ABSTRACT
Secondary metabolites are an important source of pharmaceuticals and key modulators of microbe-microbe interactions. The bacterium Serratia marcescens is part of the Enterobacteriaceae family of eubacteria and produces a number of biologically active secondary metabolites. In this study, we screened for novel regulators of secondary metabolites synthesized by a clinical isolate of S. marcescens and found mutations in a gene for an uncharacterized UmoB/IgaA family member here named gumB. Mutation of gumB conferred a severe loss of the secondary metabolites prodigiosin and serratamolide. The gumB mutation conferred pleiotropic phenotypes, including altered biofilm formation, highly increased capsular polysaccharide production, and loss of swimming and swarming motility. These phenotypes corresponded to transcriptional changes in fimA, wecA, and flhD. Unlike other UmoB/IgaA family members, gumB was found to be not essential for growth in S. marcescens, yet igaA from Salmonella enterica, yrfF from Escherichia coli, and an uncharacterized predicted ortholog from Klebsiella pneumoniae complemented the gumB mutant secondary metabolite defects, suggesting highly conserved function. These data support the idea that UmoB/IgaA family proteins are functionally conserved and extend the known regulatory influence of UmoB/IgaA family proteins to the control of competition-associated secondary metabolites and biofilm formation.
IMPORTANCE IgaA/UmoB family proteins are found in members of the Enterobacteriaceae family of bacteria, which are of environmental and public health importance. IgaA/UmoB family proteins are thought to be inner membrane proteins that report extracellular stresses to intracellular signaling pathways that respond to environmental challenge. This study introduces a new member of the IgaA/UmoB family and demonstrates a high degree of functional similarity between IgaA/UmoB family proteins. Moreover, this study extends the phenomena controlled by IgaA/UmoB family proteins to include the biosynthesis of antimicrobial secondary metabolites.
KEYWORDS: secondary metabolite, competition, flagella, biofilm, motility, surfactant, capsular polysaccharide
INTRODUCTION
Microorganism-derived secondary metabolites include important interspecies competitive factors and are a major potential source of therapeutic agents (1–6). The bacterium Serratia marcescens is found in a wide range of ecological niches and is known for the production of antibiotic secondary metabolites, including the peptide antibiotic althiomycin, the red pigment prodigiosin, and the biosurfactant serratamolide, also known as serrawettin W1 (7–11).
Both prodigiosin and serratamolide are reported to have broad-range antimicrobial activity, and serratamolide promotes swarming motility, a behavior that may allow the bacterium to become less susceptible to antibiotics and move into more favorable microenvironments (8, 12–17). Serratamolide from S. marcescens is hemolytic and was demonstrated to enable bacteria of different genera to evade phagocytosis by neutrophils in vitro, together suggesting that it may be a virulence factor and impact polymicrobial infections (18, 19). Regulation of secondary metabolites by S. marcescens occurs at the transcriptional level by several transcription factors, including CopA, cAMP receptor protein (CRP), EepR, HexS, PigP, RssAB, SmaI, and SpnR (19–27).
The goal of this study was to characterize the pleiotropic phenotypes of a newly identified secondary metabolite regulator. The mutations in mutants with reduced prodigiosin and serratamolide in S. marcescens mapped to an uncharacterized gene predicted to code for a protein of the IgaA/UmoB family. IgaA/UmoB family proteins were first described from the highly motile bacterium Proteus mirabilis by the Hughes group in 1998, where mutation of the umoB gene conferred a downregulation of the flhDC flagellar master regulator operon (28). Since then, most studies on this family of proteins have been performed using Salmonella enterica serovar Typhimurium. The igaA gene was found to be a regulator of intracellular growth in S. enterica and was later shown to regulate capsular polysaccharide production and pathogenesis (29–33). In Escherichia coli, mutations in the IgaA/UmoB-encoding gene, yrfF, were found to be enriched following serial coculture with macrophages and were found in a keratitis isolate (34, 35). The igaA and yrfF genes are essential for growth in Salmonella and E. coli bacteria (30), but umoB is not required for Proteus spp. to survive (28). The essential nature of igaA and yrfF allowed identification of suppressor mutations of the lethality phenotype that mapped to the Rcs regulatory system (30, 31, 36). Subsequent studies using bacteria from several genera indicate that IgaA/UmoB controls the Rcs phosphorelay signaling system that controls hundreds of genes (32, 37–39). Data suggest that IgaA/UmoB, together with RcsF, regulates the Rcs system in response to envelope stress and surface interactions by controlling the phosphorylation state of histidine kinase protein RcsC through an unknown mechanism (39–41). S. marcescens has an Rcs phosphorelay system that has been implicated in the regulation of outer membrane vesicle production and could be activated by a mutation of genes involved in biosynthesis of the enterobacterial common antigen outer membrane component (42, 43).
Because igaA and yrfF are essential genes in E. coli and S. enterica, there is incomplete knowledge about the range of biological processes that are mediated through this conserved family of proteins. This study characterized pleiotropic phenotypes of a mutant involving the S. marcescens IgaA/UmoB family gene, gumB, and found novel roles for IgaA/UmoB family proteins in their influence over secondary metabolite prodigiosin and biofilm production.
RESULTS
Isolation of an IgaA/UmoB family gene in S. marcescens.
Transposon (Tn) mutagenesis was used to identify genes that regulate secondary metabolite production in a clinical keratitis isolate of S. marcescens, strain K904 (25). Two mutants with severely reduced red pigmentation and hemolysis were isolated from nonsaturating mutagenesis of ∼6,000 mutants (Fig. 1A and B). The coloration of the mutant colonies ranged from white (24 h) to pink over time (≥48 h); similarly, over time, the colonies became elastic (gummy) and recalcitrant to manipulation with a toothpick and developed a rugose colony morphology (Fig. 1A).
FIG 1.
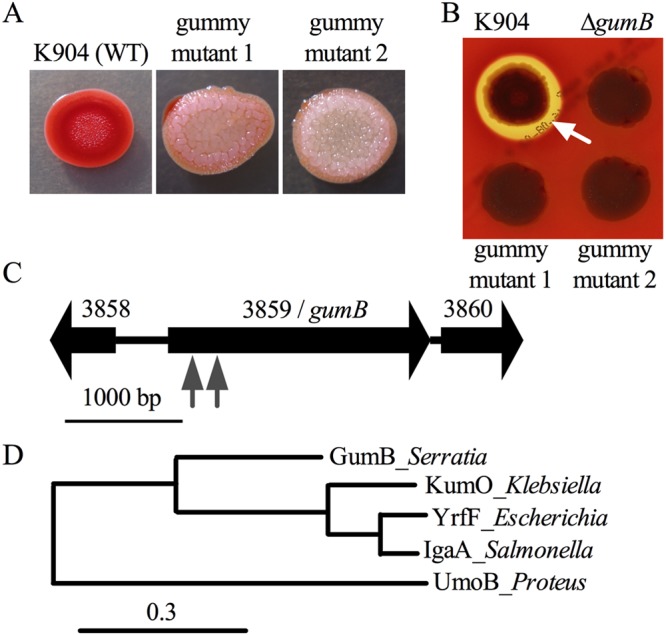
Isolation of mutants with secondary metabolite biosynthesis defects and a distinct colony morphology. (A) Representative images of the wild-type (WT) strain K904 and the two gummy mutants. The colonies were top-lit and demonstrate a lack of prodigiosin and altered colony morphology. (B) Representative images show a hemolysis zone (white arrow) on blood agar plates around K904 and a lack of hemolysis zones around two gumB transposon mutants and a defined gumB mutant. The plate was lighted from below to highlight the hemolysis zone, obscuring colony color. (C) Genetic map of the gumB gene and surrounding genes. Transposon insertion sites are noted by gray arrow points. (D) Phylogram of amino acid sequences of known and predicted IgaA/UmoB family proteins. The dendrogram was made using the “one-click” mode of the tree rendering program from www.Phylogeny.fr, using default settings. The number of substitutions per site are proportional to the branch length.
The mutations mapped to an uncharacterized gene in the S. marcescens genome, corresponding to open reading frame (ORF) SMDB11_3859 of sequenced strain Db11 (44). The mutations mapped to bp 381 relative to the start codon for gummy mutant 1 and bp 168 for gummy mutant 2 (Fig. 1C). Other mutants defective in both prodigiosin and serratamolide production mapped to the previously identified eepS gene (27) and an uncharacterized transcription factor of the GntR family that will be described elsewhere.
