Significance
Mitochondria generate the cellular fuel ATP to sustain complex life. Production of ATP depends on the oxidation of energy-rich compounds to produce the proton motive force (pmf), a chemical potential difference for protons, across the inner membrane. The pmf drives the ATP synthase to synthesize ATP via a mechanical rotary mechanism. The structures and functions of the protein components of this molecular machine, especially those involved directly in the catalytic formation of ATP, are widely conserved in metazoans, fungi, and eubacteria. Here we show that the proposal that this conservation does not extend to the ATP synthase from Trypanosoma brucei, a member of the euglenozoa and the causative agent of sleeping sickness in humans, is incorrect.
Keywords: ATP synthase, Trypanosoma brucei, p18 subunit, catalytic domain, structure
Abstract
The structures and functions of the components of ATP synthases, especially those subunits involved directly in the catalytic formation of ATP, are widely conserved in metazoans, fungi, eubacteria, and plant chloroplasts. On the basis of a map at 32.5-Å resolution determined in situ in the mitochondria of Trypanosoma brucei by electron cryotomography, it has been proposed that the ATP synthase in this species has a noncanonical structure and different catalytic sites in which the catalytically essential arginine finger is provided not by the α-subunit adjacent to the catalytic nucleotide-binding site as in all species investigated to date, but rather by a protein, p18, found only in the euglenozoa. A crystal structure at 3.2-Å resolution of the catalytic domain of the same enzyme demonstrates that this proposal is incorrect. In many respects, the structure is similar to the structures of F1-ATPases determined previously. The α3β3-spherical portion of the catalytic domain in which the three catalytic sites are found, plus the central stalk, are highly conserved, and the arginine finger is provided conventionally by the α-subunits adjacent to each of the three catalytic sites found in the β-subunits. Thus, the enzyme has a conventional catalytic mechanism. The structure differs from previous described structures by the presence of a p18 subunit, identified only in the euglenozoa, associated with the external surface of each of the three α-subunits, thereby elaborating the F1-domain. Subunit p18 is a pentatricopeptide repeat (PPR) protein with three PPRs and appears to have no function in the catalytic mechanism of the enzyme.
The ATP synthases, also known as F-ATPases or F1Fo-ATPases, are multisubunit enzyme complexes found in energy-transducing membranes in eubacteria, chloroplasts, and mitochondria (1, 2). They make ATP from ADP and phosphate under aerobic conditions using a proton-motive force (pmf), generated by respiration or photosynthesis, as a source of energy. To date, studies of the subunit compositions, structures, and mechanism of the ATP synthases have been confined mainly to the vertebrates, especially humans and bovines, and to various fungi, eubacteria, and chloroplasts of green plants. These studies have established the conservation of the central features of these rotary machines. They are all membrane-bound assemblies of multiple subunits organized into membrane-intrinsic and membrane-extrinsic sectors.
The membrane-extrinsic sector, known as F1-ATPase, is the catalytic part in which ATP is formed from ADP and inorganic phosphate. It can be detached experimentally from the membrane domain in an intact state, and retains the ability to hydrolyze, but not synthesize, ATP. The membrane intrinsic sector, sometimes called Fo, contains a rotary motor driven by pmf and is connected to the extrinsic domain by a central stalk and a peripheral stalk. The enzyme’s rotor constitutes the central stalk and an associated ring of c-subunits in the membrane domain. The central stalk lies along an axis of sixfold pseudosymmetry and penetrates into the α3β3-domain, where the catalytic sites of the enzyme are found at three of the interfaces of α- and β-subunits. The penetrant region of the central stalk is an asymmetric α-helical coiled coil, and its rotation inside the α3β3-domain takes each catalytic site through a series of conformational changes that lead to the binding of substrates and the formation and release of ATP.
During ATP hydrolysis in the experimentally detached F1-domain, the direction of rotation, now driven by energy released from the hydrolysis of ATP, is opposite to the synthetic sense. Extensive structural analyses, mostly by X-ray crystallography at atomic resolution, have shown that the F1-domains of the enzymes from bovine (3–23) and yeast (24–30) mitochondria, chloroplasts (31, 32), and eubacteria (33–39) are highly conserved. Not only is there conservation of the subunit compositions of the α3β3-domain and the central stalk (γ1ε1 in eubacteria and chloroplasts, and γ1δ1 plus an additional unique subunit, confusingly called ε, attached to the δ-subunit in mitochondria orthologs), but also the sequences of subunits are either highly conserved or absolutely conserved in many key residues. This extensive conservation includes residues in catalytic interfaces and in the catalytic sites themselves. In the β-subunits, they include a hydrophobic pocket where the adenine ring of ADP (or ATP) is bound; a P-loop sequence that interacts with the α-, β-, and γ-phosphates of ATP and provides residues involved either directly or indirectly via water molecules in the binding of a hexa-coordinate magnesium ion; and, in the adjacent α-subunit, an “arginine finger” residue, which senses whether ADP or ATP is bound to the catalytic site. Indeed, these catalytic features are common to a wide range of NTPases (40, 41), and together with conserved structural features are characteristic of the canonical ATP synthase.
