Significance
Proteins usually fold to their thermodynamically stable states. The serine protease inhibitors (serpins) modulate several biological processes such as blood clotting and inflammation. These proteins are unusual because their functionally active state, and consequently the state that they need to fold to and populate, is significantly less stable than an alternate latent conformation. This metastability facilitates conformational conversion to the inactive state which irreversibly traps the protease. However, it also makes the serpins prone to disease-linked polymerization. Here, we investigate the structural basis for this conformational choice using folding simulations of the serpin, α1-antitrypsin. We find that the structure of the protease-binding reactive center loop is determined early during folding and gates kinetic accessibility to the latent conformation.
Keywords: α1-antitrypsin, reactive center loop, protein folding simulations, kinetic trap, alternate folded conformations
Abstract
For successful protease inhibition, the reactive center loop (RCL) of the two-domain serine protease inhibitor, α1-antitrypsin (α1-AT), needs to remain exposed in a metastable active conformation. The α1-AT RCL is sequestered in a β-sheet in the stable latent conformation. Thus, to be functional, α1-AT must always fold to a metastable conformation while avoiding folding to a stable conformation. We explore the structural basis of this choice using folding simulations of coarse-grained structure-based models of the two α1-AT conformations. Our simulations capture the key features of folding experiments performed on both conformations. The simulations also show that the free energy barrier to fold to the latent conformation is much larger than the barrier to fold to the active conformation. An entropically stabilized on-pathway intermediate lowers the barrier for folding to the active conformation. In this intermediate, the RCL is in an exposed configuration, and only one of the two α1-AT domains is folded. In contrast, early conversion of the RCL into a β-strand increases the coupling between the two α1-AT domains in the transition state and creates a larger barrier for folding to the latent conformation. Thus, unlike what happens in several proteins, where separate regions promote folding and function, the structure of the RCL, formed early during folding, determines both the conformational and the functional fate of α1-AT. Further, the short 12-residue RCL modulates the free energy barrier and the folding cooperativity of the large 370-residue α1-AT. Finally, we suggest experiments to test the predicted folding mechanism for the latent state.
Sequences of proteins that undergo conformational transitions encode two structurally distinct conformations, only one of which is usually functionally active (1–3). When the same protein sequence encodes two or more structurally distinct conformations, it is imperative that the protein fold to and populate the functionally relevant conformation. This poses a problem for those unusual proteins whose functionally relevant conformation is significantly less stable than the alternative conformation [e.g., serine protease inhibitors (4), α-lytic protease (5), hemagglutinin (6)]. An elegant solution to this problem is to limit the kinetic accessibility of folding to the more stable conformation relative to folding to the less stable but functionally relevant conformation (7–9). Here, we study a serine protease inhibitor (serpin) to identify the structural basis for how the kinetic accessibility to fold to a metastable, active conformation is increased to avoid folding to the more stable, latent conformation.
Serine protease inhibitors, or serpins, are a large family of multidomain proteins that have similar structures but perform diverse functions in different organisms (10). In serpins, the metastability of the active conformation facilitates capture of the protease and conformational conversion to a stable state in which the protease is also inactivated (4). In humans, serpins are involved in regulating the function of several proteases such as elastase in lung tissue, thrombin and tissue plasminogen activator in blood, etc. Mutations to the serpins which cause loss of function or induce aberrant polymerization can lead to disease conditions such as liver cirrhosis, dementia, emphysema, thrombosis, and hemorrhagic diathesis (10). The underlying cause of these diseases seems to be the metastable nature of the active functional form of serpins, which makes serpins susceptible to misfolding and polymerization (10). Understanding the structural basis of how the serpins fold to the metastable form while avoiding folding to the stable latent form could also help in understanding the mechanistic basis of this polymerization.
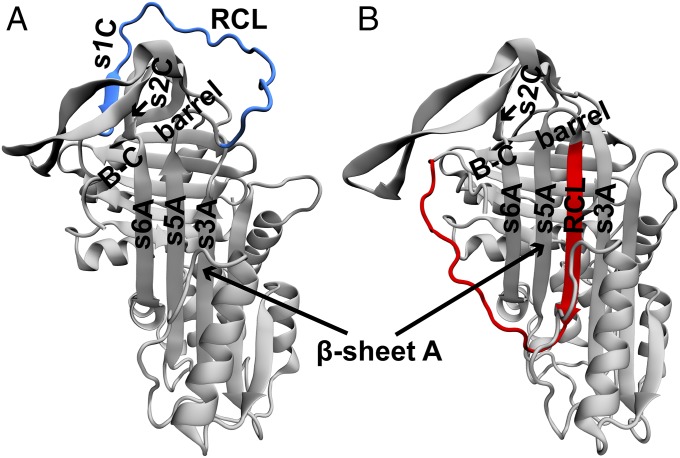
Human α1-antitrypsin (α1-AT) is an archetypal serpin which folds to an active, metastable, native conformation (4) (Figs. 1A and 2A). The active conformation of α1-AT has an exposed loop termed as the reactive center loop (RCL) which gets attacked by the protease (Fig. 1A). The protease proceeds to cleave a peptide bond in the RCL, and, in this process, it forms a covalently bound acyl-protease intermediate with the cleaved RCL. The α1-AT inhibits the protease before enzyme turnover by undergoing a large conformational transition (4). This conformational transition involves insertion of the cleaved RCL into the already existing β-sheet A between strands s5A and s3A (Fig. 1A). Consequently, the covalently bound protease is translocated to the opposite pole of α1-AT. The final conformation of α1-AT with an inserted RCL has two features: (i) It is inactive because the RCL is not exposed and is unable to bind another protease (4), and (ii) it is thermodynamically more stable than the active conformation. The midpoint of a thermal melt for active α1-AT is at 59 °C, while that for its cleaved, inactive conformation is at 124 °C (11). Thus, a higher thermodynamic stability is afforded by the insertion of the RCL into β-sheet A. A structurally equivalent latent conformation in which an uncleaved RCL is inserted into β-sheet A is also known to exist (12). It should be noted that this X-ray crystal structure (shown in Figs. 1B and 2B) was obtained by making several mutations to α1-AT (13). In summary, to be functionally active, a newly synthesized α1-AT must fold to the less stable conformation and avoid folding to the more stable, latent conformation.
