Significance
Spontaneous fMRI signals are used to understand human brain organization throughout the life span and in disease states. Spontaneous fMRI signals contain many artifacts, and removing these artifacts is vital to properly studying neurobiological signals. We report successful removal of a major artifact, spatially focal motion artifact, from resting state fMRI signals via multiecho imaging techniques. By removing motion artifact, we isolate a second kind of motion-associated signal, a respiratory signal, that occurs across the entire brain. We illustrate several techniques that remove this respiratory artifact, yielding fMRI data free of motion-related influences. These two kinds of motion-related signals have distinct physical and spatial bases, and each can strongly and differentially influence signal patterns in fMRI data.
Keywords: fMRI, multiecho, functional connectivity, motion artifact, respiration
Abstract
“Functional connectivity” techniques are commonplace tools for studying brain organization. A critical element of these analyses is to distinguish variance due to neurobiological signals from variance due to nonneurobiological signals. Multiecho fMRI techniques are a promising means for making such distinctions based on signal decay properties. Here, we report that multiecho fMRI techniques enable excellent removal of certain kinds of artifactual variance, namely, spatially focal artifacts due to motion. By removing these artifacts, multiecho techniques reveal frequent, large-amplitude blood oxygen level-dependent (BOLD) signal changes present across all gray matter that are also linked to motion. These whole-brain BOLD signals could reflect widespread neural processes or other processes, such as alterations in blood partial pressure of carbon dioxide (pCO2) due to ventilation changes. By acquiring multiecho data while monitoring breathing, we demonstrate that whole-brain BOLD signals in the resting state are often caused by changes in breathing that co-occur with head motion. These widespread respiratory fMRI signals cannot be isolated from neurobiological signals by multiecho techniques because they occur via the same BOLD mechanism. Respiratory signals must therefore be removed by some other technique to isolate neurobiological covariance in fMRI time series. Several methods for removing global artifacts are demonstrated and compared, and were found to yield fMRI time series essentially free of motion-related influences. These results identify two kinds of motion-associated fMRI variance, with different physical mechanisms and spatial profiles, each of which strongly and differentially influences functional connectivity patterns. Distance-dependent patterns in covariance are nearly entirely attributable to non-BOLD artifacts.
Resting state functional magnetic resonance imaging (fMRI) has become a major technique for investigating human and nonhuman brain organization. In a typical experiment, a subject lies quietly in the scanner for 5 min or longer while spontaneous fMRI signals are recorded. The feature of interest in the data is usually covariance between time series in different parts of the brain. A large body of work indicates that covariance in spontaneous fMRI signals can serve as an index of functional relatedness between brain regions (reviewed in refs. 1, 2).
In addition to reflecting neural activity, fMRI signals reflect a variety of nonneurobiological processes, including thermal noise and multiple sources of structured noise. Examples of structured noise sources include head motion, pulsation of brain tissue during cardiac cycles, scanner hardware artifacts, and artifacts caused by lung expansion during respiration (other sources exist; reviewed in ref. 3). Multiple studies of human fMRI signals indicate that neurobiological signals are a relatively small proportion of signal variance, on the order of 5–20% of the recorded signal (4, 5).
A critical element of any functional connectivity analysis is thus to distinguish between neurobiological and nonneurobiological signals in the fMRI time series. Failure to make these distinctions can lead to false conclusions because sources of structured noise, and thus structured covariance, often scale with factors of interest. For example, children move more than young adults, and it is now recognized that many functional connectivity properties initially reported to be related to development were actually properties of motion artifacts that were more prevalent in younger subjects (6, 7).
One way to separate neurobiological signals from many nonneurobiological signals is to characterize fMRI signal decay properties. fMRI signal at a voxel can be approximated as a monoexponential decay following the formula , where S0 is the initial signal intensity upon radiofrequency excitation at t = 0 and T2* is the time constant of signal decay (signal decay rate can also be expressed as R2* = 1/T2*). Most fMRI datasets are single-echo datasets in which signal is acquired once at a delay [echo time (TE)] of ∼30–35 ms following excitation. In single-echo data, if signal changes at a voxel from volume to volume, it cannot be determined whether the change is due to a changed decay rate or a changed starting intensity. Multiecho sequences acquire signals multiple times per excitation, allowing investigators to disambiguate S0 from R2* effects (SI Appendix, Fig. S1).
Separation of S0 from R2* effects can be helpful in fMRI data because neural activity principally changes R2* but not S0, whereas many sources of structured noise (e.g., head motion) principally alter S0 but not R2*. fMRI signals sensitive to changes in R2* are commonly called blood oxygen level-dependent (BOLD) signals, because a major influence on R2* is the amount of deoxygenated hemoglobin in a voxel. This amount is affected by vascular responses to recent neural activity (a signal of interest) and by the overall oxygen and carbon dioxide content of incoming arterial blood (contingent on lung ventilation, not a signal of interest).
The multiecho analyses in this paper were motivated by the fact that it has been difficult to remove the influence of motion artifact from single-echo fMRI data (reviewed in ref. 8). The importance of identifying and removing motion artifact has been amply demonstrated in the developmental, clinical, and aging literature in recent years (e.g., refs. 9, 10), but this issue is vital beyond these domains. Recent analyses of neurotypical adults in the NIH-sponsored Human Connectome Project have indicated that numerous behavioral (e.g., working memory) and physiological (e.g., obesity) variables correlate with head motion and with distance-dependent patterns of covariance in fMRI data that are caused by head motion (11, 12). In addition, patterns of dynamics in fMRI signals are now reported to be largely explained by head motion (13). The existence of possible genetic predispositions to motion (14), and therefore combined neural and artifactual correlates of motion, creates an additional, challenging set of influences to untangle during data analysis. Progress in functional connectivity studies of all kinds is therefore contingent on convincing separation of motion artifact from neurobiological signals.
Several studies have provided evidence that denoising techniques based on multiecho decay can reduce motion artifact (e.g., refs. 15–17), but the extent of motion artifact removal is incompletely known (reviewed in ref. 18). We thus set out to determine whether multiecho- independent component analysis (ME-ICA) eliminated motion artifacts in fMRI data. Surprisingly, we found that functional connectivity properties became more strongly tied to motion after multiecho denoising, despite the fact that motion artifact was clearly being removed from the data. This result led to the realization that global BOLD signals commonly accompany motion in fMRI data, and that many of these global BOLD signals are due to changes in respiration (rather than motion, per se). We present and compare several data-driven methods to remove such signals, yielding time series essentially free of the influence of motion.
Results
Separation of S0 from R2* Effects Reveals Prominent, Brain-Wide BOLD Signals.