Based on a structural model at 32.5-Å resolution derived by electron cryotomography (ECT), it has been suggested recently that the structure of the F1-catalytic domain and its catalytic mechanism in the ATP synthase from Trypanosoma brucei have diverged extensively from the canonical complex in an unprecedented manner (42). It has been proposed that the structure of this F1-domain is much more open than those described in other species, and that the “arginine finger” is provided not by the α-subunit, but rather by an additional p18-subunit found only in the euglenozoa (43–49). Here we examine this proposal in the context of a structure of the F1-domain of the T. brucei ATP synthase determined by X-ray crystallography at 3.2-Å resolution.
Results and Discussion
Structure Determination.
The crystals of the T. brucei F1-ATPase have the unit cell parameters a = 124.2 Å, b = 206.4 Å, and c = 130.2 Å, with α = γ = 90.0° and β = 104.9°, and they belong to space group P21, with one F1-ATPase in the asymmetric unit. Data processing and refinement statistics are presented in Table S1. The final model of the complex contains the following residues: αE, 20–125, 137–416, and 423–560; αTP, 22–127, 137–414, and 421–560; αDP, 22–125, 137–416, and 424–560; βE, 6–492; βTP, 7–494; βDP, 8–488; γ, 2–58 and 66–285; δ, 5–16 and 32–165; ε, 1–66; and three copies of p18, residues 6–169, 6–167, and 6–170, attached to the αTP-, αDP-, and αE-subunits, respectively (see below). An ADP molecule and a magnesium ion are bound to each of the three α-subunits and to the βTP- and βDP-subunits, whereas the βE-subunit has a bound ADP molecule without a magnesium ion. A similar nucleotide occupancy of catalytic and noncatalytic sites has been reported in the bovine F1-ATPase crystallized in the presence of phosphonate (20) and in the F1-ATPase from Caldalkalibacillus thermarum (38). These structures are interpreted as representing a posthydrolysis state in which the ADP molecule has not been released from the enzyme.
An unusual feature of the T. brucei F1-ATPase is that the diphosphate catalytic interface is more open than the triphosphate catalytic interface, similar to the F1-ATPase from Saccharomyces cerevisiae (24), whereas the converse is observed in all other structures (Table S2). As usual, the empty interface is the most open of the three catalytic interfaces (Table S2). The rotational position of the γ-subunit (determined by superposition of crown regions of structures) is +23.1° relative to the bovine phosphate release dwell, which is at or close to the catalytic dwell at +30° in the rotary catalytic cycle (6).
Structure of the F1-ATPase from T. brucei.
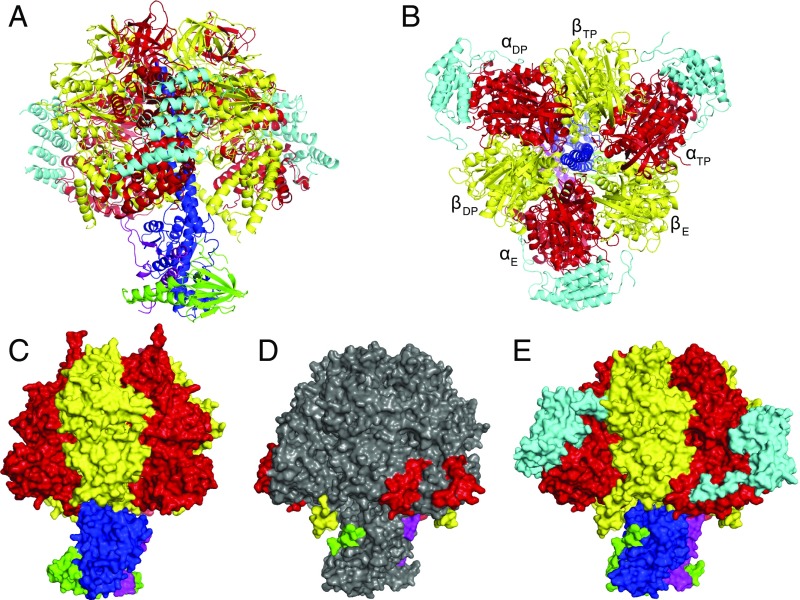
The structure consists of an α3β3-complex with α- and β-subunits arranged in alternation around an antiparallel α-helical coiled coil in the γ-subunit (Fig. 1). The rest of the γ-subunit sits beneath the α3β3-complex and is associated with the δ- and ε-subunits. Together, these three subunits form the central stalk. Thus, the overall structure of this catalytic domain of the ATP synthase complex is very close to structures of canonical F1-ATPases determined in the mitochondria of other species, and in eubacteria and chloroplasts. For example, in a comparison of backbone atoms with the bovine F1-ATPase crystallized in the presence of phosphonate (20), the rmsd is 3.24 Å. As in these other canonical structures, each of the α- and β-subunits in the T. brucei F1-ATPase has three domains. The N-terminal domain (residues 1–103 and 1–88 in α- and β-subunits, respectively) consists of a six-stranded β-barrel in both α- and β-subunits, and these six β-domains are associated in a stable annulus known as the “crown”. The central domain (residues 104–389 and 89–365 in α- and β-subunits, respectively) provides the nucleotide-binding sites (Fig. S1). The C-terminal domain consists of a bundle of seven and four α-helices in α- and β-subunits, respectively. The crown stabilizes the entire F1-domain, and, during rotary catalysis, the rest of the α- and β-subunits swing from this crown in response to the rotation of the asymmetrical α-helical coiled-coil region of the γ-subunit.
Fig. 1.