At the amino acid level, the identified gene from strain K904 is predicted to code for a protein that is 99% identical (701/710 amino acids) to the corresponding SMDB11_3859-encoded predicted protein. This protein consists of a Pfam PF07095/IgaA domain and is 42% identical to UmoB from Proteus mirabilis, 56% identical to YrfF of Escherichia coli, and 57% identical to IgaA from Salmonella enterica serovar Typhimurium. The family of proteins is conserved in the Enterobacteriaceae, including a putative protein coded by an uncharacterized ORF in K. pneumoniae predicted to be 54% identical to GumB at the amino acid level (see Fig. S1 in the supplemental material). Phylogenetic analysis suggests that the SMDB11_3859-encoded protein is structurally intermediate in relatedness between UmoB and the highly similar IgaA and YrfF proteins (Fig. 1D). In silico analysis of the GumB primary sequence suggests that it is in the inner membrane with three transmembrane helices (Fig. S1). Published models propose that IgaA/UmoB sits in the inner membrane and responds to RcsF localization or conformation to transmit envelop stress signals, and this is accomplished through IgaA/UmoB control of the Rcs transcription factor system (39–41).
Since umoB was the first gene of the IgaA/UmoB family with experimentally defined function, we here refer to the S. marcescens SMDB11_3859 as the gummy homolog of umoB, gumB.
Unlike igaA and yrfF, the gumB gene is not essential for viability.
The gumB open reading frame appears to be monocistronic (Fig. 1C), although there is a predicted operon 98 bp downstream of the stop codon of gumB. The adjacent operon composed of ORFs SMDB11_3960 and SMDB11_3961 is predicted to code for a GMP/IMP nucleotidase and ribosome-associated heat shock protein Hsp15, respectively. As a first step to test whether the observed mutant phenotypes were due to a loss of GumB activity or a polar effect on an adjacent gene, an in-frame deletion mutation of gumB was introduced into the K904 chromosome by two-step allelic replacement. Colonies of the resulting mutant strain were similar to the transposon mutants with respect to a lack of pigmentation and the development of an elastic and highly textured colony morphology (Fig. 2A and Movie S1 in the supplemental material). The isolation of transposon insertion mutations in gumB and the generation of a gumB deletion mutation suggest that unlike what was observed with igaA from S. enterica and yrfF from E. coli, the gumB gene of S. marcescens is not essential for growth.
FIG 2.
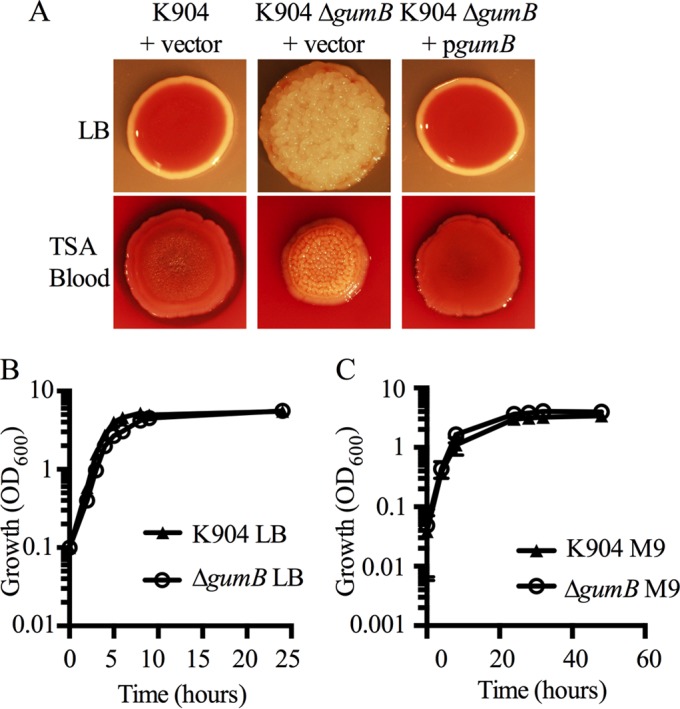
Complementation of gumB mutant defects and growth analysis. (A) The gumB mutant colony and color phenotypes are complemented by wild-type gumB on a plasmid under the control of the Plac promoter. Representative images are of the strain K904 and the ΔgumB mutant with vector control plasmid pMQ132 and complementation plasmid pgumB (pMQ480). Bacteria were incubated for 48 h at 30°C on LB or blood agar plates and lighted from above. (B and C) Growth curves of the wild-type strain K904 and the ΔgumB mutant in LB (B) or M9 minimal medium supplemented with glucose (C). Means ± standard deviation (SD) are shown, n ≥ 3.
Complementation analysis of the ΔgumB mutation with wild-type gumB on a multicopy plasmid further supports that the observed mutant defects, loss of pigmentation, and rugose phenotypes are due to a mutation of gumB and not another mutation or a polar effect on adjacent genes (Fig. 2A).
Growth analysis in rich medium demonstrates the slightly retarded growth of the ΔgumB mutant in log phase between 4 and 8 h; however, the gumB mutant achieved a final optical density similar to that of the wild type by stationary phase (∼9 h) (Fig. 2B). Given the slight growth reduction of the gumB mutant in rich medium, we tested whether it was defective in primary metabolite production by assessing growth in M9 minimal medium. The ΔgumB mutant grew slightly better than the wild type in M9 medium supplemented with glucose (Fig. 2C). These data indicate that despite the pleiotropic phenotypes imparted by a mutation of gumB, the mutant strain is able to generate all of the primary metabolites necessary for growth in minimal medium.
ΔgumB mutant phenotypes are complemented by the igaA gene from S. enterica, yrfF from E. coli, and kumO from K. pneumoniae.
To genetically test whether the GumB protein shares functionality with the IgaA/UmoB family proteins noted above, genes from E. coli (yrfF), K. pneumoniae (kumO), and S. enterica (igaA) were cloned and introduced into the ΔgumB mutant strain on a multicopy plasmid. The pigment and colony morphology phenotypes of the ΔgumB mutant were restored to those of the wild type by the expression of igaA and yrfF (Fig. 3A). The K. pneumoniae gene complemented the colony morphology phenotype and partially complemented the pigment phenotype that is clear in the larger colonies (Fig. 3B). These results support the idea that the UmoB/IgaA family proteins are highly conserved among opportunistic pathogens from the Enterobacteriaceae.
FIG 3.
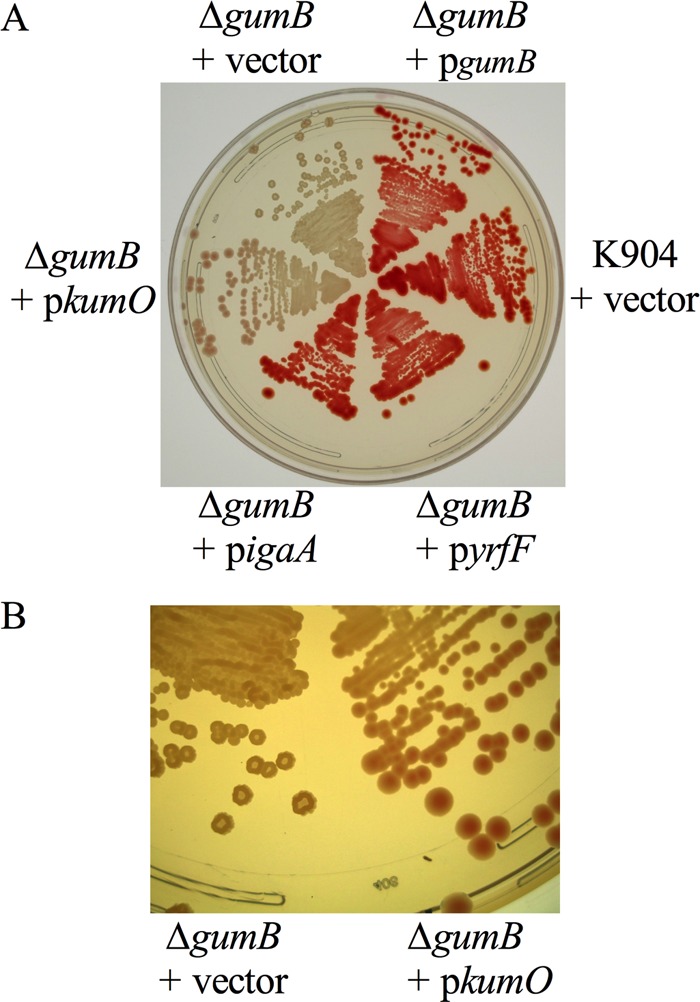
Complementation of gumB mutant defects by IgaA/UmoB family genes from several genera. (A) Image of the ΔgumB mutant with vector alone (pMQ132) or pMQ132 with various IgaA/UmoB family genes under the control of Plac to test for complementation. (B) Zoomed-in view of panel A to highlight the colony morphology complementation of the gumB mutant with a predicted IgaA/UmoB gene from K. pneumoniae (kumO).