Fig. 1.
Structures of the active and latent conformations of α1-AT. Cartoons of the X-ray crystal structures of the (A) active and (B) latent conformations of α1-AT drawn from PDBs 1QLP and 1IZ2, respectively. The structures were aligned to each other using the STAMP algorithm in Visual Molecular Dynamics (VMD 1.8.7) (46). The segments which align well with each other are colored gray, while those which do not are colored (A) blue and (B) red. These regions correspond to the conformation of the RCL, which is (A) solvent exposed while it is (B) inserted into β-sheet A. Additional structural features of α1-AT such as strands s3A, s5A, s6A, s1c, and s2c and the B-C β-barrel are also marked. The domains of α1-AT in both conformations are shown in Fig. 2, and the key interactions distinguishing the two conformations are shown in Fig. 3. All structures in this manuscript are drawn in VMD 1.8.7 (46).
Fig. 2.
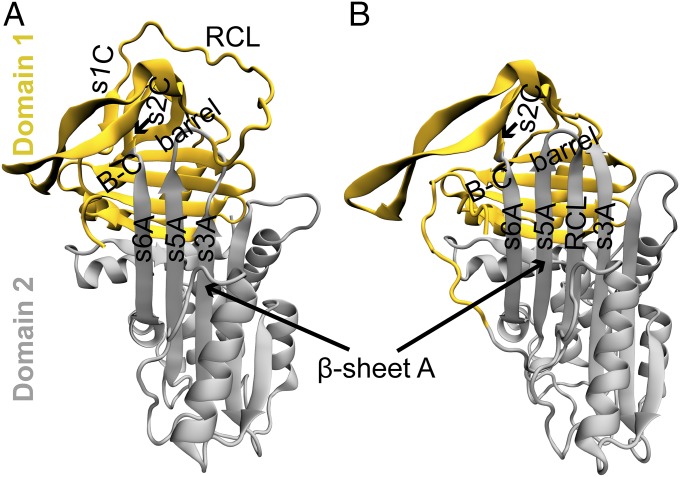
Domain definitions of the active and latent conformations of α1-AT. The structures of the (A) active and (B) latent conformations of α1-AT are colored according to their domains; α1-AT has two domains, according to the CATH database (23). Domains 1 and 2 are colored yellow and gray, respectively, in both conformations (see SI Appendix for definitions, and see SI Appendix, Fig. S1). Some of the key structural elements such as the RCL and strands s3A, s5A, and s6A are also marked. The RCL is in domain 1 in the active conformation, while it is a part of domain 2 in the latent conformation.
Previous studies have hypothesized that large free energy barriers must prevent folding to the more stable, latent conformation, and that the active conformation is a kinetically trapped folding intermediate whose subsequent folding to the more stable, latent conformation is coupled to its inhibitory function (4). Extensive experimental studies on the mechanism of folding to the less stable, active conformation of α1-AT revealed that the insertion of the RCL into β-sheet A is prevented in the early stages of folding (14, 15). By monitoring the formation of backbone structure during its folding by pulsed-labeling hydrogen−deuterium exchange coupled to mass spectrometry (HX-MS), it was observed that the B-C β-barrel folds early, allowing the s1C−s2C interaction to anchor one end of the RCL in an exposed conformation (14). On the other hand, the formation of side-chain packing interactions was monitored by hydroxyl-labeling mass spectrometry, and it was observed that early hydrophobic collapse in the core of β-sheet A prevents the insertion of RCL between strands s5A and s3A (15). Although these experiments identify the mechanisms by which the RCL is maintained in its exposed conformation during folding, a comparison with the folding mechanism of the latent conformation is not available. Here, we perform folding simulations of both the active and the latent conformations of α1-AT using structure-based (or Gō) models (16, 17) to understand the structural basis of the hypothesized larger kinetic barrier to fold to the latent conformation.
Results
Folding Simulations Capture the Mechanism of Folding of WT α1-AT to the Active Conformation.
Structure-based models (SBMs) are models based on the energy landscape theory of protein folding (16, 17) and have been successful at capturing the folding mechanisms and free energy barriers of diverse proteins (18–20). We constructed an SBM which encodes the structure of the active conformation of α1-AT as the global minimum of its potential energy function. Each residue of the protein is coarse-grained and represented by a single C-α atom (with native contacts shown in Fig. 3). We then performed simulations at the folding temperature (Tf) to obtain the folding free energy profile (FEP; see SI Appendix, SI Methods for details). The Tf is that temperature at which folding and unfolding is equally likely.
Fig. 3.