Structure of the F1-ATPase from T. brucei. The α-, β-, γ-, δ-, ε-, and p18-subunits are shown in red, yellow, blue, green, magenta, and cyan, respectively. (A and B) Side (A) and top (B) views in cartoon representation. (C–E) Side views in surface representation rotated 180° relative to A. (C) The bovine enzyme (12). (D and E) The T. brucei enzyme. In D, p18 has been omitted, and only additional regions not found in the bovine enzyme are colored; the rest of the structure is gray. The two additional sections in the α-subunit (red) interact with the p18-subunit. (E) p18 is present and is shown interacting with the α-subunit.
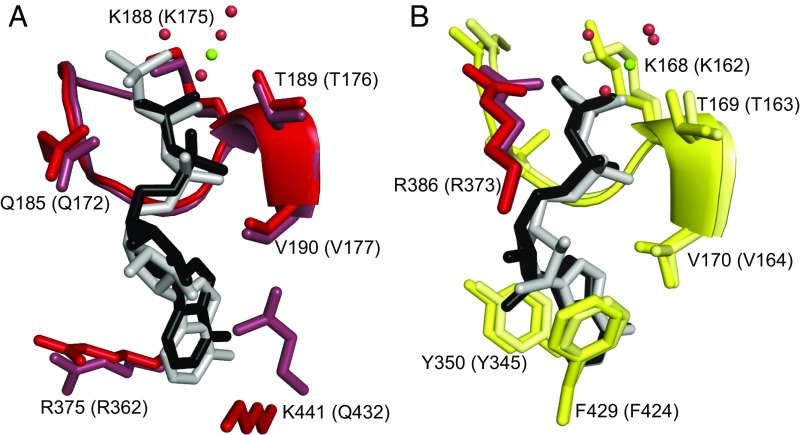
The six bound ADP molecules occupy nucleotide-binding sites that are very similar in structure to those in other ATP synthases. They retain the conventional features of a hydrophobic pocket to bind the adenine ring, and a characteristic P-loop sequence (GDRQTGKT in the α-subunit, residues 182–189; GGAGVGKT in the β-subunit, residues 162–169) interacting with the α- and β-phosphates of ADP or ATP (Fig. 2). The five magnesium ions are hexacoordinated by a threonine residue (residues 189 and 169 in α- and β-subunits, respectively) and four water molecules in each case. In the canonical enzymes, the nucleotides bound to the β-subunits participate in catalysis and exchange during a catalytic cycle, whereas those bound to the α-subunits are permanently bound to the enzyme and do not participate in catalysis. The close similarity of the structures of the T. brucei and bovine F1-ATPases suggests strongly that the α- and β-subunits in the T. brucei enzyme have the same, or very similar, roles to those in the bovine enzyme. Thus, the nucleotide-binding sites in the β-subunits are part of the catalytic sites of the enzyme, the other important catalytic feature being αArg-386, the arginine finger residue, which is positioned in the catalytic site in the βDP-subunit from T. brucei, for example, in exactly the same position occupied by the equivalent residue, αArg-373, in the bovine enzyme (Fig. 2).
Fig. 2.
Conservation of the noncatalytic and catalytic nucleotide-binding sites in the F1-ATPase from T. brucei. (A) The noncatalytic site in the αDP-subunit superposed onto the equivalent site in the bovine enzyme (12). (B) The catalytic site in the βDP-subunit superposed onto the equivalent site in the bovine enzyme. Residue αR386 is the catalytically essential arginine finger (equivalent to αR373 in the bovine protein). Residues contributed by α- and β-subunits are shown in red and yellow, respectively (with the bovine residues in muted colors), and the bound ADP molecules are in black in the T. brucei enzymes and in gray in the bovine enzymes. The green and red spheres represent magnesium ions and water molecules, respectively (in T. brucei only). The residue numbers in parentheses denote the equivalent bovine residues.
Despite the general conservation of the structure and mechanism of the T. brucei F1-ATPase, the euglenozoan enzyme is elaborated relative to the bovine enzyme, for example. First, the α-subunit in T. brucei is cleaved in vivo by proteolysis at two adjacent sites, removing residues 128–135 (Fig. S2) (50). The cleavage of α-subunits has been noted in other euglenozoan ATP synthases as well (48, 51–53), although the sites of cleavage have not been characterized precisely. In the bovine enzyme, the equivalent region (residues 117–123) forms an external loop (Fig. S2). These cleavages have no evident impact on the stability of either the α-subunit or the F1-ATPase complex itself. Second, the α-, β-, δ-, and ε-subunits of the T. brucei enzyme have additional surface features that are not found in the known structures of other F1-ATPases (Fig. 1). The most extensive are residues 483–498 and 536–560 in the C-terminal region of the α-subunit, and their significance is discussed below. The additional surface features in the β-, δ-, and ε-subunits are residues 485–499, 1–17, and 39–50, respectively. Those in the β- and ε-subunits have no obvious functions. The resolved residues of the additional sequence in the δ-subunit increases its area of interaction with the γ-subunit from 1,000 Å2 to 1,700 Å2. The C-terminal region of the γ-subunit from residues 286–304, although not resolved in the structure, is 19 residues longer than in the bovine enzyme, for example, and in the intact ATP synthase it could extend beyond the crown region, possibly making contacts, permanently or transiently, during rotary catalysis with the oligomycin sensitivity conferral protein (OSCP), a component of the peripheral stalk. In other species, the OSCP is bound to the F1-domain by the N-terminal regions of the three α-subunits (19, 29, 37, 54).