Motility defects were conferred by mutation of gumB.
Tests were done to determine whether GumB shared function with IgaA and UmoB in the regulation of flagellum-based motility. The ΔgumB mutant did not produce a swimming zone when measured at 24 h, unlike the wild type (Fig. 4A). GumB was also required for surface swarming motility (Fig. 4A). The gumB mutant swarming defects (Fig. 4B) and swimming defects (Fig. 4C) were complemented by the wild-type gumB gene on a plasmid.
FIG 4.
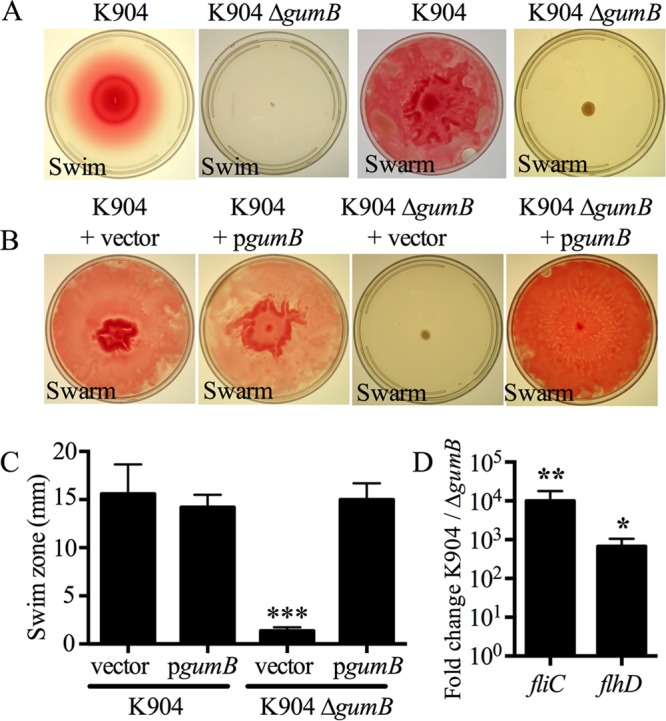
S. marcescens GumB is necessary for swimming and swarming motility. (A) Representative images of the swimming and swarming defects of the ΔgumB mutant. (B) Representative images of complementation of the ΔgumB swarming defect. (C) Complementation of the ΔgumB swimming phenotype. n ≥ 5 independent plates per genotype from 2 separate days. Asterisks indicate a significant difference from all of the other genotypes (P < 0.001). Mean and SD are shown. Vector, pMQ132; pgumB, pMQ480. (D) Fold change of both flagellum-related gene transcript levels. qRT-PCR of fliC and flhD gene expression, n ≥ 4. Mean and SD are shown. *, P < 0.05; **, P < 0.01.
To test whether expression of the flagellar master regulator operon flhDC was altered in the ΔgumB mutant, quantitative-reverse transcriptase PCR (qRT-PCR) was employed. The transcript level of the flhD gene was reduced (P < 0.05, Mann-Whitney test) by several orders of magnitude in the ΔgumB mutant compared to the wild-type K904 strain when measured at an optical density at 600 nm (OD600) of 3 (Fig. 4D). This reduction in flhDC operon expression correlated with an ∼10,000-fold reduction of flagellar gene (fliC) expression when measured by qRT-PCR (Fig. 4D). Transmission electron microscopy (TEM) analysis of the ΔgumB mutant cells for stationary-phase liquid cultures also demonstrated a loss of flagella (Fig. S2), with 71% of the wild-type cells having a flagellum, and <1% of ΔgumB mutant cells with a flagellum (n = 105 cells for each genotype; P < 0.001, Fisher's exact test). Together, these data suggest that GumB plays a role in flagellum-based motility through positive control of the flhDC flagellum master regulator operon.
Secondary metabolite defects of the gumB mutant.
The red pigment prodigiosin is a secondary metabolite made by a subset of S. marcescens strains and is thought to be involved in competition between microbes and to modulate energy levels during stationary phase. IgaA family proteins have not been implicated in the control of secondary metabolites, so the obvious defect in pigmentation of the ΔgumB mutant was quantified. Prodigiosin was extracted and measured from stationary-phase cultures of the wild-type and ΔgumB mutant strains with complementation plasmid, pMQ480, or vector negative control. Prodigiosin levels were significantly lower in the ΔgumB mutant and could be complemented in trans (Fig. 5A).
FIG 5.
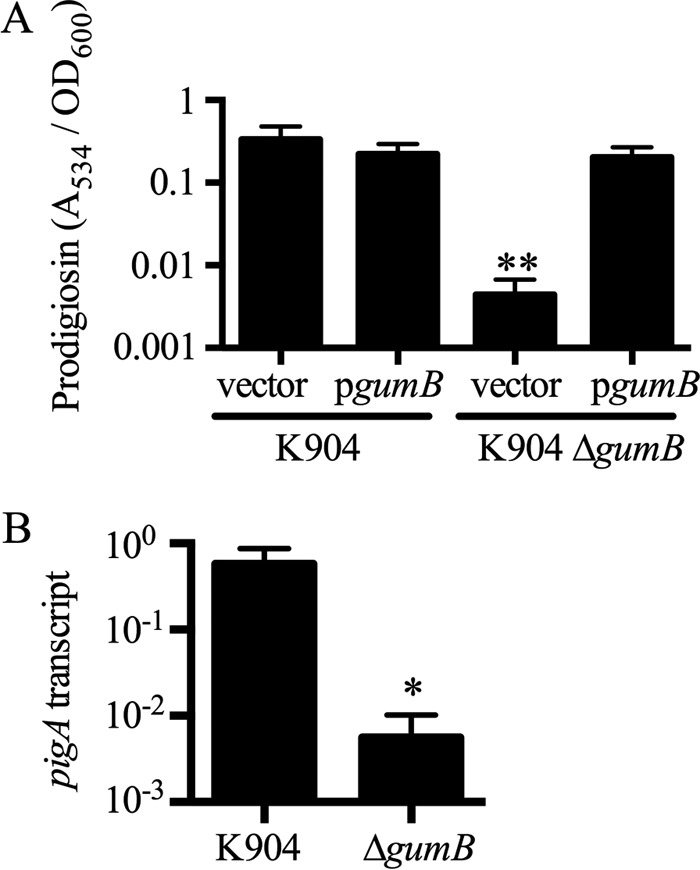
Prodigiosin pigmentation is severely reduced in a ΔgumB mutant. (A) The gumB mutant grown in LB medium is defective in prodigiosin production (75-fold reduction compared to strain K904; n = 3, P < 0.01). Extracted prodigiosin was measured by absorbance, and the defect could be complemented by wild-type gumB in trans. The vector control plasmid is pMQ132, and pgumB is pMQ480. (B) qRT-PCR analysis revealed reduced expression from the prodigiosin biosynthetic locus; pigA gene expression in the ΔgumB mutant was down 103-fold compared to the wild type (P = 0.029, n = 4). Means and standard deviations are shown. *, P < 0.05; **, P < 0.01.
This effect could stem from reduced transcriptional expression of the prodigiosin biosynthesis operon. Reverse transcriptase PCR (RT-PCR) analysis was used on stationary-phase cultures, as prodigiosin is produced during stationary phase (OD600, 3). A clear ∼100-fold reduction in the pigA transcript level was observed in the ΔgumB mutant, suggesting that the gumB mutant pigment defect is largely at the transcriptional level (Fig. 5B).