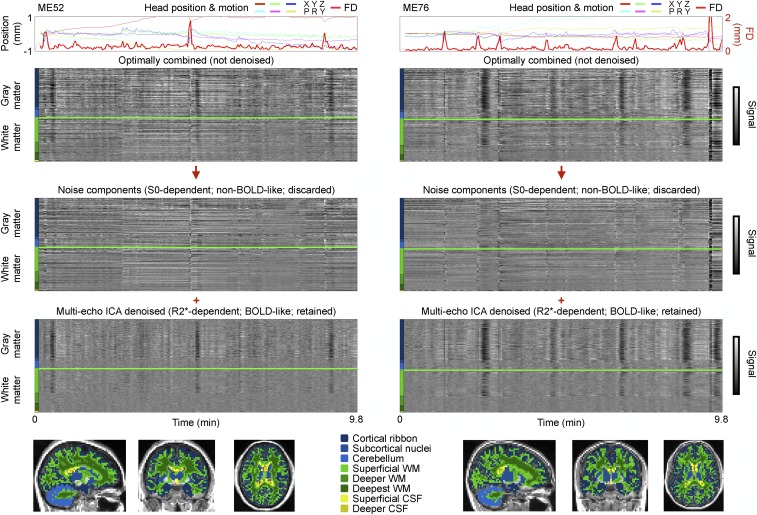
ME-ICA denoising was applied to 89 four-echo datasets [these datasets were first published by Kundu et al. (16) and will be referred to as ME datasets]. ME-ICA first identifies spatially structured signals in the datasets via ICA and categorizes the signals according to how strongly they reflect S0 versus R2* modulation over time. ME-ICA then discards (most) S0-dependent signals and retains (most) R2*-dependent signals following automated criteria (i.e., without training or user intervention). We are interested in the variance before denoising and the variance retained and discarded by ME-ICA. Data before denoising are illustrated via the “optimally combined” image, which is a weighted (by T2*) average of the multiple images, following the nomenclature of Kundu et al. (16). Data from two subjects are presented in Fig. 1 to illustrate how signals are separated by ME-ICA (these scans are representative; such plots for each ME subject are shown in Online Movie 1 (movies can be seen or downloaded at www.jonathanpower.net/paper-multiecho.html).
Fig. 1.
ME-ICA denoising separates non–BOLD-like signals from BOLD-like signals. (Top) ME-ICA results for two ME subjects are shown. (Bottom) Colored masks denote FreeSurfer-derived brain compartments used to organize the voxels in heat maps (colored bars). CSF, cerebrospinal fluid; WM, white matter.
Two aspects of signal separation, visually evident in Fig. 1, will be explored in this report. First, in the discarded S0-dependent (non–BOLD-like) components, signals are often related to head motion, or to sustained shifts in head position. These signals are usually not similar across the brain (vertically, there are few uniform bands of black or white; instead, a “salt and pepper” appearance is typical). The characteristics of this variance will be explored later in this report. Second, in the retained R2*-dependent (BOLD-like) components, vertical black and white bands representing signal increases and decreases across all gray matter are conspicuous. We will use the word “global” to refer to signals present across most or all of gray matter, and we will use the mean signal across cortical voxels as a global signal [across ME subjects, this mean cortical signal correlates at r = 0.99 ± 0.01 with the signal of all gray matter (cortex, cerebellum, and subcortical nuclei) and at r = 0.99 ± 0.01 with the mean signal of the whole brain (all gray matter, white matter, and ventricles)]. The separation of nonglobal versus global variance into non–BOLD-like versus BOLD-like components is typical of every subject of the ME cohort (Online Movie 1 and SI Appendix, Table S1; the global signals in optimally combined and ME-ICA denoised images correlate at r = 0.95 ± 0.08 across subjects).
ICA signal separation techniques, which optimize spatial independence among signals, do not separate spatially widespread signals from spatially specific signals in these data [SI Appendix, Supplemental Results (for elaboration) and SI Appendix, Fig. S2]. A corollary is that multiple BOLD-like ICA components (and virtually no non–BOLD-like components) include the global signal and, further, that there is no way to sort components to separate global from focal BOLD-like signals (SI Appendix, Supplemental Results). Given these considerations, to ensure that the global BOLD signals identified by ME-ICA were truly R2* signals, we also fit the multiecho data to the equation to yield S0 and R2* estimates at each voxel and time point. This procedure, which we term “FIT,” produced similar signal separation as ME-ICA and clearly indicated that global signals were BOLD signals (SI Appendix, Fig. S3 and Table S1; the global signals in ME-ICA denoised and FIT R2* images correlate at r = 0.96 ± 0.06 across subjects).
Prominent, Brain-Wide BOLD Signals Are Related to Breathing Patterns.
Brain-wide BOLD signals may represent widespread neural processes (19–23). On the other hand, brain-wide BOLD modulation can also occur via “nonneural” mechanisms involving changes in arterial partial pressure of carbon dioxide (pCO2) or partial pressure of oxygen (24–27). For example, hypoventilation maneuvers like breath-holding cause transient hypercapnia, which increases cerebral blood flow, which increases BOLD signal throughout the brain. In contrast, hyperventilation maneuvers reduce pCO2 and cause an opposite set of effects on BOLD signals. Spontaneous changes in respiratory rates in quietly resting subjects could therefore also cause global modulations of BOLD signals.
To help disambiguate whether neural versus respiratory processes cause global signal modulations, we obtained three-echo resting state fMRI data from 12 subjects (two runs per subject) while monitoring breathing patterns with a respiratory belt and heart rate with a pulse oximeter. These data are a subset of scans from a larger task fMRI study on object naming (we call these NA scans). Data from these NA subjects underwent ME-ICA and FIT procedures. The multiecho fits to NA data are noisier than the ME data because fewer echoes were acquired (three vs. four) and because the NA voxels are smaller (27 vs. 62 mm3) (there is therefore a smaller ratio of “signal” to thermal noise in the NA data). Nineteen of the 24 NA scans completed ME-ICA and FIT (ME-ICA failed in several scans; SI Appendix, Supplemental Methods).
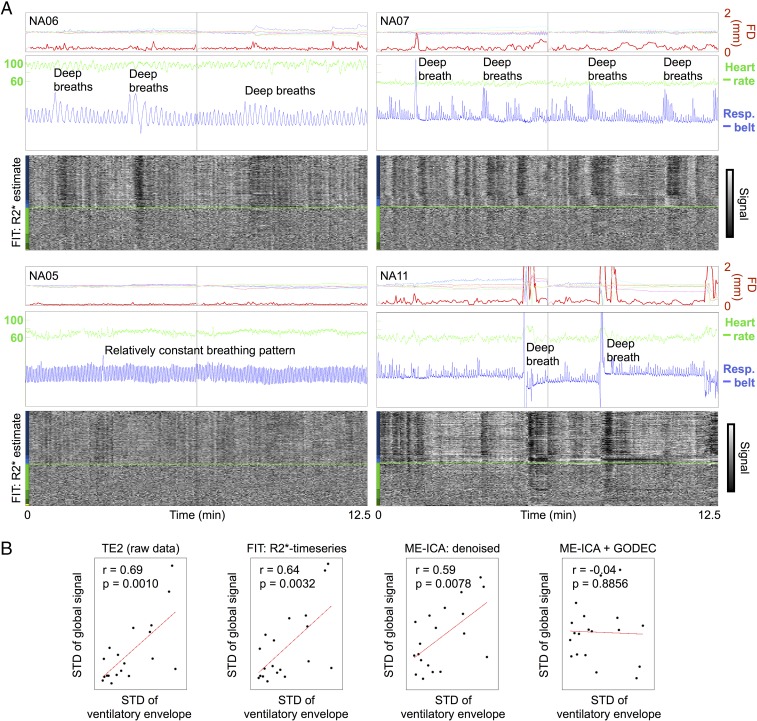
The breathing patterns in NA subjects differed across subjects, as can be seen in the blue respiratory belt traces of Fig. 2. In subjects with variable respiratory patterns, such as deep breaths or periods of relatively shallow breathing, prominent, widespread changes in BOLD signals are seen under both ME-ICA and FIT procedures (Fig. 2A). In contrast, in subjects with relatively constant respiratory rhythms (Fig. 2A, Bottom Left), global modulations of signals are much less pronounced. Plots similar to those in Fig. 2A (but also including the ME-ICA time series) can be seen for each NA subject in Online Movie 2.
