Significance
While the critical role of RIPK1 kinase activity in mediating necroptosis and RIPK1-dependent apoptosis has been established, we still know little about how the nonkinase domains of RIPK1 regulate its kinase activity. Establishing the role of RIPK1-death domain (DD) in mediating RIPK1 activation and formation of complex II provides an important insight into the molecular mechanism by which RIPK1 is activated during the transition from complex I to complex II. These results suggest that RIPK1-DD self-association may provide an amplification mechanism to promote the activation of RIPK1 kinase activity for mediating signal transduction to lead to cell death. Our results also suggest that the activation of RIPK1 may be regulated by its concentration as increased expression of RIPK1 under pathological conditions may promote its dimerization and activation.
Keywords: RIPK1, death domain, dimerization, necroptosis, RIPK1-dependent apoptosis
Abstract
RIPK1 is a critical mediator of cell death and inflammation downstream of TNFR1 upon stimulation by TNFα, a potent proinflammatory cytokine involved in a multitude of human inflammatory and degenerative diseases. RIPK1 contains an N-terminal kinase domain, an intermediate domain, and a C-terminal death domain (DD). The kinase activity of RIPK1 promotes cell death and inflammation. Here, we investigated the involvement of RIPK1-DD in the regulation of RIPK1 kinase activity. We show that a charge-conserved mutation of a lysine located on the surface of DD (K599R in human RIPK1 or K584R in murine RIPK1) blocks RIPK1 activation in necroptosis and RIPK1-dependent apoptosis and the formation of complex II. Ripk1K584R/K584R knockin mutant cells are resistant to RIPK1 kinase-dependent apoptosis and necroptosis. The resistance of K584R cells, however, can be overcome by forced dimerization of RIPK1. Finally, we show that the K584R RIPK1 knockin mutation protects mice against TNFα-induced systematic inflammatory response syndrome. Our study demonstrates the role of RIPK1-DD in mediating RIPK1 dimerization and activation of its kinase activity during necroptosis and RIPK1-dependent apoptosis.
RIPK1 is a critical mediator of cell death and inflammation downstream of TNFR1 upon stimulation by TNFα, a potent proinflammatory cytokine involved in a multitude of human inflammatory and degenerative diseases (1–3). TNFα may promote the activation of apoptosis or necroptosis, mediated by TNFR1 through intracellular signaling processes involving the formation of sequential protein complexes. Activation of TNFR1 by TNFα leads to the rapid formation of a transient complex termed complex I, or TNF-RSC, associated with the intracellular death domain (DD) of TNFR1. The components of complex I include TRADD and RIPK1, which are both DD-containing proteins that interact with TNFR1 via homotypic DD interaction (4). In apoptosis-competent cells, complex I transitions into complex IIa, which includes RIPK1, FADD, and caspase-8, to promote apoptosis (5). When apoptosis is inhibited, necroptosis may be activated by the formation of complex IIb, consisting of RIPK1, FADD, caspase-8, and RIPK3, which in turn promotes the phosphorylation and oligomerization of MLKL and the execution of necrosis (6–9).
RIPK1 is composed of an N-terminal serine/threonine kinase domain, an intermediate domain, and a C-terminal DD (10). The kinase activity encoded by the N-terminal kinase domain is essential for necroptosis and RIPK1-dependent apoptosis induced by TNFα (11–13). The intermediate domain is involved in mediating NF-κB and MAPK activation through ubiquitination at K377 by cIAP1 and binding with TRAF2, NEMO, and TAK1 (14). The RIP homotypic interaction motif (RHIM) in the intermediate domain regulates necroptosis by interaction with RIPK3, as mutating IQIG in the core RHIM motif of RIPK1 to AAAA disrupts the interaction of RIPK1 and RIPK3 (15). On the other hand, the C-terminal DD is known to be involved in the recruitment of RIPK1 to a death receptor signal complex, such as TNFR1, upon the stimulation of its cognitive ligand TNFα. The DD of RIPK1 is known to mediate the binding to other DD-containing adaptor proteins, e.g., TRADD and FADD, for its recruitment into complex I and to mediate apoptosis (16, 17). However, the functional role of RIPK1-DD in regulating the activation of its N-terminal kinase domain has not been investigated.
The DD superfamily has emerged as a prime mediator of cell death and inflammation signal transmission. DD-containing proteins usually form homodimers or oligomers based on homo- or hetero-association among subfamily members (18). However, the role of DD-mediated homo- or heterodimerization in enzymatic activities that may be encoded by other parts of the molecules has rarely been investigated. In this study, we investigated the involvement of RIPK1-DD in the activation of its kinase activity. We show that mutating K599 in human RIPK1, or its conserved residue K584 in murine RIPK1, a lysine located on the surface of the death domain to arginine, blocks RIPK1 homodimerization, kinase activation, and the formation of complex II in necroptosis and RIPK1-dependent apoptosis (RDA). Ripk1K584R/K584R knockin mutant cells are resistant to RIPK1-dependent apoptosis and necroptosis. The resistance of Ripk1K584R/K584R mutant cells, however, can be overcome by forced dimerization of RIPK1. Finally, we show that the K584R mutation protects mice against TNFα-induced systematic inflammatory response syndrome (SIRS). Our study demonstrates the role of RIPK1-DD in mediating RIPK1 dimerization and activation.
Results
K599R Mutation Blocks RIPK1-DD Interaction.