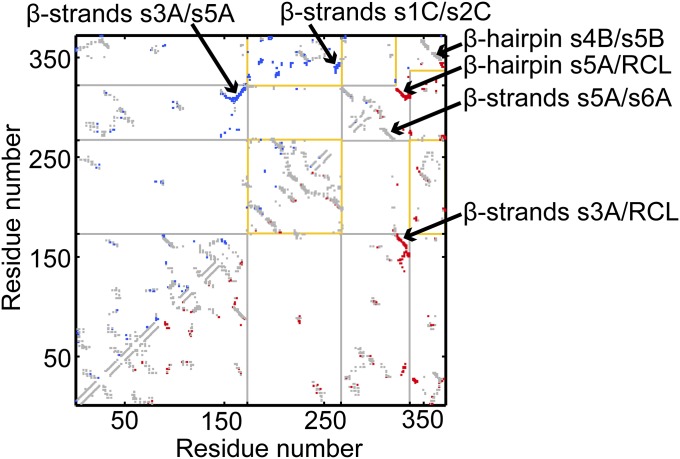
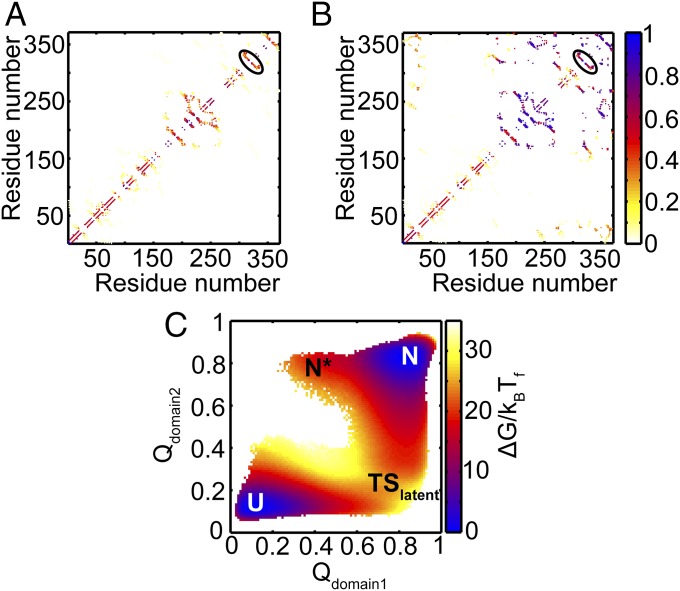
Native contact map of the active and latent conformations of α1-AT. In this map, a native contact between residues i and j is marked at (i, j), with residue numbers as the X and Y axes. The native contact maps of the active conformation and the latent conformation are shown in the upper and lower triangles, respectively. Common contacts are in gray, while those unique to the active and latent conformations are in blue and red, respectively. For example, interactions between strands s3A and s5A are unique to the active conformation, while those between strands s5A and RCL, and those between s3A and RCL, are unique to the latent conformation. The latter two sets of contacts form as a result of the insertion of the RCL between s3A and s5A. Contacts of domain 1 are bounded by yellow lines, while those of domain 2 are bounded by gray lines. Contacts which are bounded by both yellow and gray lines are interdomain contacts. The domain boundaries near the RCL are different for the active and the latent conformations and are represented by the incomplete bounding boxes near the C-terminal region of the contact map.
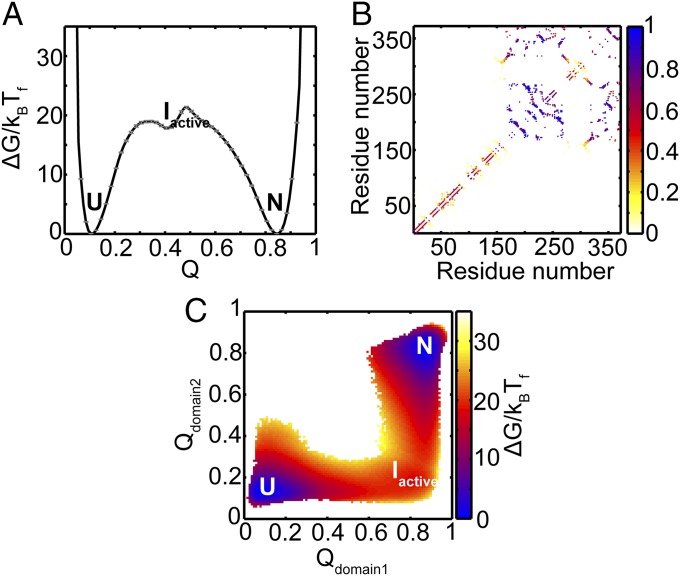
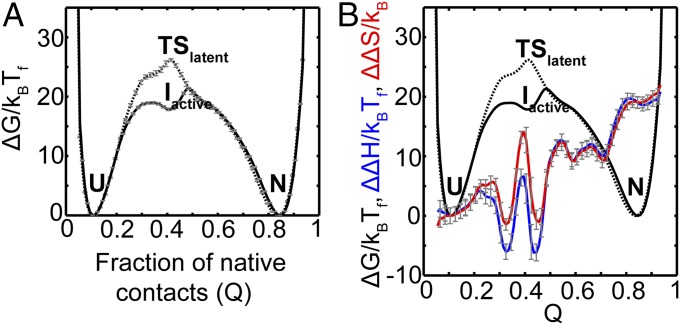
The FEP at Tf for α1-AT to fold to its active conformation is shown in Fig. 4A as a function of the fraction of native contacts formed (Q). Native contacts are nonbonded interactions between those residues that are close in the native structure. These interactions are then mapped onto the corresponding C-α atoms of the residues (Fig. 3). The reaction coordinate Q measures the extent to which these native contacts are formed during folding and therefore represents the extent to which the protein has folded. Q has previously been used to understand the progress of protein folding (19, 21, 22). In Fig. 4A, the value of Q is 0.1 when the protein is unfolded (U), and is 0.84 when it is in the native state (N). Two large free energy barriers of ∼19 and 22 kBTf are observed at Q ≈ 0.31 and 0.49, respectively. An intermediate (termed Iactive) is also populated in between these two barriers at Q ≈ 0.4 with ΔGIactive−U ≈ 18 kBTf.
Fig. 4.