Fig. 2.
Global BOLD signal changes often reflect respiratory patterns. (A) Motion and physiological traces are shown for four NA subjects, as are FIT R2* time series, following conventions of earlier figures. The blue trace is the raw respiratory belt trace, and the green trace is the heart rate derived from peaks in the pulse oximeter trace. Resp., Respiratory. (B) Correlation between variance in respiratory traces and variance in global fMRI signals, shown for the raw fMRI data at TE2, for FIT R2* time series, for ME-ICA denoised time series, and for ME-ICA + GODEC time series. Each point is a scan. Respiratory variance (x axis) is defined as the SD of the envelope of the normalized respiratory belt waveform.
Statistically, there is a strong relationship across subjects between variability in respiratory patterns and fluctuations in global fMRI signals (Fig. 2B). To capture the variability of the respiratory patterns, the respiratory belt waveforms (which are in arbitrary units) were z-scored, the envelope of the resulting waveform was calculated, and the SD of this envelope was then calculated. Those measures correlated with the SD of the global fMRI signal at r = 0.69, r = 0.64, and r = 0.59 in TE2, FIT R2*, and ME-ICA denoised data, respectively, which are all significant correlations (P < 0.001, P < 0.003, and P < 0.008, respectively). In contrast, there is no significant correlation between variability in heart rate and fluctuations in global fMRI signals: r = 0.07, r = 0.03, and r = −0.16 are the respective correlations in TE2, FIT R2*, and ME-ICA denoised data.
To further link fMRI signal changes to respiratory patterns, we examined the temporal relationship between breathing and global fMRI signals. We modeled the mean gray matter signal after single deep breaths in NA subjects and compared these signal modulations with those seen in the literature after instructed deep breaths or after spontaneous breaths. The timing and duration of signal modulation in the NA data (e.g., NA11) match those found in other reports (SI Appendix, Fig. S4): a signal peak around 5–7 s after the breath, followed by a long trough with a nadir between 15 and 20 s postbreath.
These results indicate that respiration-induced changes in pCO2 are responsible for a major portion of global BOLD fluctuations in the NA data. The characteristics of respiratory signals in the NA data are that they are slow, large-amplitude changes in gray matter signals that attenuate with depth in the white matter (by depth, we mean distance from gray matter, not the cranium). Many global signals in the ME datasets share these characteristics. For example, in the NA data, several black bands in Fig. 2 are preceded by head motion and large inspirations. Such patterns (minus the respiratory trace) are also seen in the ME data in Fig. 1. These patterns are consistent with deep breaths that (i) move the head and (ii) transiently alter blood gases to produce prolonged, brain-wide decreases in BOLD signal. We will show below that both respiratory variability (in NA data) and global covariance (in NA and ME data) are strongly linked to head motion, findings that are consistent with respiration causing a substantial portion of head motion.
Two Methods to Remove Brain-Wide Signals.
Multiecho techniques can separate many artifacts from BOLD signals in resting state fMRI data, but multiecho techniques cannot distinguish between respiratory signals and neurobiological signals because they arise from the same BOLD mechanism. On first principles, global respiratory signals of the kind demonstrated here will (i) spuriously increase covariance in time series and (ii) mimic dynamics in time series. It is thus essential to remove (or control for) such respiratory effects to draw valid neurobiological conclusions from BOLD data.
It has recently been reported that global respiratory signals persist through several common denoising approaches, including models that attempt to approximate respiratory variance from respiratory belt traces, models that utilize nuisance compartment signals, and ICA (28). The NA data corroborate this report: Brain-wide signals time-locked to respiratory changes persist after ME-ICA or FIT (Fig. 2), after regression of multiple models of respiratory variance (SI Appendix, Fig. S5), and after regression of motion estimates and nuisance compartment signals (SI Appendix, Figs. S6 and S7; see Online Movie 3 for the same model applied to each ME subject). The correlation between respiratory signal variability and global signal variability remains strong and significant after these processing streams (SI Appendix, Fig. S8). The NA data thus show that global respiratory variance is both present and inadequately removed by all of the above denoising strategies. In a dataset such as the ME dataset, there is, at present, no way to determine which subset of global fluctuations is respiratory (this is also the situation for most fMRI datasets); thus, the only way to remove respiratory global fluctuations is to remove all global fluctuations.
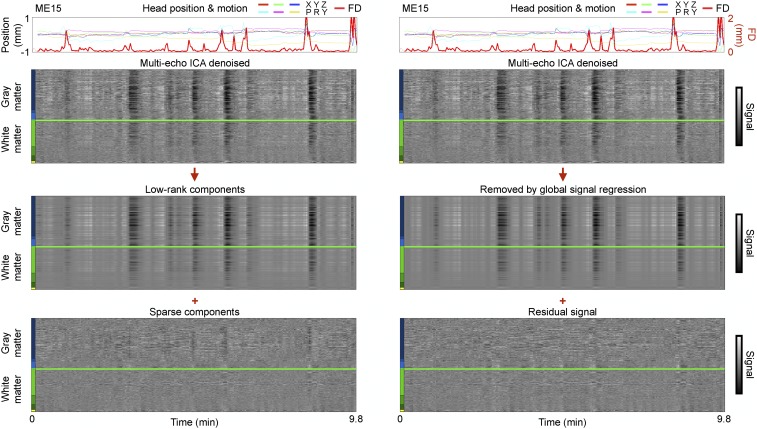
Two methods capable of removing global fluctuations in ME-ICA denoised data are illustrated in Fig. 3 (others are discussed below and in SI Appendix). Go Decomposition (GODEC) is a recently developed multivariate technique that can separate spatially widespread (low-rank) signals from spatially focal (sparse) signals in an fMRI dataset (29). A univariate approach to removing global fluctuations in fMRI data is to regress the global signal from all time series (here, we use the mean cortical signal as the global signal, which, as mentioned above, is essentially identical to the whole-brain signal that is often used for such regression purposes). The principal difference between these approaches is that the mean gray matter time series includes both the “true” global signal as well as a mixture of more focal signals present only in particular brain regions. In contrast, GODEC is intended to capture only the true global signal without incorporating the more focal signals. It is visually apparent that both GODEC and mean cortical signal regression identify and remove similar global fluctuations in BOLD signals (Fig. 3 and SI Appendix, Fig. S9; Online Movie 4 shows both techniques in each ME subject, and Online Movie 5 shows all versions of variance separation in time on cortical surfaces for each subject). Across ME subjects, the variance removed by GODEC and mean signal regression correlates at 0.87 ± 0.16. GODEC sometimes does not fully remove global fluctuations (e.g., ME13, ME20; visible in Online Movie 4). Both mean signal regression and GODEC abolish the relationship between respiratory pattern variability and global signal variability (Fig. 2B).
Fig. 3.
Low-rank signals are similar to global signals. For an ME scan shown in Fig. 1, ME-ICA denoised data are shown undergoing GODEC and global signal regression.