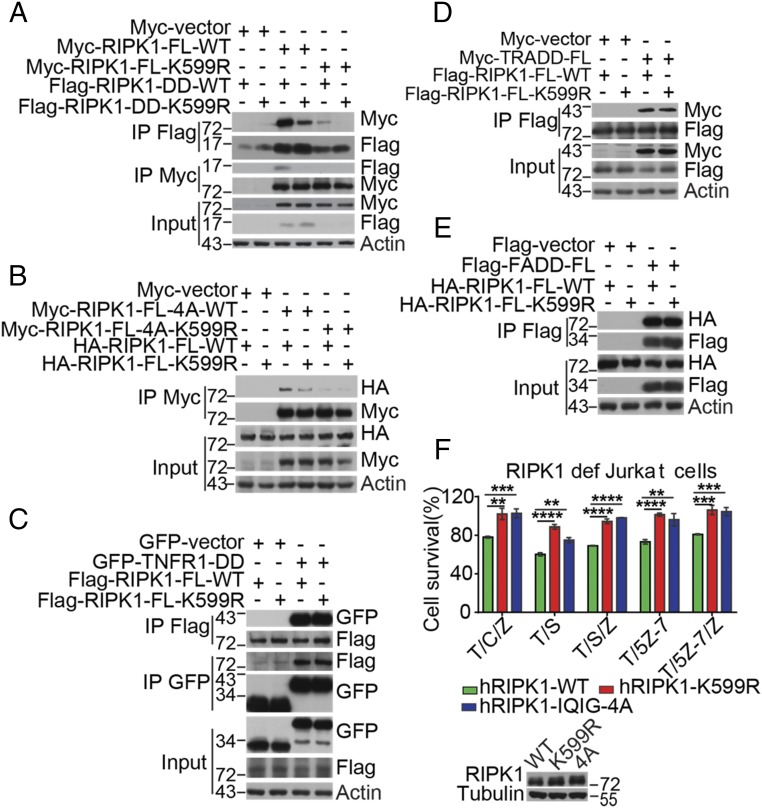
All members of the DD superfamily show a conserved 6–α-helical bundle structural fold (19). However, they also contain distinguishing sequence and structural features that differentiate them from each other. To date, the structure of RIPK1-DD has not been resolved as the purified protein is prone to form aggregates by self-association. Mutagenesis studies of TNFR1-DD have shown that the α2, α3, and α4 helixes contain residues that may be involved in mediating homodimerization as well as hetero-association with other DDs (20, 21). K599 of RIPK1 is one of the conserved residues on the surface of DD formed by α2–α3, so we hypothesized that it might be important for mediating RIPK1-DD homodimerization. We experimentally tested this possibility first in 293T cells and found that the K599R mutation in RIPK1-DD dramatically reduced the binding between RIPK1-DD and full-length RIPK1 (Fig. 1A and Fig. S1A).
Fig. 1.
The hRIPK1 K599R mutation affects RIPK1 dimerization and cell death. (A) HEK293T cells were cotransfected with Myc-tagged full-length RIPK1, Flag-tagged RIPK1-DD WT, or Flag-tagged RIPK1-DD K599R expression plasmids as indicated for 24 h. The cells were lysed with 0.5% Nonidet P-40 buffer and divided equally into two parts. The cell lysates were immunoprecipitated with anti-Flag antibody-conjugated agarose and anti-Myc antibody-conjugated agarose. respectively. (B) HEK293T cells were cotransfected with Myc-tagged full-length RIPK1-4A, Myc-tagged full-length RIPK1-4A-K599R, HA-tagged full-length RIPK1-WT, and HA-tagged full-length RIPK1-K599R expression plasmids as indicated. The cell lysates were immunoprecipitated with anti-Myc–conjugated agarose. (C–E) HEK293T cells were cotransfected with full-length RIPK1-WT or RIPK1-K599R expression plasmids and GFP-tagged TNFR1 death domain (C) or Myc-tagged full-length TRADD (D) or Flag-tagged full-length FADD expression plasmids (E) for 24 h. The cells were then lysed with 0.5% Nonidet P-40 buffer. The cell lysates were immunoprecipitated with anti-Flag antibody-conjugated agarose or anti-GFP antibody as indicated. (A–E) Immunocomplexes were analyzed by Western blotting using indicated antibodies. (F) RIPK1-deficient Jurkat cells were infected with retrovirus encoding HA-tagged hRIPK1 WT, HA-tagged hRIPK1 K599R, and HA-tagged hRIPK1 IQIG-4A by the Tet-On Advanced Inducible Expression System. Reconstituted Jurkat cells were treated with 1 μg/mL doxycycline for 48 h to induce the expression of RIPK1. The cells were then pretreated with 100 nM SM-164, 100 nM 5Z-7, and 1 μg/mL CHX for 2 h and treated with 100 ng/mL TNFα and 50 μM zVAD.fmk for an additional 8 h. The cell survival was measured by CellTiterGlo. (**P < 0.01; ***P < 0.001; ****P < 0.0001.) (Lower) Western blot analysis of RIPK1 expression levels in RIPK1-deficient Jurkat cells infected with retrovirus encoding HA-tagged hRIPK1 WT, K599R, and 4A.
The RHIM motif in the intermediate domain can mediate strong oligomerization of RIPK1 in the 293T overexpression system (22). To investigate the contribution of DD in the context of full-length RIPK1, we neutralized the RHIM by introducing IQIG-4A. We found that the binding of K599R FL RIPK1 with IQIG-4A FL RIPK1 was reduced compared with that of WT FL RIPK1 with IQIG-4A FL RIPK1 (Fig. 1B). Furthermore, the K599R+IQIG-4A double-mutated RIPK1 lost the ability to bind with other RIPK1 (Fig. 1B), suggesting that both DD and RHIM were involved in mediating RIPK1-RIPK1 homo-interaction.
Interestingly, while K599R RIPK1 blocked the binding with RIPK1-DD, it had no significant influence on its hetero-dimerization with TNFR1-DD (Fig. 1C), TRADD-DD (Fig. 1D), or FADD-DD (Fig. 1E). This result suggests that, although the members of the DD family share a similar core 3D structure, subtle sequence differences on the surface of DDs may determine the specificity of their interactions.
K599R RIPK1 Mutation Blocks Necroptosis and RDA of Human Cells.
We next analyzed the contribution of K599 in human RIPK1 to TNFα-induced cell death by complementation of RIPK1-deficient cells. We induced necroptosis and apoptosis with different combinations of TNFα, pan-caspase inhibitor zVAD.fmk, cycloheximide (CHX), small-molecule IAP antagonist SM-164 or TAK1 inhibitor (5Z)-7-oxozeaenol (5Z-7) and found that, in contrast to that of WT RIPK1 complementation, RIPK1-deficient Jurkat cells reconstituted with K599R or IQIG-4A RIPK1 were highly resistant to necroptosis induced by TNFα/CHX/zVAD.fmk, TNFα/SM-164/zVAD.fmk, or TNFα/5Z-7/zVAD.fmk (Fig. 1F). Interestingly, K599R showed better suppression than IQIG-4A in RIPK1-dependent apoptosis triggered by TNFα/SM-164 or TNFα/5Z-7. On the other hand, K599R RIPK1 did not affect RIPK1-kinase–independent apoptosis induced by TNFα/CHX treatment (Fig. S1B). These results suggest that the DD-mediated RIPK1 interaction regulates the cell death in both RIPK1-dependent apoptosis and necroptosis, but not in RIPK1-independent apoptosis, while the RHIM may be primarily involved in mediating its interaction with the RHIM of RIPK3 in necroptosis as previously reported (15).
Ripk1K584R Knockin Mutation Blocks Necroptosis and RIPK1-Dependent Apoptosis.