Folding FEP and average contact map of the intermediate ensemble of active α1-AT. (A) FEP plotted as a function of the fraction of native contacts (Q) at the folding temperature (Tf). The FEP and the error bars represent the average and SD from four independent replicates. The native ensemble, N, is at Q ≈ 0.84; the intermediate ensemble, Iactive, is at Q ≈ 0.4; and the unfolded ensemble, U, is at Q ≈ 0.1. (B) The average contact map of the intermediate, Iactive, shows that domain 1 is mostly folded, while domain 2 is unfolded. The colors represent the probability of contact formation. The average structure of the Iactive intermediate is shown in SI Appendix, Fig. S2. (C) The average 2DFES as a function of Qdomain2 and Qdomain1 shows that domain 1 folds before domain 2. The colors show the height of the free energy in kBTf with the same scale as that shown in A. The errors on the 2DFES are shown in SI Appendix, Fig. S3.
According to the CATH database, α1-AT is defined to have two domains (Fig. 2) (23). The average contact map of the Iactive intermediate (Fig. 4B) reveals that the region corresponding to the B-C β-barrel in domain 1 is significantly folded, while domain 2 is not. The 2D free energy surface (2DFES) along the fraction of native contacts formed in domain 2 (Qdomain2) versus those formed in domain 1 (Qdomain1) also shows that domain 1 folds first, followed by domain 2 (Fig. 4C).
Previous studies have shown, using both far-UV circular dichroism and fluorescence spectroscopies, that the reversible equilibrium unfolding transition of WT α1-AT is observed to be three-state, with the presence of an intermediate at 1 M to 1.5 M Guanidium chloride (13, 24, 25). Fluorescence from site-specific probes (24) and FRET dyes (25) suggests that the region at the base of β-sheet A (part of domain 2; see Fig. 2A) is less stable and unfolds first, while β-sheet B (part of domain 1) is more stable and unfolds last. Kinetics experiments also suggest that folding and unfolding of α1-AT involves the formation of an intermediate species and therefore occurs in two distinct kinetic phases (14, 26). Pulsed-labeling HX-MS−based refolding kinetics of α1-AT suggests that polypeptide segments in domain 1 fold first (no lag phase, τfast ≈ 1,000 s), followed by segments in domain 2 (have a lag phase, τslow ≈ 1,500 s) (14). Further, unfolding kinetics experiments on WT and mutants of α1-AT monitored using bis-ANS fluorescence showed that β-strand s5A and helix hI (both are present in domain 2) unfold in the first phase that leads to the formation of the intermediate (26). This is followed by unfolding of the rest of the protein (i.e., regions in domain 1) in the second phase. Taken together, both equilibrium and kinetics studies of folding and unfolding of α1-AT suggest that domain 2 unfolds first, followed by domain 1, and that the order is reversed during refolding. However, refolding kinetics monitored by side-chain hydroxyl-labeling mass spectrometry shows an early hydrophobic collapse in the core of β-sheet A (15). This mechanism suggests that domain 2 folds first, followed by domain 1, and is not captured in our simulations. The presence of side-chains in a model implies a heterogeneity in the number of native and nonnative interactions that a residue can make, as well as in the excluded volume. It is likely that the absence of both nonnative interactions and side-chains in the present C-α SBM precludes the observation of hydrophobic collapse phenomena in our simulations (27–29). Finally, an intermediate is not observed in calorimetric measurements of specific heat capacity versus temperature (11), while it is observed as an equilibrium intermediate in chemical denaturation studies (11, 13, 24, 25). The marginal stability of Iactive in the thermally driven folding simulations presented here (Fig. 4A) may be the reason for the absence of this intermediate in the calorimetric data. However, we find that the contact pattern of the high-energy intermediate observed in our simulations closely resembles the intermediate observed in chemical denaturation studies. Thus, it is possible that the same intermediate ensemble, Iactive, is stabilized by chemical denaturants in experiments. In summary, we find that the structural features of the folding landscape of a C-α SBM of active α1-AT agrees well with far-UV CD [equilibrium studies (13, 24)], pulsed-labeling HX-MS [refolding kinetics study (14)], and bis-ANS fluorescence [unfolding kinetics study (26)] experiments.
Additionally, the refolding free energy barrier computed from our simulations suggests a folding time constant of τ ≈ 1,000 s (see SI Appendix, SI Results for the calculation), which is in good agreement with the experimental folding times of τfast ≈ 1,000 s and τslow ≈ 1,500 s (14). The correct prediction of the folding mechanism of the active conformation of α1-AT, the agreement of folding rates with experiment, and the absence of multiple routes and extensively populated off-pathway intermediates in Fig. 4C suggest that Q is an appropriate reaction coordinate for the folding of the active conformation. Here, we also use Q as a reaction coordinate because previous studies have shown its appropriateness in protein folding simulations (19, 21, 22). In the next section, we compare the folding FEP of the active conformation with that of the folding FEP of the latent conformation.
The Barrier to Fold to the Latent Conformation of α1-AT Is Larger than the Barrier to Fold to the Active Conformation.
Previous studies on serpins have hypothesized that folding to the metastable active conformation must have a lower free energy barrier compared with folding to the more stable latent conformation (4). To test this hypothesis, we constructed a C-α SBM of the latent conformation and obtained its folding FEP at its Tf (see Methods).