Motion Indexes Both Focal S0 and Global R2* Signals.
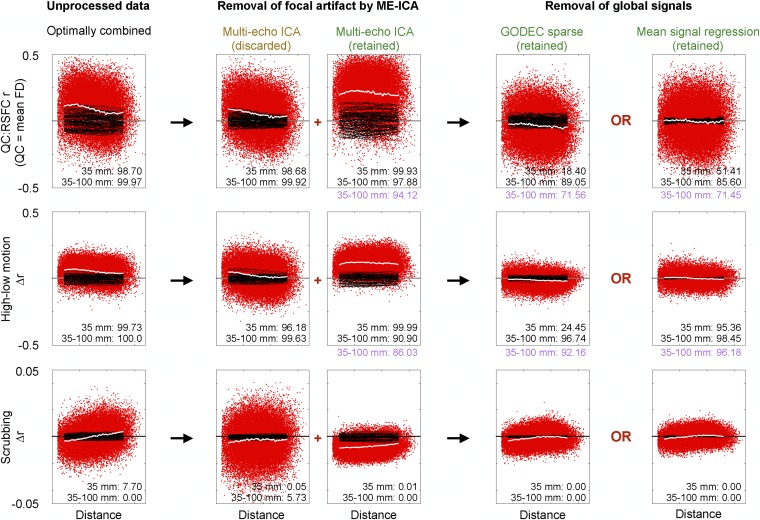
One of the best-established findings in functional connectivity MRI studies is that motion tends to elevate short-distance signal covariance more than long-distance covariance (6, 7, 10, 30). A major goal of this study is to determine the extent to which multiecho methods remove motion artifact in functional connectivity data. Accordingly, we present three analyses designed to identify the presence of motion artifact: QC:RSFC (quality control:resting state functional connectivity) plots, high–low-motion differences, and scrubbing plots (analyses reviewed in ref. 8).
We describe the logic of the analyses in Fig. 4 before examining the results. In Fig. 4, Top, QC:RSFC plots show how subject motion correlates, across subjects, with functional connectivity measures. In each subject’s data, we calculate some 35,000 pairwise correlations between signals at a set of 264 regions of interest (ROIs) that span the brain. Then, the RSFC measures are correlated, across subjects, with the mean motion [mean framewise displacement (FD)] displayed by each subject. Positive QC:RSFC correlations indicate that RSFC values are higher in higher motion subjects. The resulting QC:RSFC correlations are plotted as a function of the distance separating the ROIs that generate the correlations. The red points show each of the QC:RSFC values obtained, and a white smoothing curve summarizes the behavior of the data. Ten thousand permutations of subject motion (mean FD) generate random expectations for QC:RSFC correlations, and black smoothing curves from 50 of these permutations are shown for visual comparison. The white smoothing curve in Fig. 4, Left, is an inverted sigmoid, higher at the left (short distances) and less high at the right (long distances), indicating that subjects with higher motion tend to have higher functional connectivity correlations, especially among nearby regions of the brain. The entire smoothing curve is also above zero, indicating that all correlations tend to be increased in higher motion subjects. The high–low-motion plot in Fig. 4, Center, follows a similar presentation format, except that the computations are a median split of the cohort by motion (mean FD), followed by subtracting mean low-motion values from mean high-motion values at all correlations. Ten thousand permutations of mean FD yield random expectations. The smoothing curve is, again, an inverted sigmoid and is also elevated above zero, yielding the same kinds of conclusions as the QC:RSFC analyses.
Fig. 4.
Spatial interpretation of denoising steps in ME data. Distance-dependent motion-related artifact is assayed with three kinds of analyses at several stages of denoising (columns): QC:RSFC analyses (Top), differences in correlations between high- and low-motion subjects (Middle), and scrubbing analyses (Bottom). The red points and the white smoothing curve display actual data, and the black smoothing curves depict 50 of the 10,000 conducted permutations of mean FD (Top and Middle) or censored volumes (Bottom). The inset numbers are the percentiles of observed data among permutations, in terms of smoothing curves at 35 mm (to index all motion-related signals) and the difference between smoothing curves at 35 and 100 mm (to index distance dependence). Purple permutation ranks are drawn from SI Appendix, Fig. S10 and reflect smoothing curve values after censoring volumes with FD > 0.2 mm. Nuisance regression in the plot at the far right contains a single regressor: the mean cortical signal. (Top) RMS values of the plots (from left to right) are 0.16, 0.13, 0.25, 0.13, and 0.13. (Middle) RMS values of the plots (from left to right) are 0.08, 0.09, 0.13, 0.05, and 0.04. (Top) Mean values are 0.09, 0.05, 0.21, −0.03, and 0.00. (Middle) Mean values are 0.05, 0.02, 0.11, −0.01, and 0.00. Slopes of linear fits to the QC:RSFC curves in optimally combined and ME-ICA denoised data are −0.009 and −0.002, and corresponding fits to the high–low data are −0.005 and −0.001. Δr, change in correlation.
Motion-associated variance can be thought of as a superposition of focal (different signals in different parts of the brain) and global (similar signals throughout the brain) effects associated with motion (8). Global effects associated with motion will tend to elevate the dependence on motion of correlations at all distances. However, focal effects associated with motion will cause differential effects across distance, namely, to increase dependence on motion of correlations at short distances (where focal effects are shared) but to decrease such dependence at longer distances (where different focal effects occur). In terms of the white smoothing curves, global effects elevate the curve at all distances, whereas focal effects should elevate the curve at short distances and drive it toward zero or negative values at long distances, causing it to be sloped in some way. We measure two properties of the smoothing curves to capture these phenomena: the value at 35 mm (short distance) captures both focal and global effects, and the difference between values at 100 mm and 35 mm captures the differential across distance caused by focal effects. We measure these properties for all permutation curves, and report the rank of the observed curve among the permutation curves. To return to the examples studied above, the QC:RSFC smoothing curve of the first column (Fig. 4, Top Left) is elevated relative to random expectations [rank 9,870 of 10,000 at 35 mm (i.e., P < 0.026), indexing focal and global effects] and also shows more differential over distance than would be randomly expected [rank 9,997 of 10,000 (i.e., P < 0.0006), indexing focal effects]. The ranks of the high–low-split curve are significant in a similar way (P < 0.0054 and P < 0.0001, respectively). We may thus surmise the existence of both global- and focal motion-associated effects in the optimally combined data.
The final kind of motion-related analysis is a scrubbing analysis, shown in Fig. 4, Bottom. Whereas the QC:RSFC analyses and high–low analyses show across-subject or across-scan differences contingent on motion, the scrubbing analysis is a within-scan analysis. It shows the properties of particular volumes of scans, which, in this case, are the properties of volumes displaying detectable motion (FD > 0.2 mm). We calculate in each subject the same ∼35,000 correlations as above, first including all volumes (as in the above analyses) and then censoring (scrubbing) the suspicious high-motion volumes. For each subject, we subtract the original from the scrubbed correlation values, and then plot the mean values seen across subjects. In this way, the properties of the exact volumes being censored are demonstrated. In Fig. 4, Left, the properties of this plot are inverted from the QC:RSFC and high–low plots: When high-motion volumes are withheld, correlations are reduced at short distances and are elevated at longer distances, and the white line is approximately zero-centered. The rank of the 35- to 100-mm difference is 0/10,000, indicating that the distance-dependent effects are highly significant (i.e., P < 0.0001). The direct interpretation is that the exact volumes censored (here, those acquired during times of motion) contain distance-dependent covariance (i.e., focal motion-associated effects).