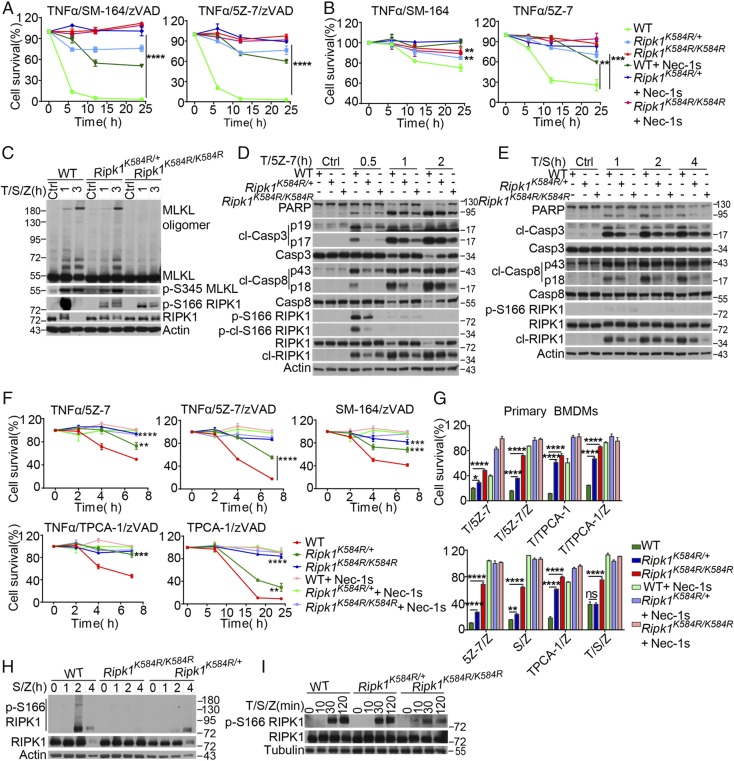
K584 in murine RIPK1 corresponds to K599 in human RIPK1 (Fig. S1C). To further investigate the importance of K584-mediated RIPK1-DD homodimerization, we generated Ripk1K584R knockin mutant mice by mutating the conserved lysine (K) at position 584 to arginine (K584R) (Fig. S2A). Ripk1K584R/K584R mice were born with expected Mendelian ratios (Fig. S2B), showed no developmental defects, and were normal as adults. We obtained primary mouse embryo fibroblasts (MEFs) derived from littermates of E14 WT Ripk1, Ripk1K584R/+, and Ripk1K584R/K584R embryos. We found that Ripk1K584R/K584R MEFs showed strong resistance to necroptosis and RDA, comparable to Nec-1s treatment, when induced by different stimuli, whereas Ripk1K584R/+ MEFs showed partial but statistically significant protection (Fig. 2 A and B and Fig. S2C). p-S166 RIPK1—a biomarker of RIPK1 kinase activation (23) in MEFs stimulated by treatments that induced necroptosis and RDA—was strongly reduced by heterozygous or homozygous K584R mutation (Fig. 2 C and D). Phosphorylation and oligomerization of MLKL was blocked in Ripk1K584R/K584R MEFs and delayed in Ripk1K584R/+ MEFs under necroptotic conditions (Fig. 2C). The activation of RIPK1 kinase activity as well as the cleavage of caspase-3, caspase-8, PARP-1, and RIPK1 in RDA induced by TNFα/5Z-7 or TNFα/SM164 were inhibited in both Ripk1K584R/K584R MEFs and Ripk1K584R/+ MEFs (Fig. 2 D and E). On the other hand, Ripk1K584R/K584R MEFs and Ripk1K584R/+ MEFs showed no resistance to RIPK1-independent apoptosis induced by TNFα/CHX, but were protected against necroptosis induced by TNFα/CHX/zVAD.fmk (Fig. S2 D–F).
Fig. 2.
Ripk1K584R/K584R mutant cells are resistant to necroptosis and RDA. (A) WT, Ripk1K584R/+, and Ripk1K584R/K584R immortalized MEFs were pretreated with 25 nM SM-164 or 100 nM 5Z-7 for 2 h as indicated and then treated with 20 ng/mL TNFα and 50 μM zVAD.fmk for 3, 16, and 24 h. (B) WT, Ripk1K584R/+, and Ripk1K584R/K584R immortalized MEFs were pretreated with 25 nM SM-164 or 100 nM 5Z-7 for 2 h as indicated and then treated with 20 ng/mL TNFα for 3, 16, and 24 h. (C) WT, Ripk1K584R/+, and Ripk1K584R/K584R immortalized MEFs were pretreated with 100 nM SM-164 (S) for 2 h, and then 100 ng/mL TNFα (T) and 50 μM zVAD.fmk (Z) were added for 1 and 3 h. The cells were lysed with 0.5% Nonidet P-40 buffer and analyzed by Western blotting with anti-MLKL antibody after a nonreducing SDS/PAGE or with anti–p-S345-MLKL, p-S166 RIPK1, and RIPK1 antibodies in normal SDS/PAGE. (D) WT, Ripk1K584R/+, and Ripk1K584R/K584R immortalized MEFs were pretreated with 500 nM 5Z-7 with for 1 h, and then 100 ng/mL TNFα (T) was added for 0.5, 1, and 2 h. The cells were lysed with 0.5% Nonidet P-40 buffer and analyzed by Western blotting with indicated antibodies. (E) WT, Ripk1K584R/+, and Ripk1K584R/K584R immortalized MEFs were pretreated with 100 nM SM-164 for 2 h and then treated with 100 ng/mL TNFα as indicated for 1, 2, and 4 h. The cells were lysed with 0.5% Nonidet P-40 buffer and analyzed by Western blotting with indicated antibodies. (F) WT, Ripk1K584R/+, and Ripk1K584R/K584R primary peritoneal macrophages were treated with TNFα/5Z-7, TNFα/5Z-7/zVAD, SM-164/zVAD, or TNFα/TPCA-1/zVAD as indicated for 2, 4, and 7 h. The cells were treated with TPCA-1/zVAD as indicated for 7, 18, and 24 h. (G) WT, Ripk1K584R/+, and Ripk1K584R/K584R primary bone marrow derived macrophages were treated with different combination of TNFα, 5Z-7, TPCA-1, SM-164 and zVAD as indicated for 16 h. (H) WT, Ripk1K584R/+ and Ripk1K584R/K584R primary bone marrow-derived macrophages were treated with 100 nM SM-164 and 50 μM zVAD.fmk for 1 h, 2 h and 4 h. The cells were lysed with 0.5% Nonidet P-40 buffer and analyzed by Western blotting with the indicated antibodies. (I) WT, Ripk1K584R/+, and Ripk1K584R/K584R primary bone marrow-derived macrophages were pretreated with 100 nM SM-164 for 2 h, and then 100 ng/mL TNFα and 50 μM zVAD.fmk were added for an additional 10, 30, and 120 min. The cells were lysed with 0.5% Nonidet P-40 buffer and analyzed by Western blotting with indicated antibodies. The cell survival in A, B, F, and G was measured by CellTiterGlo. Results shown are averages of triplicates ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
We also analyzed the responses of peritoneal macrophages (PMs) and bone-marrow–derived primary macrophages (BMDMs) to a variety of pronecroptosis and pro-RDA stimuli, triggered by a combination of TNFα, 5Z-7, SM-164, or TPCA-1 (IKKβ inhibitor) and zVAD.fmk. We found that Ripk1K584R/K584R PMs (Fig. 2F) and BMDMs (Fig. 2G) also showed strong resistance to necroptosis and RDA induced by different stimuli, whereas heterozygous Ripk1K584R/+ PMs and BMDMs showed partial resistance (Fig. 2 F and G). BMDMs can undergo necroptosis in response to SM-164/zVAD.fmk alone due to low levels of autocrine TNFα signaling (7, 24). The activation of RIPK1, as shown by p-S166 RIPK1, was reduced in Ripk1K584R/K584R BMDMs under necroptosis conditions induced by SM-164/zVAD.fmk as well as TNFα/SM-164/zVAD.fmk (Fig. 2 H and I). On the other hand, Ripk1K584R/+ BMDMs showed resistance and defective RIPK1 activation in response to SM-164/zVAD.fmk but not TNFα/SM-164/zVAD.fmk. Correspondingly, the activation of RIPK1 in Ripk1K584R/+ BMDMs was blocked when induced by SM-164/zVAD.fmk but not TNFα/SM-164/zVAD.fmk (Fig. 2 H and I).
