Abstract
Ultimately, viral evolution is a consequence of mutations that arise within and spread between infected hosts. The transmission bottleneck determines how much of the viral diversity generated in one host passes to another during transmission. It therefore plays a vital role in linking within-host processes to larger evolutionary trends. Although many studies suggest that transmission severely restricts the amount of genetic diversity that passes between individuals, there are important exceptions to this rule. In many cases, the factors that determine the size of the transmission bottleneck are only beginning to be understood. Here, we review how transmission bottlenecks are measured, how they arise, and their consequences for viral evolution.
Keywords: virus, bottleneck, transmission, diversity, evolution
Main Text
Many viral pathogens exist as diverse populations within infected hosts. The diversity present in this “mutant swarm” provides the raw material on which selection can act. Although populations within a host may reach as high as 1014 virions [1], viruses are frequently subject to bottleneck events as they spread within and between hosts [2]. These bottlenecks drastically reduce the size of the population and, consequently, its genetic diversity. Because the population that develops after a genetic bottleneck is derived from a small sample of the ancestral population, this process can dramatically alter the relative frequency of mutations in the population.
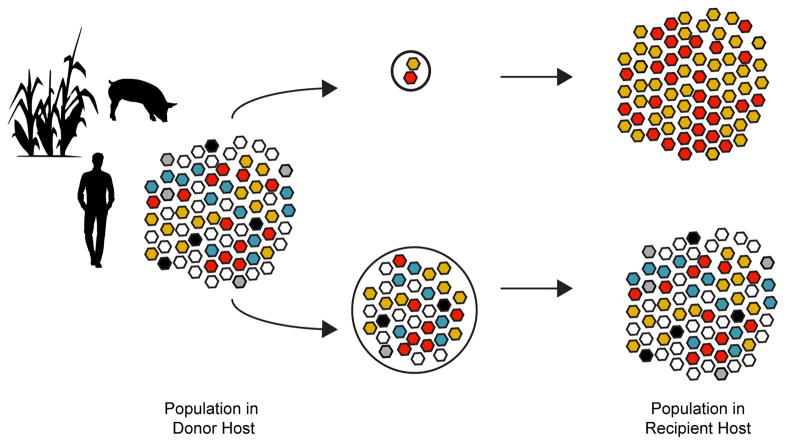
The stringency of the transmission bottleneck plays an important role in linking within-host processes to a pathogen’s larger evolutionary dynamics. Stringent, or tight, transmission bottlenecks limit the diversity of the founding population in the recipient and alter the mutational composition of the population in the recipient relative to that in the donor (Figure 1, top). However, if the transmission bottleneck is loose, transmission does not significantly impact variant frequencies and the composition of the founding population in the recipient more closely matches that present in the donor at the time of transmission (Figure 1, bottom).
Figure 1.
The effect of transmission bottlenecks on viral diversity. In a variety of hosts (e.g. humans, pigs, plant shown here), stringent bottlenecks (top) limit the size and diversity of a population and drastically alter their composition. The large populations that pass through loose bottlenecks (bottom) allow for transmission of rare variants. As a result the diversity of the population in the recipient approximates that of the donor.
Although transmission bottlenecks play an important role in viral evolution, relatively little is known about their size and determinants. Many quantitative studies suggest that bottlenecks are tight [3,4]; however, there are exceptions and even conflicting reports for viruses with similar transmission pathways. Importantly, the factors that determine the stringency of the transmission bottleneck are poorly understood. Here, we briefly review how transmission bottlenecks are measured, how they arise, and their impact on viral evolution across biological scales. For a more comprehensive review of bottlenecks, including those found at the within-host and cellular scale, we direct the reader to reference [3].