The biosurfactant serratamolide (45), also called serrawettin W1 (8), is antimicrobial and required for the hemolytic activity of S. marcescens strain K904 on blood agar plates and for growth inhibition of Staphylococcus aureus and other microbes (14, 19). The ΔgumB mutant was defective in hemolysis and antistaphylococcal inhibition zones, suggesting a serratamolide defect (Fig. 6A and B). The ΔgumB mutant was correspondingly defective in the biosynthesis of serratamolide when relative amounts were measured by mass spectrometry (Fig. 6C) and in expression of the serratamolide biosynthesis gene swrW at an OD600 of 3.0 (Fig. 6D). Hemolysis, an indirect measurement of serratamolide production, was used in a complementation analysis, and gumB on a plasmid was able to restore serratamolide production to the ΔgumB mutant (Fig. 6A).
FIG 6.
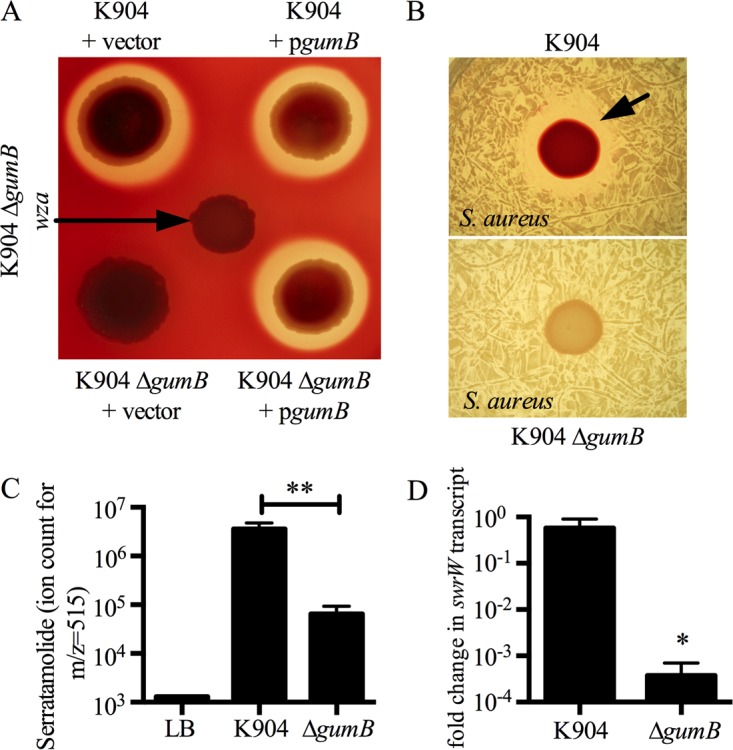
Serratamolide is severely reduced in a ΔgumB mutant. (A) Image of blood agar plate, lit from below. Serratamolide is responsible for hemolysis zones generated by strain K904. The ΔgumB mutant was defective for hemolysis and could be complemented by wild-type gumB in trans. The vector control plasmid is pMQ132, and pgumB is pMQ480. (B) Representative top-lit image. Inhibition of S. aureus bacterial growth (black arrow) by S. marcescens is serratamolide dependent and is eliminated in the ΔgumB mutant. (C) Mass spectrometry reveals a >50-fold significant reduction of serratamolide in the ΔgumB mutant (P < 0.01, n = 3). (D) qRT-PCR analysis demonstrates reduced expression of the serratamolide biosynthetic gene swrW, which was reduced >1,000-fold in the relative transcript levels for the mutant compared to the wild type (P = 0.029, n = 4). Mean and SD are shown. *, P < 0.05; **, P < 0.01.
The quantitative RT-PCR-generated expression levels of pswP, a gene necessary for both prodigiosin and serratamolide production (46), were similar for the K904 (0.0014 ± 0.0027) and the K904 ΔgumB (0.0012 ± 0.0024) strains (P = 0.84, Student's t test). Similarly, multicopy expression of pswP did not alter the pigment or serratamolide defects of the ΔgumB mutant, suggesting that the gumB secondary metabolite defect was PswP independent.
GumB inhibits exopolysaccharide biosynthesis.
To determine the mechanism by which GumB-deficient mutant colonies become elastic, suppressor mutant analysis was performed. Transposon mutagenesis was performed on the ΔgumB mutant strain, and colonies that had lost the rugose colony morphology were chosen for analysis. Multiple independent transposon mutations in the gumB deletion mutant strain yielded smooth nongummy colonies (Fig. 7A). These transposons mapped to adjacent genes in the formerly described capsular polysaccharide/enterobacterial common antigen biosynthetic locus, which has recently been shown to be important for virulence (47, 48). Five mutations were in the wza polysaccharide exporter gene, and two others were found upstream of the wecA exopolysaccharide biosynthesis gene; these genes correspond to strain Db11 ORFs SMDB11_2151 and SMDB11_2152, respectively. TEM analysis of bacteria grown on plates had staining consistent with excessive extracellular polysaccharide surrounding ΔgumB mutant bacteria that is absent in the ΔgumB wza double mutant (Fig. S2). These data suggested that capsular polysaccharide transcription in the ΔgumB mutant was increased.
FIG 7.
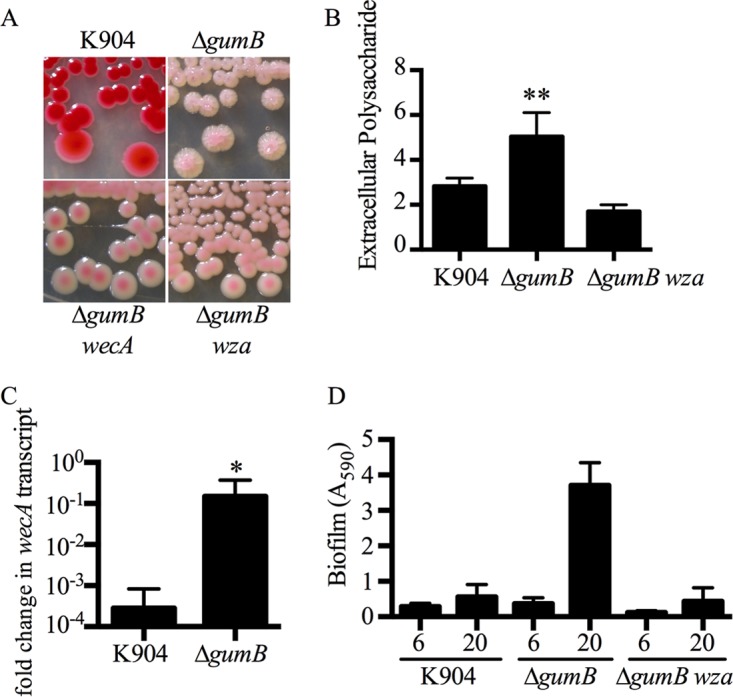
Gummy phenotype of the ΔgumB mutant requires the capsular polysaccharide operon. (A) Suppressor mutants of the rugose colony morphology phenotype of the ΔgumB mutant map to and wza genes of the capsular polysaccharide operon. (B) Extracellular polysaccharide (EPS) was extracted and measured from liquid cultures. Mean and SD are shown, n = 4. Asterisks indicate a significant increase relative to other groups (ANOVA, Tukey's posttest, P < 0.01). (C) qRT-PCR analysis revealed elevated relative wecA gene expression levels in the ΔgumB mutant (P = 0.016, n = 5). Mean and SD are shown. *, P < 0.05. (D) Biofilm formation in LB medium on borosilicate glass tubes grown under high-sheer conditions for 6 and 20 h and stained with crystal violet. The ΔgumB mutant had an elevated biofilm that was absent in a ΔgumB wza mutant. Mean and SD are shown, n = 12 biological replicates.
Total extracellular polysaccharide production was measured from the liquid cultures of the wild-type K904 strain, the ΔgumB mutant, and the ΔgumB wza double mutant. The amount of extracellular polysaccharide extracted from the ΔgumB mutant was ∼2-fold higher than that from the wild type and the ΔgumB wza double mutant (Fig. 7B). Dry weight polysaccharide measurements showed a similar trend, with 3.2 ± 0.8 mg/ml derived from K904, 5.6 ± 0.7 mg/ml from the ΔgumB mutant, and 2.1 ± 0.3 mg/ml from the ΔgumB wza mutant.
Transcriptional analysis of a gene from the capsular polysaccharide biosynthesis operon, wecA, supports the prediction that there is a deregulation of capsular polysaccharide production in the ΔgumB mutant. There was an approximate 100-fold increase in the wecA transcript level in the ΔgumB mutant strain compared to the K904 strain, at an OD600 of 3 (P < 0.05; Fig. 7C). These data suggest that the gummy and rugose colony phenotype of the gumB mutant is a result of deregulated capsular polysaccharide production.