Ripk1K584R Mutation Disrupts the Formation of Complex II.
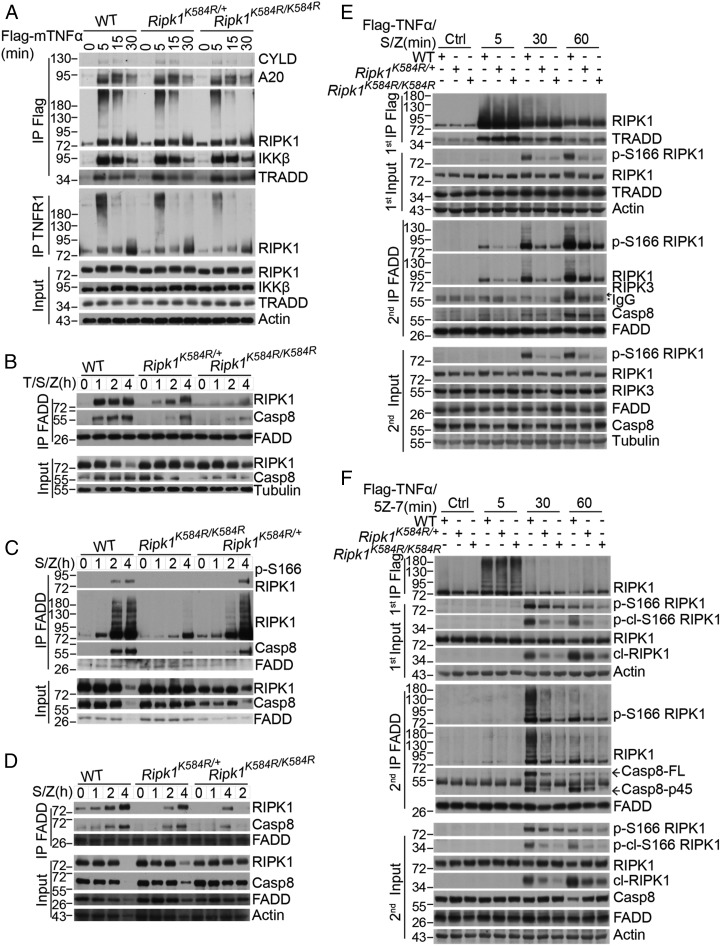
In TNFα-stimulated cells, RIPK1 is recruited rapidly to TNFR1 to form the TNF-signaling complex (TNF-RSC, or complex I) along with other components such as TRADD and CYLD. To determine the effects of RIPK1 K584R on complex I, we compared the formation of complex I using primary WT Ripk1, Ripk1K584R/+, and Ripk1K584R/K584R MEFs generated from the same pregnant female. We found that the recruitment and ubiquitination of RIPK1 in complex I of TNFα-stimulated Ripk1K584R/+ and Ripk1K584R/K584R MEFs decreased slightly and that of TRADD increased slightly compared with WT MEFs, while that of the deubiquitinase CYLD (cylindromatosis), A20 (tumor necrosis factor α-induced protein 3), and IKKβ (inhibitor of nuclear factor κ-B kinase subunit β) were not affected (Fig. 3A). Similarly, no strong difference was found in the recruitment of RIPK1, TRADD, or p-IKKα/β to complex I in WT Ripk1 and Ripk1K584R/+ MEFs, and a partial effect in complex I was found in Ripk1K584R/K584R MEFs stimulated by TNFα/SM-164, TNFα/SM-164/zVAD.fmk, TNFα/5Z-7, or TNFα/5Z-7/zVAD.fmk (Fig. S3 A and B). A similar result was found in complex I of RIPK1-deficient Jurkat cells reconstituted with hRIPK1 WT or K599R induced by Flag-TNFα (Fig. S3C). In addition, no difference in the recruitment of Sharpin, a key component of the M1-ubiquitinating LUBAC complex, which suppresses RIPK1 activation (25), was found in complex I of WT Ripk1 and Ripk1K584R/K584R MEFs (Fig. S3D). Similarly, the early activation profile of IKKα/β, JNK, ERK, p38, p-IκBα, or degradation of IκBα was not changed by K584R mutation (Fig. S3E).
Fig. 3.
The Ripk1K584R mutation disrupts the formation of complex IIa. (A) WT, Ripk1K584R/+, and Ripk1K584R/K584R primary MEFs were treated for 100 ng/mL Flag-TNFα for indicated time points. The cells were lysed with 0.5% Nonidet P-40 buffer and divided equally into two parts for immunoprecipitation with anti-Flag and anti-TNFR1 antibody, respectively. All immunoprecipitated complexes and whole-cell lysates were analyzed by Western blotting with the indicated antibodies. (B–D) WT, Ripk1K584R/+, and Ripk1K584R/K584R primary MEFs (B), primary BMDMs (C), or primary peritoneal macrophages (D) were pretreated with 50 nM SM-164 for 2 h, and then TNFα and zVAD were added as indicated for 1, 2, and 4 h. The cells were lysed with 0.5% Nonidet P-40 buffer and immunoprecipitated with anti-FADD antibody. The immunoprecipitated complex was then analyzed by Western blotting with caspase-8, TRADD, FADD, and RIPK1 antibodies. (E and F) WT, Ripk1K584R/+, and Ripk1K584R/K584R immortalized MEFs were pretreated with 100 nM SM-164 (E) or 500 nM 5Z-7 (F) for 2 h and 50 μM zVAD for 30 min, and then 100 ng/mL TNFα was added for 5, 30, and 60 min. The cells were lysed with 0.5% Nonidet P-40 buffer. The cell lysates were collected and sequentially immunoprecipitated with anti-Flag and anti-FADD antibody. First immunoprecipitation (IP): The TNFR1 complex I was immunoprecipitated using anti-Flag antibody. Second IP: the supernatants after the first IP were then immunoprecipitated with anti-FADD antibody. The immunoprecipitated complexes and whole-cell lysates were analyzed by Western blotting with the indicated antibodies.