Measuring transmission bottlenecks
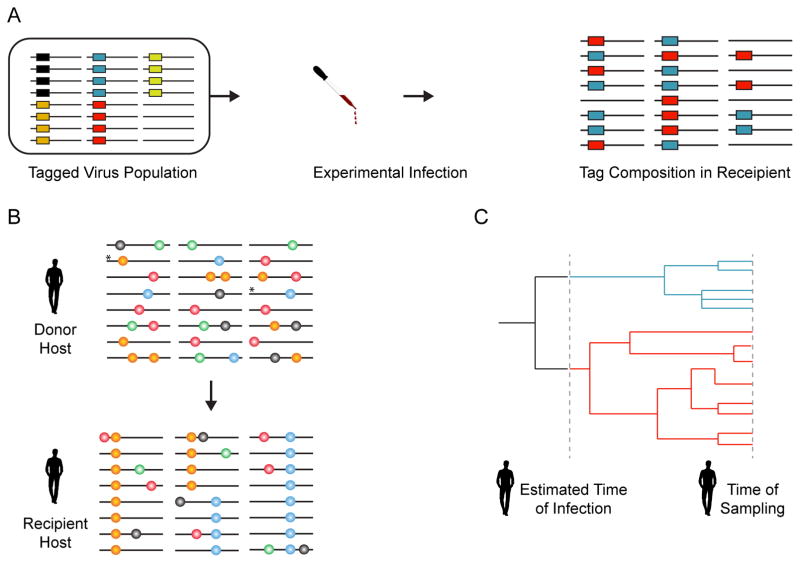
Transmission bottlenecks are measured by their effect on viral diversity. In experimental systems, within-host diversity can be approximated using a defined population of viruses that are tagged with genetic markers. If the markers are selectively neutral, the number of distinct markers that pass from donor to recipient reflects the sampling event of the bottleneck as opposed to selection within either host (Figure 2A). This technique has been used to qualitatively estimate a stringent bottleneck for aphid transmission of cucumber mosaic virus (an average of 3 of 12 markers were transmitted) [5] and aerosol transmission of influenza in ferrets and guinea pigs (2–5 of 100 sequence tags were transmitted) [6**]. In a particularly elegant experiment, Moury and colleagues artificially inoculated aphid vectors with mixtures of 2 Potato Y virus mutants prior to feeding the aphids on pepper plants [7*]. By modeling the number of plants exposed to only one of the mutants, Moury et al. found that aphid transmission imposes a bottleneck of 0.5–3.2 virions on Potato Y virus.
Figure 2.
Measuring transmission bottlenecks. (A) The number of donor-derived, neutral markers detected in the recipient is an indication of the stringency of the transmission bottleneck. Here, 3 of the 6 markers were transmitted suggesting a stringent bottleneck. (B) Shared diversity data from natural systems can be used to estimate a bottleneck. In the example, only two donor genotypes, denoted with *, were transmitted to the recipient suggesting a stringent bottleneck. Other de novo mutations arise on these backgrounds after transmission. (C) Coalescent models allow one to work backward from the time of sampling and estimate the number of genotypes that could plausibly give rise to the observed diversity. In this case, the two lineages are traced back to two genetically distinct variants present at transmission.
Because natural systems do not offer the opportunity for a barcoding approach, early studies characterized the transmission bottleneck qualitatively based on the degree of shared diversity found within transmission pairs (Figure 2B). Clonal sequencing of influenza virus isolates from swine and equine transmission chains found transmission pairs shared minority variants [8–10]. Studies of aphid, mechanical, and vertical transmission of Zucchini Yellow Mosaic Virus found similar results [11,12]. These studies suggest that transmission bottlenecks are sometimes sufficiently loose to allow for the transmission of low-frequency mutations.
More quantitative approaches can also be employed to estimate the transmission bottleneck from shared diversity data. In these models, the transmission process is assumed to be a random sampling of the donor population and individual variants are assumed to be transmitted independently of one another. The probability that a variant is transmitted is derived from a binomial distribution and is positively correlated with its frequency in the donor and the size of the bottleneck. More complexity can be incorporated into these models to tease apart the relative impact of within- and between-host processes (see ref [13**] for a thorough discussion and comparison of common models). One such model has been used to estimate a loose bottleneck of roughly 200 genomes in a recent study of human transmission of influenza virus [13**,14]. This estimate is much larger than that provided by the barcode experiments previously discussed. The large discrepancy in these studies highlights the need for a more complete understanding of the viral, host, and environmental factors that determine transmission bottleneck sizes.
When only one member of a transmission pair is available, the diversity present in the infected host can be used to estimate the number of genotypes in the founding population. Coalescent theory works backward in time, tracing the evolutionary history of the current population back to common ancestors [15]. Coalescent models based on the current diversity, the viral evolutionary rate, and the estimated time of infection can be used to determine how many genotypes were present in the founding population (Figure 2C). Phylogenetic analysis of HIV evolution suggests that most infections derive from small founding populations of only one genotype [16,17]. A similar approach has been used to estimate a stringent transmission bottleneck for HCV [18*–21].