Mutation of the capsule gene wza in the ΔgumB mutant did not alter hemolysis, motility, or pigmentation phenotypes, suggesting that the excessive capsular polysaccharide of the ΔgumB mutant is not responsible for most of its phenotypes (Fig. 6A, 7A, and S3).
Because extracellular polysaccharides can promote biofilm formation, we tested whether the gumB mutant had altered biofilm formation. Biofilms were formed on borosilicate glass under high-sheer conditions (rotated at ∼62 rpm) in LB medium and then stained with crystal violet. At 6 h, the ΔgumB mutant had a minor increase in biofilm formation compared to K904, whereas the ΔgumB wza::Tn double mutant had a 2.3-fold reduction in biofilm formation compared to K904 (Fig. 7D). At 20 h, the ΔgumB wza::Tn double mutant was defective (6.5-fold; P < 0.01, analysis of variance [ANOVA] with Tukey's posttest) in biofilm formation compared to the ΔgumB mutant, suggesting that under the tested conditions, the extracellular polysaccharide regulated by GumB contributes to biofilm formation (Fig. 7D).
GumB mutant phenotypes are not strain specific.
To test whether ΔgumB mutant phenotypes were specific to strain K904, the gumB gene was deleted in S. marcescens strains Db11, a nonpigmented insect pathogen, and CHASM, a pigmented environmental isolate. These deletions conferred a similar pigment phenotype in CHASM (Fig. S4A) and similar swarming defects in both CHASM and Db11 (Fig. S4B), indicating that the role of GumB in regulating secondary metabolite production and motility is not strain specific.
DISCUSSION
This study, designed to identify new regulators of secondary metabolism in S. marcescens, reports a new member of the IgaA/UmoB family of proteins, named GumB. Mutation of the gumB gene introduced pleiotropic phenotypes, most notably a severe reduction in secondary metabolite production.
The genetic results presented here indicate that IgaA/UmoB family genes from several genera of Enterobacteriaceae, including an uncharacterized ortholog from K. pneumoniae, are functionally and structurally conserved. Therefore, it was somewhat surprising that the gumB gene is not essential for viability in S. marcescens as yrfF and igaA are in E. coli and S. enterica, respectively. Analysis of the S. marcescens genome suggests that there is not another obvious IgaA/UmoB gene, so the essential requirement for IgaA/UmoB family proteins found in E. coli and S. enterica must be downstream of GumB and absent or redundant in S. marcescens. Similar to the situation in S. marcescens, the umoB gene in P. mirabilis is not necessary for viability (28). While gumB is not essential, there was a slight growth delay in the ΔgumB mutant grown in LB medium compared to the wild type, although the final culture densities were the same. This growth delay may be due to altered expression of cell division genes in Rcs system and IgaA/UmoB gene mutants, as seen in E. coli and S. enterica (36, 49–51).
The polysaccharide phenotype of the gumB mutant is similar to those of S. enterica with partial-function igaA mutations, where increased activation of the Rcs system is required for the highly mucoid phenotype (29–33). This hypercapsule phenotype conferred by mutation of IgaA/UmoB genes is predicted to be the reason why E. coli cells with nonlethal mutations in yrfF had a selective advantage in a model in which they were serially cocultured with a phagocytic cell line (34). The hypercapsule suggests that IgaA/UmoB proteins may negatively regulate virulence; however, studies with S. enterica bearing nonlethal mutations in igaA indicate that IgaA, at least, is required for virulence in a rodent infection model (31). Importantly, data presented indicated that, other than biofilm formation and colony morphology, excess polysaccharide production is not responsible for the other gumB mutant phenotypes.
This study provides evidence for polysaccharides contributing to S. marcescens biofilm formation, whereas previous studies suggest a dominant role for type I fimbriae (52, 53). This is a new role for IgaA/UmoB family proteins, although it has been demonstrated that the Rcs system can control biofilms (54), and it is likely that GumB works through regulation of the Rcs system in S. marcescens. This is predicted because known IgaA/UmoB proteins, such as IgaA and YrfF, regulate the Rcs system, and here, we showed that the igaA and yrfF genes could complement gumB defects, implying structural and functional similarity between GumB, IgaA, and YrfF. In-progress genetic studies further support that gumB mutant phenotypes require the S. marcescens Rcs system genes (N. A. Stella and R. M. Q. Shanks, unpublished data).
Whereas GumB shares function with known IgaA/UmoB family proteins with respect to flagellar and polysaccharide biosynthesis, it was unknown whether this family of proteins mediates secondary metabolite function. Here, we measured a severe reduction in both prodigiosin and serratamolide. Given the ability of these two compounds to prevent the growth of other microbes, promote motility, promote resistance to antimicrobials via swarming motility, provide resistance to phagocytosis, and cause lysis of mammalian cells, a gumB mutant of S. marcescens would be expected to be at a competitive disadvantage in many niches. A loss of secondary metabolite production was observed when the gumB gene was mutated in three different strains. This is important, as a given gene does not necessarily confer the same phenotypes when mutated in different strains, as has been reported for cyclic-AMP-regulated pathways in S. marcescens (27).
IgaA family proteins are thought to regulate the Rcs system in response to cellular stress (39, 42, 55, 56). Based on this model, GumB likely senses extracellular stress and modifies the activity of the RcsC and or RcsD histidine kinases that in turn influence the expression of many genes by the RcsB response regulator. One may thus consider a ΔgumB mutant cell to be in a constitutively “stressed-out” state. The production of excess capsular polysaccharide is thought to relieve outer membrane stress (57) and promote biofilm formation, both of which may enhance survival. In this stressed state, the bacteria increase the expression of capsular polysaccharide genes but stop the production of flagella and competition-promoting secondary metabolites, perhaps to conserve energy and reduce activation of host immune systems through eliminating the production of pathogen-associated molecular patterns. How GumB interacts with other regulators is unknown at this time; however, it is notable that regulation of pigment, serratamolide, and flagellum by GumB follows a directly opposite trend from that of cAMP-CRP (19, 25, 53, 58), so it could be that GumB-associated phenotypes are mediated through control of intracellular cAMP levels and the corresponding activity of the CRP. Indeed, there is evidence in P. mirabilis that the Rcs system regulates CRP expression (54).
In summary, this report describes a new regulator of secondary metabolites, motility, and biofilm formation in the bacterium S. marcescens. The GumB gene is predicted to be highly important for S. marcescens strains to compete in a variety of environments, given the biological functions that it controls. These could include the use of S. marcescens as a biological control agent, where its GumB-regulated behaviors would be necessary, and in the human gut, where successful competition of S. marcescens is associated with Crohn's disease (59). Last, this study extends the known role of the conserved IgaA/GumB family proteins to the control of secondary metabolites and biofilm formation.
MATERIALS AND METHODS
Strains and growth medium used in this study.
Bacteria were grown in lysogeny broth (LB) (60) or M9 minimal medium (61) supplemented with glucose at 0.4% (wt/vol). Saccharomyces cerevisiae was grown in yeast extract-peptone-dextrose (YPD) broth or synthetic complete (SC)-uracil medium (62). Swimming and swarming agars consisted of LB medium supplemented with 0.3% and 0.6% (wt/vol) agar, respectively. Cultures were grown with aeration using a TC-7 tissue culture roller. All incubations were performed at 30°C. The strains used are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| SM10 λpir | E. coli conjugation strain | 70 |
| S17-1 λpir | E. coli conjugation strain | 70 |
| EC100D | E. coli cloning strain, pir-116 version | Epicentre |
| K746 | E. coli keratitis isolate | 71 |
| LT2 | S. enterica serovar Typhimurium | 72 |
| 883 | K. pneumoniae, clinical isolate | 73 |
| CHASM | S. marcescens, environmental isolate | 25 |
| K904 | S. marcescens, keratitis isolate | 25 |
| Db11 | S. marcescens, insect isolate | 74 |
| CMS2265 | K904 gummy mutant 1, K904 gumB::Tn | This study |
| CMS4001 | K904 ΔgumB | This study |
| CMS4124 | K904 ΔgumB wza::Tn | This study |
| CMS4225 | K904 fimC-kan | 75 |
| CMS4619 | K904 ΔgumB wecA::Tn | This study |
| CMS4320 | K904 gummy mutant 2, K904 gumB::Tn | This study |
| CMS4462 | Db11 ΔgumB | This study |
| CMS4662 | CHASM ΔgumB | This study |
| Plasmids | ||
| pBT20 | Transposon delivery plasmid | 63 |
| pSC189 | Transposon delivery plasmid | 64 |
| pMQ132 | pBBR1-replicon shuttle vector, aacC-1 | 68 |
| pMQ200 | oriR6K plasmid with PBAD promoter, nptII | 68 |
| pMQ460 | Allelic replacement vector | 76 |
| pMQ480 | pMQ132 plus gumB from S. marcescens strain K904 | This study |
| pMQ507 | pMQ460 plus ΔgumB | This study |
| pMQ530 | pMQ132 plus igaA from S. enterica strain LT2 | This study |
| pMQ529 | pMQ132 plus kumO from K. pneumoniae strain 883 | This study |
| pMQ531 | pMQ132 plus yrfF from E. coli strain K746 | This study |
Genetic manipulations and plasmid construction.
