cell Rep. 2017;20:1229–41. doi: 10.1016/j.celrep.2017.07.025. PMID:28768205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aufderheide A, Unverdorben P, Baumeister W, Forster F. Structural disorder and its role in proteasomal degradation. FEBS Lett. 2015;589:2552–60. doi: 10.1016/j.febslet.2015.07.034. PMID:26226424 [DOI] [PubMed] [Google Scholar]
- 105.Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. Codon usage influences the local rate of translation elongation to regulate Co-translational protein folding. Mol Cell. 2015;59:744–54. doi: 10.1016/j.molcel.2015.07.018. PMID:26321254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol Cell. 2013;49:411–21. doi: 10.1016/j.molcel.2013.01.020. PMID:23395271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. PMID:11932736 [DOI] [PubMed] [Google Scholar]
- 108.Singh G, Pratt G, Yeo GW, Moore MJ. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu Rev Biochem. 2015;84:325–54. doi: 10.1146/annurev-biochem-080111-092106. PMID:25784054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zonta E, Bittencourt D, Samaan S, Germann S, Dutertre M, Auboeuf D. The RNA helicase DDX5/p68 is a key factor promoting c-fos expression at different levels from transcription to mRNA export. Nucleic Acids Res. 2013;41:554–64. doi: 10.1093/nar/gks1046. PMID:23143267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Le Hir H, Sauliere J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol. 2016;17:41–54. PMID:26670016 [DOI] [PubMed] [Google Scholar]
- 111.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–68. doi: 10.1016/j.cell.2013.04.028. PMID:23663783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–26. doi: 10.1038/nsmb.1461. PMID:18641664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9:800–5. PMID:12389039 [DOI] [PubMed] [Google Scholar]
- 114.Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29:271–8. doi: 10.1016/j.molcel.2007.11.035. PMID:18243121 [DOI] [PubMed] [Google Scholar]
- 115.Koga M, Hayashi M, Kaida D. Splicing inhibition decreases phosphorylation level of Ser2 in Pol II CTD. Nucleic Acids Res. 2015;43:8258–67. doi: 10.1093/nar/gkv740. PMID:26202968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lemieux B, Blanchette M, Monette A, Mouland AJ, Wellinger RJ, Chabot B. A function for the hnRNP A1/A2 proteins in transcription elongation. PLoS One. 2015;10:e0126654. doi: 10.1371/journal.pone.0126654. PMID:26011126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chorev M, Carmel L. The function of introns. Front Genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. PMID:22518112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gallegos JE, Rose AB. The enduring mystery of intron-mediated enhancement. Plant Sci. 2015;237:8–15. doi: 10.1016/j.plantsci.2015.04.017. PMID:26089147 [DOI] [PubMed] [Google Scholar]
- 119.Merkin JJ, Chen P, Alexis MS, Hautaniemi SK, Burge CB. Origins and impacts of new mammalian exons. Cell Rep. 2015;10:1992–2005. doi: 10.1016/j.celrep.2015.02.058. PMID:25801031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell. 2014;53:779–90. doi: 10.1016/j.molcel.2014.01.017. PMID:24560925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.de Almeida SF, Carmo-Fonseca M. Reciprocal regulatory links between cotranscriptional splicing and chromatin. Semin Cell Dev Biol. 2014;32:2–10. doi: 10.1016/j.semcdb.2014.03.010. PMID:24657193 [DOI] [PubMed] [Google Scholar]
- 122.Moehle EA, Braberg H, Krogan NJ, Guthrie C. Adventures in time and space: Splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014;11:313–9. doi: 10.4161/rna.28646. PMID:24717535 [DOI] [PMC free article] [PubMed] [Google Scholar]