Since the Ripk1K584R mutation, especially in heterozygosity, resulted in minimal difference in complex I but could strongly block the activation of caspases in RDA, which is mediated by complex II, we next analyzed its effect on binding of RIPK1 with FADD, a critical step in the formation of complex II. We stimulated MEFs with TNFα/SM-164/zVAD.fmk, where zVAD.fmk was used to prevent the cleavage of RIPK1, and examined the interaction between RIPK1 and FADD. We found that both homozygous and heterozygous K584R mutations in MEFs strongly inhibited the interaction between RIPK1 and FADD as well as between FADD and caspase-8 (Fig. 3B). Similarly, the RIPK1 K584R mutation also blocked SM-164/zVAD.fmk-induced complex II formation in primary BMDMs (Fig. 3C) and PMs (Fig. 3D). Both the interactions of RIPK1 with FADD, and of FADD with caspase-8, were inhibited by both heterozygous and homozygous K584R mutations in primary BMDMs and PMs treated with SM-164/zVAD.fmk. On the other hand, in RIPK1-independent apoptosis induced by TNFα/CHX, RIPK1 was not recruited into the FADD-associated complex IIa at all, which explained why K599R/K584R mutants did not affect this type of cell death (Fig. S3F).
We next compared the time course of RIPK1 activation during necroptosis by analyzing the profiles of RIPK1 in three cellular compartments, cytosol, associated with complex I, and associated with complex II, by sequential immunoprecipitation with Flag-TNFα and FADD, respectively. We found that in MEFs stimulated by TNFα/SM-164/zVAD.fmk for 5 min, a substantial amount of RIPK1 was already associated with complex I (Fig. 3E). However, the K584R mutation, in either homozygous or heterozygous MEFs, had no effect on the recruitment of RIPK1 in complex I. On the other hand, the pool of activated RIPK1 in the cytosol, detectable mainly after 30 min TNFα/SM-164/zVAD.fmk stimulation, was strongly inhibited in both Ripk1K584R/+ and Ripk1K584R/K584R MEFs. Furthermore, the activated RIPK1 was found to be associated mainly with FADD after 30 and 60 min of TNFα/SM-164/zVAD.fmk stimulation in WT MEFs, which was also strongly inhibited by both homozygous and heterozygous K584R mutation. The interaction of activated RIPK1 with RIPK3 was detected in WT MEFs at 60 min, but not at 30 min, after the stimulation, which was also reduced by the K584R mutation. Thus, RIPK1-DD is predominantly required for RIPK1 activation during the transition from complex I to complex II in necroptosis.
We also analyzed the time course of RIPK1 activation in RDA using sequential immunoprecipitation. In MEFs stimulated by TNFα/5Z-7 for 5 min, highly ubiquitinated RIPK1 was found to be associated with complex I, which was not changed by the K584R mutation (Fig. 3F). Activated RIPK1, both in the cytosol and in association with FADD in complex IIa, was detected at 30 min after stimulation in WT MEFs, which was strongly inhibited by both heterozygous and homozygous K584R mutation. Thus, residue K584 in RIPK1-DD is important for promoting the activation of RIPK1 and is required for the formation of complex IIa in RDA as well.
RIPK1 K584R Mutation Does Not Directly Block RIPK1 Kinase Activity.
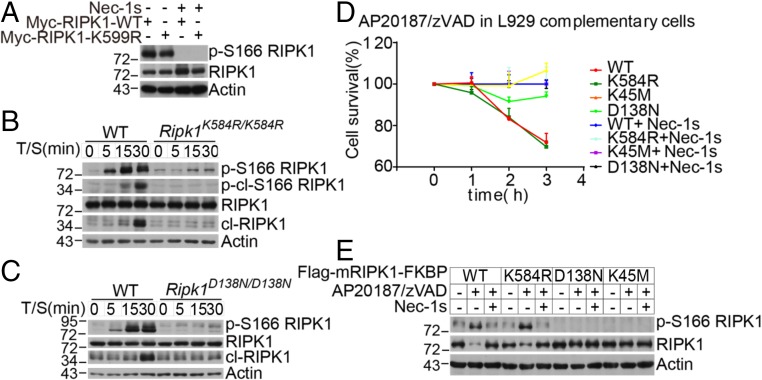
To determine if K599R mutation in RIPK1 might directly block RIPK1 kinase activity, we overexpressed hRIPK1 WT and hRIPK1 K599R in RIPK1 knockout 293T cells. We found that the levels of p-S166 RIPK1 in cells expressing WT and K599R RIPK1 were comparable and that both were effectively inhibited by Nec-1s (Fig. 4A and Fig. S4A). In addition, we purified recombinant death domain proteins of WT and K599R RIPK1 and compared their thermal stability. We found that the thermal stabilities of WT and K599R death domains were very similar (Fig. S4 B and C), suggesting that K599R mutation did not destabilize the DD structure, which is consistent with its protein surface location.
Fig. 4.
The RIPK1 K584R mutation does not directly block RIPK1 kinase activity. (A) RIPK1 KO 293T cells were transfected with Myc-tagged RIPK1 WT or K599R in the absence or presence of 10 μM Nec-1s. The cell lysates were collected 24 h after transfection and analyzed by Western blotting with the indicated antibodies. (B and C) WT and Ripk1K584R/K584R (B) and WT and Ripk1D138N/D138N (C) immortalized MEFs were pretreated with 100 nM SM-164 2 h and then treated with 100 ng/mL TNFα for 5, 15, and 30 min. The cells were lysed with 0.5% Nonidet P-40 buffer and analyzed by Western blotting analysis of p-S166 RIPK1 and RIPK1 antibodies as indicated. (D and E) RIPK1 KO L929 cells were infected with retrovirus encoding Flag-tagged mRIPK1 WT-FKBP, K584R-FKBP, K45M-FKBP, and D138N-FKBP by the Tet-On Advanced Inducible Expression System. RIPK1-reconstituted L929 cells were treated with 1 μg/mL doxycycline for 48 h to induce the expression of RIPK1. The cells were then pretreated with 10 μM Nec-1s for 30 min and then treated with 1 nM AP20187 and 50 μM zVAD.fmk. The cell survival was measured with CellTiterGlo (D). The RIPK1 kinase activity was analyzed by Western blotting with the indicated antibodies (E).