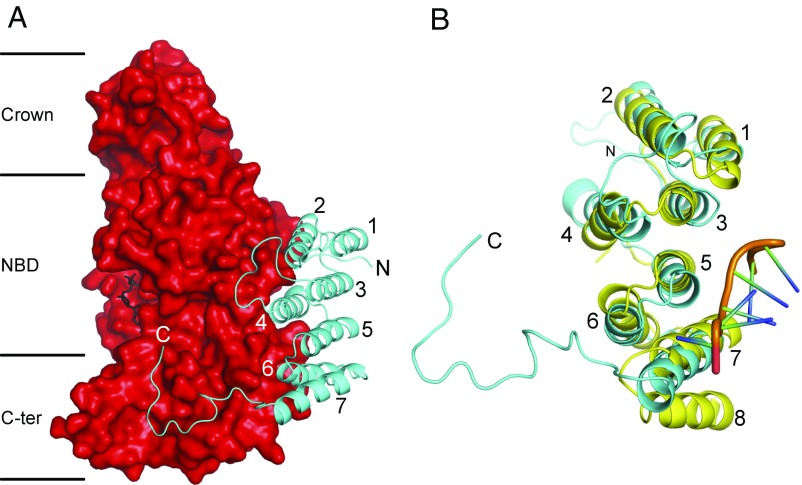
Third, and most significantly from a structural standpoint, the T. brucei F1-ATPase has an additional p18-subunit bound to each of its three α-subunits (50). The buried surface areas of interaction of the p18-subunits with their partner αE-, αTP-, and αDP-subunits are 2,500, 2,600, and 2,500 Å2, respectively. All three p18-subunits are folded into seven α-helices, H1–H7, with an unstructured C-terminal region from residues 151–170. The subunit is bound via H2 and H4 to the surface of the nucleotide-binding domain of an α-subunit and via H5 and H6 to the surface of its C-terminal domain. H7 is not in contact with the α-subunit (Fig. S2) but is bound to H6. The unstructured C-terminal tail interacts with the C-terminal domain of the α-subunit, traveling toward, but not entering, the noncatalytic interface with the adjacent β-subunit (Figs. 1 and 3 and Fig. S3). In this region, the extended C-terminal element of the p18-subunit interacts with the two additional segments of sequence (residues 483–498 and 536–560) found in the T. brucei α-subunit (Fig. S3). The first additional segment is largely extended, starting with one α-helical turn (residues 483–485). The second additional segment starts with one α-helical turn (residues 536–539), is followed by an extended region (residues 540–544), and terminates with an α-helix (residues 546–558) that doubles back into the noncatalytic interface and interacts with the extreme C-terminal end of the p18-subunit.
Fig. 3.
Structure of the p18-subunit of the F1-ATPase from T. brucei, and its relation to a PPR protein. (A) A p18 subunit (cyan) in cartoon representation, folded into α-helices H1–H7, with an extended C-terminal region from residues 151–170, bound to the αDP-subunit in solid representation (red). The N-terminal, nucleotide-binding, and C-terminal domains of the α-subunit are indicated by Crown, NBD, and C-ter, respectively; the bound ADP molecule is in black. (B) Comparison of the p18-subunit with an example PPR protein, the PPR10 protein from Z. mays (yellow) (57). PPR10 has 18 PPRs; the structures of PPRs 11–14 are shown (Fig. S3). The orange region represents the backbone of an eight-residue ribonucleotide bound to PPR10. The three PPRs in the p18-subunit correspond to H1 plus H2 (residues 20–28 and 33–45), H3 plus H4 (residues 52–64 and 78–93), and H5 plus H6 (residues 99–112 and 115–126). PPR10 has an additional α-helix, labeled 8, which together with α-helix 7 constitutes a fourth PPR.
Role of the p18-Subunit.
As noted previously, the sequence of the p18-subunit is related to the pentatricopeptide repeat (PPR) proteins (55), which are found in association with RNA molecules primarily in mitochondria and chloroplasts, as well as in some bacterial species. These proteins are characterized by a 35-aa degenerate sequence motif related to, but distinct from, the motif in the tetratricopeptide repeat (TPR) proteins (56). The PPR repeat is folded into a helix-turn-helix motif, and PPR proteins usually contain several tandem repeats associated into a superhelix, with a concave groove on one face that serves as a binding surface for RNA ligands. The p18-subunit of the F1-ATPase from T. brucei is predicted to be a PPR protein with three PPRs, whereas it was previously thought to have two PPRs (50, 55). Although the probability score (49%) is rather low, as reflected in the weak correspondence of the sequences of the three predicted PPRs to the PPR consensus (Fig. S4), the topography of p18 closely follows the topographies of other well-predicted and well-established PPR proteins, such as the RNA-binding PPR protein PPR10 from Zea mays (57) (Fig. S4).
This structural comparison (rmsd 2.3 Å) illustrates that, as predicted, the p18-subunit has three PPRs, consisting of H1 plus H2, H3 plus H4, and H5 plus H6. H7 could be a relic of the first element of a fourth PPR, in which H8 has evolved into the extended C-terminal tail region of the p18-subunit (Fig. 3B). However, p18 does not have a site equivalent to the RNA-binding site in PPR10, and other residues required for RNA binding in α-helix-7 of PPR10 have been substituted in H7 of p18. Therefore, there is no evidence suggesting that p18 has any role in binding an RNA molecule, and its role in the T. brucei F1-ATPase remains obscure, although its presence is essential for assembly of the enzyme (50). The sequences of p18-subunits, including the PPR repeats, are highly conserved across the euglenozoa, suggesting that the structure and the mode of interaction of the various p18-proteins with their cognate F1-ATPases are conserved as well (Fig. S5).