Transposon mutagenesis was performed as previously described (52) using transposon delivery plasmids pBT20 (63) and pSC189 (64). Insertion sites were mapped by marker rescue (64) or arbitrary PCR (65) followed by sequencing. Mutants of S. marcescens strain K904 were plated on LB or blood agar and were visually screened for a loss of pigmentation and hemolysis zones, as previously described (27).
Allelic replacement of the gumB open reading frame (ORF) was performed as previously described but using plasmid pMQ507 (all plasmids used are listed in Table 1) (66). The mutation deletes 2,044 bp of the 2,133-bp gene, starting at the ninth codon. To make pMQ507, 470 bp of DNA upstream of gumB and 662 bp of DNA downstream of gumB were amplified with primers that have additional sequence such that the amplicons can recombine with each other and with allelic replacement vector pMQ460. This and other plasmids were made by homologous recombination using yeast in vivo recombination (67, 68). The oligonucleotide primers used to make pMQ507 were primers 3419 to 3422 and are listed in Table 2. Plasmids were verified by PCR and sequencing of cloned junctions, and deletion mutations were verified by PCR.
TABLE 2.
Oligonucleotide primers used in this study
| Primer no. | Purposea | Sequence (5′ to 3′)b |
|---|---|---|
| 1055 | fimA-RT | ACTACACCCTGCGTTTCGAC |
| 1056 | fimA-RT | GCGTTAGAGTTTGCCTGACC |
| 2638 | 16S-RT | AACTGGAGGAAGGTGGGGAT |
| 2639 | 16S-RT | AGGAGGTGATCCAACCGCA |
| 2891 | pswP-RT | CGTGACATCGTCACCTTCACG |
| 2892 | pswP-RT | GCCAAAGAGAGCCTGTTCAAG |
| 2911 | pigA-RT | GGAGCGAACTGACCTTCAAC |
| 2912 | pigA-RT | CTGTTCCAGACGCAGTTTCA |
| 3417 | gumB cloning | ggccagtgccaagcttgcatgcctgcaggtcgactctaTTGAAGCAGCTGTCGTAGTAAC |
| 3418 | gumB cloning | tgtgagcggataacaatttcacacaggaaacagctATGAGCACAATAGTGTTGATATTGG |
| 3419 | gumB deletion | cggccagtgccaagcttgcatgcctgcaggtcgactctaGATTGCCTCAAAGAGGTTACC |
| 3420 | gumB deletion | gaagcagctgtcgtagtaacgctggCCAATATCAACACTATTGTGCTCAT |
| 3421 | gumB deletion | atgagcacaatagtgttgatattggCCAGCGTTACTACGACAGCTGCTTC |
| 3422 | gumB deletion | attgtgagcggataacaatttcacacaggaaacagctGGTTGAACCCGGTGTGCTGTTGC |
| 3570 | wecA-RT | ACCGAGCATCACTTCCTGAT |
| 3571 | wecA-RT | GTACCTGACGCTGATGCTGA |
| 3575 | flhD-RT | AATGTTTCGCCTGGGTATTG |
| 3576 | flhD-RT | ATAGCAAAATGCCGGTATGG |
| 3582 | swrW-RT | AATTAGGCGAGATCGAGCAA |
| 3583 | swrW-RT | AACAGGACGGCACCATAAAG |
| 3655 | yrfF cloning | aaattctgttttatcagaccgcttctgcgttctgatTTATTCGATAAGGCTTTCTGAAGG |
| 3656 | yrfF cloning | gtgagcggataacaatttcacacaggaaacagctATGAGCACCATTGTGATTTTTTTAGC |
| 3658 | igaA cloning | aattctgttttatcagaccgcttctgcgttctgatTCAGATGAGATTTTCCGGAGAGACG |
| 3659 | igaA cloning | gtgagcggataacaatttcacacaggaaacagctATGAGCACCATTCTGATTTTTATAGC |
| 3661 | kumO cloning | caaattctgttttatcagaccgcttctgcgttctgatTTAATCTTGCGAGTCAGATGAGG |
| 3662 | kumO cloning | aattgtgagcggataacaatttcacacaggaaacagctATGGGCACCTTTCTGATATTCC |
| 3809 | fliC-RT | GTATCTCTCTGGCGCAGACC |
| 3810 | fliC-RT | ATGGTTTCACCGTCGTTAGC |
“-RT” indicates that primers are used for qRT-PCR.
Uppercase letters indicate sequences that prime amplification of the desired DNA, whereas lowercase letters target homologous recombination with a plasmid.
To make complementation plasmids, the igaA-umoB family genes from E. coli (yrfF), Klebsiella pneumoniae (kumO), S. marcescens (gumB), and S. enterica (igaA) were amplified with primers 3655/3656, 3661/3662, 3417/3418, and 3658/3659, respectively. These ORFs were placed under transcriptional control of the E. coli Plac promoter on pMQ132 (68).
Transmission electron microscopy.
TEM of bacteria was performed using cultures grown overnight in LB medium for 18 to 20 h or taken from agar plates after 48 h of growth, washed in phosphate-buffered saline (PBS), and processed and observed as previously described (53). At least four independent preparations were made for each group.
Secondary metabolite assays.
Prodigiosin was extracted from cultures grown for 18 to 20 h in LB medium using acidified ethanol and the absorbance measured, as previously described (25).
Serratamolide was measured by mass spectrometry from cultures grown for 20 h in LB medium and normalized to an OD600 of 2.0, as previously described (27). Zones of growth inhibition for Staphylococcus aureus were measured as previously described (14).
Extracellular polysaccharide quantitation.
S. marcescens polysaccharides were isolated as described by Anderson et al. (48) and Masuko et al. (69). Briefly, bacteria were grown overnight in LB medium for 16 to 20 h and pelleted by centrifugation at 25,000 × g and 15°C for 15 min. The supernatants were discarded, and the bacterial pellets were suspended in 30 ml of PBS and 6 ml of 1% Zwittergent 3-14 (EMD Millipore) in 100 mM citric acid (pH 2) and incubated at 50°C for 20 min. Following incubation, the bacteria were pelleted by centrifugation at 25,000 × g and 15°C for 30 min, and the cell-free supernatants were transferred to 250-ml centrifuge bottles. Four volumes of cold (−20°C) ethanol were added to each and the samples placed at −20°C overnight. After overnight precipitation, the samples were centrifuged at 27,000 × g and 4°C for 45 min, and the supernatants were discarded. The precipitated polysaccharides were allowed to air-dry in a chemical fume hood. The polysaccharides were weighed, and the total carbohydrate content was quantitated using a microplate phenol-sulfuric assay (48, 69).
Biofilm assay.
Biofilm formation on borosilicate glass was performed as previously described (52, 53). Biofilms were stained with crystal violet (0.1% [wt/vol]) and solubilized with glacial acetic acid (33%), and the absorbance at 590 nm was measured using a plate reader (BioTek Synergy 2). Biofilms were generated for 6 and 20 h in LB medium in tubes rotated on a TC-7 tissue culture roller (New Brunswick Scientific).
qRT-PCR and RNA preparation.
RNA preparation from cultures with an OD600 of 3.0 and qRT-PCR were performed as previously described (27). Briefly, cultures were treated with RNAprotect reagent (Qiagen), and RNA was purified using an RNeasy kit (Qiagen) and concentrated using a spin column (RNA Clean and Concentrator; Zymo Research). Two rounds of DNase treatment were performed (10 units for 15 min at room temperature on column with Qiagen DNase I, and 1 unit for 30 min at 37°C with Promega RQ1 DNase after purification and prior to the concentration step). Controls for cDNA preparation included no-reverse transcriptase reactions for each RNA sample, included to validate the absence of chromosomal DNA in RNA samples; any samples with detectable chromosomal DNA contamination were excluded prior to experimentation. The coefficient of variance was less than 10% between experiments.