We next compared the activation of RIPK1 in WT Ripk1 and Ripk1K584R/K584R MEFs stimulated with TNFα/SM-164 or TNFα/5Z-7 for a short time (15–30 min). The activation of RIPK1, as indicated by p-S166 RIPK1, was strongly inhibited by K584R mutation (Fig. 4B and Fig. S4D). The extent of RIPK1 inhibition by K584R mutation in this assay was comparable to that of D138N RIPK1 mutation, which directly inhibits the catalytic kinase activity of RIPK1 (Fig. 4C and Fig. S4E).
Taken together, our results revealed that, while the K584R mutation did not directly inhibit RIPK1 kinase activity or affect the biophysical properties of RIPK1-DD, it strongly inhibited the activation of RIPK1 and formation of complex II in RDA and necroptosis.
Defect of K584R Mutation Can Be Overcome by Forced Dimerization.
To test if the K584R mutation blocks the activation of RIPK1 in necroptosis by preventing homodimerization, we engineered RIPK1 knockout L929 cells to express an inducible and dimerizable RIPK1 construct with fused FKBP at the C terminus of RIPK1. The addition of AP20187 and zVAD.fmk effectively induced the activation of RIPK1 and necroptosis, which was blocked by genetic inactivation of RIPK1 using the D138N or K45M mutation or by pharmacological inhibition of RIPK1 using Nec-1s. Interestingly, while D138N RIPK1 and K45M RIPK1 remained inactive upon forced dimerization, K584R mutation in this forced dimerization system was unable to prevent the cell death or RIPK1 activation as shown by p-S166 of RIPK1 (Fig. 4 D and E). This result suggests that K584/K599 in the DD regulates the kinase activity of RIPK1 by mediating RIPK1 dimerization, which can be bypassed upon forced dimerization.
Ripk1K584R Mice Are Resistant to TNFα-Induced SIRS.
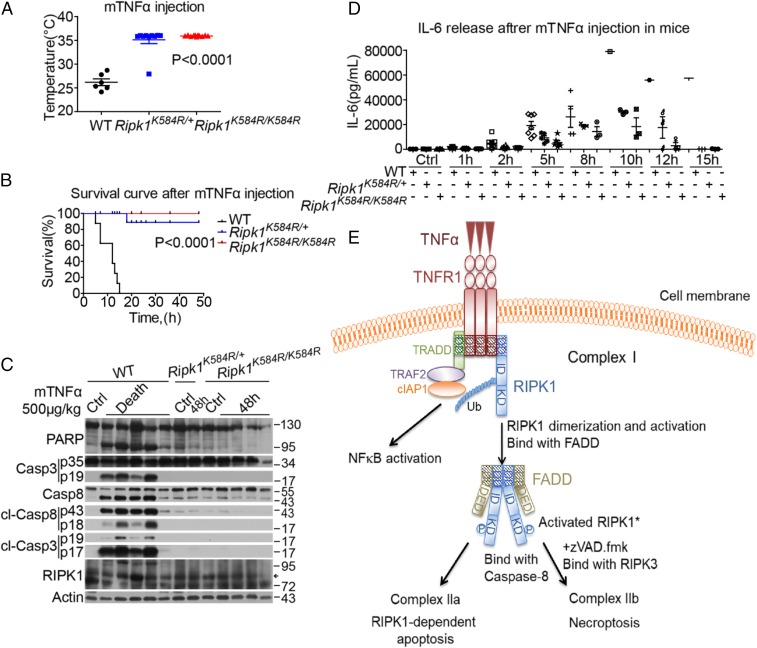
SIRS is a serious condition characterized by cytokine storm, organ failure, and lethality (26). SIRS can be modeled in mice by i.v. delivery of TNFα. Both necroptosis and RDA are known to be critically involved in mediating TNFα-induced SIRS (27, 28). WT Ripk1, Ripk1K584R/+, and Ripk1K584R/K584R mouse littermates at 6–8 wk of age were injected i.v. with mTNFα to induce SIRS. We found that, while WT mice experienced hypothermia and died by 16 h, Ripk1K584R/+ mice exhibited dramatically less hypothermia and improved survival, and Ripk1K584R/K584R mice were completely resistant to hypothermia and TNFα-induced lethality (Fig. 5 A and B).
Fig. 5.
Ripk1K584R mice are protected against TNFα-induced SIRS. (A) Body temperatures of 6 male WT, 10 male Ripk1K584R/+, and 11 male Ripk1K584R/K584R mice (6–8 wk old) were measured 10 h after i.v. injection with 500 μg/kg mTNFα (∼10 μg/mouse). (B) Survival curves of 8 male WT and 9 male Ripk1K584R/+ and 12 male Ripk1K584R/K584R mice (6–8 wk old) injected with mTNFα intravenously. (C) Five WT, one Ripk1K584R/+, and four Ripk1K584R/K584R mice (8–10 wk old) were injected with mTNFα. A WT mouse, a Ripk1K584R+ mouse, and a Ripk1K584R/K584R mouse (8–10 wk) were injected with vehicle PBS as control. Thymi were harvested from five WT mice immediately after they died from mTNFα injection, or from other mice 48 h after injection. The tissues were lysed and analyzed by Western blotting with corresponding antibodies. (D) Serum concentrations of IL6 were demonstrated using ELISA after mTNFα injection at the indicated time points. Eight male WT, six male Ripk1K584R/+, and seven male Ripk1K584R/K584R mice (6–8 wk old) were analyzed. (E) Model showing that, in cells stimulated by TNFα, RIPK1 is rapidly recruited into TNFR1 with TRADD, TRAF2, cIAP1/2, etc., to form complex I, which exists for 5–15 min. During the transition of complex I to complex II, RIPK1 undergoes dimerization, which leads to its activation. Activated RIPK1* then binds to FADD. In apoptosis-competent cells, the RIPK1*/FADD complex in turn binds to caspase-8 (complex IIa) to mediate caspase activation and apoptosis. When caspase-8 fails to be activated, the RIPK1*/FADD/caspase-8 complex binds with RIPK3 to form complex IIb, which executes necroptosis by mediating the activation and oligomerization of MLKL.