Determinants of bottleneck size
Most transmission studies suggest tight bottlenecks and small founding populations (see tables in [3] and [4]). However, as mentioned above, these estimates can vary significantly depending on the virus, host, route of transmission, and experimental design. Understanding the factors that determine the size of the transmission bottleneck is vital to interpreting the effect transmission has on viral evolution. Work in Tobacco etch virus (TEV) suggests that the size of the bottleneck is dose dependent, with higher exposure doses corresponding to larger founding populations [22]. Evidence from mixed infections of influenza virus in a guinea pig model is consistent with a dose dependence model [23]. Further support comes from experimental infections with tagged influenza clones in ferret and guinea pig models, which indicate that the more limiting exposure dose of aerosol transmission imposes a significantly more stringent bottleneck than contact transmission [6**]. Additionally, coinfection by other pathogens, which can limit innate defenses and modulate the immune response, has been correlated with loose bottlenecks in HIV and HCV [24,25,26]. Taken together these data suggest that the transmission bottleneck is not constant, but rather a complex function of both viral and host factors.
Complicating matters is the observation that segregation of the viral population within the donor can also restrict the amount of diversity transmitted to the recipient. Work in animal models of influenza virus suggest that populations in the upper respiratory tract seed transmission and can be distinct from populations at other sites of infection [6**,27]. The stringent bottleneck associated with aphid transmission of cucumber mosaic virus [28] is likely the result of extreme viral segregation within the donor. Most plant cells are infected by only one genotype, and aphids are unlikely to feed on many donor cells prior to transmission [29]. Other vector-transmitted viruses undergo an additional bottlenecking event within the vector. Smith et al. used fluorescent Venezuelan equine encephalitis virus (VEEV) replicons to show that an average of 28 midgut cells in the mosquito are initially infected by the virus [30]. This small population size is consistent with observations of Dengue virus in infected mosquitos [31] and likely contributes to the stringent bottleneck observed during mosquito-mediated transmission of VEEV in a mouse model [32*].
To this point we have focused on stochastic bottlenecks that randomly sample the donor host population during transmission. In some cases, however, selection within the donor and/or recipient hosts can impose selective sweeps that decrease the diversity of founding populations. While HIV-infected individuals can harbor highly diverse viral populations, phylogenetic approaches indicate that often only one donor genotype contributes to founding the recipient population [17,33]. This trend is often observed even when physical barriers to infection are bypassed, as is the case among intravenous drug users [34,35]. Comparisons between the populations present in the donor genital tract and recipient blood stream suggest that minor variants in the genital tract are preferentially transmitted [36**]. This biased transmission implies that the bottleneck event is not a random sample of the donor population. Larger cohort studies have shown that transmitted viruses are most similar to those present early in the donor infection [37] and are better suited to spread between hosts [38*]. Transmitted viruses are characterized by CCR5 receptor preference [39–42], lower levels of glycosylation on the surface envelope protein [43,44], and lower susceptibility to type 1 IFN [45,46*] than the majority of variants present in chronically infected hosts. Taken together, these data suggest that selective pressures in naïve hosts impact the stringency of the HIV transmission bottleneck. For more detailed reviews of HIV transmission, we direct the reader to references [33,47,48].
Evolutionary consequences of transmission bottlenecks
Transmission bottlenecks determine the extent to which within-host diversity contributes to evolutionary trends at higher scales. While the relatively high mutation rates and large population sizes of many viruses may allow these pathogens to rapidly adapt to their host, the rate of adaptation is not unlimited. In particular, the rate depends on the effective population size [49*]. The effective population size can be roughly thought of as the number of viruses that replicate and contribute genomes to the next generation. It is usually smaller than the census population (see [50] for a thorough review and discussion of effective population size). In large effective populations, selection is efficient, deleterious mutations are purged, and beneficial mutations steadily increase in frequency over time [51]. However, alleles in small populations are subject to sampling error known as random genetic drift. Drift introduces noise so that selection does not efficiently fix beneficial mutations or purge deleterious ones [52]. Stringent transmission bottlenecks reduce the effective population size of viral pathogens between hosts, increase genetic drift, and decrease the efficiency of selection.