The FEP versus Q of latent α1-AT is shown in Fig. 5A (dotted line). The unfolded (U) and native (N) states are folded similarly to the corresponding states in the active α1-AT SBM and are present at Q ≈ 0.1 and Q ≈ 0.84, respectively. A single free energy barrier of ∼26 kBTf separates the native and the unfolded ensembles. Unlike the FEP of the active conformation (Fig. 4A), this free energy barrier is sharper (Fig. 5A) and an intermediate population is not observed. Such a barrier is in agreement with denaturant-induced equilibrium unfolding experiments on the latent conformation which show a single cooperative transition (13). The extent of increased stability of the latent conformation of WT α1-AT is not available from experiment. Nevertheless, to assess the effect of this stability on the barrier height, we reweighted the FEP of the latent conformation to that temperature below Tf where the height of the folding barrier is the same as that of the active conformation. We find that, as long as the latent state is stable up to a free energy of ∼13 kBT units more than the active conformation (SI Appendix, Fig. S12), α1-AT will find it easier to access the active conformation. This equals an excess stability of about 7.7 kcal/mol at 25 °C (without including any empirical model related corrections). The stability of the active conformation at 25 °C value is 11 kcal/mol (24). Therefore, as long as the stability of the latent conformation is less than ∼18.7 kcal/mol (11 + 7.7 kcal/mol), the active conformation will have a smaller folding barrier than the latent conformation.
Fig. 5.
Folding FEP of latent α1-AT compared with that of active α1-AT. (A) The FEP plotted as a function of the fraction of native contacts (Q) at the folding temperature (Tf) is shown as a dotted line. The FEP and the error bars represent the average and SD from four independent replicates. The FEP of active α1-AT is reproduced from Fig. 4A (solid black line) for comparison. The native ensemble, N, is at Q ≈ 0.84; the transition state ensemble, TSlatent, is at Q ≈ 0.4; and the unfolded ensemble, U, is at Q ≈ 0.1. (B) The average FEPs from A are reproduced. The relative changes in enthalpy, ΔΔH (active−latent), and entropy ΔΔS (active−latent), between the folding of active and latent α1-AT, plotted versus Q are shown in blue and red, respectively. This plot shows that ΔΔS at Q ≈ 0.4 is higher than ΔΔH at Q ≈ 0.4. The error bars (gray) represent the SD from four independent replicates.
The position of the transition state (TSlatent) at Q ≈ 0.4 is similar to the position of the intermediate Iactive in the FEP of the active conformation, implying that both TSlatent and Iactive are folded to a similar extent in terms of the fraction of native contacts formed. However, Iactive is more stable than TSlatent by ΔΔG (active–latent) ≈ −8kBTf. The corresponding ΔΔH (active–latent) and ΔΔS (active–latent) at Q ≈ 0.4 are observed to be ∼6 kBTf and ∼14 kB, respectively (Fig. 5B, blue and red lines). Therefore, folding to the latent conformation has a higher folding free energy barrier compared with folding to the active conformation of α1-AT, and this is due to the relatively lower entropy of TSlatent compared with Iactive. In the next section, we compare the folding mechanism of the active conformation to that of the latent conformation to understand the structural reasons behind the differences in their free energy barriers.
Early Formation of the s5A-RCL Contacts Distinguishes the Folding to the Latent Conformation.
The folding mechanism of the active conformation suggests that domain 1 is already folded in Iactive, and therefore the RCL is already in the exposed conformation (Fig. 4 B and C). The average contact map of the near-unfolded ensemble at Q ≈ 0.2 from the FEP of the latent conformation is shown in Fig. 6A. It shows that the native contacts between s5A and RCL are already formed, as they are contacts between residues that are nearby in sequence (i.e., local contacts). Further folding to the transition state ensemble involves the folding of domain 1 (Fig. 6B). Even though this ensemble is folded to a similar extent as Iactive (both ensembles are at Q ≈ 0.4), the TSlatent ensemble also has partial formation of several long-range contacts, i.e., contacts between residues that are farther apart in sequence (Fig. 6B). These are both interdomain contacts as well as contacts of domain 2. Finally, domain 2 folds completely to form the latent conformation (Fig. 6C). Therefore, despite the overall similarity of the 2DFESs shown in Figs. 4C and 6C, and the partial contact maps shown in Figs. 4B and 6B, an important distinguishing feature in folding to the latent conformation is the early formation of the contacts between s5A and RCL. These contacts stabilize the RCL in its inserted conformation early during folding to the latent conformation. In contrast, contacts within the B-C β-barrel, not present in the latent state simulations, form early in the active state simulations and help to keep the RCL exposed. In the next section, we discuss the implications of the folding mechanisms of the active and the latent conformations on the function of α1-AT. A discussion about what these folding mechanisms might mean for disease-linked polymerization is given in SI Appendix, SI Discussion.
Fig. 6.
Folding mechanism of latent α1-AT. (A) The average contact map of the near-unfolded ensemble at Q ≈ 0.2 shows that the contacts between β-strand s5A and RCL are already formed (black ellipse). The colors represent the probability of contact formation. (B) The average contact map of the transition state ensemble at Q ≈ 0.4 shows that domain 1 and the contacts between β-strand s5A and RCL (black ellipse) are formed. In addition, several contacts between the N and C termini which are also interdomain contacts are also partially formed. (C) The average 2DFES plotted as a function of Qdomain2 and Qdomain1 shows that domain 1 folds before domain 2 during folding. The colors show the height of the free energy in kBTf with the same scale as that shown in Fig. 5A. The errors on the 2DFES are shown in SI Appendix, Fig. S4.
Discussion
Function and Folding Fate Are both Encoded by the Conformation of the RCL.