Consistent with their resistance to TNFα injection, the thymus of WT mice, but not Ripk1K584R/+ and Ripk1K584R/K584R mice, showed significant caspase activation (Fig. 5C). Inhibition of RIPK1 activation by K584R mutation also reduced inflammatory response as shown by reduction of interleukin-6 (IL6) release upon TNFα administration in Ripk1K584R/+ and Ripk1K584R/K584R mice compared with that in WT mice (Fig. 5D). Thus, the K584R RIPK1 mutation protects mice against TNFα-induced SIRS, similarly to inhibiting the kinase activity of RIPK1 by D138N knockin mutation or Nec-1s (28, 29).
Discussion
In this paper, we demonstrate that K599 (human)/K584 (mouse) in the C-terminal DD of RIPK1 distal from its N-terminal kinase domain is critically involved in mediating the activation of RIPK1 kinase in both necroptosis and RIPK1-dependent apoptosis (Fig. 5E). Our results suggest that RIPK1-DD–mediated dimerization is critical in promoting the activation of RIPK1 during the transition from complex I to complex II in TNFα-stimulated cells under both necroptosis and RDA conditions. Furthermore, we show that the K599R/K584R mutation in RIPK1-DD is sufficient to block RIPK1 activation in the cytosol and the formation of complex II, but has a minimal effect on its recruitment or ubiquitination in complex I. Since the K599R mutation blocks the DD–DD interaction of RIPK1 but not with the DDs of TNFR1, TRADD, or FADD, K599/K584 is a residue important for RIPK-1 DD homodimerization, but less so for heterodimerization. Furthermore, since K599R/K584R affects the dimerization and activation of RIPK1, but does not affect the recruitment of RIPK1 into complex I, our results suggest that the heterodimerization of RIPK1 with other DDs is distinguishable from its homodimerization.
While artificial dimerization of the truncated RIPK1 kinase domain has long been known to be able to activate its kinase activity (30), the molecular mechanism that may promote the dimerization of RIPK1 under physiological conditions has been unclear until now. We propose that K599/K584 is directly involved in mediating RIPK1-DD homodimerzation and RIPK1 kinase activation in necroptosis and RDA induced by TNFα in vitro and in vivo. K599/K584 is localized in α2–α3 helices of the RIPK1-DD, a region in the DDs of other family members, including Fas, FADD, and TNFR1, which are directly involved in homo- and hetero-association of DDs in structural studies (21, 31, 32). Our study does not rule out that K599/K584 might be subject to modification, e.g., ubiquitination, acetylation, or methylation. However, since the defect of K584R can be overcome by forced dimerization, the effect of such modification, if it exists, is likely to affect RIPK1 dimerization. Our study suggests that a subtle change in a key residue involved in protein–protein interaction can be sufficient to selectively block homodimerization but not heterodimerization.
We show that RIPK1 DD dimerization-mediated activation of RIPK1 is essential for its interaction with FADD to form complex II. Furthermore, we show that the interaction of activated RIPK1 with FADD occurs temporally first under necroptosis conditions, before the binding with RIPK3 is detected. This is consistent with apoptosis being the primary mechanism in apoptosis-competent cells; unless the interaction of activated RIPK1 with FADD fails to launch the activation of caspase-8, RIPK3 may not be recruited to promote the activation of necroptosis.
Consistent with the dimerization model in mediating the activation of RIPK1, the control of RIPK1 may be regulated in a RIPK1 concentration-dependent manner. For example, while the heterozygous K584R mutation effectively blocks RIPK1 kinase activation in MEFs, which express lower concentrations of RIPK1, it is not sufficient to block the activation of RIPK1 in TNFα/SM-164/zVAD.fmk-stimulated BMDMs, which express higher levels of RIPK1 than that of MEFs. Thus, the regulation of RIPK1 activation may exhibit cell-type specificity based on its expression levels and modification. Furthermore, we predict that, under pathological conditions when RIPK1 expression is induced, the control of its activation might be different from that of WT cells under control conditions. Understanding the dynamic control of RIPK1 kinase activation is critical to developing effective therapies for the treatment of human inflammatory and degenerative diseases.
Materials and Methods
Reagents and Antibodies.
The following commercial antibodies and reagents were used in this study: RIPK1 (Cell Signaling Technology 3493); p-S166 RIPK1 (Cell Signaling Technology 31122); TNFR1 (R&D Systems AF-425-PB); Myc (Sigma Aldrich C3956); GFP (Santa Cruz sc-9996); FADD [Abcam ab124812 and Santa Cruz (6036)]; α-Tubulin (Sigma-Aldrich T9026); β-actin (Santa Cruz 81178); Flag (Cell Signaling Technology 2368); p-S345 MLKL (Abcam ab196436); IκBα, p- IκBα, p-p38, caspase-8, caspase-3, cl-caspase-8, and cl-caspase-3 (Cell Signaling Technology); Flag, Myc antibody-conjugated agarose (Sigma); SYRO Orange Protein gel stain (Life Technologies s6651); 5Z-7 (Sigma-Aldrich O9890); and Flag-TNFα (Enzo, Life Sciences).
Cell Culture.
HEK293T, MEFs, L929, and BMDM cells were cultured in DMEM (Gibco) with 10% (vol/vol) FBS (Gibco) and 100 units/mL penicillin/streptomycin. Jurkat cells were cultured in RPMI-1640 (Gibco) with 10% (vol/vol) FBS (Gibco) and 100 units/mL penicillin/streptomycin. All cells were cultured in 37 °C with 5% CO2.
Statistics.
Data are expressed as the mean ± SEM. Error bars indicate SEM. Pairwise comparisons between two groups were performed using the Student’s t test. Differences were considered statistically significant if *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, or not significant (ns). At least three independent biological repeats were included in each data point. Each experiment was repeated at least three times.
Mice.
Ripk1K584R/K584R knock-in mice were generated by mutating the conserved lysine (AAA) at position 584 to arginine (AGG) via CRISPR/Cas9 system. The sgRNA and donor constructs were injected into embryonic stem cells derived from C57BL/6J mice. sgRNA (5′-CTCAGTGAAGCCCAGCTTGC-3′)-targeted exon 11 directed Cas9 endonuclease cleavage of the Ripk1 gene and created a double-stranded break. Such breaks are repaired by donor vector-mediated homology-directed repair to result in mutation (codon 584K to 584R). To disrupt the sgRNA recognize site, we also introduced an amino acid synonymous mutation at the A587 site (GCC to GCA) (Fig. S2A). The Ripk1K584R/K584R mice were backcrossed with C57BL/6J for eight generations.