Stringent transmission bottlenecks may therefore pose a significant barrier to adaptive evolution. Because most mutations are deleterious, repeated bottleneck events fix deleterious mutations and decrease viral fitness over time. This process, known as Muller’s ratchet, opposes purifying selection and contributes to the deleterious load often observed at the tips of viral phylogenetic trees [53,54]. Although the fixation of deleterious mutations decreases fitness along a single transmission chain, it is unlikely to drastically decrease a virus’ overall fitness at a global scale. Competition at the interhost level, can serve to maintain viral fitness [55,56*]. Notably, populations with low fitness are not as susceptible to Muller’s ratchet as well-adapted populations with high fitness [57,58]. We speculate that the fixation of deleterious mutations during transmission is less likely to affect the evolution of recently emerged viruses, which might have lower fitness in their new host species. However, emerging viruses face an alternative problem as stochastic bottlenecks may purge beneficial mutations before they reach fixation.
While transmission bottlenecks are expected to slow adaptive evolution, they may provide potential advantages to evolving pathogens. Stringent bottlenecks purge the population of defective interfering particles, which limit viral replication [59]. Bottlenecks also increase genetic drift and provide a mechanism for virus populations to traverse potential fitness valleys and escape local fitness maxima [60].
Concluding thoughts
The available data suggest that transmission frequently imposes a stringent bottleneck that dramatically reduces the level of diversity in the founding population. In many cases, however, transmission bottlenecks appear to be sufficiently wide to transmit minority variants. Defining the host and viral factors that determine the transmission bottleneck is an important step in developing strategies to limit viral transmission. A more complete understanding of viral transmission bottlenecks is also necessary to link within-host population dynamics to larger evolutionary trends.
Highlights.
Bottlenecks restrict the transmission of genetic diversity between hosts
A variety of methods can be used to estimate bottleneck size in experimental and natural infections
Some bottlenecks are selective, but most appear to be stochastic in nature
Bottlenecks link within host processes to larger evolutionary dynamics
Acknowledgments
We thank members of our laboratory for helpful discussion and a critical reading of the manuscript. This work was supported by grants from the Burroughs Wellcome Fund (PATH 1016763) and NIH (R01 AI118886) to ASL. JTM was supported by the Michigan Predoctoral Training Program in Genetics (T32 GM007544).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Russell CA, Fonville JM, Brown AEX, Burke DF, Smith DL, James SL, Herfst S, van Boheemen S, Linster M, Schrauwen EJ, et al. The Potential for Respiratory Droplet-Transmissible A/H5N1 Influenza Virus to Evolve in a Mammalian Host. 2012;336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeiffer JK, Kirkegaard K. Bottleneck-mediated quasispecies restriction during spread of an RNA virus from inoculation site to brain. Proceedings of the National Academy of Sciences. 2006;103:5520–5525. doi: 10.1073/pnas.0600834103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zwart MP, Elena SF. Matters of Size: Genetic Bottlenecks in Virus Infection and Their Potential Impact on Evolution. Annual Review of Virology. 2015;2:161–179. doi: 10.1146/annurev-virology-100114-055135. [DOI] [PubMed] [Google Scholar]
- 4.Gutiérrez S, Michalakis Y, Blanc S. Virus population bottlenecks during within-host progression and host-to-host transmission. Current Opinion in Virology. 2012;2:546–555. doi: 10.1016/j.coviro.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Ali A, Li H, Schneider WL, Sherman DJ, Gray S, Smith D, Roossinck MJ. Analysis of genetic bottlenecks during horizontal transmission of Cucumber mosaic virus. J Virol. 2006;80:8345–8350. doi: 10.1128/JVI.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Varble A, Albrecht RA, Backes S, Crumiller M, Bouvier NM, Sachs D, García-Sastre A, tenOever BR. Influenza A virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe. 2014;16:691–700. doi: 10.1016/j.chom.2014.09.020. Study in which 100 neutral barcodes were used to measure and compare the influenza transmission bottleneck imposed by airborne and contact mediated transmission in ferret and guinea pig models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Moury B, Fabre F, Senoussi R. Estimation of the number of virus particles transmitted by an insect vector. Proceedings of the National Academy of Sciences. 