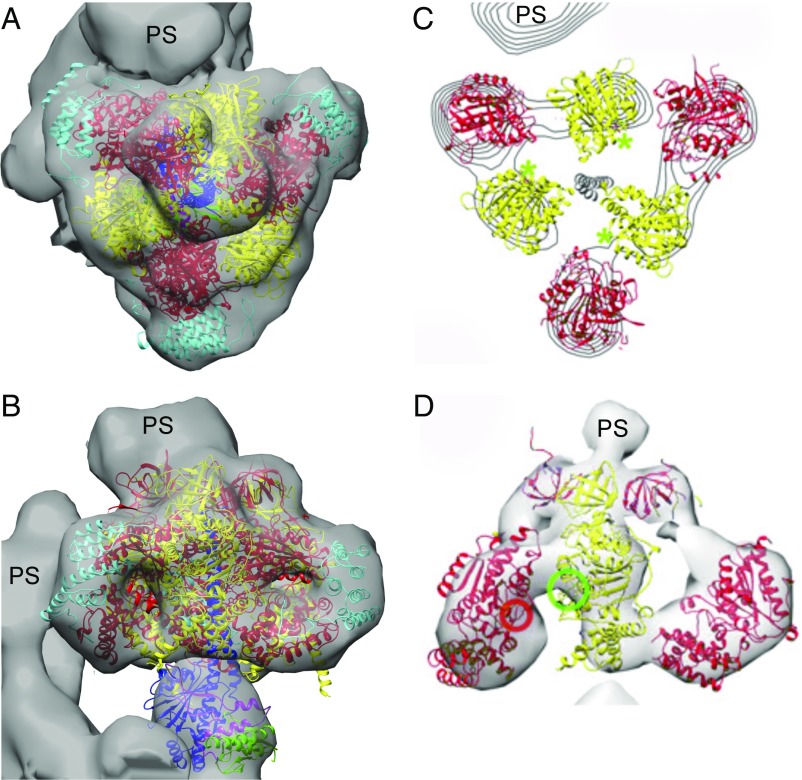
Structure of the T. brucei F1-ATPase and the ECT Map.
The structure of the F1-ATPase from T. brucei described above at 3.2-Å resolution was docked into the map of the ATP synthase complex from the same organism at 32.5-Å resolution derived by ECT of mitochondrial membranes (Fig. 4). The structure of the catalytic domain described herein closely fits the region of the map with the mushroom shape, characteristic of the catalytic F1-domain of other ATP synthases. Thus, this correspondence is also consistent with the T. brucei ATP synthase having a canonical catalytic domain elaborated by the attachment of the three p18-subunits. It does not support the proposal in Fig. 4 C and D)), where the map has been interpreted as having a catalytic domain in which the nucleotide-binding and C-terminal domains of the α-subunits are displaced outward away from the central stalk, and the role of the p18-subunit, bound in an unspecified position, is to provide the catalytically essential arginine finger residue (42).
Fig. 4.
Relationship of the crystallographic structure of the F1-domain of the ATP synthase from T. brucei to an ECT map of the intact ATP synthase in situ in mitochondrial membranes from T. brucei. The subunits of the F1-domain are colored as in Fig. 1. (A and B) Top (A) and side (B) views of the ECT map (gray), determined independently at 32.5-Å resolution with the crystallographic structure of the F1-domain determined at 3.2-Å resolution docked manually inside the ECT map, with subunits αDP and βTP proximal to the peripheral stalk. (C and D) A published interpretation of the same ECT map proposing a structure of T. brucei F1-ATPase in which the α-subunit is opened away from the central stalk, with the p18-subunit (not shown) contributing to the catalytic sites by providing the arginine finger residue (red circle) in D (42). The catalytic sites are indicated by green asterisks in C and by a green circle in D. PS, peripheral stalk of the enzyme. C and D modified from ref. 42.
Two other features in the ECT map can also be interpreted in terms of the characterized structures of canonical ATP synthases. First, the uninterpreted region of density above the F1-domain in Fig. 4 corresponds to the upper part of the peripheral stalk in other ATP synthases. This region is occupied by the OSCP in the eukarya, and by the orthologous δ-subunit in eubacterial and chloroplast enzymes. As the ATP synthase in T. brucei and other euglenozoa that have been examined contain orthologs of the OSCP (49, 58, 59), it is highly probable that the T. brucei OSCP provides this feature in the ECT map and that, as in the well- characterized ATP synthases, it is attached to the F1-domain via interactions with the N-terminal regions of the three α-subunits, which extend from the “top” of the crown domain. The role of the peripheral stalk in ATP synthases is to provide the stator of the enzyme with integrity by connecting the α3β3-domain to the essential ATP6 (subunit a in eubacteria, subunit IV in chloroplasts) in the membrane domain. ATP6 and orthologs, together with the c-ring in the rotor, provide the transmembrane pathway for protons (2). To maintain the integrity of this pathway, and to keep ATP synthesis coupled to proton motive force, the static ATP6 and the rotating c-ring must be kept in contact by the action of the peripheral stalk.