Statistical analysis.
Experiments were done at least twice, with a minimum of three biological replicates. The GraphPad Prism software was used to perform Student's t tests, Mann-Whitney U tests, Fisher's exact test, and one-way ANOVA with Tukey's posttest. Significance was set at a P value of <0.05.
Accession number(s).
The sequences of the gumB gene from S. marcescens K904 and the kumO gene from K. pneumoniae were deposited in GenBank under accession numbers KY098906 and KY098907, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vaughn Cooper, Kelly Hughes, and Ming-Hong Nguyen for the kind gifts of bacterial strains, and Marissa Aston, James Fender, and Kristin Hunt for technical assistance.
This study was supported by NIH grants EY027331 (to R.M.Q.S.), EY017271 and EY024785 (to K.M.B.), and EY08098 (Core Grant for Vision Research), the Eye and Ear Foundation of Pittsburgh, and unrestricted funds from Research to Prevent Blindness.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02575-17.
REFERENCES
- 1.Demain AL. 1992. Microbial secondary metabolism: a new theoretical frontier for academia, a new opportunity for industry. Ciba Found Symp 171:3–16, discussion 16–23. [DOI] [PubMed] [Google Scholar]
- 2.Coulthurst SJ, Barnard AM, Salmond GP. 2005. Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat Rev Microbiol 3:295–306. doi: 10.1038/nrmicro1128. [DOI] [PubMed] [Google Scholar]
- 3.Ou X, Zhang B, Zhang L, Zhao G, Ding X. 2009. Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl Environ Microbiol 75:2158–2165. doi: 10.1128/AEM.02209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbeva P, Silby MW, Raaijmakers JM, Levy SB, Boer W. 2011. Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J 5:973–985. doi: 10.1038/ismej.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. 2014. Metabolic potential of endophytic bacteria. Curr Opin Biotechnol 27:30–37. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tralau T, Sowada J, Luch A. 2015. Insights on the human microbiome and its xenobiotic metabolism: what is known about its effects on human physiology? Expert Opin Drug Metab Toxicol 11:411–425. doi: 10.1517/17425255.2015.990437. [DOI] [PubMed] [Google Scholar]
- 7.Wasserman HH, Keggi JJ, McKeon JE. 1961. Serratamolide, a metabolic product of Serratia. J Am Chem Soc 83:4107–4108. doi: 10.1021/ja01480a046. [DOI] [Google Scholar]
- 8.Matsuyama T, Sogawa M, Nakagawa Y. 1989. Fractal spreading growth of Serratia marcescens which produces surface active exolipids. FEMS Microbiol Lett 52:243–246. doi: 10.1111/j.1574-6968.1989.tb03630.x. [DOI] [PubMed] [Google Scholar]
- 9.Williamson NR, Fineran PC, Leeper FJ, Salmond GP. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4:887–899. doi: 10.1038/nrmicro1531. [DOI] [PubMed] [Google Scholar]
- 10.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerc AJ, Song L, Challis GL, Stanley-Wall NR, Coulthurst SJ. 2012. The insect pathogen Serratia marcescens Db10 uses a hybrid non-ribosomal peptide synthetase-polyketide synthase to produce the antibiotic althiomycin. PLoS One 7:e44673. doi: 10.1371/journal.pone.0044673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overhage J, Bains M, Brazas MD, Hancock RE. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai S, Tremblay J, Deziel E. 2009. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ Microbiol 11:126–136. doi: 10.1111/j.1462-2920.2008.01747.x. [DOI] [PubMed] [Google Scholar]
- 14.Kadouri DE, Shanks RM. 2013. Identification of a methicillin-resistant Staphylococcus aureus inhibitory compound isolated from Serratia marcescens. Res Microbiol 164:821–826. doi: 10.1016/j.resmic.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapenda JC, Silva PA, Vicalvi MC, Sena KX, Nascimento SC. 2015. Antimicrobial activity of prodigiosin isolated from Serratia marcescens UFPEDA 398. World J Microbiol Biotechnol 31:399–406. doi: 10.1007/s11274-014-1793-y. [DOI] [PubMed] [Google Scholar]
- 16.Danevčič T, Boric Vezjak M, Tabor M, Zorec M, Stopar D. 2016. Prodigiosin induces autolysins in actively grown Bacillus subtilis cells. Front Microbiol 7:27. doi: 10.3389/fmicb.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suryawanshi RK, Patil CD, Koli SH, Hallsworth JE, Patil SV. 2016. Antimicrobial activity of prodigiosin is attributable to plasma-membrane damage. Nat Prod Res 31:572–577. doi: 10.1080/14786419.2016.1195380. [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki Y, Oka S, Hara-Hotta H, Yano I. 1993. Stimulation and inhibition of polymorphonuclear leukocytes phagocytosis by lipoamino acids isolated from Serratia marcescens. FEMS Immunol Med Microbiol 6:265–271. doi: 10.1111/j.1574-695X.1993.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 19.Shanks RMQ, Stella NA, Lahr RM, Wang S, Veverka TI, Kowalski RP, Liu X. 2012. Serratamolide is a hemolytic factor produced by Serratia marcescens. PLoS One 7:e36398. doi: 10.1371/journal.pone.0036398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horng YT, Deng SC, Daykin M, Soo PC, Wei JR, Luh KT, Ho SW, Swift S, Lai HC, Williams P. 2002. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol Microbiol 45:1655–1671. doi: 10.1046/j.1365-2958.2002.03117.x. [DOI] [PubMed] [Google Scholar]
- 21.Coulthurst SJ, Williamson NR, Harris AK, Spring DR, Salmond GPC. 2006. Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology 152:1899–1911. doi: 10.1099/mic.0.28803-0. [DOI] [PubMed] [Google Scholar]
- 22.Tanikawa T, Nakagawa Y, Matsuyama T. 2006. Transcriptional downregulator HexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens. Microbiol Immunol 50:587–596. doi: 10.1111/j.1348-0421.2006.tb03833.x. [DOI] [PubMed] [Google Scholar]
- 23.Williamson NR, Simonsen HT, Harris AK, Leeper FJ, Salmond GP. 2006. Disruption of the copper efflux pump (CopA) of Serratia marcescens ATCC 274 pleiotropically affects copper sensitivity and production of the tripyrrole secondary metabolite, prodigiosin. J Ind Microbiol Biotechnol 33:151–158. doi: 10.1007/s10295-005-0040-9. [DOI] [PubMed] [Google Scholar]
- 24.Horng YT, Chang KC, Liu YN, Lai HC, Soo PC. 2010. The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens. Int J Med Microbiol 300:304–312. doi: 10.1016/j.ijmm.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Kalivoda EJ, Stella NA, Aston MA, Fender JE, Thompson PP, Kowalski RP, Shanks RM. 2010. Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res Microbiol 161:158–167. doi: 10.1016/j.resmic.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanks RM, Lahr RM, Stella NA, Arena KE, Brothers KM, Kwak DH, Liu X, Kalivoda EJ. 2013. A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One 8:e57634. doi: 10.1371/journal.pone.0057634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stella NA, Lahr RM, Brothers KM, Kalivoda EJ, Hunt KM, Kwak DH, Liu X, Shanks RM. 2015. Serratia marcescens cyclic AMP-receptor protein controls transcription of EepR, a novel regulator of antimicrobial secondary metabolites. J Bacteriol 197:2468–2478. doi: 10.1128/JB.00136-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dufour A, Furness RB, Hughes C. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol Microbiol 29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- 29.Cano DA, Martinez-Moya M, Pucciarelli MG, Groisman EA, Casadesus J, Garcia-Del Portillo F. 2001. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect Immun 69:6463–6474. doi: 10.1128/IAI.69.10.6463-6474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa CS, Pettinari MJ, Mendez BS, Anton DN. 2003. Null mutations in the essential gene yrfF (mucM) are not lethal in rcsB, yojN or rcsC strains of Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett 222:25–32. doi: 10.1016/S0378-1097(03)00221-0. [DOI] [PubMed] [Google Scholar]