2007;104:17891–17896. doi: 10.1073/pnas.0702739104. An elegant study that used phenotypic differences in virulence (as opposed to sequence-based approaches) to model and quantitatively measure the number of infectious virions transmitted by aphids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murcia PR, Hughes J, Battista P, Lloyd L, Baillie GJ, Ramirez-Gonzalez RH, Ormond D, Oliver K, Elton D, Mumford JA, et al. Evolution of an Eurasian Avian-like Influenza Virus in Naïve and Vaccinated Pigs. PLoS Pathog. 2012;8:e1002730. doi: 10.1371/journal.ppat.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murcia PR, Baillie GJ, Stack JC, Jervis C, Elton D, Mumford JA, Kellam P, Grenfell BT, Holmes EC, Wood JLN. Evolution of equine influenza virus in vaccinated horses. J Virol. 2013;87:4768–4771. doi: 10.1128/JVI.03379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes J, Allen RC, Baguelin M, Hampson K, Baillie GJ, Elton D, Newton JR, Kellam P, Wood JLN, Holmes EC, et al. Transmission of Equine Influenza Virus during an Outbreak Is Characterized by Frequent Mixed Infections and Loose Transmission Bottlenecks. PLoS Pathog. 2012;8:e1003081. doi: 10.1371/journal.ppat.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons HE, Dunham JP, Stack JC, Dickins BJA, Pagán I, Holmes EC, Stephenson AG. Deep sequencing reveals persistence of intra- and inter-host genetic diversity in natural and greenhouse populations of zucchini yellow mosaic virus. J Gen Virol. 2012;93:1831–1840. doi: 10.1099/vir.0.042622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons HE, Dunham JP, Zinn KE, Munkvold GP, Holmes EC, Stephenson AG. Zucchini yellow mosaic virus (ZYMV, Potyvirus): vertical transmission, seed infection and cryptic infections. Virus Research. 2013;176:259–264. doi: 10.1016/j.virusres.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Sobel Leonard A, Weissman DB, Greenbaum B, Ghedin E, Koelle K. Transmission Bottleneck Size Estimation from Pathogen Deep-Sequencing Data, with an Application to Human Influenza A Virus. J Virol. 2017;91:e00171–17. doi: 10.1128/JVI.00171-17. An in-depth analysis of transmission bottlenecks in naturally occurring human infections, as well as a detailed discussion of the models that can be applied to deep sequencing data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon LLM, Song T, Rosenfeld R, Lin X, Rogers MB, Zhou B, Sebra R, Halpin RA, Guan Y, Twaddle A, et al. Quantifying influenza virus diversity and transmission in humans. Nat Genet. 2016;48:195–200. doi: 10.1038/ng.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingman JFC. On the Genealogy of Large Populations. Journal of Applied Probability. 1982;19:27. [Google Scholar]
- 16.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci US A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards CTT, Holmes EC, Wilson DJ, Viscidi RP, Abrams EJ, Phillips RE, Drummond AJ. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evol Biol. 2006;6:28. doi: 10.1186/1471-2148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, Cameron B, Maher L, Dore GJ, White PA, et al. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 2011;7:e1002243. doi: 10.1371/journal.ppat.1002243. Here the authors present a longitudinal study of early-phase HCV infection. They use coalescent approaches to elucidate population dynamics beginning with the transmission bottleneck and find evidence for a secondary, selective bottleneck that takes place later during infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho CKY, Raghwani J, Koekkoek S, Liang RH, Van der Meer JTM, Van Der Valk M, De Jong M, Pybus OG, Schinkel J, Molenkamp R. Characterization of Hepatitis C Virus (HCV) Envelope Diversification from Acute to Chronic Infection within a Sexually Transmitted HCV Cluster by Using Single-Molecule, Real-Time Sequencing. J Virol. 2017;91:e02262–16. doi: 10.1128/JVI.02262-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Arienzo V, Moreau A, D’Alteroche L, Gissot V, Blanchard E, Gaudy-Graffin C, Roch E, Dubois F, Giraudeau B, Plantier J-C, et al. Sequence and functional analysis of the envelope glycoproteins of hepatitis C virus variants selectively transmitted to a new host. J Virol. 2013;87:13609–13618. doi: 10.1128/JVI.02119-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, Learn GH, Hraber P, Goepfert PA, Saag MS, et al. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS Pathog. 2012;8:e1002880. doi: 10.1371/journal.ppat.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwart MP, Daròs J-A, Elena SF. One Is Enough: In Vivo Effective Population Size Is Dose-Dependent for a Plant RNA Virus. 2011;7:e1002122–12. doi: 10.1371/journal.ppat.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao H, Steel J, Lowen AC. Intrahost Dynamics of Influenza Virus Reassortment. J Virol. 2014;88:7485–7492. doi: 10.1128/JVI.00715-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sagar M, Lavreys L, Baeten JM, Richardson BA, Mandaliya K, Ndinya-Achola JO, Kreiss JK, Overbaugh J. Identification of modifiable factors that affect the genetic diversity of the transmitted HIV-1 population. AIDS. 2004;18:615. doi: 10.1097/00002030-200403050-00005. [DOI] [PubMed] [Google Scholar]
- 25.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, et al. Inflammatory Genital Infections Mitigate a Severe Genetic Bottleneck in Heterosexual Transmission of Subtype A and C HIV-1. PLoS Pathog. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fauteux-Daniel S, Larouche A, Calderon V, Boulais J, Béland C, Ransy DG, Boucher M, Lamarre V, Lapointe N, Boucoiran I, et al. Vertical transmission of hepatitis C virus: variable transmission bottleneck and evidence of mid-gestation in utero infection. J Virol. 2017 doi: 10.1128/JVI.01372-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, et al. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature. 2015;526:122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betancourt M, Fereres A, Fraile A, García-Arenal F. Estimation of the effective number of founders that initiate an infection after aphid transmission of a multipartite plant virus. J Virol. 2008;82:12416–12421. doi: 10.1128/JVI.01542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeshita M, Shigemune N, Kikuhara K, Furuya N, Takanami Y. Spatial analysis for exclusive interactions between subgroups I and II of Cucumber mosaic virus in cowpea. Virology. 2004;328:45–51. doi: 10.1016/j.virol.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 30.Smith DR, Adams AP, Kenney JL, Wang E, Weaver SC. Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology. 2008;372:176–186. doi: 10.1016/j.virol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lequime S, Fontaine A, Ar Gouilh M, Moltini-Conclois I, Lambrechts L. Genetic Drift, Purifying Selection and Vector Genotype Shape Dengue Virus Intra-host Genetic Diversity in Mosquitoes. PLoS Genet. 2016;12:e1006111. doi: 10.1371/journal.pgen.1006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Forrester NL, Guerbois M, Seymour RL, Spratt H, Weaver SC. Vector-borne transmission imposes a severe bottleneck on an RNA virus population. PLoS Pathog. 2012;8:e1002897. doi: 10.1371/journal.ppat.1002897. Neutral sequence tags were used to show that the observed transmission bottleneck in a mouse model of VEEV is the result of intrahost bottlenecks in the mosquito vector. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw GM, Hunter E. HIV transmission. Cold Spring Harb Perspect Med. 2012;2:a006965–a006965. doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masharsky AE, Dukhovlinova EN, Verevochkin SV, Toussova OV, Skochilov RV, Anderson JA, Hoffman I, Cohen MS, Swanstrom R, Kozlov AP. A substantial transmission bottleneck among newly and recently HIV-1-infected injection drug users in St Petersburg, Russia. J Infect Dis. 2010;201:1697–1702. doi: 10.1086/652702. [DOI] [PubMed] [Google Scholar]
- 35.Bar KJ, Li H, Chamberland A, Tremblay C, Routy JP, Grayson T, Sun C, Wang S, Learn GH, Morgan CJ, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84:6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Boeras DI, Hraber PT, Hurlston M, Evans-Strickfaden T, Bhattacharya T, Giorgi EE, Mulenga J, Karita E, Korber BT, Allen S, et al. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proc Natl Acad Sci US A. 2011;108:E1156–63. doi: 10.1073/pnas.1103764108. Study that provides strong evidence for a selective HIV bottleneck by noting transmitted genotypes were not the predominant genotype present in the donor genital tract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redd AD, Collinson-Streng AN, Chatziandreou N, Mullis CE, Laeyendecker O, Martens C, Ricklefs S, Kiwanuka N, Nyein PH, Lutalo T, et al. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J Infect Dis. 2012;206:1433–1442. doi: 10.1093/infdis/jis503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, et al. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345:1254031. doi: 10.1126/science.1254031. Sequence data from a large HIV transmission cohort were used to show preferential transmission of amino acids predicted to increase fitness in naïve hosts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. The Journal of Experimental Medicine. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scarlatti G, Tresoldi E, Björndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 41.Long EM, Rainwater SMJ, Lavreys L, Mandaliya K, Overbaugh J. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res Hum Retroviruses. 2002;18:567–576. doi: 10.1089/088922202753747914. [DOI] [PubMed] [Google Scholar]
- 42.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. The Journal of Experimental Medicine. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, Lapedes AS, Shen T, Gaschen B, Krishnamoorthy M, Li H, et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 2011;7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 45.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology. 2013;10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci US A. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. Here the authors use a substantial number of infectious molecular clones to experientially elucidate several phenotypic differences between transmitted/founder viruses and those present during chronic infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronen K, Sharma A, Overbaugh J. HIV transmission biology: translation for HIV prevention. AIDS. 2015;29:2219–2227. doi: 10.1097/QAD.0000000000000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kariuki SM, Selhorst P, Ariën KK, Dorfman JR. The HIV-1 transmission bottleneck. Retrovirology. 2017;14:22. doi: 10.1186/s12977-017-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Rouzine IM, Rodrigo A, Coffin JM. Transition between stochastic evolution and deterministic evolution in the presence of selection: general theory and application to virology. Microbiology and Molecular Biology Reviews. 2001;65:151–185. doi: 10.1128/MMBR.65.1.151-185.2001. A detailed but accessible review of the population genetics at play in viral systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 51.Willi Y, Van Buskirk J, Hoffmann AA. Limits to the Adaptive Potential of Small Populations. Annu Rev Ecol Evol Syst. 2006;37:433–458. [Google Scholar]
- 52.Robertson A. A Theory of Limits in Artificial Selection. Proceedings of the Royal Society B: Biological Sciences. 1960;153:234–249. [Google Scholar]
- 53.Pybus OG, Rambaut A, Belshaw R, Freckleton RP, Drummond AJ, Holmes EC. Phylogenetic Evidence for Deleterious Mutation Load in RNA Viruses and Its Contribution to Viral Evolution. Molecular Biology and Evolution. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 54.Koelle K, Rasmussen DA. The effects of a deleterious mutation load on patterns of influenza A/H3N2’s antigenic evolution in humans. eLife. 2015;4:e07361. doi: 10.7554/eLife.07361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergstrom CT, McElhany P, Real LA. Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proceedings of the National Academy of Sciences. 1999;96:5095–5100. doi: 10.1073/pnas.96.9.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Elena SF, Sanjuan R, Bordería AV, Turner PE. Transmission bottlenecks and the evolution of fitness in rapidly evolving RNA viruses. INFECTION, GENETICS AND EVOLUTION. 2001;1:41–48. doi: 10.1016/s1567-1348(01)00006-5. An experimental comparison of the fitness effects of stringent bottlenecks on vertical and horizontal transmission pathways. [DOI] [PubMed] [Google Scholar]
- 57.Novella IS, Dutta RN, Wilke CO. A linear relationship between fitness and the logarithm of the critical bottleneck size in vesicular stomatitis virus populations. J Virol. 2008;82:12589–12590. doi: 10.1128/JVI.01394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novella IS, Elena SF, Moya A, Domingo E, Holland JJ. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visser PB, Brown DJ, Brederode FT, Bol JF. Nematode transmission of tobacco rattle virus serves as a bottleneck to clear the virus population from defective interfering RNAs. Virology. 1999;263:155–165. doi: 10.1006/viro.1999.9901. [DOI] [PubMed] [Google Scholar]
- 60.Rozen DE, Habets MGJL, Handel A, de Visser JAGM. Heterogeneous Adaptive Trajectories of Small Populations on Complex Fitness Landscapes. 2008;3:e1715–6. doi: 10.1371/journal.pone.0001715. [DOI] [PMC free article] [PubMed] [Google Scholar]