In Fig. 4, the QC:RSFC and high–low analyses are performed on all volumes, with scrubbing effects reported (Fig. 4, Bottom) to illustrate the properties of volumes acquired during motion. In SI Appendix, Fig. S10, the QC:RSFC and high–low analyses are repeated while withholding scrubbed volumes, and the distant-dependent rankings of those analyses are shown in purple in Fig. 4 to illustrate additional improvements made possible by censoring high-motion volumes.
We are now positioned to examine these analyses across stages of processing. QC:RSFC plots demonstrate strong distance dependence in optimally combined data (Fig. 4, Left) and in S0 data [Fig. 4, Center (column 2)], but attenuated distance dependence in BOLD-like data, whether ME-ICA denoised data [Fig. 4, Center (column 3)] or data after removal of global signals by either GODEC or mean signal regression [Fig. 4, Right (columns 4 and 5)] are considered. Distance dependence is reduced to insignificant levels in the BOLD-like data, especially when motion-exhibiting volumes are withheld (Fig. 4, purple values). Importantly, ME-ICA unmasks a strong global motion-associated influence in R2* data [Fig. 4, Center (column 3)]: Average QC:RSFC values rise from 0.08 in optimally combined data to 0.21 in ME-ICA denoised data. A straightforward interpretation is that ME-ICA removes many focal S0 artifacts whose influence had initially obscured the influence of global signals at all distances. The QC:RSFC smoothing curves become approximately zero-centered after removing global signals by either method. The high–low plots corroborate the QC:RSFC patterns. The scrubbing plots indicate that motion-exhibiting volumes contain distance-dependent covariance at all stages of processing, effects that are highly significant [e.g., P < 0.0001 or P < 0.0002 for both curve measurements (35 mm and 35–100 mm) in all BOLD-like images].
The critical points of these analyses are: (i) variance in both non–BOLD-like and BOLD-like signals scales with motion; (ii) motion-related variance in S0 signals is mostly focal, and thus distance-dependent; (iii) motion-related variance in R2* signals is mostly global, and thus distance-independent; and (iv) some focal artifact (and thus distance-dependent covariance) is present in motion-exhibiting volumes at all stages of denoising.
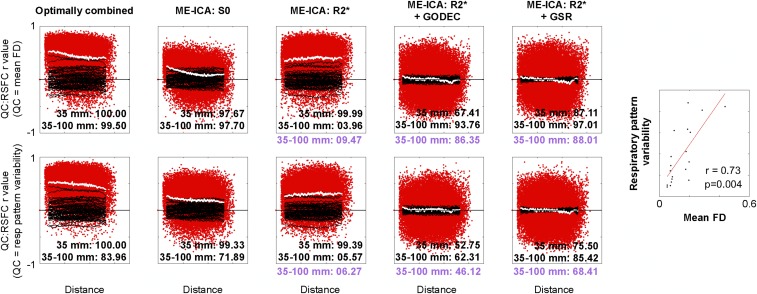
It is instructive to revisit the NA data in light of these analyses. Fig. 5, Top shows QC:RSFC plots using mean FD as the QC vector, as in Fig. 4, whereas Fig. 5, Bottom uses respiratory pattern variability as the QC vector. The rows show similar patterns because mean FD and respiratory pattern variability correlate at r = 0.73 (P = 0.004). These NA results mirror those of the ME data and confirm the existence of two distinct types of motion-associated variance with different spatial profiles, one composed of S0 signals and the other of R2* signals.
Fig. 5.
Replication in NA data of two kinds of motion-associated signals with different spatial profiles and distinct physical bases. QC:RSFC plots are formed for NA data as they were for the ME data in Fig. 4. Both S0 and R2* variance is contingent on subject motion but with different spatial patterns. Two kinds of QC:RSFC plot are shown for the NA subjects: using mean FD as the QC measure (Top; as in Fig. 4) and variability in the standardized respiratory (resp) belt envelope as the QC measure (Bottom; the same measure used in Fig. 2B). The plots show similar phenomena because, as the plot at the far right shows, mean motion and respiratory pattern variability are highly correlated. Purple values reflect permutation ranks after motion censoring as in Fig. 4.
Discussion
Signal denoising is a central concern in human neuroimaging, and multiecho fMRI sequences are attractive because they contain information, not present in typical single-echo sequences, that can be used for denoising purposes. This information is especially valuable in resting state fMRI datasets, in which there is no task timing to help isolate signal from “noise.” In this study, we examined how motion-related variance manifested and could be removed from multiecho data. Our principal findings are: (i) there are two major and separable kinds of motion-associated fMRI signals, respiratory T2* signals and artifactual S0 signals, (ii) motion-associated respiratory T2* signals are essentially whole-brain signals, (iii) motion-associated S0 signals are mostly spatially focal, (iv) motion-dependent covariance patterns in fMRI data are superpositions of these two kinds of motion-associated signals, (v) ME-ICA techniques remove the spatially focal S0 signals but not the spatially widespread T2* signals, and (vi) multiple techniques can identify and remove spatially widespread signals in fMRI data to yield times series nearly free of motion-dependent characteristics.
Elaborations on a Spatial Framework for Motion-Related Variance.
The results of this study fit well into a spatial framework for motion-related variance outlined by Power et al. (8). The framework derives from the fact that in all large datasets examined for motion artifact (to the authors’ knowledge), before denoising, signal correlations are increased in higher motion subjects, with the largest increases occurring at short distances (e.g., refs 6, 7, 10, 30–32). The same pattern is observed in the ME and NA data. This pattern may be viewed as a superposition of focal and nonfocal effects, both indexed by motion: Focal signal changes ought to enhance nearby correlations and cause distant correlations to approach zero or negative values, whereas nonfocal signal changes ought to increase correlations at all distances.
By splitting fMRI signals into S0 and R2* components, multiecho denoising techniques demonstrate that different physical mechanisms tend to cause the focal versus nonfocal element of motion-associated variance. The focal element is mostly caused by S0 modulation, and the nonfocal element is mostly caused by R2* modulation. These two kinds of motion-associated signal (focal and global) can compete and partially mask one another’s presence in motion-related analyses. For example, Fig. 4, Center (column 3) (ME-ICA denoised data) shows that motion increases all correlations at all distances, but the strong influence seen at a distance is really only appreciated once it is “unmasked” by the removal of focal artifact during the denoising process (Fig. 4, Center (column 2)].
Most S0 signal changes are focal, but there are exceptions. For example, the pan-brain signal decrease occurring during a large motion at the end of scan ME076 (Fig. 1) is identified as S0 artifact by both ME-ICA and FIT procedures (SI Appendix, Fig. S3). Immediate transient pan-brain signal decreases have been reported previously at times of motion (33). Such pan-brain changes are characteristic of some large motions, but most smaller motions tend to cause a variety of focal signal changes that manifest as the more typical salt and pepper appearance of S0 signals in Fig. 1 and Online Movie 1.