The structures of the active and latent conformations of α1-AT show that an exposed RCL is necessary for protease inhibition and that the insertion of the RCL into β-sheet A makes the serpin inactive (4, 13). The presence of an exposed RCL versus an inserted RCL is the main structural difference between the active and latent conformations of α1-AT (Fig. 1). SBMs for the active and latent conformations of α1-AT incorporate each of the two different conformations of RCL separately. Folding simulations of these SBMs reveal that early formation of an inserted RCL conformation imposes a larger free energy barrier relative to the early formation of an exposed RCL conformation (Fig. 5A). While the exposed RCL is wholly part of domain 1, the inserted RCL conformation has native contacts with both domains (Fig. 2). We suggest that this causes the RCL to act as a “pin” that structurally links both the domains in the latent conformation. When the inserted conformation of the RCL forms early, the folding free energy barrier for further folding is larger because the inserted RCL couples folding of the two domains and causes them to fold in tandem (Fig. 5A). The inserted RCL involves the formation of a β-hairpin with β-strand s5A (the required interactions are local in sequence; Fig. 3), and we therefore predict that this β-hairpin will fold early in WT α1-AT experiments where the protein sequence likely encodes both the active and the latent conformations. As the barrier to further folding to the latent state is larger, this β-hairpin must unfold before the protein can fold to the active conformation. This phenomenon arises when geometrical constraints in the protein make it harder for some of the near-unfolded states to fold further and is known as backtracking (30). On the other hand, when the exposed conformation of the RCL forms early, the folding free energy barrier is lower, because the RCL does not couple the folding of domain 2 to domain 1. This leads to the formation of an entropically stabilized intermediate Iactive and folding to the active conformation (Fig. 5). This interpretation is consistent with the observation of a three-state unfolding transition for the active α1-AT, compared with the two-state unfolding transition of latent α1-AT (13). In summary, the RCL, when in its inserted conformation, creates an interface between the two domains of α1-AT. This, in turn, creates a larger barrier to folding and makes folding more cooperative. On the other hand, the RCL in its exposed conformation cannot do the same, and the two domains of α1-AT fold semiindependently with a folding intermediate and a lower barrier (by ∼8kBTf) to folding. The conformation of the RCL adopted early during folding tunes the strength of the interdomain interactions to modulate the folding free energy barrier, and this is consistent with observations made earlier on repeat proteins (31). Thus, even a small loop, here the 12-residue RCL, can modulate the folding cooperativity or the all-or-nothing folding of a large protein such as the 370-residue α1-AT.
Destabilizing mutations in the B-C β-barrel [V364A, F366A/L286A, V364A/L288A, V364A/F366A, and I229A/V364A (13); see SI Appendix, Fig. S5] likely switch the stability of the RCL-exposed and RCL-inserted conformations along the folding route and open a trapdoor (1) to facilitate folding to the latent conformation instead (13). We propose that the structural differences between RCL conformations adopted early during folding determine the folding barrier, the final state to which the protein folds to, and therefore the functional fate of α1-AT. The similarity between the structures of the active conformations, the latent conformations, the mechanism of protease inhibition (4, 32), and the folding mechanism (14, 33) of serpins suggests that the conformation of the RCL is key to both folding and function. This is in contrast to several proteins where there is a tradeoff between functional residues and folding (34). The early folding of the RCL implies that the conformation of α1-AT is decided early during folding and “locked” in by the folding of the rest of the large α1-AT. We hypothesize that this locking in could be the cause of the large barrier to conformational transition that is seen in α1-AT (1).
The focus of the current simulations is to understand the structural basis by which α1-AT gates kinetic accessibility to the active conformation using single structure encoding SBMs. However, to understand the competition between folding to the active conformation versus the latent conformation as well as the conformational transition between the two conformations, a dual SBM, encoding both structures in the potential energy function (35), is required, and we plan to construct and simulate such an SBM in the future. In the next section, we describe an experimental strategy to confirm several results from our current simulations.
Folding to the Latent Conformation Could Be Captured by Engineering Disulfide Bonds into the RCL.
While the folding kinetics of the active conformation has been measured experimentally (14, 15), it is not possible to measure the folding kinetics of the latent conformation, because WT α1-AT always folds to the active conformation. Multiple destabilizing mutations in the B-C β-barrel are necessary to facilitate folding to the latent conformation (13). The folding kinetics of this multiple mutant α1-AT may not report on the actual folding rate of the latent conformation unless the effects of these destabilizing mutations are accounted for. Instead, we propose the following strategy to measure the folding kinetics of the active and latent conformations using a single protein sequence. Folding simulations of the SBM of latent α1-AT show that the RCL forms a β-hairpin with β-strand s5A very early during folding (Fig. 6A). We propose that a disulfide bond can be engineered into WT α1-AT to lock the two ends of the s5A/RCL β-hairpin together. This is expected to stabilize the inserted RCL conformation and facilitate folding to the latent conformation. A potential location for this disulfide bond is between the N terminus of β-strand s5A and the C terminus of the RCL. This could be achieved by the triple mutation Cys232Ala/Val333Cys/Phe352Cys [residue numbering according to Protein Data Bank (PDB) 1QLP] in α1-AT. The Cys232Ala mutation is necessary to replace the native Cys residue (36), while the mutation at residues Val333 and Phe352 could encode the engineered disulfide bond. We also suggest Lys335Cys/Ala350Cys as an alternative location to engineer the disulfide bond between strands s5A and the RCL. In the past, engineered disulfide bonds have been used to successfully stabilize specific conformations in serpins (36, 37), and to map folding pathways, alter folding rates (38), or capture alternative native conformations in other proteins (39, 40). Folding kinetics of the disulfide-bonded α1-AT under oxidizing conditions are expected to report on the folding rate of the latent conformation. On the other hand, folding kinetics under reducing conditions, i.e., conditions under which the disulfide bond will not be formed, are expected to report on the folding rate of the active conformation. Our simulations (Fig. 5A) predict that folding to the latent conformation will be slower than folding to the active one. We note that engineering the disulfide bond is not expected to change the present latent state α1-AT simulations, because the SBM contains only the latent conformation and the RCL already inserts early in these simulations. However, the effects of engineering an appropriate disulphide bond can be tested in the context of a dual SBM of α1-AT.