The peripheral stalk is the most divergent of the essential features of ATP synthases (2). Apart from the OSCP and orthologous δ-subunits, the subunit compositions, sequences, and structures of the related and structurally simpler eubacterial and chloroplast peripheral stalks differ greatly from the more complex structurally characterized peripheral stalks in mitochondrial enzymes, although they are all dominated by approximately parallel, antiparallel, and apparently rigid long α-helical structures connecting the OSCP to the ATP6 subunit (and orthologs) running alongside the catalytic domains. The peripheral stalks of ATP synthases in the mitochondria of euglenozoa (59) and in the green alga, Polytomella (60), appear to be even more divergent than those in characterized mitochondrial enzymes. Their subunit compositions are more complex, and as is evident in the map feature to the left of the F1-domain in Fig. 4 and in other published images, they are thicker and apparently more robust than structurally characterized peripheral stalks. More details are likely to emerge in the near future, most likely from the application of cryo-EM imaging of individual particles of these enzymes. These endeavors are driven by the imperative to use knowledge of the structure of the ATP synthase from T. brucei (61, 62) to aid the development of new drugs to treat patients with sleeping sickness by finding selective inhibitors of its activity.
Materials and Methods
Crystallization of F1-ATPase from T. brucei.
The F1-ATPase purified from T. brucei (50) was crystallized at 4 °C by the microbatch method under paraffin oil. The enzyme was dissolved at a protein concentration of 9.0 mg/mL in buffer consisting of 20 mM Tris⋅HCl pH 7.5, 100 mM NaCl, 10 mM MgSO4 and 1 mM ADP. This protein solution was mixed in wells in microbatch plates with an equal volume of 7.7% (wt/vol) PEG 10,000 dissolved in a buffer containing 100 mM 2-(N-morpholino)-ethanesulfonic acid pH 6.0 under a layer of paraffin oil. The plates were kept at 4 °C. Crystals appeared after 48 h, and were harvested 8 d later. They were cryoprotected by the addition of 15 µL of a solution containing 10 mM Tris⋅HCl pH 8.0, 10 mM NaCl, 5 mM MgSO4, 0.5 mM ADP, 50 mM 2-(N-morpholino)-ethanesulfonic acid pH 6.0, 5% (wt/vol) PEG 12,000, and 30% (vol/vol) glycerol to each well. After 5 min, the crystals were harvested with a MicroLoop (MiTeGen), flash-frozen, and stored in liquid nitrogen.
Data Collection and Structure Determination.
X-ray diffraction data were collected at 100 K from cryoprotected crystals with a PILATUS3 2M detector (Dectris) at a wavelength of 0.966 Å at the European Synchrotron Radiation Facility, Grenoble, France, using the MXPressE automated screening protocol (63, 64). Diffraction images were integrated with iMOSFLM (65), and the data were reduced with AIMLESS (66). Anisotropic correction was applied using STARANISO (staraniso.globalphasing.org). Molecular replacement using the α3β3-domain from the structure of the ground state structure of bovine F1-ATPase [Protein Data Bank (PDB) ID code 2JDI] was carried out with PHASER (67). Nucleotides, magnesium ions, and water molecules were removed from the model. Rigid body refinement and restrained refinement were performed with REFMAC5 (68). Manual rebuilding was performed with Coot (69), alternating with refinement performed with REFMAC5. For calculations of Rfree, 5% of the diffraction data were excluded from the refinement. Additional electron density features, adjacent to the α-subunits, were attributed to p18. Initially, poly-Ala α-helices were fitted into this additional density with Coot (69), and the assignment of the direction of the α-helices was guided by secondary structure predictions performed with PSIPRED (70). This prediction also detected structural homology of p18 with PPR10 from Z. mays (PDB ID code 4M59). Stereochemistry was assessed with MolProbity (71), and images of structures and electron density maps were prepared with PyMOL (72). Structural comparisons of T. brucei F1-ATPase with bovine F1-ATPase inhibited with dicyclohexylcarbodiimide (PDB ID code 1E79) (12), bovine F1-ATPase crystallized in the presence of phosphonate (PDB ID code 4ASU) (20), bovine F1-ATPase inhibited with ADP-AlF4 (PDB ID code 1H8E) (16), and the ground state structure of yeast F1-ATPase (PDB ID code 2HLD) (24) and of the p18-subunit from T. brucei with PPR10 from Z. mays (PDB ID code 4M59) (57) were done with Coot (69) and PyMOL (72). The p18-subunit was assessed for the presence of PPR and TPR sequences with TPRpred (73), and α-helices were assigned according to PyMOL.
Supplementary Material
Acknowledgments
We thank the staff at beamline ID30A-1 MASSIF-1 at the European Synchrotron Radiation Facility for their help. This work was supported by the Medical Research Council UK by Grants MC_U105663150 and MR/M009858/1 (to J.E.W.) and MC_U105184325 (to A.G.W.L.); the Czech Republic Ministry of Education, Youth, and Sports (European Research Council CZ Grant LL1205, to A.Z.); the Postdok_BIOGLOBE project, cofinanced by the European Social Fund and the Czech Republic (Grant CZ.1.07/2.3.00/30.0032, to A.Z. and O.G.); and European Molecular Biology Organization (Short-Term Fellowship ASTF 81-2016, to O.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6F5D).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720940115/-/DCSupplemental.