It is thus a useful heuristic to think of motion-related changes as focal S0 changes, so long as it is remembered that S0 changes can also be global. Similarly, it is a useful heuristic to think of motion-associated R2* changes as global, so long as it is remembered that there is spatial weighting to this global BOLD signal: Studies of ventilation-induced BOLD changes demonstrate that although changes are present throughout all gray matter, signal changes are most prominent in a “sensorimotor” pattern emphasizing occipital and peri-Rolandic cortex (28, 34–36).
Global fMRI Signals Are Largely Respiratory-Related BOLD R2* Signals.
In the NA cohort, variability in respiratory patterns is strongly linked to variability in the global signal (Fig. 2); in many instances, there are obvious temporal links between deep breaths and global decreases (Fig. 2, vertical black bands) in BOLD signals (SI Appendix, Fig. S4). Recent reports show that other breathing patterns, such as hypopnea or hyperpnea, coincide with prolonged increases and decreases in fMRI signals, respectively (28). Although such findings do not exclude a neurobiological component to global signals, they do indicate that a large portion of the variance in the global signal is modulated along with respiratory patterns, which have a well-established mechanism of globally modulating BOLD signal through ventilatory effects on pCO2.
The global BOLD signals observed on deep inspiration are not consistent with a typical “evoked” neural signal due to motor activity. Typical BOLD motor responses (i) are (multi)focal and (ii) exhibit maximal signal change ∼6 s after the motor activity, with resolution of signal changes by ∼15–20 s. In contrast, the global BOLD modulations seen here span all of gray matter and reach their maximal change ∼15–20 s after an inspiration, with resolution in ∼30–40 s (SI Appendix, Fig. S4). These characteristics are in accord with prior reports on the time course of pCO2-related changes in BOLD signal, such as those seen with instructed breaths (35, 37, 38).
There are strong correlations between breathing pattern variability, subject motion, and global signal variability. A likely explanation for many transient, prominent global signal changes (Fig. 2, black bands) is that subjects move their heads as they breathe deeply and pCO2 alteration then causes the global signal changes seen over the next 40 s. Not all respiratory variance will follow this single-deep-breath pattern, and other patterns, such as more rapid or slower breathing, would also produce global BOLD signal changes, although with other temporal features (e.g., more sustained global signal changes, different signs in change according to hyperventilation vs. hypoventilation). Other respiratory changes can and do occur with inspiration (immediate motion artifact as well as B0 shifts from lung expansion), but these will transpire over a shorter time scale and by different mechanisms.
At present, it is difficult to measure respiration and effectively model respiratory fMRI signals. This study and others (e.g., ref. 28) have shown that existing respiratory models using measures derived from respiratory belts often fall short of adequately modeling respiratory signals. The measure we used, the variability of the respiratory belt envelope, was highly predictive of global signal variability and may have some utility in future modeling efforts.
Multiple Approaches to Removing Global Artifacts.
If respiratory signals constitute much of the global signal variance but are either unidentifiable (if no respiratory records are present) and/or difficult to remove (since neither ICA nor common nuisance regressions adequately remove them), then, at present, the only way to remove those signals from individual fMRI scans is to remove all global signals. Multiple approaches can be used to remove global signals, and we next review the approaches we used and others that could achieve comparable results.
One simple and reliable way to remove global signals is to calculate and remove the mean signal of an image or, nearly identically, the mean gray matter or cortical signal. These signals are all similar because (i) gray matter has large signal amplitude relative to white matter and (ii) gray matter, especially the cortex, constitutes large portions of the brain. However, regression of mean signals has been criticized on multiple grounds (39, 40). The central objection is that nonglobal signals will enter the mean signal calculation, and thus regression of the mean signal will change the covariance structure between nonglobal signals beyond just removing an additive global signal. The effect size of this argument can range from undetectable to marked, depending on the dimensionality of the data, and the relevant dimensionality of fMRI data is not well understood (36). A limitation of using a single global signal for this regression is that the vascular response to arterial pCO2 may vary depending on the location of a neurovascular unit in the vascular tree (e.g., proximal vs. distal branching from the arterial supply), and a single regressor cannot fully capture such lags.
Another approach to removing global fMRI signals is multivariate signal partitioning of gray matter signals, which may avoid the above criticisms by decomposing the data into (typically independent) global and nonglobal elements. Signal partitioning algorithms have undergone much development in the past decade in response to demand for automated separation of background (global, low-rank) from sparse elements in visual images. The most common version of signal partitioning is robust principal component analysis (PCA), which is related to GODEC and can yield similar signal separation as GODEC (SI Appendix, Supplemental Methods and Supplemental Results). We focused on GODEC because it is computationally efficient in comparison to robust PCA. For illustrative purposes, SI Appendix, Fig. S12 shows robust PCA and GODEC producing similar signal partitioning in a single dataset (Online Movie 6 shows such plots for all ME subjects). Quantitatively, group correlation matrices after GODEC and robust PCA are highly similar (SI Appendix, Fig. S13).
Although GODEC and robust PCA work well in most scans to fully identify global signals, global signals do visibly persist occasionally in the sparse partition (Fig. 3 and Online Movie 6). Some user oversight is thus needed for these techniques. Additionally, these approaches are tunable and can incorporate variable numbers of signals as part of the low-rank solution. When too few signals are included, global signals may persist in the sparse space as noted just above, but when too many signals are included, “resting state network” signals that span substantial portions of the brain can be included in the background. In our data, we saw this effect when approximately five or more signals entered the background (SI Appendix, Fig. S14). We thus chose tunings that typically identified one to four signals per subject as the background space. The desired dimensionality of partitioning is likely to vary with scan characteristics (i.e., likely higher with higher spatial and/or temporal resolution).
A related approach to multivariate partitioning of gray matter signals is to define nuisance regressors via PCA performed on nuisance compartment (i.e., not gray matter) signals, an approach called CompCor in fMRI studies (41). The global fMRI signal can readily be identified by this approach without explicitly saying so, since PCA performed on voxels in white matter yields nuisance signals correlated at nearly r = 1 with the global fMRI signal. Such high correlations can be obtained even when the nuisance voxels are located considerably away from the gray matter (SI Appendix, Fig. S11). Further, voxels outside the brain proper (e.g., in sinuses) contain the global signal (28). If such voxels are included in the “nuisance” signals that undergo PCA, the global signal or something very like it will be identified and removed. SI Appendix, Fig. S12 shows CompCor operating on a single dataset. Corresponding group correlation matrices are shown in SI Appendix, Fig. S13.
The correlation matrices we obtained from GODEC and robust PCA are very similar (SI Appendix, Fig. S13), and both matrices are also similar to matrices produced by mean signal regression. The principal difference between these techniques is that GODEC and robust PCA yielded slightly higher values at negative correlations and slightly lower values at high correlations. These differences may be attributable to some combination of incomplete removal of global artifacts in some subjects by robust PCA and GODEC, the tendency of mean signal regression to cause or enhance negative correlations, and the possibility that real signals of interest entered the background partition in robust PCA and GODEC. The correlation values obtained by CompCor are broadly similar but slightly higher than those obtained by the other methods, a result that can be attributed to the fact that global signals are often visibly remaining (although attenuated) in the data. All of these methods remove global signals to an extent that zero-centers QC:RSFC curves, thus removing group-level dependence of global covariance on motion (SI Appendix, Fig. S13).