Conclusions
The serine protease inhibitor, α1-AT, is a two-domain protein which folds to a metastable, active conformation that can convert into a thermodynamically stable latent conformation. Folding simulations of both conformations using separate SBMs revealed that it is harder to fold to the latent conformation. The RCL, which houses the protease binding site, is exposed in the active conformation, while it is inserted into a β-sheet in the latent conformation. The early formation of native contacts between the β-strand s5A and the RCL constrains latent α1-AT to fold with an inserted RCL. An inserted RCL couples the folding of the two domains of α1-AT and causes several long-range interdomain interactions to form in the transition state ensemble of the latent state. This imposes a larger entropic penalty on the transition state and increases the free energy barrier to fold to the latent state. In contrast, the exposed conformation of the RCL in the active α1-AT is stabilized in an on-pathway intermediate, Iactive, and this reduces the free energy barrier by ∼8 kBTf and facilitates faster folding to the active conformation. Thus, the RCL, a small 12-amino acid loop, determines the free energy barrier and, in turn, the all-or-nothing folding or folding cooperativity of the large 370-amino acid α1-AT. It should also be noted that this difference between the large folding free energy barriers of the two conformations (26 − 18 = 8 kBTf) is larger than the entire folding free energy barriers of most single-domain proteins. Further, in contrast to what is seen in several proteins where regions that drive folding are separate from functional regions, the RCL and its conformation determine the barrier to folding to the two alternative conformations as well as the functional fate of α1-AT. We propose that engineering a disulfide bond which locks the RCL in the inserted conformation under oxidizing conditions will likely cause α1-AT to fold to the latent conformation without the need for destabilizing mutations as were required earlier. Experimental measurement of folding kinetics and routes under oxidizing and reducing conditions will allow for testing our result that folding to the latent conformation is slower than folding to the active conformation. The metastability of the active conformation has been hypothesized to facilitate disease-causing polymerization. Using the structural ensembles populated in our simulations, such as the intermediate ensemble, Iactive, we are also able to propose mechanisms of serpin polymerization (SI Appendix, SI Discussion and Fig. S13).
Methods
We simulated the α1-AT conformations using C-α SBMs in which each residue is represented by a bead at its C-α atom (18). The present procedure of using C-α SBMs of alternative conformations to understand their differing barriers is similar to that used for determining the folding barriers of the syn- and anti-ROP dimers (41). We obtained the native structures of active and latent conformations of α1-AT from PDB IDs 1QLP and 1IZ2, respectively (Fig. 1). Native contact maps for both conformations were calculated using the CSU software (42). The coordinates of the C-α atoms from the PDB file and its corresponding native contact map were given as inputs to the SMOG webserver (43) to separately generate the C-α SBM for the two conformations. Molecular dynamics simulations were performed using GROMACS (44) and replica exchange umbrella sampling (REUS) at the folding temperature (Tf) (45). The resulting simulations were reweighted using the weighted histogram analysis method to obtain the unbiased distribution of conformational states. The negative logarithm of this distribution is plotted as the folding FEP at Tf. Further details on the proteins, SBMs, REUS simulations, and simulation analyses are given in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
S.G. thanks the Government of India, Department of Atomic Energy for core funding and the Government of India, Department of Science and Technology for the Ramanujan Fellowship (Grant SR/S2/RJN-63/2009, 5 y, with effect from April 15, 2010).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708173115/-/DCSupplemental.
References
- 1.Schug A, Whitford PC, Levy Y, Onuchic JN. Mutations as trapdoors to two competing native conformations of the Rop-dimer. Proc Natl Acad Sci USA. 2007;104:17674–17679. doi: 10.1073/pnas.0706077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amemiya T, Koike R, Fuchigami S, Ikeguchi M, Kidera A. Classification and annotation of the relationship between protein structural change and ligand binding. J Mol Biol. 2011;408:568–584. doi: 10.1016/j.jmb.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 3.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whisstock JC, Bottomley SP. Molecular gymnastics: Serpin structure, folding and misfolding. Curr Opin Struct Biol. 2006;16:761–768. doi: 10.1016/j.sbi.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Sohl JL, Jaswal SS, Agard DA. Unfolded conformations of alpha-lytic protease are more stable than its native state. Nature. 1998;395:817–819. doi: 10.1038/27470. [DOI] [PubMed] [Google Scholar]
- 6.Carr CM, Chaudhry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinner AR, Karplus M. A metastable state in folding simulations of a protein model. Nat Struct Biol. 1998;5:236–241. doi: 10.1038/nsb0398-236. [DOI] [PubMed] [Google Scholar]
- 8.Honeycutt JD, Thirumalai D. Metastability of the folded states of globular proteins. Proc Natl Acad Sci USA. 1990;87:3526–3529. doi: 10.1073/pnas.87.9.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C, Park SH, Lee MY, Yu MH. Regulation of protein function by native metastability. Proc Natl Acad Sci USA. 2000;97:7727–7731. doi: 10.1073/pnas.97.14.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gettins PG, Olson ST. Inhibitory serpins. New insights into their folding, polymerization, regulation and clearance. Biochem J. 2016;473:2273–2293. doi: 10.1042/BCJ20160014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaslik G, et al. Effects of serpin binding on the target proteinase: Global stabilization, localized increased structural flexibility, and conserved hydrogen bonding at the active site. Biochemistry. 1997;36:5455–5464. doi: 10.1021/bi962931m. [DOI] [PubMed] [Google Scholar]