Genotyping of K584R mice was conducted with mouse-tail DNAs by PCR (95 °C, 5 min; 95 °C, 30 s; 58 °C, 30 s; 72 °C, 45 s; 72 °C, 5 min; 10 °C, hold, 35 cycles) and confirmed by sequencing analysis. The PCR primers used for genotyping were the following: forward primer—5′-GTACTGAAGAAGGAATGGAGCTGAG-3′; reverse primer—5′-AGTTTATTGGGCACAGGGAACAGTG-3′; and sequencing primer—5′-AATCCAAGTCCCAGAGAGACAAAGG-3′. Mice were housed and cared for in a specific pathogen-free environment, and all animal procedures were performed according to the protocols approved by the Standing Animal Care Committee at Interdisciplinary Research Center of Biology and Chemistry, Shanghai Institute of Organic Chemistry.
Viability Assays.
General cell survival was measured by the ATP luminescence assay CellTiterGlo (Promega). The percentage of viability was normalized to readouts of untreated cells of each genotype.
Lentivirus Production and Infection.
pTet-On puro vector carrying hRIPK1-WT, hRIPK1-K599R, hRIPK1-IQIG-4A, mRIPK1-WT-FKBP, mRIPK1-K45M-FKBP, and mRIPK1-WT-D138N-FKBP was transfected individually into 293T cells with VSVG and GAG plasmids for 48 h. Harvested supernatant media from transfected 293T cells was filtered with a 45-μM membrane. Filtered media containing retrovirus particles was used to infect target cells.
Immunoprecipitation.
Cells were lysed with Nonidet P-40 buffer (120 mM NaCl, 10 mM Tris⋅HCl at pH 7.4, 1 mM EDTA, 0.2% Nonidet P-40, 10% glycerol) supplemented with 1 mM PMSF, 1× protease inhibitor mixture (Roche), 10 mM β-glycerophosphate, 5 mM NaF, and 1 mM Na3VO4. Debris was precipitated, and the lysate was incubated with an antibody overnight at 4 °C. The immunocomplex was captured by protein A/G agarose (Life Technologies) with the appropriate antibodies for 1–2 h at 4 °C. Beads were washed four times, and the immunocomplex was eluted from beads by loading buffer.
ELISA.
The blood obtained from mouse tail veins was centrifuged to remove cell debris. The levels of IL6 were determined using ELISA kits according to the manufacturer’s instructions (R&D Systems).
Protein Purification.
The human RIPK1 death domain (583–671 aa) was expressed as N-terminal Trx-6*His fusion proteins in Escherichia coli BL21 (DE3) following overnight induction at 4 °C and purified by Ni2+ affinity resin (GE Healthcare). Proteins were eluted from a Ni-NTA column by a buffer of 25 mM Tris (pH 7.5), 500 mM NaCl, and 300 mM imidazole. The eluted proteins were purified by a Superdex 200 column using the buffer containing 25 mM Tris (pH 7.5), 300 mM NaCl, and 1 mM DTT.
Thermal Shift Assay.
Recombinant human RIPK1 WT and mutant K599R death domains (4 μM, 6 μM) were mixed with the dye in a final reaction volume of 10 μL. The protein thermal stability was analyzed by differential scanning calorimetry on a 7500 Fast Real-Time PCR System according to the manufacturer’s recommendations using a melt protocol of 25 °C to 95 °C at a 1% ramp rate (equivalent to 0.015 °C/s). Data were analyzed using the Protein Thermal Shift Software.
Supplementary Material
Acknowledgments
We thank Dr. Palak Amin for critical reading and helpful comments on the manuscript and Professor Jiahuai Han (Xiamen University) for providing Ripk1 knockout L929 cells and RIPK1 knockout 293T cells. This work was supported in part by China National Natural Science Foundation Grant 31530041; National Key R&D Program of China Grant 2016YFA0501900; the Chinese Academy of Sciences; National Institute of Neurological Disorders and Stroke Grant 1R01NS082257 and National Institute on Aging Grants 1R01AG047231 and RF1AG055521 (to J.Y.); and Natural Science Foundation of Shanghai Grant 16ZR1443900 (to B.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722013115/-/DCSupplemental.
References
- 1.Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol. 2014;76:129–150. doi: 10.1146/annurev-physiol-021113-170259. [DOI] [PubMed] [Google Scholar]
- 2.Ito Y, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 2016;353:603–608. doi: 10.1126/science.aaf6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol. 2013;14:727–736. doi: 10.1038/nrm3683. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Goeddel DV. TNF-R1 signaling: A beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 5.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 6.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 10.Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: A novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 11.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dondelinger Y, et al. NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol Cell. 2015;60:63–76. doi: 10.1016/j.molcel.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Geng J, et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun. 2017;8:359. doi: 10.1038/s41467-017-00406-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 15.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 17.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 18.Park HH, et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park YH, Jeong MS, Park HH, Jang SB. Formation of the death domain complex between FADD and RIP1 proteins in vitro. Biochim Biophys Acta. 2013;1834:292–300. doi: 10.1016/j.bbapap.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Sukits SF, et al. Solution structure of the tumor necrosis factor receptor-1 death domain. J Mol Biol. 2001;310:895–906. doi: 10.1006/jmbi.2001.4790. [DOI] [PubMed] [Google Scholar]
- 21.Telliez JB, et al. Mutational analysis and NMR studies of the death domain of the tumor necrosis factor receptor-1. J Mol Biol. 2000;300:1323–1333. doi: 10.1006/jmbi.2000.3899. [DOI] [PubMed] [Google Scholar]
- 22.Wu XN, et al. Distinct roles of RIP1-RIP3 hetero- and RIP3-RIP3 homo-interaction in mediating necroptosis. Cell Death Differ. 2014;21:1709–1720. doi: 10.1038/cdd.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ofengeim D, et al. Activation of necroptosis in multiple sclerosis. Cell Rep. 2015;10:1836–1849. doi: 10.1016/j.celrep.2015.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christofferson DE, et al. A novel role for RIP1 kinase in mediating TNFα production. Cell Death Dis. 2012;3:e320. doi: 10.1038/cddis.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei R, et al. SPATA2 regulates the activation of RIPK1 by modulating linear ubiquitination. Genes Dev. 2017;31:1162–1176. doi: 10.1101/gad.299776.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balk RA. Systemic inflammatory response syndrome (SIRS): Where did it come from and is it still relevant today? Virulence. 2014;5:20–26. doi: 10.4161/viru.27135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton K, et al. RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ. 2016;23:1565–1576. doi: 10.1038/cdd.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duprez L, et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35:908–918. doi: 10.1016/j.immuni.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 29.Polykratis A, et al. Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J Immunol. 2014;193:1539–1543. doi: 10.4049/jimmunol.1400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 31.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 32.Jeong EJ, et al. The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. J Biol Chem. 1999;274:16337–16342. doi: 10.1074/jbc.274.23.16337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.