The central point we wish to emphasize is that multiple methods can (i) visibly remove global signals in gray plots (Fig. 3), (ii) eliminate motion-dependent patterns in data (Figs. 4 and 5), and (iii) eliminate the association of respiratory pattern variability with global variance (Figs. 2B and 5). Further work, likely involving simulations, will be needed to determine the precise differences between GODEC, mean signal regression, and other methods of global signal removal.
Steps Needed to Eliminate the Influence of Motion from fMRI Time Series.
Our original purpose in this study was to investigate the success of ME-ICA for removing motion artifact. Our results indicate that ME-ICA does an excellent job of identifying and removing focal S0-related artifacts, many of which are motion-related (Figs. 1, 4, and 5). In two respects, however, the data are not completely denoised. First, the analyses reported in this paper indicate that much of the remaining R2*-related variance is linked to motion and is likely related to respiration. In this sense, ME-ICA has not finished denoising the data, and techniques that remove global signals (e.g., GODEC, mean signal regression, others) are needed to eliminate most of the remaining motion dependence. In theory, if multiple sources of global signals exist, techniques that selectively removed the motion-associated signals (e.g., respiration) would suffice to remove motion dependence from the data. Second, the scrubbing analyses indicate that at no stage of denoising was focal artifact completely removed from the data: Censoring volumes with FD > 0.2 mm produces decreases in short-distance correlations more than long-distance correlations (Fig. 4) at all stages of denoising, including after ME-ICA + GODEC. This result stems from the fact that nonfocal signal-removing techniques (e.g., GODEC) will do little for focal artifacts, and it is already evident from prior reports that ME-ICA alone does not fully restore signals acquired during motion to normalcy (e.g., figure 1 and figure S5 of ref. 16). The failure to completely remove focal (and/or global) artifacts during motion is shared across a wide variety of examined denoising techniques (reviewed in ref. 8). Thus, something needs to be done to address the volumes irreparably corrupted by subject motion. In this study, censoring those volumes reduced the remaining small influence of motion to negligible levels.
Two findings of this study need to be emphasized. First, full removal of the influence of motion on fMRI time series requires at least two, and likely three, elements: (i) removal of focal artifacts that are immediately produced by motion, (ii) removal of global signals due to ventilation that scale with head motion, and (iii) excision (or downweighing or replacement) of the time points that are beyond repair in i. The combination of these three elements can eliminate the unwanted influences of motion within an fMRI dataset.
The second finding deserving emphasis is that distant-dependent changes in signal covariance indexed by motion are mainly attributable to non-BOLD artifact. In Figs. 4 and 5, discarded S0 signals exhibit strong distance dependence, whereas the retained R2* signals are nearly distance-independent with respect to motion. If we consider that ME-ICA has likely not perfectly separated S0 and R2* effects (seen by the occasional splitting of global BOLD signals into both retained and discarded components) and also that signals during motion remain somewhat corrupted (as shown in the scrubbing analyses of Fig. 4), it seems likely that some S0 signals are still in the retained R2* components, which may account for the slight distance dependence that remains in the retained data (before censoring). Future refinements of multiecho procedures may improve such signal separation. Regardless, it is clear that motion-related distance dependence is mostly, if not entirely, a product of non-BOLD artifacts in fMRI time series.
Materials and Methods
Full descriptions of the data and methods are provided in SI Appendix.
In brief, 89 multiecho datasets previously published in a study by Kundu et al. (16) are examined; these fMRI data are from adults from whom informed consent was obtained in a study approved by the Local Research Ethical Committee at the University of Cambridge (LREC 11/EE/0198). These datasets are four-echo datasets (TE = 12 ms, 28 ms, 44 ms, and 60 ms) and are called the ME cohort; each subject contributed a single 9.8-min scan. An additional set of multiecho scans was obtained from 12 subjects at the NIH with simultaneous monitoring of respiration and heart rhythms via respiratory belt and pulse oximeter. These subjects gave written consent and were part of a larger study approved by the National Institute of Mental Health Institutional Review Board (Protocol 10-M-0027). These are three-echo datasets (TE = 12 ms, 24.5 ms, and 37 ms) and are called the NA cohort; each subject contributed two 6.25-min scans. Details of the sequences and datasets are listed in SI Appendix, Supplemental Methods and Table S2.
All scans underwent standard preprocessing, including motion estimation, slice time correction, and transformation to a target atlas. All scans underwent multiecho denoising designed to isolate S0 from R2* influences in the data. To create a single image for a scan, signals from all echoes were optimally combined using weighting by estimated T2* of each voxel as in the study by Kundu et al. (16). ME-ICA denoising was performed as in the study by Kundu et al. (16), and we examine undenoised images (optimally combined), denoised images (BOLD-like, R2*-modulating components), and the discarded signals (non–BOLD-like, S0-modulating components). All multiecho scans also underwent a procedure we term FIT, in which S0 and R2* were fit to a monoexponential decay at each voxel and time point by least squares, and we examine denoised images (R2* estimates) and discarded signals (S0 estimates).
The plots in Figs. 1–3 are described by Power et al. (42). Briefly, a T1-weighted image in register with an echoplanar imaging (EPI) image was used to derive brain compartments via FreeSurfer; the mean and linear trend terms of the EPI image were removed at each voxel; and the time series of all voxels in the image are shown as a grayscale heat map, ordered by tissue compartment. Several kinds of traces are shown above these heat maps. Head position estimates are from realignment algorithms. The red FD trace is based on the sum of the differentiated position estimates, and thus indexes head motion. Mean FD was the average value of the FD trace. Blue respiratory belt traces are raw data, and green heart rate traces are derived from the intervals between peaks in the pulse oximeter trace. All physiological traces and derived measures were visually checked in their entirety. Respiratory pattern variability was defined as the SD of the envelope of the z-scored respiratory belt waveform. Compartment signals, such as the mean gray matter signal, are derived from appropriate FreeSurfer compartments. When various denoising models are used subsequent to ME-ICA (e.g., ME-ICA and then mean cortical signal regression), the models are applied to the ME-ICA denoised image. GODEC is a multivariate technique that can separate nonfocal signals from focal signals in a dataset via techniques related to robust PCA decomposition.
The plots shown in Figs. 4 and 5 reflect standard analyses developed in the field to assay fMRI data for motion artifact, as reviewed by Power et al. (8). Briefly, signals were extracted in each subject from 264 10-mm-diameter spherical ROIs, as reported by Power et al. (43), that span much of the brain. Pairwise correlations were calculated in each subject’s data and were Fisher z-transformed. In QC:RSFC analyses, mean FD was correlated across subjects with each pairwise correlation to yield a 264 × 264 matrix, which was then plotted as a function of the distance separating ROIs. In high–low-motion differences, a median split of subjects by mean FD was performed, the mean of each subgroup was calculated, and the difference between subgroups was plotted as a function of distance. In scrubbing analyses, signal correlations were calculated using all time points and then again without high-motion (FD > 0.2 mm) time points, the difference in resulting correlations was calculated, and the mean of this difference across subjects was plotted as a function of distance. The red points and the white smoothing curves display actual data, and the black smoothing curves depict 50 of the 10,000 conducted permutations of mean FD (QC:RSFC and high–low analyses) or of censored volumes (scrubbing analyses). Smoothing curves were generated via sliding boxcars of 1,000 points. The inset numbers are the percentiles of observed data among permutations, in terms of smoothing curve values at 35 mm (to index all motion-related signals, focal and nonfocal) and the difference between smoothing curve values at 35 and 100 mm (to index distance dependence, mainly due to focal signals).