- 12.Lomas DA, Elliott PR, Chang WS, Wardell MR, Carrell RW. Preparation and characterization of latent alpha 1-antitrypsin. J Biol Chem. 1995;270:5282–5288. doi: 10.1074/jbc.270.10.5282. [DOI] [PubMed] [Google Scholar]
- 13.Im H, Woo M-S, Hwang KY, Yu M-H. Interactions causing the kinetic trap in serpin protein folding. J Biol Chem. 2002;277:46347–46354. doi: 10.1074/jbc.M207682200. [DOI] [PubMed] [Google Scholar]
- 14.Tsutsui Y, Dela Cruz R, Wintrode PL. Folding mechanism of the metastable serpin α1-antitrypsin. Proc Natl Acad Sci USA. 2012;109:4467–4472. doi: 10.1073/pnas.1109125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stocks BB, Sarkar A, Wintrode PL, Konermann L. Early hydrophobic collapse of α1-antitrypsin facilitates formation of a metastable state: Insights from oxidative labeling and mass spectrometry. J Mol Biol. 2012;423:789–799. doi: 10.1016/j.jmb.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 17.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 19.Chavez LL, Onuchic JN, Clementi C. Quantifying the roughness on the free energy landscape: Entropic bottlenecks and protein folding rates. J Am Chem Soc. 2004;126:8426–8432. doi: 10.1021/ja049510+. [DOI] [PubMed] [Google Scholar]
- 20.Hyeon C, Thirumalai D. Capturing the essence of folding and functions of biomolecules using coarse-grained models. Nat Commun. 2011;2:487. doi: 10.1038/ncomms1481. [DOI] [PubMed] [Google Scholar]
- 21.Cho SS, Levy Y, Wolynes PG. P versus Q: Structural reaction coordinates capture protein folding on smooth landscapes. Proc Natl Acad Sci USA. 2006;103:586–591. doi: 10.1073/pnas.0509768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best RB, Hummer G, Eaton WA. Native contacts determine protein folding mechanisms in atomistic simulations. Proc Natl Acad Sci USA. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sillitoe I, et al. CATH: Comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015;43:D376–D381. doi: 10.1093/nar/gku947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James EL, Whisstock JC, Gore MG, Bottomley SP. Probing the unfolding pathway of alpha1-antitrypsin. J Biol Chem. 1999;274:9482–9488. doi: 10.1074/jbc.274.14.9482. [DOI] [PubMed] [Google Scholar]
- 25.Liu L, Werner M, Gershenson A. Collapse of a long axis: Single-molecule Förster resonance energy transfer and serpin equilibrium unfolding. Biochemistry. 2014;53:2903–2914. doi: 10.1021/bi401622n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knaupp AS, et al. The roles of helix I and strand 5A in the folding, function and misfolding of α1-antitrypsin. PLoS One. 2013;8:e54766. doi: 10.1371/journal.pone.0054766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarrine-Afsar A, et al. Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding. Proc Natl Acad Sci USA. 2008;105:9999–10004. doi: 10.1073/pnas.0801874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T, Chan HS. Native contact density and nonnative hydrophobic effects in the folding of bacterial immunity proteins. PLOS Comput Biol. 2015;11:e1004260. doi: 10.1371/journal.pcbi.1004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, et al. 2017 All-atom simulations predict how single point mutations promote serpin misfolding. Available at https://arxiv.org/abs/1707.05019. Accessed October 29, 2017.
- 30.Gosavi S, Whitford PC, Jennings PA, Onuchic JN. Extracting function from a beta-trefoil folding motif. Proc Natl Acad Sci USA. 2008;105:10384–10389. doi: 10.1073/pnas.0801343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagai T, Azia A, Trizac E, Levy Y. Modulation of folding kinetics of repeat proteins: Interplay between intra- and interdomain interactions. Biophys J. 2012;103:1555–1565. doi: 10.1016/j.bpj.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman GA, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 33.Chandrasekhar K, et al. Cellular folding pathway of a metastable serpin. Proc Natl Acad Sci USA. 2016;113:6484–6489. doi: 10.1073/pnas.1603386113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giri Rao VVH, Gosavi S. Using the folding landscapes of proteins to understand protein function. Curr Opin Struct Biol. 2016;36:67–74. doi: 10.1016/j.sbi.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Whitford PC, Sanbonmatsu KY, Onuchic JN. Biomolecular dynamics: Order-disorder transitions and energy landscapes. Rep Prog Phys. 2012;75:076601. doi: 10.1088/0034-4885/75/7/076601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamasaki M, Sendall TJ, Pearce MC, Whisstock JC, Huntington JA. Molecular basis of α1-antitrypsin deficiency revealed by the structure of a domain-swapped trimer. EMBO Rep. 2011;12:1011–1017. doi: 10.1038/embor.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamasaki M, Li W, Johnson DJD, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 38.Abkevich VI, Shakhnovich EI. What can disulfide bonds tell us about protein energetics, function and folding: Simulations and bioninformatics analysis. J Mol Biol. 2000;300:975–985. doi: 10.1006/jmbi.2000.3893. [DOI] [PubMed] [Google Scholar]
- 39.Clarke J, Fersht AR. Engineered disulfide bonds as probes of the folding pathway of barnase: Increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 40.Cho SS, Levy Y, Onuchic JN, Wolynes PG. Overcoming residual frustration in domain-swapping: The roles of disulfide bonds in dimerization and aggregation. Phys Biol. 2005;2:S44–S55. doi: 10.1088/1478-3975/2/2/S05. [DOI] [PubMed] [Google Scholar]
- 41.Levy Y, Cho SS, Shen T, Onuchic JN, Wolynes PG. Symmetry and frustration in protein energy landscapes: A near degeneracy resolves the Rop dimer-folding mystery. Proc Natl Acad Sci USA. 2005;102:2373–2378. doi: 10.1073/pnas.0409572102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 43.Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN. SMOG@ctbp: Simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. 2010;38:W657–W661. doi: 10.1093/nar/gkq498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murata K, Sugita Y, Okamoto Y. Free energy calculations for DNA base stacking by replica-exchange umbrella sampling. Chem Phys Lett. 2004;385:1–7. [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.