References
- 1.Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 2.Walker JE. Structure, mechanism, and regulation of ATP synthases. In: Wikström M, editor. Mechanisms of Primary Energy Transduction in Biology. Royal Society of Chemistry; London: 2017. pp. 338–373. [Google Scholar]
- 3.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8-Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 4.Abrahams JP, et al. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc Natl Acad Sci USA. 1996;93:9420–9424. doi: 10.1073/pnas.93.18.9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bason JV, Montgomery MG, Leslie AGW, Walker JE. Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase. Proc Natl Acad Sci USA. 2014;111:11305–11310. doi: 10.1073/pnas.1411560111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bason JV, Montgomery MG, Leslie AGW, Walker JE. How release of phosphate from mammalian F1-ATPase generates a rotary substep. Proc Natl Acad Sci USA. 2015;112:6009–6014. doi: 10.1073/pnas.1506465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci USA. 2006;103:8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9-Å resolution. J Biol Chem. 2007;282:14238–14242. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- 9.Braig K, Menz RI, Montgomery MG, Leslie AGW, Walker JE. Structure of bovine mitochondrial F1-ATPase inhibited by Mg(2+) ADP and aluminium fluoride. Structure. 2000;8:567–573. doi: 10.1016/s0969-2126(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 10.Cabezón E, Montgomery MG, Leslie AGW, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Biol. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- 11.Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons C, Montgomery MG, Leslie AGW, Walker JE. The structure of the central stalk in bovine F1-ATPase at 2.4-Å resolution. Nat Struct Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 13.Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. How the regulatory protein, IF1, inhibits F1-ATPase from bovine mitochondria. Proc Natl Acad Sci USA. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc Natl Acad Sci USA. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kagawa R, Montgomery MG, Braig K, Leslie AGW, Walker JE. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menz RI, Walker JE, Leslie AGW. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 17.Menz RI, Leslie AGW, Walker JE. The structure and nucleotide occupancy of bovine mitochondrial F1-ATPase are not influenced by crystallisation at high concentrations of nucleotide. FEBS Lett. 2001;494:11–14. doi: 10.1016/s0014-5793(01)02302-x. [DOI] [PubMed] [Google Scholar]
- 18.Orriss GL, Leslie AGW, Braig K, Walker JE. Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: The structure provides further support for a rotary catalytic mechanism. Structure. 1998;6:831–837. doi: 10.1016/s0969-2126(98)00085-9. [DOI] [PubMed] [Google Scholar]
- 19.Rees DM, Leslie AGW, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rees DM, Montgomery MG, Leslie AGW, Walker JE. Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc Natl Acad Sci USA. 2012;109:11139–11143. doi: 10.1073/pnas.1207587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Raaij MJ, Abrahams JP, Leslie AGW, Walker JE. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc Natl Acad Sci USA. 1996;93:6913–6917. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watt IN, Montgomery MG, Runswick MJ, Leslie AGW, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou A, et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. Elife. 2015;4:e10180. doi: 10.7554/eLife.10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabaleeswaran V, Puri N, Walker JE, Leslie AGW, Mueller DM. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 2006;25:5433–5442. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabaleeswaran V, et al. Asymmetric structure of the yeast F1 ATPase in the absence of bound nucleotides. J Biol Chem. 2009;284:10546–10551. doi: 10.1074/jbc.M900544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 27.Robinson GC, et al. The structure of F1-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF1. Open Biol. 2013;3:120164. doi: 10.1098/rsob.120164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arsenieva D, Symersky J, Wang Y, Pagadala V, Mueller DM. Crystal structures of mutant forms of the yeast F1 ATPase reveal two modes of uncoupling. J Biol Chem. 2010;285:36561–36569. doi: 10.1074/jbc.M110.174383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vinothkumar KR, Montgomery MG, Liu S, Walker JE. Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy. Proc Natl Acad Sci USA. 2016;113:12709–12714. doi: 10.1073/pnas.1615902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn A, et al. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groth G, Pohl E. The structure of the chloroplast F1-ATPase at 3.2-Å resolution. J Biol Chem. 2001;276:1345–1352. doi: 10.1074/jbc.M008015200. [DOI] [PubMed] [Google Scholar]
- 32.Groth G. Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc Natl Acad Sci USA. 2002;99:3464–3468. doi: 10.1073/pnas.052546099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shirakihara Y, et al. The crystal structure of the nucleotide-free α3β3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer. Structure. 1997;5:825–836. doi: 10.1016/s0969-2126(97)00236-0. [DOI] [PubMed] [Google Scholar]
- 34.Cingolani G, Duncan TM. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat Struct Mol Biol. 2011;18:701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy A, Hutcheon ML, Duncan TM, Cingolani G. Improved crystallization of Escherichia coli ATP synthase catalytic complex (F1) by introducing a phosphomimetic mutation in subunit ε. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:1229–1233. doi: 10.1107/S1744309112036718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirakihara Y, et al. Structure of a thermophilic F1-ATPase inhibited by an ε-subunit: Deeper insight into the ε-inhibition mechanism. FEBS J. 2015;282:2895–2913. doi: 10.1111/febs.13329. [DOI] [PubMed] [Google Scholar]