- 31.Domínguez-Bernal G, Pucciarelli MG, Ramos-Morales F, Garcia-Quintanilla M, Cano DA, Casadesus J, Garcia-del Portillo F. 2004. Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol Microbiol 53:1437–1449. doi: 10.1111/j.1365-2958.2004.04213.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Zhao Y, McClelland M, Harshey RM. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J Bacteriol 189:8447–8457. doi: 10.1128/JB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariscotti JF, García-del Portillo F. 2009. Genome expression analyses revealing the modulation of the Salmonella Rcs regulon by the attenuator IgaA. J Bacteriol 191:1855–1867. doi: 10.1128/JB.01604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miskinyte M, Sousa A, Ramiro RS, de Sousa JA, Kotlinowski J, Caramalho I, Magalhaes S, Soares MP, Gordo I. 2013. The genetic basis of Escherichia coli pathoadaptation to macrophages. PLoS Pathog 9:e1003802. doi: 10.1371/journal.ppat.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Tyne D, Ciolino JB, Wang J, Durand ML, Gilmore MS. 2016. Novel phagocytosis-resistant extended-spectrum beta-lactamase-producing Escherichia coli from keratitis. JAMA Ophthalmol 134:1306–1309. doi: 10.1001/jamaophthalmol.2016.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cano DA, Dominguez-Bernal G, Tierrez A, Garcia-Del Portillo F, Casadesus J. 2002. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tierrez A, Garcia-del Portillo F. 2004. The Salmonella membrane protein IgaA modulates the activity of the RcsC-YojN-RcsB and PhoP-PhoQ regulons. J Bacteriol 186:7481–7489. doi: 10.1128/JB.186.22.7481-7489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mariscotti JF, Garcia-Del Portillo F. 2008. Instability of the Salmonella RcsCDB signalling system in the absence of the attenuator IgaA. Microbiology 154:1372–1383. doi: 10.1099/mic.0.2007/015891-0. [DOI] [PubMed] [Google Scholar]
- 39.Morgenstein RM, Rather PN. 2012. Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J Bacteriol 194:669–676. doi: 10.1128/JB.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho SH, Szewczyk J, Pesavento C, Zietek M, Banzhaf M, Roszczenko P, Asmar A, Laloux G, Hov AK, Leverrier P, Van der Henst C, Vertommen D, Typas A, Collet JF. 2014. Detecting envelope stress by monitoring beta-barrel assembly. Cell 159:1652–1664. doi: 10.1016/j.cell.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 41.Laloux G, Collet JF. 2017. Major Tom to ground control: how lipoproteins communicate extracytoplasmic stress to the decision center of the cell. J Bacteriol 199:e00216-. doi: 10.1128/JB.00216-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castelli ME, Vescovi EG. 2011. The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens. J Bacteriol 193:63–74. doi: 10.1128/JB.00839-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon KJ, Castelli ME, Garcia Vescovi E, Feldman MF. 2012. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J Bacteriol 194:3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, Katsura K, Kurokawa K, Oshima K, Hattori M, Parkhill J, Sebaihia M, Coulthurst SJ, Gotoh N, Thomson NR, Ewbank JJ, Hayashi T. 2014. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol 6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wasserman HH, Keggi JJ, McKeon JE. 1962. The structure of serratamolide. J Am Chem Soc 84:2978–2982. doi: 10.1021/ja00874a028. [DOI] [Google Scholar]
- 46.Sunaga S, Li H, Sato Y, Nakagawa Y, Matsuyama T. 2004. Identification and characterization of the pswP gene required for the parallel production of prodigiosin and serrawettin W1 in Serratia marcescens. Microbiol Immunol 48:723–728. doi: 10.1111/j.1348-0421.2004.tb03597.x. [DOI] [PubMed] [Google Scholar]
- 47.Castelli ME, Fedrigo GV, Clementin AL, Ielmini MV, Feldman MF, Garcia Vescovi E. 2008. Enterobacterial common antigen integrity is a checkpoint for flagellar biogenesis in Serratia marcescens. J Bacteriol 190:213–220. doi: 10.1128/JB.01348-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson MT, Mitchell LA, Zhao L, Mobley HLT. 2017. Capsule production and glucose metabolism dictate fitness during Serratia marcescens bacteremia. mBio 8:e00740-. doi: 10.1128/mBio.00740-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gervais FG, Phoenix P, Drapeau GR. 1992. The rcsB gene, a positive regulator of colanic acid biosynthesis in Escherichia coli, is also an activator of ftsZ expression. J Bacteriol 174:3964–3971. doi: 10.1128/jb.174.12.3964-3971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carballès F, Bertrand C, Bouche JP, Cam K. 1999. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol Microbiol 34:442–450. doi: 10.1046/j.1365-2958.1999.01605.x. [DOI] [PubMed] [Google Scholar]
- 51.Costa CS, Anton DN. 2001. Role of the ftsA1p promoter in the resistance of mucoid mutants of Salmonella enterica to mecillinam: characterization of a new type of mucoid mutant. FEMS Microbiol Lett 200:201–205. doi: 10.1111/j.1574-6968.2001.tb10716.x. [DOI] [PubMed] [Google Scholar]
- 52.Shanks RMQ, Stella NA, Kalivoda EJ, Doe MR, O'Dee DM, Lathrop KL, Guo FL, Nau GJ. 2007. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J Bacteriol 189:7262–7272. doi: 10.1128/JB.00859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalivoda EJ, Stella NA, O'Dee DM, Nau GJ, Shanks RM. 2008. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl Environ Microbiol 74:3461–3470. doi: 10.1128/AEM.02733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howery KE, Clemmer KM, Rather PN. 2016. The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence. Curr Genet 62:775–789. doi: 10.1007/s00294-016-0579-1. [DOI] [PubMed] [Google Scholar]
- 55.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 56.Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. 2014. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proc Natl Acad Sci U S A 111:E4350–E4358. doi: 10.1073/pnas.1417138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gottesman S, Stout V. 1991. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol 5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 58.Stella NA, Kalivoda EJ, O'Dee DM, Nau GJ, Shanks RM. 2008. Catabolite repression control of flagellum production by Serratia marcescens. Res Microbiol 159:562–568. doi: 10.1016/j.resmic.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoarau G, Mukherjee PK, Gower-Rousseu C, Hager C, Chandra J, Retueto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn's disease. mBio 7:e01250-. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams MH. 1959. Bacteriophages. Interscience Publishers, Inc., New York, NY. [Google Scholar]
- 62.Burke D, Dawson D, Stearns T. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 63.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol 55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 64.Chiang SL, Rubin EJ. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179–185. doi: 10.1016/S0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 65.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. 1999. Genetic approaches to the study of biofilms. Methods Enzymol 310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 66.Kalivoda EJ, Horzempa J, Stella NA, Sadaf A, Kowalski RP, Nau GJ, Shanks RM. 2011. New vector tools with a hygromycin resistance marker for use with opportunistic pathogens. Mol Biotechnol 48:7–14. doi: 10.1007/s12033-010-9342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shanks RM, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S, Lee YC. 2005. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem 339:69–72. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 70.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalivoda EJ, Brothers KM, Stella NA, Schmitt MJ, Shanks RM. 2013. Bacterial cyclic AMP-phosphodiesterase activity coordinates biofilm formation. PLoS One 8:e71267. doi: 10.1371/journal.pone.0071267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neidhardt FC. 1996. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 73.Olonisakin TF, Li H, Xiong Z, Kochman EJ, Yu M, Qu Y, Hulver M, Kolls JK, St Croix C, Doi Y, Nguyen MH, Shanks RM, Mallampalli RK, Kagan VE, Ray A, Silverstein RL, Ray P, Lee JS. 2016. CD36 provides host protection against Klebsiella pneumoniae intrapulmonary infection by enhancing LPS responsiveness and macrophage phagocytosis. J Infect Dis 214:1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flyg C, Kenne K, Boman HG. 1980. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J Gen Microbiol 120:173–181. [DOI] [PubMed] [Google Scholar]
- 75.Shanks RM, Stella NA, Brothers KM, Polaski DM. 2016. Exploitation of a “hockey-puck” phenotype to identify pilus and biofilm regulators in Serratia marcescens through genetic analysis. Can J Microbiol 62:83–93. doi: 10.1139/cjm-2015-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shanks RM, Stella NA, Hunt KM, Brothers KM, Zhang L, Thibodeau PH. 2015. Identification of SlpB, a cytotoxic protease from Serratia marcescens. Infect Immun 83:2907–2916. doi: 10.1128/IAI.03096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