Supplementary Material
Acknowledgments
We thank Kevin Tran, David Godlove, and the NIH Helix/Biowulf staff for their computing support. This work was supported by the Intramural Research Program, National Institute of Mental Health/NIH (Grants ZIAMH002920 and NCT01031407), and by the Sackler Institute for Developmental Psychobiology (J.D.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited on OpenfMRI, https://openfmri.org/dataset (accession no. ds000258).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720985115/-/DCSupplemental.
References
- 1.Buckner RL, Krienen FM, Yeo BT. Opportunities and limitations of intrinsic functional connectivity MRI. Nat Neurosci. 2013;16:832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- 2.Power JD, Schlaggar BL, Petersen SE. Studying brain organization via spontaneous fMRI signal. Neuron. 2014;84:681–696. doi: 10.1016/j.neuron.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy K, Birn RM, Bandettini PA. Resting-state fMRI confounds and cleanup. Neuroimage. 2013;80:349–359. doi: 10.1016/j.neuroimage.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianciardi M, et al. Sources of functional magnetic resonance imaging signal fluctuations in the human brain at rest: A 7 T study. Magn Reson Imaging. 2009;27:1019–1029. doi: 10.1016/j.mri.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcus DS, et al. WU-Minn HCP Consortium Human connectome project informatics: Quality control, database services, and data visualization. Neuroimage. 2013;80:202–219. doi: 10.1016/j.neuroimage.2013.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satterthwaite TD, et al. Impact of in-scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Power JD, Schlaggar BL, Petersen SE. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage. 2015;105:536–551. doi: 10.1016/j.neuroimage.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyszka JM, Kennedy DP, Paul LK, Adolphs R. Largely typical patterns of resting-state functional connectivity in high-functioning adults with autism. Cereb Cortex. 2014;24:1894–1905. doi: 10.1093/cercor/bht040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel JS, et al. Data quality influences observed links between functional connectivity and behavior. Cereb Cortex. 2016;27:4492–4502. doi: 10.1093/cercor/bhw253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgson K, et al. Shared genetic factors influence head motion during MRI and body mass index. Cereb Cortex. 2016;27:5539–5546. doi: 10.1093/cercor/bhw321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laumann TO, et al. On the stability of BOLD fMRI correlations. Cereb Cortex. 2016;27:4719–4732. doi: 10.1093/cercor/bhw265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng LL, et al. Neurobiological basis of head motion in brain imaging. Proc Natl Acad Sci USA. 2014;111:6058–6062. doi: 10.1073/pnas.1317424111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bright MG, Murphy K. Removing motion and physiological artifacts from intrinsic BOLD fluctuations using short echo data. Neuroimage. 2013;64:526–537. doi: 10.1016/j.neuroimage.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kundu P, et al. Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proc Natl Acad Sci USA. 2013;110:16187–16192. doi: 10.1073/pnas.1301725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dipasquale O, et al. Comparing resting state fMRI de-noising approaches using multi- and single-echo acquisitions. PLoS One. 2017;12:e0173289. doi: 10.1371/journal.pone.0173289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundu P, et al. Multi-echo fMRI: A review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage. 2017;154:59–80. doi: 10.1016/j.neuroimage.2017.03.033. [DOI] [PubMed] [Google Scholar]
- 19.Schölvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci USA. 2010;107:10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong CW, Olafsson V, Tal O, Liu TT. Anti-correlated networks, global signal regression, and the effects of caffeine in resting-state functional MRI. Neuroimage. 2012;63:356–364. doi: 10.1016/j.neuroimage.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong CW, Olafsson V, Tal O, Liu TT. The amplitude of the resting-state fMRI global signal is related to EEG vigilance measures. Neuroimage. 2013;83:983–990. doi: 10.1016/j.neuroimage.2013.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong CW, DeYoung PN, Liu TT. Differences in the resting-state fMRI global signal amplitude between the eyes open and eyes closed states are related to changes in EEG vigilance. Neuroimage. 2016;124:24–31. doi: 10.1016/j.neuroimage.2015.08.053. [DOI] [PubMed] [Google Scholar]
- 23.Matsui T, Murakami T, Ohki K. Transient neuronal coactivations embedded in globally propagating waves underlie resting-state functional connectivity. Proc Natl Acad Sci USA. 2016;113:6556–6561. doi: 10.1073/pnas.1521299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci USA. 1990;87:9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stillman AE, Hu X, Jerosch-Herold M. Functional MRI of brain during breath holding at 4 T. Magn Reson Imaging. 1995;13:893–897. doi: 10.1016/0730-725x(95)00037-h. [DOI] [PubMed] [Google Scholar]
- 26.Poulin MJ, Liang PJ, Robbins PA. Dynamics of the cerebral blood flow response to step changes in end-tidal PCO2 and PO2 in humans. J Appl Physiol (1985) 1996;81:1084–1095. doi: 10.1152/jappl.1996.81.3.1084. [DOI] [PubMed] [Google Scholar]
- 27.Kastrup A, Li TQ, Takahashi A, Glover GH, Moseley ME. Functional magnetic resonance imaging of regional cerebral blood oxygenation changes during breath holding. Stroke. 1998;29:2641–2645. doi: 10.1161/01.str.29.12.2641. [DOI] [PubMed] [Google Scholar]
- 28.Power JD, Plitt M, Laumann TO, Martin A. Sources and implications of whole-brain fMRI signals in humans. Neuroimage. 2017;146:609–625. doi: 10.1016/j.neuroimage.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou TTD. 2011 GoDec: Randomized low-rank and sparse matrix decomposition in noisy case. Proceedings of the Twenty-Eighth International Conference on Machine Learning. Available at https://opus.lib.uts.edu.au/bitstream/10453/19145/1/2011001843OK.pdf. Accessed January 24, 2018.
- 30.Power JD, Plitt M, Kundu P, Bandettini PA, Martin A. Temporal interpolation alters motion in fMRI scans: Magnitudes and consequences for artifact detection. PLoS One. 2017;12:e0182939. doi: 10.1371/journal.pone.0182939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muschelli J, et al. Reduction of motion-related artifacts in resting state fMRI using aCompCor. Neuroimage. 2014;96:22–35. doi: 10.1016/j.neuroimage.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess GC, et al. Evaluation of denoising strategies to address motion-correlated artifacts in resting-state functional magnetic resonance imaging data from the human connectome project. Brain Connect. 2016;6:669–680. doi: 10.1089/brain.2016.0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satterthwaite TD, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 35.Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 36.Power JD, Laumann TO, Plitt M, Martin A, Petersen SE. On global fMRI signals and simulations. Trends Cogn Sci. 2017;21:911–913. doi: 10.1016/j.tics.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: The temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage. 2008;40:644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage. 2009;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: Are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saad ZS, et al. Trouble at rest: How correlation patterns and group differences become distorted after global signal regression. Brain Connect. 2012;2:25–32. doi: 10.1089/brain.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Power JD. A simple but useful way to assess fMRI scan qualities. Neuroimage. 2017;154:150–158. doi: 10.1016/j.neuroimage.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Power JD, et al. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.