- 37.Morales-Rios E, Montgomery MG, Leslie AGW, Walker JE. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0-Å resolution. Proc Natl Acad Sci USA. 2015;112:13231–13236. doi: 10.1073/pnas.1517542112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferguson SA, Cook GM, Montgomery MG, Leslie AGW, Walker JE. Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum. Proc Natl Acad Sci USA. 2016;113:10860–10865. doi: 10.1073/pnas.1612035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobti M, et al. Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. Elife. 2016;5:e21598. doi: 10.7554/eLife.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakrishnan C, Dani VS, Ramasarma T. A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 2002;15:783–798. doi: 10.1093/protein/15.10.783. [DOI] [PubMed] [Google Scholar]
- 42.Mühleip AW, Dewar CE, Schnaufer A, Kühlbrandt W, Davies KM. In situ structure of trypanosomal ATP synthase dimer reveals a unique arrangement of catalytic subunits. Proc Natl Acad Sci USA. 2017;114:992–997. doi: 10.1073/pnas.1612386114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balabaskaran Nina P, et al. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8:e1000418. doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balabaskaran Nina P, et al. ATP synthase complex of Plasmodium falciparum: Dimeric assembly in mitochondrial membranes and resistance to genetic disruption. J Biol Chem. 2011;286:41312–41322. doi: 10.1074/jbc.M111.290973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cardol P, et al. The mitochondrial oxidative phosphorylation proteome of Chlamydomonas reinhardtii deduced from the genome sequencing project. Plant Physiol. 2005;137:447–459. doi: 10.1104/pp.104.054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Lis R, Mendoza-Hernández G, Groth G, Atteia A. New insights into the unique structure of the F0F1-ATP synthase from the chlamydomonad algae Polytomella sp. and Chlamydomonas reinhardtii. Plant Physiol. 2007;144:1190–1199. doi: 10.1104/pp.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaidya AB, Mather MW. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 2009;63:249–267. doi: 10.1146/annurev.micro.091208.073424. [DOI] [PubMed] [Google Scholar]
- 48.Perez E, et al. The mitochondrial respiratory chain of the secondary green alga Euglena gracilis shares many additional subunits with parasitic Trypanosomatidae. Mitochondrion. 2014;19 Pt B:338–349. doi: 10.1016/j.mito.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Zíková A, Schnaufer A, Dalley RA, Panigrahi AK, Stuart KD. The F0F1-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 2009;5:e1000436. doi: 10.1371/journal.ppat.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gahura O, et al. The F1-ATPase from Trypanosoma brucei is elaborated by three copies of an additional p18-subunit. FEBS J. 2017 doi: 10.1111/febs.14364. [DOI] [PubMed] [Google Scholar]
- 51.Speijer D, et al. Characterization of the respiratory chain from cultured Crithidia fasciculata. Mol Biochem Parasitol. 1997;85:171–186. doi: 10.1016/s0166-6851(96)02823-x. [DOI] [PubMed] [Google Scholar]
- 52.Nelson RE, Aphasizheva I, Falick AM, Nebohacova M, Simpson L. The I-complex in Leishmania tarentolae is an uniquely-structured F1-ATPase. Mol Biochem Parasitol. 2004;135:221–224. doi: 10.1016/j.molbiopara.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Dean S, Gould MK, Dewar CE, Schnaufer AC. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci USA. 2013;110:14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carbajo RJ, et al. How the N-terminal domain of the OSCP subunit of bovine F1F0-ATP synthase interacts with the N-terminal region of an alpha subunit. J Mol Biol. 2007;368:310–318. doi: 10.1016/j.jmb.2007.02.059. [DOI] [PubMed] [Google Scholar]
- 55.Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol Cell Biol. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small ID, Peeters N. The PPR motif A TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 57.Yin P, et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504:168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- 58.Cano-Estrada A, et al. Subunit–subunit interactions and overall topology of the dimeric mitochondrial ATP synthase of Polytomella sp. Biochim Biophys Acta. 2010;1797:1439–1448. doi: 10.1016/j.bbabio.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 59.Yadav KNS, et al. Atypical composition and structure of the mitochondrial dimeric ATP synthase from Euglena gracilis. Biochim Biophys Acta. 2017;1858:267–275. doi: 10.1016/j.bbabio.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Allegretti M, et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521:237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 61.Šubrtová K, Panicucci B, Zíková A. ATPaseTb2, a unique membrane-bound F0F1-ATPase component, is essential in bloodstream and dyskinetoplastic trypanosomes. PLoS Pathog. 2015;11:e1004660. doi: 10.1371/journal.ppat.1004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panicucci B, Gahura O, Zíková A. Trypanosoma brucei TbIF1 inhibits the essential F1-ATPase in the infectious form of the parasite. PLoS Negl Trop Dis. 2017;11:e0005552. doi: 10.1371/journal.pntd.0005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svensson O, Malbet-Monaco S, Popov A, Nurizzo D, Bowler MW. Fully automatic characterization and data collection from crystals of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 2015;71:1757–1767. doi: 10.1107/S1399004715011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bowler MW, et al. MASSIF-1: A beamline dedicated to the fully automatic characterization and data collection from crystals of biological macromolecules. J Synchrotron Radiat. 2015;22:1540–1547. doi: 10.1107/S1600577515016604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schrödinger LLC. 2015. The PyMOL Molecular Graphics System, version 1.8. (Schrödinger, LLC, New York)
- 73.Alva V, Nam SZ, Söding J, Lupas AN. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016;44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.